Abstract
The 11-kb sequence encompassing the alkylbenzene upper pathway in Pseudomonas putida 01G3, a psychrotrophic strain able to degrade alkylbenzenes at low temperatures, was characterized. Together with a potential regulator (EbdR), six putative enzymes (EbdAaAbAcAdBC) were identified, and they exhibited highly significant similarities with enzymes implicated in the equivalent pathway in P. putida RE204. ebd genes appeared to be preferentially induced by ethylbenzene. Multiple-alignment data and growth rate measurements led us to classify 01G3 and closely related strains in two groups with distinct substrate specificities. Close to identified genes, remnants of IS5-like elements provided insight into the evolution of catabolic sequences through rearrangements from a less complex ancestral cluster.
To some extent, the xyl, tod, and bph operons in the aromatic compound-degrading strains Pseudomonas putida PaW1 (27) and F1 (28) and Pseudomonas pseudoalcaligenes KF707 (22), respectively, can be considered the prototypes of meta-aromatic compound degradative pathways. Both tod and bph operons initiate the degradation steps with an aromatic oxidation catalyzed by a multicomponent dioxygenase. As is the case for strains F1 and KF707, most of the degradative pseudomonads that have been studied are mesophilic. Since much of our planet is often exposed to temperatures below 5°C, it is likely that psychrotrophic and psychrophilic strains play an important role in bioremediation of polluted cold environments. We previously isolated P. putida 01G3 for its ability to metabolize toluene at 4°C (5). In this study, using an insertional mutagenesis approach, we performed molecular characterization of the alkylbenzene catabolic pathway responsible for this low-temperature degradation. Comparisons with previously identified pathways led us to distinguish two subfamilies of pathways. Possible implications for regulation of an adjacent cloned gene are discussed.
Isolation and characterization of mini-Tn5lacZ1 mutants.
Using a P. putida 01G3 spontaneous Smr mutant strain (01G3S) as the recipient and the transposon delivery vector pUTmini-Tn5lacZ1 (Kmr) (7) harbored by Escherichia coli S17-1 (24), we performed a random mutagenesis experiment (9) to generate clones affected in the alkylbenzene catabolic pathway. Approximately 25,000 tranposon insertion mutants were grown on selective MMO medium (18) and screened for both reductive dioxygenase and catechol 2,3-dioxygenase activities by using indigo (Ind) (14) and catechol (Cat) tests (26). Fifteen clones (Ind− and/or Cat−) were unable to grow on MMO medium supplemented with toluene (0.1%, vol/vol) and were kept and used for further analysis.
A 0.9-kb HindIII fragment of the kanamycin resistance gene was used as a probe to subclone two genomic fragments, A1 (a 7.8-kb SacI fragment from G3A1, a Ind− Cat+ mutant) and A32 (a 6.7-kb BamHI fragment from G3A32, a Ind− Cat− mutant), both containing transposon insertion breakpoints, into the pBluescript SKII+ phagemid, and the fragments were sequenced. A1 contained a putative open reading frame (ORF) truncated by a transposon insertion whose deduced translation product exhibited the highest level of similarity (98% identity, 104 of 106 amino acids) with the C-terminal portion of the α-subunit of isopropylbenzene dioxygenase (IpbAa) encoded by plasmid pRE4 in P. putida RE204 (8, 9). A32 also contained a truncated ORF whose deduced protein sequence exhibited 88% identity (120 of 136 amino acids) with the C-terminal part of IpbR, the ipb positive regulator encoded by the same plasmid.
Cloning alkylbenzene catabolism genes.
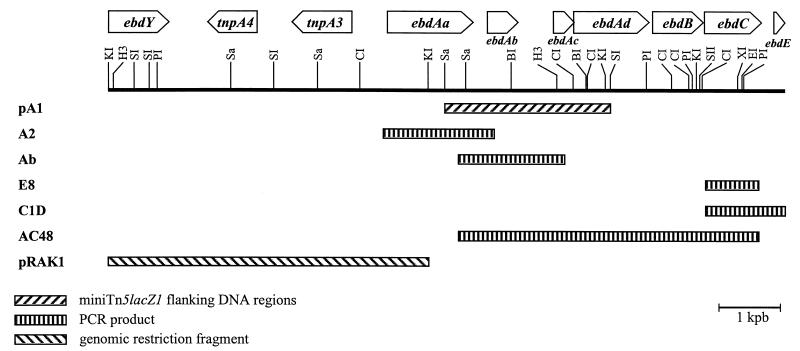
An extensive search for plasmid DNA in 01G3 by various extraction methods was negative, suggesting that the genes encoding the alkylbenzene catabolic pathway in 01G3 are located on the chromosome. This practically excludes the possibility that P. putida 01G3 and RE204 represent close variants of the same strain. Nevertheless, on the basis of the high level of similarity between 01G3 partial sequences and RE204 partial sequences, the following primers were designed to generate overlapping 01G3 DNA amplicons: forward primers P1 (5′-GGGTGAGAAACTGGTCTTCG-3′; positions 7571 to 7590), P2 (5′-CCTTTTTGTGCTCAGATGGGGGTCG-3′; positions 8745 to 8770), and P5 (5′-GCATGGAGATCTTTCCTCTC-3′; positions 12758 to 12777); and reverse primers P3 (5′-CTCCATCGCCTTGTTTCGGG-3′; positions 9320 to 9339), P4 (5′-GGTGCTGCTTTTATCTTGCCCGTGC-3′; positions 10481 to 10505), P6 (5′-TGACCCCACATATCGATCCG-3′; positions 13586 to 13605), and P7 (5′-CTGGTGACGCCGATTTTCTTGC-3′; positions 14047 to 14068) (numbering based on the sequence deposited under accession no. AF006691). Four amplicons (A2 [1.77 kb] obtained with primers P1 and P3, Ab [1.70 kb] obtained with primers P2 and P4, C1D [1.31 kb] obtained with primers P5 and P7, and AC48 [4.86 kb] obtained with primers P2 and P6) were cloned into the pGEM-T vector (Promega) and sequenced which allowed us to reconstitute a unique data set covering 6,381 bp (Fig. 1).
FIG. 1.
Cloning and sequencing strategy for the genes encoding catabolism of alkylbenzene in P. putida 01G3 (11,023-bp fragment). The directions of transcription are indicated by arrowheads. The pBluescriptII SK-derived plasmids (pA1 and pRAK1) and amplicons (A2, Ab, E8, C1D, and AC48) used for sequencing are shown. Abbreviations: CI, ClaI; BI, BamHI; EI, EcoRI; H3, HindIII; KI, KpnI; PI, PstI; SI, SacI; SII, SacII; Sa, SalI; XI, XhoI; kpb, kilobase pair.
Protein sequence analysis of the pathway.
When the sequence was analyzed, six putative proteins could be directly related to alkylbenzene catabolism. Four of them (EbdAa, EbdAb, EbdAc, and EbdAd) exhibited the highest levels of identity with the ipb dioxygenase components of RE204 (98, 88, 87, and 94% with IpbAa, IpbAb, IpbAc, and IpbAd, respectively [Table 1]). The other proteins (EbdB and EbdC) were found to be very similar to RE204 IpbB (cis-dihydrodiol dehydrogenase) and IpbC (3-isopropylcatechol-2,3-dioxygenase) (96 and 95% identity, respectively). The sequence identified was presumed to encode all the enzymes necessary for alkylbenzene upper pathway degradation in 01G3. Another ORF, designated ebdE, which may encode the 54 N-terminal amino acids of a 2-hydroxypenta-2,4-dienoate hydratase, was also detected further downstream (Fig. 1). The previously reported (19) internal gene orf3, whose functionality remains to be demonstrated, was not found in P. putida 01G3. The general organization of the ebd ORFs along the chromosome was found to be identical to the organization observed in the ipb operon in RE204 (9), as well as the organizations observed in other aromatic compound catabolism operons (1, 10, 19, 20, 22, 28).
TABLE 1.
Catabolic genes of P. putida 01G3 involved in biotransformation of alkylbenzene to corresponding catechol derivatives
|
P. putida 01G3 ebd upper pathway
|
Similar sequences
|
||||
|---|---|---|---|---|---|
| Gene | Location (positions) | G+C content (%) | Mol wt (amino acid residues)a | Sequence | % Identityb |
| ebdAa | 4596–5969 | 52.1 | 52,283 (459) | RE204 IpbAa | 98 |
| JR1 IpbA1 | 90 | ||||
| ebdAb | 6202–6696 | 49.1 | 19,467 (166) | RE204 IpbAb | 85 |
| JR1 IpbA2 | 76 | ||||
| ebdAc | 7266–7593 | 48.8 | 11,688 (109) | RE204 IpbAc | 87 |
| JR1 IpbA3 | 74 | ||||
| ebdAd | 7592–8821 | 59.1 | 43,390 (411) | RE204 IpbAd | 93 |
| JR1 IpbA4 | 79 | ||||
| ebdB | 8868–9695 | 54.8 | 29,093 (276) | RE204 IpbB | 95 |
| JR1 IpbB | 85 | ||||
| ebdC | 9703–10638 | 51 | 34,693 (312) | RE204 IpbC | 94 |
| JR1 IpbC | 81 | ||||
Predicted molecular weight (number of amino acid residues) of the corresponding protein based on the gene sequence data.
Sequence and growth rate comparison.
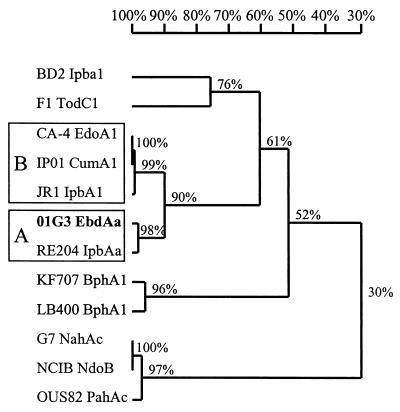
Using the DNAMAN analysis package (Lynnon Biosoft), we aligned α-subunit sequences from aromatic ring-hydroxylating dioxygenases (16) with the EbdAa sequence (Fig. 2). EbdAa appeared to be more closely related (98% identity) to the RE204 dioxygenase α-subunit IpbAa than to any other sequence (group A). The α-subunits of Pseudomonas sp. strain JR1 (19) and Pseudomonas fluorescens IP01 (1) isopropylbenzene dioxygenases (IpbA1 and CumA1, respectively), as well as of P. fluorescens CA-4 ethylbenzene dioxygenase (EdoA1) (6), formed a distinct group (group B) exhibiting 99% identity. The level of identity between groups A and B was 90%. Similar results were obtained with all other proteins (data not shown). Surprisingly, when β-subunits were compared, the EbdAb sequence lacked 20 amino acids (positions 210 to 229 in IpbAa). This deletion, however, did not seem to affect the mineralizing activity of 01G3 (Table 2). When data from all six proteins were taken into account, the group A and B peptide sequences exhibited 93 and 97.6% identity, respectively, on average, and the level of identity between groups A and B was only 80%. At the DNA level (6,045 bp; positions 4596 to 10640 in 01G3) 01G3 and RE204 (group A) exhibited only 77% identity. The level of identity between IP01 and JR1 (group B) was 98%. For the same region, groups A and B exhibited only 63% identity. This means that 01G3 and RE204 could not represent variants of the same strain and shows that group B is more homogeneously conserved than group A.
FIG. 2.
Relationships of 01G3 α-subunit (EbdAa) with α-subunits of related multicomponent dioxygenases. The values at the nodes are pairwise alignment identity values. The following proteins and bacterial strains (GenBank accession numbers) are included: BD2 IpbA1, isopropylbenzene dioxygenase from R. erythropolis BD2 (U24277); F1 TodC1, toluene dioxygenase from P. putida F1 (J04996); CA-4 EdoA1, ethylbenzene dioxygenase from P. fluorescens CA-4 (AF049851); IP01 CumA1, cumene dioxygenase from P. fluorescens IP01 (D37828); JR1 IpbAa, isopropylbenzene dioxygenase from Pseudomonas sp. strain JR1 (U53507); 01G3 EbdAa, alkylbenzene dioxygenase from P. putida 01G3; RE204 IpbAa, isopropylbenzene dioxygenase from P. putida RE204 (AF006691); KF707 BphA1, biphenyl dioxygenase from P. pseudoalcaligenes KF707 (AF049345); LB400 BphA1, biphenyl dioxygenase from Burkholderia sp. strain LB400 (M86348); G7 NahAc, naphthalene dioxygenase from P. putida G7 (M83949); NCIB NdoB, naphthalene dioxygenase from P. putida NCIB 9816 (M23914); OUS82 PahAc, polycyclic aromatic hydrocarbon dioxygenase from P. putida OUS82 (D16629).
TABLE 2.
Growth rates of alkylbenzene-degrading Pseudomonas strains on different substrates
| Strain | Growth rate (h−1) ona:
|
||
|---|---|---|---|
| Toluene | Ethylbenzene | Isopropylbenzene | |
| P. putida 01G3 | 0.38 ± 0.03 | 0.44 ± 0.01 | 0.18 ± 0.03 |
| P. putida RE204 | 0.43 ± 0.02 | 0.49 ± 0.02 | 0.19 ± 0.01 |
| P. fluorescens IP01 | 0.50 ± 0.07 | 0.28 ± 0.03 | 0.32 ± 0.01 |
| Pseudomonas sp. strain JR1 | 0.36 ± 0.04 | 0.30 ± 0.05 | 0.26 ± 0.02 |
Kinetics were determined at 30°C in MMO medium (40 ml) supplemented with an aromatic compound (0.1%, vol/vol) in the gas phase. The results are means based on at least three distinct experiments.
The capacities of groups A and B to metabolize various potential substrates were compared by measuring the growth rates at 30°C (Table 2). Strains were grown on MMO medium, and toluene, ethylbenzene, or isopropylbenzene was supplied in the gas phase at a concentration of 0.1% (vol/vol) as the sole source of carbon and energy. All strains were able to metabolize the substrates tested. The highest growth rates were observed with ethylbenzene for group A strains and with toluene for group B strains. These results confirmed the dichotomy observed with the multiple-sequence alignments. The finding that two groups of closely related proteins apparently discriminated between closely related substrates is intriguing and needs to be confirmed biochemically.
Characterization of ebd promoter.
Using a 900-bp DNA fragment from ebdAa as a probe, we cloned a KpnI 5.5-kb fragment overlapping the 611-bp ebdAa ORF (pRAK1) (Fig. 1). Three other ORFs, tnpA3, tnpA4, and ebdY (ebdY was interrupted at a KpnI cloning site), were detected in this fragment. Compared to sequences from protein databases, the TnpA3 sequence (326 amino acids) showed 95% identity with transposases described for P. putida H (accession no. AF052751), Pseudomonas stutzeri AN10 (2), and P. putida RE204. The TnpA4 sequence (271 amino acids) exhibited only 63% identity with a putative transposase of Pseudomonas syringae (accession no. M14366). The ebdY partial translation product exhibited 54% identity (181 of 325 amino acids) with NahY, a protein implicated in naphthalene chemotaxis in P. putida G7 (13) and with no equivalent described for RE204.
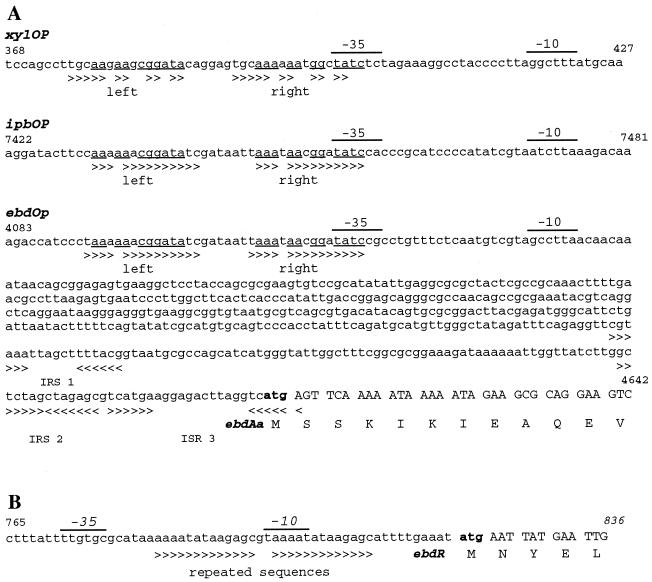
By analogy with putative −35 and −10 boxes of XylOmOp (11) and IpbOp (8), putative −35 and −10 boxes were identified upstream of ebdAa (positions 471 and 448 upstream of ATG, respectively [Fig. 3A]). In addition, an almost perfect (14 of 15 bp) direct repeat motif separated by 6 bp was found to overlap the −35 box. An identical motif (36 of 36 bp) has been described previously at the same position in the ipb operator-promoter region (ipbOP) in RE204 (8), and a closely related motif (22 of 36 bp) has been found upstream of xyl genes (xylOmOp) in P. putida PaW1 (15). No putative XylS activation-related sequence (11) could be detected in ebdOp or in ipbOp. This supports the apparent conservation noticed previously in the two clusters. No other promoter sequences could be detected upstream of the other ORFs, indicating that the ebd genes may be expressed as a single operon. This finding does not exclude the possibility that there are cryptic alternative internal initiations.
FIG. 3.
Regulatory elements of the alkylbenzene catabolism operon. (A) Comparison of the ebd upstream region with outstanding elements harbored by the TOL (pWWO) and pRE4 plasmids. (B) Promoter region of the potential regulator gene ebdR. Putative −35 and −10 boxes are indicated. Regions where potential ebd, ipb, and xyl operators are identical are underlined. The greater-than symbols indicate conserved bases in tandem repeats, and the greater-than and less-than symbols together indicate conserved bases in inverted repeat sequences (IRS). The translation starting points for ebdAa and ebdR are indicated by boldface type.
Cloning and sequence analysis of an upper pathway putative regulator, ebdR.
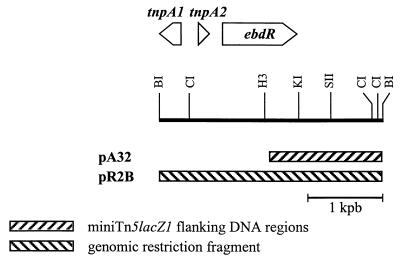
Since the A32 translation product is a good candidate for a regulator of the ebd pathway, a 700-bp SacII subfragment of pA32 was used as a probe to clone a single approximately 3-kb BamHI fragment (pR2B) containing the complete ebdR ORF (Fig. 4). EbdR exhibited 90% identity (299 of 332 amino acids) with the IpbR regulatory protein from RE204 and 67% identity (222 of 332 amino acids) with its equivalent (IpbR) recently identified in JR1 (accession no. AF155505). Multiple-sequence alignments (data not shown) confirmed the division into two groups described above. At the DNA level, identity was restricted to the sole ebdR coding sequence. EbdR was also found to be similar (50% identity on average) to many regulatory proteins of the XylS-AraC family, all of which have been implicated in regulation of aromatic compound catabolic pathways (12) and all of which have the helix-turn-helix motifs involved in DNA binding (4).
FIG. 4.
Cloning and sequencing strategy for the genes encoding catabolism of alkylbenzene in P. putida 01G3 (3,013-bp fragment). The directions of translation are indicated by arrowheads. pA32 and pR2B are pBluescriptII SK+ derivatives used for sequencing. Abbreviations: CI, ClaI; BI, BamHI; H3, HindIII; KI, KpnI; SII, SacII; kpb, kilobase pair.
Analysis of ebdR upstream sequence.
Analysis of the 822-bp upstream region of the ebdR gene did not reveal any significant similarity with the corresponding region in ipbR or xylS, apart from the putative −35 and −10 boxes (Fig. 3B). A 13-bp perfect direct repeat separated by only 2 bp was found to partially overlap the putative −10 element. No equivalent feature was found either in ipbR or in xylS upstream sequences. However, we cannot rule out the possibility that this site may represent a new type of regulator binding site.
Two incomplete transposase-like genes (tnpA1 and tnpA2) apparently transcribed in opposite directions were detected. The TnpA1 protein exhibited 94% identity (90 of 96 amino acids) with TnpA3 encoded upstream of ebdAa. A similar value was obtained for the 96 N-terminal residues of the three transposases encoded upstream of the naphthalene-degrading operon in P. stutzeri AN10 (2, 3). TnpA2, interrupted by an EbdR sequence, was only 48 amino acids long and thus was considered nonfunctional. No significant similarities with a putative transposase described for P. putida RE204 could be found.
Isolation of an ebdC mutant.
The promoterless lacZ gene of miniTn5lacZ1 was introduced into the ORF of ebdC together with the kanamycin marker. Briefly, a 830-bp fragment containing part of ebdC was isolated from a PCR product (E8 amplicon) obtained with primers P5 and P6 by using BglII and ClaI restriction enzymes (corresponding to the underlined bases in the primer sequences in the “Cloning alkylbenzene catabolism genes” section) and subcloned. The plasmid was linearized with EcoRI at an ebdC internal EcoRI site and ligated with a 5.2-kb EcoRI fragment from pUTmini-Tn5lacZ1. The construct was excised from the vector as a XbaI-PstI fragment (5.8 kbp) and subcloned in the pME3087 (Tcr) mobilizable vector (21). The resulting pME8-17 plasmid was then transfected into the S17-1 donor strain (24) prior to conjugation. Integration of the plasmid into the 01G3S recipient yielded Kmr Tcr Smr transconjugants. All LacZ+ merodiploids lacked the characteristic yellow color after catechol spraying, thus showing inactivation of ebdC after insertional recombination and eliminating the possibility of a duplication. All clones still exhibited reductive dioxygenase activity, as determined with the indole test (14). Subcloning and sequencing of the G3D24 mutant DNA insert clearly showed that the intact lacZ gene was effectively inserted 618 bp downstream of the ebdC initiation site (data not shown).
Influence of aromatic substrate on ebd expression level.
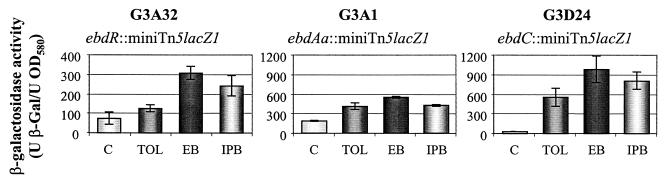
We showed previously that 17°C is a hinge temperature for 01G3 growth (5). G3A32, G3A1, and G3D24 mutants (inactivated in ebdR, ebdAa, and ebdC) were grown at 17°C in selective Luria-Bertani medium supplemented with toluene, ethylbenzene, and isopropylbenzene as potential inducers. β-Galactosidase activity was directly assayed in cell lysates (17). All mutants displayed a marked increase in β-galactosidase activity for all compounds tested compared to a nonsupplemented control (Fig. 5). This result supports the hypothesis that positive regulation occurs. For each mutant, although ebd cluster expression appeared to be enhanced by various related compounds, ethylbenzene was the preferential inducer.
FIG. 5.
Activity of the lacZ reporter gene in ebd insertional mutants. Cells were grown at 17°C in selective Luria-Bertani medium supplemented with toluene (TOL), ethylbenzene (EB), or isopropylbenzene (IPB) until the end of exponential growth. C, unsupplemented control. The results are means based on triplicate determinations. OD580, optical density at 580 nm.
Catabolic sequence evolution.
It has been suggested previously that random recruitment and assembly of preexisting sequences may lead to acquisition or variegation of catabolic pathways (2, 23, 25). Recurrent remnants of IS5-like elements were considered to be supporting evidence that transposition events occur in the actual nah operon structure (2, 3, 8). The presence close to the ebd degradative cluster of four putative transposases exhibiting an average level of identity of 49% with the IS5 functional enzyme supports this hypothesis. One of these transposases, TnpA2, appeared to be truncated by a fragment encoding TnpA1 and EbdR. This may be interpreted as a relic of either the imbrication of two successive transpositions or a failed transposition process. Whatever the mechanism, our data support the idea that like the nah pathway genes of P. stutzeri AN10, the alkylbenzene degradation pathway genes of P. putida 01G3 may have evolved from a less complex ancestral cluster. Bosch et al. have recently suggested that the nah pathway could represent an evolutionary step towards specificity initiated with a large-spectrum catabolic pathway (2). Given this, the ipb pathway carried by plasmid pRE4 in RE204 could be a good candidate for an earlier plasmid pathway that led to emergence of the genomic ebd cluster in 01G3 after chromosomal integration.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the EMBL database under accession numbers AJ293587 and AJ293588.
Acknowledgments
This work was supported by the Contrat de Plan Inter Régional “Grand Bassin Parisien.” P. Chablain and A. Zgoda were recipients of a fellowship from the Ministère de l'Education Nationale, de la Recherche et de la Technologie.
We are grateful to C. Regeard, Université de Rouen, for his help with mutagenesis techniques. We are indebted to R. Eaton, U.S. Environmental Protection Agency, B. Averhoff, Universität Göttingen, Göttingen, Germany, and T. Omori, University of Tokyo, Tokyo, Japan, for supplying bacterial strains RE204, JR1, and IP01.
REFERENCES
- 1.Aoki H, Kimura T, Habe H, Yamane H, Kodama T, Omori T. Cloning, nucleotide sequence and characterization of the genes encoding the enzymes involved in the degradation of cumene to 2-hydroxy-6-oxo-7-methylocta-2,4-dienoic acid in Pseudomonas fluorescens IP01. J Ferment Bioeng. 1996;81:187–196. [Google Scholar]
- 2.Bosch R, Garcia-Valdes E, Moore E R. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation lower pathway from Pseudomonas stutzeri AN10. Gene. 2000;245:65–74. doi: 10.1016/s0378-1119(00)00038-x. [DOI] [PubMed] [Google Scholar]
- 3.Bosch R, Garcia-Valdes E, Moore E R. Genetic characterization and evolutionary implications of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene. 1999;236:149–157. doi: 10.1016/s0378-1119(99)00241-3. [DOI] [PubMed] [Google Scholar]
- 4.Brennan R G, Matthews B W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989;264:1903–1906. [PubMed] [Google Scholar]
- 5.Chablain P A, Philippe G, Groboillot A, Truffaut N, Guespin-Michel J F. Isolation of a soil psychrotrophic toluene-degrading Pseudomonas strain: influence of temperature on the growth characteristics on different substrates. Res Microbiol. 1997;148:153–161. doi: 10.1016/S0923-2508(97)87646-2. [DOI] [PubMed] [Google Scholar]
- 6.Corkery D M, Dobson A D. Reverse transcription-PCR analysis of the regulation of ethylbenzene dioxygenase gene expression in Pseudomonas fluorescens CA-4. FEMS Microbiol Lett. 1998;166:171–176. doi: 10.1111/j.1574-6968.1998.tb13886.x. [DOI] [PubMed] [Google Scholar]
- 7.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaton R W, Selifonova O V, Gedney R M. Isopropylbenzene catabolic pathway in Pseudomonas putida RE204: nucleotide sequence analysis of the ipb operon and neighboring DNA from pRE4. Biodegradation. 1998;9:119–132. doi: 10.1023/a:1008386221961. [DOI] [PubMed] [Google Scholar]
- 9.Eaton R W, Timmis K N. Characterization of a plasmid-specified pathway for catabolism of isopropylbenzene in Pseudomonas putida RE204. J Bacteriol. 1986;168:123–131. doi: 10.1128/jb.168.1.123-131.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson B D, Mondello F J. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J Bacteriol. 1992;174:2903–2912. doi: 10.1128/jb.174.9.2903-2912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallegos M T, Marques S, Ramos J L. Expression of the TOL plasmid xylS gene in Pseudomonas putida occurs from a ς70-dependent promoter or from ς70- and ς54-dependent tandem promoters according to the compound used for growth. J Bacteriol. 1996;178:2356–2361. doi: 10.1128/jb.178.8.2356-2361.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallegos M T, Michan C, Ramos J L. The XylS/AraC family of regulators. Nucleic Acids Res. 1993;21:807–810. doi: 10.1093/nar/21.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimm A C, Harwood C S. NahY, a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J Bacteriol. 1999;181:3310–3316. doi: 10.1128/jb.181.10.3310-3316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins R O, Dalton H. The use of indigo as a spectrophotometric assay substrate for toluene dioxygenase. FEMS Microbiol Lett. 1985;30:227–231. [Google Scholar]
- 15.Kessler B, Timmis K N, de Lorenzo V. The organization of the Pm promoter of the TOL plasmid reflects the structure of its cognate activator protein XylS. Mol Gen Genet. 1994;244:596–605. doi: 10.1007/BF00282749. [DOI] [PubMed] [Google Scholar]
- 16.Mason J R, Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 18.Palleroni N J, Doudoroff M. Some properties and taxonomic subdivisions of the genus Pseudomonas. Annu Rev Phytopath. 1972;10:73–100. [Google Scholar]
- 19.Pflugmacher U, Averhoff B, Gottschalk G. Cloning, sequencing, and expression of isopropylbenzene degradation genes from Pseudomonas sp. strain JR1: identification of isopropylbenzene dioxygenase that mediates trichloroethene oxidation. Appl Environ Microbiol. 1996;62:3967–3977. doi: 10.1128/aem.62.11.3967-3977.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon M J, Osslund T D, Saunders R, Ensley B D, Suggs S, Harcourt A, Suen W C, Cruden D L, Gibson D T, Zylstra G J. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene. 1993;127:31–37. doi: 10.1016/0378-1119(93)90613-8. [DOI] [PubMed] [Google Scholar]
- 21.Simon R, Priefer U, Pehle A. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 22.Taira K, Hirose J, Hayashida S, Furukawa K. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J Biol Chem. 1992;267:4844–4853. [PubMed] [Google Scholar]
- 23.van der Meer J R, de Vos W M, Harayama S, Zehnder A J. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voisard C, Bull C, Keel C, Laville J, Mautofer M, Schnider U, Defago G, Haas D. Biocontrol of root disease by Pseudomonas fluorescens CHAO: current concepts and experimental approaches. In: O'Gara F, Dowling D N, Boesten B, editors. Molecular ecology of rhizosphere microorganisms. Weinheim, Germany: VCH; 1994. pp. 67–89. [Google Scholar]
- 25.Williams P A, Sayers J R. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation. 1994;5:195–217. doi: 10.1007/BF00696460. [DOI] [PubMed] [Google Scholar]
- 26.Winstanley C, Morgan J A, Pickup R W, Saunders J R. Use of a xylE marker gene to monitor survival of recombinant Pseudomonas putida populations in lake water by culture on nonselective media. Appl Environ Microbiol. 1991;57:1905–1913. doi: 10.1128/aem.57.7.1905-1913.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Worsey M J, Williams P A. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]