Abstract
Bourbon virus (BRBV) is an emerging tick-borne orthomyxovirus that causes severe febrile illness in humans. There are no specific treatments for BRBV disease currently available. Here, we developed a highly accessible and robust, quantitative fluorescence-based BRBV minigenome (MG) system and applied it to high-throughput antiviral drug screening. We demonstrated that human dihydroorotate dehydrogenase (DHODH) inhibitors, hDHODH-IN-4 and brequinar, efficiently reduced BRBV RNA synthesis, and validated these findings using infectious BRBV in vitro. The DHODH inhibitors also exhibited high potency in inhibiting MG activities of other orthomyxoviruses with emerging zoonotic potential, including bat influenza A virus, swine influenza D virus, and Thogoto virus. Our newly developed MG system is a powerful platform for antiviral drug screening across the Orthomyxoviridae family, enabling rapid development and deployment of antivirals against future emerging orthomyxoviruses.
Keywords: Bourbon virus, orthomyxoviruses, emerging zoonotic viruses, antivirals, minigenome
1. Introduction
Bourbon virus (BRBV) belongs to the genus Thogotovirus in the family Orthomyxoviridae (Kosoy et al., 2015), which is comprised of seven virus genera including Alphainfluenzavirus, Betainfluenzavirus, Gammainfluenzavirus, Deltainfluenzavirus, Isavirus, Quaranjavirus, and Thogotovirus (International Committee on Taxonomy of Viruses (ICTV) Reports, 2020). The genome of BRBV consists of six segments of single-stranded, negative-sense RNA in which each segment is known to encode for a single viral protein: segments 1–3 and 5 encode for the subunits of the viral RNA-dependent RNA polymerase (RdRp) complex (PB2, PB1, and PA) and nucleoprotein (NP) respectively, and segments 4 and 6 encode for the glycoprotein (GP) and matrix (M) protein, respectively (Lambert et al., 2015). Members of the family Orthomyxoviridae have a wide host range and broad geographic distribution, and thus have been recognized as important pathogens threatening public health via zoonotic transmission to humans (Asha and Kumar, 2019; Kumar et al., 2018).
BRBV is a tick-borne virus transmitted by the Lone Star tick - Amblyomma americanum (Godsey et al., 2021; Savage et al., 2017; Savage et al., 2018). BRBV was first isolated from a patient in Bourbon County, Kansas in the United States of America (USA) in 2014 (Kosoy et al., 2015). The patient developed febrile illness with a maculopapular rash, and died 11 days after symptom onset from complications related to thrombocytopenia, leukopenia, renal dysfunction, acute respiratory distress syndrome, and shock. To date, several additional cases of BRBV infection, including a second fatal case identified in Missouri, USA in 2017, have been reported in the central and southcentral United States (Bricker et al., 2019; Centers for Disease Control and Prevention).
While documented cases of human disease caused by BRBV infection are limited, serological surveys of wild and domesticated animals demonstrated the presence of BRBV-neutralizing antibodies in wide variety of species, including the domesticated dog (Canis lupus familiaris), eastern cottontail (Sylvilagus floridanus), horse (Equus caballus), raccoon (Procyon lotor), and white-tailed deer (Odocoileus virginianus) (Jackson et al., 2019; Komar et al., 2020). These accumulating field data clearly indicate that both wild and domesticated animals are frequently exposed to BRBV, and suggest the possibility of more human cases of BRBV infection that might have gone unnoticed or been misdiagnosed.
Despite the prevalent risk of BRBV exposure in both humans and animals, there are no specific treatments for BRBV disease currently available. In this study, we developed a fluorescence-based minigenome (MG) system for BRBV and performed high-throughput drug screening targeting viral RNA replication/transcription. Additionally, we established MG systems for other orthomyxoviruses which have potential as emerging zoonotic pathogens and applied these systems for testing the compound’s antiviral efficacy. Here, we provide a highly efficient, pan-orthomyxovirus drug screening platform that enables us to promptly respond to the emergence of novel pathogenic orthomyxoviruses.
2. Materials and Methods
2.1. Chemical compounds
Antiviral Compound Library (HY-L027), hDHODH-IN-4 (HY-128787; CAS No. 1644156-56-8), brequinar (HY-108325; CAS No. 96187-53-0), and sofosbuvir (HY-15005; CAS No. 1190307-88-0) were purchased from MedChemExpress. Favipiravir (T-705) (A11590; CAS No. 259793-96-9) was purchased from AdooQ. Compounds were resuspended in dimethyl sulfoxide (DMSO) and stored at −80°C.
2.2. Cells and viruses
HEK293 (CRL-1573), HEK293T/17 (ATCC, CRL-11268), Huh7 (a kind gift from Yoshiharu Matsuura, Osaka University), and Vero E6 cells (ATCC, CRL-1586) were maintained in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin (PS). BHK-21 cells (ATCC, CCL-10) were maintained in EMEM supplemented with 10% Tryptose phosphate broth, 5% FBS, and 1% PS.
BRBV-Original isolate and Thogoto virus (THOV) IIA isolate were kindly provided by Brandy J. Russell of the Centers for Disease Control and Prevention (CDC). BRBV and THOV stocks were prepared in Huh7 and BHK-21 cells, respectively, by passaging the original virus once.
BRBV titer was determined by focus-forming assay. Briefly, Vero E6 cells were seeded in 24-well plate (1.5 × 105 cells/well) or 96-well plate (2 × 104 cells/well) 1 day before infection. Cells were infected with 10-fold serial dilutions of virus and overlayed with 1.2% carboxymethylcellulose. After 2 days post-infection (dpi), cells were fixed with 10% neutral buffered formalin. Cells were permeabilized using 1:1 ratio of methanol:acetone for 10 min at room temperature followed by blocking in 1% BSA for 1 hour at room temperature. Affinity purified rabbit polyclonal antibody was produced against peptide antigen Cys-EDEQRDLWLEEVTRQLNTLTPVIRG from BRBV M protein by Biomatik and used as a primary antibody. Primary antibody was added to the cells at concentration of 1:1000 in PBS containing 1% BSA and incubated at 4°C overnight. Visualization of viral foci was accomplished by using goat anti-rabbit AlexaFluor 488 (ThermoFisher Scientific) at concentration of 1:1000.
2.3. Viral genome sequencing
BRBV-Original and THOV IIA were passaged once in Vero E6 and BHK-21 cells, respectively. Viral RNA was extracted and purified using QIAamp Viral RNA Mini kit (QIAGEN). Full-length viral genome sequences were determined by 3’ and 5’RACE and Sanger sequencing. All sequence information is available from the Genbank database (accession numbers OL989462–OL989467).
2.4. Generation of minigenome plasmids and helper plasmids
Human RNA polymerase I (hPolI)-driven BRBV MG plasmids were generated based on BRBV segment 2. Briefly, BRBV segment 2 was amplified by RT-PCR from viral genomic RNA and the cDNA was cloned into a cloning vector. The PB1 open reading frame (ORF) was replaced with either the nanoluciferase (NLuc) (Hall et al., 2012) or the ZsGreen fluorescent protein gene (Matz et al., 1999), while the 3’ and 5’ non-coding regions (NCRs), which are crucial for initiation of transcription, and replication of viral RNA via recognition by the viral RdRp, were left intact. The cDNA consisting of the segment 2 3’ and 5’ NCRs flanking a reporter gene was then cloned into a pPolI vector which possesses the hPolI promoter and terminator sequences as previously described (Flick et al., 2003).BRBV MG plasmids based on segment 1, 4, and 6 were also generated by the same cloning methods described above. To generate BRBV helper plasmids, the PB2, PB1, PA, and NP ORFs were amplified and the cDNA was cloned into a pCAGGs vector possessing the cytomegalovirus enhancer fused to the chicken beta-actin promoter (Niwa et al., 1991).
THOV MG and helper plasmids were generated by the same cloning methods described above but using genomic THOV IIA RNA as a template for RT-PCR. Influenza A virus (IAV) MG and helper plasmids were generated based on identified sequences of human A(H1N1) pandemic 2009 strain, A/Mexico/InDRE4487/2009 (Tsuda et al., 2017). Bat IAV and swine influenza D virus (IDV) MG systems were constructed based on sequences of A/flat-faced bat/Peru/033/2010 (H18N11) (CY125942.1, CY125943.1, CY125944.1, CY125946.1) and D/swine/Oklahoma/1334/2011 (LC522351.1, LC522352.1, LC522353.1, LC522355.1).
2.5. Minigenome reporter assays
For the luciferase-based MG assay, HEK293 cells (2 × 105 cells/well) were seeded in 12-well plates 1 day before transfection. Cells were transfected using Transit-LT1 with a MG-NLuc plasmid (0.5 μg) together with helper plasmids PB2, PB1, PA, and NP (0.5 μg each). A plasmid encoding firefly luciferase under the control of an SV40 promoter (10 ng, pGL2-control; Promega) was also included as a transfection control (Hoenen et al., 2010). Cells were lysed using Passive Lysis Buffer (Promega), and luciferase activity was measured by NanoGlo-Dual Luciferase Reporter Assay System (Promega). For the fluorescence-based MG assay, HEK293T/17 cells (1.5 × 105 cells/well) were seeded in 24-well plate 1 day before transfection. Cells were transfected using Transit-LT1 with a MG-ZsGreen plasmid (0.25 μg) together with helper plasmids PB2, PB1, PA, and NP (0.1 μg each). Fluorescence signal was measured by Glomax Discover Microplate Reader (Promega) with emission filter 500–550/excitation filter Blue 475nm. Fluorescence images were obtained using a ZOE Fluorescent Cell Imager (Bio-Rad).
The fluorescence-based MG assay was further optimized to a 96-well plate using HEK293T/17 cells (6 × 104 cells/well). Optimized plasmid amounts were: 125 ng of MG and 1.56 ng of each helper plasmids for BRBV, THOV, and IAV MG systems; 62.5 ng of MG and 3.125 ng of each helper plasmids for bat IAV MG system; and 125 ng of MG and 0.78 ng of each helper plasmids for IDV MG system.
2.6. Compound screening using fluorescence-based MG systems
The Antiviral Compound Library was screened at the dose of 50 nM using the aforementioned optimized fluorescence-based BRBV MG system in 96-well plate. Similarly, the inhibitory effects of hDHODH-IN-4, brequinar, sofosbuvir, and favipiravir on MG activity were tested using optimized fluorescence-based orthomyxovirus MG systems in 96-well plates. Compounds were added to the cells after 8 hours post-transfection (hpt), and fluorescence signals were measured at 3 days post-transfection (dpt) followed by cell viability assay using CellTiter Glo Luminescent Cell Viability Assay (Promega).
2.7. Virus growth kinetics
Huh7 cells (3 × 105 cells/well) were seeded in 6-well plate 1 day before infection. Cells were infected with BRBV at a multiplicity of infection (MOI) of 0.1. After 1 hour adsorption with tilting every 15 minutes, cells were washed 3 times with serum-free DMEM and 3.5 ml of DMEM supplemented with 2% FBS was added to the cells. Immediately after adding medium, 0.5 ml of supernatants were harvested as a sample of 0 dpi. Up to 3 dpi, 0.5 ml of supernatants were harvested at the indicated timepoints and replaced with an equal volume of fresh medium supplemented with 2% FBS. All supernatant samples were stored at −80 °C until use for titration.
2.8. Evaluation of compound efficacy on BRBV replication
Huh7 cells (8 × 104 cells/well) were seeded in 24-well plate 1 day before infection and infected with BRBV at an MOI of 0.1. After 1 hour adsorption with tilting every 15 minutes, cells were washed 3 times with serum-free DMEM and 0.5 ml of DMEM supplemented with 2% FBS and compounds or DMSO vehicle was added to the cells. Supernatants were harvested at 3 dpi and stored at −80°C until use for titration.
2.9. Cell viability assay
Huh7 cells (2 × 104 cells/well) were seeded in 96-well plate 1 day before compound treatment. After 3 days post-treatment, cell viability was measured by CellTiter Glo Luminescent Cell Viability Assay (Promega).
2.10. Biosafety
All experiments using infectious BRBV and THOV were performed in a biosafety level 3 (BSL-3) and biosafety level 2+ (BSL-2+) facilities, respectively, at Mayo Clinic in accordance with approval and guidelines provided by the Mayo Clinic Institutional Biosafety Committee (IBC). Sample inactivation and removal from the facility was performed in accordance with standard operating protocols approved by the IBC.
2.11. Statistical analyses
Experiments were performed as three biological replicates. Statistical analyses were performed using the t test and one-way ANOVA with GraphPad Prism 9 version 9.1.0.
3. Results
3.1. Generation of human polymerase I-driven BRBV minigenome system
BRBV-Original isolate was passaged once in our lab and genome sequences were determined by Sanger sequencing. We confirmed that our verified sequences of segment 3 (OL989464) and 4 (OL989465) were identical to those previously deposited in Genbank (segment 3: KP657748.2, segment 4: KU708255.1), however some nucleotide differences were found in our verified sequences of segment 1 (OL989462), 2 (OL989463), 5 (OL989466), and 6 (OL989467) (Fig. 1A). Nucleotide differences identified in ORF regions of segment 2 and 5 resulted in a cluster of amino acid (aa) changes, totaling 4 and 5 amino acid differences in PB1 and NP, respectively. Notably, aa 573–577 in PB1 and aa 67–71 in NP in other BRBV isolates, including BRBV-STL, -KS15–1735, and -MO13–2499 (kindly provided by Brandy J. Russell of the CDC and passaged once in Huh7 cells in our lab), were confirmed to be all identical to those of our passaged BRBV-Original (data not shown).
Fig. 1:
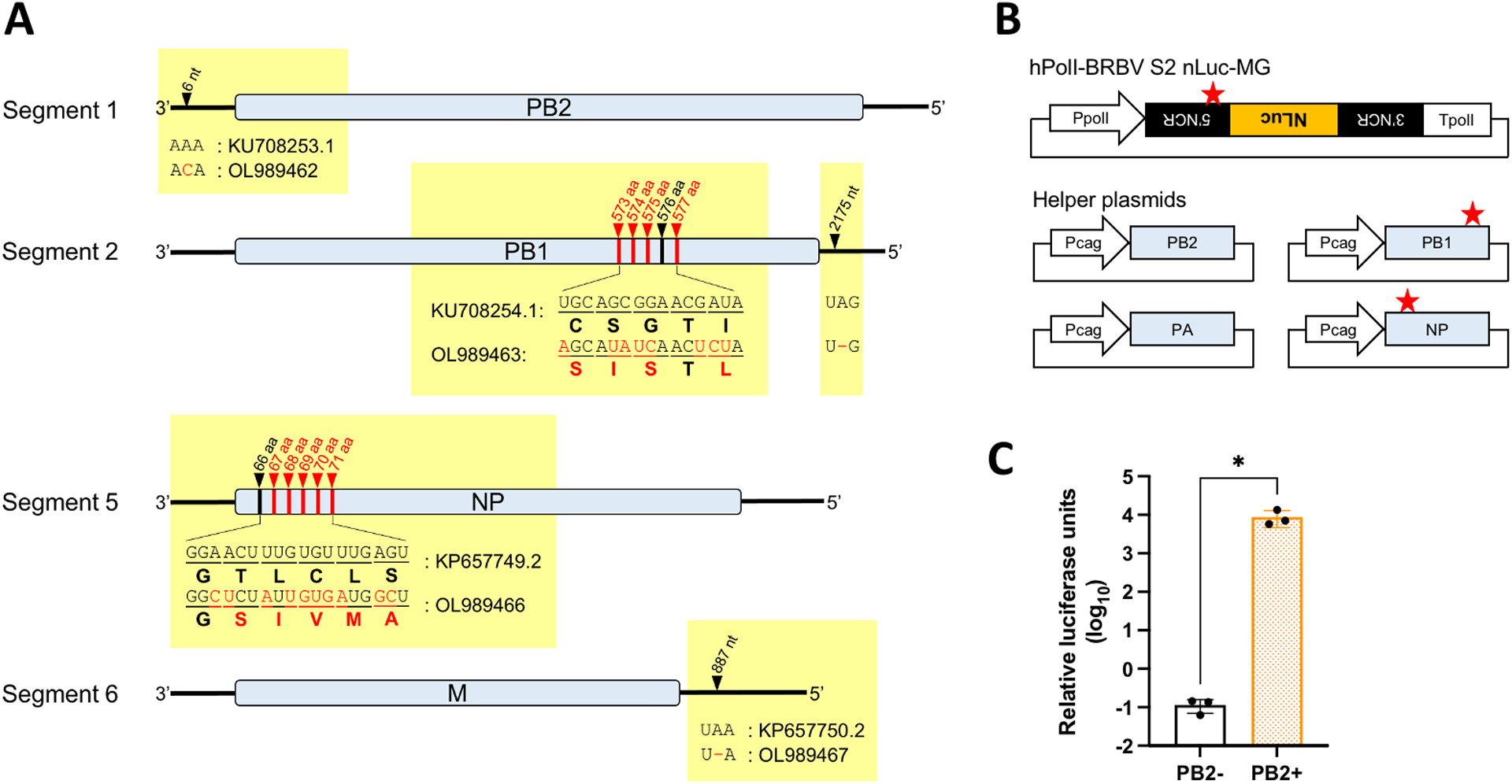
Luminescence-based BRBV MG system in HEK293 cells. A) Nucleotide and amino acid differences between previously Genbank-deposited BRBV sequences (segment 1: KU708253.1, segment 2: KU708254.1, segment 5: KP657749.2, segment 6: KP657750.2) and our verified sequences (segment 1: OL989462, segment 2: OL989463, segment 5: OL989466, segment 6: OL989467). Differences are shown in red. B) Schematics of the segment 2-based BRBV MG plasmid encoding nanoluciferase (NLuc) gene and helper plasmids encoding PB2, PB1, PA, and NP. The sequence differences in the 5’ non-coding region (5’ NCR) of segment 2, as well as the PB1 and NP ORFs, found in our verified sequences are indicated by red stars. PpolI: human RNA polymerase I promoter, TpolI: human RNA polymerase I terminator, Pcag: cytomegalovirus enhancer fused to the chicken beta-actin promoter. C) BRBV MG activity shown by relative luciferase units (NLuc activity normalized by firefly luciferase activity) with or without supplementation of PB2 helper plasmid at 48 hours post-transfection. HEK293 cells were seeded in 12-well plates and transfected with a MG-NLuc plasmid (0.5 μg) together with helper plasmids (0.5 μg each) and a firefly luciferase-expressing plasmid (10 ng). All data are representative of the average of three independent experiments. *: p < 0.05.
To establish a BRBV MG system, we constructed helper protein-expressing plasmids based on our verified sequences, together with a MG encoding NLuc gene in segment 2 (Fig. 1B). Using this system, we observed an approximately 5-log induction of MG luciferase activity relative to PB2(−) negative control at 48 hpt (Fig. 1C), indicating that our verified sequences are functional in driving BRBV MG activity.
3.2. Generation and optimization of a fluorescence-based BRBV minigenome system for the use of compound screening
To develop a fluorescence-based BRBV MG system for use in high-throughput antiviral compound screening, we first optimized our system using the Glomax Discover Microplate Reader, which can quantify fluorescence signals. Among all conditions tested, we found that the expression of ZsGreen in HEK293T/17 cells exhibited the highest quantified fluorescence signal, consistent with our fluorescence microscopy images (Fig. S1A–C). According to this data, we constructed a BRBV segment 2 MG encoding ZsGreen (Fig. 2A) and performed the MG assay in HEK293T/17 cells in 24-well plate format. The MG was successfully transcribed as evidenced by the significantly higher fluorescence signal compared to the PB2(−) negative control (Fig. 2B–C). MG activities gradually increased over time and showed the highest activity at 72 hpt (Fig. 2D). Fluorescent activity using MGs based on segment 2 and 6 were relatively higher than those of segment 1 and 4, although there was no statistical significance (Fig. S2A–C).
Fig. 2:

Fluorescence-based BRBV MG system in HEK293T/17 cells. A) Schematic of the segment 2-based BRBV MG plasmid encoding ZsGreen gene. PpolI: human RNA polymerase I promoter, TpolI: human RNA polymerase I terminator, NCR: non-coding region. BRBV MG activity is shown by B) fluorescence signals and C) fluorescence intensity with or without supplementation of PB2 helper plasmid at 72 hours post-transfection. D) Time course of BRBV MG activity at 24, 48, and 72 hours post-transfection. Fold induction of BRBV MG activity was obtained by normalizing PB2+ fluorescence intensity with PB2-. For B)-D), HEK293T/17 cells were seeded in 24-well plates and transfected with a MG-ZsGreen plasmid (0.25 μg) together with helper plasmids (0.1 μg each). E) BRBV MG activities induced by transfection of serially diluted helper plasmids at 72 hours post-transfection. HEK293T/17 cells were seeded in 96-well plates and transfected with a MG-ZsGreen plasmid (125 ng) together with helper plasmids (25 ng each as the initial amount which were further diluted for :2, :8, :16, :32, and :64). Data are representative of the average of three independent experiments or triplicate in one experiment. ***: p < 0.0005, ****: p < 0.00005.
Since it was suggested that saturation of helper proteins in the cells due to transfection of excessive amount of helper plasmids may mask the functionality of compound targeting viral RNA synthesis (Mendoza et al., 2021), we assessed whether we could reduce the amount of helper plasmids while retaining the optimum MG activity. Using a 96-well plate format, we demonstrated that helper plasmids diluted 16-fold from the original condition still induced a comparable level of MG activity (Fig. 2E). Notably, we saw significantly higher MG activity in absolute value by downsizing of the plate format to 96-well (Fig. 2E) compared to 24-well format (Fig. 2C). Thus, this optimized platform was chosen for further experiments to evaluate compound efficacy in inhibiting BRBV MG activity.
3.3. High-throughput compound screening assay using BRBV minigenome system
To identify compound(s) that efficiently inhibit BRBV RNA synthesis, we utilized an Antiviral Compound Library (MedChemExpress), a collection of 596 bioactive anti-virus compounds including 126 small molecules that target influenza virus. By utilizing an optimized BRBV MG system for compound screening (Z’factor = 0.8), we found that 5 compounds reduced MG activity more than 40% compared to the vehicle control at the 50 nM dose: hDHODH-IN-4, NMS-873, harringtonine, brequinar, and floxuridine (Fig. 3A, Table S1). Cells treated with 50 nM of hDHODH-IN-4, harringtonine, brequinar, and floxuridine maintained 90% of cell viability relative to the vehicle control, while NMS-873 showed significant cytotoxicity. Broad-spectrum viral RdRp inhibitors such as favipiravir, ribavirin, and GS-441524 (a predominant metabolite of remdesivir) (Tian et al., 2021a; Tian et al., 2021b) were not efficacious in reducing BRBV MG activity under the conditions tested (Table S1).
Fig. 3:
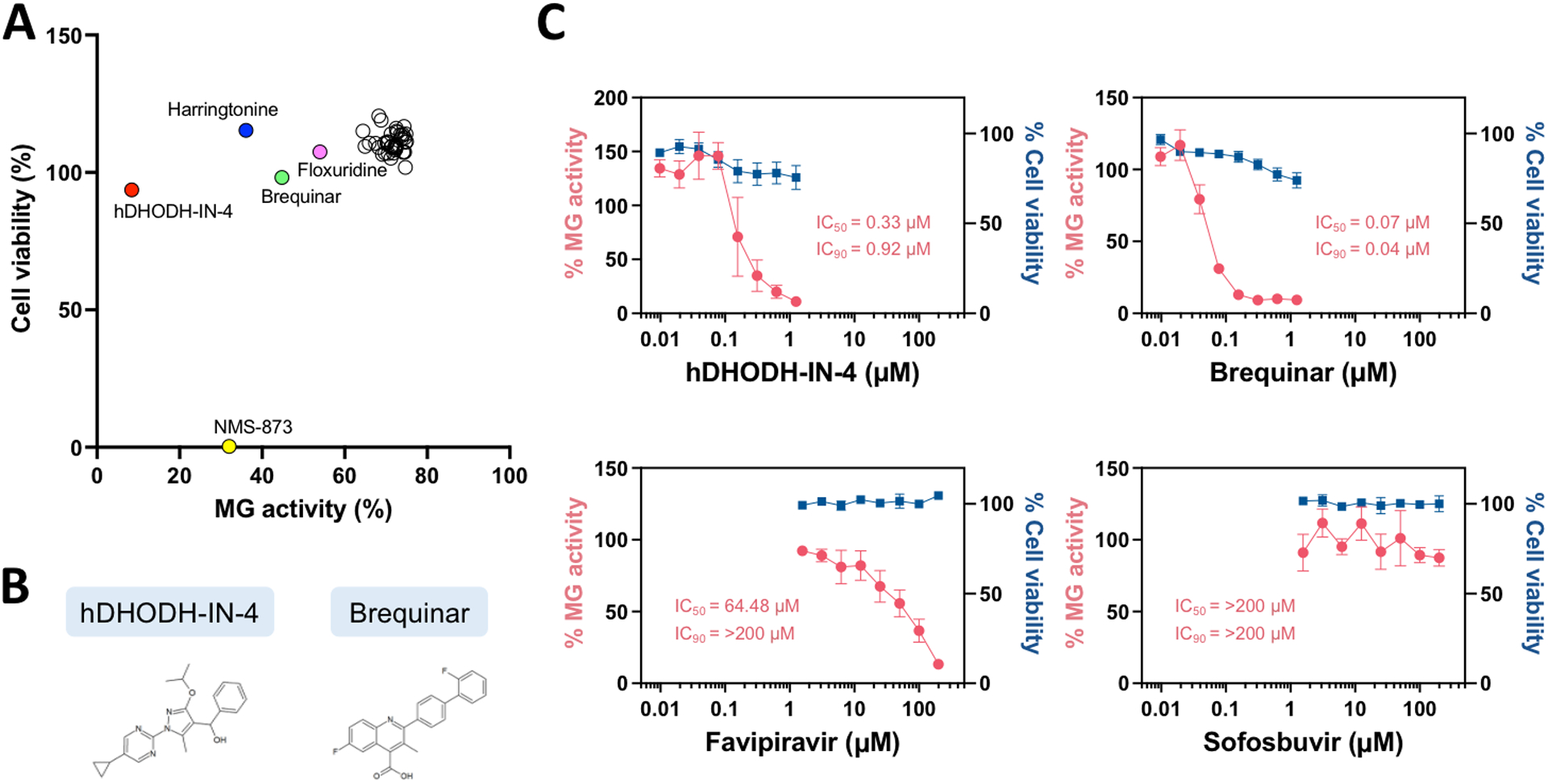
Evaluation of compound efficacies on BRBV MG activity using an optimized, fluorescence-based BRBV MG system. A) High-throughput antiviral compound screening was performed once. HEK293T/17 cells were seeded in 96-well plates and transfected with a MG-ZsGreen plasmid (125 ng) together with helper plasmids (1.56 ng each). Compounds (50 nM) were added to the cells after 8 hours post-transfection, and fluorescence signals were measured at 72 hours post-transfection followed by cell viability assay. Percent reduction of BRBV MG activity and cell viability in compound-treated cells are calculated relative to DMSO vehicle control-treated cells. Compounds that inhibited BRBV MG activity more than 25% relative to the DMSO vehicle control were included in the plot. B) Chemical structures of hDHODH-IN-4 and brequinar. C) IC50 and IC90 of hDHODH-IN-4, brequinar, favipiravir, and sofosbuvir are determined by using serially diluted compounds with BRBV MG system. HEK293T/17 cells were seeded in 96-well plates and transfected with a MG-ZsGreen plasmid (125 ng) together with helper plasmids (1.56 ng each). Compounds were added to the cells after 8 hours post-transfection, and fluorescence signals were measured at 72 hours post-transfection followed by cell viability assay. Data are representative of the average of three independent experiments.
The IC50 and IC90 values were further determined for two dihydroorotate dehydrogenase (DHODH) inhibitors, hDHODH-IN-4 and brequinar (Fig. 3B), along with favipiravir in our BRBV MG platform. We found that hDHODH-IN-4 and brequinar exhibited IC50 and IC90 values in the nanomolar range, which were significantly lower than those of favipiravir (Fig. 3C, Table 1). Additionally, we included sofosbuvir, an FDA-approved hepatitis C virus RdRp inhibitor which has been shown to have no effect on negative-strand RNA viruses in our MG assay as a negative control (Bhatia et al., 2014; Deval et al., 2015). As expected, sofosbuvir did not show any inhibition of BRBV MG activity (Fig. 3C).
Table 1.
IC50 values of each compounds on inhibition of various orthomyxovirus MG activities (μM)
| hDHODH-IN-4 | Brequinar | Favipiravir | Sofosbuvir | |
|---|---|---|---|---|
| IAV MG | 0.32 | 0.12 | 196.23 | >200 |
| Bat IAV MG | 0.22 | 0.07 | 108.71 | >200 |
| IDV MG | 0.23 | 0.05 | 92.67 | >200 |
| THOV MG | 0.12 | 0.04 | 111.66 | >200 |
| BRBV MG | 0.33 | 0.07 | 64.48 | >200 |
3.4. DHODH inhibitors counteract BRBV replication in vitro
Next, we evaluated antiviral efficacy of newly identified compounds, hDHODH-IN-4 and brequinar, against BRBV in Huh7 cells, which support BRBV replication with the highest titer reached at 3 dpi at an MOI of 0.1 (Fig. 4). We found that hDHODH-IN-4 and brequinar are remarkably potent in inhibiting BRBV replication, with IC50 and IC90 in the nanomolar range (Fig. 5A–B). Favipiravir also was capable of inhibiting BRBV replication, albeit with an IC50 lower than that previously reported (0.54 μM in our study; 310 μM in Bricker et al., 2019), and with significantly less inhibition than hDHODH-IN-4 and brequinar. These results clearly demonstrated a high potency of DHODH inhibitors to counteract BRBV replication in vitro.
Fig. 4:
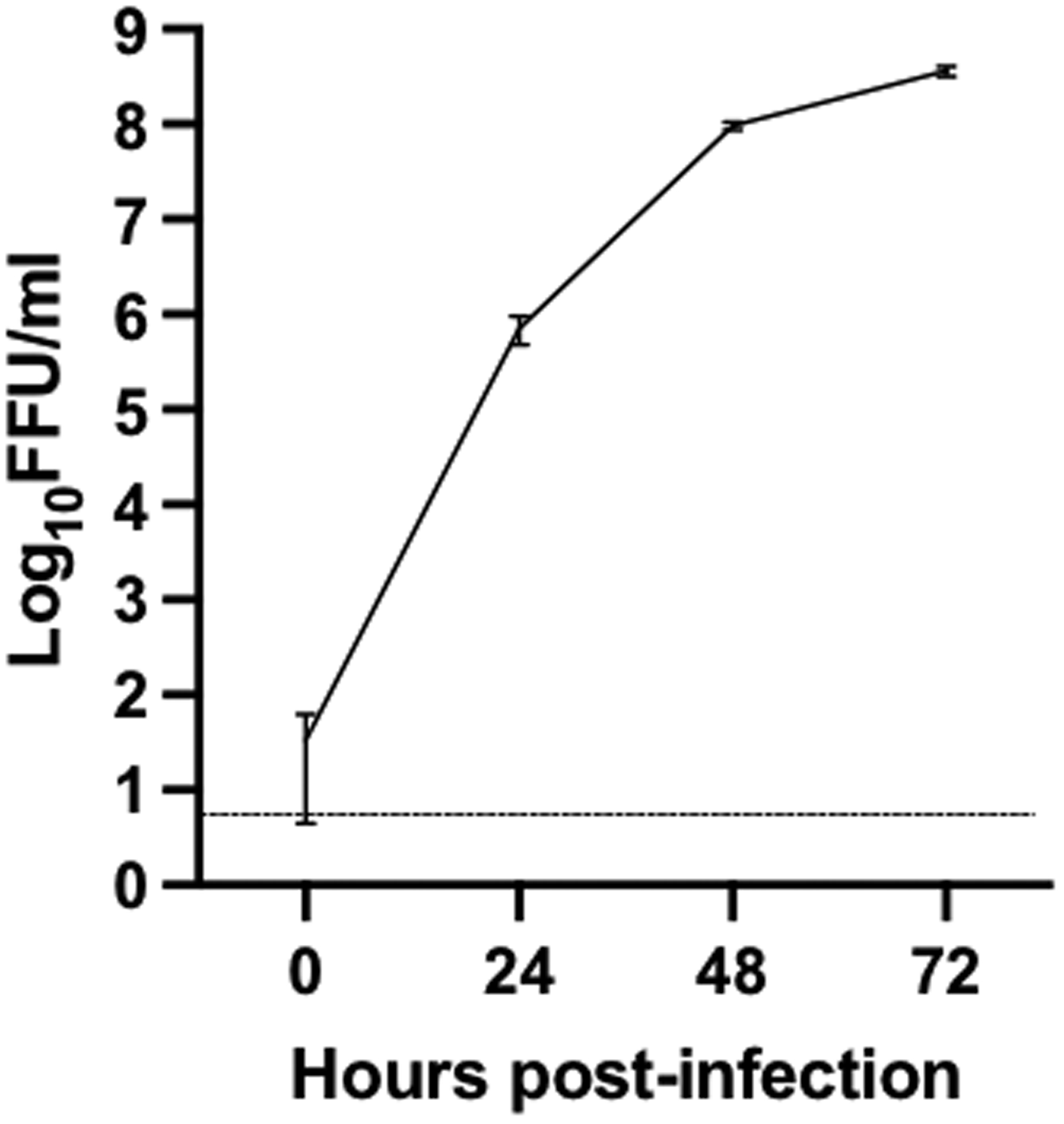
BRBV growth kinetics in Huh7 cells at 0, 24, 48, and 72 hours post-infection. Huh7 cells were seeded in 6-well plate and infected with BRBV at a multiplicity of infection of 0.1. Viral titers in culture supernatant are represented as log10 focus-forming unit (FFU)/ml. The dotted line represents the limit of detection. Data are representative of the average of three independent experiments.
Fig. 5:
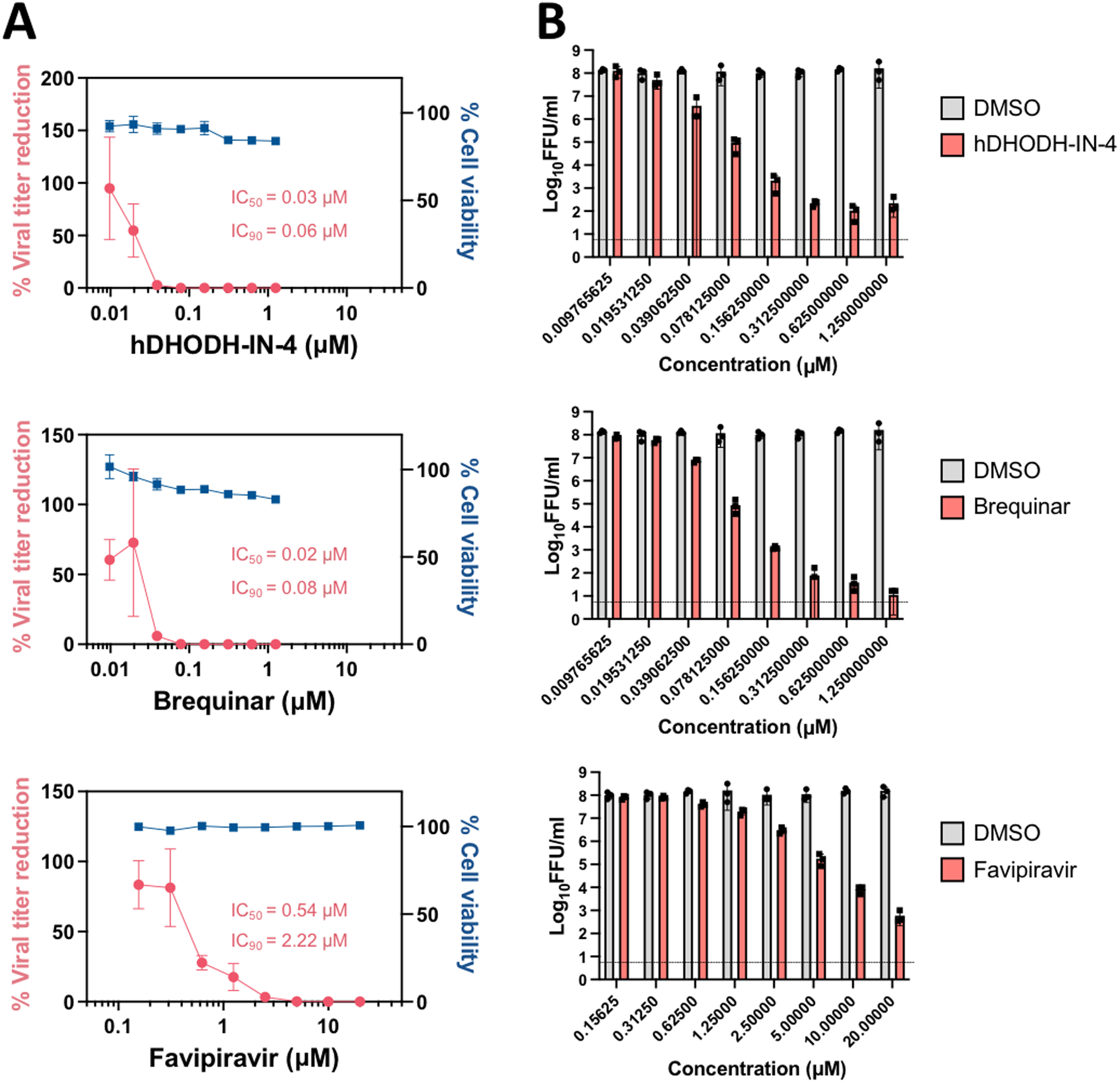
Evaluation of compound efficacies on BRBV replication in Huh7 cells. Huh7 cells were seeded in 24-well plate and infected with BRBV at a multiplicity of infection of 0.1 and treated with serially diluted compounds, hDHODH-IN-4, brequinar, and favipiravir. Supernatants were harvested at 72 hours post-infection and used for titration. Data are shown as A) percent viral titer reduction and cell viability in compound-treated cells relative to DMSO vehicle control-treated cells with IC50 and IC90 values and B) log10 focus-forming unit (FFU)/ml. Data are representative of the average of three independent experiments.
3.5. Broad spectrum efficacy of DHODH inhibitors on various orthomyxovirus MG activities
In recent decades, a number of novel orthomyxoviruses have been identified worldwide (Allison et al., 2015; Campos et al., 2019; Chiapponi et al., 2016; Ducatez et al., 2015; Ejiri et al., 2018; Ellis J.C., 2010; Ferguson et al., 2015; Flynn et al., 2018; Hause et al., 2013; Jiang et al., 2014; Kessell et al., 2012; Mitra et al., 2016; Murakami et al., 2016; Tong et al., 2012; Tong et al., 2013; Yang et al., 2021). To validate whether our novel fluorescence-based MG system can be applied to a high-throughput screening scheme for pan-orthomyxovirus antiviral drugs, we generated additional MG systems that cover a broad range of orthomyxoviruses with emerging zoonotic potential, including bat influenza A virus (bat IAV: A/flat-faced bat/Peru/033/2010 (H18N11)) (Tong et al., 2013), swine influenza D virus (swine IDV: D/swine/Oklahoma/1334/2011) (Hause et al., 2013), and Thogoto virus (THOV: prototype IIA), along with a human influenza A (H1N1) virus pandemic strain (IAV: A/Mexico/InDRE4487/2009) (Tsuda et al., 2017) as a control (Fig. 6A), and tested compound’s inhibitory effects on their MG activities. All MGs were successfully transcribed, with the THOV MG platform having the highest activity (Fig. 6B–C, S3A–B). Helper plasmids were diluted 16-fold for IAV, bat IAV and THOV MG systems, and 32-fold for IDV MG system from the original condition according to our plasmid titration assay to optimize MG activity (Fig. 6D) and used for further experiments. We demonstrated that hDHODH-IN-4 and brequinar have a pan-inhibitory effect on all orthomyxovirus MG activities in the nanomolar range, whereas the IC50 and IC90 of favipiravir were in the micromolar range (Table 1, Fig. S4A–D). No inhibitory activity occurred with sofosbuvir treatment in any orthomyxovirus MG system. In summary, we showed that our fluorescence-based MG system is a robust platform to identify antiviral drugs targeting orthomyxoviruses. Our data all strongly suggest that DHODH inhibitors could be an effective treatment approach across the Orthomyxoviridae family, including potential emerging zoonotic orthomyxoviruses.
Fig. 6:
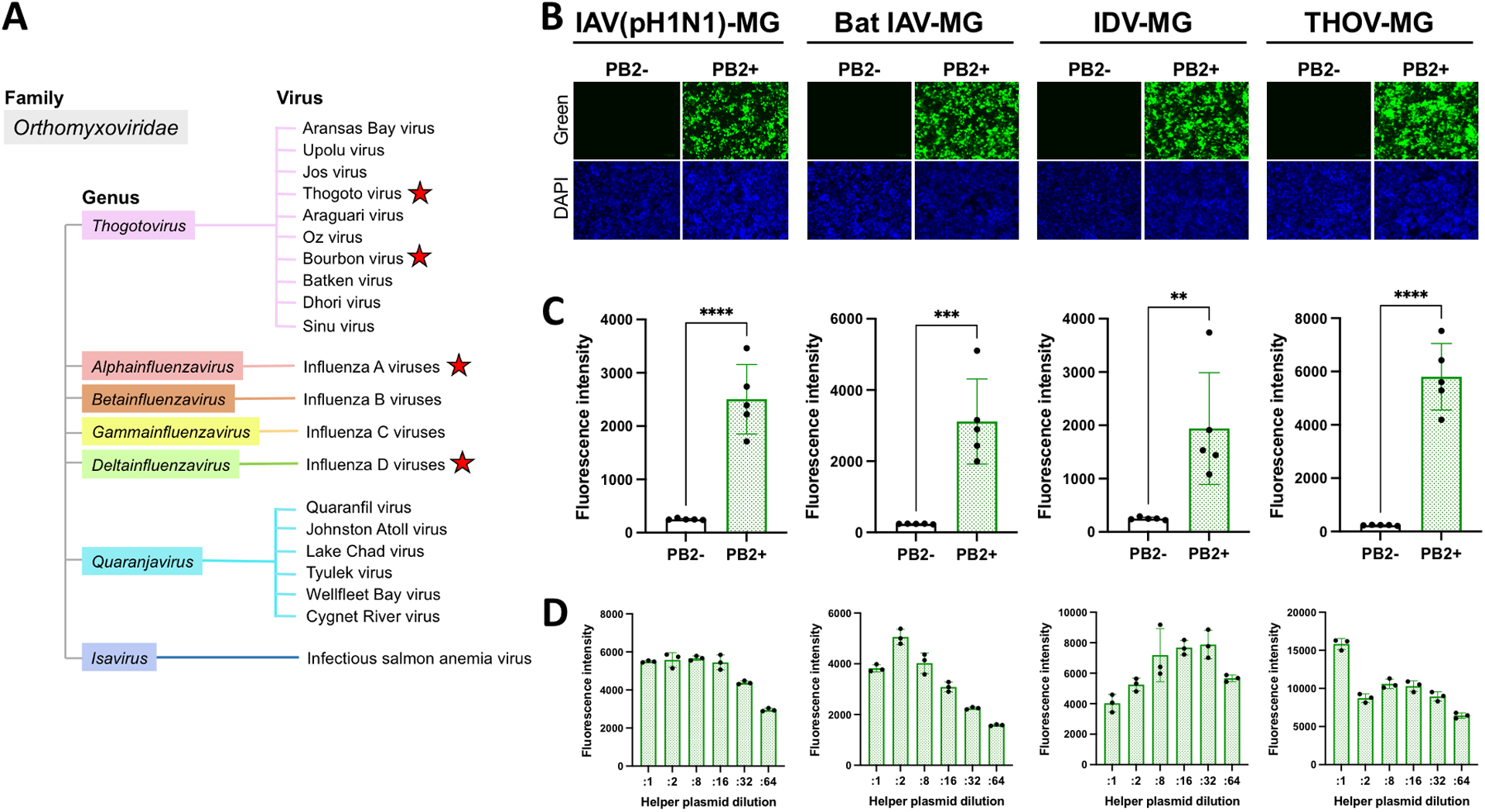
Fluorescence-based orthomyxovirus MG systems in HEK293T/17 cells. A) Classification of viruses belonging to the family Orthomyxoviridae. The names of viruses which have been isolated are shown. MG systems were generated for viruses labeled with red stars. MG activities are shown by B) fluorescence signals and C) fluorescence intensity with or without supplementation of PB2 helper plasmid at 72 hours post-transfection. For B) and C), HEK293T/17 cells were seeded in 24-well plates and transfected with a MG-ZsGreen plasmid (0.25 μg) together with helper plasmids (0.1 μg each). D) MG activities induced by transfection of serially diluted helper plasmids. HEK293T/17 cells were seeded in 96-well plates and transfected with an IAV (pH1N1) MG-ZsGreen, IDV MG-ZsGreen, and THOV MG-ZsGreen plasmid (125 ng) or bat IAV MG-ZsGreen plasmid (62.5 ng) together with helper plasmids (25ng each as the initial amount which were further diluted for :2, :8, :16, :32, and :64). Data are representative of the average of three independent experiments or triplicate in one experiment. **: p < 0.005, ***: p < 0.0005, ****: p < 0.00005.
4. Discussion
In the past few decades, numerous novel orthomyxoviruses such as BRBV (Bricker et al., 2019; Kosoy et al., 2015), bat-associated IAVs (Campos et al., 2019; Tong et al., 2012; Tong et al., 2013; Yang et al., 2021), IDVs (Chiapponi et al., 2016; Ducatez et al., 2015; Ferguson et al., 2015; Flynn et al., 2018; Hause et al., 2013; Jiang et al., 2014; Mitra et al., 2016; Murakami et al., 2016), Wellfleet Bay virus (Allison et al., 2015; Ellis et al., 2010), Cygnet River virus (Kessell et al., 2012), and Oz virus (Ejiri et al., 2018) have been identified worldwide. Among them, BRBV is known to be pathogenic to humans. In this study, we established a highly accessible pan-orthomyxovirus drug screening platform based on a fluorescent protein reporter system. While luminescence-based assays have a high detection sensitivity, they require multiple steps to obtain results including removal of cell supernatants, washing, lysis, and mixture of cellular lysates with luciferase substrate. In contrast, our fluorescence-based orthomyxovirus MG systems do not require any downstream manipulation, which enabled us to perform high-throughput compound screening in a simple, rapid, and inexpensive way with quantitative output using a multimode plate reader. Importantly, we demonstrated that the efficacy of compounds for inhibition of BRBV RNA synthesis in the MG system was consistent with that with infectious virus (Fig. 3C, 5A–B), indicating a good reliability of our MG-based compound screening platform. We expect that our quantitative MG system can be promptly applied to newly emerged orthomyxoviruses to evaluate antivirals, if the need arises.
In this study, we identified that two DHODH inhibitors, hDHODH-IN-4 and brequinar, efficiently inhibit BRBV replication in vitro. The IC50 of hDHODH-IN-4 and brequinar are 25.7 nM and 17.9 nM, respectively (Fig. 5A), which are significantly lower than that of favipiravir (IC50: 540 nM; 310 μM (Bricker et al., 2019)) and myricetin (IC50: 4.6 μM (Hao et al., 2020)), the compounds previously found to counteract BRBV infection. DHODH is a mitochondrial enzyme that catalyzes the conversion of dihydroorotate to orotic acid in the de novo pyrimidine biosynthesis pathway (Barnes et al., 1993; Boukalova et al., 2020; Evans and Guy, 2004). The mechanism of antiviral action of DHODH inhibitors is by the inhibition of de novo pyrimidine synthesis that results in depletion of nucleotides required for virus replication (Coelho and Oliveira, 2020; Hoffmann et al., 2011; Zhou et al., 2021). Mammalian cells possess two pyrimidine synthesis pathways: salvage and de novo. While the salvage pathway supplies basal level of pyrimidine in normal proliferative cells, the de novo pathway supplies additional amounts of pyrimidine to meet the enormous demand for nucleotides in metabolically activated cells (e.g., virus infected cells). Because the DHODH inhibitors only targets the de novo pathway but leaves the salvage pathway intact, this host-targeting strategy can be effectively applied to counteract virus infection with minimum damage on cell viability. In addition to this main antiviral action, DHODH inhibitors have been shown to amplify innate immunity, which may also contribute to their antiviral activities (Lucas-Hourani et al., 2013; Luthra et al., 2018; Yang et al., 2018). The pathway of host pyrimidine metabolism, especially via DHODH, has become of great interest as a target of antiviral as well as cancer therapy in recent years (Boukalova et al., 2020; Kaur et al., 2021; Madak et al., 2019; Vyas and Ghate, 2011; Zhou et al., 2021). To date, there are 3 DHODH inhibitors approved by the US FDA: leflunomide (Arava; approved for use in rheumatoid arthritis and psoriatic arthritis), teriflunomide (Aubagio; approved for use in relapsing forms of multiple sclerosis), and Atovaquone (Mepron; approved for use in Pneumocystis jiroveci induced pneumonia). A number of DHODH inhibitors with different structures have been reported, and some of them have shown broad-spectrum activity against negative-strand RNA viruses including influenza A and B viruses, positive-strand RNA viruses, and DNA viruses (Bonavia et al., 2011; Hahn et al., 2020; Hoffmann et al., 2011; Xiong et al., 2020; Yang et al., 2018). Brequinar, one of the identified compounds in this study, has been evaluated in phase I/II clinical trials against COVID-19 (ClinicalTrials.gov Identifier: NCT04575038 and NCT04425252) and may also be a good candidate to treat BRBV disease. While DHODH inhibitors have been successfully used for a range of diseases, some adverse events including fever, headache, diarrhea, nausea, or difficulty breathing may occur in some patients during the use of FDA-approved DHODH inhibitors (Aly et al., 2017; Fan et al., 2022). Continued studies will be therefore required to carefully evaluate in vivo efficacies and adverse effects of DHODH inhibitors to apply them for infectious viral diseases.
We observed a large disparity in the IC50 values of favipiravir in inhibition of BRBV replication in vitro between our study and the previous study (IC50 = 0.54 μM in our study; 310 μM in Bricker et al., 2019). One possible reason behind this disparity could be due to the different of virus isolates used for each study: BRBV-STL was used in the previous study, whereas we used BRBV-Original. Interestingly, we found two amino acid differences in the PB1 polymerase subunit at aa 81 and aa 558 between these two BRBV isolates, suggesting that these amino acids may be responsible for differential impact of favipiravir. In future studies, we will characterize the potential differences in favipiravir sensitivity seen among BRBV isolates.
With our orthomyxovirus MG systems, we found a unique characteristic of IDV genome replication/transcription: unlike other orthomyxovirus MG systems, IDV MG activity increased by reducing the amount of helper proteins (Fig. 6D). This insight suggests the interference of RNP function by either excessive amount of helper proteins or possibly by unknown viral protein(s) encoded by the ORF sequence of IDV segment 1, 2, 3, or 5. Indeed, PB2-S1, a novel viral protein encoded by a spliced mRNA from IAV PB2 segment, has been shown to interfere with viral polymerase activity due to its PB1-binding capability (Yamayoshi et al., 2016). In addition to compound screening, our established orthomyxovirus MG systems will also serve as a great tool to perform head-to-head comparison of viral RNA synthesis among various orthomyxoviruses, facilitating a better understanding of orthomyxovirus biology.
Supplementary Material
Fig. S1: Selection of fluorescent reporter protein and cell line for efficient detection of fluorescence signal. HEK293 or HEK293T/17 cells were seeded in 96-well plate and transfected with empty vector, GFP-, or ZsGreen-expressing pCAGGs plasmid (100 ng). Data are shown by A) fluorescence signals and B) fluorescence intensity at 72 hours post-transfection. C) Fold induction of ZsGreen intensity is obtained by normalizing fluorescence intensity measured in the ZsGreen-expressing cells with empty vector-transfected cells. Data are representative of the average of triplicate in one experiment. **: p < 0.005, ****: p < 0.00005.
Fig. S2: Comparison of activities of BRBV MGs constructed based on segment 1, 2, 4, or 6. A) The nucleotide sequences of the 3’ and 5’ non-coding regions for BRBV segment 1 (S1), 2 (S2), 4 (S4), and 6 (S6). B) Fluorescence intensity with or without supplementation of PB2 helper plasmid at 72 hours post-transfection. HEK293T/17 cells were seeded in 24-well plate and transfected with a MG-ZsGreen plasmid (0.25 μg) together with helper plasmids (0.1 μg each). C) Fold induction of BRBV MG activity was obtained by normalizing PB2+ fluorescence intensity with PB2-. Data are representative of the average of three independent experiments. *: p < 0.05, **: p < 0.005, ***: p < 0.0005, ns: p > 0.05.
Fig. S3: Comparison of MG activities among different orthomyxovirus MG systems. A) Fluorescence intensity with or without supplementation of PB2 helper plasmid at 72 hours post-transfection. HEK293T/17 cells were seeded in 24-well plate and transfected with an IAV (pH1N1) MG-ZsGreen, bat IAV MG-ZsGreen, IDV MG-ZsGreen, and THOV MG-ZsGreen plasmid (0.25 μg) together with helper plasmids (0.1 μg each). B) Fold induction of MG activity was obtained by normalizing PB2+ fluorescence intensity with PB2-. Data are representative of the average of three independent experiments. **: p < 0.005, ***: p < 0.0005, ****: p < 0.00005, ns: p > 0.05.
Fig. S4: Evaluation of compound efficacies on orthomyxovirus MG activities using optimized, fluorescence-based A) IAV (pH1N1) MG, B) bat IAV MG, C) IDV MG, and D) THOV MG systems. HEK293T/17 cells were seeded in 96-well plates and transfected with an IAV MG-ZsGreen and THOV MG-ZsGreen plasmid (125 ng) together with helper plasmids (1.56 ng each) or bat IAV MG-ZsGreen plasmid (62.5 ng) together with helper plasmids (3.125 ng each) or IDV MG-ZsGreen plasmid (125 ng) together with helper plasmids (0.78 ng each). Compounds were added to the cells after 8 hours post-transfection, and fluorescence signals were measured at 72 hours post-transfection followed by cell viability assay. IC50 and IC90 of hDHODH-IN-4, brequinar, favipiravir, and sofosbuvir are determined by using serially diluted compounds with all MG systems. Percent reduction of MG activity and cell viability in compound-treated cells are calculated relative to DMSO vehicle control-treated cells. Data are representative of the average of three independent experiments.
Table S1: Summary of high-throughput antiviral drug screening using BRBV MG system
Highlights.
We developed a minigenome-based high-throughput antiviral screening system for BRBV, an emerging pathogenic orthomyxovirus
DHODH inhibitors inhibited BRBV minigenome activity and viral replication in vitro with IC90/EC90 in the nanomolar range
The system was applied for other orthomyxoviruses with emerging zoonotic potential, including bat IAV, IDV, and THOV
We present a highly accessible platform for high-throughput antiviral screening across the Orthomyxoviridae family
Acknowledgement
We would like to thank Brandy J. Russell (Division of Vector-Borne Diseases at the Centers for Disease Control and Prevention [CDC]) for providing viruses including BRBV-Original, -STL, -KS15-1735, -MO13-2499, and THOV IIA. We also would like to thank Dr. Michael A. Barry (Department of Infectious Diseases at Mayo Clinic) for providing scientific input and lab resources.
Funding
H.E. and S.Y. were supported in part by the Mayo Clinic internal start up-budget.
Footnotes
Declaration of competing interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
References
- Allison AB, Ballard JR, Tesh RB, Brown JD, Ruder MG, Keel MK, Munk BA, Mickley RM, Gibbs SE, Travassos da Rosa AP, Ellis JC, Ip HS, Shearn-Bochsler VI, Rogers MB, Ghedin E, Holmes EC, Parrish CR, Dwyer C, 2015. Cyclic avian mass mortality in the northeastern United States is associated with a novel orthomyxovirus. J Virol 89, 1389–1403. 10.1128/JVI.02019-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly L, Hemmer B, Korn T, 2017. From Leflunomide to Teriflunomide: Drug Development and Immunosuppressive Oral Drugs in the Treatment of Multiple Sclerosis. Curr Neuropharmacol 15, 874–891. 10.2174/1570159X14666161208151525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asha K, Kumar B, 2019. Emerging Influenza D Virus Threat: What We Know so Far! J Clin Med 8. 10.3390/jcm8020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes T, Parry P, Hart I, Jones C, Minet M, Patterson D, 1993. Regional mapping of the gene encoding dihydroorotate dehydrogenase, an enzyme involved in UMP synthesis, electron transport, and superoxide generation, to human chromosome region 16q22. Somat Cell Mol Genet 19, 405–411. 10.1007/BF01232751. [DOI] [PubMed] [Google Scholar]
- Bhatia HK, Singh H, Grewal N, Natt NK, 2014. Sofosbuvir: A novel treatment option for chronic hepatitis C infection. J Pharmacol Pharmacother 5, 278–284. 10.4103/0976-500X.142464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonavia A, Franti M, Pusateri Keaney E, Kuhen K, Seepersaud M, Radetich B, Shao J, Honda A, Dewhurst J, Balabanis K, Monroe J, Wolff K, Osborne C, Lanieri L, Hoffmaster K, Amin J, Markovits J, Broome M, Skuba E, Cornella-Taracido I, Joberty G, Bouwmeester T, Hamann L, Tallarico JA, Tommasi R, Compton T, Bushell SM, 2011. Identification of broad-spectrum antiviral compounds and assessment of the druggability of their target for efficacy against respiratory syncytial virus (RSV). Proc Natl Acad Sci U S A 108, 6739–6744. 10.1073/pnas.1017142108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukalova S, Hubackova S, Milosevic M, Ezrova Z, Neuzil J, Rohlena J, 2020. Dihydroorotate dehydrogenase in oxidative phosphorylation and cancer. Biochim Biophys Acta Mol Basis Dis 1866, 165759. 10.1016/j.bbadis.2020.165759. [DOI] [PubMed] [Google Scholar]
- Bricker TL, Shafiuddin M, Gounder AP, Janowski AB, Zhao G, Williams GD, Jagger BW, Diamond MS, Bailey T, Kwon JH, Wang D, Boon ACM, 2019. Therapeutic efficacy of favipiravir against Bourbon virus in mice. PLoS Pathog 15, e1007790. 10.1371/journal.ppat.1007790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos ACA, Goes LGB, Moreira-Soto A, de Carvalho C, Ambar G, Sander AL, Fischer C, Ruckert da Rosa A, Cardoso de Oliveira D, Kataoka APG, Pedro WA, Martorelli LFA, Queiroz LH, Cruz-Neto AP, Durigon EL, Drexler JF, 2019. Bat Influenza A(HL18NL11) Virus in Fruit Bats, Brazil. Emerg Infect Dis 25, 333–337. 10.3201/eid2502.181246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Bourbon virus (accessed 5 January 2022).
- Chiapponi C, Faccini S, De Mattia A, Baioni L, Barbieri I, Rosignoli C, Nigrelli A, Foni E, 2016. Detection of Influenza D Virus among Swine and Cattle, Italy. Emerg Infect Dis 22, 352–354. 10.3201/eid2202.151439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho AR, Oliveira PJ, 2020. Dihydroorotate dehydrogenase inhibitors in SARS-CoV-2 infection. Eur J Clin Invest 50, e13366. 10.1111/eci.13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval J, Hong J, Wang G, Taylor J, Smith LK, Fung A, Stevens SK, Liu H, Jin Z, Dyatkina N, Prhavc M, Stoycheva AD, Serebryany V, Liu J, Smith DB, Tam Y, Zhang Q, Moore ML, Fearns R, Chanda SM, Blatt LM, Symons JA, Beigelman L, 2015. Molecular Basis for the Selective Inhibition of Respiratory Syncytial Virus RNA Polymerase by 2’-Fluoro-4’-Chloromethyl-Cytidine Triphosphate. PLoS Pathog 11, e1004995. 10.1371/journal.ppat.1004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatez MF, Pelletier C, Meyer G, 2015. Influenza D virus in cattle, France, 2011–2014. Emerg Infect Dis 21, 368–371. 10.3201/eid2102.141449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejiri H, Lim CK, Isawa H, Fujita R, Murota K, Sato T, Kobayashi D, Kan M, Hattori M, Kimura T, Yamaguchi Y, Takayama-Ito M, Horiya M, Posadas-Herrera G, Minami S, Kuwata R, Shimoda H, Maeda K, Katayama Y, Mizutani T, Saijo M, Kaku K, Shinomiya H, Sawabe K, 2018. Characterization of a novel thogotovirus isolated from Amblyomma testudinarium ticks in Ehime, Japan: A significant phylogenetic relationship to Bourbon virus. Virus Res 249, 57–65. 10.1016/j.virusres.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Ellis JC, C.S.J., Shearn-Boschler VI, 2010. Cyclic mass mortality of common eiders at Cape Cod, MA: an ongoing puzzle., In: Abstracts of the 37th Pacific Seabird Group Annual Meeting. Long Beach (CA): Pacific Seabird Group; 2010., pp. 17–18. [Google Scholar]
- Evans DR, Guy HI, 2004. Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. J Biol Chem 279, 33035–33038. 10.1074/jbc.R400007200. [DOI] [PubMed] [Google Scholar]
- Fan JH, Fogt F, Berger JR, 2022. Gastrointestinal pathologic findings of teriflunomide associated diarrhea. Mult Scler Relat Disord 58, 103506. 10.1016/j.msard.2022.103506. [DOI] [PubMed] [Google Scholar]
- Ferguson L, Eckard L, Epperson WB, Long LP, Smith D, Huston C, Genova S, Webby R, Wan XF, 2015. Influenza D virus infection in Mississippi beef cattle. Virology 486, 28–34. 10.1016/j.virol.2015.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick R, Flick K, Feldmann H, Elgh F, 2003. Reverse genetics for crimean-congo hemorrhagic fever virus. J Virol 77, 5997–6006. 10.1128/jvi.77.10.5997-6006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn O, Gallagher C, Mooney J, Irvine C, Ducatez M, Hause B, McGrath G, Ryan E, 2018. Influenza D Virus in Cattle, Ireland. Emerg Infect Dis 24, 389–391. 10.3201/eid2402.170759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsey MS, Rose D, Burkhalter KL, Breuner N, Bosco-Lauth AM, Kosoy OI, Savage HM, 2021. Experimental Infection of Amblyomma americanum (Acari: Ixodidae) With Bourbon Virus (Orthomyxoviridae: Thogotovirus). J Med Entomol 58, 873–879. 10.1093/jme/tjaa191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn F, Wangen C, Hage S, Peter AS, Dobler G, Hurst B, Julander J, Fuchs J, Ruzsics Z, Uberla K, Jack HM, Ptak R, Muehler A, Groppel M, Vitt D, Peelen E, Kohlhof H, Marschall M, 2020. IMU-838, a Developmental DHODH Inhibitor in Phase II for Autoimmune Disease, Shows Anti-SARS-CoV-2 and Broad-Spectrum Antiviral Efficacy In Vitro. Viruses 12. 10.3390/v12121394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV, 2012. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol 7, 1848–1857. 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Ning K, Wang X, Wang J, Cheng F, Ganaie SS, Tavis JE, Qiu J, 2020. Establishment of a Replicon Reporter of the Emerging Tick-Borne Bourbon Virus and Use It for Evaluation of Antivirals. Front Microbiol 11, 572631. 10.3389/fmicb.2020.572631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, Armien A, Kaplan B, Chakravarty S, Hoppe AD, Webby RJ, Simonson RR, Li F, 2013. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog 9, e1003176. 10.1371/journal.ppat.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T, Jung S, Herwig A, Groseth A, Becker S, 2010. Both matrix proteins of Ebola virus contribute to the regulation of viral genome replication and transcription. Virology 403, 56–66. 10.1016/j.virol.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Hoffmann HH, Kunz A, Simon VA, Palese P, Shaw ML, 2011. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc Natl Acad Sci U S A 108, 5777–5782. 10.1073/pnas.1101143108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Committee on Taxonomy of Viruses (ICTV) Reports, 2020. Virus Taxonomy: 2020 Release, Orthomyxoviridae, pp. EC 52, Online meeting, October 2020. (accessed 22 December 2021).
- Jackson KC, Gidlewski T, Root JJ, Bosco-Lauth AM, Lash RR, Harmon JR, Brault AC, Panella NA, Nicholson WL, Komar N, 2019. Bourbon Virus in Wild and Domestic Animals, Missouri, USA, 2012–2013. Emerg Infect Dis 25, 1752–1753. 10.3201/eid2509.181902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WM, Wang SC, Peng C, Yu JM, Zhuang QY, Hou GY, Liu S, Li JP, Chen JM, 2014. Identification of a potential novel type of influenza virus in Bovine in China. Virus Genes 49, 493–496. 10.1007/s11262-014-1107-3. [DOI] [PubMed] [Google Scholar]
- Kaur H, Sarma P, Bhattacharyya A, Sharma S, Chhimpa N, Prajapat M, Prakash A, Kumar S, Singh A, Singh R, Avti P, Thota P, Medhi B, 2021. Efficacy and safety of dihydroorotate dehydrogenase (DHODH) inhibitors “leflunomide” and “teriflunomide” in Covid-19: A narrative review. Eur J Pharmacol 906, 174233. 10.1016/j.ejphar.2021.174233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessell A, Hyatt A, Lehmann D, Shan S, Crameri S, Holmes C, Marsh G, Williams C, Tachedjian M, Yu M, Bingham J, Payne J, Lowther S, Wang J, Wang LF, Smith I, 2012. Cygnet River virus, a novel orthomyxovirus from ducks, Australia. Emerg Infect Dis 18, 2044–2046. 10.3201/eid1812.120500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar N, Hamby N, Palamar MB, Staples JE, Williams C, 2020. Indirect Evidence of Bourbon Virus (Thogotovirus, Orthomyxoviridae) Infection in North Carolina. N C Med J 81, 214–215. 10.18043/ncm.81.3.214. [DOI] [PubMed] [Google Scholar]
- Kosoy OI, Lambert AJ, Hawkinson DJ, Pastula DM, Goldsmith CS, Hunt DC, Staples JE, 2015. Novel thogotovirus associated with febrile illness and death, United States, 2014. Emerg Infect Dis 21, 760–764. 10.3201/eid2105.150150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B, Asha K, Khanna M, Ronsard L, Meseko CA, Sanicas M, 2018. The emerging influenza virus threat: status and new prospects for its therapy and control. Arch Virol 163, 831–844. 10.1007/s00705-018-3708-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AJ, Velez JO, Brault AC, Calvert AE, Bell-Sakyi L, Bosco-Lauth AM, Staples JE, Kosoy OI, 2015. Molecular, serological and in vitro culture-based characterization of Bourbon virus, a newly described human pathogen of the genus Thogotovirus. J Clin Virol 73, 127–132. 10.1016/j.jcv.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas-Hourani M, Dauzonne D, Jorda P, Cousin G, Lupan A, Helynck O, Caignard G, Janvier G, Andre-Leroux G, Khiar S, Escriou N, Despres P, Jacob Y, Munier-Lehmann H, Tangy F, Vidalain PO, 2013. Inhibition of pyrimidine biosynthesis pathway suppresses viral growth through innate immunity. PLoS Pathog 9, e1003678. 10.1371/journal.ppat.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra P, Naidoo J, Pietzsch CA, De S, Khadka S, Anantpadma M, Williams CG, Edwards MR, Davey RA, Bukreyev A, Ready JM, Basler CF, 2018. Inhibiting pyrimidine biosynthesis impairs Ebola virus replication through depletion of nucleoside pools and activation of innate immune responses. Antiviral Res 158, 288–302. 10.1016/j.antiviral.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madak JT, Bankhead A 3rd, Cuthbertson CR, Showalter HD, Neamati N, 2019. Revisiting the role of dihydroorotate dehydrogenase as a therapeutic target for cancer. Pharmacol Ther 195, 111–131. 10.1016/j.pharmthera.2018.10.012. [DOI] [PubMed] [Google Scholar]
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA, 1999. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol 17, 969–973. 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- Mendoza CA, Yamaoka S, Tsuda Y, Matsuno K, Weisend CM, Ebihara H, 2021. The NF-kappaB inhibitor, SC75741, is a novel antiviral against emerging tick-borne bandaviruses. Antiviral Res 185, 104993. 10.1016/j.antiviral.2020.104993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra N, Cernicchiaro N, Torres S, Li F, Hause BM, 2016. Metagenomic characterization of the virome associated with bovine respiratory disease in feedlot cattle identified novel viruses and suggests an etiologic role for influenza D virus. J Gen Virol 97, 1771–1784. 10.1099/jgv.0.000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Endoh M, Kobayashi T, Takenaka-Uema A, Chambers JK, Uchida K, Nishihara M, Hause B, Horimoto T, 2016. Influenza D Virus Infection in Herd of Cattle, Japan. Emerg Infect Dis 22, 1517–1519. 10.3201/eid2208.160362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J, 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199. 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Reports, I.C.o.T.o.V.I., 2020. Virus Taxonomy: 2020 Release, Orthomyxoviridae, pp. EC 52, Online meeting, October 2020.
- Savage HM, Burkhalter KL, Godsey MS Jr., Panella NA, Ashley DC, Nicholson WL, Lambert AJ, 2017. Bourbon Virus in Field-Collected Ticks, Missouri, USA. Emerg Infect Dis 23, 2017–2022. 10.3201/eid2312.170532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage HM, Godsey MS Jr., Panella NA, Burkhalter KL, Manford J, Trevino-Garrison IC, Straily A, Wilson S, Bowen J, Raghavan RK, 2018. Surveillance for Tick-Borne Viruses Near the Location of a Fatal Human Case of Bourbon Virus (Family Orthomyxoviridae: Genus Thogotovirus) in Eastern Kansas, 2015. J Med Entomol 55, 701–705. 10.1093/jme/tjx251. [DOI] [PubMed] [Google Scholar]
- Tian D, Liu Y, Liang C, Xin L, Xie X, Zhang D, Wan M, Li H, Fu X, Liu H, Cao W, 2021a. An update review of emerging small-molecule therapeutic options for COVID-19. Biomed Pharmacother 137, 111313. 10.1016/j.biopha.2021.111313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Qiang T, Liang C, Ren X, Jia M, Zhang J, Li J, Wan M, YuWen X, Li H, Cao W, Liu H, 2021b. RNA-dependent RNA polymerase (RdRp) inhibitors: The current landscape and repurposing for the COVID-19 pandemic. Eur J Med Chem 213, 113201. 10.1016/j.ejmech.2021.113201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO, 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A 109, 4269–4274. 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO, 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog 9, e1003657. 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda Y, Weisend C, Martellaro C, Feldmann F, Haddock E, 2017. Pathogenic analysis of the pandemic 2009 H1N1 influenza A viruses in ferrets. J Vet Med Sci 79, 1453–1460. 10.1292/jvms.16-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas VK, Ghate M, 2011. Recent developments in the medicinal chemistry and therapeutic potential of dihydroorotate dehydrogenase (DHODH) inhibitors. Mini Rev Med Chem 11, 1039–1055. 10.2174/138955711797247707. [DOI] [PubMed] [Google Scholar]
- Xiong R, Zhang L, Li S, Sun Y, Ding M, Wang Y, Zhao Y, Wu Y, Shang W, Jiang X, Shan J, Shen Z, Tong Y, Xu L, Chen Y, Liu Y, Zou G, Lavillete D, Zhao Z, Wang R, Zhu L, Xiao G, Lan K, Li H, Xu K, 2020. Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly-emerged coronavirus SARS-CoV-2. Protein Cell 11, 723–739. 10.1007/s13238-020-00768-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamayoshi S, Watanabe M, Goto H, Kawaoka Y, 2016. Identification of a Novel Viral Protein Expressed from the PB2 Segment of Influenza A Virus. J Virol 90, 444–456. 10.1128/JVI.02175-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CF, Gopula B, Liang JJ, Li JK, Chen SY, Lee YL, Chen CS, Lin YL, 2018. Novel AR-12 derivatives, P12–23 and P12–34, inhibit flavivirus replication by blocking host de novo pyrimidine biosynthesis. Emerg Microbes Infect 7, 187. 10.1038/s41426-018-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Schountz T, Ma W, 2021. Bat Influenza Viruses: Current Status and Perspective. Viruses 13. 10.3390/v13040547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Tao L, Zhou X, Zuo Z, Gong J, Liu X, Zhou Y, Liu C, Sang N, Liu H, Zou J, Gou K, Yang X, Zhao Y, 2021. DHODH and cancer: promising prospects to be explored. Cancer Metab 9, 22. 10.1186/s40170-021-00250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: Selection of fluorescent reporter protein and cell line for efficient detection of fluorescence signal. HEK293 or HEK293T/17 cells were seeded in 96-well plate and transfected with empty vector, GFP-, or ZsGreen-expressing pCAGGs plasmid (100 ng). Data are shown by A) fluorescence signals and B) fluorescence intensity at 72 hours post-transfection. C) Fold induction of ZsGreen intensity is obtained by normalizing fluorescence intensity measured in the ZsGreen-expressing cells with empty vector-transfected cells. Data are representative of the average of triplicate in one experiment. **: p < 0.005, ****: p < 0.00005.
Fig. S2: Comparison of activities of BRBV MGs constructed based on segment 1, 2, 4, or 6. A) The nucleotide sequences of the 3’ and 5’ non-coding regions for BRBV segment 1 (S1), 2 (S2), 4 (S4), and 6 (S6). B) Fluorescence intensity with or without supplementation of PB2 helper plasmid at 72 hours post-transfection. HEK293T/17 cells were seeded in 24-well plate and transfected with a MG-ZsGreen plasmid (0.25 μg) together with helper plasmids (0.1 μg each). C) Fold induction of BRBV MG activity was obtained by normalizing PB2+ fluorescence intensity with PB2-. Data are representative of the average of three independent experiments. *: p < 0.05, **: p < 0.005, ***: p < 0.0005, ns: p > 0.05.
Fig. S3: Comparison of MG activities among different orthomyxovirus MG systems. A) Fluorescence intensity with or without supplementation of PB2 helper plasmid at 72 hours post-transfection. HEK293T/17 cells were seeded in 24-well plate and transfected with an IAV (pH1N1) MG-ZsGreen, bat IAV MG-ZsGreen, IDV MG-ZsGreen, and THOV MG-ZsGreen plasmid (0.25 μg) together with helper plasmids (0.1 μg each). B) Fold induction of MG activity was obtained by normalizing PB2+ fluorescence intensity with PB2-. Data are representative of the average of three independent experiments. **: p < 0.005, ***: p < 0.0005, ****: p < 0.00005, ns: p > 0.05.
Fig. S4: Evaluation of compound efficacies on orthomyxovirus MG activities using optimized, fluorescence-based A) IAV (pH1N1) MG, B) bat IAV MG, C) IDV MG, and D) THOV MG systems. HEK293T/17 cells were seeded in 96-well plates and transfected with an IAV MG-ZsGreen and THOV MG-ZsGreen plasmid (125 ng) together with helper plasmids (1.56 ng each) or bat IAV MG-ZsGreen plasmid (62.5 ng) together with helper plasmids (3.125 ng each) or IDV MG-ZsGreen plasmid (125 ng) together with helper plasmids (0.78 ng each). Compounds were added to the cells after 8 hours post-transfection, and fluorescence signals were measured at 72 hours post-transfection followed by cell viability assay. IC50 and IC90 of hDHODH-IN-4, brequinar, favipiravir, and sofosbuvir are determined by using serially diluted compounds with all MG systems. Percent reduction of MG activity and cell viability in compound-treated cells are calculated relative to DMSO vehicle control-treated cells. Data are representative of the average of three independent experiments.
Table S1: Summary of high-throughput antiviral drug screening using BRBV MG system


