Abstract
Microbial pathogens employ signaling systems through cyclic (di-) nucleotide monophosphates serving as second messengers to increase fitness during pathogenesis. However, signaling schemes via second messengers in Porphyromonas gingivalis, a key Gram-negative anaerobic oral pathogen, remain unknown. Here, we report that among various ubiquitous second messengers, P. gingivalis strains predominantly synthesize bis-(3′,5′)-cyclic di-adenosine monophosphate (c-di-AMP), which is essential for their growth and survival. Our findings demonstrate an unusual regulation of c-di-AMP synthesis in P. gingivalis. P. gingivalis c-di-AMP phosphodiesterase (PDE) gene (pdepg) positively regulates c-di-AMP synthesis and impedes a decrease in c-di-AMP concentration despite encoding conserved amino acid motifs for phosphodiesterase activity. Instead, the predicted regulator gene cdaR, unrelated to the c-di-AMP PDE genes, serves as a potent negative regulator of c-di-AMP synthesis in this anaerobe. Further, our findings reveal that pdepg and cdaR are required to regulate the incorporation of ATP into c-di-AMP upon pyruvate utilization, leading to enhanced biofilm formation. We show that shifts in c-di-AMP signaling change the integrity and homeostasis of cell envelope, importantly, the structure and immunoreactivity of the lipopolysaccharide layer. Additionally, microbe–microbe interactions and the virulence potential of P. gingivalis were modulated by c-di-AMP. These studies provide the first glimpse into the scheme of second messenger signaling in P. gingivalis and perhaps other Bacteroidetes. Further, our findings indicate that c-di-AMP signaling promotes the fitness of the residents of the oral cavity and the development of a pathogenic community.
Subject terms: Pathogens, Dentistry
Introduction
Periodontitis is a highly prevalent and destructive infectious, inflammatory disease of the tissues supporting the teeth that is induced by a synergistic community of virulent subgingival bacteria1,2. Among recognized pathogens, the Gram-negative anaerobe Porphyromonas gingivalis has been strongly implicated in the onset and development of periodontitis and several systemic diseases in adults upon producing an array of virulence factors, orchestrating dysbiotic inflammation, and disrupting host–microbial homeostasis even at low abundance2–12. Yet, how P. gingivalis can persist in the subgingival niche, induce ecological changes, and promote polymicrobial infection remain obscure. P. gingivalis has long been described as a highly proteolytic and asaccharolytic pathogen. It utilizes the type IX secretion system (T9SS) and T9SS cargo proteins, such as potent proteases known as gingipains, to provide proteinaceous nutrients while enabling the manipulation of immune components4,13–15. Our recent work revealed that the metabolic plasticity of P. gingivalis allows the consumption of exogenous pyruvate as an important source of carbon and energy16,17. Utilization of pyruvate significantly rewires the central metabolism of P. gingivalis toward de novo biosynthesis of phosphorylated and non-phosphorylated carbohydrates, promotes the expression of fimbrial adhesins, and subsequently enhances biofilm formation, co-colonization of P. gingivalis and other accessory species, and invasion16,17. Metabolomics data16 suggested that cyclic (di-) nucleotide monophosphates may broadcast metabolic signals in P. gingivalis. Still, the scheme of signaling through second messengers and their biological roles in this anaerobe and other Bacteroidetes remain largely unknown. Signaling systems via cyclic (di-) nucleotide monophosphate second messengers have been long described as fundamental regulatory mechanisms by which bacterial pathogens can promptly respond to specific stimuli and control appropriate biological outputs and pathoadaptive mechanisms, including phenotypic changes, the motility-sessility switch, host colonization, biofilm formation, virulence, stress tolerance, and antibiotic resistance18–21. Paradoxically, Chaudhuri et al.22 reported the presence of bis-(3′,5′)-cyclic di-guanosine monophosphate (c-di-GMP) signaling in P. gingivalis while pioneering works indicated that neither c-di-GMP metabolizing enzymes nor c-di-GMP receptors exist in Bacteroidetes23,24. Such inconsistent findings justify further investigation of signaling systems in P. gingivalis and other periodontopathogens.
In this study, we sought to reveal the scheme of nucleotide-based signaling in P. gingivalis strains using direct analytical evidence and approaches, and to determine biological significance during pathogenesis. Our data demonstrate that bis-(3′,5′)-cyclic di-adenosine monophosphate (c-di-AMP) is the primary and essential signaling system in P. gingivalis, which controls cell growth, cell wall integrity, biofilm formation, and virulence. Since c-di-AMP signaling does not exist in mammals, it is potentially a novel druggable target for controlling pathogens belonging to the phylum Bacteroidetes.
Results
Analysis of nucleotide-based signaling molecules in P. gingivalis
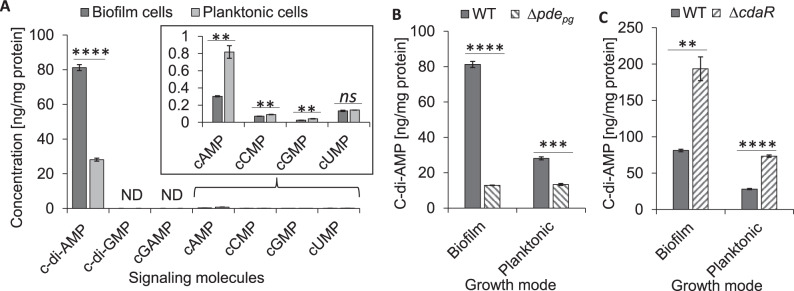
It is noteworthy to mention that genetic and phenotypic variability, e.g., fimbriation vs. encapsulation, exists among clinically prevalent strains of P. gingivalis, which significantly impacts host–pathogen interactions and virulence potential25,26. Here we sought to provide a first glimpse into signaling schemes of two phenotypically different strains 381 (hyperfimbriated/non-encapsulated) and W50 (encapsulated/fimbriated), with emphasis on strain 381. Strain 381 is a robust biofilm former exhibiting high invasion efficiency, which induces proinflammatory cytokines and periodontal bone loss27,28. We analyzed ubiquitous bacterial cyclic nucleotide monophosphates (cNMPs) and cyclic di-nucleotide monophosphates (c-di-NMPs) in metabolic extracts of biofilm- and planktonic-grown cells of strain 381. We found that c-di-AMP is the most abundant signaling molecule in planktonic and biofilm cells of P. gingivalis (Fig. 1A). Other c-di-NMPs, i.e., c-di-GMP and 2′3′-cyclic GMP-AMP (cGAMP) were not detected. Among detected cNMPs, the cellular concentration of cAMP was less than 0.9 ng per mg protein in strain 381. In contrast, we quantified cCMP, cGMP, and cUMP concentrations in the low-nanogram range (<0.15 ng per mg protein). Planktonic cells of strain 381 displayed higher concentrations of cNMPs than biofilm cells; the cellular concentration of cAMP in planktonic cells was almost 2.7 times higher than biofilm cells (Fig. 1A). Similar to strain 381, the cellular concentration of c-di-AMP in strain W50 (a weak biofilm former) was the greatest among detected molecules. However, it was almost 7.8 times less than strain 381 (Supplemental Fig. 1A). Similarly, the concentration of cNMPs in strain W50 was in the low-nanogram range, and the cAMP concentration in strain W50 was about two times higher than strain 381 (Supplemental Fig. 1B), indicating c-di-AMP and cAMP may distinctively associate with specific phenotypes and strains in this species. Overall, our data demonstrate that P. gingivalis strains do not possess c-di-GMP signaling; instead, they predominantly harness c-di-AMP signaling, which positively correlates with the potential for biofilm formation. Supplementary Table 1 represents the mean values of cNMPs, c-di-NMPs, and other molecules in pmol/mg protein.
Fig. 1. Comprehensive analysis of cyclic mono- and di-nucleotide second messengers in P. gingivalis 381.
A Cellular concentration of ubiquitous bacterial second messengers in the cells of P. gingivalis in biofilm or planktonic modes of growth. Mononucleotide second messengers are in the low-nanogram range (inside box). B c-di-AMP in P. gingivalis ∆pdepg. C c-di-AMP in P. gingivalis ∆cdaR. Graphs represent the mean ± SE (three biological replicates) of nucleotide second messengers which were analyzed with a student’s t test (**P < 0.01; ***P < 0.001; ****P < 0.0001; ns not significant). Scatter plots in Supplementary Fig. 9 display the data distribution. Standard curves are presented in Supplementary Fig. 10. ND not detected, WT wild type.
C-di-AMP turnover in P. gingivalis in response to biologically relevant stimuli
Given that c-di-AMP was the predominant second messenger in P. gingivalis strains, we sought to investigate the molecular mechanism of c-di-AMP signaling, as determined by highly conserved c-di-AMP-metabolizing enzymes in prokaryotes, i.e., di-adenylate cyclases (DACs) and the DHH/DHHA1- or PgpH/HD (or His-Asp)-type domain-containing phosphodiesterases (PDEs)29,30. In general, these enzymes collectively control the delicate equilibrium between the synthesis of c-di-AMP from two molecules of ATP or its hydrolysis into 5′-phosphadenylyl-adenosine (pApA) or two molecules of adenosine monophosphate (AMP) in response to perceived stimuli. Subsequently, a specific cellular concentration of c-di-AMP must engage specific receptors/effectors to control desired biological outputs20,21,31,32. Using NCBI BLAST and the Phyre2 Protein Fold Recognition Server33, we found that the products of two nearby genes PGN_0523 and PGN_0521 have 36% and 38% structural homology with Listeria monocytogenes DAC and PgpH/HD-type PDE enzymes, respectively. We did not find a DHH-DHHA1 domain-containing c-di-AMP PDEs in P. gingivalis. In addition, multiple sequence alignments of these proteins predicted highly conserved motifs for mediating c-di-AMP binding and fulfilling synthesis or degradation processes (Supplemental Fig. 2). Hence, we posited that deletion of PGN_0523 (∆dacpg) or PGN_0521 (∆pdepg) would change the cellular concentration of c-di-AMP significantly. Our attempt to generate a dacpg deletion mutant was unsuccessful, indicating that dacpg is essential for P. gingivalis growth. This finding is consistent with previous reports demonstrating that c-di-AMP synthases are necessary for the growth of several bacterial species34 and dacpg was identified among inherently essential genes in P. gingivalis based on transposon sequencing libraries35–37. Strikingly, deletion of the pdepg gene, which is typically expected to elevate c-di-AMP concentration, significantly reduced it by 2.1- and 6.3-fold in planktonic- and biofilm-grown cells, respectively, when compared to the wild type (Fig. 1B and Supplemental Fig. 3A). While in trans complementation of the pdepg mutant was not achievable, in cis complementation with pdepg yielded increased c-di-AMP concentration to the same concentration of wild-type c-di-AMP (Supplemental Fig. 3B). To the best of our knowledge, this is the first c-di-AMP PDE enzyme that positively regulates the cellular concentrations of c-di-AMP among bacteria.
Upon further investigation to find a gene candidate acting as a negative regulator of c-di-AMP turnover, the STRING database38 suggested PGN_1486 encoding a putative YbbR- or CdaR-like protein may be functionally associated with the c-di-AMP signaling network in P. gingivalis. Interestingly, deletion of PGN_1486 (∆cdaR) significantly increased c-di-AMP concentrations by about 2.6- and 2.4-fold, in planktonic and biofilm cells, respectively (Fig. 1C and Supplemental Fig. 3A). cis complementation of ∆cdaR with cdaR reduced c-di-AMP concentrations to the same concentration as wild-type c-di-AMP (Supplemental Fig. 3B), which rules out polar effects on nearby genes.
A comparison of the cellular concentration of cNMPs in the mutants showed that they do not significantly change between ∆pdepg and ∆cdaR in biofilm mode, although both mutants contained much less cAMP concentration than the wild type (Supplemental Fig. 3C). However, in planktonic mode, the deletion of cdaR almost doubled the cellular concentrations of cCMP, cGMP, and cUMP and impeded the reduction of cAMP (Supplemental Fig. 3D). Overall, these data suggest crosstalk between the c-di-AMP signaling pathway and other signaling systems in P. gingivalis only in the planktonic mode of growth.
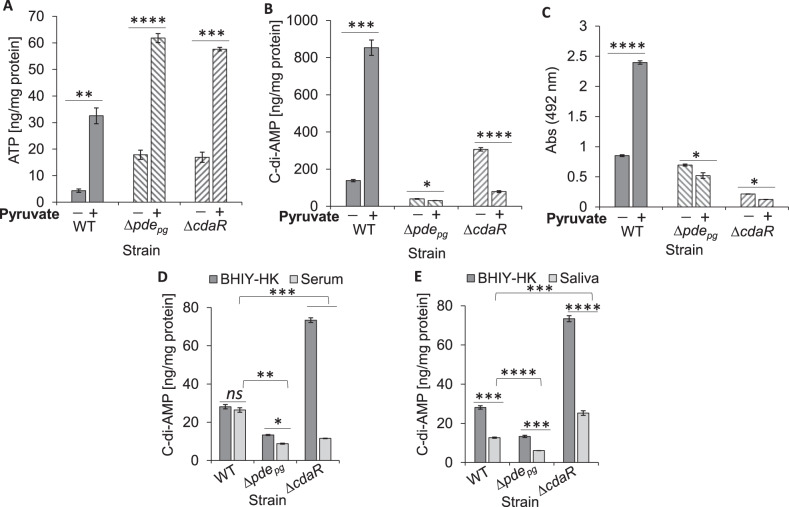
In order to provide more insights into the mechanism of c-di-AMP turnover, the products of c-di-AMP degradation, i.e., pApA or AMP, were quantified in the wild-type and mutants. Similar to c-di-AMP quantification data, ∆pdepg and ∆cdaR cells contained lower and higher pApA and/or AMP concentrations respectively, compared to wild type (Supplemental Fig. 3E, F). However, alterations in pApA or AMP concentrations in the mutants were disproportionate to quantified c-di-AMP concentrations, indicating that c-di-AMP degradation may not typically occur in P. gingivalis. Therefore, we hypothesize that the availability of ATP may regulate c-di-AMP signaling at the synthesis level in this organism. Quantification of cellular ATP showed that ∆pdepg and ∆cdaR mutants significantly accumulate ATP three to four times more than the wild type (Supplemental Fig. 4). These data suggest that PDEpg and CdaR are critical for determining c-di-AMP equilibrium through controlling the synthesis process rather than hydrolysis. As a proof of concept, we used pyruvate as a source of carbon and energy for P. gingivalis16,17 to provide a permissive condition for in situ provision of ATP followed by simultaneous quantification of ATP and c-di-AMP. Intriguingly, pairwise comparisons showed that although pyruvate utilization increased ATP concentrations significantly in ∆pdepg and ∆cdaR mutants compared to the wild-type (Fig. 2A), it elevated the c-di-AMP concentration significantly only in the wild-type and decreased it in the mutants (Fig. 2B). Consistent with these findings, pyruvate addition greatly enhanced biofilm formation of the wild-type while reducing the mutants’ ability to form a biofilm (Fig. 2C). These data indicate that coexistence of PDEpg and CdaR is required to regulate the incorporation of ATP into c-di-AMP synthesis in P. gingivalis and regulate biofilm formation through a yet to be determined mechanism.
Fig. 2. Both pdepg and cdaR genes are required for the synthesis of c-di-AMP and biofilm formation while they regulate differentially the cellular level of c-di-AMP in P. gingivalis in response to biologically relevant stimuli.
A, B Cellular levels of ATP and c-di-AMP in biofilm cells in response to pyruvate availability. C Monospecies biofilm formation with WT, ∆pdepg, and ∆cdaR; in the presence or absence of pyruvate. D, E Differential regulation of c-di-AMP levels in the WT, ∆pdepg, and ∆cdaR mutants in response to 10% human serum (D) or 10% saliva (E). Graphs represent the mean ± SE (three biological replicates) of c-di-AMP concentrations which were analyzed with a Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns not significant). Scatter plots in Supplementary Fig. 9 display the data distribution. Standard curves are presented in Supplementary Fig. 10. WT wild-type.
To assess the response of c-di-AMP signaling to biologically relevant stimuli, we applied human serum or saliva as primary constituents of subgingival and supragingival environments, respectively. To this end, Brain Heart Infusion (supplemented with yeast extract, hemin, and menadione (BHIYHK))-grown cells in exponential phase (OD600: 0.65; 5 × 108 CFU) were incubated in 10% human serum or saliva for 30 min followed by extraction and quantification of molecules. The cellular concentration of c-di-AMP remained unchanged in the wild type in response to serum but was reduced in both mutants (Fig. 2D). Saliva reduced the concentration of c-di-AMP in the wild-type and mutants (Fig. 2E). Overall, these data suggest that c-di-AMP signaling is regulated differently in response to different stimuli in the oral cavity, and that both PDEpg and CdaR are necessary for maintaining optimal concentrations of c-di-AMP, particularly in the subgingival environment. Supplementary Table 1 represents the mean values of cNMPs, c-di-NMPs, and other molecules in pmol/mg protein.
Cell growth and biofilm formation are regulated by c-di-AMP turnover in P. gingivalis
We first assessed the colony morphology and growth rate of the ∆pdepg and ∆cdaR mutants. In general, strain 381 displays a smooth appearance on BHIYHK plates but colonies of ∆pdepg displayed a glossier appearance, while ∆cdaR appeared as rough colonies (Supplemental Fig. 5A). Given that P. gingivalis intrinsically grows slowly but tends to auto-aggregate progressively in bovine serum albumin (BSA)-only medium over short periods16,39, we used this condition to assess cell surface properties of ∆pdepg and ∆cdaR mutants in static liquid cultures. ∆cdaR cells demonstrated strong cohesiveness and auto-aggregation, whereas ∆pdepg cells showed less auto-aggregation compared to the parental strain (Supplemental Fig. 5B). Both mutants were unaffected in their ability to form black-pigmented colonies on blood agar plates due to heme accumulation on the cell surface. However, the mutants did not display hemolytic activity on blood agar (Supplemental Fig. 5C). These findings suggest that c-di-AMP signaling in P. gingivalis can impact cell surface properties.
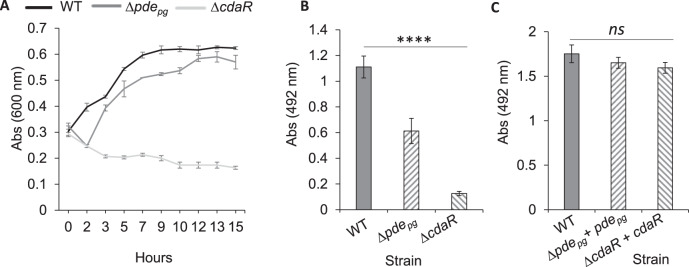
In chronic periodontal infection, P. gingivalis interacts with a complex microbial community in the subgingival crevice, between the tooth surface and the adjacent epithelium where a serum exudate (known as gingival crevicular fluid) constitutes the primary nutritional flow5,40. Our previous findings demonstrated that P. gingivalis could efficiently utilize serum components which play an ancillary role in developing polymicrobial biofilms upon establishing physical and cross-feeding networks with accessory species16. Here, we applied similar experimental conditions, i.e., the use of human serum albumin (HSA)- or human serum-based media that offer biologically relevant model systems for studying proteolytic activity, biofilm formation, and in situ interactions between different oral species16,17,39. Analysis of planktonic growth rate showed that ∆pdepg, and to a much greater degree, ∆cdaR displayed a significant growth defect in chemically defined medium supplemented with human serum albumin (CDM-HSA-HK) (Fig. 3A). In monospecies biofilm accumulation in microtiter plates, the ∆pdepg and ∆cdaR mutants formed almost two- and six-fold less biofilm than the wild type, while the biofilm phenotype was restored to parental concentrations in cis-complemented strains (Fig. 3B, C). Consistent with these data, quantitative image analysis of biofilms grown in the CDM-Serum-HK medium revealed that ∆pdepg forms a less developed biofilm with a lower biovolume value compared to the wild type. The ∆cdaR mutant displayed a greater reduction in the ability to form biofilms. Further, the biofilms of both mutants contained higher numbers of dead cells and displayed lower compactness values than the wild-type (Fig. 4).
Fig. 3. Comparison of planktonic growth rate and biofilm formation.
A Graph shows that ∆pdepg displays a lower rate of planktonic growth than the wild-type and ∆cdaR displays a significant growth defect. Cells were cultivated in the CDM-HSA-HK medium. B Biofilm assay using 96-well plates shows that both mutants produce less biofilms than WT in the CDM-HSA-HK medium. However, complementation of the mutants with relevant genes restored biofilm formation to the level of the wild type (C). Graphs represent mean biomass of biofilms ± SE (three biological replicates) at 48 h, as determined by safranin staining, which were analyzed using ANOVA test (Shapiro–Wilk test: P > 0.05). Asterisks indicate pairs of significantly different values (post hoc Tukey’s HSD test: ****P < 0.0001; ns not significant).
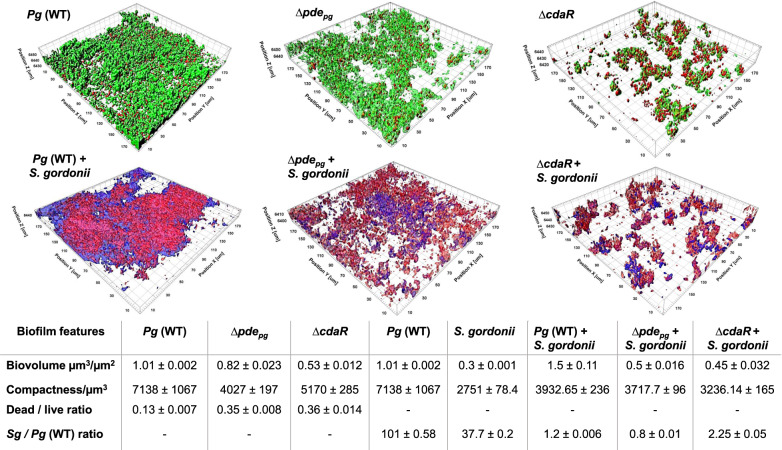
Fig. 4. Effects of defective regulation of c-di-AMP levels on biofilm formation of P. gingivalis WT, ∆pdepg, and ∆cdaR and its colocalization with S. gordonii.
CLSM images of mono- and dual-species biofilms were analyzed by IMARIS image analysis software. The resultant biofilm parameters are summarized in the table. The cell community dimensions are provided as µm3/µm−2 and the values of three biological replicates were calculated per unit that represent mean biovolume/unit ± SE, compactness/unit ± SE, dead/live ratio, and S. gordonii/P. gingivalis ratio, as determined by appropriate staining methods. Sg S. gordonii, Pg P. gingivalis, WT wild-type.
Given that our prior findings demonstrated that among different oral species, only P. gingivalis could efficiently exploit serum components for the development of polymicrobial biofilm with Streptococcus gordonii16, we hypothesized that the ∆pdepg and ∆cdaR mutants may display different capabilities of co-colonization with S. gordonii. Quantitative image analysis revealed an efficient co-colonization of P. gingivalis and S. gordonii to form dual-species communities with both species almost equally present (Fig. 4 and Supplemental Fig. 6). However, deletion of pdepg or cdaR significantly changed the interaction of P. gingivalis with S. gordonii, resulting in a threefold or more decrease of biofilm volume and compactness. Moreover, the S. gordonii population was almost double the population of the ∆cdaR mutant in their mixed biofilm (Fig. 4). Overall, these data indicate that changes in c-di-AMP concentrations significantly alter biofilm features and microbial interactions between P. gingivalis and accessory species.
C-di-AMP signaling controls cell wall homeostasis and stress tolerance
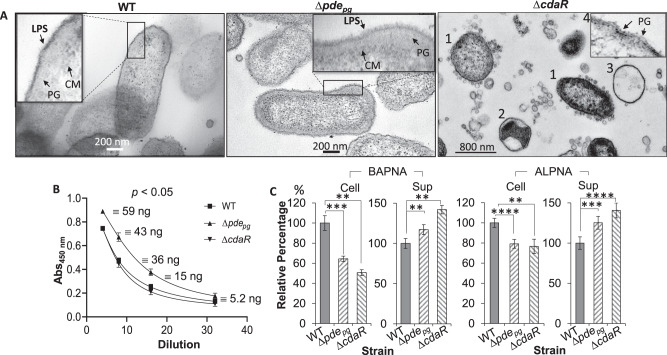
To examine directly if c-di-AMP signaling impacts cell surface and membrane structures, we performed transmission electron microscopy (TEM) on biofilm cells of the wild type and mutants. TEM images revealed a network of macromolecules extending much further from the less uniform outer leaflet of the outer membrane (OM) of ∆pdepg cells compared to the wild type (Fig. 5A and Supplemental Fig. 7A, B). We posited that this network of macromolecules comprises lipopolysaccharides (LPSs) since the sample preparation procedure predominantly stains phosphoryl and carboxyl groups of lipids and lipopolysaccharides, the outer face of the OM is primarily composed of LPSs, and strain 381 does not produce capsular polysaccharides41–43. Intriguingly, the cells of ∆cdaR mutant displayed a significant heterogeneity in cell shape and cell envelope as follows: (1) rod- and round-shaped cells with fully or partially intact envelopes that are overloaded with outer membrane vesicles (OMVs) varying in shape and size; (2) round cells with an intact monolayer of membrane encompassing agglomerated cytoplasmic materials with void spaces in the cells; (3) entirely void round-shaped structures with an intact monolayer of membrane; and (4) cells without recognizable OM layers which display a bare peptidoglycan layer (Fig. 5A and Supplemental Fig. 7C, D). Such heterogeneity was also distinguishable in confocal laser microscopy images (Supplemental Fig. 7E), supporting our notion that observed structural heterogeneity in cells and their envelope was caused by the deletion of the cdaR gene, rather than the sample preparation procedure.
Fig. 5. Effects of defective regulation of the cellular c-di-AMP level on P. gingivalis cell envelope, the immunoreactivity of LPS, and gingipain activities.
A Cells were stained with osmium tetroxide and uranyl acetate and imaged by transmission electron microscopy. Cells of ∆cdaR mutant displayed a significant shape and cell envelope heterogeneity represented by rod- and round-shaped cells with fully or partially intact cell envelopes that are overloaded with OMVs varying in shape and size (1); cells with an intact monolayer of membrane encompassing agglomerated cytoplasmic materials (2) or entirely void round-shaped structures with an intact monolayer of the membrane (3), cells displaying bare peptidoglycan layers (4). B ELISA assays of cell lysates using anti-P. gingivalis LPS monoclonal antibody. The graph represents the mean ± SE of the immunoreactivity of cell lysates (three biological replicates). The standard curve is presented in Supplementary Fig. 10. C Gingipain-dependent proteolytic activities of P. gingivalis strains cells. Graphs represent the mean ± SE (three biological replicates) of the activity of arginine (BAPNA) and lysine (ALPNA) gingipains which were analyzed with a Student’s t test (**P < 0.01; ***P < 0.001; ****P < 0.0001). Scatter plots in Supplementary Fig. 9 display the data distribution. Sup cell-free supernatant, BAPNA N-α-benzoyl-l-arginine-p-nitroanilide, ALPNA N-α-acetyl-l-lysine-p-nitroanilide.
To supplement the TEM-based observations, we performed an ELISA assay using anti-P. gingivalis LPS monoclonal antibody on cell lysates of planktonic cells from the exponential phase of growth. ∆pdepg cells exhibit higher immunoreactivity to the antibody than the wild type, while ∆cdaR cells show slightly less (P < 0.05) (Fig. 5B).
In general, changes in LPS composition affect cell wall homeostasis, hence the sensitivity of Gram-negative bacteria to antibiotics and environmental stresses44–46. To assess if c-di-AMP-dependent changes in the cell envelope impact P. gingivalis susceptibility to stresses, we measured the minimum inhibitory concentration (MIC) of amoxicillin and sensitivity to sodium dodecyl sulfate (SDS) for each strain. The MIC test showed that deletion of pdepg or cdaR significantly increased P. gingivalis susceptibility to amoxicillin by about fivefold and tenfold, respectively, compared to the wild-type (Supplemental Fig. 8A). Similarly, the ∆pdepg and ∆cdaR mutants displayed greater sensitivity to SDS and were completely lysed in the presence of 0.001% SDS (Supplemental Fig. 8B). Overall, these data indicate that c-di-AMP signaling regulates the composition of LPS in P. gingivalis directly or indirectly and plays a significant role in cell envelope homeostasis and protection against stresses.
C-di-AMP signaling controls the virulence potential of P. gingivalis
Since A-LPS variant of P. gingivalis is employed for post-translational modification of gingipains and anchorage on the cell surface47,48, we hypothesized that such heterogeneity in LPS structure may impact the association of gingipains on the surface of the mutants. A gingipain assay49 showed that the cell-free supernatants of both mutants show more gingipain activity, and cell surfaces have less activity, compared to the wild-type (Fig. 5C).
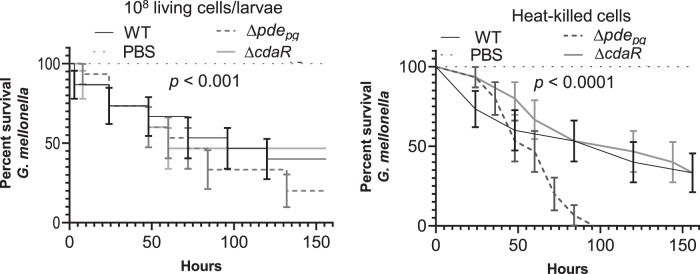
To assess the virulence potential of the mutants, we applied the Galleria mellonella infection model, an increasingly popular model for determining the virulence of bacterial pathogens, including periodontopathogens, because of similarities with the mammalian innate immune system50–52. Living cells of the ∆pdepg mutant exhibited higher virulence with only 20% larval survival after 156 h of infection (Fig. 6). On the other hand, living cells of the ∆cdaR mutant were slightly less virulent than the wild type. Further, the release of metabolites and lipid-based contents by heat treatment increased the toxicity of the ∆pdepg mutant (0% larval survival after 100 h of infection) (Fig. 6). Heat-killed cells of the ∆cdaR mutant showed less toxicity than that of the wild-type within 60 h of the assay, but both resulted in 33% larval survival after 156 h (Fig. 6). Overall, our data indicate that the c-di-AMP signaling pathway is associated with the virulence potential of P. gingivalis, and pdepg and cdaR act oppositely in the determination of c-di-AMP concentration in the cells and virulence potential.
Fig. 6. Survival of G. mellonella larvae injected with ∼108 CFU/ml of P. gingivalis WT, ∆pdepg, and ∆cdaR mutants.
The control group was injected with sterile PBS. Survival data were plotted using the Log-rank (Mantel–Cox) test. Graph shows the average of two biological replicates and the standard error (n = 15 larvae per group).
Discussion
The results presented in this study reveal the schemes of nucleotide-based second messenger signaling in P. gingivalis. This anaerobe is a key player in the etiology of periodontitis and may be an important paradigm for signaling mechanisms via nucleotide-based second messengers in Bacteroidetes. We found that, among ubiquitous cNMPs and c-di-NMPs, P. gingivalis strains predominantly synthesize c-di-AMP, which is essential for their growth and survival. Consistent with previous reports indicating that the c-di-GMP signaling is absent in Bacteroidetes23,24, we could not detect c-di-GMP or cGAMP in P. gingivalis strains. To date, the essential role of c-di-AMP signaling for bacterial growth, and their role in controlling a wide range of cellular and biological processes, including biofilm formation, central metabolism, cell wall homeostasis, and osmoregulation, have been addressed primarily in Gram-positive species20,34,53–61 and our understanding of its biological significance in the pathogenesis of Gram-negative pathogens remain largely limited. Emerging data indicate that the c-di-AMP signaling mechanism follows the same general principle as for other second messengers in which cognate metabolizing enzymes control the equilibrium between synthesis and degradation of c-di-AMP molecules, which are necessary for engaging specific protein or riboswitch receptors to act as direct effectors for regulating desired biological outputs62,63. Here, we show that as opposed to this general principle, as determined by DAC and the DHH/DHHA1- or PgpH/HD domain-containing PDE enzymes, respectively, the concentration of c-di-AMP is positively regulated by pdepg in P. gingivalis. In addition, low cellular concentrations of pApA or AMP (c-di-AMP degradation products) and their disproportionate relationship to c-di-AMP concentrations suggest that, at least under our experimental condition, PDEpg may not act as a c-di-AMP phosphodiesterase in P. gingivalis despite displaying conserved amino acid motifs with functionally related proteins. Instead, PDEpg impedes a decrease in c-di-AMP concentrations. However, its exact function(s) or the necessity of specific stimuli to induce a phosphodiesterase activity or the existence of an alternative mechanism or gene for c-di-AMP degradation in this species awaits further investigation. Intriguingly, we found that the cdaR gene, unrelated to the c-di-AMP PDE enzymes, serves as a potent negative regulator of c-di-AMP synthesis. In general, Ybbr domain-containing proteins (also known as CdaA regulatory protein CdaR in Gram-positive species) have been characterized as membrane proteins with sensory domains for an unknown signal59,64. It has been noted that cdaR and dac genes are often clustered together in one operon, and their products interact to regulate the c-di-AMP synthesis, and hence downstream cellular pathways58,65. However, different CdaR proteins have been found to act oppositely on the c-di-AMP synthesis in different species, such as Bacillus subtilis and L. monocytogenes59,66. In P. gingivalis, these two genes are located in two different genomic regions. Our findings indicate that simultaneous involvement of dacpg, pdepg, and cdaR is not only necessary for regulation of c-di-AMP synthesis and cell homeostasis but also essential for growth, biofilm formation, and fitness in different microenvironments. Interestingly, the high cellular concentration of c-di-AMP, achieved upon pyruvate supplementation, did not perturb wild-type cell hemostasis, but instead enhanced cell proliferation and biofilm biomass. Hence, we can conclude that the proposed toxicity of excess c-di-AMP in bacteria may not be the case in P. gingivalis, likely because the counterparts DACpg, PDEpg, and CdaR may exert other activities that mitigate directly or indirectly such an adverse effect. Previous studies have addressed the necessity of c-di-AMP signaling for cell wall synthesis and metabolic homeostasis in the Gram-positive Firmicutes to cope with stresses and damages20,31,55,67, and our data indicate that c-di-AMP signaling and/or the activities of cognate protein counterparts can regulate the structure of LPS and cell envelope integrity of Gram-negative species, hence susceptibility to membrane-targeting antimicrobials. Notably, the absence of cdaR significantly dysregulates the integrity of the cell envelope and cellular homeostasis, leading to a membrane with numerous OMV-like and unusual membranous structures. P. gingivalis can produce a copious amount of OMVs loaded with various virulence factors, including LPS and gingipains, for dissemination and distal impacts39,68,69. However, the mechanistic basis of OMV biogenesis remains unknown. Our findings suggest CdaR is a strong candidate in the regulation of OMV biogenesis, and further investigation is ongoing to understand its exact function.
It is plausible that P. gingivalis has evolved c-di-AMP signaling for appropriate responses and enhanced fitness in the surrounding environment, which is a complex polymicrobial community with serum (subgingival) or saliva (supragingival) fluid phases. We found that c-di-AMP signaling is differentially regulated in response to human saliva and serum, indicating distinct functions in different microenvironments. Previous studies, including ours, demonstrated co-colonization and physical interaction of P. gingivalis with other potential pathogens, as well as the ancillary role of P. gingivalis in the establishment of metabolic crosstalk within the polymicrobial community upon the utilization of human serum components2,16,70,71. Further, a mixed community of P. gingivalis and S. gordonii is more pathogenic than P. gingivalis alone72,73. Our findings demonstrate that a c-di-AMP signaling pathway is required for the optimal interaction of P. gingivalis and S. gordonii. Our work also provided the first glimpse into the c-di-AMP-dependent variation of LPS that subsequently affects the association of T9SS cargo proteins such as gingipains to the cell surface. Both LPS and gingipains are notable virulence factors playing prominent roles in the pathological outcome of periodontitis by being involved in the stimulation of proinflammatory responses, tissue destruction, host-defense perturbation, and bone resorption74–76. Previous studies have addressed LPS heterogeneity in P. gingivalis in response to certain stimuli77,78 but the mechanistic basis of the shifts in the LPS profile remains obscure. Here, we revealed that P. gingivalis LPS profile and virulence potential change concomitant with c-di-AMP signaling alterations. The control of virulence potential via shifting the structure and immunostimulatory properties of LPS is an essential aspect of bacterial pathoadaptation by which pathogens can disguise themselves from host detection, regulate virulence and immunostimulatory effects to promote persistence in the host or protect themselves from host-defense mechanisms, antimicrobials, and environmental stresses79–81. Hence, we consider the c-di-AMP-dependent LPS heterogeneity in P. gingivalis as a novel mechanism for regulating pathogenesis and virulence. This notion is further supported by our findings that P. gingivalis displays a higher level of LPS immunoreactivity, as well as virulence and toxicity in the absence of pdepg in our infection model. However, further investigations are required to reveal the mechanistic basis of the regulation of cellular pathways by c-di-AMP signaling and their biological importance in the onset and development of periodontitis by using different infection models.
Overall, to our knowledge, the unusual nature of c-di-AMP turnover in P. gingivalis and its involvement in regulating LPS profile and cell envelope integrity is unique to this species. Since the c-di-AMP signaling and cognate regulatory proteins are absent in mammals, they can be attractive antimicrobial drug targets for treating chronic infections such as periodontitis. However, further understanding of the role of c-di-AMP signaling in microbial dysbiosis, host–pathogen interaction, and periodontal inflammation is required to develop more precise and informed therapeutic interventions.
Methods
Ethics statement
Saliva collection was approved by the University of Louisville IRB, Protocol # 21.0925 and categorized as a prospective collection of biological specimens for research purposes by noninvasive means. Written informed consent was obtained from all participants.
Bacterial strains, growth conditions, and chemicals
Bacteria used in this study were P. gingivalis strains 381 (with isogenic mutants described below) and W50, Escherichia coli strain DH5-α (New England BioLabs GmbH), and Streptococcus gordonii DL-1. Bacto™ Trypticase Soy Broth (BD Biosciences) supplemented with 5 μg/ml hemin and 1 μg/ml menadione (TSBHK) or Bacto™ Brain Heart Infusion Broth (BHI) without sucrose (BD Biosciences) supplemented with 0.5% yeast extract (Fisher Scientific), hemin, and menadione (BHIYHK), or TSBHK supplemented with 5% defibrinated sheep blood (BAPHK) were used for cultivation of P. gingivalis. Bacto™ Agar was used for solid media. BHI medium was used for the cultivation of S. gordonii. For chemically defined media (CDM), bovine serum albumin (BSA; Alfa Aesar), human serum albumin (HSA, Alfa Aesar), or normal human serum (Merck; Millipore) were dissolved in basal salts (BS) buffer (14 mM Na2HPO4, 10 mM KCl, 10 mM MgCl2, pH 7.3), then filtered (0.22 µm) and supplemented with hemin (H)/menadione (K). For pyruvate-relevant assays, CDM was supplemented with 0.5% pyruvate (Sigma). P. gingivalis or S. gordonii were incubated at 37 °C anaerobically (5% hydrogen, 10% carbon dioxide, and 85% nitrogen). When necessary, 10% human serum (Merck; Millipore) or 10% saliva in water were prepared and then filter-sterilized (0.22-µm). Saliva from healthy individuals was collected 2 h after food consumption. After chewing a walnut-size ball of sterile parafilm for 10 min, saliva was expectorated into a container on ice until approximately 10 ml of saliva was collected. Following collection, dithiothreitol (DTT) was added to a final concentration of 2.5 mM. Samples were incubated on ice for 10 min followed by removal of particles and debris using centrifugation at 13,000 × g for 10 min. Clarified saliva was diluted to 10% with distilled water and then filter-sterilized (0.22-µm). Aliquots of saliva were stored at −20 °C until needed82. Enzymes for genetic manipulations and cloning were purchased from New England BioLabs, Ipswich, MA, USA, and all chemicals were purchased from Sigma-Aldrich unless otherwise indicated. Solvents used for the analysis of nucleotide second messengers were HPLC grade.
Construction of mutants and complementation
Deletions of pdepg (PGN_0521) and cdaR (PGN_1486) were generated through homologous recombination using linear DNA fragments17,83. Linear fragments carrying 1-kb flanking regions of the target genes, as well as a promoterless erythromycin resistance gene (ermF), were obtained from P. gingivalis strain 381 genome and plasmid pVA2198 as a template, respectively, using primers listed in Supplementary Table 2. The PCR was performed using Phusion Hot Start Flex 2X Master Mix (New England BioLabs; cat# M0536S) and consisted of 25 cycles with a temperature profile of 30 s at 98 °C, 30 seconds at 57 °C for annealing, and 10 min at 72 °C for the extension. The NEBuilder HiFi DNA assembly cloning kit (New England BioLabs; cat# E5520S) was used as described in the manufacturer’s protocol for assembling DNA fragments into linear cassettes consisting of the 1 kb upstream region of the gene-ermF-1-kb downstream region of the gene. Competent cells were prepared using 15 ml of the actively growing culture of P. gingivalis strain 381 (OD600 = 0.7). Cells were pelleted and washed twice with electroporation buffer (10% glycerol and 1 mM MgCl2; filter-sterilized; stored at 4 °C). The pellet was resuspended in 0.5 ml of electroporation buffer and distributed into 100-μl aliquots. Electroporation was performed using a 100-μl sample of cells to which ~2 μg of the assembled DNA fragments was added and placed in a sterile electrode cuvette (0.2-cm gap). The cells were pulsed with a Bio-Rad gene pulser at 2.5 kV for 5.5 ms and then incubated on ice for 5 min. The cell suspension was then added to 1 ml of TSBHK and incubated for ~16 h. Transformants were selected on BHIYHK plates supplemented with antibiotic followed by further confirmation via genomic DNA isolation using a Wizard® Genomic DNA Purification Kit (Promega) and amplification of upstream/downstream regions of the deleted gene followed by DNA sequencing (Eurofins Genomics)84. For in cis complementation, the abovementioned strategy for the generation of linear fragments was employed but the linear fragment contained 1-kb flanking regions of the genes, the relevant gene under its native promoter, and the tetracycline resistance gene (tetQ) obtained from plasmid pT-COW85. Confirmation of gene insertion was by colony selection on antibiotic plates, genome isolation, and amplification of the gene.
Growth rate analysis and biofilm assays
For growth rate analysis, P. gingivalis strains were grown anaerobically on BAPHK for 2–3 days and inoculated in TSBHK or BHIYHK, then incubated for 18 h anaerobically. Cultures grown overnight were centrifuged and the pellets were washed with pre-reduced BS buffer and then suspended in pre-reduced working medium. Bacterial growth was then monitored by measuring the optical density at 600 nm (OD600). To estimate the number of viable bacteria, planktonic growth cultures were subsequently prepared at dilutions for plating on BAPHK plates and inoculated anaerobically for 4–5 days. The numbers of colony-forming unit (CFU) of growth culture at different time points were determined.
Biofilm assays were performed in 96-well flat-bottom plates (FALCON®, Corning Inc., USA)16. To this end, 4 ml of overnight culture was pelleted inside an anaerobic chamber using a mini centrifuge and washed with pre-reduced BS buffer and pelleted. Bacterial cells were resuspended in the working medium supplemented with 5 μg/ml hemin and 1 μg/ml menadione to reach an optical density at OD600 = 0.1 (~5 × 107 cells/ml). A total volume of 200 μl of the cells was applied in each well and anaerobically incubated for 48 h. The plates were washed twice by immersing in distilled water to remove free cells, and air-dried. A total volume of 200 μl of 0.1% safranin in water was used to stain each well for 30 min. The plates were washed by immersing twice in distilled water, and air-dried. The extraction of the stain from biofilms was performed using 200 μl of 90% ethanol containing 1% sodium dodecyl sulfate for 30 min followed by reading the absorbance at 492 nm.
For biofilm analysis by confocal laser scanning microscopy, the inoculum was prepared as described above but diluted to OD600 = 0.2 and 1 ml of the cells was added to each well of a 12‐well plate (Greiner Bio‐one) containing a circular glass coverslip16. Plates were incubated anaerobically for 24 h. Glass coverslips were gently washed with distilled water to remove free cells and stained with either the Invitrogen Live/Dead BacLight Bacterial Viability Kit (Invitrogen) or ViaGramTM Red+ Bacterial Gram Stain and Viability Kit (Molecular Probes, Invitrogen) as described by the manufacturer. Stained coverslips were gently placed face down on a glass microscope slide, and biofilms were visualized on a Leica SP8 confocal microscope (Leica Microsystems Inc.), using 507/545 nm (SYTO 9), 614/745 nm (PI), 410/547 nm (DAPI), and 590/750 nm (Texas Red) spectra, and ×63 objective. Images were analyzed using IMARIS image analysis software (Bitplane AG).
Analysis of nucleotide second messengers
High-performance liquid chromatography coupled with tandem mass spectrometry (HPLC-MS/MS) was employed to perform a comprehensive analysis on various nucleotide second messengers and relevant derivatives86. Various internal standards, including 13C2015N10-c-di-GMP and 13C2015N10-c-di-AMP, were used in this analysis. To this end, biofilm or planktonic cells were prepared as described above. Cells were incubated on ice for 20 min and then harvested by centrifugation at 4700 × g, 4 °C for 10 min. Washed cells were resuspended with 300 μL ice-cold extraction solvent containing acetonitrile/methanol/water (2/2/1, v/v/v) and incubated on ice for 15 min. Suspensions were heated at 95 °C for 10 min and cooled again on ice. Following centrifugation at 20,800 × g and 4 °C for 10 min, supernatants were collected, and pellets were subjected to extraction twice with solvent without heating. Combined supernatants of three extraction steps were stored at −20 °C overnight for further protein precipitation followed by centrifugation for protein precipitant separation. All residual pellets were collected for protein quantification using the Bicinchoninic Acid (BCA) Kit (Thermo Scientific) and supernatants were dried in a Speed-Vac (Thermo Scientific) at 40 °C. Dried samples were dissolved in 200 μL sample solvent and vortexed for 30 s. After centrifugation, 40 μL of sample solvent with internal standards were directly added in each MS vial with an insert and 40 μL of supernatant were transferred to a MS vial with insert and mixed three times by pipetting. Analysis was performed using liquid chromatography-mass spectrometry (LC-MS).
Detection and quantification of cNMPs and c-di-NMPs
For the determination of cNMPs, the resuspended samples were mixed 1:2 with an aqueous internal standard solution (100 mg/mL tenofovir). This solution was transferred to measurement vials and analyzed by liquid chromatography (LC) coupled tandem mass spectrometry (5500QTRAP; ABSciex, Framingham, Massachusetts)87,88. Cyclic nucleotide monophosphates were separated by reversed-phase chromatography on a C18 column (Zorbax eclipse XCB-C18 50 mm × 4.6 mm; 1.8 µm; Agilent, Santa Clara, California, USA). Solvent A was methanol–water (3:97, v/v), solvent B was methanol–water (97:3, v/v), with each containing 50 mM ammonium acetate and 0.1 vol.% acetic acid. A gradient was used that started with 0% B. Within 5 min, the content of solvent B was increased to 50%. During this time, the elution of the cNMPs was completed. Re-equilibration to the starting conditions was achieved within 3 min. The total run time of the analysis was 8 min at a flow rate of 500 µL/min. Mass spectrometric analysis was performed on a tandem mass spectrometer (QTRAP; MA, USA) using selected reaction monitoring (SRM) in positive ionization mode.
Chromatographic separation of c-di-NMPs was performed by reversed-phase chromatography (RP chromatography) on a C18 column (Nucleodur Pyramid C18 3 µ 50 × 3 mm; Macherey-Nagel; Germany), using water containing 10 mM NH4Ac and 0.1% HAc as eluent A, and pure methanol as eluent B. The following gradient was applied: 0 to 4 min, 0% B, 4 to 7.3 min 0–10% B, 7.3–8.3 min 10–30% B and 11–13 min 0% B. The flow rate was set at 600 µL/min. Mass spectrometric analysis was performed on a tandem mass spectrometer (API4000; MA, USA) performing selected reaction monitoring (SRM)89. The mass spectrometer was equipped with an electrospray ionization source (ESI) and ionization was performed in positive mode for all analytes.
ELISA quantification of c-di-AMP and ATP
C-di-AMP was also analyzed using a c-di-AMP Enzyme-Linked Immunosorbent Assay (ELISA) Kit (Cayman Chemical, USA). Briefly, the extraction of metabolites was performed as described above for HPLC-MS/MS. Each sample was assayed at a minimum of two dilutions, each in triplicates. A 50 µl sample was added to each relevant well followed by the addition of 50 µl tracer. Within 15 min, 50 µl of the antibody was added. The plate was covered and incubated for 2 h at room temperature on an orbital shaker. After washing the wells five times with the Wash Buffer of the kit, wells were developed with the addition of 175 µl of TMB (3,3’,5,5’-tetramethylbenzidine) Substrate Solution (Sigma-Aldrich, St. Louis, MO, USA). The reaction was stopped upon adding 75 µl of the kit’s HRP Stop Solution and the plate was read at 450 nm. The standard plot was calculated based on %Bound/Maximum Bound (%B/B0) versus serial dilutions of c-di-AMP using a four-parameter logistic fit. The %B/B0 value for each sample was also calculated and the concentration of c-di-AMP was identified on the standard curve.
ATP quantification assay was performed using ATPliteTM Luminescence Assay System (https://www.perkinelmer.com) on the same samples explained above. Briefly, a 50 µl of each sample was added to microtiter plate wells in triplicate. 50 µl of the kit’s lysis buffer and 60 µl of the kit’s reconstituting buffer were added and mixed on a rotational shaker (700 rpm) for 5 min. 50 µl of the reconstituted enzyme was added to the wells and mixed for 3 min and immediately the luminescence was measured at 578 nm. The ATP concentration for each sample was calculated on the standard curve which was set up in the same plate using the kit’s ATP standard.
LPS ELISA assay
For LPS ELISAs, autoclaved extracts of cells were used. Actively growing cells (OD600 = 0.7) were pelleted, diluted in sterile distilled water, and normalized to an OD600 of 0.5. 1.5 ml of the suspension was pelleted, and half the supernatant was removed. The cells were resuspended in the final 0.75 ml and autoclaved at 120 °C for 30 min. Once cooled, the extracts were centrifuged, and the supernatants were stored for ELISA assay83. Autoclaved extracts were diluted in 50 mM carbonate/bicarbonate buffer, pH 9.6, and serially diluted in a 96-well plate which was incubated at 4 °C overnight90. Plates were washed with PBS containing 0.05% Tween 20. A solution of 5% milk powder in PBS was used to block wells for 1 h at ambient temperature. After repeated washing, wells were incubated for 1 h at 37 °C with 0.2 µg/ml anti-Porphyromonas gingivalis LPS monoclonal antibody (Sigma; SAB4200834). Wells were washed and then incubated for 1 h at 37 °C with a goat anti-mouse IgG2b cross-adsorbed secondary-HRP antibody (Invitrogen; Cat# M32407) diluted at 1:2000 in PBS containing 0.1% Tween 20 and 0.1% bovine serum albumin. After a final wash, wells were incubated with TMB until sufficient color appeared. The reaction was stopped with an equal portion of 1 M HCl, and the absorbance was measured at OD450. Pure lipopolysaccharide from P. gingivalis (InvivoGen; Cat#tlrl-pglps) was used to plot the standard curve.
The MIC and SDS sensitivity assays
For MIC test and SDS sensitivity assay, P. gingivalis strains were grown anaerobically on BAPHK for 2–3 days and inoculated in TSBHK overnight anaerobically. Cultures grown overnight were centrifuged and the pellets were washed with pre-reduced medium and then suspended in pre-reduced working TSBHK medium to reach OD600 = 0.35. Various concentrations of amoxicillin or SDS were added to the cultures. Cultures were incubated anaerobically, and their growth rate was monitored for 24 h in the MIC test and for 8 h in the SDS sensitivity assay. The lowest concentration of antibiotic preventing bacterial growth was considered the MIC (the minimal concentration that inhibited growth by ≥95%).
Transmission electron microscopy (TEM)
Biofilm cells were harvested and washed with ice-cold PBS three times, and fixation solution (0.1 M phosphate buffer, 2% paraformaldehyde, 2% glutaraldehyde in 20 ml of MilliQ water, pH: 7.4) was added without resuspending the bacteria pellet. The pellet was then refrigerated at 4 °C for at least 48 h. The pellet was washed three times in 0.1 M phosphate buffer for 20 s each wash. 1% osmium tetroxide solution was added and allowed to incubate for 2 h at room temperature. After the osmium solution was removed, the pellets were washed four times (10 min each wash) in 0.1 M phosphate buffer, and the pellets held at 4 °C overnight. A Durcopan resin solution was first mixed up using Durcopan A, B, C, and D components. To a special flask, a 50/50 solution of Durcopan resin and 200 Proof EMS-grade ethanol was made and was set aside for the completion of the pellet dehydration step. Samples were then moved to glass Wheaton sample jars (VWR; Cat# 225536). The 0.1 M phosphate buffer was removed from the sample pellets, and 50%, 70%, 95%, and 100% ethanol were sequentially added, swirled, allowed to sit for 15 min, and removed. Treatment with 100% ethanol was repeated for two more times. The pellet pieces were then added to the 50/50 resin/ethanol mixture and placed on a rotator wheel for 1 h. After the mixture was removed, a 3:1 resin/100% ethanol mixture was added to the pellet samples, and then the samples were placed on a rotator wheel for 1 h, then it replaced with pure resin. The samples were then placed in a compression chamber and allowed to sit overnight. The pellet pieces were then placed into plastic TEM sample capsules, filled completely with resin, and placed into a compression chamber for 1 h (this was done to remove any air bubbles remaining in the resin). The samples were removed, and then placed in an oven set at 60 °C for 2 days for polymerization. The capsules (containing the bacteria pellets) were sectioned using a Leica EM UC7 model ultramicrotome at a thickness of 70 nm. The sections generated were placed on copper EM grids for staining. The sample grids were placed on top of drops of 8% aqueous solution of uranyl acetate for 5 min. Each grid was washed via ten dips in three DI H2O-filled beakers. The grids were then allowed to dry for 1 h, and then placed on top of drops of 3% stabilized solution of lead citrate for 2 min. The grids were then washed via ten dips in three DI H2O beakers, and then allowed to dry for 1 h. The grids were visualized on a Hitachi HT7700 Transmission Electron Microscope.
Gingipain assay
To assess the activity of arginine and lysine gingipains, cells were grown anaerobically in 10 ml of BHIYHK to mid-exponential phase. Planktonic cultures were pelleted and normalized to an OD600: 1.0, and 1 ml of each culture was centrifuged at 15,000 × g for 10 min at 4 °C. The supernatants were transferred to separate tubes while the pellets were resuspended in the gingipain assay buffer containing 200 mM Tris, 5 mM CaCl2, 150 mM NaCl, and 10 mM l-cysteine, pH 7.6. The cells and supernatants were diluted 1:10 in the gingipain assay buffer and then each sample was assayed at a minimum of two dilutions in the gingipain assay buffer, each in triplicates. The initial absorbance at 405 nm was recorded and the plates were then incubated at 37 °C for 10 min. The reagents N-α-benzoyl-l-arginine-p-nitroanilide (BAPNA) or N-α-acetyl-l-lysine-p-nitroanilide (ALPNA) was added to the relevant wells at a final concentration of 1 mM and the plates were incubated for 2 h at 37 °C49. The final absorbance (405 nm) of the wells was measured and the difference between the initial and final absorbance was determined.
Galleria mellonella infection model
Larvae of G. mellonella were used to assess virulence of P. gingivalis strain 381 and c-di-AMP relevant mutants as described previously51,91. To this end, BHIYHK-grown bacteria were transferred to fresh medium to OD600: 0.3 and incubated anaerobically to mid-exponential phase. Bacteria were centrifuged and washed with PBS. The pellets were resuspended in PBS and then normalized to OD600: 1.0. Groups of 15 larvae with optimal weight, ranging from 200 to 300 mg, were injected with 5 μl of bacterial inoculum containing ~8 × 108 CFU using a Hamilton syringe. Injection was carried out into the hemocoel of each larva via the last left proleg. After injection, larvae were kept at 37 °C, and survival was recorded at select intervals for up to 160 h.
Statistical analysis
All experiments were conducted using at least three biological replicates. GraphPad Prism version 9.2.0. was applied for performing Student’s t test in pairwise comparisons. For other data, the Shapiro–Wilk test was used to evaluate the normality of the distribution of the data. Parametric data were analyzed by ANOVA followed by post hoc Tukey’s honestly significant difference (HSD) test for pairwise comparisons, using GraphPad Prism version 9.2.0. Differences in the data were considered significant when the probability value was < 5.0% (P value < 0.05). The results of virulence assessment using Galleria mellonella infection model were analyzed with GraphPad Prism 9.2.0. software by using the Mantel–Cox log-rank test. Experiments were performed independently at least two times.
Reporting summary
Further information on experimental and research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We are thankful to Ken Brittian (Electron Microscopy Core, KY-INBRE, University of Louisville) for performing TEM analysis. We gratefully acknowledge the support of Dr. Mary Ellen Davey (University of Florida) to M.F.M in this work. This work was supported by start-up funding from the Department of Oral Immunology and Infectious Diseases, School of Dentistry, University of Louisville, NIH/NIDCR: R03DE031854, and NIH/NIGMS: GM125504 awarded to M.F.M.
Author contributions
M.F.M. conceived the project, designed the experiments, and wrote the manuscript. M.F.M. and S.G. contributed to data acquisition. H.B. and R.S. contributed to determining the concentrations of cNMPs and c-di-NMPs using LC-MS. R.J.L. and D.A.S. contributed to conceiving the project and performed critical review of the manuscript. All authors reviewed and edited the manuscript.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper, and its Supplementary Information files or from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-022-00316-w.
References
- 1.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral. Microbiol. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat. Rev. Microbiol. 2018;16:745–759. doi: 10.1038/s41579-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinberg A, Belton CM, Park Y, Lamont RJ. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 1997;65:313–316. doi: 10.1128/iai.65.1.313-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potempa J, Banbula A, Travis J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol 2000. 2000;24:153–192. doi: 10.1034/j.1600-0757.2000.2240108.x. [DOI] [PubMed] [Google Scholar]
- 5.Socransky SS, Haffajee AD, Smith C, Dibart S. Relation of counts of microbial species to clinical status at the sampled site. J. Clin. Periodontol. 1991;18:766–775. doi: 10.1111/j.1600-051X.1991.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 6.Uematsu H, Hoshino E. Predominant obligate anaerobes in human periodontal pockets. J. Periodontal Res. 1992;27:15–19. doi: 10.1111/j.1600-0765.1992.tb02080.x. [DOI] [PubMed] [Google Scholar]
- 7.Mysak J, et al. Porphyromonas gingivalis: major periodontopathic pathogen overview. J. Immunol. Res. 2014;2014:476068. doi: 10.1155/2014/476068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cugini, C., Klepac-Ceraj, V., Rackaityte, E., Riggs, J. E. & Davey, M. E. Porphyromonas gingivalis: keeping the pathos out of the biont. J. Oral Microbiol. 5, 19804 (2013). [DOI] [PMC free article] [PubMed]
- 9.How KY, Song KP, Chan KG. Porphyromonas gingivalis: an overview of periodontopathic pathogen below the gum line. Front. Microbiol. 2016;7:53. doi: 10.3389/fmicb.2016.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen I, Lambris JD, Hajishengallis G. Porphyromonas gingivalis disturbs host-commensal homeostasis by changing complement function. J. Oral. Microbiol. 2017;9:1340085. doi: 10.1080/20002297.2017.1340085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 1998;62:1244–1263. doi: 10.1128/MMBR.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajishengallis G, Lamont RJ. Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur. J. Immunol. 2014;44:328–338. doi: 10.1002/eji.201344202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y, Nguyen KA, Potempa J. Dichotomy of gingipains action as virulence factors: from cleaving substrates with the precision of a surgeon’s knife to a meat chopper-like brutal degradation of proteins. Periodontol 2000. 2010;54:15–44. doi: 10.1111/j.1600-0757.2010.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grenier D, et al. Effect of inactivation of the Arg- and/or Lys-gingipain gene on selected virulence and physiological properties of Porphyromonas gingivalis. Infect. Immun. 2003;71:4742–4748. doi: 10.1128/IAI.71.8.4742-4748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slaney JM, Gallagher A, Aduse-Opoku J, Pell K, Curtis MA. Mechanisms of resistance of Porphyromonas gingivalis to killing by serum complement. Infect. Immun. 2006;74:5352–5361. doi: 10.1128/IAI.00304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moradali, M. F. & Davey, M. E. Metabolic plasticity enables lifestyle transitions of Porphyromonas gingivalisnpj Biofilms Microbiomes7, 1–13 (2021). [DOI] [PMC free article] [PubMed]
- 17.Moradali MF, Ghods S, Angelini TE, Davey ME. Amino acids as wetting agents: surface translocation by Porphyromonas gingivalis. ISME J. 2019;13:1560–1574. doi: 10.1038/s41396-019-0360-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gürsoy UK, Gürsoy M, Könönen E, Sintim HO. Cyclic dinucleotides in oral bacteria and in oral biofilms. Front. Cell Infect. Microbiol. 2017;7:273. doi: 10.3389/fcimb.2017.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson JW, et al. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat. Chem. Biol. 2013;9:834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stülke J, Krüger L. Cyclic di-AMP signaling in bacteria. Annu. Rev. Microbiol. 2020;74:159–179. doi: 10.1146/annurev-micro-020518-115943. [DOI] [PubMed] [Google Scholar]
- 21.Yin W, et al. A decade of research on the second messenger c-di-AMP. FEMS Microbiol Rev. 2020;44:701–724. doi: 10.1093/femsre/fuaa019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaudhuri S, et al. Identification of a diguanylate cyclase and its role in Porphyromonas gingivalis virulence. Infect. Immun. 2014;82:2728–2735. doi: 10.1128/IAI.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pesavento C, Hengge R. Bacterial nucleotide-based second messengers. Curr. Opin. Microbiol. 2009;12:170–176. doi: 10.1016/j.mib.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Enersen M, Olsen I, Kvalheim Ø, Caugant DA. fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis. J. Clin. Microbiol. 2008;46:31–42. doi: 10.1128/JCM.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Igboin CO, Griffen AL, Leys EJ. Porphyromonas gingivalis strain diversity. J. Clin. Microbiol. 2009;47:3073–3081. doi: 10.1128/JCM.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chastain-Gross, R. P. et al. Genome Sequence of Porphyromonas gingivalis Strain 381. Genome Announc5, e01467-16 (2017). [DOI] [PMC free article] [PubMed]
- 28.Rodrigues PH, et al. Porphyromonas gingivalis strain specific interactions with human coronary artery endothelial cells: a comparative study. PLoS ONE. 2012;7:e52606. doi: 10.1371/journal.pone.0052606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang F, et al. Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase. Biochem J. 2018;475:191–205. doi: 10.1042/BCJ20170739. [DOI] [PubMed] [Google Scholar]
- 30.Commichau, F. M., Heidemann, J. L., Ficner, R. & Stülke, J. Making and breaking of an essential poison: the cyclases and phosphodiesterases that produce and degrade the essential second messenger cyclic di-AMP in bacteria. J. Bacteriol. 201, e00462-18 (2019). [DOI] [PMC free article] [PubMed]
- 31.Hengge R, et al. Recent advances and current trends in nucleotide second messenger signaling in bacteria. J. Mol. Biol. 2019;431:908–927. doi: 10.1016/j.jmb.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 32.He J, Yin W, Galperin MY, Chou SH. Cyclic di-AMP, a second messenger of primary importance: tertiary structures and binding mechanisms. Nucleic Acids Res. 2020;48:2807–2829. doi: 10.1093/nar/gkaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fahmi, T., Port, G. C. & Cho, K. H. c-di-AMP: an essential molecule in the signaling pathways that regulate the viability and virulence of Gram-positive bacteria. Genes8, 197 (2017). [DOI] [PMC free article] [PubMed]
- 35.Hutcherson JA, et al. Comparison of inherently essential genes of Porphyromonas gingivalis identified in two transposon-sequencing libraries. Mol. Oral. Microbiol. 2016;31:354–364. doi: 10.1111/omi.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein BA, Duncan MJ, Hu LT. Defining essential genes and identifying virulence factors of Porphyromonas gingivalis by massively parallel sequencing of transposon libraries (Tn-seq) Methods Mol. Biol. 2015;1279:25–43. doi: 10.1007/978-1-4939-2398-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein BA, et al. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics. 2012;13:578. doi: 10.1186/1471-2164-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vermilyea DM, Moradali MF, Kim HM, Davey ME. PPAD activity promotes outer membrane vesicle biogenesis and surface translocation by Porphyromonas gingivalis. J. Bacteriol. 2021;203:e00343–20. doi: 10.1128/JB.00343-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loesche WJ, Grossman NS. Periodontal disease as a specific, albeit chronic, infection: diagnosis and treatment. Clin. Microbiol. Rev. 2001;14:727–752. doi: 10.1128/CMR.14.4.727-752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aduse-Opoku J, et al. Identification and characterization of the capsular polysaccharide (K-antigen) locus of Porphyromonas gingivalis. Infect. Immun. 2006;74:449–460. doi: 10.1128/IAI.74.1.449-460.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen T, et al. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J. Bacteriol. 2004;186:5473–5479. doi: 10.1128/JB.186.16.5473-5479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham LL, Harris R, Villiger W, Beveridge TJ. Freeze-substitution of gram-negative eubacteria: general cell morphology and envelope profiles. J. Bacteriol. 1991;173:1623–1633. doi: 10.1128/jb.173.5.1623-1633.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamaki S, Sato T, Matsuhashi M. Role of lipopolysaccharides in antibiotic resistance and bacteriophage adsorption of Escherichia coli K-12. J. Bacteriol. 1971;105:968–975. doi: 10.1128/jb.105.3.968-975.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajagopal S, Sudarsan N, Nickerson KW. Sodium dodecyl sulfate hypersensitivity of clpP and clpB mutants of Escherichia coli. Appl. Environ. Microbiol. 2002;68:4117–4121. doi: 10.1128/AEM.68.8.4117-4121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, et al. LPS remodeling is an evolved survival strategy for bacteria. Proc. Natl Acad. Sci. USA. 2012;109:8716–8721. doi: 10.1073/pnas.1202908109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paramonov N, et al. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol. Microbiol. 2005;58:847–863. doi: 10.1111/j.1365-2958.2005.04871.x. [DOI] [PubMed] [Google Scholar]
- 48.Rangarajan M, et al. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J. Bacteriol. 2008;190:2920–2932. doi: 10.1128/JB.01868-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veillard F, Potempa B, Poreba M, Drag M, Potempa J. Gingipain aminopeptidase activities in Porphyromonas gingivalis. Biol. Chem. 2012;393:1471–1476. doi: 10.1515/hsz-2012-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pereira, M. F., Rossi, C. C., da Silva, G. C., Rosa, J. N. & Bazzolli, D. M. S. Galleria mellonella as an infection model: an in-depth look at why it works and practical considerations for successful application. Pathog. Dis.78, ftaa056 (2020). [DOI] [PubMed]
- 51.Kim HM, Davey ME. Synthesis of ppGpp impacts type IX secretion and biofilm matrix formation in Porphyromonas gingivalis. npj Biofilms Microbiomes. 2020;6:5. doi: 10.1038/s41522-020-0115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dos Santos JD, et al. Immunomodulatory effect of photodynamic therapy in Galleria mellonella infected with Porphyromonas gingivalis. Micro. Pathog. 2017;110:507–511. doi: 10.1016/j.micpath.2017.07.045. [DOI] [PubMed] [Google Scholar]
- 53.Devaux L, et al. Cyclic di-AMP regulation of osmotic homeostasis is essential in Group B Streptococcus. PLoS Genet. 2018;14:e1007342. doi: 10.1371/journal.pgen.1007342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bai Y, et al. Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-AMP. PLoS ONE. 2012;7:e35206. doi: 10.1371/journal.pone.0035206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dengler V, et al. Mutation in the C-di-AMP cyclase dacA affects fitness and resistance of methicillin resistant Staphylococcus aureus. PLoS ONE. 2013;8:e73512. doi: 10.1371/journal.pone.0073512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gundlach J, et al. An essential poison: synthesis and degradation of cyclic di-AMP in Bacillus subtilis. J. Bacteriol. 2015;197:3265–3274. doi: 10.1128/JB.00564-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo Y, Helmann JD. Analysis of the role of Bacillus subtilis σ(M) in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol. Microbiol. 2012;83:623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mehne FM, et al. Cyclic di-AMP homeostasis in Bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J. Biol. Chem. 2013;288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. C-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12:594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng X, Zhang Y, Bai G, Zhou X, Wu H. Cyclic di-AMP mediates biofilm formation. Mol. Microbiol. 2016;99:945–959. doi: 10.1111/mmi.13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao A, Serganov A. Structural insights into recognition of c-di-AMP by the ydaO riboswitch. Nat. Chem. Biol. 2014;10:787–792. doi: 10.1038/nchembio.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krüger L, et al. A meet-up of two second messengers: the c-di-AMP receptor DarB controls (p)ppGpp synthesis in Bacillus subtilis. Nat. Commun. 2021;12:1210. doi: 10.1038/s41467-021-21306-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Corrigan RM, Gründling A. Cyclic di-AMP: another second messenger enters the fray. Nat. Rev. Microbiol. 2013;11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 65.Zhu Y, et al. Cyclic-di-AMP synthesis by the diadenylate cyclase CdaA is modulated by the peptidoglycan biosynthesis enzyme GlmM in Lactococcus lactis. Mol. Microbiol. 2016;99:1015–1027. doi: 10.1111/mmi.13281. [DOI] [PubMed] [Google Scholar]
- 66.Rismondo J, et al. Phenotypes associated with the essential diadenylate cyclase CdaA and its potential regulator CdaR in the human pathogen Listeria monocytogenes. J. Bacteriol. 2016;198:416–426. doi: 10.1128/JB.00845-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Witte CE, et al. Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. mBio. 2013;4:e00282–13. doi: 10.1128/mBio.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Z, Liu D, Liu S, Zhang S, Pan Y. The role of Porphyromonas gingivalis outer membrane vesicles in periodontal disease and related systemic diseases. Front. Cell Infect. Microbiol. 2020;10:585917. doi: 10.3389/fcimb.2020.585917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farrugia C, Stafford GP, Murdoch C. Porphyromonas gingivalis outer membrane vesicles increase vascular permeability. J. Dent. Res. 2020;99:1494–1501. doi: 10.1177/0022034520943187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuboniwa M, et al. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol. Microbiol. 2006;60:121–139. doi: 10.1111/j.1365-2958.2006.05099.x. [DOI] [PubMed] [Google Scholar]
- 71.Kuboniwa M, et al. Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nat. Microbiol. 2017;2:1493–1499. doi: 10.1038/s41564-017-0021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hajishengallis G, Lamont RJ. Dancing with the stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. 2016;24:477–489. doi: 10.1016/j.tim.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daep CA, Novak EA, Lamont RJ, Demuth DR. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect. Immun. 2011;79:67–74. doi: 10.1128/IAI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hausmann E, Weinfeld N, Miller WA. Effects of lipopolysaccharides on bone resorption in tissue culture. Calcif. Tissue Res. 1972;9:272–282. doi: 10.1007/BF02061967. [DOI] [PubMed] [Google Scholar]
- 75.Millar SJ, Goldstein EG, Levine MJ, Hausmann E. Modulation of bone metabolism by two chemically distinct lipopolysaccharide fractions from Bacteroides gingivalis. Infect. Immun. 1986;51:302–306. doi: 10.1128/iai.51.1.302-306.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Steffen MJ, Holt SC, Ebersole JL. Porphyromonas gingivalis induction of mediator and cytokine secretion by human gingival fibroblasts. Oral. Microbiol. Immunol. 2000;15:172–180. doi: 10.1034/j.1399-302x.2000.150305.x. [DOI] [PubMed] [Google Scholar]
- 77.Al-Qutub MN, et al. Hemin-dependent modulation of the lipid A structure of Porphyromonas gingivalis lipopolysaccharide. Infect. Immun. 2006;74:4474–4485. doi: 10.1128/IAI.01924-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Olsen I, Singhrao SK. Importance of heterogeneity in Porhyromonas gingivalis lipopolysaccharide lipid A in tissue specific inflammatory signalling. J. Oral. Microbiol. 2018;10:1440128. doi: 10.1080/20002297.2018.1440128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matsuura M. Structural modifications of bacterial lipopolysaccharide that facilitate Gram-Negative bacteria evasion of host innate immunity. Front. Immunol. 2013;4:109. doi: 10.3389/fimmu.2013.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mazgaeen, L. & Gurung, P. Recent advances in lipopolysaccharide recognition systems. Int. J. Mol. Sci.21, 379 (2020). [DOI] [PMC free article] [PubMed]
- 81.Moradali MF, Rehm BHA. Polymers produced by bacteria: from pathogenesis to advanced materials. Nat. Rev. Microbiol. 2020;18:195–210. doi: 10.1038/s41579-019-0313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Q, Wright CJ, Dingming H, Uriarte SM, Lamont RJ. Oral community interactions of Filifactor alocis in vitro. PLoS ONE. 2013;8:e76271. doi: 10.1371/journal.pone.0076271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moye ZD, Valiuskyte K, Dewhirst FE, Nichols FC, Davey ME. Synthesis of sphingolipids impacts survival of Porphyromonas gingivalis and the presentation of surface polysaccharides. Front. Microbiol. 2016;7:1919. doi: 10.3389/fmicb.2016.01919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davey ME, Duncan MJ. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J. Bacteriol. 2006;188:5510–5523. doi: 10.1128/JB.01685-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valentine PJ, Shoemaker NB, Salyers AA. Mobilization of bacteroides plasmids by bacteroides conjugal elements. J. Bacteriol. 1988;170:1319–1324. doi: 10.1128/jb.170.3.1319-1324.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bähre H, Kaever V. Mass spectrometric analysis of non-canonical cyclic nucleotides. Handb. Exp. Pharm. 2017;238:293–306. doi: 10.1007/164_2016_5001. [DOI] [PubMed] [Google Scholar]
- 87.Bähre H, Kaever V. Measurement of 2’,3’-cyclic nucleotides by liquid chromatography-tandem mass spectrometry in cells. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014;964:208–211. doi: 10.1016/j.jchromb.2014.02.046. [DOI] [PubMed] [Google Scholar]
- 88.Beste KY, Burhenne H, Kaever V, Stasch JP, Seifert R. Nucleotidyl cyclase activity of soluble guanylyl cyclase α1β1. Biochemistry. 2012;51:194–204. doi: 10.1021/bi201259y. [DOI] [PubMed] [Google Scholar]
- 89.Bähre H, Kaever V. Identification and quantification of cyclic di-guanosine monophosphate and its linear metabolites by reversed-phase LC-MS/MS. Methods Mol. Biol. 2017;1657:45–58. doi: 10.1007/978-1-4939-7240-1_5. [DOI] [PubMed] [Google Scholar]
- 90.Bainbridge BW, Hirano T, Grieshaber N, Davey ME. Deletion of a 77-base-pair inverted repeat element alters the synthesis of surface polysaccharides in Porphyromonas gingivalis. J. Bacteriol. 2015;197:1208–1220. doi: 10.1128/JB.02589-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gaca AO, Abranches J, Kajfasz JK, Lemos JA. Global transcriptional analysis of the stringent response in Enterococcus faecalis. Microbiology. 2012;158:1994–2004. doi: 10.1099/mic.0.060236-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper, and its Supplementary Information files or from the corresponding author upon request.