Abstract
A molecular approach was used to investigate a recently described candidate division of the domain Bacteria, TM7, currently known only from environmental 16S ribosomal DNA sequence data. A number of TM7-specific primers and probes were designed and evaluated. Fluorescence in situ hybridization (FISH) of a laboratory scale bioreactor using two independent TM7-specific probes revealed a conspicuous sheathed-filament morphotype, fortuitously enriched in the reactor. Morphologically, the filament matched the description of the Eikelboom morphotype 0041-0675 widely associated with bulking problems in activated-sludge wastewater treatment systems. Transmission electron microscopy of the bioreactor sludge demonstrated that the sheathed-filament morphotype had a typical gram-positive cell envelope ultrastructure. Therefore, TM7 is only the third bacterial lineage recognized to have gram-positive representatives. TM7-specific FISH analysis of two full-scale wastewater treatment plant sludges, including the one used to seed the laboratory scale reactor, indicated the presence of a number of morphotypes, including sheathed filaments. TM7-specific PCR clone libraries prepared from the two full-scale sludges yielded 23 novel TM7 sequences. Three subdivisions could be defined based on these data and publicly available sequences. Environmental sequence data and TM7-specific FISH analysis indicate that members of the TM7 division are present in a variety of terrestrial, aquatic, and clinical habitats. A highly atypical base substitution (Escherichia coli position 912; C to U) for bacterial 16S rRNAs was present in almost all TM7 sequences, suggesting that TM7 bacteria, like Archaea, may be streptomycin resistant at the ribosome level.
It is now well recognized that microbial diversity is greatly underestimated by cultivation studies because most microorganisms observable in nature typically cannot be cultivated by standard techniques (2). One result of this limited access has been the extremely narrow focus of microbiology on a very few microbial species. In fact, approximately 65% of worldwide research in microbiology from 1991 to 1997 was dedicated to only eight species of bacteria, representing only three main bacterial lineages (divisions) (14). The advent of culture-independent rRNA-based molecular methods has greatly enhanced our understanding of the extent and character of bacterial diversity (22). Over 40 bacterial divisions are now recognized, only half of which have representatives obtained in pure culture (16). Consequently, environmental-clone sequences comprise a significant portion of the rRNA sequences present in public databases.
Candidate division TM7 is one of several newly described bacterial divisions exclusively characterized by environmental sequence data (16). TM7 was originally proposed based on partial 16S ribosomal DNA (rDNA) sequences obtained from PCR clonal studies of a peat bog (one sequence, after which the division was named [24]), a mature forest soil (two sequences [6]), and two sequencing batch reactor sludges (seven sequences [5]). Therefore, the initial aim of the present study was to fully sequence a number of the original 16S rDNA clones from the study of Bond et al. (5) and complete a more rigorous phylogenetic analysis of the division. From these and other full-length sequences, TM7-specific oligonucleotide primers and probes were designed and evaluated. We report the characterization of a conspicuous TM7 morphotype, fortuitously enriched in a laboratory scale bioreactor, and discuss the possible phylogenetic implications for this candidate bacterial division.
MATERIALS AND METHODS
Sample collection and processing.
Sludge samples (50 ml) were obtained from a laboratory scale sequencing batch reactor operating under enhanced biological phosphorus removal (EBPR) conditions (8) and two full-scale biological nutrient removal wastewater treatment plants, Loganholme (used to seed the laboratory scale reactor) and Noosa wastewater treatment plants. A 50-g sample of garden topsoil was collected at the University of Queensland. Aliquots (0.5 ml or 0.5 g) of the samples were fixed in paraformaldehyde as previously described (4) and stored at 4°C. DNA was extracted from sample aliquots (1 ml or 1 g) using the FastDNA SPIN kit (BIO 101, Inc.), purified through Chroma Spin + TE-1000 columns (Clontech Laboratories Inc.) according to the manufacturer's instructions, and stored at −70°C.
PCR clone libraries.
PCR was performed on bulk DNA extracted from the three activated-sludge samples and the garden topsoil sample. Each 25-μl reaction mixture contained (as final concentrations) 1× Tth plus reaction buffer (Biotech International), 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate 0.3 U of Tth plus DNA polymerase (Biotech International), 10 to 50 ng of genomic DNA, and 50 ng of each forward and reverse primer. Thermal cycling was carried out on a Perkin-Elmer DNA Thermal Cycler 480 with an initial denaturation step of 96°C for 10 min, followed by 28 cycles at 94°C for 1 min, 60°C for 1 min, and 72°C for 2 min; cycling was completed by a final elongation period of 5 min at 72°C. In all reactions, the broad-specificity reverse primer 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) was used combined with a TM7-specific forward primer, either TM7314F or TM7580F (Table 1). An optimal annealing temperature of 60°C was determined empirically to remove nonspecific amplification products. Clone libraries of the TM7314F-1492R PCR products from the two full-scale activated-sludge DNAs were prepared and screened by restriction fragment length polymorphism analysis, and representatives were sequenced as previously described (17). rDNA clones from clone libraries previously prepared in our laboratory were resuscitated from glycerol storage at −70°C on Luria broth-ampicillin plates, and inserts were reamplified with cloning vector primers and sequenced. All sequencing was performed by the Australian Genome Research Facility, University of Queensland, on Applied Biosystems 377 automated sequencers.
TABLE 1.
TM7-specific PCR primers and FISH probes and optimized conditions for use
| Oligonucleotide | E. coli no. | Sequence (5′-3′) | Length (nt) | Tm (°C)a | % GC content | Optimized conditions
|
|
|---|---|---|---|---|---|---|---|
| Annealing temp (°C)b | % Formamidec | ||||||
| Primers | |||||||
| TM7314F | 295–314 | GAGAGGATGATCAGCCAG | 20 | 54 | 50 | 60 | |
| TM7580F | 563–580 | AYTGGGCGTAAAGAGTTGC | 19 | 58 | 50 | 60 | |
| Probes | |||||||
| TM7305 | 305–322 | GTCCCAGTCTGGCTGATC | 18 | 56 | 61 | 30 | |
| TM7905 | 905–926 | CCGTCAATTCCTTTATGTTTTA | 22 | 56 | 32 | 20 | |
| TM7522 | 522–537 | CGTATGACCGCGGCTG | 16 | 61 | 69 | NA | |
| TM7567 | 567–585 | CCTACGCAACTCTTTACGCC | 20 | 60 | 55 | NA | |
Nearest-neighbor melting temperature.
Annealing temperature of PCR using 1492R as reverse primer.
46°C hybridization temperature and 48°C wash temperature. NA, not applicable (no specific hybridization signal was obtained).
Phylogenetic analyses.
Sequences were compiled using the SeqEd software package (Applied Biosystems) and compared to available databases by use of the Basic Local Alignment Search Tool (BLAST) (1) to determine approximate phylogenetic affiliations and to indicate the presence of any existing sequences that might be included in the novel divisions which had not previously been identified. All sequences were examined for chimera formation as previously described (17). The compiled sequences were aligned using the ARB software package (http://www.mikro.biologie.tu-muenchen.de/), and the alignments were refined manually. Phylogenetic trees based on comparative analysis of the 16S rRNA genes were constructed by distance and parsimony methods, with and without corrections for rate variation and GC bias, using PAUP∗ version 4.0b2a (written by David L. Swofford) as described previously (11). The robustness of the tree topology was tested by bootstrap resampling under a range of outgroup configurations (9).
Two TM7-specific PCR primers and four TM7-specific fluorescence in situ hybridization (FISH) probes were designed using the probe design tool of the ARB software package (see above). Based on comparative analysis of all sequences in the database, the program selected regions within the TM7 sequences which allowed the construction of primers and probes specific for all or part of the TM7 division. The design parameters used where possible were centralized nontarget mismatches for probes, 3′-end nontarget mismatches for primers, a nearest-neighbor melting temperature of >56°C for probes (calculated using 50 mM NaCl and 50 μM oligonucleotide), and Escherichia coli ribosome relative probe accessibility of >20% (13). Primer and probe sequences were subsequently confirmed for specificity using BLAST. Selected parameters of the primers and probes are listed in Table 1, and their target specificities are shown in Fig. 1.
FIG. 1.
Evolutionary-distance dendrogram of candidate division TM7 and other bacterial division level groups based on comparative analyses of 16S rDNA data. Division and subdivision designations are bracketed on the right. Specificities of primers and probes designed to target the TM7 division (detailed in Table 1) are indicated in italics outside the brackets. Branch points supported (bootstrap values, >74%) by all inference methods used are indicated by solid circles, and those supported by most inference methods are indicated by open circles. Branch points without circles were not resolved (bootstrap values, <75%) as specific groups in different analyses and at the division level were collapsed back to the next significant node. Clones sequenced in the present study are in boldface. Partial-length TM7 sequences (<600 nt) were inserted into the tree using the parsimony insertion tool of ARB to show their approximate positions and are indicated by dashed line segments. Archaeal outgroups (not shown) for the tree were Methanococcus vannielii (M36507) and Sulfolobus acidocaldarius (D14876). The bar represents 10% estimated sequence divergence.
FISH microscopy.
Probes were commercially synthesized and 5′ labeled with the fluorochrome fluorescein isothiocyanate (FITC), CY3, or CY5 (Genset, Paris, France, or Interactiva, Ulm, Germany). Paraformaldehyde-fixed samples (see above) were dual or triple hybridized with one or two TM7-specific probes (Table 1) and probe EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′), which targets most bacteria, at empirically determined optimal stringencies (Table 1) (see Results) using a standard FISH protocol (21). Sphingomonas sp. strain BF14 (Australian Collection of Microorganisms (ACM) 4962) and Micrococcus luteus (ACM 975) were grown in pure culture on R2A agar medium (23) (2 days at 28°C) as negative controls for TM7305 and TM7905 probe optimization, respectively. A Zeiss LSM510 or Bio-Rad MRC 1024 confocal laser scanning microscope was used for visualization of FISH preparations. Red and green fields were registered in Adobe Photoshop 4.0 and annotated and compiled in Microsoft PowerPoint 98.
Bright-field microscopy.
Gram, methylene blue (for polyanions), and Sudan black (for lipophilic inclusions, including poly-β-hydroxybutyrate [PHB]) stains were prepared for the activated-sludge samples as previously described (4). All bright-field microscopy, including phase-contrast microscopy, was conducted on a Zeiss Axiophot2 microscope.
Transmission electron microscopy (TEM).
A sample of the laboratory scale sequencing batch reactor sludge was fixed in 3% glutaraldehyde in 0.1 M cacodylate buffer for 2 h at room temperature. The fixed biomass was washed twice in 0.1 M cacodylate buffer and embedded in 2% agarose. The sample was subsequently postfixed in 1% osmium tetroxide in 0.1 M cacodylate for 1 h and washed again in buffer prior to being dehydrated through a graded acetone series and embedded in Epon. Thin sections were stained with 5% uranyl acetate in 50% methanol and Reynolds lead citrate and viewed in a JEOL 1010 transmission electron microscope.
Nucleotide sequence accession numbers.
The 36 rDNA clones sequenced in this study have the GenBank accession numbers AF268992 to AF269027.
RESULTS
Phylogeny of the TM7 division.
Thirteen partially sequenced 16S rDNA clones (<500 nucleotides [nt]) from previous published (clone prefix SBR [5]) and unpublished (clone prefixes SBRH and GC) studies of activated sludges in our laboratory were selected for complete sequencing because of their apparent affiliation with candidate division TM7 or unresolved phylogenetic affiliations. Comparative analysis of the 13 fully sequenced clones enabled all clones, with the exception of SBRH63 and SBR1093, to be phylogenetically resolved into recognized divisions or recently described candidate divisions in the domain Bacteria (16). No chimeric sequences were detected. Seven clone sequences were unambiguously affiliated with TM7, and one clone was affiliated with each of the green nonsulfur, termite group I, TM6, and NKB19 (27) groups (Fig. 1). SBRH63 forms a monophyletic group with clone sequences from deep-sea sediments and soils (BD group [Fig. 1]), and SBR1093, together with its closest relative, sludge clone 1959, likely constitutes the nucleus of a new division level group in the domain Bacteria. Monophyly of the groups was established by bootstrap resampling and varying the division level outgroup composition. All groups presented in Fig. 1 were reproducibly monophyletic and unassociated with other recognized bacterial divisions, supporting their candidacy as bacterial divisions (9, 16). Based on sequence data from this study (see below) and the public databases, three subdivisions can presently be resolved in the TM7 division (Fig. 1), with a maximum intradivision sequence divergence of 17% (using the Lane mask [18; http://www.mikro.biologie.tu-muenchen.del). For reference, the 16S rDNA sequences of the TM7 division are 22 to 25% and 21 to 24% dissimilar to those of E. coli (Proteobacteria) and Bacillus subtilis (low-G+C gram-positive bacteria) 16S rDNAs, respectively, also using the Lane mask.
TM7-specific PCR clone libraries and FISH.
PCR primers and FISH probes specific for candidate division TM7 were designed based on comparative analysis of available TM7 16S rDNA sequences (Table 1). TM7-specific PCR using TM7314F-1492R and TM7580F-1492R, indicated the presence of members of the division in laboratory scale and full-scale activated sludges and a garden topsoil. Clone libraries were prepared from the TM7314F-1492R PCR products of the two full-scale activated sludges. Following screening by restriction fragment length polymorphism, 5 and 18 clones were fully sequenced from the Loganholme (TM7LH clones) and Noosa (NoosaAW clones) wastewater treatment plants, respectively. The phylogenetic positioning of these clones within the TM7 division (Fig. 1) confirms the specificity of the PCR and substantially expands the known diversity of the TM7 division. No chimeric sequences were detected.
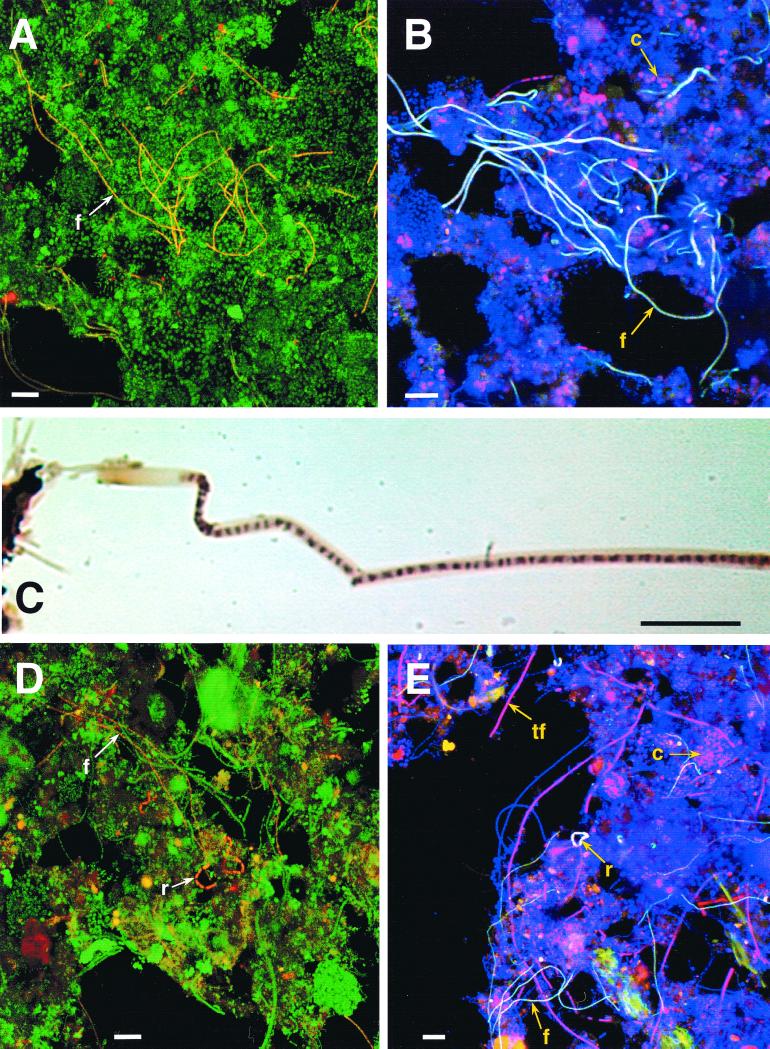
The activated-sludge samples were selected for initial FISH studies because their extracted DNAs yielded strong TM7-specific PCR products (data not shown). Probe TM7305, which targets most of TM7 subdivision 1 (Table 1 and Fig. 1), revealed a conspicuous filament morphotype, occurring in long chains, in the laboratory scale sludge (Fig. 2A). Probe TM7905, designed subsequently and targeting nearly the entire division (Table 1 and Fig. 1), when used in combination with TM7305 confirmed that the filament morphotype belonged to the TM7 division (Fig. 2B). Presumably, the filament morphotype belongs to TM7 subdivision 1, as that is the only subdivision targeted by TM7305 (Fig. 1). In addition, TM7905 hybridized to cocci (Fig. 2B) that may be representatives of TM7 subdivisions 2 and/or 3, which are not targeted by TM7305 (Fig. 1). However, this observation requires confirmation with additional TM7 division-specific probes. Virtually all filaments in the laboratory scale sludge specifically bound both the TM7305 and TM7905 probes (Fig. 2B), ensuring that filaments observed in this sludge by bright-field microscopy likely were members of the TM7 division (Fig. 2C) (see below).
FIG. 2.
Light microscopy of the microbial communities of laboratory scale and full-scale activated-sludge samples. All bars are 10 μm, except for that in panel C, which is 5 μm. (A and D) Confocal laser scanning microscopy (CLSM) images of a laboratory scale EBPR sludge (A) and Loganholme wastewater treatment plant (WTP) sludge (D) dual hybridized with FITC-labeled EUB338 and CY3-labeled TM7305. TM7 morphotypes appear yellow-orange, and other bacteria appear green. (C) Bright-field micrograph of a Gram stain of the sheathed-filament morphotype in the laboratory scale sludge. Note the sheath and prominent cuff. (B and E) CLSM images of the laboratory scale EBPR sludge (B) and Noosa WTP sludge (E) triple hybridized with FITC-labeled TM7305, CY3-labeled TM7905, and CY5-labeled EUB338. TM7 morphotypes which bind both TM7-specific probes and EUB338 appear white, morphotypes which bind only TM7905 and EUB338 appear pink, and other non-TM7 bacteria appear blue. Selected TM7 morphotypes are highlighted with arrows. f, filament; c, cocci; r, rod; tf, thick filament.
FISH analysis of two full-scale wastewater treatment plant sludges, with probes TM7305 and TM7905, indicated the presence of a number of TM7 morphotypes (Fig. 2D and E). This is consistent with the greater complexity of full-scale sludge microbial communities relative to the laboratory scale sludge which had been selectively enriched for a phenotype phosphate removal. Two morphotypes a sheathed filament and a sausage-shaped rod occurring in short chains (Fig. 2E), hybridized both TM7-specific probes. Additional morphotypes, a thick filament and cocci (Fig. 2E), bound TM7905 only, consistent with the broader phylogenetic specificity of the probe compared to TM7305.
Two additional TM7-specific probes (TM7522 and TM7567) did not result in any specific hybridization signal, as is often the case for untested FISH probes (2), and were not pursued further. However, their sequences are included in Table 1 because they should prove useful in ex situ methods, such as slot blot hybridization. The optimal stringencies for the FISH probes were determined by varying the formamide concentration in the hybridization buffer as described previously (8). Since no pure cultures of TM7 have yet been obtained, optimized conditions were inferred when specific fluorescence decreased sharply above 30% formamide for TM7305 and 20% for TM7905. The nontarget cultivated organisms closest to the probes were used as negative controls to corroborate the optimized stringency values. Both Sphingomonas sp. strain BF14 (accession no. Z23157; one mismatch to TM7305) and M. luteus (accession no. M38242; two mismatches to TM7905) showed no specific hybridization signal to the probes at 0, 10, 20, or 30% formamide.
Bright-field microscopy and TEM of TM7 filament morphotype.
All three sludge samples were studied by bright-field microscopy using the Gram, methylene blue, and Sudan black stains. The last two stains are used routinely in the study of sludges which remove phosphorus to determine organisms which accumulate polyphosphate and PHB, respectively (8). The TM7 filament morphotype was easily discernible in bright-field preparations of the laboratory scale sludge sample due to its distinctive morphology and presence in relatively high numbers. It was nonmotile and nonbranching and consisted of gram-variable to -positive cells surrounded by a prominent sheath which stained pink with the safranin counterstain (Fig. 2C). Attached growth was generally absent (Fig. 2A and B). In the laboratory scale reactor, the filaments grew to several hundred micrometers in length and were intertwined in the flocs (Fig. 2A). The filament stained negatively for both polyanions and PHB (data not shown), suggesting it does not play a role in typical phosphorus removal processes (see Discussion). TM7 morphotypes observed by FISH (Fig. 2D and E) could not be readily distinguished in bright-field microscopy preparations due to their low numbers and the complexity of the full-scale sludge communities.
Fortuitously, the TM7 sheathed-filament morphotype was virtually the only filament type observed in the laboratory scale reactor sludge and was present in substantial numbers (Fig. 2A). Therefore, it was decided to prepare sections of the reactor sludge for TEM to investigate the ultrastructure of these conspicuous filaments. Approximately 200 filaments were observed by TEM, and the following parameters represent an average of these observations. Each filament comprised tens to hundreds of closely appressed cells, each completely surrounded along its length by a pronounced sheath (Fig. 3A). The diameter and length of individual cells varied between 0.3 to 0.5 and 0.6 to 1.7 μm, respectively. The cell envelope had an ultrastructure typical of gram-positive bacteria, consisting of a cytoplasmic membrane and an outer wall (Fig. 3B and C). The wall was commonly 20 nm thick but was as thin as 12 nm in places, and it had a trilaminar appearance (Fig. 3B, C, and D). The outer layer of the wall displayed an unusual series of fine ridges that ran longitudinally along the cell (Fig. 3D). The sheath varied between 25 and 50 nm in thickness and had a close association with the cell wall at points along its length (Fig. 3A and D). The sheath was often observed to end in a cuff which extended beyond the cells (Fig. 3E).
FIG. 3.
Transmission electron micrographs of TM7 sheathed-filament morphotype. (A and B) Longitudinal sections of filament showing pronounced sheath (S), attached to the filament at one point (A), and septum (SE). Bars = 0.5 μm (A) and 100 nm (B). (C) High-magnification image of a cross section of a cell showing typical gram-positive cell envelope, cytoplasmic membrane (CM), and outer wall (W). Note the trilaminar appearance of the wall. Bar = 50 nm. (D) Cross section of a filament showing a series of fine ridges (R) running longitudinally along the cells, the sheath (S), and close association of the sheath with the filament (A). Bar = 100 nm. (E) Filament terminus showing a sheath cuff extending beyond the cells. Bar = 0.5 μm.
DISCUSSION
TM7 sheathed-filament morphotype.
A conspicuous sheathed-filament morphotype was highlighted in a laboratory scale activated sludge using two independent FISH probes (TM7305 and TM7905) specific for candidate division TM7 (Fig. 2A and B). Nearly all filaments observed in the laboratory scale sludge were targeted by both probes, indicating that the vast majority of filaments in the sludge were members of the TM7 division. However, the filament morphotype may comprise more than one species or even genus of the TM7 division, since the probes target a broad phylogenetic group (Fig. 1) and morphology alone usually is a poor indicator of phylogenetic conformity (28). TEM of the same laboratory scale sludge sample agreed with the epifluorescence microscopy in that a sheathed filament was the dominant morphotype observed. Over 200 filaments were examined, and all but 2 had a distinctive sheathed morphotype with a typical gram-positive cell envelope (Fig. 3). The possibility of the dominant filament type seen by TEM not being a member of the TM7 division is extremely remote, since almost all filaments observed by epifluorescence microscopy in the laboratory scale sludge bound both TM7-specific FISH probes (Fig. 2A and B). Therefore, we conclude that candidate division TM7 is only the third major lineage of the domain Bacteria to possess members with bona fide gram-positive cell envelopes (together with the Actinobacteria and low-G+C gram-positive divisions). Deinococcus species, of the Thermus-Deinococcus lineage, are known to stain gram positive, but this is due to a thick peptidoglycan layer in an otherwise gram-negative cell envelope type (http://206.67.72.215:6336/contents/).
Although sheathed filaments are relatively uncommon among isolated and characterized bacteria, particularly gram-positive organisms (15), they are observed frequently in activated sludges and have been classified by morphology according to the system of Eikelboom (12). The TM7 filament most closely resembled Eikelboom type 0041, a gram-positive sheathed filament frequently displaying attached growth (7, 25). Type 0041 has been implicated in bulking problems and is commonly observed in foams of activated sludges (25). A notable increase in TM7 filament numbers was recorded in the Noosa sludge during a foaming episode (unpublished observation), suggesting their involvement in this problem. However, type 0041 cannot be reliably distinguished from type 0675, a morphologically similar gram-positive sheathed filament (25), highlighting the limitations of a morphology-based microbial classification system. Therefore, it is quite likely that type 0041-0675 represents more than one organism, of which the TM7 filament is one example.
The TM7 filaments did not accumulate polyphosphate or PHB in the laboratory scale sequencing batch reactor operated under EBPR conditions (8). This suggests that the filament is not involved in standard physiological transformations thought to occur in EBPR sludge.
Candidate division TM7.
Sequence data from published and unpublished culture-independent PCR clonal studies suggest that members of candidate division TM7 are widely distributed in the environment. TM7 sequences have been detected in a range of chemically and geographically diverse habitats, including terrestrial (soils, rhizosphere, and peat bog), aquatic (groundwater, freshwater, seawater, and deep-sea sediments), and clinical (human oral cavity and mouse feces) locales (Fig. 1). No representatives have yet been identified in thermophilic environments, although this may be a sampling artifact. PCR clone library and FISH analyses indicate that the division is well represented in activated sludges (Fig. 1 and 2) and suggest TM7 bacteria may be common components of these systems. Several different morphotypes were observed in full-scale activated sludges using TM7 division-specific probes, including various filaments, rods, and cocci (Fig. 2D and E). This is not surprising, since the division probably comprises hundreds to thousands of species, which might be expected to have a variety of morphotypes. Design and application of a FISH probe targeting a subdivision of the Acidobacteria lineage revealed a similar diversity of morphotypes in environmental samples (19).
Candidate division TM7 has a relatively modest intradivision 16S rDNA sequence divergence of 17% compared to a range of 13 to 33% for a broad sampling of bacterial divisions (10). This is approximately the same degree of divergence recognized in the division Actinobacteria (10), which comprises at least 35 formally described families (26). However, we feel it is too early to predict what (if anything) 16S rDNA sequence divergence may indicate in terms of ecological and physiological diversity at the division level.
The finding of a typical gram-positive cell envelope in the TM7 sheathed-filament morphotype raises the question of the occurence of this fundamental structure in the TM7 division. The gram-positive cell envelope is a largely unifying taxonomic character of the two well-known gram-positive bacterial lineages, the Actinobacteria and low-G+C gram-positive divisions (28). Therefore, it is quite possible that the same feature is widespread throughout the TM7 division. It also will be of interest to determine if components characteristic of gram-positive cell envelopes, such as teichoic acids (20), are present in TM7 bacteria.
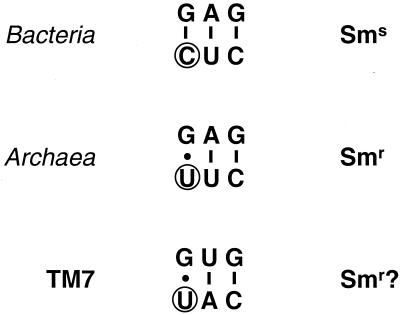
The basis for the specificity of one TM7-specific probe (TM7905) is positions 911 and 912 (E. coli numbering). All TM7 sequences, with the exceptions of clones K20-12, GC1, and SBR2060 (Fig. 1), have adenosine (A) and uracil (U) residues at positions 911 and 912, respectively, which are highly atypical for members of the domain Bacteria (Fig. 4). All other bacteria sequenced to date have a U at 911 and cytosine (C) at 912 (as do K20-12 and SBR2060). Archaea also have a U at 911 but have a U in common with the TM7 division at 912 (Fig. 4). It is the latter residue which is primarily responsible for resistance to the aminoglycoside antibiotic streptomycin in Archaea and streptomycin sensitivity in Bacteria (3). Therefore, TM7 bacteria may be resistant to streptomycin at the ribosome level. This hypothesis awaits experimental verification. Unfortunately, streptomycin resistance was not useful for selective enrichment of TM7 bacteria, since plasmid-borne streptomycin resistance is widespread in the environment and allows rapid overgrowth of enrichment cultures by fast-growing organisms, such as representatives of the division Proteobacteria (unpublished observations).
FIG. 4.
Consensus sequences of part of 16S rRNA stem 30 (according to the ARB numbering system) in Bacteria (except TM7), Archaea, and TM7. The residue at position 912 (circled) is primarily responsible for streptomycin resistance (Smr) and sensitivity (Sms) in different domains (3). Canonical and noncanonical base pairing between residues are indicated by lines and dots, respectively.
In conclusion, TM7 is a phylogenetically independent division level lineage in the domain Bacteria that is widespread in the environment and comprises representatives with noteworthy characteristics. The probes and primers described in this paper should provide useful tools for further study of this novel group.
ACKNOWLEDGMENTS
We thank Brigitte Pertschy and Rebecca Smith for preparing and screening TM7-specific clone libraries, Paul Burrell for fully sequencing a number of sludge clones, and Greg Crocetti for operating the sequencing batch reactors. We also thank Gavin Symonds and Erin Collins for assistance with confocal laser scanning microscopy and Ed DeLong for pointing out the molecular basis of streptomycin resistance.
P.H. and G.W.T. are funded by the Cooperative Research Centre for Waste Management and Pollution Control Ltd., a center established and supported under the Australian Government's Cooperative Research Centres Program.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amils R, Cammarano P, Londei P. Translation in archaea. In: Kates M, Kushner D J, Matheson A T, editors. The biochemistry of archaea (archaebacteria) Vol. 26. Amsterdam, The Netherlands: Elsevier; 1993. pp. 393–438. [Google Scholar]
- 4.Bond P L, Erhart R, Wagner M, Keller J, Blackall L L. Identification of some of the major groups of bacteria in efficient and nonefficient biological phosphorus removal activated sludge systems. Appl Environ Microbiol. 1999;65:4077–4084. doi: 10.1128/aem.65.9.4077-4084.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brand P A J, Tiedt L R, Hamilton-Attwell V L. Some observations on the morphology and the anatomy of filament type 0041. Water S Afr. 1987;13:1–6. [Google Scholar]
- 8.Crocetti G R, Hugenholtz P, Bond P L, Schuler A, Keller J, Jenkins D, Blackall L L. Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl Environ Microbiol. 2000;66:1175–1182. doi: 10.1128/aem.66.3.1175-1182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalevi, D., P. Hugenholtz, and L. L. Blackall. A multiple-outgroup approach to resolving division-level phylogenetic relationships using 16S rDNA data. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 10.Dojka M A, Harris J K, Pace N R. Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria. Appl Environ Microbiol. 2000;66:1617–1621. doi: 10.1128/aem.66.4.1617-1621.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dojka M A, Hugenholtz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eikelboom D H. Filamentous organisms observed in activated sludge. Water Res. 1975;9:365–388. [Google Scholar]
- 13.Fuchs B M, Wallner G, Beisker W, Schwippl I, Ludwig W, Amann R. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 1998;64:4973–4982. doi: 10.1128/aem.64.12.4973-4982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galvez A, Maqueda M, Martinez-Bueno M, Valdivia E. Publication rates reveal trends in microbiological research. ASM News. 1998;64:269–275. [Google Scholar]
- 15.Holt J G, Hirsch P, Mulder E G. Sheathed bacteria. In: Staley J T, Bryant M P, Pfennig N, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 3. Baltimore, Md: Williams & Wilkins; 1989. pp. 1994–2009. [Google Scholar]
- 16.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- 19.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K-H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 20.Madigan M T, Martinko J M, Parker J. Brock biology of microorganisms. 9th ed. London, United Kingdom: Prentice-Hall, Inc.; 2000. [Google Scholar]
- 21.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 22.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 23.Reasoner D S, Geldreich E E. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rheims H, Rainey F A, Stackebrandt E. A molecular approach to search for diversity among bacteria in the environment. J Ind Microbiol. 1996;17:159–169. [Google Scholar]
- 25.Seviour E M, Seviour R J, Lindrea K C. Descriptions of the filamentous bacteria causing bulking and foaming in activated sludge plants. In: Seviour R J, Blackall L L, editors. The microbiology of activated sludge. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 301–348. [Google Scholar]
- 26.Stackebrandt E, Rainey F A, Ward-Rainey N L. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–491. [Google Scholar]
- 27.von Wintzingerode F, Landt O, Ehrlich A, Göbel U B. Peptide nucleic acid-mediated PCR clamping as a useful supplement in the determination of microbial diversity. Appl Environ Microbiol. 2000;66:549–557. doi: 10.1128/aem.66.2.549-557.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]