Abstract
The viability of the human probiotic strains Lactobacillus paracasei NFBC 338 and Bifidobacterium sp. strain UCC 35612 in reconstituted skim milk was assessed by confocal scanning laser microscopy using the LIVE/DEAD BacLight viability stain. The technique was rapid (<30 min) and clearly differentiated live from heat-killed bacteria. The microscopic enumeration of various proportions of viable to heat-killed bacteria was then compared with conventional plating on nutrient agar. Direct microscopic enumeration of bacteria indicated that plate counting led to an underestimation of bacterial numbers, which was most likely related to clumping. Similarly, LIVE/DEAD BacLight staining yielded bacterial counts that were higher than cell numbers obtained by plate counting (CFU) in milk and fermented milk. These results indicate the value of the microscopic approach for rapid viability testing of such probiotic products. In contrast, the numbers obtained by direct microscopic counting for Cheddar cheese and spray-dried probiotic milk powder were lower than those obtained by plate counting. These results highlight the limitations of LIVE/DEAD BacLight staining and the need to optimize the technique for different strain-product combinations. The minimum detection limit for in situ viability staining in conjunction with confocal scanning laser microscopy enumeration was ∼108 bacteria/ml (equivalent to ∼107 CFU/ml), based on Bifidobacterium sp. strain UCC 35612 counts in maximum-recovery diluent.
Probiotics are described as “living micro-organisms, which upon ingestion in certain numbers exert health benefits beyond inherent basic nutrition” (12). Accumulating clinical evidence supporting the health-promoting characteristics of Lactobacillus and Bifidobacterium intestinal isolates (for reviews see references 24 and 29) has led to increased commercial interest in developing novel probiotic food products. Those products which have received the most attention as probiotic carriers include fermented milks, unfermented milks with cultures added, ice cream, frozen yogurt, and various cheeses (for reviews see references 18, 31, 32, and 34). It has been suggested that probiotic products should contain at least 107 CFU per ml or g (15).
Bacterial viability is typically assessed by plate counting on a suitable growth medium. However, there are a number of disadvantages associated with this approach. For example, plate counting is time-consuming, often requiring 2 to 3 days of incubation, microorganisms may be unevenly distributed in the product, and bacteria may occur in chains and/or clumps, resulting in underestimation of the true bacterial count (6). In addition, oxidative killing of anaerobic microorganisms such as Bifidobacterium during plating may also contribute to an underestimation of bacterial numbers. A more direct approach may be the use of a microscopic technique; however, this requires differentiation of live and dead bacteria. Direct epifluorescent counting has been described as a suitable method for enumeration of total bacteria in environmental samples” (17). Fluorescence microscopy has the advantage of allowing a rapid and direct assessment of cell viability (17, 22), although particular strains cannot be identified. Fluorescent indicators of viability may be based on membrane integrity, enzyme activity, membrane potential, respiration, or pH gradient (9, 20, 21, 23, 27, 33). The LIVE/DEAD BacLight viability kit (Molecular Probes Inc., Eugene, Oreg.) was developed to differentiate live and dead bacteria based on plasma membrane permeability and has been used to monitor growth of bacterial populations (38). This kit comprises two fluorescent nucleic acid stains: SYTO9 and propidium iodide. SYTO9 (excitation and emission maxima, 480 and 500 nm) penetrates both viable and nonviable bacteria (Handbook of Fluorescent Probes and Research Chemicals, 6th ed., Molecular Probes, Inc.), while propidium iodide (excitation and emission maxima, 490 and 635 nm) penetrates bacteria with damaged plasma membranes only (16, 21), quenching the green SYTO9 fluorescence. Thus, bacterial cells with compromised membranes fluoresce red and those with intact membranes fluoresce green.
Confocal scanning laser microscopy (CSLM) has been used extensively in cell biology (39) and was used to study viability of Escherichia coli and Salmonella where rhodamine 123 and propidium iodide were employed to differentiate viable from nonviable bacteria based on membrane potential and integrity (19). Conventional epifluorescence microscopy may be used for viability staining of liquid samples such as milk (26). However, the optical sectioning capability of CSLM has the advantages of increased sensitivity and reduced out-of-focus blur, enabling observation of subsurface structures of foods in situ (3, 13). In addition, digital acquisition of images by CSLM enables rapid enumeration of bacteria by image analysis (4).
In this study, in situ LIVE/DEAD BacLight bacterial viability staining in conjunction with CSLM was compared with standard plate counting for enumeration and viability assessment of bacteria in various probiotic dairy products, including reconstituted skim milk (RSM), fermented milk, full-fat cheddar cheese, and spray-dried probiotic milk powder.
MATERIALS AND METHODS
Bacterial strains and media.
The potentially probiotic strains Lactobacillus paracasei subsp. paracasei NFBC 338, Bifidobacterium sp. strain UCC 35612, and Bifidobacterium sp. strain UCC 401 were isolated from the human gastrointestinal tract (5, 25) and were obtained from University College Cork, Cork, Ireland, under a restricted-materials transfer agreement. The Lactobacillus strain was cultured as described previously (10), while the Bifidobacterium strain was cultured in MRS broth (Difco Laboratories, Detroit, Mich.) supplemented with 0.05% (wt/vol) cysteine HCl (Sigma-Aldrich Ireland, Dublin, Ireland) (7). For cheddar cheese manufacture, cheesemaking starters Lactococcus lactis subsp. cremoris strains 223 and 227 and an adjunct culture of Bifidobacterium lactis Bb-12 were obtained from C. Hansen Laboratories (Little Island, Cork, Ireland) in the form of freeze-dried pellets. L. paracasei NFBC 338 was enumerated in milk and dairy products by pour plating on MRS agar (Difco Laboratories). Tryptone-phytone-yeast extract agar containing NPNL selective solution (neomycin sulfate [20 mg/liter], paromomycin sulfate [40 mg/liter], nalidixic acid [3 mg/liter], lithium chloride [600 mg/liter]) (30, 37) was used for selective enumeration of bifidobacteria from fermented milk and cheddar cheese, while MRS agar supplemented with 0.05% (wt/vol) cysteine HCl was used for enumeration from RSM suspensions. All dilutions were performed using maximum-recovery diluent (MRD) (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) and plates were incubated under anaerobic conditions at 37°C for 3 days for both lactobacilli and bifidobacteria.
Plate count enumeration of probiotic bacteria in milk and dairy products.
Cells from 100 ml of stationary-phase cultures of L. paracasei NFBC 338 or Bifidobacterium sp. strain UCC 35612 were concentrated by centrifugation at 3,640 × g for 10 and 20 min, respectively. The resultant cells were then resuspended by vortex mixing for 10 s in 100 ml of RSM (10% wt/vol) that had previously been sterilized at 121°C for 5 min. A 10-ml sample of the RSM containing resuspended cells was then incubated at 90°C for 5 min. Unheated and heat-treated RSM cell suspensions were vortex mixed in various proportions to give mixtures of live and dead bacteria in which the proportion of live bacteria varied in 10% increments from 0 to 100% (vol/vol). Bacteria in mixtures containing 0, 10, 50, 90, and 100% (vol/vol) live bacteria were enumerated as outlined above.
Pasteurized whole milk supplemented with skim milk powder (16.5% total solids) was heat treated at 90°C for 15 min. After cooling to 37°C, the milk was inoculated (2% vol/vol) with an overnight broth culture of Bifidobacterium sp. strain UCC 401 and incubated at 37°C for 24 h until a pH of 4.8 was reached. Duplicate samples of fermented milk were emulsified in sterile 2% (wt/vol) trisodium citrate, and serial dilutions in MRD were pour plated as described above.
A pilot-scale cheesemaking trial was performed according to the experimental protocol described by Gardiner et al. (10). A control vat contained a 1.5% (vol/vol) inoculum of starter cultures only and the experimental vat contained an additional culture of Bifidobacterium sp. strain Bb12, added as an adjunct to the starter culture to yield ∼108 CFU bifidobacteria per ml of cheesemilk. Cheeses were sampled at 1 and 3 months and bifidobacteria were enumerated by plate counting as described for probiotic fermented milk above.
To manufacture a probiotic-containing spray-dried powder, cells from an overnight MRS broth culture of L. paracasei NFBC 338 (200 ml) were resuspended in 1,500 ml of RSM (25% wt/vol), which had been previously heat treated at 90°C for 30 min. The suspension was spray dried in a Buchi B191 mini-spray dryer (Buchi Labortechnik AG, Flawil, Switzerland) as previously described (11). The inlet air temperature was set at 160°C and outlet air temperatures ranging from 71 to 78°C were used, yielding skim milk powders containing viable lactobacilli at ∼1010 CFU/g. Lactobacilli were enumerated in duplicate following 2 months of storage at 4°C.
In situ viability staining and CSLM imaging.
All microscopy work was performed using an LSM310 confocal scanning laser microscope (Carl Zeiss Ltd., Welwyn Garden City, Herts., United Kingdom) using the method involving LIVE/DEAD BacLight viability staining essentially as previously described (11). Randomly selected areas of each sample were imaged using a ×63 magnification objective with a numerical aperture of 1.4. Confocal illumination was provided by a Kr/Ar laser (488-nm laser excitation) fitted with a long-pass 514-nm emission filter. A 580-nm beam splitter was used together with a long-pass 520-nm filter (green fluorescence signal) and long-pass 590-nm filter (red fluorescence signal). Simultaneous dual-channel imaging using pseudocolor was used to display green and red fluorescence. The confocal pinhole was set to give an x-y resolution of 0.2 μm and an axial resolution of 1.0 μm. Red-green-blue images (24 bit), 512 by 512 pixels, were acquired using a zoom factor of 2.0, giving a final pixel resolution of 0.2 μm/pixel and representing a volume of 1.05 × 10−8 ml per field of view. Thus, for direct enumeration of bacteria per milliliter, a microscopic factor of 1.05 × 108 was used. For triple-channel imaging, a transmitted photodetector was used in conjunction with interference contrast optics and the transmitted image was colored blue. Image analysis was performed on CSLM images using a Kontron KS400 image analysis system (Imaging Associates Ltd., Thame, Oxfordshire, United Kingdom). Images of stained bacteria were segmented using color thresholding to separate the red and green fluorescence signals. Two parameters were then measured: (i) green fluorescence as a percentage of total green and red fluorescence and (ii) numbers of individual green fluorescing bacteria. To separate clusters of bacteria, erosion-dilation algorithms included in the image analysis software were used. Direct microscopic counts were normalized to take into account the dilution effect caused by adding the viability stain to the sample. To simultaneously visualize the structure of the spray-dried particles and the red- and green-fluorescing bacteria, triple-channel imaging was used. To confirm that the glycerol-based staining mixture did not affect viability staining, live and heat-killed L. paracasei NFBC 338 microorganisms in RSM were prepared as described above. When mixed with the glycerol-based stain at a ratio of 1:1, live cells fluoresced green and heat-killed cells fluoresced red.
(i) Minimum detection limit of the in situ viability staining and CSLM enumeration method using bifidobacteria suspended in MRD.
To establish the sensitivity of the in situ viability staining technique, an actively growing broth culture of Bifidobacterium sp. strain UCC 35612 was diluted in MRD to yield an approximate log dilution series of 105 to 109 CFU/ml. Plate count enumeration of the broth culture and in situ staining with CSLM enumeration of the dilution series were performed as described for RSM. Bacteria from 50 microscopic fields were counted using image analysis, and results were expressed as numbers of bacteria per milliliter.
(ii) In situ viability staining and CSLM enumeration of lactobacilli and bifidobacteria in RSM.
The LIVE/DEAD BacLight viability stain, prepared according to the manufacturer's instructions, was incubated with equal volumes of milk containing 0 to 100% live bacteria, prior to CSLM imaging. The specificity of the two individual LIVE/DEAD BacLight staining components was verified in the milk by adding 5 μl of each staining component to separate 100-μl samples of milk inoculated with either a live culture or a heat-treated culture of L. paracasei NFBC 338. CSLM imaging confirmed that SYTO9 stained both live and dead bacteria green, whereas propidium iodide stained only heat-killed bacteria red (data not shown). Results from in situ viability staining were obtained within 30 min of sampling the milk.
(iii) In situ viability staining and CSLM enumeration of bacteria in fermented milk, cheddar cheese, and spray-dried probiotic milk powder.
Equal volumes of probiotic fermented milk (pH 5.6) and LIVE/DEAD BacLight viability stain were vortex mixed for 1 min and green fluorescent bacteria from 20 fields were enumerated. LIVE/DEAD BacLight viability stain (25 μl) was also added to freshly cut sections of 2-month-old cheddar cheese, and a coverslip was placed on top. CSLM images were obtained ∼10 μm below the level of the coverslip after 20 min of incubation in the dark at room temperature. CSLM imaging data from dry powders were compared with data from the reconstituted product (10% [wt/vol]). To prevent dissolution of the spray-dried powder particles during in situ viability staining, a glycerol-based staining mixture was prepared from the LIVE/DEAD BacLight staining components, as follows. SYTO9 and propidium iodide were each dissolved in separate 1-ml samples of distilled water to give final concentrations of 60 and 300 mM, respectively. Seventy-five microliters of SYTO9 solution and 25 μl of propidium iodide solution were then added, with vortex mixing, to 400 μl of glycerol (Sigma-Aldrich Ireland). This ratio of SYTO9 to propidium iodide was found to be optimal for the production of an adequate green fluorescence signal. Spray-dried probiotic milk powder (∼10 μg) was gently mixed with 10 μl of this staining mixture on a microscope slide.
Statistical analysis.
The significance of the difference between the means obtained by direct microscopic counting and plate count enumeration was determined by a one-tailed Student t test (20 df).
RESULTS AND DISCUSSION
Minimum detection limit of the in situ viability staining and CSLM enumeration method using Bifidobacterium sp. strain UCC 35612.
In order to relate in situ viability staining and CSLM enumeration to plate count data, it was first necessary to establish the minimum detection limit of the in situ CSLM technique. Results of CSLM enumeration of LIVE/DEAD BacLight-stained bifidobacteria in MRD indicated a minimum detection limit of ∼108 bacteria/ml or ∼107 CFU/ml from plating (Table 1). These data suggest that plate counting underestimates the actual viable cell population by a factor of at least 10, confirming reports by other researchers (6, 17, 28). Results further indicate that LIVE/DEAD BacLight viability staining may be a suitable means of assessing in situ the viability of bacteria in probiotic foods, given that the recommended minimum number of probiotic bacteria in such food products is approximately 107 CFU/ml (15). It should be noted, however, that the sensitivity of the viability staining method could be greatly increased by filtration and/or centrifugation to concentrate the recovered cells (17, 26).
TABLE 1.
Direct microscopic counts by in situ viability staining and CSLM enumeration and plate counts of serially diluted Bifidobacterium sp. strain UCC 35612 in maximum-recovery diluent
| Plate count (CFU/ml)a | Direct microscopic count of green (viable) fluorescent bacteria
|
|
|---|---|---|
| Mean of 50 fields | No. of bacteria/ml, normalized | |
| 1.25 × 105 | 0 | 0 |
| 1.25 × 106 | 0.1 | 2.1 × 107 |
| 1.25 × 107 | 2.3 | 4.8 × 108 |
| 1.25 × 108 | 14.6 | 3.1 × 109 |
| 1.25 × 109b | 172.8 | 3.6 × 1010 |
Estimated from serial dilutions of broth culture.
Mean of triplicate analyses.
In situ viability staining and CSLM enumeration of probiotic bacteria in RSM.
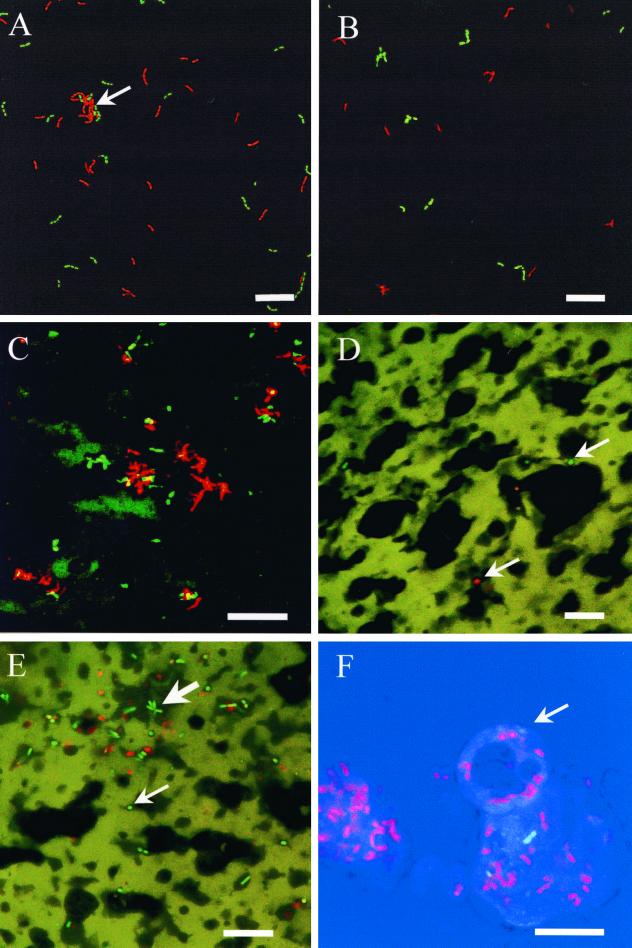
The specificity of the LIVE/DEAD BacLight stain was then assessed using known ratios of live to dead (heat-killed) bacteria in RSM. Both L. paracasei NFBC 338 and Bifidobacterium sp. strain UCC 35612 produced a strong red or green fluorescence depending on whether the cultures were dead or live, respectively (Fig. 1A and B). Background fluorescence from milk proteins was low, enabling clear discrimination of viable and nonviable bacterial cells. CSLM observations indicated that cells of L. paracasei NFBC 338 were often in short chains of four cells in addition to larger clumps of up to 200 microorganisms. In contrast, Bifidobacterium sp. strain UCC 35612 appeared as individual cells or small clumps of <4 cells. A good correlation was obtained between percentages of live bacteria and green cells (green fluorescence expressed as a percentage of red and green fluorescence), as measured by analysis of CSLM images for L. paracasei NFBC 338 (R2 = 0. 99) and for Bifidobacterium sp. strain UCC 35612 (R2 = 0.98). This indicates that in situ viability staining and CSLM imaging constitute a valid quantitative technique for estimating the proportion of viable to dead bacterial cells in milk.
FIG. 1.
CSLM images of dairy products stained with LIVE/DEAD BacLight viability stain. (A and B) L. paracasei NFBC 338 and Bifidobacterium sp. strain UCC 35612, respectively, suspended in reconstituted skim milk. Dual-channel CSLM images represent a 50:50 mixture of live and heat-treated bacteria. Live bacteria are green; dead bacteria are red. Note the clumping of lactobacilli in panel A (arrow). Bar = 10 μm. (C) Dual-channel CSLM projection (z depth, 15 μm) of probiotic milk fermented with Bifidobacterium sp. strain UCC 401. Note extensive clumping of bacteria. Bar = 25 μm. (D and E) Dual-channel CSLM images of control (D) and probiotic (E) cheddar cheese. Note the star-shaped cluster of rod-shaped cells characteristic of some Bifidobacterium strains (E, large arrow), short rods and cocci of presumptive nonstarter lactic acid bacteria (small arrows), and background green fluorescence of protein matrix with dark spaces containing fat. Bar = 25 μm. (F) Probiotic skim milk powder stained in situ with glycerol-based LIVE/DEAD BacLight viability stain. Triple-channel CSLM image shows live and dead lactobacilli (green and red, respectively) and transmitted image (blue). The arrow indicates a powder particle. Bar = 5 μm.
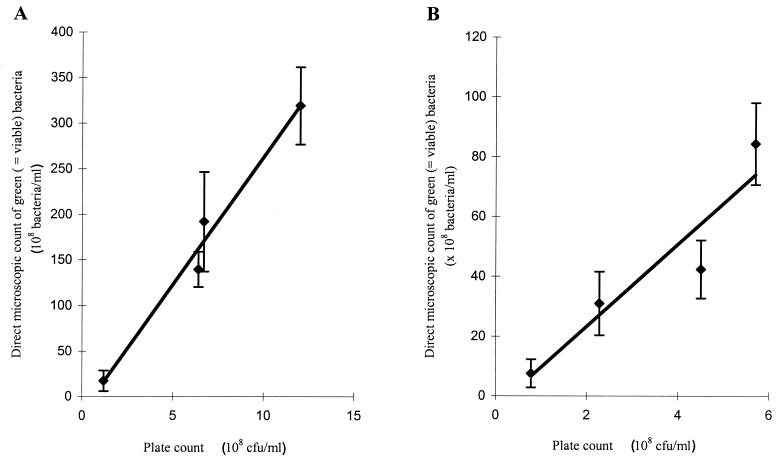
Data obtained by direct CSLM enumeration of bacteria were then compared with those obtained by plate counting for L. paracasei NFBC 338 and Bifidobacterium sp. strain UCC 35612 in RSM. The correlations of 0.98 and 0.89 for various ratios of live to dead L. paracasei NFBC 338 and Bifidobacterium sp. strain UCC 35612, respectively, confirm the quantitative capability of in situ viability staining and CSLM enumeration (Fig. 2). Relative to the direct microscopic counts, however, L. paracasei NFBC 338 and Bifidobacterium sp. strain UCC 35612 plate counts were approximately 20-fold and 10-fold lower, respectively. These results are consistent with the greater degree of clumping exhibited by L. paracasei NFBC 338 (Fig. 1) compared with Bifidobacterium sp. strain UCC 35612.
FIG. 2.
Correlation between direct microscopic counts (mean of six replicates) of green-fluorescing bacteria stained with LIVE/DEAD BacLight viability stain and plate counts (mean of duplicates) of L. paracasei NFBC 338 (R2 = 0.98) (A) and Bifidobacterium sp. strain UCC 35612 (R2 = 0.89) (B) in reconstituted skim milk. The error bars represent 95% confidence intervals.
In situ viability staining and CSLM enumeration of bifidobacteria in probiotic fermented milk.
In situ LIVE/DEAD BacLight staining showed red- and green-fluorescing bacteria occurring singly or in small clumps of up to 20 cells in the probiotic fermented milk (Fig. 1C). Some background fluorescence was present in the green channel. Bacterial cells were irregularly shaped rods with occasional branching, a morphologic characteristic of some Bifidobacterium sp. (30). Enumeration by CSLM indicated a viable count equivalent to 3.2 × 108 bacteria/ml, comparing favorably to the plate count of 2.3 × 108 CFU/ml (Table 2). The higher count (P < 0.001) obtained by direct microscopic enumeration was most likely due to clumping of bacteria and killing on media selective for bifidobacteria.
TABLE 2.
Comparison of LIVE/DEAD BacLight viability staining and direct CSLM microscopic enumeration with plate counting for probiotic bacteria in dairy products
| Product | Sample | Microscopic count (viable bacteria/ml or g)a | Plate count (CFU/ml or g)b | P |
|---|---|---|---|---|
| Fermented milk | Bifidobacterium sp. strain UCC 401 | 3.2 × 108 (1.6 × 107) | 2.3 × 108 (1.1 × 108) | <0.001 |
| Cheddar cheese | Control | 7.0 × 107 (1.0 × 107) | 3.3 × 105 (4.2 × 105) | <0.001 |
| Bifidobacterium sp. strain Bb12 | 1.5 × 108 (5.6 × 107) | 3.6 × 108 (2.8 × 107) | <0.001 | |
| Spray-dried powder | L. paracasei NFBC 338 | 6.3 × 108 (3.6 × 108) | 1.1 × 109 (6.4 × 107) | <0.05 |
Mean of 20 fields. Values in parentheses are standard deviations.
Mean of duplicate analyses. Values in parentheses are standard deviations.
In situ viability staining and CSLM enumeration in probiotic cheddar cheese.
The in situ viability staining and CSLM imaging technique was used to enumerate total viable bacteria directly from cheddar cheese ripened for 2 months and compared with plate counts for enumeration of viable bifidobacteria (Table 2). Enumeration by CSLM indicated a total viable count equivalent to 1.5 × 108 bacteria/g, which was lower than the bifidobacterial plate count of 3.6 × 108 CFU/g. The probiotic cheddar cheese contained approximately twice as many viable bacteria as the control cheese, as determined by the in situ CSLM method. If higher bacterial counts in the probiotic cheese were exclusively due to bifidobacteria, in situ viability staining indicated a count of approximately 7.5 × 107 bifidobacteria/g. Positive identification of bifidobacteria in situ would require an alternative approach such as fluorescent in situ hybridization or immunofluorescent labeling (14). Therefore, it was not possible to distinguish bifidobacteria in the cheese from nonstarter lactic acid bacteria; rather, it is the comparison with total numbers in the control cheese that is given. However, the probiotic cheddar cheese contained several star-shaped clusters of rod-shaped bacteria (Fig. 1E) typical of some Bifidobacterium strains (30), including Bifidobacterium lactis Bb-12. These clusters were not present in the control Cheddar cheese (Fig. 1D). Cell morphology was confirmed by adjusting the focal plane of the CSLM. Bacteria were not homogeneously distributed but frequently occurred in clumps of up to 20 cells. Some background fluorescence of the protein matrix was seen in the green channel, although this was at a lower intensity than bacterial fluorescence. Fat globules appeared as dark rounded regions by negative contrast as observed in a previous study (8). The homogeneous staining of the protein matrix with SYTO9 was most likely due to nonspecific binding of the stain to milk proteins. Small (<2-μm) patches of diffuse red fluorescence, possibly due to exogenous microbial nucleic acids, were also observed in both probiotic and control cheddar cheese samples (data not shown).
In situ viability staining and CSLM enumeration of spray-dried L. paracasei NFBC 338 in skim milk powder.
The counts of live bacteria in spray-dried form, as determined by image analysis of CSLM images and plate counts, are shown in Table 2. CSLM enumeration indicated a viable count of 6.3 × 108 bacteria/g in the rehydrated powder, which was significantly lower (P < 0.05) than that obtained by plate counting (1.1 × 109 CFU/g). Triple channel imaging using the glycerol-based mixture of propidium iodide and SYTO9 enabled in situ observation of both red- and green-fluorescing L. paracasei NFBC 338 cells within the powder particles (Fig. 1F). A low level of background fluorescence from the milk powder was observed in the green channel. Serial CSLM optical sections indicated that bacteria were encapsulated within the spray-dried powder particles, confirming earlier work (11). Higher numbers of bacteria fluoresced green in the rehydrated than in the dry powder. The low number of green-fluorescing bacteria (<1 bacterium/field) in the dry powder compared with that in the rehydrated powder suggested that the bacterial plasma membrane was compromised in the dehydrated state, as expected (1, 35), but recovered somewhat when rehydrated. Spray drying has been shown to result in cell membrane damage, as indicated by the increased sensitivity of L. paracasei NFBC 338 to NaCl following drying (11). It is possible that reversible melting of membrane lipids at temperatures of ∼50°C (36) and/or removal of bound water from cell wall proteins during the drying process (1) may be responsible. It has been reported that slow rehydration procedures can increase the viability of spray-dried L. bulgaricus (35). For more detailed study of the effect of sublethal stress on bacterial viability, other fluorescent viability indicators, such as esterase activity, membrane potential, or respiratory activity, may be more suitable than techniques based on membrane permeability (2, 19).
Conclusions.
The results of this study indicate that in situ LIVE/DEAD BacLight viability staining and CSLM enumeration may be of value for the rapid estimation of viable bacteria in some dairy products, which could take over 3 days to achieve by plate counting. The data demonstrate that microscopic viability counting of probiotic milk and fermented milk yield consistently higher counts (up to 20-fold for milk) than plate counting. This may be expected given the high degree of clumping observed with some of the strains and the possible killing of cells by selective media. Microscopic counts were lower than plate counts for cheese products and spray-dried cultures, highlighting the need for further work to establish the effect of environmental factors such as pH, ionic profile, and water activity on viability staining.
ACKNOWLEDGMENTS
This work was supported by the European Research and Development Fund and by the European Union (SM&T-CT98–2235). G.E.G. and S.J.M. were supported by Teagasc Walsh Fellowships.
REFERENCES
- 1.Brennan M, Wanismail B, Johnson M C, Ray B. Cellular damage in dried Lactobacillus acidophilus. J Food Prot. 1986;49:47–53. doi: 10.4315/0362-028X-49.1.47. [DOI] [PubMed] [Google Scholar]
- 2.Breuwer P. Assessment of viability of micro-organisms employing fluorescence techniques. Ph.D. thesis. Wageningen, The Netherlands: Aula vab de Landbouwuniversiteit te Wageningen; 1996. [Google Scholar]
- 3.Brooker B E. Imaging food systems by confocal scanning laser microscopy. In: Dickenson E, editor. New physico-chemical techniques for the characterisation of complex food systems. London, United Kingdom: Blackie Academic and Professional; 1995. pp. 53–68. [Google Scholar]
- 4.Caldwell D E, Korber D R, Lawrence J R. Confocal laser microscopy and digital image analysis in microbial ecology. Adv Microb Ecol. 1992;12:1–67. [Google Scholar]
- 5.Collins J K, Thornton G, O'Sullivan G. Selection of probiotic strains for human applications. Int Dairy J. 1998;8:487–490. [Google Scholar]
- 6.Daley R J. Direct epifluorescence enumeration of native aquatic bacteria: uses, limitations and comparative accuracy. In: Costerton J W, editor. Native aquatic bacteria: enumeration, activity and ecology. ASTM STP 605. Philadelphia, Pa: American Society for Testing and Materials; 1979. pp. 29–45. [Google Scholar]
- 7.de Man J C, Rogosa M, Sharpe M E. A medium for the cultivation of lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 8.Everett D W, Ding K, Olson N F, Gunasekaran S. Applications of confocal microscopy to fat globule structure in cheese. In: Malin E L, Tunick M H, editors. Chemistry of structure-function relationships in cheese. New York, N.Y: Plenum Press; 1995. [DOI] [PubMed] [Google Scholar]
- 9.Gant V A, Warnes G, Phillips I, Savidge G F. The application of flow cytometry to the study of bacterial responses to antibiotics. J Med Microbiol. 1993;39:147–154. doi: 10.1099/00222615-39-2-147. [DOI] [PubMed] [Google Scholar]
- 10.Gardiner G E, Ross R P, Collins J K, Fitzgerald G F, Stanton C. Development of a probiotic cheddar cheese containing human-derived Lactobacillus paracasei strains. Appl Environ Microbiol. 1998;64:2192–2199. doi: 10.1128/aem.64.6.2192-2199.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardiner G E, O'Sullivan E, Kelly J, Auty M A E, Fitzgerald G F, Collins J K, Ross R P, Stanton C. Comparative survival of human-derived Lactobacillus paracasei and L. salivarius strains during heat treatment and spray drying. Appl Environ Microbiol. 2000;66:2605–2612. doi: 10.1128/aem.66.6.2605-2612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guarner F, Schaafsma G J. Probiotics. Int J Food Microbiol. 1998;39:237–238. doi: 10.1016/s0168-1605(97)00136-0. [DOI] [PubMed] [Google Scholar]
- 13.Heertje I, van der Vlist P, Blonk J C G, Hendrickx H A C, Brackenhof G J. Confocal scanning laser microscopy in food research: some observations. Food Microstruct. 1987;6:115–120. [Google Scholar]
- 14.Hugenholtz J, Veldkamp H, Konings W N. Detection of specific strains and variants of Streptococcus cremoris in mixed cultures by immunofluorescence. Appl Environ Microbiol. 1987;53:149–155. doi: 10.1128/aem.53.1.149-155.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishibashi N, Shimamura S. Bifidobacteria: research and development in Japan. Food Technol. 1993;47:126–135. [Google Scholar]
- 16.Jepras T I, Carter J, Pearson S C, Paul F E, Wilkinson M J. Development of a robust flow cytometry assay for determining numbers of viable bacteria. Appl Environ Microbiol. 1995;61:2696–2701. doi: 10.1128/aem.61.7.2696-2701.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kepner R J, Pratt J R. Use of fluorochromes for direct enumeration of total bacteria in environmental samples: past and present. Microbiol Rev. 1994;58:603–615. doi: 10.1128/mr.58.4.603-615.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y K, Salminen S. The coming of age of probiotics. Trends Food Sci Technol. 1995;6:241–245. [Google Scholar]
- 19.López-Amorós R, Castel S, Comas-Riu J, Vives-Rego J. Assessment of E. coli and Salmonella viability and starvation by confocal laser microscopy and flow cytometry using rhodamine 123, DiBAC4(3), propidium iodide and CTC. Cytometry. 1997;29:298–305. doi: 10.1002/(sici)1097-0320(19971201)29:4<298::aid-cyto6>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 20.Mason D J, Allman R, Lloyd D. Uses of membrane potential dyes with bacteria. In: Lloyd D, editor. Flow cytometry in microbiology. London, United Kingdom: Springer-Verlag; 1993. pp. 67–81. [Google Scholar]
- 21.Mason D J, Allman R, Sark J M, Lloyd D. Rapid estimation of antibiotic susceptibility with flow cytometry. J Microsc. 1994;176:8–16. doi: 10.1111/j.1365-2818.1994.tb03494.x. [DOI] [PubMed] [Google Scholar]
- 22.McFeters G A, Feiping P Y, Pyle B H, Stewart P S. Physiological assessment of bacteria using fluorochromes. J Microbiol Methods. 1995;21:1–13. doi: 10.1016/0167-7012(94)00027-5. [DOI] [PubMed] [Google Scholar]
- 23.Molenaar D, Abee T, Konings W N. Continuous measurement of the cytoplasmic pH in Lactococcus lactis with a fluorescent pH indicator. Biochim Biophys Acta. 1991;1115:75–83. doi: 10.1016/0304-4165(91)90014-8. [DOI] [PubMed] [Google Scholar]
- 24.Naidu A S, Bidlack W R, Clemens R A. Probiotic spectra of lactic acid bacteria (LAB) Crit Rev Food Sci Nutr. 1999;38:13–126. doi: 10.1080/10408699991279187. [DOI] [PubMed] [Google Scholar]
- 25.O'Riordan K, Fitzgerald G F. Evaluation of bifidobacteria for the production of antimicrobial compounds and assessment of performance in cottage cheese at refrigeration temperature. J Appl Microbiol. 1998;85:104–114. doi: 10.1046/j.1365-2672.1998.00474.x. [DOI] [PubMed] [Google Scholar]
- 26.Pettipher G L, Mansell R, McKinnon C H, Cousins C. Rapid membrane filtration epifluorescent microscopy technique for direct enumeration of bacteria in raw milk. Appl Environ Microbiol. 1980;39:423–429. doi: 10.1128/aem.39.2.423-429.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez G G, Phipps D, Ishiguro K, Ridgway H F. Use of a fluorescent redox probe for visualization of actively respiring bacteria. Appl Environ Microbiol. 1992;58:1801–1808. doi: 10.1128/aem.58.6.1801-1808.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roszak D B, Colwell R R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salminen S, Ouwehand A G, Isolauri E. Clinical applications of probiotic bacteria. Int Dairy J. 1998;8:563–572. [Google Scholar]
- 30.Scardovi V. Genus Bifidobacterium Orla-Jensen 1924, 472AL. In: Sneath P H, Nair N S, Sharpe M E, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 2. Baltimore, Md: Williams and Wilkins; 1986. pp. 1418–1434. [Google Scholar]
- 31.Shah N P. Bifidobacteria: characteristics and potential for application in fermented milk products. Milchwissenschaft. 1997;52:16–20. [Google Scholar]
- 32.Stanton C, Gardiner G E, Lynch P B, Collins J K, Fitzgerald G F, Ross R P. Probiotic cheese. Int Dairy J. 1998;8:491–496. [Google Scholar]
- 33.Stubberfield L C F, Shaw P J A. A comparison of tetrazolium reduction and FDA hydrolysis with other methods of microbial activity. J Microbiol Methods. 1990;12:151–162. [Google Scholar]
- 34.Tamime A Y, Marshall V M, Robinson R K. Microbiological and technological aspects of milks fermented by bifidobacteria. J Dairy Res. 1995;62:151–187. doi: 10.1017/s002202990003377x. [DOI] [PubMed] [Google Scholar]
- 35.Teixeira P, Castro H, Kirby R. Spray drying as a method for preparing concentrated cultures of Lactobacillus bulgaricus. J Appl Bacteriol. 1995;78:456–462. [Google Scholar]
- 36.Teixeira P, Castro H, Mohácsi-Farkas C, Kirby R. Identification of sites of injury in Lactobacillus bulgaricus during heat stress. J Appl Microbiol. 1997;83:219–226. doi: 10.1046/j.1365-2672.1997.00221.x. [DOI] [PubMed] [Google Scholar]
- 37.Teraguchi S, Uehara M, Ogasa K, Mitsuoka T. Enumeration of bifidobacteria in dairy products. Jpn J Bacteriol. 1978;33:753–761. [PubMed] [Google Scholar]
- 38.Virta M, Lineri S, Kankaanpää P, Karp M, Peltonen K, Nuutila J, Lilius E-M. Determination of complement-mediated killing of bacteria by viability staining and bioluminescence. Appl Environ Microbiol. 1998;64:515–519. doi: 10.1128/aem.64.2.515-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright S J, Centonze V E, Stricker S A, DeVries P J, Paddock S W, Schatten G. Introduction to confocal microscopy and three-dimensional reconstruction. Methods Cell Biol. 1993;38:1–45. doi: 10.1016/s0091-679x(08)60998-x. [DOI] [PubMed] [Google Scholar]