Abstract
Hyperphosphorylation of the neuronal tau protein is a hallmark of neurodegenerative tauopathies such as Alzheimer’s disease. A central unanswered question is why tau becomes progressively hyperphosphorylated. Here, we show that tau phosphorylation is governed by interdependence— a mechanistic link between initial site-specific and subsequent multi-site phosphorylation. Systematic assessment of site interdependence identified distinct residues (threonine-50, threonine-69, and threonine-181) as master sites that determine propagation of phosphorylation at multiple epitopes. CRISPR point mutation and expression of human tau in Alzheimer’s mice showed that site interdependence governs physiologic and amyloid-associated multi-site phosphorylation and cognitive deficits, respectively. Combined targeting of master sites and p38α, the most central tau kinase linked to interdependence, synergistically ablated hyperphosphorylation. In summary, our work delineates how complex tau phosphorylation arises to inform therapeutic and biomarker design for tauopathies.
Master sites drive global tau hyperphosphorylation in Alzheimer’s disease.
INTRODUCTION
Alzheimer’s disease (AD) is a fatal neurodegenerative disease, for which no cure exists (1, 2). AD is characterized by extracellular β-amyloid (Aβ) plaques and the intraneuronal accumulation of neurofibrillary tangles consisting of tau, a microtubule-associated neuronal protein that regulates the cytoskeleton of all neurons (3–5). Tau pathology, the progressive deposition of tau, correlates with cognitive decline in AD (3, 5). Thus, tau pathology is predictive of disease progression, supporting a central role of tau (6), which makes tau a prime target of AD diagnostic and therapeutic developments (4). Pathologic tau occurs in a hyperphosphorylated state, i.e., excessive modification with phospho-groups at multiple amino acid side chains simultaneously (7, 8).
Tau has a large potential for combinatorial phosphorylation (7, 9). Phosphorylation at serine (S) or threonine (T) residues followed by proline (P) is the most common form of phospho-epitope in tau, particularly within the proline-rich region (PRR) and C-terminal region (CTR) of tau (8), and includes 17 distinct SP or TP phosphorylation sites, many of which are prominently phosphorylated in aggregated tau in neurofibrillary tangles (5, 7). These sites are targeted by proline-directed SP/TP kinases, the main kinases involved in hyperphosphorylation and therefore pathologic tau (7, 10, 11). Tau phosphorylation is linked to aggregation into multimers and fibrils (12) and modulates cognitive deficits induced by pathogenic tau or by Aβ (13, 14). Despite these clear associations of phosphorylated tau with disease, phosphorylated tau also occurs in physiologic states (15), and site-specific tau phosphorylation mediates functions other than disease progression, including modulation of signaling complexes or lowering of microtubule binding affinity of tau (13, 16). It remains an unresolved question how site-specific and hyperphosphorylation of tau are mechanistically linked.
To fill this gap in knowledge, we screened tau phosphorylation with a focus on SP/TP sites, the most common type of tau phospho-epitope, for mechanistic relations between site-specific and multi-site phosphorylation. We identify central sites, which we term “master sites,” whose phosphorylation governs overall multi-site tau phosphorylation, and we validate these master sites in experiments in vitro and in mouse brain. Thus, tau phosphorylation is regulated by an intrinsic mechanism that modulates relations between phospho-sites and determines how tau arrives at a multi-site phosphorylated state. Overall, this study builds a hitherto unknown basis for the mechanistic connection between site-specific and hyperphosphorylation of tau with broad implications for tauopathies and tau-mediated neurologic disorders.
RESULTS
We propose that tau phosphorylation is mechanistically interdependent, i.e., initial phosphorylation at one site modulates the phosphorylation state at another site (Fig. 1A). We propose and define the term “interdependence” as the mechanistic relationship that modulates tau phosphorylation at a given site by prior phosphorylation at another site.
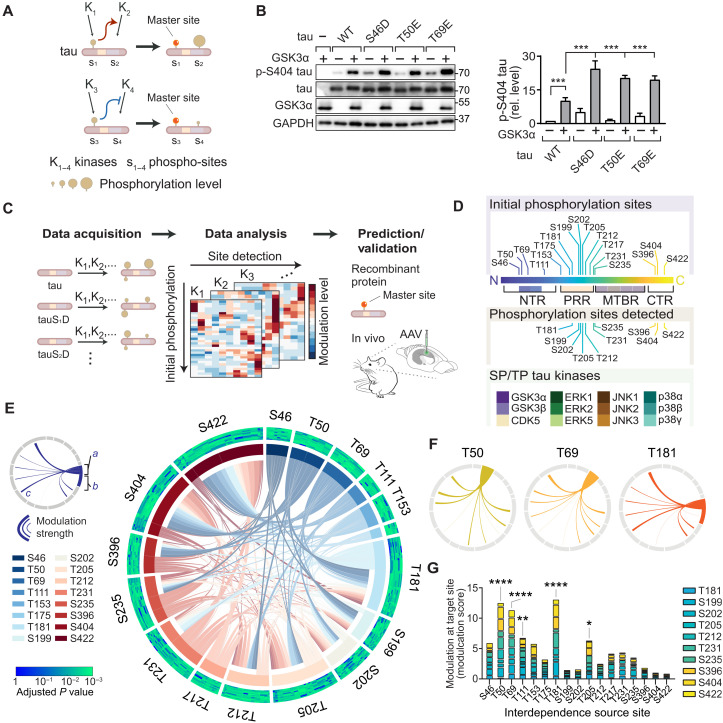
Fig. 1. Interdependence defines master sites integral to tau multi-site phosphorylation.
(A) Schematic of site interdependence. Phosphorylation at one site (e.g., S1) by kinase K1 disproportionately affects phospho-state at another site (e.g., S2). (B) Site interdependence enhances distal tau phosphorylation. Immunoblots from 293T cells expressing human wild-type tau (WT) or indicated variants ± glycogen synthase kinase 3α (GSK3α). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), loading control. n = 3. Note that positive interdependence promotes higher pS404 levels in tau variants. (C) Mapping tau SP/TP phosphorylation interdependence. Expression of tauWT or variants with phospho-mimetic conversions, representing initial phosphorylation state, in cells ± kinases (K1, K2,…). Phospho-status is quantified with epitope-specific antibodies and mapped onto multidimensional dataset. Data analysis identifies interdependence patterns and master sites, followed by validation. (D) Seventeen initial phosphorylation sites, 10 sites detected, and 12 SP/TP kinases in screen. (E) Integrated plot of tau SP/TP phosphorylation interdependence for 10 target and 17 source sites. Modulation a, at source site; b, at source site; c, toward target site. Adjusted P values for modulation are plotted for each site (Spearman’s correlation). Cumulative modulation is represented by sector width. (F) Interdependence for master sites. (G) Cumulative modulation scores (factor of correlation coefficient and relative phosphorylation level). Means ± SEM. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05 [analysis of variance (ANOVA)].
To systematically explore interdependence for SP/TP phosphorylation of tau, we set up a site interdependence screen based on a tractable experimental paradigm in cultured cells that combined initial phosphorylation, increased kinase activity, and assessment of resulting tau phosphorylation at specific epitopes. We used 293T cells expressing tau variants defining a site-specific initial phosphorylation state (based on phospho-mimicking side chains, i.e., serine → aspartate; threonine → glutamate). We raised activity of one tau kinase by concomitant expression of a SP/TP kinase (Fig. 1B and fig. S1). Controls without expression of kinase (i.e., tau alone) served to determine the effect that fixed phosphomimetic residue has on modifications at other SP/TP sites (Fig. 1B and fig. S1). Mimicking initial phosphorylation at distal SP/TP sites resulted in instances of significant modulation of site-specific tau phosphorylation, i.e., significantly different levels of detectable phospho-tau epitopes, which were quantified through densitometric analysis of immunoblots and normalized to total tau and loading control [glyceraldehyde-3-phosphate dehydrogenase (GAPDH)] (Fig. 1B and fig. S1). Integrating these experimental conditions facilitated data acquisition across three parameters: (i) pre-SP/TP site phosphorylation, (ii) increased SP/TP kinase activity, and (iii) SP/TP site phosphorylation status (Fig. 1C). Resulting multidimensional data built the framework for definition of tau phosphorylation interdependence, to be further validated in vivo and explored mechanistically (Fig. 1C and fig. S2). Our experimental setup based on SDS–polyacrylamide gel electrophoresis (PAGE) and immunoblot for detection was superior to dot blot detection of phosphorylated tau (fig. S3). Validated phospho-epitope–specific tau antibodies were used for detection (fig. S4 and table S1). Phospho-mimicking tau variants did not induce aggregation or unfolded stress response (see Materials and Methods and fig. S5). Analogous tau expression in cultured cells served previously to identify tau kinase and phosphorylation-dependent mechanisms relevant to neurons in vitro and in vivo (16, 17).
Expanding this approach across 17 SP/TP initial phosphorylation sites and 12 kinases produced a multidimensional dataset of steady-state levels of tau phosphorylation at 10 different phospho-sites in relation to different kinase activities and initial phosphorylation (Fig. 1, C and D, fig. S2, and table S1). Notably, multi-site phosphorylation exhibits distinct steady-state forms for tau (18) and other proteins (19). Global data analysis across all 12 SP/TP kinases demonstrated interdependence across individual sites and different SP/TP kinases (fig. S6), exhibiting predominantly positive modulation, likely due to enhanced kinase activities in our experimental approach or limited effects of the kinases studied on tau phosphatase activity (20–22). Clustering the data along initial phosphorylation and kinase activity showed that sites T181, T205, T212, S404, and S422 were hotspots within the PRR and CTR for modulation by tau phosphorylation interdependence (fig. S6). Thus, SP/TP initial phosphorylation differentially and distally modulates phosphorylation across tau. These results identify site interdependence as a key feature of tau phosphorylation in living cells and show that interdependence effects are quantifiable in direction, strength, and extent.
Comparing data from different SP/TP kinases revealed similarities, as well as unique patterns in site interdependence (fig. S7). Mitogen-activated protein (MAP) kinase p38α showed most extensive interdependence effects (i.e., numerous SP/TP site modulation) (fig. S7), whereas few interdependence effects were apparent in extracellular signal–regulated kinase 1 (ERK1) and p38γ activities toward tau (fig. S7). Thus, initial phosphorylation can alter the site-specific activity of kinases toward tau and site interdependence is notably kinase specific.
We aimed to determine positional characteristics and magnitude of interdependence in tau. To visualize phosphorylation interdependence, we plotted modulation effects along the primary sequence of tau, based on the parameters of initial phosphorylation and target site modulation, including directionality and strength of effects (fig. S8). This showed proximal and distal interdependence effects for each tau kinase between phosphorylation events in the N-terminal region (NTR), PRR, and CTR (fig. S8).
To identify significant interdependence links between tau phosphorylation sites, we analyzed levels of epitope phosphorylation by cross-correlation depending on (i) the phosphorylation site targeted, (ii) initial phosphorylation, or (iii) increased kinase activity in our experimental system (fig. S9, A and B). This permitted effect comparisons of individual phosphorylation events for all and for individual SP/TP kinases to identify significantly similar or dissimilar behavior (fig. S9, A and B). Resulting Spearman’s correlation coefficients enabled calculation and ranking of modulation scores to determine most impactful initial phosphorylation sites and performing hierarchical clustering of sites or kinases to visualize dis/similarities. Integrated analysis for strength, target, and significance of modulation produced an overview of the complexity of tau phosphorylation within paradigm parameters (i.e., 17 initial phosphorylation, 12 SP/TP kinases, and 10 phosphorylation detection sites) and revealed relations of broadest impact across SP/TP phosphorylation events and kinases (Fig. 1E). NTR sites T50 and T69 presented with numerous significant interdependence connections onto other tau phosphorylation sites (Fig. 1, F and G). T181 and T205 within tau’s PRR had multiple high-significance relations with other sites across most kinases tested (Fig. 1, F and G). Notably, CTR sites (i.e., S396, S404, and S422) contributed significantly less to tau phosphorylation interdependence across all kinases tested when compared to sites within the NTR and the PRR (Fig. 1G and fig. S10). Sites markedly governing a broad range of tau SP/TP epitopes were termed master sites. Initial modification at master sites was a strong predictor of subsequent tau phosphorylation (Fig. 1, E and G, and fig. S10). Across all SP/TP kinases, master sites in the NTR (T50 and T69) as well as PRR’s T181 and T205 formed clusters of similar modulation relations (fig. S9D). S396 and S404 clustered separately with PRR sites (e.g., T231 and S235) and with CTR site S422, respectively, suggesting that proximity within tau’s primary sequence is not a dominant driver of site interdependence in tau phosphorylation (fig. S9D). Ranking all phospho-sites by impact and significance of modulation revealed cumulative modulation by prior phosphorylation status at T181, S199, and S422 (fig. S9C). These sites are prominent epitopes of pathological hyperphosphorylated tau detected in human brain samples and mouse models of AD and other tauopathies (15, 23, 24). Modulation of S422, a hallmark of advanced tau pathology (25), was prominently associated with prior phosphorylation and kinase activities of p38α, glycogen synthase kinase 3β (GSK3β), and c-Jun N-terminal kinase 3 (JNK3) (fig. S9C). Identified master sites T50, T69, T181, and, to a lesser extent, T111 and T205 made significant contributions to initial phosphorylation-dependent modulation of tau phosphorylation across tau kinases tested (Fig. 1G and fig. S10). These results suggest that a critical contribution to sequential tau phosphorylation arises from initial phosphorylation at specific master sites in the NTR and PRR.
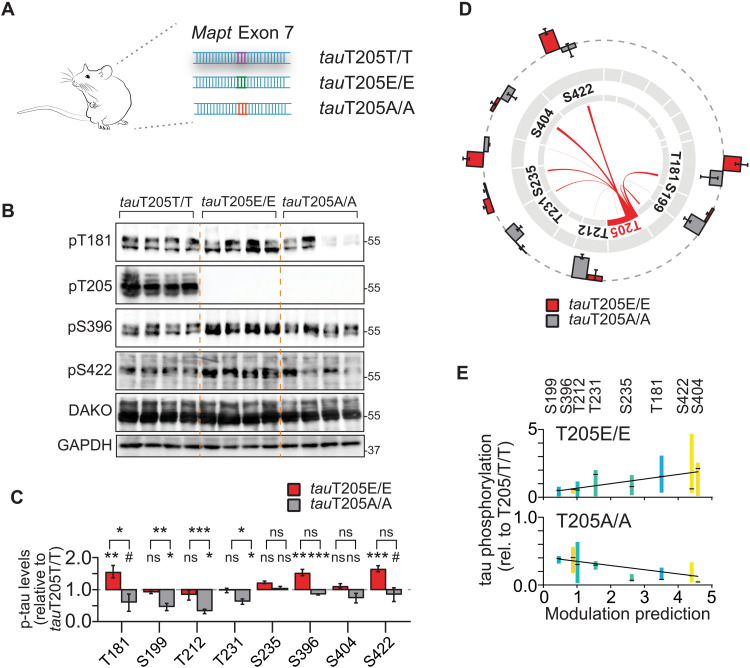
We next confirmed predictions of master sites by proof-of-concept experiments in systems of progressive complexity. In cultured cells, tau phosphorylation by kinases GSK3β and p38α was modulated by T50, T69, T181, and T205 (fig. S11). We wondered whether tau phospho-site interdependence is a physiologic mechanism that occurs during healthy brain function. To test this idea, we focused on master site T205. T205 is of interest because (i) it is a critical residue within the AT8 epitope of pathological tau, a hallmark histological marker of Alzheimer’s tauopathy (26–28) and (ii) T205 phosphorylation of endogenous tau conveys site-specific functions (13, 16). To address the physiologic relation of T205 phosphorylation and multi-site tau phosphorylation in vivo, we used mice, in which the endogenous Mapt gene was CRISPR-Cas9–edited at the T205 codon to a phospho-deficient [threonine → alanine (A)] or phospho-mimetic side chain [→ glutamate (E)] at this position (Fig. 2A) (13). CRISPR tauT205A and tauT205E mice allow for testing for requirement and sufficiency of this master site in physiologic phosphorylation in murine tau, respectively. Brains of tauT205E/E, tauT205A/A and tauT205T/T control mice showed markedly different levels of tau phosphorylation at multiple epitopes including T181, S199, T212, T231, S396, and S422 in tauT205E/E and tauT205A/A as compared with tauT205T/T controls, despite overall similar total tau levels (Fig. 2, B and C, and fig. S12). This was confirmed in immunoprecipitated murine tau followed by phospho-peptide detection (fig. S12). Differential tau phosphorylation in tauT205E/E and tauT205A/A brains significantly correlated with predicted modulation by T205 phosphorylation in vitro, particularly for T181 and S422 (Fig. 2, D and E). These results show that modification of endogenous tau at T205 modulates physiologic tau phosphorylation at multiple epitopes and that predicted site interdependence in cultured cells applies to endogenous tau in brain tissue.
Fig. 2. Master site interdependence controls endogenous tau phosphorylation in vivo.
(A) CRISPR-Cas9 genome-edited mice with conversion of codon in Mapt gene encoding for master site T205 to phospho-deficient alanine (tauT205A) or phospho-mimicking (tauT205E) codon. (B and C) Gene editing of master site T205 in mice modulates levels of site-specific tau phosphorylation of endogenous murine tau predicted by interdependence analysis. Levels of indicated phospho-tau levels in cortex of tauT205A/A, tauT205E/E relative to control tauT205T/T mice, and total tau levels. n = 5 to 6. (D) Visual correlation of T205 interdependence determined in cultured cells with tau phosphorylation levels at different epitopes in CRISPR T205 mice. Differences in tau phosphorylation in tauT205A/A and tauT205E/E as compared with tauT205T/T cortex mapped onto T205-dependent modulation at SP/TP sites. n = 5 to 6. (E) Regression analysis of T205-dependent modulation strength predicted at indicated sites in tau with differences in cortical phosphorylation levels of tauT205A/A and tauT205E/E mice. Values are means ± 10 to 90 percentile relative to tauT205T/T. tauT205A/A (n = 12), P < 0.0001; tauT205E/E (n = 11), P = 0.0002 (linear regression with F test). Data are presented as means ± SEM unless indicated otherwise. ***P < 0.001; **P < 0.01; *P < 0.05; ns, not significant (ANOVA).
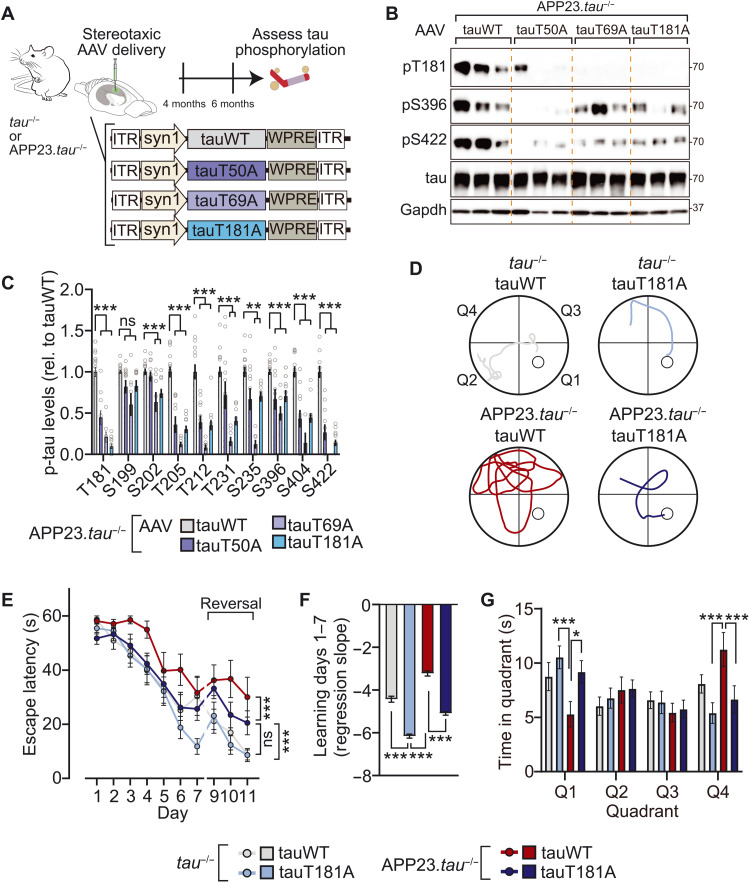
Murine and human tau share a high degree of phospho-epitope identity (15, 29). This suggests that master site interdependence is transferable to human tau, including in the context of AD. Notably, tau phosphorylation—at multiple and individual sites—is involved in tau-dependent AD mouse models, including downstream of Aβ in Aβ precursor protein (APP) transgenic models (13, 16, 30, 31). We investigated master sites T50, T69, and T181 in an AD mouse model by expressing adeno-associated virus (AAV)–delivered wild-type human tau (human tauWT) or phospho-deficient tau variants in the brains of APP23.tau−/− mice, in which the influence of endogenous tau is precluded (Fig. 3A) (13, 32). We addressed master site–dependent tau phosphorylation in this APP transgenic mouse model because tau is critical for phenotypes and signaling downstream of Aβ (33, 34). The reconstitution approach by AAV-mediated neuronal expression of tau variants permitted assessment of site interdependence solely in human tau in living brain tissue, including in the context of a disease model. We focused in these experiments on assessing requirement of these sites for multi-site phosphorylation of human tau as an approach complementary to the phospho-mimicking tauT50E, tauT69E, and tauT181E variants used in cultured cell experiments (fig. S11). AAV-mediated expression of phospho-deficient tau variants showed that T181 and, to a lesser extent, T50 and T69 control multi-site phosphorylation of human tau in the context of high Aβ levels in AD mice (Fig. 3, B and C). Notably, levels of phosphorylation of T205, S396, S404, S422 and others were markedly lower when tau with ablated T50, T69, or T181 site was expressed in APP mice, confirming predictions of phosphorylation modulation by these master sites (Fig. 3, B and C, and fig. S13). These results were supported by mass spectrometry and phospho-peptide enrichment of tau immunoprecipitated from APP23.tau−/− and tau−/− mice expressing tau variants, which also indicated that occurrence of non-SP/TP phosphorylation was lower in tauT50A, tauT69A, or tauT181A as compared with tauWT (fig. S13). To assess the consequence of master site ablation on cognitive phenotypes of AD mice, APP23.tau−/− mice expressing tauWT or tauT181A variant after intracranial AAV delivery performed spatiotemporal learning paradigm (Morris water maze) (Fig. 3, D to G, and fig. S14). TauWT-expressing APP23.tau−/− mice showed strong impairment of learning and cognitive flexibility in the acquisition and reversal phase, respectively, as well as of memory in the probe trails, while tauT181A-expressing APP23.tau−/− mice showed significantly better learning and memory. All experimental groups had comparable swim speeds and visual acuity (fig. S14). To control for effects on locomotor activity, mice performed open field tests, which did not show differences between tauWT and tauT181A-expressing APP23.tau−/− nor tau−/− mice (fig. S14). These results show that Aβ-associated phosphorylation of human tau at multiple pathologically relevant epitopes is regulated by master sites in line with our interdependence predictions. Furthermore, ablation of master site T181 results in modulation of Aβ-associated cognitive deficits.
Fig. 3. Master site interdependence governs human tau phosphorylation and Aβ-tau–induced cognitive impairment in vivo.
(A) Schematic of AAV-mediated expression of tauWT or indicated phospho-deficient tau variants in tau−/− or APP23.tau−/− (B and C) Master site–governed SP/TP phosphorylation of human tau in an AD mouse model. Tau-deficient human APPswe transgenic mice (APP23.tau−/−) were injected intracerebrally with AAV-driving neuronal expression of tauWT or indicated tau variants (tauT50A, tauT69A, and tauT181A). Phosphorylation levels of tau variant or tauWT-expressing mice relative to total tau levels were assessed by immunoblot. n = 8 to 16. (D to G) Morris water maze spatiotemporal learning paradigm with APP23.tau−/− and tau−/− mice injected intracerebrally with AAV-driving neuronal expression of tauWT or tauT181A. (D) Morris water maze escape paths on day 7. Quadrants as indicated with hidden platform in Q1. (E) Escape latency during acquisition phase (days 1 to 7) and reversal phase (days 9 to 11). (F) Acquisition phase learning curve slopes. (G) Probe trials on day 8. Q1, target quadrant. Data are presented as means ± SEM unless indicated otherwise. ***P < 0.001; **P < 0.01; *P < 0.05 (ANOVA).
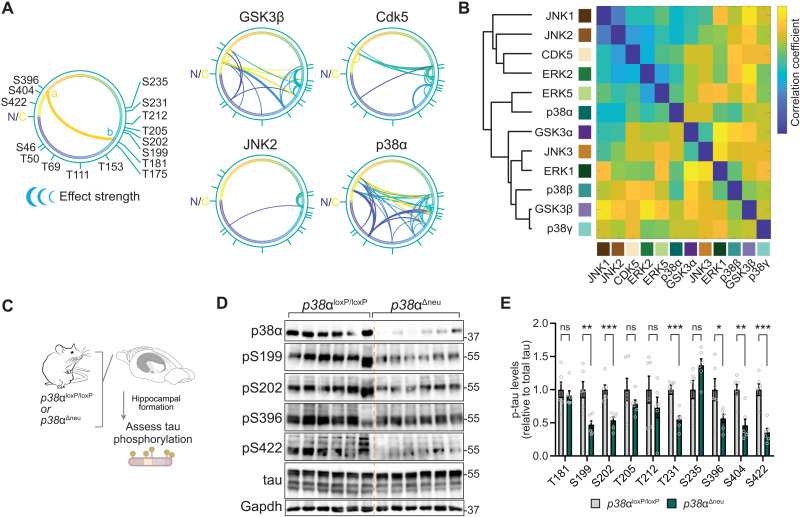
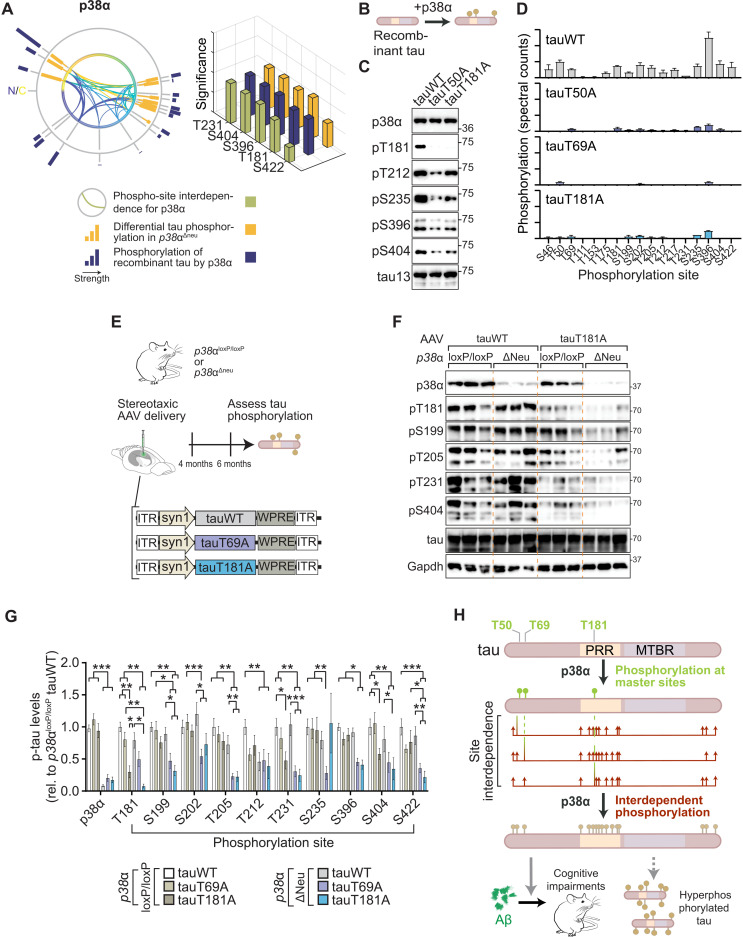
We next asked which tau SP/TP kinases are governed most extensively by interdependence. Plotting modulation by initial tau phosphorylation for kinases individually corroborated kinase-specific interdependence in cultured cells (Fig. 4A and fig. S15). Cross-correlation of all phosphorylation events depending on initial phosphorylation returned varying degrees of similarity and dissimilarity of tau kinases in terms of interdependence of tau phospho-sites (Fig. 4B). Hierarchical clustering of tau kinases placed p38α in proximity with ERK2 and ERK5 and removed from p38 family members p38β and p38γ (Fig. 4B), despite a high degree of sequence identity between p38 kinases (35). We next addressed whether p38α made critical contributions to multi-site tau phosphorylation in vivo. We probed for murine tau phosphorylation at a range of epitopes in neuron-specific p38α knockout (p38αΔneu) and control p38αloxP/loxP mouse hippocampus (Fig. 4C), a brain region with high neuronal p38α expression (36). We found a marked reduction in tau phosphorylation at multiple sites in p38αΔneu as compared with p38αloxP/loxP hippocampus (Fig. 4, D and E, and fig. S16). Tau phosphorylation in p38β−/− and p38γ−/− mice was comparable to control mice for almost all sites tested (fig. S16), suggesting that within the p38 family, p38α rather than p38β or p38γ is a high-impact, multi-epitope tau kinase in vivo. These results show that p38α is a hippocampal tau master kinase in neurons in vivo, likely due to interdependence mechanisms.
Fig. 4. MAP kinase p38α is a multi-site tau kinase in vivo.
(A) Tau SP/TP phosphorylation interdependence is kinase specific. Directionality of interdependence relations between initial phosphorylation (a) and modulated phosphorylation (b). Connecting line thickness indicates significance and modulation strength. Kinases with extensive high-significance interdependence (GSK3β and p38α) and kinases with sparse relations (Cdk5 and JNK2) are shown. n = 4. N, N terminus; C, C terminus of tau (441 amino acids) primary sequence. (B) Correlation matrix of tau phosphorylation interdependence relations associated with 12 SP/TP kinases (n = 4), including hierarchical clustering by pair-wise Pearson’s correlations. Note that tau kinase similarity differs from evolutionary relation of kinase families for GSK3 and p38 kinases. (C to E) Phosphorylation of endogenous tau is reduced at multiple epitopes in hippocampus of mice with neuron-restricted p38α deficiency (p38αΔneu) as compared with control p38αloxP/loxP mice. (D) Representative immunoblots probed with antibodies for pS199 tau, pS202 tau, pS396 tau, pS404 tau, pS422 tau, tau, and p38α are shown. n = 6. Residual p38α in p38αΔneu is due to expression in glial cells (36). (E) Immunoblots in (D) were quantified relative to Gapdh and tau. Data are expressed as means ± SEM. ***P < 0.001; **P < 0.01; *P < 0.05. Note that most phosphorylation sites tested show markedly lower levels in p38αΔneu as compared with control p38αloxP/loxP mice.
Interdependence in tau phosphorylation can arise from intrinsic (i.e., solely between substrate and kinase) or extrinsic mechanisms (i.e., requirement for additional factors). To explore intrinsic mechanisms of interdependence, we performed in vitro kinase reactions (16). We focused on comparisons of p38α with the prominent tau kinases GSK3β (37) and Cdk5 (38, 39). p38α showed extensive phosphorylation of recombinant tau in vitro at multiple sites (fig. S17A and table S5). Phosphorylation of recombinant tau when incubated with CDK5 was limited to specific sites, which correlated with interdependence relations predicted for this kinase (fig. S17B and table S5). Direct tau phosphorylation by GSK3β did not correlate with interdependence identified for this kinase (fig. S17B), suggesting that GSK3β requires extrinsic factors in the cellular context such as engagement of priming kinases for further tau phosphorylation (40). Levels and significance of recombinant tau phosphorylation and differential phosphorylation in p38α-deficient mice correlated with interdependence predicted for p38α (Fig. 5A and fig. S17B).
Fig. 5. Removal of single master site and kinase p38α abolishes tau multi-site hyperphosphorylation.
(A) Mapping of tau site interdependence and differential levels of p38α-mediated tau phosphorylation. Levels (circular graph) and significance (bar chart) of differential tau phosphorylation in brains of p38α-deficient mice (p38αΔneu) and of recombinant tau by p38α correlates with predicted interdependence for p38α. (B) Schematic of in vitro kinase reactions of recombinant tauWT, tauT50A, tauT69A, or tauT181A with p38α to delineate intrinsic basis of tau phosphorylation interdependence. (C) Immunoblots (n = 6) and (D) phospho-proteomic quantification (n = 3) of kinase reactions of p38α with indicated tau variants showing markedly lower phosphorylation of recombinant tau variants at multiple epitopes. (E to G) Combination of master sites T50 and T181 and master kinase p38α governs phosphorylation of human tau. (E) Schematic of stereotaxic delivery of AAVs expressing human tau (tauWT) or indicated tau variants in hippocampus of p38αΔneu and control p38αloxP/loxP mice. (F) Representative immunoblots for tauWT- and tauT181A-expressing mice probed with phospho-tau epitope, tau, and p38α antibodies. n = 10 to 12. (G) Immunoblots were quantified relative to total tau and tauWT. Note that the absence of master sites T69 and T181 in human tau ablates the majority of tau phosphorylation in p38αΔneu compared with p38αloxP/loxP controls. (H) Hypothetical model of tau multi-site hyperphosphorylation governed by site interdependence and mediated by p38α. Initial phosphorylation by p38α at master sites, T50, T69, or T181, promotes subsequent multi-site phosphorylation by p38α. Thus, interdependence between master sites and other phospho-sites may promote tau hyperphosphorylation and contribute to Aβ-induced cognitive deficits. MTBR, microtubule-binding region. Data are presented as means ± SEM unless indicated otherwise. ***P < 0.001; **P < 0.01; *P < 0.05 (ANOVA).
To explore master sites in intrinsic p38α-tau mechanisms, we phosphorylated master site–deficient tau (i.e., tauT50A, tauT69A, and tauT181A) or WT tau with p38α (Fig. 5B and fig. S18). Ablation of T50, T69, or T181 rendered tau less amenable to phosphorylation by p38α at multiple sites (Fig. 5, C and D, fig. S18, and table S6), showing that modulation depending on these sites contributes to direct tau phosphorylation by p38α due to an intrinsic kinase-substrate mechanism. T181 phosphorylation by p38α preceded subsequent multi-site phosphorylation at PRR, and CTR epitopes in kinase reactions stopped at different time points (fig. S19). We next addressed whether these sites were decisive for multi-site phosphorylation by p38α specifically in neurons in vivo. While AAV-mediated delivery of human tauWT to p38αΔNeu brains did not result in lowered tau phosphorylation, likely due to presence of murine tau (13, 16), expression of human tauT69A or tauT181A in p38αΔNeu resulted in markedly lower phosphorylation at multiple sites, with pronounced synergism of kinase and master site ablation (Fig. 5, E to G, and fig. S20). In summary, phosphorylation of master sites acting in synergy with p38α is required for extensive and interdependent tau phosphorylation at multiple epitopes, accelerates aggregation of human tau in vitro, and contributes to Aβ-induced cognitive deficits in vivo (Fig. 5H and fig. S21).
DISCUSSION
In summary, our work details how interdependence mechanistically links site-specific tau phosphorylation to hyperphosphorylation and defines master sites that govern phosphorylation across the entire tau molecule. Site interdependence is a physiologic mechanism and is, notably, intrinsic to the tau protein. This may not only govern physiologic behavior of tau but may also promote hyperphosphorylation and gain of pathologic function. Moreover, it provides a concept for synergism in posttranslational modifications with specific master sites critically controlling global changes in multi-site substrates.
Our screen in 293T cells based on expression of human tau phospho-site variants and SP/TP kinases revealed a complex network of site interdependence. This proposes a hierarchy of tau phosphorylation events that includes long-range effects between sites in distal regions of tau. It surpasses simple directional priming of neighboring sites (41, 42). Long-range site interdependence between phospho-sites in the NTR, PRR, and CTR of tau differs from priming phosphorylation in tau and other multi-site kinase substrates (43, 44), as priming phosphorylation is limited by both proximity and directionality (45). Our results implicate T50, T69, T181, and T205 as key sites that drive multi-site phosphorylation of human tau. Site interdependence is deterministic of tau multi-site phosphorylation and, therefore, of critical interest for diagnostic and therapeutic approaches. Our study purposefully focusses on SP/TP phosphorylation of tau—the most abundant form of tau modification—to delineate the concept of site interdependence. However, detection limitations of phospho-tau immunoblots due to unavailability of antibodies for all SP/TP sites and of mass spectrometry due to low peptide abundance will entail future studies about interdependence of N-terminal SP/TP and non–SP/TP sites. Moreover, our work does not extend directly to tau kinases other than the main proline-directed kinases or to tau phosphatase, which both likely contribute to multi-site phosphorylated tau states (5, 7).
The principal concept of site interdependence applies to physiologic tau phosphorylation. Results from tauT205A and tauT205E mice present T205 as a physiologic master site of tau phosphorylation. They also show that site interdependence is a driving mechanism in multi-site phosphorylation of murine tau. Notably, T205 phosphorylation dampens neurotoxic Aβ signaling and maintains cognitive function (13, 16). However, engaging T205 phosphorylation may eventually promote physiologic multi-site and potentially hyperphosphorylation through site interdependence. It remains unclear, however, why interdependence mechanisms exist that promote tau multi-site modification in the healthy brain.
Aβ-induced cognitive deficits are dependent on tau (33, 34) and reconstituting tau reinstates cognitive deficits (13). APP23.tau−/− mice expressing tau lacking master site T181 did not show significant cognitive deficits and resulted in learning performance comparable to nontransgenic mice. A site-specific function of T181 cannot be excluded at this point. However, lack of master site T181 in human tau was clearly associated with reduced Aβ-induced impact on learning and memory and correlates with lower multi-site phosphorylation of tau at SP/TP and non–SP/TP sites. These results suggest that multi-site phosphorylation, as well as concomitant negative impact on cognitive function, can be alleviated by “breaking” individual master sites within tau.
Synergism of master sites and kinase activity converts p38α into a “master” tau kinase in vitro with isolated tau and in vivo in hippocampal neurons. Normally, p38α targets substrates at stringent consensus epitopes (46) in conjunction with docking motifs (47), neither of which are present in tau. Notably, removal of a single master site was sufficient to ablate p38α-mediated multi-site tau phosphorylation in isolated protein and brain. Thus, site interdependence can direct protein kinase activities toward multiple epitopes in multi-site substrates, such as tau. p38 kinases are highly similar, sharing ~60 to 70% sequence identity. However, they act differently toward tau in vivo (this study) and in short-term kinase reactions [(16) and this study]. This is unexpected because p38 kinases can phosphorylate conventional substrates with equal efficiency (48, 49). We compared tau phosphorylation patterns in p38αΔneu mice with those from p38β and p38γ knockout mice. Genetic deletion of p38β and p38γ does not markedly reduce multi-site tau phosphorylation, but rather at individual sites. This and our data on master site ablation in p38α kinase assays and in the living brain support that interdependence is a driving mechanism for p38α. Other prominent tau SP/TP kinases GSK3β and Cdk5 appear to differ in their mechanism of tau phosphorylation, potentially due to priming site effects by non–SP/TP kinases (50). Future studies will be needed to resolve the relation of p38α, GSK3β, and Cdk5 in tau phosphorylation.
Notably, expression of tau master site variants in p38αloxP/loxP control brains did not results in pronounced reduction of tau phosphorylation at multiple epitopes, unlike expression in p38αΔNeu or in APP23.tau−/− brains. This is likely due to presence of murine tau in the tau-competent genetic backgrounds and its impact on transgenic human tau (51). Mouse tau follows interdependence principles comparably to human tau as our results from tauT205 CRISPR mice indicate. Influence of murine tau on tau phosphorylation of human tau in tau-competent mice also raises the possibility that both intra- and intermolecular mechanisms determine interdependence and govern tau phosphorylation of a pool of tau proteins, at least in living cells.
Site interdependence can create stable species of multi-phosphorylation substrates, similar to other proteins with multi-site modification (19). Notably, multi-site phosphorylation of isolated tau by p38α strictly required master sites T50, T69, or T181 individually. However, these sites may not act independently in vivo as ablation of T205 in tauT205 CRISPR mice and expression of tau variants result in reciprocal changes in their phosphorylation abundance (determined by phospho-peptide mass spectrometry). T50 is not present in murine tau and may therefore promote multi-site phosphorylation specifically in human tau. Our results in cells and with isolated tau indicate that tau phosphorylation at master sites is mutually subject to interdependence effects by prior phosphorylation. This is also the case for T181 in human tau in vivo based on immunoblot and mass spectrometry data. In isolated tau, T181 phosphorylation preceded multi-site phosphorylation, indicating that master site phosphorylation may be an early event under experimental conditions with reduced parameters. Detection of phospho-T50 and phospho-T69 epitopes is limited by low levels in mass spectrometry data to conclude on interdependence between these two sites that specifically could not be determined. In recombinant phosphorylation of tau by p38α, T50 and T69 phosphorylation is reduced in tauT181A, indicating that under isolated reaction conditions, these sites are mutually interdependent.
Site interdependence may explain the abundance of specific tau phospho-epitopes in pathological tau within and outside the central nervous system (24), providing a novel rationale for phospho-tau–based biomarker design in tauopathies. Master site T181 phosphorylation bears strong potential as diagnostic biomarker (24). Ablating T181 resulted in marked reduction of tau multi-site phosphorylation. Thus, T181 in tau may serve as both a diagnostic and therapeutic target. Targeting tau master sites is likely applicable to neurological disorders beyond AD, such as frontotemporal dementia, Parkinson’s disease, stroke, and chronic traumatic encephalitis, in which pathologically phosphorylated tau is abundant (4, 52).
Overall, site interdependence may answer a central question in Alzheimer’s research: Why and how does the neuronal tau protein get progressively hyperphosphorylated? This raises the exciting possibility that therapeutic inhibition of master sites will block intrinsic augmenting mechanisms of tau phosphorylation, decrease levels of pathologic tau, and, thus, maintain healthy cellular function in AD and other tauopathies.
MATERIALS AND METHODS
Mice
Mice with targeted alleles for p38α (Mapk14) [p38αloxP;B6.129S4(Cg)-Mapk14tm1.2Lex], for p38β (Mapk11) [p38β−; B6.129S4(Cg)-Mapk11tm1.2Lex], for p38γ (Mapk12) [p38γ−; B6.129S4(Cg)-Mapk12tm1.2Lex], tau-deficient mice [tauKOeGFP; B6.129S4(Cg)-Mapttm1(EGFP)Klt], APP transgenic APP23 mice [B6.Cg-Tg(Thy1-APP)3Somm], transgenic mice expressing Cre recombinase in neurons including in the cortex and hippocampus [B6.FVB/N-Tg(Thy1-cre)1Vln] (16), and mice with murine Mapt gene targeted by CRISPR-Cas9 to exchange the codon corresponding to T194 (i.e. T205 in human tau) were previously described (13). All mouse strains were backcrossed and maintained on a C57Bl/6 background. Mice were housed in individually vented cage systems (Tecniplast, Italy) with access to food (Gordon Specialty Feeds, standard rodent chow) and water ad libitum. Lighting was adjusted to a circadian day-night cycle. Regular health checks were carried out by animal facility staff and researchers, and only animals with normal health parameters (body weight and coat condition) were included in experimentation. All animal experimentation and husbandry were approved by the Animal Ethics Committees of Flinders University and Macquarie University and followed Institutional Animal Care and Use Committee guidelines. Mice were genotyped by polymerase chain reaction (PCR) using isopropanol-precipitated DNA as template and OneTaq DNA polymerase (New England Biolabs, (NEB), #M0480) as described previously (53). TauT205A and tauT205E mice were genotyped using isopropanol-precipitated DNA from tail biopsies as template for tetra-primer amplification-refractory mutation system PCR (13). Oligonucleotide primers (desalted, Macrogen, Korea) for genotyping targeted alleles and transgenes by PCR are described in table S2.
Behavior and memory testing
Morris water maze
Water maze tests were performed as previously described (16). Briefly, the water maze (145 cm in diameter, white opaque high-density polyethylene, custom-built, Acrilix) surrounded by white screens was filled with opacified room-temperature water until 30 cm below outer edge. A hidden platform (12.6 cm in diameter, 1 cm below water surface, ConductScience) was placed in the quadrant opposite of entry quadrant. Four cue boards were affixed at perimeter of tank. Movements were recorded with a digital camera mounted above the maze. Entry position in quadrant 1 was semirandomized at four positions. Mice were gently lowered into the water facing the maze wall. Acquisition phase (days 1 to 7) included four consecutive 60-s trials per test day, with 30-s pause once the mouse reached the hidden platform. Two probe trials (30 s) were performed after the hidden platform was removed on day 8. For the reversal learning phase (days 9 to 11), platform was moved to a different quadrant and cue boards were randomized. A high-contrast marker was attached to the hidden platform on day 12 for visual cued trials. Automated movement tracking was performed using ANY-maze software (version 7.10, Stoelting). Escape was defined as finding and ascending the hidden platform. Acquisition curves (days 1 to 7) were analyzed with linear regression to determine learning curve slopes (GraphPad Prism).
Open-field test
Open-field paradigm was done as previously described (36). Briefly, tests were performed in 44-cm2 square gray opaque Perspex boxes. Mice were placed in the arena facing the center and allowed 10 min for spontaneous exploration. Automated movement tracking was performed with ANY-maze (Stoelting). Distance traveled by mice in inner and outer zones of the open-field arena was quantified.
Tau variant library and protein kinase expression constructs
A human cDNA clone encoding the longest isoform of human tauWT (2N4R, 441 amino acids) was used as the starting WT sequence. Specific amino acid substitutions within proline-directed phosphorylation epitopes from SP or TP to aspartic acid–proline (DP) or glutamic acid–proline (EP), respectively, were made using Q5 site–directed mutagenesis kit (NEB, #0554) or had been described previously (16). Oligonucleotides (Macrogen, Korea) used for mutagenic PCRs to generate phospho-mimetic and phospho-deficient (serine/threonine to alanine) variants are listed in table S4. All tau variants were cloned into pcDNA3.2/V5 (Invitrogen, #12489019) propagated and amplified in Escherichia coli DH5α (NEB, NEB5-alpha, #C2988). Plasmids were isolated using PureLink HiPure plasmid Maxiprep kit (Invitrogen, #K210016). Constructs for expression of human protein kinase were obtained from Addgene for GSK3 alpha pMT2 (Addgene, #15896, a gift from J. Woodgett), hemagglutinin (HA) GSK3 beta S9A pcDNA3 (Addgene, #14754, a gift from J. Woodgett), pcDNA3-cdk5GFP (Addgene, #1346, a gift from L.-H. Tsai), pFLAG-CMV-hErk1 (Addgene, #49328, a gift from M. Cobb), pcDNA3-HA-ERK2 WT (Addgene, #8974, a gift from J. Blenis), pCI-HA-ERK5-FL (Addgene, #31817, a gift from A. Winoto), pCDNA3 Flag Jnk1a2 (Addgene, #13751, a gift from R. Davis), pCDNA3 Flag Jnk2a2 (Addgene, #13755, a gift from R. Davis), and pCDNA3 Flag Jnk3a2 (Addgene, #13759, a gift from R. Davis). Constructs for expression of active human p38 kinases p38α, p38β, and p38γ (54) were cloned into pcDNA3.1(+) (Invitrogen) including a HA tag at the start of the open reading frame as previously described (16). All kinase constructs were cloned into pcDNA3.1(+) (Invitrogen), propagated and amplified in E. coli DH5α (New England Biolabs, NEB5-alpha, NEB#C2988). All sequences were verified by sequencing (Macrogen, South Korea) and sequence analysis (CodonCode Aligner). All tau constructs were tested for expression of full-length tau in mammalian cells (293T) by plasmid DNA transfection and Western blot.
Recombinant tau protein
Recombinant human tau proteins were generated, expressed, and purified as described previously (16). Briefly, coding sequences of human tauWT (441 amino acids) or of phospho-deficient variants tauT50A, tauT69A, or tauT181A or murine tau WT (430 amino acids) or murine tauT69A or tauT181A were cloned between Nde I and Bam HI restriction sites in the pHIS17 vector using restriction-ligation cloning using NdeI-HF (NEB, #R0111), BamHI-HF (NEB#R3136), and T4 DNA ligase (NEB#M2020). Sequence-verified clones were transformed into E. coli BL21(DE3)pLys (Promega, #L1195) of Shuffle T7 express (NEB#C3030), and recombinant protein expression was induced by addition of isopropyl-β-d-1-thiogalactopyranoside (1 mM; Meridian Bioscience, #BIO-37036) within the exponential growth phase (at an optical density at 600 nm of 0.6) at 37°C for 3 to 4 hours. Expression of recombinant proteins was confirmed on 8% SDS-PAGE gel and by visualization with Coomassie brilliant blue stain (Sigma-Aldrich). Bacterial cells containing recombinant protein were lysed by sonication (5 min, 1-min pulses at 30% duty cycle) on ice in bacterial lysis buffer [50 mM sodium phosphate buffer (pH 7.4), 300 mM sodium chloride, lysozyme (0.2 mg/ml; Sigma-Aldrich), deoxyribonuclease (20 μg/ml; NEB#0303), 1 mM magnesium chloride, and 1 mM Pefabloc SC (Sigma-Aldrich)]. Bacterial lysates were cleared by centrifugation at 16,000g for 15 min at 4°C. After confirmation of neutral pH, recombinant proteins were purified by batch purification on cobalt–immobilized metal affinity chromatography resin (His GraviTrap TALON, Cytiva), washed with low imidazole buffer [50 mM sodium phosphate buffer (pH 7.4), 300 mM sodium chloride, and 5 mM imidazole], and eluted twice with high imidazole buffer [50 mM sodium phosphate buffer (pH 7.4), 300 mM sodium chloride, and 150 mM imidazole]. Recombinant tau proteins were buffer exchanged by dialysis against a tris buffer [50 mM tris (pH 7.4) and 300 mM NaCl]. Purity and integrity of recombinant proteins were confirmed on Coomassie brilliant blue–stained SDS-PAGE and by immunoblot using tau13 antibody. Protein concentration of recombinant proteins was determined by protein assay (Bio-Rad).
Cell culture
Human embryonic kidney 293T cells (American Type Culture Collection, #CRL3216) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco), supplemented with 10% fetal bovine serum (FBS; Gibco, #10099), 1% (v/v) penicillin/streptomycin (Gibco, #10378016), and 2 mM GlutaMAX (l-alanyl-l-glutamine dipeptide supplement; Gibco, #35050061) in controlled atmosphere containing 5% CO2 at 37°C. Passage numbers of cultured 293T cells were maintained within the range of 3 to 10 passages. Cells were passaged when reaching 70 to 80% confluence. Twenty-four hours before transfection with tau and/or protein kinase expression constructs, cells were seeded at 8 × 104 density into a 24-well plate in fresh medium. Two hours before transfection, culture medium was exchanged, and cells were transfected at 70% confluency using calcium precipitation as described previously (36). Briefly, plasmid DNA (1 μg of total plasmid amount) was mixed with calcium chloride (2 M; Sigma-Aldrich, #499609) while vortexing. Plasmid DNA–calcium precipitated was distributed into culture plate wells after 10 min. Forty-eight hours after transfection, cells were washed in 1× phosphate-buffered saline (PBS), harvested, and lysed in 1× SDS–Laemmli sample buffer [60 mM tris-Cl (pH 6.8), 2% (w/v) SDS (Sigma-Aldrich, #28312), 10% glycerol (Sigma-Aldrich, #G5516), and 0.01% bromophenol blue (Sigma-Aldrich, #B0126)] supplemented with 5% β-mercaptoethanol and 10 mM sodium fluoride (NaF). The supernatant was collected, and samples were sonicated before being snap-frozen in liquid nitrogen and stored at −80°C.
Production of recombinant AAV
Packaging of AAV vectors was performed as previously (13, 16). The following procedure is described in (55). For packaging of AAV particles, 293T cells were seeded in complete DMEM (Gibco, #11965175), supplemented with 10% FBS (Gibco, #10099), 1% (v/v) penicillin/streptomycin (Gibco, #10378016), and 2 mM GlutaMAX (l-alanyl-l-glutamine dipeptide supplement, Gibco, #35050061) at 70 to 80% confluence. Three hours before transfection, cell culture medium was replaced with Iscove’s modified Dulbecco’s medium (Gibco, #12240053) with 5% FBS (Gibco, #10099). Transfection included viral genome-containing plasmid, pFΔ6 as helper plasmid, and AAV-PHP.B plasmid, which encodes rep and cap sequences (56), and was performed with polyethylenimine-Max (PEI-Max; Polysciences, #24765-1) transfection reagent at a ratio of plasmid DNA:PEI of 3:1. Seventy-two hours after transfection, cells and supernatant were collected. Cell supernatants were clarified by addition of 40% PEG 8000/2.5 M sodium chloride to a final concentration of 8% PEG 8000/0.5 M sodium chloride and incubated at 4°C for at least 2 hours. Cleared supernatant was spun at 2000g for 30 min at 4°C. Combined cell pellet and precipitate containing viral particles were treated with sodium deoxycholate (0.5% final concentration; Sigma-Aldrich, #D6750) and benzonase (~500 U; Sigma-Aldrich, E8263) at 37C for 40 min. Following addition of sodium chloride, incubation at 56°C for 40 min and cycles of freeze-thawing, solution containing viral particles was spun for 30 min at 5000g and 4°C. Purification of supernatants was performed by iodixanol (OptiPrep, Sigma-Aldrich, #D1556) gradient ultracentrifugation (475,900g for 2 hours at 18°C). Viral particles were concentrated and buffer-exchanged into PBS (pH 7.4) with 100-kDa centrifugation filter units (Millipore, #ACS510024) at 5000g and 4°C. Upon titration of viral particles with quantitative PCR (qPCR), aliquots were stored at −80°C. AAV titration was performed by qPCR of serial dilutions of purified AAV concentrates using Luna Universal qPCR mix (NEB#M3003). Oligonucleotide primers for AAV titration purpose were forward inverted terminal repeat (ITR) primer, 5′-GGAACCCCTAGTGATGGAGTT, and reverse ITR primer, 5′-CGGCCTCAGTGAGCGA (57). Determined AAV titers were (expressed as viral genomes per milliliter) AAV-PHP.B-syn1-tauWT (8.42 × 1013), AAV-PHP.B-syn1-tauT50A (1.49 × 1014), AAV-PHP.B-syn1-tauT69A (1.68 × 1014), and AAV-PHP.B-syn1-tauT181A (1.70 × 1014). For application by stereotaxic injection, concentrated purified AAV solutions were diluted 1:1 with sterile saline.
Stereotaxic injection
Delivery of AAV particles by stereotaxic injection was performed following our previously published method (35). Mice assigned to AAV delivery by stereotaxic injection were 2 to 4 months of age. Mice were anesthetized by injection of ketamine/xylazine (80/8 mg/kg of body weight, ip). General anesthesia was confirmed, and the scalp was shaved and disinfected with alcoholic chlorhexidine. Subsequently, mice were mounted onto a stereotaxic frame (Kopf Instruments, Tujunga, USA, catalog nos. 940 and 926). Midline scalp incision was made, skull surface was cleared, and the stereotaxic frame was centered on bregma as reference position. Stereotaxic coordinates were obtained from the mouse brain atlas (Paxinos and Franklin, fifth edition). Bur holes were formed with a microdrill (Stoelting, #58610v) fitted with a 0.05-mm-diameter tungsten carbide drill head (Fine Science Tools, Meisinger #310104001) to infuse at coordinates for hippocampus [lateral (L), ±1.85; anterior-posterior (AP), −2.0; dorso-ventral (DV), −2.0] and cortex (L, ±1.85; AP, −2.0; DV, −0.5). For stereotaxic injections, 400 nl (1 × 1010 viral particles) of AAV particle solution was infused at two positions bilaterally into the hippocampus and cortex of ketamine-anesthetized mice. The injection needle (34 gauges; World Precision Instruments, Saratosa, USA) remained in position for 2 min before infusion at 200 nl/min using a microinjection syringe fitted with a 34-gauge needle under control of a nanoliter pump system (World Precision Instruments, Saratosa, USA, catalog no. UMP3). After volume delivery, the injection needle was left in position for additional 5 min before retraction.
Mouse brain lysates
Lysates were prepared as previously described (36, 55). Briefly, murine cortical and/or hippocampal tissue was extracted upon transcardial perfusion with PBS (pH 7.4) (Gibco, #10010023). Extracted tissue were maintained on ice and either processed immediately or snap-frozen in liquid nitrogen for subsequent storage at −80°C. Tissue material was weighed before addition of ice-cold radioimmunoprecipitation assay (RIPA) buffer [20 mM tris (pH 8.0), 150 mM sodium chloride, 1 mM sodium EDTA, 1 mM activated sodium orthovanadate (Na3VO4), 1 mM NaF, 1 mM glycerophosphate, 2.5 mM sodium pyrophosphate (Na2H2P2O7), 1 mM phenylmethylsulfonyl fluoride, protease inhibitors (cOmplete, Roche, catalog no. 11697498001), 1% NP-40 substitute (Sigma-Aldrich, Merck, Munich, Germany, catalog no. 11754599001), 0.1% SDS, and 0.5% sodium deoxycholate] was added at a ratio of 10 μl of buffer per milligram tissue weight, and tissue was homogenized with a dounce homogenizer (10 strokes, 650 rpm; Heidolph, Schwabach, Germany) on ice. Resulting lysates were sonicated (2 × 2 s; 20% amplitude; Qsonica) before centrifugation at 16,000g for 10 min at 4°C. Protein concentrations were determined with colorimetric assay kit [DC Protein Assay (Bio-Rad, Hercules, USA, catalog no. 5000111) for samples in RIPA buffer and Pierce 660-nm protein assay (Thermo Fisher Scientific, catalog no. 22662) for samples in 1× SDS–Laemmli sample buffer).
Immunoblotting
To analyze protein levels in lysates, Western blotting was performed as described previously (16, 32). Briefly, cell or tissue lysates were separated by SDS-PAGE (8 or 10%), transferred to nitrocellulose filter membranes (Immobilon-NC, Merck Millipore; catalog no. HATF00010). Membranes were blocked in 3% bovine serum albumin (BSA) (Sigma-Aldrich, catalog no. 9048-46-8) in tris-buffered saline buffer with Tween 20 (TBS-T; 0.5%) and incubated with primary antibodies diluted in 3% BSA in TBS-T overnight at 4°C. After three washes for 10 min in TBS-T, membranes were incubated at ambient temperature for 1 hour with secondary antibodies. Subsequently, membranes were washed three times for 5 min before development of enhanced chemiluminescence reaction (Merck Millipore, #WBLUR0500). Chemiluminescence signals were imaged on a ChemiDoc MP (Bio-Rad) digital system. We assessed specificity of antibodies for designated individual phospho-epitopes in tau by expression of phospho-deficient tau variants and immunoblot. We selected site-specific phospho-tau antibodies with no or minimal effects of overlapping phospho-epitopes in tau (see fig. S4 and table S1). Primary antibodies used in this study were anti-human tau (tau13) (1:5000; Santa Cruz Biotechnology, #sc-21796), tau (1:5000; DAKO, A0024), tau D5D8N (1:1000; Cell Signaling Technology, #43894), anti–phospho–threonine-181 tau (1:1000; Abcam, #ab75679,), anti–phospho–serine-199 (1:1000; Abcam, #ab4749), anti–phospho–serine-202 (1:1000; Abcam, #ab108387), anti–phospho–threonine-205 (1:2000; Abcam, #ab4841), anti–phospho–threonine-212 (1:1000; Abcam, #ab4842), anti–phospho–threonine-231 (1:1000; Abcam, #ab194815), anti–phospho–serine-235 (1:1000; Abcam, #ab306640), anti–phospho–serine-396 (1:2000; Abcam, #ab109390), anti–phospho–serine-404 (1:1000; Abcam, #ab92676), anti–phospho–serine-422 (1:1000; Abcam, #ab79415), anti-GAPDH (1:5000; Millipore, #ab2302), anti-p38α (1:1000; Cell Signaling Technology, #9212), anti-p38β (1:1000; Novus Biologicals, OTI1C2), anti-p38γ (1:500; R&D Systems), anti-GSK3 (Abcam, #15314), anti-GRP78 (1:1000; Abcam, #ab21685), anti-Hsp70 (1:1000; Santa Cruz Biotechnology, #sc-32239), anti–green fluorescence protein (GFP; 1:1000; Abcam, #ab290), and anti-HA7 (1:5000; Sigma-Aldrich, #H3663). Horseradish peroxidase (HRP)–coupled secondary antibodies used include HRP-coupled secondary antibodies goat–anti-rabbit (1:5000; Santa Cruz Biotechnology, #sc-2004) and goat–anti-mouse (1:5000; Santa Cruz Biotechnology, #sc-2005).
Immunoblot quantification
Densitometric analysis of immunoblot images was done as previously described (16). Briefly, densitometric quantification of immunoblot results was performed using ImageJ 2.0.0-rc-49/1.51d. The rectangle analysis tool in ImageJ 2.0.0-rc-49/1.51d was used to select the length and width of the lane to be measured, beginning at the first lane, and using the identical frame across all lanes. Following this, the density of a band was defined by measurement of the total intensity peak area within the reduced dimension representation of the lane. A numerical value for each lane was generated, which was deducted from the background value of the immunoblot. Following this, all blots were normalized to the loading control (GAPDH) and tau expression (antibodies tau13 or DAKO for human or mouse tau, respectively). Subsequently, all values were analyzed relative to the control condition. For phospho-site interdependence screening in cultured cells, levels of tau phosphorylated at specific epitopes were expressed relative to total tau signals, normalized to GAPDH. Protein kinase expression was quantified normalized to ensure that similar amounts of kinase expression were achieved across conditions. Changes between WT and phospho-mimicking tau variants in the absence of kinase overexpression were included by normalizing phospho-epitope signals to either tauWT or the respective tau variant expression without kinase expression. Each dataset is an average of at least three to four repeats. For plotting in clustergrams (hierarchical clustering analysis data represented as heatmaps; MATLAB function: clustergram; see figs. S6 and S7), relative modulation was calculated from normalized relative densitometric data averaged and weighted on the basis of probability values [P < 0.05, false discovery rate (FDR) corrected by the Benjamini-Hochberg method] (58, 59). Validity of weighted probabilities in multiple comparisons by P values density in pairwise comparisons of phosphorylation levels between tauWT and tau phospho-mimicking variants and by q value distribution upon positive FDR (pFDR) analysis was confirmed using MATLAB’s pFDR function (60).
In vitro kinase assays
In vitro kinase assay was performed as previously published (16). All human recombinant kinase assays were purchased from Promega and include CDK5/p25 (V3231), ERK1 (V1951), ERK2 (V1961), GSK3α (V3051), GSK3β (V1991), JNK1 (V4070), JNK3 (V3821), p38α (V2701), p38β (V4154), and p38γ (V3371). Briefly, 0.5 μg of recombinant kinase was incubated with 1 μg of recombinant human tau in kinase reaction buffer containing 40 mM tris (pH 7.5), 20 mM magnesium chloride, BSA (0.1 mg/ml), and 50 μM dithiothreitol (Promega). Reactions were supplemented with 50 μM adenosine triphosphate (ATP; Sigma-Aldrich, #A2383) to initiate protein kinase reaction and substrate phosphorylation. Negative control conditions contained reaction buffer without addition of ATP. Samples were incubated at 30°C for 30 min, before being stored at −80°C until phospho-peptide enrichment and mass spectrometry or analysis by SDS-PAGE, followed by immunoblot. To resolve tau phosphorylation at different time points of kinase reactions, assays were stopped by addition of SDS sample buffer including β-mercaptoethanol.
Immunoprecipitation
Tau protein from brain lysates (cortex or hippocampus) was immunoprecipitated as described previously (16). Briefly, brain tissue lysates in RIPA buffer including phosphatase and protease inhibitors with normalized concentration (300 μg of total protein/250 μl of total volume) were incubated with either tau13 (1:250; B11E8, Abcam, #ab19030) or with tau D5D8N (1:200; Cell Signaling Technology, #43894) for precipitation of human or mouse tau, respectively. Samples were rotated at 4°C at 6 rpm for 3 hours. Buffer-equilibrated protein G magnetic beads (New England Biolabs, #1430) were added at 25 μl of bead slurry per sample and incubated further for 45 to 60 min. Beads were washed three times at 4°C for 10 min with RIPA buffer and rinsed once with ammonium bicarbonate (100 mM) before on-beads digest (1:100 trypsin:protein ratio) and subsequent phospho-peptide enrichment, and sample preparation for mass spectrometry. A 5% fraction was withheld for immunoblot.
Phospho-peptide enrichment and mass spectrometry
Phospho-peptide mapping of tau was done as previously described (16). Protein samples were reduced with 3 mM tris(2-carboxyethyl)phosphine (56°C for 10 min), followed by alkylation using 6 mM iodoacetamide (Sigma-Aldrich) at ambient temperature for 30 min. Samples were buffer exchanged and concentrated using 100 mM ammonium bicarbonate and 3-kDa spin-filter concentrators (Amicon Ultra-4 Centrifugal Filters, Merck, catalog no. UFC800324). Samples were trypsin digested [1:25 (w/w) trypsin:protein ratio for 16 hours at 37°C; sequencing grade trypsin, Promega, catalog no. V5111). A portion of the material was enriched for phospho-peptides using Titansphere Phos-TiO kit, with TiO2 Spin tips (GL Sciences), following a standard protocol provided by the manufacturer. Phospho-peptide–enriched and nonenriched samples were analyzed by liquid chromatography with tandem mass spectrometry (LC-MS/MS) using Orbitrap mass spectrometers [LTQ-Orbitrap Velos with collision-induced dissociation (CID) and electron-transfer dissociation (ETD) activation modes and higher energy collisional dissociation (HCD) on the Q Exactive Plus, both from Thermo Electron] to maximize phospho-peptide identification. Chromatography was carried out by nano-LC (Dionex UltiMate 3000 HPLC, Thermo Fisher Scientific) with autosampler system (Dionex). Peptides (1 to 7 μl injected) were first captured on a C18 cartridge (Acclaim PepMap 100, 5 μm, 100 Å, Thermo Scientific Dionex), switching to a capillary column (10 cm) containing C18 reverse phase packing (Reprosil-Pur, 1.9 μm, 200 Å, Dr. Maisch GmbH), supported within a column heater (45°C; Sonation GmbH). Peptides were eluted using a 40-min gradient of buffer A (H2O:CH3CN of 98:2 containing 0.1% formic acid) to 45% buffer B (H2O:CH3CN of 20:80 containing 0.1% formic acid) at 200 nl/min, with high voltage applied at the column inlet. Mass spectrometer settings were electrospray voltage (2000 V), capillary temperature (275° to 300°C), positive ion mode, data-dependent acquisition mode with a survey scan acquired (mass/charge ratio, 375 to 1750) and up to 10 multiply charged ions (charge state ≥ 2+) isolated for MS/MS fragmentation (counts > 2500 for CID, >5000 for ETD, and intensity threshold of 8.0 × 104 for HCD). Nitrogen was used as HCD collision gas and fluoranthene anion reagent for ETD. Peak lists were generated from the raw data using MASCOT Distiller (Matrix Science, London, England) and searched using the MASCOT search engine (version 2.5, Matrix Science) and the NCBInr database (downloaded 24-10-15) using Homo sapiens taxonomy for peptide annotation. Search parameter settings for MASCOT search were (i) peptide tolerance of ±4 parts per million and MS/MS tolerances of ±0.4 Da for CID and ETD or ±0.05 Da for HCD (61); (ii) variable modifications were carbamidomethyl-cysteine, methionine oxidation, phospho-serine/phospho-threonine, and phospho-tyrosine; (iii) peptide charge of 2+, 3+, and 4+; and (iv) enzyme specificity trypsin with up to three missed cleavages allowed. Label-free, semiquantitative determination of abundance was done by spectral count analysis, defined as total number of spectra recorded for a phospho-epitope as performed previously (16). Quantitation of phospho-peptides from immunoprecipitated tau was normalized to the relative amount of precipitated tau as determined by immunoblot and densitometric measurements (ImageJ). Purity of isolated recombinant tau proteins confirmed by LC-MS/MS protein identification of nonenriched in vitro kinase reaction samples and standardized TiO2-enrichment of phospho-peptides supported the validity of label-free abundance comparison using spectral counts (62).
Statistical analysis of phosphorylation site modulation
Data were processed and analyzed on the basis of Spearman’s dissimilarity statistic implemented with a custom-built script in MATLAB (R019b, MathWorks, MA, USA). Spearman’s rank-order correlation is a nonparametric measure of associations that allows the quantification and hypothesis testing of correlations between pairwise variables. First, for each site, linear correlations between three experimental variables/parameters (tau initial phosphorylation site, tau phospho-site detection levels, and SP/TP tau kinase) were computed keeping one variable constant (e.g., either the initial phosphorylation or phospho-epitope detected was kept constant) depending on the pairwise analysis conducted. Then, for each comparison, random permutation testing was implemented using Spearman’s dissimilarity metric. Spearman’s rank correlation coefficient measures the strength and direction of an association between two parameters. Use of Spearman’s rank correlation was implicated because of data demonstrating a monotonic relationship. Calculation of Spearman’s rank-order correlation was based on
Where ρ is the Spearman’s rank correlation coefficient, d2 is the squared difference between ranks, and n is the number of measurements. Spearman’s rank-order correlations were analyzed for statistically significant positive and negative correlations across all initial phosphorylation, detected phosphorylation levels, and tau kinases expressed using a data randomization and multiple comparisons approach. The random permutation model was generated using n = 100 random rearrangements of the data for each kinase, again ensuring constant variables of initial phosphorylation and detection site for pairwise comparisons. Pairwise null distributions were plotted for each comparison along the Spearman’s dissimilarity metric with 0 representing complete positive correlation, 1 being random nonassociation, and 2 being complete negative correlation. Probability values were obtained for each pairwise correlation using a Fisher’s exact testing to reject the null hypothesis. Multiple comparisons were corrected for pFDR using the Benjamini-Hochberg method in MATLAB. Significance threshold for individual interactions was set at P < 0.05. Correlation coefficient matrices were plotted by clustergram function in MATLAB including hierarchical clustering. Spearman’s rank correlations coefficients and cophenetic distances were used to generate dendrograms for initial/source site and tau kinase (dis)similarities based on quantified tau phosphorylation interdependence (see fig. S9D and Fig. 4B, respectively).
Graphical representation of results
A custom-built MATLAB (R2019b, MathWorks) script was used for clustergram representation of data. Data were displayed using the clustergram function with hierarchical clustering, which is on the basis of Euclidean distance between data pairs. For plotting of site interdependence relations along the tau molecule in circular or linear representations of the tau primary sequence, data were standardized with a logistic weighting function across the data dimension based on probability values (P < 0.05, FDR corrected by the Benjamini-Hochberg method) (58, 59). Data were plotted on double-linear or circular graphs representing the primary sequence of tau. Notably, neither double-linear nor circular representation necessarily distinguish between intra- or intermolecular mechanisms of interdependence between phosphorylation sites. Double-linear graphs to represent site interdependence relations between initial and subsequent phosphorylation in tau from cultured cells for each of 12 SP/TP kinases (see fig. S8) were plotted with a custom-built MATLAB script.
For integration of site interdependence across conditions of increased kinase activity of all 12 kinases (see Fig. 1E and fig. S10), data were plotted on a circular representation of phosphorylation sites summarizing site interdependence within the sector for each phospho-site–based including both at source and at target of interdependence relation, thus defining the width of the sector edge. Probability values were plotted for each interdependence relation across all protein kinases on the outer perimeter of the circular graph. Modulation score for individual source sites of interdependence relations (see Fig. 1G) is a factor of Spearman’s correlation rank coefficient (Fisher’s exact test, Benjamini-Hochberg multiple comparisons adjustment) and the averaged relative phosphorylation level at each target site for the source site.
For generation of circular graphs representing site interdependence relations along the primary sequence of tau (see Fig. 4A and fig. S15), a custom-built MATLAB script is based on circularGraph function (Paul Kassebaum, 2020). The circularGraph function (www.github.com/paul-kassebaum-mathworks/circularGraph, GitHub) was used to plot statistically significant modulation of phosphorylation along tau. Data pertaining to phospho-peptide spectral counts or differential phosphorylation levels from mouse brain were plotted along the perimeters of circularGraph.
Statistical analysis
For statistical analysis, GraphPad Prizm (v7.0c) was used. Comparisons of two experimental groups for protein/phosphorylation level data were performed with unpaired, two-tailed Student’s t test. For comparisons for more than two experimental groups, analysis of variance (ANOVA) was performed with multiple comparisons using the Holm-Sidak method. Data are expressed as means ± SEM. unless stated otherwise in the figure legend.
Acknowledgments
We thank the technical staff of the Dementia Research Centre (T. Butler, J. Spathos, and Y. Lin) and of the Biomedical Engineering, Flinders Medical Centre, and the staff of the Macquarie Animal Research Services of Macquarie University and of the College of Medicine and Public Health Animal Facility of Flinders University for animal husbandry. We furthermore thank G. R. White (Indiana University, Bloomington, US), T. Mueller (Macquarie University, Sydney, Australia), and L. M. Ittner (Macquarie University, Sydney, Australia) for discussions on concept and data analysis, as well as Y. L. Chew (Flinders University, Australia) for critical reading of the manuscript.
Funding: This work was supported by funding from the National Health and Medical Research Council (grant nos. 1143978 and 1176628); the Australian Research Council (grant nos. DP170100843, DP200102396, and DP220101900); the Dementia Australia Research Foundation; the Flinders Foundation, Flinders University, and Macquarie University; and the BrightFocus Foundation (grant no. A2022022F to K.S.) is a Scientia Professor Henry Brodaty Post-doctoral Fellow of the Dementia Australia Research Foundation. A.I. is a National Health and Medical Research Council Emerging Leadership 2 fellow (grant no. 1176628).
Author contributions: A.I. conceived of the original study concept and led the study. A.I. and K.S. designed overarching research strategy and the specific research plan of all experiments and obtained funding. K.S. and A.I. generated materials, performed experiments, analyzed data, and prepared figures. M.G. analyzed data, wrote analysis scripts, and prepared figures. H.I.A. and A.V. provided critical assistance with generation of materials. A.R.P.T. and P.R.A. provided important technical assistance. A.P. conducted proteomic analysis. A.I. supervised and contributed to experiments. K.S., A.P., and A.I. wrote the manuscript. All authors contributed to the final version of the manuscript.
Competing interests: A.I. is a coinventor on a patent application related to targeting p38γ and T205 tau in AD and other neurodegenerative diseases (Australian patent number APA#2016900764). The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Data containing raw immunoblots for experiments summarized in Fig. 1 can be found through Dryad doi.org/10.5061/dryad.bvq83bkc0.
Supplementary Materials
This PDF file includes:
Figs. S1 to S21
Tables S1 to S6
References
REFERENCES AND NOTES
- 1.Scheltens P., Blennow K., Breteler M. M. B., de Strooper B., Frisoni G. B., Salloway S., van der Flier W. M., Alzheimer’s disease. Lancet 388, 505–517 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D. J., Hardy J., The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 8, 595–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busche M. A., Hyman B. T., Synergy between amyloid-β and tau in Alzheimer’s disease. Nat. Neurosci. 23, 1183–1193 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Chang C. W., Shao E., Mucke L., Tau: Enabler of diverse brain disorders and target of rapidly evolving therapeutic strategies. Science 371, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Mandelkow E., Tau in physiology and pathology. Nat. Rev. Neurosci. 17, 5–21 (2016). [DOI] [PubMed] [Google Scholar]
- 6.La Joie R., Visani A. V., Baker S. L., Brown J. A., Bourakova V., Cha J., Chaudhary K., Edwards L., Iaccarino L., Janabi M., Lesman-Segev O. H., Miller Z. A., Perry D. C., O’Neil J. P., Pham J., Rojas J. C., Rosen H. J., Seeley W. W., Tsai R. M., Miller B. L., Jagust W. J., Rabinovici G. D., Prospective longitudinal atrophy in Alzheimer’s disease correlates with the intensity and topography of baseline tau-PET. Sci. Transl. Med. 12, eaau5732 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noble W., Hanger D. P., Miller C. C., Lovestone S., The importance of tau phosphorylation for neurodegenerative diseases. Front. Neurol. 4, 83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ittner A., Ittner L. M., Dendritic tau in Alzheimer’s disease. Neuron 99, 13–27 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Neddens J., Temmel M., Flunkert S., Kerschbaumer B., Hoeller C., Loeffler T., Niederkofler V., Daum G., Attems J., Hutter-Paier B., Phosphorylation of different tau sites during progression of Alzheimer’s disease. Acta Neuropathol. Commun. 6, 52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaherty D. B., Soria J. P., Tomasiewicz H. G., Wood J. G., Phosphorylation of human tau protein by microtubule-associated kinases: GSK3β and cdk5 are key participants. J. Neurosci. Res. 62, 463–472 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Qi H., Prabakaran S., Cantrelle F. X., Chambraud B., Gunawardena J., Lippens G., Landrieu I., Characterization of neuronal tau protein as a target of extracellular signal-regulated kinase. J. Biol. Chem. 291, 7742–7753 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Despres C., Byrne C., Qi H., Cantrelle F. X., Huvent I., Chambraud B., Baulieu E. E., Jacquot Y., Landrieu I., Lippens G., Smet-Nocca C., Identification of the Tau phosphorylation pattern that drives its aggregation. Proc. Natl. Acad. Sci. U.S.A. 114, 9080–9085 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ittner A., Asih P. R., Tan A. R. P., Prikas E., Bertz J., Stefanoska K., Lin Y., Volkerling A. M., Ke Y. D., Delerue F., Ittner L. M., Reduction of advanced tau-mediated memory deficits by the MAP kinase p38γ. Acta Neuropathol. 140, 279–294 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Wang X., Liu Q., Li X. G., Zhou Q. Z., Wu D. Q., Li S. H., Liu Y. C., Wang J. Z., T217-phosphorylation exacerbates tau pathologies and tau-induced cognitive impairment. J. Alzheimers Dis. 81, 1403–1418 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Morris M., Knudsen G. M., Maeda S., Trinidad J. C., Ioanoviciu A., Burlingame A. L., Mucke L., Tau post-translational modifications in wild-type and human amyloid precursor protein transgenic mice. Nat. Neurosci. 18, 1183–1189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ittner A., Chua S. W., Bertz J., Volkerling A., van der Hoven J., Gladbach A., Przybyla M., Bi M., van Hummel A., Stevens C. H., Ippati S., Suh L. S., Macmillan A., Sutherland G., Kril J. J., Silva A. P. G., Mackay J. P., Poljak A., Delerue F., Ke Y. D., Ittner L. M., Site-specific phosphorylation of tau inhibits amyloid-β toxicity in Alzheimer’s mice. Science 354, 904–908 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Hoover B. R., Reed M. N., Su J., Penrod R. D., Kotilinek L. A., Grant M. K., Pitstick R., Carlson G. A., Lanier L. M., Yuan L. L., Ashe K. H., Liao D., Tau mislocalization to dendritic spines mediates synaptic dysfunction independently of neurodegeneration. Neuron 68, 1067–1081 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stepanov A., Karelina T., Markevich N., Demin O., Nicholas T., A mathematical model of multisite phosphorylation of tau protein. PLOS ONE 13, e0192519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomson M., Gunawardena J., Unlimited multistability in multisite phosphorylation systems. Nature 460, 274–277 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guillonneau M., Paris F., Dutoit S., Estephan H., Bénéteau E., Huot J., Corre I., Oxidative stress disassembles the p38/NPM/PP2A complex, which leads to modulation of nucleophosmin-mediated signaling to DNA damage response. FASEB J. 30, 2899–2914 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Leslie S. N., Nairn A. C., cAMP regulation of protein phosphatases PP1 and PP2A in brain. Biochim. Biophys. Acta Mol. Cell Res. 1866, 64–73 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D., Kanthasamy A., Yang Y., Anantharam V., Kanthasamy A., Protein kinase Cδ negatively regulates tyrosine hydroxylase activity and dopamine synthesis by enhancing protein phosphatase-2A activity in dopaminergic neurons. J. Neurosci. 27, 5349–5362 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blennow K., Hampel H., Weiner M., Zetterberg H., Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat. Rev. Neurol. 6, 131–144 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Barthelemy N. R., Li Y., Joseph-Mathurin N., Gordon B. A., Hassenstab J., Benzinger T. L. S., Buckles V., Fagan A. M., Perrin R. J., Goate A. M., Morris J. C., Karch C. M., Xiong C., Allegri R., Mendez P. C., Berman S. B., Ikeuchi T., Mori H., Shimada H., Shoji M., Suzuki K., Noble J., Farlow M., Chhatwal J., Graff-Radford N. R., Salloway S., Schofield P. R., Masters C. L., Martins R. N., O’Connor A., Fox N. C., Levin J., Jucker M., Gabelle A., Lehmann S., Sato C., Bateman R. J., Dade E. M.; Dominantly Inherited Alzheimer Network , A soluble phosphorylated tau signature links tau, amyloid and the evolution of stages of dominantly inherited Alzheimer’s disease. Nat. Med. 26, 398–407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grueninger F., Bohrmann B., Czech C., Ballard T. M., Frey J. R., Weidensteiner C., von Kienlin M., Ozmen L., Phosphorylation of Tau at S422 is enhanced by Aβ in TauPS2APP triple transgenic mice. Neurobiol. Dis. 37, 294–306 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Braak E., Braak H., Mandelkow E. M., A sequence of cytoskeleton changes related to the formation of neurofibrillary tangles and neuropil threads. Acta Neuropathol. 87, 554–567 (1994). [DOI] [PubMed] [Google Scholar]
- 27.Goedert M., Jakes R., Vanmechelen E., Monoclonal antibody AT8 recognises tau protein phosphorylated at both serine 202 and threonine 205. Neurosci. Lett. 189, 167–170 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Clinton L. K., Blurton-Jones M., Myczek K., Trojanowski J. Q., LaFerla F. M., Synergistic interactions between Aβ, tau, and α-synuclein: Acceleration of neuropathology and cognitive decline. J. Neurosci. 30, 7281–7289 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trushina N. I., Bakota L., Mulkidjanian A. Y., Brandt R., The evolution of tau phosphorylation and interactions. Front. Aging Neurosci. 11, 256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otth C., Concha I. I., Arendt T., Stieler J., Schliebs R., González-Billault C., Maccioni R. B., AβPP induces cdk5-dependent tau hyperphosphorylation in transgenic mice Tg2576. J. Alzheimers Dis. 4, 417–430 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Regan P., Piers T., Yi J. H., Kim D. H., Huh S., Park S. J., Ryu J. H., Whitcomb D. J., Cho K., Tau phosphorylation at serine 396 residue is required for hippocampal LTD. J. Neurosci. 35, 4804–4812 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stefanoska K., Volkerling A., Bertz J., Poljak A., Ke Y. D., Ittner L. M., Ittner A., An N-terminal motif unique to primate tau enables differential protein-protein interactions. J. Biol. Chem. 293, 3710–3719 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ittner L. M., Ke Y. D., Delerue F., Bi M., Gladbach A., van Eersel J., Wölfing H., Chieng B. C., Christie M. D. J., Napier I. A., Eckert A., Staufenbiel M., Hardeman E., Götz J., Dendritic function of tau mediates amyloid-q toxicity in Alzheimer’s disease mouse models. Cell 142, 387–397 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Roberson E. D., Scearce-Levie K., Palop J. J., Yan F., Cheng I. H., Wu T., Gerstein H., Yu G. Q., Mucke L., Reducing endogenous tau ameliorates amyloid β-induced deficits in an Alzheimer’s disease mouse model. Science 316, 750–754 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Asih P. R., Prikas E., Stefanoska K., Tan A. R. P., Ahel H. I., Ittner A., Functions of p38 MAP kinases in the central nervous system. Front. Mol. Neurosci. 13, 570586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stefanoska K., Bertz J., Volkerling A. M., van der Hoven J., Ittner L. M., Ittner A., Neuronal MAP kinase p38α inhibits c-Jun N-terminal kinase to modulate anxiety-related behaviour. Sci. Rep. 8, 14296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hooper C., Killick R., Lovestone S., The GSK3 hypothesis of Alzheimer’s disease. J. Neurochem. 104, 1433–1439 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patrick G. N., Zukerberg L., Nikolic M., de la Monte S., Dikkes P., Tsai L. H., Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature 402, 615–622 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Seo J., Kritskiy O., Watson L. A., Barker S. J., Dey D., Raja W. K., Lin Y. T., Ko T., Cho S., Penney J., Silva M. C., Sheridan S. D., Lucente D., Gusella J. F., Dickerson B. C., Haggarty S. J., Tsai L. H., Inhibition of p25/Cdk5 attenuates tauopathy in mouse and iPSC models of frontotemporal dementia. J. Neurosci. 37, 9917–9924 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho J. H., Johnson G. V., Glycogen synthase kinase 3β phosphorylates tau at both primed and unprimed sites: Differential impact on microtubule binding. J. Biol. Chem. 278, 187–193 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Lee V. M., Brunden K. R., Hutton M., Trojanowski J. Q., Developing therapeutic approaches to tau, selected kinases, and related neuronal protein targets. Cold Spring Harb. Perspect. Med. 1, a006437 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Domise M., Didier S., Marinangeli C., Zhao H., Chandakkar P., Buée L., Viollet B., Davies P., Marambaud P., Vingtdeux V., AMP-activated protein kinase modulates tau phosphorylation and tau pathology in vivo. Sci. Rep. 6, 26758 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hao B., Oehlmann S., Sowa M. E., Harper J. W., Pavletich N. P., Structure of a Fbw7-Skp1-cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26, 131–143 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Koivomagi M., Valk E., Venta R., Iofik A., Lepiku M., Balog E. R. M., Rubin S. M., Morgan D. O., Loog M., Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature 480, 128–131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koivomagi M., Örd M., Iofik A., Valk E., Venta R., Faustova I., Kivi R., Balog E. R. M., Rubin S. M., Loog M., Multisite phosphorylation networks as signal processors for Cdk1. Nat. Struct. Mol. Biol. 20, 1415–1424 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knight J. D., Tian R., Lee R. E. C., Wang F., Beauvais A., Zou H., Megeney L. A., Gingras A. C., Pawson T., Figeys D., Kothary R., A novel whole-cell lysate kinase assay identifies substrates of the p38 MAPK in differentiating myoblasts. Skelet. Muscle 2, 5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheridan D. L., Kong Y., Parker S. A., Dalby K. N., Turk B. E., Substrate discrimination among mitogen-activated protein kinases through distinct docking sequence motifs. J. Biol. Chem. 283, 19511–19520 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang Y., Li Z., Schwarz E. M., Lin A., Guan K., Ulevitch R. J., Han J., Structure-function studies of p38 mitogen-activated protein kinase: Loop 12 influences substrate specificity and autophosphorylation, but not upstream kinase selection. J. Biol. Chem. 272, 11096–11102 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Cuenda A., Cohen P., Buee-Scherrer V., Goedert M., Activation of stress-activated protein kinase-3 (SAPK3) by cytokines and cellular stresses is mediated via SAPKK3 (MKK6); comparison of the specificities of SAPK3 and SAPK2 (RK/p38). EMBO J. 16, 295–305 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertrand J., Plouffe V., Senechal P., Leclerc N., The pattern of human tau phosphorylation is the result of priming and feedback events in primary hippocampal neurons. Neuroscience 168, 323–334 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Ando K., Leroy K., Héraud C., Yilmaz Z., Authelet M., Suain V., de Decker R., Brion J. P., Accelerated human mutant tau aggregation by knocking out murine tau in a transgenic mouse model. Am. J. Pathol. 178, 803–816 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Irwin D. J., Lee V. M., Trojanowski J. Q., Parkinson’s disease dementia: Convergence of α-synuclein, tau and amyloid-β pathologies. Nat. Rev. Neurosci. 14, 626–636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ittner A., Block H., Reichel C. A., Varjosalo M., Gehart H., Sumara G., Gstaiger M., Krombach F., Zarbock A., Ricci R., Regulation of PTEN activity by p38δ-PKD1 signaling in neutrophils confers inflammatory responses in the lung. J. Exp. Med. 209, 2229–2246 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Avitzour M., Diskin R., Raboy B., Askari N., Engelberg D., Livnah O., Intrinsically active variants of all human p38 isoforms. FEBS J. 274, 963–975 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Asih P. R., Stefanoska K., Prikas E., Ittner A., High level forebrain expression of active tau kinase p38γ exacerbates cognitive dysfunction in aged APP-transgenic Alzheimer’s mice. Neuroscience 484, 53–65 (2022). [DOI] [PubMed] [Google Scholar]
- 56.Chan K. Y., Jang M. J., Yoo B. B., Greenbaum A., Ravi N., Wu W. L., Sánchez-Guardado L., Lois C., Mazmanian S. K., Deverman B. E., Gradinaru V., Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aurnhammer C., Haase M., Muether N., Hausl M., Rauschhuber C., Huber I., Nitschko H., Busch U., Sing A., Ehrhardt A., Baiker A., Universal real-time PCR for the detection and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum. Gene Ther. Methods 23, 18–28 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Genovese C. R., Lazar N. A., Nichols T., Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage 15, 870–878 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Ignatiadis N., Klaus B., Zaugg J. B., Huber W., Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods 13, 577–580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Storey J., The positive false discovery rate: A Bayesian interpretation and the q-value. Ann. Stat. 31, 2013–2035 (2003). [Google Scholar]
- 61.Watson J. B., Szijan I., Coulter P. M. II, Localization of RC3 (neurogranin) in rat brain subcellular fractions. Brain Res. Mol. Brain Res. 27, 323–328 (1994). [DOI] [PubMed] [Google Scholar]
- 62.Lundgren D. H., Hwang S. I., Wu L., Han D. K., Role of spectral counting in quantitative proteomics. Expert Rev. Proteomics 7, 39–53 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Sumara G., Formentini I., Collins S., Sumara I., Windak R., Bodenmiller B., Ramracheya R., Caille D., Jiang H., Platt K. A., Meda P., Aebersold R., Rorsman P., Ricci R., Regulation of PKD by the MAPK p38δ in insulin secretion and glucose homeostasis. Cell 136, 235–248 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heinrichsdorff J., Luedde T., Perdiguero E., Nebreda A. R., Pasparakis M., p38α MAPK inhibits JNK activation and collaborates with IκB kinase 2 to prevent endotoxin-induced liver failure. EMBO Rep. 9, 1048–1054 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ercan E., Eid S., Weber C., Kowalski A., Bichmann M., Behrendt A., Matthes F., Krauss S., Reinhardt P., Fulle S., Ehrnhoefer D. E., A validated antibody panel for the characterization of tau post-translational modifications. Mol. Neurodegener. 12, 87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao L., Liang Y., Liu Y., Xu Y., Wan W., Zhu C., Pseudo-phosphorylation at AT8 epitopes regulates the tau truncation at aspartate 421. Exp. Cell Res. 370, 103–115 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Martorell A. J., Paulson A. L., Suk H.-J., Abdurrob F., Drummond G. T., Guan W., Young J. Z., Kim D. N.-W., Kritskiy O., Barker S. J., Mangena V., Prince S. M., Brown E. N., Chung K., Boyden E. S., Singer A. C., Tsai L.-H., Multi-sensory gamma stimulation ameliorates Alzheimer’s-associated pathology and improves cognition. Cell 177, 256–271.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qi B., Yang Y., Cheng Y., Sun D., Wang X., Khanna R., Ju W., Nasal delivery of a CRMP2-derived CBD3 adenovirus improves cognitive function and pathology in APP/PS1 transgenic mice. Mol. Brain 13, 58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seo J. S., Lee S., Shin J. Y., Hwang Y. J., Cho H., Yoo S. K., Kim Y., Lim S., Kim Y. K., Hwang E. M., Kim S. H., Kim C. H., Hyeon S. J., Yun J. Y., Kim J., Kim Y., Alvarez V. E., Stein T. D., Lee J., Kim D. J., Kim J. I., Kowall N. W., Ryu H., McKee A. C., Transcriptome analyses of chronic traumatic encephalopathy show alterations in protein phosphatase expression associated with tauopathy. Exp. Mol. Med. 49, e333 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Foidl B. M., Humpel C., Differential hyperphosphorylation of Tau-S199, -T231 and -S396 in organotypic brain slices of Alzheimer mice. A model to study early tau hyperphosphorylation using okadaic acid. Front. Aging Neurosci. 10, 113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moosecker S., Gomes P., Dioli C., Yu S., Sotiropoulos I., Almeida O. F. X., Activated PPARγ abrogates misprocessing of amyloid precursor protein, tau missorting and synaptotoxicity. Front. Cell Neurosci. 13, 239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ittner A., Bertz J., Suh L. S., Stevens C. H., Götz J., Ittner L. M., Tau-targeting passive immunization modulates aspects of pathology in tau transgenic mice. J. Neurochem. 132, 135–145 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S21
Tables S1 to S6
References