Abstract
Molecular tests are highly reliable and sensitive but lack portability and are not simple to use; conversely, easy-to-use antigenic tests still lack high performance. BioScreen combines single-molecule sensitivity and outstanding reliability with ultraportability and simplicity of use. This digital platform is capable of artificial intelligence–based binary classification at the limit of identification of a single marker/virus in 0.1 ml. The diagnostic sensitivity, specificity, and accuracy reach 99.2% as validated through 240 assays, including a pilot clinical trial. The versatile immunometric system can detect the SARS-CoV-2 virus, spike S1, and immunoglobulin G antigen proteins in saliva, blood serum, and swab. BioScreen has a small footprint comprising a disposable cartridge and a handheld electronic reader connected to a smart device. The sample handling is minimal, and the assay time to result is 21 min. Reliable and sensitive self-testing with an ultraportable and easy-to-use diagnostic system operated directly by a patient holds the potential to revolutionize point-of-care testing and early diagnosis.
POC antigenic testing is unreliable, but BioScreen performs like a molecular test and is also ultraportable, fast, and disposable.
INTRODUCTION
The ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, with its over 450 million infected and 6 million deaths (as of March 2022), is still threatening the world, while noncommunicable diseases, e.g., cardiovascular syndromes, cancers, and neurodegenerative pathologies, are stably among the top 10 causes of death in higher-income countries (1). Point-of-care (POC) testing devices can offer early diagnostic with fast time to results at a low cost. This can limit the pathogens spreading in the case of pandemics (2). In progressive noncommunicable diseases, early detection and/or continuous monitoring can improve treatment efficacy and quality of life. However, until very recently, typical POC testing drawbacks included limit of detections (LOD) (3) higher than 104 markers per milliliter, corresponding to femtomolar (10−15 M) concentrations (4), and a high incidence of false negatives (5).
Because of the coronavirus disease 2019 (COVID-19) pandemic, POC testing has been recognized as a top priority by the World Health Organization (WHO) (6). The scientific community responded with an incredible effort that has come up with the study and development of several innovative POC diagnostic tests. The best-performing ones are the platforms based on the reverse transcription, loop-mediated isothermal amplification (RT-LAMP) (7) and on the CRISPR-Cas Abbot ID NOW (8). These are both molecular assays, namely, they involve the detection of the viral genome. These approaches have been proven capable of assaying SARS-CoV-2 viral RNA in nasal, nasopharyngeal, or throat swabs at an LOD of 10 to 100 copies per milliliter (5). Similar figures of merit have also been demonstrated in POC tests such as the CRISPR-Cas–based smartphone-read saliva tests (9) or, to a less high-performance level, in the RT-LAMP Harmony COVID-19 (10). While being quantitative with a wide dynamic range, molecular-based POC tests still have not fully addressed key aspects such as portability, simplicity of use, and independence from resource/infrastructure. These require minimizing assay steps and manual operation in sample preparation as well as full integration with automated and ultraportable devices. On the other hand, no such improvements in the performance level of immunometric later flow antigenic tests can be reported. Still, the convenient, easy to use one-step, disposable, and very fast lateral flow immunometric assays result in LODs reaching 108 entities per milliliter and as yet hold an unacceptably high incidence of false negatives (11–13). These features still make them unreliable for a diagnosis.
Bioelectronic sensors are, in principle, ideal for field-deployable diagnostic as they can be produced at a low cost and can provide an electronic response for direct digital data communication (14–20). State-of-the-art bioelectronic technologies operating at the lowest possible LOD of 1 entity in 0.1 ml have been based so far on organic (21–34) or graphene (16) bioelectronic field-effect transistors (FETs). These have triggered a great deal of attention as they offer label-free ultrahigh sensitive detection along with versatility; viruses (26) and their capsid proteins (25, 31) as well as protein (22–24, 27, 29, 30, 32–34) and amino acid–based (28) markers can be detected. High selectivity is assured by the biofunctionalization of the sensing gate with a densely packed layer of capturing antibodies (22–27, 29–34). These technologies, however, focus on the ultralow detection limits and high selectivity, but key aspects such as easy and user-friendly assay procedures, miniaturization and portable readout equipment, and, most relevantly, reliability, i.e., a low incidence of false positives and false negatives, are still overlooked.
Here, the handheld, ultraportable, fully integrated, single-molecule BioScreen binary platform, comprising a disposable bioelectronic cartridge driven by a handheld readout circuit connected via Bluetooth to a smart device, is proposed. Notably, the platform is based on antigenic immunometric detection; hence, it is highly versatile as, by just changing the capturing antibody, it can be successfully tuned to address many different applications. The modular system is proven capable of detecting the SARS-CoV-2 virus as well as the SARS-CoV-2 spike S1 capsid (S1) and immunoglobulin G (IgG) proteins directly in patients’ saliva as well as in blood serum and swab. Digital immunometric detections and artificial intelligence (AI)–based binary classifications of a single entity are accomplished beyond limit of identification (LOI; granting false-positive and false-negative random errors below 1%) (3), granting extremely high reliability (diagnostic sensitivity, diagnostic specificity, and diagnostic accuracy of 99.2%). The performance level is validated through 240 label-free assays carried out directly in biofluids, including those from 18 COVID-19 patients. Selectivity is achieved by very easily integrating into the cartridge a layer of capturing antibodies stably physisorbed on a millimeter-wide gate electrode, exhibiting a shelf life of at least 2 weeks. The assay operation is highly user-friendly, and it encompasses a few easy steps taking, overall, 21 min.
RESULTS AND DISCUSSION
The handheld BioScreen system is displayed in Fig. 1A, along with an illustration of a typical POC ambient (Fig. 1B). An assay of a saliva sample self-collected by a patient is shown in an explanatory video (section S1) that evidences how fast and easy it is to perform a whole BioScreen assay. The procedure is rapid and user-friendly, resulting in a straightforward delivery of the assay outcome directly to an end-user that can read it on a smartphone or on a tablet.
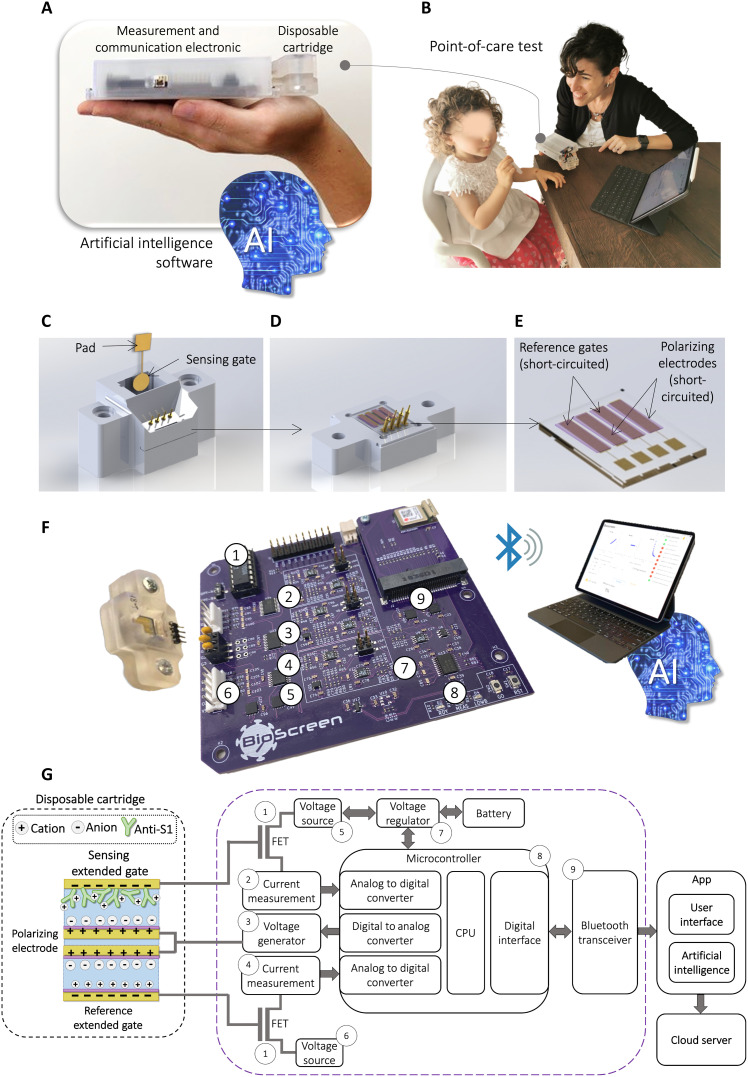
Fig. 1. The BioScreen POC platform.
(A) Pictures of the ultraportable BioScreen platform, showing the disposable fluidic cartridge and the ultraportable reader that sends the data to a smart device via Bluetooth; the output data are analyzed with an ad hoc designed AI software. (B) Picture of a typical operational environment where the saliva is self-collected and put in the disposable fluidic cartridge. (C) Rendering of the disposable bioelectronic cartridge enclosing a fluidic device embedding four gate electrodes (not visible in this panel) and the sensing gate. The latter comprises a gold sensing area and a contact pad. The circular area is biofunctionalized with a stably physisorbed layer of capturing antibodies. The sensing gate is easily and stably inserted into the fluidic system. (D) Details of the bottom of the fluidic device comprising four electrodes. (E) Zoomed view into the four gold electrodes that are coated with the antifouling proprietary layer (violet). Two of them serve as reference gates, while the other two serve as counter electrodes. (F) Actual three-dimensional–printed cartridge and the reusable electronic driving/readout circuit connected via Bluetooth to a smart device. The acquired data are handled via a dedicated app, while the AI-based binary analysis output sorts out the samples. (G) On the left, a schematic illustration of the electrodes in the disposable cartridge is displayed. A representation of the device operation principle is also provided showing the details of the capacity-coupled charge double layers forming at the interfaces between the electrodes’ surfaces and the electrolyte. The electrodes in the cartridge are connected to the electronic system as extended gates of the FETs (shown on the right). The main building blocks of the reader are displayed including the digital-to-analog converters, the microcontroller unit, the Bluetooth communication, and the energy management unit for low-power battery operation.
The core of the BioScreen platform is the disposable bioelectronic cartridge comprising a fluidic device filled with the gating electrolyte or with the sample to be assayed (Fig. 1C). Embedded in the fluidic device is the sensing array, encompassing four gold electrodes and the sensing gate (Fig. 1D). This is stably inserted into the fluidics after its biofunctionalization with a layer of capturing antibodies [e.g., anti–SARS-CoV-2 spike S1 (anti-S1)] for the selective immunometric recognition of the analyte (e.g., the SARS-CoV-2 virion via the binding of its capsid protein S1). The biofunctionalization protocol involves the physisorption of the capturing antibodies directly on the gold sensing gate (see Materials and Methods). Surface plasmon resonance (SPR) characterization (see Materials and Methods) assesses that a very stable adherent layer encompassing (1.13 ± 0.04) × 1012 capturing proteins per square centimeter is attached to the electrode. After 45 hours of storage in phosphate-buffered saline (PBS) solution and after several washing steps, the gate is still uniformly covered by the physisorbed biolayer (section S2). Moreover, a physisorbed anti-S1 layer can capture the S1 proteins even after overnight storage in deionized (DI) water or after storage for 2 weeks in PBS (section S3). These data prove that (i) a physisorbed layer of capturing antibodies is stable and suitable to carry out our immunometric electronic sensing; (ii) the operation or even overnight storage of the biolayer in water does not harm the capturing antibodies’ biological functionality; and (iii) the biofunctionalized sensing gate holds a shelf life of at least 3 weeks; therefore, it can be likely fabricated along with the cartridge. This very important occurrence implies that no biofunctionalization or coating process is needed to be performed right before the assay. Therefore, in contrast to the state-of-the-art single-molecule sensors (table S1), the biofunctionalization process time (taking at least 2 hours) does not add to the time to results.
The other four electrodes (Fig. 1E) are gold strips coated with an antifouling conducting layer, proven to prevent nonspecific adsorption from the analyte solution (section S4). Two of the four electrodes serve as reference gates, enabling us to assess whether any kind of spurious surface potential shift (instabilities) occurs at any stage of the bioelectronic sensing measurements. The other two electrodes enable the polarization of the electrolyte solution that fills the fluidic cell. Low ionic strength DI water serves as the electrolyte that maximizes the Debye length, leaving the electrostatic changes associated with the biochemical specific binding unscreened (35). Pictures of the disposable cartridge and the reusable driving/readout electronic Bluetooth connected to a smart device are shown in Fig. 1F. In Fig. 1G (left), a schematic representation of the cartridge and the device operation principle is provided. The sensing and reference gates work as extended electrodes connected to the gate terminals of two FETs. Together, they form the extended gate transistor (EGT)–based BioScreen architecture (36) comprising the sensing EGT and the reference EGT. Each gate is capacitively coupled to the polarizing electrode, acting as the counter electrode that is connected to the signal generator. Upon biasing, the ions from the solution form charge double layers that couple each gate (sensing and reference) to the corresponding EGT. Self-dissociation of water produces a low concentration (ca. 5 μM) of positive and negative ions in the electrolyte that can gate a transistor (37). The selective binding at the biofunctionalized sensing gate shifts its surface potential, and an adjustment of the charge double layer takes place, producing a threshold voltage (VT) shift in the sensing EGT that is read as an output drain current (ID) change (38). By contrast, no selective binding occurs at the reference gates and, hence, no current variation is read on the reference EGT. A schematic layout of the electrodes’ connections to the circuit is provided in the right part of Fig. 1G along with all the components for data digitalization and communication. A Bluetooth connection with a smart device endowed with an app along with an ad hoc developed BioScreen AI-based binary classifier completes the system. The data analysis is done offline, but it can be prospectively performed via a connection to a cloud so that the classifier output is displayed, through the app, directly back on the smart device.
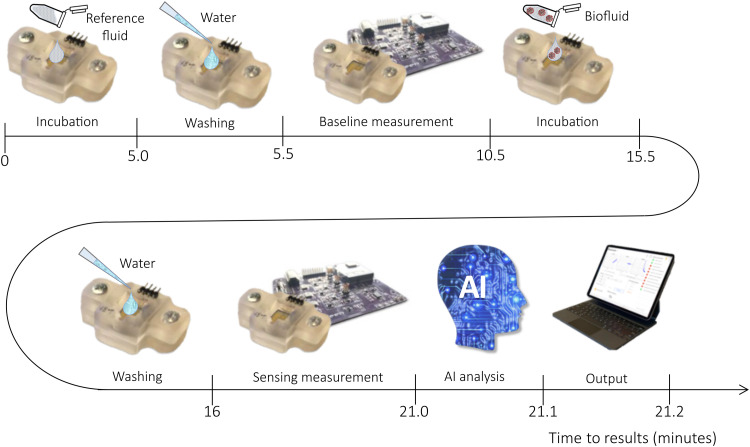
The BioScreen platform operation is user-friendly and fast, as illustrated in Fig. 2. Here, the main steps of the assay shown in the video are schematically provided (section S1). The set of operations to be performed is extremely simple and can be executed by an untrained operator. It could even be performed directly by the patient at home following simple instructions provided by the app. The overall time to result is 21.2 min. The assay foresees, after installing and opening the app, a first step when the disposable fluidic device is filled with 100 μl of the reference fluid, in which the sensing gate electrode (not shown) is incubated for 5 min. The fluidic element is washed and filled with DI water, and the baseline current is measured. This step involves the automatic reiterated acquisition of 30 electrical transfer characteristics, addressed as cycling (vide infra), that lasts for 5 min. The DI water is discarded, the cartridge is filled with 100 μl of the sample to be analyzed, for instance, the patient’s self-sampled saliva, and a second incubation follows. A further washing step and cycling of the sensing gate is performed before the BioScreen AI-based binary classifier output is released.
Fig. 2. Fast BioScreen assay—The sensing steps.
The disposable cartridge is the fluidic device, and the biofunctionalized sensing gate is specific for a given application. For the sake of clearness, the sensing gate is not shown. The cartridge is connected to the electronic reader, and the app is installed on a tablet. In step 1, the cartridge is loaded with 100 μl of the specific sample diluting solution used as reference fluid where the biofunctionalized sensing gate is incubated for 5 min. The reference fluid is discarded, and the fluidic device is washed and (step 2) filled with 100 μl of DI water (0.5 min). The measurement of the baseline current is run (step 3) by the system for 5 min by reiteratively measuring the EGT output current curves. This is addressed as the cycling and is detailed in Materials and Methods. The sample to be analyzed (e.g., 100 μl of a patient’s saliva) is loaded in the fluidic device and incubated for 5 min (step 4). The fluidic device is washed and filled with DI water (0.5 min), and a final set of EGT curves is run (step 5). This is the measurement of the sensing signal taking 5 min. Then, 0.2 min is approximately needed for the data to be encrypted, transferred to the app, and analyzed by the BioScreen AI-based binary classifier to provide the positive/negative response (step 6). Overall, the assay takes 21.2 min.
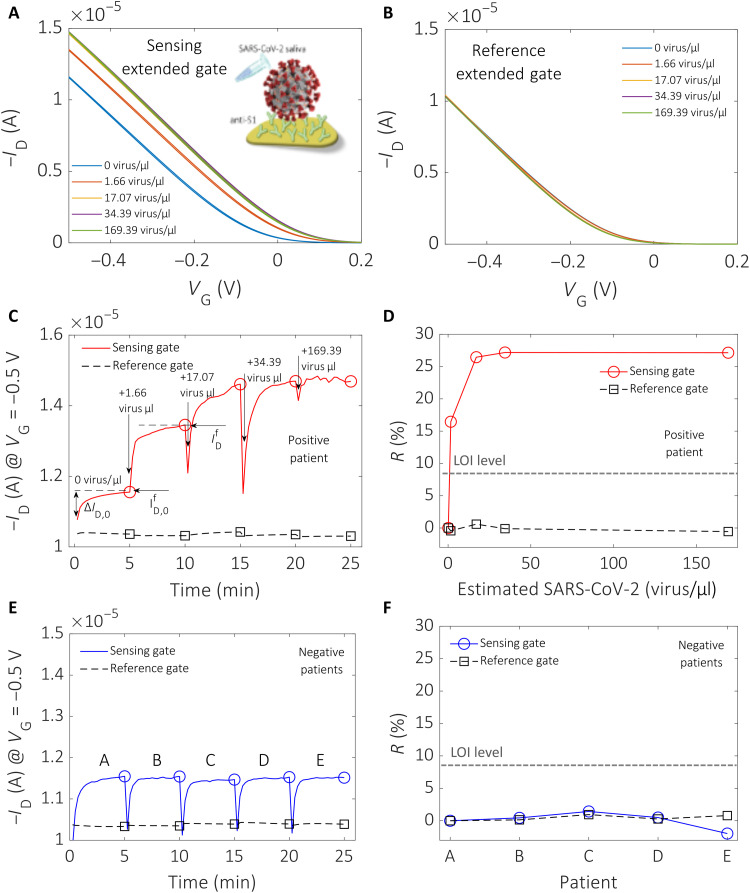
Raw data taken with the BioScreen platform directly in the self-sampled saliva of positive and negative patients are given in Fig. 3. The sensing gate is functionalized with the anti-S1 antibody and incubated for 5 min in the saliva to be assayed. The gate is measured in DI water by acquiring 30 subsequent transfer characteristics, ID (drain current) versus VG (gate voltage) at VD (drain voltage) = −0.1 V, with the source terminal connected to the ground. This measurement run is addressed as the cycling. The 30th transfer characteristics measured with the sensing EGT and the reference EGT are shown in Fig. 3, A and B, respectively. The gate voltage range is chosen between 0.2 and −0.5 V to ensure that Faradaic leakage currents, connected with redox reactions detrimental to the electrode stability, are negligible (section S5). The viral concentrations assayed are as follows: 0 virus/μl (blue line; this is the baseline measured in the saliva diluent serving as reference fluid, namely, Pipes), 1.66 virus/μl (red line), 17.07 virus/μl (yellow line), 34.39 virus/ml (purple line), and 169.39 virus/μl (green line). The original positive sample contained a viral concentration of 169.39 virus/μl. This sample was progressively diluted with the saliva diluent. The clinical samples were assessed as positive by real-time reverse transcription polymerase chain reaction (rRT-PCR) and quantified by fully automated chemiluminescent enzyme immunoassay. Detailed virus quantification procedures and dilution protocols are given in section S6. A change in current is measured when passing from 0 viral concentration (baseline) to concentrations up to 169.39 virus/μl (Fig. 3A). Conversely, the ID measured on the reference gate is stable, and no appreciable change is seen at any of the concentrations assayed (Fig. 3B). This proves the excellent stability of the system in the absence of a specific immunometric binding reaction. The measurements displayed in Fig. 3C are the transient ID values at VG = −0.5 V and VD = −0.1 V measured from the 1st to the 30th cycle. The red trace is measured on the sensing gate, while the dashed black one is taken on the reference gate while both are immersed in the fluidic cell loaded with DI water. The red trace data evidence the ID dynamics occurring when the sensing gate is cycled, which is typically seen in biofunctionalized electrolyte-gated transistors (27). ID, measured on the sensing gate incubated in the sample to be assayed, reaches a final value, , after full cycling in DI water at each assayed concentration. For the reference fluid (zero analyte concentration), ID reaches a final value, , while the current overall change during the cycling is addressed as ∆ID,0. Figure 3D shows the percentage of the relative variation of the current measured at the end of the cycling, , relative to the corresponding baseline level, . This is taken as the sensing response given in Eq. 1
| (1) |
Fig. 3. Detection of SARS-CoV-2 virus in untreated saliva clinical samples.
(A) Last (30th cycle) transfer characteristics, ID versus VG at VD = −0.1 V, are shown as a function of the SARS-CoV-2 virion concentration in saliva, assessed with an anti-S1 biofunctionalized sensing EGT. Five different concentrations are assayed, and the corresponding current changes are shown. (B) Last transfer curves (30th cycle) as a function of the SARS-CoV-2 concentration measured with the reference EGT. No shift in the current is measured after assaying all five saliva samples. (C) Transient ID values [extracted from the transfer curves in (A)] at VG = −0.5 V and VD = −0.1 V, at each cycle from the 1st to the 30th, upon assaying saliva samples at increasing virion concentration, are provided. The red line is relevant to the ID current values measured with the sensing gate, and the hollow dots mark the 30th cycle for each sample. The current measured on the reference gate is also shown as a dashed black line. The hollow squares mark the end of the measuring cycle. (D) The red line is the R (%) value measured on a positive patient’s sample with the sensing gate, while the dashed black trace is taken, contemporarily, on the reference gate. (E) Five saliva samples collected from negative patients (A, B, C, D, and E) measured following the protocol described in (C). The blue line shows the data from the sensing gate, while the dashed black trace is measured with the reference gate. The hollow symbols mark the end of the cycling. (F) R (%) estimated from the data of (E); the blue line shows the data from the sensing gate, while the black trace is measured with the reference gate. The hollow symbols mark the end of the measuring cycle. The limit-of-identification (LOI) level is given by the dashed black line.
At the lowest concentration, equal to 1.66 virus/μl, an R (%) value as high as 16.5% is measured with the sensing EGT. The signal saturates at R = 27.2% when a 34.39 virus/μl concentration is assayed. As a comparison, the response obtained with the reference EGT is lower than 0.6%, proving the extremely high electronic stability of the EGT-based BioScreen system.
To assess the selectivity of the BioScreen platform, negative control experiments were performed assaying, with an anti-S1 functionalized sensing EGT, several saliva samples self-collected by healthy (negative) patients. The status for each patient was assessed by at least three subsequent negative rRT-PCR tests in 6 weeks. The blue trace in Fig. 3E shows the ID (VG = −0.5 V, VD = −0.1 V) transient signal measured with an anti-S1 sensing EGT after incubation in the saliva samples of the A, B, C, D, and E healthy patients. The negative status of these patients was assessed by rRT-PCR and three subsequent follow-up tests. The measurements show extremely reproducible values and current dynamics, both on the sensing EGT (blue trace) and on the reference EGT (black trace). The R (%) values from Eq. 1 for each patient measured with the sensing EGT and reference EGT are displayed in Fig. 3F. The maximum response of the reference EGT on these negative patients is less than 1.43%, and the sensing EGT responses are almost perfectly overlapped. This proves the extremely high reproducibility of the BioScreen system when operated directly in the saliva of a patient, along with the high selectivity. The LOI level, estimated as the average of the signal measured on the assays of the negative patients, plus six times its SD (section S7), is as low as 8.5%. This means that even the lowest concentration assayed, namely, 1.66 virus/μl (166 ± 13 viruses in 100 μl), holds a high level of confidence that can grant false-positive and false-negative random errors lower than 1% (section S7). While this is compelling proof of BioScreen ability to sense a few viruses directly in saliva, reliable single-entity detection is not yet proven.
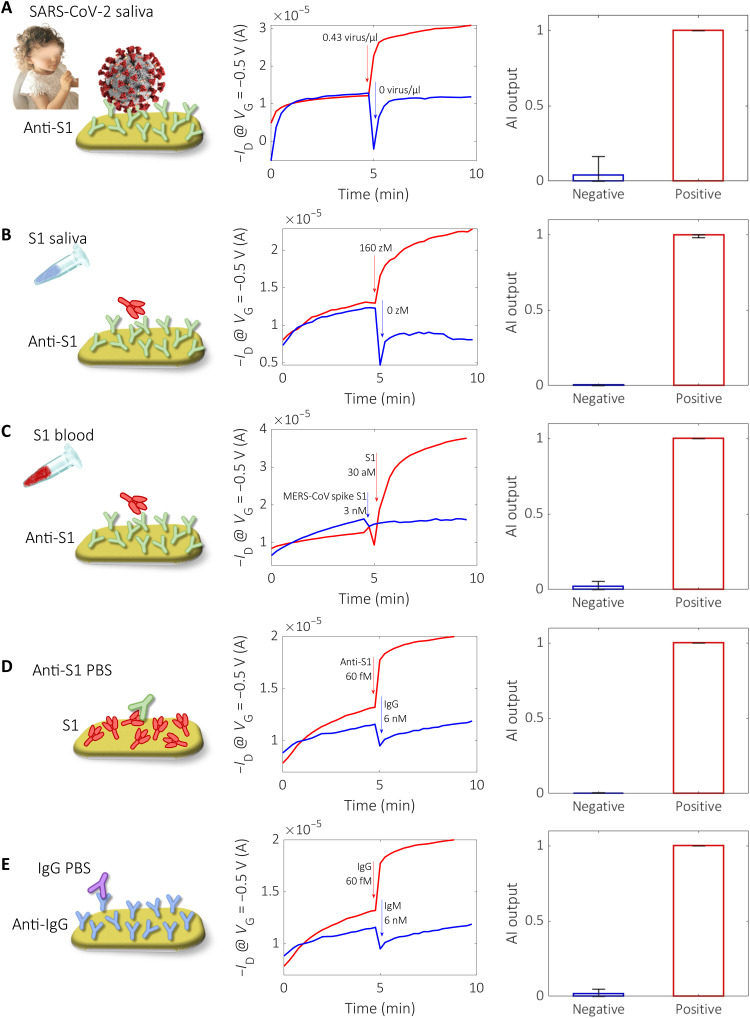
The BioScreen system performance levels are validated by analyzing 240 samples, including 18 COVID-19 patients. The generality of the approach is exemplified in Fig. 4, where the sensing and reference transient traces for each of the different classes of samples assayed are provided, namely, the detection of the SARS-CoV-2 virus in real patients’ saliva (Fig. 4A), its capsid protein S1 in saliva (Fig. 4B) and in human whole blood (Fig. 4C), its capturing antibody anti-S1 in PBS (Fig. 4D), and its IgG in PBS (Fig. 4E). Only the capturing protein, specific for the target species to be detected, changes from one assay to the other. For each assay, a negative control experiment is also provided.
Fig. 4. General applicability of the binary BioScreen platform.
In the first column, the different types of assays carried out with the BioScreen platform are schematically shown. The second column shows the measured transient current traces for the positive (red curves) and negative samples (blue curves). The first 5 min are relevant to cycles 1 to 30, measured in DI water after incubation in the reference fluid; in the subsequent 5 min (cycles 31 to 60), the same gate is measured in DI water after incubation in the sample to be assayed. The third column shows the output of the AI binary classifier. The different assays represented are (A) SARS-CoV-2 virus detected in a patient’s saliva (0.43 virus/μl; 43 ± 7 viruses per 100 μl); (B) S1 protein added in human saliva (160 zM; 10 ± 3 in 100 μl) detected with anti-S1 capturing antibody (the reference fluid is 0 zM Pipes); (C) S1 protein added in whole human blood (30 aM; 1.8 × 103 in 100 μl) detected with anti-S1; (D) PBS anti-S1 solution (60 fM; 3.6 × 106 in 100 μl), with the S1 protein serving as capturing antibody [the reference fluid is PBS added with IgG (6 nM; 4 × 1011 in 100 μl)]; and (E) PBS solution of IgG (60 fM; 3.6 × 106 in 100 μl), with anti-IgG as capturing antibody [the reference fluid is PBS, and the negative sample is a PBS solution of IgM (6 nM; 4 × 1011 in 100 μl)].
The different classes of assays are exemplified by a cartoon in the first column of Fig. 4. The transient ID measurements (current value at VG = −0.5 V, VD = −0.1 V) for each assay are given in the central column as red traces for the positive samples/patients and blue traces for the negative samples/patients. After incubating the sensing electrode in the reference fluid, the electrode is washed in DI water and a cycling run, namely, 30 transfer characteristics (ID − VG) are recorded (1st to 30th), in DI water is carried out. Then, the same gate is incubated into the sample to be assayed, and after washing, another set of 30 ID − VG characteristics is also acquired (31st to 60th) in DI water. The third column shows the output of the AI-based binary BioScreen classifier (section S8); error bars are the incidences of false positive and false negative on the output responses.
In more detail, the different assays represented are as follows: SARS-CoV-2 virus detected in a patient’s saliva (0.43 virus/μl; 43 ± 7 viruses per 100 μl) [the assay involves the anti-S1 biofunctionalized sensing gate immersed in the saliva diluent (0.5 M Pipes, pH 6.8) and afterward in the saliva samples of a healthy (blue trace) or diseased (red curve) patients] (Fig. 4A); SARS-CoV-2 capsid spike S1 (S1) protein added in human saliva (160 zM; 10 ± 3 in 100 μl), with anti-S1 serving as capturing antibody (the reference fluid is 0 zM Pipes) (Fig. 4B); S1 protein added in whole human blood (30 aM; 1.8 × 103 in 100 μl), with anti-S1 serving as capturing antibody [the reference fluid is whole blood, and the negative control is performed with whole blood added with the nonbinding Middle East respiratory syndrome coronavirus (MERS-CoV) spike S1 (3 nM, 2 × 1011 in 100 μl)] (Fig. 4C); PBS anti-S1 solution (60 fM; 3.6 × 106 in 100 μl), with the S1 protein serving as capturing antibody [the reference fluid is PBS added with IgG (6 nM; 4 × 1011 in 100 μl)] (Fig. 4D); and PBS solution of IgG (60 fM; 3.6 × 106 in 100 μl), with anti-IgG serving as capturing antibody [the reference fluid is PBS, and the negative sample is a PBS solution of IgM (6 nM; 4 × 1011 in 100 μl)] (Fig. 4E).
Very relevantly, a clear response is seen in Fig. 4A, where the detection of the whole SARS-CoV-2 virion directly in saliva is shown as already pointed out in Fig. 3. Interesting is the assay in Fig. 4B where a concentration of the SARS-CoV-2 capsid protein S1 of 160 × 10−21 M (zM) in 100 μl is assayed directly in untreated human saliva; this sample encompasses as low as 10 ± 3 spike S1 proteins. The specificity of the assay is also extremely high as, for instance, in the S1/anti-S1 assay, is carried out in whole human blood (Fig. 4C). Here, the analyte is detected at a low concentration of 30 × 10−18 M, while the response to MERS-CoV-2 spike S1 at a much higher concentration of 3 × 10−9 M (control experiment) results in a clearly negative output. Relevantly, despite the eight orders of magnitude higher concentration of the MERS-CoV-2 spike S1 solution as compared to the SARS-CoV-2 spike S1 one, MERS-CoV-2 spike S1 does not nonspecifically bind with the anti–SARS-CoV-2 spike S1 (anti-S1) layer. This explains why a stable sensing signal is measured when directly assaying with the BioScreen platform untreated patients’ saliva or human whole blood. On the other hand, the very high specificity of the assay is fully confirmed by the trace curves in Fig. 4 (D and E), where the assay of anti-S1 captured by a layer of S1 and the IgG captured by anti-IgG are shown, respectively.
More generally, the data of the middle panels of Fig. 4 show how the blue traces can be different from the red traces when the sensing signal is analyzed, namely, when the traces measured between the 31st cycle and the 60th cycle are compared. Despite the sample-to-sample differences, no such differences can be qualitatively appreciated when comparing the red and blue traces acquired after incubation of the gate in the reference fluid (first 30 cycles). A more quantitative comparison between those features that are connected with the current dynamic rate of the biofunctionalized gates will be on the basis of the AI analysis of the whole set of data provided in the following.
While at this point we have shown that the BioScreen already reaches an LOI of 10 ± 3 viruses in 0.1 ml along with outstanding selectivity, diagnostic reliability still needs to be assessed. An elementary approach to sort the whole set of 240 assays into binary zero (negative)/one (positive) responses would be to use the R sensing response (Eq. 1) as an AI feature (39). This too simplistic approach leads, however, to a very erroneous and unreliable output affected by 2% of false-positive and 11% of false-negative random errors. This leads to a diagnostic sensitivity, defined as true positives/(true positives + false negatives), of 89.9%, which is comparable to those shown by the already available POC immunometric systems (see table S3 in section S9). Hence, choosing feature R as the only sorting parameter leads to a misclassification of the data. An example of this misclassification is given in fig. S8 of section S8, for the negative sample R = 0.042, while the positive one is lower being R = 0.022. The dispersion of the whole 240 dataset is given in fig. S9 of section S8, which evidences the highly populated confusion region where positive and negative outputs overlap.
To improve the binary sorting reliability, the use of an AI-based classifier can be the right choice (40, 41). To this end, a BioScreen binary classifier was designed, which is able to directly read raw data like those in Figs. 3 and 4 and produces an output attributing 0 to the negative samples (N) and 1 to the positive ones (P). Before entering into the details of the ad hoc designed classifier, by virtue of an example, the AI outputs relevant to the data of Fig. 4 are shown in the figure panels on the right. As it is apparent, a clear and very pronounced differentiation of the negative samples from the positive ones is always achieved.
The first step to develop the BioScreen AI-based binary classifier was to consider a further feature that, along with R, allows for better profiling of the measured signals. The more uncorrelated the two features, the better the classifier. Besides the R response, another very characteristic aspect in bioelectronics is the current dynamic upon cycling, which deeply characterizes the sensing output as can be seen in fig. S8, Fig. 3 (A to C), and Fig. 4 (middle panels). The dynamic processes, being correlated to the formation of the charge double layer at the electrode/electrolyte interface when the gate potential is applied, are highly specific to a given gate. In general, it can depend on elements such as (i) the chemical and mechanical nature of the layer of the capturing antibody, (ii) the biochemical process involved in the assay, and (iii) the fluid in which the assay is carried out. The sensing gate, after being incubated in a high ionic strength solution (the sample to be assayed or in the reference fluid), is then washed and cycled in a low ionic strength DI water electrolyte. The current dynamics is hence typical of the characteristic features of each biofunctionalized electrode and can be affected by the processes by which the gate adjusts to the new environment characterized by a much lower ionic strength. Hence, the cycling of the gate in the reference fluid, leading to stabilization within 30 cycles or not, allows unveiling the rate of the stabilization process, which is a key characteristic feature of a given gate. For instance, the current dynamics in the reference fluid (cycles 1 to 30) in fig. S8A is quite slow being far from the steady state even at the last 30th cycle. Conversely, fig. S8B shows a case where the dynamic is much faster, as the curve measured in the reference fluid is closer to its asymptote at the end of the cycling. Featuring this characteristic aspect of a given gate is hence very much useful. A simple feature, named RD, representing the dynamic behavior of a given gate, can be given as Eq. 2
| (2) |
where the current change, ∆ID,0, is computed between one of the first cycles (e.g., the 5th cycle to minimize the impact of the adjustment to the low ionic strength environment) and the last one (30th) measured in the reference fluid (see Fig. 3C). As anticipated, the first ID points are affected by the change of the ionic strength of the liquid, while after few initial cycles the sensor characteristic dynamic shows a more consistent trend. For a given , a slower (faster) dynamic results in a smaller (larger) RD. Sorting all the 240 data against the RD feature leads to an error of 2% false positives and 4% false negatives. While improved, these are still high. However, they are already good enough to correctly classify the data in fig. S8, as with RD a comparably larger output for the positive (0.875) sample is measured compared to the negative one (0.343). Moreover, RD increases consistently compared to R.
A second step of designing an AI-based reliable binary classifier is to combine the information encoded in the R and RD features by plotting the distribution of the positive and negative samples in the 2D feature space (fig. S10A, negative cases in blue and positive cases in red). A certain degree of correlation between the two features exists because R and RD both rely on the current change compared to the baseline. However, the normalization factor is completely different, and this leads to a better discrimination of the positive and negative samples in the central region of the feature space where the confusion region can fall. As it will be better clarified later in the text, the very good quality of the dataset measured by the BioScreen sensing technology is evidenced by the fact that positive and negative clusters form concave enclosures with a very low overlap as shown in fig. S10B. This occurrence enabled us to choose a classifier architecture involving just one output node comprising perceptrons that do not need to be separately trained for the recognition of positive and negative samples, without compromising its generality and the quality of its recognition ability.
The third step in the design of the BioScreen binary classifier can be accomplished with a multilayer perceptron network (MLPn) (42) architecture based on perceptron (neuron) nodes belonging to layers connected to previous/following ones through a defined pattern. Each perceptron linearly combines the input values and gives its output after filtering through a sigmoid function (e.g., the logistic function for binary classification), which compresses the output values into the 0 to 1 range. The MLPn used in this study, involving three perceptrons in the first layer, is shown in fig. S11.
The network is fed by the R and RD features’ values extracted from each of the 240 raw data. Each perceptron node in the first layer (1, 2, 3) produces a linear combination of the chosen features, whose general equation is v = Rw + RDwD + b, where w and wD are the weights of the features R and RD, respectively, and b is the bias. The geometric meaning of the first layer is in the definition of the three dotted black lines given in fig. S10B, each of which divides the 2D feature space into two half-planes. The resulting v values are then passed to a sigmoidal function. Here, they are bounded between 0 and 1 values depending on which half-plane a given sample belongs to. These are the binary values assigned as output to each perceptron. The segments of the dotted black lines overlapping with the negative or positive clusters (solid black segments) separate the assigned positive from the negative ones. The output of nodes 1, 2, and 3 feed nodes A and B, forming the subsequent layer (hidden layer) (39) that combines the half-planes into convex regions (A and B of fig. S10C), namely, regions where every two points can be connected with a segment that is totally within the region itself. Lines 1 and 2 concur to form the convex region A (at the right of line 1 and above line 2) including part of the positive samples. Lines 2 and 3 form another convex region B (above line 2 and at the right of line 3), which partially overlaps region A but includes the remaining positive samples. The third layer combines these regions by merging or intersecting them and creates more sophisticated structures. The actual merging of regions A and B creates a concave area including all positive (P) samples, except for 1 positive sample (false negative) and 1 negative sample (false positive). All negative samples (N) outside the merged A and B regions are correctly classified as negatives. The information of how the half-planes combine to define convex clusters is included in the weights of nodes A and B. Last, the output node in the classification layer (CL) gives the final answer by combining the convex clusters into the concave P and N regions.
The quality, wide applicability, and robustness of the classifier here presented rely on the topology achieved by careful data analysis along with the use of a validated dataset of 240 samples including many different biochemical assays carried out in various biofluids, namely, SARS-CoV-2 virus as well as the SARS-CoV-2 spike S1 (S1) and IgG proteins, directly in patients’ saliva as well as in blood serum and swab. The validated dataset is built by discarding those bioelectronic cartridges (fluidics) that showed instabilities in the trace of the reference electrode and/or a noisy dynamic in the assay of the reference biofluid with the sensing gate. Relevantly, the decision of discarding a gate can be taken already after just 10.5 min from the beginning of the assay (Fig. 2). The AI output can also give information on the level of reliability of a given output. If a nonreliable output (inconclusive) is obtained for an assay, that sample can be assayed again. To this end, a figure of merit that automatically assesses the quality of each output can be provided as described later in the text. This largely contributes to gather a very high quality dataset with the BioScreen platform.
It is worth noting that the high quality and the nonoverfitting nature of the binary classifier here designed rely on the following facts: (i) The number of parameters used in this study is 18, which is very low, resulting from three independent parameters (w, wD, and b) for each of the three nodes in the first layer (1, 2, and 3) and the three parameters for nodes A, B, and CL, respectively, and (ii) a set of 75% samples randomly chosen out of the 240 available is used for training the network, while the remaining 25% is used for the testing. This makes the numerosity of the training set (180 samples) 10 times larger than the number of parameters that by itself rule out the risk of overfitting. Moreover, repeating the random sample selection many times leads to final parameters that are very similar because the positive and negative clusters almost do not overlap. This is due to the very high quality sensing technology on which BioScreen relies.
As the assessment of the inconclusive outputs is concerned, a figure of merit that automatically assesses the quality of each output is also provided. The output of the BioScreen classifier is 0 for clearly negative events, 1 for positive events, and something in between around the clusters separating border where decision making is inconclusive. This transition from 0 to 1 can be defined by means of an output quality (OQ) function defined as OQN = (0.5 – N)/0.5 for negatives identified by N < 0.5, and OQP = (P – 0.5)/0.5 for positives identified by P > 0.5. OQ is 1 when N is close to 0 or when P is close to 1. When OQ is close to 0, the system output is around 0.5, meaning that a univocal class identification is not possible. In these inconclusive cases, the OQ value gives a warning so that the assay can be repeated.
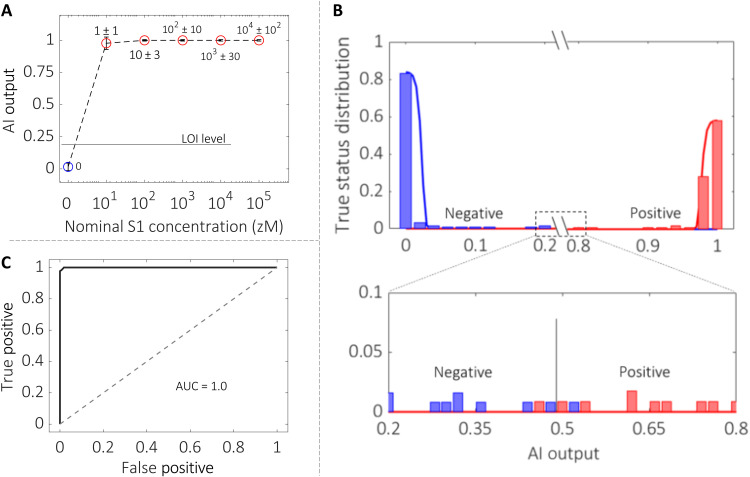
The classifier was tested at first on the saliva samples from healthy donors to which the SARS-CoV-2 capsid S1 protein was added, spanning the 16 zM to 160 aM concentration range corresponding to 1 ± 1 to 104 ± 102 proteins in 100 μl. Figure 5A shows the AI-based BioScreen classifier output plotted versus the S1 protein concentration. An LOI level of 0.22 is estimated from the negative data (section S7); hence, it is well below the AI signal output at 16 zM, namely, 1 ± 1 protein in 100 μl. This shows that a single protein can be detected at the LOI, meaning that this output holds a statistical confidence level such that false positives and false negatives are lower than 1% (3). Figure 5B plots the sample class distributions against the AI classifier output for the complete samples’ dataset of 240 data. The blue bars refer to the negative cases, and the red bars indicate the positive ones. Continuous lines show the Gaussian distribution of the negative and positive samples. The two distributions have mean values of 0 for negatives and 1 for positives, respectively. As it is clear, the two sets of output data are very well separated and the magnification of the central region where confusions can occur shows that we have six samples whose assigned value is very close to 0.5. Among those, one red point falls in the blue field and one blue point falls in the red field. Those are a single false negative over 121 negative outputs populating the blue field, and a false positive over 119 positive outputs populating the red field. The final error rate of the BioScreen classifier is, hence, 0.83%, corresponding to one negative misclassification and one positive misclassification. This is perfectly in line with the level of confidence expected based on the LOI level chosen. If the OQ function is applied to the six points in 0.5 AI output region (Fig. 5B, bottom panel), an OQ factor very close to 0 comes out (fig. S12), giving a clear indication that replicates of these analyses would have been beneficial for achieving an even better sorting.
Fig. 5. The AI binary classifier.
(A) The BioScreen AI classifier zero/one output for the assay of the SARS-CoV-2 virus capsid S1 protein added to the saliva of healthy volunteers is shown as a function of the S1 concentration ranging from 16 zM to 160 aM; this corresponds to 100 μl in a number of capsid proteins ranging from 1 ± 1 to 104 ± 102. The reference fluid is the saliva diluent (0.5 M Pipes, pH 6.8). Overall, 96 samples were assayed. The error bars are evaluated as asymmetric one SD over the 36 negative and 60 positive assayed samples. The LOI level, estimated as the average value of the negative control experiments plus six times the SD (section S7), is 0.22. This is marked as a horizontal dotted black line in the graph. (B) Histograms of the true sample status (positive or negative) versus the BioScreen AI-based binary classifier output for all of the 240 samples assayed. Blue bars stand for negative outputs, while red ones address positive answers. Continuous lines are Gaussian distributions, with a mean of 0 for negative data and 1 for the positive data. (C) Receiver operating characteristic (ROC) curve, featuring the true positives (239) versus the false positives (1), of the BioScreen AI binary classifier for all of the 240 samples. The area under the curve (AUC) is nearly one given the almost ideal ROC curve.
The output of the classifier for all the 240 samples is 238 samples that are correctly classified, 1 false negative, and 1 false positive. This leads to a false-positive and false-negative error of 0.42% and an overall error of 0.83%. Therefore, diagnostic sensitivity, given by true positives/(true positives + false negatives), diagnostic specificity, given by true negatives/(true negatives + false positives), and diagnostic accuracy, given by (true positives + true negatives)/(true negatives + false positives + false negatives + true positives), equal to 99.2% are obtained. These are excellent figures of merit that confirm the high quality and robustness of the classifier proposed. As already mentioned, the small number of parameters as compared to the training dataset (18 versus 180) also leads to good generalization ability (no overfitting) of the network.
Figure 5C shows the receiver operating characteristic (ROC) curve. The ROC curve is a standard quality–related curve of binary classifiers. It shows the relationship between true positives and false positives. When the classifier just “flips a coin” giving an output of random result, the dashed line is seen. In this case, accepting false-positive results of 0.1 results in a true-positive answer of 0.1. When a classifier is working perfectly, on the other hand, 0 false positives along with 1 (or 100%) true positives are observed (42). Our classifier is almost perfect on the studied dataset, so the ROC curve has the expected ideal shape. ROC comprises an ideal vertical segment at 0 false positive, varying the true-positive values from 0 to 1, and a horizontal segment at 1 true positive, varying the false-positive values from 0 to 1. The system answer on the entire dataset has only two errors, so there is only a slight deviation from the ideal behavior in the top-left region of the ROC curve. This extremely high reliability over 240 assays is mirrored in the area under the curve (AUC) being almost one.
On the basis of the data shown, we can state that the ultraportable, low-cost BioScreen POC platform with a disposable cartridge can reliably assess in 21 min, if a 100-μl sample of a self-collected biofluid such as saliva contains or not at least one virus or protein marker. This enables the screening of a population against a given disease, quickly identifying the subset of the diseased individuals with an error (false positives or false negatives) below 1%. Such a platform outperforms all the available POC systems (2, 4, 7–13, 16, 26, 27, 43) (section S9) and fully complies with the latest WHO recommendations for POC devices (6). REASSURED is the acronym adopted by WHO to characterize a POC device ideally encompassing real-time connection, ease specimen collection, be affordable, sensitive, specific, user-friendly, rapid, robust, equipment-free, and allow for a straightforward delivery to end-users. It is a fact that, to date, no single-molecule sensing platform combines all the features encoded in the REASSURED acronym. BioScreen platform outperforms state-of-the-art technologies, and it uniquely fully complies with all the REASSURED requirements for a POC system. In the following, a better-detailed explanation of this occurrence is given: Real-time connection: The BioScreen platform holds a reliable real-time connection via an ad hoc designed app that connects directly with the smart device. Data could also be transferred to the cloud for further processing on a standard network connection taking a few seconds (Fig. 1F). Ease specimen collection: Assays can be performed on self-sampled biofluids such as saliva. We have proven that the BioScreen platform can very well detect one virion of SARS-CoV-2 in saliva (Fig. 5A). Sensitive: The SARS-CoV-2 virus capsid protein is detected at an LOI of 1 ± 1 entity in 0.1 ml (10 to 20 zM). Hence, the BioScreen immunoassay is performed at the lowest possible limit. If the fluid contains at least one target molecule, the binary response is positive; if there are none, the response is negative. As an example, for the whole virus in patients’ saliva samples, the AI-based binary BioScreen classifier output is comparable to that of the rRT-PCR qualitative test (Fig. 5A). Selective, versatile, and reliable: The assay can highly selectively detect the SARS-CoV-2 virus and its capsid protein, antigen, and IgG in the nasal swab, in human whole blood, and in saliva. The analytical selectivity is proven against negative control assays. The number of different types of assays also proves the versatility of the BioScreen platform (Table 1). The extremely high reliability is proven by an incidence of false-positive and false-negative random errors below 1% along with diagnostic sensitivity, diagnostic specificity, and diagnostic accuracy of 99.2%. Rapid: The overall time to response to carry out the procedure described above is ca. 21 min. This is the effective time from the cartridge connection to the electronics to the delivery of the results. The electrodes in the disposable cartridge are stable over at least 2 weeks and are foreseen to be fabricated with the cartridge, meaning that no biofunctionalization or coating process is needed to be performed right before the assay. Therefore, in contrast to the state-of-the-art single-molecule sensors (table S1), the biofunctionalization process time (taking at least 2 hours) does not add to the time to results. User-friendly: The system operation is straightforward. The disposable cartridge is connected to the electronic reader, the app is opened (after having been installed) on a smart device (phone or tablet), and the user can start the analysis. At first, a set of reference measurements is automatically run by the system with the reference fluid comprising no analyte. From this dataset, the baseline current level is extracted. Then, the disposable cartridge is filled with 0.1 ml of saliva from a patient (or with the sample to be analyzed), and after 5 min of incubation and washing, a new set of measurements is run. These data provide the sensing signal level for a given sample. Then, the data are encrypted, transferred to the app, and analyzed by the AI algorithm, providing the response (Fig. 2 and explanatory video in section S1). Affordable and deliverable to end-users: The BioScreen platform developed up to this stage at the technology readiness level 5 (component and/or breadboard validated in simulated or real-place environment) is based on standard silicon-based driving/readout/communication electronics comprising a disposable three-dimensional (3D)–printed cartridge. Both components are built with assessed technologies that can be produced in large scale and at low cost. The prototype system is 9 cm large, 11 cm wide, and 2.5 cm thick, so it is handheld (Fig. 1) and ultraportable. The operation steps can all be prospectively performed by an untrained operator, and the proposed platform is ideal to be used right away by the end-user directly at home sampling his/her own saliva.
Table 1. Detailed list of all the samples assayed in the present work.
UTM, universal transport media for viruses.
|
Analyte:
Protein/virus |
Physisorbed
capturing antibody |
Biofluid/reference
fluid (diluent) |
Type of sample |
Number of assayed
samples |
Patients |
| S1 added | Anti-S1 | Saliva/Pipes* diluent | Positive | 60 | |
| S1 added | Anti-S1 | Saliva/Pipes diluent | Negative | 36 | 1 |
| Virus real sample | Anti-S1 | Saliva/Pipes diluent | Positive | 17 | 5 |
| Virus real sample | Anti-S1 | Saliva/Pipes diluent | Negative | 36 | 5 |
| Virus real sample | Anti-S1 | Swab/UTM diluent | Positive | 9 | 4 |
| Virus real sample | Anti-S1 | Swab/UTM diluent | Negative | 3 | 1 |
| S1 added | Anti-S1 | Whole blood | Positive | 8 | |
| S1 added | Anti-S1 | Whole blood | Negative | 10 | 1 |
| S1 added | Anti-S1 | PBS† diluent | Positive | 8 | |
| S1 added | Anti-S1 | PBS diluent | Negative | 14 | |
| Anti-S1 added | S1 | Whole blood | Positive | 9 | |
| Anti-S1 added | S1 | Whole blood | Negative | 10 | 1 |
| Anti-S1 added | S1 | PBS diluent | Positive | 4 | |
| Anti-S1 added | S1 | PBS diluent | Negative | 8 | |
| IgG added | Anti-IgG | PBS diluent | Positive | 4 | |
| IgG added | Anti-IgG | PBS diluent | Negative | 4 | |
| Total: 240 | Total: 18 (9 positive + 9 negative) |
*Pipes: 0.5 M, pH 6.8.
†PBS: 162 mM, pH 7.4.
In conclusion, the BioScreen handheld, easy-to-operate digital platform, returning a binary response with a threshold at a single molecule in 21 min, is shown to outperform available technologies fully complying with all the WHO’s requirements for POC testing. BioScreen, operating directly in a relevant environment, can assess whether a 0.1-ml self-collected sample of saliva contains or not at least a single virus or protein marker, enabling the quick identification of the subset of diseased asymptomatic patients, with false-positive and false-negative errors below 1%. This was possible by combining an extremely sensitive and selective bioelectronic device with an AI-based binary classifier that encodes features characteristic of a bioelectronic sensing output. The BioScreen general applicability, low cost, and convenient digital data handing provide breakthroughs in early and widespread diagnostic with foreseen applications in public health managing, precision medicine, and advanced biomedical research.
MATERIALS AND METHODS
Materials
SARS-CoV-2 (2019-nCov) spike S1 recombinant monoclonal antibody (anti-S1) expressed from human embryonic kidney (HEK) 293 cells, SARS-CoV-2 (2019-nCov) spike S1 His Recombinant Protein, MW 76.5 kDa, and MERS-CoV spike S1 protein S1 Subunit (AFS88936.1) (Met1-Glu725) fused with a polyhistidine tag at the C terminus, MW 79.9 kDa, were purchased from Sino Biological and used with no further purification. The anti-human IgG (anti-IgG) is an Fc-specific antibody affinity produced in a goat (molecular weight, ~144 kDa), while the human IgG (~150 kDa) affinity ligand and the human IgM (~950 kDa) ligand were extracted from human serum. All the Igs used are polyclonal antibodies and were purchased from Sigma-Aldrich and readily used. PBS (Sigma-Aldrich) solution presents osmolality and ion concentrations matching those of the human body (isotonic). One tablet of PBS was dissolved in 0.2 ml of water [high-performance liquid chromatography (HPLC) grade], resulting in 0.01 M phosphate buffer, 0.0027 M potassium chloride, and 0.137 M sodium chloride (pH 7.4) at 25°C. Water (HPLC-grade) was purchased from Sigma-Aldrich and used with no further purification. Piperazine-N,N′-bis(2-ethanesulfonic acid), piperazine-1,4-bis(2-ethanesulfonic acid), and 1,4-piperazinediethanesulfonic acid (Pipes) (0.5 M) buffer solution (pH 6.8) was purchased from Thermo Fisher Scientific. Human blood produced from a healthy donor aged between 18 and 60, screened and tested negative for HIV and hepatitis B and C, was purchased from Research Donors Ltd. (London, UK).
Disposable cartridge fabrication
The bioelectronic cartridges comprising a fluidic device (Fig. 1B) filled with the gating electrolyte (DI water) and with the sample to be assayed or with the reference fluid were fabricated by stereolithographic 3D printing (FormLab 2, clear resin). The gate array, encompassing four gold electrodes (area = 0.1 cm2 each), is embedded in the fluidic device. These are fabricated by sputtering and patterned through a shadow mask. The sensing gate is fabricated on a PEN (polyethylene naphthalate) foil by shadow mask lithography and e-beam evaporation of Ti/Au (5 nm/50 nm) films, with the circular area being about 0.2 cm2. The sensing gate is biofunctionalized with a layer of capturing antibodies, while the other four electrodes (Fig. 1C) are coated with an antifouling layer. This is proven to prevent spurious adsorption of the solutions to be assayed, even when a whole real biofluid such as saliva, is involved (section S4). The reference gates enable to assess whether spurious surface potential shifts occur during the sensing measurements. The other two electrodes enable the polarization of the electrolyte solution, in which all electrodes are immersed. The polarizing and the reference electrodes comprise two short-circuited elements (Fig. 1E) to assure a uniform biasing of the electrolyte solution. During the reading, DI water serves as the electrolyte to maximize the Debye length on the sensing gate and hence to leave the electrostatic effects associated with the biochemical specific binding unscreened (35).
Biofunctionalization
The biofunctionalization protocol involves the physisorption of the capturing proteins directly on the gold sensing gate. This was cleaned and put in contact with 70 μl of the PBS (ionic strength is 163 mM and pH 7.4) of the capturing proteins (50 μg/ml). SPR characterization (section S2) assesses that a layer (ca. 10 nm thick) with trillions of proteins is attached to the electrode. This is relevant as a highly densely packed layer of recognition elements has been used in single-molecule detection at a millimeter-wide electronic interface (26, 27). Moreover, the SPR analysis proves the extremely high stability of these gates: After 45 hours of storage in PBS solution (ionic strength, is = 163 mM and pH 7.4) and after several washing steps, the gate is still uniformly covered by a layer comprising (1.13 ± 0 0.04) × 1012 antibodies/cm2. The physisorbed anti-S1 (anti–SARS-CoV-2 spike S1) layer can capture the S1 proteins, even after overnight storage in DI water (ionic strength, is = 5 μM) or after storage for 2 weeks in PBS (section S3).
Handheld low-cost reader electronic circuit
The self-collected saliva is placed in the disposable bioelectronic cartridge that is connected to the ultraportable (9 cm × 11 cm × 2.5 cm) electronic reader that acquires, digitalizes, and transmits the data to a smart device. The app installed on the smart device enables the data handling, while the analysis is performed with the AI-based BioScreen binary classifier. The electronic circuit board (Fig. 1, F and G) is based on a mixed-signal approach integrating analog and digital functions as well as the power management unit for battery operation and voltage regulation. The biosensor array is fully managed by the microcontroller, which generates the voltage waveforms applied to the gate electrode and reads the output currents of the transistors connected to the sensing and reference gates. A front-end analog interface has been designed for proper signal conditioning, an integrated circuit with 24-bit sigma-delta analog-to-digital converters is used for data acquisition, and a 16-bit digital-to-analog converter is embedded into the microcontroller. This reduces the power consumption and cost, providing a portable, battery-powered, and small-footprint system. A Bluetooth module is used for transferring the measurements to a smart device, such as a phone or tablet, which displays the curves. All the FETs used are p-type accumulation-mode transistors; hence, the gate voltage is swept toward more negative potentials.
Sensing protocol
The fluidic device is filled with the reference or diluent fluid, namely, Pipes for saliva, Universal transport medium (UTM) for swab, or PBS (pH 7.4 and is = 162 mM); here, the sensing gate, covered with a physisorbed layer of capturing antibodies, is incubated for 5 min. The reference fluid is discarded, and the fluidic device is washed and filled in with DI water (is = 5 μM). In DI water, a set of 30 transfer characteristics (1st to 30th), ID (drain current) versus VG (gate voltage) at VD (drain voltage) = −0.1 V, is measured contemporarily with the sensing EGT and the reference EGT. This step, addressed as cycling, takes 5 min. The transient ID current traces comprise the current values of the transfer characteristics at VG = −0.5 V. The reference fluid is discarded and the fluidic device is filled with the sample to be assayed, the electrode is incubated for 5 min, the sample is discarded, and the fluidic device is washed and filled with DI water. A second cycling step comprising the measurement of other 30 transfer characteristics (31st to 60th) is performed. The gate voltage range spans between 0.2 and −0.5 V, as this ensures that Faradaic leakage currents, connected with redox reactions detrimental for the electrode stability, are negligible (section S5). This is mirrored also by the occurrence that the transfer characteristics, always measured in the forward and backward mode, show a negligible hysteresis as proven by the curves in Fig. 3A and fig. S7. As it can be seen, while the curves are all measured in the forward and reverse mode, no hysteresis is observed on the ID curves and a very negligible hysteresis is seen in the low IG leakage current. This rules out that any detrimental faradaic current is flowing through the gate and the polarizing counter electrode (Fig. 1G), as expected in a stably operated bioelectronic transistor (44). A very characteristic aspect in bioelectronics is the current dynamics that deeply affect the sensing output as can be seen in fig. S8 and Figs. 3 and 4.
The viral concentration in real samples has been estimated from the S1 antigen level quantitatively assessed with the LUMIPULSE SARS-CoV-2 antigen test (Fujirebio Inc., Tokyo, Japan). A viral concentration of 1 virus/μl (100 ± 10 viruses per 100 μl) has been estimated with an S1 antigen concentration of 0.15 pg/ml (2 × 10−15 M, 2 fM, 1.2 × 105 antigens in 100 μl). Saliva samples were diluted 1:2 in piperazine-N,N′-bis(2-ethanesulfonic acid), piperazine-1,4-bis(2-ethanesulfonic acid), and Pipes diluent, vortexed, transferred into a sterile tube, and centrifuged at 1000g for 15 min. Aliquots (100 μl) of the supernatant were used for testing on the LUMIPULSE G1200 system and with the BioScreen platform. In particular, the saliva samples at different virion concentrations ranging from 1.66 to 169.39 virus/μl have been prepared for the BioScreen assay by standard serial dilutions in Pipes saliva diluent from a positive saliva sample with an antigen level of 25 pg/ml (327 fM; 2 × 107 in 100 μl; section S6). The sensing measurements on the real samples were carried out, bringing the BioScreen platform to the hygiene and preventive medicine department—COVID-19 epidemiological surveillance laboratory of the University of Bari in compliance with all the needed safety rules.
Assays performed
The BioScreen platform was used to assay 240 samples, 119 positives and 121 negatives. The large samples’ ensemble includes a pilot clinical study on nine patients positive to COVID-19 and nine negatives showing the feasibility of performing the BioScreen assays directly in saliva in a clinical environment, namely, the hygiene and preventive medicine department—COVID-19 epidemiological surveillance laboratory at the University of Bari. The other samples tested were real human fluids (whole blood and saliva) from healthy volunteers or PBS solutions spiked with the targeted markers. The measurement (cycling) of the sensing gate in the reference fluid with the associated ID current dynamic curve was important to assess the reliability of the sensing gate. The high-quality dataset of 240 samples analyzed was validated by the careful screening of the reference electrode trace stability and the ID current dynamics evaluated in the reference fluid and eventually also in the sample. When either one of these traces showed an overall unstable behavior, the electrode and the related dataset were discarded. A total of five sensing gates were discarded, and the corresponding current values were not included in the dataset. For some of the real sample assayed, a dilution up to 1:8 with the reference fluid was needed. In Table 1, a detailed list with all the samples assayed is given.
The AI-based BioScreen binary classifier
R and RD features given as Eqs. 1 and 2, respectively, are taken as the AI features. An MLPn architecture based on six perceptron nodes (42) is designed for the binary zero/one classification. Each perceptron node associates a linear equation v = Rw + RD wD + b, where w and wD are weights of features R and RD, respectively, and b is the bias. Eighteen independent parameters need to be computed with a set of 180 samples randomly chosen out of the 240 available to train the network, while the remaining 60 are used for testing. This makes the numerosity of the training set 10 times the number of parameters adding generality to the approach. The quality of the BioScreen classifier output is assessed by the OQ function. OQ is 1 when N is close to 0 or when P is close to 1. OQ is close to 0 when the system output is around 0.5, meaning that a clear class identification is not possible. So, in this case, the OQ value gives a warning (after just 10.5 min) that the assay has to be repeated.
Acknowledgments
We thank A. Sallustio and C. Di Franco for support in performing the COVID-19 assays and for the metallization of the sensing gates, respectively.
Funding: The project BIOSCREEN (POR FESR 2014-2020, ID number 1831459, CUP E81B20000320007 funded by the European Commission, the Italian Government, and Regione Lombardia) and ERCStg 2021 “A binary sensor with single-molecule digit to discriminate biofluids enclosing zero or at least one biomarker” (NoOne) (Grant Agreement ID 101040383) are acknowledged for partial financial support.
Author contributions: F.T. and L.T. conceived the idea and designed the experiments. E.M. contributed to the design of the experiments and executed all the sensing experiments. SPR experiments were performed by L.S. The activities at the hygiene and preventive medicine department—COVID-19 epidemiological surveillance laboratory of the University of Bari were supervised by M.C., assisted by D.L. F.T. and L.T. conceived and designed the fluidic device. Z.M.K.-V. designed the AI tool and performed the data analysis. F.T. conceived and designed the system architecture. M.R. designed the specific circuit topologies and developed the app. F.T. and L.T. wrote the paper, and all authors provided feedback.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The datasets generated and analyzed during the current study are available in the ReCaS (https://recascloud.ba.infn.it/index.php/s/ECsOKmgD2DXdpn1) repository, University of Bari. Approval by the Ethics Committee was waived because the samples were anonymized. All procedures were performed in accordance with the Declaration of Helsinki, as revised in 2013, for research involving human subjects.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S12
Tables S1 to S3
Sections S1 to S9
References
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.World Health Organization, The Top 10 Causes of Death—Factsheet (World Health Organization, 2020), pp. 1–9. [Google Scholar]
- 2.Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z., Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 19, 171–183 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson M., Ellison S. L. R., Wood R., Harmonized guidelines for single-laboratory validation of methods of analysis (IUPAC Technical Report). Pure Appl. Chem. 74, 835–855 (2002). [Google Scholar]
- 4.Boehringer H. R., O’Farrell B. J., Lateral flow assays in infectious disease diagnosis. Clin. Chem. 68, 52–58 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnaout R., Lee R. A., Lee G. R., Callahan C., Cheng A., Yen C. F., Smith K. P., Arora R., Kirby J. E., The limit of detection matters: The case for benchmarking severe acute respiratory syndrome coronavirus 2 testing. Clin. Infect. Dis. 73, e3042–e3046 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Land K. J., Boeras D. I., Chen X. S., Ramsay A. R., Peeling R. W., REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 4, 46–54 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dao Thi V. L., Herbst K., Boerner K., Meurer M., Kremer L. P. M., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M. L., Boulant S., Klein S., Chlanda P., Khalid D., Miranda I. B., Schnitzler P., Kräusslich H. G., Knop M., Anders S., A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 12, eabc7075 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbot ID NOW, www.abbott.com/IDNOW.html?CID=OUS_OK.
- 9.Ning B., Yu T., Zhang S., Huang Z., Tian D., Lin Z., Niu A., Golden N., Hensley K., Threeton B., Lyon C. J., Yin X. M., Roy C. J., Saba N. S., Rappaport J., Wei Q., Hu T. Y., A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Sci. Adv. 7, eabe3703 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panpradist N., Kline E. C., Atkinson R. G., Roller M., Wang Q., Hull I. T., Kotnik J. H., Oreskovic A. K., Bennett C., Leon D., Lyon V., Gilligan-Steinberg S. D., Han P. D., Drain P. K., Starita L. M., Thompson M. J., Lutz B. R., Harmony COVID-19: A ready-to-use kit, low-cost detector, and smartphone app for point-of-care SARS-CoV-2 RNA detection. Sci. Adv. 7, eabj1281 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mertens P., De Vos N., Martiny D., Jassoy C., Mirazimi A., Cuypers L., Van den Wijngaert S., Monteil V., Melin P., Stoffels K., Yin N., Mileto D., Delaunoy S., Magein H., Lagrou K., Bouzet J., Serrano G., Wautier M., Leclipteux T., Van Ranst M., Vandenberg O., Development and potential usefulness of the COVID-19 Ag Respi-Strip diagnostic assay in a pandemic context. Front. Med. 7, 225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh N. K., Ray P., Carlin A. F., Magallanes C., Morgan S. C., Laurent L. C., Aronoff-Spencer E. S., Hall D. A., Hitting the diagnostic sweet spot: Point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. Biosens. Bioelectron. 180, 113111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broughton J. P., Deng X., Yu G., Fasching C. L., Servellita V., Singh J., Miao X., Streithorst J. A., Granados A., Sotomayor-gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C., Guevara H., Wadford D. A., Chen J. S., Chiu C. Y., CRISPR – Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 38, 870–874 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkula V., Berto M., Diacci C., Patrahau B., Di Lauro M., Kovtun A., Liscio A., Sensi M., Samorì P., Greco P., Bortolotti C. A., Biscarini F., Harnessing selectivity and sensitivity in electronic biosensing: A novel lab-on-chip multigate organic transistor. Anal. Chem. 92, 9330–9337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rani D., Singh Y., Salker M., Vu X. T., Ingebrandt S., Pachauri V., Point-of-care-ready nanoscale ISFET arrays for sub-picomolar detection of cytokines in cell cultures. Anal. Bioanal. Chem. 412, 6777–6788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seo G., Lee G., Kim M. J., Baek S. H., Choi M., Ku K. B., Lee C. S., Jun S., Park D., Kim H. G., Kim S. J., Lee J. O., Kim B. T., Park E. C., Il Kim S., Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 14, 5135–5142 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Liu Q., Zhao C., Chen M., Liu Y., Zhao Z., Wu F., Li Z., Weiss P. S., Andrews A. M., Zhou C., Flexible multiplexed In2O3 nanoribbon aptamer-field-effect transistors for biosensing. iScience 23, 101469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko J. W., Woo J. M., Jinhong A., Cheon J. H., Lim J. H., Kim S. H., Chun H., Kim E., Park Y. J., Multi-order dynamic range DNA sensor using a gold decorated SWCNT random network. ACS Nano 5, 4365–4372 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Liang Y., Xiao M., Wu D., Lin Y., Liu L., He J., Zhang G., Peng L. M., Zhang Z., Wafer-scale uniform carbon nanotube transistors for ultrasensitive and label-free detection of disease biomarkers. ACS Nano 14, 8866–8874 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Hajian R., Balderston S., Tran T., deBoer T., Etienne J., Sandhu M., Wauford N. A., Chung J. Y., Nokes J., Athaiya M., Paredes J., Peytavi R., Goldsmith B., Murthy N., Conboy I. M., Aran K., Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 3, 427–437 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacChia E., Torricelli F., Bollella P., Sarcina L., Tricase A., Di Franco C., Österbacka R., Kovács-Vajna Z. M., Scamarcio G., Torsi L., Large-area interfaces for single-molecule label-free bioelectronic detection. Chem. Rev. 122, 4636–4699 (2022). [DOI] [PubMed] [Google Scholar]
- 22.Macchia E., Romele P., Manoli K., Ghittorelli M., Magliulo M., Kovács-Vajna Z. M., Torricelli F., Torsi L., Ultra-sensitive protein detection with organic electrochemical transistors printed on plastic substrates. Flex. Print. Electron. 3, 034002 (2018). [Google Scholar]
- 23.Wang X., Wang Y., Wu Y., Xiao Y., A highly sensitive and versatile chiral sensor based on a top-gate organic field effect transistor functionalized with thiolated β-cyclodextrin. Analyst 144, 2611–2617 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Wu Y., Wang X., Li X., Xiao Y., Wang Y., Cyclodextrin derivatives functionalized highly sensitive chiral sensor based on organic field-effect transistor. Chin. Chem. Lett. 31, 99–102 (2019). [Google Scholar]
- 25.Sailapu S. K., Macchia E., Merino-Jimenez I., Esquivel J. P., Sarcina L., Scamarcio G., Minteer S. D., Torsi L., Sabaté N., Standalone operation of an EGOFET for ultra-sensitive detection of HIV. Biosens. Bioelectron. 156, 112103 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Guo K., Wustoni S., Koklu A., Díaz-Galicia E., Moser M., Hama A., Alqahtani A. A., Ahmad A. N., Alhamlan F. S., Shuaib M., Pain A., McCulloch I., Arold S. T., Grünberg R., Inal S., Rapid single-molecule detection of COVID-19 and MERS antigens via nanobody-functionalized organic electrochemical transistors. Nat. Biomed. Eng. 5, 666–677 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Macchia E., Manoli K., Holzer B., Di Franco C., Ghittorelli M., Torricelli F., Alberga D., Mangiatordi G. F., Palazzo G., Scamarcio G., Torsi L., Single-molecule detection with a millimetre-sized transistor. Nat. Commun. 9, 3223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macchia E., Manoli K., Di Franco C., Picca R. A., Österbacka R., Palazzo G., Torricelli F., Scamarcio G., Torsi L., Organic field-effect transistor platform for label-free, single-molecule detection of genomic biomarkers. ACS Sens. 5, 1822–1830 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Macchia E., Tiwari A., Manoli K., Holzer B., Ditaranto N., Picca R. A., Cioffi N., Di Franco C., Scamarcio G., Palazzo G., Torsi L., Label-free and selective single-molecule bioelectronic sensing with a millimeter-wide self-assembled monolayer of anti-immunoglobulins. Chem. Mater. 31, 6476–6483 (2019). [Google Scholar]
- 30.Macchia E., Manoli K., Holzer B., Di Franco C., Picca R. A., Cioffi N., Scamarcio G., Palazzo G., Torsi L., Selective single-molecule analytical detection of C-reactive protein in saliva with an organic transistor. Anal. Bioanal. Chem. 411, 4899–4908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macchia E., Sarcina L., Picca R. A., Manoli K., Di Franco C., Scamarcio G., Torsi L., Ultra-low HIV-1 p24 detection limits with a bioelectronic sensor. Anal. Bioanal. Chem. 412, 811–818 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Picca R. A., Manoli K., Macchia E., Sarcina L., Di Franco C., Cioffi N., Blasi D., Österbacka R., Torricelli F., Scamarcio G., Torsi L., Ultimately sensitive organic bioelectronic transistor sensors by materials and device structure design. Adv. Funct. Mater. 30, 1904513 (2020). [Google Scholar]
- 33.Macchia E., Sarcina L., Driescher C., Gounani Z., Tewari A., Osterbacka R., Palazzo G., Tricase A., Kovács-Vajna Z. M., Viola F., Modena F., Caironi M., Torricelli F., Esposito I., Torsi L., Single-molecule bioelectronic label-free assay of both protein and genomic markers of pancreatic mucinous cysts’ in whole blood serum. Adv. Electron. Mater. 7, 2100304 (2021). [Google Scholar]
- 34.Macchia E., Manoli K., Di Franco C., Scamarcio G., Torsi L., New trends in single-molecule bioanalytical detection. Anal. Bioanal. Chem. 412, 5005–5014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu C. H., Sarangadharan I., Regmi A., Chen Y. W., Hsu C. P., Chang W. H., Lee G. Y., Chyi J. I., Chen C. C., Shiesh S. C., Bin Lee G., Wang Y. L., Beyond the Debye length in high ionic strength solution: Direct protein detection with field-effect transistors (FETs) in human serum. Sci. Rep. 7, 5256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaisti M., Detection principles of biological and chemical FET sensors. Biosens. Bioelectron. 98, 437–448 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Kergoat L., Herlogsson L., Braga D., Piro B., Pham M. C., Crispin X., Berggren M., Horowitz G., A water-gate organic field-effect transistor. Adv. Mater. 22, 2565–2569 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Torricelli F., Adrahtas D. Z., Biscarini F., Bonfiglio A., Bortolotti C. A., Frisbie C. D., Mcculloch I., Macchia E., Malliaras G. G., Electrolyte- gated transistors for enhanced performance bioelectronics. Nat. Rev. Methods Primers 1, 66 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elemento O., Leslie C., Lundin J., Tourassi G., Artificial intelligence in cancer research, diagnosis and therapy. Nat. Rev. Cancer 21, 747–752 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Gupta A., Anjum, Gupta S., Katarya R., InstaCovNet-19: A deep learning classification model for the detection of COVID-19 patients using chest x-ray. Appl. Soft Comput. 99, 106859 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X., Faes L., Kale A. U., Wagner S. K., Fu D. J., Bruynseels A., Mahendiran T., Moraes G., Shamdas M., Kern C., Ledsam J. R., Schmid M. K., Balaskas K., Topol E. J., Bachmann L. M., Keane P. A., Denniston A. K., A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis. Lancet Digit. Health 1, e271–e297 (2019). [DOI] [PubMed] [Google Scholar]
- 42.R. M. Hristev, The ANN Book (Computational Systems Biology Group, 1998).
- 43.Fabiani L., Saroglia M., Galatà G., De Santis R., Fillo S., Luca V., Faggioni G., D’Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F., Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: A reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 171, 112686 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macchia E., Picca R. A., Manoli K., Di Franco C., Blasi D., Sarcina L., Ditaranto N., Cioffi N., Österbacka R., Scamarcio G., Torricelli F., Torsi L., About the amplification factors in organic bioelectronic sensors. Mater. Horiz. 7, 999–1013 (2020). [Google Scholar]
- 45.Kretschmann E., Raether H., Notizen: Radiative decay of non radiative surface plasmons excited by light. Z. Naturforsch. A 23, 2135–2136 (1968). [Google Scholar]
- 46.E. T. Gedig, in Handbook of Surface Plasmon Resonance, R. B. M. Schasfoort, Ed. (The Royal Society of Chemistry, ed. 2, 2017), pp. 173–220. [Google Scholar]
- 47.Homola J., Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 108, 462–493 (2008). [DOI] [PubMed] [Google Scholar]
- 48.De Feijter J. A., Benjamins J., Veer F. A., Ellipsometry as a tool to study the adsorption behavior of synthetic and biopolymers at the air–water interface. Biopolymers 17, 1759–1772 (1978). [Google Scholar]
- 49.Ball V., Ramsden J. J., Buffer dependence of refractive index increments of protein solutions. Biopolymers 46, 489–492 (1998). [Google Scholar]
- 50.Miyazaki C. M., Shimizu F. M., Ferreira M., Surface plasmon resonance (SPR) for sensors and biosensors. Nanocharacterization Tech. , 183–200 (2017). [Google Scholar]
- 51.Winspall 3.02 software, p. 3.
- 52.Refractive Index reference value; https://refractiveindex.info/.
- 53.Tan Y. H., Liu M., Nolting B., Go J. G., Gervay-Hague J., Liu G. Y., A nanoengineering approach for investigation and regulation of protein immobilization. ACS Nano 2, 2374–2384 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D. Male, J. Brostoff, D. B. Roth, I. Roitt, Immunology (Elsevier, ed. 8, 2013), vol. 4. [Google Scholar]
- 55.A. K. Abbas, A. H. Lichtman, S. Pillai, Cellular and Molecular Immunology (Saunders, ed. 5, 2003), pp. 189–214. [Google Scholar]
- 56.Hirotsu Y., Maejima M., Shibusawa M., Amemiya K., Nagakubo Y., Hosaka K., Sueki H., Hayakawa M., Mochizuki H., Tsutsui T., Kakizaki Y., Miyashita Y., Omata M., Prospective study of 1308 nasopharyngeal swabs from 1033 patients using the LUMIPULSE SARS-CoV-2 antigen test: Comparison with RT-qPCR. Int. J. Infect. Dis. 105, 7–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K., Sueki H., Hayakawa M., Mochizuki H., Tsutsui T., Kakizaki Y., Miyashita Y., Yagi S., Kojima S., Omata M., Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int. J. Infect. Dis. 99, 397–402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bahadır E. B., Sezgintürk M. K., Lateral flow assays: Principles, designs and labels. Trends Anal. Chem. 82, 286–306 (2016). [Google Scholar]
- 59.Mahmoudi T., de la Guardia M., Shirdel B., Mokhtarzadeh A., Baradaran B., Recent advancements in structural improvements of lateral flow assays towards point-of-care testing. Trends Anal. Chem. 116, 13–30 (2019). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S12
Tables S1 to S3
Sections S1 to S9
References
Movie S1