Abstract
Background and aim
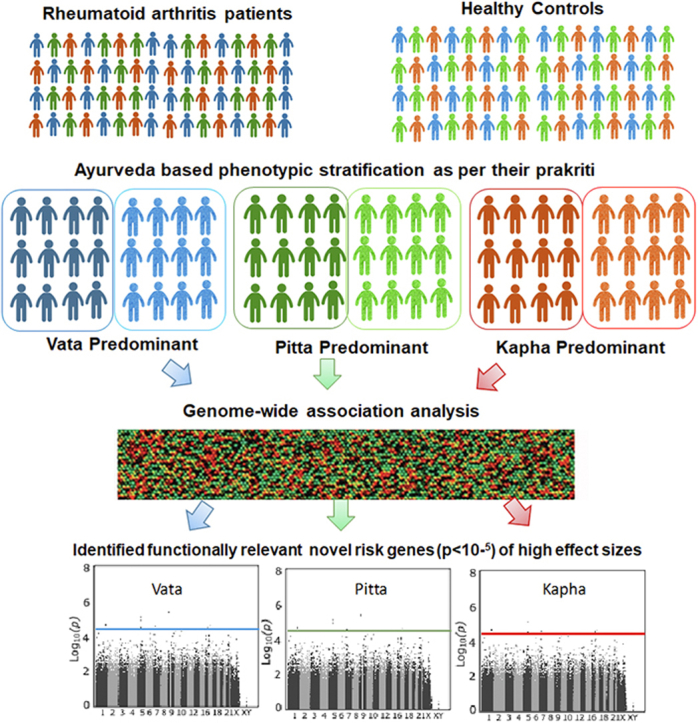
Genome wide association studies have scaled up both in terms of sample size and range of complex disorders investigated, but these have explained relatively little phenotypic variance. Of the several reasons, phenotypic heterogeneity seems to be a likely contributor for missing out genetic associations of large effects. Ayurveda, the traditional Indian system of medicine is one such tool which adopts a holistic deep phenotyping approach and classifies individuals based on their body constitution/prakriti. We hypothesized that Ayurveda based phenotypic stratification of healthy and diseased individuals will allow us to achieve much desired homogeneous cohorts which would facilitate detection of genetic association of large effects. In this proof of concept study, we performed a genome wide association testing of clinically diagnosed rheumatoid arthritis patients and healthy controls, who were re-phenotyped into Vata, Pitta and Kapha predominant prakriti sub-groups.
Experimental procedure
Genotypes of rheumatoid arthritis cases (Vata = 49; Pitta = 117; Kapha = 78) and controls (Vata = 33; Pitta = 175; Kapha = 85) were retrieved from the total genotype data, used in a recent genome-wide association study performed in our laboratory. A total of 528461 SNPs were included after quality control. Prakriti-wise genome-wide association analysis was employed.
Results and conclusion
This study identified (i) prakriti-specific novel disease risk genes of high effect sizes; (ii) putative candidates of novel therapeutic potential; and (iii) a good correlation between genetic findings and clinical knowledge in Ayurveda. Adopting Ayurveda based deep phenotyping may facilitate explaining hitherto undiscovered heritability in complex traits and may propel much needed progress in personalized medicine.
Keywords: Rheumatoid arthritis, Ayurgenomics, Ayurveda, Genome-wide association study, P4 medicine
Abbreviations: GWAS, Genome-wide association study; RA, Rheumatoid arthritis; P4 medicine, Predictive, Preventive, Personalized and Participatory medicine; SNPs, Single nucleotide polymorphisms; Trx, Thioredoxin; Cul3, Cullin E3 ubiquitin ligase; lncRNA, Long intergenic non-coding RNA; MAF, Minor allele frequency; HWE, Hardy-Weinberg equilibrium; QC, Quality control
Graphical abstract
1. Introduction
The inherent goal of Predictive, Preventive, Personalized and Participatory (P4) Medicine is to shift the paradigm in medicine from reactive and generalized to proactive and personalized and hence from disease to wellness. This transformation in healthcare can be achieved by (i) predicting an individual's predisposition to a disease; (ii) stratifying patients to facilitate potential personalized nutritional and drug treatment strategies; (iii) reducing adverse drug reactions; (iv) identifying new druggable targets and their development; and (v) reducing the time, cost, and also failure rate of clinical trials for new therapies. Past decade witnessed explosion of genome-wide association studies (GWASs) of clinically defined cases against non-compromised individuals as controls with a hope that discovering risk genes in large cohorts would provide insights into patient stratification and further aid towards achieving P4 medicine goal. Despite the apparent success of this approach in complex traits, this technology-driven GWAS strategy which primarily relied on large sample sizes has witnessed serious limitations such as non-identification of disease associated genes of large effects which contribute to missing heritability and also non-replication of observed associations across studies. This limitation has been largely due to the inherent heterogeneity owing to endogenous and exogenous factors involved in complex disorders, and by increasing sample sizes, led to inadvertent scaling up of heterogeneity in parallel and hence reduced the statistical power to detect real associations. Different strategies are now being used to overcome the GWAS related limitations and these include turning the emphasis on to rare variants, epigenetic modifications, miRNA etc [1]. Phenotype resolution however seems a likely major determinant of the success or failure of GWAS to date. The importance of accurate phenotyping over increasing sample size to detect true associations has been addressed in a recent study. The authors used both simulated and GWAS data for Type I and Type II diabetes and demonstrated that statistical power to detect real association was reduced when the study cohort was heterogeneous. In other words, if GWAS were carried out in more homogeneous sample sets the magnitude of risk conferred by the marginally/modestly significant risk variants would have been larger [2].
Phenotype definitions in modern medicine largely depend on quantifiable parameters and ignore the underlying heterogeneity in disease pathogenesis. Therefore, we believe it is time to revisit and adopt newer non-conventional phenotyping approaches which may be able to capture molecular variability underlying the disease. Similar to personalized medicine, Ayurveda is not a 'one-size-fits-all' approach but on the contrary addresses inter-individual variability effectively. It adopts a holistic approach toward healthy living on the basis of the concepts of Tridosha and Prakriti. According to Ayurveda, all matter is comprised of the five basic elements or building blocks of nature: Earth, Air, Water, Fire, and Space. Varying combinations of these elements form the three basic humors/forces of the human body namely Vata Dosha, Pitta Dosha and Kapha Dosha collectively called as Tridoshas [3]. Each of these Doshas have distinct properties and when there is balance between the constituents, they work in harmony through the body to maintain homeostasis. In other words, according to Ayurveda, maintenance of this balance is health and imbalance is disease. The innate proportion of Doshas that a zygote acquires at the time of conception determines its Prakriti, and this represents a summed-up phenotype or basic constitution type of an individual [3]. Prakriti defines physical, physiological, and psychological traits of an individual and is the template for individualized diet, lifestyle counselling and disease treatment. A few salient Prakriti specific phenotypes are dryness, lightness, coldness (physical), low strength, irregular digestion (physiological), unstable/confused mind (psychological) for Vata Prakriti [4]; Burning sensation, excessive sweating (physical), excessive appetite, intolerance to heat (psychological), bold, short tempered, irritable (psychological) for Pitta Prakriti [5]; and heaviness in the body, over weight (physical), lethargy, slow movements (physiological), calm, relaxed mind (psychological) for Kapha Prakriti [6]. To this extent one's Prakriti may be considered as the Ayurvedic equivalent of describing the unique genetic constitution (genome) of each individual in modern biology. However, Ayurveda doctrines go further and according to Tridosha theory, depending upon the individual or combinatorial proportions of Tridosha in each person, there are seven possible Prakriti types namely Vata, Pitta, Kapha, Vata-Pitta, Pitta-Kapha, Vata-Kapha and Vata-Pitta-Kapha contributing to wide phenotypic diversity highlighting their practice of deep phenotyping. Furthermore, according to its doctrines and practice, each of these Prakriti types is the determinant of its own characteristic features such as metabolic profiles, disease predisposition, and natural history in individuals with respective Prakriti [7,8]. This Dosha type is also postulated to be responsible for disease characteristics such as severity, therapeutic recommendations, and treatment outcome in individuals.
An empirical validation of this concept has been provided by our previous pilot study on candidate gene associations with rheumatoid arthritis (RA; termed Amavata in Ayurveda), among three subgroups that were based on Ayurveda based phenotyping [9]. This study revealed association of inflammatory genes with RA among Vata predominant Prakriti subjects whereas oxidative stress genes were associated with Pitta subgroup. Further, severity of RA in the patients of Vata Prakriti was significantly higher compared to Pitta and Kapha predominant subgroups which is in agreement with what is known for Amavata in Ayurveda literature.
Significant correlations have also been established between Prakriti and single nucleotide polymorphisms (SNPs) in genes such as HLA in healthy individuals and EGLN1 in patients suffering from high-altitude pulmonary edema [10,11]. Apropos to the above evidence, differences in genome wide expression and biochemical profiles have been observed between the three extreme Prakriti types [12]. Interestingly, a genome-wide analysis has revealed 52 Prakriti differentiating SNPs in healthy individuals across the three predominant Prakriti groups [13]. Of note, another recent study utilized modelling methods using the phenotyping data and was able to classify healthy individuals into three distinct clusters, which matched with the extreme Prakriti groups as classified by clinicians [14].
With this background, we hypothesize that GWAS carried out on homogeneous but small cohorts obtained by deep phenotyping based on Ayurveda doctrines will be more insightful since Ayurveda has comprehensive criteria to stratify not only controls but also cases. This approach would facilitate identification of genetic associations of large effect sizes which may enable filling the present knowledge gap in complex disease biology. In the present study, an Ayurgenomics approach was adopted wherein we carried out a GWAS on RA patients and healthy controls who were re-phenotyped for their Prakriti type. Genome-wide analysis of Prakriti matched RA cases and controls revealed potential Prakriti-specific genetic associations. We believe this Ayurgenomics approach offers itself as a novel tool to perform Prakriti based deep phenotyping of study cohorts prior to their inclusion in contemporary GWASs to obtain homogeneous cohorts and consequently identify true genetic associations with large effect sizes.
2. Materials and methods
2.1. Study cohort
244 RA cases [49 Vata; 117 Pitta; 78 Kapha] and 293 controls [33 Vata; 175 Pitta; 85 Kapha] sub-grouped based on their predominant Prakriti type (briefly described below) were recruited from Department of Ayurveda, Holy family hospital, New Delhi. Both cases and controls were matched for age [18–50 years], gender and ethnicity. DNA from venous blood drawn from study subjects was extracted according to routine phenol-chloroform protocol. This cohort has been used in two previous studies [9,15]. Institutional ethics committee clearance was obtained from Ayurveda Department, Holy Family Hospital and Department of Genetics, University of Delhi South Campus, New Delhi, prior to initiation of this study and the protocol were carried out in accordance with the approved guidelines. Written informed consent was obtained from each participant.
2.2. Ayurveda based phenotyping
The Prakriti of each subject was assessed independently by two Ayurveda physicians using a validated questionnaire based on physical, physiological and psychological characteristics recommended by the Central Council for Research in Ayurveda and Siddha, Department of AYUSH, Ministry of health and family welfare, Government of India, New Delhi (http://www.ccras.nic.in/). Physique, skin texture, hunger, thirst, digestive capacity, temperament and memory are some of the major attributes evaluated to determine an individual's Prakriti. Predominant Prakriti was allotted if ≥ 70% dominance of a single Dosha score was obtained. Only individuals with predominance of either Vata, Pitta or Kapha Doshas were included in the study as described previously [9]. Objective parameters like height, weight, body mass index, blood pressure, swelling, blood/serum examination, X-rays and magnetic resonance imaging were used in the clinical assessment of cases. In addition, visual analogue scale [16] was used for most of the subjective features like pain, swelling, burning sensation, heaviness etc. Blood was drawn and used for analysis of haemoglobin%, erythrocyte sedimentation rate, rheumatoid factor and anti-cyclic citrullinated peptide antibody levels as described elsewhere [9]. RA diagnosis of all the patients thus enrolled were independently confirmed by an orthopaedic surgeon.
2.3. Genotyping and quality control
Genotype information of above mentioned total cohort (n = 244 cases, 293 controls) was retrieved from the total genotype data generated on Illumina Human660W Quad BeadChip v1.C (655 216 markers) genotyping platform, as described before [15]. Briefly, DNA isolated from venous blood of the study cohort was genotyped at a commercial facility. Genome studio software was used to perform initial quality control (QC). Genomic data was then filtered using standard quality control steps on PLINK (v1.9beta3) [17]. Samples with a call rate of <90%, individuals with discordant/ambiguous sex, putative inbreeding/contaminated samples (heterozygosity rate >4 ± standard deviation), ethnic outliers, cryptic relatedness (pi_hat>0.25) were removed. Cryptic relatedness was determined by identity-by-descent analysis using PLINK V.1.07.18. Before SNP QC, markers for which no intensity peaks were detected, copy number variations and non-autosomal chromosomes were excluded from the analysis. Inclusion criteria comprised of call rate >90%; minor allele frequency (MAF) ≥0.01; Hardy–Weinberg equilibrium (HWE) tests (p ≤ 5 E-06) in controls.
2.4. Statistical analysis
Association was tested for total cohort as well as for three separate Prakriti matched case–control samples. Allele frequencies between cases and controls in each of these categories were compared using Fisher's exact test on PLINK. A p-value threshold of 1 E-05 and a Bonferroni-corrected p-value threshold of 9.46 E-08 were considered suggestive and genome-wide significant, respectively. To identify potential link between the novel genetic variants identified with RA subgroups in this study and RA based on literature evidence, p-values were retrieved from Open Targets platform [18]. Biological interactions between genes were inferred using GeneMANIA prediction server which provides interactive functional association network [19].
3. Results
Power analysis performed with Quanto software showed >80% power in Vata, Pitta, and Kapha sub-groups.
3.1. Association findings
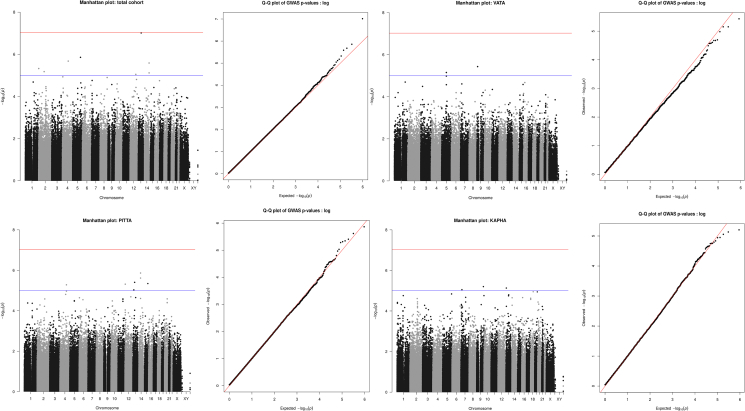
A total of 444 individuals [229 cases (45 Vata, 113 Pitta, 71 Kapha) and 215 controls (24 Vata, 131 Pitta, 60 Kapha)] and 528461 SNPs remained for downstream analysis after QC. A total number of SNPs and individuals removed after each QC step has been described in Supplementary Fig. 1. GWA analysis was performed for the total study cohort as well as for the three predominant Prakriti groups separately. Novel SNPs with suggestive p-value 1 E-05 [Table 1] were found to be associated with the three RA sub-groups. However, none of the SNPs surpassed Bonferroni-corrected p-value. Manhattan and Q–Q plots are shown in Fig. 1. It was notable that all the associated markers/genes were unique to each of the groups. Broad function of the gene/nearest gene identified in Prakriti-wise analysis are also described below.
Table 1.
Genome-wide suggestive significant (p-value < 1 E-05) variants associated with RA in total study cohort and Prakriti specific RA groups. The significance status of these SNPs in other groups are also shown. Positions refer to assembly GRCh37.
| SNP | Chr | Position | Location | Gene/Nearest gene | Risk allele | PGWASTotal | OR (95% CI)* | PGWASVata | OR (95% CI)* | PGWASPitta | OR (95% CI)* | PGWASKapha | OR (95% CI)* |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total study cohort | |||||||||||||
| rs340575 (C>A) | 13 | 72876358 | intergenic | MZT1 | C | 9.707E-08 | 6.52 (2.91-14.62) | 0.21 | 2E-05 | 0.003 | |||
| rs10056189 (C>T) | 5 | 145804118 | intergenic | TCERG1 | C | 1.394E-06 | 3.41 (2.02-5.78) | 0.05 | 0.0008 | 0.01 | |||
| rs4956041 (C>T) | 4 | 109113914 | intron | LEF1-AS1 | T | 2.105E-06 | 1.92 (1.47-2.51) | 0.01 | 0.01 | 0.006 | |||
| rs4983599 (G>A) | 14 | 105011436 | intron | TMEM179 | A | 2.591E-06 | 2.13 (1.55-2.93) | 0.05 | 0.0005 | 0.06 | |||
| rs2867116 (C>A) | 2 | 682363 | upstream | TMEM18 | C | 4.734E-06 | 4.56 (2.26-9.23) | 0.0001 | 0.02 | 0.02 | |||
| rs7556762 (T>G) | 2 | 101765922 | intron | TBC1D8 | T | 6.651E-06 | 2.41 (1.63-3.55) | 0.01 | 0.002 | 0.06 | |||
| rs4548807 (G>A) | 14 | 105040988 | intron | TMEM179 | A | 7.633E-06 | 2.12 (1.53- 2.96) | 0.003 | 0.007 | 0.06 | |||
| rs3782690 (T>G) | 12 | 106464063 | intron | NUAK1 | T | 8.948E-06 | 1.84 (1.41-2.42) | 0.2 | 0.005 | 0.0008 | |||
| Vata | |||||||||||||
| rs1953175 (G>T) | 9 | 13505851 | intergenic | RP11-536O18.1 | T | 0.2 | 3.684E-06 | 6.41 (2.87-14.34) | 0.2 | 0.3 | |||
| rs4352629 (C>T) | 5 | 87756821 | intron | CTC-498M16.4 | T | 0.3 | 7.028E-06 | 5.64 (2.63-12.08) | 0.7 | 0.1 | |||
| rs7448716 (A>G) | 5 | 87752695 | intron | CTC-498M16.4 | G | 0.2 | 7.028E-06 | 5.64 (2.63-12.08) | 0.7 | 0.1 | |||
| Pitta | |||||||||||||
| rs11625685 (T>C) | 14 | 52931884 | intron | TXNDC16 | C | 5E-05 | 0.4 | 1E-06 | 4.50 (2.35-8.61) | 0.2 | |||
| rs11623917 (A>G) | 14 | 52921896 | intron | TXNDC16 | G | 0.0001 | 0.5 | 2E-06 | 4. 37 (2.28-8.38) | 0.3 | |||
| rs9527038 (A>G) | 13 | 53503775 | intergenic | PCDH8 | G | 0.003 | 0.5 | 4E-06 | 2.48 (1.69-3.64) | 0.8 | |||
| rs4620912 (C>T) | 15 | 86389516 | intergenic | KLHL25 | T | 4E-05 | 0.5 | 5E-06 | 2.80 (1.8-4.37) | 0.5 | |||
| rs10849264 (A>G) | 12 | 5531307 | intergenic | NTF3 | G | 0.0003 | 0.3 | 5E-06 | 2.35 (1.62-3.39) | 0.7 | |||
| rs1390079 (T>C) | 4 | 125523042 | intergenic | RP11-93I21.3 | C | 0.007 | 0.5 | 5E-06 | 13.32 (3.09-57.45) | 0.3 | |||
| rs7323558 (C>T) | 13 | 37250048 | intron | SERTM1 | T | 0.002 | 0.9 | 9E-06 | 2.305 (1.60-3.33) | 0.7 | |||
| Kapha | |||||||||||||
| rs3120029 (G>A) | 9 | 129649356 | downstream | ZBTB34 | A | 0.2 | 0.4 | 0.2 | 6E-06 | 4.243 (2.21-8.16) | |||
| rs9512378 (A>G) | 13 | 27363688 | intergenic | GPR12 | A | 0.01 | 0.8 | 0.7 | 7E-06 | 4.60 (3.31-9.15) | |||
| rs11762117 (C>A) | 7 | 20391383 | intron | ITGB8 | C | 0.02 | 0.3 | 0.6 | 9E-06 | 8.74 (2.93-26.03) | |||
*Odds Ratio (OR) shown for genome-wide suggestively significant SNPs.
Fig. 1.
Manhattan plots depicting SNP associations with the (a) total RA cohort and (b–d) Prakriti specific RA sub-groups in the north Indian population, and respective Q–Q plots. On the Manhattan plot, all SNPs are plotted according to their position on each chromosome on x-axis, against their association (-log10 (p-value)) on y-axis. The red line represents the Bonferroni-corrected threshold (p-value = 9.46 E-08), while the blue line represents the suggestive association threshold (p = 1E-05). The inset Q–Q plots show the observed (y-axis) against the expected (x-axis) distribution of GWAS p-values under the null hypothesis for the total RA cohort and Prakriti sub-groups.
3.1.1. Total cases vs controls
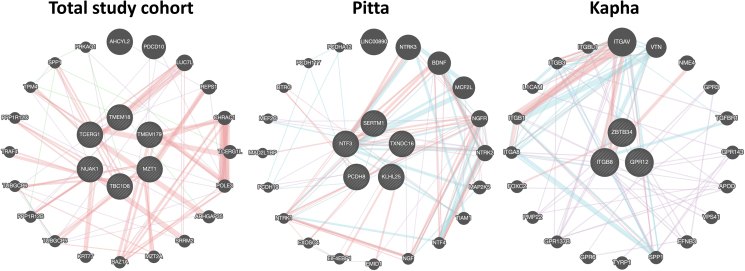
TMEM179, TMEM18, NUAK1, TBC1D8, and LEF1-AS1, together with MZT1 (downstream of rs340575) and TCERG1 (downstream of rs10056189) were identified to be associated when analyzing total RA cases and controls. All six genes, except for the RNA gene LEF1-AS1, were associated with musculoskeletal system disease (Open Targets, p-value 0.02), which includes conditions that affect joints, such as osteoarthritis, rheumatoid arthritis, psoriatic arthritis, gout, ankylosing spondylitis among others. Of note, we also found that all these six genes physically and genetically interact with each other and are co-expressed (Fig. 2). Furthermore, LEF1-AS1, NUAK1, and TBC1D8 were seen to be associated with systemic juvenile idiopathic arthritis (Open Targets, p-value 0.002), and a genomic marker located at C14orf180 (overlapped with TMEM179), rs4264325, was also seen to be significant for RA susceptibility. Of note, all these genes were also seen to be associated in all the three subgroups but at lower significance level [Table 1].
Fig. 2.
GeneMANIA network showing potential interactions with novel RA genes identified in total study cohort and in Prakriti-specific RA sub-groups. The genes of interest are represented in the middle with striped circles. Pink lines represent physical interactions; green lines represent genetic interactions; purple lines represent co-expression; blue lines represent pathways. Thickness of the lines represents interaction strength.
3.1.2. Vata sub-group
Test of association between RA cases and controls categorized under Vata predominant Prakriti identified one SNP (rs1953175) in RP11-536O18.1 and two SNPs (rs4352629 and rs7448716) in CTC-498M16.4 [Table 1]. None of these three Vata-specific SNPs showed association in the total group, or in Pitta and Kapha cohorts [Table 1].
3.1.3. Pitta sub-group
In Pitta group, we found six genes namely TXNDC16 (rs11625685; rs11623917), PCDH8 (rs9527038), KLHL25 (rs4620912) NTF3 (rs10849264), RP11-93I21.3 (rs1390079), and SERTM1 (rs7323558) significantly associated [Table 1] and all of which were functionally relevant. Additionally, physical interaction, co-expression or their presence in common pathways were found among these genes (Fig. 2, Supplementary Table 1).
3.1.4. Kapha sub-group
Three genes namely ZBTB34 (rs3120029), ITGB8 (rs11762117), and GPR12 (rs9512378) were found to be significantly associated in Kapha group but not in Vata and Pitta [Table 1]. All three genes were found to be physically interacting or were part of common pathways [Fig. 2, Supplementary Table 1]. Genomic markers in all three genes were associated with scoliosis (Open Targets, p-value 0.002), fat body mass (Open Targets, p-value 0.01), and also rheumatic disease (Open Targets, p-value 0.01).
4. Discussion
RA is a chronic inflammatory joint disease affecting synovial tissue in multiple joints but with poorly uncovered etiology. It is a clinically and biologically heterogeneous disease with respect to both disease course and treatment outcome suggesting distinct molecular mechanisms contributing to RA in different patients. For instance, differences in the activation of the STAT1 pathway between rheumatoid tissues confirm etiological heterogeneity [20]. Continuous efforts are being made to sub-classify clinically diagnosed RA on the basis of molecular criteria/signatures using OMICS or more recently using phenome-wide association study approach [21]. While we still await their deliverables, exploring non-conventional phenotyping approaches and providing scientific validation for their utility as an adjunct could accelerate the progress in this endeavour. In this proof-of-concept study, we performed a GWAS of RA patients and healthy controls who were phenotypically sub-classified into three Prakriti groups namely Vata, Pitta and Kapha predominant by employing the ancient deep phenotyping principles practiced in Ayurveda system [7,8]. Despite small numbers in each of the three subgroups, this novel Ayurgenomics approach identified Prakriti-specific genes of high effect sizes [Table 1], most of them not hitherto identified for RA across multiple GWASs performed in large cohorts or even in meta-analyses data [22]. The relevance of these novel susceptibility genes in the context of RA are briefly discussed below.
4.1. Vata sub-group
Although no direct link was found between RA and the two Vata specific genes, CTC-498M16.4 has been shown to be associated with attention deficit/hyperactivity disorder (ADHD), rs4916723 (GWAS, p-value 2.67 E-05) being the lead SNP conditioning the gene [23]. This is important considering that though ADHD is currently conceptualized as a neurodevelopmental disorder, recent findings [24] have shown genetic connection between ADHD and immune alterations/autoimmune disorders. Further, the infective component in RA etiology indicates that there could be shared risk pathways between RA and ADHD with pleiotropic genetic effects.
4.2. Pitta sub-group
TXNDC16 encodes for Thioredoxin (Trx) Domain Containing 16, which is an endoplasmic reticulum-associated glycoprotein and is believed to have putative redox activity [25]. A substantial number of studies have demonstrated the role of oxidative stress in RA pathogenesis [26]. It has been reported that cytosolic Trx system has a role in RA and Trx1 has shown to be significantly increased in the synovial fluid of RA patients [25].
PCDH8 is a protocadherin involved in neural development and function and it is shown to be dysregulated in several types of cancers and playing a critical role in tumor progression. Notably, a global gene expression profiling of chondrogenic tissues during in vivo development in mice showed involvement of Pcdh8 in chondrogenesis [27], a process by which cartilage is formed.
KLHL25 belongs to Kelch family of proteins that function as substrate-specific adaptors for Cullin E3 ubiquitin ligase (Cul3), a core component of the ubiquitin-proteasome system to regulate the protein turnover. It is important to mention here that our earlier study has shown an association between CUL1 haplotype and methotrexate response in a north Indian population [28]. Similar findings were also observed in a RA cohort of Japanese origin [29]. Our findings lend further support to the role of ubiquitin pathway in autoimmunity and inflammation. Recent studies have identified mutations of several Kelch proteins in skeletal muscle disorders [30]. Though, no direct role of KLHL25 has been implicated in RA, a recent study has proposed that increase in inflammatory processes and reactive oxygen species production leads to skeletal muscle deterioration [31] which in turn contributes to a vicious cycle of disease activity, muscle inflammatory signalling and disrupted remodelling, physical inactivity, and disability in patients with RA [32].
Protein (NT-3) encoded by NTF3 is a member of the neurotrophin family which are essential for the development and maintenance of the vertebrate nervous system. Neutrophins and their receptors are shown to be expressed in the non-neuronal cells [33] supporting the role of neurotrophins beyond neurogenesis. A study has shown that LPS-treated mouse macrophages resulted in up-regulation of NT-3 leading to overproduction of nitric oxide, suggesting that NT-3 may play important roles in the function of macrophages during inflammatory responses and in tissue repair [34]. The role of NT-3 in RA has been empirically demonstrated by recent studies wherein over-expression of NT-3 in serum of RA patients [35] and high expression of NTF-3 in RA synovial fibroblasts compared with healthy synovial fibroblasts under normoxic conditions has been observed [36]. In a recent study, NT-3 and its high affinity receptor TrkC were found to be highly induced at the injury site and endogenous NT-3 was found to promote bone repair [37]. In addition, NT-3 has also been implicated in neuropathic pain which is often poorly alleviated by first- and second-line medications due to lack of efficacy and/or dose-limiting side-effects [38]. Notably, neuropathic pain in substantial number of RA patients has been associated with vitamin D deficiency [39] or with high disease activity and weight [40]. These observations are of clinical relevance since a better understanding of neuropathic pain mechanisms will provide a more targeted approach to pain treatment in RA.
RP11-93I21.3 is a long intergenic non-coding RNA (lncRNA). Though there is no report suggesting direct functional involvement of RP11-93I21.3 in RA pathogenesis, several lncRNAs are shown to be dysregulated in RA and are correlated with disease activity [41].
SERTM1, is a serine rich and transmembrane domain containing 1 gene and has shown to be downregulated in psoriasis patients, thereby likely to play an important role in inflammation associated with RA patients [42].
4.3. Kapha sub-group
ZBTB34, a nuclear protein, is a new member of the BTB/POZ zinc finger protein family. Although exact role of ZBTB34 is not known, some of the proteins of this family critically regulate development of specific lineages in the immune system, promote oncogenesis and maintain stem cells. It has also been suggested that ZBTB34 might function as a transcriptional repressor [43]. In zebrafish, ZBTB is predicted to be involved in negative regulation of transcription by RNA polymerase II; regulation of cytokine production; and regulation of immune system process [44]. ZBTB34 has been shown to be overexpressed in the whole blood of axial spondylarthritis/ankylosing spondylitis patients compared to healthy controls [45]. Of note, our previous GWAS has shown significant association of a different SNP (rs561041) closest to ZBTB34 with RA [15].
GPR12 is classified as an orphan G protein-coupled receptor. Disruption of Gpr12 gene in mice has shown to provoke changes in both lipid and carbohydrate metabolism resulting in dyslipidemia and obesity [46] and therefore considered to be involved in regulating energy expenditure and important for future drugs that target this receptor. Of note, GPR12 has been identified as a novel target of Cannabidiol, which is shown to have therapeutic potential for arthritis pain-related behaviors and inflammation without evident side-effects [47].
ITGB8 is a member of the integrin beta chain family and has been involved in angiogenesis deregulation in systemic sclerosis, a chronic autoimmune rheumatic disorder [48]. Furthermore, importance of ITGB8 in chondrogenesis has been previously established [49], suggesting its direct involvement in RA pathology, as progressive loss of cartilage due to inflammatory response is one of the disease characteristics. This derives further support from a gene expression study wherein ITGB8 was shown to be highly expressed in RA synovial fibroblasts compared with healthy synovial fibroblasts under normoxic conditions suggesting its role in chronic synovitis [36].
It is important to mention here that even the interacting partners of genes associated with Pitta and Kapha subgroups such as BDNF, NGFR, ITGAV [Fig. 2, Supplementary Table 1] were also found to be very relevant in disease pathogenesis [50]. Besides these, the involvement of some of the genes evidenced in different pathways strengthens our findings. For example, KLH25 in Class I MHC mediated antigen processing and presentation pathway; NTF3 and ITGB8 in Apoptotic pathways in synovial fibroblasts.
Our results also highlight the striking difference in the genetic associations identified in the total group versus those in each of the three Prakriti based subgroups [Table 1]. These findings imply that even small sample size of tightly defined cases and controls or just precise phenotyping may have led to comparatively more genetically homogeneous groups which were sufficient to maximize the detection of common alleles conferring high risk and minimize statistical noise. That P-values of genes are largely driven by sample size is further exemplified by HLA associations which are more significant in the total cohort (n = 229 cases and 215 controls) and Pitta subgroup (n = 117 cases and n = 175 controls) compared to Vata and Kapha in our study [Table 2]. This may also explain limited genetic associations that we observed in the Vata group in this study compared to Pitta and Kapha subgroups. Nevertheless, a recent report wherein a locus with genome-wide significance was identified near the gene encoding parathyroid hormone-like hormone in a GWAS performed in a cohort of only 40 patients with peripartum cardiomyopathy [51] supports the power of sample homogeneity in association studies.
Table 2.
List of SNPs in and around HLA region which surpassed genome-wide significance (p < 10−5) in our previous RA GWAS [13] and their significance status in the three Ayurveda sub-groups and total cohort.
| SNP | Chr | Position | Gene | Location | Minor allele | PGWASTotal | OR (95% CI) Total | PGWASVata | OR (95% CI) Vata | PGWAS Pitta | OR (95% CI) Pitta | PGWAS Kapha | OR (95% CI) Kapha |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs9275406 | 6 | 32669955 | HLA-DQB1 | flanking_5UTR | T | 0.004 | 1.66 (1.18–2.34) | 0.09 | 2.75 (0.87–8.66) | 0.008 | 1.88 (1.19–2.97) | 0.55 | 1.23 (0.68–2.23) |
| rs9275388 | 6 | 32669084 | HLA-DQB1 | flanking_5UTR | C | 0.004 | 1.65 (1.18–2.32) | 0.09 | 2.75 (0.87–8.66) | 0.01 | 1.84 (1.17–2.90) | 0.46 | 1.26 (0.70–2.27) |
| rs9275390 | 6 | 32669156 | HLA-DQB1 | flanking_5UTR | C | 0.004 | 1.65 (1.18–2.32) | 0.09 | 2.75 (0.87–8.66) | 0.01 | 1.84 (1.17–2.90) | 0.46 | 1.26 (0.70–2.27) |
| rs9275407 | 6 | 32670037 | HLA-DQB1 | flanking_5UTR | T | 0.003 | 1.67 (1.19–2.35) | 0.09 | 2.75 (0.87–8.66) | 0.008 | 1.88 (1.19–2.97) | 0.46 | 1.26 (0.70–2.27) |
| rs9275393 | 6 | 32669439 | HLA-DQB1 | flanking_5UTR | A | 0.005 | 1.63 (1.16–2.29) | 0.14 | 2.63 (0.83–8.34) | 0.01 | 1.82 (1.16–2.88) | 0.46 | 1.26 (0.70–2.27) |
| rs9275425 | 6 | 32670874 | HLA-DQB1 | flanking_5UTR | A | 0.005 | 1.63 (1.16–2.29) | 0.09 | 2.75 (0.87–8.66) | 0.01 | 1.77 (1.12–2.81) | 0.46 | 1.28 (0.71–2.32) |
| rs9275439 | 6 | 32671521 | HLA-DQB1 | flanking_5UTR | C | 0.003 | 1.67 (1.19–2.35) | 0.09 | 2.75 (0.87–8.66) | 0.01 | 1.84 (1.17–2.90) | 0.46 | 1.31 (0.72–2.36) |
| rs9275427 | 6 | 32670915 | HLA-DQB1 | flanking_5UTR | T | – | – | – | – | – | – | – | – |
| rs9275374 | 6 | 32668526 | HLA-DQB1 | flanking_5UTR | T | 0.004 | 1.65 (1.18–2.32) | 0.09 | 2.75 (0.87–8.66) | 0.01 | 1.84 (1.17–2.90) | 0.46 | 1.26 (0.70–2.27) |
| rs9275418 | 6 | 32670244 | HLA-DQB1 | flanking_5UTR | G | 0.005 | 1.64 (1.17–2.30) | 0.14 | 2.63 (0.83–8.34) | 0.01 | 1.84 (1.17–2.90) | 0.46 | 1.26 (0.70–2.27) |
| rs9275428 | 6 | 32670978 | HLA-DQB1 | flanking_5UTR | G | 0.007 | 1.62 (1.15–2.27) | 0.09 | 2.75 (0.87–8.66) | 0.01 | 1.79 (1.13–2.83) | 0.55 | 1.23 (0.68–2.23) |
| rs9275424 | 6 | 32670576 | HLA-DQB1 | flanking_5UTR | G | 0.008 | 1.57 (1.13–2.19) | 0.06 | 2.94 (0.94–9.22) | 0.04 | 1.62 (1.04–2.54) | 0.46 | 1.30 (0.72–2.34) |
| rs9275371 | 6 | 32668296 | HLA-DQB1 | flanking_5UTR | C | – | – | – | – | – | – | – | – |
| rs9275595 | 6 | 32681355 | HLA-DQA2 | flanking_5UTR | C | 0.007 | 1.63 (1.14–2.34) | 0.20 | 2.38 (0.75–7.57) | 0.01 | 1.94 (1.19–3.16) | 0.75 | 1.14 (0.62–2.12) |
| rs9275582 | 6 | 32680070 | HLA-DQA2 | flanking_5UTR | T | 0.007 | 1.63 (1.14–2.34) | 0.20 | 2.38 (0.75–7.57) | 0.01 | 1.94 (1.19–3.16) | 0.75 | 1.14 (0.62–2.12) |
| rs2856725 | 6 | 32666738 | HLA-DQB1 | flanking_5UTR | C | 0.09 | 0.78 (0.59–1.04) | 0.14 | 0.57 (0.28–1.18) | 0.77 | 0.93 (0.63–1.37) | 0.10 | 0.62 (0.36–1.07) |
| rs2647012 | 6 | 32664458 | HLA-DQB1 | flanking_5UTR | T | 0.08 | 0.77 (0.58–1.02) | 0.13 | 0.55 (0.27–1.14) | 0.70 | 0.91 (0.62–1.34) | 0.11 | 0.63 (0.37–1.07) |
| rs2858305 | 6 | 32670464 | HLA-DQB1 | flanking_5UTR | G | 0.08 | 0.78 (0.58–1.03) | 0.13 | 0.55 (0.27–1.14) | 0.69 | 0.92 (0.62–1.35) | 0.11 | 0.64 (0.38–1.08) |
| rs2856717 | 6 | 32670308 | HLA-DQB1 | flanking_5UTR | A | 0.10 | 0.78 (0.59–1.04) | 0.13 | 0.55 (0.27–1.14) | 0.77 | 0.94 (0.64–1.36) | 0.11 | 0.63 (0.37–1.07) |
| rs10484561 | 6 | 32665420 | HLA-DQB1 | flanking_5UTR | G | 0.004 | 1.99 (1.24–3.2) | 0.06 | 6.54 (0.82–52.32) | 0.00 | 2.5 (1.31–4.78) | 1.00 | 1.06 (0.48–2.37) |
| rs7774434 | 6 | 32657578 | HLA-DQB1 | flanking_5UTR | T | 0.02 | 0.72 (0.55–0.94) | 0.05 | 0.47 (0.23–0.97) | 0.23 | 0.79 (0.55–1.14) | 0.11 | 0.65 (0.40–1.07) |
| rs13192471 | 6 | 32671103 | HLA-DQB1 | flanking_5UTR | C | 0.01 | 1.74 (1.12–2.70) | 0.03 | 7.94 (1.01–62.64) | 0.03 | 1.89 (1.06–3.34) | 1.00 | 1.06 (0.48–2.37) |
| rs9275572 | 6 | 32678999 | HLA-DQA2 | flanking_5UTR | A | 0.09 | 0.78 (0.59–1.03) | 0.20 | 0.6 (0.30–1.26) | 0.63 | 0.90 (0.62–1.32) | 0.11 | 0.63 (0.37–1.07) |
| rs9275596 | 6 | 32681631 | HLA-DQA2 | flanking_5UTR | C | 0.01 | 0.65 (0.47–0.9) | 0.01 | 0.36 (0.16–0.80) | 1.00 | 0.97 (0.63–1.52) | 0.00 | 0.38 (0.20–0.71) |
| rs13209234 | 6 | 32415975 | HLA-DRA | flanking_3UTR | A | 0.06 | 1.43 (0.99–2.06) | 0.14 | 2.56 (0.81–8.10) | 0.16 | 1.45 (0.88–2.39) | 0.75 | 1.12 (0.59–2.12) |
| rs7755224 | 6 | 32652317 | HLA-DQB1 | flanking_5UTR | G | 0.006 | 1.93 (1.21–3.08) | 0.06 | 6.54 (0.82–52.32) | 0.01 | 2.35 (1.25–4.44) | 1.00 | 1.06 (0.48–2.37) |
| rs9391858 | 6 | 32341398 | C6orf10 | flanking_5UTR | G | 0.06 | 1.42 (0.99–2.03) | 0.09 | 2.75 (0.87–8.66) | 0.26 | 1.35 (0.83–2.20) | rs9391858 | 1.20 (0.64–2.27) |
Taken together, the novel study findings lend credence that association studies conducted on homogeneous subgroups obtained by Ayurveda based deep phenotyping enable identification of disease specific genes of major effect size. Of the genes identified in the different subgroups in our study, NTF3, KLH25, TXNDC16, PCDH8, ITGB8, and GPR12 [Table 1] are promising and may provide a new perspective and prompt us to explore their active involvement and therapeutic potential in chronic inflammatory arthritis. However, with the limited data on the functions of most of the genes, it may be difficult to establish direct genotype and Prakriti-associated phenotype (physical/physiological/psychological traits) correlations; and to also comment on Prakriti-specific disease mechanism. Yet Ayurveda wisdom (explained briefly below) supports utility of Ayurgenomics approach to understand heterogeneity in disease biology to some extent.
4.4. Insights into RA biology from Ayurveda
Stratifying healthy individuals into seven constitution types or Prakriti for predicting Prakriti-specific disease susceptibilities and clinical outcomes such as treatment response forms the basis of Ayurveda medical practice and also explains inter-individual variability. To elaborate this further, individuals with Vata Prakriti are more predisposed to RA and are the most difficult group to treat compared to the Pitta subgroup who are less prone, manifest mild to moderate symptoms and are also easier to treat with better outcome [52]. Furthermore, disease severity is more pronounced in RA patients with Vata Prakriti, who suffer severe throbbing pain, which worsens in cold weather; Pitta patients experience burning sensation, redness, swelling, and inflammation, which worsens with hot weather; and Kapha patients show loss of movement, itching, joint swelling and edema (without inflammation), with other symptoms including dullness, heaviness and aches [53].
As for treatment, Ayurveda believes that RA (Amavata) is a problem of the gut, or in other words, a metabolic disorder, and therefore improving the digestive capacity which varies according to individual's Prakriti is the primary focus of its treatment regime. This is in line with the emerging role of gut microbiome dysbiosis in RA. According to Ayurveda, Mandagni [hypofunctioning] of Agni/digestive power (corresponding to enzymes, chemicals, hormones, neurotransmitters and cytokines known to modern medicine) results in impaired digestion and absorption of food, which leads to the formation of immunologic and toxic substances called “Ama” [54]. This Ama when circulates in the body lodges in the joints and leads to inflammation. Of note, Agni which is responsible for metabolism, absorption, etc is believed to be Prakriti specific with best/strong digestive/metabolic power in Pitta followed by Kapha and then Vata sub-groups [55]. Therefore, the treatment of Amavata focuses primarily on improving the digestive capacity, and removal of Ama which in other words is treating the cause of the disease. Reducing the pain/inflammation is a secondary treatment based on the disease symptoms and as mentioned above, this treatment is also Prakriti specific.
5. Conclusions
Identification of novel Prakriti specific and more importantly, functionally relevant susceptibility genes of intermediate/high effect size for RA, suggest that Ayurveda based deep phenotyping could be an effective approach to achieve the highly desirable sample homogeneity in complex trait genetics. This may propel i) a better development of multi-omics signature based prognostic and diagnostic markers and ii) allow Prakriti specific nutritional and therapeutic intervention strategies. Further, such homogeneous cohorts will catalyse rare variant identification as the focus of genetic studies turns from common to rare variants. We strongly believe that using non-conventional phenotyping approaches practiced in complementary systems of medicine such as Ayurveda, Unani, and Chinese traditional medicine along with modern medicine diagnostic/therapeutic knowledge will broaden our horizon of disease biology and provide insights into disease genetics, which remains an urgent unmet need to break ground in complex traits and fulfil the P4 medicine goal. However, these novel findings endorse replication in independent cohorts.
Sources(s) of funding
This work was supported by Central Council for Research in Ayurvedic Sciences, Ministry of AYUSH, New Delhi, India [grant number F.No. Z.31018/18/2006-R&P]; and Department of Biotechnology, New Delhi, India [grant number #BT/01/COE/07/UDSC/2008 (Phase I)].
Author contributions
Garima Juyal: Methodology/Study design, Formal analysis, Investigation, Data curation, Writing- Original draft, Writing-review and editing, Visualization, Supervision; Anuj Pandey: Formal analysis, Data Curation, Writing-review and editing; Sara L. Garcia: Software, Formal analysis, Data curation, Writing-review and editing, Visualization; Sapna Negi: Data curation, Writing-review and editing; Ramneek Gupta: Software, Formal analysis, Data curation, Writing-review and editing; Uma Kumar: Methodology/Study design, Investigation, Resources, Writing-review and editing, Funding acquisition; Bheema Bhat: Conceptualization, Methodology/Study design, Investigation, Resources, Data curation, Writing-review and editing, Visualization, Supervision, Project administration, Funding acquisition; Ramesh C Juyal: Conceptualization, Methodology/Study design, Resources, Writing-review and editing, Project administration, Funding acquisition; Thelma B K: Conceptualization, Methodology/Study design, Formal analysis, Investigation, Resources, Data curation, Writing- Original draft, Writing-review and editing, Visualization, Supervision, Project administration, Funding acquisition.
Consent to participate
Written informed consent was obtained from all subjects participating in this study.
Conflict of interest
None.
Acknowledgments
Gratefully acknowledge Dr. Preeti Wakhode, Ayurveda physician, for independent assessment of the study cohort for their Prakriti using the questionnaire; and Apoorva Anand, Anuroop Venkateswaran and Neha Rana for their help with initial data collation. Infrastructure support provided to the Department of Genetics, University of Delhi South Campus, by the University Grants Commission, New Delhi under the Special Assistance Programme and Department of Science and Technology, New Delhi under FIST and DU-DST PURSE programmes; are gratefully acknowledged. Senior research fellowship to AP by Council of Scientific and Industrial Research, New Delhi, India and Financial support from Idella Foundation to SLG is greatly acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Peer review under responsibility of Transdisciplinary University, Bangalore.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jaim.2022.100578.
Contributor Information
Garima Juyal, Email: garimajuyal@gmail.com.
B.K. Thelma, Email: thelmabk@south.du.ac.in, thelmabk@gmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Seyhan A.A., Carini C. Are innovation and new technologies in precision medicine paving a new era in patients centric care? Open Access Journal of Translational Medicine. J Transl Med. 2019;17:114. doi: 10.1186/s12967-019-1864-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manchia M., Cullis J., Turecki G., Rouleau G.A., Uher R., Alda M. The impact of phenotypic and genetic heterogeneity on results of genome wide association studies of complex diseases. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakraborty R. 2017. Genesis of personalized medicine: relevance of Ayurveda in the present millennium. [DOI] [Google Scholar]
- 4.Charak Samhita of Charak, Vimansthaan; Rogbhishakjitiya vimanadhyay: Chapter 8, verse 98. Chaukhamba Bharati Academy; Varanasi: 2001. pp. 773–774. [Google Scholar]
- 5.Charak Samhita of Charak, Vimansthaan; Rogbhishakjitiyavimanadhyay: Chapter 8, verse 97. Chaukhamba Bharati Academy; Varanasi: 2001. p. 773. [Google Scholar]
- 6.Charak Samhita of Charak, Vimansthaan; Rogbhishakjitiyavimanadhyay: Chapter 8, verse 96. Chaukhamba Bharati Academy; Varanasi: 2001. p. 772. [Google Scholar]
- 7.Charak Samhita of Charak, Vimansthaan; Rogbhishakjitiyavimanadhyay: Chapter 8, verse 100. Chaukhamba Bharati Academy; Varanasi: 2001. p. 774. [Google Scholar]
- 8.Charak Samhita of Charak, Vimana Sthana; Rogabhishakjitiya vimanadhyay: Chapter 8, verse 99. Chaukhamba Bharati Academy; Varanasi: 2001. p. 774. [Google Scholar]
- 9.Juyal R.C., Negi S., Wakhode P., Bhat S., Bhat B., Thelma B.K. Potential of Ayurgenomics approach in complex trait research: leads from a pilot study on rheumatoid arthritis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0045752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhushan P., Kalpana J., Arvind C. Classification of human population based on HLA gene polymorphism and the concept of Prakriti in Ayurveda. J Alternative Compl Med. 2005;11:349–353. doi: 10.1089/acm.2005.11.349. [DOI] [PubMed] [Google Scholar]
- 11.Aggarwal S., Negi S., Jha P., Singh P.K., Stobdan T., Pasha M.A.Q., et al. EGLN1 involvement in high-altitude adaptation revealed through genetic analysis of extreme constitution types defined in Ayurveda. Proc Natl Acad Sci U S A. 2010;107 doi: 10.1073/pnas.1006108107. –6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prasher B., Negi S., Aggarwal S., Mandal A.K., Sethi T.P., Deshmukh S.R., et al. Whole genome expression and biochemical correlates of extreme constitutional types defined in Ayurveda. J Transl Med. 2008;6:48. doi: 10.1186/1479-5876-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Govindaraj P., Nizamuddin S., Sharath A., Jyothi V., Rotti H., Raval R., et al. Genome-wide analysis correlates Ayurveda prakriti. Sci Rep. 2015;5 doi: 10.1038/srep15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiwari P., Kutum R., Sethi T., Shrivastava A., Girase B., Aggarwal S., et al. Recapitulation of Ayurveda constitution types by machine learning of phenotypic traits. PLoS One. 2017;12 doi: 10.1371/journal.pone.0185380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negi S., Juyal G., Senapati S., Prasad P., Gupta A., Singh S., et al. A genome-wide association study reveals ARL15, a novel non-HLA susceptibility gene for rheumatoid arthritis in North Indians. Arthritis Rheum. 2013;65:3026–3035. doi: 10.1002/art.38110. [DOI] [PubMed] [Google Scholar]
- 16.Melzack R. The McGill Pain Questionnaire: major properties and scoring methods. Pain. 1975;1:277–299. doi: 10.1016/0304-3959(75)90044-5. [DOI] [PubMed] [Google Scholar]
- 17.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvalho-Silva D., Pierleoni A., Pignatelli M., Ong C.K., Fumis L., Karamanis N., et al. Open Targets Platform: new developments and updates two years on. Nucleic Acids Res. 2019;47:D1056–D1065. doi: 10.1093/nar/gky1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van der Pouw Kraan T.C.T.M., Van Gaalen F.A., Kasperkovitz P.V., Verbeet N.L., Smeets T.J.M., Kraan M.C., et al. Rheumatoid arthritis is a heterogeneous disease: evidence for differences in the activation of the STAT-1 pathway between rheumatoid tissues. Arthritis Rheum. 2003;48:2132–2145. doi: 10.1002/art.11096. [DOI] [PubMed] [Google Scholar]
- 21.Liao K.P., Sparks J.A., Hejblum B.P., Kuo I.H., Cui J., Lahey L.J., et al. Phenome-wide association study of autoantibodies to citrullinated and noncitrullinated epitopes in rheumatoid arthritis. Arthritis Rheumatol. 2017;69:742–749. doi: 10.1002/art.39974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buniello A., Macarthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/NAR/GKY1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao C., Laporte A.D., Spiegelman D., Akçimen F., Joober R., Dion P.A., et al. Transcriptome-wide association study of attention deficit hyperactivity disorder identifies associated genes and phenotypes. Nat Commun. 2019;10 doi: 10.1038/s41467-019-12450-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tylee D.S., Sun J., Hess J.L., Tahir M.A., Sharma E., Malik R., et al. Genetic correlations among psychiatric and immune-related phenotypes based on genome-wide association data. Am J Med Genet Part B Neuropsychiatr Genet. 2018;177:641–657. doi: 10.1002/ajmg.b.32652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanschmann E.M., Godoy J.R., Berndt C., Hudemann C., Lillig C.H. Thioredoxins, glutaredoxins, and peroxiredoxins-molecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxidants Redox Signal. 2013;19:1539–1605. doi: 10.1089/ars.2012.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fonseca LJS Da, Nunes-Souza V., Goulart M.O.F., Rabelo L.A. Oxidative stress in rheumatoid arthritis: what the future might hold regarding novel biomarkers and add-on therapies. Oxid Med Cell Longev. 2019;2019 doi: 10.1155/2019/7536805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cameron T.L., Belluoccio D., Farlie P.G., Brachvogel B., Bateman J.F. Global comparative transcriptome analysis of cartilage formation in vivo. BMC Dev Biol. 2009;9 doi: 10.1186/1471-213X-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Negi S., Kumar A., Thelma B.K., Juyal R.C. Association of Cullin1 haplotype variants with rheumatoid arthritis and response to methotrexate. Pharmacogenetics Genom. 2011;21:590–593. doi: 10.1097/FPC.0b013e3283492af7. [DOI] [PubMed] [Google Scholar]
- 29.Kawaida R., Yamada R., Kobayashi K., Tokuhiro S., Suzuki A., Kochi Y., et al. CUL1, a component of E3 ubiquitin ligase, alters lymphocyte signal transduction with possible effect on rheumatoid arthritis. Gene Immun. 2005;6:194–202. doi: 10.1038/sj.gene.6364177. [DOI] [PubMed] [Google Scholar]
- 30.Gupta V.A., Beggs A.H. Kelch proteins: emerging roles in skeletal muscle development and diseases. Skeletal Muscle. 2014;4:11. doi: 10.1186/2044-5040-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oyenihi A.B., Ollewagen T., Myburgh K.H., Powrie Y.S.L., Smith C. Redox status and muscle pathology in rheumatoid arthritis: insights from various rat hindlimb muscles. Oxid Med Cell Longev. 2019;2019 doi: 10.1155/2019/2484678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huffman K.M., Jessee R., Andonian B., Davis B.N., Narowski R., Huebner J.L., et al. Molecular alterations in skeletal muscle in rheumatoid arthritis are related to disease activity, physical inactivity, and disability. Arthritis Res Ther. 2017;19:12. doi: 10.1186/s13075-016-1215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nockher W.A., Renz H. Neurotrophins in clinical diagnostics: pathophysiology and laboratory investigation. Clin Chim Acta. 2005;352:49–74. doi: 10.1016/j.cccn.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Barouch R., Appel E., Kazimirsky G., Brodie C. Macrophages express neurotrophins and neurotrophin receptors: regulation of nitric oxide production by NT-3. J Neuroimmunol. 2001;112:72–77. doi: 10.1016/S0165-5728(00)00408-2. [DOI] [PubMed] [Google Scholar]
- 35.Panezai J., Ali A., Ghaffar A., Benchimol D., Altamash M., Klinge B., et al. Upregulation of circulating inflammatory biomarkers under the influence of periodontal disease in rheumatoid arthritis patients. Cytokine. 2020;131 doi: 10.1016/j.cyto.2020.155117. [DOI] [PubMed] [Google Scholar]
- 36.Del Rey M.J., Izquierdo E., Usategui A., Gonzalo E., Blanco F.J., Acquadro F., et al. The transcriptional response of normal and rheumatoid arthritis synovial fibroblasts to hypoxia. Arthritis Care Res. 2010;62:3584–3594. doi: 10.1002/art.27750. [DOI] [PubMed] [Google Scholar]
- 37.Su Y.W., Chung R., Ruan C.S., Chim S.M., Kuek V., Dwivedi P.P., et al. Neurotrophin-3 induces BMP-2 and VEGF activities and promotes the bony repair of injured growth plate cartilage and bone in rats. J Bone Miner Res. 2016;31:1258–1274. doi: 10.1002/jbmr.2786. [DOI] [PubMed] [Google Scholar]
- 38.Khan N., Smith M.T. Neurotrophins and neuropathic pain: role in pathobiology. Molecules. 2015;20:10657–10688. doi: 10.3390/molecules200610657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yesil H., Sungur U., Akdeniz S., Gurer G., Yalcin B., Dundar U. Association between serum vitamin D levels and neuropathic pain in rheumatoid arthritis patients: a cross-sectional study. Int J Rheum Dis. 2018;21:431–439. doi: 10.1111/1756-185X.13160. [DOI] [PubMed] [Google Scholar]
- 40.Ito S., Kobayashi D., Murasawa A., Narita I., Nakazono K. An analysis of the neuropathic pain components in rheumatoid arthritis patients. Intern Med. 2018;57:479–485. doi: 10.2169/internalmedicine.9235-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lao M.X., Xu H.S., Guo L.S. Involvement of long non-coding RNAs in the pathogenesis of rheumatoid arthritis. Chin Med J (Engl) 2020;133:941–950. doi: 10.1097/CM9.0000000000000755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn R., Yan D., Chang H.W., Lee K., Bhattarai S., Huang Z.M., et al. RNA-seq and flow-cytometry of conventional, scalp, and palmoplantar psoriasis reveal shared and distinct molecular pathways. Sci Rep. 2018;8:11368. doi: 10.1038/s41598-018-29472-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi J., Zhang X., Zhang H.K., Yang H.M., Zhou Y.B., Han Z.G. ZBTB34, a novel human BTB/POZ zinc finger protein, is a potential transcriptional repressor. Mol Cell Biochem. 2006;290:159–167. doi: 10.1007/s11010-006-9183-x. [DOI] [PubMed] [Google Scholar]
- 44.ZFIN the Zebrafish Information Network n.d. https://zfin.org/(accessed October 25, 2020).
- 45.Park R., Kim T.-H., Ji J.D. Gene expression profile in patients with axial spondyloarthritis: meta-analysis of publicly accessible microarray datasets. J Rheum Dis. 2016;23:363. doi: 10.4078/jrd.2016.23.6.363. [DOI] [Google Scholar]
- 46.Bjursell M., Gerdin A.K., Jönsson M., Surve V.V., Svensson L., Huang X.F., et al. G protein-coupled receptor 12 deficiency results in dyslipidemia and obesity in mice. Biochem Biophys Res Commun. 2006;348:359–366. doi: 10.1016/j.bbrc.2006.07.090. [DOI] [PubMed] [Google Scholar]
- 47.Hammell D.C., Zhang L.P., Ma F., Abshire S.M., McIlwrath S.L., Stinchcomb A.L., et al. Transdermal cannabidiol reduces inflammation and pain-related behaviours in a rat model of arthritis. Eur J Pain (United Kingdom) 2016;20:936–948. doi: 10.1002/ejp.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giusti B., Margheri F., Rossi L., Lapini I., Magi A., Serratì S., et al. Correction: desmoglein-2-integrin Beta-8 interaction regulates actin assembly in endothelial cells: deregulation in Systemic sclerosis. PLoS One. 2013;8(7) doi: 10.1371/annotation/b41766f2-c23d-455e-8d6e-e4bce5ae1d80. doi: 10.1371/journal.pone.0068117). PLoS One 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.LaPointe V.L.S., Verpoorte A., Stevens M.M. The changing integrin expression and a role for integrin β8 in the chondrogenic differentiation of mesenchymal stem cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0082035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai N.S., Yu H.C., Huang Tseng H.Y., Hsu C.W., Huang H Bin, Lu M.C. Increased serum levels of brain-derived neurotrophic factor contribute to inflammatory responses in patients with rheumatoid arthritis. Int J Mol Sci. 2021;22:1–15. doi: 10.3390/IJMS22041841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horne B.D., Rasmusson K.D., Alharethi R., Budge D., Brunisholz K.D., Metz T., et al. Genome-wide significance and replication of the chromosome 12p11.22 locus near the PTHLH gene for peripartum cardiomyopathy. Circ Cardiovasc Genet. 2011;4:359–366. doi: 10.1161/CIRCGENETICS.110.959205. [DOI] [PubMed] [Google Scholar]
- 52.Charaka Samhita of Agnivesha, Sutrasthaan; Mahachatuspada adhyaya; Chapter10, verse 11, Varanasi: Chaukhamba Sanskrit series. 2002. pp. 197–198. [Google Scholar]
- 53.Madhava Nidana of Madhavakara, Amavatanidana [Chapter 25], verse 1, Varanasi: Chaukhambha Orientalia seventh edition. 2005. p. 95. [Google Scholar]
- 54.Madhava Nidana of Madhavakara, Amavatanidana [Chapter 25], verse 11, Chaukhambha Orientalia 7th ed. 2005. p. 96. [Google Scholar]
- 55.Charaka Samhita of Agnivesha, Vimanasthana; Raganikavimana adhyaya; Chapter 6, verse 12, Varanasi. Chaukhamba Sanskrit series; 2003. p. 189. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.