Abstract
We have developed a xylose-dependent expression system for tight and modulated expression of cloned genes in Bacillus subtilis. The expression system is contained on plasmid pSWEET for integration at the amyE locus of B. subtilis and incorporates components of the well-characterized, divergently transcribed xylose utilization operon. The system contains the xylose repressor encoded by xylR, the promoter and 5′ portion of xylA containing an optimized catabolite-responsive element, and intergenic xyl operator sequences. We have rigorously compared this expression system to the isopropyl-β-d-thiogalactopyranoside-induced spac system using a thermostable β-galactosidase reporter (BgaB) and found the xyl promoter-operator to have a greater capacity for modulated expression, a higher induction/repression ratio (279-fold for the xyl system versus 24-fold with the spac promoter), and lower levels of expression in the absence of an inducer. We have used this system to probe an essential function in wall teichoic acid biosynthesis in B. subtilis. Expression of the teichoic acid biosynthesis gene tagD, encoding glycerol-3-phosphate cytidylyltransferase, from the xylose-based expression system integrated at amyE exhibited xylose-dependent complementation of the temperature-sensitive mutant tag-12 when grown at the nonpermissive temperature. Plasmid pSWEET thus provides a robust new expression system for conditional complementation in B. subtilis.
Manipulated gene expression in bacteria is of fundamental importance to understanding the effects of expression or depletion of gene products on bacterial physiology. Such investigations ideally require the controlled expression of a cloned gene from a tightly regulated, inducible promoter. This is particularly true in studies of indispensable genes in bacteria where targeted deletions of these genes require expression from complementing copies of the genes. The dispensability of such genes and the phenotypes associated with their loss are most rigorously examined through conditional expression of the complementing genes. Especially challenging in such studies are the need for extremely tight regulation of expression and the tendency for the majority of regulated expression systems to “leak” in the absence of an inducer.
The state of the art in controlled expression in Bacillus subtilis is the spac expression system, which is based on the application of the lac repressor-operator control system from Escherichia coli. In B. subtilis, the system uses a constitutive penicillinase promoter to express the lac repressor. Isopropyl-β-d-thiogalactopyranoside (IPTG)-dependent expression of the target gene is from a hybrid promoter-operator consisting of the SPO1 bacteriophage promoter and lac operator sequence (35). Despite its broad use for conditional expression in B. subtilis, the spac system has been widely recognized for its capacity to allow significant expression in the absence of an inducer. Indeed, only relatively recently has there been a systematic study of expression from the spac promoter, indicating demonstrable uninduced expression of lacZ from the spac promoter of the pMUTIN vector system (33).
The xylose operon has emerged as a well-characterized B. subtilis regulatory system with the potential for particularly tight transcriptional regulation (5, 6, 12, 13, 16–19). Xylose utilization in B. subtilis requires the production of xylose isomerase (XylA) and xylulose kinase (XylB) and is regulated at the level of transcription by a xylose-responsive repressor protein encoded by xylR and by catabolite repression. Genes xylR and xylAB are divergently transcribed from a common intergenic region containing xyl operator sequences which are bound by XylR in the absence of an inducer. Transcription of the xyl operon is also catabolite repressed through the cis-acting catabolite-responsive element (CRE) located in the coding sequence of xylA.
Cell wall teichoic acids are a diverse group of phosphate-rich polymers which are covalently linked to peptidoglycan and constitute a substantial portion of the cell wall of gram-positive bacteria. Teichoic acids have been implicated as virulence factors in a variety of gram-positive bacterial infections (14, 22, 30), and growing evidence indicates that teichoic acid biosynthesis is indispensable for the growth of B. subtilis (2, 21, 23). Conditional lethal mutations in the poly(glycerol phosphate) teichoic acid gene cluster (tag) of B. subtilis 168 have been mapped to a number of genes, including tagD (27), encoding glycerol-3-phosphate cytidylyltransferase (25).
In this work, we have taken advantage of a notable depth of knowledge of the xylose utilization operon to develop a xylose-dependent promoter-operator system for tight and modulated expression of cloned genes in B. subtilis. As a point of reference, we have rigorously compared this expression system to the IPTG-induced spac system for efficiency of regulation and modulation of expression. Finally, we have put expression of tagD under control of the xylose-based expression system described here and have for the first time shown trans complementation of a teichoic acid mutant.
MATERIALS AND METHODS
General methods.
Strains, plasmids, and primers used are listed in Tables 1 and 2. B. subtilis strains were grown in rich (Luria-Bertani [LB]) or minimal [15 mM (NH4)2SO4, 80 mM K2HPO4, 44 mM KH2PO4, 3.4 mM sodium citrate, 1 mM MgSO4 (pH 7.4)] medium plus arabinose (0.2%). (Arabinose was previously found not to exert a catabolite-repressive effect on the xylose operon [18].) The following concentrations of antibiotics were used for selection: 50 μg of ampicillin per ml, 10 μg of chloramphenicol (CHL) per ml, and 1 μg of erythromycin per ml. Unless otherwise stated, glucose was added to 0.2%, xylose was added to 2%, IPTG was added to 1 mM, and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was added to 80 μg/ml. Cloning was performed in E. coli strain Novablue (Novagen) according to established methods (29). Transformation of these cells was performed according to the manufacturer's instructions. Restriction enzymes were purchased from New England Biolabs (Beverly, Mass.) and used according to the manufacturer's instructions. Transformation of B. subtilis organisms was performed according to procedures previously described (4); derivatives of pDG364 were targeted to amyE via double recombination as linear DNA, using approximately 1 μg of PstI-digested plasmid. All other chemicals were purchased from Sigma (Mississauga, Ontario, Canada).
TABLE 1.
List of strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| B. subtilis | ||
| EB161 | trpC2 pheA1 amyE::ery | Bacillus Genetic Stock Center 1A717 |
| EB106 | trpC2 pheA1 amyE::xylR PxylA cat86 | This work |
| EB107 | trpC2 pheA1 amyE::xylR PxylA bgaB cat86 | This work |
| EB105 | trpC2 pheA1 amyE::Pspac lacI cat86 | This work |
| EB104 | trpC2 pheA1 amyE::Pspac bgaB cat86 | This work |
| EB103 | trpC2 pheA1 amyE::Pspac bgaB lacI cat86 | This work |
| EB6 | hisA1 argC4 metC3 | L5087 (2) |
| EB4a | hisA1 argC4 metC3 tag-12 φ29r | L6602 (2) |
| EB123 | hisA1 argC4 metC3 tag-12 φ29ramyE::xylR PxylA tagD cat86 | This work |
| EB127 | hisA1 argC4 metC3 tag-12 φ29ramyE::Pspac tagD lacI cat86 | This work |
| E. coli | ||
| Novablueb | endA1 hsdR17 (rK12− mK12+) supE44 thi-1 recA1 gyrA96 relA1 lac[F′ proA+B+ lacIqZΔM15::Tn10(TcR)] | Novagen |
| Plasmids | ||
| pDG364 | B. subtilis vector for integration at amyE | Bacillus Genetic Stock Center (3) |
| pSWEET | pDG364 with xyl expression system | This work |
| pSWEET-bgaB | pSWEET with bgaB from B. stearothermophilus and polylinker | This work |
| pSWEET-tagD | pSWEET with wild-type tagD from B. subtilis 168 | This work |
| pDR67 | Plasmid containing spac expression system for integration at amyE | 15 |
| pSPAC-bgaB ΔlacI | pDR67 with bgaB from B. stearothermophilus without lacI gene | This work |
| pSPAC-bgaB | pDR67 with bgaB from B. stearothermophilus | This work |
| pSPAC-tagD | pDR67 with tagD from B. subtilis 168 | This work |
Temperature-sensitive mutant in teichoic acid biosynthesis ascribed to tagD.
Cloning strain.
TABLE 2.
List of primers
| Oligonucleotide | Sequenceab | Restriction site(s) |
|---|---|---|
| AB01 | 5′-GGCCGGATCCGAATTACATTTTAACGATATC-3′ | BamHI |
| AB02c | 5′-GCGCTTAATTAAACCACTTTGTTAACGCTTACAAAATAGTT-3′ | PacI |
| AB03 | 5′-GCGCTTAATTAACATTAGGAAGGACGCTTTCTT-3′ | PacI |
| AB04 | 5′-CCGGATCCGCTTATAAACCAGCAATTTCCTC-3′ | BamHI |
| AB05 | 5′-GGAAGGATCCGCGCTTAATTAAACCACTTTGTTAACGCTT-3′ | BamHI/PacI |
| AB06 | 5′-GGAAAGATCTTTACATTTTAACGATATCTAGAAAAT-3′ | BglII |
| AB07d | 5′-CCTGCCCGGG/TTAATTAACATTAGGAAGGAGCGTTTCTTTAAATGAATGTGTTATCCTCAATTTGTTACGG-3′ | SmaI/PacI |
| AB08e | 5′-GGAAGGATCC/AAGCTT/GCGGCCGC/CCGGGC/TAGCCTAAACCTTCCCGGCTTCATCATGCTCTCT-3′ | BamHI/HindIII/ NotI/SrfI/NheI |
| AB09 | 5′-GGCGGATCC/AGATCTCTAAACCTTCCCGG-3′ | BamHI/BglII |
Restriction sites are indicated in italics.
Multiple restriction sites are separated by a forward slash (/).
Underlined text indicates the optimized CRE site, and bold text indicates mismatches from the xylA CRE site.
Underlined text indicates the ribosome binding site, and bold text indicates a 9-bp spacer between the ribosome binding site and the initiation codon.
Underlined text indicates bases shared by two restriction sites.
Plasmid construction.
Plasmid pSWEET-tagD was constructed using the following strategy. The xylR gene, the xylA promoter, and the first 58 nucleotides of the xylA gene (including the CRE site) were amplified as a single product from B. subtilis W23 chromosomal DNA, using the primers AB01 and AB02. Primer AB02 incorporated two mutations in the CRE site, G→T and A→T at positions 3 and 10, respectively (18, 34). Gene tagD, from −24 to stop (including its native ribosome binding site), was amplified from B. subtilis 168 using the primers AB03 and AB04. The above PCR products were digested with PacI and ligated together. The ligation product was reamplified with primers AB01 and AB04, digested with BamHI, and ligated into BamHI-digested pDG364.
To create pSWEET-bgaB the xylR-xylA fragment was reamplified from pSWEET-tagD with primers AB05 and AB06. The amplified fragment was digested with BamHI and BglII and ligated into BamHI-digested pDG364 to create plasmid pSWEET. Plasmid pKL4 was used as a template for amplification of bgaB (31) using primers AB07 and AB08. Primer AB07 incorporated nucleotides −24 to −1 of B. subtilis tagD so that the ribosome binding sites and their contexts were identical in pSWEET-tagD and pSWEET-bgaB. The bgaB PCR product was digested with PacI and BamHI and ligated into pSWEET digested with the same enzymes.
To facilitate a rigorous comparison of the xylose-based expression system in pSWEET with that of the commonly used spac system, we constructed clones of bgaB and tagD under the control of the spac promoter in pDR67 (15) with ribosome binding sites and contexts identical to those used with pSWEET (i.e., nucleotides −24 to −1 of tagD, as described above). Those plasmids were named pSPAC-bgaB and pSPAC-tagD, respectively. In addition, we constructed a lacI deletion of pSPAC-bgaB, designated pSPAC-bgaB ΔlacI, in order to test the importance of lacI to the regulation of the spac promoter. Gene bgaB was amplified from pKL4 using primers AB07 and AB09. To create pSPAC-bgaB, the amplified product was digested with SmaI and BglII and ligated to SmaI- and BglII-digested pDR67. For pSPAC-bgaB ΔlacI, the amplified product was digested with SmaI and BamHI and ligated to SmaI- and BamHI-digested pDR67. To place tagD under the control of the spac promoter, tagD was excised from pSWEET-tagD with PacI and BamHI and ligated into PacI- and BamHI-digested pSPAC-bgaB.
β-Galactosidase activity assay.
An assay of thermostable β-galactosidase has been described previously (31, 32). In brief, cells were grown to an optical density at 600 nm (OD600) of 0.5, pelleted, and resuspended in an equivalent volume of buffer B (25 mM potassium phosphate, 50 mM KCl, and 1 mM MgSO4 [pH 6.4]). Sample aliquots ranging from 0.1 to 0.5 ml were diluted to 0.8 ml in buffer B and lysed for 30 min at 28°C with the addition of 16 μl of lysozyme (10 mg/ml). Lysis was followed by the addition of 40 μl of 10% Triton X-100. Subsequently, a preincubation at 60°C for 15 min was used to inactivate endogenous β-galactosidase. The assay was initiated with the addition of 0.2 ml of o-nitrophenyl-β-d-galactopyranoside (ONPG) at a concentration of 4 mg/ml and quenched by the addition of 0.5 ml of 1 M Na2CO3. Absorbance was recorded at 420 nm in a Spectramax Plus spectrophotometer (Molecular Devices).
RESULTS
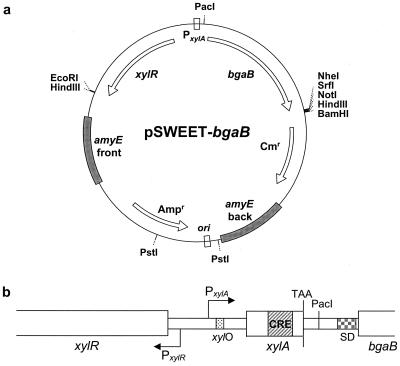
We chose to construct a xylose-based expression system by incorporating components of the xylose operon from strain W23 of B. subtilis. The xylose utilization machinery of this B. subtilis strain has been the subject of extensive characterization (5, 6, 12, 13, 16–19). The xyl expression system developed in this work includes the xylose repressor encoded by xylR, intergenic xyl operator sequences, the xylA promoter, and the 5′ portion of xylA containing an optimized CRE (Fig. 1). The system is contained on plasmid pSWEET, a derivative of pDG364 (3), for ready integration of the expression system in the B. subtilis chromosome at amyE.
FIG. 1.
Map of plasmid pSWEET-bgaB. (a) Significant features of the xylose-based expression system. Plasmid pSWEET-bgaB is a derivative of pDG364 (3), which allows integration into the B. subtilis chromosome at amyE via double recombination and selection with CHL (10 μg/ml). The plasmid also has an E. coli origin of replication, denoted ori, and an ampicillin resistance cassette (50 μg/ml) for routine cloning steps. On the outside of the plasmid map, restriction sites of interest are highlighted, including two PstI sites for convenient plasmid linearization, a PacI site (eight-base recognition sequence TTAATTAA) upstream of bgaB, and a polylinker downstream of bgaB (HindIII is not unique). (b) Close-up of key elements of the xylose expression system (not to scale). Shown are xylR, encoding the xylose repressor; the xyl intergenic region, including promoters for xylR (PxylR), xylA (PxylA), and xyl operator sequences (xylO); translationally truncated xylA (first 58 nucleotides followed by an in-frame TAA), including an optimized CRE in xylA (see Materials and Methods and reference 18); PacI 5′ cloning site; ribosome binding site (SD) native to B. subtilis tagD; and gene bgaB, encoding a thermostable β-galactosidase from B. stearothermophilus.
Transcriptional regulatory sequences in the xyl operon include tandem overlapping operator sequences downstream of the xylA transcriptional start site (5) and a CRE located 36 nucleotides into the coding sequence of the xylose isomerase gene (xylA) (18). To ensure that the xyl system was maximally subjected to catabolite repression, we included the first 58 nucleotides of the gene xylA and introduced two mismatches in the CRE site to perfectly match the consensus sequence established previously (18, 34). An in-frame stop codon was placed in the xylA gene following the 58th nucleotide, effectively truncating the XylA protein. Downstream of that translational stop, we constructed pSWEET-bgaB so that PacI (eight-base recognition sequence TTAATTAA) and one of several polylinker enzymes could be used to replace bgaB and its associated ribosome binding site with any sequences of interest.
Schrogel and Allmansberger (31) have optimized the use of heat-stable β-galactosidase, bgaB from B. stearothermophilus, as a reporter gene in B. subtilis, which permits inactivation of endogenous background β-galactosidase activity. We employed this reporter system in order to describe the lower limits of transcriptional control afforded by the xyl system relative to the spac system. For an unbiased comparison of the xyl and spac expression systems, we constructed pSPAC-bgaB and pSWEET-bgaB so that their ribosome binding sites and their contexts were identical (see Materials and Methods).
Figure 2a and b demonstrate expression of bgaB from the xyl and spac expression systems (induced with xylose and IPTG, respectively) as indicated by X-Gal hydrolysis in LB agar (EB107 and EB103, respectively). Strain EB104 (spac system without lacI) also showed significant X-Gal hydrolysis. Strains EB106 (xyl) and EB105 (spac) were negative-control strains, containing the corresponding expression system but lacking the bgaB gene, and showed no evidence of X-Gal hydrolysis. Strains containing both expression systems were plated on X-Gal in the absence of any inducer to assess transcriptional control (i.e., the leakiness of the expression system). Figure 2c shows that the spac system (EB103) produced clearly discernible levels of BgaB as revealed by the extent of X-Gal hydrolysis. This cleavage is not due to endogenous β-galactosidase genes, as demonstrated by the lack of color development in the negative-control strain (EB105). The strain lacking lacI (EB104) served to illustrate that while the lac repressor allowed significant transcriptional control, it did not limit expression beyond the detection of this assay. In contrast, the xyl expression system (EB107) showed no discernible expression in the absence of an inducer and showed no significant deviation from the negative-control strain (EB106).
FIG. 2.
Detection of reporter gene expression by X-Gal hydrolysis on solid media. (a) Strains EB106 (pSWEET) and EB107 (pSWEET-bgaB) were plated on LB–CHL–X-Gal in the presence of 2% xylose. (b) Strains EB103 (pSPAC-bgaB), EB104 (pSPAC-bgaB ΔlacI), and EB105 (pSPAC) were plated on LB–CHL–X-Gal in the presence of 1 mM IPTG. (c) All five strains were plated on LB–CHL–X-Gal in the absence of an inducer. Strains were grown overnight at 37°C and then incubated at 55°C for color development (up to 36 h).
To better characterize the xyl and spac expression systems, B. subtilis strains carrying transcriptional fusions of bgaB to Pxyl and Pspac, respectively, were assayed for β-galactosidase activity after growth in liquid media. Figure 3 shows an example (β-galactosidase activities of EB104 under inducing conditions) of our efforts to ensure that the assay was linear both with time and with the volume of cells. Under the conditions used in this work, o-nitrophenol production remained linear for 240 min and was directly dependent on the amount of sample added to the assay. Having established parameters for a linear response in the assay of thermostable β-galactosidase, EB103 and EB107 were grown in both rich (LB) and minimal media (plus 0.2% arabinose) in the presence or absence of an inducer. The xyl expression system showed a significantly higher ratio of induction to repression than the spac system (ratios of 279 versus 24 in rich media, respectively), which can be attributed to higher levels of expression under inducing conditions (Table 3). Our analysis of the spac system indicates a somewhat lower induction/repression ratio than that previously recorded (140-fold) for the comparable spac system of pMUTIN1 (33), perhaps due to differences in the genetic contexts of each system. Overall, the xyl system demonstrated a 16-fold increase in expression relative to the spac system (β-galactosidase units of 12,824 and 814 in rich media for the xyl and spac systems, respectively). Under noninducing conditions, however, there was no significant difference between the xyl and spac expression systems using this assay (β-galactosidase units of 46 ± 21 [mean ± standard deviation] and 34 ± 20 in rich medium, respectively). Indeed, significant noise in this assay, which was particularly troublesome at the lower limits of detection, may have precluded our detection of otherwise significant differences between these two expression systems.
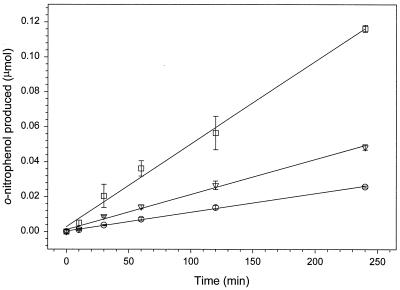
FIG. 3.
Linearity of heat-stable β-galactosidase assay. Strain EB104 was grown (LB-CHL with 1 mM IPTG) to mid-log phase (OD600 = 0.5) and assayed for BgaB activity (see Materials and Methods) at different sample volumes (○, ▿, and □ denote 0.1, 0.2 and 0.5 ml of culture, respectively) for up to 4 h. β-Galactosidase activity was defined as micromoles of o-nitrophenol released after 0, 10, 30, 60, 120, and 240 min. Slopes for 0.1, 0.2, and 0.5 ml of culture were 0.11, 0.20, and 0.47 nmol/min. Errors are standard deviations from three separate experiments.
TABLE 3.
Induction and repression of xyl and spac expression systemsa
| Expression system | Medium | β-Gal activity (mean ± SD)
|
Induction/ repression ratio | ||
|---|---|---|---|---|---|
| Inducedb | Uninduced | Repressed | |||
| xyl (EB107) | Minimal | 12,795 ± 858 | 89 ± 19 | 52 ± 17c | 246 |
| xyl (EB107) | Rich | 12,824 ± 658 | 46 ± 21 | NDd | 279 |
| spac (EB103) | Minimal | 953 ± 43 | 111 ± 22 | ND | 9 |
| spac (EB103) | Rich | 814 ± 48 | 34 ± 20 | ND | 24 |
Saturated cultures EB107 (pSWEET-bgaB) and EB103 (pSPAC-bgaB) were diluted (100-fold) into fresh rich LB or minimal medium, grown to mid-log phase, and assayed as described in Materials and Methods. Errors are standard deviations from at least three separate experiments. β-Gal, β-galactosidase.
Medium supplemented with 2% xylose (EB107) or 1 mM IPTG (EB103).
Medium supplemented with 0.2% glucose.
ND, not determined.
The role of the CRE in catabolite repression of the xyl operon and its capacity for optimization through mutation has been well documented (13, 16, 18, 34). Furthermore, glucose-specific, XylR-mediated repression has also been reported (18). In Table 3, we report about a 250-fold difference in reporter gene expression, comparing growth on minimal medium with xylose (2%) to growth on minimal medium plus glucose (0.2%). This result is consistent with previous studies of the xyl regulon in which induction/repression ratios as high as 260-fold have been reported (13). We also examined reporter gene expression after growth in minimal medium with xylose (0.2%) and one of three sugars (glucose, fructose, or glycerol) at a level of 0.2%. Regardless of the type of sugar added, we observed about a 100-fold decrease in reporter gene expression (data not shown). These findings are also consistent with a previous study in which the CRE sequence used in our studies (G→T and A→T at positions 3 and 10, respectively) resulted in similar levels of catabolite-mediated repression and masked to a large extent the glucose-specific effect seen with wild-type CREs (18). Kim et al. (17) have described a xylose-inducible integration vector using xyl regulatory sequences from Bacillus megaterium. Induction/repression ratios ranging from 150- to 200-fold were reported for that system, which lacked the CRE sequence and was not subject to catabolite repression. Eichenbaum et al. have also published a comparison study of a plasmid-based B. subtilis xylose expression system with spac, lac, and nisA promoters (9). In that work, the xyl and spac systems demonstrated similar induction/repression ratios in B. subtilis (11- and 16-fold, respectively), though few details were published regarding the construction of that xylose-based expression system.
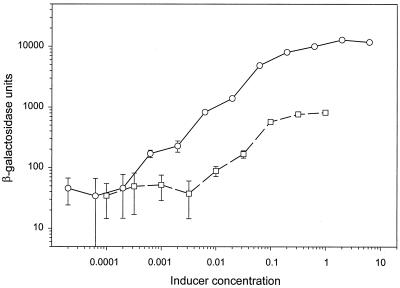
Having determined the extremes of expression, we wanted to assess the ability of each system to modulate expression in response to an inducer. Strains EB107 and EB103 were grown in various concentrations of inducers (xylose and IPTG, respectively) in rich liquid media, and expression levels were assessed using the β-galactosidase assay (Fig. 4). The xylose-induced expression system showed a particular capacity for modulation, as β-galactosidase activity varied from 30 units to about 11,000 units over an inducer concentration range of 3.5 log units (0.0002 to 0.63% xylose). In contrast, the spac system was modulated over an inducer concentration range of only 1.5 log units (0.003 to 0.1 mM IPTG).
FIG. 4.
Modulation of the xyl and spac expression systems. Saturated cultures of EB107 (pSWEET-bgaB) and EB103 (pSPAC-bgaB) were inoculated (1/100) into fresh LB-CHL media supplemented with increasing concentrations of inducer, i.e., 0.00002 to 6.3% xylose (○) and 100 nM to 1 mM IPTG (□). Cultures were grown to mid-log phase and assayed for BgaB activity (see Materials and Methods). β-Galactosidase units are picomoles of o-nitrophenol released per minute per milliliter of culture at an OD600 of 0.5. Errors are standard deviations from at least three separate experiments.
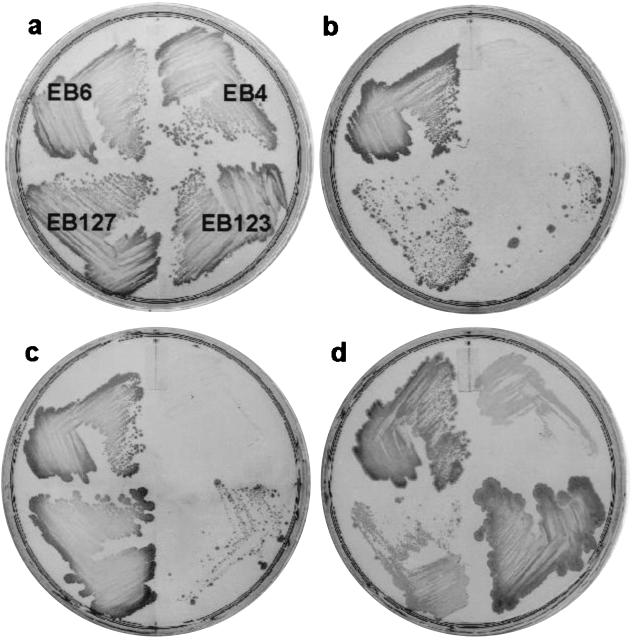
As a test case for the capacity of the xyl expression system to provide conditional complementation of essential functions, we attempted to complement a temperature-sensitive mutant (tag-12) previously attributed to B. subtilis tagD (23), which encodes glycerol-3-phosphate cytidylyltransferase. Again, for comparative purposes, tagD was cloned into both pSWEET and pSPAC in order to create EB123 and EB127, which expressed wild-type tagD in the temperature-sensitive background under xyl and spac control, respectively. We assessed the ability of each system to complement the temperature-sensitive mutant at the nonpermissive temperature. As a control, we included the parental tag-12 temperature-sensitive strain (EB4) and the isogenic tag+ strain (EB6) in these experiments. As expected, EB6 grew well at both the permissive (30°C) and nonpermissive (47°C) temperatures, while EB4 showed almost no growth at the nonpermissive temperature (Fig. 5a and b). Both EB123 and EB127 were able to complement the mutant at the nonpermissive temperature in the presence of their respective inducers (Fig. 5c and d). In the absence of added inducer, the spac expression system (EB127) showed substantial growth at the nonpermissive temperature, while the xyl system (EB123) showed only very slight growth relative to the control EB4 (Fig. 5b). Interestingly, we have consistently noted that in the absence of inducer and with a heavy inoculum, a mixed population of large and small colonies is evident with strains EB123 and EB127 (e.g., Fig. 5b). These large colonies subsequently demonstrate a temperature-insensitive and inducer-independent growth phenotype and may have arisen from recombination of the temperature-sensitive copy of tagD at the tag locus with the wild-type copy of tagD resident at amyE.
FIG. 5.
Xylose-dependent complementation of a temperature-sensitive mutant in teichoic acid biosynthesis. Strains were plated on LB agar in the presence or absence of an inducer and incubated at 30°C (permissive temperature) or 47°C (nonpermissive temperature). Strains EB4 (tag-12), EB6 (tag+), EB123 (tag-12 pSWEET-tagD), and EB127 (tag-12 pSPAC-tagD) were plated at 30°C (a), plated at 47°C (b), supplemented with 1 mM IPTG at 47°C (c), and supplemented with 2% xylose at 47°C (d). Strains shown in panels b through d were plated as indicated in panel a.
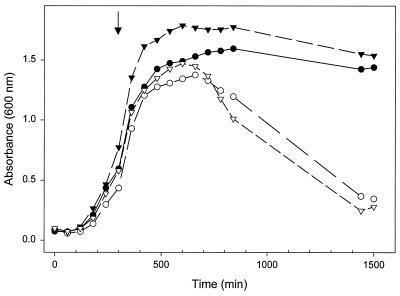
To further assess the ability of the xyl system to conditionally complement the tag-12 mutant, a growth curve was examined using strain EB123 (expression of tagD under xyl control in the tag-12 background) grown in the presence and absence of the inducer xylose (0.2%) (Fig. 6). Again, the parental strains EB4 and EB6 were included as controls. The cultures were initially grown at the permissive temperature (30°C) for 300 min and were then shifted to the nonpermissive temperature (47°C) and monitored for up to 25 h. The tag-12 mutant (EB4) showed a lytic phenotype upon temperature shift, as indicated by the steady decrease in OD after 660 min. In the absence of xylose, strain EB123 duplicated the growth and lysis exhibited by EB4. In the presence of the inducer, EB123 showed slightly better growth than EB6, likely due to the presence of a rich carbon source (xylose). Nevertheless, we are certain that this complementation was due to the expression of tagD and not the presence of xylose, since, when grown at the nonpermissive temperature and in the presence of xylose, EB4 showed only very slight growth (Fig. 5d). We also tested EB127 (expression of tagD under spac control in the tag-12 background) in an analogous experiment and found that the strain exhibited a growth curve which was indistinguishable from that seen for EB123 (data not shown).
FIG. 6.
Growth profile of the B. subtilis tag-12 mutant and conditional complementation with tagD under xylose control. Overnight cultures (LB) of EB4 (tag-12), EB6 (tag+), and EB123 (tag-12 pSWEET-tagD) were inoculated (1/100) into fresh medium (LB) and grown for 25 h, with periodic monitoring of growth by OD600. Growth curves for EB4 and EB6 are represented by the symbols ○ and ●, respectively. EB123 was grown in the presence (▾) and absence (▿) of 0.2% xylose. Growth was at 30°C (permissive temperature for tag-12) until mid-log phase, when cultures were shifted to 47°C (restrictive temperature for tag-12) at 300 min (denoted by the arrow).
DISCUSSION
We have undertaken the development and characterization of a system for efficient expression in B. subtilis in order to characterize putatively indispensable functions in teichoic acid biosynthesis. To that end, we have taken advantage of an extensive body of work on the xylose utilization operon of B. subtilis W23, which has described in detail the machinery for xylose induction and catabolite repression in that system (5, 6, 12, 13, 16, 18, 19). Accordingly, we chose to construct a xylose-based expression system by incorporating components of the xylose operon from strain W23. The expression system contains the xylose repressor encoded by xylR, intergenic xyl operator sequences, the xylA promoter, and the 5′ portion of xylA containing an optimized CRE. These components are present on plasmid pSWEET, a derivative of pDG364 (3), for integration into the B. subtilis chromosome at amyE.
We have made significant contributions to the improvement of xylose-based systems (9, 17) with the inclusion of xyl sequences important in catabolite repression (CREs) and with an extensive characterization of the expression system constructed. The xyl expression system developed was effective in achieving very low levels of induction in the absence of an inducer, was capable of a wide range of induction levels, and had the capacity for modulated expression over a broad scale of inducer concentrations. We have carefully compared the performance of the xyl expression system to that of the widely used spac system as a point of reference and found it to be superior to spac in each of these characteristics.
The tight control of expression afforded by the xyl expression system is an attractive feature in the study of null mutations, where strongly regulated expression of a complementing copy of the gene of interest can lead to an unambiguous interpretation of phenotype. Indeed, this expression system has particular utility in the study of null mutations in essential genes. In the studies detailed here, the temperature-sensitive mutant tag-12 showed little or no growth at the restrictive temperature. Significant growth was seen, however, at the nonpermissive temperature for the tag-12 mutant with spac-tagD at amyE in the absence of IPTG, whereas only very slight growth was evident with the xyl-tagD complementation system in the absence of xylose. Were this a tagD null mutant, complementation experiments with the spac expression system might incorrectly indicate that a phenotype of impaired growth, not lethality, is associated with a tagD knockout.
The particular capacity for modulation of gene expression with the xyl system is arguably a very attractive feature for the deliberate expression of cloned genes in B. subtilis. While the spac system modulated expression over a narrow range of inducer concentrations (1.5 log units), the xyl system controlled expression over a >3.5-log span of xylose concentrations. When combined with the large induction/repression ratio characteristic of the xyl system, this broad response to inducer concentration should facilitate exploration of the effects of expression level of a cloned gene on phenotype. Indeed, protein expression levels can be important to phenotypic analyses in bacteria for a wide variety of reasons, including an exquisite sensitivity, in some cases, to protein stoichiometry, such as that observed for the E. coli proteins FtsZ and FtsA (7, 8).
Despite the apparent superiority of the xyl promoter-operator system over that of spac, both systems showed considerable capacity for control of expression in B. subtilis. Together, these two systems may be particularly useful in instances where more than one induction system may be warranted. Plasmid pMUTIN, for example, provides the means for facile inactivation of a target gene while placing downstream genes under the control of the spac promoter to test for polar effects (33). Complementation of the inactivated gene by placing a copy under spac transcriptional control would not provide the means to distinguish the effects of transcription of downstream genes from those of complementation. Use of pSWEET for this purpose would facilitate an unequivocal analysis of any phenotype(s) associated with the null mutant by placing the target gene in trans under the control of the xylose promoter.
The tag genes are responsible for the synthesis of poly(glycerol phosphate), the predominant cell wall-linked polyanionic polymer of B. subtilis strain 168. A considerable body of work indicates an essential role in this strain for poly(glycerol phosphate) synthesis (2, 23, 24, 26, 27). Paradoxically, two other wall polymers, poly(glucose N-acetylgalactosamine phosphate) and teichuronic acid, are also produced and are capable of at least partially substituting for the predominant polymer (10, 11). In fact, certain strains of B. subtilis have been reported to completely replace wall teichoic acid with phosphate-free teichuronic acid under phosphate-limiting conditions (20). Therefore, while teichoic acid biosynthesis may have great potential as a therapeutic drug target in gram-positive bacterial physiology, a clear understanding of the putatively essential role for this polymer, even in the model organism B. subtilis 168, has remained elusive.
An essential role for tagD, encoding glycerol-3-phosphate cytidylyltransferase (25), was indicated by the localization of two thermosensitive mutations, tag-11 and tag-12, to tagD (23). In addition, glycerol-3-phosphate cytidylyltransferase activity in extracts of the tag-11 mutant was shown to be thermolabile and significantly reduced relative to that of wild-type extracts (27). In the work reported here, we noted a pronounced growth defect in the tag-12 mutant, such that this strain showed little or no growth at the restrictive temperature and demonstrated a significant drop in OD soon after a temperature shift from 30 to 47°C. The decrease in cell density upon shifting to the nonpermissive temperature is remarkable in its similarity to the lytic response normally reserved for defects in peptidoglycan biosynthesis and is consistent with previous studies detailing gross morphological changes associated with teichoic acid mutants (1, 28). Using the xyl expression system developed here, we have demonstrated, for the first time, trans complementation of a teichoic acid biosynthesis mutant. Rescue of the tag-12 mutant at the restrictive temperature with tagD under xyl control was xylose dependent, unequivocally indicating a role for tagD in this growth defect. This work sets the stage for further analysis of teichoic acid biosynthesis genes in B. subtilis through the construction of null mutants therein and conditional complementation using pSWEET.
ACKNOWLEDGMENTS
We thank Dimitri Karamata for tag and tag-12 B. subtilis strains, Oliver Schrogel for pKL4, Petra Levin for pDR67, and Christopher Murphy for fruitful discussions. We also thank Justin Nodwell, Gerry Wright, and Janet Wood for offering comments on the manuscript.
This work was supported by an operating grant and scholarship from the Medical Research Council of Canada to E.D.B. and by a postgraduate fellowship to A.P.B. from the Natural Sciences and Engineering Research Council of Canada.
REFERENCES
- 1.Boylan R J, Mendelson N H, Brooks D, Young F E. Regulation of the bacterial cell wall: analysis of a mutant of Bacillus subtilis defective in biosynthesis of teichoic acid. J Bacteriol. 1972;110:281–290. doi: 10.1128/jb.110.1.281-290.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briehl M, Pooley H M, Karamata D. Mutants of Bacillus subtilis 168 thermosensitive for growth and wall teichoic acid biosynthesis. J Gen Microbiol. 1989;135:1325–1334. [Google Scholar]
- 3.Cutting S M, Horn P B V. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Toronto, Canada: John Wiley and Sons; 1990. pp. 27–61. [Google Scholar]
- 4.Cutting S M, Youngman P. Gene transfer in gram-positive bacteria. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 348–364. [Google Scholar]
- 5.Dahl M K, Degenkolb J, Hillen W. Transcription of the xyl operon is controlled in Bacillus subtilis by tandem overlapping operators spaced by four base-pairs. J Mol Biol. 1994;243:413–424. doi: 10.1006/jmbi.1994.1669. [DOI] [PubMed] [Google Scholar]
- 6.Dahl M K, Schmiedel D, Hillen W. Glucose and glucose-6-phosphate interaction with Xyl repressor proteins from Bacillus spp. may contribute to regulation of xylose utilization. J Bacteriol. 1995;177:5467–5472. doi: 10.1128/jb.177.19.5467-5472.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dewar S J, Begg K J, Donachie W D. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992;174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichenbaum Z, Federle M J, Marra D, de Vos W M, Kuipers O P, Kleerebezem M, Scott J R. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl Environ Microbiol. 1998;64:2763–2769. doi: 10.1128/aem.64.8.2763-2769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellwood D C, Tempest D W. Influence of culture pH on the content and composition of teichoic acids in the walls of Bacillus subtilis. J Gen Microbiol. 1972;73:395–402. doi: 10.1099/00221287-73-2-395. [DOI] [PubMed] [Google Scholar]
- 11.Estrela A I, Pooley H M, de Lencastre H, Karamata D. Genetic and biochemical characterization of Bacillus subtilis 168 mutants specifically blocked in the synthesis of the teichoic acid poly(3-O-beta-D-glucopyranosyl-N-acetylgalactosamine 1-phosphate): gneA, a new locus, is associated with UDP-N-acetylglucosamine 4-epimerase activity. J Gen Microbiol. 1991;137:943–950. doi: 10.1099/00221287-137-4-943. [DOI] [PubMed] [Google Scholar]
- 12.Gärtner D, Degenkolb J, Ripperger J A, Allmansberger R, Hillen W. Regulation of the Bacillus subtilis W23 xylose utilization operon: interaction of the Xyl repressor with the xyl operator and the inducer xylose. Mol Gen Genet. 1992;232:415–422. doi: 10.1007/BF00266245. [DOI] [PubMed] [Google Scholar]
- 13.Gärtner D, Geissendörfer M, Hillen W. Expression of the Bacillus subtilis xyl operon is repressed at the level of transcription and is induced by xylose. J Bacteriol. 1988;170:3102–3109. doi: 10.1128/jb.170.7.3102-3109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heumann D, Barras C, Severin A, Glauser M P, Tomasz A. Gram-positive cell walls stimulate synthesis of tumor necrosis factor alpha and interleukin-6 by human monocytes. Infect Immun. 1994;62:2715–2721. doi: 10.1128/iai.62.7.2715-2721.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ireton K, Rudner D Z, Siranosian K J, Grossman A D. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- 16.Jacob S, Allmansberger R, Gärtner D, Hillen W. Catabolite repression of the operon for xylose utilization from Bacillus subtilis W23 is mediated at the level of transcription and depends on a cis site in the xylA reading frame. Mol Gen Genet. 1991;229:189–196. doi: 10.1007/BF00272155. [DOI] [PubMed] [Google Scholar]
- 17.Kim L, Mogk A, Schumann W. A xylose-inducible Bacillus subtilis integration vector and its application. Gene. 1996;181:71–76. doi: 10.1016/s0378-1119(96)00466-0. [DOI] [PubMed] [Google Scholar]
- 18.Kraus A, Hueck C, Gärtner D, Hillen W. Catabolite repression of the Bacillus subtilis xyl operon involves a cis element functional in the context of an unrelated sequence, and glucose exerts additional xylR-dependent repression. J Bacteriol. 1994;176:1738–1745. doi: 10.1128/jb.176.6.1738-1745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreuzer P, Gärtner D, Allmansberger R, Hillen W. Identification and sequence analysis of the Bacillus subtilis W23 xylR gene and xyl operator. J Bacteriol. 1989;171:3840–3845. doi: 10.1128/jb.171.7.3840-3845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang W K, Glassey K, Archibald A R. Influence of phosphate supply on teichoic acid and teichuronic acid content of Bacillus subtilis cell walls. J Bacteriol. 1982;151:367–375. doi: 10.1128/jb.151.1.367-375.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazarevic V, Karamata D. The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol Microbiol. 1995;16:345–355. doi: 10.1111/j.1365-2958.1995.tb02306.x. [DOI] [PubMed] [Google Scholar]
- 22.Matsuura T, Miyake Y, Nakashima S, Komatsuzawa H, Akagawa Y, Suginaka H. Isolation and characterization of teichoic acid-like substance as an adhesin of Staphylococcus aureus to HeLa cells. Microbiol Immunol. 1996;40:247–254. doi: 10.1111/j.1348-0421.1996.tb03341.x. [DOI] [PubMed] [Google Scholar]
- 23.Mauel C, Young M, Karamata D. Genes concerned with synthesis of poly(glycerol phosphate), the essential teichoic acid in Bacillus subtilis strain 168, are organized in two divergent transcription units. J Gen Microbiol. 1991;137:929–941. doi: 10.1099/00221287-137-4-929. [DOI] [PubMed] [Google Scholar]
- 24.Mauel C, Young M, Margot P, Karamata D. The essential nature of teichoic acids in Bacillus subtilis as revealed by insertional mutagenesis. Mol Gen Genet. 1989;215:388–394. doi: 10.1007/BF00427034. [DOI] [PubMed] [Google Scholar]
- 25.Park Y S, Sweitzer T D, Dixon J E, Kent C. Expression, purification, and characterization of CTP:glycerol-3-phosphate cytidylyltransferase from Bacillus subtilis. J Biol Chem. 1993;268:16648–16654. [PubMed] [Google Scholar]
- 26.Pooley H M, Abellan F-X, Karamata D. CDP-glycerol:poly(glycerophosphate) glycerophosphotransferase, which is involved in the synthesis of the major wall teichoic acid in Bacillus subtilis 168, is encoded by tagF (rodC) J Bacteriol. 1992;174:646–649. doi: 10.1128/jb.174.2.646-649.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pooley H M, Abellan F-X, Karamata D. A conditional-lethal mutant of Bacillus subtilis 168 with a thermosensitive glycerol-3-phosphate cytidylyltransferase, an enzyme specific for the synthesis of the major wall teichoic acid. J Gen Microbiol. 1991;137:921–928. doi: 10.1099/00221287-137-4-921. [DOI] [PubMed] [Google Scholar]
- 28.Prayitno N R, Archibald A R. The effects of growth conditions on cell wall composition and cell morphology in a temperature-sensitive tagB mutant of Bacillus subtilis. World J Microbiol Biotechnol. 1997;13:207–217. [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schneider O, Michel U, Zysk G, Dubuis O, Nau R. Clinical outcome in pneumococcal meningitis correlates with CSF lipoteichoic acid concentrations. Neurology. 1999;53:1584–1587. doi: 10.1212/wnl.53.7.1584. [DOI] [PubMed] [Google Scholar]
- 31.Schrogel O, Allmansberger R. Optimisation of the BgaB reporter system: determination of transcriptional regulation of stress responsive genes in Bacillus subtilis. FEMS Microbiol Lett. 1997;153:237–243. doi: 10.1111/j.1574-6968.1997.tb10488.x. [DOI] [PubMed] [Google Scholar]
- 32.Shimotsu H, Henner D J. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene. 1986;43:85–94. doi: 10.1016/0378-1119(86)90011-9. [DOI] [PubMed] [Google Scholar]
- 33.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 34.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yansura D G, Henner D J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]