Abstract
Most mitochondrial proteins are synthesized as precursor proteins (preproteins) in the cytosol and imported into mitochondria. The translocator of the outer membrane (TOM) complex functions as a main entry gate for the import of mitochondrial proteins. The TOM complex is a multi-subunit membrane protein complex composed of a β-barrel channel Tom40 and six single-pass membrane proteins. Recent cryo-EM studies have revealed high-resolution structures of the yeast and human TOM complexes, which enabled us to discuss the mechanism of protein import at an amino-acid residue level. The cryo-EM structures show that two Tom40 β-barrels are surrounded by two sets of small Tom subunits to form a dimeric structure. The intermembrane space (IMS) domains of Tom40, Tom22, and Tom7 form a binding site for presequence-containing preproteins in the middle of the dimer to achieve their efficient transfer of to the downstream translocase, the TIM23 complex. The N-terminal segment of Tom40 spans the channel from the cytosol to the IMS to interact with Tom5 at the periphery of the dimer, where downstream components of presequence-lacking preproteins are recruited. Structure-based biochemical analyses together with crosslinking experiments revealed that each Tom40 channel possesses two distinct paths and exit sites for protein translocation of different sets of mitochondrial preproteins. Here we summarize the current knowledge on the structural features, protein translocation mechanisms, and remaining questions for the TOM complexes, with particular emphasis on their determined cryo-EM structures. This article is an extended version of the Japanese article, Structural basis for protein translocation by the translocase of the outer mitochondrial membrane, published in SEIBUTSU BUTSURI Vol. 60, p. 280-283 (2020).
Keywords: mitochondria, protein translocation, TOM complex preprotein, Cryo-EM
Significance
The TOM complex in the mitochondrial outer membrane functions as the main entry gate for the import of ~1,000 different mitochondrial proteins synthesized in the cytosol into mitochondria. Recent progress in cryo-electron microscopy with single particle analyses allowed us to determine high-resolution structures of the TOM complex, which, in combination with structure-based biochemical analyses, led to revealing significant insight into the molecular mechanisms of the precise as well as efficient import of mitochondrial proteins with divergent chemical and physical properties.
Introduction
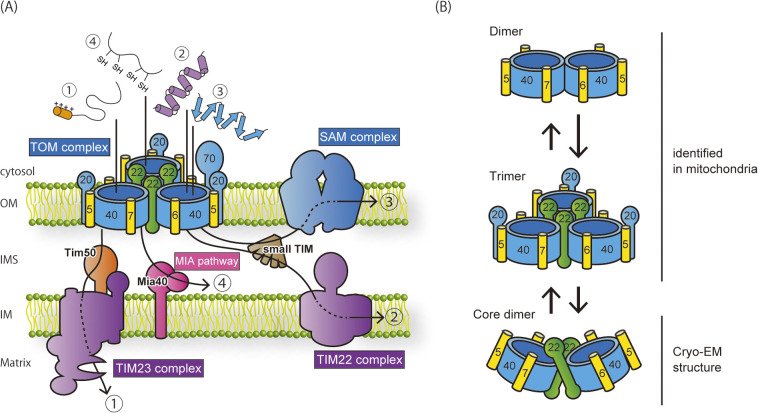
Mitochondria are essential organelles that play key roles in numerous cellular processes such as energy production, signal transduction, and metabolic reactions. To fulfill these functions, around 1,000 different resident proteins work in mitochondria. Mitochondria evolved from aerobic bacteria that were engulfed by ancestral eukaryotic cells and developed into their present shapes as endosymbionts. Today’s mitochondria consist of two membranes, the outer (OM) and inner membranes (IM), and two aqueous compartments, the intermembrane space (IMS) between the outer and inner membranes and the matrix surrounded by the inner membrane. Mitochondria have their own genomic DNA in their matrix, yet it encodes only about ten different proteins. Nearly all mitochondrial proteins are encoded by the nuclear genome, synthesized in the cytosol as precursor proteins (preproteins), and imported back into mitochondria. Therefore, eukaryotic cells have a dedicated system consisting of targeting signals encrypted in mitochondrial proteins and import machineries that decode their targeting signals and import them into mitochondria [1–3]. The imported mitochondrial preproteins are further sorted to different mitochondrial sub-compartments. Today, four major translocators, the TOM and SAM complexes in the OM and the TIM22 and TIM23 complexes in the IM, working on four different import and sorting pathways are known (Fig. 1A).
Figure 1 .
Import pathways of mitochondrial proteins. (A) Four major import pathways and translocators are shown. Upon translocation into the IMS through the TOM complex in the OM, the import pathway is branched to four different pathways. Pathway 1: presequence-containing preproteins are inserted into the IM or transported to the matrix through the TIM23 complex. Pathway 2: polytopic inner-membrane proteins, such as carrier-family proteins, are transferred to the IMS chaperones, small TIM proteins, and inserted into the IM by the TIM22 complex. Pathway 3: β-barrel proteins are transferred to the small TIM proteins and inserted into the OM by the SAM complex. Pathway 4: small soluble IMS proteins are transferred to Mia40 and undergo oxidative folding in the IMS. This schematic diagram is modified from Ref. 4. (B) Different assembly forms of the TOM complex. In intact mitochondria, the TOM complex exists in a dynamic exchange between a major form of the trimer and a minor form of the dimer lacking Tom22 and Tom20. The purified TOM complex in detergent micelles is a mixture of the trimer and the dimer, which contains Tom22 but not Tom20, named the core dimer.
Mitochondrial preproteins can be classified into several different classes according to their sorting pathways after crossing the OM through the TOM complex. Most matrix-targeted preproteins use an N-terminal positively charged amphipathic helix-forming sequence as a targeting signal (presequence) to move through the TOM complex and are passed onto the translocase of the inner mitochondrial membrane (TIM23) complex in the IM (Fig. 1A, pathway 1) [4–6]. Inner-membrane polytopic membrane proteins like carrier proteins and outer-membrane β-barrel proteins are synthesized without an N-terminal presequence and transferred from the TOM complex to the small TIM chaperones in the IMS, and then to the TIM22 complex in the IM and the SAM complex in the OM, respectively (Fig. 1A pathway 2, 3) [1–4,7].
Soluble IMS proteins with specific cysteine-rich motifs are transferred from the TOM complex to the mitochondrial IMS import and assembly (MIA) system (Fig. 1A pathway 4) [8–11].
Each class of newly synthesized preproteins are initially recognized by the receptors of the TOM complex on the mitochondrial surface, cross the OM through the TOM channel, and then transferred to the downstream components, which can be recruited to the outlet of the TOM channel. Therefore, the TOM complex plays a central role in the intramitochondrial sorting as well as in the OM translocation of preproteins.
Structure of the TOM Complex
The TOM complex is a membrane protein complex consisting of a β-barrel channel protein Tom40 and six α-helical single-pass membrane proteins including the receptor subunits Tom70, Tom22, and Tom20, and the auxiliary subunits Tom7, Tom6, and Tom5 (Fig. 1A). These subunits are well conserved from yeast to human, yet sequence analysis across the eukaryotic kingdoms suggests that the ancestral TOM complex could consist of Tom40, Tom22, and Tom7 [12].
The first structural characterization of the TOM complex was conducted by the electron microscopy (EM) analysis in 1998, which revealed that the solubilized and purified TOM complex from Neurospora crassa is a mixture of the trimer containing three pores and dimer with two pores [13]. Afterward, a low-resolution 3D structure of the trimeric TOM complex was reported at 18 Å resolution in 2008 [14], and a mid-resolution dimeric structure was elucidated at 6.8 Å resolution in 2017 by single particle analysis using cryo-EM (Fig. 1B) [15]. Our previous systematic crosslinking analyses revealed that the TOM complex functions as a trimer in intact mitochondria; we could construct a trimer model of the yeast TOM complex based on the structural mapping by in vivo site-specific photocrosslinking, in which three Tom40 channels are sandwiched by the transmembrane helices of the Tom22 subunits, resulting in a triangular form of the entire complex (Fig. 1B) [7]. By using this information, the mid-resolution cryo-EM studies could suggest the spatial arrangement of each subunit, although the cryo-EM structure was a dimeric complex, not a trimeric complex [15]. A previous study also indicated that a fraction of the TOM complex forms a dimer with two Tom40 channels, but lacking the Tom22 subunit in intact mitochondria (Fig. 1B) [16]. The trimeric TOM complex and dimeric one likely handle different subsets of mitochondrial precursor proteins for translocation across the outer membrane [16]. However, it was difficult to discuss the precise mechanisms of the TOM-mediated protein translocation at the level of amino acid residues for more than two decades due to the lack of the high-resolution structures of the TOM complex.
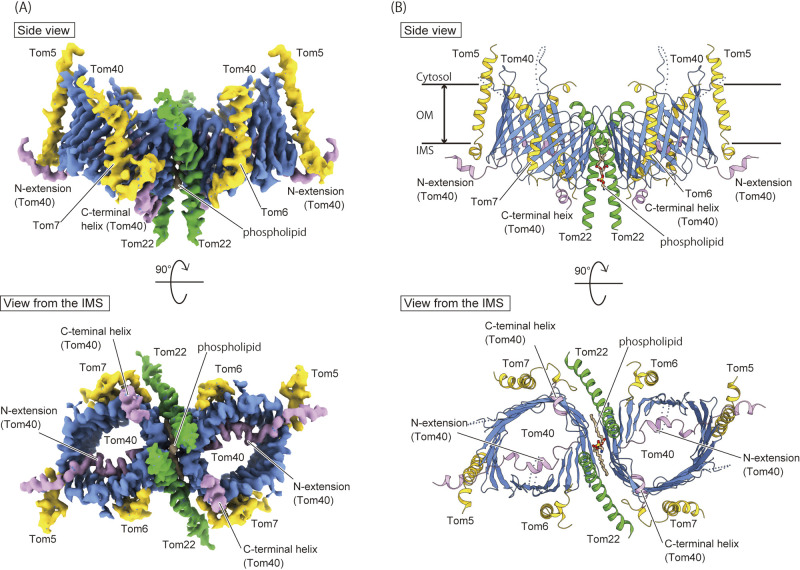
In 2019, our group and Tucker et al. independently determined long-awaited high-resolution structures of the TOM complex of Saccharomyces cerevisiae by cryo-EM single particle analysis (Fig. 2A) [17,18]. The determined cryo-EM structures showed that the TOM complex is a dimeric form, like the mid-resolution cryo-EM structure [15], in which Tom40 forms a 19-strand transmembrane β-barrel channel, and is surrounded by the transmembrane α-helices of Tom22, Tom5, Tom6, and Tom7 (Fig. 2B). The first and last β-strands (β1 and β19) of Tom40 associate with each other, forming a parallel β-sheet, to close the β-barrel. The receptor subunit Tom22 forms a kinked transmembrane helix in the middle of the molecule, whereas its N-terminal receptor domain on the cytosolic side is partially disordered (Fig. 2B and 3A). Although another receptor subunit Tom20 was contained in the purified TOM complex, the determined cryo-EM dimeric structure lacked EM density corresponding to Tom20, suggesting that Tom20 easily dissociates from the TOM dimer [13,19].
Figure 2 .
The cryo-EM structure of the TOM complex from Saccharomyces cerevisiae at 3.8 Å resolution. (A) Cryo-EM density map of the core dimer of the TOM complex (EMDB-9851). The β-barrel channels of the Tom40 are colored blue, the N-extension and the C-terminal helix are colored in pink, the Tom22 subunit in green, and the small Tom subunits such as Tom5, Tom6 and Tom7 in yellow. The phospholipid molecule inserted between the Tom40 channels is colored in brown. (B) The ribbon presentation of the overall structure of the core dimer of the TOM complex (PDB ID: 6JNF). The coloring scheme is the same as in (A). The ribbon presentations are modified from Ref. 4. Molecular graphics were illustrated with Cuemol 2.0 (http://www.cuemol.org) and UCSF chimeraX (http://www.cgl.ucsf.edu/chimerax/).
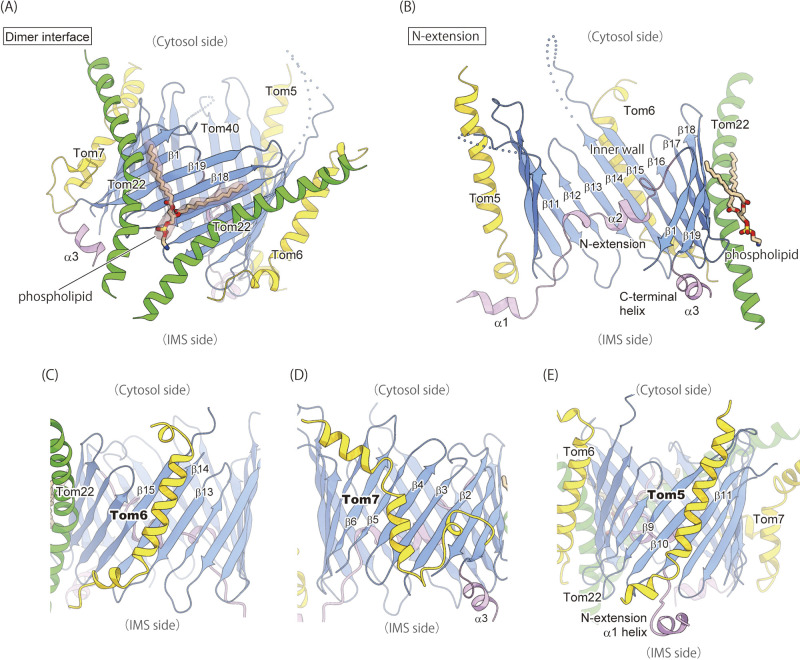
Figure 3 .
The detailed structures of the TOM complex. The coloring scheme is the same as in Figure 2. (A) A close-up view of the dimer interface. Two Tom22 molecules tether the two Tom40 β-barrels. The phospholipid molecule bound to the interface is shown by ball-and-stick model and the EM density is shown by surface representation. (B) A cut-away view of the Tom40 channel. The N-extension and the inner wall are visible because of the removal of strands β2–β8. (C–E) The structures of the transmembrane helices of Tom6 (C), Tom7 (D), and Tom5 (E). Molecular graphics were illustrated with Cuemol 2.0 (http://www.cuemol.org). (A) and (B) are modified from Ref. 4.
The dimeric structure determined by cryo-EM analysis is likely different from the “Tom22-lacking dimer” found in intact mitochondria in the previous biochemical study [16], so that it is referred to as a “core dimer” (Tom22-containing dimer) (Fig. 1B). The core dimer is only found in the structural analysis in the presence of detergent, but observation of the detergent-solubilized TOM complex by high-speed atomic force microscopy (HS-AFM) shows that the trimer is easily converted to a dimer, whose dimension is close to the core dimer rather than the Tom22-lacking dimer, suggesting that the core dimer forms a stable structural unit in the trimer in the mitochondrial membrane [17].
The two Tom40 β-barrels come in contact with each other only at the cytosolic edge and have two molecules of Tom22 inserted between the barrels in the bilayer membrane core (Fig. 2B and 3A). The transmembrane helices of Tom22 and the Tom40 β-barrels form a cavity with opening on the IMS side, and one phospholipid molecule is inserted into the cavity with its head group facing the IMS (Fig. 3A). The presence of the kinked Tom22 helices with a phospholipid molecule explains the tilt of the Tom40 β-barrels, which causes the distorted lipid bilayer (Fig. 2B and 3A). Tom40 has a long N-terminal segment composed of 82 amino-acid residues (N-extension), preceding the first β-strand (β1), and this N-extension penetrates into the β-barrel pore from the cytosolic side all the way to the IMS (Fig. 3B). Two α-helices, α1 and α2, are formed in the N-extension; the α1 helix is located in the IMS and likely associates with the IMS part of Tom5. In contrast, the following α2 helix in the N-extension tightly binds to the inner wall of the Tom40 channel (β11–β17) through hydrophilic interactions (Fig. 3B). On the other hand, the C-terminal segment of Tom40 following the last β-strand (β19) forms a short α-helix (α3) with a flexible loop on the IMS side (Fig. 3B).
Tom5, Tom6, and Tom7, small auxiliary subunits called “small TOM subunits”, bind to the outer wall of the Tom40 β-barrels mainly through hydrophobic interactions (Fig. 2B). The TM helix of Tom6 interacts with the outer wall of the Tom40 β-barrel through interactions with the β13–β15 strands, which are bound by the N-extension of Tom40 on the other side inside the β-barrel (Fig. 3C). On the opposite side to Tom6 of the β-barrel, Tom7 is bound to strands β1–β6 with an unusual “z-shape” topology, in which a short hydrophobic helix spans the membrane toward the IMS, the subsequent hydrophilic loop runs through the IMS, and the hydrophobic helix goes back into the membrane core (Fig. 3D). Tom5 is located in the distal position of the dimer and its long helix binds to strands β9–β11 of Tom40 (Fig. 3E). The long helix of Tom5 is kinked on the IMS side to expose its C-terminal segment to the IMS, which interacts with the α1 helix of the N-extension of Tom40 (Fig. 3B, E). Our biochemical analysis showed that a double mutant of the N-terminal deletion of Tom40 and Tom5 knockout exhibits a synthetic growth defect [17], suggesting the functional cooperation between Tom5 and the N-extension of Tom40.
Protein Translocation Mechanism
Based on the precise structural information, we can now discuss the mechanism of mitochondrial protein import in detail. Upon translocation across the outer membrane through the TOM complex, most mitochondrial proteins are sorted to different sub-compartments via one of the four major protein translocation pathways (Fig. 1). About 60% of mitochondrial proteins possess an N-terminal presequence and are imported into the matrix through the TIM23 pathway (Fig. 1A Pathway 1). Functional studies previously suggested that a trans presequence-binding site is formed on the IMS side of the TOM complex, where Tom7 [20] and the IMS regions of Tom22 [21–23] and Tom40 [23,24] are involved.
Upon translocation through the Tom40 channel, presequence-containing preproteins are transferred to Tim50, an essential subunit of the translocase of inner mitochondrial membrane (TIM23) complex [5,6]. In the cryo-EM structure, the hydrophilic loop between the Tom7 helices α2 and α3, the disordered C-terminal IMS domain of Tom22, and the C-terminal α3 helix of Tom40 (Fig. 4A) are in the close vicinity in the middle of the dimeric structure, which is consistent with the above-mentioned biochemical studies. Furthermore, in vivo crosslinking study indicated that the downstream receptor Tim50 is recruited to the C-terminal IMS domains of Tom22 and Tom40 [7,17]. Thus, the combination of structural and biochemical analyses revealed the presence of the translocation exit site for presequence-containing preproteins in the middle of the dimeric TOM complex for efficient transfer to the recruited downstream component, Tim50 (Fig. 4A).
Figure 4 .
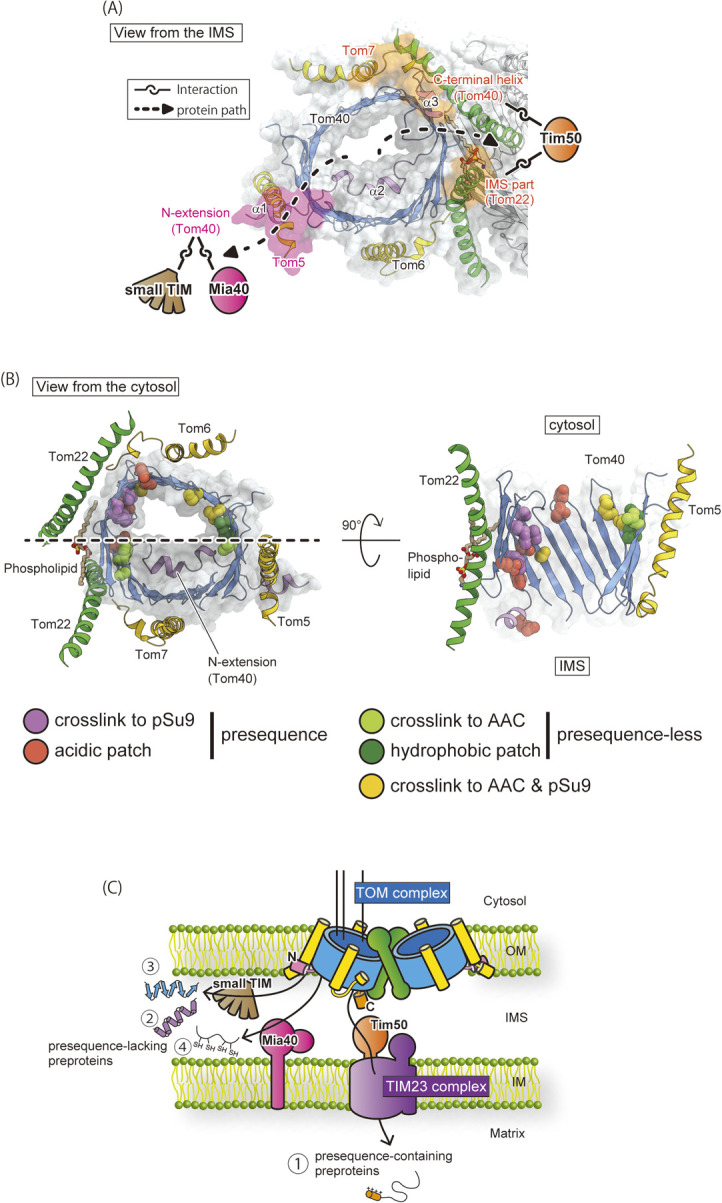
Protein paths and exits of the Tom40 import channel in the TOM complex. (A) Two exit sites are shown on the structure of the TOM complex viewed from the IMS. The exit sites of the presequence-containing and presequece-lacking preproteins are highlighted in orange and pink, respectively. Protein paths are shown by broken lines. The crosslinked downstream components (Tim50, Mia40 and small TIM) are indicated. (B) The protein paths in the Tom40 channel. (Left) The Tom40 channel and surrounding Tom22, Tom7, Tom6, and Tom5 subunits viewed from the cytosol; (right) cross section of the inner wall opposite to the N-extension with a section plane indicated by broken line. For the path for presequence-containing preproteins, the glutamate residues are shown by space-filling form in pink and the residues crosslinked to the model substrate, pSu9-DHFR, by space-filling form in orange. These residues contribute to forming the protein path in the proximity of Tom22. For the path for presequence-lacking preproteins, the hydrophobic residues are shown by space-filling form in green, and the residues crosslinked to the model substrate, AAC-DHFR, by space-filling form in yellowish green. These residues form a protein path leading to Tom5. The residues crosslinked to both pSu9-DHFR and AAC-DHFR are colored in yellow. (C) Schematic model of the protein import and sorting pathways of the TOM complex. The coloring scheme is the same as in Figures 1 and 2. Molecular graphics were illustrated with Cuemol 2.0 (http://www.cuemol.org). (A) and (C) are modified from Ref. 4, and (B) is modified from Ref. 25.
On the other hand, inner-membrane carrier proteins, outer-membrane β-barrel proteins, and small cysteine-rich IMS proteins are synthesized as preproteins lacking a cleavable N-terminal presequence (Fig. 1A pathways 1, 2, and 3). A previous site-specific crosslinking study revealed that the small TIM chaperones are crosslinked to the IMS-exposed part (residues 11–44) of the Tom40 N-extension [7]. The N-extension of Tom40 likely forms an exit site for presequence-less inner-membrane carrier proteins and outer-membrane β-barrel proteins in cooperation with the IMS part of Tom5 for efficient protein transfer to the small TIM chaperones (Fig. 4A). Supporting this model, mitochondria with Tom40 variant lacking the N-terminal 57 residues of Tom40 showed defective in vitro import of carrier and β-barrel precursors, but not of presequence-containing preproteins [7]. We also found that Mia40/Tim40 of the MIA pathway [8–11] was also crosslinked to the disordered region of the Tom40 N-extension including residues 34 and 44 [17], suggesting that the N-extension recruits Mia40 as well as the small TIM chaperones to the IMS outlet of the Tom40 channel (Fig. 4A). Interestingly, in vitro import of MIA substrates, small TIM chaperones, depended on the presence of Tom5 [17], but in the presence of Tom5, the N-terminal deletion of Tom40 up to residue 59 including the α1 helix impaired the in vitro import of Tim9 and Tim12 via the MIA pathway [17]. These results indicate that the import of the MIA substrates requires Tom5 and the N-extension of Tom40 (Fig. 4A). Therefore, presequence-lacking preproteins, the carrier and β-barrel precursors and MIA substrates leave the Tom40 import channel from the exit site formed by Tom5 and the N-terminal extension of Tom40 at the periphery of the dimeric TOM complex, which is on the opposite side of the Tom22/Tom7/Tom40 trans site as an exit for presequence-containing precursors. The IMS-exposed portion of the N-terminal extension of Tom40 recruits Mia40 (for MIA substrates) and small TIM chaperones (for carrier and β-barrel precursors) to ensure the efficient transfer of presequence-lacking preproteins (Fig. 4A).
In addition to the separated translocation exits in the Tom40 channel for different classes of mitochondria preproteins, the high-resolution cryo-EM structures of the TOM complex showed that the inner surface of the Tom40 import channel is optimized for guiding preproteins to their exit sites through distinct paths for efficient transfer to distinct subsequent downstream components. That is, the site-specific crosslinking indicated that presequence-containing and presequence-lacking (hydrophobic) preproteins move along the two separate paths on the inner wall of the Tom40 channel (Fig. 4B) [7,25]. Analysis of electric potential surface shows that the inner wall of the Tom40 channel is negatively charged overall, thereby promoting protein translocation of positively charged presequences [18], yet a closer look at the structure reveals the non-uniform distribution of charged residues. Acidic residues are aligned from the cytosolic side to the IMS side near the dimer interface, forming an acidic path for smooth interactions with preproteins with a positively charged presequence, such as pSu9-DHFR, as revealed by site-specific crosslinking [7,17]. These aligned acid patches are further connected to the C-terminal helix of Tom40 on the IMS side, which forms a trans presequence-binding site (Fig. 4B). The inner wall of the Tom40 channel also contains aligned hydrophobic patches, which include the residues crosslinked to a carrier precursor substrate, like AAC-DHFR (Fig. 4B) [7,17]. Among them, one hydrophobic patch is close to the periphery of the dimer, where Tom5 binds from the outside. This suggests that presequence-lacking preproteins are all guided to the exit site formed by Tom5 and the N-extension of Tom40 (Fig. 4B). The functional importance of acidic and hydrophobic patches was demonstrated by verifying the growth of the cells with mutant Tom40s, in which the acidic/hydrophobic residues of the aligned patches are replaced with basic/hydrophilic residues, respectively [18].
In summary, the Tom40 import channel of the TOM complex possesses two distinct protein paths and exits (Fig. 4C). Presequence-containing preproteins are guided to the exit formed by Tom7, Tom22, and the C-terminal helix of Tom40 in the middle of the dimer. Presequence-lacking preproteins are guided to the exit involving the N-extension with the aid of Tom5 at the dimer periphery (Fig. 4C). Separation of the exits at the Tom40 import channel with the dimension of ~40 Å × 30 Å could contribute to the recruitment of distinct downstream components of the import pathways without steric crowding. Distortion of the bilayer membrane around the TOM complex also contributes to the separation of the outlet of the two Tom40 channels. Thus, the TOM complex can achieve the efficient transport of around 1,000 different mitochondrial preproteins with divergent structural and chemical properties by equipping different translocation routes.
Remaining Questions and Perspectives
Although high-resolution structures of the TOM complexes in the form of the core dimer became available, it remains unexplained how the core dimer is related to the functional trimer and (Tom22-lacking) dimer identified in intact mitochondria. A previous study showed that the trimeric form is a major assembly form of the TOM complex in intact mitochondria, and the trimer undergoes a dynamic conversion into a minor form of the dimer lacking Tom22, and this exchange could contribute to the efficient replacement of a defective subunit with a newly synthesized subunit. (Fig. 1B) [7]. As the cryo-EM analysis led to the successful determination of only the dimeric form containing Tom22, this core dimer could represent a stable assembly unit to function as an assembly platform for developing into a trimeric form [15,17,18], (Fig. 1B). HS-AFM is a powerful tool to analyze dynamic behaviors of protein complex structures at a nm spatial resolution [26]. Our HS-AFM analysis indicated that the purified TOM complex is indeed a mixture of a trimer and a dimer, and the trimer tends to dissociate into a dimer and a monomer, and the dimeric form exhibits a similar dimension to the core dimer revealed by the cryo-EM analyses [17]. These structural observations suggest that the trimeric TOM complex is unstable after solubilization and could be converted to the core dimer. Interestingly, since the geometrical arrangement of each subunit in the core dimer is not compatible with the crosslinking results for the TOM complex in intact mitochondria, a significant conformational rearrangement would be required for the conversion of the trimer into the core dimer form. To further understand the physiological significance and molecular mechanism of such a conformational change, the structural determination of the mature trimeric form of the TOM complex is essential.
To obtain a high-resolution structure of the substrate-bound form of the TOM complex is also a challenging subject. For example, since the TOM complex exists in a dynamic equilibrium, HS-AFM could be used for direct visualization of the conversion between different substrate-bound forms. Besides, as the TOM complex can work in cooperation with other components along the import pathways, such as the TIM 23 complex, the SAM complex, small TIM chaperones, and Mia40, structural analyses of the super-complex between the TOM complex and these components/protein complexes should be also attractive as well as essential subjects. We thus expect more high-resolution structures involving the TOM complex to come, which will provide detailed mechanical insight into the operation of the TOM complex-mediated mitochondrial protein import.
Conflict of Interest
The authors declare no conflicts of interest.
Author Contributions
Y.A. and T.E. wrote the paper.
Acknowledgements
This work was supported by JSPS KAKENHI to Y.A. (20H02583), to T.E. (15H05705 and 2222703). YA was supported by Leading Initiative for Excellent Young Researchers, MEXT, Japan.
References
- [1].Endo, T., Yamano, K.. Multiple pathways for mitochondrial protein traffic. Biol. Chem. 390, 723–730 (2009). https://doi.org/10.1515/BC.2009.087 [DOI] [PubMed] [Google Scholar]
- [2].Neupert, W. A perspective on transport of proteins into mitochondria: A myriad of open questions. J. Mol. Biol. 427, 1135–1158 (2015). https://doi.org/10.1016/j.jmb.2015.02.001 [DOI] [PubMed] [Google Scholar]
- [3].Pfanner, N., Warscheid, B., Wiedemann, N.. Mitochondrial proteins: From biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 20, 267–284 (2019). https://doi.org/10.1038/s41580-018-0092-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Araiso, Y. Structural basis for protein translocation by the translocase of the outer mitochondrial membrane. SEIBUTSU BUTSURI 60, 280–283 (2020). https://doi.org/10.2142/biophys.60.280 [Google Scholar]
- [5].Yamamoto, H., Esaki, M., Kanamori, T., Tamura, Y., Nishikawa, S., Endo, T.. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell 111, 519–528 (1999). https://doi.org/10.1016/s0092-8674(02)01053-x [DOI] [PubMed] [Google Scholar]
- [6].Mokranjac, D., Paschen, S. A., Kozany, C., Prokisch, H., Hoppins, S. C., Nargang F. E., et al. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 22, 816–825 (2003). https://doi.org/10.1093/emboj/cdg090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shiota, T., Imai, K., Qiu, J., Hewitt, V. L., Tan, K., Shen, H. H., et al. Molecular architecture of the active mitochondrial protein gate. Science 349, 1544–1548 (2015). https://doi.org/10.1126/science.aac6428 [DOI] [PubMed] [Google Scholar]
- [8].Gornicka, A., Bragoszewski, P., Chroscicki, P., Wenz, L. S., Schulz, C., Rehling, P., et al. A discrete pathway for the transfer of intermembrane space proteins across the outer membrane of mitochondria. Mol. Biol. Cell 25, 3999–4009 (2014). https://doi.org/10.1091/mbc.E14-06-1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Naoe, M., Ohwa, Y., Ishikawa, D., Ohshima, C., Nishikawa, S., Yamamoto, H., et al. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J. Biol. Chem. 279, 47815–47821 (2004). https://doi.org/10.1074/jbc.M410272200 [DOI] [PubMed] [Google Scholar]
- [10].Chacinska, A., Pfannschmidt, S., Wiedemann, N., Kozjak, V., Sanjuán Szklarz, L. K., Schulze-Specking, A., et al. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 23, 3735–3746 (2004). https://doi.org/10.1038/sj.emboj.7600389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mesecke, N., Terziyska, N., Kozany, C., Baumann, F., Neupert, W., Hell, K., et al. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121, 1059–1069 (2005). https://doi.org/10.1016/j.cell.2005.04.011 [DOI] [PubMed] [Google Scholar]
- [12].Fukasawa, Y., Oda, T., Tomii, K., Imai, K.. Origin and evolutionary alteration of the mitochondrial import system in eukaryotic lineages. Mol. Biol. Evol. 35, 1574–1586 (2017). https://doi.org/10.1093/molbev/msx096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Künkele, K. P., Heins, S., Dembowski, M., Nargang, F. E., Benz, R., Thieffry, M., et al. The preprotein translocation channel of the outer membrane of mitochondria. Cell 93, 1009–1019 (1998). https://doi.org/10.1016/s0092-8674(00)81206-4 [DOI] [PubMed] [Google Scholar]
- [14].Model, K., Meisinger, C., Kühlbrandt, W.. Cryo-electron microscopy structure of a yeast mitochondrial preprotein translocase. J. Mol. Biol. 383, 1049–1057 (2008). https://doi.org/10.1016/j.jmb.2008.07.087 [DOI] [PubMed] [Google Scholar]
- [15].Bausewein, T., Mills, D. J., Langer, J. D., Nitschke, B., Nussberger, S., Kühlbrandt, W.. Cryo-EM structure of the TOM core complex from Neurospora crassa. Cell 170, 693–700 (2017). https://doi.org/10.1016/j.cell.2017.07.012 [DOI] [PubMed] [Google Scholar]
- [16].Sakaue, H., Shiota, T., Ishizaka, N., Kawano, S., Tamura, Y., Tan, K. S., et al. Porin associates with Tom22 to regulate the mitochondrial protein gate assembly. Mol. Cell 73, 1044–1055 (2019). https://doi.org/10.1016/j.molcel.2019.01.003 [DOI] [PubMed] [Google Scholar]
- [17].Araiso, Y., Tsutsumi, A., Qiu, J., Imai, K., Shiota, T., Song, J., et al. Structure of mitochondrial import gate reveals distinct preprotein paths. Nature 575, 395–401 (2019). https://doi.org/10.1038/s41586-019-1680-7 [DOI] [PubMed] [Google Scholar]
- [18].Tucker, K., Park, E.. Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nat. Struct. Mol. Biol. 26, 1158–1166 (2019). https://doi.org/10.1038/s41594-019-0339-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Model, K., Prinz, T., Ruiz, T., Radermacher, M., Krimmer, T., Kühlbrandt, W., et al. Protein translocase of the outer mitochondrial membrane: Role of import receptors in the structural organization of the TOM complex. J. Mol. Biol. 316, 657–666 (2002). https://doi.org/10.1006/jmbi.2001.5365 [DOI] [PubMed] [Google Scholar]
- [20].Esaki, M., Shimizu, H., Ono, T., Yamamoto, H., Kanamori, T., Nishikawa, S., et al. Mitochondrial protein import. Requirement of presequence elements and TOM components for precursor binding to the TOM complex. J. Biol. Chem. 279, 45701–45707 (2004). https://doi.org/10.1074/jbc.M404591200 [DOI] [PubMed] [Google Scholar]
- [21].Court, D. A., Nargang, F. E., Steiner, H., Hodges, R. S., Neupert, W., Lill, R.. Role of intermembrane-space domain of the preprotein receptor Tom22 in protein import into mitochondria. Mol. Cell Biol. 16, 4035–4042 (1996). https://doi.org/10.1128/MCB.16.8.4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Moczko, M., Bömer, U., Kübrich, M., Zufall, N., Hönlinger, A., Pfanner, N.. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol. Cell Biol. 17, 6574–6584 (1997). https://doi.org/10.1128/MCB.17.11.6574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kanamori, T., Nishikawa, S., Nakai, M., Shin, I., Schultz P. G., Endo, T.. Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane. Proc. Natl. Acad. Sci. U.S.A. 96, 3634–3639 (1999). https://doi.org/10.1073/pnas.96.7.3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rapaport, D., Neupert, W., Lill, R.. Mitochondrial protein import. Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J. Biol. Chem. 272, 18725–18731 (1997). https://doi.org/10.1074/jbc.272.30.18725 [DOI] [PubMed] [Google Scholar]
- [25].Araiso, Y., Imai, K., Endo, T. Role of the TOM complex in protein import into Mitochondria: Structural views. Annu. Rev. Biochem. 91, Online ahead of print (2022). https://doi.org/10.1146/annurev-biochem-032620-104527 [DOI] [PubMed]
- [26].Uchihashi, T., Kodera, N., Ando, T.. Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nat. Protoc. 7, 1193–1206 (2012). https://doi.org/10.1038/nprot.2012.047 [DOI] [PubMed] [Google Scholar]