Abstract
Genes for subunits of acetyl coenzyme A carboxylase (ACC), which is the enzyme that catalyzes the first step in the synthesis of fatty acids in Lactobacillus plantarum L137, were cloned and characterized. We identified six potential open reading frames, namely, manB, fabH, accB, accC, accD, and accA, in that order. Nucleotide sequence analysis suggested that fabH encoded β-ketoacyl-acyl carrier protein synthase III, that the accB, accC, accD, and accA genes encoded biotin carboxyl carrier protein, biotin carboxylase, and the β and α subunits of carboxyltransferase, respectively, and that these genes were clustered. The organization of acc genes was different from that reported for Escherichia coli, for Bacillus subtilis, and for Pseudomonas aeruginosa. E. coli accB and accD mutations were complemented by the L. plantarum accB and accD genes, respectively. The predicted products of all five genes were confirmed by using the T7 expression system in E. coli. The gene product of accB was biotinylated in E. coli. Northern and primer extension analyses demonstrated that the five genes in L. plantarum were regulated polycistronically in an acc operon.
Strains of Lactobacillus plantarum form a group of industrially important lactic acid bacteria that are widely used as starters to stimulate malolactic fermentation in wine and lactic acid fermentation in meat and vegetables. We previously isolated L. plantarum from a fermented fish and rice food that is produced in the Philippines, and one of the strains, L137, which can hydrolyze starch and contained 15 plasmids, was studied in detail (32). Using a small plasmid, pLTK2, we developed a host-vector system for L. plantarum (15). In general, growth of lactic acid bacteria requires various amino acids, vitamins (including biotin), and lipids or fatty acids, and growth is stimulated in the presence of Tween 80. Although lactic acid bacteria are industrially important gram-positive bacteria, the mechanisms involved in lipid biosynthesis in these bacteria have not been well characterized. To our knowledge, only genes for biotin carboxylase in Lactococcus lactis have been sequenced (accession no. X76191). Recent studies suggest that fatty acids might act as signaling molecules that are important for cellular differentiation in gram-positive bacteria (1, 43). Acetyl coenzyme A (acetyl-CoA) carboxylase (ACC) is an enzyme that is essential for the first step in biosynthesis of fatty acids, and this enzyme belongs to the group of carboxylases that use biotin as a cofactor and bicarbonate as a source of the carboxyl group. ACC catalyzes the addition of CO2 to acetyl-CoA to generate malonyl-CoA. The ACC of both eukaryotic and prokaryotic organisms, such as Escherichia coli (17, 22, 23), Bacillus (27), Pseudomonas (4), Mycobacterium (30), Corynebacterium (14), and Anabaena sp. (9) strains, have been studied. In each of these organisms, the organization of the genes for each subunit of ACC is different even though the amino acid sequence around the active sites of each ACC is well conserved. We have been interested in the genes that are involved in fatty acid biosynthesis in L. plantarum, and previously we have described cloning of the accC gene for a subunit of biotin carboxylase (P. Kiatpapan, H. Kobayashi, M. Sakaguchi, H. Ono, Y. Kaneko, and Y. Murooka, Abstr. IX Congr. Bacteriol. Appl. Microbiol., p. 62, 1999).
In this report, we describe the complete nucleotide sequences, organization, and details of expression of the genes for β-ketoacyl acyl carrier protein (ACP) synthase III and ACC in L. plantarum.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
Fragments of the chromosomal DNA of L. plantarum L137 (32) were cloned in pT7Blue (Novagen, Madison, Wis.), pUC18/19 (49), pBluescript II KS+ (Stratagene, La Jolla, Calif.), and pTWV228 (Takara Shuzo Co. Ltd., Shiga, Japan). In T7 expression experiments, we used pVEX11 which had been provided by Y. Yamada (Yamaguchi University, Yamaguchi, Japan). L. plantarum L137 (32) and E. coli JM109 (49) and BL21(DE3) (39) were used as host strains. Lactic acid bacteria were grown at 30°C in De Man-Rogosa-Sharpe medium (Difco Laboratories, Detroit, Mich.). E. coli was grown in Luria-Bertani medium (39), M9 medium (39), or YT broth (39) at 37°C. E. coli L8 (11) containing a temperature-sensitive lesion in the biotin carboxyl carrier protein (BCCP), accB22(Ts) [fabE22(Ts)], and E. coli LA1-6 (24), which contains a temperature-sensitive lesion in the β subunit of the carboxyltransferase, accD6(Ts) fab-6(Ts) (E. coli Genetic Stock Center, Yale University, New Haven, Conn.), were cultured at 30°C.
Restriction enzymes and T4 DNA ligase, obtained from Takara Shuzo Co. Ltd. or from Toyobo Co. Ltd. (Osaka, Japan), were used as suggested by the suppliers. [35S]methionine was purchased from Amersham Pharmacia Biotech (Buckinghamshire, United Kingdom). Reagent grade chemicals were obtained from Nacalai Tesque Inc. (Kyoto, Japan) or Sigma Chemical Co. (St. Louis, Mo.).
Manipulation of DNA.
Chromosomal DNA was prepared from L. plantarum as described previously (15). Preparation of plasmid DNA and genetic manipulations of L. plantarum were performed as described by Kaneko et al. (15). Plasmid DNA was transferred to L. plantarum by electroporation with a Gene Pulser (Bio-Rad Laboratories, Hercules, Calif.). Preparation of DNA and genetic manipulation of E. coli were performed by standard methods (39).
Cloning of the acc genes.
Degenerate oligonucleotide primers BC1 (5′-CA[T/C]CCIGGITA[T/C]GGITT[T/C][T/C]TIGC-3′) and BC2 (5′-CICC[A/G]TG[T/C]TCIAC[T/C]TGIA[A/G]IC-3′) were designed by reference to the conserved amino acid sequences AIHPGYGFLA/SENAD/NFA and YFM/IEMNTRI/VQVEH, respectively. PCR amplification was performed with 2.5 U of Taq polymerase. The parameters used for PCR were as follows: 30 cycles of denaturation at 94°C for 30 s, annealing at 45°C for 45 s, and elongation at 72°C for 60 s after an initial 5-min denaturation step; and a final 10-min extension step. The PCR product was purified from an agarose gel and ligated to the pT7Blue vector. DNA-DNA hybridization was performed by using the standard protocol (42) with a digoxigenin (DIG) chemiluminescence detection kit (Roche Diagnostics GmbH, Mannheim, Germany). The DNA probe was labeled with DIG-High Prime (Roche Diagnostics) by using a DIG labeling kit from Boehringer (Mannheim, Germany).
DNA sequencing.
Nucleotide sequences of both strands were determined by the dideoxy chain termination method (40) with ABI PRISM dye terminator cycle sequencing Ready Reaction kits (PE Biosystems) and an AutoRead 1000 sequencing kit (Amersham Pharmacia Biotech, Uppsala, Sweden). Gaps in sequences were eliminated by using customized oligonucleotide primers. Sequences were assembled and analyzed by using the GENETYX-MAC program, version 8 (Software Development, Tokyo, Japan). A homology search was carried out with the BLAST (Basic Local Alignment Search Tool) program (2) by using the DNA Data Bank of Japan (DDBJ) database.
Synthesis of products of genes from L. plantarum in E. coli.
The fragment containing fabH and accBCDA was amplified by PCR with pACC as the template. Forward primer 5′-CATATGATGCCAACTTATAC-3′ and reverse primer 5′-GAATTCGAAGCGCATCCG-3′ were used to create an NdeI restriction site at the initiation codon of fabH and an EcoRI site downstream of the accA termination codon. The PCR was performed in an automated temperature-controlled system (PC700; Astec Co., Fukuoka, Japan). The 100-μl reaction mixture contained 10 ng of template, 50 pmol of each primer, 1 U of Taq polymerase, and each deoxynucleoside triphosphate at a concentration of 0.2 mM in 10 mM Tris-HCl buffer (pH 8.3). A 40-μl layer of mineral oil was used to prevent evaporation. The sample was first incubated at 95°C for 5 min and then subjected to 30 cycles of denaturation (20 s at 95°C), annealing (60 s at 60°C), and extension (90 s at 72°C). The PCR product was eluted from an agarose gel after electrophoresis with a GFX PCR DNA and Gel Band purification kit (Amersham Pharmacia Biotech) and cloned into the pT7Blue vector. The NdeI-EcoRI fragment was isolated from the resulting plasmid and subcloned into pVEX-11 expression vector that had been digested with NdeI and EcoRI. The resulting plasmid, pT7ACC, which included a 4.6-kb fragment, was used to transform E. coli BL21(DE3). Ampicillin-resistant colonies were selected, and cells from individual colonies were picked and cultured in M9 medium supplemented with glucose (0.4%) and amino acids (100 μg/ml each, except for methionine) until the absorbance at 600 nm (optical density at 600 nm [OD600]) reached 0.7. Synthesis of T7 RNA polymerase was induced by 30 min of incubation with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and then cells were treated with rifampin (200 μg/ml) for 30 min. Cellular proteins were labeled for 10 min with [35S]methionine (37 Bq/ml). Cell pellets were washed with 10 mM HEPES buffer (pH 7.4), and proteins were separated by sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel electrophoresis (PAGE) (20). The gel was fixed and dried overnight, and then labeled proteins were visualized by autoradiography.
Construction of pTACO-1.
The promoter region and the 5′ end of the open reading frame (ORF) of fabH (nucleotides [nt] 664 to 960) were amplified by PCR with primers 5′-TCTAGAGGTGTTTGCGCAGAAAGTCC-3′ (forward primer) and 5′-GATCCAACATATTGTCCCATGGCG-3′ (reverse primer). The amplified fragment obtained after PCR was cloned to the pT7Blue vector to obtain pTPac. The gene for cholesterol oxidase (choA′) from pCO117 (29) was amplified by PCR by using primers 5′-GCATGACTGCACAACAGC-3′ and 5′-CGACTAGTTGGTGCGTTCCTTC-3′ as the forward and reverse primers, respectively, and a SpeI site was generated at the 3′ end of the gene. An NdeI restriction site at the 5′ end was generated after the amplified fragment was subcloned into the pT7Blue vector. The NdeI-SpeI fragment containing the choA′ gene was ligated into plasmid pTPac that had been digested with NdeI and SpeI to generate pTACO-1. E. coli cells carrying plasmid pTACO-1 were used for preparation of total RNA.
Preparation of RNA.
The methods used for preparation of RNA from E. coli and primer extension were based on the method described by Kashima et al. (16), with some modifications. Cells were grown in 40 ml of 2× YT broth and incubated until the OD600 reached 0.7. Cells were harvested and suspended in 1 ml of lysis buffer, which contained 0.5% SDS, 20 mM sodium acetate (pH 5.5), and 1 mM EDTA. The lysate was extracted for 5 min at 65°C with 1 ml of phenol that had been saturated with 20 mM sodium acetate (pH 5.5). After centrifugation, the aqueous phase was extracted once with an equal volume of a mixture of chloroform and isoamyl alcohol (24:1, vol/vol). The RNA was precipitated in 3 volumes of absolute ethanol. The RNA pellet was treated with 20 U of DNase (RNase-free) and 25 U of RNase inhibitor in 50 μl of distilled water at 37°C for 15 min. Total RNA from L. plantarum L137 was prepared with a Fast RNA Blue kit (Bio 101, Inc., Calif.) by using a 40-ml culture with an OD600 of 0.4.
Northern blot analysis.
L137 RNA (approximately 20 μg) was electrophoretically separated on 1.2% (wt/vol) agarose gels containing 0.66 M formaldehyde. After electrophoresis, the gels were treated with 0.05 N NaOH for 30 min. The RNA was transferred to a nylon membrane (Hybond-N; Amersham) by standard methods (39). Specific DNA fragments used as hybridization probes were labeled with DIG-High Prime (Boehringer Mannheim, Indianapolis, Ind.). The DraI-NheI and NruI-HindIII DNA fragments (Fig. 1) were used as probes that were specific for fabH and accC, respectively. Prehybridization was performed at 42°C for 2 h in hybridization buffer containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 2% blocking solution, 0.1% lauryl sarcosine, 7.0% SDS, and 50% formamide in 50 mM phosphate buffer. The probe was heated to 95°C and then added to the prehybridization mixture at a final concentration of about 100 ng/ml. Hybridization was continued at 42°C overnight. The blots were washed once with 2× SSC–0.1% SDS for 10 min at room temperature and twice with 0.1× SSC–0.1% SDS for 20 min at 55°C. The probes were visualized on MXJB-1 film (Eastman Kodak, Rochester, N.Y.) by using a DIG-High Prime DNA labeling and detection starter kit II (Boehringer Mannheim) according to the manufacturer's instructions along with the chemiluminescence substrate CSPD.
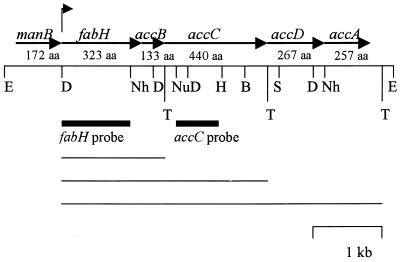
FIG. 1.
Restriction map and organization of the manB, fabH, accB, accC, accD, and accA genes in L. plantarum. The six ORFs identified are indicated by arrows. The arrow above the map indicates the major transcriptional start site identified. T, potential terminator site. The probes used for Northern blot analysis are indicated below the map by thick lines. Potential transcripts (from the start to the terminators) are indicated below the probes by thin lines. Selected restriction sites are indicated. Restriction site abbreviations: B, BalI; D, DraI; E, EcoRI; H, HindIII; Nh, NheI; Nu, NruI; S, SalI. aa, amino acids.
Primer extension analysis.
The primer extension reaction was performed as follows. A 0.2-pmol portion of fluorescein isothiocyanate (FITC)-labeled oligonucleotide primer for L. plantarum (5′-FITC-ATTATCAACAACGCGTCCCG-3′) or for E. coli (5′-FITC-ACAGATGCTGTTGTGCAGTC-3′) was mixed with 30 μg of RNA in 20 μl (total volume) of distilled water, and then the mixture was heated at 60°C for 1.5 h. After slow cooling to room temperature for 2 h, 6.4 μl of 5× transcriptase buffer, 5.0 μl of a mixture containing each deoxynucleoside triphosphate at a concentration of 2.5 mM, and 100 U of reverse transcriptase (Promega, Madison, Wis.) were added. After incubation at 37°C for 1 h and then at 42°C for 30 min, the transcript was precipitated in ethanol, redissolved in 10 to 20 μl of formamide dye (39), and then denatured by boiling for 2 min. The initiation site of transcription was determined by electrophoresis of the product of primer extension on a 6% polyacrylamide gel that contained 8 M urea next to the products of DNA sequencing reactions generated with the same primer and the ALF DNA sequencing system (Amersham Pharmacia Biotech).
Complementation test and biotinylation of AccB.
A DNA fragment containing the fabH and accBCDA genes was amplified by PCR performed with synthesized oligonucleotide primers in order to generate SacI and EcoRV sites at the 5′ and 3′ ends, respectively. The PCR fragment was subcloned into plasmid pTWV228 digested with SacI and HincII and gave rise to pWACC. Plasmid pWACC was transferred to E. coli L8 [accB22(Ts)] and E. coli LA1-6 [accD6(Ts)]. The transformants obtained were grown at 42°C to confirm that complementation of E. coli mutant strains L8 and LA1-6 by L. plantarum accB and accD, respectively, occurred. A biotinylation experiment was performed by using E. coli carrying pWACC. Cells were grown in Luria-Bertani medium until the OD600 reached 0.6, IPTG was added to a final concentration of 1 mM, and the cells were grown for 2 h. The cultured cells were harvested and lysed by boiling them for 2 to 3 min in sample buffer (39), and the proteins were separated by SDS-PAGE (ready gel; 10 to 20% polyacrylamide; Bio-Rad) and electrophoretically transferred to a nitrocellulose membrane. Biotinylating protein was detected by using streptavidin conjugated with alkaline phosphatase (Bethesda Research Laboratories Inc.) as described previously (4).
Nucleotide sequence accession number.
The nucleotide sequence reported here has been deposited in the DDBJ database under accession no. AB025973.
RESULTS
Isolation and cloning of acc genes.
To clone the genes that encode subunits of ACC, we designed oligonucleotides for use as primers by using the conserved sequences in biotin carboxylase (accC) genes of various organisms (4, 9, 17, 22, 27). An approximately 600-bp amplified product was obtained when the genomic DNA of L. plantarum L137 was used as the template. Sequence analysis confirmed that this product encoded a putative protein that was highly homologous to the biotin carboxylase subunit of ACC from various organisms. Since the organizations or orders of the genes for subunits of ACC differ markedly in different prokaryotes, we attempted to isolate the entire complement of ACC genes from L. plantarum L137. Southern hybridization analysis performed with the DIG-labeled 600-bp fragment as the probe and chromosomal DNA that had been digested with EcoRI, BamHI, SalI, or XhoI yielded a 5.5-kbp EcoRI fragment which was cloned into pBluescript KS+ to generate plasmid pACC. Plasmid pACC was used for analysis of nucleotide sequences.
Nucleotide sequence upstream and downstream of the accC gene.
Analysis of the 5.5-kbp sequence revealed six potential ORFs which were encoded on the same DNA strand (DDBJ accession no. AB025973). A database search identified these ORFs as homologs of the following genes in E. coli: manB, fabH, accB, accC, accD, and accA. Therefore, we tentatively gave the same designations to the genes from L. plantarum in that order (Fig. 1). Each of the ORFs had an ATG initiation codon, as well as a sequence with reasonable homology to the consensus ribosome-binding site of E. coli (41) and Lactobacillus (46).
ORFs upstream of the accC gene.
The first ORF encoded a putative protein containing 172 amino acids. There were high levels of homology at the amino acid level (61 to 67%) between this ORF and the manB gene that encodes the phosphotransferase IIB component of the phosphotransferase system (PTS) in E. coli (5) and Bacillus subtilis (18) and enzyme element II B (manB) of the mannose PTS in Lactobacillus curvatus (47). The manB gene was followed by a flanking region at nucleotides 756 to 918 and then by the fabH gene.
The putative fabH gene encoded a 34.0-kDa protein that exhibited significant homology (56 to 61%) to FabH from E. coli (45), B. subtilis (18), and Streptomyces glaucescens (44). The FabH protein corresponds to β-ketoacyl ACP synthase III, which catalyzes the first condensation reaction in the biosynthesis of fatty acids (10, 26). The consensus amino acid sequence AACAGF at the active site of the condensing enzyme was found in the FabH protein of L. plantarum at amino acids 112 to 117 (accession no. AB025973).
The third ORF encoded a putative 133-amino-acid protein that was 54 to 64% homologous to BCCP (encoded by accB), one of the subunits of ACC in E. coli (22), Pseudomonas aeruginosa (4), B. subtilis (27), and Anabaena sp. strain PCC7120 (9). The lysine residue that serves as a biotin-binding site in the biotin-dependent carboxylase in B. subtilis (27) was also found in AccB of L. plantarum at position 96 (accession no. AB025973). The amino acid sequence surrounding the biotin-binding site was MKLF and was identical to that in B. subtilis AccB.
Downstream of the accB gene of L. plantarum, we found the accC gene, which encoded a homolog of biotin carboxylase. This gene was immediately downstream of the termination codon of the accB gene. The deduced amino acid sequence of AccC was 79 to 84% homologous to the sequences of E. coli and B. subtilis. The amino acid sequence included a predicted ATP-binding site, GGGGKG, located at positions 161 to 166, with strong similarity to similar sequences in other organisms (4, 9, 22, 27). Several palindromic sequences, which served as transcriptional terminators, were found in the 3′ regions of accB and accC.
ORFs downstream of the accC gene.
We found two more ORFs downstream of the accC gene of L. plantarum. The amino acid compositions and amino acid sequences deduced from these two ORFs were very similar to those of the β and α subunits of the carboxyltransferase of E. coli, which are encoded by accD and accA. One base of the ORF of the putative accD sequence overlapped with the last codon of the accC gene. The putative accA gene of L. plantarum encoded a protein containing 257 amino acids. The amino acid residues at the predicted carboxybiotin-binding site (GGARMQE) in the β subunit of the carboxyltransferase and the putative acyl-CoA-binding site (SGGAL) in the α subunit were identical to those in E. coli (23) and B. subtilis (21). However, the genes for these two subunits in E. coli are located at sites on the E. coli chromosome different from the sites on the chromosome of L. plantarum. The gene for the α subunit is located downstream of the polC gene (4.3 min on the genetic map of E. coli), whereas the gene for the β subunit is located within an ORF with an unknown function designated dedB and usg (50 min on the genetic map of E. coli [23]). By contrast, the putative accD and accA genes of L. plantarum were located downstream of fabH, accB, and accC and overlapped. The accA gene of B. subtilis is also located downstream of the accD (or yttI) gene (21).
The 3′ nontranslated region of the accA gene of L. plantarum was followed by a palindromic structure that was bordered by five A residues. This region should be able to form a 20-bp imperfect stem-loop structure with a calculated ΔG value of 118.3 kJ/mol (25°C). These features are typical of a rho-independent signal for termination of transcription in E. coli (34).
Expression of the fabH, accB, accC, accD, and accA genes of L. plantarum.
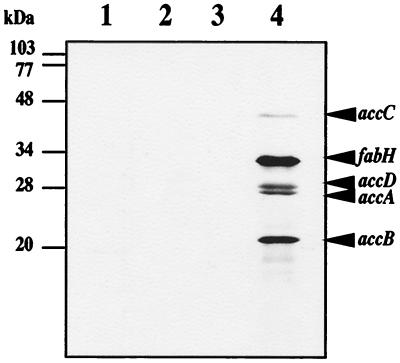
To confirm expression of proteins from the fabH-accBCDA gene cluster of L. plantarum L137, we subcloned the five ORFs into an expression vector, pVEX11, under control of the T7 promoter to generate pT7ACC. We transferred pT7ACC into E. coli BL21(DE3) and analyzed the gene products by SDS-PAGE. The estimated molecular masses of the proteins encoded by the fabH, accB, accC, accD, and accA genes were 35, 14, 47, 28, and 27 kDa, respectively. Five [35S]methionine-labeled proteins with molecular masses of 47, 34, 27, 25, and 17 kDa were recognized after SDS-PAGE (Fig. 2). The molecular mass of each protein, as estimated from its mobility during SDS-PAGE, was identical to that calculated from the deduced amino acid sequence, with one exception. AccB yielded a slightly larger molecular mass after SDS-PAGE than the molecular mass calculated from the amino acid sequence. Thus, in E. coli, the fabH, accB, accC, accD, and accA genes from L. plantarum were overexpressed from a single operon under control of the T7 promoter.
FIG. 2.
Expression of products of the L. plantarum fabH, accB, accC, accD, and accA genes under the T7 promoter. Proteins were separated by electrophoresis on an SDS-polyacrylamide gel, and [35S]methionine-labeled proteins were visualized by autoradiography. Cells were prepared from strain BL21 carrying pVEX11 (control) (lanes 1 and 2) or pT7ACC (lanes 3 and 4). Lanes 1 and 3, no IPTG induction; lanes 2 and 4, IPTG induction. The arrowheads indicate the positions of the expression products obtained. The corresponding genes coding for the proteins are indicated. The numbers on the left indicate the molecular masses of the standards.
Transcription of fabH and accBCDA.
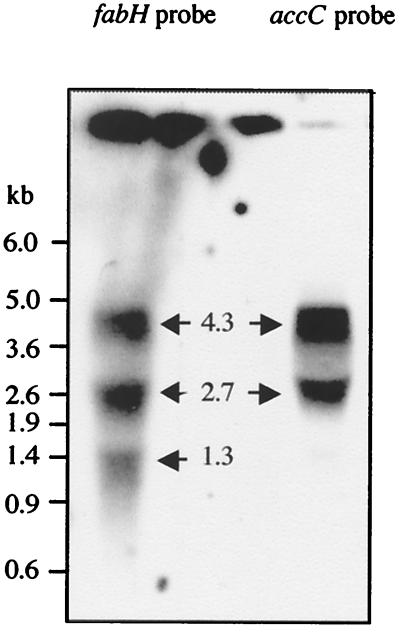
To confirm that cotranscription of the fabH and accBCDA genes occurred in L. plantarum, RNA blot hybridization was studied. Hybridization was performed by using logarithmically growing strain L137 cells with either a probe specific for fabH or a probe specific for accC (Fig. 1). Autoradiography results revealed that at least three species of RNA were transcribed. Transcripts that were 4.3, 2.7, and 1.3 kb long (Fig. 3) were observed; the 4.3-kb transcript hybridized to the two probes, and its size was identical to that of the theoretical transcript from the site of initiation of transcription of mRNA for fabH to a predicted termination site for accA which is located 12 bp downstream from the TTA termination codon of accA (Fig. 1). The 2.7-kb transcript bound to the two probes, whereas the 1.3-kb transcript bound to the fabH probe but not to the accC probe, indicating that the 2.7- and 1.3-kb transcripts may start at the initiation site in the fabH gene and may stop at the predicted termination sites in the accC and accB genes (Fig. 1), respectively, although their functions are unknown.
FIG. 3.
Northern blot analysis of transcripts from the fabH-accBCDA regions. Total RNA was isolated from L. plantarum L137. Two probes specific for fabH (DraI-NheI DNA fragment in fabH) and accC (NruI-HindIII DNA fragment in accC) were used. The sizes of mRNA species that hybridized with the probes (arrows) are indicated. The positions of size standards are indicated on the left.
Analysis of the promoter region upstream of the fabH gene.
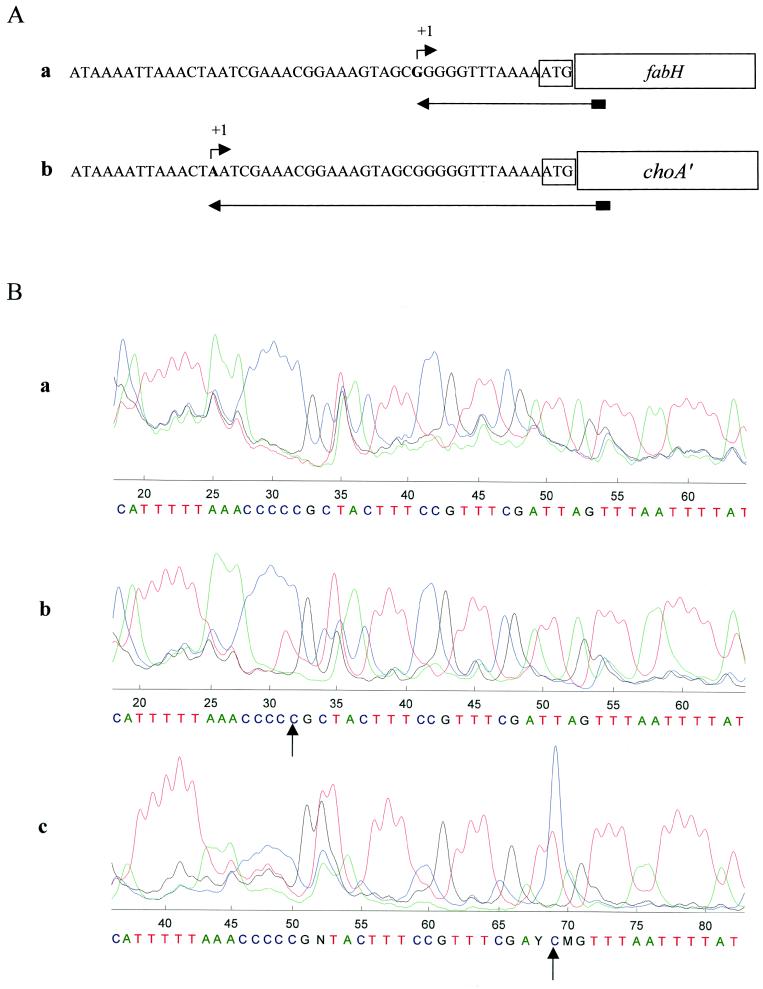
Since the genes fabH and accBCDA genes were separated from the first ORF (manB), we searched for a potential promoterlike sequence that resembled the −35 and −10 sequences of E. coli and lactobacilli (7). A −35 sequence (TTGACG) from nt 854 to 859 and two potential −10 sequences from nt 876 to 881 (TAAAAT) and from nt 878 to 882 (AAAATT) were very similar to −35 and −10 sequences. A −35 sequence (TTTTAT) from nt 867 to 872 and a −10 sequence (TCGAAA) from nt 890 to 895 were similar to the sequences of promoters in lactobacilli (8). To identify the site of initiation of transcription in the putative promoter region, we performed a primer extension analysis with total RNA prepared from L. plantarum L137. We identified the initiation site by comparing the data obtained from nucleotide sequencing of the products of the primer extension reaction with the results of a thymidine terminating reaction (Fig. 4Bb). The +1 site guanine residue was located 12 bp upstream of the initiation codon of fabH found in L. plantarum (nt 907, accession no. AB025973). This site is located at the site of a possible ribosome-binding site and 1 bp upstream, as reported for the rec operon of Streptococcus pneumoniae (33). We also performed a primer extension reaction analysis with RNA from E. coli(pTACO-1) and loaded the FITC-labeled primer together with the products of a cytosine terminating reaction. The +1 site adenine residue, which was 31 bp upstream of the initiation codon of fabH (nt 888, accession no. AB025973), was identified as the site of initiation of transcription (Fig. 4Bc). Differences between the sites of initiation of transcription in E. coli and lactic acid bacteria have been reported for several operons (3, 8, 33).
FIG. 4.
Mapping of the 5′ end of fabH by primer extension analysis by using a 5′-end FITC-labeled primer. See text for details. (A) Primer extensions with RNA from L. plantarum (a) and RNA from E. coli that harbored pTACO-1 (b). The arrows indicate the transcript determined by primer extension. +1 indicates the site of initiation of transcription. (B) Chromatography of the products of sequencing reactions. Green, blue, black, and red lines indicate the products of adenine, cytosine, guanine, and thymine sequencing reactions, respectively. The arrow indicates the site of initiation of transcription (+1) in the primer extension reaction. Reaction products were loaded together with the products of thymine (red) (b) or cytosine (blue) (c) sequencing reactions. (a) DNA sequence used as a control; (b) RNA prepared from L. plantarum L137; (c) RNA prepared from E. coli that harbored pTACO-1.
The promoter upstream of the fabH gene was able to mediate expression of a reporter enzyme, cholesterol oxidase (3 U/mg of protein), in L. plantarum that harbored a plasmid in which the promoter was fused to the choA′ gene (data not shown). Together, our results indicate that fabH and accBCDA is an operon in L. plantarum, which we designated the acc operon.
Functional analysis of accB and accD by complementation and biotinylation experiments.
We tested the ability of the L. plantarum accB and accD genes to complement E. coli L8 [accB22(Ts)] (11) and E. coli LA1-6 [accD6(Ts)] (24). Both mutants are viable at 30°C but not at higher temperatures. Plasmid pWACC carrying the fabH-accBCDA genes was transferred to E. coli L8 and LA1-6 at 30°C. The transformants were incubated at 42°C. The transformants of strains L8 and LA1-6 carrying pWACC could grow at 42°C, but the strains without the plasmid could not. These results indicated that the L. plantarum accB and accD genes were able to complement the BCCP in E. coli L8 and the β subunit of carboxyltransferase in E. coli LA1-6, respectively.
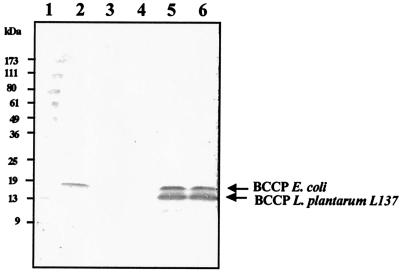
Furthermore, biotinylation of the gene product from L. plantarum accB was tested by performing a Western blot analysis with streptavidin conjugated with alkaline phosphatase. Two biotin-containing proteins were detected in E. coli carrying plasmid pWACC as bands at 14 and 17 kDa (Fig. 5, lane 5). The size of the 14-kDa protein corresponded to the size of L. plantarum BCCP estimated from the amino acid sequence. The 17-kDa protein is the E. coli biotin-binding BCCP. IPTG induction was observed only in the biotin-binding BCCP of L. plantarum (Fig. 5, lane 6). However, a low level of induction was observed due to the low copy number of plasmid pTWV228 used in construction of pWACC. When cells prepared from E. coli L8 grown at 30, 37, and 42°C were used, the biotinylating protein was observed only at 30°C, and this protein was 17 kDa long (Fig. 5, lanes 2 through 4). These results suggest that the L. plantarum BCCP has the biotinylation function in E. coli.
FIG. 5.
Western blot analysis of biotinylation of BCCP of L. plantarum L137 in E. coli. Lane 1 contained protein molecular mass standards. E. coli mutant strain L8 was grown at 30°C (lane 2), 37°C (lane 3), or 42°C (lane 4). E. coli carrying plasmid pWACC without IPTG induction (lane 5) or with IPTG induction (lane 6) was also included. The positions of the streptavidin-binding biotinylating BCCP in E. coli and in L. plantarum are indicated by arrows. The numbers on the left indicate the molecular masses of the standards.
DISCUSSION
ACC, the first enzyme in the fatty acid biosynthetic pathway, has been extensively studied by gene cloning and enzyme characterization in E. coli. The first reaction consists of two half-reactions: carboxylation of biotin which is bound to BCCP with bicarbonate, which is catalyzed by biotin carboxylase; and subsequent transfer of the carboxyl group from carboxybiotin to acetyl-CoA to obtain malonyl-CoA, which is catalyzed by carboxyltransferase. Moreover, it has been reported that the first step in the elongation of fatty acid chains is catalyzed by β-ketoacyl ACP synthase III, which is encoded by fabH (6, 10, 26).
Since the first enzymatic step in a metabolic pathway is often rate limiting, we attempted to isolate and characterize the genes for ACC in a lactic acid bacterium. In this study, we cloned and characterized the homologs of the manB, fabH, accB, accC, accD, and accA genes in L. plantarum.
The enzymes of the PTS in L. curvatus are encoded by four genes, manABCD. These proteins are similar to the PTS transporters EIIA, EIIB, EIIC, and EIID of the mannose class, and it has been proposed that EIIB, encoded by manB, might play a regulatory role in L. curvatus (47). However, the manB gene in L. plantarum seems to be organized differently than the manB gene in L. curvatus. The arrangements of genes involved in phosphorylation of sugar or PTSs are different in different species.
The fabH and accBCDA genes appear to form a single operon, although we found three sizes of transcripts in L. plantarum. In E. coli (22), B. subtilis (27), and P. aeruginosa (4), the accB and accC genes are transcribed together. The accD and accA genes in E. coli are located at different positions (4.3 and 50 min, respectively) (23), whereas in B. subtilis the accA and accD (yttI) genes are located in the rrnB-dnaB region (21). In P. aeruginosa, the accA and accD genes are neither immediately upstream nor immediately downstream of the accBC operon (4). The fabH gene encodes β-ketoacyl ACP synthase III, which catalyzes the condensation of acetyl-CoA with malonyl-ACP to initiate chain elongation in the biosynthesis of fatty acids. This gene is located in the plsX-fab gene cluster in E. coli (50). A study of the regulation of expression of the fab gene cluster in E. coli suggested that a promoter in the plsX coding sequence might allow read-through of downstream fab genes (35). Full-length and short transcripts were generated depending on the amount of ACP, a small protein containing fewer than 90 amino acids that plays a key role in lipid biosynthesis and is encoded by the acpP gene. The fabH gene in Streptomyces glaucescens is located in the fab gene cluster, in which the genes are arranged in the order fabD, fabH, fabC, fabB, which implies that there is a functional connection between the metabolism of fatty acids and biosynthesis of polyketide (44). In E. coli and Salmonella typhimurium, the fabH gene is organized in a cluster with the g30k, rmpF, and plsX genes and other fab genes (namely, fabD, fabG, acpP, and fabF) (31, 36, 45). In B. subtilis, the two putative fabH genes, fabH1 and fabH2 (equivalent to yjaX and yhfB), are also located outside the fab cluster (18, 28). The fabH1 (yjaX), yjaY, and fabH2 (yhfB) genes encode the subunits of 3-oxoacyl ACP synthase. In P. aeruginosa, the fabH gene is also not part of the major cluster of fab genes (19). Unlike the fabH gene in Streptomyces, E. coli, and B. subtilis, the fabH gene of L. plantarum is not located in the same transcript as the acpP or plsX gene. It seems likely that the promoter upstream of the fabH gene in L. plantarum is a major promoter responsible for transcription of the fabH and accBCDA genes together, although we found three sizes of transcripts. In the T7 RNA polymerase expression system, the L. plantarum fabH-accBCDA genes are cotranscribed as a single transcript (Fig. 2). The T7 RNA polymerase may read through two potential terminators. The Northern blot analysis in which the fabH and accC probes were used suggested that the transcripts started from the fabH promoter seem to stop at the termination signals of accB, accC, and accA (Fig. 3). Such heterologous transcripts of one operon have been found in gram-positive bacteria (12, 13, 48). However, we have not ruled out the possibility that there might be a minor promoter(s) located within the acc gene cluster.
The complementation test showed that the L. plantarum BCCP containing the Met-Lys-Leu biotinylation motif was able to complement the accB mutation in E. coli. The highly conserved MKM motif for biotinylation was studied previously in E. coli, and no biotinylation of MAK or KAM mutants was observed (37). Only the lysine residue in its native position in the hairpin turn, KKM or MKK, was found to be biotinylated. Our results, together with the biotinylation test results, also support this conclusion. The L. plantarum BCCP, which contains the MKL motif, was biotinylated in E. coli even though the L. plantarum BCCP lacks Thr-94, which was suggested to form a protruding polypeptide “thumb” (38). The putative L. plantarum accD gene was also able to complement the accD mutation in E. coli.
In bacteria, fatty acids are primarily the precursors of phospholipids rather than storage fuel, and thus ACC activity is coordinated with cell growth and division (1, 25, 26). The requirement for biotin and lipids in most lactobacilli suggests that expression of the acc operon might play an important role in growth of the bacteria. Lactic acid bacteria living under fatty conditions, such as meat or milk, might be expected to lose the ability to synthesize lipids or fatty acids. However, the bacteria retain these genes in an operon, and the genes are efficiently expressed to allow synthesis of fatty acids upon addition of biotin. The differences in gene organization and coregulation of expression of ACC subunits among microorganisms might reflect differences in control of growth under various environmental conditions.
ACKNOWLEDGMENTS
We thank Y. Yamada of Yamaguchi University for providing pVEX11 and Mary Berlyn of the E. coli Genetic Stock Center for providing E. coli mutant strains L8 and LA1-6.
This work was supported in part by Monbusho, Japan (Kiban B; grant 10556019). P.K. was supported by the Ronpaku Program of the Japan Society for the Promotion of Science.
REFERENCES
- 1.Allen S D, Siders J A, Riddell M J, Fill J A, Wegener W S. Cellular fatty acid analysis in the differentiation of Clostridium in the clinical microbiology laboratory. Clin Infect Dis Suppl. 1995;2:198–201. doi: 10.1093/clinids/20.supplement_2.s198. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Stephen F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araya-Kojima T, Ishibashi N, Shimamura S, Tanaka K, Takahashi H. Identification and molecular analysis of Lactococcus lactis rpoD operon. Biosci Biotechnol Biochem. 1995;59:73–77. doi: 10.1271/bbb.59.73. [DOI] [PubMed] [Google Scholar]
- 4.Best E A, Knauf V C. Organization and nucleotide sequences of the genes encoding the biotin carboxyl carrier protein and biotin carboxylase protein of Pseudomonas aeruginosa acetyl coenzyme A carboxylase. J Bacteriol. 1993;175:6881–6889. doi: 10.1128/jb.175.21.6881-6889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Choi K H, Heath R J, Rock C O. β-Ketoacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J Bacteriol. 2000;182:365–370. doi: 10.1128/jb.182.2.365-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Djordjevic G, Bojovic B, Banina A, Topissirovic L. Cloning of promoter-like sequences from Lactobacillus paracasei subsp. paracasei CG11 and their expression in Escherichia coli, Lactococcus lactis, and Lactobacillus reuteri. Can J Microbiol. 1994;40:1043–1050. doi: 10.1139/m94-165. [DOI] [PubMed] [Google Scholar]
- 8.Fremaux C, Ahn C, Klaenhammer T R. Molecular analysis of the lactacin F operon. Appl Environ Microbiol. 1993;59:3906–3915. doi: 10.1128/aem.59.11.3906-3915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gornicki P, Scappino L A, Haselkorn R. Genes for two subunits of acetyl coenzyme A carboxylase of Anabaena sp. strain PCC 7120: biotin carboxylase and biotin carboxyl carrier protein. J Bacteriol. 1993;175:5268–5272. doi: 10.1128/jb.175.16.5268-5272.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han L, Lobo S, Reynolds K A. Characterization of β-ketoacyl-acyl carrier protein synthase III from Streptomyces glaucescens and its role in initiation of fatty acid biosynthesis. J Bacteriol. 1998;180:4481–4486. doi: 10.1128/jb.180.17.4481-4486.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harder M E, Beacham I R, Cronan J E, Jr, Beacham K, Honeger J L, Silbert D F. Temperature-sensitive mutants of Escherichia coli requiring saturated and unsaturated fatty acid for growth: isolation and properties. Proc Natl Acad Sci USA. 1972;69:3105–3109. doi: 10.1073/pnas.69.11.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopwood D A, Bibb M J, Chater K F, Janssen G R, Malpartida F, Smith C P. Regulation of gene expression in antibiotic-producing Streptomyces. In: Booth I R, Higgins C F, editors. Regulation of gene expression—25 years on. Cambridge, United Kingdom: SGM; 1986. pp. 251–276. [Google Scholar]
- 13.Horii M, Ishizaki T, Paik S-Y, Manome T, Murooka Y. An operon containing the genes for cholesterol oxidase and a cytochrome P-450-like protein from a Streptomyces sp. J Bacteriol. 1990;172:3644–3653. doi: 10.1128/jb.172.7.3644-3653.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jager W, Peter-Wendisch P G, Kalinowski J, Puhler A. A Corynebacterium glutamicum gene encoding a two-domain protein similar to biotin carboxylases and biotin-carboxyl-carrier proteins. Arch Microbiol. 1996;166:76–82. doi: 10.1007/s002030050359. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko Y, Kobayashi H, Kiatpapan P, Nishimoto T, Napitupulu R, Ono H, Murooka Y. Development of a host-vector system for Lactobacillus plantarum L137 isolated from a traditional fermented food produced in the Philippines. J Biosci Bioeng. 2000;89:62–67. doi: 10.1016/s1389-1723(00)88051-2. [DOI] [PubMed] [Google Scholar]
- 16.Kashima Y, Nakajima Y, Nakano T, Tayama K, Koizumi Y, Udaka S, Yanagida F. Cloning and characterization of ethanol-regulated esterase genes in Acetobacter pasteurianus. J Biosci Bioeng. 1999;87:19–27. doi: 10.1016/s1389-1723(99)80003-6. [DOI] [PubMed] [Google Scholar]
- 17.Kondo H, Shiratsuchi K, Yoshimoto T, Masuda T, Kitazono A, Tsuru D, Anai M, Sekiguchi M, Tanabe T. Acetyl-CoA carboxylase from Escherichia coli: gene organization and nucleotide sequence of the biotin carboxylase subunit. Proc Natl Acad Sci USA. 1991;88:9730–9733. doi: 10.1073/pnas.88.21.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunst F, Ogasawara N, Moszer I, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 19.Kutchma A J, Hoang T T, Schweizer H P. Characterization of a Pseudomonas aeruginosa fatty acid biosynthetic gene cluster: purification of acyl carrier protein (ACP) and malonyl-coenzymeA:ACP transacylase (FabD) J Bacteriol. 1999;181:5498–5504. doi: 10.1128/jb.181.17.5498-5504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lapidus A, Galleron N, Sorokin A, Ehrlich S D. Sequencing and functional annotation of the Bacillus subtilis genes in 200 kb rrnB-dnaB region. Microbiology. 1997;143:3431–3441. doi: 10.1099/00221287-143-11-3431. [DOI] [PubMed] [Google Scholar]
- 22.Li S J, Cronan J E., Jr The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-coA carboxylase. J Biol Chem. 1992;267:855–863. [PubMed] [Google Scholar]
- 23.Li S J, Cronan J E., Jr The genes encoding the two carboxyltransferase subunits of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992;267:16841–16847. [PubMed] [Google Scholar]
- 24.Li S J, Rock C O, Cronan J E., Jr The dedB (usg) open reading frame of Escherichia coli encodes a subunit of acetyl-coenzyme A carboxylase. J Bacteriol. 1992;174:5755–5757. doi: 10.1128/jb.174.17.5755-5757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S J, Cronan J E., Jr Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first step of lipid biosynthesis. J Bacteriol. 1993;175:332–340. doi: 10.1128/jb.175.2.332-340.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magnuson K, Jackowski S, Rock C O, Cronan J E. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marini P, Li S J, Gardiol D, Cronan J E, de Mendoza D. The genes encoding the biotin carboxyl carrier protein and biotin carboxylase subunits of Bacillus subtilis acetyl coenzyme A carboxylase, the first enzyme of fatty acid synthesis. J Bacteriol. 1995;177:7003–7006. doi: 10.1128/jb.177.23.7003-7006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morbidoni H R, de Mendoza D, Cronan J E., Jr Bacillus subtilis acyl carrier protein is encoded in a cluster of lipid biosynthesis genes. J Bacteriol. 1996;178:4794–4800. doi: 10.1128/jb.178.16.4794-4800.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nomura N, Choi K P, Yamashita M, Yamamoto H, Murooka Y. Genetic modification of the Streptomyces cholesterol oxidase gene for expression in Escherichia coli and development of promoter-probe vectors for use in enteric bacteria. J Ferment Bioeng. 1995;79:410–416. [Google Scholar]
- 30.Norman E, De Smet K A, Stoker N G, Ratledge C, Wheeler P R, Dale J W. Lipid synthesis in mycobacteria: characterization of the biotin carboxyl carrier protein genes from Mycobacterium leprae and M. tuberculosis. J Bacteriol. 1994;176:2525–2531. doi: 10.1128/jb.176.9.2525-2531.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh W, Larson T J. Physical locations of genes in the me(ams)-rpk PlsX-fab/plsX-fab region of the Escherichia coli K-12 chromosome. J Bacteriol. 1992;174:7873–7874. doi: 10.1128/jb.174.23.7873-7874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olympia M, Fukuda H, Ono H, Kaneko Y, Takano M. Characterization of starch hydrolyzing lactic acid bacteria isolated from a fermented fish and rice food, Burong Isda, and its amylolytic enzyme. J Ferment Bioeng. 1995;80:193–197. [Google Scholar]
- 33.Pearce B J, Naughton A M, Campbel E A, Masure H R. The rec locus, a competence-induced operon in Streptococcus pneumoniae. J Bacteriol. 1995;177:86–93. doi: 10.1128/jb.177.1.86-93.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Platt T. Transcription termination and regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- 35.Podkovyrov S M, Larson T J. Identification of promoter and stringent regulation of transcription of the fabH, fabD and fabG genes encoding fatty acid biosynthetic enzymes of Escherichia coli. Nucleic Acids Res. 1996;24:1747–1752. doi: 10.1093/nar/24.9.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rawlings M, Cronan J E. The gene encoding Escherichia coli acyl carrier protein lies within a cluster of fatty acid biosynthetic genes. J Biol Chem. 1992;267:5751–5754. [PubMed] [Google Scholar]
- 37.Reche P, Li Y L, Fuller C, Eichhorn K, Perham R N. Selectivity of post-translational modification in biotinylated proteins: the carboxy carrier protein of the acetyl-CoA carboxylase of Escherichia coli. Biochem J. 1998;329:589–596. doi: 10.1042/bj3290589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts E L, Shu N, Howard M J, Broadhurst R W, Chapman-Smith A, Wallace J C, Morris T, Cronan J E, Jr, Perham R N. Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy. Biochemistry. 1999;38:5045–5053. doi: 10.1021/bi982466o. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shine J, Dalgarno L. The 3-terminal sequence of Escherichia coli 16S ribosomal RNA: complementary to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Southern E M. Detection of specific sequences among DNA fragment separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 43.Stuible H P, Wagner C, Andreou I, Huter G, Haselmann J, Schweizer E. Identification and functional differentiation of two type I fatty acid synthases in Brevibacterium ammoniagenes. J Bacteriol. 1996;178:4787–4793. doi: 10.1128/jb.178.16.4787-4793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Summer R G, Ari A, Shen B, Wessel W A, Hutchinson A R. Malonyl-coenzyme A:acyl carrier protein acyltransferase of Streptomyces glaucescens: a possible link between fatty acid and polyketide biosynthesis. Biochemistry. 1995;34:9389–9402. doi: 10.1021/bi00029a015. [DOI] [PubMed] [Google Scholar]
- 45.Tsay J-T, Oh W, Larson T J, Jackowski S, Rock C O. Isolation and characterization of the β-ketoacyl-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12. J Biol Chem. 1992;267:6807–6814. [PubMed] [Google Scholar]
- 46.Van de Guchte M, Kok J, Venema G. Gene expression in Lactococcus lactis. FEMS Microbiol Rev. 1992;88:73–92. doi: 10.1111/j.1574-6968.1992.tb04958.x. [DOI] [PubMed] [Google Scholar]
- 47.Veyrat A, Gorsalbes M J, Perez-Martinez G. Lactobacillus curvatus has a glucose transport system homologous to the mannose family of phosphoenolpyruvate-dependent phosphotransferase systems. Microbiology. 1996;142:3469–3477. doi: 10.1099/13500872-142-12-3469. [DOI] [PubMed] [Google Scholar]
- 48.Westpheling J M, Brawner M. Two transcribing activities are involved in expression of the Streptomyces galactose operon. J Bacteriol. 1989;171:1355–1361. doi: 10.1128/jb.171.3.1355-1361.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of M13 mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Cronan J E., Jr Transcriptional analysis of essential genes of the Escherichia coli fatty acid biosynthesis gene cluster by functional replacement with the analogous Salmonella typhimurium gene cluster. J Bacteriol. 1998;180:3295–3303. doi: 10.1128/jb.180.13.3295-3303.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]