Abstract
Although advanced age and presence of comorbidities significantly impact the variation observed in the clinical symptoms of COVID-19, it has been suggested that genetic variants may also be involved in the disease. Thus, the aim of this study was to perform a systematic review with meta-analysis of the literature to identify genetic polymorphisms that are likely to contribute to COVID-19 pathogenesis. Pubmed, Embase and GWAS Catalog repositories were systematically searched to retrieve articles that investigated associations between polymorphisms and COVID-19. For polymorphisms analyzed in 3 or more studies, pooled OR with 95% CI were calculated using random or fixed effect models in the Stata Software. Sixty-four eligible articles were included in this review. In total, 8 polymorphisms in 7 candidate genes and 74 alleles of the HLA loci were analyzed in 3 or more studies. The HLA-A*30 and CCR5 rs333Del alleles were associated with protection against COVID-19 infection, while the APOE rs429358C allele was associated with risk for this disease. Regarding COVID-19 severity, the HLA-A*33, ACE1 Ins, and TMPRSS2 rs12329760T alleles were associated with protection against severe forms, while the HLA-B*38, HLA-C*6, and ApoE rs429358C alleles were associated with risk for severe forms of COVID-19. In conclusion, polymorphisms in the ApoE, ACE1, TMPRSS2, CCR5, and HLA loci appear to be involved in the susceptibility to and/or severity of COVID-19.
Introduction
Coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was identified in China near the end of 2019, and progressed to a pandemic condition in March 2020, resulting in a major public health problem worldwide due to its social and economic burdens [1]. As of February 1, 2022, COVID-19 affected more than 370 million people, and caused more than 5,658,702 deaths (https://www.who.int/publications/m/item/weekly-operational-update-on-covid-19—1-february-2022).
Clinical manifestations of COVID-19 vary from an asymptomatic infection, dry cough, sore throat, fever, shortness of breath, fatigue, muscle pain, headache, loss of taste or smell, vomiting, diarrhea, to acute respiratory distress syndrome. Approximately 15% of patients develop the severe form, which can progress to pneumonia, respiratory failure, kidney injury, multiorgan dysfunction, and death [2, 3]. The variation in symptoms and severity of COVID-19 is partially explained by known risk factors, including advanced age, male gender, and presence of comorbidities, such as diabetes, obesity, hypertension, and heart disease [4, 5]. However, severe outcomes have also been observed in young and healthy patients, suggesting that other risk factors, such as genetic predisposition, may increase the risk to and/or severity of this disease [6–8].
It is well known that host genetic polymorphisms play a key role in the susceptibility or resistance to different viral infections [9, 10]. Taking into account the main role of host genes in the entry and replication of SARS-CoV-2 in cells and in mounting the immune response, it seems that a combination of multiple genes might be involved in COVID-19 pathogenesis [9]. Accordingly, to date, numerous studies have been conducted on the association between genetic polymorphisms and COVID-19 [6, 7, 9–11]. Some studies have indicated that polymorphisms in genes related to innate and adaptive immune response [toll-like receptors (TLRs), human leukocyte antigen (HLA) class I and II, and cytokines/chemokines] and in genes involved in viral binding and entry into host cells (angiotensin converting enzyme-2 –ACE2, and transmembrane serine protease–TMPRSS) are associated with COVID-19 development and/or severity [6–8, 12]. However, it is still unclear which and to what degree specific polymorphisms contribute to the susceptibility for this disease [6].
Thus, aiming to identify the genetic factors that may influence COVID-19 susceptibility and severity, we conducted a comprehensive and updated systematic review of the literature on the subject followed by meta-analyses of those polymorphisms analyzed in three or more studies. Even though few systematic reviews have been published regarding the association between polymorphisms in different genes and COVID-19 [6, 7, 10, 12].
Materials and methods
Literature search strategy and eligibility criteria
This comprehensive and updated systematic review was performed and written according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA), Meta-analysis of Observational Studies in Epidemiology (MOOSE) statements and guideline for Systematic Reviews of Genetic Association Studies [13–15], and it was registered at PROSPERO (http://www.crd.york.ac.uk/PROSPERO) under the CRD42021248091 number. We performed a search at PubMed and Embase repositories for all English, Portuguese, and Spanish language original articles that analyzed potential associations between genetic polymorphisms and susceptibility/severity for COVID-19, up to July, 2021. For this, the following MeSH terms were used: (SARS-CoV-2 OR COVID-19 OR severe acute respiratory syndrome OR SARS virus) AND (polymorphism, genetic OR polymorphism, single nucleotide OR polymorphism, single-stranded conformational OR polymorphism, restriction fragment length OR DNA copy number variations OR amplified fragment length polymorphism analysis OR mutation OR mutation rate OR INDEL mutation OR mutation, missense OR point mutation OR frameshift mutation OR codon, nonsense). In addition, studies of interest were also searched in the GWAS Catalog (https://www.ebi.ac.uk/gwas).
Two independent investigators (C.D and L.A.B) screened and evaluated the eligibility of each study retrieved from the online repositories by reviewing titles and abstracts. When abstracts did not provide adequate information, the full texts of the extracted articles were also reviewed, as previously reported by our group [16, 17]. Discrepancies between the two investigators were settled by debate between them and, when necessary, a third reviewer (D.C.) was consulted. All observational human studies that compared frequencies of at least one polymorphism between patients with and without COVID-19 or between COVID-19 patients with different degrees of severity were included in this systematic review. Moreover, reference lists coming from the articles fulfilling our eligibility criteria were manually searched to identify other potentially relevant citations.
The exclusion criteria were: 1) articles without enough data to estimate an OR with 95% CI; 2) duplicated studies (in this case, the most complete study was chosen for inclusion); and 3) non-human studies.
Data extraction and quality evaluation
Necessary information from each study was individually extracted by C.D. and L.A.B. using a standardized form [16, 17]. Agreement was pursued in all evaluated items of this form; however, when an agreement could not be reached, divergences in data extraction were solved by referring to the original article or by consulting another investigator (D.C.). Data retrieved from each study were as follows: 1) characteristics of the studies and samples (including publication year, name of first author, number of subjects in each analyzed group, mean age, gender, country, and ethnicity); and 2) data of the polymorphisms of interest [including their identification, allele/genotype frequencies, and OR (95% CI)]. When data were not available in the article, the authors were contacted by email for the necessary information, but only part of them answered.
The Clark-Baudouin Score (CBS) was used to evaluate the quality of the included studies [18]. This score applies pre-defined criteria to assess each publication, highlighting quality issues in the conduction of studies and interpretation of results. Using a 10-point scoring sheet, investigators can evaluate sections of the articles related to reproducibility, selection of subjects, statistical analyses, and genotyping methods.
Statistical analyses for meta-analysis
Those polymorphisms analyzed in three or more studies were submitted to meta-analyses using the Stata 15.0 software (StataCorp, College Station, TX, USA). Goodness-of-fitness χ2 tests were used to evaluate whether genotype frequencies were in conformity with the Hardy-Weinberg Equilibrium (HWE) in the control groups. Associations between individual polymorphisms and COVID-19 susceptibility and/or severity were analyzed using OR (95% CI) calculations for the allele contrast, dominant, recessive, and additive inheritance models, categorized as suggested by a previous publication [19]. For the HLA allelic analysis, frequency was calculated as the number of cases or controls harbouring at least one positive event (one allele type) divided by the total number of chromosomes included in each of the corresponding groups [20]. Inter-studies heterogeneity was tested using χ2-based Cochran’s Q statistic, while inconsistency was quantified with the I2 metric [21, 22]. When P < 0.10 (Q statistic) and/or I2 > 50%, heterogeneity was considered statistically relevant. In this case, the DerSimonian and Laird random effect model (REM) was used to calculate OR (95% CI) for each study and for the pooled effect. In the lack of significant inter-studies heterogeneity, the fixed effect model (FEM) was used for this calculation.
Results
Literature search
Fig 1 shows the flow diagram illustrating the strategy used to identify and select studies for inclusion in our systematic review and meta-analyses. A total of 2936 articles were retrieved after searching PubMed, Embase, and GWAS Catalog resources, and 2727 of them were excluded during the review of titles and abstracts due to disagreements with our defined eligibility criteria. Two hundred and nine articles remained to be full text evaluation. Nevertheless, after carefully analyzing the full texts, another 145 studies were excluded, and a total of 64 articles were included in this systematic review (Table 1 and Fig 1). Among them, 30 studies, where the same SNP was evaluated in at least 3 articles and frequency data was available, were included in the meta-analyses.
Fig 1. Flowchart illustrating the search strategy used to identify association studies between genetic polymorphisms and COVID-19 disease.
Table 1. Characteristics of studies included in the systematic review.
| Reference | Population | Sample (case/control) | Gene | Results |
|---|---|---|---|---|
| Agwa et al., 2021 [23] | Egyptian | 141 cases / 100 controls | INFλ, TLL1, DDR1 |
Disease susceptibility: The IFN-λ rs12979860 C/C, TLL1 rs17047200 A/A and the DDR1 rs4618569 A/A genotypes were associated with COVID-19 (P = 0.011, P = 0.012, and P = 0.026, respectively). Severity: The DDR1 rs4618569 A/G was associated with COVID-19 severity (P = 0.007). |
| Alghamdi et al., 2021 [24] | Saudi | 880 cases | IFITM3 |
Disease susceptibility: The rs12252 G allele was associated with risk for hospital admission (OR = 1.65, 95% CI 1.01–2.70, P = 0.04). Severity: The rs12252 G allele conferred risk for mortality (OR = 2.2, 95% CI 1.16–4.20, P = 0.01). |
| Amodio et al., 2020 [25] | Italian | 381 cases | IFNL3, IFNL4 | Severity: The IFNL4 rs368234815 DelG/DelG genotype was associated with risk for higher viral loads in COVID-19 patients (OR = 1.24, 95% CI 1.09–1.40). |
| Amoroso et al., 2021 [26] | Italian | 219 cases /40,685 controls | HLA-A, -B, -DRB1 |
Disease susceptibility: The HLA-DRB1*08 allele was associated with risk for COVID-19 (OR = 1.9, 95% CI 1.2–3.1, P = 0.003) Severity: The HLA-DRB1*08 allele conferred risk for death (OR = 2.9, 95% CI 1.15–7.21, P = 0.023). |
| Avendaño-Félix et al., 2021 [27] | Mexican | 193 cases | IL-10 | Severity: The rs1800871 and rs1800872 polymorphisms were not associated with COVID-19 severity (P = 0.286 and P = 0.235, respectively) and related-outcomes (P = 0.499 and P = 0.531). |
| Benetti et al., 2020 [28] | Italian | 131 cases /258 controls | WES | Disease susceptibility: ACE2 allelic variability was higher in control group compared to the patient cohort, detected from a cumulative analysis of the identified variants (P <0.029). |
| Benetti et al., 2020 [29] | Italian | 35 cases / 150 controls | WES | Disease susceptibility: Through the gene burden test, mutations in PRKRA and LAPTM4B genes were identified as being risk factors, while mutations in OR4C5 and NDU-FAF7 genes represented protective factors for COVID-19. |
| Bernas et al., 2021 [30] | German | 4758 cases /10,5008 controls | CCR5 |
Disease susceptibility: The CCR5 Δ32 polymorphism was not associated with COVID-19 (OR = 0.96, 95% CI 0.89–1.03, P = 0.21). Severity: The CCR5 Δ32 polymorphism did not differ significantly between individuals with or without symptomatic infection (OR = 1.13, 95% CI 0.88–1.45, P = 0.32), severe respiratory tract infection (OR = 1.03, 95% CI 0.88–1.22, P = 0.68) or respiratory hospitalization (OR = 1.16, 95% CI 0.79–1.69, P = 0.45). |
| Cabrera-Marante et al., 2020 [31] | Latin-american, Spanish, Polish | 22 cases | PRF1 | Severity: Two of 22 patients showed PRF1 A91V mutation in heterozygosis (allele frequency = 0.045). These 2 A91V-positive patients had higher fever associated with respiratory symptoms and died. |
| Cafiero et al., 2021 [32] | Italian | 104 cases | ACE1, ACE2, AGT, AGTR1 | Severity: The ACE2 rs2074192 T, ACE1 Del, and AGT rs699 C alleles were more frequent in symptomatic patients vs. asymptomatic (P = 0.001, P <0.001, and P = 0.033, respectively). |
| Calabrese et al., 2020 [33] | Italian | 68 cases / 222 controls | ACE1 | Severity: The frequency of ACE1 Del/Del genotype was higher in COVID-19 patients with pulmonary embolism (PE) than patients without PE (72 vs. 46.5%; P = 0.048). |
| Cantalupo et al., 2021 [34] | Italy | 202 cases /929 controls (rs35951367) 221 cases/1084 controls (rs3441865) 147 cases / 1095 controls (rs333) | WES | Disease susceptibility: The CCR5 rs35951367 C allele was associated with risk for COVID-19 (OR = 1.307, 95% CI 1.01–1.70, P = 0.043). The CCR5 rs34418657 G/T genotype was more frequent in patients with COVID-19 than controls (OR = 3.978, 95% CI 1.060–14.933, P = 0.027). No association was found between the CCR5 Δ32 (rs333) polymorphism and COVID-19 (P = 0.99). |
| Coto et al., 2021 [35] | Spanish | 318 cases / 350 controls | ABO | Disease susceptibility: The rs8176719 polymorphism was not associated with risk for COVID-19 or disease severity. |
| Cuesta-Llavona et al., 2021 [36] | Spanish | 801 cases / 650 controls | CCR5 | Disease susceptibility: Homozygosis for the CCR5 Δ32 deletion (rs333) conferred protection against COVID-19 (OR = 0.66, 95% CI 0.49–0.88, P = 0.01). |
| Del Ser et al., 2021 [37] | Spanish | 62 cases / 851 controls | APOE | Disease susceptibility: The APOE ε4 allele was associated with the presence of symptoms of COVID-19 (OR = 1.85, 95% CI 1.13–2.88, P = 0.010). |
| Dite et al., 2021 [38] | British | 1582 casesa | Array | Severity: A score of 64 SNPs was associated with risk for COVID-19 severity (OR = 1.19, 95% CI 1.15–1.22, P <0.001). A model incorporating this score and clinical risk factors showed 111% better discrimination of disease severity than a model with just age and gender. |
| Ellinghaus et al., 2020 [39] | Italian, Spanish | 835 cases / 1255 controls 775 cases/ 950 controls | GSA | Severity: The 3p21.31 cluster was identified as a susceptibility locus in patients with COVID-19 with respiratory failure (OR = 1.77, 95% CI 1.48–2.11; P = 1.15×10−10). |
| Gavriilaki et al., 2021 [40] | Greek | 97 cases | NGS | Severity: Patients carrying the THBD rs1042580 C and CFH rs800292 G alleles did not require ICU hospitalization (vs. patients carrying the other alleles). Polymorphisms in ADAMTS13, C3 and CFH genes were associated with risk for ICU hospitalization (P = 0.022). |
| Gómez et al., 2020 [41] | Spanish | 204 cases / 536 controls | ACE1, ACE2 | Severity: The ACE1 Del/Del genotype was associated with severe COVID-19 (P = 0.049). The ACE2 rs2285666 polymorphism was not associated with disease severity. |
| Gómez et al., 2021 [42] | Spanish | 311 cases / 440 controls | IFITM3 | Disease susceptibility: The IFITM3 rs12252 C allele was associated with risk for COVID-19 hospitalization after adjustment by age and gender (OR = 2.02, 95%CI 1.19–3.42, P = 0.01). |
| Grimaudo et al., 2021 [43] | Italian | 383 cases | MERTK, INFL4, PNPLA3, TLL1 | Severity: In patients younger than 65 years, the PNPLA3 rs738409 G/G (OR = 4.69, 95% CI 1.01–22.04, P = 0.049) and TLL1 rs17047200 T/T (OR = 9.1, 95% CI 1.45–57.3, P = 0.018) genotypes were associated with risk for disease severity. |
| Gunal et al., 2021 [44] | Turkish | 90 cases | ACE1 | Severity: The ACE1 Ins/Ins genotype conferred protection against severe COVID-19 (OR = 0.323, 95% CI 0.112–0.929, P = 0.036). |
| Hamet et al., 2021 [45] | British | 1644 cases / 15962 controlsa | Array | Severity: The ACE2 rs2074192 T allele was associated with more severe outcomes of COVID-19 in obese smoking males of 50 years or older (OR = 4.07, P = 0.036). |
| Hubacek et al., 2021 [46] | Czech | 416 cases / 2404 controlsd | CCR5 | Severity: The frequency of CCR5 Δ32 allele was higher in COVID-19 asymptomatic patients (23.8%) than COVID-19-symptomatic patients (16.7%) (P = 0.03). |
| Hubacek et al., 2021 [47] | Czech | 408 cases / 2559 controlsd | ACE1 | Disease susceptibility: The frequency of ACE1 Ins/Ins genotype was higher in COVID-19 patients vs. controls (26.2% vs. 21.2%; OR = 1.55, 95% CI 1.17–2.05, P = 0.02). |
| Hubacek et al., 2021 [46] | Czech | 408 cases / 2606 controlsd | APOE |
Disease susceptibility: The frequency of the APOE4 allele did not differ between the group of SARS-CoV-2-positive subjects and the control population (P = 0.11). Severity: The presence of least one APOE4 allele was higher in symptomatic COVID-19 subjects than controls (OR = 1.43, 95% CI 1.05–1.95, P = 0.03). Genotype frequencies were almost identical in COVID-19-asymptomatic subjects and in the control group population (P = 0.86). |
| Karakas Çelik et al., 2021 [48] | Turkish | 155 cases | ACE1, ACE2 | Severity: ACE1 Ins/Del and ACE2 rs2106809 and rs2285666 polymorphisms were not associated with COVID‐19 severity. |
| Kerget et al., 2021 [49] | Turkish | 70 cases | IL-6 | Severity: The IL-6 rs2074192 G/G genotype was associated with COVID-19 severity (P = 0.002). |
| Kolin et al., 2020 [50] | British | 968 cases / 1734 controlsa | Array | Disease susceptibility: Genome-wide association analysis did not show any significant loci in the meta-analysis (P >0.050). |
| Kuo et al., 2020 [51] | British | 622 cases / 322326 controlsa | Array |
Disease susceptibility: The ApoE ε4ε4 genotype was associated with risk of COVID-19 positivity (OR = 2.24, 95% CI 1.72–2.93, P = 3.24 × 10−9) vs. e3e3 genotype. Severity: The presence of the ApoE ε4ε4 genotype conferred risk for mortality (OR = 4.29, 95% CI 2.38–7.72, P = 1.22 × 10−6) vs. e3e3 genotype. |
| Latini et al., 2020 [52] | Italian | 131 cases / Controlse | WES | Disease susceptibility: Furin rs769208985 A and TMPRSS2 rs114363287 A alleles were more frequent in COVID-19 than GnomAD controls (P = 0.005 and P = 0.016, respectively). TMPRSS2 rs75603675 T and rs12329760 A alleles were less frequent in COVID-19 patients than GnomAD (P = 0.0446 and P = 0.023, respectively). |
| Lehrer et al., 2021 [53] | British | 688 casesa | S1R | Severity: The S1R rs17775810 T/T genotype was associated with the lowest death rate (0%, P = 0.020). |
| Lehrer et al., 2021 [54] | British | 712 cases / 9265 controlsa | GWAS-Chr9 | Disease susceptibility: No association was found between the rs657252 polymorphism in Chr9 and COVID-19. |
| Littera et al., 2020 [55] | Italian | 182 cases / 619 controls | HLA-A, -B, -C, -DRB1 |
Disease susceptibility: The haplotype HLA-A*02:05, B*58:01, C*07:01, DRB1*03:01 protected against SARS-CoV-2 infection. HLA-C*04:01 allele and the haplotype HLA-A*30:02, B*14:02, C*08:02 (OR = 3.8, 95% CI 1.8–8.1, P = 0.025) were more frequent in patients than controls. Severity: HLA-DRB1*08:01 allele was only present in hospitalized patients (OR >2.5, 95% CI 2.7–220.6, P = 0.024). |
| Lorente et al., 2020 [56] | Spanish | 72 cases / 3,886 controls | HLA-A, -B, -C, -DRB1, -DQB1 | Severity: The HLA-A*11, HLA-C*01 and HLA-DQB1*04 alleles were associated with higher mortality due to COVID-19 (OR = 7.69, 95% CI 1.06–55.65, P = 0.040; OR = 11.18, 95% CI 1.05–118.70, P = 0.040; and OR = 9.96, 95% CI 1.23–80.36, P = 0.030; respectively). |
| Malaquias et al., 2020 [57] | Brazilian | 6 cases / 11 controls | MBL2 | Disease susceptibility: The rs180040 A/A, rs1800451 G/G and rs5030737 C/C genotypes had a higher prevalence in the COVID-19 group. |
| Martínez-Sanz et al., 2021 [58] | Spanish | 39 cases / 28 controls | Array | Disease susceptibility: The ACE2 rs2106806 A (OR = 3.75, 95% CI 1.23–11.43, P = 0.015) and rs6629110 T (OR = 3.39, 95% CI 1.09–10.56, P = 0.028) alleles were associated with risk for COVID-19. |
| Medetalibeyouglu et al., 2021 [59] | Turkish | 284 cases / 100 controls | MBL2 | Disease susceptibility: The B/B genotype of the codon 54 A/B (Gly54Asp: rs1800450) variant in the MBL2 gene was more frequent in COVID-19 cases vs. controls (10.9% vs. 1.0%; OR = 12.1, 95% CI 1.6–90.1, P = 0.001). |
| Möhlendick et al., 2021 [60] | Germany | 297 cases / 253 controls | ACE1, ACE2 |
Disease susceptibility: The ACE2 rs2285666 G/G genotype was associated with risk for COVID-19 (OR = 1.91, 95% CI 1.13–3.24, P = 0.02). No association was found between the ACE1 rs1799752 polymorphism and COVID-19. Severity: The ACE2 rs2285666 G/G genotype confer risk for serious course of COVID-19 compared to moderate course (OR = 3.04, 95% CI 1.47–6.27, P = 0.002) and is also associated with mortality (OR = 2.69, 95% CI 1.02–7.11, P = 0.05). |
| Monticelli et al., 2021 [61] | Italian | 1177 casesb | WES | Severity: The TMPRSS2 rs2298659 A and the rs12329760 T alleles were more frequent among mild cases of COVID-19 than severe cases (P = 0.004 and P = 0.029, respectively). |
| Naemi et al., 2021 [62] | Asian | 95 cases | HLA-A, -B, -C, -DRB1, -DQA1, -DQB1 | Severity: No association was found between these HLA genotypes and COVID-19 severity. |
| Novelli et al., 2020 [63] | Italian | 131 cases / 1000 Controlse | WES | Disease susceptibility: No association was found between ACE2 polymorphisms (rs140312271, rs2285666 and rs41303171) and COVID-19. |
| Novelli et al., 2020 [64] | Italian | 99 cases / 1017 controls | NGS | Disease susceptibility: The frequencies of three HLA alleles were higher in cases vs. controls: HLA B*27:07 (2.02% vs. 0.10%; P = 0.004), DRB1*15:01 (10.10% vs. 4.62%, P = 0.048), and DQB1*06:02 (7.58% vs. 3.64%, P = 0.016). |
| Pairo-Castineira et al., 2021 [65] | 2244 casesc | GWAS | Severity: Polymorphisms in Chr 12q24.13 (rs10735079, P = 1.65 × 10−8, near to OAS1, OAS2 and OAS3 genes), Chr 19p13.2 (rs74956615, P = 2.3 × 10−8, near TYK2), Chr 19p13.3 (rs2109069, P = 3.98 × 10−12, in DPP9), and Chr 21q22.1 (rs2236757, P = 4.99 × 10−8, in IFNAR2) were associated with COVID-19 severity. | |
| Petrazzuolo et al., 2020 [66] | French | 140 cases | FPR1 | Severity: No association was found between the FPR1 rs5030880 and rs867228 polymorphisms and COVID-19 severity. |
| Posadas-Sánchez et al., 2021 [67] | Mexican | 90 cases / 263 controls | DPP4 | Disease susceptibility: The DPP4 rs3788979 T/T genotype was associated with risk for COVID-19 (OR = 4.28, 95% CI 2.12–8.62, P = 4.7 × 10−5; recessive model). |
| Ravikanth et al., 2021 [68] | Indian | 510 cases / 500 controls | WES | Severity: The TMPRSS2 rs12329760 A allele was less frequent in patients with mild-to-moderate (P = 0.004) or severe disease (P = 0.010) vs. asymptomatic patients. |
| Russo et al., 2021 [69] | Italian | 500 cases / 283 controls | WES | Severity: The TNFRSF13 rs61756766 C allele was more frequent in severe cases vs. non-severe (OR = 11.5, 95% CI 1.3–100, P = 0.010) and asymptomatic patients (OR = 3.7, 95% CI 1.3–10.6, P = 0.020). |
| Saleh et al., 2021 [70] | Egyptian | 900 cases / 184 controls | TNFA | Disease susceptibility: The A/A genotype of the TNF G308A polymorphism was associated with risk for COVID-19 (OR = 3.06, 95% CI 1.26–7.44, P = 0.019). |
| Salem Hareedy et al., 2021 [71] | Egyptian | 46 cases / 14 controls | CYP2D6*4, CYP2D6*2XN, CYP3A4*1B, CYP3A5*3 | Disease susceptibility: Carriers of the CYP2D*2XN C/C genotype had the lower risk for a positive anti-COVID-19 IgG or IgM. The CYP3A4*1B A/A genotype conferred protection against positive anti-COVID-19 IgM (vs. G/G genotype). |
| Schönfelder et al., 2021 [72] | Germany | 239 cases / 253 controls | IFITM3 |
Disease susceptibility: The IFITIM3 rs12252 and rs34481144 polymorphisms were not associated with COVID-19 development (OR = 1.37, 95% CI 0.73–2.58, P = 0.340; OR = 0.96, 95% CI 0.65–1.41, P = 0.840; respectively). Severity: The IFITIM3 rs12252 and rs34481144 polymorphisms did not confer risk to COVID-19 severity (OR = 0.89, 95% CI 0.35–2.25, P = 1.00; OR = 1.77, 95% CI 0.94–3.32, P = 0.100; respectively). |
| Schönfelder et al., 2021 [73] | Germany | 239 cases / 253 controls | TMPRSS2 | Disease susceptibility: The TMPRSS2 rs383510 C/C genotype was associated with risk for COVID-19 infection (OR = 1.73, 95% CI 1.15–2.59, P = 0.010). The rs2070788 and rs12329760 polymorphisms were not associated with COVID-19. |
| Scutt et al., 2021 [74] | British | 705 cases / 471506 controlsa | Array | Disease susceptibility: The INK4A/ARF rs10757278 G/A genotype was associated with lower risk of hospital admission for COVID-19 in non-Caucasian patients (A/A + G/G vs. A/G; OR = 0.56, 95% CI 0.37–0.85, P = 0.006). |
| Shikov et al., 2020 [75] | Russian | 37 cases /21 controls | ACE2, ACE1 | Disease susceptibility: No association was found between ACE2 and ACE1 polymorphisms and COVID-19. |
| Shkunikov et al., 2021 [76] | Russian | 111 cases / 428 controls | NGS | Disease susceptibility: The HLA-A*01:01 allele was associated with risk for COVID-19, while the HLA-A*02:01 and HLA-A*03:01 alleles conferred protection. |
| Torre-Fuentes et al., 2021 [77] | Spanish | 4 cases / 71 controls | WES | Disease susceptibility: No association was found between ACE2, TMPRSS2 and FURIN polymorphisms and COVID-19. |
| Valenti et al., 2021 [78] | Spanish | 72 cases | Chr3 | Severity: The rs11385942 G/A genotype was associated with COVID-19 severity. |
| Verma et al., 2021 [79] | Indian | 269 cases | ACE1 | Severity: The ACE1 Del/Del genotype was associated with risk for severe COVID-19 (OR = 3.69, 95% CI 1.612–8.431, P = 0.002). |
| Vietzen et al., 2021 [80] | 361 cases / 260 controls | HLA-E, KLRC2 |
Disease susceptibility: The KLRC2 Del allele conferred risk for hospitalization (OR = 2.6, P = 0.0006) and hospitalization in ICU (OR = 7.1, P <0.0001) vs. non-hospitalized patients and controls. Severity: The HLA-E*0101 allele was also associated with risk for hospitalization (OR = 2.1, P = 0.010) and hospitalization in ICU (OR = 2.7, P = 0.010). |
|
| Wang et al., 2020 [81] | Chinese | 332 cases | GWAS* / HLA-A, -B, -C, -DRB1, -DQB1, -DPB1, -DQA1 | Severity: The TMEM189–UBE2V1 rs6020298 A allele was more frequent in patients with severe COVID-19 than non-severe patients (0.59 vs. 0.45) and conferred risk for mild + severe disease (OR = 1.2, P = 4.1 x 10−6). The TMPRSS2 rs12329760 minor allele was less frequent among patients with severe COVID-19 vs. mild symptomatic patients. HLA-A* 11:01, B*51:01, and C*14:02 alleles were associated with risk for severe COVID-19. |
| Wang et al., 2020 [82] | Chinese | 82 cases / 3548 controls | NGS | Disease susceptibility: HLA-B*15:27 and HLA-C*07:29 were associated with risk for COVID-19 disease (OR = 3.59; 95% CI 1.72–7.50, P = 0.030; and OR = 130.20, 95% CI 5.28–3211, P = 0.025, respectively). |
| Wulandari et al., 2021 [83] | Indonesian | 95 cases | TMPRSS2 | Severity: No association was found between the rs12329760 polymorphism and COVID-19 severity. |
| Zhang et al., 2020 [84] | China | 80 cases | IFITM3 | Severity: The IFITM3 rs12252 C/C genotype was associated with disease severity in an age-dependent manner (OR = 6.37, P <0.001). |
| Zhou et al., 2020 [85] | British | 1091 cases / 2793 controlsa | TMPRSS2, ACE2 | Disease susceptibility: After analyzing 17 and 31 tag SNPs of ACE2 and TMPRSS2 genes, respectively, the rs7282236 SNP in TMPRSS2 gene was the only one associated with risk of COVID-19 disease (OR = 1.33, 95% CI 1.14–1.54, P = 2.31 × 10−4). |
Chr: chromosome; GSA: Global Screening Array; GWAS: Genome-wide Association Study; ICU: intensive care unit, NGS: next-generation sequencing; WES: Whole exome sequencing
adata from UK biobank
bdata from GEN-COVID Multicenter Study
cdata from GenOMICC database
dcontrols data from post-MONICA study
econtrols data from GnomAD database.
Qualitative synthesis of studies that analyzed associations of SNPs and COVID-19
Table 1 shows the compiled main data of the 64 eligible studies included in this systematic review. More than 200 polymorphisms and 50 genes/loci were studied regarding their associations with COVID-19 susceptibility or severity of this disease. Most of the studies compared polymorphism frequencies in patients who tested positive for COVID-19 compared to negative controls. Twenty-three studies evaluated polymorphisms in COVID-19 patients categorized according to different degrees of disease severity. S1 Table shows the quality of all studies included in this systematic review, which was evaluated using the CBS as described in the Methods Section. Considering a score system that ranges from 0 to 10 points according to the adherence to pre-defined criteria, none of the studies reached 9 points. However, the majority of the studies (70.1%) were classified as presenting good quality since they were awarded 6 to 8 points. The remaining articles were awarded with less than 6 points.
More information regarding the COVID-19 diagnostic criteria, definition of severity degrees, age, ethnicity, gender, and genotyping techniques are described in S2 Table. The most studied candidate genes/loci were: HLA, ABO, ACE1, ACE2, APOE, CCR5, TMPRSS2, and IFITM3. In total, 8 polymorphisms in 7 candidate genes and 74 alleles of the HLA loci (A, B, C, DRB1, DQA1, and DQB1) were analyzed in ≥3 studies and subsequently included in the meta-analyses.
Meta-analyses of ACE2, ACE1, and TMPRSS2 polymorphisms
Two polymorphisms in the ACE2 gene were included in meta-analyses (Table 2). The pooled data of 3 studies for the rs41303171 (T/C) polymorphism [28, 63, 77] and 3 studies for the rs2285666 (C/T) polymorphism [41, 58, 63] indicated no association between them and the risk for COVID-19.
Table 2. Meta-analyses of the association between polymorphisms in candidate genes and COVID-19 development and severity.
| Polymorphism | Localization/Position | Inheritance model | Studies | I2 | Model | OR (95% CI) |
|---|---|---|---|---|---|---|
| COVID-19 infection vs. Control | ||||||
| ACE2 rs2285666 | chrX:15592225 / Intron | Dominant | 3 | 64.1% | Random | 0.95 (0.57–1.56) |
| ACE2 rs41303171 | chrX:15564175 / Exon | Allele | 3 | 66.3% | Random | 1.52 (0.24–9.61) |
| Dominant | 3 | 67.8% | Random | 1.36 (0.20–9.20) | ||
| ACE1 Ins/Del | chr17:63488530–63488543 / Intron | Allele | 4 | 61.7% | Random | 1.00 (0.82–1.22) |
| Dominant | 4 | 64.1% | Random | 0.95 (0.70–1.28) | ||
| Recessive | 4 | 64.2% | Random | 0.93 (0.64–1.37) | ||
| Additive | 4 | 72.3% | Random | 0.89 (0.55–1.46) | ||
| TMPRSS2 rs12329760 | chr21:41480570 / Exon | Allele | 3 | 12.6% | Fixed | 1.08 (0.92–1.27) |
| Dominant | 3 | 0% | Fixed | 1.18 (0.96–1.45) | ||
| CCR5 rs333 | chr3:46373453–46373487 / Exon | Allele | 3 | 44.6% | Fixed | 0.80 (0.68–0.96)* |
| Dominant | 3 | 40.3% | Fixed | 0.82 (0.68–0.98)* | ||
| ApoE ε4 | chr19:44908684 and chr19:44908822† / Exon | Allele | 3 | 41.8% | Fixed | 1.32 (1.20–1.45)* |
| Dominant | 3 | 58.2% | Random | 1.38 (1.09–1.75)* | ||
| Recessive | 3 | 28.2% | Fixed | 1.94 (1.50–2.50)* | ||
| Additive | 3 | 27.1% | Fixed | 2.05 (1.58–2.65)* | ||
| ABO rs8176719 | chr9:133257521–133257522 / Exon | Allele | 3 | 80.7% | Random | 1.22 (0.99–1.49) |
| COVID-19 mild/moderate vs. severe | ||||||
| ACE1Ins/Del | chr17:63488530–63488543 / Intron | Allele | 5 | 45.4% | Fixed | 0.67 (0.56–0.82)* |
| Dominant | 5 | 41.4% | Fixed | 0.62 (0.47–0.83)* | ||
| Recessive | 5 | 0% | Fixed | 0.69 (0.50–0.95)* | ||
| Additive | 5 | 0% | Fixed | 0.49 (0.33–0.72)* | ||
| TMPRSS2 rs12329760 | chr21:41480570 / Exon | Allele | 5 | 0% | Fixed | 0.77 (0.66–0.91)* |
| Dominant | 5 | 0% | Fixed | 0.74 (0.61–0.90)* | ||
| Recessive | 5 | 0% | Fixed | 0.71 (0.44–1.15) | ||
| Additive | 5 | 0% | Fixed | 0.65 (0.40–1.06) | ||
| CCR5 rs333 | chr3:46373453–46373487 / Exon | Allele | 3 | 67.2% | Random | 0.83 (0.59–1.16) |
| Dominant | 3 | 67.4% | Random | 0.83 (0.58–1.18) | ||
| IFITM3 rs12252 | chr11:320772 / Exon | Allele | 4 | 65.6% | Random | 1.04 (0.62–1.75) |
| Dominant | 4 | 65.8% | Random | 0.97 (0.53–1.77) | ||
| Recessive | 4 | 22.5% | Fixed | 1.04 (0.44–2.46) | ||
| Additive | 4 | 34.3% | Fixed | 0.78 (0.31–1.91) | ||
| ApoE ε4 | chr19:44908684 and chr19:44908822† / Exon | Allele | 3 | 0% | Fixed | 1.36 (1.07–1.73)* |
| Dominant | 3 | 0% | Fixed | 1.30 (0.97–1.72) | ||
| ABO rs8176719 | chr9:133257521–133257522 / Exon | Allele | 4 | 0% | Fixed | 0.94 (0.85–1.05) |
OR: odds ratio; CI: confidence interval.
* Indicates a significant association at P <0.05.
† Location of the two polymorphisms (rs429358 and rs7412) that generated the ApoE ε4 haplotype.
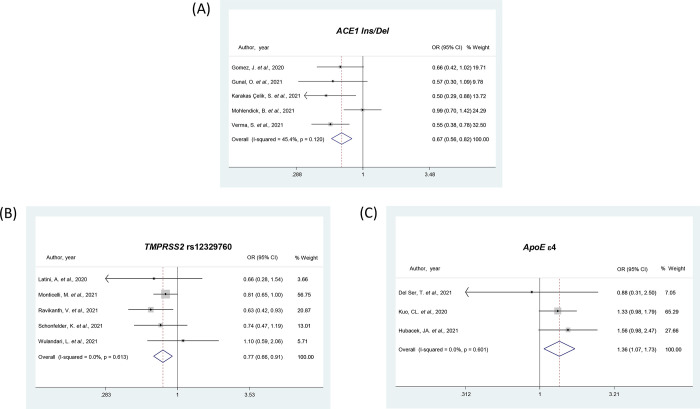
The rs1799752 (Ins/Del) polymorphism in the ACE1 gene was analyzed in 4 studies [33, 41, 47, 60] and the meta-analysis indicated no association between the Ins allele and the risk for COVID-19 (Table 2). Regarding COVID-19 severity, 8 studies [32, 33, 44, 47, 48, 58, 60, 79] were included. However, we analyzed the pooled data from 5 studies [41, 44, 48, 60, 79] that included severe COVID-19 patients compared to other degrees of severity (moderate, mild and/or asymptomatic). The meta-analysis of these studies showed an association between the ACE1 rs1799752 Ins allele and protection against the most severe form of COVID-19, in all inheritance models (OR = 0.67, 95% CI 0.56–0.82, Table 2 and Fig 2A for the allele model). Hubacek et al. [47] and Cafiero et al. [32] studies only compared asymptomatic vs. symptomatic patients, while the study be Calabrese et al. [33] compared groups according to the presence of thromboembolism in patients with severe COVID-19. Of note, when we included all the 8 studies in the meta-analysis, the Ins allele remained associated with protection against severe COVID-19 (OR = 0.60, 95% CI 0.39–0.94, for the allele model).
Fig 2.
Forest plots showing individual and pooled ORs (95% CIs) for the associations between the ACE1 Ins/Del (A), TMPRSS2 rs12329760 (B), and ApoE ε4 (C) polymorphisms and COVID-19 severity, under the allele contrast model.
The TMPRSS2 rs12329760 (C/T) polymorphism was analyzed in 3 studies regarding COVID-19 infection [52, 68, 73, 77] and 5 studies investigating disease severity [61, 68, 73, 83] (Table 2). Although the rs12329760 polymorphism was not associated with the risk of COVID-19, this meta-analysis showed that the T allele of this polymorphism confers protection for the most severe form of COVID-19 when considering both allele (OR = 0.77, 95% CI 0.66–0.91; Fig 2B) and dominant model (OR = 0.74, 95% CI 0.61–0.90) models (Table 2).
Meta-analyses of HLA alleles
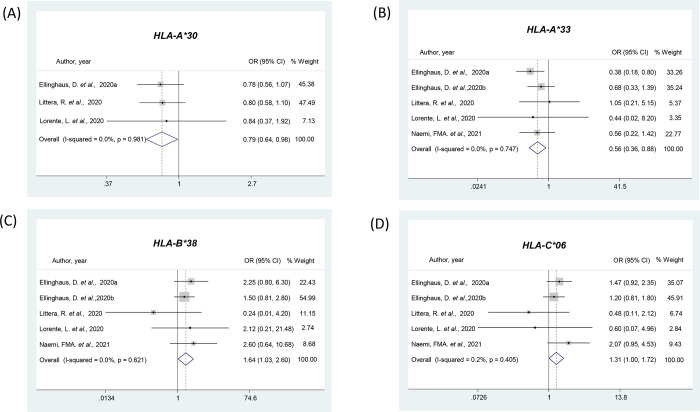
The A, B, C, DRB1, DQB1, and DQA1 alleles of the HLA were analyzed according to the risk of COVID-19 (S3 Table) or the severity of the disease (S4 Table). The HLA-A*30 allele was analyzed in 3 studies [39, 55, 56], and the pooled analysis showed this allele confers protection against COVID-19 (OR = 0.79, 95% CI 0.64–0.98; S3 Table and Fig 3A).
Fig 3. Forest plots showing individual and pooled ORs (95% CIs) for the associations between HLA alleles and COVID-19 presence or severity.
(A) Forest plot for HLA-A*30 and COVID-19 presence. (B) Forest plot for HLA-A*33 and COVID-19 severity. (C) Forest plot for HLA-B*38 and COVID-19 severity. (D) Forest plot for HLA-B*06 and COVID-19 severity. a Data from an Italian population; b Data from a Spanish population.
Regarding COVID-19 severity, the pooled data of 4 articles (5 studies) [39, 55, 56, 62] showed the association between the HLA-A*33 allele and protection for the most severe form of disease (OR = 0.56, 95% CI 0.36–0.88; S4 Table and Fig 3B). In contrast, the HLA-B*38 and HLA-C*06 alleles, both analyzed in the same 4 articles (5 studies) [39, 55, 56, 62], were associated with risk for the most severe form of COVID-19 (OR = 1.64, 95% CI 1.03–2.60 and OR = 1.31, 95% CI 1.00–1.72, respectively; S4 Table and Fig 3C and 3D). Our meta-analyses demonstrated that the other 70 alleles of the A, B, C, DRB1, DQB1, and DQA1 loci were not associated with COVID-19 development or severity (S3 and S4 Tables).
Meta-analyses of CCR5 and IFITM3 polymorphisms
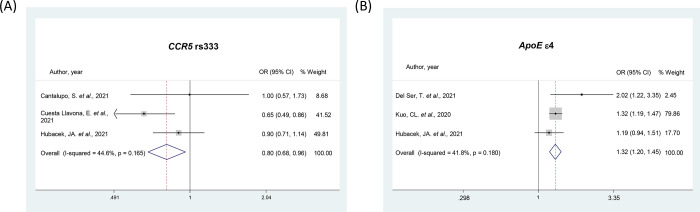
Three studies were included in the meta-analyses of CCR5 rs333 (Ins/Del) polymorphism regarding the risk of COVID-19 and its severity [30, 36, 46] (Table 2). The Del allele was associated with protection for COVID-19 infection considering both allele (OR = 0.80, 95% CI 0.68–0.96; Fig 4A) and dominant (OR = 0.82, 95% CI 0.68–0.98) models; however, this polymorphism was not associated with the severity of the disease (Table 2).
Fig 4.
Forest plots showing individual and pooled ORs (95% CIs) for the associations between the CCR5 rs333 (A) and ApoE ε4 (B) polymorphisms and COVID-19 presence, both under the allele contrast model.
For the IFITM3 rs12252 (T/C) polymorphism, the pooled analyses of 4 studies [24, 42, 72, 84] indicated no association of this polymorphism and different degrees of COVID-19 severity, for all tested genetic models (Table 2).
Meta-analyses of ApoE and ABO polymorphisms
The ApoE ε4 genotype was analyzed in 3 studies [37, 46, 51] regarding both COVID-19 infection and severity (Table 2). Meta-analyses showed the ε4 allele was associated with risk for COVID-19 presence in all genetic models (OR = 1.32, 95% CI 1.20–1.45, Fig 4B for the allele model). The ε4 allele was also associated with risk for the most severe form of COVID-19 when considering the allele model (OR = 1.36, 95% CI 1.07–1.73, Fig 2C).
The rs8176719 (-/C) polymorphism in the ABO gene was evaluated in 3 studies (2 articles) [38, 39] about COVID-19 development and 4 studies (3 articles) regarding disease severity [35, 38, 39] (Table 2). The pooled analyses indicated the Ins C allele is not associated with COVID-19 presence or severity in the allele model.
Discussion
Elucidating the genetic determinants of SARS-CoV-2 infection is essential for understanding the pathophysiology of COVID-19 and the inter-individual variability in its severity; thus, contributing to the development of updated vaccines and new antivirals. Hence, in this systematic review, we summarized the results of 64 eligible articles that analyzed the association between genetic polymorphisms and risk for infection or severity of COVID-19. Moreover, data regarding polymorphisms in 8 genes (HLA, ABO, ACE1, ACE2, APOE, CCR5, TMPRSS2, and IFITM3) were meta-analyzed in relation to the risk of infection and severity of COVID-19. Pooled results demonstrated that polymorphisms in the ApoE, ACE1, TMPRSS2, CCR5, and HLA genes appear to be involved in the susceptibility to and/or severity of COVID-19.
Angiotensin-converting enzyme 2 (ACE2) and type II transmembrane serine protease (TMPRSS2) are candidate genes for susceptibility for SARS-CoV-2 infection since SARS-CoV-2 uses the ACE2 receptor for cell entry, while the serine protease TMPRSS2 is required for priming of the viral spike (S) protein [86, 87]. ACE2 and ACE1, together with renin and angiotensin, constitute the renin angiotensin aldosterone system (RAAS), which is a complex system involved in multiple biological process that regulated blood pressure homeostasis and extracellular volume, and inflammation, which is closely related to COVID-19 morbidity and mortality, as it affects bradykinin production [88, 89]. Following the viral entry, ACE2 is down-regulated, causing an ACE1/ACE2 imbalance and contributing to RAAS overactivation and pulmonary shutdown. The consequent increased ACE1 activity and reduced ACE2 expression increase the risk of pulmonary diseases by increasing the lung vascular permeability; thus, leading to lung damage [90–92]. Accordingly, studies have reported the association between polymorphisms in ACE1, ACE2, and TMPRSS2 genes and SARS-CoV-2 infection [28, 32, 33, 41, 44, 48, 52, 58, 60, 61, 63, 68, 73, 77, 83]; however, the results are still contradictory. In the present meta-analysis, two ACE2 polymorphisms (rs2285666 and rs41303171) were analyzed, but no association with COVID-19 was found. Nevertheless, we demonstrated an association between the T allele of the TMPRSS2 rs12329760 polymorphism and protection against the most severe form of COVID-19.
Regarding the ACE1 gene, the insertion/deletion (Ins/Del) of 287-bp in the Alu-sequence of intron 16, represented by four individual SNPs (rs4646994, rs1799752, rs4340 and rs13447447), modulates ACE1 expression [93–95]. This Ins/Del variant results in alternative splicing, leading to protein shortening and loss of the catalytically active domain in ACE1 Ins allele carriers [92]. Moreover, the ACE1 Ins/Del variant explains about 60% of variability in ACE1 levels in the general population since ACE1 levels in Ins/Ins carriers are approximately half of that of Del/Del carriers [39, 93, 96]. In the context of SARS-CoV-2 infection, studies have reported variations in COVID-19 recovery and prevalence rates are associated to ACE1 Ins/Del frequency and geographical variations of this variant [97, 98]. Here, we showed an association between the ACE1 Ins allele and protection against severe COVID-19.
Major histocompatibility complex genes (MHC, known as Human Leukocyte Antigens, HLA) play a critical role in immune response [99]. The HLA system is a remarkably polymorphic region and genetic variants of HLA have been reported to affect the clinical course of patients infected with different viruses [100], including SARS-CoV-1 [101]. A specific set of HLA will present the peptides of the degraded virus to receptors on T cells, thus eliciting an immune response for virus eradication [102]. The set of HLA alleles inherited by an individual will determine the immune responses to viruses according to the selected peptides that can bind to the peptide‐binding groove [102]. Studies in different populations have shown associations between some HLA class I (A, B, and C) and class II (DRB1, DQA1, and DQB1) alleles and COVID-19 susceptibility and/or severity [82, 103]. Our meta-analyses did not confirm the results of previous individual studies; however, we identified new HLA alleles associated with COVID-19: the HLA-A*30 and HLA-A*33 were associated with protection against COVID-19 infection and the most severe form of this disease, respectively. Besides, the HLA-B*38 and HLA-C*06 alleles were associated with risk for severe COVID-19.
The interferon-induced transmembrane 3 (IFITM3) is an IFN-stimulated gene (ISG) essentially expressed on endosomes and lysosomes [104]. IFITM3 is part of an ISG family (IFITM) responsible for inhibiting the fusion between viral and cellular membranes of many viruses, such as influenza A H1N1 virus, dengue virus, and SARS-CoV [104]. On the other hand, it was recently shown that IFITM proteins are cofactors for efficient SARS-CoV-2 infection in human cells [105], reaffirming a key role of this gene in the susceptibility to COVID-19. Nevertheless, here, the IFITM3 rs12252 polymorphism was not associated with COVID-19 severity. Of note, we did not analyze this polymorphism regarding COVID-19 infection susceptibility due to lack of studies. Although this SNP in IFITM3 gene was not associated with COVID-19, it is noteworthy that type I IFN (IFN-I)-stimulated immunity has been shown to influence COVID-19 severity. Inborn errors of IFN-I pathway and pre-existing auto-antibodies neutralizing IFN-I appear to be strong determinants of critical COVID-19 pneumonia in about 15–20% of patients [106]. Asano et al., [107] reported that deleterious X-linked TLR7 mutations were observed in 16 male subjects from a cohort of 1202 patients with unexplained critical COVID-19 pneumonia. The patients’ blood plasmacytoid dendritic cells (pDCs) produced low levels of IFN-I in response to SARS-CoV-2. Human TLR7 and pDCs are essential for protective IFN-I immunity against SARS-CoV-2 in the respiratory tract. Moreover, Zhang et al., [108] showed that inborn errors of TLR3- and IRF-7 dependent IFN-I immunity can cause life-threatening COVID-19 pneumonia in patients with no prior severe infection.
Chemokines act attempting to maintain the immune homeostasis and to defend the body against harmful stimuli, such as SARS-CoV-2 infection [109]. CCR5 encodes a chemokine receptor expressed in macrophages and T cells, and its upregulation has been confirmed in COVID-19 patients [110]. Furthermore, an anti-CCR5 treatment has been shown to relieve the symptoms and the cytokine storm in COVID-19 patients who are critically ill [109]. The CCR5 gene is located at 3p21.31, a gene cluster region associated with severe COVID-19 courses [39]. The most studied CCR5 polymorphism regarding COVID-19 susceptibility is the Δ32 Ins/Del (rs333) [30, 34, 36, 46]. The CCR5 rs333 Del allele results in loss of function of the protein; being a major determinant of the resistance to HIV infection since the CCR5 protein serves as one of the gateways for the HIV virus [111]. Accordingly, our meta-analysis showed the CCR5 rs333 Del allele was associated with protection against COVID-19 infection [34, 36, 46].
A Genome-Wide Association Study (GWAS) carried out by the Severe COVID-19 GWAS Group [39] reported that one of the 2 strongest signals associated with severe COVID-19 was located within the ABO blood-group system. The involvement of ABO blood groups in COVID-19 susceptibility has been reported in both genetic and non-genetic studies. The blood group O was previously associated with a lower risk of acquiring COVID-19 when compared to subjects with non-O blood groups, whereas the blood A group was associated with a higher risk for this disease than non-A blood groups [39]. One of the assumptions is that the A-antigen causes P-selectin and intercellular cell adhesion molecule 1 binding to endothelial cells, increasing the probability of cardiovascular disease. Another explanation is that individuals with blood group O have decreased levels of von Willebrand factor, lowering the thrombotic disease risk [reviewed in [103]]. The rs8176719 polymorphism is the main determinant of the O blood group and has been investigated as a potential marker of COVID-19 susceptibility. However, some studies did not confirm these findings [35, 38]. In our meta-analysis, we demonstrated that the ABO rs8176719 - /C SNP was not associated with COVID-19 infection neither with different stages of severity.
The ApoE ε4 genotype was investigated in the UK Biobank Cohort, being associated with COVID-19 severity and mortality [51]. This finding was replicated in other studies [37, 46]. Apolipoprotein E (ApoE) is broadly expressed in human tissues and has an essential role in lipid transport, which has a key role in many functions, including immunity [112]. The most studied polymorphisms in ApoE are the rs429358 (ApoE4, C/T) and rs7412 (ApoE2, C/T), both located at exon 4. Three haplotypes are generated from these two polymorphisms (ε2, ε3 and ε4), codifying 3 protein isoforms (E2, E3 and E4). Moreover, these haplotypes can combine in 6 different variants: ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε3, ε3/ε4, and ε4/ε4 [112]. Among them, the ancestral ApoE ε4/ε4, generally considered deleterious, is a significant risk factor for Alzheimer’s disease and other human pathologies, including type 2 diabetes and cardiovascular disease, which are known risk factors for worst outcomes of COVID-19 [112–114]. In the present meta-analysis, the pooled data of three studies confirmed the association of the ε4 allele with both risk to COVID-19 presence and severe outcomes of the disease. It has been hypothesized that elevated cholesterol and oxidized lipoprotein levels, linked to the effects of ApoE ε4/ε4 variant, is associated with increased pneumocyte susceptibility to infection and to exaggerated lung inflammation [112]. Moreover, the frequency of the ε4 allele is higher in African-Americans who had increased mortality due to COVID-19 compared to Caucasian populations [115].
The results of the present meta-analysis should be interpreted within the context of a few limitations. Inter-studies heterogeneity is common in meta-analyses of genetic association studies and it should be cautiously interpreted. Some included studies did not test the control groups for COVID-19 or included controls derived from previous databank or ecological studies without COVID-19 information. Moreover, the COVID-19 severity criteria varied among the studies. Particular studies had included asymptomatic patients while others only included patients with at least a given symptom. Due to the presence of more than 2 groups of COVID-19 severity stages (mild, moderate and severe), we have categorized the patients regarding COVID-19 severity in different ways; however, it was more rational to show the data categorizing the most severe group against the others groups (asymptomatic and/or mild plus moderate). It was not possible to evaluate the association with mortality, as only few studies presented data comparing COVID-19 survivors and non-survivors. Furthermore, the impact of gender and age, which may influence the COVID-19 predisposition, could not be assessed due to the small number of studies for each SNP. Genetic background among different populations may significantly influence COVID-19 susceptibility, and the studies included in the present meta-analysis comprised different ethnicities. However, due to the small number of studies for each ethnicity, we were not able to analyze the impact of genetic background on the results. Finally, we cannot be sure that small negative studies were overlooked since we could not perform the publication bias analysis due to the small amount of studies for each SNP.
The infection with SARS-CoV-2 and its clinical course are dependent on the complex relationship between the virus and the host immune system. In this meta-analysis, we identified, for the first time, that four alleles of the HLA class I loci (A*30, A*33, B*38 and C*06) are associated with COVID-19. Moreover, we confirmed the association between COVID-19 susceptibility and polymorphisms in the ApoE, ACE1, TMPRSS2, and CCR5 genes. These findings will guide further epidemiological studies on host genetics as well as the development of innovative treatments. Considering that specific genetic polymorphisms might lead to severe COVID-19 outcomes, it is of extreme importance to use individual genetic data to employ personalized therapeutics and improve the COVID-19 prognostic.
Supporting information
(DOCX)
(XLSX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was partially supported by grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, grant numbers 401610/2020-9 and 425579/2018-2), Fundo de Incentivo à Pesquisa e Eventos (FIPE) at Hospital de Clínicas de Porto Alegre (grant number: 2020-0218), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). D.C., C.B.L. and N.E.L are recipients of a scholarship from CNPq, while C.D. is a recipient of scholarship from CAPES.
References
- 1.Guo G, Ye L, Pan K, Chen Y, Xing D, Yan K, et al. New Insights of Emerging SARS-CoV-2: Epidemiology, Etiology, Clinical Features, Clinical Treatment, and Prevention. Front Cell Dev Biol. 2020;8:410. doi: 10.3389/fcell.2020.00410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alshoabi SA, Alhazmi FH, Abdulaal OM, Gameraddin MB, Algaberi AK, Hamid AM, et al. Frequent clinical and radiological manifestations of the Novel SARS-CoV-2: A review article. J Family Med Prim Care. 2021;10(1):122–6. doi: 10.4103/jfmpc.jfmpc_1985_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atzrodt CL, Maknojia I, McCarthy RDP, Oldfield TM, Po J, Ta KTL, et al. A Guide to COVID-19: a global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS J. 2020;287(17):3633–50. doi: 10.1111/febs.15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakeshbandi M, Maini R, Daniel P, Rosengarten S, Parmar P, Wilson C, et al. The impact of obesity on COVID-19 complications: a retrospective cohort study. Int J Obes (Lond). 2020;44(9):1832–7. doi: 10.1038/s41366-020-0648-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–62. doi: 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anastassopoulou C, Gkizarioti Z, Patrinos GP, Tsakris A. Human genetic factors associated with susceptibility to SARS-CoV-2 infection and COVID-19 disease severity. Hum Genomics. 2020;14(1):40. doi: 10.1186/s40246-020-00290-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elhabyan A, Elyaacoub S, Sanad E, Abukhadra A, Elhabyan A, Dinu V. The role of host genetics in susceptibility to severe viral infections in humans and insights into host genetics of severe COVID-19: A systematic review. Virus Res. 2020;289:198163. doi: 10.1016/j.virusres.2020.198163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grolmusz VK, Bozsik A, Papp J, Patocs A. Germline Genetic Variants of Viral Entry and Innate Immunity May Influence Susceptibility to SARS-CoV-2 Infection: Toward a Polygenic Risk Score for Risk Stratification. Front Immunol. 2021;12:653489. doi: 10.3389/fimmu.2021.653489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debnath M, Banerjee M, Berk M. Genetic gateways to COVID-19 infection: Implications for risk, severity, and outcomes. FASEB J. 2020;34(7):8787–95. doi: 10.1096/fj.202001115R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos-Lopez O, Daimiel L, Ramirez de Molina A, Martinez-Urbistondo D, Vargas JA, Martinez JA. Exploring Host Genetic Polymorphisms Involved in SARS-CoV Infection Outcomes: Implications for Personalized Medicine in COVID-19. Int J Genomics. 2020;2020:6901217. doi: 10.1155/2020/6901217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozturk R, Tasova Y, Ayaz A. COVID-19: pathogenesis, genetic polymorphism, clinical features and laboratory findings. Turk J Med Sci. 2020;50(SI-1):638–57. doi: 10.3906/sag-2005-287 [DOI] [PubMed] [Google Scholar]
- 12.SeyedAlinaghi S, Mehrtak M, MohsseniPour M, Mirzapour P, Barzegary A, Habibi P, et al. Genetic susceptibility of COVID-19: a systematic review of current evidence. Eur J Med Res. 2021;26(1):46. doi: 10.1186/s40001-021-00516-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283(15):2008–12. doi: 10.1001/jama.283.15.2008 [DOI] [PubMed] [Google Scholar]
- 15.Sagoo GS, Little J, Higgins JP. Systematic reviews of genetic association studies. Human Genome Epidemiology Network. PLoS Med. 2009;6(3):e28. doi: 10.1371/journal.pmed.1000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Souza BM, Brondani LA, Boucas AP, Sortica DA, Kramer CK, Canani LH, et al. Associations between UCP1 -3826A/G, UCP2 -866G/A, Ala55Val and Ins/Del, and UCP3 -55C/T polymorphisms and susceptibility to type 2 diabetes mellitus: case-control study and meta-analysis. PLoS One. 2013;8(1):e54259. doi: 10.1371/journal.pone.0054259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brondani LA, Assmann TS, de Souza BM, Boucas AP, Canani LH, Crispim D. Meta-analysis reveals the association of common variants in the uncoupling protein (UCP) 1–3 genes with body mass index variability. PLoS One. 2014;9(5):e96411. doi: 10.1371/journal.pone.0096411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark MF, Baudouin SV. A systematic review of the quality of genetic association studies in human sepsis. Intensive Care Med. 2006;32(11):1706–12. doi: 10.1007/s00134-006-0327-y [DOI] [PubMed] [Google Scholar]
- 19.Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J. The choice of a genetic model in the meta-analysis of molecular association studies. Int J Epidemiol. 2005;34(6):1319–28. doi: 10.1093/ije/dyi169 [DOI] [PubMed] [Google Scholar]
- 20.Castano-Rodriguez N, Diaz-Gallo LM, Pineda-Tamayo R, Rojas-Villarraga A, Anaya JM. Meta-analysis of HLA-DRB1 and HLA-DQB1 polymorphisms in Latin American patients with systemic lupus erythematosus. Autoimmun Rev. 2008;7(4):322–30. doi: 10.1016/j.autrev.2007.12.002 [DOI] [PubMed] [Google Scholar]
- 21.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58. doi: 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 22.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60. doi: 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agwa SHA, Kamel MM, Elghazaly H, Abd Elsamee AM, Hafez H, Girgis SA, et al. Association between interferon-lambda-3 rs12979860, tll1 rs17047200 and ddr1 rs4618569 variant polymorphisms with the course and outcome of sars-cov-2 patients. Genes. 2021;12(6). doi: 10.3390/genes12060830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alghamdi J, Alaamery M, Barhoumi T, Rashid M, Alajmi H, Aljasser N, et al. Interferon-induced transmembrane protein-3 genetic variant rs12252 is associated with COVID-19 mortality. Genomics. 2021;113(4):1733–41. doi: 10.1016/j.ygeno.2021.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amodio E, Pipitone RM, Grimaudo S, Immordino P, Maida CM, Prestileo T, et al. SARS-CoV-2 viral load, ifnλ polymorphisms and the course of COVID-19: An observational study. Journal of Clinical Medicine. 2020;9(10):1–9. doi: 10.3390/jcm9103315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amoroso A, Magistroni P, Vespasiano F, Bella A, Bellino S, Puoti F, et al. HLA and AB0 Polymorphisms May Influence SARS-CoV-2 Infection and COVID-19 Severity. Transplantation. 2021;105(1):193–200. doi: 10.1097/TP.0000000000003507 [DOI] [PubMed] [Google Scholar]
- 27.Avendaño-Félix M, Ochoa-Ramírez LA, Ramos-Payán R, Aguilar-Medina M, Ayala-Ham A, Rendón-Aguilar H, et al. Lack of Effects of the Genetic Polymorphisms of Interleukin-10 in Clinical Outcomes of COVID-19. Viral immunology. 2021. doi: 10.1089/vim.2021.0022 [DOI] [PubMed] [Google Scholar]
- 28.Benetti E, Tita R, Spiga O, Ciolfi A, Birolo G, Bruselles A, et al. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur J Hum Genet. 2020;28(11):1602–14. doi: 10.1038/s41431-020-0691-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benetti E, Giliberti A, Emiliozzi A, Valentino F, Bergantini L, Fallerini C, et al. Clinical and molecular characterization of COVID-19 hospitalized patients. PLoS ONE. 2020;15(11 November). doi: 10.1371/journal.pone.0242534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernas SN, Baldauf H, Wendler S, Heidenreich F, Lange V, Hofmann JA, et al. CCR5Δ32 mutations do not determine COVID-19 disease course. International Journal of Infectious Diseases. 2021;105:653–5. doi: 10.1016/j.ijid.2021.02.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabrera-Marante O, de Frías ER, Pleguezuelo DE, Allende LM, Serrano A, Laguna-Goya R, et al. Perforin gene variant A91V in young patients with severe COVID-19. Haematologica. 2020;105(12):2844–6. doi: 10.3324/haematol.2020.260307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cafiero C, Rosapepe F, Palmirotta R, Re A, Ottaiano MP, Benincasa G, et al. Angiotensin system polymorphisms’ in sars-cov-2 positive patients: Assessment between symptomatic and asymptomatic patients: A pilot study. Pharmacogenomics and Personalized Medicine. 2021;14:621–9. doi: 10.2147/PGPM.S303666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calabrese C, Annunziata A, Coppola A, Pafundi PC, Guarino S, Di Spirito V, et al. ACE Gene I/D Polymorphism and Acute Pulmonary Embolism in COVID19 Pneumonia: A Potential Predisposing Role. Frontiers in Medicine. 2020;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cantalupo S, Lasorsa VA, Russo R, Andolfo I, D’alterio G, Rosato BE, et al. Regulatory noncoding and predicted pathogenic coding variants of ccr5 predispose to severe covid-19. International Journal of Molecular Sciences. 2021;22(10). doi: 10.3390/ijms22105372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coto E, Albaiceta GM, Clemente MG, Gómez J. Lack of association between SNPsrs8176719 (O blood group) and COVID-19: Data from Spanish age matched patients and controls. Transfusion. 2021;61(2):654–6. doi: 10.1111/trf.16206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuesta-Llavona E, Gómez J, Albaiceta GM, Amado-Rodríguez L, García-Clemente M, Gutiérrez-Rodríguez J, et al. Variant-genetic and transcript-expression analysis showed a role for the chemokine-receptor CCR5 in COVID-19 severity. International Immunopharmacology. 2021;98. doi: 10.1016/j.intimp.2021.107825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Del Ser T, Fernández-Blázquez MA, Valentí M, Zea-Sevilla MA, Frades B, Alfayate E, et al. Residence, Clinical Features, and Genetic Risk Factors Associated with Symptoms of COVID-19 in a Cohort of Older People in Madrid. Gerontology. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dite GS, Murphy NM, Allman R. An integrated clinical and genetic model for predicting risk of severe COVID-19: A population-based case-control study. PLoS One. 2021;16(2):e0247205. doi: 10.1371/journal.pone.0247205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, et al. Genomewide association study of severe covid-19 with respiratory failure. New England Journal of Medicine. 2020;383(16):1522–34. doi: 10.1056/NEJMoa2020283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gavriilaki E, Asteris PG, Touloumenidou T, Koravou EE, Koutra M, Papayanni PG, et al. Genetic justification of severe COVID-19 using a rigorous algorithm. Clinical Immunology. 2021;226. doi: 10.1016/j.clim.2021.108726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gómez J, Albaiceta GM, García-Clemente M, López-Larrea C, Amado-Rodríguez L, Lopez-Alonso I, et al. Angiotensin-converting enzymes (ACE, ACE2) gene variants and COVID-19 outcome. Gene. 2020;762. doi: 10.1016/j.gene.2020.145102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gómez J, Albaiceta GM, Cuesta-Llavona E, García-Clemente M, López-Larrea C, Amado-Rodríguez L, et al. The Interferon-induced transmembrane protein 3 gene (IFITM3) rs12252 C variant is associated with COVID-19. Cytokine. 2021;137:155354. doi: 10.1016/j.cyto.2020.155354 [DOI] [PubMed] [Google Scholar]
- 43.Grimaudo S, Amodio E, Pipitone RM, Maida CM, Pizzo S, Prestileo T, et al. PNPLA3 and TLL-1 Polymorphisms as Potential Predictors of Disease Severity in Patients With COVID-19. Frontiers in Cell and Developmental Biology. 2021;9. doi: 10.3389/fcell.2021.627914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gunal O, Sezer O, Ustun GU, Ozturk CE, Sen A, Yigit S, et al. Angiotensin-converting enzyme-1 gene insertion/deletion polymorphism may be associated with COVID-19 clinical severity: a prospective cohort study. Ann Saudi Med. 2021;41(3):141–6. doi: 10.5144/0256-4947.2021.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamet P, Pausova Z, Attaoua R, Hishmih C, Haloui M, Shin J, et al. SARS-CoV-2 Receptor ACE2 Gene Is Associated with Hypertension and Severity of COVID 19: Interaction with Sex, Obesity, and Smoking. American journal of hypertension. 2021;34(4):367–76. doi: 10.1093/ajh/hpaa223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hubacek JA, Dlouha L, Dusek L, Majek O, Adamkova V. Apolipoprotein E4 Allele in Subjects with COVID-19. Gerontology. 2021;67(3):320–2. doi: 10.1159/000516200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hubacek JA, Dusek L, Majek O, Adamek V, Cervinkova T, Dlouha D, et al. ACE I/D polymorphism in Czech first-wave SARS-CoV-2-positive survivors. Clinica Chimica Acta. 2021;519:206–9. doi: 10.1016/j.cca.2021.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karakaş Çelik S, Çakmak Genç G, Pişkin N, Açikgöz B, Altinsoy B, Kurucu İşsiz B, et al. Polymorphisms of ACE (I/D) and ACE2 receptor gene (Rs2106809, Rs2285666) are not related to the clinical course of COVID-19: A case study. Journal of Medical Virology. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kerget F, Kerget B. Frequency of interleukin-6 rs1800795 (-174G/C) and rs1800797 (-597G/A) polymorphisms in COVID-19 patients in Turkey who develop macrophage activation syndrome. Japanese journal of infectious diseases. 2021. [DOI] [PubMed] [Google Scholar]
- 50.Kolin DA, Kulm S, Christos PJ, Elemento O. Clinical, regional, and genetic characteristics of Covid-19 patients from UK Biobank. PLoS ONE. 2020;15(11 November). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuo CL, Pilling LC, Atkins JL, Masoli JAH, Delgado J, Kuchel GA, et al. APOE e4 Genotype Predicts Severe COVID-19 in the UK Biobank Community Cohort. J Gerontol A Biol Sci Med Sci. 2020;75(11):2231–2. doi: 10.1093/gerona/glaa131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Latini A, Agolini E, Novelli A, Borgiani P, Giannini R, Gravina P, et al. COVID-19 and Genetic Variants of Protein Involved in the SARS-CoV-2 Entry into the Host Cells. Genes (Basel). 2020;11(9). doi: 10.3390/genes11091010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lehrer S, Rheinstein PH. Homozygosity for rs17775810 Minor Allele Associated With Reduced Mortality of COVID-19 in the UK Biobank Cohort. In Vivo. 2021;35(2):965–8. doi: 10.21873/invivo.12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lehrer S, Rheinstein PH. ABO blood groups, COVID-19 infection and mortality. Blood Cells Mol Dis. 2021;89:102571. doi: 10.1016/j.bcmd.2021.102571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Littera R, Campagna M, Deidda S, Angioni G, Cipri S, Melis M, et al. Human Leukocyte Antigen Complex and Other Immunogenetic and Clinical Factors Influence Susceptibility or Protection to SARS-CoV-2 Infection and Severity of the Disease Course. The Sardinian Experience. Frontiers in Immunology. 2020;11. doi: 10.3389/fimmu.2020.605688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lorente L, Martín MM, Franco A, Barrios Y, Cáceres JJ, Solé-Violán J, et al. HLA genetic polymorphisms and prognosis of patients with COVID-19. Medicina Intensiva. 2020. doi: 10.1016/j.medin.2020.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malaquias MAS, Gadotti AC, Motta-Junior JDS, Martins APC, Azevedo MLV, Benevides APK, et al. The role of the lectin pathway of the complement system in SARS-CoV-2 lung injury. Translational Research. 2020. doi: 10.1016/j.trsl.2020.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martínez-Sanz J, Jiménez D, Martínez-Campelo L, Cruz R, Vizcarra P, Sánchez-Conde M, et al. Role of ACE2 genetic polymorphisms in susceptibility to SARS-CoV-2 among highly exposed but non infected healthcare workers. Emerging Microbes and Infections. 2021;10(1):493–6. doi: 10.1080/22221751.2021.1902755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Medetalibeyoglu A, Bahat G, Senkal N, Kose M, Avci K, Sayin GY, et al. Mannose binding lectin gene 2 (rs1800450) missense variant may contribute to development and severity of COVID-19 infection. Infect Genet Evol. 2021;89:104717. doi: 10.1016/j.meegid.2021.104717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Möhlendick B, Schönfelder K, Breuckmann K, Elsner C, Babel N, Balfanz P, et al. ACE2 polymorphism and susceptibility for SARS-CoV-2 infection and severity of COVID-19. Pharmacogenetics and genomics. 2021. doi: 10.1097/FPC.0000000000000436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monticelli M, Mele BH, Benetti E, Fallerini C, Baldassarri M, Furini S, et al. Protective role of a tmprss2 variant on severe covid-19 outcome in young males and elderly women. Genes. 2021;12(4). doi: 10.3390/genes12040596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naemi FMA, Al-Adwani S, Al-Khatabi H, Al-Nazawi A. Association between the HLA genotype and the severity of COVID-19 infection among South Asians. J Med Virol. 2021;93(7):4430–7. doi: 10.1002/jmv.27003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Novelli A, Biancolella M, Borgiani P, Cocciadiferro D, Colona VL, D’Apice MR, et al. Analysis of ACE2 genetic variants in 131 Italian SARS-CoV-2-positive patients. Hum Genomics. 2020;14(1):29. doi: 10.1186/s40246-020-00279-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Novelli A, Andreani M, Biancolella M, Liberatoscioli L, Passarelli C, Colona VL, et al. HLA allele frequencies and susceptibility to COVID-19 in a group of 99 Italian patients. HLA. 2020;96(5):610–4. doi: 10.1111/tan.14047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591(7848):92–8. doi: 10.1038/s41586-020-03065-y [DOI] [PubMed] [Google Scholar]
- 66.Petrazzuolo A, Le Naour J, Vacchelli E, Gaussem P, Ellouze S, Jourdi G, et al. No impact of cancer and plague-relevant FPR1 polymorphisms on COVID-19. OncoImmunology. 2020;9(1). doi: 10.1080/2162402X.2020.1857112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Posadas-Sánchez R, Sánchez-Muñoz F, Guzmán-Martín CA, Hernández-Díaz Couder A, Rojas-Velasco G, Fragoso JM, et al. Dipeptidylpeptidase-4 levels and DPP4 gene polymorphisms in patients with COVID-19. Association with disease and with severity. Life Sci. 2021;276:119410. doi: 10.1016/j.lfs.2021.119410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ravikanth V, Sasikala M, Naveen V, Latha SS, Parsa KVL, Vijayasarathy K, et al. A variant in TMPRSS2 is associated with decreased disease severity in COVID-19. Meta Gene. 2021;29. doi: 10.1016/j.mgene.2021.100930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Russo R, Andolfo I, Lasorsa VA, Cantalupo S, Marra R, Frisso G, et al. The TNFRSF13C H159Y Variant Is Associated with Severe COVID-19: A Retrospective Study of 500 Patients from Southern Italy. Genes (Basel). 2021;12(6). doi: 10.3390/genes12060881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saleh A, Sultan A, Elashry MA, Farag A, Mortada MI, Ghannam MA, et al. Association of TNF-α G-308 a Promoter Polymorphism with the Course and Outcome of COVID-19 Patients. Immunological Investigations. 2020. doi: 10.1080/08820139.2020.1851709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salem Hareedy M, Rashad SM, Hetta HF, Hassanien SM, Abdellatif H, Hassanien M. CYP2D6 and CYP3A4 variants influence the risk and outcome of COVID-19 infection among rheumatoid arthritis patients maintained on hydroxychloroquine. Drug Metabolism and Personalized Therapy. 2021;36(2):99–111. doi: 10.1515/dmdi-2020-0164 [DOI] [PubMed] [Google Scholar]
- 72.Schönfelder K, Breuckmann K, Elsner C, Dittmer U, Fistera D, Herbstreit F, et al. The influence of IFITM3 polymorphisms on susceptibility to SARS-CoV-2 infection and severity of COVID-19. Cytokine. 2021;142:155492. doi: 10.1016/j.cyto.2021.155492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schönfelder K, Breuckmann K, Elsner C, Dittmer U, Fistera D, Herbstreit F, et al. Transmembrane serine protease 2 Polymorphisms and Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Type 2 Infection: A German Case-Control Study. Frontiers in Genetics. 2021;12. doi: 10.3389/fgene.2021.667231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scutt DG, Overall DA. Single nucleotide polymorphisms in key aging pathways, and phenotypic markers of frailty are associated with increased odds of hospital admission with COVID-19. Journal of Infection. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shikov AE, Barbitoff YA, Glotov AS, Danilova MM, Tonyan ZN, Nasykhova YA, et al. Analysis of the Spectrum of ACE2 Variation Suggests a Possible Influence of Rare and Common Variants on Susceptibility to COVID-19 and Severity of Outcome. Frontiers in Genetics. 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shkurnikov M, Nersisyan S, Jankevic T, Galatenko A, Gordeev I, Vechorko V, et al. Association of HLA Class I Genotypes With Severity of Coronavirus Disease-19. Frontiers in Immunology. 2021;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Torre-Fuentes L, Matías-Guiu J, Hernández-Lorenzo L, Montero-Escribano P, Pytel V, Porta-Etessam J, et al. ACE2, TMPRSS2, and Furin variants and SARS-CoV-2 infection in Madrid, Spain. Journal of Medical Virology. 2021;93(2):863–9. doi: 10.1002/jmv.26319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valenti L, Griffini S, Lamorte G, Grovetti E, Uceda Renteria SC, Malvestiti F, et al. Chromosome 3 cluster rs11385942 variant links complement activation with severe COVID-19. J Autoimmun. 2021;117:102595. doi: 10.1016/j.jaut.2021.102595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Verma S, Abbas M, Verma S, Khan FH, Raza ST, Siddiqi Z, et al. Impact of I/D polymorphism of angiotensin-converting enzyme 1 (ACE1) gene on the severity of COVID-19 patients. Infect Genet Evol. 2021;91:104801. doi: 10.1016/j.meegid.2021.104801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vietzen H, Zoufaly A, Traugott M, Aberle J, Aberle SW, Puchhammer-Stöckl E. Deletion of the NKG2C receptor encoding KLRC2 gene and HLA-E variants are risk factors for severe COVID-19. Genetics in Medicine. 2021. doi: 10.1038/s41436-020-01077-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang F, Huang S, Gao R, Zhou Y, Lai C, Li Z, et al. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discovery. 2020;6(1). doi: 10.1038/s41421-020-00231-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang W, Zhang W, Zhang J, He J, Zhu F. Distribution of HLA allele frequencies in 82 Chinese individuals with coronavirus disease-2019 (COVID-19). HLA. 2020;96(2):194–6. doi: 10.1111/tan.13941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wulandari L, Hamidah B, Pakpahan C, Damayanti NS, Kurniati ND, Adiatmaja CO, et al. Initial study on TMPRSS2 p.Val160Met genetic variant in COVID-19 patients. Hum Genomics. 2021;15(1):29. doi: 10.1186/s40246-021-00330-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y, Qin L, Zhao Y, Zhang P, Xu B, Li K, et al. Interferon-Induced Transmembrane Protein 3 Genetic Variant rs12252-C Associated With Disease Severity in Coronavirus Disease 2019. J Infect Dis. 2020;222(1):34–7. doi: 10.1093/infdis/jiaa224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou J, Liu C, Sun Y, Huang W, Ye K. Cognitive disorders associated with hospitalization of COVID-19: Results from an observational cohort study. Brain, Behavior, and Immunity. 2020. doi: 10.1016/j.bbi.2020.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181(4):894–904 e9. doi: 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–80 e8. doi: 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scialo F, Daniele A, Amato F, Pastore L, Matera MG, Cazzola M, et al. ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung. 2020;198(6):867–77. doi: 10.1007/s00408-020-00408-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rysz S, Al-Saadi J, Sjostrom A, Farm M, Campoccia Jalde F, Platten M, et al. COVID-19 pathophysiology may be driven by an imbalance in the renin-angiotensin-aldosterone system. Nat Commun. 2021;12(1):2417. doi: 10.1038/s41467-021-22713-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hu Y, Liu L, Lu X. Regulation of Angiotensin-Converting Enzyme 2: A Potential Target to Prevent COVID-19? Front Endocrinol (Lausanne). 2021;12:725967. doi: 10.3389/fendo.2021.725967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Magrone T, Magrone M, Jirillo E. Focus on Receptors for Coronaviruses with Special Reference to Angiotensin- Converting Enzyme 2 as a Potential Drug Target—A Perspective. Endocr Metab Immune Disord Drug Targets. 2020;20(6):807–11. doi: 10.2174/1871530320666200427112902 [DOI] [PubMed] [Google Scholar]
- 92.Yamamoto N, Ariumi Y, Nishida N, Yamamoto R, Bauer G, Gojobori T, et al. SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene. 2020;758:144944. doi: 10.1016/j.gene.2020.144944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gemmati D, Bramanti B, Serino ML, Secchiero P, Zauli G, Tisato V. COVID-19 and Individual Genetic Susceptibility/Receptivity: Role of ACE1/ACE2 Genes, Immunity, Inflammation and Coagulation. Might the Double X-chromosome in Females Be Protective against SARS-CoV-2 Compared to the Single X-Chromosome in Males? Int J Mol Sci. 2020;21(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gemmati D, Tisato V. Genetic Hypothesis and Pharmacogenetics Side of Renin-Angiotensin-System in COVID-19. Genes (Basel). 2020;11(9). doi: 10.3390/genes11091044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhong WG, Wang Y, Zhu H, Zhao X. Meta analysis of angiotensin-converting enzyme I/D polymorphism as a risk factor for preeclampsia in Chinese women. Genet Mol Res. 2012;11(3):2268–76. doi: 10.4238/2012.May.21.1 [DOI] [PubMed] [Google Scholar]
- 96.Delanghe JR, Speeckaert MM, De Buyzere ML. COVID-19 infections are also affected by human ACE1 D/I polymorphism. Clin Chem Lab Med. 2020;58(7):1125–6. doi: 10.1515/cclm-2020-0425 [DOI] [PubMed] [Google Scholar]
- 97.Delanghe JR, Speeckaert MM, De Buyzere ML. The host’s angiotensin-converting enzyme polymorphism may explain epidemiological findings in COVID-19 infections. Clin Chim Acta. 2020;505:192–3. doi: 10.1016/j.cca.2020.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hatami N, Ahi S, Sadeghinikoo A, Foroughian M, Javdani F, Kalani N, et al. Worldwide ACE (I/D) polymorphism may affect COVID-19 recovery rate: an ecological meta-regression. Endocrine. 2020;68(3):479–84. doi: 10.1007/s12020-020-02381-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. 2013;14:301–23. doi: 10.1146/annurev-genom-091212-153455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dutta M, Dutta P, Medhi S, Borkakoty B, Biswas D. Polymorphism of HLA class I and class II alleles in influenza A(H1N1)pdm09 virus infected population of Assam, Northeast India. J Med Virol. 2018;90(5):854–60. doi: 10.1002/jmv.25018 [DOI] [PubMed] [Google Scholar]
- 101.Lin M, Tseng HK, Trejaut JA, Lee HL, Loo JH, Chu CC, et al. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet. 2003;4:9. doi: 10.1186/1471-2350-4-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dendrou CA, Petersen J, Rossjohn J, Fugger L. HLA variation and disease. Nat Rev Immunol. 2018;18(5):325–39. doi: 10.1038/nri.2017.143 [DOI] [PubMed] [Google Scholar]
- 103.Deng H, Yan X, Yuan L. Human genetic basis of coronavirus disease 2019. Signal Transduct Target Ther. 2021;6(1):344. doi: 10.1038/s41392-021-00736-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13(1):46–57. doi: 10.1038/nri3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Muller JA, Gross R, Conzelmann C, Kruger J, Merle U, Steinhart J, et al. SARS-CoV-2 infects and replicates in cells of the human endocrine and exocrine pancreas. Nat Metab. 2021;3(2):149–65. doi: 10.1038/s42255-021-00347-1 [DOI] [PubMed] [Google Scholar]
- 106.Zhang Q, Bastard P, Effort CHG, Cobat A, Casanova JL. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature. 2022;603(7902):587–98. doi: 10.1038/s41586-022-04447-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Asano T, Boisson B, Onodi F, Matuozzo D, Moncada-Velez M, Maglorius Renkilaraj MRL, et al. X-linked recessive TLR7 deficiency in ~1% of men under 60 years old with life-threatening COVID-19. Sci Immunol. 2021;6(62). doi: 10.1126/sciimmunol.abl4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370(6515). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zeng Z, Lan T, Wei Y, Wei X. CCL5/CCR5 axis in human diseases and related treatments. Genes Dis. 2021. doi: 10.1016/j.gendis.2021.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ray PR, Wangzhou A, Ghneim N, Yousuf MS, Paige C, Tavares-Ferreira D, et al. A pharmacological interactome between COVID-19 patient samples and human sensory neurons reveals potential drivers of neurogenic pulmonary dysfunction. Brain Behav Immun. 2020;89:559–68. doi: 10.1016/j.bbi.2020.05.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, et al. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272(5270):1955–8. doi: 10.1126/science.272.5270.1955 [DOI] [PubMed] [Google Scholar]
- 112.Gkouskou K, Vasilogiannakopoulou T, Andreakos E, Davanos N, Gazouli M, Sanoudou D, et al. COVID-19 enters the expanding network of apolipoprotein E4-related pathologies. Redox Biol. 2021;41:101938. doi: 10.1016/j.redox.2021.101938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mahley RW. Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders. J Mol Med (Berl). 2016;94(7):739–46. doi: 10.1007/s00109-016-1427-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.El-Lebedy D, Raslan HM, Mohammed AM. Apolipoprotein E gene polymorphism and risk of type 2 diabetes and cardiovascular disease. Cardiovasc Diabetol. 2016;15:12. doi: 10.1186/s12933-016-0329-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Inal J. Biological Factors Linking ApoE epsilon4 Variant and Severe COVID-19. Curr Atheroscler Rep. 2020;22(11):70. doi: 10.1007/s11883-020-00896-y [DOI] [PMC free article] [PubMed] [Google Scholar]