Abstract
The intestinal epithelium represents the most regenerative tissue in the human body, located in proximity to the dense and functionally diverse microbial milieu of the microbiome. Episodes of tissue injury and incomplete healing of the intestinal epithelium are a prerequisite for immune reactivation and account for recurrent, chronically progressing phenotypes of inflammatory bowel diseases (IBD). Mitochondrial dysfunction and associated changes in intestinal epithelial functions are emerging concepts in the pathogenesis of IBD, suggesting impaired metabolic flexibility of epithelial cells affects the regenerative capacity of the intestinal tissue. Next to rendering the intestinal mucosa susceptible to inflammatory triggers, metabolic reprogramming of the epithelium is implicated in shaping adverse microbial environments. In this review, we introduce the concept of “metabolic injury” as a cell autonomous mechanism of tissue wounding in response to mitochondrial perturbation. Furthermore, we highlight epithelial metabolism as intersection of microbiome, immune cells and epithelial regeneration.
Introduction - from energy deficiency to chronic inflammation
Inflammatory bowel diseases (IBD) are a paradigm for the complex interplay of gene-environment interactions in the development and progression of immune-mediated pathologies, and together with metabolic disorders, their incidence and prevalence are increasing, following a pattern of industrialization1,2. Crohn’s disease and ulcerative colitis are the two main clinical phenotypes3 and dysbiotic changes of the intestinal microbiome emerged as a common link between host genetic susceptibility (>240 variant alleles)4,5 and environmental cues6,7. Crohn’s disease is characterized by discontinuous and transmural inflammation predominantly affecting the ileo-colonic part of the intestine, while ulcerative colitis is restricted to the colonic mucosa. These anatomically and functionally distinct areas of the intestinal tract create spatially adapted microbial habitats, likely contributing to the heterogeneity of disease phenotypes in IBD. The intestinal epithelium represents the frontline of the complex pathogenesis, lying at the interface of luminal inflammatory triggers such as the microbiome and host immune cells, and a breach of this well-structured barrier is suggested as cornerstone of chronic inflammation. Consequently, episodes of tissue injury and incomplete healing of the intestinal epithelium are a prerequisite for immune reactivation and account for recurrent, chronically progressing phenotypes of IBD. Injury-associated stem cells are imperative in orchestrating tissue regeneration, and dynamic adaptations of mitochondrial metabolism in the intestinal stem cell (ISC) niche are essential to ensure tissue homeostasis. Thus, these results support the concept in which a reduced metabolic flexibility of IECs affects the regenerative capacity of the epithelium and renders the intestinal mucosa towards increased susceptibility to inflammatory triggers8. Mitochondria are increasingly recognized as site of microbial signal-integration, and microbiome-derived metabolic signals emerge as an important player in determining the ability for mucosal healing9–11. Intermittent flares of mucosal inflammation are associated with rapid individual changes in the microbiome of Crohn’s disease patients, and relapsing as well as remitting Crohn’s disease phenotypes are transmissive via fecal transfer in germ-free mouse models12, supporting the hypothesis that functional alterations in the microbiome not only contribute to the progression but also to the remission of disease.
W.E. Roediger proposed in 1980 that ulcerative colitis is an energy deficiency disease of the intestinal epithelium defined by diminished butyrate oxidation leading to alterations in mucus synthesis, and crypt cell maturation13. In the inflamed tissue microenvironment of IBD patients, infiltrating immune cells, together with the energy requirements of resident epithelial and stroma cells limit the available oxygen14, and together with a reduced blood supply, these changes contribute to hypoxic conditions in chronic inflammation15. Interestingly, mitochondrial dysfunction and associated changes in intestinal epithelial functions are suggested as early event in the pathogenesis of IBD, preceding inflammatory tissue aberrations8,16–21. In particular, impaired mitochondrial function in intestinal epithelial cells (IEC) is associated with reduced stemness and Paneth cell dysfunction18,20,22. However, even under inflammation-associated hypoxia, the remaining oxygen in the intestinal tissue is sufficient to support mitochondrial oxidative phosphorylation (OXPHOS)15, and these changes cannot explain the complex and early (before onset of histological inflammation) alterations in epithelial metabolism observed in intestinal inflammation13,18,20. It seems an intriguing hypothesis that alterations in epithelial cell oxidative metabolism based on disturbed mitochondrial functions is not only a consequence, but rather a causal factor in the development of chronic intestinal inflammation23–25. First hints towards this idea came from genome-wide association studies mapping IBD genetic risk loci to mitochondrial function-associated genes26–28, implying mitochondrial disturbances as underlying mechanism in IBD pathogenesis.
In this review, we introduce the concept of “metabolic injury” as a cell autonomous mechanism of organ and tissue wounding in response to mitochondrial perturbation (Fig. 1). By accounting for unresolved tissue injury, metabolic injury might play a key role in the pathogenesis of inflammatory and tumorigenic disorders of the digestive tract. Furthermore, we highlight epithelial metabolism as intersection of microbiome, immune cells and epithelial regeneration.
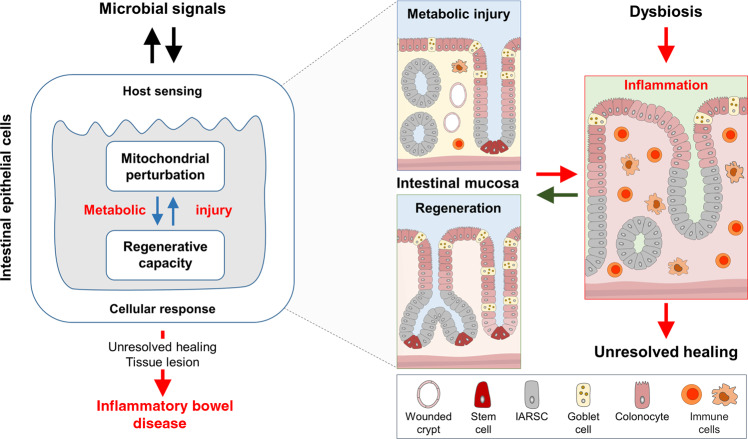
Fig. 1. The novel concept of metabolic injury as underlying mechanism of recurrent inflammation in patients with inflammatory bowel diseases (IBD).
Mitochondrial perturbation of the colonic epithelium e.g., caused by a combination of genetic predisposition and environmental triggers results in tissue injury (referred to as metabolic injury). Unresolved metabolic injury and incomplete tissue regeneration take place at the intersection of mitochondrial stress signaling (UPRmt) and changes of epithelial metabolism driving microbial dysbiosis and chronic inflammation in patients with IBD. UPRmt mitochondrial unfolded protein responses, IARSC injury-associated regenerative stem cells.
Intestinal epithelial architecture and metabolism
The intestinal epithelium is a multicellular interface located in close proximity to a complex and dense microbial milieu. Simultaneously intestinal epithelial cells absorb nutrients, and form a physical and immune-mediated barrier against adverse components of the luminal environment. The intestinal epithelium represents the most regenerative tissue in the human body highlighted by the fact that self-renewal of this single cell-layered interface (30–40 m2) is completed every 3–5 days, most likely as a protective mechanism against injuries and infections. Under homeostatic conditions of self-renewal but also in response to injury, crypt base columnar cells (CBCs) expressing the leucine-rich repeat-containing G protein-coupled receptor 5 (Lgr5) feed the transit amplifying zone of the epithelium, and progenitor cells finally differentiate into mature secretory (e.g., Paneth cells, goblet cells, enteroendocrine cells) and absorptive cells (columnar shaped enterocytes)29. Olfactomedin 4 (Olfm4), is an alternative marker for highly proliferative intestinal stem cells, co-expressed with, but not restricted to Lgr5+ stem cells. Numbers of Olfm4 positive cells expands during mucosal healing processes30, and are implicated in colorectal cancer31. The Olfm4 gene shows an abnormal methylation and expression patterns in ulcerative colitis32, and encodes a putative secreted glycoprotein. Yet, Olfm4 is also involved in the regulation of mitochondrial respiration and cellular ATP level33. Paneth cells contribute to stem cell homeostasis and anti-microbial defence exclusively in the small intestine, while goblet cells are spread throughout the entire epithelial lining of the digestive tract, and specifically in the colon, these secretory cells produce a thick and well-structured mucus layer responsible for the sequestration of bacteria from the host34. Loss of Lgr5+ stem cells in response to severe (often transmural) injury requires additional regenerative capacities in the intestine, involving quiescent slow-cycling cells located at the +4 position in the crypt, de-differentiated epithelial cells and reserve stem cells35. A breach in barrier integrity is associated with inflammatory pathologies36, and requires immediate action to restore barrier function and to induce mucosal healing37.
Metabolism, long perceived as mere supplier of ATP, is increasingly appreciated to reflect and determine cellular phenotypes. The central metabolic pathways, including mitochondrial OXPHOS, glycolysis, tricarboxylic acid (TCA) cycle, pentose phosphate pathway, fatty acid oxidation, fatty acid synthesis and amino acid metabolism, are tightly interrelated and essentially contribute to the availability of biosynthetic precursors and energy. Beyond constituting “building blocks”, central components of intermediary metabolism are co-factors or co-substrates of chromatin-modifying enzymes, and metabolic enzymes are directly involved in the control of gene expression38. Thus, metabolism and gene expression employ a regulatory interface, and consequently, IEC differentiation requires distinct metabolic identities, characterized by highly regulated changes in mitochondrial activity. Crypt cells including the stem cell niche and transit amplifying cells mainly rely on glycolysis for ATP generation, whereas differentiation and maturation of intestinal epithelial cells is accompanied by increased dependence on mitochondrial OXOHOS to meet their energetic needs. This metabolic gradient along the crypt-villus axis and is reflected by the mitochondrial content of the cells, which also determines cellular levels of reactive oxygen species (ROS). The balance of ROS scavenger systems and ROS produced by the mitochondrial respiratory chain control the activation of mitochondria-dependent apoptotic cascades in senescent epithelial cells, hence mitochondrial functions steer and coordinate epithelial renewal and cell shedding39. Not only mitochondrial metabolism, but also mitochondrial proteostasis and the associated mitochondrial unfolded protein response (UPRmt) contribute to these processes and are involved in intestinal pathologies by controlling self-renewal and the proliferative capacity of the epithelium22,40. This is not only critical to maintain epithelial homeostasis, but also crucial during inflammation and for dedifferentiation processes associated with wound healing. Both requires a tight regulation of cell proliferation and cell death programs and hence, mitochondrial functions. Furthermore, mitochondrial metabolism is a driving force in the generation of wound-associated epithelial cells41 a cell type required for efficient barrier restoration upon injury. Thus, shifts in epithelial metabolism under inflammatory conditions might partly represent changes in IEC subtype composition and differentiation state and therefore, be secondary to inflammation. Vice versa, genetic risk factors affecting cellular metabolism might render particularly IECs sensitive to environmental triggers of inflammation, and thereby impair the regenerative capacity of the epithelium.
In the light of these findings, the functional plasticity of the intestinal epithelium and its regenerative response to injury are modulated or even controlled by mitochondrial metabolism, supporting the hypothesis that mitochondrial exhaustion contributes to the functional perturbation of the epithelium in IBD (referred to as metabolic injury).
Mitochondrial signaling in the epithelium
Mitochondria play a profound role as platforms sensing the cellular environment and eliciting appropriate responses to cope with physiological disturbances. For example, mitochondria contribute to inflammatory processes by production of ROS42, and mitochondrial DNA can act as damage-associated molecular pattern (DAMP), promoting inflammation through toll-like receptor 9-dependent mechanisms43. Yet, also mitochondrial metabolic activity itself and the associated signaling pathways constitute critical checkpoints for ensuring epithelial integrity in the intestine39. Unfolded protein responses (UPR) are cornerstones in the homeostatic regulation of organelle function44, first described for the endoplasmic reticulum and implicated in the pathogenesis of IBD45,46. Mitochondrial UPR is essential in maintaining cellular metabolism and function47, but under conditions of intestinal inflammation, the consequences of sustained mitochondrial stress in the epithelium are completely unclear. Highlighting the importance of mitochondrial proteostasis for intestinal homeostasis, overexpression of prohibitin 1, a mitochondrial chaperone for proteins belonging to the electron transport chain in intestinal epithelial cells48, or therapeutic delivery of prohibitin 1 via nanoparticles49 protected mice from chemically induced colitis. Vice versa, conditional deletion of the mitochondrial chaperone heat shock protein 60 (Hsp60) in intestine22 and in liver50 gave rise to the global concept that cell autonomous perturbation of mitochondrial function requires paracrine mediators to cause tissue injury. Using mouse models in which loss of Hsp60 causes UPRmt activation and subsequent mitochondrial dysfunction specifically in intestinal stem cells (ISC) or IECs, we illustrated the crucial role of mitochondrial metabolism for intestinal stemness, Paneth cell functionality and the proliferative capacity of IECs18,22,51. Deficiency in mitochondrial OXPHOS upon deletion of Hsp60 causes loss of Lgr5+ ISCs and emergence of non-granular, dysfunctional Paneth cells, accompanied by tissue aberrations reminiscent of mucosal wound healing18,22. Importantly, mitochondrial-dysfunction associated aberrances of the intestinal stem cell niche were not only observed in mouse models of intestinal inflammation, but also in a cohort of Crohn’s disease patients18. In this prospective patient cohort, presence of mitochondrial dysfunction-associated markers in the intestinal stem cell niche in non-inflamed tissue margins could stratify the risk of disease recurrence18, suggesting that metabolic perturbations in the crypt epithelium precede inflammatory tissue lesions.
A number of IBD-related genetic risk loci map to mitochondrial function-associated genes, supporting the idea that mitochondrial perturbations and limited metabolic flexibility sensitize the intestinal epithelium to additional insults26–28. For example, SLC22A5 encodes the carnitine transporter OCTN2 involved in mitochondrial fatty acid oxidation52. IRGM affects mitochondrial fission53, a process crucial for the dynamic adaptation of mitochondrial function to physiological cellular changes54. Another IBD risk allele involved in the regulation of PPIF (encoding Cyclophilin D)4 affects cell death mechanisms by controlling mitochondrial membrane potential and the mitochondrial permeability transition pore (MPTP)55. Mitophagy removes dysfunctional mitochondria and several genetic risk variants are associated with this cellular process. Among these are PARK756, SMURF157, LRRK258, but also prominent IBD-related genes including ATG16L159,60 and NOD261,62. Overall, a recent analysis found that 5% of IBD susceptibility genes identified have direct roles in regulating mitochondrial homeostasis56. Of note, also mitochondrial-encoded DNA polymorphisms are associated with ulcerative colitis63,64, and experiments using conplastic mouse strains (possessing identical nuclear DNA but distinct mitochondrial DNA) suggest that increased mitochondrial OXPHOS and ATP levels protect from experimental colitis65. In summary, changed expression of mitochondrial genes and proteins, mitochondriopathy, perturbed mitochondrial dynamics, mitochondrial dysfunction, and activation of mitochondrial stress signaling were observed in IBD patients39,66,67, and added new facets to the old “energy deficiency” hypothesis (reviewed in9,68). Consequently, mitochondrial metabolism and signaling including the UPRmt have been implicated in integrating nutrient- and microbiota-derived signals in the intestinal stem cell niche8, critically affecting the regenerative capacity of the epithelium under homeostatic and disease conditions.
Metabolic circuits between the microbiome and the intestinal epithelium
The human digestive tract harbors a complex array of microorganisms, including bacteria, archaea, viruses and fungi69. Bacteria colonize the compartmentalized gastrointestinal tract in a spatially structured manner following a gradient from the proximal to the distal part of the intestine, reaching highest density and functional diversity in the colon70. In comparison to the small intestine, the motility of the colon is substantially slowed down leading to a prolonged retention of luminal content (20–50 h) and the accumulation of biologically active metabolites71. In this context, the term microbiome describes the “theatre of activity” including the complex physio-chemical characteristics of the microbial communities within the niche shaped by the host72. Disruption of this microbiome-host symbiosis contributes to the initiation and progression of immune and metabolic diseases, such as IBD73, graft-versus-host diseases (GvHD)74, and type 2 diabetes75, underlining the proposition that microbe-host interactions are critically important for human health76.
Several pathogens and their toxins specifically target and disrupt mitochondrial function77, and vice versa, mitochondrial stress in IECs increases bacterial translocation78. Yet, the mucosal interface not only responds to infections, but also to non-pathogenic bacteria79–81. This finding seeded the idea that intestinal tissue homeostasis requires active engagement of multifaceted microbial (such as pattern recognition receptors)82 and chemical sensors83 (including olfactory receptors and purinergic receptors (ATP receptors)84–87) and implicated the microbiome not only contributing to IBD progression but also to confer protective mechanisms. A main paradigm in IBD pathogenesis is that mucosal tolerance towards the “normal” microbiota is lost, and interestingly, mitochondrial dysfunction impairs the ability of the intestinal epithelium to be tolerant to commensal bacteria62. On the other hand, the microbiome provides metabolic support for the epithelium, including butyrate13, lactate88, and purines89 (Fig. 2). Thus, the microbiome can modulate ISC niche function and host metabolism through direct contact or release of products/ metabolites90,91. Studies in germ-free mice demonstrated a profound effect of the microbiome on amino acid, glutathione and overall energy metabolism92,93, as well as on IEC-maturation and differentiation94,95. Of note, expression of the lactate and butyrate transporter solute carrier family 16 member 1 (SLC16A1/ MCT1) is reduced in ulcerative colitis, along with genes encoding enzymes of the mitochondrial β-oxidation pathway96, indicating that reduced availability of substrates might arise from bacterial alterations as well as cellular disturbances. Consequently, loss of metabolic circuits is associated with the development of chronic inflammation18,21,40,66,67,97 and implicate a mitochondrial (genetic) susceptibility-plus-microbiome function interaction in IBD pathogenesis. Intestinal epithelial mitochondria might be key to initiation of inflammatory processes, as they are integral to epithelial homeostasis and at the same time, are exposed to beneficial and detrimental luminal factors. In line, several microbiome-derived mediators have been identified that enhance epithelial regeneration by sustaining cellular energetics and are depleted under inflammatory conditions.
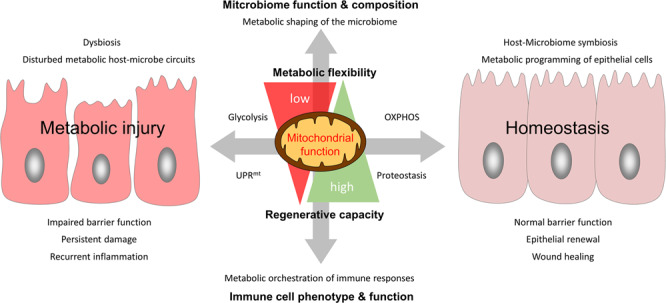
Fig. 2. Intestinal epithelial mitochondrial function as intersection of microbiome, immune cells and epithelial regeneration.
The intestinal epithelium senses the microbial environment via pattern recognition receptors and receptors sensing metabolites. Left: Under homeostatic conditions, intestinal epithelial cell (IEC) mitochondrial function contributes to the selection of a beneficial microbiome by maintaining low luminal oxygen concentration through oxidative phosphorylation (OXPHOS) and supporting production of antimicrobial peptides (AMPs). The microbiome provides metabolic support of epithelial cells by fermentation products such as short chain fatty acids (SCFAs), lactate, purines, and carnitines, thereby promoting cellular energetics and metabolic flexibility of IECs. As the ability to adapt mitochondrial functionality to the cellular demand determines the epithelial regenerative capacity, perturbations of mitochondrial metabolism result in metabolic injury of the epithelium (right). Shifting cellular metabolism away from OXPHOS to glycolysis (leading to elevated O2 levels) and impaired AMP production might result in dysbiosis, in turn aggravating the pro-inflammatory environment by reducing beneficial metabolites/ increasing disadvantageous microbial functions. IECs suffering from mitochondrial perturbation are exposed to high levels of reactive oxygen species (ROS) and activate mitochondrial stress signaling pathways such as mitochondrial unfolded protein response (UPRmt). Host genetics impact the selection of microbiota (left) and mitochondrial functions (right). IEC metabolism in conjunction with microbiota-derived metabolites likely controls mucosal immune cell recruitment and differentiation, thus orchestrating healing responses. Vice versa, immune cell-derived factors such as cytokines steer epithelial responses by targeting mitochondrial functions and metabolism. IL interleukin, TNF tumor necrosis factor.
Microbiome-derived metabolic signals and mucosal healing
Fecal stream diversion in patients with active Crohn’s disease and fecal microbial transplantation98–100 provided first clinical evidence to support the hypothesis that the intestinal microbial milieu contributes to disease recurrence101,102. Microbiome alterations are evident in the initiation and progression of disease activity, and considering the high energy demand of the inflamed mucosa, the (disturbed) interplay of epithelial metabolic functions and the microbial milieu might be of particular relevance in IBD67.
Disturbed metabolic circuits between microbiome and host, including butyrate13, carnitine103, purine89 and tryptophan metabolism104, are involved in the regulation of epithelial regenerative capacity and the development chronic inflammation, supporting the idea that mitochondrial perturbations contribute to IBD pathogenesis18,21,40,66,67,97. IECs are highly adapted to their anatomical location. Hence, small intestinal enterocytes predominantly utilize glucose and glutamine for energy generation, while microbiota-derived short-chain fatty acids (SCFA) represent the major energy source for colonocytes39.
There have been numerous reports indicating reductions in SCFA-producing bacteria in IBD105. However, substantial inter-individual variations are reported for SCFA levels106, and it is unlikely that the reductions in SCFAs observed under inflammatory conditions result in a primary energy deficiency of colonocytes. Under healthy conditions colonic concentrations of butyrate range from 10 to 20 mM107 (70–100 mM for the SCFAs acetate, propionate and butyrate combined108), and portal vein plasma concentrations range from 14–20 µM109,110, despite metabolization rates of 70–90% given for butyrate in colonocytes108,111. Plasma concentrations of acetate and propionate are even higher than those of butyrate110, indicating a spillover of SCFAs into the blood stream and suggesting SCFA availability to exceed the physiological energy demand of colonocytes.
Furthermore, SCFAs regulate PGC1α, a major transcription factor of mitochondrial biogenesis, and other genes involved in energy metabolism while promoting IEC growth112–114. Consistently, colonocytes from germ-free mice show a metabolic shift from OXPHOS to glycolysis93 and concomitantly, diminished cell cycle progression115. These effects were associated with perturbed pyruvate dehydrogenase (PDH) function, and could be rescued by supplementation of butyrate115. Of note, the metabolic effects were attributed to butyrate serving as energy substrate and not butyrate acting as inhibitor of HDAC activity93, which has been reported to inhibit proliferation of colonic ISCs via Foxo3116. Propionate and acetate are additional substrates for the TCA cycle and ameliorate metabolic and intestinal diseases by activating intestinal gluconeogenesis (like microbiome-derived succinate117)118 and by enhancing innate immune responses via free fatty acid receptor 2 (FFAR2)-signaling119.
Similarly, lactate serves as energy substrate fueling OXPHOS, thereby enhancing intestinal stemness120. Microbiome-derived lactate furthermore accelerates colonocyte turnover121 and by activating Gpr81 either on Paneth cells or on stromal cells, augments Wnt factor-production, resulting in Lgr5+ ISCs expansion and protection from acute intestinal damage88. Recently, also microbiome-derived purines have been identified as checkpoint metabolites, critically modulating cellular energetics, proliferation, and epithelial barrier function122. Purines like microbiome-derived hypoxanthine allow efficient biosynthesis of nucleotides and IECs can improve their energy balance by preferentially salvaging exogenous purines for ATP biosynthesis123. The need for nucleotide substrates increases substantially during inflammation, injury and wound healing, and consistently, disease severity correlated with loss of hypoxanthine in the epithelium in a chemically induced murine model of colitis123. In line, supplementation of hypoxanthine or selective colonization with purine-producing bacteria improved cellular energetics, promoted mucus generation and epithelial barrier function, resulting in enhanced wound healing and protection from colitis122. Under normal conditions, the intestinal microbiome produces and releases purines at high levels123, yet meeting the energy demand required for mucosal regeneration seems to be a bottle neck during active inflammation. Thus, supplementation of purines has been suggested as therapeutic approach to promote wound healing and remission in IBD patients122,123.
Metabolism of aromatic amino acids, particularly tryptophan, is another example for the tight interplay of microbiome and host metabolism124. Bacterial as well as host catabolism of the essential amino acid tryptophan gives rise to metabolites such as indoles and kynurenines, that are sensed by the aryl hydrocarbon receptor (Ahr)87,125. Ahr is a ligand-activated transcription factor located in the cytosol as well as inner mitochondrial membrane126 and impacts mitochondrial respiration and other cellular metabolic pathways125,127. Exerting various effects on epithelial and immune cells, Ahr-signaling orchestrates the key players of mucosal healing processes128. Activation of Ahr for example improves intestinal barrier function by modulating notch signaling129 and maintaining ISC homeostasis130,131. Concomitantly, Ahr activation induces regulatory T cells132, sustains IL-22 production in innate lymphoid cell (ILC) 3133–135 and enhances interleukin (IL) 10 receptor expression on epithelial cells136. These pathways are essential for intestinal wound healing and particularly important in the resolution of tissue damage128. Diets enriched in tryptophan137 or tryptophan-derived metabolites131,136 as well as enhanced bacterial tryptophan metabolism135,138 are shown to protect from inflammatory damage and accelerate wound healing. Conversely, microbiome-derived indole derivatives are diminished in IBD104, and this metabolic trait has been linked to reduced mucosal barrier integrity and disturbed immune balance under disease conditions124, suggesting these metabolites for prophylactic and therapeutic treatments131.
Reversely, microbiota-host interactions also contribute to disease susceptibility and progression. For instance, the bacterial metabolite hydrogen sulfide (H2S) is potentially harmful for IECs by inducing genotoxic damage and impairing mitochondrial OXPHOS139,140. In IBD, increased abundance of sulfate-reducing bacteria (i.e., H2S-producers) and in parallel, decreased expression of mitochondrial proteins involved in H2S- detoxification on host side have been reported67 and a recent integrated microbiota and metabolite profile-analysis linked Crohn’s disease activity to bacterial sulfur metabolism12 (Fig. 3). Thus, not only loss-of-function but also gain-of-function of the microbiome is crucial for IBD pathogenesis.
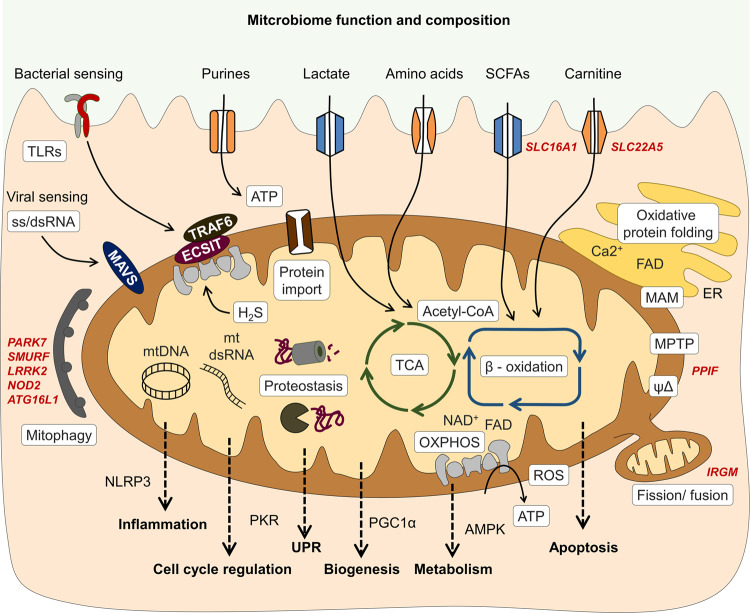
Fig. 3. Intestinal epithelial cell mitochondria serve as metabolic signaling platform translating microbiome-derived signals into mucosal responses.
Intestinal epithelial cells (IECs) sense the microbial environment via pattern recognition receptors including toll-like receptors (TLR) and take up diet and microbiota-derived metabolites. Activation of TLR signaling can impact the electron transport chain (ETC) and oxidative phosphorylation (OXPHOS) via TNF receptor associated factor (TRAF) 6 and ECSIT (Evolutionarily conserved signaling intermediate in Toll pathway), altering production of reactive oxygen species (ROS). Viral sensing involves mitochondrial antiviral-signaling protein (MAVS) and initiates inflammatory responses i.e., activating NFκB pathway. Microbiota-derived metabolites feed the tricarboxylic acid (TCA) cycle and mitochondrial beta oxidation or can enhance cellular energetics through salvage pathways (purines). Hydrogen sulfide (H2S) produced by bacteria can as electron donor for the ETC, but at high concentrations, inhibit Complex IV activity and other mitochondrial proteins. Mitochondria are embedded in an organelle network, in particular, an exchange of calcium, FAD, and ATP with the endoplasmic reticulum (ER) occurs at mitochondria-associated membranes (MAM) and is important for ER oxidative protein folding. Proteostasis, depending on protein import, chaperone activity, and proteases, is crucial to sustain mitochondrial functions, and disturbances of mitochondrial proteostasis are signaled by the mitochondrial unfolded protein response (UPRmt). Release of mitochondrial DNA and double-stranded (ds) mitochondrial RNA184 under stress conditions promotes inflammatory signaling and regulates cell cycle progression. Fission and fusion events as well as mitophagy are critical regulators of mitochondrial homeostasis and prevent accumulation of dysfunctional mitochondria and excess ROS production. Thus, mitochondria integrate environmental signals into metabolism, downstream employing various signaling pathways to contribute to cell fate decisions and determine cellular phenotypes. Gene names of known IBD risk variants involved in mitochondrial functions are given in dark red. AMPK AMP-activated protein kinase, NLRP3 NLR family pyrin domain containing 3, PGC1α Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, PKR double-stranded RNA-activated protein kinase, SCFAs short chain fatty acids.
Epithelial metabolic shaping of the microbiome
Several metabolic pathways are shared between microbiome and host, and common metabolites might originate from microbial or host metabolism. Hence, the question arises whether alterations in metabolite levels observed in the intestinal lumen under inflammatory conditions are due to microbiome alterations or mucosal dysfunction141. Tryptophan metabolism is a paradigm for shared and tightly entangled metabolic functions.
Intriguing aspects of host-microbiome interaction converging on tryptophan metabolism have been highlighted in Card9-deficient mice104, REG3A transgenic mice142 and Ido1-deficient mice135. Caspase recruitment domain family member 9 (CARD9) is a susceptibility gene for IBD and functions in the immune response against microorganisms. Host deficiency in Card9 or carrying CARD9 risk alleles results in a microbiome with reduced capability to metabolize tryptophan into Ahr ligands, resulting in increased colitis susceptibility in the mouse model104. Vice versa, mice transgenic for the human secreted antimicrobial peptide REG3A, display altered composition of their microbiota, favoring L-Ornithine-producing lactobacilli. In turn, microbiome-derived L-Ornithine promotes generation of the Ahr ligand L-kynurenine in IECs, increasing ILC3 cell numbers and intestinal mucus formation142. Similarly, increased tryptophan availability in mice deficient in tryptophan-metabolizing indoleamine 2,3 dioxygenase-1 (Ido1) leads to an expansion of intestinal lactobacilli which use tryptophan instead of sugar as energy source. Lactobacilli produce the Ahr ligand indole-3-aldehyde and thereby contribute to mucosal IL-22 expression and colonization resistance to the fungus Candida albicans135. These findings exemplify the bi-directional cross-talk between host genetics/host-derived factors, and microbiome composition/function, and its relevance to disease.
Despite enormous efforts in cataloguing microbiome alterations143, the functional specificity and cause of dysbiosis is not well understood144,145. An intriguing new concept suggests that metabolic reprogramming of the intestinal epithelium contributes to the dysbiotic adaptation of microbial communities146, highlighting the bi-directional metabolic interaction of microbiome and host at the intestinal interface.
The underlying hypothesis is that epithelial metabolism is integral to the mechanisms used by the host to shape the microbiome for its own benefit147. To achieve this, differentiated colonocytes are supposed to preferentially oxidize butyrate and other fatty acids and employ OXPHOS, with oxidation of SCFAs accounting for approximately 70% of oxygen consumption in colonocytes108. As a result of high epithelial oxygen consumption, the epithelial surface is kept in a hypoxic state, favoring a microbiota dominated by obligate anaerobic bacteria. Thereby, oxygen depletion fosters bacteria converting fiber into fermentation products and making an otherwise non-usable energy source accessible to the host146. This mutualism is disrupted upon injury or inflammation, potentially causing a shift in colonocyte metabolism away from fatty acid oxidation, thereby reducing epithelial oxygen consumption. Parallel to the increase in oxygen emanating from the epithelial surface, luminal availability of by-products of the host inflammatory response such as nitrate, favors the growth of facultative anaerobes from the family of Enterobacteriaceae146,148,149. As outlined before, metabolic alterations cause proportional shifts in epithelial subtypes of the epithelium. Crypt hyperplasia, a common feature of IBD150,151, results from an expansion of transit amplifying cells and concomitantly involves a reduction of terminally differentiated epithelial cells, such as goblet cells152. These changes can be regarded as an excessive epithelial repair program. In comparison to terminally differentiated colonocytes, metabolism of transit amplifying cells is characterized by low oxygen consumption153, resembling alterations observed in host epithelial metabolism under inflammatory conditions.
Consistently, metabolic reprogramming of IECs offers new promising treatment options for IBD patients, targeting both, epithelial restitution and restoration of host-microbiome symbiosis.
Epithelial – immune cell metabolic circuits
As mentioned above, tryptophan is catabolized by Ido1 in IECs as well as lamina propria immune cells, generating immunoregulatory kynurenine-based metabolites and resulting in tryptophan depletion. Under inflammatory conditions, such as IBD and GvHD but also in colorectal cancer (CRC), expression of Ido1 is strongly elevated154,155. Notably, the serum kynurenine to tryptophan ratio as a measure of Ido1 activity might be used as a disease biomarker156. The role of Ido1 and tryptophan metabolism, together with arginine metabolizing enzymes, has been extensively studied in the context of metabolic reprogramming of immune cells for balancing immunoregulatory and pro-inflammatory phenotypes157–159, thus being critical to wound healing, neoplasia, autoimmunity and the rejection of transplanted tissues155,160–162. Effects of Ido1 activity include promotion of T cell- mediated tolerance and antimicrobial effects154 and Ido1 might also act at the site of expression to decrease T-cell proliferation and survival, diminishing colonic inflammation and reducing disease severity155. Hence, the induction of Ido1 in intestinal epithelial cells under inflammatory conditions most likely represents an attempt to dampen immune cell activity and lower inflammation-associated tissue injury. In line, a subset of Paneth cells expressing Ido1 has recently been identified that might be involved in controlling immune responses towards the intestinal microbiome163. Comparing epithelial Ido1 expression with the extensive data available for the role of Ido1 in immune cells161 and considering the concept of competition for nutrients in the control of immune responses164, IECs potentially engage their metabolism to actively orchestrate mucosal immune responses. Determining the availability of metabolites in the lamina propria in conjunction with the microbiome, IECs might coordinate the recruitment of immune cells and guide immune cell functions to ensure efficient wound healing and barrier restitution upon mucosal injury.
Of note, immune cells and their cytokines reciprocally regulate IECs metabolism and function. For example, the pro-inflammatory cytokines TNF and IL-6, but also regulatory cytokines like IL-22 and IL-10 have been demonstrated to regulate mitochondrial metabolism165–167 as well as ISC activation and proliferation168–170. Moreover, Ido1 is induced by the pro-inflammatory cytokine interferon gamma, whereas anti-inflammatory cytokines including IL-10 and transforming growth factor (TGF) β inhibit Ido1, thus, activity of Ido1 in IECs reflects the balance between pro- and anti-inflammatory signals171. Collectively, this highlights IECs metabolism and mitochondria as interface of microbiome and immune cell-derived signals and underlines the potential of epithelial metabolism as therapeutic target.
Attempts to modify IEC metabolism, using P110, a small peptide inhibitor of mitochondrial fission172, or olaparib, a clinically applied PARP inhibitor improving mitochondrial function173, already succeeded in reducing chemically induced colitis in mice. Additionally, established drugs like 5-amino salicylic acid, that alters mitochondrial metabolism174, might already be efficient by targeting epithelial metabolism. Of note, metformin, first-line medication for the treatment of type 2 diabetes, strongly affects intestinal microbiome composition and function175, and in parallel, alters IEC metabolism176. With regard to the host-microbiome symbiosis outlined above, identification of specific microbiome-derived metabolites conferring to epithelial homeostasis could be a promising aim of future research.
Open questions and conclusion
Metabolic fitness emerged as new frontier in intestinal epithelial homeostasis and disease pathogenesis, and multiple extrinsic as well as intrinsic factors converge at this junction. Mitochondrial dysfunction is an early event in IBD pathogenesis, preceding inflammatory tissue aberrations8,18–21. Yet, it still needs to be clarified of whether these alterations are cause or consequence in response to injury and inflammation. These findings and insights gained in the field of host-microbiome symbiosis added new dimensions to the old hypothesis of IBD being an “energy deficiency disease” of the intestinal epithelium, evolving the idea of epithelial metabolism as central gatekeeper of barrier integrity and mucosal tolerance.
We propose that intrinsic defects in cellular metabolism cause epithelial dysfunction (metabolic injury) evoking attempts of the ISC niche to reconstitute normal tissue architecture and function. Metabolic injury may cause aberrant tissue responses reminiscent of intestinal reconstitution and, failure to resolve metabolic injuries leads to incomplete tissue healing and persistence of focal inflammatory lesions, predisposing IBD patients to remitting disease phenotypes or even tumor formation. The underlying causes of metabolic injury are most likely highly individual, and comprise an interrelated portfolio of genetic susceptibility and triggers from the luminal environment, including diet and the microbiome10,177–180. Functional adaptations of the epithelium and in particular, the ISC niche are initiated by extrinsic signals affecting cellular metabolism. It remains largely elusive which factors targeting mitochondrial function control epithelial cell regeneration in response to metabolic disruption, and how these signals contribute to either healing and tissue homeostasis, or favor chronic inflammation or tumorigenesis.
More research is needed to elucidate the metabolic program of the intestinal interface, and its pathogenic role in the etiology of inflammatory and tumorigenic disorders. Next to metabolites involved in host-microbiome metabolic communication discussed in this review, there is a vast number of additional candidate pathways like vitamin B181 and lipid metabolism182 that converge on epithelial function183. Thus, a better molecular understanding of signals and mediators in regenerative tissue responses and resolution of metabolic injuries is critical to develop clinically-relevant therapeutic interventions focusing on the enforcement of epithelial regenerative capacity by improving metabolic fitness, a novel strategy for combating intestinal diseases.
Acknowledgements
This review was supported by Collaborative Research Center (CRC) 1371 funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation – project number 395357507).
Author contributions
E.R. and D.H. wrote the manuscript
Funding
Open Access funding enabled and organized by Projekt DEAL.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ng SC, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2018;390:2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 2.Piovani D, et al. Environmental risk factors for inflammatory bowel diseases: an umbrella review of meta-analyses. Gastroenterology. 2019;157:647–659 e644. doi: 10.1053/j.gastro.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 3.Chang JT. Pathophysiology of Inflammatory Bowel Diseases. N. Engl. J. Med. 2020;383:2652–2664. doi: 10.1056/NEJMra2002697. [DOI] [PubMed] [Google Scholar]
- 4.Nasser J, et al. Genome-wide enhancer maps link risk variants to disease genes. Nature. 2021;593:238–243. doi: 10.1038/s41586-021-03446-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Lange KM, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yadav P, et al. Genetic factors interact with tobacco smoke to modify risk for inflammatory bowel disease in humans and mice. Gastroenterology. 2017;153:550–565. doi: 10.1053/j.gastro.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urbauer E, Rath E, Haller D. Mitochondrial metabolism in the intestinal stem cell niche-sensing and signaling in health and disease. Front. Cell Dev. Biol. 2020;8:602814. doi: 10.3389/fcell.2020.602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho, G. T. & Theiss, A. L. Mitochondria and inflammatory bowel diseases: toward a stratified therapeutic intervention. Annu. Rev. Physiol.84, 435–459 (2022). [DOI] [PMC free article] [PubMed]
- 10.Jackson DN, Theiss AL. Gut bacteria signaling to mitochondria in intestinal inflammation and cancer. Gut Microbes. 2020;11:285–304. doi: 10.1080/19490976.2019.1592421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng CW, et al. Ketone body signaling mediates intestinal stem cell homeostasis and adaptation to diet. Cell. 2019;178:1115–1131 e1115. doi: 10.1016/j.cell.2019.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metwaly A, et al. Integrated microbiota and metabolite profiles link Crohn’s disease to sulfur metabolism. Nat. Commun. 2020;11:4322. doi: 10.1038/s41467-020-17956-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roediger WE. The colonic epithelium in ulcerative colitis: an energy-deficiency disease? Lancet. 1980;2:712–715. doi: 10.1016/S0140-6736(80)91934-0. [DOI] [PubMed] [Google Scholar]
- 14.Colgan SP, Taylor CT. Hypoxia: an alarm signal during intestinal inflammation. Nat. Rev. Gastroenterol. Hepatol. 2010;7:281–287. doi: 10.1038/nrgastro.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colgan SP, Curtis VF, Campbell EL. The inflammatory tissue microenvironment in IBD. Inflamm. Bowel Dis. 2013;19:2238–2244. doi: 10.1097/MIB.0b013e31828dcaaf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santhanam S, et al. Mitochondrial electron transport chain complex dysfunction in the colonic mucosa in ulcerative colitis. Inflamm. Bowel Dis. 2012;18:2158–2168. doi: 10.1002/ibd.22926. [DOI] [PubMed] [Google Scholar]
- 17.Sifroni KG, et al. Mitochondrial respiratory chain in the colonic mucosal of patients with ulcerative colitis. Mol. Cell Biochem. 2010;342:111–115. doi: 10.1007/s11010-010-0474-x. [DOI] [PubMed] [Google Scholar]
- 18.Khaloian S., et al. Mitochondrial impairment drives intestinal stem cell transition into dysfunctional Paneth cells predicting Crohn’s disease recurrence. Gut69, 1939–1951 (2020). [DOI] [PMC free article] [PubMed]
- 19.VanDussen KL, et al. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology. 2014;146:200–209. doi: 10.1053/j.gastro.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson DN, et al. Mitochondrial dysfunction during loss of prohibitin 1 triggers Paneth cell defects and ileitis. Gut. 2020;69:1928–1938. doi: 10.1136/gutjnl-2019-319523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alula K. M., et al. Targeting mitochondrial damage as a therapeutic for ileal Crohn’s disease. Cells10, 1349 (2021). [DOI] [PMC free article] [PubMed]
- 22.Berger E, et al. Mitochondrial function controls intestinal epithelial stemness and proliferation. Nat. Commun. 2016;7:13171. doi: 10.1038/ncomms13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beltran B, et al. Mitochondrial dysfunction, persistent oxidative damage, and catalase inhibition in immune cells of naive and treated Crohn’s disease. Inflamm. Bowel Dis. 2010;16:76–86. doi: 10.1002/ibd.21027. [DOI] [PubMed] [Google Scholar]
- 24.Fukushima K, Fiocchi C. Paradoxical decrease of mitochondrial DNA deletions in epithelial cells of active ulcerative colitis patients. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;286:G804–G813. doi: 10.1152/ajpgi.00398.2003. [DOI] [PubMed] [Google Scholar]
- 25.Roediger WE, Nance S. Metabolic induction of experimental ulcerative colitis by inhibition of fatty acid oxidation. Br. J. Exp. Pathol. 1986;67:773–782. [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez A, et al. Association of the organic cation transporter OCTN genes with Crohn’s disease in the Spanish population. Eur. J. Hum. Genet. 2006;14:222–226. doi: 10.1038/sj.ejhg.5201529. [DOI] [PubMed] [Google Scholar]
- 27.Lamhonwah AM, Tein I. Novel localization of OCTN1, an organic cation/carnitine transporter, to mammalian mitochondria. Biochem Biophys. Res Commun. 2006;345:1315–1325. doi: 10.1016/j.bbrc.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 28.Yu X, et al. Association of UCP2 −866 G/A polymorphism with chronic inflammatory diseases. Genes Immun. 2009;10:601–605. doi: 10.1038/gene.2009.29. [DOI] [PubMed] [Google Scholar]
- 29.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 30.van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 31.Liu W, et al. Olfactomedin 4 deletion induces colon adenocarcinoma in Apc(Min/+) mice. Oncogene. 2016;35:5237–5247. doi: 10.1038/onc.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taman H, et al. Genome-wide DNA methylation in treatment-naive ulcerative colitis. J. Crohns Colitis. 2018;12:1338–1347. doi: 10.1093/ecco-jcc/jjy117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W, et al. Olfactomedin 4 deletion improves male mouse glucose intolerance and insulin resistance induced by a high-fat diet. Endocrinology. 2018;159:3235–3244. doi: 10.1210/en.2018-00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurashima Y, Kiyono H. Mucosal ecological network of epithelium and immune cells for gut homeostasis and tissue healing. Annu Rev. Immunol. 2017;35:119–147. doi: 10.1146/annurev-immunol-051116-052424. [DOI] [PubMed] [Google Scholar]
- 35.Rees WD, Tandun R, Yau E, Zachos NC, Steiner TS. Regenerative intestinal stem cells induced by acute and chronic injury: the saving grace of the epithelium? Front. Cell Dev. Biol. 2020;8:583919. doi: 10.3389/fcell.2020.583919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akdis, C. A. Does the epithelial barrier hypothesis explain the increase in allergy, autoimmunity and other chronic conditions? Nat. Rev. Immunol.21, 739–751 (2021). [DOI] [PubMed]
- 37.Quiros M, Nusrat A. Contribution of wound-associated cells and mediators in orchestrating gastrointestinal mucosal wound repair. Annu. Rev. Physiol. 2019;81:189–209. doi: 10.1146/annurev-physiol-020518-114504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Knaap JA, Verrijzer CP. Undercover: gene control by metabolites and metabolic enzymes. Genes Dev. 2016;30:2345–2369. doi: 10.1101/gad.289140.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rath E, Moschetta A, Haller D. Mitochondrial function - gatekeeper of intestinal epithelial cell homeostasis. Nat. Rev. Gastroenterol. Hepatol. 2018;15:497–516. doi: 10.1038/s41575-018-0021-x. [DOI] [PubMed] [Google Scholar]
- 40.Rath E, et al. Induction of dsRNA-activated protein kinase links mitochondrial unfolded protein response to the pathogenesis of intestinal inflammation. Gut. 2012;61:1269–1278. doi: 10.1136/gutjnl-2011-300767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyoshi H, et al. Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. EMBO J. 2017;36:5–24. doi: 10.15252/embj.201694660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West AP, et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyapati RK, et al. Mitochondrial DNA is a pro-inflammatory damage-associated molecular pattern released during active IBD. Inflamm. Bowel Dis. 2018;24:2113–2122. doi: 10.1093/ibd/izy095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grootjans J, Kaser A, Kaufman RJ, Blumberg RS. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016;16:469–484. doi: 10.1038/nri.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shkoda A, et al. Interleukin-10 blocked endoplasmic reticulum stress in intestinal epithelial cells: impact on chronic inflammation. Gastroenterol. 2007;132:190–207. doi: 10.1053/j.gastro.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 46.Kaser A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shpilka T, Haynes CM. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 2018;19:109–120. doi: 10.1038/nrm.2017.110. [DOI] [PubMed] [Google Scholar]
- 48.Theiss AL, et al. Prohibitin is a novel regulator of antioxidant response that attenuates colonic inflammation in mice. Gastroenterology. 2009;137:199–208. doi: 10.1053/j.gastro.2009.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Theiss AL, et al. Nanoparticle-based therapeutic delivery of prohibitin to the colonic epithelial cells ameliorates acute murine colitis. Inflamm. Bowel Dis. 2011;17:1163–1176. doi: 10.1002/ibd.21469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan D, et al. Kupffer cell-derived Tnf triggers cholangiocellular tumorigenesis through JNK due to chronic mitochondrial dysfunction and ROS. Cancer Cell. 2017;31:771–789 e776. doi: 10.1016/j.ccell.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waldschmitt N, et al. C/EBP homologous protein inhibits tissue repair in response to gut injury and is inversely regulated with chronic inflammation. Mucosal. Immunol. 2014;7:1452–1466. doi: 10.1038/mi.2014.34. [DOI] [PubMed] [Google Scholar]
- 52.Lin Z, et al. OCTN1 variant L503F is associated with familial and sporadic inflammatory bowel disease. J. Crohns Colitis. 2010;4:132–138. doi: 10.1016/j.crohns.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 53.Prescott NJ, et al. Independent and population-specific association of risk variants at the IRGM locus with Crohn’s disease. Hum. Mol. Genet. 2010;19:1828–1839. doi: 10.1093/hmg/ddq041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elrod JW, Molkentin JD. Physiologic functions of cyclophilin D and the mitochondrial permeability transition pore. Circ. J. 2013;77:1111–1122. doi: 10.1253/circj.CJ-13-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ho GT, et al. MDR1 deficiency impairs mitochondrial homeostasis and promotes intestinal inflammation. Mucosal. Immunol. 2018;11:120–130. doi: 10.1038/mi.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Franke A, et al. Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL) Nat. Genet. 2010;42:292–294. doi: 10.1038/ng.553. [DOI] [PubMed] [Google Scholar]
- 58.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/ng.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 60.Iida T, Onodera K, Nakase H. Role of autophagy in the pathogenesis of inflammatory bowel disease. World J. Gastroenterol. 2017;23:1944–1953. doi: 10.3748/wjg.v23.i11.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 62.Saxena A, Lopes F, Poon KKH, McKay DM. Absence of the NOD2 protein renders epithelia more susceptible to barrier dysfunction due to mitochondrial dysfunction. Am. J. Physiol. Gastrointest. Liver Physiol. 2017;313:G26–G38. doi: 10.1152/ajpgi.00070.2017. [DOI] [PubMed] [Google Scholar]
- 63.Rosa A, et al. Ulcerative colitis is under dual (mitochondrial and nuclear) genetic control. Inflamm. Bowel Dis. 2016;22:774–781. doi: 10.1097/MIB.0000000000000694. [DOI] [PubMed] [Google Scholar]
- 64.Dankowski T, et al. Male-specific association between MT-ND4 11719 A/G polymorphism and ulcerative colitis: a mitochondria-wide genetic association study. BMC Gastroenterol. 2016;16:118. doi: 10.1186/s12876-016-0509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bar F, et al. Mitochondrial gene polymorphisms that protect mice from colitis. Gastroenterology. 2013;145:1055–1063 e1053. doi: 10.1053/j.gastro.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 66.Haberman Y, et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat. Commun. 2019;10:38. doi: 10.1038/s41467-018-07841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mottawea W, et al. Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn’s disease. Nat. Commun. 2016;7:13419. doi: 10.1038/ncomms13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Denson LA. Mitochondrial networks: a new therapeutic target in colitis. Cell Mol. Gastroenterol. Hepatol. 2020;10:426–427. doi: 10.1016/j.jcmgh.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gilbert JA, et al. Current understanding of the human microbiome. Nat. Med. 2018;24:392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 71.Donia MS, Fischbach MA. HUMAN MICROBIOTA. Small molecules from the human microbiota. Science. 2015;349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berg G, et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome. 2020;8:103. doi: 10.1186/s40168-020-00875-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lloyd-Price J, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peled JU, et al. Microbiota as predictor of mortality in allogeneic hematopoietic-cell transplantation. N. Engl. J. Med. 2020;382:822–834. doi: 10.1056/NEJMoa1900623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reitmeier S, et al. Arrhythmic gut microbiome signatures predict risk of type 2 diabetes. Cell Host Microbe. 2020;28:258–272 e256. doi: 10.1016/j.chom.2020.06.004. [DOI] [PubMed] [Google Scholar]
- 76.Metwaly, A., Reitmeier, S. & Haller, D. Microbiome risk profiles as biomarkers for inflammatory and metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. (2022). 10.1038/s41575-022-00581-2. Online ahead of print. [DOI] [PubMed]
- 77.He D, et al. Clostridium difficile toxin A triggers human colonocyte IL-8 release via mitochondrial oxygen radical generation. Gastroenterology. 2002;122:1048–1057. doi: 10.1053/gast.2002.32386. [DOI] [PubMed] [Google Scholar]
- 78.Lewis K, et al. Enhanced translocation of bacteria across metabolically stressed epithelia is reduced by butyrate. Inflamm. Bowel Dis. 2010;16:1138–1148. doi: 10.1002/ibd.21177. [DOI] [PubMed] [Google Scholar]
- 79.Haller D, et al. Non-pathogenic bacteria elicit a differential cytokine response by intestinal epithelial cell/leucocyte co-cultures. Gut. 2000;47:79–87. doi: 10.1136/gut.47.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hooper LV, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 81.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 82.Burgueno JF, Abreu MT. Epithelial Toll-like receptors and their role in gut homeostasis and disease. Nat. Rev. Gastroenterol. Hepatol. 2020;17:263–278. doi: 10.1038/s41575-019-0261-4. [DOI] [PubMed] [Google Scholar]
- 83.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020;17:223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 84.Inami A., Kiyono H., Kurashima Y. ATP as a pathophysiologic mediator of bacteria-host crosstalk in the gastrointestinal tract. Int. J. Mol. Sci. 19, 2371 (2018). [DOI] [PMC free article] [PubMed]
- 85.Zietek T, Rath E. Inflammation meets metabolic disease: gut feeling mediated by GLP-1. Front Immunol. 2016;7:154. doi: 10.3389/fimmu.2016.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kotlo K, et al. The olfactory G protein-coupled receptor (Olfr-78/OR51E2) modulates the intestinal response to colitis. Am. J. Physiol. Cell Physiol. 2020;318:C502–C513. doi: 10.1152/ajpcell.00454.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marinelli L, et al. Identification of the novel role of butyrate as AhR ligand in human intestinal epithelial cells. Sci. Rep. 2019;9:643. doi: 10.1038/s41598-018-37019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee YS, et al. Microbiota-derived lactate accelerates intestinal stem-cell-mediated epithelial development. Cell Host Microbe. 2018;24:833–846 e836. doi: 10.1016/j.chom.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 89.Cader MZ, et al. FAMIN is a multifunctional purine enzyme enabling the purine nucleotide cycle. Cell. 2020;180:278–295 e223. doi: 10.1016/j.cell.2019.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ocansey DKW, et al. Mesenchymal stem cell-gut microbiota interaction in the repair of inflammatory bowel disease: an enhanced therapeutic effect. Clin. Transl. Med. 2019;8:31. doi: 10.1186/s40169-019-0251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Everard A, et al. Intestinal epithelial MyD88 is a sensor switching host metabolism towards obesity according to nutritional status. Nat. Commun. 2014;5:5648. doi: 10.1038/ncomms6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mardinoglu A, et al. The gut microbiota modulates host amino acid and glutathione metabolism in mice. Mol. Syst. Biol. 2015;11:834. doi: 10.15252/msb.20156487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Donohoe DR, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Macpherson AJ, McCoy KD. Standardised animal models of host microbial mutualism. Mucosal Immunol. 2015;8:476–486. doi: 10.1038/mi.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wichmann A, et al. Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host Microbe. 2013;14:582–590. doi: 10.1016/j.chom.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 96.De Preter V, et al. Impaired butyrate oxidation in ulcerative colitis is due to decreased butyrate uptake and a defect in the oxidation pathway. Inflamm. Bowel Dis. 2012;18:1127–1136. doi: 10.1002/ibd.21894. [DOI] [PubMed] [Google Scholar]
- 97.Delpre G, Avidor I, Steinherz R, Kadish U, Ben-Bassat M. Ultrastructural abnormalities in endoscopically and histologically normal and involved colon in ulcerative colitis. Am. J. Gastroenterol. 1989;84:1038–1046. [PubMed] [Google Scholar]
- 98.van Lier Y. F., et al. Donor fecal microbiota transplantation ameliorates intestinal graft-versus-host disease in allogeneic hematopoietic cell transplant recipients. Sci. Transl. Med.12, eaaz8926 (2020). [DOI] [PubMed]
- 99.Paramsothy S., et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet389, 1218–1228 (2017). [DOI] [PubMed]
- 100.Kootte RS, et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 2017;26:611–619 e616. doi: 10.1016/j.cmet.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 101.Rutgeerts P, et al. Effect of faecal stream diversion on recurrence of Crohn’s disease in the neoterminal ileum. Lancet. 1991;338:771–774. doi: 10.1016/0140-6736(91)90663-A. [DOI] [PubMed] [Google Scholar]
- 102.D’Haens GR, et al. Early lesions of recurrent Crohn’s disease caused by infusion of intestinal contents in excluded ileum. Gastroenterol. 1998;114:262–267. doi: 10.1016/S0016-5085(98)70476-7. [DOI] [PubMed] [Google Scholar]
- 103.Smith S. A., et al. Mitochondrial dysfunction in inflammatory bowel disease alters intestinal epithelial metabolism of hepatic acylcarnitines. J. Clin. Invest. 131, e133371 (2021). [DOI] [PMC free article] [PubMed]
- 104.Lamas B, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Caruso R, Lo BC, Nunez G. Host-microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 2020;20:411–426. doi: 10.1038/s41577-019-0268-7. [DOI] [PubMed] [Google Scholar]
- 106.Bloemen JG, et al. Short chain fatty acids exchange across the gut and liver in humans measured at surgery. Clin. Nutr. 2009;28:657–661. doi: 10.1016/j.clnu.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 107.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cook SI, Sellin JH. Review article: short chain fatty acids in health and disease. Aliment Pharm. Ther. 1998;12:499–507. doi: 10.1046/j.1365-2036.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- 109.van der Beek CM, et al. Hepatic uptake of rectally administered butyrate prevents an increase in systemic butyrate concentrations in humans. J. Nutr. 2015;145:2019–2024. doi: 10.3945/jn.115.211193. [DOI] [PubMed] [Google Scholar]
- 110.Peters SG, Pomare EW, Fisher CA. Portal and peripheral blood short chain fatty acid concentrations after caecal lactulose instillation at surgery. Gut. 1992;33:1249–1252. doi: 10.1136/gut.33.9.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Boets E, et al. Systemic availability and metabolism of colonic-derived short-chain fatty acids in healthy subjects: a stable isotope study. J. Physiol. 2017;595:541–555. doi: 10.1113/JP272613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lukovac, S. et al. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. mBio5, e01438-14 (2014). [DOI] [PMC free article] [PubMed]
- 113.Park JH, et al. Promotion of intestinal epithelial cell turnover by commensal bacteria: role of short-chain fatty acids. PLoS ONE. 2016;11:e0156334. doi: 10.1371/journal.pone.0156334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tan J, et al. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 115.Donohoe DR, Wali A, Brylawski BP, Bultman SJ. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PLoS ONE. 2012;7:e46589. doi: 10.1371/journal.pone.0046589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kaiko GE, et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;165:1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.De Vadder F, et al. Microbiota-Produced Succinate Improves Glucose Homeostasis via Intestinal Gluconeogenesis. Cell Metab. 2016;24:151–157. doi: 10.1016/j.cmet.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 118.De Vadder F, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 119.Fachi J. L., et al. Acetate coordinates neutrophil and ILC3 responses against C. difficile through FFAR2. J. Exp. Med. 217, jem.20190489 (2020). [DOI] [PMC free article] [PubMed]
- 120.Rodriguez-Colman MJ, et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature. 2017;543:424–427. doi: 10.1038/nature21673. [DOI] [PubMed] [Google Scholar]
- 121.Okada T, et al. Microbiota-derived lactate accelerates colon epithelial cell turnover in starvation-refed mice. Nat. Commun. 2013;4:1654. doi: 10.1038/ncomms2668. [DOI] [PubMed] [Google Scholar]
- 122.Lee JS, et al. Microbiota-sourced purines support wound healing and mucous barrier function. iScience. 2020;23:101226. doi: 10.1016/j.isci.2020.101226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee JS, et al. Hypoxanthine is a checkpoint stress metabolite in colonic epithelial energy modulation and barrier function. J. Biol. Chem. 2018;293:6039–6051. doi: 10.1074/jbc.RA117.000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu Y, Hou Y, Wang G, Zheng X, Hao H. Gut microbial metabolites of aromatic amino acids as signals in host-microbe interplay. Trends Endocrinol. Metab. 2020;31:818–834. doi: 10.1016/j.tem.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 125.Brinkmann V, Ale-Agha N, Haendeler J, Ventura N. The aryl hydrocarbon receptor (AhR) in the aging process: another puzzling role for this highly conserved transcription factor. Front. Physiol. 2019;10:1561. doi: 10.3389/fphys.2019.01561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Casado FL. The aryl hydrocarbon receptor relays metabolic signals to promote cellular regeneration. Stem Cells Int. 2016;2016:4389802. doi: 10.1155/2016/4389802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hwang HJ, et al. Mitochondrial-targeted aryl hydrocarbon receptor and the impact of 2,3,7,8-tetrachlorodibenzo-p-dioxin on cellular respiration and the mitochondrial proteome. Toxicol. Appl. Pharm. 2016;304:121–132. doi: 10.1016/j.taap.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Leoni G, Neumann PA, Sumagin R, Denning TL, Nusrat A. Wound repair: role of immune-epithelial interactions. Mucosal. Immunol. 2015;8:959–968. doi: 10.1038/mi.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu Z, et al. Aryl hydrocarbon receptor activation maintained the intestinal epithelial barrier function through Notch1 dependent signaling pathway. Int J. Mol. Med. 2018;41:1560–1572. doi: 10.3892/ijmm.2017.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Metidji A, et al. The environmental sensor AHR protects from inflammatory damage by maintaining intestinal stem cell homeostasis and barrier integrity. Immunity. 2018;49:353–362 e355. doi: 10.1016/j.immuni.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Scott SA, Fu J, Chang PV. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc. Natl Acad. Sci. USA. 2020;117:19376–19387. doi: 10.1073/pnas.2000047117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cervantes-Barragan L, et al. Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8alphaalpha(+) T cells. Science. 2017;357:806–810. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li S, Bostick JW, Zhou L. Regulation of innate lymphoid cells by aryl hydrocarbon receptor. Front. Immunol. 2017;8:1909. doi: 10.3389/fimmu.2017.01909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tan, J., Ni, D., Ribeiro, R. V., Pinget, G. V. & Macia, L. How changes in the nutritional landscape shape gut immunometabolism. Nutrients13, 823 (2021). [DOI] [PMC free article] [PubMed]
- 135.Zelante T, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 136.Lanis JM, et al. Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal. Immunol. 2017;10:1133–1144. doi: 10.1038/mi.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Islam J, et al. Dietary tryptophan alleviates dextran sodium sulfate-induced colitis through aryl hydrocarbon receptor in mice. J. Nutr. Biochem. 2017;42:43–50. doi: 10.1016/j.jnutbio.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 138.Wlodarska M, et al. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe. 2017;22:25–37 e26. doi: 10.1016/j.chom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Saint-Georges-Chaumet Y, Edeas M. Microbiota-mitochondria inter-talk: consequence for microbiota-host interaction. Pathog. Dis. 2016;74:ftv096. doi: 10.1093/femspd/ftv096. [DOI] [PubMed] [Google Scholar]
- 140.Ijssennagger N, van der Meer R, van Mil SWC. Sulfide as a mucus barrier-breaker in inflammatory bowel disease? Trends Mol. Med. 2016;22:190–199. doi: 10.1016/j.molmed.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 141.Roediger WE, Lawson MJ, Nance SH, Radcliffe BC. Detectable colonic nitrite levels in inflammatory bowel disease–mucosal or bacterial malfunction? Digestion. 1986;35:199–204. doi: 10.1159/000199368. [DOI] [PubMed] [Google Scholar]
- 142.Qi H, et al. Lactobacillus maintains healthy gut mucosa by producing L-Ornithine. Commun. Biol. 2019;2:171. doi: 10.1038/s42003-019-0424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schirmer M, Garner A, Vlamakis H, Xavier RJ. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019;17:497–511. doi: 10.1038/s41579-019-0213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017;14:573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Metwaly A, Haller D. Multi-omics in IBD biomarker discovery: the missing links. Nat. Rev. Gastroenterol. Hepatol. 2019;16:587–588. doi: 10.1038/s41575-019-0188-9. [DOI] [PubMed] [Google Scholar]
- 146.Litvak Y., Byndloss M. X., Baumler A. J. Colonocyte metabolism shapes the gut microbiota. Science362, eaat9076 (2018). [DOI] [PMC free article] [PubMed]
- 147.Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S. The evolution of the host microbiome as an ecosystem on a leash. Nature. 2017;548:43–51. doi: 10.1038/nature23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Winter SE, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lupp C, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 150.Arato A, Savilahti E, Paszti I. Crypt hyperplasia related to increased lymphocyte activation in the rectal mucosa of children with ulcerative colitis. Z. Gastroenterol. 1994;32:483–487. [PubMed] [Google Scholar]
- 151.Kilgore SP, Sigel JE, Goldblum JR. Hyperplastic-like mucosal change in Crohn’s disease: an unusual form of dysplasia? Mod. Pathol. 2000;13:797–801. doi: 10.1038/modpathol.3880138. [DOI] [PubMed] [Google Scholar]
- 152.Strugala V, Dettmar PW, Pearson JP. Thickness and continuity of the adherent colonic mucus barrier in active and quiescent ulcerative colitis and Crohn’s disease. Int J. Clin. Pr. 2008;62:762–769. doi: 10.1111/j.1742-1241.2007.01665.x. [DOI] [PubMed] [Google Scholar]
- 153.Fan YY, et al. A bioassay to measure energy metabolism in mouse colonic crypts, organoids, and sorted stem cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;309:G1–G9. doi: 10.1152/ajpgi.00052.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ciorba MA. Indoleamine 2,3 dioxygenase in intestinal disease. Curr. Opin. Gastroenterol. 2013;29:146–152. doi: 10.1097/MOG.0b013e32835c9cb3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jasperson LK, et al. Indoleamine 2,3-dioxygenase is a critical regulator of acute graft-versus-host disease lethality. Blood. 2008;111:3257–3265. doi: 10.1182/blood-2007-06-096081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gupta NK, et al. Serum analysis of tryptophan catabolism pathway: correlation with Crohn’s disease activity. Inflamm. Bowel Dis. 2012;18:1214–1220. doi: 10.1002/ibd.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dowling JK, et al. Mitochondrial arginase-2 is essential for IL-10 metabolic reprogramming of inflammatory macrophages. Nat. Commun. 2021;12:1460. doi: 10.1038/s41467-021-21617-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rodriguez PC, Ochoa AC, Al-Khami AA. Arginine metabolism in myeloid cells shapes innate and adaptive immunity. Front Immunol. 2017;8:93. doi: 10.3389/fimmu.2017.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mondanelli G, Iacono A, Allegrucci M, Puccetti P, Grohmann U. Immunoregulatory interplay between arginine and tryptophan metabolism in health and disease. Front. Immunol. 2019;10:1565. doi: 10.3389/fimmu.2019.01565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Szondi DC, Wong JK, Vardy LA, Cruickshank SM. Arginase signalling as a key player in chronic wound pathophysiology and healing. Front. Mol. Biosci. 2021;8:773866. doi: 10.3389/fmolb.2021.773866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mellor AL, Lemos H, Huang L. Indoleamine 2,3-dioxygenase and tolerance: where are we now? Front. Immunol. 2017;8:1360. doi: 10.3389/fimmu.2017.01360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mbongue JC, et al. The role of indoleamine 2, 3-dioxygenase in immune suppression and autoimmunity. Vaccines. 2015;3:703–729. doi: 10.3390/vaccines3030703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pflugler S, et al. IDO1(+) Paneth cells promote immune escape of colorectal cancer. Commun. Biol. 2020;3:252. doi: 10.1038/s42003-020-0989-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kedia-Mehta N, Finlay DK. Competition for nutrients and its role in controlling immune responses. Nat. Commun. 2019;10:2123. doi: 10.1038/s41467-019-10015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hahn WS, et al. Proinflammatory cytokines differentially regulate adipocyte mitochondrial metabolism, oxidative stress, and dynamics. Am. J. Physiol. Endocrinol. Metab. 2014;306:E1033–E1045. doi: 10.1152/ajpendo.00422.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hu M, Lin H, Yang L, Cheng Y, Zhang H. Interleukin-22 restored mitochondrial damage and impaired glucose-stimulated insulin secretion through down-regulation of uncoupling protein-2 in INS-1 cells. J. Biochem. 2017;161:433–439. doi: 10.1093/jb/mvw084. [DOI] [PubMed] [Google Scholar]
- 167.Ip WKE, Hoshi N, Shouval DS, Snapper S, Medzhitov R. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science. 2017;356:513–519. doi: 10.1126/science.aal3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bradford EM, et al. Epithelial TNF receptor signaling promotes mucosal repair in inflammatory bowel disease. J. Immunol. 2017;199:1886–1897. doi: 10.4049/jimmunol.1601066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jeffery V, Goldson AJ, Dainty JR, Chieppa M, Sobolewski A. IL-6 signaling regulates small intestinal crypt homeostasis. J. Immunol. 2017;199:304–311. doi: 10.4049/jimmunol.1600960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zha JM, et al. Interleukin 22 expands transit-amplifying cells while depleting Lgr5(+) stem cells via inhibition of wnt and notch signaling. Cell Mol. Gastroenterol. Hepatol. 2019;7:255–274. doi: 10.1016/j.jcmgh.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Badawy AA. Kynurenine pathway of tryptophan metabolism: regulatory and functional aspects. Int J. Tryptophan Res. 2017;10:1178646917691938. doi: 10.1177/1178646917691938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mancini NL, et al. Perturbed mitochondrial dynamics is a novel feature of colitis that can be targeted to lessen disease. Cell Mol. Gastroenterol. Hepatol. 2020;10:287–307. doi: 10.1016/j.jcmgh.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kovacs D, et al. Olaparib: a clinically applied PARP inhibitor protects from experimental Crohn’s disease and maintains barrier integrity by improving bioenergetics through rescuing glycolysis in colonic epithelial cells. Oxid. Med. Cell Longev. 2021;2021:7308897. doi: 10.1155/2021/7308897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lee JY, et al. High-fat diet and antibiotics cooperatively impair mitochondrial bioenergetics to trigger dysbiosis that exacerbates pre-inflammatory bowel disease. Cell Host Microbe. 2020;28:273–284 e276. doi: 10.1016/j.chom.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mueller NT, et al. Metformin affects gut microbiome composition and function and circulating short-chain fatty acids: a randomized trial. Diabetes Care. 2021;44:1462–1471. doi: 10.2337/dc20-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yang M, et al. Inhibition of mitochondrial function by metformin increases glucose uptake, glycolysis and GDF-15 release from intestinal cells. Sci. Rep. 2021;11:2529. doi: 10.1038/s41598-021-81349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Potdar AA, et al. Ileal gene expression data from Crohn’s disease small bowel resections indicate distinct clinical subgroups. J. Crohns Colitis. 2019;13:1055–1066. doi: 10.1093/ecco-jcc/jjz021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Liu TC, et al. Interaction between smoking and ATG16L1T300A triggers Paneth cell defects in Crohn’s disease. J. Clin. Invest. 2018;128:5110–5122. doi: 10.1172/JCI120453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Liu TC, et al. Western diet induces Paneth cell defects through microbiome alterations and farnesoid X receptor and type I interferon activation. Cell Host Microbe. 2021;29:988–1001 e1006. doi: 10.1016/j.chom.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gruber L, Lichti P, Rath E, Haller D. Nutrigenomics and nutrigenetics in inflammatory bowel diseases. J. Clin. Gastroenterol. 2012;46:735–747. doi: 10.1097/MCG.0b013e31825ca21a. [DOI] [PubMed] [Google Scholar]
- 181.Yoshii K, Hosomi K, Sawane K, Kunisawa J. Metabolism of dietary and microbial Vitamin B family in the regulation of host immunity. Front Nutr. 2019;6:48. doi: 10.3389/fnut.2019.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lamichhane S., et al. Linking gut microbiome and lipid metabolism: moving beyond associations. Metabolites11, 55 (2021). [DOI] [PMC free article] [PubMed]
- 183.Hennequart M, et al. The impact of physiological metabolite levels on serine uptake, synthesis and utilization in cancer cells. Nat. Commun. 2021;12:6176. doi: 10.1038/s41467-021-26395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kim Y, et al. PKR senses nuclear and mitochondrial signals by interacting with endogenous double-stranded RNAs. Mol. Cell. 2018;71:1051–1063 e1056. doi: 10.1016/j.molcel.2018.07.029. [DOI] [PubMed] [Google Scholar]