Antinucleocapsid antibodies (anti-N Abs) are currently being used to diagnose prior SARS-CoV-2 infection in individuals and to determine community seroprevalence of SARS-CoV-2. In a large, randomized, double-blind, placebo-controlled vaccine efficacy trial of the mRNA-1273 vaccine, anti-N Ab status was determined in participants who had prior SARS-CoV-2 infection confirmed by polymerase chain reaction or anti-N Abs.
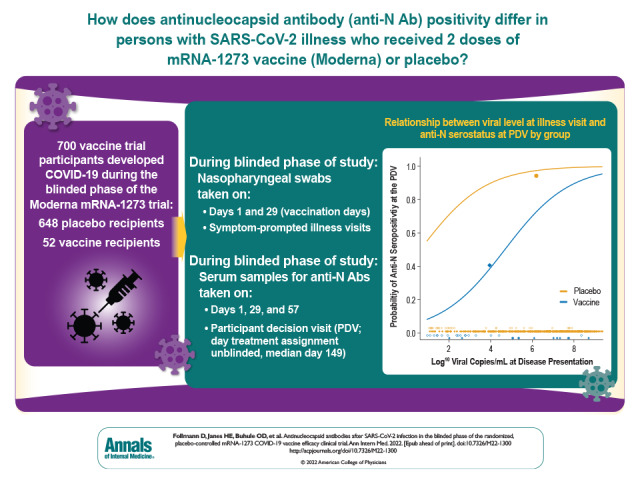
Visual Abstract. Antinucleocapsid Antibodies After SARS-CoV-2 Infection in COVE.
Antinucleocapsid antibodies (anti-N Abs) are currently being used to diagnose prior SARS-CoV-2 infection in individuals and to determine community seroprevalence of SARS-CoV-2. In a large, randomized, double-blind, placebo-controlled vaccine efficacy trial of the mRNA-1273 vaccine, anti-N Ab status was determined in participants who had prior SARS-CoV-2 infection confirmed by polymerase chain reaction or anti-N Abs.
Abstract
Background:
Immunoassays for determining past SARS-CoV-2 infection have not been systematically evaluated in vaccinated persons in comparison with unvaccinated persons.
Objective:
To evaluate antinucleocapsid antibody (anti-N Ab) seropositivity in mRNA-1273 (Moderna) vaccinees with breakthrough SARS-CoV-2 infection.
Design:
Nested substudy of a phase 3 randomized, double-blind, placebo-controlled vaccine efficacy trial. (ClinicalTrials.gov: NCT04470427)
Setting:
99 sites in the United States, July 2020 through March 2021.
Participants:
Participants were aged 18 years or older, had no known history of SARS-CoV-2 infection, and were at risk for SARS-CoV-2 infection or severe COVID-19. Substudy participants were diagnosed with SARS-CoV-2 infection during the trial's blinded phase.
Intervention:
2 mRNA-1273 or placebo injections 28 days apart.
Measurements:
Nasopharyngeal swabs from days 1 and 29 (vaccination days) and from symptom-prompted illness visits were tested for SARS-CoV-2 via polymerase chain reaction (PCR). Serum samples from days 1, 29, and 57 and the participant decision visit (PDV, when participants were informed of treatment assignment; median day 149) were tested for anti-N Abs by the Elecsys immunoassay.
Results:
Among 700 participants with PCR-confirmed COVID-19 illness during the blinded phase of the trial (through March 2021), seroconversion to anti-N Abs (median of 53 days after diagnosis) occurred in 21 of 52 mRNA-1273 vaccinees (40% [95% CI, 27% to 54%]) versus 605 of 648 placebo recipients (93% [CI, 92% to 95%]). Each 1-log increase in SARS-CoV-2 viral copies at diagnosis was associated with 90% higher odds of anti-N Ab seroconversion (odds ratio, 1.90 [CI, 1.59 to 2.28]).
Limitation:
The scope was restricted to mRNA-1273 vaccinees and the Elecsys assay, the sample size was small, data on Delta and Omicron infections were lacking, and the analysis did not address a prespecified objective of the trial.
Conclusion:
Vaccination status should be considered when interpreting seroprevalence and seropositivity data based solely on anti-N Ab testing.
Primary Funding source: National Institute of Allergy and Infectious Diseases of the National Institutes of Health.
A reliable diagnostic tool to identify recent and remote infection with SARS-CoV-2 is needed to enable population-based seroprevalence studies, ascertain infection status in vaccine clinical trials, and diagnose postinfectious complications of COVID-19, such as multisystem inflammatory syndrome. Antibodies against the nucleocapsid protein (anti-N Abs) are not elicited by COVID-19 vaccines that target the spike protein, including all vaccines currently used in the United States, and assays that measure anti-N Abs have demonstrated high sensitivity and specificity in many studies when used at least 14 days after infection (1). In addition, the anti-N Ab response in unvaccinated persons has been reported to be durable, with half-life estimates ranging from 68 to 283 days (2, 3). The determination of previous SARS-CoV-2 infections in participants in a pivotal phase 3 vaccine clinical trial (4, 5) and in seroprevalence studies done early in the pandemic (6) relied on detection of antispike or anti-N Abs via chemiluminescence immunoassays in unvaccinated persons. However, it is unknown how vaccine-induced immunity may affect seroconversion to nonspike proteins and whether immunoassays measuring nonspike seroconversion would have similar performance in seroprevalence studies of previous SARS-CoV-2 infection in regions of the world with high vaccination coverage. To answer this question, we evaluated vaccine and placebo recipients in the mRNA-1273 (Moderna) phase 3 clinical trial (ClinicalTrials.gov: NCT04470427) (4, 5) in whom SARS-CoV-2 infection was detected during the blinded, placebo-controlled phase of the trial. Serum samples from these participants were assayed for anti-N Abs through about 5 months after enrollment.
Methods
Study Design and Population
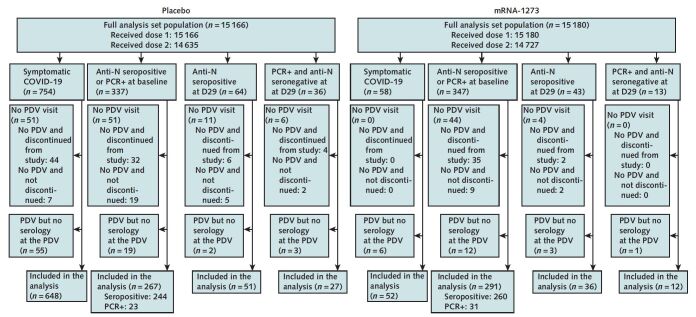
Between 27 July and 23 October 2020, the COVE (Coronavirus Efficacy) trial enrolled 30 420 U.S. adults aged 18 years or older at appreciable risk for SARS-CoV-2 infection or high risk for severe COVID-19 at 99 sites across the United States. Participants were randomly assigned 1:1 to receive two 100-μg doses of mRNA-1273 vaccine or placebo, given at day 1 (baseline) and day 29. Participant characteristics, study procedures, and primary efficacy results are described elsewhere (4, 5). Infections with SARS-CoV-2 were detected on the basis of both symptom-prompted testing and planned testing at specific study visits (Figure 1). Blood was collected for anti-N Ab serology at days 1, 29, and 57 and at the participant decision visit (PDV), when participants were informed about their blinded randomization assignment and offered mRNA-1273 vaccination if previously assigned to placebo. Nasopharyngeal swabs for SARS-CoV-2 reverse transcriptase polymerase chain reaction (RT-PCR) testing were collected at day 1, day 29, and the PDV in all participants. The PDVs occurred a median of 149 days (IQR, 133 to 164 days) after enrollment. In addition, participants meeting predefined clinical criteria for suspected COVID-19 during follow-up had an illness visit for collection of a nasopharyngeal swab for SARS-CoV-2 PCR testing. Nasal swabs or saliva samples could be submitted if a clinic or home visit could not be completed. Symptom severity, including body temperature and oxygen saturation, was collected daily via telemedicine visit for 14 days (or until symptoms resolved, whichever was longer). The present analysis is based on data obtained through the completion of the blinded phase of the study with a data cutoff date of 26 March 2021. The Appendix Figure) shows the flow of participants with detected SARS-CoV-2 infection from enrollment through inclusion in the analysis.
Figure 1. Method of SARS-CoV-2 infection determination and sampling schedule.

D = day; N = nucleocapsid; NP = nasopharyngeal; PCR = polymerase chain reaction; PDV = participant decision visit.
Appendix Figure. Study flow diagram.
Flow of participants with detected SARS-CoV-2 infection from enrollment through to inclusion in the analysis. Data cutoff date: 26 March 2021. Symptomatic COVID-19 indicates an illness visit due to eligible symptoms (4, 5) and positive SARS-CoV-2 reverse transcriptase PCR (RT-PCR) within 14 d after dose 2 and before the PDV in participants with no evidence of previous SARS-CoV-2 infection. Anti-N seropositive indicates positive binding antibodies against SARS-CoV-2 nucleocapsid protein as defined by a positive Elecsys (Roche Diagnostics International) test result, in the absence of a previous positive Elecsys result. PCR+ indicates a nasopharyngeal swab positive for SARS-CoV-2 by RT-PCR in the absence of both symptoms and binding antibodies against SARS-CoV-2 nucleocapsid protein by Elecsys at the same time point, in the absence of a previous positive PCR result. D29 and D57 denote the day 29 and the day 57 study visits, respectively. D = day; N = nucleocapsid; PDV = participant decision visit; PCR = polymerase chain reaction.
* Six participants with major protocol deviations and 2 who were randomly assigned twice were excluded from the full analysis set.
The mRNA-1273-P301 study is being conducted in accordance with the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use, Good Clinical Practice guidelines, and applicable government regulations. The Central Institutional Review Board approved the mRNA-1273-P301 protocol and consent forms. All participants provided written informed consent before enrollment. Central institutional review board services for the study were provided by Advarra.
Laboratory Assays
Anti-N Abs were detected using the Elecsys Anti-SARS-CoV-2 immunoassay (Roche Diagnostics International) (7) on samples from days 1, 29, and 57 and the PDV. The Elecsys assay has a binary interpretation, “nonreactive” or “reactive,” for the presence of anti-N Abs. Day 57 50% inhibitory dilution neutralizing antibody titers, measured by an assay using SARS-CoV-2 spike-pseudotyped virus, were previously reported (8). Nasopharyngeal swabs, nasal swabs, and saliva samples were tested for SARS-CoV-2 via RT-PCR (Viracor, Eurofins Clinical Diagnostics). In COVID-19 cases, viral level was assessed at the illness visit and on days 3, 5, 7, 9, 14, 21, and 28 after the illness visit by SARS-CoV-2 quantitative RT-PCR, and cycle threshold values were converted to a number of viral genome copies (9). Sample collection was by swab at the illness visit and by saliva on other illness days.
Definitions
In the full analysis set of participants who were randomly assigned and received at least 1 dose of vaccine or placebo, the outcome of anti-N seropositivity at the PDV was compared between mRNA-1273 vaccine and placebo recipients who had SARS-CoV-2 infection detected before the PDV. The analysis was stratified by method and timing of SARS-CoV-2 infection detection; 4 mutually exclusive groups of infections were defined as follows. The first group of infections is primary end point COVID-19 cases detected at an illness visit at least 14 days after dose 2 and up to the PDV. Cases were defined as occurring when participants in the per protocol population were SARS-CoV-2–negative at baseline and had at least 2 symptoms from among fever (temperature ≥38 °C), chills, myalgia, headache, sore throat, or new olfactory or taste disorder, or as occurring in those who had at least 1 respiratory sign or symptom (including cough, shortness of breath, or clinical or radiographic evidence of pneumonia) and at least 1 nasopharyngeal swab, nasal swab, or saliva sample (or respiratory sample, if the participant was hospitalized) that was positive for SARS-CoV-2 by RT-PCR (4, 5). The second group of infections comprises participants who were anti-N seropositive by Elecsys assay or had a nasopharyngeal swab positive by RT-PCR at baseline (day 1). Participants in the third infection group were anti-N seropositive at day 29 without prior evidence of infection by serology or RT-PCR. The fourth group of infections consists of participants with a nasopharyngeal swab positive by RT-PCR at day 29 who were seronegative at day 29 without prior evidence of infection by serology or RT-PCR.
Statistical Analysis
Analyses to assess anti-N seropositivity at the PDV by infection group and randomization group were prespecified before analysis but were not prespecified in the study protocol. The outcome of PDV anti-N seropositivity was compared between infection groups and randomization groups using Fisher exact tests. The association of PDV anti-N seropositivity with time since COVID-19 diagnosis and other factors was assessed using logistic regression. A Wilcoxon rank-sum test was used to compare antispike antibody responses in those who were anti-N seropositive versus seronegative at the PDV. A Wald test of interaction was used to assess whether the effect of illness visit viral level on PDV anti-N seropositivity differed between randomization groups. Receiver-operating characteristic curves were compared using a nonparametric approach (10). All tests are 2-sided with a type I error rate equal to 0.05 and no adjustment for multiplicity.
Role of the Funding Source
The National Institutes of Health and National Institute of Allergy and Infectious Diseases had no role in the study design, data collection, or analysis; the decision to publish; or the preparation of the manuscript. The mRNA-1273-P301 study is sponsored by Moderna, but no other funding or support was provided for this nested substudy; Moderna authors reviewed and approved the analysis plan and manuscript.
Results
Infection with SARS-CoV-2 before the PDV was detected in 4 mutually exclusive groups. A total of 812 participants who were SARS-CoV-2–negative at baseline (754 placebo recipients and 58 vaccine recipients) (Supplement Table 1) had illness 14 or more days after full receipt of study vaccine or placebo and were diagnosed via a PCR swab obtained at an illness visit associated with that event. Median times between second dose and illness were 77 and 73 days for the vaccine and placebo groups, respectively. For the other 3 groups, SARS-CoV-2 infection was first detected through planned specimen collection at baseline or day 29. Infection was first detected at baseline by anti-N seropositivity or nasopharyngeal swab RT-PCR positivity in 684 participants (337 placebo recipients and 347 vaccine recipients) (Supplement Table 2). Postbaseline infections were first detected by anti-N seropositivity at day 29 in 107 participants (64 placebo recipients and 43 vaccine recipients) and by PCR positivity at day 29 in 49 participants (36 placebo recipients and 13 vaccine recipients) (Supplement Tables 3 and 4). For each of these 4 groups, Supplement Tables 1 to 4 also present baseline characteristics by group.
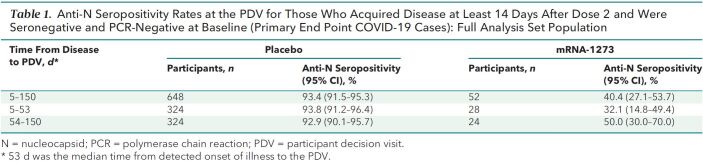
Table 1 shows anti-N seropositivity at the PDV for participants with COVID-19 detected at an illness visit, which differed substantially between groups: 40.4% (21 of 52) in vaccine recipients versus 93.4% (605 of 648) in placebo recipients (P < 0.001). The median time from illness to the PDV was 53 days, with a range of 5 to 150 days. The large between-group difference was apparent both among COVID-19 cases diagnosed within 53 days of the PDV and among cases diagnosed longer than 53 days from the PDV. Anti-N seropositivity at the PDV was not significantly associated with days between illness and PDV or with age, sex, body mass index, race/ethnicity, or “at risk for severe COVID-19” status at baseline in either vaccine or placebo recipients (Supplement Table 5). Of the 52 vaccine recipients, 36 had their 50% inhibitory dilution antispike neutralizing antibody titer measured at day 57 as part of the case-cohort immunogenicity and immune correlates assessment (8) and also had serostatus measured at the PDV. Among these 36 persons, the median day 57 titers were not significantly different between the 20 who were anti-N seronegative and the 16 who were anti-N seropositive at the PDV (200 vs. 158 IU50/mL; P = 0.39). However, anti-N seropositivity was positively associated with days between second vaccination and illness among vaccine recipients (P = 0.048) but not among placebo recipients (P = 0.95), suggesting that absence of anti-N seroconversion among vaccinated persons may be a lesser issue the farther the illness is from last vaccination. There was also a trend toward higher odds of anti-N seropositivity for vaccine recipients with higher total COVID-19 symptom scores (P = 0.077; P = 0.55 for placebo recipients).
Table 1.
Anti-N Seropositivity Rates at the PDV for Those Who Acquired Disease at Least 14 Days After Dose 2 and Were Seronegative and PCR-Negative at Baseline (Primary End Point COVID-19 Cases): Full Analysis Set Population
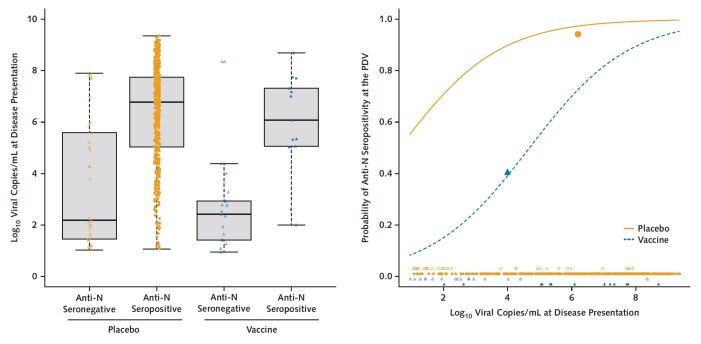
We assessed whether a higher SARS-CoV-2 viral level at the illness visit was associated with anti-N seropositivity at the PDV. The median viral level at the day 1 illness visit was significantly higher in placebo recipients who were anti-N seropositive at the PDV (6.8 log10 copies/mL) than in placebo recipients who were anti-N seronegative at the PDV (2.2 log10 copies/mL) (P < 0.001). A similar result was seen in vaccine recipients, with analogous viral levels of 6.1 log10 copies/mL and 2.4 log10 copies/mL, respectively (P < 0.001) (Figure 2, left). Using logistic regression with terms for group and viral level, we found that an increase in illness visit viral level of 1 log10 nearly doubled the odds of anti-N seropositivity at the PDV (odds ratio, 1.90 [95% CI, 1.59 to 2.28] per 1-log10 increase). Yet, viral level at the illness visit did not fully explain the large difference in PDV seropositivity between groups: For any given viral level, the odds of anti-N seropositivity were 13.67 times higher (CI, 5.17 to 36.16 times higher) for the placebo group than the vaccine group. For example, a vaccine recipient with 2.0 log10 viral copies/mL at the illness visit has an estimated probability of PDV anti-N seropositivity of 0.15, whereas a placebo recipient with the same viral level at the illness visit has an estimated probability of 0.71 (Figure 2, right). We also compared the overall prediction of PDV seropositivity using viral level at the day 1 illness visit versus the average overall illness visits; the latter measure is influenced by both magnitude and duration of viral replication. Viral level at the day 1 illness visit performed better, with an area under the receiver-operating characteristic curve of 0.84 versus 0.78 (P = 0.042).
Figure 2. Relationship between viral level at disease presentation and anti-N serostatus at the PDV, by group.
Closed and open shapes denote anti-N seropositive and anti-N seronegative cases, respectively. Left. Log10 viral copies/mL at disease presentation for placebo (orange circles) and vaccine (blue triangles) groups, by PDV anti-N serostatus. Right. Predicted probability of PDV anti-N seropositivity, by group. The 2 large closed shapes are plotted at the mean viral level and the mean PDV anti-N seropositivity rates, respectively, by group. N = nucleocapsid; PDV = participant decision visit.
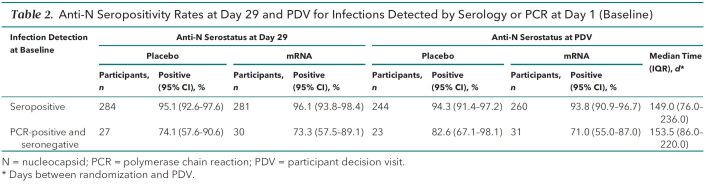
Table 2 presents the anti-N seropositivity rates at various study time points for infections detected at baseline, before receipt of study product. We found similar anti-N seropositivity rates between the 2 groups: Rates at day 29 were 95.1% (270 of 284) for the placebo group and 96.1% (270 of 281) for the mRNA-1273 group. These high rates were maintained through the PDV a median of 149 days later. For participants who were PCR-positive and anti-N–seronegative for SARS-CoV-2 at baseline, the day 29 anti-N seropositivity rates were 74.1% (20 of 27) for the placebo group and 73.3% (22 of 30) for the mRNA-1273 group. These rates were similar at the PDV: 82.6% (19 of 23) and 71.0% (22 of 31), respectively. Within each group, the anti-N seropositivity rates at day 29 and the PDV were significantly lower for participants who were PCR-positive at baseline than for those who were anti-N–seropositive at baseline (P < 0.001 for each group).
Table 2.
Anti-N Seropositivity Rates at Day 29 and PDV for Infections Detected by Serology or PCR at Day 1 (Baseline)
Table 3 presents seropositivity rates for infections detected at day 29 among participants who were SARS-CoV-2–negative at baseline; these rates are after dose 1 and thus subject to partial vaccine effects. For participants who were anti-N–seropositive on day 29, the anti-N seropositivity rates at day 57 were 86.9% (53 of 61) and 84.6% (33 of 39) for the placebo and mRNA-1273 groups, respectively. The rates were similar at the PDV, a median of 118 days later: 84.3% (43 of 51) and 86.1% (31 of 36), respectively. These rates were not significantly different at day 57 versus the PDV (P = 1.00 for mRNA-1273 and P = 0.79 for placebo) or by group (P = 0.77 at day 57 and P = 1.00 at PDV). For those who were newly RT-PCR–positive and anti-N–seronegative at day 29, the analogous seropositivity rates were 60.0% (18 of 30) and 38.5% (5 of 13) at day 57 and 70.4% (19 of 27) and 50.0% (6 of 12) at the PDV. These rates did not differ significantly at day 57 versus the PDV (P = 0.70 for mRNA-1273 and P = 0.58 for placebo) or by group (P = 0.32 at day 57 and P = 0.29 at PDV). Consistent with the effects seen among baseline infections, the anti-N seropositivity rates at day 57 and the PDV were significantly lower for participants who were RT-PCR–positive and anti-N–seronegative at day 29 than for those who were anti-N–seropositive at day 29 (P < 0.001 and P = 0.011, respectively).
Table 3.
Anti-N Seropositivity Rates at Day 57 and PDV for Infections Detected by Serology or PCR at Day 29 (Dose 2)
Discussion
These data show that, among the participants with PCR-confirmed COVID-19, anti-N Ab seropositivity at a median of 53 days after diagnosis occurred in 40% of the mRNA-1273 vaccine recipients versus 93% of the placebo recipients. Given the short time frame, the likely explanation is a vaccine-induced reduction in seroconversion. Anti-N seropositivity correlated with SARS-CoV-2 viral level at the illness visit, with each log increase in viral level nearly doubling the odds of anti-N seropositivity at the PDV. Because the viral level on the day of the illness visit in mRNA-1273 recipients with COVID-19 has been shown to be 100-fold lower than that in placebo recipients with COVID-19 (9), the lower anti-N seropositivity in the mRNA-1273 recipients could be partly explained by their reduced exposure to N-antigen. However, strong vaccine effects remain; at 2.0 log10 copies/mL, the predicted probability of seroconversion was 0.15 for vaccinated persons with COVID-19, compared with 0.71 for placebo recipients with COVID-19. This may be due to a difference in the live virus replication between vaccine and placebo recipients, which cannot be differentiated by the RT-PCR test. Another potential explanation is that the vaccine has much larger effects on reducing replication outside the nose, as was shown in a study evaluating the mRNA-1273 vaccine against SARS-CoV-2 challenge in a nonhuman primate model (11). There may be other features of the initial course of infection that influence anti-N Ab seroconversion and are affected by vaccination. Of note, the average viral level across post–COVID-19 illness visits was found to be a worse predictor of PDV seroconversion.
Infection with SARS-CoV-2 results in a wide spectrum of clinical outcomes from subclinical to fatal. Symptomatic COVID-19 of any severity and severe COVID-19 are of clear medical relevance. Although subclinical SARS-CoV-2 infection is of less direct medical importance, asymptomatic acquisition and carriage are important factors in household, institutional, and community transmission (12–15). Anti-N Ab seroconversion has been to date the major focus for defining subclinical and past COVID-19 detection, both on the individual and population levels (16–18).
Our study has shown that anti-N seropositivity as evidence of previous SARS-CoV-2 infection is complex and may be subject to large vaccine effects. Multiple studies have reported that some fraction of PCR-confirmed SARS-CoV-2 infections are not accompanied by seroconversion; estimates range from 5% to 36% (19–23). Although a large seroprevalence study in Ireland (24) found low rates of anti-N Ab seroconversion in hospital health care workers who were fully vaccinated (BNT162b2), our study provides the first evidence to our knowledge from a randomized, placebo-controlled trial with systematic surveillance for infection. This effect has consequences for interpretation of end points in vaccine trials, observational studies, and serosurveys and for monitoring and responding to the ongoing pandemic. The interpretation of anti-N Ab–based diagnostics of long COVID and multisystem inflammatory syndrome in adults and children with prior vaccination requires an understanding of the limitations and variability of the test performance by vaccination status and time since vaccination, among other variables.
Even with frequent serosampling, serosurveys that rely on antibodies to the N protein may underestimate within-community transmission dynamics. A meta-analysis showed that of the studies that reported the assay used for diagnosing previous infection, 59% used N-targeting tests (25). In our study, the seroconversion rate was about 40% at a median of 53 days after diagnosis for fully vaccinated persons who developed COVID-19. After 1 dose, the seroconversion rates for those who were PCR-positive at day 29 were 38.5% at day 57 and 50.0% at the PDV (median of 112 days after diagnosis); the seroconversion rates after 2 doses for those who were PCR-positive could not be evaluated in our study but are likely lower. Although anti-N Ab seroconversion is high for unvaccinated persons, the proportion of people immunologically naive to SARS-CoV-2 is diminishing as the global population acquires immunity through infection and vaccination (26–28). Hence, conclusions about the prevalence and incidence of SARS-CoV-2 infection based on serologic assays need to be weighed in the context of our results.
We found that participants with infection detected before vaccination, and those with infection diagnosed via serology before full vaccination, remained seropositive for the period of observation (to the PDV). Reductions in seroconversion rates were most evident in those who met COVID-19 case criteria for the primary end point—that is, became ill 14 or more days after full vaccination. Whether PCR-positive persons with asymptomatic breakthrough infections seroconvert at reduced rates will require study in cohorts with systematic asymptomatic testing. Vaccine efficacy trials for COVID-19 (5, 29–31) use anti-N Ab seroconversion as a secondary or exploratory end point to assess vaccine efficacy against asymptomatic SARS-CoV-2 infection, or as part of a secondary end point to assess vaccine efficacy against SARS-CoV-2 infection of any severity. Because we found that infections in the placebo group were about twice as likely to manifest through anti-N Ab seroconversion as those in the vaccine group, our results suggest that caution is needed when interpreting vaccine estimates against such end points. One solution may be to define the end point of interest as an infection that is accompanied by seroconversion. This seroconversion end point would exclude about 60% of COVID-19 cases and an unknown percentage of PCR-positive cases in fully vaccinated persons. The effect would likely be to reduce the overall sensitivity of the SARS-CoV-2 infection case definition. Whether infected persons who do not seroconvert after vaccination are less likely to transmit to others is also unknown. An additional approach that may reflect transmission potential would be to contrast the mean viral loads over all persons at a single point in time (or over the course of the acute illness), a metric known as vaccine efficacy on prevalent viral load (32, 33). Although the lack of anti-N seroconversion may indicate that the breakthrough infection was immunologically “silent,” other immunologic markers may be useful to determine these infections. These include SARS-CoV-2–specific cellular responses against the N and M proteins (34). These tests are at present expensive and often require special sampling and processing techniques that make implementation for large-scale seroprevalence studies difficult, but they would be important to explore in a research setting.
This analysis has a few limitations. First, our participants received the mRNA-1273 vaccine exclusively. Whether vaccination with BNT162b2 or Ad26.COV2.S interferes with anti-N seropositivity after infection to the same magnitude as mRNA-1273 is unknown. In a study of 31 persons, most of whom received BNT162b2 and then became infected with SARS-CoV-2, anti-N Abs were detected in 68% at 5 weeks after infection using enzyme-linked immunosorbent assay (35). Second, we used only the Elecsys assay, which has some of the highest sensitivity and specificity data, and whether other immunoassays will have different sensitivity for detecting recent or past infections in vaccinated persons is unknown. Third, our sample is small, with 52 fully vaccinated persons with breakthrough COVID-19 assessed for seroconversion a median of 53 days after diagnosis; thus, transient anti-N seropositivity events could go undetected. However, the close follow-up and uniformity in obtaining the swabs and serum samples increase confidence in and interpretability of our findings. Fourth, our vaccinated COVID-19 case patients exhibited illness a median of 77 days after their second dose, and most (86%) had mild to moderate disease (4). Whether anti-N seroconversion rates are higher for persons who are farther from second vaccination or have more severe disease, and lower for persons after a third or fourth dose, deserves further investigation. Fifth, postvaccination SARS-CoV-2 infections that are caused by the Delta or Omicron variant have been associated with higher viral level on diagnosis and more rapid clearance (36, 37). Whether breakthrough infections due to these variants are associated with different rates of seroconversion cannot be ascertained using these data. Finally, this analysis did not address a prespecified objective of the COVE study. Although the statistical analysis plan was prespecified, the COVE study was not designed to assess the sensitivity or specificity of the Elecsys assay in vaccinees versus placebo recipients.
In summary, these data suggest that assay limitations may exist in detecting anti-N Abs in persons recently vaccinated with mRNA-1273. Only an estimated 40% of mRNA-1273 vaccine recipients, versus 93% of placebo recipients, were anti-N–positive a median of 53 days after COVID-19 diagnosis. Determining population-level SARS-CoV-2 infections via serosurveillance and diagnosing past SARS-CoV-2 infection in the era of COVID-19 vaccination require further research on detection of recent or remote SARS-CoV-2 infection in vaccinated persons. Vaccination status should be taken into account when interpreting population and individual-level seroprevalence and seropositivity data.
Supplementary Material
Footnotes
This article was published at Annals.org on 5 July 2022.
* Drs. Follmann and Janes contributed equally to this work.
† Drs. El Sahly and Baden contributed equally to this work.
References
- 1. Tan SS , Saw S , Chew KL , et al. Comparative clinical evaluation of the Roche Elecsys and Abbott severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) serology assays for coronavirus disease 2019 (COVID-19). Arch Pathol Lab Med. 2021;145:32-38. [PMID: ] doi: 10.5858/arpa.2020-0499-SA [DOI] [PubMed] [Google Scholar]
- 2. Gallais F , Gantner P , Bruel T , et al. Evolution of antibody responses up to 13 months after SARS-CoV-2 infection and risk of reinfection. EBioMedicine. 2021;71:103561. [PMID: ] doi: 10.1016/j.ebiom.2021.103561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dan JM , Mateus J , Kato Y , et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371. [PMID: ] doi: 10.1126/science.abf4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. El Sahly HM , Baden LR , Essink B , et al; COVE Study Group. Efficacy of the mRNA-1273 SARS-CoV-2 vaccine at completion of blinded phase. N Engl J Med. 2021;385:1774-1785. [PMID: ] doi: 10.1056/NEJMoa2113017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baden LR , El Sahly HM , Essink B , et al; COVE Study Group. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403-416. [PMID: ] doi: 10.1056/NEJMoa2035389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bajema KL , Wiegand RE , Cuffe K , et al. Estimated SARS-CoV-2 seroprevalence in the US as of September 2020. JAMA Intern Med. 2021;181:450-460. [PMID: ] doi: 10.1001/jamainternmed.2020.7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roche Diagnostics. Elecsys Anti-SARS-CoV-2. Updated 11 February 2022. Accessed at https://diagnostics.roche.com/us/en/products/params/elecsys-anti-sars-cov-2.html on 11 February 2022.
- 8. Gilbert PB , Montefiori DC , McDermott AB , et al; Immune Assays Team. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43-50. [PMID: ] doi: 10.1126/science.abm3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pajon R , Paila YD , Girard B , et al; COVE Trial Consortium. Initial analysis of viral dynamics and circulating viral variants during the mRNA-1273 phase 3 COVE trial. Nat Med. 2022;28:823-830. [PMID: ] doi: 10.1038/s41591-022-01679-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DeLong ER , DeLong DM , Clarke-Pearson DL . Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837-45. [PMID: ] [PubMed] [Google Scholar]
- 11. Corbett KS , Nason MC , Flach B , et al. Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science. 2021;373:eabj0299. [PMID: ] doi: 10.1126/science.abj0299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Subramanian R , He Q , Pascual M . Quantifying asymptomatic infection and transmission of COVID-19 in New York City using observed cases, serology, and testing capacity. Proc Natl Acad Sci U S A. 2021;118. [PMID: ] doi: 10.1073/pnas.2019716118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Emery JC , Russell TW , Liu Y , et al; CMMID COVID-19 Working Group. The contribution of asymptomatic SARS-CoV-2 infections to transmission on the Diamond Princess cruise ship. Elife. 2020;9. [PMID: ] doi: 10.7554/eLife.58699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilmes P , Zimmer J , Schulz J , et al. SARS-CoV-2 transmission risk from asymptomatic carriers: results from a mass screening programme in Luxembourg. Lancet Reg Health Eur. 2021;4:100056. [PMID: ] doi: 10.1016/j.lanepe.2021.100056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johansson MA , Quandelacy TM , Kada S , et al. SARS-CoV-2 transmission from people without COVID-19 symptoms. JAMA Netw Open. 2021;4:e2035057. [PMID: ] doi: 10.1001/jamanetworkopen.2020.35057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones JM , Stone M , Sulaeman H , et al. Estimated US infection- and vaccine-induced SARS-CoV-2 seroprevalence based on blood donations, July 2020-May 2021. JAMA. 2021;326:1400-1409. [PMID: ] doi: 10.1001/jama.2021.15161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Z , Lewis B , Berney K , et al. Social vulnerability and rurality associated with higher SARS-CoV-2 infection-induced seroprevalence: a nationwide blood donor study, United States, July 2020 - June 2021. Clin Infect Dis. 2022. [PMID: ] doi: 10.1093/cid/ciac105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stone M , Di Germanio C , Wright DJ , et al; NHLBI Recipient Epidemiology and Donor Evaluation Study-IV-Pediatric (REDS-IV-P). Use of US blood donors for national serosurveillance of severe acute respiratory syndrome coronavirus 2 antibodies: basis for an expanded national donor serosurveillance program. Clin Infect Dis. 2022;74:871-881. [PMID: ] doi: 10.1093/cid/ciab537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oved K , Olmer L , Shemer-Avni Y , et al. Multi-center nationwide comparison of seven serology assays reveals a SARS-CoV-2 non-responding seronegative subpopulation. EClinicalMedicine. 2020;29:100651. [PMID: ] doi: 10.1016/j.eclinm.2020.100651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pathela P , Crawley A , Weiss D , et al; NYC Serosurvey Team. Seroprevalence of severe acute respiratory syndrome coronavirus 2 following the largest initial epidemic wave in the United States: findings from New York City, 13 May to 21 July 2020. J Infect Dis. 2021;224:196-206. [PMID: ] doi: 10.1093/infdis/jiab200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu W , Russell RM , Bibollet-Ruche F , et al. Predictors of nonseroconversion after SARS-CoV-2 infection. Emerg Infect Dis. 2021;27:2454-2458. [PMID: ] doi: 10.3201/eid2709.211042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thiruvengadam R , Chattopadhyay S , Mehdi F , et al; DBT India Consortium for COVID 19 Research. Longitudinal serology of SARS-CoV-2-infected individuals in India: a prospective cohort study. Am J Trop Med Hyg. 2021;105:66-72. [PMID: ] doi: 10.4269/ajtmh.21-0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nasrallah GK , Dargham SR , Shurrab F , et al. Analytic comparison between three high-throughput commercial SARS-CoV-2 antibody assays reveals minor discrepancies in a high-incidence population. Sci Rep. 2021;11:11837. [PMID: ] doi: 10.1038/s41598-021-91235-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Allen N , Brady M , Carrion Martin AI , et al. Serological markers of SARS-CoV-2 infection; anti-nucleocapsid antibody positivity may not be the ideal marker of natural infection in vaccinated individuals [Letter]. J Infect. 2021;83:e9-e10. [PMID: ] doi: 10.1016/j.jinf.2021.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen X , Chen Z , Azman AS , et al. Serological evidence of human infection with SARS-CoV-2: a systematic review and meta-analysis. Lancet Glob Health. 2021;9:e598-e609. [PMID: ] doi: 10.1016/S2214-109X(21)00026-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Our World in Data. Coronavirus (COVID-19) vaccinations. Updated 22 February 2022. Accessed at https://ourworldindata.org/covid-vaccinations on 23 February 2022.
- 27. Madhi SA, Kwatra G, Myers JE, et al. South African population immunity and severe Covid-19 with Omicron variant. medRxiv. Preprint posted online 27 January 2022. doi: 10.1101/2021.12.20.21268096 [DOI]
- 28. Bergeri I, Whelan M, Ware H, et al. Global epidemiology of SARS-CoV-2 infection: a systematic review and meta-analysis of standardized population-based seroprevalence studies, Jan 2020-Dec 2021. medRxiv. Preprint posted online 14 February 2022. doi: 10.1101/2021.12.14.21267791 [DOI]
- 29. Heath PT , Galiza EP , Baxter DN , et al; 2019nCoV-302 Study Group. Safety and efficacy of NVX-CoV2373 Covid-19 vaccine. N Engl J Med. 2021;385:1172-1183. [PMID: ] doi: 10.1056/NEJMoa2107659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sadoff J , Gray G , Vandebosch A , et al; ENSEMBLE Study Group. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N Engl J Med. 2021;384:2187-2201. [PMID: ] doi: 10.1056/NEJMoa2101544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Falsey AR , Sobieszczyk ME , Hirsch I , et al; AstraZeneca AZD1222 Clinical Study Group. Phase 3 safety and efficacy of AZD1222 (ChAdOx1 nCoV-19) Covid-19 vaccine. N Engl J Med. 2021;385:2348-2360. [PMID: ] doi: 10.1056/NEJMoa2105290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Follmann DA , Fay MP . Vaccine efficacy at a point in time. Biostatistics. 2022. [PMID: ] doi: 10.1093/biostatistics/kxac008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kennedy-Shaffer L , Kahn R , Lipsitch M . Estimating vaccine efficacy against transmission via effect on viral load. Epidemiology. 2021;32:820-828. [PMID: ] doi: 10.1097/EDE.0000000000001415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zuo J , Dowell AC , Pearce H , et al. Robust SARS-CoV-2-specific T cell immunity is maintained at 6 months following primary infection. Nat Immunol. 2021;22:620-626. [PMID: ] doi: 10.1038/s41590-021-00902-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bates TA , McBride SK , Leier HC , et al. Vaccination before or after SARS-CoV-2 infection leads to robust humoral response and antibodies that effectively neutralize variants. Sci Immunol. 2022;7:eabn8014. [PMID: ] doi: 10.1126/sciimmunol.abn8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Singanayagam A , Hakki S , Dunning J , et al; ATACCC Study Investigators. Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. Lancet Infect Dis. 2022;22:183-195. [PMID: ] doi: 10.1016/S1473-3099(21)00648-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Puhach O, Adea K, Hulo N, et al. Infectious viral load in unvaccinated and vaccinated patients infected with SARS-CoV-2 WT, Delta and Omicron. medRxiv. Preprint posted online 11 January 2022. doi: 10.1101/2022.01.10.22269010 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.