Abstract
Objective:
Genome-wide association studies (GWAS) have identified a number of genetic variants associated with the susceptibility of bladder cancer (BC) in European and Chinese populations. Here, we assessed the association of two of these variants, rs9642880 and rs710521 in an Egyptian patients and also examined the expression of c-Myc.The basis was due to the absence of studies on Egyptian patients to determine the association between rs9642880& rs710521 and bladder cancer risk, particularly due to the known role of the variant (rs9642880) in the Progression and development of bladder cancer.
Methods:
Urine samples were collected from onehundred and fiftybladder cancer patients under particular standards and fifty healthy controls. Genomic DNA was extracted, rs9642880 G>T and rs710521 A>G polymorphisms were amplified, assessed via restriction fragment length polymorphism(RFLP) and sequenced. Urine retrieved results were compared to the histopathological diagnosis of tissue biopsies and to the results of C-Myc immunohistochemistry. Data were statistically analyzed using Microsoft Excel 2016, association between significant genotypes of the two studied variables and bladder cancer risk was performed.
Results:
We found that the TT genotype of rs9642880 G>T was strongly associated with the risk of bladder cancer, andfor rs710521 A>G, AG genotype was also identified to has an association with bladder cancer risk.All 150 tumor sections showed positive immunoreactivity for c-Myc in the nucleus and the cytoplasm.
Conclusion:
Identifying the association to risk of bladder cancer using genetic analysis will help in the early detection of the disease.
Key Words: RFLP, bladder cancer, genotype, genome, wide association study
Introduction
Bladder cancer is ranked 9th in terms of commonality amongst both men and women worldwide, with an annual incidence of around 430,000 cases (Martin et al., 2018), with a rank of 13th in terms of cancer-related mortality annually (Cumberbatch et al., 2019). It occurs in both sexes, there is a male predominance, and it ranked 7th in terms of occurrence for men(Rampias et al., 2014). Bladder cancer varies among countries, with the highest incidence rates in Europe, the United States and Egypt, whereas the lowest rates in Asia and Africa, it is known of a cancer of industrialized nations, with higher incidence in high resource versus low resource countries, determined as high as triple fold (Altunkol, et al., 2018). Cigarette smoke and occupational exposures are examples of two well-established factors in the development of bladder cancer. However, the basis of genetic susceptibility to bladder cancer is largely unknown(Chang et al., 2009). In Egypt, urinary bladder cancer incidence is among the highest worldwide. Chronic bladder infection with Schistosoma haematobium, the trematode causing urinary schistosomiasis as well as smoking are the most important risk factors for bladder cancer in Egypt (Samah, et al., 2015).
Urothelial BC (UBC) is the most common subtype. Approximately 75% of patients present with non–muscle-invasive disease, confined to the bladder mucosa/submucosa. This stage is usually managed with local treatment and surveillance, and has a particularly high prevalence due to the nonaggressive natural history of this disease (Allory et al., 2014).
It is well known that single nucleotide polymorphisms (SNPs) contribute to interindividual differences in cancer susceptibility(Dudek et al., 2013). DNA from different individuals is identical for most base positions; however, variants are observed approximately every 500 bases. If a variant occurs in more than 1% of the population, it is defined as a single nucleotide polymorphism. Today, up to 900,000 SNPs and 900,000 copy number variations can be determined in a single analysis on SNP chips(Klaus et al., 2011).Single nucleotide polymorphisms are the most common type of variation in genetic sequences. Accumulating evidence suggests that SNPs are closely related to the occurrence and development of tumors (Tengyun, et al., 2021).
Genome-wide association studies have become a powerful approach for identifying disease susceptibility variants. The first of GWAS for bladder cancer was conducted in 2008, by Kiemeney et al., they reported it on a European population and two common variants associated with BC—rs9642880 G>T upstream of MYC on chromosome 8q24.21 and rs710521 A>G located near TP63 on chromosome 3q28. Since then most GWAS for bladder cancer were mostly for European and Chinese populations (Wang et al., 2009). This approachencouraged us to evaluate the association between rs710521 at 3q28 and bladder cancer risk in Egyptian population. Most of the variants reported by GWAS are merely associated markers and the underlying molecular bases of these associations are unknown. Therefore, we hypothesized that if the 3q28& 8q24. region plays a role in susceptibility to bladder cancer, there must be a casual risk variant that might be related with some regulated elements controlling the expression of the key genes. To localize the causal variant at 3q28 & 8q24 associated with bladder cancer susceptibility, an additional attempt to decrease invasivenessinvolved extracting genomic DNA from urine samples rather than blood.Using molecular diagnostic markers for detecting several diseases is one of the most growing areasin the world, as reported by (Gu et al., 2011) that the diagnosis of bladder cancer includes urine-based Markers Cystoscopy, cytology, and biopsy. This study aims to assess the cumulative effect of some single nucleotide polymorphism (rs964880 G>T and rs710521 A>G) in measuring the genetic susceptibility of bladder cancer in Egyptian patients by genotyping using RFLP and sequencing techniques.
c-Myc has been reported to be the master regulator of metabolic processes involved in cell proliferation. c-Myc is deregulated in many human cancers in which it triggers tumorigenesis through the transcriptional modulation of many genes (Hammam et al., 2017). In fact, it has been recently proposed that c-Myc is a “general” transcription factor, in the sense that high levels of c-Myc in tumor cells produce elevated levels of transcripts from the existing gene expression program of tumor cells (Charles et al., 2012). As a master regulator of many of the processes associated with cancer, c-Myc is an attractive therapeutic target(Miller et al., 2012).
Materials and Methods
Selection of study population
This study was conducted on150 bladder cancer patients, and 50 healthy volunteers from (Theodor Bilharz Research Institute) at the beginning of 2020. It included a patient criterion of diagnosed bladder cancer patients who did not receive any type of therapy, all cases were histologically diagnosed directly after removal the tissue specimens from the patients and stored at -80°C and used when necessary. All cases were processed routinely into paraffin sections and pathologically diagnosed for tumor stage and grade. According to the health organization 1973 grading of the urothelial carcinoma: well-differentiated (grade1, G1), moderately differentiated (grade2, G2) or poorly differentiated (grade3, G3). The exclusion criteria included previous cancer, metastasized cancer from other or unknown origins and previous radiotherapy or chemotherapy. Before recruitment, a signed informed consent was obtained fromall participants. The research protocol was approved by the institutional review board ofthe Ethics Committee of Theodor Bilharz Research Institute, in accordance with the institutional guidelines.Twenty ml urine samples from 150 BC patients and 50 healthy volunteers were collected, centrifuged at 4000 rpm at 25°C for 10 min, samples were processed immediately when received, aliquoted and stored at -20°C till use.
DNA Extraction from Urine Samples
DNA extraction was carried out using Qiagen Kits (Hilden, Germany) as per manufacturer instructions, the purified DNA was dissolved in 50 µl of water, measured on a Nanodrop ND-2000c (Thermo Scientific, Waltham, MA, USA), and stored at −20°C for further analyses.
PCR of Rs9642880 & Rs710521 genomic DNA fragments
The rs9642880 G>T and rs710521 A>G polymorphisms were amplified by the polymerase chain reaction using the following primers:
5-CCACCACTCTCAGCCTTTTC-3 (forward) and 5-TGGGATTACAAGTGTGAACCTG-3 (reverse) for rs9642880 G>T polymorphism and 5-TTATCAGTGCTTATTCATTTTTGAAG-3 (forward) and 5-TTTCCAAGAGATTTCAGTGTACAAC-3 (reverse) for rs710521 A>G polymorphism(Wang et al, 2009). These primers generated PCR products of 203 bp and 131 bp respectively. The reaction was conducted at Bio-RAD T100Thermal cycler as follows:initial denaturation at 95°C for 3 min,then amplification was done using 35 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 1 min, followed by final extension at 72°C for 5 min. Seven μL from completed PCR product were loaded on 3% agarose gel and samples were run for 45 min at 100V on a Bio-RAD electrophoresis chamber, with 5 μL of 100-1,000 bp DNA ladder RTU used as a marker.
RFLP analysis and Molecular Detection via Sequencing
An aliquot of 5 μl of each PCR product was subjected to restriction digestion at 37 °C for 30 min in a final volume of 20 μl of digestion buffer. Thermo Scientific FastDigestStyI was used to digest rs9642880, whileMboII was used to digest rs710521.Then 7μl of the digested samples were resolved on 3% agarose gel for electrophoresis and were run for 45 min at 100V to detect the fragments allele in RFLP patterns (Wang et al., 2009). Gel results obtained using a UV imaging system and selected bands were purified from the gel and sequenced using Applied Biosystems 3500/3500xL Genetic Analyzer at Colors for Research, Medical Laboratories, Egypt. The results of the sequence were analyzed, revised, lined up alongside the NCBI samples using BLAST.
Histopathological study
Thisstudy included 150 formalin-fixed and paraffin-embedded sections of bladder biopsiesobtained from patients who had undergone radical cystectomy with pelvic lymphadenectomy at urology department of Theodor Bilharz Research Institute, Cairo, Egypt, during the period between January 2019 and December 2019 and also 50 non-malignant specimen were included as a control. All specimens were processed at Pathology Department ofTheodor Bilharz Research Institute. Clinical data were collected from the medical records and included: age, gender, lymph node andstage grouping. All patients in this study did not receive neoadjuvant chemotherapy or radiotherapy.
All hematoxylin and eosin (H and E) stained sections were re-evaluated independently by two pathologists to determine the variant and divergent differentiation of UC, presence of bilharziasis, grading and pathological TNM stage(Kyritsi et al., 2018). The tumors were staged according to American Joint Committee on Cancer (AJCC) staging system, 8th edition (Magers et al., 2019) and histological variants were classified according to World Health Organization (WHO) classification of tumors of the urinary tract (Humphrey et al., 2016).
c-Myc immunohistochemistry
REFERENCE (Aboushousha T, Hammam O, Aref A, Kamel A, Badawy M, Abdel Hamid A. Tissue Profile of CDK4 and STAT3 as Possible Innovative Therapeutic Targets in Urinary Bladder Cancer. Asian Pac J Cancer Prev. 2020 Feb 1;21(2):547-554. doi: 10.31557/APJCP.2020.21.2.547. PMID: 32102537; PMCID: PMC7332140.
Formalin-fixed and paraffin-embedded tissue sections, each of 4-μm-thick, were cut from each block and stained immunohistochemically with c-Myc antibody ((A-14): sc-789, rabbit polyclonal affinity purified antibody raised against c-terminus of c-Myc protein of human origin, Santa Cruz Biotechnology, Santa Cruz, CA, USA) using the manufacturer’s protocol. Tissue sections were first incubated in the oven for 1 hour at 95°C, dewaxed, rehydrated and submitted for antigen retrieval by incubating the sections in PT-link od DAKO autoimmunostainer (Dako EnVisionTM FLEX). Target Retrieval Solution, Citrate buffer, Low PH 6.1 (50x) (Code DM829). After cooling at room temperature, sections were washed in distilled water for 2 minutes. The endogenous peroxidase was blocked using Dako EnVisionTM FLEX Peroxidase Blocking Reagent (Code SM801) for 5 minutes, then rinsed twice with diluted phosphate buffered saline (PBS) using Dako EnVisionTM FLEX Wash Buffer (20x) (Code DM831). The samples were incubated overnight (~20 hours) at 4°C with the primary antibody at dilution of 1:100 (optimum dilution). On the next day, the slides were washed with PBS twice. The slides were then treated with a secondary antibody applied for 20 min at room temperature using Dako EnVisionTM FLEX HRP (Horseradish peroxidase) (Code SM802) and again washed twice with PBS. Finally, the sections were visualized by diaminobenzidine staining using Dako EnVisionTM FLEX DAB and Chromogen (Code DM827) and Substrate Buffer (Code SM803), then the sections were counterstained with ready to use hematoxylin using Dako EnVisionTM FLEX Hematoxylin (Code SM806), dehydrated with gradient ethanol, cleared with xylene and sealed for microscopic examination. Burkitt lymphoma tissue sections served as positive and negative (By omitting the primary antibody) controls.(Aboushousha et al., 2020)
Evaluation of c-Myc immunostaining
The expression of c-Myc in each tumor section was assessed independently by 2 pathologists who had no previous knowledge of clinical data. Staining expression was assessed with regard to percentage of positive cells, cellular location and intensity. Histoscore (H-score) system was applied to quantify c-Myc expression with a range of 0-300. H-score was achieved by semi-quantitative assessment of both the intensity, classified as negative (0), weak (1+), moderate (2+) and strong (3+) and the percentage of positive cells according to the following formula (Elwy et al., 2019).
H-score = 1 x (% cells 1+) + 2 x (% cells 2+) + 3 x (% cells 3+).
H-score was calculated for each case and the mean score was calculated for each group of cases.
Statistical analysis
The data were analyzedusing Microsoft Excel 2016 and statistical package for social science ‘IBM SPSS Statistics for Windows, version 26 (IBM Corp., Armonk, N.Y., USA)’. Continuous normally distributed variables were represented as mean±SD. with 95% confidence interval and using the frequencies and percentage for categorical variables; a P value < 0.05 was considered statistically significant. To compare the means of normally distributed variables between groups, the Student’s t test was performed, while χ2 test was used to determine the distribution of categorical variables between groups. Logistic regression analysis was done to identify predictor genotype associated with the risk of bladder cancer. In addition, Hardy-Weinberg equilibrium (HWE) was used to assess independence between the alleles inherited from the parents.
Haplotype analysis
The data were analyzed using HAPLOTYPE ANALYSIS software v1.05 is a software written in Visual Basic for Applications (VBA) within Excel (Eliades N-G and Eliades D, 2009). It is a new software for analysis of data from organell which based on the frequency of haplotypes. Population genetic structure from the population samples (inter-population analysis) is computed, utilizing: Nei’s minimum genetic distance (Nei, 1987) and genetic differentiation among the populations and contribution of each of them to the total diversity (Finkeldey and Murillo, 1999).
Results
The present study included 150 bladder cancer subjects and 50 unaffected controls. Demographic, clinical characteristics of the study participants and their histopathological data are represented in (Table 1). Patients were confirmed bladder cancer by histopathological examination of bladder mass biopsies, without radiation, chemotherapy or had a history of cancer, metastasized cancer from other organs. Controls were selected from individuals participating in a community-based screening program for non-infectious diseases conducted during the same period when the cases were recruited. All of the participants were genetically unrelated Egyptian individuals, and they each donated approximately 20 ml of urine. Smoking information were collected through face-to-face interviewed by trained interviewers and those who had smoked less than an average of one cigarette per day and < 1 year in their lifetime were defined as nonsmokers; while others were considered as smokers (Salim et al., 2012).
Table 1.
Demographic, Clinical and Histopathological Data of Bladder Cancer Patients
| Control | Cases | P- value | ||
|---|---|---|---|---|
| N=50 | N=150 | |||
| Age | Mean±SD | 44.5±12.4 | 59.4±8.6 | 0.001** |
| Min - Max | 22.0 - 68.0 | 26.0 - 73.0 | ||
| Sex | Female | 17 (34.0%) | 21 (14.0%) | 0.1 |
| Male | 33 (66.0%) | 129 (86.0%) | ||
| Bilhariziasis | Negative | 50 (100.0%) | 33 (22.0%) | 0.001** |
| Positive | 0 (0.0%) | 117 (78.0%) | ||
| Smoker | No | 50 (100.0%) | 39 (26.0%) | 0.001** |
| Yes | 0 (0.0%) | 111 (74.0%) | ||
| Clinical Ex. | Negative | 50 (100.0%) | 0 (0.0%) | N.A |
| T1 | 0 (0.0%) | 21 (14.0%) | ||
| T2 | 0 (0.0%) | 75 (50.0%) | ||
| T3 | 0 (0.0%) | 54 (36.0%) | ||
| Cytology | Negative | 50 (100.0%) | 54 (36.0%) | 0.001** |
| Positive | 0 (0.0%) | 96 (64.0%) | ||
| Histopathological characteristics | Negative | 50 (100.0%) | 0 (0.0%) | N.A |
| Squamous cell carcinoma | 0 (0.0%) | 30 (20.0%) | ||
| Urothelial carcinoma | 0 (0.0%) | 120 (80.0%) | ||
| Stage of Tumor | Negative for malignancy | 50 (100.0%) | 0 (0.0%) | N.A |
| Non muscle invasive bladder cancer(NMIBC) | 0 (0.0%) | 12 (8.0%) | ||
| Muscle invasive bladder cancer(MIBC) | 0 (0.0%) | 123 (82.0%) | ||
| Superficial bladder cancer | 0 (0.0%) | 15 (10.0%) | ||
| Serum creatinine | Normal | 50 (100.0%) | 123 (82.0%) | 0.001** |
| High | 0 (0.0%) | 27 (18.0%) | ||
| Lymph node | Negative for malignancy | 50 (100.0%) | 75 (50.0%) | 0.001** |
| Positive | 0 (0.0%) | 75 (50.0%) | ||
| Gradesof Tumor | Negative | 50 (100.0%) | 0 (0.0%) | N.A |
| GI | 0 (0.0%) | 15 (10.0%) | ||
| GII | 0 (0.0%) | 39 (26.0%) | ||
| GIII | 0 (0.0%) | 96 (64.0%) |
Age is represented as Mean ± SD; the data were analyzed by student t test. While Sex, Bilhariziasis, Smoker, Clinical Ex., Cytology, Histopathological characteristics, Stage of Tumor, Serum creatinine, Lymph node, and Grades of Tumor are represented as F (%) frequency and percent; the data were analyzed by X2 test. * P-value <0.05 is significant, ** P. value <0.01 is highly significant.
Molecular detection with RFLP technology and Sequencing
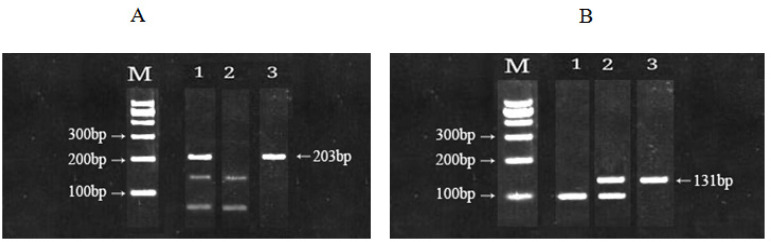
The results of the molecular genotypepatterns of rs9642880 G>T and rs710521 A>G are presented in (Fig.1, A&B) which showing electrophoresis of PCR products in the polymorphism on 3% agarose gel for 45 min at 100 V. Figure 1 (A), the first column represents the size marker 100-1000 bp, followed by the genotype patterns of rs9642880 GT, TT and GG. while Figure 2 (B), the first column represents the size marker 100-1000 bp., followed by the genotype patterns of rs710521 AA, AG and GG. The molecular genotyping was performed using RFLP technique to identify the genetic diversity. The genotype distributions were significantly different between patients and controls.
Figure 1.
A, Electrophoresis in 3% agarose gel showed genotype patterns of rs9642880, M, DNA marker, genotypes (GT) ( TT) and(GG) in columns 1,2 and 3 respectively; B, The genotype patterns of rs710521, M, DNA marker, genotypes (AA) (AG) and (GG) in columns 1,2 and 3 respectively
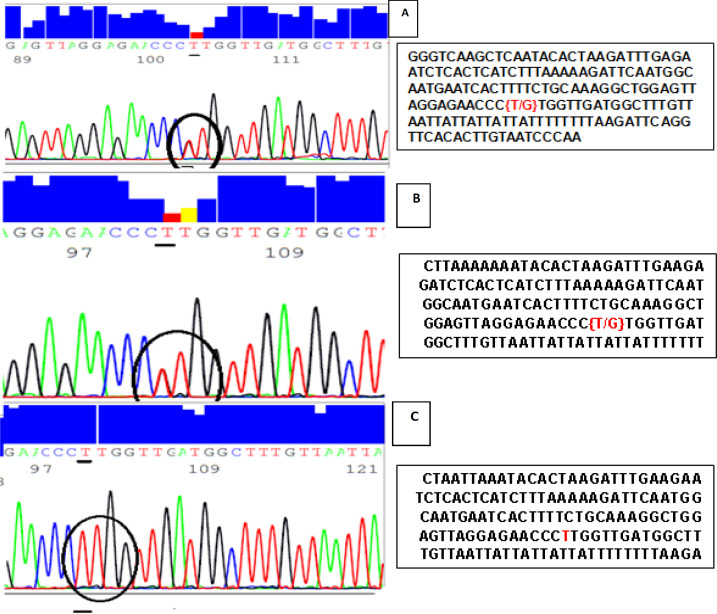
Figure 2.
Forward Sequencing Peaks for PCR Amplified rs9642880 Fragment for three Different Bladder Cancer Patients. Graph A and B indicates that these two patients have GT genotype while graph C indicates that this patient has TT genotype. Related sequences in the region of rs9642880 polymorphism was highlighted in the sequence text, circled and underlined in the graph
The samples were analyzed using Applied Biosystems 3500/3500xL Genetic Analyzer at Colors for Research, Medical Laboratories, Egypt. Figure 2 illustrates peaks of 3 forward sequenced samples for 3 different patients. Graph A and B indicates that these two patients have GT genotype while graph C indicates that this patient has TT genotype.
Hardy-Weinberg Equilibrium(HWE)
Hardy-Weinberg equilibrium (HWE) is useful to assess independence between the alleles inherited from the parents.In this study, tests of HWE were carried out for rs9642880 G/T upstream of MYC on chromosomes 8q24 and rs710521 A/G located near TP63 on chromosome 3q28 among controls and bladder cancer patients independently.
The genotype distribution of rs9642880 and rs710521 polymorphisms was checked in the controls and cases, the results showed consistency with HWE in the control group (P. value =0.69 and 0.81, respectively) and for cases (P. value =0.43 and 0.80, respectively). Therefore, our controls and selected bladder cancer subjects were a typical Mendelian population (Table 2).
Table 2.
Observed and Expected Variables Genotypes for Controls and Cases
| SNPs Genotype | N=50 | Observed results | Expected results | p-value | ||
|---|---|---|---|---|---|---|
| (Current Study) | (H-W equilibrium) | |||||
| Control | rs9642880 | GG | 27 | 0.54 | 0.196 | 0.69 |
| GT | 15 | 0.3 | 0.494 | |||
| TT | 8 | 0.16 | 0.31 | |||
| rs710521 | AA | 6 | 0.12 | 0.025 | 0.81 | |
| AG | 17 | 0.34 | 0.265 | |||
| GG | 27 | 0.54 | 0.71 | |||
| Total | 50 | 1 | 1 | |||
| Cases | rs9642880 | GG | 90 | 0.6 | 0.153 | 0.43 |
| GT | 9 | 0.06 | 0.477 | |||
| TT | 51 | 0.34 | 0.37 | |||
| rs710521 | AA | 30 | 0.2 | 0.06 | 0.8 | |
| AG | 69 | 0.46 | 0.37 | |||
| GG | 51 | 0.34 | 0.57 | |||
| Total | 150 | 1 | 1 |
H-W equilibrium, Hardy-Weinberg equilibrium; n, Number; P. Value is depending on the X2 test; *P. value <0.05 is significant,; ** P. value <0.01 is highly significant.
In the present study, genotype distribution and allele frequency were analyzed in rs9642880 and rs710521, aiming to elucidate their association with bladder cancer risk.
By comparing the distribution of rs9642880 polymorphism genotypes in bladder cancer with healthy controls, it revealed a statistically higher significant frequency of GT genotype in controls than cases (P. value = 0.001) with protection rate from bladder cancer [OR (95% C.I) =0.18(0.071 - 0.457), (P. value =0.001)], while TT genotype showed a statistically higher significant in cases more than controls (P. value = 0.01) with higher risk development to the bladder cancer [OR (95% C.I) =1.912(0.809 - 4.522), (P. value =0.03)]. Interestingly, it was found that the results at the allele level G/T upstream of c-Myc on chromosomes 8q24 were non-significant (Table 3).
Table 3.
Genotyping Distribution of the Studied Variables
| SNPs | Genotype /Allele | Control | Cases | aP. value | OR | 95% C.I | bP. value |
|---|---|---|---|---|---|---|---|
| N=50 | N=150 | ||||||
| rs9642880 | GG | 27 (54.0%) | 90 (60.0%) | 0.2 | 1(reference) | ||
| GT | 15 (30.0%) | 9 (6.0%) | 0.001** | 0.18 | 0.071 - 0.457 | 0.001** | |
| TT | 8 (16.0%) | 51 (34.0%) | 0.01* | 1.912 | 0.809 - 4.522 | 0.03* | |
| G Allele | 69 (0.690) | 189 (0.630) | 0.3 | 1 (reference) | |||
| T Allele | 31 (0.310) | 111 (0.370) | 1.307 | 0.805 - 2.122 | 0.3 | ||
| rs710521 | AA | 6 (12.0%) | 30 (20.0%) | 0.06 | 1 (reference) | ||
| AG | 17 (34.0%) | 69 (46.0%) | 0.02* | 1.812 | 0.291 - 3.262 | 0.04* | |
| GG | 27 (54.0%) | 51 (34.0%) | 0.001** | 0.378 | 0.140 - 1.020 | 0.04* | |
| A Allele | 29 (0.290) | 129 (0.430) | 0.01* | 1.847 | 1.133 - 3.011 | 0.01* | |
| G Allele | 71 (0.710) | 171 (0.570) | 1 (reference) |
Genotype /Allele are represented as F (%) frequency and percent; the data were analyzed by X2 test; * P. value <0.05 is significant, ** P. value <0.01 is highly significant; OR, Odds Ratio; CI, Confidence Interval; *P value ≤ 0.05 significant; **P value ≤ 0.01 highly significant; aP. Value is depending on the X2 test, while bP. Value is depending onLogistic Regression analysis.
For to rs710521 polymorphism genotypes, the results showed a statistically higher significant frequency of AG genotype in cases than controls (P. value = 0.02) with higher risk development to bladder cancer [OR (95% C.I) =1.812(0.291 - 3.262), (P. value =0.04)], while GG genotype showed a statistically higher significant in controls more than cases (P. value = 0.001) with protection rate from bladder cancer [OR (95% C.I) =0.378(0.140 - 1.020), (P. value =0.04)].It was striking to notice that, the A allele is more significant in cases than controls and on the contrary for the G allele (P. value = 0.01), with higher risk development to bladder cancer [OR (95% C.I) =1.847(1.133 - 3.011), (P. value =0.01)] (Table 3).
According to the association study between TT genotype of rs9642880 polymorphism with the studied parameters, a significant association was found with T3 regarding clinical examination (P. value =0.001) and susceptibility to bladder cancer is 8.0 times (95% C.I =2.409 - 26.567, P. value = 0.001). Also, with the cases which have positive cytology results to the bladder cancer (P. value =0.03) and susceptibility to bladder cancer is 4.8 times (95% C.I =1.059 - 21.753, P. value = 0.04) (Table 4).
Table 4.
The Association between the Significant Genotypes of “rs9642880” with Bladder Cancer Risk
| rs9642880 | Associations | Risk assessment | |||||
|---|---|---|---|---|---|---|---|
| GT | TT | aP. value | OR | 95% C.I | bP. value | ||
| n=9 | n=51 | ||||||
| Sex | Female | 3 (33.3%) | 12 (23.5%) | 0.02* | 1.09 | 0.969 - 1.226 | 0.1 |
| Male | 6 (66.7%) | 39 (76.5%) | |||||
| Bilhariziasis | Negative | 3 (33.3%) | 18 (35.3%) | 0.6 | 0.917 | 0.204 - 4.109 | 0.9 |
| Positive | 6 (66.7%) | 33 (64.7%) | |||||
| Smoker | No | 3 (33.3%) | 15 (29.4%) | 0.5 | 1.2 | 0.265 - 5.438 | 0.8 |
| Yes | 6 (66.7%) | 36 (70.6%) | |||||
| Clinical Ex. | T1 | 0 (0.0%) | 6 (11.8%) | 0.2 | - | - | - |
| T2 | 6 (66.7%) | 21 (41.2%) | 0.01* | 3.5 | 1.413 - 8.672 | 0.01* | |
| T3 | 3 (33.3%) | 24 (47.1%) | 0.001** | 8 | 2.409 - 26.567 | 0.001** | |
| Cytology | Negative | 6 (66.7%) | 15 (29.4%) | 0.03* | 4.8 | 1.059 - 21.753 | 0.04* |
| Positive | 3 (33.3%) | 36 (70.6%) | |||||
| Histopathological characteristics |
Scc | 3 (33.3%) | 15 (29.4%) | 0.5 | 1.2 | 0.265 - 5.438 | 0.8 |
| UC | 6 (66.7%) | 36 (70.6%) | |||||
| Stage | NMIBC | 0 (0.0%) | 3 (5.9%) | 0.4 | - | - | - |
| MIBC | 9 (100.0%) | 42 (82.4%) | 0.02* | 1.667 | 2.272 - 7.587 | 0.1 | |
| Superficial bladder cancer | 0 (0.0%) | 6 (11.8%) | 0.2 | - | - | - | |
| Serum Creatinine | Normal | 6 (66.7%) | 39 (76.5%) | 0.4 | 0.615 | 0.133 - 2.841 | 0.5 |
| High | 3 (33.3%) | 12 (23.5%) | |||||
| Lymph Node | Negative | 0 (0.0%) | 27 (52.9%) | 0.03* | 0.111 | 0.013 - 0.954 | 0.06 |
| Positive | 9 (100.0%) | 24 (47.1%) | |||||
| Tumor Grade | GI | 0 (0.0%) | 6 (11.8%) | 0.2 | - | - | - |
| GII | 3 (33.3%) | 15 (29.4%) | 0.4 | 1.035 | 1.448 - 10.271 | 0.5 | |
| GIII | 6 (66.7%) | 30 (58.8%) | 0.1 | 1.123 | 2.081 - 8.013 | 0.2 | |
SCC, squamous cell carcinoma; TCC, non-papillary transitional cell carcinoma; Ta, Superficial bladder cancer; NMIBC, Non muscle invasive bladder cancer; MIBC, Muscle invasive bladder cancer; OR, Odds Ratio; CI, Confidence Interval; *P value ≤ 0.05 significant; **P value ≤ 0.01 highly significant; aP. Value is depending on the X2 test, while bP. Value is depending onLogistic Regression analysis.
With regard to the association between AG genotype of rs710521 polymorphism, a significant association was observed in patients withBilharziasis (P. value =0.02) and susceptibility to bladder cancer is 2.36 times (95% C.I =1.143 - 7.907, P. value = 0.03). In addition, the patients with T2 and T3 regarding clinical examination (P. value= 0.01 and 0.04) respectively and susceptibility to bladder cancer is 4.0 times (95% C.I =1.129 - 14.175, P. value = 0.03) in T2 and 1.9 times (95% C.I =0.958 - 3.384, P. value = 0.05) in T3 (Table 5).
Table 5.
The Association between the Significant Genotypes of “rs710521” with Bladder Cancer Risk
| rs710521 | Associations | Risk assessment | |||||
|---|---|---|---|---|---|---|---|
| AG | GG | aP. value | OR | 95% C.I | bP. value | ||
| n=69 | n=51 | ||||||
| Sex | Female | 12 (17.4%) | 6 (11.8%) | 0.3 | 1.579 | 0.550 - 4.535 | 0.4 |
| Male | 57 (82.6%) | 45 (88.2%) | |||||
| Bilhariziasis | Negative | 9 (13.0%) | 15 (29.4%) | 0.02* | 2.36 | 1.143 - 7.907 | 0.03* |
| Positive | 60 (87.0%) | 36 (70.6%) | |||||
| Smoker | No | 30 (43.5%) | 6 (11.8%) | 0.1 | 0.769 | 0.174 - 1.307 | 0.1 |
| Yes | 39 (56.5%) | 45 (88.2%) | |||||
| Clinical Ex. | T1 | 30 (43.5%) | 33 (64.7%) | 0.01* | 1.1 | 0.671 - 1.804 | 0.7 |
| T2 | 12 (17.4%) | 3 (5.9%) | 0.01* | 4 | 1.129 - 14.175 | 0.03* | |
| T3 | 27 (39.1%) | 15 (29.4%) | 0.04* | 1.9 | 0.958 - 3.384 | 0.05* | |
| Cytology | Negative | 21 (30.4%) | 15 (29.4%) | 0.5 | 1.05 | 0.476 - 2.316 | 0.9 |
| Positive | 48 (69.6%) | 36 (70.6%) | |||||
| Histopathological characteristics |
Scc | 15 (21.7%) | 9 (17.6%) | 0.4 | 1.296 | 0.517 - 3.251 | 0.6 |
| UC | 54 (78.3%) | 42 (82.4%) | |||||
| Stage | NMIBC | 3 (4.3%) | 3 (5.9%) | 0.5 | 1 | 0.202 - 4.955 | 0.9 |
| MIBC | 57 (82.6%) | 42 (82.4%) | 0.9 | 1.357 | 0.911 - 2.022 | 0.1 | |
| Superficial bladder cancer | 9 (13.0%) | 6 (11.8%) | 0.9 | 1.5 | 0.534 - 4.214 | 0.4 | |
| Serum Creatinine | Normal | 57 (82.6%) | 36 (70.6%) | 0.09 | 1.979 | 0.832 - 4.706 | 0.1 |
| High | 12 (17.4%) | 15 (29.4%) | |||||
| Lymph Node | Negative | 30 (43.5%) | 24 (47.1%) | 0.5 | 0.865 | 0.418 - 1.791 | 0.7 |
| Positive | 39 (56.5%) | 27 (52.9%) | |||||
| Tumor Grade | GI | 9 (13.0%) | 6 (11.8%) | 0.9 | 1.5 | 0.534 - 4.214 | 0.4 |
| GII | 18 (26.1%) | 12 (23.5%) | 0.7 | 1.5 | 0.723 - 3.114 | 0.3 | |
| GIII | 42 (60.9%) | 33 (64.7%) | 0.5 | 1.273 | 0.807 - 2.008 | 0.3 | |
SCC, squamous cell carcinoma; TCC, non-papillary transitional cell carcinoma; Ta, Superficial bladder cancer; NMIBC, Non muscle invasive bladder cancer; MIBC, Muscle invasive bladder cancer; OR, Odds Ratio; CI, Confidence Interval; *P value ≤ 0.05 significant; **P value ≤ 0.01 highly significant; aP. Value is depending on the X2 test, while bP. Value is depending onLogistic Regression analysis.
Haplotype association analysis
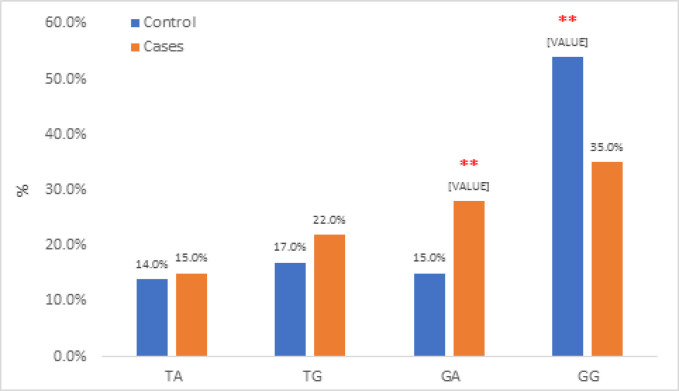
In addition to the individual statistical analysis of each polymorphism, the association analysis of haplotypes was also performed. The collection of alleles at all variable sites in a chromosome region is called haplotype. Haplotype analysis involves different combinations of the two studied variables (rs9642880 and rs710521), producing 4 different haplotypes (TA, TG, GA and GG) whichwere included in the statistical analysis (ّFigure 3, Table 6).
Figure 3.
Total Haplotype Association between Controls and Cases
Table 6.
Summary of Haplotype Association Analysis between Controls and Cases
| Haplotype association | Control | Cases | aP. value | OR | 95% CI | bP. value |
|---|---|---|---|---|---|---|
| N (%) | N (%) | |||||
| (1 0) TA | 14 (14.0%) | 45 (15.0%) | 0.8 | 1(reference) | ||
| (1 1) TG | 17 (17.0%) | 66 (22.0%) | 0.2 | 1.208 | 0.541 - 2.695 | 0.6 |
| (0 0) GA | 15 (15.0%) | 84 (28.0%) | 0.001** | 5.6 | 3.233 - 9.700 | 0.001** |
| (0 1) GG | 54 (54.0%) | 105 (35.0%) | 0.001** | 0.514 | 0.370 - 0.714 | 0.001** |
The data represented as frequency and percent for the studied variants associations; The sequence of alleles depending on the following variable order respectively: rs9642880, rs710521; Binary haploid data observed for RFLP- PCR (0 if absent, 1 if Present); OR, Odds Ratio; CI, Confidence Interval; *P value ≤ 0.05 significant; **P value ≤ 0.01 highly significant; aP. Value is depending on the X2 test, while bP. Value is depending onLogistic Regression analysis
Haplotype frequencies for the two studied variables combination were estimated for bladder cancer cases and controls separately and the risk of haplotype was also estimated (Table 6). The results showed that the most significant prevalent haplotype sequence is GA in cases = 84(28.0%) while in controls = 15(15.0%), (P. value =0.001) with the risk susceptibility to bladder cancer is 5.6 times (95% C.I =3.233 - 9.700, P. value = 0.001). In contrast, the GG haplotype increased in controls than cases with (P. value = 0.001) with the protection susceptibility to bladder cancer is 0.514 times (95% C.I =0.370 - 0.714, P. value = 0.001) (Table 6).
Immunohistochemical profile of c-Myc
Based on knowledge that c-Myc is a transcription factor protein, so its nuclear localization in tumor cells was expected. The cytoplasmic localization for some unknown reason and of a controversial issue was also detected. Therefore, in this study we evaluated both expression patterns collectively and correlated both of them with different histopathological parameters.
Normal urothelium adjacent to tumor showed weak to moderate cytoplasmic expression and focal nuclear staining. Stromal and endothelial cells showed focal cytoplasmic and nuclear expression. All 150 tumor sections showed positive immunoreactivity for c-Myc in the nucleus and the cytoplasm. Meanwhile, tumor cells showed variability in nuclear expression according to different histopathological parameters that were studied (Figure 4).
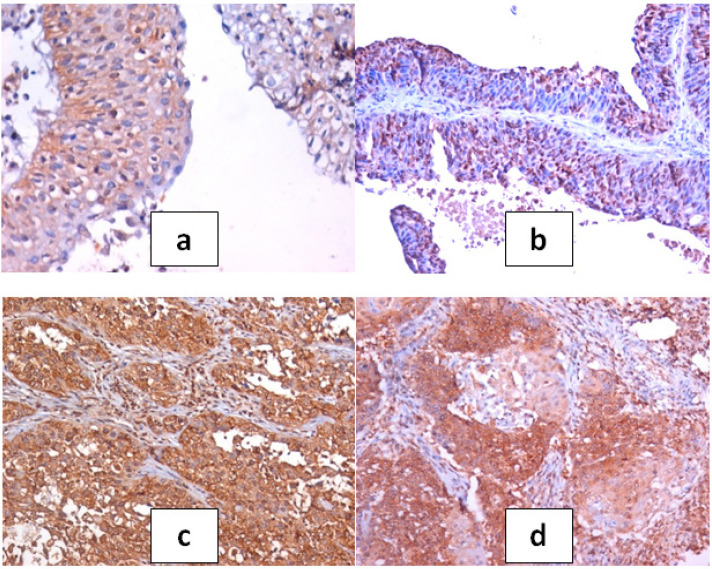
Figure 4.
a, Section in urinary bladder tissue of mild case of cystitis showing discrete nuclear and cystoplasmic spots of c-Myc positivity within the urothelium (c-Myc IHC, DAB, x400); b, Moderate nuclear positivity in papillary low grade TCC (c-Myc IHC, DAB, x200); c, Dense diffuse nuclear and cytoplasmic positivity in case of MIBC of high grade TCC (c-Myc IHC, DAB, x200); d, Dense patchy positivity in case of MIBC of SCC type (c-Myc IHC, DAB, x200)
H-scores of c-Myc expression in tumor tissue are summarized in (Table 7). Comparison between median H-scores of c-Myc nuclear expression of non-malignant and malignant cases was statistically significant (p<0.001). The highest median H-score was noted with MIBC of high grade anaplasia and positive lymph node metastasis especially of SCC type of bladder cancer and the lowest one was noted with the superficial bladder cancer of PTCC of low grade anaplasia with negative lymph node metastasis. Comparison between median H-scores of bilharzial associated and non-associated bladder cancer showed higher values of mean H-scores on bilharzial associated cases, however this could not be taken as a fact because it may influenced by the type of tumor, grade and stage of invasion.
Table 7.
Differential Expression of c-Myc in Relation to Variable Histopathological Parameters of Bladder Cancer
| Histopathological Parameter |
N | Mean H-score |
Std. Deviation |
|---|---|---|---|
| Nonmalignant & cystitis | 50 | 44.17 | 41.17 |
| UC (all) | 122 | 176.41## | 97.36 |
| PTCC | 41 | 162.5 | 115.51A |
| TCC | 81 | 177.50$ | 84.97 |
| SCC | 28 | 220.03##* | 64.49 |
| BILHARZIASIS | |||
| POSITIVE | 119 | 183.44 | 96.36 |
| NEGATIVE | 31 | 130.85** | 64.37 |
| GRADE | |||
| G1 | 21 | 82.5 | 58.77 |
| G2 | 34 | 221.11^^ | 60.28 |
| G3 | 95 | 285.00^^$$ | 17.32 |
| STAGE | |||
| Ta | 14 | 42.5 | 8.66 |
| NMIBC | 14 | 160.47@@ | 82.46 |
| MIBC | 122 | 241.43@@"" | 63.35 |
| Histopathological Parameter |
N | Mean H-score |
Std. Deviation |
| LYMPH NODES | |||
| -ve | 75 | 151.36 | 96.36 |
| +ve | 75 | 231.67&& | 63.37 |
| Total | 200 | 144.35 | 102.29 |
UC, urothelial carcinoma; PTCC, Papillary transitional cell carcinoma; TCC, non-papillary transitional cell carcinoma; SCC, squamous cell carcinoma; Ta, Superficial bladder cancer; NMIBC, Non muscle invasive bladder cancer; MIBC, Muscle invasive bladder cancer; ##, High significant difference with non-malignant cases; $, Significant difference with PTCC; *, Significant difference with UC; **, High significant difference with positive bilharzial cases; ^^, High significant difference with G1; $$, High significant difference with G2; @@, High significant difference with Ta; "", High significant difference with NMIBC; &&, High significant difference with +ve LN.
Discussion
Most patients with advanced bladder cancer present with painless gross hematuria sometimes accompanied by the sensation of urgently needing to urinate, and even urge incontinence. When a patient is suspected of having bladder cancer, the initial assessment involves cystoscopy and imaging of the urinary tract through techniques such as sonography and computed tomography urography. However, tiny papillary tumors and carcinoma in situ are difficult to detect using standard white-light cystoscopy or urinary tract imaging, which may account for the misdiagnosis or early recurrence of bladder cancer(Lin, et al., 2021). This difficulty has led scientists to identify new numerous noninvasive biomarkers in urine that can serve as diagnostic indicators of bladder cancer.
Bladder cancer in Egypt is still a major problem where patients presented with advanced stage of the disease. Also, the survival rate is still low. Early detection is needed to detect the disease at early stage. Many advances have been made to discover a reliable marker that may detect the disease in its early stages (Samah et al., 2015).Genome Wide Association study identified two common variants on different chromosomes that confer susceptibility to bladder cancer. The two variants are single nucleotide polymorphisms rs9642880 G>T on 8q24and rs710521 A>G on 3q28 (Wang et al., 2009). The aim of this study is to genotype rs9642880 G>T and rs710521 A>G and to evaluate the association of these variants to the risk of bladder cancer in Egyptian population using PCR- RFLP and sequencing.
In the present study, it was found that the increased risk with bladder cancer in the rs9642880 G>T polymorphism was more pronounced in old men, smokers and bilharzial patients.
Regarding the genotype distribution in bladder cancer patients, (Table3) shows the results of patients and controls,for the rs9642880 variant,60% of the patients were genotyped with GG, 34% with genotype TT while the genotype GT is only6%. As for the rs710521, results show 69% of the patients were genotyped with AG,51% for GG and only 30% for the AA genotype.
According to Wang et al., (2009) a similar study that was carried out on Chinese population with results compatible to our study. It shows difference in that rs9642880 –GT/TT genotypes- indicate association to a higher risk of bladder cancer while in our study on the Egyptian population the genotype TT in rs9642880 is the only one that indicates high risk development to bladder cancer but GT indicates protection from bladder cancer.
Regarding rs710521 genotypes in Egyptian patients, the genotype GG indicates protection from bladder cancer, but AG genotype is the only one which indicates high risk to bladder cancer while no genotype was associated with bladder cancer risk in the Chinese population.
The allele frequency of the two variants was analyzed in the Egyptian bladder cancer patients, concerning rs9642880,it was found that the results at the allele level G/T upstream of c-MYC on chromosomes 8q24 were non-significant,this result is opposite to that of the Chinese patients of Wang et al., 2009 who reported that the T allele is associated with bladder cancer risk and also against the result of another study for Cortessis et al., 2010 thatreported a high association of rs9642880 allele T with increased risk of bladder cancer.
Our results have been clarified that the A allele frequency for the variant rs710521is more significant in cases than controls with higher risk development to bladder cancer and this is in contrast to the results of the Chinese patients which reported that no significant association with bladder cancer risk was observed for rs710521(Wang et al., 2009).
Our study is the first study carried out for genotyping the two variants rs9642880 G>T and rs710521 A>G to validate the association with the risk of bladder cancer in Egyptian population. Our results are consistent with the results of Tang et al., (2016) who reported an obvious relationship between rs9642880 polymorphism and increased risk of bladder cancer that was detected in all models and they added that their results pointed to the T allele of rs9642880 confers susceptibility to bladder cancerand this contradicts our results.
Rs710521 is located near TP63 on chromosome 3q28. TP63 has a strong homology with the well-known tumor suppressor protein TP53 (Wen, et al., 2019). However, the significant association between the A allele of rs710521 and bladder cancer risk was first described by Kiemeney et al., 2008 was not confirmed in the study belongs toWang et al., 2009. In order to clarify this discrepancy, we analyzed the allele frequency of rs710521 A>G in 150 urinary bladder cancer patients and 50 controls. Our results clearly showed that the A allele of rs710521 is indeed significantly associated with higher bladder cancer risk in Egyptian population. Analysis of the (A/G) substitution (rs710521) on chromosome 3q28, position and differentiation between the homozygous (A/A), homozygous (G/G) and the heterozygous (A/G) was detected via sequencing.
Several studies reported the association of other multiple genetic variants within the chromosome 8q24 region with the risk of bladder cancer; however, the mechanism still indefinable due to some known genes’ deaths at this locus and further research are being carried out (Wang et al., 2009).
Based on knowledge that c-Myc is a transcription factor protein, so its nuclear localization in tumor cells was expected (Jawad et al., 2019). The cytoplasmic localization for some unknown reason and of a controversial issue was also detected. The highest median H-score was noted with MIBC of high grade anaplasia and positive lymph node metastasis especially of SCC type of bladder cancer and the lowest one was noted with the superficial bladder cancer of PTCC of low grade anaplasia with negative lymph node metastasis. Previous investigation of infiltrating bladder tumors revealed c-Myc overexpression in 56% of T1 tumors and 59% of muscle-infiltrating tumors (Schmitz-Dräger et al., 1997).
The data indicate that elevated mRNA expression as a consequence of increases in c-Myc gene copy number often underlies c-Myc protein over-expression in bladder cancer. This increase may be a consequence of, most frequently, chromosome 8q gain and, occasionally, gene amplification, while in some tumors deregulation of mRNA expression occurs without evident changes in the c-Myc gene copy number.(Cristoph et a;, 1999).
Comparison between median H-scores of c-Myc in bilharzial associated and non-associated bladder cancer showed higher values of mean H-scores on bilharzial associated cases, however this could not be taken as a fact because it may be influenced by the type of tumor, grade and stage of invasion. Experimental studies on ras mutations in vitro have also shown a cumulative effect with other genetic alterations, including those of c-erbB-2 and c-Myc, leading to more aggressive and invasive tumor properties in bilharzial associated bladder cancer (Kroft S H, Oyasu R. 1994).The GWAS approach has been successful in the identification of UBC risk genetic variants. Furthermore, it pointed towards new genes involved in bladder cancer development, possibly leading to the development of new therapies in the future. Until now, individual genetic risk assessment using the risk variants has not been clinically relevant, however, it cannot be excluded that, with increasing numbers of identified variants, genetic counseling of high risk groups, based on GWAS results, will become possible . GWAS results are very promising in bringing us closer to the era of personalized medicine. However, they still pose a particular challenge in the interpretation of their functional significance.
As a conclusion, bladder cancer risk being expected by histopathological grading and staging as well as immunohistologically using c-Myc is associated specifically with TT genotype of rs9642880 on the 8q24 and with the AG genotype of re710521 on 3q28. Identifying the association to risk of bladder cancer using genetic analysis will help in the early detection of the disease. These findings provide new insights into bladder cancer etiology, and the causal variants associated with these risk regions need to be identified by subsequent fine-mapping and functional studies.
Author Contribution Statement
all authors were contributing equally in all aspects of the study.
Acknowledgements
None.
References
- Aboushousha T, Hammam O, Aref A, et al. Tissue Profile of CDK4 and STAT3 as possible innovative therapeutic targets in urinary bladder cancer. Asian Pac J Cancer Prev. 2020;21:547–54. doi: 10.31557/APJCP.2020.21.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allory Y, Beukers W, Sagrera , et al. Telomerase reverse transcriptase promoter mutations in bladder cancer: High frequency across stages, detection in urine, and lack of association with outcome. Eur Urol. 2014;65:360–6. doi: 10.1016/j.eururo.2013.08.052. [DOI] [PubMed] [Google Scholar]
- Altunkol A, Savaş M, Dilmeç F, et al. Detection of CYP1A1 and GSTP1 gene polymorphisms in bladder cancer patients in a Turkish population using a polymerase chain reaction-restriction fragment length polymorphism method. Turk J Urol. 2018;44:125–31. doi: 10.5152/tud.2018.23571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Chiu , C , Wang H, et al. Significant association of ERCC6 single nucleotide polymorphisms with bladder cancer susceptibility in Taiwan. Anticancer Res. 2009;29:5121–4. [PubMed] [Google Scholar]
- Charles Y, Jakob L, Peter B, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortessis V, Yuan J, Berg D, et al. Risk of urinary bladder cancer is associated with 8q24 variantrs 9642880[T] in multiple Racial/Ethnic Groups: Results from the Los Angeles–Shanghai Case–Control Study. Cancer Epidemiol Biomarkers Prev. 2010;19:3150–6. doi: 10.1158/1055-9965.EPI-10-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumberbatch M, Noon A. Epidemiology, aetiology and screening of bladder cancer. Transl Androl Urol. 2019;8:5–11. doi: 10.21037/tau.2018.09.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek A, Grotenhuis A, Vermeulen S, Kiemeney L, Verhaegh G. Urinary bladder cancer susceptibility markers What do we know about functional mechanisms? Int J Mol Sci. 2013;14:12346–66. doi: 10.3390/ijms140612346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwy A, Tarek M, Ahmed R, Mahmoud I. Prognostic value of c-Myc immunohistochemical expression in muscle invasive urothelial carcinoma of the urinary Bladder: A retrospective study. Asian Pac J Cancer Prev. 2019;20:3735–46. doi: 10.31557/APJCP.2019.20.12.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkeldey R, Murillo O. Contributions of subpopulations to total gene diversity. Theoretical Appl Genet. 1999;98:664–8. [Google Scholar]
- Frank C, Bettina S, Bernd J, Schmitz-D , Wolfgang A. Over-expression and amplification of the c-myc gene in human urothelial carcinoma. Int J Cancer. 1999;84:169–73. doi: 10.1002/(sici)1097-0215(19990420)84:2<169::aid-ijc13>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Gu J, Wu X. Genetic susceptibility to bladder cancer risk and outcome. Per Med. 2011;8:365–74. doi: 10.2217/PME.11.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammam O, Aboushousha T, El-Hindawi A, et al. Expression of FGFR3 protein and gene amplification in urinary bladder lesions in relation to schistosomiasis. Open Access Maced J Med Sci. 2017;5:160–6. doi: 10.3889/oamjms.2017.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey A, Holger M, Antonio L, Thomas M, Victor E. The 2016 WHO classification of tumours of the urinary system and male genital organs-part B: Prostate and Bladder Tumours. Eur Urol. 2016;70:106–19. doi: 10.1016/j.eururo.2016.02.028. [DOI] [PubMed] [Google Scholar]
- Jawad Z, Mohammed Z, Jeddoaa A. Molecular detection of MTHFR gene. J Pharm Sci Res. 2019;11:584–9. [Google Scholar]
- Kiemeney L, Thorlacius S, Sulem P, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–12. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus G, Silvia S, Marie-Louise L, et al. Genetic variants in urinary bladder cancer: collective power of the “wimp SNPs”. Arch Toxicol. 2011;85:539–54. doi: 10.1007/s00204-011-0676-3. [DOI] [PubMed] [Google Scholar]
- Kroft S, Oyasu R. Urinary bladder cancer: mechanism of development and progression. Lab Invest. 1994;71:158–74. [PubMed] [Google Scholar]
- Kyritsi F, Loffredo C, Zheng Y, Philips G, Amr S. Urinary bladder cancer in Egypt: Are there gender differences in its histopathological presentation? Adv Urol. 2018;2018:1–7. doi: 10.1155/2018/3453808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Tsai K. Circulating miRNAs Act as Diagnostic Biomarkers for Bladder Cancer in Urine. Int J Mol Sci. 2021;22:4278. doi: 10.3390/ijms22084278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magers M, Lopez-Beltran A, Montironi R, et al. Staging of bladder cancer. Histopathology. 2019;74:112–34. doi: 10.1111/his.13734. [DOI] [PubMed] [Google Scholar]
- Martin C, Franklin D, Colette L, et al. The global epidemiology of bladder cancer: a joinpoint regression analysis of its incidence and mortality trends and projection. Sci Rep. 2018;8:1129. doi: 10.1038/s41598-018-19199-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D, Thomas S, Islam A, Muench D, Sedoris K. C-Myc and cancer metabolism. Clin Cancer Res. 2012;18:5546–53. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York. 1987;Columbia University:Press, pp 219–20. [Google Scholar]
- Rampias T, Vgenopoulou P, Avgeris M, et al. A new tumor suppressor role for the Notch pathway in bladder cancer. Nat Med. 2014;20:1199–205. doi: 10.1038/nm.3678. [DOI] [PubMed] [Google Scholar]
- Salim E. Mutation analysis and expression of fibroblast growth factor receptor 3 in Bilharzial urinary bladder carcinomas from Egyptian patients. Egypt J Exp Biol. 2012;8:145–53. [Google Scholar]
- Samah M, Ayman M, Ahmed M, et al. Bat-26 is associated with clinical stage and lymph node status in schistosomiasis associated bladder cancer. Am J Biochem. 2015;5:15–21. [Google Scholar]
- Schmitz-Dräger B, Schulz W, Jürgens B, et al. c-myc in bladder cancer Clinical findings and analysis of mechanism. Urol Res. 1997;25:45–9. doi: 10.1007/BF00942047. [DOI] [PubMed] [Google Scholar]
- Tang J, Li X, Jiang X, et al. The association between rs9642880 gene polymorphism and bladder cancer risk: A meta-analysis. Int J Clin Exp Med. 2016;8:20236–44. [PMC free article] [PubMed] [Google Scholar]
- Tengyun L, Xiaoni L, Guofei Z, et al. Single nucleotide polymorphism mutation related genes in bladder cancer for the treatment of patients: a study based on the TCGA database. Biotechnol Biotechnol Equipment. 2021:35. [Google Scholar]
- Wang M, Wang M, Zhang W, et al. Common genetic variants on 8q24 contribute to susceptibility to bladder cancer in a Chinese population. Carcinogenesis. 2009;30:991–6. doi: 10.1093/carcin/bgp091. [DOI] [PubMed] [Google Scholar]
- Wen Y, Zhu C, Li N, et al. Fine mapping in chromosome 3q28 identified two variants associated with lung cancer risk in asian population. J Cancer. 2019;10:1862–9. doi: 10.7150/jca.28379. [DOI] [PMC free article] [PubMed] [Google Scholar]