Abstract
Bacteria commonly form aggregates in a range of coral species [termed coral-associated microbial aggregates (CAMAs)], although these structures remain poorly characterized despite extensive efforts studying the coral microbiome. Here, we comprehensively characterize CAMAs associated with Stylophora pistillata and quantify their cell abundance. Our analysis reveals that multiple Endozoicomonas phylotypes coexist inside a single CAMA. Nanoscale secondary ion mass spectrometry imaging revealed that the Endozoicomonas cells were enriched with phosphorus, with the elemental compositions of CAMAs different from coral tissues and endosymbiotic Symbiodiniaceae, highlighting a role in sequestering and cycling phosphate between coral holobiont partners. Consensus metagenome-assembled genomes of the two dominant Endozoicomonas phylotypes confirmed their metabolic potential for polyphosphate accumulation along with genomic signatures including type VI secretion systems allowing host association. Our findings provide unprecedented insights into Endozoicomonas-dominated CAMAs and the first direct physiological and genomic linked evidence of their biological role in the coral holobiont.
Coral-associated bacterial aggregates may play a role in phosphate sequestration inside corals.
INTRODUCTION
Endosymbiotic microbial aggregates are a common feature within tissues of many animals and often demonstrate tight mutualistically beneficial symbiotic roles. These structures (named bacteriocytes, bacteriomes, trophosomes, “bacterial aggregates,” “cyst-like aggregates,” or “intracellular colonies of bacteria”) have been widely reported in terrestrial eukaryotes such as plants [e.g., (1, 2)], insects [e.g., (3–5)], and freshwater single-celled eukaryotes (6) in addition to many marine phyla such as gutless oligochaetes (7), deep-sea tubeworms (8), sponges (9), sea anemones (10), ascidians (11), mollusks (12, 13), and corals (14). Many of these microbial symbionts live in specific compartments of their host, maintaining an obligate relationship that facilitates holobiont fitness through metabolic interactions, nutrient exchange, and defense mechanisms (1–7, 9, 15, 16). However, for many marine invertebrates such as corals, sea anemones, ascidians, and mollusks, the identity and function of the bacteria within these aggregates are currently poorly understood despite being prevalent.
Numerous 16S ribosomal RNA (rRNA) amplicon-based studies have reported Endozoicomonas (Gammaproteobacteria)–affiliated taxa as the dominant members of the coral microbiome, with diverse phylotypes associated with individual species and across taxonomically dispersed hosts (11, 17–20). Endozoicomonas strains have been isolated from corals, and their metabolic potential was inferred from derived genomic studies on the isolates and metagenomes assembled from culture-independent studies, suggesting integrated metabolic links facilitating nutrient acquisition and provision and therefore potentially important roles in host health (14, 21–24). Some Endozoicomonas have been reported to locate within aggregates of the coral tissues (14) and other marine invertebrates (10, 11, 13), although other bacterial taxa including Rickettsiales (Alphaproteobacteria) and Chlamydia (Chlamydiae) have also been reported to form aggregations in corals (23, 25) and mollusks (26, 27); hence, these coral-associated microbial aggregates (CAMAs) could be constructed with polymorphous types of bacteria (28, 29). However, the spatial localization of these symbionts is still largely overlooked, particularly with respect to individual phylotypes and populations across bacterial lineages. In addition, the linkages between bacterial physiology and metabolic roles within the coral holobiont are unknown.
Here, we provide an unprecedented characterization of CAMAs in the coral Stylophora pistillata, visualizing the in situ distribution of CAMAs in the three-dimensional (3D) space of the coral polyp and estimating the numbers of bacteria in respective CAMAs. Furthermore, we identified and characterized bacterial phylotypes within CAMAs from coral samples from two geographic locations and confirmed their phylogenetic relationships and putative functions through metagenomic approaches. Last, we mapped elemental distribution within the CAMAs and elucidated the putative ecological functions of CAMAs in the coral holobiont (see more details in the workflow of our research in fig. S1).
RESULTS
High-resolution characterization of CAMAs within coral tissues
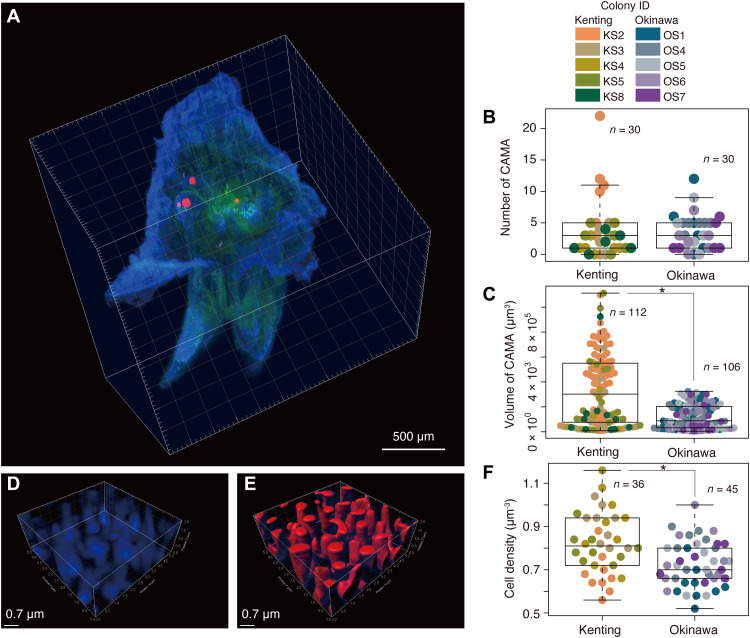
We successfully determined the number and volume of CAMAs in single S. pistillata polyps (n = 6) derived from each of five colonies sampled from Kenting Peninsula, Taiwan, and Okinawa Island, Japan (Fig. 1A and figs. S1A and S2A) by the newly established method in combination with lightsheet microscopic and confocal microscopic observations. A total of 218 CAMAs were characterized with an average of 3.73 ± 4.68 (SD) and 3.53 ± 2.83 per polyp from Kenting and Okinawa samples, respectively (Fig. 1B), with the maximum in one polyp being 22 from a Kenting sample (Fig. 1B) under the lightsheet microscopy. Only a few polyps did not contain CAMAs (five from Kenting and three from Okinawa samples). The average volume of CAMAs (estimated as a minimum by three auto-threshold algorithms; see fig. S2C and Supplementary Methods) was significantly different between Kenting and Okinawa samples being 337,321 ± 287,620 μm3 and 120,641 ± 95,478 μm3, respectively (P < 0.01, Wilcoxon test; Fig. 1C). With observation in the confocal microscopy, we found that the higher CAMA volume was also reflected in the average cell density [0.82 ± 0.14 cells/μm3 in Kenting samples (n = 36) versus 0.72 ± 0.10 cells/μm3 in Okinawa samples (n = 45)] (P < 0.001, t test; Fig. 1, D to F) in addition to the estimated total cells housed in CAMAs within a single polyp being approximately 1.03 × 106 for Kenting and 0.31 × 106 for Okinawa samples.
Fig. 1. Estimation of bacterial populations within CAMAs of single coral polyps.
(A) Representative 3D image using a lightsheet microscope demonstrating the localization of four CAMAs in the tentacles of a single polyp, merged with signals from fluorescence in situ hybridization (FISH) with EUB338mix probe (for all bacteria) labeled with Cy3 (red) and autofluorescence of coral tissue (blue) and Symbiodiniaceae (green) using the lightsheet microscope system. Dot plots showing (B) the number of CAMAs and (C) the volume of each CAMA. For estimation of bacterial cell density in single CAMAs, (D) 4′,6-diamidino-2-phenylindole signals (blue) are shown in a 3D image from a confocal microscope and (E) its surface rendering image (red) processing in Imaris software. (F) Dot plot showing the cell density within CAMAs. Asterisks indicate significant differences between Kenting and Okinawa samples (n = 5 colonies each): P = 2.12 × 10−8, Wilcoxon test (C), and P = 5.12 × 10−4, t test (F), respectively.
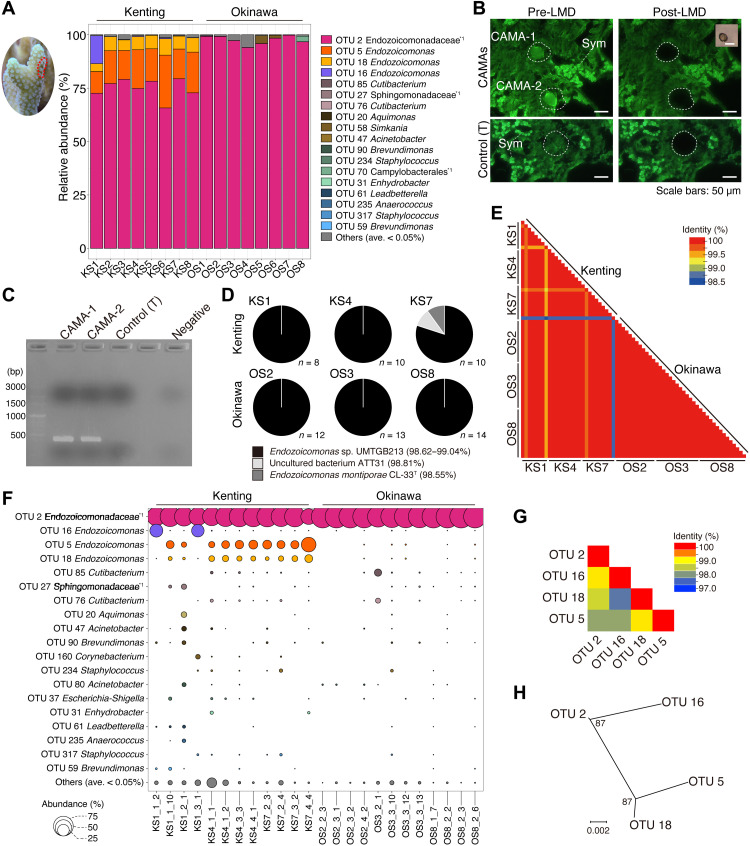
Bacterial community profiling of DNA derived from S. pistillata tissues (n = 8 from each location; V6-V8 regions of the 16S rRNA gene) retrieved 669,773 sequences that were clustered at 99.5% sequence identity into 102 operational taxonomic units (OTUs). Across all samples, sequences affiliated with the Endozoicomonadaceae family were dominant with OTU 2 (assigned to Endozoicomonadaceae unclassified) ranging from 65.8 to 99.8% of the total sequences retrieved from coral tissues from both locations (Fig. 2A). Two other Endozoicomonas OTUs (5 and 18) represented, on average, 16 and 6% of sequences retrieved from Kenting samples, although they were only in low relative abundance (between 0.013 and 0.053%) in Okinawa samples (Fig. 2A). OTU 16 demonstrated high relative abundance (13.1%) in only Kenting colony KS1 (Fig. 2A). The relative abundance of other taxa was low across all samples with Simkania (OTU 58, 5.1%) and Campylobacterales unclassified (OTU 70, 2.9%) being most noteworthy.
Fig. 2. Bacterial community patterns of CAMAs within the tissues of the coral S. pistillata.
(A) Bar plots showing the relative abundance of 16S rRNA reads from coral tissues sampled from Kenting (KS) and Okinawa (OS) (note the example tissue region sampled shown in coral image surrounded by the red dotted line). (B) Representative images of pre- and post-LMD collections of individual CAMAs (top) and control coral tissues without CAMAs (bottom) closely located with Symbiodiniaceae (Sym). (C) Representative image of electrophoresis showing 16S rRNA amplified gene bands (V6-V8 regions, expected size: 424 bp), from CAMA-1 and CAMA-2 and control [T (coral tissue)]. (D) Pie charts showing identified 16S rRNA gene bacterial hits (V6-V8 regions) from BLAST searches derived from 67 individual CAMAs (obtained through LMD) and sourced from three colonies each from Kenting and Okinawa sampling sites using direct Sanger sequencing. (E) Heatmap demonstrating similarities among sequences retrieved from 67 individual CAMAs derived from Kenting and Okinawa samples. (F) Bubble chart showing the bacterial composition of four individual CAMAs randomly selected from three Kenting and Okinawa samples and subjected to direct Sanger sequencing. (G) Heatmap of similarities and (H) phylogenetic tree representing genomic differences of four dominant OTUs belonging to Endozoicomonadaceae.
Laser microdissection (LMD) selectively excised 67 individual CAMAs (area from 516.5 to 6424.5 μm2; average, 2053.1 μm2) from three colonies in each location and was successfully combined with 16S rRNA gene (V6-V8 regions) sequencing of extracted DNA to profile the CAMA-specific microbial community (Fig. 2, B and C). Regions of coral tissue with no CAMAs were similarly processed with no amplified polymerase chain reaction (PCR) signal obtained (Fig. 2C). DNAs derived from the individual CAMAs were of sufficient quality to perform direct Sanger sequencing. Sequences were obtained from 65 of 67 individual CAMAs affiliated to Endozoicomonas sp. UMTGB213 [BLAST sequence identity, 98.6 to 99.0% over 415 to 437 base pairs (bp); GenBank accession no. MG896199 originating from marine tunicates; Fig. 2D]. Two individual CAMAs from Kenting colony KS7 showed the highest sequence identity to an uncultured bacterium ATT31 and Endozoicomonas montiporae CL-33T retrieved from Acropora and Montipora coral samples (BLAST sequence identity, 98.8 and 98.6% over 419 and 415 bp; GenBank accession nos. FJ809552 and CP013251, respectively). All sequences from the individual CAMAs demonstrated ≥98.5% sequence identity to each other (Fig. 2E).
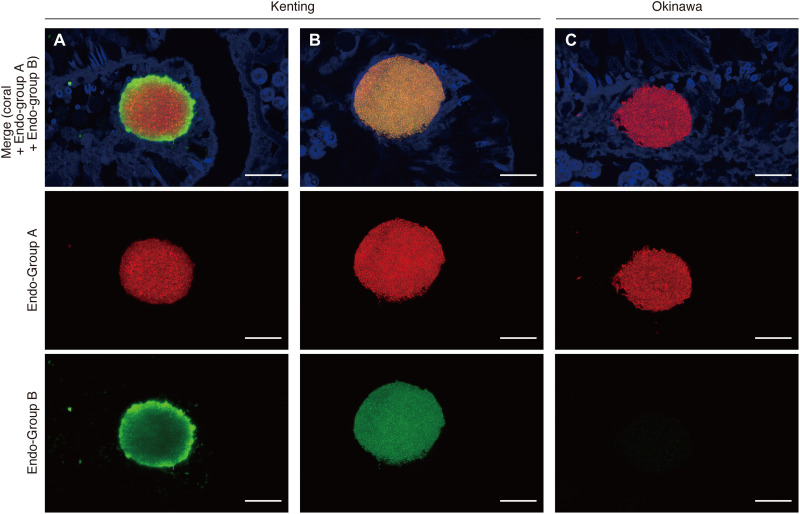
We also used 16S rRNA metabarcoding analysis (V6-V8 regions) to profile the bacterial communities of 24 individual CAMAs (a subset of the 67 sampled CAMAs detailed previously), obtaining 492,018 bacterial reads that clustered (99.5%) into 151 OTUs (table S1). OTU 2 (Endozoicomonadaceae unclassified) was again dominant, representing 71.7 and 97.7% of sequences retrieved from CAMAs derived from each of three Kenting and Okinawa colonies, respectively (Fig. 2F). All individual CAMAs from Okinawa samples were monopolized by OTU 2 (Fig. 2F). For Kenting samples, OTU 2 coexisted with Endozoicomonas OTUs 5 and 18 (average relative abundances of 11.6 and 4.1% respectively), although in two individual CAMAs on the colony KS1 (see KS1_1_2 and KS1_3_1 in Fig. 2F), OTUs 2 and 16 coexisted at the exclusion of OTUs 5 and 18. OTUs 5 and 18 were never retrieved from the same aggregation as OTU 16, and this pattern was also consistent with bacterial profiles generated from the bulk tissue analysis (Fig. 2, A and F). The four Endozoicomonadaceae OTUs showed high sequence identity (≥97.7%) (Fig. 2G) and were grouped into two phylogenetic groups (OTUs 2 and 16 and OTUs 5 and 18; Fig. 2H), indicating that different phylotypes are in the same CAMA. Fluorescent in situ hybridization (FISH) confirmed sequencing patterns, with two 16S rRNA gene probes (Endo-Group A targeting OTUs 2 and 16 and Endo-Group B targeting OTUs 5 and 18; details in Supplementary Methods) demonstrating visible individual bacterial cells within CAMAs, although only Endo-Group A probe hybridized to aggregates of the Okinawa samples (Fig. 3 and fig. S3). In Kenting samples, binding and fluorescent signals from both probes were detected and colocalized in single aggregations, although the patterns of probe hybridization were different in different CAMAs (Fig. 3). For example, strong Endo-group B probe binding is observed on the periphery of some CAMAs, while in others, hybridization of both probes is evenly distributed across the aggregation, confirming that phylotypes are colocated in the same CAMA. Other bacterial 16S rRNA gene sequences were also retrieved from the individual CAMAs, although their average relative abundance was <0.5% of reads and were affiliated to Cutibacterium (OTUs 76 and 85), Sphingomonadaceae unclassified (OTU 27), Aquimonas (OTU 20), and Acinetobacter (OTU 47). Because of low relative abundance, it is unsure whether these reads are derived from taxa that are colocated within the CAMAs or are contaminant reads that are known to appear in samples that have low concentration of extracted DNA (30).
Fig. 3. Visualization of Endozoicomonas phylotypes within individual CAMAs in the tissues of S. pistillata.
Representative confocal images depicting the hybridization of two Endozoicomonadaceae-specific probes (Endo-Group A probe targeting OTUs 2 and 16 and Endo-Group B probe targeting OTUs 5 and 18) labeled with Cy3 (red) and Cy5 (green), respectively. Two different patterns are observed with the Endo-group B probe binding prominent in the CAMA periphery (A): Homogeneous binding of both Endo-group B and Endo-group A is observed in other CAMAs (B) in Kenting samples, while the Endo-Group A probe is exclusively visible in Okinawa samples (C). Scale bars, 20 μm.
Functional signatures of Endozoicomonas taxa within CAMAs
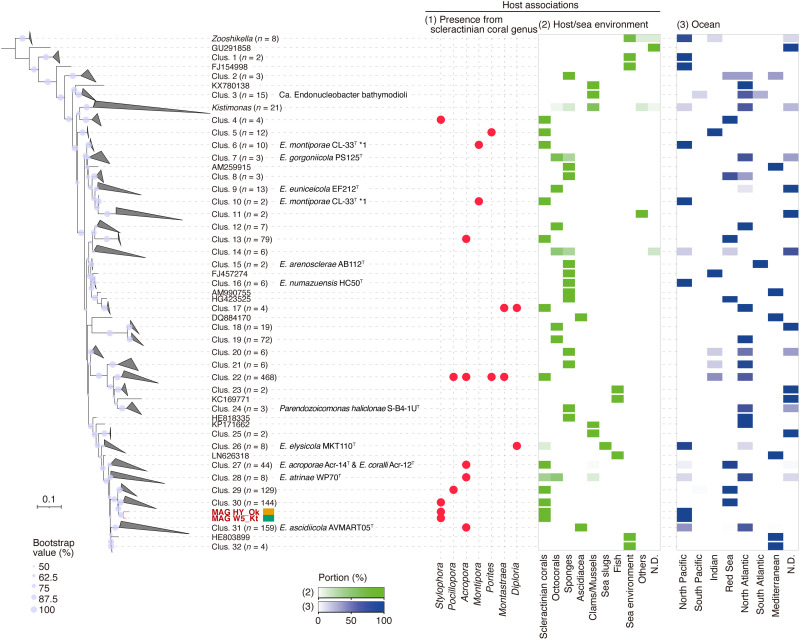
Two metagenome-assembled genomes (MAGs) of Endozoicomonadaceae (W5_Kt and HY_Ok; Supplementary Methods) generated from pooled DNA derived from individual CAMAs cut by LMD (n = 84 from Kenting and n = 57 from Okinawa) and whole genome–amplified DNA were estimated to be 3.43 and 3.93 Mbp in size, 89 and 91% complete with 1.8 and 1.9% contamination, respectively (see more details in table S2). Full-length 16S rRNA genes from each MAG confirmed high sequence identity to each other (98.5%) and the four Endozoicomonadaceae OTUs derived from bacterial community analysis (identities ranged from 98.18 to 99.55%; table S3). We generated the maximum likelihood (ML) phylogenetic tree with our MAG sequences and 1289 near-complete Endozoicomonadaceae 16S rRNA sequences (>1400 bp) from the SILVA SSU r138 database (31) and included representatives of the genus Endozoicomonas, Kistimonas, and Zooshikella. Both MAG-generated 16S sequences were most closely related to sequences (n = 144) within cluster 30 that included Endozoicomonas taxa derived from S. pistillata corals of the Red Sea (Fig. 4). The sequences in the other monophylogenetic branching cluster (Clus. 29; Fig. 4) were derived from Pocillopora damicornis from the Red Sea (n = 127 sequences) and the Great Barrier Reef (n = 2 sequences), which is based on host phylogenetics and is closely related to S. pistillata (32). Other closely placed clusters (i.e., clusters 27 to 32) were characterized by sequences, which included type strains of Endozoicomonas acroporae Acr-14T, Endozoicomonas coralli Acr-12T, Endozoicomonas atrinae WP70T, and Endozoicomonas ascidiicola AVMART05T retrieved from marine invertebrates sampled across different oceans and including the coral genus Acropora (n = 43 of 44 in Clus. 27, n = 3 of 8 in Clus. 28, and n = 1 of 159 in Clus. 31) (Fig. 4). We also calculated the in silico genome-genome distances among the available Endozoicomonadaceae genomes using average nucleotide identity (ANI), and our two MAGs showed closest genetic relatedness with the type strains E. acroporae Acr-14T, E. atrinae WP70T, and E. ascidiicola AVMART05T (ANI values ranged from 72.85 to 75.49%) and other Endozoicomonas genomes Endozoicomonas elysicola MKT110T and DSM22380 and Endozoicomonas sp. G2-1 (ANI values ranging from 73.93 to 75.20%). However, our two MAGs (ANI value 99.2% to each other) were still clearly distinguished from the other genomes (fig. S4).
Fig. 4. ML phylogenetic tree illustrating the placement of the 16S rRNA gene sequences from two bacterial MAGs (W5_Kt and HY_Ok) derived from S. pistillata CAMAs among the family Endozoicomonadaceae.
The dataset included 1289 bacterial 16S rRNA gene sequences reference sequences (>1400 bp) from the SILVA database. The tree is rooted to the genus Zooshikella. Bootstrap values greater than 50% are shown at the nodes (based on 1000 replications). Dot plot and heatmaps showing the relative abundance of sequences based on the following categories: (1) derived from scleractinian corals, (2) derived from marine hosts or seawater environments, and (3) derived from ocean geographic locations. In the category of host/sea environment, others are included green algae, marine annelid, marine nemertean, starfish, and zoanthid. ND, no data.
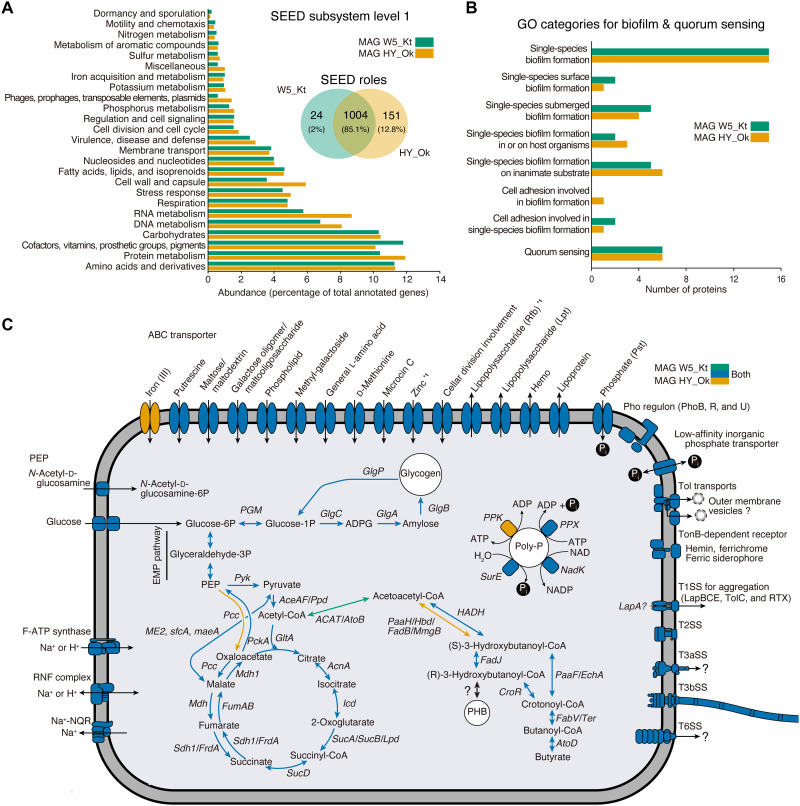
The functional features of our two Endozoicomonas MAGs (W5_Kt 3,181 CDS and HY_Ok 3,561 CDS), classified using the SEED genome annotation subsystems (at level 1), were both dominated by an abundance of amino acids and derivatives (11.3%), protein metabolism (11.1%), cofactors, vitamins, prosthetic groups, pigments (11.0%), and carbohydrates (10.3%), although only ~85.1% of the SEED role categories were overlapped across the two MAGs (Fig. 5A). The MAGs were interrogated for genomic signatures related to biofilm formation and quorum sensing, which potentially facilitate aggregation formation in coral tissues, and on the basis of Gene Ontology (GO) categories, 37 predicted proteins in each MAG were identified, including single-species biofilm formation (GO:0044010, 15 proteins each) and quorum sensing (GO:0009372, six proteins each) (Fig. 5B and table S4). Furthermore, we screened for eukaryotic repeat domains that may facilitate host-symbiont interactions and identified 26 and 13 predicted proteins encoding ankyrin and tetratricopeptide repeats, respectively, in our MAGs (table S5).
Fig. 5. Genomic features of two MAGs derived from CAMAs.
(A) Bar plot and Venn diagram representing the percentage of the two MAGs (W5_Kt and HY_Ok) annotated to the SEED category level 1 and both number and percentage of proteins shared or specific for the MAGs based on SEED category role, respectively. (B) Bar plot depicting the number of the proteins related to biofilm formations and quorum sensing identified in the MAGs and annotated in the GO database. (C) Schematic representation of inferred metabolic capabilities of the MAGs annotated to KEGG orthology. Colors (see top right side) indicate the presence of the enzyme from only W5_Kt (green), only HY_Ok (brown), or both (blue). *1 indicates that only one block is missing in ABC transporters (zinc in W5_Kt and Rfb in HY_Ok). ATP, adenosine triphosphate.
Several secretory systems were nearly complete components, including the general Tat (twin-arginine protein translocation) and type I-III and VI secretion systems (SSs) (table S6 and fig. S5). In T3SS, it was estimated that approximately 40% of effectors were enzymes based on the annotation with the SEED category (table S7). T6SS was positioned within a gene cluster and on a single contig of the HY_OK MAGs [based on rapid annotations using subsystems technology (RAST) and the domain search]; and unexpectedly, each locus contained a Tat pathway signal (with nondomain) next to the VGR-related protein (fig. S5). To comprehensively identify the array of the T6SS gene cluster in the two MAGs, we analyzed 18 available genomes from Endozoicomonadaceae, which included six representatives obtained from corals. The presented ortholog [based on Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology] phylogenetic positions were analyzed with close relative sequences from the National Center for Biotechnology Information (NCBI) nr database. While the T6SS were absent in some genomes derived from marine organisms such as corals, ascidians, and sea slug, it was distributed in Endozoicomonas genomes retrieved from sponges [Endozoicomonas arenosclerae (AB112 and E-MC227), Endozoicomonas numazuensis HC50, Parendozoicomonas haliclonae (ERS1628946), moss (Endozoicomonas sp. AB1-5), mollusk (E. atrinae WP70), starfish (Kistimonas asteriae KMD001), and marine sediment (Endozoicomonas sp. SM1973); fig. S6A]. The presence or absence of T6SS gene signatures among the genomes appeared host specific (fig. S6A) except for our two MAGs, which were unique in displaying the T6SS gene cluster in the coral-associated Endozoicomonas genomes. In addition, the phylogeny based on TssB and TssC orthologs (form the tail sheath) in T6SS showed the phylogenetic independence of bacterial taxonomy and host association and likely shared the ancestor with the families Oceanospirillaceae, Vibrionaceae, and several families from the order Alteromonadales (fig. S6B). The TssB and TssC orthologs in our two MAGs were closest to E. atrinae WP70 (fig. S6B), supporting the 16S rRNA gene phylogeny (Fig. 4).
On the basis of the KEGG orthology, both MAGs displayed nearly all genes required for the tricarboxylic acid cycle, glycolysis mediated by glucose transporters (PtsG and Crr) in the phosphotransferase system (phosphoenolpyruvate), Ton and Tol system, and adenosine triphosphate synthases (Fig. 5C and table S8). The MAGs also revealed the potential to store organic carbon in the form of polyhydroxybutyrate and phosphorous in the form of polyphosphates (note the lack of PPK gene in W5_Kt bin-genome) (Fig. 5C). We identified three genes (PPX, SurB, and NadK) involved in the utilization of polyphosphates in the MAGs (Fig. 5C). Two phosphonate transport systems were also predicted [high-affinity phosphate transporter PstSCAB (including ABC transporter) under PhoP regulon PhoBRU and the low-affinity system]. In addition to the phosphate ABC transporter, we also identified several ABC transporters for putrescine, maltose/maltodextrin, galactose oligomer/maltooligosaccharide, phospholipid, general l-amino acid, d-methionine, zinc, and lipopolysaccharides (Fig. 5C).
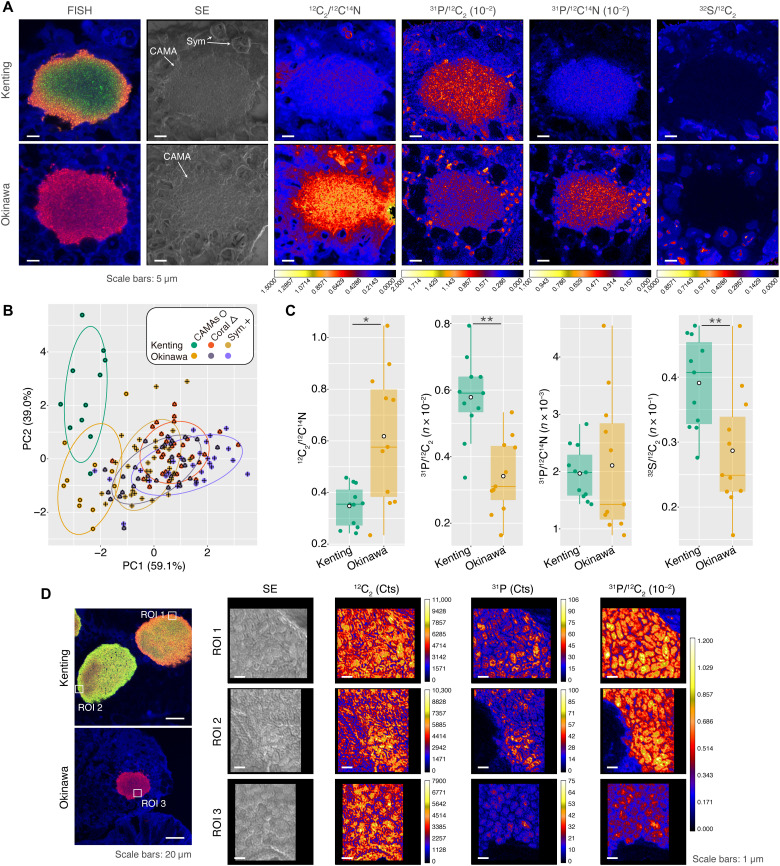
FISH-NanoSIMS (nanoscale secondary ion mass spectrometry) identified the elemental mapping of carbon, nitrogen, phosphorus, and sulfur around and inside CAMAs derived from coral samples from Kenting and Okinawa. As a precaution, those elements from volatile, water- and ethanol-soluble compounds might not be detected due to multiple steps of the sample preparation in this study. The elemental abundance assigned from three regions (CAMAs, coral tissues, and Symbiodiniaceae) (Fig. 6, A to C, and figs. S7 and S8) demonstrated that the C/N, P/C, P/N, and S/C elemental ratios of CAMAs were distinct from that of coral tissues and Symbiodiniaceae [F = 0.293, P = 0.001, two-way permutational multivariate analysis of variance (PERMANOVA); CAMAs against coral tissues and Symbiodiniaceae in the post hoc pairwise test, Padjusted ≤ 0.05 in all combinations; Fig. 6B]. Elemental distributions were also mapped at the single-cell level, demonstrating that phosphorus enrichment was intracellular and heterogeneous among the different cells in the CAMAs, whereas the phosphorus of the coral tissues was at a relatively low concentration (except for enriched phosphorus vesicle-like structures in the coral tissues surrounding the CAMAs) (Fig. 6D and fig. S7). The enrichment in phosphorus indicates a role of CAMAs in sequestering and cycling phosphate between the coral holobiont partners, which supports the genomic evidence of the MAGs in storing and using polyphosphates and transporting phosphonates. In addition, C/N, P/N, and S/C ratios at the single-cell level were variable across the CAMAs (fig. S9). Although we were not able to clarify the differences between Endozoicomonas phylotypes within the individual CAMAs, those results demonstrate that individual bacterial cells modulated divergent elemental inclusions.
Fig. 6. FISH-NanoSIMS profiling of the natural elemental distribution of CAMAs in coral tissues.
In representative FISH-NanoSIMS images, (A) the confocal merge images representing the signals from FISH [hybridized with Endo-Group A probe (red, Cy3) and Endo-Group B probe (green, Cy5)] and coral autofluorescence (blue). Other columns depicting NanoSIMS ion maps of secondary electron (SE) and the ratios with 12C2/12C14N (C/N), 31P/12C2 (P/C), 31P/12C14N (P/N), and 32S/12C2 (S/C). Sym, Symbiodiniaceae. (B) Biplot of principal components (PC) analysis of natural elemental ratios from three regions: CAMAs (n = 11 CAMAs from both Kenting and Okinawa samples), coral tissues (n = 32 regions from Kenting samples and n = 29 regions from Okinawa samples), and Symbiodiniaceae (n = 33 cells from Kenting samples and n = 28 cells from Okinawa samples). Different colors distinguish points from Kenting and Okinawa and sample types. (C) Dot and box plots representing comparisons within CAMA regions among the ratios C/N, P/C, P/N, and S/C from Kenting and Okinawa samples. Asterisk indicates statistically significant differences (*P < 0.05 and **P < 0.01, Wilcoxon signed-rank test). (D) Representative FISH-NanoSIMS confocal images showing two CAMAs (periphery probe binding and homogeneous types) from Kenting and Okinawa samples. Three regions of interest (ROIs; two regions from Kenting and one region from Okinawa) in FISH images corresponding to NanoSIMS maps of SE, single-element measurement (12C2 and 31P), and P/C ratio.
DISCUSSION
This study provides an unprecedented, high-resolution characterization of CAMAs within the polyps of S. pistillata, revealing an average bacterial density of 0.77 cells/μm3 within the structures and an estimated 0.67 × 106 bacterial cells housed within the aggregates in a single polyp. CAMAs were dominated by Endozoicomonas, although individual CAMAs have multiple phylotypes and interrogation of recovered MAGs, linked with cell-level elemental mapping, and indicated a capability of sequestering phosphate and potentially cycling within the coral holobiont. Phosphorus enrichment was higher in CAMAs than in the surrounding coral tissues and Symbiodiniaceae and, at the cell level, could be observed in the intracellular spaces of Endozoicomonas cells within the CAMAs. Coral tissues fluctuate in the levels of oxygen with oxygenic photosynthesis driven by the Symbiodiniaceae creating hyperoxic conditions during the day and night time respiration, resulting in low or even anaerobic conditions (33). Consistent with the phosphorus enrichments at the single-cell level by NanoSIMS imaging, the reconstructed MAGs identified the capability of Endozoicomonas to synthesize polyphosphate via the putative polyphosphate kinase (PPK) protein (with aerobic conditions during daytime) and release the phosphate via the polyphosphatase enzyme PPX (with low oxygen or anaerobic conditions during the night), with associated transporters (high- and low-affinity phosphate transporters) corresponding to phosphate uptake and release. Phosphate enrichment can reduce Symbiodiniaceae photosynthesis, which, in turn, lowers calcification processes and subsequent coral growth [e.g., (34, 35)]. Sequestering of polyphosphate within the CAMAs may therefore represent a buffering mechanism to modulate phosphate internally within the coral tissues, facilitating efficient photosynthesis by the Symbiodiniaceae endosymbionts and optimizing coral growth during daytime. Previous studies have suggested that the frequency of CAMAs in coral tissues could be influenced by dissolved nutrients (29), and therefore, high P levels in surrounding seawater could facilitate the development of CAMAs in the coral tissues. A role for bacteria in phosphate cycling within coral reef marine invertebrate hosts might be common. Zhang et al. (36) detected a group of uncharacterized cyanobacterial symbionts in three sponges that enabled polyphosphate synthesis and suggested that those bacteria play a role in phosphorus sequestration and recycling in the environment.
An intriguing genomic feature of the two recovered MAGs in this study was the near-complete set of T6SS structure genes, in contrast to other genomes of Endozoicomonas retrieved from corals that did not carry either any or only one of the T6SS-like genes, although they are commonly detected in Endozoicomonas species from noncoral hosts. Bacterial T6SS is a common secretion system, detected in approximately 25% of all Gram-negative bacteria (37) and important for virulence traits in pathogenesis (38), bacterial communication (39), cell aggregate formations (40), and biofilm formation (41). Many plant pathogenic bacteria process the T6SS, producing the virulent effectors to eliminate their antagonists and suppress plant defenses, thereby allowing bacterial colonization in host tissues (42). This process resembles the symbiosis establishment of Vibrio fischeri in the marine bobtail squid, in which T6SS is used to eradicate their competitors (43). In the plant, recent evidence demonstrated a beneficial role for the rhizobial T6SS, promoting the formation of root nodules and subsequent plant growth (44). Given these findings in other hosts, the T6SS system in Endozoicomonas may facilitate and maintain CAMA formation in the host coral. The predicted T6SS proteins showed phylogenetic relatedness to members within the families Oceanospirillaceae, Vibrionaceae, and several families from the order Alteromonadales from marine ecosystems, which may have been acquired horizontally. A recent comparative genomic analysis of culturable coral-associated bacteria has revealed that the Vibrionales genomes have a greater number of T6SS-related domains than other bacterial taxa, and pathogenic Vibrio strains in addition have more than nonpathogenic Vibrio strains (20). The specific roles of T6SS-related domains in the Endozoicomonas are not known, although potentially important in establishing their dominance in CAMAs. Whether the Endozoicomonas are parasitic, commensal, or mutualistic is also still to be resolved.
CAMAs derived from two sampling locations (Okinawa, Japan and Kenting, Taiwan) were characterized via 16S rRNA gene sequencing of bulk tissues and laser dissection excision of individual CAMAs (n = 67), with Okinawa samples particularly dominated by one Endozoicomonas phylotype (OTU 2). Kenting samples displayed higher dispersion in Endozoicomonas phylotypes, and although OTU 2 was still the most dominant, three other Endozoicomonas OTUs were characterized, although OTUs 5 and 18 were never retrieved with the OTU 16 phylotype across any of the samples, which may indicate competition exclusion. Closely related organisms that occupy overlapping niches have been documented to compete for resources (45), which can allow for symbiotic bacteria to monopolize a host niche (46, 47). However, why a single phylotype (OTU 2) is found in CAMAs of samples from Okinawa while multiple related Endozoicomonas phylotypes coexist in Kenting samples is still to be elucidated. The Endozoicomonas genus displays high taxonomic diversity being a host generalist, although specific subclades have been identified across coral taxa (48). Previous studies have also identified that S. pistillata harbors geographically distinct genotypes of Endozoicomonas (14), and taxonomic comparisons from our study demonstrated phylotypes detected here clustered into this S. pistillata–specific subclade. These sequences also shared a basal clade with P. damicornis–associated Endozoicomonas, which along with S. pistillata belong in the Pocilloporidae family and are commonly recognized as also having prevalent CAMAs within their tissues (14, 25, 49). These close host-bacterial phylogenetic relationships provide further evidence for patterns of phylosymbiosis across coral species and the potential for cophylogeny (48, 50, 51).
Endosymbiotic microbial aggregates are critical to the biological success of many higher organisms. CAMAs have been widely documented, although understanding their distribution in coral polyps, geographic specificity, genetic divergence, and functions that underpin coral health has remained elusive. This study provides unprecedented insights into these tissue structures and identifies dominant Endozoicomonas phylotypes and their genomic and metabolic signatures that are integrated into the coral holobiont.
MATERIALS AND METHODS
Sample collections
We collected a total of 16 S. pistillata coral colonies from two locations (Kenting, Taiwan, and Okinawa, Japan; fig. S1) in September and October 2017. Three nubbins (approximately 20 mm in length) from each colony were fixed in either modified methacarn (MM) solution (MM samples), 4% paraformaldehyde (PFA samples), or 100% ethanol (EtOH samples). Subsequently, the MM samples and PFA samples were decalcified with 14% EDTA and Morse’s solution, respectively (further details in Supplementary Methods).
Molecular characterization of bacteria associated with coral tissues and individual CAMAs
Coral tissues were removed from EtOH stored samples (n = 8 from each of Kenting and Okinawa samples) using a sterile scalpel blade, and the total genomic DNA was extracted using the DNeasy PowerBiofilm Kit (QIAGEN) according to the manufacturer’s protocol. MM-fixed samples (n = 3 from each of Kenting and Okinawa samples) were embedded in optimal cutting temperature compound after decalcification and sectioned to a thickness of 10 μm in a cryostat Leica CM3050 S (Leica). Intact individual CAMAs that were stained with SYBR gold [0.2% (v/v), Invitrogen] were excised from coral tissues using an LMD microscope system (PALM MicroBeam, Zeiss) in addition to areas of coral tissues without CAMAs as negative controls. The total genomic DNA from individual microdissected CAMAs (n = 67) and coral tissues (n = 29) was extracted using the QIAamp DNA Micro Kit (QIAGEN) according to the manufacturer’s protocol (further details in Supplementary Methods).
DNA extracted from the individual CAMAs were subjected to PCR amplification of the V6-V8 hypervariable region of the 16S rRNA gene using universal primers 968F (52) and 1391R (53), with positive samples confirmed by gel electrophoresis. All control tissue samples (without CAMAs) resulted in no amplified products. Direct Sanger sequencing of amplified products was conducted with resulting sequences manually edited and classified by a BLASTN search against the nr/nt database in NCBI (further details in Supplementary Methods). 16S rRNA gene amplicon sequencing was also performed on DNA derived from individual CAMAs from the samples subjected to the direct Sanger sequencing (n = 4 CAMAs from three colonies sampled from both Kenting and Okinawa; n = 24 total) and genomic DNA derived from the tissue of corals (n = 8 from both Kenting and Okinawa; n = 16 total), again targeting the V6-V8 regions with universal primer pair 968F and 1391R on the Illumina MiSeq platform (Illumina).
Sequences were quality filtered and then clustered into zero-radius OTUs (zOTUs) using USEARCH v11 (54, 55) and MOTHUR v.1.39.5 (56) pipelines (further details in Supplementary Methods). The zOTU sequences were further reclustered into OTUs at a 99.5% similarity level and assigned to known taxonomic groups by mapping to the SILVA SSU r138.1 database (www.arb-silva.de/) using MOTHUR with a cutoff value of 80. Sequences identified as unknown, eukaryote, and chloroplasts were removed. A total of 1,161,791 reads belonging to 187 OTUs were obtained from all samples (the individual samples ranging from 3754 to 49,742). Phylogenetic relationships among individual OTUs were inferred from pairwise comparisons and construction of ML trees using MEGA 7.0.26 (57) with 1000 bootstrap replicates.
DNA derived from individual CAMAs was pooled (between 8 and 48 CAMAs) for subsequent metagenome sequencing. Because of the low DNA concentration derived from individual CAMAs, whole-genome amplification (WGA) was performed using the REPLI-g FFPE Kit (QIAGEN) without the random ligation step. The resulting DNA was a product of 84 individual CAMAs from Kenting and n = 57 individual CAMAs from Okinawa. The WGA libraries were sequenced on the Illumina HiSeq (pair-end 150 bp) platform. Two consensus MAGs from the samples were reconstructed and annotated from the de novo assemblies (see more details in Supplementary Methods). Following the MAG reconstructions, we analyzed phylogeny and functional features that included overall metabolisms, biofilm formation, quorum sensing, predicted metabolic pathway, secretion systems (especially T3SS and T6SS), and protein with eukaryotic repeat domains (further details in Supplementary Methods).
FISH characterization of CAMAs
A total of 60 dissected PFA-fixed S. pistillata polyps (n = 6 polyps from five colonies from Kenting and Okinawa) were processed through a methanol dehydration series before rehydration following the protocol of Liu et al. (58), which facilitates decolorizing and permeabilization of the tissues. QuickHCR-FISH (59) was undertaken using rRNA-targeted oligonucleotide probes [EUB338mix (60), initiatorH adapter for most bacteria and Non338 (61) and initiatorC adapter for a negative control] and corresponding amplifier probes (H-amplifier probes labeled with Cy3 and C-amplifier probes labeled with Alexa Fluor 488). Hybridized polyps were embedded in acrylamide gel and treated with refractive index matching for observations. A Lightsheet Z.1 with ZEN (black edition) software was used for the acquisition of 3D images of whole polyps. We reconstructed the 3D image of the polyp to detect CAMA distribution and their number and volume within the individual polyps using Imaris software 8.0.2 (Bitplane Inc.) and ImageJ 1.53d (further details in Supplementary Methods) (62). Statistical analyses were computed using R v.3.6.1 (63) with the package car v.3.0-11 (64). The comparisons in the number and volume of CAMA from two Kenting and Okinawa were conducted using a Wilcoxon rank-sum test. Differences between cell densities derived from tissues from the two sampling locations were tested using a Student’s t test.
PFA-fixed and paraffin-embedded samples were sectioned to a thickness of 10 μm and subjected to quickHCR-FISH as previously detailed although also stained with 4′,6-diamidino-2-phenylindole (DAPI). The 3D CAMA structures were scanned with a ×63 objective in Z-stack mode (0.1-μm interval) using a confocal laser scanning microscope Zeiss LSM 880 with Airyscan (Zeiss) with Zen SP2.3 software (Zeiss). Image processing allowed quantification of visualized individual cells on DAPI signals using ImageJ 1.53d (62) and Imaris software 8.0.2 (Bitplane Inc.; further details in Supplementary Methods).
Two specific 16S rDNA probes were designed specifically targeting the V6-V8 regions of 16S rRNA genes of OTUs 2 and 16 (Endo-group A) and OTUs 5 and 18 (Endo-group B) using the web-based tool Design Probes in DECIPHER (65). In addition, two competitor probes for each targeted region were designed for eliminating potential nonspecific binding (further details in Supplementary Methods). The paraffin-embedded and PFA-fixed samples were sectioned to a thickness of 5 μm and subjected to quickHCR-FISH (59) with the rRNA-targeted oligonucleotide probes (Endo-Group A, initiatorH and Endo-Group B, initiatorR) with corresponding competitor probes (Comp-Endo-Group A and Comp-Endo-Group B) and amplifier probes (H-amplifier probes labeled with Cy3 and R-amplifier probes labeled with Cy5). The sections were observed using the Zeiss LSM 880 (Zeiss) confocal laser scanning microscope (further details in Supplementary Methods).
NanoSIMS imaging of CAMAs and surrounding coral tissues
To reveal the elemental distributions of CAMA, Symbiodiniaceae, and coral tissue, we applied a FISH-NanoSIMS technique. Two serial sections of one colony (KS2 and OS6) were sectioned to a thickness of 5 μm. For one serial section, quickHCR-FISH (59) with the specific probes was performed according to the methods detailed previously and images acquired by the confocal microscopy. Another serial section was mounted onto indium tin oxide glass coated with poly-l-lysine solution (Sigma-Aldrich), dewaxed by xylene, and air-dried overnight for subsequent NanoSIMS imaging. The elemental distribution of carbon, nitrogen, phosphorus, and sulfur of CAMAs within coral tissue, regions of the coral tissue, and Symbiodiniaceae cells was mapped using a NanoSIMS 50L (Cameca-Ametek, Gennevilliers; further details in Supplementary Methods).
Principal components analysis was performed on the elemental ratios (12C2−/12C14N−, 31P−/12C2−, 31P−/12C14N−, and 32S−/12C2−) detected from individual NanoSIMS images using the function “prcomp” in statistical software R v.4.0.1 (63) and evaluated by two-way PERMANOVA with 999 permutations based on Bray-Curtis dissimilarities using the package vegan v.2.5-7 (66) after normalization with function “decostand” in the vegan package in the R. Post hoc pairwise comparisons were also performed for the regions and sample sites using the package pairwiseAdonis v.0.0.1 (67). For the locational comparison of each ratio in CAMAs, coral tissues, and Symbiodiniaceae, the comparisons were tested using the Wilcoxon rank-sum test in the statistical software R.
Acknowledgments
This project was supported by the Academia Sinica, Taiwan. We thank four imaging facilities at Academia Sinica, as follows: the Imaging Core Facility at the ICOB, the Neuroscience Core Facility (AS-CFII-110-101), the Academia Sinica Advanced Optics Microscope Core Facility (AS-CFII-108-116), and the IPMB Live-Cell-Imaging Core Lab for the technical support in microscopic imaging and analysis. We thank The NGS Core Facility at Biodiversity Research Center, Academia Sinica for technical support of the high-throughput sequencing. We are also grateful to R. Machida and summer intern M. Chen for advice and help in the phylogenetic analysis.
Funding: Financial support for this study came from Academia Sinica.
Author contributions: N.W., D.G.B., and S.-L.T. conceived the project. N.W., S.-P.Y., C.-Y.L., and H.Y. collected the samples. N.W., M.-T.H., Y.-C.T., X.T., and B.-C.C. performed the FISH experiments, all observations, and data analyses. N.W., K.T., H.-J.C., Y.-H.C., P.-W.C, and Y.-J.C. conducted genomic experiments and analyses. N.W., S.S.-Y.H., and D.-C.L. carried out FISH-NanoSIMS experiment and analyses. N.W., D.G.B., and S.-L.T. had major contributions to manuscript writing and figure making. All authors critically reviewed all contents.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All sequencing data produced in this study have been deposited in NCBI under BioProject accession number PRJNA783437 at www.ncbi.nlm.nih.gov/bioproject/. In addition, direct Sanger sequencing data have appeared in the GenBank nucleotide sequence database under accession numbers OL957474 to OL957540 at www.ncbi.nlm.nih.gov/genbank/.
Supplementary Materials
This PDF file includes:
Supplementary Methods
Figs. S1 to S11
Tables S2, S3, S5, S7, and S9
References
Other Supplementary Material for this manuscript includes the following:
Tables S1, S4, S6, and S8
REFERENCES AND NOTES
- 1.Elbeltagy A., Nishioka K., Sato T., Suzuki H., Ye B., Hamada T., Isawa T., Mitsui H., Minamisawa K., Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl. Environ. Microbiol. 67, 5285–5293 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kondorosi E., Mergaert P., Kereszt A., A paradigm for endosymbiotic life: Cell differentiation of rhizobium bacteria provoked by host plant factors. Annu. Rev. Microbiol. 67, 611–628 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Stouthamer R., Breeuwer J. A. J., Hurst G. D. D., Wolbachia pipientis: Microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53, 71–102 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Kuechler S. M., Renz P., Dettner K., Kehl S., Diversity of symbiotic organs and bacterial endosymbionts of Lygaeoid bugs of the families Blissidae and Lygaeidae (Hemiptera: Heteroptera: Lygaeoidea). Appl. Environ. Microbiol. 78, 2648–2659 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balmand S., Lohs C., Aksoy S., Heddi A., Tissue distribution and transmission routes for the tsetse fly endosymbionts. J. Invertebr. Pathol. 112 Suppl, S116–S122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graf J. S., Schorn S., Kitzinger K., Ahmerkamp S., Woehle C., Huettel B., Schubert C. J., Kuypers M. M. M., Milucka J., Anaerobic endosymbiont generates energy for ciliate host by denitrification. Nature 591, 445–450 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubilier N., Mülders C., Ferdelman T., de Beer D., Pernthaler A., Klein M., Wagner M., Erséus C., Thiermann F., Krieger J., Giere O., Amann R., Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature 411, 298–302 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Felbeck H., Chemoautotrophic potential of the hydrothermal vent tube worm, Riftia pachyptila Jones (Vestimentifera). Science 213, 336–338 (1981). [DOI] [PubMed] [Google Scholar]
- 9.Tianero M. D., Balaich J. N., Donia M. S., Localized production of defence chemicals by intracellular symbionts of Haliclona sponges. Nat. Microbiol. 4, 1149–1159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuett C., Doepke H., Grathoff A., Gedde M., Bacterial aggregates in the tentacles of the sea anemone Metridium senile. Helgol. Mar. Res. 61, 211–216 (2007). [Google Scholar]
- 11.Schreiber L., Kjeldsen K. U., Funch P., Jensen J., Obst M., López-Legentil S., Schramm A., Endozoicomonas are specific, facultative symbionts of sea squirts. Front. Microbiol. 7, 1042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponnudurai R., Kleiner M., Sayavedra L., Petersen J. M., Moche M., Otto A., Becher D., Takeuchi T., Satoh N., Dubilier N., Schweder T., Markert S., Metabolic and physiological interdependencies in the Bathymodiolus azoricus symbiosis. ISME J. 11, 463–477 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cano I., Ryder D., Webb S. C., Jones B. J., Brosnahan C. L., Carrasco N., Bodinier B., Furones D., Pretto T., Carella F., Chollet B., Arzul I., Cheslett D., Collins E., Lohrmann K. B., Valdivia A. L., Ward G., Carballal M. J., Villalba A., Marigómez I., Mortensen S., Christison K., Kevin W. C., Bustos E., Christie L., Green M., Feist S. W., Cosmopolitan distribution of Endozoicomonas-like organisms and other intracellular microcolonies of bacteria causing infection in marine mollusks. Front. Microbiol. 11, 577481 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neave M. J., Rachmawati R., Xun L., Michell C. T., Bourne D. G., Apprill A., Voolstra C. R., Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J. 11, 186–200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engebrecht J., Nealson K., Silverman M., Bacterial bioluminescence: Isolation and genetic analysis of functions from Vibrio fischeri. Cell 32, 773–781 (1983). [DOI] [PubMed] [Google Scholar]
- 16.Miyamoto N., Yoshida M.-A., Koga H., Fujiwara Y., Genetic mechanisms of bone digestion and nutrient absorption in the bone-eating worm Osedax japonicus inferred from transcriptome and gene expression analyses. BMC Evol. Biol. 17, 17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiu J.-H., Ding J.-Y., Tseng C.-H., Lou S.-P., Mezaki T., Wu Y.-T., Wang H.-I., Tang S.-L., A newly designed primer revealed high phylogenetic diversity of Endozoicomonas in coral reefs. Microbes Environ. 33, 172–185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glasl B., Smith C. E., Bourne D. G., Webster N. S., Disentangling the effect of host-genotype and environment on the microbiome of the coral Acropora tenuis. PeerJ 7, e6377 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller N., Maneval P., Manfrino C., Frazer T. K., Meyer J. L., Spatial distribution of microbial communities among colonies and genotypes in nursery-reared Acropora cervicornis. PeerJ 8, e9635 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sweet M., Villela H., Keller-Costa T., Costa R., Romano S., Bourne D. G., Cárdenas A., Huggett M. J., Kerwin A. H., Kuek F., Medina M., Meyer J. L., Müller M., Pollock F. J., Rappé M. S., Sere M., Sharp K. H., Voolstra C. R., Zaccardi N., Ziegler M., Peixoto R., Insights into the cultured bacterial fraction of corals. mSystems 6, e0124920 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding J.-Y., Shiu J.-H., Chen W.-M., Chiang Y.-R., Tang S.-L., Genomic insight into the host–endosymbiont relationship of Endozoicomonas montiporae CL-33T with its coral host. Front. Microbiol. 7, 251 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cano I., van Aerle R., Ross S., Verner-Jeffreys D. W., Paley R. K., Rimmer G. S. E., Ryder D., Hooper P., Stone D., Feist S. W., Molecular characterization of an Endozoicomonas-like organism causing infection in the king scallop (Pecten maximus L.). Appl. Environ. Microbiol. 84, e00952-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klinges J. G., Rosales S. M., McMinds R., Shaver E. C., Shantz A. A., Peters E. C., Eitel M., Wörheide G., Sharp K. H., Burkepile D. E., Silliman B. R., Vega Thurber R. L., Phylogenetic, genomic, and biogeographic characterization of a novel and ubiquitous marine invertebrate-associated Rickettsiales parasite, Candidatus Aquarickettsia rohweri, gen. nov., sp. nov. ISME J. 13, 2938–2953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tandon K., Lu C.-Y., Chiang P.-W., Wada N., Yang S.-H., Chan Y.-F., Chen P.-Y., Chang H.-Y., Chiou Y.-J., Chou M.-S., Chen W.-M., Tang S.-L., Comparative genomics: Dominant coral-bacterium Endozoicomonas acroporae metabolizes dimethylsulfoniopropionate (DMSP). ISME J. 14, 1290–1303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Work T., Aeby G., Microbial aggregates within tissues infect a diversity of corals throughout the Indo-Pacific. Mar. Ecol. Prog. Ser. 500, 1–9 (2014). [Google Scholar]
- 26.Harshbarger J. C., Chang S. C., Otto S. V., Chlamydiae (with phages), mycoplasmas, and rickettsiae in Chesapeake Bay bivalves. Science 196, 666–668 (1977). [DOI] [PubMed] [Google Scholar]
- 27.Cruz-Flores R., Cáceres-Martínez J., Rickettsiales-like organisms in bivalves and marine gastropods: A review. Rev. Aquac. 12, 2010–2026 (2020). [Google Scholar]
- 28.Costa P. M., Carreira S., Lobo J., Costa M. H., Molecular detection of prokaryote and protozoan parasites in the commercial bivalve Ruditapes decussatus from southern Portugal. Aquaculture 370-371, 61–67 (2012). [Google Scholar]
- 29.Wada N., Ishimochi M., Matsui T., Pollock F. J., Tang S.-L., Ainsworth T. D., Willis B. L., Mano N., Bourne D. G., Characterization of coral-associated microbial aggregates (CAMAs) within tissues of the coral Acropora hyacinthus. Sci. Rep. 9, 14662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karstens L., Asquith M., Davin S., Fair D., Gregory W. T., Wolfe A. J., Braun J., McWeeney S., Controlling for contaminants in low-biomass 16S rRNA gene sequencing experiments. mSystems 4, e00290-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F. O., The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cunning R., Bay R. A., Gillette P., Baker A. C., Traylor-Knowles N., Comparative analysis of the Pocillopora damicornis genome highlights role of immune system in coral evolution. Sci. Rep. 8, 16134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson H. R., Altieri A. H., Oxygen: The universal currency on coral reefs. Coral Reefs 38, 177–198 (2019). [Google Scholar]
- 34.Ferrier-Pagès C., Gattuso J.-P., Dallot S., Jaubert J., Effect of nutrient enrichment on growth and photosynthesis of the zooxanthellate coral Stylophora pistillata. Coral Reefs 19, 103–113 (2000). [Google Scholar]
- 35.Koop K., Booth D., Broadbent A., Brodie J., Bucher D., Capone D., Coll J., Dennison W., Erdmann M., Harrison P., Hoegh-Guldberg O., Hutchings P., Jones G. B., Larkum A. W. D., O’Neil J., Steven A., Tentori E., Ward S., Williamson J., Yellowlees D., ENCORE: The effect of nutrient enrichment on coral reefs. Synthesis of results and conclusions. Mar. Pollut. Bull. 42, 91–120 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Zhang F., Blasiak L. C., Karolin J. O., Powell R. J., Geddes C. D., Hill R. T., Phosphorus sequestration in the form of polyphosphate by microbial symbionts in marine sponges. Proc. Natl. Acad. Sci. U.S.A. 112, 4381–4386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cianfanelli F. R., Monlezun L., Coulthurst S. J., Aim, load, fire: The type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24, 51–62 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Bernard C. S., Brunet Y. R., Gueguen E., Cascales E., Nooks and crannies in type VI secretion regulation. J. Bacteriol. 192, 3850–3860 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallique M., Bouteiller M., Merieau A., The type VI secretion system: A dynamic system for bacterial communication? Front. Microbiol. 8, 1454 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cassan F. D., Coniglio A., Amavizca E., Maroniche G., Cascales E., Bashan Y., de-Bashan L. E., The Azospirillum brasilense type VI secretion system promotes cell aggregation, biocontrol protection against phytopathogens and attachment to the microalgae Chlorella sorokiniana. Environ. Microbiol. 23, 6257–6274 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Linares D., Jean N., Van Overtvelt P., Ouidir T., Hardouin J., Blache Y., Molmeret M., The marine bacteria Shewanella frigidimarina NCIMB400 upregulates the type VI secretion system during early biofilm formation. Environ. Microbiol. Rep. 8, 110–121 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Ryu C.-M., Against friend and foe: Type 6 effectors in plant-associated bacteria. J. Microbiol. 53, 201–208 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Speare L., Cecere A. G., Guckes K. R., Smith S., Wollenberg M. S., Mandel M. J., Miyashiro T., Septer A. N., Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc. Natl. Acad. Sci. U.S.A. 115, E8528–E8537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salinero-Lanzarote A., Pacheco-Moreno A., Domingo-Serrano L., Durán D., Ormeño-Orrillo E., Martínez-Romero E., Albareda M., Palacios J. M., Rey L., The Type VI secretion system of Rhizobium etli Mim1 has a positive effect in symbiosis. FEMS Microbiol. Ecol. 95, fiz054 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Elton C., Competition and the structure of ecological communities. J. Anim. Ecol. 15, 54–68 (1946). [Google Scholar]
- 46.Bongrand C., Ruby E. G., Achieving a multi-strain symbiosis: Strain behavior and infection dynamics. ISME J. 13, 698–706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itoh H., Jang S., Takeshita K., Ohbayashi T., Ohnishi N., Meng X.-Y., Mitani Y., Kikuchi Y., Host–symbiont specificity determined by microbe–microbe competition in an insect gut. Proc. Natl. Acad. Sci. U.S.A. 116, 22673–22682 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pollock F. J., McMinds R., Smith S., Bourne D. G., Willis B. L., Medina M., Thurber R. V., Zaneveld J. R., Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 9, 4921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bayer T., Neave M. J., Alsheikh-Hussain A., Aranda M., Yum L. K., Mincer T., Hughen K., Apprill A., Voolstra C. R., The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl. Environ. Microbiol. 79, 4759–4762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Brien P. A., Tan S., Yang C., Frade P. R., Andreakis N., Smith H. A., Miller D. J., Webster N. S., Zhang G., Bourne D. G., Diverse coral reef invertebrates exhibit patterns of phylosymbiosis. ISME J. 14, 2211–2222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Brien P. A., Andreakis N., Tan S., Miller D. J., Webster N. S., Zhang G., Bourne D. G., Testing cophylogeny between coral reef invertebrates and their bacterial and archaeal symbionts. Mol. Ecol. 30, 3768–3782 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Chen C.-P., Tseng C.-H., Chen C. A., Tang S.-L., The dynamics of microbial partnerships in the coral Isopora palifera. ISME J. 5, 728–740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jorgensen S. L., Hannisdal B., Lanzén A., Baumberger T., Flesland K., Fonseca R., Ovreås L., Steen I. H., Thorseth I. H., Pedersen R. B., Schleper C., Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc. Natl. Acad. Sci. U.S.A. 109, E2846–E2855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar R. C., Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Edgar R. C., UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., Lesniewski R. A., Oakley B. B., Parks D. H., Robinson C. J., Sahl J. W., Stres B., Thallinger G. G., Van Horn D. J., Weber C. F., Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar S., Stecher G., Tamura K., MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu C., Cheng S. H., Lin S., Illuminating the dark depths inside coral. Cell. Microbiol. 22, e13122 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Yamaguchi T., Fuchs B. M., Amann R., Kawakami S., Kubota K., Hatamoto M., Yamaguchi T., Rapid and sensitive identification of marine bacteria by an improved in situ DNA hybridization chain reaction (quickHCR-FISH). Syst. Appl. Microbiol. 38, 400–405 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Daims H., Brühl A., Amann R., Schleifer K. H., Wagner M., The domain-specific probe EUB338 is insufficient for the detection of all bacteria: Development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22, 434–444 (1999). [DOI] [PubMed] [Google Scholar]
- 61.Wallner G., Amann R., Beisker W., Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14, 136–143 (1993). [DOI] [PubMed] [Google Scholar]
- 62.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.R Core Team, R: A Language and environment for statistical computing (R Foundations for Statistical Computing, 2020); www.R-project.org/.
- 64.J. Fox, S. Weisberg, An R Companion to Applied Regression (Sage Publications, ed. 3, 2019). [Google Scholar]
- 65.Wright E. S., Yilmaz L. S., Corcoran A. M., Ökten H. E., Noguera D. R., Automated design of probes for rRNA-targeted fluorescence in situ hybridization reveals the advantages of using dual probes for accurate identification. Appl. Environ. Microbiol. 80, 5124–5133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.J. Oksanen, F. G. Blanchet, R. Kindt, P. Legendre, R. B. O’hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, H. Wagner, Vegan: Community ecology package (2020); https://cran.r-project.org/web/packages/vegan/index.html.
- 67.P. Martinez Arbizu, pairwiseAdonis: Pairwise multilevel comparison using adonis. R package version 0.0.1 (2017).
- 68.E. Nawrocki, “Structural RNA Homology Search and Alignment Using Covariance Models,” thesis, Washington University in Saint Louis, School of Medicine (2009). [Google Scholar]
- 69.S. Andrews, FastQC: A quality control tool for high throughput sequence data (2010); www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 70.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li D., Liu C.-M., Luo R., Sadakane K., Lam T.-W., MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Uritskiy G. V., DiRuggiero J., Taylor J., MetaWRAP—A flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6, 158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu Y.-W., Simmons B. A., Singer S. W., MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607 (2015). [DOI] [PubMed] [Google Scholar]
- 74.Alneberg J., Bjarnason B. S., de Bruijn I., Schirmer M., Quick J., Ijaz U. Z., Lahti L., Loman N. J., Andersson A. F., Quince C., Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Kang D. D., Li F., Kirton E., Thomas A., Egan R., An H., Wang Z., MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., Lesin V. M., Nikolenko S. I., Pham S., Prjibelski A. D., Pyshkin A. V., Sirotkin A. V., Vyahhi N., Tesler G., Alekseyev M. A., Pevzner P. A., SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.von Meijenfeldt F. A. B., Arkhipova K., Cambuy D. D., Coutinho F. H., Dutilh B. E., Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol. 20, 217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.B. Bushnell, BBMap: A fast, accurate, splice-aware aligner (LBNL-7065E, Lawrence Berkeley National Lab, 2014); www.osti.gov/biblio/1241166.
- 80.Chaumeil P.-A., Mussig A. J., Hugenholtz P., Parks D. H., GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seemann T., Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Kanehisa M., Sato Y., Morishima K., BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Kanehisa M., Goto S., KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., Formsma K., Gerdes S., Glass E. M., Kubal M., Meyer F., Olsen G. J., Olson R., Osterman A. L., Overbeek R. A., McNeil L. K., Paarmann D., Paczian T., Parrello B., Pusch G. D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O., The RAST server: Rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marchler-Bauer A., Bryant S. H., CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nawrocki E. P., Eddy S. R., Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., Vinh L. S., UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Letunic I., Bork P., Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen I.-M. A., Chu K., Palaniappan K., Ratner A., Huang J., Huntemann M., Hajek P., Ritter S., Varghese N., Seshadri R., Roux S., Woyke T., Eloe-Fadrosh E. A., Ivanova N. N., Kyrpides N. C., The IMG/M data management and analysis system v.6.0: New tools and advanced capabilities. Nucleic Acids Res. 49, D751–D763 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoon S.-H., Ha S.-M., Lim J., Kwon S., Chun J., A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286 (2017). [DOI] [PubMed] [Google Scholar]
- 93.R. A. M. Villanueva, Z. J. Chen, ggplot2: Elegant graphics for data analysis. Interdiscip. Res. Perspect. 17, 160–167 (2019). [Google Scholar]
- 94.Eichinger V., Nussbaumer T., Platzer A., Jehl M.-A., Arnold R., Rattei T., EffectiveDB—Updates and novel features for a better annotation of bacterial secreted proteins and Type III, IV, VI secretion systems. Nucleic Acids Res. 44, D669–D674 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T. L., BLAST+: Architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Castresana J., Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- 97.Felsenstein J., Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791 (1985). [DOI] [PubMed] [Google Scholar]
- 98.The UniProt Consortium , UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Binns D., Dimmer E., Huntley R., Barrell D., O’Donovan C., Apweiler R., QuickGO: A web-based tool for Gene Ontology searching. Bioinformatics 25, 3045–3046 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fu L., Niu B., Zhu Z., Wu S., Li W., CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morse A., Formic acid-sodium citrate decalcification and butyl alcohol dehydration of teeth and bones for sectioning in paraffin. J. Dent. Res. 24, 143–153 (1945). [Google Scholar]
- 102.Huang L.-K., Wang M.-J. J., Image thresholding by minimizing the measures of fuzziness. Pattern Recogn. 28, 41–51 (1995). [Google Scholar]
- 103.Li C. H., Tam P. K. S., An iterative algorithm for minimum cross entropy thresholding. Pattern Recogn. Lett. 19, 771–776 (1998). [Google Scholar]
- 104.Glasbey C. A., An analysis of histogram-based thresholding algorithms. Graph. Models Image Process. 55, 532–537 (1993). [Google Scholar]
- 105.Yilmaz L. S., Parnerkar S., Noguera D. R., mathFISH, a web tool that uses thermodynamics-based mathematical models for in silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Appl. Environ. Microbiol. 77, 1118–1122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Figs. S1 to S11
Tables S2, S3, S5, S7, and S9
References
Tables S1, S4, S6, and S8