Abstract
Euryhaline teleosts can survive in environments with different salinities. Cortisol is an important hormone for acclimation to seawater (SW) of euryhaline teleosts. Osmotic stress transcription factor 1 (OSTF1), also called the transforming growth factor-beta stimulated clone 22 domain 3 (tsc22d3), was first reported in tilapia as an acute response gene and protein under hyperosmotic stress, and it is regulated by cortisol. To date, most studies on OSTF1 have focused on freshwater inhabitants, such as tilapia, medaka, and catadromous eel. The expression of OSTF1 and the correlation between OSTF1 and cortisol in marine inhabitant euryhaline teleosts, to our knowledge, remain unclear. This study reveals the changes in the expression levels of branchial OSTF1, plasma cortisol levels, and their correlation in the marine inhabitant milkfish with ambient salinities. The two sequences of milkfish TSC22D3 transcripts were classified as OSTF1a and OSTF1b. Both genes were expressed universally in all detected organs and tissues but were the most abundant in the liver. Similar gene expression levels of ostf1a and ostf1b were found in SW- and fresh water (FW)-acclimated milkfish gills, an important osmoregulatory organ. Within 12 hours of being transferred from FW to SW, the gene expression level of ostf1b increased significantly (4 folds) within 12 h, whereas the expression level of ostf1a remained constant. Moreover, cortisol levels increased rapidly after being transferred to a hyperosmotic environment. After an intraperitoneal injection of cortisol, the gene expression levels of ostf1a and ostf1b were elevated. However, under hyperosmotic stress, ostf1a gene expression remained stable. Overall, the results revealed that ostf1b was the primary gene in milkfish responding to hypertonic stress, and cortisol concentration increased after the transfer of milkfish from FW to SW. Furthermore, cortisol injection increased the expression of ostf1a and ostf1b. As a result, factors other than cortisol may activate ostf1b in milkfish gills in response to an environmental salinity challenge.
Introduction
Euryhaline teleosts can adapt to fluctuations in environmental salinity. In 2005, the osmotic stress transcription factor 1 (OSTF1) was first identified in the primary culture of gill epithelial cells of the Mozambique tilapia (Oreochromis mossambicus) upon hyperosmotic stress [1]. It is thought to be an early and transiently upregulated gene and protein in fresh water (FW) inhabitant euryhaline teleosts [1–3]. Further studies on Japanese medaka (Oryzias latipes) revealed two transcripts of tsc22d3. ostf1a with high level of similarity with tilapia ostf1 [4]. Meanwhile, both gene expression was shown to be upregulated under hyperosmotic stress. Moreover, ostf1b modulates the expression of aquaporin 1 (AQP1), cystic fibrosis transmembrane conductance regulator (CFTR), and sodium hydrogen antiporter 3 (NHE3) for osmoregulation [4]. OSTF1 acts as a multifunctional transcription factor in fish. It participates in different downstream functions in different species, such as osmoregulation in tilapia [1], medaka [4], and Japanese eel (Anguilla japonica) [3], embryogenesis in zebrafish (Danio rerio) [5], and sex change in orange-spotted grouper (Epinephelus coioides) [6].
According to bioinformatics analysis, OSTF1 is a transcription factor characterized by a leucine zipper domain. It is a member of the transforming growth factor-beta stimulated clone 22 (tsc22) family. OSTF1 is also known as transforming growth factor-beta stimulated clone 22 domain 3 (tsc22d3) or glucocorticoid-induced leucine zipper (GILZ) [1,2,7]. In mammals, GILZ has been found to be an anti-inflammatory regulator that binds to proinflammatory transcription factors (e.g., nuclear factor kappa light chain enhancer of activated B cells, NF-κB) [8,9]. In addition, GILZ in mammalian kidney epithelial cells interacts with several epithelial Na+ channel (ENaC) regulatory proteins (e.g., serum- and glucocorticoid-induced kinase 1, SGK1) to regulate the expression and activity of ENaC by aldosterone through the MAPK pathway [10].
Cortisol, a glucocorticoid stress hormone, enables seawater (SW) adaptation in fish [11]. Cortisol affects the development and morphology of ionocytes in fish gills. Therefore, it is involved in increasing the ability of fish to osmoregulate in SW [12–14] and in upregulating several hyperosmotic stress-responsive genes [15–19]. On the other hand, cortisol plays a critical role in the energy substrate reallocation, which is crucial for SW adaptation of fish [18]. Moreover, McGuire et al. (2010) reported upregulated ostf1 expression in fresh water (FW)- or SW-acclimated tilapia gills after cortisol injection [20].
Milkfish (Chanos chanos) is a marine euryhaline teleost distributed in tropical and subtropical regions of the world [21–23]. Milkfish is one of the most important aquaculture species in Southeast Asian countries, including Indonesia, Philippines, and Taiwan [21,24,25]. It has outstanding osmoregulation ability and can survive in environments with salinity ranging from 0 to 150‰ [26,27].
To date, most studies on fish OSTF1 have examined FW inhabitant euryhaline species (e.g. tilapia and medaka) or catadromous species (e.g. eel), revealing that the gene expression level of ostf1 increased after transfer to SW from FW [1,3,4]. There was only one study on a marine inhabitant euryhaline teleost, the black porgy (Acanthopagrus schlegeli), showing that the gene expression level of ostf1 increased after transfer to FW from SW [28]. However, little is known about the expression of OSTF1 in marine euryhaline fish after exposure to hypoosmotic or hyperosmotic environments, and the effect of cortisol, the SW-acclimation hormone, on OSTF1 expression in marine euryhaline teleosts. To better understand the potential roles and regulatory mechanisms of OSTF1 in marine euryhaline teleosts, the present study aimed to investigate (i) the expression of OSTF1 in the gills of milkfish, a marine euryhaline species, when exposed to hypoosmotic (FW) or hyperosmotic (SW) environments, and (ii) the effects of cortisol on branchial OSTF1 expression in euryhaline milkfish.
Materials and methods
Experimental fish and environments
Juvenile milkfish (Chanos chanos) with 17.09 ± 1.50 g of weight and 10.55 ± 0.35 cm of length were obtained from the DaShun fish farm in Tainan, Taiwan. Milkfish were reared in brackish water (BW; 15‰) at 28 ± 1°C for two weeks, then transferred to either fresh water (FW) or seawater (SW) at 28 ± 1°C for four-weeks acclimation. BW and SW were prepared from local tap water containing appropriate amounts of artificial sea salt (Blue Treasure Tropic Fish Sea Salt, Qingdao, China). The photoperiod in the fish room was 12 h light:12 h dark. Water was continuously circulated through fabric-floss filters, and a quarter of the water was changed every two weeks. The fish were fed commercial milkfish pellet diets once daily.
Acclimation, transfer, and cortisol injection experiments
For long-term acclimation experiments, milkfish were acclimated to FW or SW for at least four weeks before tissue sampling. For the transfer experiments, FW- and SW-acclimated milkfish were transferred from FW to SW or SW to FW. For the cortisol injection experiments, FW-acclimated milkfish were injected with cortisol or DMSO (control group). Fish were sampled at 0, 3, 6, 12, and 24 h after transfer or injection.
Ethical statement
All experiments were conducted according to the principles and procedures approved by the Institutional Animal Care and Use Committee (IACUC) of National Chung Hsing University (IACUC approval no. 108–137 granted to T.H. Lee).
Blood collecting and tissue sampling
The milkfish were fasted for one day and anesthetized with 0.05% (500 μL/L) 2-phenoxyethanol (PANREAC, Barcelona, Spain) before blood collection and sampling. Milkfish blood was collected from the caudal vein using a Li-heparinized syringe with 27 G needles. The plasma was collected after centrifugation (5,000 × g for 10 min at 4°C) of the blood and stored at -20°C before analysis. After blood collection, the milkfish were sacrificed, and all tissues were sampled. The tissues were immediately frozen in liquid nitrogen and stored at -80°C before homogenization.
Total RNA extraction and reverse transcription (RT)
Total RNA was extracted using IsoI-RNA Lysis Reagent (5 Prime, Gaithersburg, MD, USA) following the manufacturer’s instructions. The RNA pellet was dissolved in sterilized distilled water and deionized water. The intact extracted RNA was verified using 1% agarose gel (SeaKem® LE Agarose; Lonza, Basel, Switzerland) electrophoresis with SafeViewTM Classic nucleic acid stain (ABM, San Jose, CA, USA). The concentration and quality of the extracted RNA were measured using NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE, USA). Purified RNA with an A260/A280 ratio between 1.8 and 2.0, and an A260/A230 ratio over 2.0, were used for all RNA experiments. For RT, first-strand cDNA was synthesized from 1 μg of total RNA samples using the iScriptTM cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) following the manufacturer’s instructions.
Real-time PCR (RT-PCR) analysis
For RT-PCR, the primers for osft1a (XM_030764688.1), ostf1b (XM_030764689.1), and β-actin (XM_030789841.1) were designed using Primer 3 Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). The amplification efficiency from the standard curve of each primer pair used for RT-PCR analysis was within the range of 95–105% (Table 1). To analyze the gene expression of osft1, the MiniOpticon Real-Time PCR system (Bio-Rad) with SYBR green (2x KAPA SYBR® FAST qPCR Master Mix; Kapa Biosystems, Wilmington, MA, USA) was used. All data were normalized using β-actin as the reference gene. To quantify gene expression level in this experiment, the comparative Ct value method was used with the following formula: 2^- [(Ct target, n–Ct β-actin, n)—(Ct target, control–Ct β-actin, control)]. In the formula, ‘Ct’ represented the threshold cycle number in the formula and ‘n’ represented each cDNA sample used in these experiments [29].
Table 1. Primer sequences used for real-time PCR in this study.
| Genes | Primer | sequence 5’ to 3’ | Amplicon size (bp) |
Efficiency (%) |
|---|---|---|---|---|
| Ostf1a | CC_ostf1a_F544 | CCATCGGACTGGACTGCT | 189 | 96.8 |
| CC_ostf1a_R733 | TCCAGGGAGTCCTGTCTCAT | |||
| Ostf1b | CC_ostf1b_F161 | CTTCAGCAGGCTGTTCTCG | 180 | 96.6 |
| CC_ostf1b_F341 | CTTGACAACAGTGCCTCTGG | |||
| β-actin | CC_β-actin_F | CCATTGAGCACGGTATTGTCA | 82 | 103.4 |
| CC_β-actin_R |
GCAACACGCAGCTCGTTGTA
|
F, forward strand; R, reverse strand.
Cortisol injection and measurement of plasma cortisol concentration
Cortisol doses used in this study were chosen according to Hu et al. (2019) [30]. Hydrocortisone (Sigma) was dissolved in dimethyl sulfoxide (DMSO; Sigma) [11,16,30], and used for intraperitoneal injection. Milkfish were injected with DMSO and 4 μg cortisol/g body weight injection, respectively [30]. The volume of dissolved cortisol was calculated according to the average milkfish weight. Before injection, fish were anaesthetized by 0.05% (500 μL/L) 2-phenoxyethanol.
Determination of cortisol concentration
Plasma was used to detect cortisol concentrations, which were determined using a fish cortisol ELISA kit (Cusabio Biotech, Wuhan, China) following the manufacturer’s instructions.
Statistical analyses
In this study, the prediction of protein motifs of the milkfish OSTF1 sequences was analyzed by Myhits (https://myhits.isb-sib.ch/cgi-bin/motif_scan). The normality test was verified to test the distribution by the method of Shapiro-Wilk normality test, and nonparametric methods were used for data analysis. The tissue distribution and the comparison between cortisol and DMSO injection were analyzed using the Mann–Whitney test, and time-course data were analyzed using the Kruskal–Wallis test with Dunn’s multiple comparisons test. All analyses were performed using GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA). Values are expressed as mean ± standard error of the mean (SEM), p<0.05 was set as the significance level.
Results
Classification of OSTF1 in milkfish
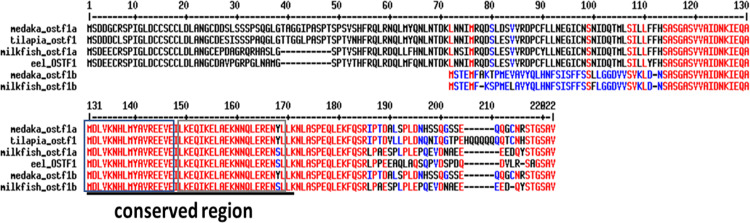
The milkfish ostf1 sequences, tsc22d3×1 (XM_030764688.1) and tsc22d3×2 (XM_030764689.1) (S1 Fig), were searched in the NCBI database. Based on the amino acid sequences similarities between milkfish TSC22D3 and medaka OSTF1a and OSTF1b, these two sequences were named OSTF1a (TSC22D3×1) and OSTF1b (TSC22D3×2), respectively. By alignment of the amino acid sequences of milkfish OSTF1 with those of the other teleosts (Fig 1), milkfish OSTF1a revealed more than 80% similarity with the sequences of the tilapia OSTF1, eel OSTF1, and medaka OSTF1a. The sequence similarity of OSTF1b was also found between milkfish and medaka. The two milkfish OSTF1 sequences shared almost 100% similarity with other teleosts in the conserved region. Based on the motif prediction, both OSTF1 sequences of milkfish have the TSC22 domain and leucine zipper (Fig 1).
Fig 1. The amino acid sequence of milkfish OSTF1a and OSTF1b was aligned with OSTF1 sequences of other teleostean species.
The identical amino acids were indicated by red residues, the low consensus amino acids were indicated by blue residues, and black residues represented the non-identical amino acids. The underline indicated the conserved region of OSTF1. The blue rectangle indicated the TSC22 domain, and the gray rectangle indicated the leucine zipper.
Tissue distribution of ostf1a and ostf1b in FW- and SW-acclimated milkfish
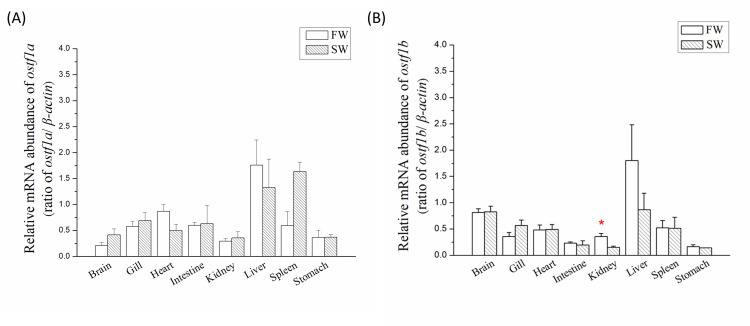
In FW- and SW-acclimated milkfish, ostf1a and ostf1b were expressed in all the tissues (Fig 2). Moreover, ostf1a was mainly expressed in the liver of FW- and SW-acclimated milkfish and in the spleen of SW-acclimated milkfish (Fig 2A), whereas the highest abundance of ostf1b was found in the liver of milkfish (Fig 2B). Meanwhile, ostf1a and ostf1b were expressed in all detected targets. Significant differences in ostf1a and ostf1b levels were found in the spleens and kidneys, respectively, between the FW and SW groups. However, in the gills, the expression levels of ostf1a (Fig 2A) and ostf1b (Fig 2B) were similar in FW- and SW-acclimated milkfish.
Fig 2. Tissue distribution of ostf1 mRNA abundance in milkfish acclimated to fresh water or seawater.
(A) ostf1a and (B) ostf1b was detected by real-time PCR. The expression has been normalized according to the expression of β-actin. Values are shown in means ± S.E.M. Gill and kidney, n = 6; other tissues, n = 3 (p < 0.05, analyzed by Mann–Whitney test). The asterisks indicated significant differences between the FW and SW group.
Gene expression of ostf1a and ostf1b in gills of milkfish upon salinity challenges
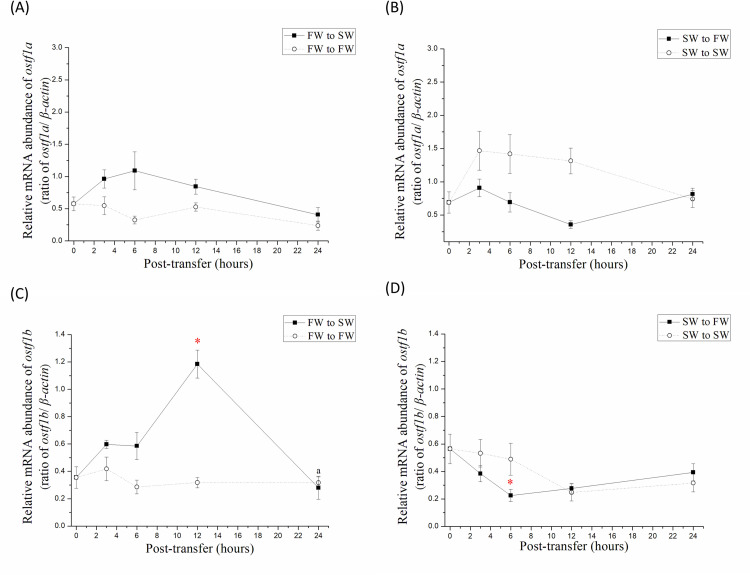
To determine whether ostf1a and ostf1b are rapid response genes to hyperosmotic or hypoosmotic stress in the gills of milkfish, one-month FW- and SW-acclimated milkfish were directly transferred to SW and FW, respectively, and set as the salinity-transfer (study) groups. Meanwhile, FW- and SW-acclimated milkfish directly transferred to FW and SW, respectively, were set as the control groups. In the SW-transfer study group, the expression level of gill ostf1a did not change significantly after transfer (Fig 3A). However, the gene expression of gill ostf1b in the SW transfer group increased significantly at 12 h post-transfer and decreased back to the original level 24 h after transfer (Fig 3C). On the other hand, in the FW-transfer study group, the expression of the ostf1a in gills was similar (Fig 3B), while branchial ostf1b expression was significantly lower at 6 h post-transfer from SW (0 h) (Fig 3D).
Fig 3. Time-course mRNA abundance of ostf1 in gills of milkfish transferred from FW to SW and SW to FW for 24 h.
(A, B) ostf1a and (C, D) ostf1b was detected by real-time PCR. The expression has been normalized according to the expression of β-actin. The time-points of sampling are 0, 3, 6, 12 and 24 h, respectively. Values were shown in means ± S.E.M. (n = 6). The asterisks indicated significant differences between each time-points and 0 h (p < 0.05, analyzed by Kruskal–Wallis test with Dunn’s multiple comparisons test).
Plasma cortisol concentration of milkfish upon salinity challenges
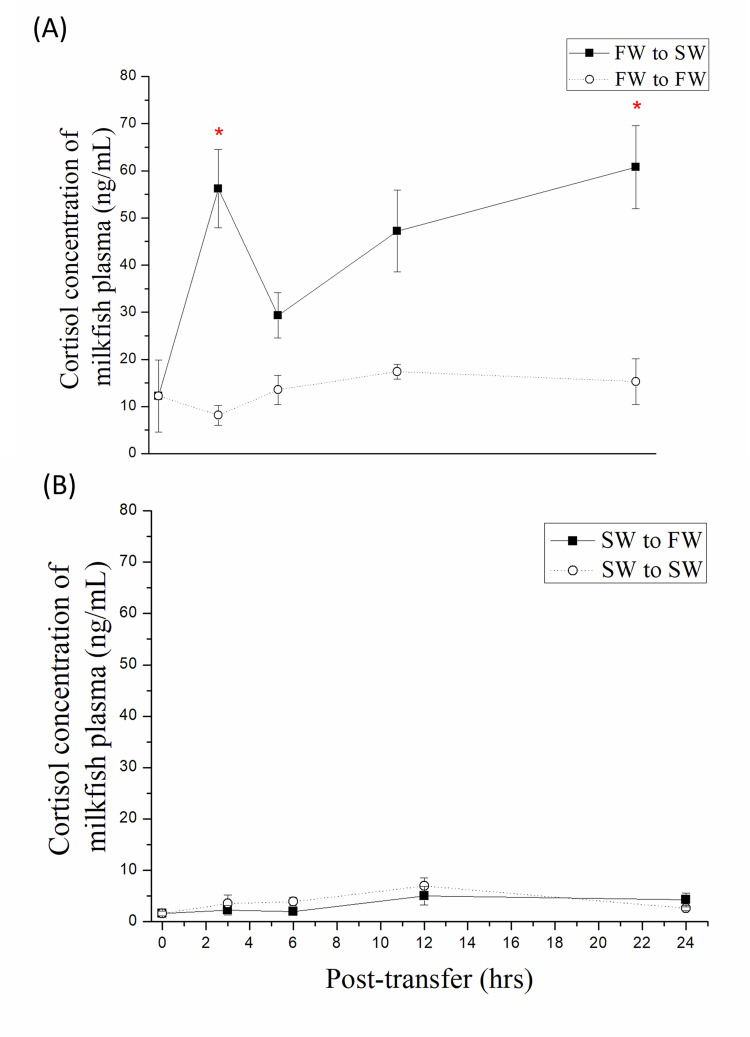
To determine the profile of cortisol concentration after salinity transfer in milkfish blood, the study and control groups of SW-transfer and FW-transfer milkfish were performed as previously described. After transfer, milkfish plasma was collected at each time point. The cortisol concentration in the plasma of the SW-transfer study group increased rapidly and significantly at 3 and 24 h post-transfer compared to FW (0 h) (Fig 4A). In contrast, plasma cortisol levels were steady in both the study and control groups of the FW transfer milkfish at 24 h post-transfer from SW (Fig 4B).
Fig 4. Plasma cortisol concentration of milkfish transferred from (A) FW to SW and (B) SW to FW for 24 h.
Values were shown in mean ± S.E.M. (n = 6). The asterisks indicated significant differences between each time-point and 0 h (p < 0.05, analyzed by Kruskal–Wallis test with Dunn’s multiple comparisons test).
Gene expression of ostf1a and ostf1b in gills of milkfish after cortisol injection
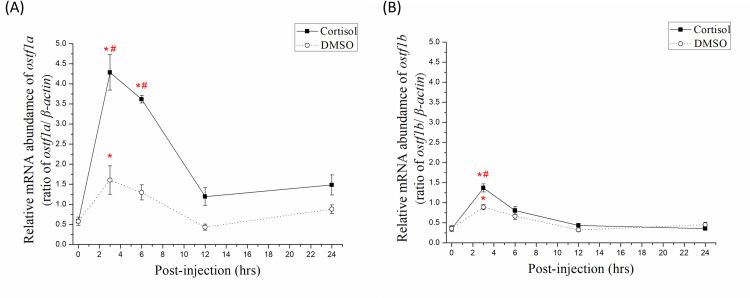
To illustrate the relationship between ostf1 and cortisol, plasma cortisol concentration in FW milkfish was measured after cortisol injection (S2 Fig), and ostf1a and ostf1b expression was detected. Comparisons between different time-points of the cortisol groups at 0 h showed that the gene expression level of ostf1a in gills of the FW milkfish increased significantly at 3 and 6 h (7–9 folds on average) and returned to the background level at 12 h after cortisol injection (Fig 5A). The gene expression level of ostf1b increased significantly at 3 h (4 folds on average) and returned to the background level after 6 h (Fig 5B). In the DMSO group, ostf1a and ostf1b expression levels increased significantly (3–4 folds on average) 3 h after injection. Comparisons between the cortisol and DMSO groups at the same time points showed that the gene expression level of ostf1a was significantly higher at 3 and 6 h (Fig 5A), while that of ostf1b was significantly higher at 3 h (Fig 5B).
Fig 5. Time-course mRNA abundance of (A) ostf1a and (B) ostf1b in gills of FW-acclimated milkfish after injection with cortisol and DMSO for 24 h.
The abundance has been normalized according to the expression of β-actin. Values were shown in mean ± S.E.M. Treatment group, n = 6; control group, n = 4. The asterisks indicated significant differences between each time-point and 0 h (p < 0.05, analyzed by Kruskal–Wallis test with Dunn’s multiple comparisons test). The sharps indicated significant differences between the cortisol group and DMSO group at the same time-point (p < 0.05, analyzed by Mann–Whitney test).
Discussion
OSTF1 was first identified in the gills of Mozambique tilapia and as a transcription factor with a leucine zipper domain [1]. Subsequently, studies on ostf1 in medaka gills were conducted on its expression and possible participation in the JNK pathway [4]. In medaka, two tsc22d3 transcripts have been named ostf1a and ostf1b [4]. This study revealed that in the seawater (SW) inhabitant milkfish, two tsc22d3 transcripts were found in the NCBI database. Based on the similarity of amino acid sequences between tilapia and medaka, the milkfish TSC22D3 was renamed as OSTF1a or OSTF1b.
Most fish studies focused on the expression and functions of ostf1 in gills [1,3,4,28]. There is limited understanding of ostf1 expression in other tissues. To our knowledge, this study is the first to reveal the tissue distribution of ostf1 in fish. In milkfish, both ostf1a and ostf1b are expressed in all tissues. The ostf1a was highly expressed in the liver and spleen of SW-acclimated milkfish, whereas ostf1b was highly expressed in the liver, and significantly higher in the kidney of FW milkfish. The gill and kidney are important organs for osmoregulation, and the liver and spleen are crucial for immune functions in teleosts [31,32]. In Atlantic salmon (Salmo salar), microarray analysis showed that the gene expression levels of ostf1 decreased in the liver, muscle, and head kidney after bacterial injection [33]. In mammals, tsc22d3 is expressed in different organs and tissues, and it participates in adipogenesis, immunity, and renal sodium transport [34]. In zebrafish, ostf1 may participate in embryonic development [5]. Taken together, ostf1 in fish may be a multifunctional transcription factor for maintaining osmoregulation, as well as playing other physiological roles.
In the gills of freshwater (FW) euryhaline fish, e.g., tilapia, medaka, and eel, ostf1 was found to be a rapid response gene to salinity challenge. The gene expression level of ostf1(b) increased 4–10 folds in 2–6 h after transfer from FW to SW, but later returned to the background level [1–4]. In the gills of milkfish transferred from FW to SW, the expression level of ostf1b elevated and reached its highest level at 12 h, and then returned to the background level at 24 h. Therefore, similar expression levels of branchial ostf1b were found in FW- and SW-acclimated milkfish. On the other hand, the expression level of ostf1b decreased significantly after 6 h of transfer from SW to FW. Accordingly, branchial ostf1b expression in marine euryhaline milkfish was found to respond rapidly when exposed to hypoosmotic environments, although it responded later than medaka, tilapia, and eels upon hyperosmotic challenge. Moreover, ostf1a expression in medaka gills increased significantly after SW exposure [4], whereas ostf1a expression in milkfish gills did not change significantly after exposure to hyperosmotic SW. Hence, milkfish ostf1b might be the major osmotic challenge, while milkfish ostf1a might be responsible for other physiological roles. In addition, when exposed to hyperosmotic stress, ostf1 of marine euryhaline milkfish showed similar responses to other FW euryhaline teleosts.
Cortisol plays an important role in the SW adaption of euryhaline teleosts [17–19]. Cortisol is critical for SW tolerance maintenance in the larval stage of the summer flounder (Paralichthys dentatus) [11]. In Asian sea bass (Lates calcarifer), cortisol can improve tolerance and survival in hypersaline environments, but not in hyposaline environments [35]. In long-term FW- and SW-acclimated milkfish, plasma cortisol levels were higher in FW than in SW milkfish [30]. In tilapia, the plasma cortisol levels were similar between FW and SW after one-month acclimation [36]. However, the plasma cortisol concentration in tilapia increases rapidly when exposed to hyperosmotic environments [37]. As in tilapia, this study revealed that after transfer from FW to SW, the plasma cortisol concentration of milkfish increased immediately. However, cortisol concentration was stable when milkfish were transferred from SW to FW. Furthermore, compared with Hu et al. (2019) [30], cortisol concentration might decrease after the transfer of milkfish from FW to SW in one day. Accordingly, in marine euryhaline milkfish, cortisol was found to be an important hormone in response to acute salinity challenge; in this regard, the marine euryhaline milkfish was similar to other FW euryhaline teleosts.
McGurie et al. (2010) reported that ostf1 expression was induced by cortisol in Mozambique tilapia [20]. In this study, after intraperitoneal injection of cortisol, two ostf1 genes identified in milkfish reached their highest expression levels at 3 h, related to changes in plasma cortisol levels after cortisol injection. As a result, cortisol can upregulate the gene expression of milkfish ostf1a and ostf1b. In addition, the increase in ostf1a level was greater than that in ostf1b level. Meanwhile, ostf1a was not found to be the primary gene when milkfish were exposed to hyperosmotic stress. We assumed that cortisol was not the primary factor in activating milkfish ostf1b in response to salinity stress, and that other factors contributed to ostf1b upregulation but did not increase ostf1a expression in response to salinity challenge. Furthermore, after injection with DMSO (the control group), the gene expression levels of ostf1a and ostf1b also increased in a short time but were 2–3 folds lower than those in the cortisol-treated group. Therefore, it will be intriguing to clarify whether ostf1b responds to hypertonic stress in milkfish gills and whether ostf1a responds to other unknown functions. In the present study, cortisol upregulated the gene expression of ostf1b in marine euryhaline milkfish, similar to freshwater euryhaline tilapia [20].
Conclusion
In summary, the two milkfish TSC22D3 sequences were classified as OSTF1a and OSTF1b. Both ostf1a and ostf1b were expressed in all examined tissues and were most abundant in the liver, but not in the gill or other osmoregulatory organs. In milkfish gills, however, only ostf1b expression was salinity-dependent upon osmotic stress. In addition, cortisol levels were enhanced after milkfish were transferred from FW to SW, and gene expression levels of both ostf1a and ostf1b increased after cortisol treatment. Taken together, we assumed that other factors, but not cortisol, would activate milkfish ostf1b in response to hyperosmotic challenge. In addition to osmoregulation, ostf1a and ostf1b may play other roles in physiological homeostasis maintenance. More research is needed to understand underlying functions of OSTF1 and the relationship between cortisol and OSTF1 in milkfish.
Supporting information
The identical amino acids were indicated by red residues, the low consensus amino acids were indicated by blue residues, and black residues represented the non-identical amino acids.
(TIF)
Values were mean ± S.E.M.. Treatment group, n = 6; control group, n = 4. The asterisks indicated significant differences between each time-point and 0 h (p < 0.05, analyzed by Kruskal–Wallis test with Dunn’s multiple comparisons test).
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was financially supported in part by research projects to T.-H. Lee from the Ministry of Science and Technology (MOST) of Taiwan (109-2313-B-005-005-MY3 and 109-2911-I-005-507) and in part by the iEGG and Animal Biotechnology Center from The Feature Area Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) of Taiwan (109-S-0023-F). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fiol DF, Kültz D. Rapid hyperosmotic coinduction of two tilapia (Oreochromis mossambicus) transcription factors in gill cells. Proceedings of the National Academy of Sciences. 2005;102(3), 927–932. doi: 10.1073/pnas.0408956102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiol DF, Chan SY, Kültz D. Regulation of osmotic stress transcription factor 1 (Ostf1) in tilapia (Oreochromis mossambicus) gill epithelium during salinity stress. Journal of Experimental Biology. 2006;209(16), 3257–3265. doi: 10.1242/jeb.02352 [DOI] [PubMed] [Google Scholar]
- 3.Tse WKF, Chow SC, Wong CKC. Cloning of eel osmotic stress transcription factor and the regulation of its expression in primary gill cell culture. Journal of Experimental Biology. 2008;211(12), 1964–1968. doi: 10.1242/jeb.017368 [DOI] [PubMed] [Google Scholar]
- 4.Tse WKF, Lai KP, Takei Y. Medaka osmotic stress transcription factor 1b (Ostf1b/TSC22D3-2) triggers hyperosmotic responses of different ion transporters in medaka gill and human embryonic kidney cells via the JNK signaling pathway. The international journal of biochemistry & cell biology. 2011;43(12), 1764–1775. [DOI] [PubMed] [Google Scholar]
- 5.Tse WKF, Jiang YJ, Wong CKC. Zebrafish transforming growth factor-β-stimulated clone 22 domain 3 (TSC22D3) plays critical roles in Bmp-dependent dorsoventral patterning via two deubiquitylating enzymes Usp15 and Otud4. Biochimica et Biophysica Acta (BBA)-General Subjects. 2013;1830(10), 4584–4593. doi: 10.1016/j.bbagen.2013.05.006 [DOI] [PubMed] [Google Scholar]
- 6.Kuo CE, Chen YM. Characterization of gonadal glucocorticoid-induced leucine zipper (GILZ) protein expression during sex change in the protogynous orange-spotted grouper, Epinephelus coioides. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2020;242, 110416. doi: 10.1016/j.cbpb.2020.110416 [DOI] [PubMed] [Google Scholar]
- 7.Tse WKF. The role of osmotic stress transcription factor 1 in fishes. Frontiers in zoology. 2014;11(1), 86. doi: 10.1186/s12983-014-0086-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koizume S, Takahashi T, Yoshihara M, Nakamura Y, Ruf W, Takenaka K, et al. Cholesterol starvation and hypoxia activate the FVII gene via the SREBP1-GILZ pathway in ovarian cancer cells to produce procoagulant microvesicles. Thrombosis and Haemostasis. 2019;119(07), 1058–1071. doi: 10.1055/s-0039-1687876 [DOI] [PubMed] [Google Scholar]
- 9.Ronchetti S, Migliorati G, Riccardi C. GILZ as a mediator of the anti-inflammatory effects of glucocorticoids. Frontiers in endocrinology. 2015;6, 170. doi: 10.3389/fendo.2015.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soundararajan R, Zhang TT, Wang J, Vandewalle A, Pearce D. A novel role for glucocorticoid-induced leucine zipper protein in epithelial sodium channel-mediated sodium transport. Journal of Biological Chemistry. 2005;280(48), 39970–39981. doi: 10.1074/jbc.M508658200 [DOI] [PubMed] [Google Scholar]
- 11.Veillette PA, Merino M, Marcaccio ND, Garcia MM, Specker JL, Cortisol is necessary for seawater tolerance in larvae of a marine teleost the summer flounder. General and comparative endocrinology. 2007;151, 116–121. doi: 10.1016/j.ygcen.2006.12.019 [DOI] [PubMed] [Google Scholar]
- 12.Dangé AD. Branchial Na+-K+-ATPase activity in freshwater or saltwater acclimated tilapia, Oreochromis (Sarotherodon) mossambicus: effects of cortisol and thyroxine. General and comparative endocrinology. 1986;62(2), 341–343. doi: 10.1016/0016-6480(86)90125-5 [DOI] [PubMed] [Google Scholar]
- 13.Evans DH, Piermarini PM, Choe KP. The multifunctional fish gill: dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiological reviews. 2005;85(1), 97–177. doi: 10.1152/physrev.00050.2003 [DOI] [PubMed] [Google Scholar]
- 14.McCormicks SD. Hormonal control of gill Na+, K+-ATPase and chloride cell function. In Fish physiology (Vol. 14, pp. 285–315). Academic Press; 1995. [Google Scholar]
- 15.Foskett JK, Bern HA, Machen TE, Conner M. Chloride cells and the hormonal control of teleost fish osmoregulation. Journal of experimental Biology. 1983;106(1), 255–281. doi: 10.1242/jeb.106.1.255 [DOI] [PubMed] [Google Scholar]
- 16.Kumai Y, Nesan D, Vijayan MM, Perry SF. Cortisol regulates Na+ uptake in zebrafish, Danio rerio, larvae via the glucocorticoid receptor. Molecular and cellular endocrinol. 2012;364, 113–125. doi: 10.1016/j.mce.2012.08.017 [DOI] [PubMed] [Google Scholar]
- 17.McCormick SD. Endocrine control of osmoregulation in teleost fish. American zoologist. 2001;41(4), 781–794. [Google Scholar]
- 18.Mommsen TP, Vijayan MM, Moon TW. Cortisol in teleosts: dynamics, mechanisms of action, and metabolic regulation. Reviews in Fish Biology and Fisheries. 1999;9(3), 211–268. [Google Scholar]
- 19.Vijayan MM, Pereira C, Grau EG, Iwama GK. Metabolic responses associated with confinement stress in tilapia: the role of cortisol. Comparative Biochemistry and Physiology Part C: Pharmacology, Toxicology and Endocrinology. 1997;116(1), 89–95. [Google Scholar]
- 20.McGuire A, Aluru N, Takemura A, Weil R, Wilson JM, Vijayan MM. Hyperosmotic shock adaptation by cortisol involves upregulation of branchial osmotic stress transcription factor 1 gene expression in Mozambique Tilapia. General and comparative endocrinology. 2010;165(2), 321–329. doi: 10.1016/j.ygcen.2009.07.016 [DOI] [PubMed] [Google Scholar]
- 21.FitzGerald WJ. Milkfish aquaculture in the Pacific: potential for the tuna longline fishery bait market. Noumea, New Caledonia: Secretariat of the Pacific Community; 2004. [Google Scholar]
- 22.Kumar N, Ambasankar K, Krishnani KK, Kumar P, Akhtar MS, Bhushan S, et al. Dietary pyridoxine potentiates thermal tolerance, heat shock protein and protect against cellular stress of Milkfish (Chanos chanos) under endosulfan-induced stress. Fish & Shellfish Immunology. 2016;55, 407–414. doi: 10.1016/j.fsi.2016.06.011 [DOI] [PubMed] [Google Scholar]
- 23.Swanson C. Early development of milkfish: effects of salinity on embryonic and larval metabolism, yolk absorption and growth. Journal of fish Biology. 1996;48(3), 405–421. [Google Scholar]
- 24.Systematics Bagarinao T., distribution, genetics and life history of milkfish, Chanos chanos. Environmental biology of fishes. 1994;39(1), 23–41. [Google Scholar]
- 25.Hanke I. Milkfish (Chanos chanos) under stress: Contributing to fish welfare in tropical aquaculture by identifying and quantifying potential stressors (Doctoral dissertation, Universität Bremen); 2019.
- 26.Crear D. Observations on the reproductive state of milkfish populations (Chanos chanos) from hypersaline ponds on Christmas Island (Pacific Ocean). In Proceedings of the World Mariculture Society (Vol. 11, No. 1‐4, pp. 548–556). Oxford, UK: Blackwell Publishing Ltd; 1980. [Google Scholar]
- 27.Ferraris RP, Almendras JM, Jazul AP. Changes in plasma osmolality and chloride concentration during abrupt transfer of milkfish (Chanos chanos) from seawater to different test salinities. Aquaculture. 1988;70(1–2): 145–157. [Google Scholar]
- 28.Choi CY, An KW. Cloning and expression of Na+/K+-ATPase and osmotic stress transcription factor 1 mRNA in black porgy, Acanthopagrus schlegeli during osmotic stress. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2008;149(1), 91–100. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4): 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 30.Hu YC, Chu KF, Hwang LY, Lee TH. Cortisol regulation of Na+, K+-ATPase β1 subunit transcription via the pre-receptor 11β-hydroxysteroid dehydrogenase 1-like (11β-Hsd1L) in gills of hypothermal freshwater milkfish, Chanos chanos. The Journal of steroid biochemistry and molecular biology. 2019;192, 105381. doi: 10.1016/j.jsbmb.2019.105381 [DOI] [PubMed] [Google Scholar]
- 31.Bo J, Cai L, Xu JH, Wang KJ, Au DWT. The marine medaka Oryzias melastigma–a potential marine fish model for innate immune study. Marine pollution bulletin. 2011;63.5–12: 267–276. [DOI] [PubMed] [Google Scholar]
- 32.Breves JP, et al. Dynamic gene expression of GH/PRL-family hormone receptors in gill and kidney during freshwater-acclimation of Mozambique tilapia. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 2011;158(2), 194–200. doi: 10.1016/j.cbpa.2010.10.030 [DOI] [PubMed] [Google Scholar]
- 33.Tacchi L, Bron JE, Taggart JB, Secombes CJ, Bickerdike R, Adler MA, et al. Multiple tissue transcriptomic responses to Piscirickettsia salmonis in Atlantic salmon (Salmo salar). Physiological genomics. 2011;43(21), 1241–1254. doi: 10.1152/physiolgenomics.00086.2011 [DOI] [PubMed] [Google Scholar]
- 34.Suarez PE, Rodriguez EG, Soundararajan R, Mérillat AM, Stehle JC, Rotman, S, et al. The glucocorticoid-induced leucine zipper (gilz/Tsc22d3-2) gene locus plays a crucial role in male fertility. Molecular endocrinology. 2012;26(6), 1000–1013. doi: 10.1210/me.2011-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sampath-Kumar R, Munro AD, Lee J, Lam TJ. Exogenous cortisol promotes survival of Asian seabass (Lates calcarifer) hatchlings exposed to hypersalinity but not hyposalinity shock. Aquaculture. 1993;116(2–3), 247–255. [Google Scholar]
- 36.Iwama GK, Takemura A, Takano K. Oxygen consumption rates of tilapia in fresh water, sea water, and hypersaline sea water. Journal of Fish Biology. 1997;51(5), 886–894. [Google Scholar]
- 37.Kammerer BD, Cech JJ Jr, Kültz D. Rapid changes in plasma cortisol, osmolality, and respiration in response to salinity stress in tilapia (Oreochromis mossambicus). Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 2010;157(3), 260–265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The identical amino acids were indicated by red residues, the low consensus amino acids were indicated by blue residues, and black residues represented the non-identical amino acids.
(TIF)
Values were mean ± S.E.M.. Treatment group, n = 6; control group, n = 4. The asterisks indicated significant differences between each time-point and 0 h (p < 0.05, analyzed by Kruskal–Wallis test with Dunn’s multiple comparisons test).
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.