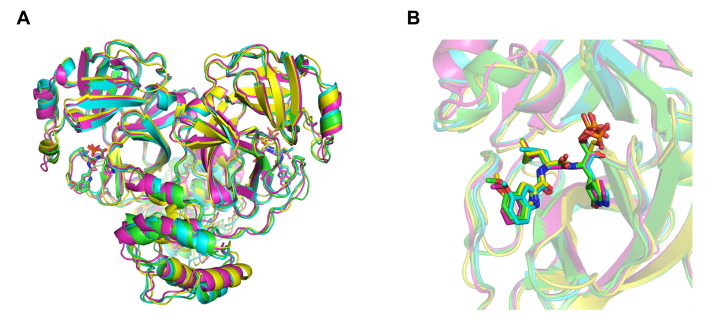
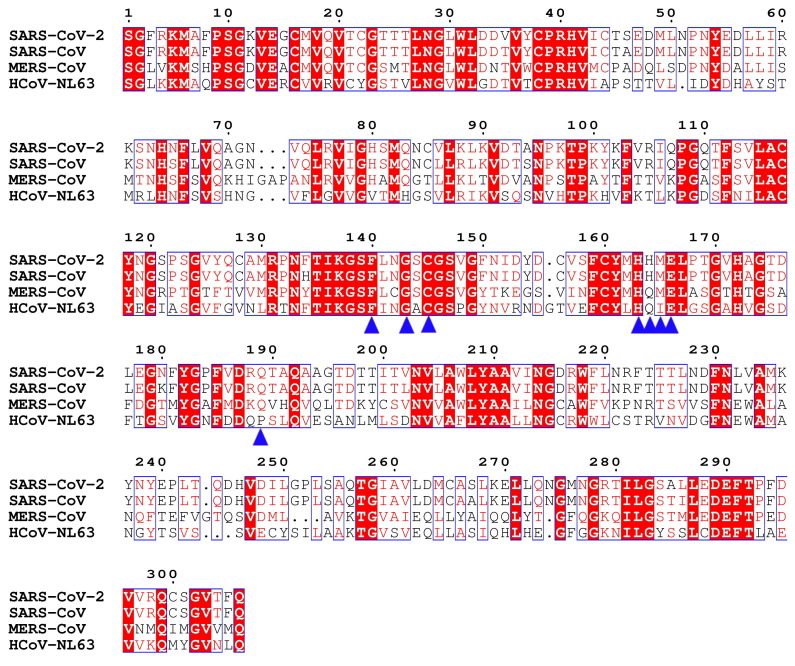
Graphical abstract
Keywords: coronavirus, main protease, PF-07304814, inhibitor, structural basis
Abstract
New variants of the severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) emerged and spread rapidly all over the world, which strongly supports the need for pharmacological options to complement vaccine strategies. Main protease (Mpro or 3CLpro) is a critical enzyme in the life cycle of SARS-CoV-2 and appears to be highly conserved among different genera of coronaviruses, making it an ideal target for the development of drugs with broad-spectrum property. PF-07304814 developed by Pfizer is an intravenously administered inhibitor targeting SARS-CoV-2 Mpro. Here we showed that PF-07304814 displays broad-spectrum inhibitory activity against Mpros from multiple coronaviruses. Crystal structures of Mpros of SARS-CoV-2, SARS-CoV, MERS-CoV, and HCoV-NL63 bound to the inhibitor PF-07304814 revealed a conserved ligand-binding site, providing new insights into the mechanism of inhibition of viral replication. A detailed analysis of these crystal structures complemented by comprehensive comparison defined the key structural determinants essential for inhibition and illustrated the binding mode of action of Mpros from different coronaviruses. In view of the importance of Mpro for the medications of SARS-CoV-2 infection, insights derived from the present study should accelerate the design of pan-coronaviral main protease inhibitors that are safer and more effective.
Introduction
Since December 2019, the pandemic of coronavirus disease 2019 (COVID-19), whose causative agent is the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has taken a tremendous toll on global health and economies1. Along with the newly identified SARS-CoV-2, a total of seven coronaviruses (CoVs) can infect human beings.2, 3 Among these, HCoV-NL63 and HCoV-229E are alphacoronaviruses, while the other five CoVs, namely HCoV-HKU1, HCoV-OC43, SARS-CoV, MERS-CoV, and SARS-CoV-2 are betacoronaviruses.3 The betacoronaviruses can be further classified into four lineages (A, B, C, and D).4, 5 HCoV-HKU1 and HCoV-OC43 belong to lineage A, while SARS-CoV and SARS-CoV-2 belong to lineage B. MERS-CoV is included in lineage C. Unlike the other four CoVs (HCoV-NL63, HCoV-HKU1, HCoV-OC43, and HCoV-229E) that cause relatively mild common colds, SARS-CoV, MERS-CoV, and SARS-CoV-2 cause deadly viral infection in human beings.4, 6, 7 Notably, despite a relatively lower mortality rate, the infection rate of SARS-CoV-2 is even higher than that of SARS-CoV and MERS-CoV.8, 9, 10 As of June 15, 2022, COVID-19 outbreak has resulted in more than 534 million confirmed cases, including more than 6 million deaths (https://covid19.who.int/). The global race to combat this pandemic has led to rapid development and a largescale administration of vaccines against SARS-CoV-2. However, the emergence of viral variants, including Delta (B.1.617.2) and Omicron (B.1.1.529), have compromised vaccine effectiveness.11, 12, 13 Given the ongoing pandemic and disruptive impact, there is still an urgent need to develop new therapeutics for the prompt and effective therapy of SARS-CoV-2 infection.
SARS-CoV-2 produces two large viral polyproteins, pp1a and pp1ab, which are processed by two cysteine proteases, the main protease (Mpro), also called 3C-like protease and the papain-like protease. In particular, Mpro, which cleaves the viral polyproteins at 11 sites to generate a series of non-structural proteins critical for virus replication and transcription, is one of the most attractive targets for numerous kinds of small molecule inhibitors against viral infections.14, 15 Mpro is highly conserved among all seven human coronaviruses and is intolerant of mutation, which makes it a broad-spectrum antiviral target.16 In addition, Mpro has no human homologs, suggesting that selective Mpro inhibitors should avoid unwanted polypharmacology and have minimal side effects.15 To date, the utilization of high-throughput Mpro inhibition assays and computer-aided drug design has led to the discovery of multiple promising Mpro inhibitors, including GC376,17, 18, 19 boceprevir,18, 19 calpain inhibitors II and XII,18 masitinib20 and several rationally redesigned noncovalent inhibitors.21, 22 Unfortunately, most of these inhibitors cannot be developed into effective drugs to treat COVID-19 due to lack of potency, toxicity, or poor metabolic properties.
The most promising inhibitors against SARS-CoV-2 Mpro with drug development potential are PF-07321332 and PF-07304814, both of which are developed by Pfizer. PF-07321332 is the first orally administered inhibitor with in vitro pan-human coronavirus antiviral activity, excellent off-target selectivity and in vivo safety profiles.23 Ritonavir can slow down the metabolic degradation of PF-07321332 in vivo when co-administrated. Currently, the emergency use of PAXLOVIDTM (PF-07321332 and ritonavir) has been authorized by the U.S. Food and Drug Administration (FDA) for the early treatment of people with mild to moderate COVID-19. Unlike PF-07321332 carrying a nitrile warhead, PF-07304814 is a hydroxymethyl ketone (HMK) based covalent inhibitor and must be administered by intravenous infusion.24 As a phosphate prodrug, PF-07304814 rapidly undergoes in vivo conversion to its active form PF-00835231, which exhibits high selectivity over human proteases and acts as a broad-spectrum coronavirus Mpro protease inhibitor.24 Recently, PF-07304814 has completed a Phase 1b study and should enter phase 2/3 study soon. Thus, PF-07304814 could be an alternative therapeutic in the future for hospitalized patients with COVID-19.
Previously, the crystal structures of SARS-CoV and SARS-CoV-2 Mpros in complex with the active moiety of PF-07304814 were solved, which revealed the structural basis of inhibition.25 In order to comprehensively assess the effectiveness and inhibition mechanism of this drug candidate, we explored the molecular basis for the small molecule inhibitor PF-07304814 targeting Mpros of coronaviruses using a structure-based approach. We solved the crystal structures of main proteases of HCoV-NL63, MERS-CoV, SARS-CoV and SARS-CoV-2 bound to PF-07304814 and revealed the architecture of the Mpro active site with bound inhibitors. A detailed analysis and structural comparison revealed the structural similarities and differences of PF-07304814 in binding different Mpros, which will provide a basis for design of broad-spectrum anti-coronaviral drugs.
Results
Broad-spectrum inhibitory activity of PF-07304814 and PF-00835231 against coronavirus Mpros
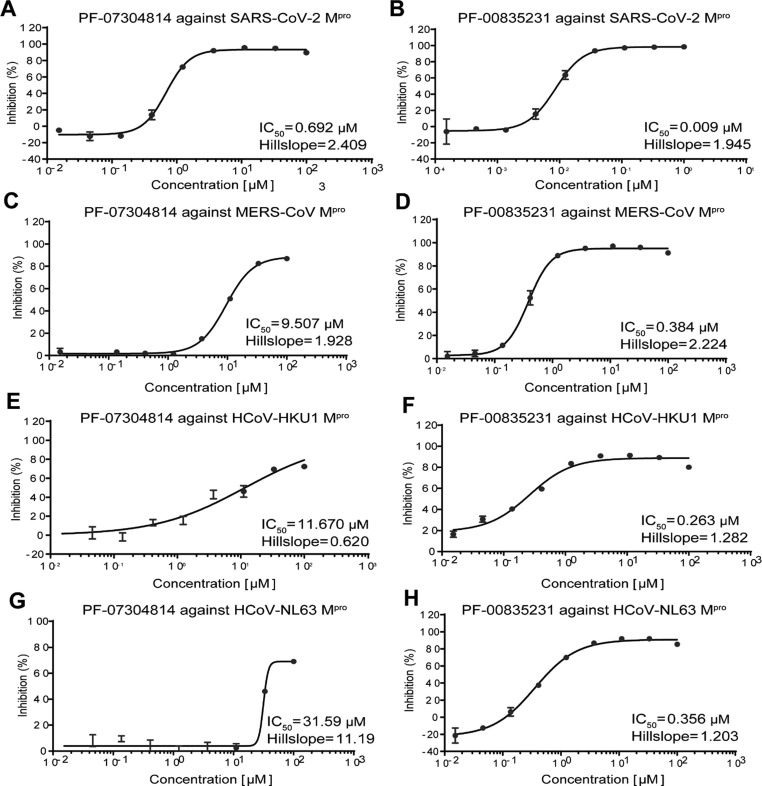
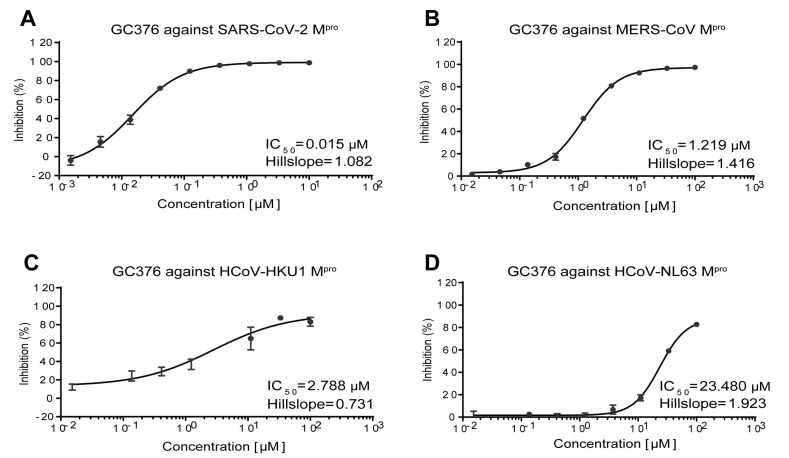
We expressed and purified Mpros of HCoV-NL63, HCoV-HKU1, SARS-CoV-2, and MERS-CoV as previously reported,16 which represent alphacoronavirus, lineage A betacoronavirus, lineage B betacoronavirus, and lineage C betacoronavirus, respectively. Fluorescence resonance energy transfer (FRET) assays were employed to determine the inhibitory activity of PF-07304814 and PF-00835231 against these Mpros. GC376 was used as a positive control and displayed IC50 values ranging from 0.015 μM to 23.48 μM (Figure S1). PF-00835231 inhibits these four Mpros with the IC50 values ranging from 0.0086 μM to 0.3839 μM (Figure 1 ), suggesting a broad-spectrum inhibitory activity. However, the inhibition efficacies of PF-07304814 against these four Mpros are much weaker than that of its active form PF-00835231, with the IC50 values ranging from 0.6921 μM to 31.59 μM.
Figure 1.
Enzymatic inhibition of PF-07304814 and PF-00835231 against main proteasesof different coronaviruses. Inhibition of PF-07304814 and PF-00835231 against main protease of SARS-CoV-2 (A, B), MERS-CoV (C, D), HCoV-HKU1 (E, F) and HCoV-NL63 (G, H). Main proteases were preincubated in the reaction buffer with various concentrations of PF-07304814 or PF-00835231 at room temperature for 30 min before reacting with the FRET substrate The IC50 values were calculated using the GraphPad Prism software.
Inhibitory mechanism of PF-07304814 against SARS-CoV-2 Mpro
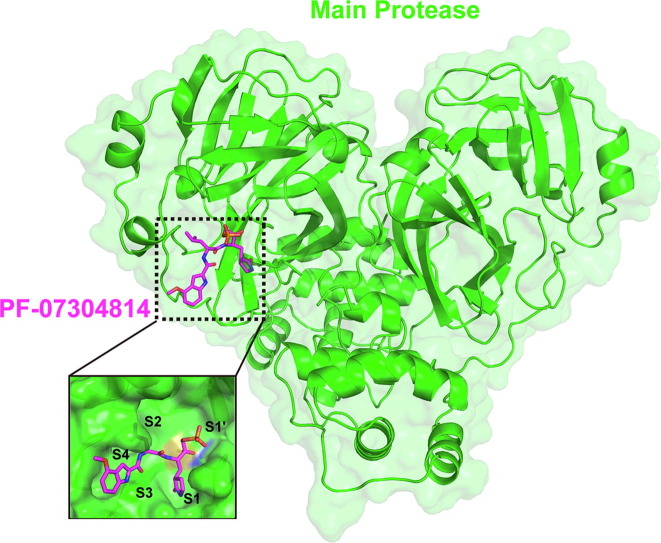
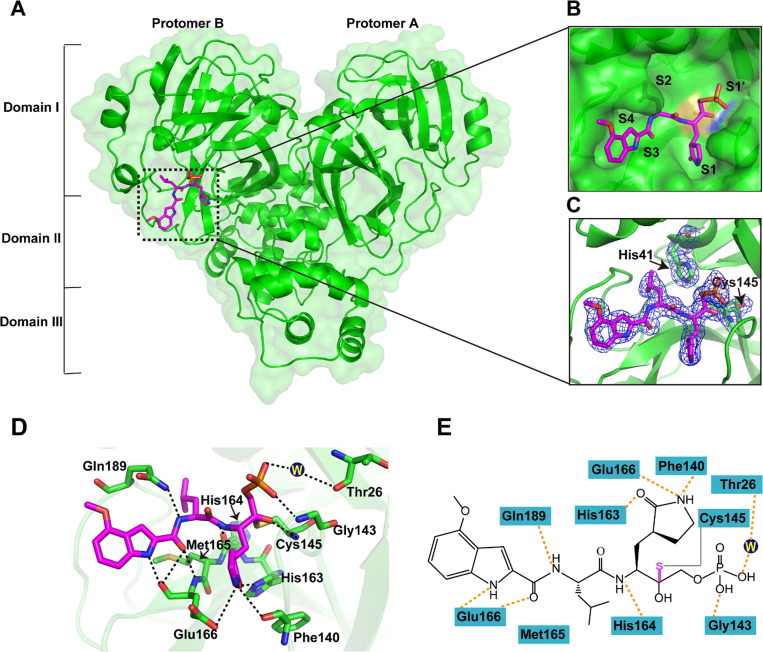
In order to figure out the inhibitory mechanisms of PF-07304814, we first determined the crystal structure of SARS-CoV-2 Mpro in complex with PF-07304814 at 1.97 Å resolution using co-crystallization method (Table 1 ). The complex structure is in space group P212121. As shown in Figure 2 , the Mpro molecule in the complex structure consists of two protomers (A and B) and each contains three distinct domains, namely domain I (residues 10–99), domain II (residues 100–184), and domain III (residues 201–303). However, PF-07304814 can be only found in protomer B (Figure 2(A)). Specifically, PF-07304814 binds to the active site situated in the cleft between domains I and II of Mpro in an extended conformation and occupies S1, S2 and partial S3 pockets of SARS-CoV-2 Mpro, but not the canonical S4 site of this protease (Figure 2(B)). The electron density map unambiguously shows that the carbonyl carbon of the HMK warhead of PF-07304814 forms a covalent bond with the sulfur atom (2.2 Å C—S bond length) of the Mpro active-site cysteine (Cys145), generating a tetrahedral carbinol complex (Figure 2(C)). Besides the typical covalent interaction, PF-07304814 forms multiple non-covalent interactions within the active site. The carbinol hydroxyl forms a hydrogen-bonding interaction with the backbone NH of Cys145. PF-07304814 consists of four moieties, namely P1′ and P1-P3. As shown in Figure 2(D) and (E), PF-07304814 has a phosphate moiety at P1′ site. One of the hydroxy groups forms a hydrogen-bonding interaction with the amide NH of Gly143, while another hydroxy group forms a hydrogen bond with Thr26 via a bridging water molecule. PF-07304814 displays a lactam ring that is deeply embedded in the S1 pocket of the protease at P1 position. The lactam carbonyl forms a strong hydrogen-bonding interaction with the Nε2 of H163, while the lactam NH forms hydrogen-bonding interactions with the side-chain oxygen of Glu166 and the backbone oxygen of Phe140. Further, a hydrogen-bonding interaction is also found between P1 NH and the main-chain carbonyl oxygen of His164. The P2 position contains a leucine moiety which inserts into the S2 pocket, which is formed by residues His41, Cys43, Met49, Pro52, Tyr54, Met165, Asp187, Arg188, and Gln189. The inhibitor P2 NH forms a hydrogen-bonding interaction with the side chain of Gln189. Met165 of SARS-CoV-2 Mpro makes several Van Der Waals interactions with the inhibitor. PF-07304814 presents an indole group at P3 position but does not fit the S3 subsite well, which is quite different with the previous reported α-ketoamide inhibitor of SARS-CoV-2 Mpro.26 The backbone carbonyl and NH of Glu166 form several hydrogen-bonding interactions with the indole fragment of PF-07304814. Therefore, PF-07304814 occupies the active site of SARS-CoV-2 Mpro by covalently binding to Cys145 and non-covalently interacting with conserved residues including Cys145, His163, His164, Glu166 and Gln189.
Table 1.
Statistics for data processing and model refinement of Mpro-PF-07304814 complexes.
| SARS-CoV-2 Mpro-PF-07304814 | SARS-CoV Mpro-PF-07304814 | MERS-CoV Mpro-PF-07304814 | HCoV-NL63 Mpro-PF-07304814 | |
|---|---|---|---|---|
| PDB code | 7VVP | 7WQI | 7WQJ | 7WQH |
| Data collection | ||||
| Synchrotron | SSRF | SSRF | SSRF | SSRF |
| Beam line | BL17U1 | BL17U1 | BL17U1 | BL17U1 |
| Wavelength (Å) | 0.97918 | 0.97918 | 0.97918 | 0.97918 |
| Space group | P212121 | P1 | P212121 | P1211 |
| a, b, c (Å) | 68.07, 102.27, 103.75 | 55.30, 61.02, 68.70 | 82.52, 92.77, 97.70 | 63.09, 82.79, 64.42 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 91.49, 101.91, 108.51 | 90.00, 90.00, 90.00 | 90.00, 109.00, 90.00 |
| Total reflections | 413,236 | 59,368 | 162,667 | 93,452 |
| Unique reflections | 49,032 | 31,785 | 19,794 | 25,391 |
| Resolution (Å) | 1.97(2.07–1.97) | 2.33(2.46–2.33) | 2.75(2.90–2.75) | 2.32(2.45–2.32) |
| R-merge (%) | 3.8(56.6) | 4.3(9.4) | 7.4(86.7) | 5.4(57.7) |
| Mean I/σ (I) | 20.6 /4.1 | 4.8 /2.5 | 9.2 /2.7 | 7.9/2.6 |
| Completeness (%) | 93.9(91.1) | 91.1(91.8) | 98.5(92.1) | 93.4(98.2) |
| Redundancy | 8.4(5.9) | 2.0(1.5) | 8.2(4.5) | 3.7(4.0) |
| Refinement | ||||
| Resolution (Å) | 34.04–1.97 | 36.15–2.33 | 67.28–2.75 | 37.02–2.32 |
| Rwork/Rfree(%) | 20.63/24.93 | 20.47/24.26 | 22.43/28.98 | 20.41/26.16 |
| Atoms | 4565 | 4531 | 4565 | 4469 |
| Mean temperature factor (Å2) | 43.0 | 39.3 | 39.5 | 44.6 |
| Bond lengths (Å) | 0.008 | 0.008 | 0.012 | 0.007 |
| Bond angles (°) | 0.943 | 0.99 | 1.05 | 0.91 |
| Preferred | 96.77 | 97.93 | 96.30 | 97.47 |
| Allowed | 3.23 | 2.07 | 3.70 | 2.53 |
| Outliers | 0 | 0 | 0 | 0 |
Figure 2.
Crystal structure of SARS-CoV-2 Mpro in complex with PF-07304814. (A) Overall structure of SARS-CoV-2 Mpro in complex with PF-07304814. Three domains and two protomers of Mpro are labeled. The substrate-binding pocket is situated within the black dotted box. PF-07304814 is shown as sticks with the carbon atoms in magentas, oxygen atoms in red, nitrogen atoms in blue, and phosphorus atoms in orange. (B) An enlarged view of the substrate-binding pocket. PF-07304814 forms a covalent bond with Cys145. The substrate-binding subsites (S1′, S1, S2, S3 and S4) are labeled. (C) A C-S covalent bond forms between Cys145 and the hydroxymethyl ketone of PF-07304814. The 2Fo–Fc density map contoured at 1.0σ is shown as blue mesh. (D) The detailed interaction in the complex structure is shown with the residues involved in inhibitor binding (within 3.5 Å) displayed as sticks. W represents the water molecule. Hydrogen-bonding interactions are shown as black dashed lines. (E) Schematic interaction between PF-07304814 and Mpro. Hydrogen-bonding interactions are shown as orange dashed lines.
Comparison of structures of SARS-CoV-2 Mpro in complex with PF-07304814 and PF-00835231
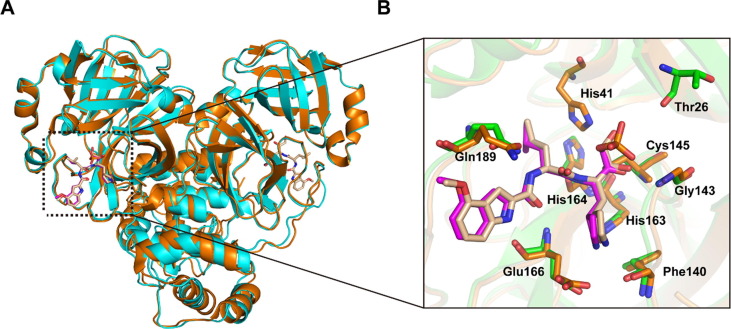
Recently, the crystal structure of PF-00835231 in complex with SARS-CoV-2 Mpro has been solved.25 PF-00835231 is the active form of PF-07304814. Unlike PF-07304814 that displays a phosphate moiety, PF-00835231 has a hydroxy group at P1′ site just after the warhead. Superimposition of Mpro-PF-00835231 and Mpro-PF-07304814 complexes reveals high similarity with the root mean square deviation (RMSD) of equivalent Cɑ positions being 1.107 Å (Figure 3 (A)). Both PF-07304814 and PF-00835231 form hydrogen-bonding interactions with residues Gln189, His163, His164, Glu166, Phe140, and Cys145 (Figure 3(B)). The largest difference was observed in interactions with the residues in S1′ pocket. The primary alcohol moiety forms a hydrogen-bonding interaction with catalytic residue His41 in the case of Mpro-PF-00835231 complex, while the phosphate group forms a hydrogen-bonding interaction with Thr26 via a water molecule in the case of Mpro-PF-07304814 complex. The difference in binding mode may contribute to the difference of inhibitory activity.
Figure 3.
Comparison of the binding modes of PF-07304814 and PF-00835231. (A) Comparison of overall structures between SARS-CoV-2 Mpro-PF-07304814 and SARS-CoV-2 Mpro-PF-00835231 (PDB ID 6XHM). Structures of SARS-CoV-2 Mpro bound to PF-07304814 and PF-00835231 are shown in cyan and orange, respectively. PF-07304814 and PF-00835231 are shown in magentas and wheat sticks, respectively. (B) A zoomed in view of substrate binding pocket of SARS-CoV-2 Mpro. The key residues of SARS-CoV-2 Mpro involved in hydrogen-bonding interaction with PF-07304814 and PF-00835231 are shown as green and orange sticks, respectively.
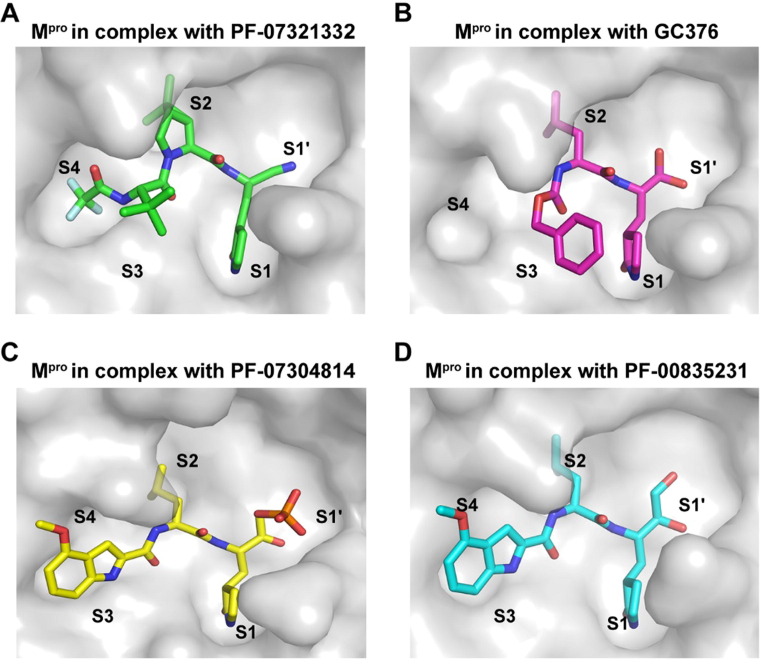
Structural comparison of PF-07304814, GC-376, and PF-07321332 in complex with SARS-CoV-2 Mpro
PF-07304814, PF-07321332, and GC-376 represent the most advanced drug candidates targeting the SARS-CoV-2 main protease. Among them, PF-07321332 was developed by modification of PF-07304814, and is an analog of the peptidomimetic compound GC376, which has been licensed to treat feline infectious peritonitis caused by a feline coronavirus. Recently, the crystal structures of GC376 and PF-07321332 in complex with SARS-CoV-2 Mpro have been solved.19, 27 All of them form a C-S covalent bond with the catalytic cysteine and display a lactam ring at the P1 position (Figure 4 ), which is suitable to occupy the S1 pocket by forming several hydrogen-bonding interactions with Phe140, His163, and Glu166. The binding pattern of PF-07304814 as well as PF-00835231 is similar to that of GC376 at the P2 position. The P3 position of these three inhibitors does not fit well with the S3 pocket, suggesting that structural optimization will be required to identify more suitable groups for this subsite. Unlike PF-07304814, PF-00835231 and GC376, PF-07321332 displays a trifluoromethyl group at the P4 position which fit the S4 subsite well. Therefore, these structures will provide molecular basis for drug development and optimization.
Figure 4.
Comparison of the binding modes of four inhibitors targeting SARS-CoV-2 Mpro. (A to D) The binding pockets of PF-07321332 (A) (PDB ID 7VLQ), GC376 (B) (PDB ID 7D1M), PF-07304814 (C) (PDB ID 7VVP), and PF-00835231 (D) (PDB ID 6XHM) bound to SARS-CoV-2 Mpro are shown. Mpros are shown as the gray surface, and the inhibitors are shown as sticks.
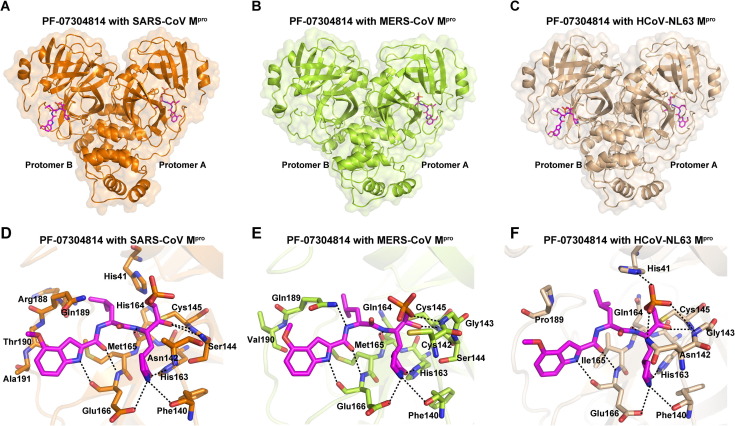
Crystal structures of PF-07304814 in complex with Mpros of SARS-CoV, MERS-CoV, and HCoV-NL63
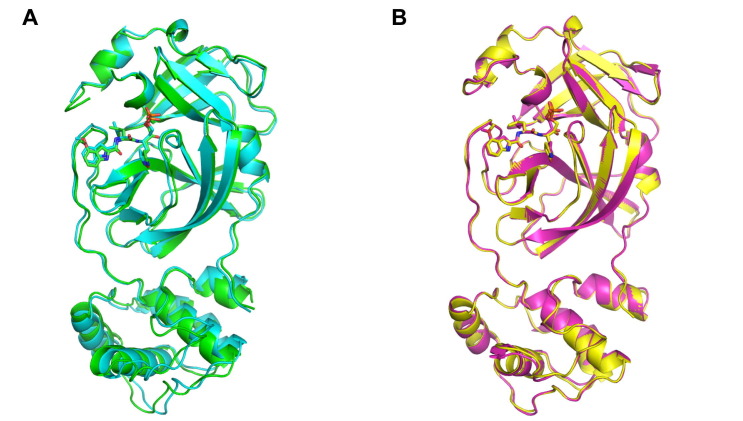
We also determined the crystal structures of PF-07304814 in complex with Mpros of SARS-CoV, MERS-CoV, and HCoV-NL63 at 2.33 Å, 2.75 Å, and 2.32 Å resolution (Table 1), respectively. The space group of these structures are P1, P212121, and P1211, respectively. By overall comparison, these three complex structures show high similarity with SARS-CoV-2 Mpro-PF-07304814 complex with the RMSD of equivalent Cɑ positions ranging from 0.851 Å to 1.435 Å. In the case of MERS-CoV Mpro-PF-07304814 complex, ligand can be only found in protomer B. In the structures of PF-07304814 in complex with SARS-CoV and HCoV-NL63 Mpros, the small molecule can be found in both protomer A and protomer B. These two protomers have similar configurations when bound to PF-07304814 (Figure S2), with the RMSD for all equivalent Ca atoms being 0.900 Å in SARS-CoV Mpro and 0.282 Å in HCoV-NL63 Mpro. As expected, PF-07304814 displays a similar conformation in the substrate-binding sites of SARS-CoV-2, SARS-CoV, MERS-CoV, and HCoV-NL63 Mpros even that the orientation of each moiety of PF-07304814 has slight differences (Figure 5 (A–C), Figure S3). Like the case with SARS-CoV-2 Mpro, PF-07304814 forms a covalent bond with the catalytic cysteine residue in Mpros of SARS-CoV, MERS-CoV, and HCoV-NL63 (Figure 5(D–F)). The key amino acid residues of different Mpros interacting with PF-07304814 are highly conserved, including Phe140, Cys145, His163, His/Gln164, Glu166, and Gln/Pro189 (Figure 5(D–F), Figure S4). Among these, Phe140, Cys145, His163, His/Gln164, and Glu166 form hydrogen-bonding interactions with the inhibitor in all the four complex structures (Figure 5(D–F)). The main differences for the four main proteases interacting with PF-07304814 were observed in the residues 142–144 and 188–192, which can be attributed to the flexibility. Specifically, the backbone NH of Ser144 in SARS-CoV Mpro form a hydrogen-bonding interaction with the carbinol hydroxyl of PF-07304814 at P1′ site (Figure 5(D)), while it is the backbone NH of Gly143 in SARS-CoV-2, MERS-CoV, and HCoV-NL63 Mpros forms hydrogen-bonding interactions with hydroxy group or backbone oxygen atom of PF-07304814 at P1′ site (Figure 2(D), Figure 5(E–F)). Furthermore, residue 189 forms van der Waals interactions with the P2 moiety of PF-07304814 in SARS-CoV-2, SARS-CoV, and HCoV-NL63 Mpros, while residue 189 in MERS-CoV Mpro forms a hydrogen-bonding interaction with the P2 NH of PF-07304814. In addition, His41 in Mpros of SARS-CoV and HCoV-NL63 is involved in the interactions with PF-07304814 (Figure 5(D), Figure 5(F)), however no interactions form between residue His41 of SARS-CoV-2 and MERS-CoV Mpros and PF-07304814 (Figure 2(A), Figure 5(E)). The imidazole ring of His41 of HCoV-NL63 Mpro flips over compared with that of SARS-CoV and forms a hydrogen-bonding interaction with the phosphate group of PF-07304814 (Figure 5(D), Figure 5(F)).
Figure 5.
Crystal structures of SARS-CoV, MERS-CoV and HCoV-NL63 Mpros in complex with PF-07304814. (A–C) Overall structures of SARS-CoV Mpro (A), MERS-CoV Mpro (B) and HCoV-NL63 Mpro (C) in complex with PF-07304814. Two protomers of Mpro are labeled. PF-07304814 is shown as sticks with the carbon atoms in magentas, oxygen atoms in red, nitrogen atoms in blue, and phosphorus atoms in orange. (D-F) The detailed interactions in the SARS-CoV Mpro-PF-07304814 complex (D), MERS-CoV Mpro-PF-07304814 complex (E), and HCoV-NL63 Mpro-PF-07304814 complex (F) are shown with the residues involved in inhibitor binding (within 3.5 Å) displayed as sticks. Hydrogen-bonding interactions are shown as black dashed lines.
These observations provide the structural basis for how PF-07304814 inhibits different coronavirus Mpros and support that PF-07304814 can be a potent inhibitor with broad-spectrum property.
Discussion
There is an urgent need to develop effective drugs as the SARS-CoV-2 pandemic continues to cause significant chaos to the international community and pose a threat to public health. Mpro is a promising drug target of SARS-CoV-2 for its vital role in viral replication and high conservation among different human-infected coronaviruses. Moreover, Mpro has no homolog in humans. Although a great number of inhibitors have been found to show potent inhibitory activity to SARS-CoV-2 Mpro until now, few have been approved clinically. PF-07304814 is a SARS-CoV-2 Mpro inhibitor developed by Pfizer and has entered into clinical trial.24 It must be administrated through intravenous injection, thus making it suitable to treat critical patients with COVID-19. In this study, we found PF-07304814 is a potent inhibitor against SARS-CoV-2 Mpro and showed broad-spectrum inhibitory activity against Mpros from multiple human-infected coronaviruses, similar with another ketoamide inhibitor boceprevir.19 With the emergence of SARS-CoV-2 variants as well as other possible human-infected coronaviruses, inhibitors targeting such a viral target with conserved and essential properties may complement the clinical use of available vaccines and antivirals.
However, we observed that the in vitro inhibitory efficacy of PF-07304814 varied among the different main proteases, with the IC50 values against main protease activities of SARS-CoV, MERS-CoV, and HCoV-NL63 less potent when compared with that against main protease activity of SARS-CoV-2. The reason could be many factors. The binding affinity differences between the inhibitor and enzyme may be one of the reasons for the different inhibitory effects of PF-07304814. In addition, we cannot rule out that differences in protein stability may contribute to the inhibition differences.
We also solved the crystal structures of PF-07304814 in complex with Mpros of four human-infected coronaviruses (HCoV-NL63, MERS-CoV, SARS-CoV, and SARS-CoV-2). The structures indicate that PF-07304814 is covalently bound to the catalytic cysteine of Mpro, and formed multiple hydrogen bonds with conserved residues within the active site. Compared with PF-00835231, PF-07304814 employs a similar binding pattern to SARS-CoV-2 Mpro but with some key differences. These data complement the previous report about the structure of PF-00835231 in complex with Mpros of SARS-CoV-2 and SARS-CoV and are beneficial to comprehensively understand the inhibitory mechanism of PF-07304814 as well as PF-00835231, thus providing structural basis and critical insight for pharmaceutical development. Further structure-based optimization of such covalent inhibitors would generate potent drugs against current COVID-19 pandemic and potential pandemic in future.
Materials and Methods
Expression and purification of Mpro from different human CoVs
The codon-optimized cDNAs for Mpro of SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-HKU1 and HCoV-NL63 were commercially synthesized. Synthesized genes were then subcloned into the pET-28a vector with a 6*His tag fused to their N terminus. The expression and purification of these main proteases were performed according to the standard method described previously by our lab.16 TEV protease was used to remove the N-terminal His tag of Mpros. Then Mpros were further purified by size-exclusion chromatography.
Enzymatic assays
Fluorogenic substrates as a donor and quencher pair were synthesized. The inhibitory activities of the PF-07304814 and PF-00835231 against main proteases of SARS-CoV-2, MERS-CoV, HCoV-HKU1, and HCoV-NL63 were measured using the FRET-based enzymatic assays. GC376 was used as a positive control. Firstly, 1 μL of main protease (200 nM) was incubated with various concentrations of GC376, PF-07304814 or PF-00835231 at room temperature for 30 min in a 384-well plate, and then FRET substrates were added into each well. The reaction was monitored for 20 min, and the data were calculated at 10 min intervals by linear regression. The IC50 values were calculated by plotting the initial velocity against various concentrations of testing inhibitor using the dose–response curve in GraphPad Prism software.
Crystallization
PF-07304814 was mixed with Mpros according to a molar ratio of 3:1 and the mixture was incubated for 30 min on ice. The mixture was then used for crystallization, which was performed using a sitting-drop vapor diffusion method at 20 °C. After 3–5 days, the complex crystals of Mpros with PF-07304814 were obtained. The crystals of SARS-CoV-2 Mpro in complex with PF-07304814 were grown in the reservoir solution comprising 0.1 M HEPES pH 7.5, 20% w/v PEG 10000. The crystals of SARS-CoV Mpro in complex with PF-07304814 were grown in the reservoir solution comprising 0.1 M HEPES pH 7.5, 10% PEG 8000, 8% Ethylene glycol. The crystals of MERS-CoV Mpro in complex with PF-07304814 were grown in the reservoir solution comprising 10% PEG 200, 0.1 M Bis-Tris-propane pH9.0, 18% PEG 8000. The crystals of HCoV-NL63 Mpro in complex with PF-07304814 were grown in the reservoir solution comprising 0.1 M Sodium citrate tribasic dihydrate pH 5.0, 18% w/v PEG 20,000.
Data collection, structure determination and refinement
The obtained crystals were cryo-protected by soaking in reservoir solution supplemented with 20% glycerol before flash-cooling in liquid nitrogen. All data sets were collected at 100 K on macromolecular crystallography beamline02U1 (BL02U1) at Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China). All collected data were handled by the HKL 2000 software package. The structures were determined by molecular replacement with PHENIX software. The program Coot was used to rebuild the initial model. Data collection and refinement statistics are summarized in Table 1. Coordinates and structure factors for SARS-CoV Mpro-PF-07304814, SARS-CoV Mpro-PF-07304814, MERS-CoV Mpro-PF-07304814 and HCoV-NL63 Mpro-PF-07304814 complexes have been deposited in the Protein Data Bank (PDB) under accession numbers 7VVP, 7WQI, 7WQJ and 7WQH, respectively.
CRediT authorship contribution statement
Jian Li: Conceptualization, Investigation, Methodology, Data curation, Writing – review & editing, Funding acquisition. Cheng Lin: Investigation. Xuelan Zhou: Investigation. Fanglin Zhong: Investigation. Pei Zeng: Investigation. Peter J. McCormick: Investigation, Writing – review & editing. Haihai Jiang: Conceptualization, Supervision, Writing – original draft, Writing – review & editing. Jin Zhang: Conceptualization, Supervision, Writing – review & editing, Funding acquisition.
Acknowledgments
Acknowledgements
J.L. was supported by Gannan Medical University (QD201910), Jiangxi natural science Foundation for distinguished young scholar (20212ACB216001), Jiangxi key research and development program (20203BBG73063) and Jiangxi “Double Thousand Plan (jxsq2019101064)''. J.Z. was supported by the Thousand Young Talents Program of China, the National Natural Science Foundation of China (grant no. 31770795; grant no. 81974514), and the Jiangxi Province Natural Science Foundation (grant no. 20181ACB20014). P.J.M was supported by the Foreign Talent project of Jiangxi Province. This work was also supported by Ganzhou COVID-19 Emergency Research Project (2020.17), Major science and technology programs of Ganzhou City (2020.67) and Ganzhou Zhanggong District COVID-19 prevention and control key research projects (2020.67).
Author contributions
J.L. H.J and J.Z. initiated and supervised the project. C.L., X.Z., F.Z. and P.Z. crystallized the protein complexes, performed soaking experiments. J.L., C.L., X.Z., F.Z., P.Z., P.J.M. and J.Z. collected X-ray data, and solved and refined structures. H.J., J.L., P.J.M and J.Z analyzed the data and wrote the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Edited by M.F. Summers
Footnotes
Supplementary material to this article can be found online at https://doi.org/10.1016/j.jmb.2022.167706.
Appendix A. Supplementary material
The following are the Supplementary material to this article:
Supplementary Figure S1.
Enzymatic inhibition of GC376 against main protease of different coronaviruses. Inhibition of GC376 against main protease of SARS-CoV-2 (A), MERS-CoV (B), HCoV-HKU1 (C), and HCoV-NL63 (D). Main proteases were preincubated in the reaction buffer with various concentrations of GC376 at room temperature for 30 min before reacting with the FRET substrate The IC50 values was calculated using the GraphPad Prism software.
Supplementary Figure S2.
Structural comparison of protomer A and B of SARS-CoV and HCoV-NL63 Mpros bound to PF-07304814. (A) Structural comparison of protomer A and protomer B of SARS-CoV Mpro bound to PF-07304814. Protomer A (cyan) and B (green) of SARS-CoV Mpro are shown as cartoon, while ligand shown as sticks. (B) Structural comparison of protomer A and protomer B of HCoV-NL63 Mpro bound to PF-07304814. Protomer A (yellow) and B (magentas) of HCoV-NL63 Mpro are shown as cartoon, while ligand shown as sticks.
Supplementary Figure S3.
Structural comparison of PF-07304814 in complex with Mpros of SARS-CoV, MERS-CoV, and HCoV-NL63. (A) Structural alignment of CoV Mpros bound to PF-07304814 with SARS-CoV-2 Mpro-inhibitor complex in green, SARS-CoV Mpro-inhibitor complex in cyan, MERS-CoV Mpro-inhibitor complex in yellow, and HCoV-NL63 Mpro-inhibitor complex in magentas. (B) An enlarged view of the substrate-binding pocket.
Supplementary Figure S4.
Sequence alignment of Mpros from SARS-CoV-2, SARS-CoV, MERS-CoV and HCoV-NL63. Amino acid sequence of Mpros from SARS-CoV-2, SARS-CoV, MERS-CoV and HCoV-NL63 are retrieved from GenBank. ClustalX (1.83) software is used for sequence alignment and ESPript 3.0 was used to generate a graphical representation. The key residues involve in interacting with inhibitors are labeled with blue triangle.
Data availability
Data will be made available on request.
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye Z.W., Yuan S., Yuen K.S., Fung S.Y., Chan C.P., Jin D.Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020;16:1686–1697. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., et al. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;28:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nature Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madewell Z.J., Yang Y., Longini I.M., Jr, Halloran M.E., Dean N.E. Household Transmission of SARS-CoV-2: A Systematic Review and Meta-analysis. JAMA Netw. Open. 2020;3:e2031756. doi: 10.1001/jamanetworkopen.2020.31756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel K.P., Vunnam S.R., Patel P.A., Krill K.L., Korbitz P.M., Gallagher J.P., Suh J.E., Vunnam R.R. Transmission of SARS-CoV-2: an update of current literature. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:2005–2011. doi: 10.1007/s10096-020-03961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M., Zhang L., Li Q., Wang B., Liang Z., Sun Y., Nie J., Wu J., et al. Reduced sensitivity of the SARS-CoV-2 Lambda variant to monoclonal antibodies and neutralizing antibodies induced by infection and vaccination. Emerg. Microbes Infect. 2022;11:18–29. doi: 10.1080/22221751.2021.2008775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ai J., Zhang H., Zhang Y., Lin K., Zhang Y., Wu J., Wan Y., Huang Y., et al. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg. Microbes Infect. 2021;22:1–24. doi: 10.1080/22221751.2021.2022440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L., Iketani S., Guo Y., Chan J.F., Wang M., Liu L., Luo Y., Chu H., et al. Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2. Nature. 2021;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 14.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses - drug discovery and therapeutic options. Nature Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of Anti-SARS Drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 16.Li J., Zhou X., Zhang Y., Zhong F., Lin C., McCormick P.J., Jiang F., Luo J., et al. Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode. Sci. Bull. (Beijing) 2021;66:661–663. doi: 10.1016/j.scib.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vuong W., Khan M.B., Fischer C., Arutyunova E., Lamer T., Shields J., Saffran H.A., McKay R.T., et al. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nature Commun. 2020;11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. CellRes. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., et al. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nature Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drayman N., DeMarco J.K., Jones K.A., Azizi S.A., Froggatt H.M., Tan K., Maltseva N.I., Chen S., et al. Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2. Science. 2021;373:931–936. doi: 10.1126/science.abg5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitamura N., Sacco M.D., Ma C., Hu Y., Townsend J.A., Meng X., Zhang F., Zhang X., et al. Expedited Approach toward the Rational Design of Noncovalent SARS-CoV-2 Main Protease Inhibitors. J. Med. Chem. 2022;65:2848–2865. doi: 10.1021/acs.jmedchem.1c00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C.H., Stone E.A., Deshmukh M., Ippolito J.A., Ghahremanpour M.M., Tirado-Rives J., Spasov K.A., Zhang S., et al. Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations. ACS Cent. Sci. 2021;7:467–475. doi: 10.1021/acscentsci.1c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owen D.R., Allerton C., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 24.Boras B., Jones R.M., Anson B.J., Arenson D., Aschenbrenner L., Bakowski M.A., Beutler N., Binder J., et al. Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19. Nature Commun. 2021;12:6055. doi: 10.1038/s41467-021-26239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman R.L., Kania R.S., Brothers M.A., Davies J.F., Ferre R.A., Gajiwala K.S., He M., Hogan R.J., et al. Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19. J. Med. Chem. 2020;63:12725–12747. doi: 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:eabb3405. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Lin C., Zhou X., Zhong F., Zeng P., Yang Y., Zhang Y., Yu B., et al. Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332. J. Virol. 2022;96:e0201321. doi: 10.1128/jvi.02013-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.