Abstract
Purpose
To develop a tool to measure the pH at the surfaces of individual cells.
Procedures
The SNARF pH-sensitive dye was conjugated to a pHLIP® peptide (pH-Low Insertion Peptide) that binds cellular membranes in tumor spheroids. A beam splitter allows simultaneous recording of two images (580 and 640 nm) by a CCD camera. The ratio of the two images is converted into a pH map resolving single spheroid cells. An average pH for each cell is calculated and a pH histogram is derived.
Results
Surface pH depends on cellular glycolytic activity, which was varied by adding glucose or deoxy-glucose. Glucose was found to decrease the surface pH relative to the pH of the bulk solution. The surface pH of metastatic cancer cells was lower than that of non-metastatic cells indicating a higher glycolytic activity.
Conclusions
Our method allows cell surface pH measurement and its correlation with cellular glycolytic activity.
Keywords: pHLIP, Warburg effect, tumor acidity, pH measurements, SNARF fluorescence
INTRODUCTION
Otto Warburg discovered that cancer cells predominately use glycolysis for ATP production even when the supply of oxygen is sufficient, and he also found that the tumor microenvironment is more acidic than that in normal tissues [1]. For many years the main tool for measuring the pH in tumor tissue was a needle pH meter, which is an invasive method that damages cells and could not distinguish between intercellular and extracellular pHs [2]. It was believed that intracellular pHi and extracellular pHe were both acidic until it was shown that pHi is rather neutral (7.2, as might have been expected) in both normal and cancer cells, while pHe is acidic in tumors, in contrast to the normal pHe found in healthy tissue [3–5]. Tumor acidosis is caused mostly by the enhancement of glycolytic metabolism and an inhibition of the phosphorylation-oxidation pathway of energy production [6–7]. To maintain normal pHi cancer cells have to pump out protons produced during glycolysis [8]. Membrane carbonic anhydrases further induce cell surface acidification by hydrating cell-generated CO2 into HCO3− and H+ at the cancer cell membrane [7]. As a result, it is expected that an “acidic layer” will be formed around cancer cells, and that a steep proton concentration gradient might exist near the surfaces of cancer cells. Thus, to assess cancer cell pH, it must be measured at the surface, and not in the bulk extracellular fluid.
We recently reported an approach for cell surface pH measurements using a pHLIP (pH-Low Insertion Peptide) conjugated with a pH-sensitive ratiometric fluorescent dye, SNARF [9]. pHLIPs can target tumors and deliver imaging and therapeutic agents to cancer cells within tumors [10–15]. When it contacts an acidic cell, pHLIP forms a transmembrane helix with its N-terminus outside the cell and its C-terminus inserted across the cell membrane. Conjugation of SNARF to the pHLIP N-terminus positions the dye near the surface of the cell, where it can measure the pH in the extracellular space near the surface [9]. The pH is measured as the ratio of two peaks in the SNARF emission at 580 nm and 640 nm, converting the ratio into pH using a calibration curve. However, this previous approach does not allow measurement of pH at a single cell level. In this report, we describe a way to measure surface pH at single cell resolution.
MATERIALS AND METHODS
Materials
D-(+)-glucose and 2-deoxy-D-glucose were purchased from Sigma-Aldrich. Matrigel® growth factor reduced basement membrane matrix and phenol red-free were obtained from Corning. The wild type (WT) pH (Low) Insertion Peptide, WT-pHLIP, was synthesized with a single Lys residue near its acetylated N-terminus (Ac-AKEQNPIYWARYADWLFTTPLLLLDLALLVDADEGT) and purified by reverse-phase chromatography by C.S. Bio. SNARF™-1 carboxylic acid, acetate, succinimidyl ester was purchased from Thermo Fisher Scientific.
Phosphate-Buffered Solutions
Phosphate-buffered solutions were prepared over the pH range of 6.0–8.0 by mixing 0.5 M dibasic and monobasic solutions (J. T. Baker). The final experimental PBS solution contained 10 mM phosphate, 150 mM NaCl (J. T. Baker), 0.2 mM MgCl2 (Sigma), and 0.2 mM CaCl2 (Sigma). Buffer solutions were sterilized by passage through a 0.2-μm filter. The final pH for each solution was measured by a Dual Star pH / ISE benchtop meter with a microelectrode (Thermo Scientific™ Orion™).
Synthesis of SNARF pHLIP
Lys-WT and SNARF-1 were dissolved in DMF (dimethylformamide, Sigma), and incubated at a ratio of 2:1 in 60% DMF (dimethylformamide), 30% 0.1 M PBS pH 9.0 and 10% pH 9.5 0.1 M sodium bicarbonate buffer for a final pH of 9.0. SNARF-1 was converted to its fluorescent form after conjugation by raising the conjugation solution’s volume by 50% with methanol and raising the solution pH to 14 with 2 M potassium hydroxide for 1 hour at room temperature. Then, pH was adjusted to pH 7.0 by adding 30% HCl. The reaction progress was monitored by reverse phase (Zorbax SB-C18 columns, 9.4 × 250 mm 5 μm, Agilent Technology) high-performance liquid chromatography (HPLC) using a gradient of 25−75% acetonitrile and water containing 0.05% of trifluoroacetic acid. The concentration of each labeled peptide in buffer was determined by SNARF-1 absorption at 548 nm, ε548=27,000 M−1 cm−1. The purity and characterization of the construct were performed by analytical HPLC and surface-enhanced laser desorption/ionization–TOF mass spectrometry.
Cell Lines
Human melanoma M4A4 and NM2C5 cell lines were obtained from the American Tissue and Culture Collection (ATCC). Cells were authenticated, stored according to supplier’s instructions, and used within 3–4 months after resuscitation of frozen aliquots. All lines were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 4.5 g/L glucose and 40 mg/L sodium pyruvate supplemented with 10% FBS (Gibco), 0.1% of 10 μg/mL ciprofloxacin·HCl (Cellgro, Voigt Global Distribution) in a humidified atmosphere of 5% CO2 at 37 °C.
Tumor Spheroids
A 2% agarose (Sigma) solution was made by dissolving in pH 7.4 PBS (Gibco). 150 μL of the solution was pipetted into each well of a 48-well flat bottom tissue culture plate (Celltreat). After the agarose gel had sufficiently settled (~1 h), 150 μL of DMEM supplemented with 10% FBS and ciprofloxacin·HCl was added to each well. The covered plate was left in a humidified atmosphere at 37°C and 5% CO2 in cell culture incubator for 24 h. On the next day, the excess medium was removed from the agarose layer. NM2C5 or M4A4 cells (10,000 cells) in 200 μL of DMEM containing 2% matrigel were added into each well and incubated for 3–4 days to allow the formation of spheroids. Matrigel was dissolved on ice overnight and added in ice cold DMEM at a concentration of 2.5% (to obtain a final concentration of 2% once added to the wells). Then the mixture was heated to 37°C before being combined with the cells.
Imaging Tumor Spheroids
Tumor spheroids of a given cell line were incubated in 50 μL of PBS buffer, pH 6.3 containing 5 μM SNARF pHLIP and either 25 mM glucose or 50 mM deoxyglucose in a humidified atmosphere of 5% CO2 at 37°C for 30 min. After treatment, the spheroids were washed three times in 1 mL of experimental PBS of the desired pH containing either 25 mM glucose or 50 mM deoxy-glucose as a control. The spheroids were then placed in a 96 well glass bottom dish for imaging.
Fluorescence images were recorded using an Olympus IX71, an inverted epifluorescence microscope, and fluorescence was excited using a FF01-531/40-25 Semrock excitation filter. The resulting fluorescence was observed using a DV2 multichannel imaging system, with FF01-579/34-25 and FF01-647/57-25 Semrock emission filters in the left and right channels, respectively, which allowed two images to be taken at the same time by a Q-imaging Retiga-SRV CCD.
Data Analysis
For each pH point, 20 spheroids were used in total: 10 spheroids were used for calibration, and 10 spheroids were used for cancer cell surface pH measurements. For calibration and pH measurements, glucose and deoxy-glucose were added to 5 sets of spheroids, respectively. At least 5 images were taken from different areas of each spheroid. All images were analyzed by our program written in Matlab R2016b. The output of the analysis included a correction curve, pH map and pH histogram for the selected cells in each image. Statistical analysis and final graphs were obtained using Origin Lab 2016.
RESULTS
To measure the pH at the surfaces of cancer cells within tumor spheroid, we used pH-senstive fluorescent dye, SNARF, (Figure 1a) conjugated to the N-terminus of pHLIP (Figure 1b). The N-terminus of the peptide stays exposed to the extracellular space after the pHLIP peptide inserts across the plasma membrane of an acidic cancer cell, positioning the SNARF near the cell surface (Figure 1c). We previously used the quenching of SNARF fluorescence by membrane impermeable Trypan Blue to show that the SNARF moiety is located in the extracellular space, where it can report the pH near the cell surface [9]. The fluorescence spectrum of SNARF exhibits two maxima, at 580 nm and 640 nm, the ratio of intensities of which correlates with the pH of the SNARF environment. The advantage of the ratiometric method is that it does not depend on the concentration of SNARF and so allows accurate measurement of pH. The intensities of the two emission maxima can be obtained simultaneously from a fluorescence spectrum recorded by a linear CCD camera in the range of 570 – 700 nm, as it was done in our previous work [9]. The spectroscopic method is accurate, however, it measures only average intensity values from the illuminated spot, which includes multiple cells and spaces between cells [9]. In the present work, we have developed a high resolution pH mapping approach based on imaging a field of cells with enough resolution to resolve each cell. SNARF fluorescence was excited using a 531±20 nm excitation filter, and the emission signal was split into two using a DV2 optical system to acquire two fluorescent images, where one image was obtained using a 579±17 nm emission filter (called 580 nm) and the other image is obtained using a 647±28.5 nm emission filter (called 640 nm). Both images originate from the same area and are acquired simultaneously by the same CCD camera, which excludes any influence of potential cell-cell intensity differences on the calculated pH values. First, correction at a pixel level is applied in order to align two selected regions from the image (Figure 2a). The intensity values of each pixel at the center (within the region of ±25 pixel units) of the selected 640 nm image region are compared with the corresponding central pixel intensity values of the 580 nm image region. The differences in intensity between 640 nm and 580 nm images are shown in Figure 2b. The two images are adjusted to minimize the intensity difference and align images for further processing. Intensity and size cutoffs were used to establish cell border outlines (red outlines in Figure 2c). The pH mapping is obtained by calculating the averaged intensity ratio for each cell and converting these ratios into pH values using calibration curves (Figure 2d). The pH histogram reflects the distribution of the surface pH for all selected cells (Figure 2e).
Figure 1. The schematic presentation of SNARF-pHLIP interaction with plasma membrane of cells.
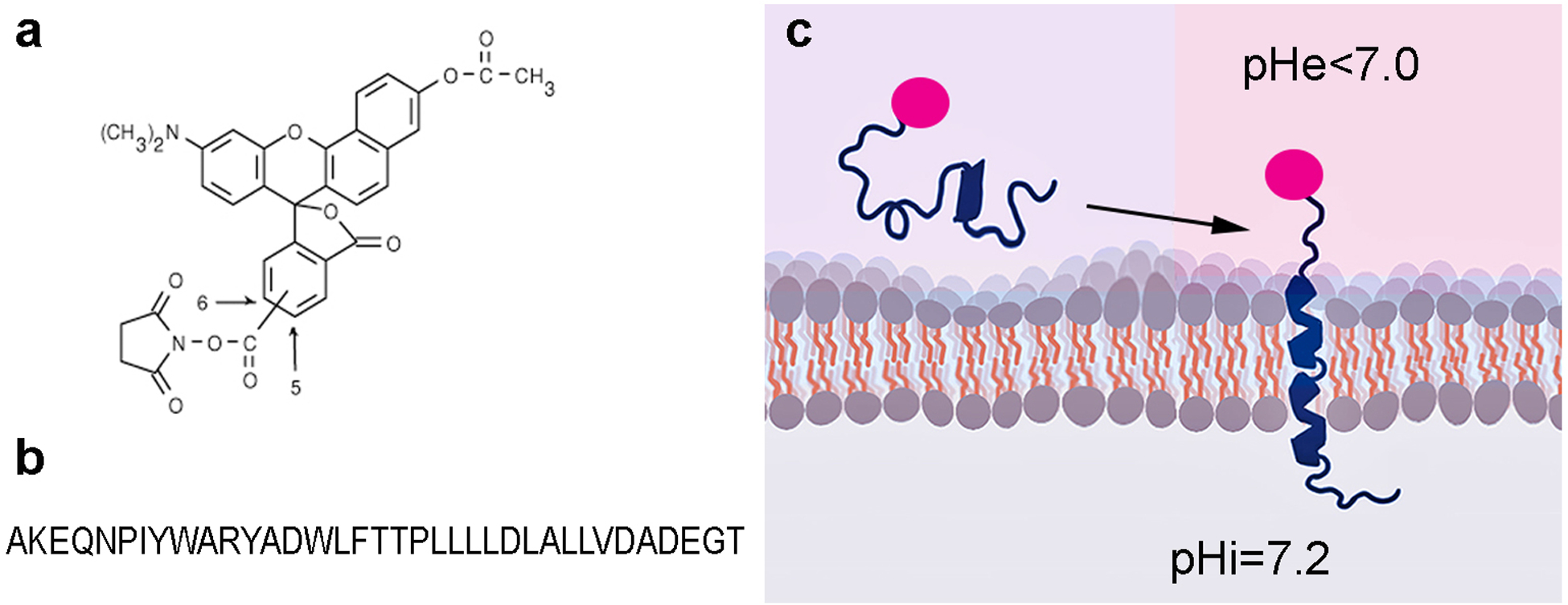
Chemical structure of pH-senstive fluorescent dye, SNARF (a) and sequence of pHLIP peptide with acytylated N-terminus (b) are shown. Schematic presentation of SNARF (shown in red) conjugated to the N-terminal part of pHLIP (shown in blue) interaction with lipid bilayer of membrane (c). pHLIP inserts into bilayer and tethers SNARF to the surface of membrane.
Figure 2. The image analysis.
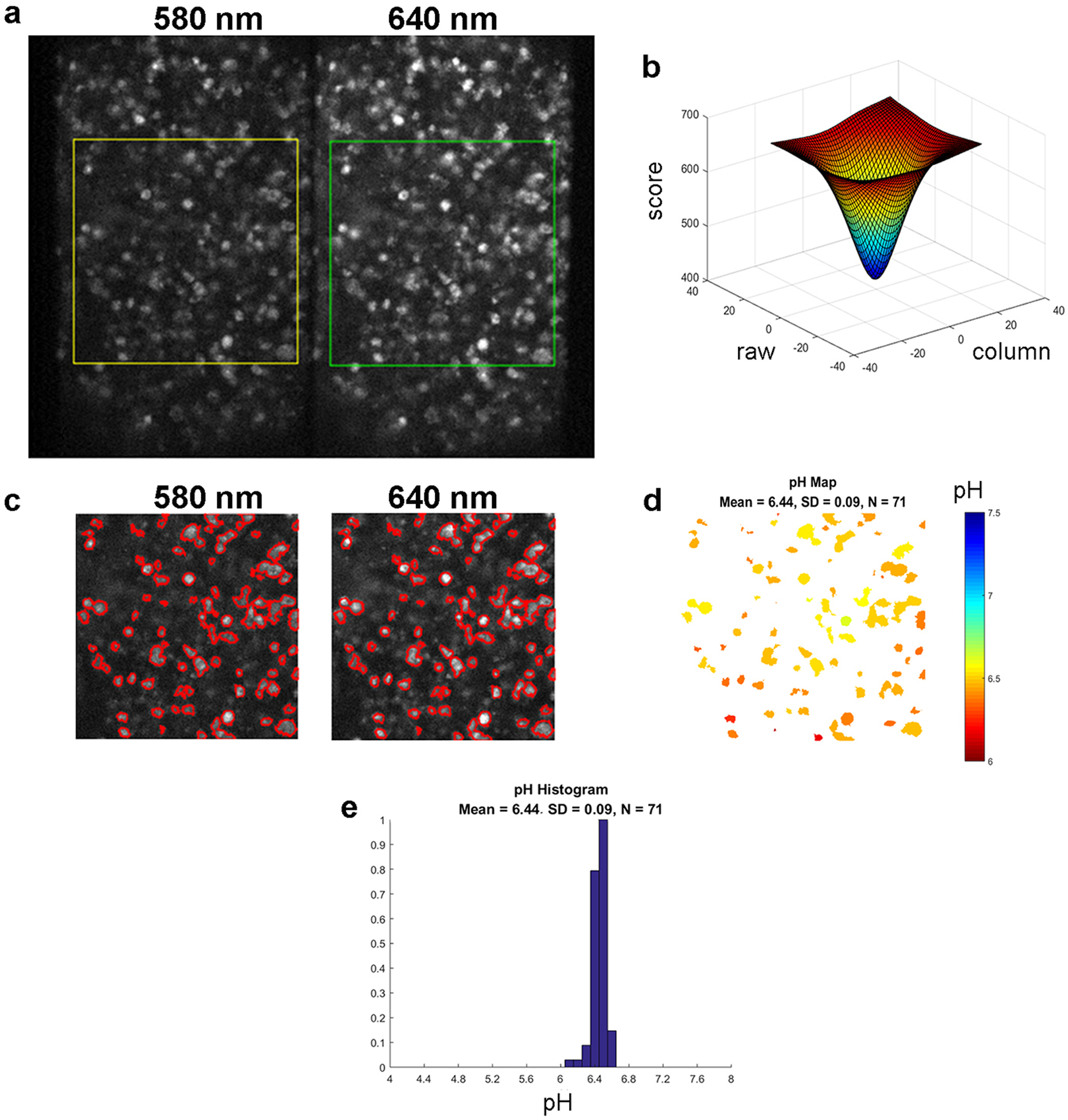
SNARF fluorescence images of N4A4 cells at 580 nm and 640 nm (a) were obtained using a DV2 beam splitting system and 579±17 nm and 647±28.5 nm emission filters, respectively. The area of interest is selected by squares. The differences in intensity values in each pixel at the center (within the region of ±25 pixels) between 640 nm and 580 nm image regions (b) were used to adjust the 580 nm image position to minimize the intensity differences (achieve the smallest score) and thus to align images for further processing. Image correction, intensity and size cutoff were used to establish the cell border outlines shown in red (c). Examples of a pH map (d), where pH values are indicated by different colors, and a pH histogram (e) for the selected region from (a) are shown.
The calibration curves were obtained for NM2C5 (Figure 3a) and M4A4 (Figure 3b) cancer cells separately. As a control, glycolysis was shut down: M4A4 or NM2C5 tumor spheroids were incubated in media with no glucose and then transferred into media containing 50 mM of a non-metabolizable analog of glucose, 2-deoxyglucose. The inhibition of glycolysis results in the shutdown of acid production and the proton flux by cells. As a result, the pH at the surface of cells is the same as in the bulk solution, allowing determination of calibration curves of the fluorescence signals with pH. We recorded numerous cell images at different buffer pHs and calculated 580/640 ratio images (RM4A4 and RNM2C5) to establish such calibration curves for each cancer cell line (Figure 3a, b):
Figure 3. Comparison of surface pH for more and less metastatic cell lines.
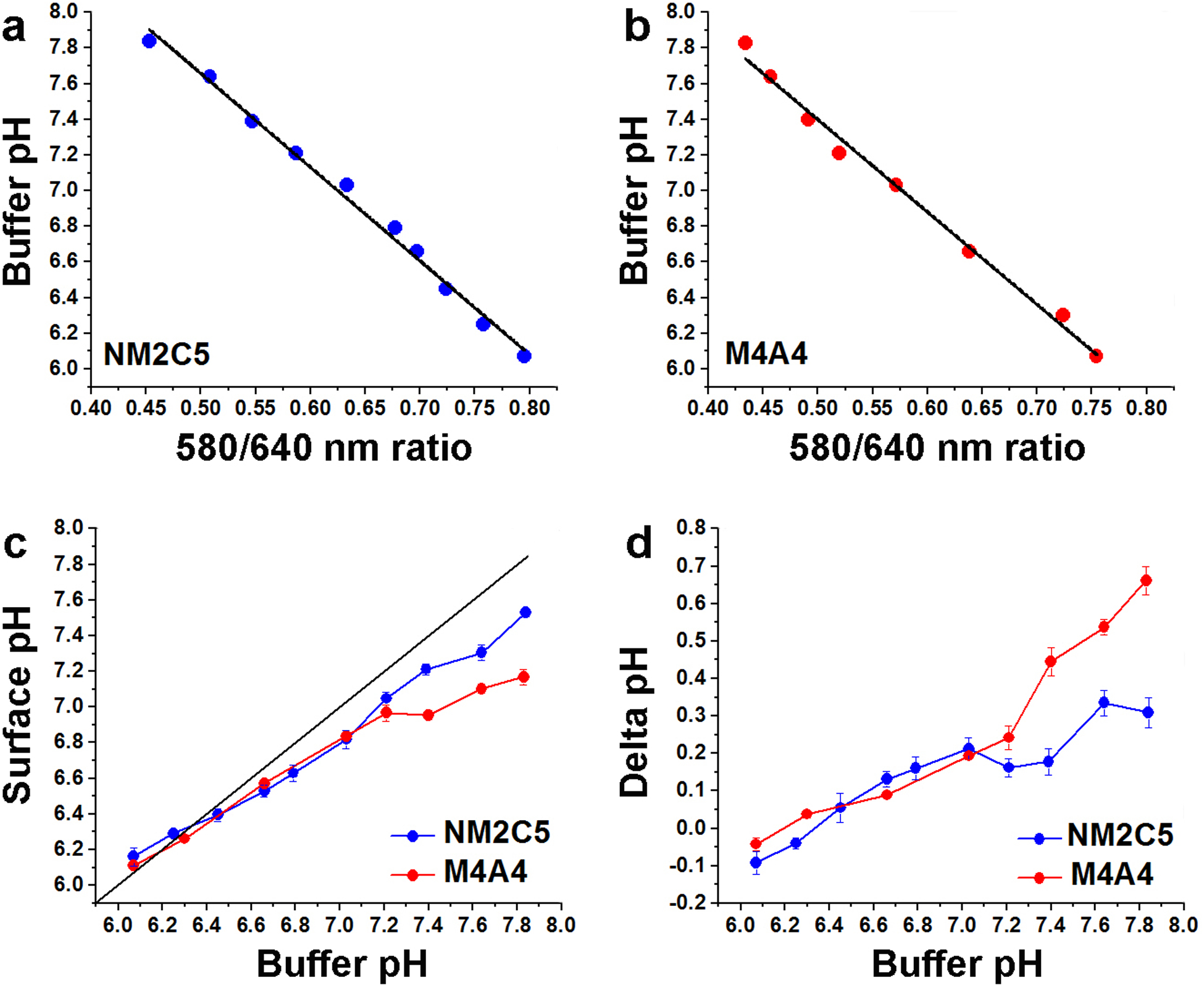
Calibration curves for NM2C5 (a) and M4A4 (b) cells grown in tumor spheroids obtained when glycolysis is suppressed are shown. A linear fit of buffer pH vs 580/640 SNARF fluorescence intensity ratio in the presence of 50 mM deoxy-glucose at various pHs of buffer gives the calibration curve data. Cell surface pH under glycolytic conditions (c) was measured using SNARF pHLIP bound to NM2C5 and M4A4 cell spheroids in the presence of 25 mM glucose at various pHs of buffer. The values were calculated using the calibration curves (a and b) obtained for each cell line to convert 580/640 nm fluorescence ratios into pH values. The cell surface pH differences (d) for non-metastatic NM2C5 and metastatic M4A4 cancer cells were calculated by comparing pH values obtained in the presence of deoxy-glucose and glucose at different buffer pHs.
The calibration curves are slightly different for M4A4 and NM2C5 cells, which might be attributed to differences in the SNARF local environments from membrane protein and/or lipid composition variation.
The calibration equations were used to calculate surface pH from the SNARF pHLIP 580/640 nm ratio imaging of spheroids of a non-metastatic cancer cell line, NM2C5, and a metastatic cancer cell line, M4A4, in the presence of 25 mM glucose, which promotes cellular metabolism. In the presence of glucose, both NM2C5 and M4A4 cells show lower pH at the surface compared with the bulk extracellular pH. Furthermore, metastatic cancer cells were slightly more acidic than non-metastatic cancer cells, especially when the pH of the medium was normal or higher than pH 7.4. When the medium pH is lower than pH 6.4, the pH at the surfaces of cancer cells equilibrates with the bulk solution pH (Figure 3a, b). The surface pH differences found in the presence of glucose versus deoxy-glucose for each type of cancer cell are shown in Figure 3c. At normal or high medium pH the metastatic cancer cells, M4A4, exhibit a significantly larger pH difference, about 0.7 pH units, compared to the non-metastatic cancer cells, NM2C5, where the pH difference is about 0.3 pH units (Figure 3d). The results are in good agreement with previously published data for these cells lines obtained by fluorescence spectroscopy [9]. The previous spectroscopic method measures the average fluorescence spectrum from an illuminated spot, which includes several cells and the area between them, and pH is calculated from the ratio of two peaks in the emission spectrum. The image-based method gives pH images of each of hundreds of individual cells in the sample and excludes the areas between the cells.
DISCUSSION
In the mid 20th century, tumor acidity was typically measured using microelectrodes directly inserted into tumor tissues. Tumors were found to be acidic, and it was believed at that time that extracellular pHe and intracellular pHi were each acidic [4]. However, microelectrodes do not directly measure the pH inside a cell. Only after the development of magnetic resonance spectroscopy (MRS) methods was it found that pHi in cancer cells is similar to or slightly more basic than that in normal cells, while pHe in tumors is more acidic than pHe in healthy tissues [3, 5]. MRS methods use pH-sensitive agents to measure pHe in tumors in vivo [16]. Those agents are distributed both in blood (where the pH is normal) and in the extracellular space. Thus, MRS provides a measure of an average pH in tumors [17–21]. Since cancer cells have to maintain pHi in the range of 7.2 – 7.4 for normal cellular functions, they have to pump out protons and lactate, which are produced in high amounts due to the enhancement of glycolysis [1]. The other by-product is CO2, which freely diffuses across the membrane and is converted into carbonic acid by membrane bound carbonic anhydrases (CAIX, CAXII), which are overexpressed in cancer cells [22–24]. Protons create a zone of high acidity near the cancer cell membrane as a result of the flux from the cell and the membrane electrochemical potential. pH increases with distance from the cancer cells as a result of protons diffusion and equilibrates with a bulk extracellular pH, which might be normal in well perfused tumor areas, or might remain lowered in poorly perfused tumor zones. Despite variations of bulk extracellular pH in tumors (documented by MRS measurements), the cell surface pH can be acidic even in areas with normal bulk pH. Acidic cell surface pH well correlates with the metastatic potential of cancer cells. Therefore, tools that measure cell surface pH should be useful for the investigation of tumor development and progression. And, it is crucial that any use of pH as a biomarker for targeting should sense acidity at the cell surfaces, as pHLIP does, directing imaging and therapeutic agents to all cancer cells within tumors.
CONCLUSIONS
In this work, we describe a new method to map cell surface pH at the level of individual cancer cells. We believe that this approach will find many applications, both in normal cell biology and in cancer biology studies. An ability to distinguish normal from cancer cells based on surface acidity might be used for the analysis of tissue samples removed in the course of needle biopsy or surgery.
ACKNOWLEDGEMENTS
We would like to acknowledge the financial support from NIH RO1 GM073857-09A1 grant to DME, OAA and YKR. We would like to thank our colleagues Anna Moshnikova and Michael Anderson for useful discussions and suggestions.
Footnotes
CONFLICT OF INTERESTS
D.M.E., O.A.A., and Y.K.R. are founders of pHLIP, Inc. They have shares in the company, but the company did not fund any part of the work reported in the paper, which was done in their academic laboratories.
REFERENCES
- 1.Warburg O (1956) On the origin of cancer cells. Science 123:309–314. [DOI] [PubMed] [Google Scholar]
- 2.Griffiths JR (1991) Are cancer cells acidic? Br J Cancer 64:425–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffiths JR, Stevens AN, Iles RA, Gordon RE, Shaw D (1981) 31P-NMR investigation of solid tumours in the living rat. Biosci Rep 1:319–325. [DOI] [PubMed] [Google Scholar]
- 4.Wike-Hooley JL, Haveman J, Reinhold HS (1984) The relevance of tumour pH to the treatment of malignant disease. Radiother Oncol 2:343–366. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Lin Y, Gillies RJ (2010) Tumor pH and its measurement. J Nucl Med 51:1167–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kroemer G, Pouyssegur J (2008) Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell 13:472–482. [DOI] [PubMed] [Google Scholar]
- 7.Damaghi M, Wojtkowiak JW, Gillies RJ (2013) pH sensing and regulation in cancer. Front Physiol 4:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiche J, Brahimi-Horn MC, Pouyssegur J (2010) Tumour hypoxia induces a metabolic shift causing acidosis: a common feature in cancer. J Cell Mol Med 14:771–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson M, Moshnikova A, Engelman DM, Reshetnyak YK, Andreev OA (2016) Probe for the measurement of cell surface pH in vivo and ex vivo. Proc Natl Acad Sci U S A 113:8177–8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreev OA, Dupuy AD, Segala M, et al. (2007) Mechanism and uses of a membrane peptide that targets tumors and other acidic tissues in vivo. Proc Natl Acad Sci U S A 104:7893–7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreev OA, Engelman DM, Reshetnyak YK (2010) pH-sensitive membrane peptides (pHLIPs) as a novel class of delivery agents. Mol Membr Biol 27:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reshetnyak YK, Yao L, Zheng S, Kuznetsov S, Engelman DM, Andreev OA (2011) Measuring tumor aggressiveness and targeting metastatic lesions with fluorescent pHLIP. Mol Imaging Biol 13:1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weerakkody D, Moshnikova A, Thakur MS, et al. (2013) Family of pH (low) insertion peptides for tumor targeting. Proc Natl Acad Sci U S A 110:5834–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreev OA, Engelman DM, Reshetnyak YK (2014) Targeting diseased tissues by pHLIP insertion at low cell surface pH. Front Physiol 5:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyatt LC, Lewis JS, Andreev OA, Reshetnyak YK, Engelman DM (2017) Applications of pHLIP Technology for Cancer Imaging and Therapy: (Trends in Biotechnology 35, 653–664, 2017). Trends Biotechnol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashim AI, Zhang X, Wojtkowiak JW, Martinez GV, Gillies RJ (2011) Imaging pH and metastasis. NMR Biomed 24:582–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moon RB, Richards JH (1973) Determination of intracellular pH by 31P magnetic resonance. J Biol Chem 248:7276–7278. [PubMed] [Google Scholar]
- 18.Gillies RJ, Liu Z, Bhujwalla Z (1994) 31P-MRS measurements of extracellular pH of tumors using 3-aminopropylphosphonate. Am J Physiol 267:C195–203. [DOI] [PubMed] [Google Scholar]
- 19.Ojugo AS, McSheehy PM, McIntyre DJ, et al. (1999) Measurement of the extracellular pH of solid tumours in mice by magnetic resonance spectroscopy: a comparison of exogenous (19)F and (31)P probes. NMR Biomed 12:495–504. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Martin ML, Herigault G, Remy C, et al. (2001) Mapping extracellular pH in rat brain gliomas in vivo by 1H magnetic resonance spectroscopic imaging: comparison with maps of metabolites. Cancer Res 61:6524–6531. [PubMed] [Google Scholar]
- 21.Rata M, Giles SL, deSouza NM, Leach MO, Payne GS (2014) Comparison of three reference methods for the measurement of intracellular pH using 31P MRS in healthy volunteers and patients with lymphoma. NMR Biomed 27:158–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wykoff CC, Beasley NJ, Watson PH, et al. (2000) Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res 60:7075–7083. [PubMed] [Google Scholar]
- 23.Griffiths JR, McIntyre DJ, Howe FA, Stubbs M (2001) Why are cancers acidic? A carrier-mediated diffusion model for H+ transport in the interstitial fluid. Novartis Found Symp 240:46–62; discussion 62–47, 152–153. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov S, Liao SY, Ivanova A, et al. (2001) Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol 158:905–919. [DOI] [PMC free article] [PubMed] [Google Scholar]


