Abstract
Fluorescence in situ hybridization (FISH) and rRNA slot blot hybridization with 16S rRNA-targeted oligonucleotide probes were used to investigate the phylogenetic composition of a marine Arctic sediment (Svalbard). FISH resulted in the detection of a large fraction of microbes living in the top 5 cm of the sediment. Up to 65.4% ± 7.5% of total DAPI (4′,6′-diamidino-2-phenylindole) cell counts hybridized to the bacterial probe EUB338, and up to 4.9% ± 1.5% hybridized to the archaeal probe ARCH915. Besides δ-proteobacterial sulfate-reducing bacteria (up to 16% 52) members of the Cytophaga-Flavobacterium cluster were the most abundant group detected in this sediment, accounting for up to 12.8% of total DAPI cell counts and up to 6.1% of prokaryotic rRNA. Furthermore, members of the order Planctomycetales accounted for up to 3.9% of total cell counts. In accordance with previous studies, these findings support the hypothesis that these bacterial groups are not simply settling with organic matter from the pelagic zone but are indigenous to the anoxic zones of marine sediments. Members of the γ-proteobacteria also constituted a significant fraction in this sediment (6.1% ± 2.5% of total cell counts, 14.4% ± 3.6% of prokaryotic rRNA). A new probe (GAM660) specific for sequences affiliated with free-living or endosymbiotic sulfur-oxidizing bacteria was developed. A significant number of cells was detected by this probe (2.1% ± 0.7% of total DAPI cell counts, 13.2% ± 4.6% of prokaryotic rRNA), showing no clear zonation along the vertical profile. Gram-positive bacteria and the β-proteobacteria were near the detection limit in all sediments.
Knowledge of the microbial diversity of marine pelagic and benthic communities has been greatly extended recently by molecular studies based on the analysis of 16S rDNA sequences (see, for example, references 9, 10, 20, 43, 44, 48, 50, 52, and 67). Numerous new 16S rDNA sequences have been retrieved both from marine sediments and from the water column, indicating that the vast majority of species has not been cultivated yet. Several studies using the cultivation-independent approach of 16S rDNA cloning have helped to elucidate common features within the microbial communities of specific habitats such as marine benthic environments (10, 35, 36, 52, 67). Furthermore, they have provided additional sequence information for the design and evaluation of nucleic acid probes for the identification and quantification of distinct bacterial populations.
While microbial diversity can be readily studied by PCR-based 16S rDNA cloning, community structure cannot be deduced from cloning studies (3) due to potential biases introduced during DNA retrieval and amplification (17, 53, 65). For reliable characterization of community structure, quantitative methods such as fluorescence in situ hybridization (FISH) or rRNA slot blot hybridization are more suitable (3). To date, a number of studies have been performed using either of these two methods to quantify different groups in marine sediments (15, 37, 38, 51, 57–59, 68). Most of these studies, however, focused on specific microbial groups such as sulfate-reducing bacteria (51, 58, 59) or Archaea (38, 68).
We describe here the community composition of a marine Arctic sediment (Smeerenburgfjorden, Svalbard) using both FISH and rRNA slot blot hybridization for quantification. The sulfate-reducing community of Smeerenburgfjorden sediment has recently been described in detail (51); sulfate reducers accounted for up to 16% of the total cell numbers and up to 29% of the prokaryotic rRNA. In the present study, we report the contribution of other major phylogenetic groups, such as the β- and γ-proteobacteria, the Cytophaga-Flavobacterium cluster, the Planctomycetales, and gram-positive bacteria, to the total microbial community along vertical gradients.
To the best of our knowledge, there is only a single previous study which has described the abundance of the different classes of proteobacteria, the Cytophaga-Flavobacterium cluster, the Planctomycetales, and gram-positive bacteria in marine sediments (37). Llobet-Brossa et al. used FISH for quantification. The present study, however, reports the first rRNA profiles of these major phylogenetic groups in marine sediments.
In addition to the quantification of these major phylogenetic groups, a new probe specific for a cluster of 16S rDNA clone sequences affiliated with free-living and endosymbiotic sulfur-oxidizing bacteria of invertebrates was developed and applied. Sequences of this group were abundant in a Svalbard sediment clone library (52) and also dominant in clone libraries from other marine sediments, e.g., different deep-sea sediments off Japan (35, 36), coastal sediments off Japan (67), and seagrass-colonized sediments from the Bassin d'Archachon (7). The potential ecological importance of this group is discussed with regard to its abundance, the stratification of its distribution, and the possible occurrence of symbiotic and free-living forms.
MATERIALS AND METHODS
Study site and sampling.
Sediment samples were collected on 28 July 1998 from Smeerenburgfjorden, Svalbard, Arctic Ocean (79°42′815"N, 11°05′189"E, “station J”). The sediment temperature was 0°C, the surface water temperature was 5°C, and the water depth was 218 m. Sediment was sampled with a Haps-corer, subsampled, and kept at in situ temperature during transport (72 h). The sediment was characterized by a soft brown silty oxidized surface (upper 2 cm) overlaying a transition zone of darker, black-streaked clayey mud. Below the transition zone (2 to 6 cm) a black sulfidic zone followed. Worm tubes as well as small shells (2 to 3 mm) were present in the sediment to a depth below 10 cm. Two parallel cores subsampled from the same Haps-corer (distance between the two core samples, ca. 10 cm) were sliced: one-half of each slice was frozen in liquid nitrogen for RNA extraction (stored at −80°C); the other half was fixed for 2 to 3 h at a final concentration of 3% formaldehyde, washed twice with 1× phosphate-buffered saline (PBS; 10 mM sodium phosphate [pH 7.2], 130 mM NaCl), and was finally stored in 1× PBS-ethanol (1:1) at −20°C.
RNA extraction and slot blot hybridization.
RNA was extracted from 1.5 ml of wet sediment (per layer) by bead beating, phenol extraction, and isopropanol precipitation as described previously (57). The quality of the RNA was checked by polyacrylamide gel electrophoresis. Approximately 50 ng of RNA was blotted onto nylon membranes (Magna Charge; Micron Separations, Westborough, Mass.) in triplicate and hybridized with radioactively labeled oligonucleotide probes as described by Stahl et al. (63). Membranes were washed at different temperatures depending on the dissociation temperature (Td) of the probe. The probes used and dissociation temperatures are given in Table 1. The dissociation temperatures of the probes were determined as described by Raskin et al. (49) with slight modifications. For Td determinations and hybridizations (probes BET42a, GAM42a, GAM660, CF319a, and PLA886), washing buffer with a lower sodium dodecyl sulfate (SDS) concentration was used (1× SSC [150 mM NaCl, 15 mM sodium citrate; pH 7.0]; 0.1% SDS). However, for hybridizations with probes Uni1390, EUB338, EUK1379, and ARCH915 a washing buffer with 1% SDS was used.
TABLE 1.
Oligonucleotide probes used in this study
| Probe | Specificity | Sequence (5′-3′) | Target RNA | Positiona (nucleotide range) | FISH (FA)b | Slot blot Td (°C)c | Source or reference |
|---|---|---|---|---|---|---|---|
| UNI1390 | Universal—all organisms | GACGGGCGGTGTGTACAA | 16S, 18S | 1390–1407 | NU | 44* | 72 |
| ARCH915 | Archaea | GTGCTCCCCCGCCAATTCCT | 16S | 915–935 | 35 | 56* | 2 |
| EUK1379 | Eucarya | TACAAAGGGCAGGGAC | 18S | 1379–1394 | NU | 42* | 28 |
| EUB338 | Bacteria | GCTGCCTCCCGTAGGAGT | 16S | 338–355 | 10 | 54* | 1 |
| NON338 | Negative control | ACTCCTACGGGAGGCAGC | 16S | 338–355 | 10 | NU | 69 |
| GP1199 | Most gram-positive bacteria | AAGGGGCATGATG | 16S | 1199–1211 | 34* | B. J. MacGregor et al.d | |
| GAM42a | γ-Proteobacteria | GCCTTCCCACATCGTTT | 23S | 1027–1043 | 35 | 60 | 40 |
| BET42a | β-Proteobacteria | GCCTTCCCACTTCGTTT | 23S | 1027–1043 | NU | 58 | 40 |
| PLA886 | Planctomycetales, some Eucarya | GCCTTGCGACCATACTCCC | 16S | 886–904 | 35 | 62 | 46 |
| CF319a | Cytophaga-Flavobacterium cluster | TGGTCCGTGTCTCAGTAC | 16S | 319–336 | 35 | 56 | 39 |
| GAM660 | 16S rDNA clone sequences affiliated with endosymbionts and some other species in the γ-proteobacteria | TCCACTTCCCTCTAC | 16S | 660–674 | 35–40 | 52 | This study |
Position in the 16S or 23S rRNA of E. coli.
FA, formamide concentrations in the hybridization buffer calculated as the percent (vol/vol). NU, not used.
*, Dissociation temperatures (Td) were determined with the washing buffer containing 1× SSC and 1% SDS.
B. J. MacGregor, S. Toze, E. W. Alm, R. Sharp, C. J. Ziemer, and D. A. Stahl, submitted for publication.
Quantification.
Hybridization signal intensity was measured with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and quantified as described previously (59). Reference rRNA isolated from pure cultures of Cytophaga lytica (DSM 7489), Pirellula marina strain 1, Methanolobus tindarius (DSM 2278), Arthrobacter strain KT1113.15, Zoogloea strain Cadagno, Halothiobacillus kellyi (DSM 13162), and sulfur-oxidizing bacterium strain OAII 2, as well as the rRNA of Saccharomyces cerevisiae and Escherichia coli (purchased from Roche, Mannheim, Germany) served as standards for hybridization with the probes given in Table 1.
FISH.
PBS-ethanol-stored samples were diluted and treated by mild sonication with an MS73 probe (Sonopuls HD70; Bandelin, Berlin, Germany) at a setting of 20 s, with an amplitude of 42 μm, and <10 W. An aliquot of 10 μl of a 1:40 dilution was filtered on 0.2-μm-pore-size GTTP polycarbonate filters (Millipore, Eschborn, Germany). Hybridization and microscopy counts of hybridized and 4′,6′-diamidino-2-phenylindole (DAPI)-stained cells were performed as described previously (62). Means were calculated from 10 to 20 randomly chosen fields on each filter section, corresponding to 800 to 1,000 DAPI-stained cells. Counting results were always corrected by subtracting signals observed with the probe NON338. Formamide concentrations are given in Table 1.
Oligonucleotides.
Oligonucleotides were purchased from Interactiva (Ulm, Germany). For FISH, oligonucleotide probes were synthesized with the fluorescent dye Cy3 at the 5′ end.
RESULTS
Total cell counts and domain-specific probing.
Total cell numbers were determined by DAPI staining. They were in the range of (2.1 to 4.7) × 109 ml of wet sediment−1 and showed little variation among two parallel sediment cores. There was no significant decrease of total cell numbers with increasing sediment depth (Table 2), even to a 19-cm depth.
TABLE 2.
Quantification of bacteria by FISH and rRNA slot blot hybridization in Smeerenburgfjorden sediments (core A/core B)
| Depth (cm) | Absolute prokaryotic cell nos (109 ml−1) | Total RNA, probe Uni1390 (ng of RNA ml−1) |
Bacteria (probe EUB338)
|
Archaea (probe ARCH915)
|
Eucarya (probe EUK1379)
|
Cytophaga-Flavobacterium (probe CF319a)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % Total DAPI cell counts | % Universal RNA | ng of RNA ml−1 | % Total DAPI cell counts | % Universal RNA | ng of RNA ml−1 | % Universal RNA | ng of RNA ml−1 | % Total DAPI cell counts | % Prokaryotic RNA | ng of RNA ml−1 | |||
| 0.25 | 2.9/2.1 | 15,658/18,429 | 57.9/72.9 | 61.3/47.8 | 9,601/9,845 | 6.4/3.3 | 1.7/0.9 | 264/169 | 19.2/26.3 | 3,010/4,841 | 9.6/12.8 | 6.1/5.4 | 606/481 |
| 0.75 | 3.5/3.3 | 14,750/22,860 | 55.9/73.6 | 66.7/59.3 | 9,845/13,564 | 1.8/1.8 | 1.3/1.2 | 191/282 | 12.4/15.9 | 1,829/3,629 | 10.3/10.4 | 5.5/5.8 | 554/799 |
| 1.25 | 3.1/3.4 | 13,943/20,448 | 56.1/65.4 | 51.5/50.8 | 7,179/10,391 | 2.9/1.8 | 1.0/1.0 | 142/206 | 25.1/30.4 | 3,500/6,225 | 7.4/6.8 | 4.1/4.1 | 299/433 |
| 1.75 | 4.2/4.2 | 11,449/13,035 | 53.1/57.7 | 58.9/62.8 | 6,744/8,183 | 2.1/1.5 | 1.2/1.1 | 135/142 | 23.9/15.2 | 2,734/1,983 | 7.2/4.8 | 4.3/3.8 | 297/315 |
| 2.25 | 4.1/3.5 | 11,276/18,647 | 57.5/48.4 | 62.3/50.5 | 7,028/9,417 | 1.8/1.5 | 1.2/1.0 | 133/188 | 18.0/20.8 | 2,026/3,883 | 7.6/3.0 | 4.2/5.4 | 299/522 |
| 2.75 | 3.7/3.3 | 7,795/8,318 | 47.9/51.2 | 63.4/69.6 | 4,944/5,791 | 1.4/0.8 | 1.2/0.9 | 93/75 | 21.4/12.0 | 1,666/995 | 8.5/7.1 | 4.3/3.6 | 217/213 |
| 3.25 | 2.7/3.2 | 8,266/9,157 | 43.7/46.6 | 56.1/69.3 | 4,638/6,344 | 1.4/1.7 | 1.0/0.8 | 86/73 | 25.8/12.0 | 2,129/1,258 | 7.6/3.9 | 4.1/3.3 | 193/215 |
| 3.75 | 3.1/3.7 | 7,896/9,342 | 42.8/50.0 | 64.9/70.8 | 5,125/6,613 | 1.4/1.7 | 1.1/0.6 | 87/58 | 12.1/13.7 | 954/1,181 | 8.2/3.3 | 3.7/3.4 | 192/229 |
| 4.25 | 3.5/3.4 | 5,964/6,494 | 43.5/47.3 | 59.2/77.3 | 3,531/5,019 | 1.4/0.4 | 1.4/0.8 | 83/52 | 18.3/12.6 | 1,093/895 | 4.3/2.8 | 4.9/3.1 | 177/159 |
| 4.75 | 3.4/3.6 | 5,559/5,952 | 28.4/42.2 | 63.4/73.5 | 3,526/4,373 | 1.4/1.1 | 1.4/0.9 | 79/54 | 15.3/13.8 | 853/811 | 4.4/1.8 | 4.2/3.2 | 152/140 |
| 5.5 | 3.5/4.6 | 3,306/4,956 | 26.0/36.0 | 77.7/80.6 | 2,570/3,997 | NDa | 1.4/1.0 | 46/49 | 8.3/15.5 | 274/769 | 1.8/2.0 | 4.0/3.4 | 106/137 |
| 6.5 | 3.7/4.7 | 3,345/4,897 | 24.1/27.4 | 87.6/90.8 | 2,929/4,448 | ND | 1.6/0.9 | 55/46 | 10.1/13.1 | 339/643 | 1.0/1.7 | 3.7/2.7 | 112/122 |
| 7.5 | 4.7/3.9 | 3,283/3,972 | 23.6/23.6 | 80.1/87.5 | 2,631/3,474 | ND | 0.9/1.2 | 31/46 | 12.4/13.5 | 408/536 | 0/0 | 3.5/3.1 | 93/110 |
| 8.5 | 2.9/3.1 | 3,599/4,331 | 28.9/22.6 | 78.5/73.9 | 2,824/3,200 | 1.0/1.7 | 1.3/1.1 | 45/46 | 11.7/12.9 | 421/559 | 0/0.5 | 3.4/3.5 | 97/112 |
| 9.5 | 3.7/3.9 | 3,199/3,589 | 22/15.7 | 71.1/79.7 | 2,273/2,861 | 1.0/0.9 | 1.1/1.0 | 35/38 | 12.6/14.9 | 402/534 | 0/0 | 3.5/3.1 | 81/90 |
| 11 | 3.9/3.4 | 1.7/0.7 | |||||||||||
| 13 | 2.7/3.4 | 0.6/0.8 | |||||||||||
| 15 | 2.5/3.4 | 0.6/1.2 | |||||||||||
| 17 | 3.3/2.7 | ||||||||||||
| 19 | 2.7/2.1 | ||||||||||||
ND, not determined.
Bacteria and Archaea were quantified by both FISH and rRNA slot blot hybridization, with domain-specific probes. FISH resulted in the detection of a large fraction of microbes living in the top 5 cm of the sediment. Up to 65.4% ± 7.5% of total DAPI cell counts hybridized to the eubacterial probe EUB338, and up to 4.9% ± 1.5% of the DAPI cell counts hybridized to the archaeal probe ARCH915 (mean of two cores along the vertical profile). The EUB338 detection rate strongly decreased by factors of 2.6 (core A) and 4.6 (core B) along a vertical profile from the sediment surface to a 10-cm depth. At a >10-cm depth, the detection rate was too low for further FISH analysis (<20% of total DAPI cell counts). Depth profiles of bacterial rRNA were in good accordance with profiles of FISH-detected cells. Recovered rRNA was highest near the surface (up to 13.6 μg ml of sediment−1) and decreased with depth to 2.3 μg ml−1.
Archaea mainly occurred only in numbers near the detection limit, set at 1% of DAPI-stained cells (Table 2). Only in the uppermost layer were Archaea found in higher numbers, with up to 6.4% of DAPI cell counts and 1.9 × 108 cells ml−1. Below the surface layer, the relative contribution of Archaea remained relatively constant at approximately 1.0 to 1.5% of total DAPI cell counts along a vertical profile. No increase of Archaea cell numbers was detected in sediment layers at depths of 11 to 15 cm. Quantification of Archaea by slot blot hybridization were in the same range (0.6 to 1.7% of total rRNA) as determined by FISH.
Eukaryotic rRNA was quantified using probe EUK1379. The highest percentages were detected in the upper layers (0- to 3.25-cm depth). The mean in this region was 20.8% ± 5.8% of total rRNA, as compared to 13.2% ± 2.3% in the layers between depths of 3.75 and 9.5 cm. rRNA detected by the bacterial, archaeal, and eukaryotic probes were 80 to 100% of total rRNA as quantified using universal probe UNI1390. Especially in the upper layers, only about 80% of total rRNA was detected with the domain-specific probes.
The two parallel cores were quite similar in total cell numbers, FISH detection rates, and recovered rRNA (Tables 2 and 3). Therefore, in the following sections the mean values of the two cores are discussed.
TABLE 3.
Further quantification of bacteria by FISH and rRNA slot blot hybridization in Smeerenburgfjorden sediments (core A/core B)
| Depth (cm) | β-Proteobacteria, probe BET42a
|
γ-Proteobacteria (probe GAM42a)
|
Clone sequences and symbionts affiliated with the γ-Proteobacteria (probe GAM660)
|
Planctomycetales (probe PLA886)
|
Gram-positive bacteria (probe GP 1199)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % Prokaryotic RNA | ng of RNA ml−1 | % Total DAPI cell counts | % Prokaryotic RNA | ng of RNA ml−1 | % Total DAPI cell counts | % Prokaryotic RNA | ng of RNA ml−1 | % Total DAPI cell counts | % Universal RNA | ng of RNA ml−1 | % Prokaryotic RNA | ng of RNA ml−1 | |
| 0.25 | 1.6/1.7 | 161/155 | 12.1/9.0 | 17.6/19.4 | 1,739/1,744 | 2.9/2.9 | 11.7/6.9 | 1,153/621 | 2.2/1.9 | 30.7/17.0 | 4,815/3,131 | 0/0.2 | 0/17 |
| 0.75 | 1.8/1.7 | 180/229 | 11.6/9.2 | 20.7/19.4 | 2,075/2,813 | 1.8/2.7 | 9.5/10.5 | 950/1,449 | 3.0/2.8 | 23.5/16.1 | 3,464/3,684 | 0/0.4 | 0/52 |
| 1.25 | 1.8/1.5 | 132/159 | 8.9/9.5 | 22.2/17.8 | 1,626/1,888 | 2.5/2.8 | 11.6/10.8 | 851/1,142 | 3.6/3.5 | 53.6/19.7 | 7,468/4,037 | 0/0.5 | 0/49 |
| 1.75 | 1.4/1.6 | 99/134 | 8.0/6.9 | 20.2/16.8 | 1,388/1,402 | 0.8/2.0 | 17.0/8.2 | 1,170/682 | 2.8/2.7 | 26.0/20.6 | 2,975/2,683 | 0.1/1.4 | 4/116 |
| 2.25 | 1.3/1.6 | 96/158 | 7.3/6.2 | 17.5/16.2 | 1,254/1,558 | 1.6/3.1 | 10.8/11.4 | 772/1,094 | 3.7/3.8 | 19.1/17.2 | 2,154/3,204 | 0.5/1.3 | 35/129 |
| 2.75 | 1.1/1.2 | 58/72 | 5.5/6.1 | 14.5/13.3 | 733/780 | 1.2/2.1 | 18.1/6.1 | 910/359 | 2.8/3.9 | 15.8/13.1 | 1,229/1,091 | 0.7/2.2 | 36/129 |
| 3.25 | 1.3/1.2 | 60/75 | 4.7/4.7 | 14.5/12.5 | 686/804 | 2.6/2.7 | 18.4/6.7 | 871/431 | 3.5/3.3 | 19.1/14.1 | 1,578/1,287 | 0.3/2.6 | 16/167 |
| 3.75 | 1.3/1.1 | 70/77 | 8.7/6.0 | 14.7/12.6 | 764/841 | 2.6/2.8 | 20.1/9.0 | 1,050/599 | 3.1/3.6 | 19.3/13.5 | 1,524/1,257 | 0.2/2.1 | 13/137 |
| 4.25 | 1.1/1.1 | 40/57 | 4.2/4.2 | 13.7/11.8 | 495/596 | 2.2/2.5 | 17.6/7.0 | 635/353 | 1.7/1.9 | 16.2/15.2 | 964/984 | 0/1.2 | 0/63 |
| 4.75 | 1.3/1.1 | 47/45 | 5.1/3.6 | 13.4/11.5 | 483/509 | 2.2/2.7 | 20.4/9.5 | 734/423 | 1.8/1.8 | 26.5/14.0 | 1,475/831 | 0/0.7 | 0/31 |
| 5.5 | 1.2/1.1 | 31/45 | 4.4/5.0 | 11.9/12.0 | 311/486 | 2.8/2.4 | 14.2/14.0 | 3,730/565 | 2.9/1.5 | 21.8/16.0 | 720/793 | 0/0.3 | 0/13 |
| 6.5 | 1.2/1.2 | 36/56 | 5.8/4.0 | 11.2/11.6 | 336/520 | 1.8/2.1 | 16.6/14.2 | 496/638 | 2.5/3.1 | 27.8/14.3 | 931/701 | 0/0.1 | 0/6 |
| 7.5 | 1.2/1.1 | 31/38 | 6.4/3.0 | 9.3/11.8 | 246/415 | 2.4/1.3 | 18.3/11.6 | 4,878/408 | 1.7/2.8 | 31.0/14.5 | 1,019/575 | 0/0 | 0/0 |
| 8.5 | 1.2/0.9 | 26/30 | 4.2/2.9 | 9.3/11.4 | 268/369 | 0.6/1.9 | 19.4/12.0 | 557/391 | 1.3/2.0 | 31.8/15.7 | 1,143/681 | 0/0.2 | 0/6 |
| 9.5 | 0.9/1.0 | 20/28 | 4.0/3.0 | 10.0/11.7 | 231/338 | 1.8/0.4 | 21.3/12.4 | 493/359 | 1.9/1.2 | 38.7/19.0 | 1,238/681 | 0/1.4 | 0/40 |
Quantification of different bacterial groups. (i) Cytophaga-Flavobacterium cluster.
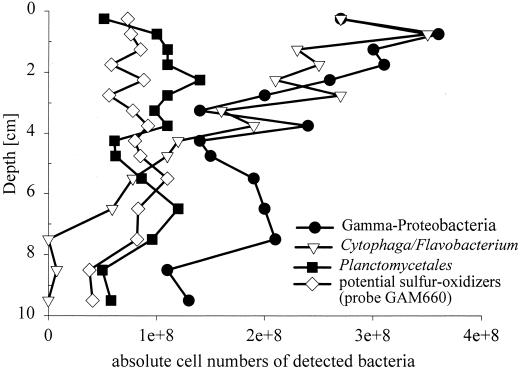
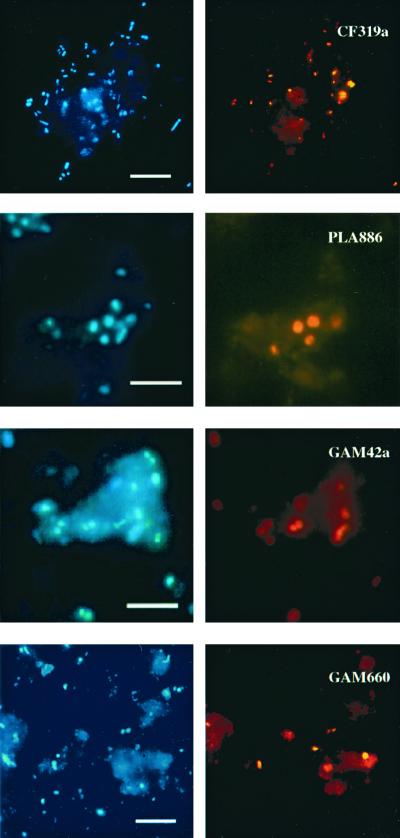
A large fraction of the microbial community could be affiliated with the Cytophaga-Flavobacterium cluster (Tables 2 and 3). Their relative abundance ranged from 11% (3.5 × 108 cells ml−1) in the uppermost layers to 3% (2.0 × 108 cells ml−1) at a 5-cm depth (Fig. 1). Below 5 cm, CF319a-target cells were near or below the detection limit. Cytophaga-Flavobacterium rRNA detection was also highest at the sediment surface (5.7% of prokaryotic rRNA) and decreased slightly to 3.3% at 9.5 cm. CF319a target cells were morphologically highly diverse and included long and short rods (0.5 to 1.5 μm in length), filaments (up to 10 μm in length) and cocci. About three-quarters of the detected cells were very small (≤0.5 μm). Several very thin filaments could barely be detected by DAPI staining. Some of the Cytophaga-Flavobacterium cells (>30%) were found attached to sediment particles or other organic matrices (Fig. 2). These cells were difficult to remove from the particles by sonication.
FIG. 1.
Vertical profiles of absolute numbers of bacteria detected. The mean of two parallel cores is shown.
FIG. 2.
Epifluorescence micrographs of bacteria in sediment samples from Smeerenburgfjorden (Svalbard). Specific hybridizations for the Cytophaga-Flavobacterium cluster (CF319a), for Planctomycetales (PLA886), the γ-proteobacteria (GAM42a), and a γ-proteobacterial subgroup which is affiliated with free-living and symbiotic sulfur-oxidizing bacteria (GAM660) and corresponding DAPI staining (same microscopic field). Bar, 5 μm (all panels).
(ii) Planctomycetales.
Probe PLA886 is specific for Pirellula spp., Planctomyces spp., Isophaera spp., and several clone sequences within the order Planctomycetales. Furthermore, the probe also binds to a wide variety of eukaryotic 18S rRNAs. For FISH analysis, this lack of specificity is not relevant because, in general, a visual differentiation of Eucarya and Bacteria is possible based on the smaller cell size of the latter. Members of the Planctomycetales made up a quantitatively important fraction of the microbial community in the Smeerenburgfjorden sediments and ranged between 1.5 and 3.7% of the total prokaryotic cell counts. There was no clear maximum visible at any specific depth. The highest detection corresponded to 1.4 × 108 cells ml of sediment−1. The cells were usually large cocci, approximately 1 μm in diameter (Fig. 2), occurring as single or rosette-forming cells or in disordered clusters of about 10 cells. All target cells showed a bright fluorescence signal. In the slot blot hybridization, the problem of hybridization of PLA886 to eukaryotic rRNA became relevant. Very high values (13.1 to 38.7% of total rRNA) were detected. A comparison of slot blot profiles for probes PLA886 and EUK1379 showed similar maxima.
(iii) γ-Proteobacteria.
γ-Proteobacteria, as detected by probe GAM42a, comprised a dominant group in the Smeerenburgfjorden sediments (Tables 2 and 3). In the upper layers, this group accounted for up to 10.5% of the total DAPI cell counts. Detection by FISH decreased slightly with increasing depth and was lowest at a 10-cm depth, with 3.5% of the total DAPI cell counts. The morphology of the GAM42a target cells was quite diverse (Fig. 2). The cell size varied, but a large fraction of detected cells was very small (size, ≤0.5 μm). The majority of target cells had a very bright FISH signal, indicating a high cellular rRNA content. The γ-proteobacterial rRNA also made up a quantitatively important fraction of the microbial-community rRNA, with up to 20.0% of prokaryotic rRNA hybridizing to GAM42a. The relative contribution to the prokaryotic rRNA decreased by a factor of approximately 2 from the surface to the 10-cm depth.
Potential sulfur-oxidizing bacteria within the γ-proteobacteria.
Probe GAM660 was designed to be specific for clone sequences affiliated with free-living and endosymbiotic sulfur-oxidizing bacteria which were abundant in a Svalbard sediment clone library (52). Because of their phylogenetic affiliation, these sequences could potentially originate from sulfur-oxidizing bacteria. In addition to our clone sequences, probe GAM660 also targets closely related (up to 97.9%) γ-proteobacterial sequences which were retrieved from other marine sediments (7, 35, 36), endosymbionts of Riftia pachyptila, other vestimentiferan tubeworms and of several bivalves (11–14, 18, 31), Thiobacillus ferrooxidans, H. kellyi and Coxiella burnetii (Table 4). A clear discrimination between target and nontarget organisms was possible with FISH as well as with rRNA slot blot hybridization. Probe GAM660 hybridized to free-living bacteria in Smeerenburgfjorden sediment samples. Up to 2.9% of the total DAPI cell counts (9.4 × 107 cells ml−1) were detected in the surface layer. In deeper layers, the detection rate remained relatively constant and varied between 0.4 and 3.1% of the total DAPI cell counts. In general, targeted cells were cocci that very often occurred as diplococci (Fig. 2). Due to their small size, it was impossible to investigate targeted cells for the presence of sulfur inclusion bodies. The FISH-detected fraction was relatively small compared with the fraction (13.2% ± 4.2% of prokaryotic RNA) detected by slot blot hybridization.
TABLE 4.
List of organisms or clone sequences targeted by GAM660
| Target | Accession no. | Target | Accession no. | |
|---|---|---|---|---|
| Solemya terraeregina gill symbiont | U62131 | Uncultured γ-proteobacterium TK99 | AB022639 | |
| Solemya pusilla gill symbiont | U62130 | Unidentified γ-proteobacterium JTB256 | AB015255 | |
| Thyasira flexuosa gill symbiont | L01575 | Unidentified γ-proteobacterium JTB255 | AB015254 | |
| Riftia pachyptila trophosome symbiont | M99451 | Unidentified γ-proteobacterium JTB23 | AB015248 | |
| Riftia pachyptila endosymbiont | U77478 | Unidentified γ-proteobacterium BD5-16 | AB015571 | |
| Lucinoma aequizonata gill symbiont | M99448 | Unidentified γ-proteobacterium BD3-6 | AB015548 | |
| Anodontia phillipiana gill symbiont | L25711 | Unidentified γ-proteobacterium BD3-1 | AB015547 | |
| Ridgeia piscesae endosymbiont | U77480 | Unidentified marine eubacterium | L10949 | |
| Lamellibrachia columna endosymbiont | U77481 | Uncultured γ-proteobacterium n7d | AF194195 | |
| Lucina pectinata gill symbiont | X84980 | Uncultured γ-proteobacterium B2M61 | AF223300 | |
| Escarpia spicata endosymbiont | U77482 | Uncultured γ-proteobacterium B2M60 | AF223299 | |
| Vestimentiferan endosymbiont Shinkai 6500 | AF65907 | Uncultured γ-proteobacterium B2M54 | AF22329 | |
| Unidentified bacterium HNSS31 | Z88573 | Uncultured γ-proteobacterium B2M28 | AF223297 | |
| Unidentified marine eubacterium | U84621 | Uncultured γ-proteobacterium B2M23 | AF223296 | |
| Uncultured γ-proteobacterium Sva1046 | AJ240991 | Uncultured marine eubacterium HstpL26 | AF159683 | |
| Uncultured γ-proteobacterium Sva0864 | AJ240988 | Uncultured bacterium clone Car16fa | AF224800 | |
| Uncultured γ-proteobacterium Sva0071 | AJ240986 | Uncultured bacterium BURTON-10 | AF142830 | |
| Uncultured γ-proteobacterium Sva0115 | AJ240974 | Thiomicrospira sp. strain Tms-MPN/Milos-CIVI | AJ247759 | |
| Uncultured γ-proteobacterium Sva0304 | AJ240971 | Halothiobacillus kellyi | AF170419 | |
| Uncultured γ-proteobacterium Sva0857 | AJ240969 | Beggiatoa sp. strain 1401-13 | L40997 | |
| Uncultured γ-proteobacterium Sva0862 | AJ240968 | Coxiella burnetii | D89799 | |
| Uncultured γ-proteobacterium Sva0120 | AJ240993 | Thiobacillus ferrooxidans | M79416 | |
| Uncultured proteobacterium clone 1605-59 | AJ007658 | Dechloromarinus chlorophilus | AF170359 | |
| Uncultured γ-proteobacterium SA28 | AB022641 | Alkalispirillum mobilis | AF114783 |
Other probes used.
Members of the β-proteobacteria and of the gram-positive bacteria were only quantified by slot blot hybridization. For probe Bet42a, maximum values of 1.7% of the total DAPI cell counts were detected at the surface, decreasing to 1% in deeper layers. The rRNA of gram-positive bacteria was barely detected in the upper and lower layers and reached a maximal mean of 1.4% prokaryotic rRNA at 3 cm.
DISCUSSION
Total cell counts in the Smeerenburgfjorden sediments were relatively constant along a vertical profile from the sediment surface to a 20-cm depth. The average abundance of (3.4 ± 0.6) × 109 ml−1 was comparable with previous reports for other marine sediments (see, for example, references 37, 57, and 70), although in contrast to our results, all other studies, including one of four other sampling sites off the coast of Svalbard (57), reported decreasing cell numbers with depth. In Svalbard sediments, Sahm et al. reported cell numbers decreased with depth by factors of 3, 7, and 9 within the first 28 cm (57). In Wadden Sea sediments, the total cell numbers decreased by a factor of 2.4 within the first 5 cm of the sediment (37). Wellsbury et al. (70) reported constant cell numbers in the uppermost layers (up to an 8-cm depth) of an estuarine sediment. They explained this rather unusual depth profile by a high tidal influence and high sediment porosity. In our case, tidal influence can be excluded. The sediment, however, was characterized by a relative high water content in the first 2 to 3 cm. Since most sediment bacteria can be found attached to particles, a higher pore water content leads to lower cell numbers per milliliter and could be one cause of the constant cell numbers throughout the profile. In contrast, the total level of RNA detectable by universal or domain-specific probes decreased markedly over the same depth. The most likely reason for the decrease in total rRNA recovered with depth is a lower cellular ribosome content with depth according to the available organic substrates. This is supported by a lower mean FISH signal of the cells in the deeper layers of the sediment (51).
As in other sediments (57, 59, 61) the recovered rRNA was mainly of bacterial and eukaryotic origin. Archaea made up only a minor part of the microbial community, with about 1 to 3% of total cells and of prokaryotic rRNA. Although a relatively large number of cells were not detected by the domain-specific probes in FISH, the lack of detection of significant amounts of archaeal rRNA in the slot blot hybridization suggests that Archaea are not a major component of this arctic sediment. To date, probe ARCH915 includes more than 95% of currently available archaeal sequences in the databases. Low Archaea counts are in accordance with previous studies from other marine sediments (37, 57).
The Cytophaga-Flavobacterium cluster and the order Planctomycetales typically contain aerobic species. Cytophaga-Flavobacterium have been shown to be abundant in the marine water column (16, 19). Recently, Llobet-Brossa et al. found significant cell numbers of both groups in Wadden Sea sediments, even in anoxic zones (37). Data from clone libraries derived from several marine sediments (20, 35, 36, 52) and a freshwater sediment (42) supported this finding. Input of complex organic substrates to anaerobic sediments resulted in a strong increase among members of the Cytophaga-Flavobacterium cluster (56). These findings indicate a potential ecological relevance of these bacteria as hydrolytic fermentative organisms in a mainly anaerobic habitat. In our study, the Cytophaga-Flavobacterium cluster, along with the γ-proteobacteria and sulfate reducers, was one of the three most abundant groups, with high numbers of more than 1.5 × 108 cells ml−1 also in the anoxic layers up to a depth of 4.75 cm. Oxygen profiles from a close station off the coast of Svalbard with a similar water depth indicated an oxygen penetration depth of about 7 mm (29). Calculations of cellular rRNA contents of Cytophaga-Flavobacterium cells made by combining FISH-detected cell numbers and the detected rRNA revealed relatively constant cellular rRNA contents with depth (range, 0.1 to 0.2 fg of rRNA per cell). Planctomycetales made up between 1.2 and 3.9% of DAPI-stained cells down to a depth of 9.5 cm, with a maximum in their proportional contribution at 2.25 cm. These data support the hypothesis that these bacterial groups are multiplying even in anoxic zones in the sediment.
A reliable quantification of Planctomycetales rRNA was not possible because of the cross-hybridization of probe PLA886 with a wide variety of Eucarya. A comparison of the slot blot profiles for probes PLA886 and EUK1379 showed similar shapes and maxima. Therefore, there was presumably a very high contribution of eukaryotic rRNA to PLA886 target rRNA. Since not all organisms targeted by EUK1379 are also targeted by PLA886, a simple subtraction of the values is not possible.
Sulfur-oxidizing bacteria isolated from marine sediments are often members of the genera Thiomicrospira (5, 6, 32) or Thiobacillus (54). In addition, Beggiatoa and Thioploca spp. have often been found in sediments and are used for ecophysiological studies (23, 30, 41). Thiomicrospira and Thiobacillus spp. were often retrieved from most-probable-number (MPN) dilution series for chemolithoautotrophic sulfur-oxidizing bacteria, but only in maximal numbers of 1.4 × 106 cells ml of sediment−1 (5, 60). In MPN dilution series of Smeerenburgfjorden sediments, the growth of chemolithoautotrophic sulfur-oxidizing bacteria was observed to 10−3 dilutions. This result contradicts the idea that they might be numerically abundant. Using the new probe GAM660 which is specific for 16S rDNA clone sequences affiliated with free-living or endosymbiotic sulfur-oxidizing bacteria retrieved from several marine sediments (7, 35, 36, 52, 67), an abundance of up to 1.1 × 108 cells ml−1 was demonstrated in Smeerenburgfjorden sediment. In Wadden Sea sediments, this group was also detected by FISH and accounted for up to 4.6 × 107 cells ml−1 sediment (up to 2.3% of total DAPI cell counts; S. Kolb and K. Ravenschlag, unpublished data). Further functional studies of GAM660 target organisms are needed to find out if these abundant bacteria are really sulfur oxidizers. Possible experiments include large-insert DNA libraries (55, 64) of GAM660 target cells for the identification of genes involved in the sulfur oxidation or the combination of microautoradiography with FISH, allowing the assignment of radiotracer uptake to specific phylogenetic groups (8, 34, 47).
In some layers, detection of the subgroup GAM660 target rRNA was even higher than the rRNA yield of total γ-proteobacterial rRNA. Due to the stringent washing temperature, hybridization with nontarget organisms having one mismatch to the probe sequence can be excluded. However, the discrepancy cannot currently be clarified, because GAM42a targets 23S rRNA. GAM660 targets mostly uncultivated organisms for which the 23S rRNA sequences are yet unknown and cannot be determined easily.
The relative contribution of GAM660 rRNA was significantly higher than for FISH-detected cells (2.4- to 32.3-fold). GAM660 also targets chemoautotrophic symbionts from several bivalve molluscs and tubeworms. Thus, the high relative percentage of GAM660-rRNA could mean a contribution of rRNA derived from endosymbiotic bacteria of bivalves or other eukaryotic hosts. Such bacteria would not have been counted in the FISH analysis due to exclusion during pipetting or sedimentation in dilution steps. The rRNA of these organisms and their hosts, however, might be included in the extracted rRNA used for slot blot hybridization. Chemoautotrophic symbionts have not yet been cultured from their hosts, nor has a free-living stage of the symbionts been isolated from the environment. There is evidence that some hosts obtain their symbionts via environmental transmission (21, 22, 33), which involves the reinfection of the new host generation from an environmental stock of free-living symbiont forms as done by, for example, Codakia orbicularis (21). GAM660 target cells could potentially represent such a free-living symbiont form. The vertical profiles of GAM660-detected rRNA, and GAM660 target cells showed no stratification as might be expected for aerobic chemoautotrophic organisms. However, nitrate respiration has been demonstrated in several endosymbionts, for example, from Solemya reidi (71), Riftia pachyptila (27), and Lucinoma aequizonata (26), as well as in the ectosymbionts of nematodes (25). For the endosymbiotic bacteria, motility of the hosts might be another explanation for the lack of zonation.
We did not screen for α-proteobacteria because the available probes, ALF1b (40) and ALF968 (45), also target a wide variety of δ-proteobacterial sequences, including sulfate-reducing bacteria and members of the genera Pelobacter, Geobacter, Desulfuromonas, Synthrophus, and Polyangium-Chondromyces. In this sediment, δ-proteobacteria contributed up to 34.5% of prokaryotic rRNA and up to 17.5% of total cell counts (51) and therefore have greatly affected the detection of α-proteobacteria.
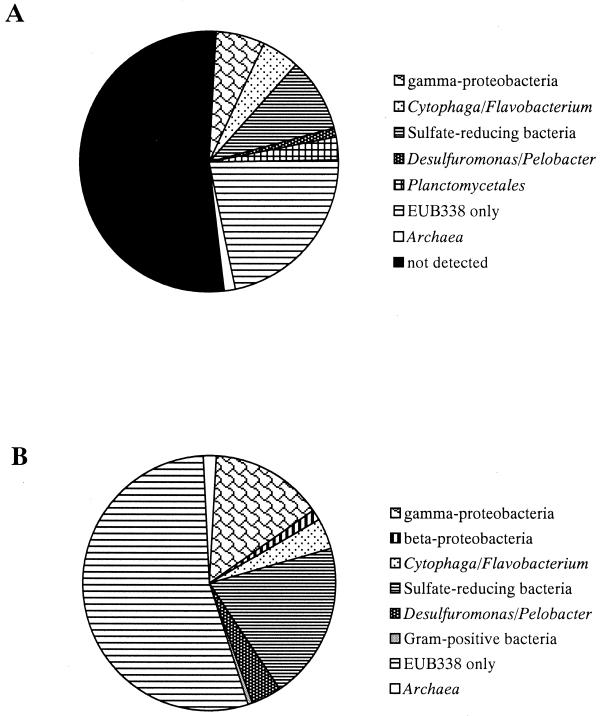
Adding up the mean detection rates along a vertical profile for the different bacterial groups (including the large fraction of sulfate-reducing bacteria (51), 57.8% ± 12.7% of total detectable bacterial cells (23.9% ± 7.5% of total DAPI cell counts), and 44.9% ± 5.5% of bacterial rRNA could be assigned to specific phylogenetic groups (Fig. 3). One explanation for the relatively large “black box” could be the limited coverage with the current probe set, which has been shown by the rapid growth of the 16S rRNA sequence database to be rather incomplete. Furthermore, there are certainly other bacterial groups which make up a quantitatively important fraction in the Smeerenburgfjorden sediments. For example, 16S rDNA sequences affiliated with the order Verrucomicrobiales (24) were repeatedly found in clone libraries from marine sediments (52, 67) or marine snow (50), and sequences related to Arcobacter spp. or other ɛ-proteobacteria were repeatedly retrieved from marine sediments (4, 35, 36). Furthermore, the genus Arcobacter accounted for up to 1.6% of total cell counts in Wadden Sea sediments (37). An ability to carry out nitrate reduction and sulfide oxidation has been reported for Arcobacter spp. (66). Further studies will be needed to investigate the quantitative contribution of Verrucomicrobium spp., Arcobacter spp., and as-yet-unknown phylogenetic groups to microbial communities of marine sediments.
FIG. 3.
Overview of the microbial community structure of Smeerenburgfjorden sediments as revealed by FISH (A) and quantitative rRNA slot blot hybridization (B). For the individual phylogenetic groups mean of the abundance along the vertical profile was calculated. Fractions shown indicate the percentage of total DAPI-stained cells (FISH) and the relative percentage of prokaryotic rRNA (slot blot), respectively. Since probe PLA886 targeting Planctomycetales is also targeting a wide variety of eukaryotes (see the text), the results are not included in part B. Due to the FISH results it can be expected that a fraction of the unidentified EUB338 target rRNA is formed by Planctomycetales.
This study reports the first rRNA profiles for major phylogenetic groups in marine sediments and compares these data with abundances determined by FISH. More combined quantitative studies of microbial community structures in marine sediments are needed to identify common benthic features. Furthermore, studies are needed to identify the organisms contributing to the large “black box.” A major goal for future work will be to combine these data with measurements of microbial activities to address the functional role of abundant phylogenetic groups in the microbial community.
ACKNOWLEDGMENTS
We thank Sebastian Behrens for assistance with FISH counting. Heike Eilers, Heinz Schlesner, and Stefan Sievert provided several strains as references. We are grateful to Carol Arnosti for her critical reading and corrections of the manuscript.
This work was supported by the Max Planck Society.
REFERENCES
- 1.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bidle K A, Kastner M, Bartlett D H. A phylogenetic analysis of microbial communities associated with methane hydrate containing marine fluids and sediments in the Cascadia margin (ODP8 site 892B) FEMS Microbiol Lett. 1999;177:101–108. doi: 10.1111/j.1574-6968.1999.tb13719.x. [DOI] [PubMed] [Google Scholar]
- 5.Brinkhoff T, Santegoeds C, Sahm K, Kuever J, Muyzer G. A polyphasic approach to study the diversity and vertical distribution of sulfur-oxidizing Thiomicrospira species in coastal sediments of the German Wadden Sea. Appl Environ Microbiol. 1998;64:4650–4657. doi: 10.1128/aem.64.12.4650-4657.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinkhoff T, Sievert S M, Kuever J, Muyzer G. Distribution and diversity of sulfur-oxidizing Thiomicrospira spp. at a shallow-water hydrothermal vent in the Aegean Sea (Milos, Greece) Appl Environ Microbiol. 1999;65:3843–3849. doi: 10.1128/aem.65.9.3843-3849.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cifuentes A, Antón J, Benlloch S, Donnelly A, Herbert R A, Rodriguez-Valera F. Prokaryotic diversity in Zostera noltii-colonized marine sediments. Appl Environ Microbiol. 2000;66:1715–1719. doi: 10.1128/aem.66.4.1715-1719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottrell M T, Kirchman D L. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–1697. doi: 10.1128/aem.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLong E F, Wu K Y, Prézelin B B, Jovine R V M. High abundance of Archaea in antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 10.Devereux R, Mundfrom G W. A phylogenetic tree of 16S rRNA sequences from sulfate-reducing bacteria in sandy marine sediment. Appl Environ Microbiol. 1994;60:3437–3439. doi: 10.1128/aem.60.9.3437-3439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Distel D L, Felbeck H, Cavanaugh C M. Evidence for phylogenetic congruence among sulfur-oxidizing chemoautotrophic bacterial endosymbionts and their bivalve hosts. J Mol Evol. 1994;38:533–542. [Google Scholar]
- 12.Distel D L, Lane D J, Olsen G J, Giovannoni S J, Pace N R, Stahl D A, Felbeck H. Sulfur-oxidizing bacterial endosymbionts analysis of phylogeny and specificity by 16S ribosomal RNA sequences. J Bacteriol. 1988;170:2506–2510. doi: 10.1128/jb.170.6.2506-2510.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Distel D L, Wood A P. Characterization of the gill symbiont of Thyasira flexuosa (Thasiridae: Bivalvia) by use of polymerase chain reaction and 16S rRNA sequence analysis. J Bacteriol. 1992;174:6319–6320. doi: 10.1128/jb.174.19.6317-6320.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durand P, Gros O, Frenkiel L, Prieur D. Phylogenetic characterization of sulfur-oxidizing bacterial endosymbionts in three tropical Lucinidae by 16S rDNA sequence analysis. Mol Mar Biol Biotechnol. 1996;5:37–42. [Google Scholar]
- 15.Edgcomb V P, McDonald J H, Devereux R, Smith D W. Estimation of bacterial cell numbers in humic acid-rich salt marsh sediments with probes directed to 16S ribosomal DNA. Appl Environ Microbiol. 1999;65:1516–1523. doi: 10.1128/aem.65.4.1516-1523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eilers H, Pernthaler J, Glöckner F O, Amann R. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol. 2000;66:3044–3051. doi: 10.1128/aem.66.7.3044-3051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldman R A, Black M B, Cary C S, Lutz R A, Vrijenhoek R C. Molecular phylogenetics of bacterial endosymbiont and their vestimentiferan hosts. Mol Mar Biol Biotechnol. 1997;6:268–277. [PubMed] [Google Scholar]
- 19.Glöckner F O, Fuchs B M, Amann R. Bacterioplancton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999c;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray J P, Herwig R P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gros O, Darasse A, Durand P, Frenkiel L, Moueza M. Environmental transmission of a sulfur-oxidizing bacterial gill endosymbiont in the tropical lucinid bivalve Codakia orbicularis. Appl Environ Microbiol. 1996;62:2324–2330. doi: 10.1128/aem.62.7.2324-2330.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gros O, Wulf-Durand P D, Frenkiel L, Moueza M. Putative environmental transmission of sulfur-oxidizing bacterial symbionts in tropical lucinid bivalves inhabiting various environments. FEMS Microbiol Lett. 1998;160:257–262. [Google Scholar]
- 23.Gundersen J K, Jørgensen B B, Larsen E, Jannasch H W. Mats of giant sulphur bacteria on deep-sea sediments due to fluctuating hydrothermal flow. Nature. 1992;360:454–456. [Google Scholar]
- 24.Hedlund B, Gosink J J, Staley J T. Verrucomicrobia div. nov., a new division of the bacteria containing three new species of Prosthecobacter. Antonie Leeuwenhoek. 1997;72:29–38. doi: 10.1023/a:1000348616863. [DOI] [PubMed] [Google Scholar]
- 25.Hentschel U, Berger E C, Bright M, Felbeck H, Ott J A. Metabolism of nitrogen and sulfur in ectosymbiotic bacteria of marine nematodes (Nematoda, Stilbonematina) Mar Ecol Prog Ser. 1999;183:149–158. [Google Scholar]
- 26.Hentschel U, Cary S C, Felbeck H. Nitrate respiration in chemoautotrophic symbionts of the bivalve Lucinoma aequizonata. Mar Ecol Prog Ser. 1993b;94:35–41. doi: 10.1128/aem.61.4.1630-1633.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hentschel U, Felbeck H. Nitrate respiration in the hydrothermal vent tubeworm Riftia pachyptila. Nature. 1993a;366:338–340. [Google Scholar]
- 28.Hicks R E, Amann R I, Stahl D A. Dual staining of natural bacterioplancton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hulth S, Blackburn T H, Hall P O J. Arctic sediments (Svalbard): consumption and microdistribution of oxygen. Mar Chem. 1994;46:293–316. [Google Scholar]
- 30.JØrgensen B B, Gallardo V A. Thioploca spp.: filamentous sulfur bacteria with nitrate vacuoles. FEMS Microbiol Ecol. 1999;28:301–313. [Google Scholar]
- 31.Krueger D M, Cavanaugh C M. Phylogenetic diversity of bacterial symbionts of Solemya hosts based on comparative sequence analysis of 16S rRNA genes. Appl Environ Microbiol. 1997;63:91–98. doi: 10.1128/aem.63.1.91-98.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuenen J G, Robertson L A, Tuovinen O H. The genera Thiobacillus, Thiomicrospira, and Thiosphaera. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. Vol. 3. New York, N.Y: Springer-Verlag; 1992. pp. 2638–2657. [Google Scholar]
- 33.Laue B E, Nelson D C. Sulfur-oxidizing symbionts have not co-evolved with their hydrothermal vent tube worm hosts: an RFLP analysis. Mol Mar Biol Biotechnol. 1997;6:180–188. [PubMed] [Google Scholar]
- 34.Lee N, Nielsen P H, Andreasen K H, Juretschko S, Nielsen J L, Schleifer K H, Wagner M. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, Kato C, Horikoshi K. Bacterial diversity in deep-sea sediments from different depths. Biodivers Conserv. 1999;8:659–677. [Google Scholar]
- 36.Li L, Kato C, Horikoshi K. Microbial diversity in sediments collected from the deepest cold-seep area, the Japan trench. Mar Biotechnol. 1999;1:391–400. doi: 10.1007/pl00011793. [DOI] [PubMed] [Google Scholar]
- 37.Llobet-Brossa E, Rossello-Mora R, Amann R. Microbial community composition of Wadden sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microbiol. 1998;64:2691–2696. doi: 10.1128/aem.64.7.2691-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacGregor B J, Moser D P, Alm E W, Nealson K H, Stahl D A. Crenarchaeota in Lake Michigan sediment. Appl Environ Microbiol. 1997;63:1178–1181. doi: 10.1128/aem.63.3.1178-1181.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 40.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 41.McHatton S C, Barry J P, Jannasch H W, Nelson D C. High nitrate concentrations in vacuolate, autotrophic marine Beggiatoa spp. Appl Environ Microbiol. 1996;62:954–958. doi: 10.1128/aem.62.3.954-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miskin I O, Farrimond P, Head I M. Identification of novel bacterial lineages as active members of microbial populations in a freshwater sediment using a rapid RNA extraction procedure and RT-PCR. Microbiology. 1999;145:1977–1987. doi: 10.1099/13500872-145-8-1977. [DOI] [PubMed] [Google Scholar]
- 43.Moyer C L, Dobbs F C, Karl D M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system. Loihi Seamount, Hawaii. Appl Environ Microbiol. 1994;60:871–879. doi: 10.1128/aem.60.3.871-879.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mullins T C, Britschgi T B, Krest R L, Gionvannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 45.Neef A. Ph.D. thesis. Munich, Germany: Technische Universität München; 1997. [Google Scholar]
- 46.Neef A, Amann R, Schlesner H, Schleifer K H. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology. 1998;144:3257–3266. doi: 10.1099/00221287-144-12-3257. [DOI] [PubMed] [Google Scholar]
- 47.Ouverney C C, Fuhrman J A. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl Environ Microbiol. 1999;65:1746–1752. doi: 10.1128/aem.65.4.1746-1752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rappé M S, Kemp P F, Giovannoni S J. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr. 1997;42:811–826. [Google Scholar]
- 49.Raskin L, Poulsen L K, Noguera D R, Rittmann B E, Stahl D A. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl Environ Microbiol. 1994;60:1241–1248. doi: 10.1128/aem.60.4.1241-1248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rath J, Wu K Y, Herndl G J, DeLong E F. High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat Microb Ecol. 1998;14:261–269. [Google Scholar]
- 51.Ravenschlag K, Sahm K, Knoblauch C, Jørgensen B B, Amann R. Community structure, cellular rRNA content and activity of sulfate-reducing bacteria in marine arctic sediments. Appl Environ Microbiol. 2000;66:3592–3602. doi: 10.1128/aem.66.8.3592-3602.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ravenschlag K, Sahm K, Pernthaler J, Amann R. High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol. 1999;65:3982–3989. doi: 10.1128/aem.65.9.3982-3989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reysenbach A-L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robertson L A, Kuenen J G. The colorless sulfur bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. Vol. 1. New York, N.Y: Springer-Verlag; 1992. pp. 385–413. [Google Scholar]
- 55.Rondon M R, August P R, Bettermann A D, Brady S F, Grossman T H, Liles M R, A. L K, Lynch B A, MacNeil I A, Minor C, Tiong C L, Gilman M, Osburne M S, Clardy J, Handelsman J, Goodman R M. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosselló-Mora R, Thamdrup B, Schaefer H, Weller R, Amann R. The response of the microbial community of marine sediments to organic carbon input under anaerobic conditions. Syst Appl Microbiol. 1999;22:237–248. doi: 10.1016/S0723-2020(99)80071-X. [DOI] [PubMed] [Google Scholar]
- 57.Sahm K, Berninger U-G. Abundance, vertical distribution, and community structure of benthic prokaryotes from permanently cold marine sediments (Svalbard, Arctic Ocean) Mar Ecol Prog Ser. 1998;165:71–80. [Google Scholar]
- 58.Sahm K, Knoblauch C, Amann R. Phylogenetic affiliation and quantification of psychrophilic sulfate-reducing isolates in marine arctic sediments. Appl Environ Microbiol. 1999;65:3976–3981. doi: 10.1128/aem.65.9.3976-3981.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sahm K, MacGregor B J, Jørgensen B B, Stahl D A. Sulfate reduction and vertical distribution of sulfate-reducing bacteria quantified by rRNA slot-blot hybridization in a coastal marine sediment. Environ Microbiol. 1999;1:65–74. doi: 10.1046/j.1462-2920.1999.00007.x. [DOI] [PubMed] [Google Scholar]
- 60.Sievert S M, Brinkhoff T, Muyzer G, Ziebis W, Kuever J. Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island (Greece) Appl Environ Microbiol. 1999;65:3834–3842. doi: 10.1128/aem.65.9.3834-3842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sievert S M, Ziebis W, Kuever J, Sahm K. Relative abundance of Archaea and Bacteria along a thermal gradient of a shallow-water hydrothermal vent quantified by rRNA slot-blot hybridization. Microbiology. 2000;146:1287–1293. doi: 10.1099/00221287-146-6-1287. [DOI] [PubMed] [Google Scholar]
- 62.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stein J L, Marsh T L, Wu K Y, Shizuya H, DeLong E F. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol. 1996;178:591–599. doi: 10.1128/jb.178.3.591-599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teske A, Sigalevich P, Cohen Y, Muyzer G. Molecular identification of bacteria from a coculture by denaturing gradient gel electrophoresis of 16S ribosomal DNA fragment as a tool for isolation in pure cultures. Appl Environ Microbiol. 1996;62:4210–4215. doi: 10.1128/aem.62.11.4210-4215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Urakawa H, Kita-Tsukamota K, Ohwada K. Microbial diversity in marine sediments from Sagami Bay and Tokyo Bay, Japan, as determined by 16S rRNA gene analysis. Microbiology. 1999;145:3305–3315. doi: 10.1099/00221287-145-11-3305. [DOI] [PubMed] [Google Scholar]
- 68.Vetriani C, Jannasch H W, MacGregor B, Sathl D A, Reysenbach A L. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl Environ Microbiol. 1999;65:4375–4384. doi: 10.1128/aem.65.10.4375-4384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganism. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 70.Wellsbury P, Herbert R A, Parkes R J. Bacterial activity and production in near-surface estuarine and freshwater sediments. FEMS Microbiol Ecol. 1996;19:203–214. [Google Scholar]
- 71.Wilmot D B, Vetter R D. Oxygen- and nitrogen-dependent sulfur metabolism in the thiotrophic clam Solemya reidi. Biol Bull. 1992;182:444–453. doi: 10.2307/1542264. [DOI] [PubMed] [Google Scholar]
- 72.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4314–4317. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]