The centrosymmetric trans-NiN4O2 coordination polyhedra of the Ni2+ ions in the complex cations and anions of the title compound are tetragonally distorted octahedra. In the crystal, O—H⋯O hydrogen bonds between the phosphonate groups of the anions result in the formation of layers oriented parallel to the bc plane, which are further arranged into a three-dimensional network due to hydrogen-bonding involving the macrocyclic di-aqua cations and water molecules.
Keywords: crystal structure, cyclam, nickel, triphosphonic acid
Abstract
The components of the title compound, [Ni(C10H24N4)(H2O)2]2[Ni(C10H24N4)(C27H24O9P3)2]·10H2O are two centrosymmetric [Ni(C10H24N4)(H2O)2]2+ dications, a centrosymmetric [Ni(C10H24N4)(C27H24O9P3)2]4− tetra-anion and five crystallographically unique water molecules of crystallization. All of the nickel ions are coordinated by the four secondary N atoms of the macrocyclic cyclam ligands, which adopt the most energetically stable trans-III conformation, and the mutually trans O atoms of either water molecules in the cations or the phosphonate groups in the anion in a tetragonally distorted NiN4O2 octahedral coordination geometry. Strong O—H⋯O hydrogen bonds between the protonated and the non-protonated phosphonate O atoms of neighboring anions result in the formation of layers oriented parallel to the bc plane, which are linked into a three-dimensional network by virtue of numerous N—H⋯O and O—H⋯O hydrogen bonds arising from the sec-NH groups of the macrocycles, phosphonate O atoms and coordinated and non-coordinated water molecules.
1. Chemical context
First-row transition-metal complexes of 14-membered cyclam-like tetraaza macrocyles (cyclam = 1,4,8,11-tetraazacyclotetradecane; C10H24N4; L) are characterized by high thermodynamic stability and kinetic inertness (Yatsimirskii & Lampeka, 1985 ▸) and are popular metal-containing building units for the construction of MOFs (Lampeka & Tsymbal, 2004 ▸; Suh & Moon, 2007 ▸; Suh et al., 2012 ▸; Stackhouse & Ma, 2018 ▸). These crystalline coordination polymers, in which oligocarboxylates are the most common bridging ligands (Rao et al., 2004 ▸), possess permanent porosity and demonstrate many promising applications in different areas (MacGillivray & Lukehart, 2014 ▸; Kaskel, 2016 ▸).
The rigid trigonal aromatic linker 1,3,5-benzenetricarboxylate, C9H3O6
3–, is widely used for the assembly of MOFs based on azamacrocyclic cations (Lampeka & Tsymbal, 2004 ▸). Its tris-monodentate coordination in the trans-axial coordination positions of the metal ions leads predominantly to the formation of two-dimensional coordination polymers with hexagonal nets of 63 topology (Alexandrov et al., 2017 ▸). Usually, the modification of this bridge through the substitution of the carboxylic groups by para-carboxybenzyl fragments (the ligand H3BTB, 4,4′,4′′-benzene-1,3,5-triyltribenzoic acid) does not affect the coordination properties of the carboxylate groups or the topological characteristics of polymeric nets but results in the enlargement of the hexagonal structural unit of the coordination polymers allowing interpenetration of the subnets (Lampeka et al., 2012 ▸; Gong et al., 2016 ▸). Compared to carboxylates, linkers with other coordinating functions, in particular oligophosphonates, have been studied to a much lesser extent (Gagnon et al., 2012 ▸; Firmino et al., 2018 ▸; Yücesan et al., 2018 ▸), though one can expect that the substitution of a mono-anionic carboxylic group by a di-anionic phosphonate group with distinct acidity, number of donor atoms and spatial directivity of coordination bonds will strongly influence the composition and topology of the coordination nets. However, except for a very recent publication (Tsymbal et al., 2022 ▸), no papers dealing with structural characterization of the complexes formed by metal azamacrocyclic cations and phosphonate ligands have been published to date.

We report here the synthesis and crystal structure of the product of the reaction of [Ni(L)](ClO4)2 with 4,4′,4′′-(1,3,5-trimethylbenzene-2,4,6-triyl)triphosphonic acid, H6Me3BTP) – the structural analogue of H3BTB, namely, bis[trans-diaqua-(1,4,8,11-tetraazacyclotetradecane-κ4 N 1,N 4,N 8,N 11)-nickel(II)] trans-{bis-[4,4′,4′′-(1,3,5-trimethylbenzene-2,4,6-triyl)tris(hydrogen phenylphosphonato-κO)-(1,4,8,11-tetraazacyclotetradecane-κ4 N 1,N 4,N 8,N 11)-nickel(II)]} decahydrate, [Ni(L)(H2O)2]2[Ni(L)(H3Me3BTP)2]·10H2O, I.
2. Structural commentary
The molecular structure of I is shown in Fig. 1 ▸. It represents a non-polymeric compound in which atom Ni1 is coordinated by two monodentate H3Me3BTP3– ligands via their phosphonate O atoms, resulting in the formation of an [Ni(L)(H3Me3BTP)2]4– complex anion, which is charge-balanced by two structurally non-equivalent [Ni(L)(H2O)2]2+ divalent cations formed by atoms Ni2 and Ni3. The coordination geometries of all the nickel ions in I have much in common: the Ni2+ ions (all with site symmetry
 ) are coordinated by the four secondary N atoms of the macrocyclic ligands L, which adopt the most energetically stable trans-III (R,R,S,S) conformation (Bosnich et al., 1965a
▸; Barefield et al., 1986 ▸) with the five-membered (N—Ni—N bite angles ≃ 85°) and six-membered (N—Ni—N bite angles ≃ 95°) chelate rings in gauche and chair conformations, respectively (Table 1 ▸). The coordination polyhedra of the metal ions can be described as tetragonally elongated trans-NiN4O2 octahedra with the Ni—N bond lengths [average value 2.068 (3) Å] slightly shorter than the Ni—O bonds which, in turn, do not display any dependence on the nature of the donor oxygen atoms. The location of the metal ions on crystallographic inversion centers enforces strict planarity of the Ni(N4) coordination moieties and the axial Ni—O bonds are nearly orthogonal to the NiN4 planes (deviations of the angles N—Ni—O from 90° do not exceed 2°).
) are coordinated by the four secondary N atoms of the macrocyclic ligands L, which adopt the most energetically stable trans-III (R,R,S,S) conformation (Bosnich et al., 1965a
▸; Barefield et al., 1986 ▸) with the five-membered (N—Ni—N bite angles ≃ 85°) and six-membered (N—Ni—N bite angles ≃ 95°) chelate rings in gauche and chair conformations, respectively (Table 1 ▸). The coordination polyhedra of the metal ions can be described as tetragonally elongated trans-NiN4O2 octahedra with the Ni—N bond lengths [average value 2.068 (3) Å] slightly shorter than the Ni—O bonds which, in turn, do not display any dependence on the nature of the donor oxygen atoms. The location of the metal ions on crystallographic inversion centers enforces strict planarity of the Ni(N4) coordination moieties and the axial Ni—O bonds are nearly orthogonal to the NiN4 planes (deviations of the angles N—Ni—O from 90° do not exceed 2°).
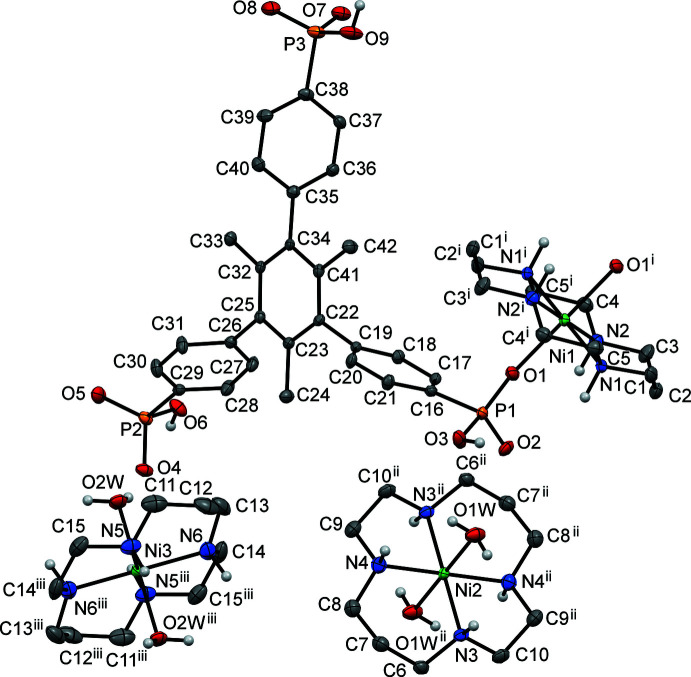
Figure 1.
The extended asymmetric unit in I showing the coordination environment of the Ni atoms and the atom-labeling scheme (displacement ellipsoids are drawn at the 30% probability level). C-bound H atoms and uncoordinated water molecules are omitted for clarity. Symmetry codes: (i) −x + 2, −y + 1, −z + 2; (ii) −x + 2, −y + 2, −z + 1; (iii) −x + 1, −y + 3, −z + 1.
Table 1. Selected geometric parameters (Å, °).
| Ni1—N1 | 2.067 (4) | Ni2—O1W | 2.105 (4) |
| Ni1—N2 | 2.064 (4) | Ni3—N5 | 2.070 (4) |
| Ni1—O1 | 2.134 (3) | Ni3—N6 | 2.056 (5) |
| Ni2—N3 | 2.072 (4) | Ni3—O2W | 2.136 (3) |
| Ni2—N4 | 2.076 (4) | ||
| N1—Ni1—N2i | 85.31 (16) | N3—Ni2—N4 | 95.34 (16) |
| N1—Ni1—N2 | 94.69 (16) | N5—Ni3—N6iii | 85.2 (2) |
| N3—Ni2—N4ii | 84.66 (16) | N5—Ni3—N6 | 94.8 (2) |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 .
.
The pendant benzene rings of the H3Me3BTP3– tri-anion in I are substantially tilted relative to the central aromatic core [average angle between the mean planes = 76 (5)°] and this feature is caused by repulsive interactions between the hydrogen atoms of the pendant rings and those of the methyl substituents of the central ring. The P—OH bond lengths [average value 1.57 (3) Å] are larger than the other P—O bonds [average value 1.501 (5) Å], thus indicating the partially delocalized character of the phosphonate groups.
3. Supramolecular features
In the crystal of I, the [Ni1(L)(H3Me3BTP)2]4– anions, [Ni2/Ni3(L)(H2O)2]2+ cations and water molecules of crystallization are linked by numerous hydrogen bonds with participation of the phosphonate groups, the secondary amino groups of the macrocycles and both the coordinated and crystalline water molecules (Table 2 ▸). A distinct lamellar structure is inherent for this compound. In particular, strong hydrogen-bonding interactions between the protonated fragments of the P1 and P3 phosphonate groups of one molecule as the donors with the non-protonated O4 and O5 atoms of the P2 group of another molecule as the acceptors [P1—O3—H3C⋯O5(x, y − 1, z), P3—O9—H9C⋯O49(x, y − 1, z + 1)], together with a weak N1—H1⋯O6 (x, y − 1, z) hydrogen bond between the secondary amino group of the macrocyclic cation [Ni1(L)] and protonated P2—O6 phosphonate fragment result in the formation of anionic layers oriented parallel to the bc plane. The distance between the parallel mean planes of the staggered by 60° trimethylbenzene rings of neighboring H3Me3BTP3– anions is 5.248 (3) Å, thus allowing us to exclude the possibility of aromatic π–π stacking interactions between them. Additionally, the negative charge of the layers are partially compensated by the incorporation within the layers of the [Ni2(L)(H2O)2]2+ cations via hydrogen bonding between the coordinated water molecules and the phosphonate O7 atom [O1W—H1WB⋯O7(x, y, z − 1)] (Fig. 2 ▸).
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O6iv | 1.00 | 2.32 | 3.196 (5) | 146 |
| N2—H2⋯O6W | 1.00 | 2.18 | 3.039 (6) | 143 |
| N3—H3⋯O7v | 1.00 | 2.13 | 3.102 (6) | 162 |
| N4—H4⋯O4W | 1.00 | 2.06 | 3.056 (6) | 173 |
| N5—H5⋯O9vi | 1.00 | 2.07 | 3.003 (6) | 155 |
| N6—H6⋯O7W ii | 1.00 | 1.98 | 2.956 (6) | 166 |
| O3—H3C⋯O5iv | 0.84 | 1.84 | 2.654 (5) | 162 |
| O6—H6C⋯O3W vii | 0.84 | 1.75 | 2.550 (5) | 159 |
| O9—H9C⋯O4viii | 0.84 | 1.74 | 2.517 (5) | 154 |
| O1W—H1WB⋯O7v | 0.87 | 1.81 | 2.679 (5) | 173 |
| O1W—H1WA⋯O4W | 0.87 | 2.45 | 3.256 (6) | 155 |
| O2W—H2WB⋯O4 | 0.86 | 1.90 | 2.729 (5) | 164 |
| O2W—H2WA⋯O7W ix | 0.86 | 1.81 | 2.675 (6) | 174 |
| O3W—H3WB⋯O2 | 0.87 | 1.81 | 2.676 (4) | 177 |
| O3W—H3WA⋯O7v | 0.85 | 1.84 | 2.689 (5) | 174 |
| O4W—H4WB⋯O3 | 0.87 | 2.26 | 3.115 (6) | 167 |
| O4W—H4WA⋯O8v | 0.87 | 1.93 | 2.796 (6) | 172 |
| O5W—H5WB⋯O5x | 0.87 | 1.98 | 2.813 (5) | 159 |
| O5W—H5WA⋯O8xi | 0.87 | 1.87 | 2.725 (5) | 168 |
| O6W—H6WB⋯O2 | 0.87 | 2.02 | 2.799 (6) | 149 |
| O6W—H6WA⋯O5W | 0.87 | 2.00 | 2.842 (5) | 164 |
| O7W—H7WB⋯O3W | 0.85 | 2.02 | 2.731 (5) | 140 |
| O7W—H7WA⋯O5W | 0.86 | 1.83 | 2.688 (5) | 173 |
Symmetry codes: (ii)
 ; (iv)
; (iv)
 ; (v)
; (v)
 ; (vi)
; (vi)
 ; (vii)
; (vii)
 ; (viii)
; (viii)
 ; (ix)
; (ix)
 ; (x)
; (x)
 ; (xi)
; (xi)
 .
.
Figure 2.
The hydrogen-bonded (dashed lines) layers in I viewed down the a axis. C-bound H atoms and macrocyclic cations formed by Ni3 have been omitted; C and N atoms of the macrocyclic cations formed by Ni2 are shown in green.
The second macrocyclic aqua cation [Ni3(L)(H2O)2]2+, due to hydrogen bonding of the coordinated water molecule with the phosphonate O4 atom (O2W—H2WB⋯O4), serves as the bridge between the layers, arranging them into a three-dimensional network (Fig. 3 ▸), which is further stabilized by numerous O—H⋯O hydrogen bonds involving the water molecules of crystallization, O3W–O7W (Table 2 ▸).
Figure 3.
The structure of I viewed down the b axis. C-bound H atoms have been omitted; C and N atoms of the macrocyclic cation formed by Ni2 and Ni3 are shown in green and violet, respectively. Water molecules of crystallization are not shown; hydrogen bonds are depicted as dashed lines.
4. Database survey
A search of the Cambridge Structural Database (CSD, version 5.43, last update March 2022; Groom et al., 2016 ▸) gave no hits related to H6Me3BTP or its complexes with metal ions, so the present work is the first structural characterization of a complex of this ligand. At the same time, several works dealing with the structures of the non-methylated analogue of the phosphonate under consideration, namely, 4,4′,4′′-benzene-1,3,5-triyl-triphosphonic acid (H6BTP), have been published. They include a methanol solvate of the free acid (CSD refcode AKEPEO; Vilela et al., 2021 ▸) and its pyridinium salt (YOLGEM; Beckmann et al., 2008 ▸), molecular complexes with solvated CoII and NiII ions (OQIZAR and OQIZEV; Pili et al., 2016 ▸) and coordination polymers formed by SrII (SOTZOR; Vaidhyanathan et al., 2009 ▸), ZnII (ISELAV02; Hermer et al., 2016 ▸), YIII (AKEPOY; Vilela et al., 2021 ▸), ZrIV (COCLIR; Taddei et al., 2014 ▸) and VIV/V (COQNAY; Ouellette et al., 2009 ▸). Interestingly, as in I, in all the metal complexes except COCLIR and ISELAV02, the ligand acts as a H3BTP3– tri-anion with three monodeprotonated phosphonate groups. On the other hand, because of the absence of methyl substituents, the molecules of the anions H n BTP(6–n)– as a whole are flatter than H3Me3BTP3– in I with a maximal tilting angle of pendant versus central benzene rings of ca 49° observed in ISELAV02. In addition, in the majority of complexes formed by H n BTP(6–n)– ligands (except AKEPOY and ISELAV02), aromatic π–π stacking interactions of different strengths are observed with centroid-to-centroid distances between the central aromatic rings ranging from 3.4 to 3.9 Å.
The Cambridge Structural Database contains also 18 hits describing the structure of the [Ni(L)(H2O)2]2+ complex cation in salts of different inorganic and organic anions as well as the charge-compensating part in anionic coordination polymers. In general, the structure of this cation in I is similar to other compounds, both from the point of view of the conformation of the macrocycle and the bond distances and angles characterizing the coordination polyhedron of the metal.
5. Synthesis and crystallization
All chemicals and solvents used in this work were purchased from Sigma–Aldrich and used without further purification. The acid H6Me3BTP was synthesized according to a procedure described previously for the preparation of H6BTP (Vaidhyanathan et al., 2009 ▸), starting from 1,3,5-trimethyl-2,4,6-tris(4′-bromophenyl)benzene instead of 1,3,5-tris(4′-bromophenyl)benzene. The complex [Ni(L)](ClO4)2 was prepared from ethanol solutions as described in the literature (Bosnich et al., 1965b ▸).
The title compound [Ni( L )(H2O)2]2[Ni( L )(H3Me3BTP)2]·10H2O, I, was prepared as follows. A solution of [Ni(L)](ClO4)2 (46 mg, 0.1 mmol) in 5 ml of water was added to 5 ml of an aqueous solution of H6Me3BTP (18 mg, 0.03 mmol) containing 2 ml of pyridine. The pink precipitate, which formed in a week, was filtered off, washed with small amounts of water, methanol and diethyl ether, and dried in air. Yield: 7 mg (10% based on acid). Analysis calculated for C84H148N12Ni3O32P6: C 45.85, H 6.78, N 7.64%. Found: C 45.73, H 6.87, N 7.51%. Single crystals of I suitable for X-ray diffraction analysis were selected from the sample resulting from the synthesis.
Caution! Perchlorate salts of metal complexes are potentially explosive and should be handled with care.
6. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. H atoms in I were placed in geometrically idealized positions and constrained to ride on their parent atoms, with C—H distances of 0.95 Å (ring H atoms), 0.98 Å (methyl H atoms), 0.99 Å (methylene H atoms), N—H distances of 1.00 Å, O—H distances of 0.84 Å (protonated phosphonate groups) and 0.87 Å (water molecules) with U iso(H) values of 1.2U eq or 1.5U eq times those of the corresponding parent atoms.
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | [Ni(C10H24N4)(H2O)2]2[Ni(C10H24N4)(C27H24O9P3)2]·10H2O |
| M r | 2200.09 |
| Crystal system, space group | Triclinic, P

|
| Temperature (K) | 160 |
| a, b, c (Å) | 9.8779 (5), 17.2467 (11), 17.6707 (11) |
| α, β, γ (°) | 61.409 (6), 77.515 (5), 77.713 (5) |
| V (Å3) | 2559.7 (3) |
| Z | 1 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.72 |
| Crystal size (mm) | 0.40 × 0.10 × 0.10 |
| Data collection | |
| Diffractometer | Rigaku Xcalibur Eos |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2020 ▸) |
| T min, T max | 0.701, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 23737, 9657, 6598 |
| R int | 0.063 |
| (sin θ/λ)max (Å−1) | 0.610 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.067, 0.161, 1.02 |
| No. of reflections | 9657 |
| No. of parameters | 629 |
| No. of restraints | 1 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.67, −0.46 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989022006624/hb8026sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022006624/hb8026Isup2.hkl
CCDC reference: 2178456
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| [Ni(C10H24N4)(H2O)2]2[Ni(C10H24N4)(C27H24O9P3)2]·10H2O | Z = 1 |
| Mr = 2200.09 | F(000) = 1166 |
| Triclinic, P1 | Dx = 1.427 Mg m−3 |
| a = 9.8779 (5) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 17.2467 (11) Å | Cell parameters from 5403 reflections |
| c = 17.6707 (11) Å | θ = 2.1–26.3° |
| α = 61.409 (6)° | µ = 0.72 mm−1 |
| β = 77.515 (5)° | T = 160 K |
| γ = 77.713 (5)° | Prism, clear light colourless |
| V = 2559.7 (3) Å3 | 0.40 × 0.10 × 0.10 mm |
Data collection
| Rigaku Xcalibur Eos diffractometer | 9657 independent reflections |
| Radiation source: fine-focus sealed X-ray tube, Enhance (Mo) X-ray Source | 6598 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.063 |
| Detector resolution: 16.1593 pixels mm-1 | θmax = 25.7°, θmin = 2.1° |
| ω scans | h = −11→12 |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2020) | k = −21→20 |
| Tmin = 0.701, Tmax = 1.000 | l = −21→21 |
| 23737 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.067 | H-atom parameters constrained |
| wR(F2) = 0.161 | w = 1/[σ2(Fo2) + (0.0527P)2 + 4.2781P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max < 0.001 |
| 9657 reflections | Δρmax = 0.67 e Å−3 |
| 629 parameters | Δρmin = −0.46 e Å−3 |
| 1 restraint |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Ni1 | 1.000000 | 0.500000 | 1.000000 | 0.0228 (2) | |
| P2 | 0.77348 (14) | 1.51832 (8) | 0.73137 (8) | 0.0273 (3) | |
| P1 | 0.86291 (13) | 0.70402 (8) | 0.82874 (8) | 0.0243 (3) | |
| P3 | 0.75955 (15) | 0.78200 (9) | 1.49060 (8) | 0.0332 (3) | |
| O5 | 0.6838 (4) | 1.5710 (2) | 0.7750 (2) | 0.0380 (9) | |
| C29 | 0.7561 (5) | 1.4015 (3) | 0.7965 (3) | 0.0241 (10) | |
| O4 | 0.7456 (4) | 1.5452 (2) | 0.6413 (2) | 0.0333 (8) | |
| O3W | 1.0380 (3) | 0.6623 (2) | 0.6411 (2) | 0.0311 (8) | |
| H3WA | 0.998703 | 0.703601 | 0.598132 | 0.047* | |
| H3WB | 1.016063 | 0.687031 | 0.675752 | 0.047* | |
| O4W | 0.6258 (4) | 0.8895 (3) | 0.6511 (3) | 0.0607 (12) | |
| H4WA | 0.622702 | 0.869018 | 0.615193 | 0.091* | |
| H4WB | 0.655562 | 0.843018 | 0.696393 | 0.091* | |
| C22 | 0.7292 (4) | 0.9758 (3) | 0.9717 (3) | 0.0193 (10) | |
| C38 | 0.7501 (5) | 0.8505 (3) | 1.3753 (3) | 0.0274 (11) | |
| C26 | 0.7310 (5) | 1.2190 (3) | 0.9041 (3) | 0.0230 (10) | |
| N1 | 0.9914 (4) | 0.4354 (3) | 0.9286 (2) | 0.0275 (9) | |
| H1 | 0.956271 | 0.481640 | 0.873064 | 0.033* | |
| O3 | 0.7242 (4) | 0.7049 (2) | 0.7985 (2) | 0.0387 (9) | |
| H3C | 0.724711 | 0.656089 | 0.798510 | 0.058* | |
| C34 | 0.7272 (4) | 0.9926 (3) | 1.1008 (3) | 0.0211 (10) | |
| N2 | 1.1919 (4) | 0.5419 (3) | 0.9365 (3) | 0.0330 (10) | |
| H2 | 1.173656 | 0.596679 | 0.881233 | 0.040* | |
| C3 | 1.2935 (5) | 0.4794 (4) | 0.9119 (4) | 0.0421 (14) | |
| H3A | 1.320107 | 0.425875 | 0.965248 | 0.051* | |
| H3B | 1.378830 | 0.507709 | 0.878884 | 0.051* | |
| O2 | 0.9773 (4) | 0.7334 (2) | 0.7517 (2) | 0.0349 (8) | |
| O9 | 0.7490 (5) | 0.6880 (2) | 1.5017 (2) | 0.0490 (11) | |
| H9C | 0.757613 | 0.649777 | 1.553173 | 0.074* | |
| C35 | 0.7357 (5) | 0.9486 (3) | 1.1963 (3) | 0.0245 (10) | |
| C41 | 0.7258 (5) | 0.9377 (3) | 1.0620 (3) | 0.0229 (10) | |
| C23 | 0.7243 (5) | 1.0683 (3) | 0.9212 (3) | 0.0229 (10) | |
| C30 | 0.6330 (5) | 1.3737 (3) | 0.8509 (3) | 0.0335 (12) | |
| H30 | 0.555907 | 1.416974 | 0.852158 | 0.040* | |
| C25 | 0.7243 (5) | 1.1221 (3) | 0.9602 (3) | 0.0216 (10) | |
| C5 | 0.8806 (6) | 0.3769 (4) | 0.9791 (3) | 0.0381 (13) | |
| H5A | 0.916444 | 0.324112 | 1.029994 | 0.046* | |
| H5B | 0.851180 | 0.356262 | 0.942372 | 0.046* | |
| C33 | 0.7319 (5) | 1.1422 (3) | 1.0923 (3) | 0.0280 (11) | |
| H33A | 0.769682 | 1.105369 | 1.147602 | 0.042* | |
| H33B | 0.637231 | 1.170079 | 1.102996 | 0.042* | |
| H33C | 0.791865 | 1.188533 | 1.053584 | 0.042* | |
| C39 | 0.6285 (5) | 0.9036 (3) | 1.3453 (3) | 0.0321 (12) | |
| H39 | 0.549541 | 0.906986 | 1.385777 | 0.039* | |
| C42 | 0.7241 (6) | 0.8387 (3) | 1.1158 (3) | 0.0333 (12) | |
| H42A | 0.818887 | 0.807974 | 1.111379 | 0.050* | |
| H42B | 0.661713 | 0.818665 | 1.094399 | 0.050* | |
| H42C | 0.690678 | 0.825159 | 1.176739 | 0.050* | |
| O7 | 0.9019 (4) | 0.7838 (2) | 1.5071 (2) | 0.0399 (9) | |
| C36 | 0.8576 (5) | 0.8993 (3) | 1.2269 (3) | 0.0283 (11) | |
| H36 | 0.938419 | 0.898840 | 1.186477 | 0.034* | |
| C19 | 0.7533 (4) | 0.9154 (3) | 0.9299 (3) | 0.0194 (10) | |
| C21 | 0.6772 (5) | 0.8176 (3) | 0.8889 (3) | 0.0282 (11) | |
| H21 | 0.603252 | 0.793241 | 0.884480 | 0.034* | |
| O8 | 0.6378 (4) | 0.8126 (3) | 1.5414 (2) | 0.0476 (10) | |
| C20 | 0.6477 (5) | 0.8787 (3) | 0.9222 (3) | 0.0287 (11) | |
| H20 | 0.553187 | 0.895677 | 0.940187 | 0.034* | |
| C2 | 1.2358 (6) | 0.4512 (4) | 0.8569 (3) | 0.0407 (14) | |
| H2A | 1.196217 | 0.505379 | 0.808660 | 0.049* | |
| H2B | 1.314402 | 0.421415 | 0.830904 | 0.049* | |
| C37 | 0.8665 (5) | 0.8500 (3) | 1.3151 (3) | 0.0306 (12) | |
| H37 | 0.952338 | 0.815768 | 1.334400 | 0.037* | |
| C32 | 0.7271 (4) | 1.0848 (3) | 1.0504 (3) | 0.0211 (10) | |
| C24 | 0.7140 (6) | 1.1079 (3) | 0.8254 (3) | 0.0314 (12) | |
| H24A | 0.660985 | 1.072081 | 0.815897 | 0.047* | |
| H24B | 0.808141 | 1.108441 | 0.793050 | 0.047* | |
| H24C | 0.666278 | 1.168989 | 0.805139 | 0.047* | |
| Ni2 | 1.000000 | 1.000000 | 0.500000 | 0.0233 (2) | |
| O1W | 0.9604 (4) | 0.8687 (2) | 0.5872 (2) | 0.0452 (10) | |
| H1WA | 0.881040 | 0.872384 | 0.618973 | 0.068* | |
| H1WB | 0.944450 | 0.844264 | 0.557414 | 0.068* | |
| N3 | 0.9605 (4) | 0.9803 (3) | 0.4004 (2) | 0.0297 (10) | |
| H3 | 0.924717 | 0.921388 | 0.427638 | 0.036* | |
| C10 | 1.0986 (5) | 0.9720 (4) | 0.3496 (3) | 0.0380 (13) | |
| H10A | 1.128892 | 1.031639 | 0.311897 | 0.046* | |
| H10B | 1.091906 | 0.945314 | 0.312116 | 0.046* | |
| N4 | 0.7940 (5) | 1.0447 (3) | 0.5321 (3) | 0.0377 (11) | |
| H4 | 0.744740 | 0.990779 | 0.568716 | 0.045* | |
| C9 | 0.7973 (6) | 1.0856 (4) | 0.5889 (3) | 0.0408 (14) | |
| H9A | 0.703665 | 1.090138 | 0.621780 | 0.049* | |
| H9B | 0.824723 | 1.146279 | 0.553175 | 0.049* | |
| C7 | 0.7183 (6) | 1.0577 (4) | 0.4022 (3) | 0.0418 (14) | |
| H7A | 0.689758 | 0.997677 | 0.440380 | 0.050* | |
| H7B | 0.646435 | 1.092349 | 0.362786 | 0.050* | |
| C6 | 0.8543 (5) | 1.0479 (3) | 0.3477 (3) | 0.0350 (12) | |
| H6A | 0.837243 | 1.030970 | 0.304733 | 0.042* | |
| H6B | 0.891090 | 1.106025 | 0.315290 | 0.042* | |
| C8 | 0.7168 (6) | 1.1019 (4) | 0.4586 (4) | 0.0434 (14) | |
| H8A | 0.758644 | 1.158069 | 0.423021 | 0.052* | |
| H8B | 0.618830 | 1.116899 | 0.480465 | 0.052* | |
| Ni3 | 0.500000 | 1.500000 | 0.500000 | 0.0280 (2) | |
| O2W | 0.5104 (4) | 1.5055 (2) | 0.6168 (2) | 0.0374 (9) | |
| H2WA | 0.451405 | 1.548647 | 0.620771 | 0.056* | |
| H2WB | 0.589347 | 1.520291 | 0.613928 | 0.056* | |
| N5 | 0.3476 (5) | 1.4161 (3) | 0.5683 (3) | 0.0436 (12) | |
| H5 | 0.333782 | 1.392139 | 0.529340 | 0.052* | |
| O7W | 1.3235 (4) | 0.6425 (3) | 0.6174 (3) | 0.0660 (14) | |
| H7WA | 1.366682 | 0.665639 | 0.637536 | 0.099* | |
| H7WB | 1.244232 | 0.671069 | 0.624896 | 0.099* | |
| N6 | 0.6644 (5) | 1.3992 (3) | 0.5280 (3) | 0.0464 (12) | |
| H6 | 0.671366 | 1.374659 | 0.486100 | 0.056* | |
| C14 | 0.7918 (6) | 1.4389 (5) | 0.5068 (4) | 0.0558 (18) | |
| H14A | 0.873557 | 1.397569 | 0.499126 | 0.067* | |
| H14B | 0.802863 | 1.450247 | 0.554899 | 0.067* | |
| O6W | 1.2369 (4) | 0.7283 (3) | 0.7946 (2) | 0.0541 (11) | |
| H6WA | 1.317921 | 0.726218 | 0.763661 | 0.081* | |
| H6WB | 1.177461 | 0.729258 | 0.764271 | 0.081* | |
| O5W | 1.4763 (3) | 0.7022 (2) | 0.6828 (2) | 0.0395 (9) | |
| H5WA | 1.538341 | 0.732237 | 0.641517 | 0.059* | |
| H5WB | 1.524801 | 0.660758 | 0.722817 | 0.059* | |
| O1 | 0.9002 (4) | 0.6178 (2) | 0.9039 (2) | 0.0326 (8) | |
| O6 | 0.9335 (4) | 1.5164 (2) | 0.7321 (2) | 0.0402 (9) | |
| H6C | 0.960853 | 1.564111 | 0.692096 | 0.060* | |
| C17 | 0.9212 (5) | 0.8304 (3) | 0.8664 (3) | 0.0229 (10) | |
| H17 | 1.015334 | 0.815323 | 0.846193 | 0.027* | |
| C16 | 0.8155 (5) | 0.7918 (3) | 0.8618 (3) | 0.0216 (10) | |
| C18 | 0.8904 (5) | 0.8913 (3) | 0.9005 (3) | 0.0238 (10) | |
| H18 | 0.964113 | 0.916614 | 0.903815 | 0.029* | |
| C40 | 0.6201 (5) | 0.9525 (3) | 1.2561 (3) | 0.0302 (12) | |
| H40 | 0.535465 | 0.988328 | 1.236326 | 0.036* | |
| C1 | 1.1247 (6) | 0.3895 (3) | 0.9048 (3) | 0.0375 (13) | |
| H1A | 1.108999 | 0.364727 | 0.867554 | 0.045* | |
| H1B | 1.157958 | 0.339182 | 0.958205 | 0.045* | |
| C28 | 0.8659 (5) | 1.3371 (3) | 0.7957 (3) | 0.0323 (12) | |
| H28 | 0.950712 | 1.353974 | 0.758385 | 0.039* | |
| C31 | 0.6206 (5) | 1.2844 (3) | 0.9032 (3) | 0.0319 (12) | |
| H31 | 0.534822 | 1.267424 | 0.939145 | 0.038* | |
| C13 | 0.6486 (8) | 1.3227 (4) | 0.6163 (4) | 0.0614 (19) | |
| H13A | 0.651043 | 1.342483 | 0.660143 | 0.074* | |
| H13B | 0.728211 | 1.275571 | 0.620745 | 0.074* | |
| C15 | 0.2154 (6) | 1.4748 (5) | 0.5758 (4) | 0.0502 (16) | |
| H15A | 0.212231 | 1.487228 | 0.625394 | 0.060* | |
| H15B | 0.133940 | 1.444182 | 0.586810 | 0.060* | |
| C11 | 0.3777 (8) | 1.3401 (5) | 0.6484 (4) | 0.0606 (19) | |
| H11A | 0.300674 | 1.302890 | 0.671624 | 0.073* | |
| H11B | 0.383358 | 1.360212 | 0.691424 | 0.073* | |
| C27 | 0.8528 (5) | 1.2485 (3) | 0.8489 (3) | 0.0315 (12) | |
| H27 | 0.930342 | 1.205552 | 0.847868 | 0.038* | |
| C12 | 0.5140 (8) | 1.2849 (4) | 0.6361 (4) | 0.065 (2) | |
| H12A | 0.519087 | 1.227524 | 0.689642 | 0.078* | |
| H12B | 0.509541 | 1.271311 | 0.588269 | 0.078* | |
| C4 | 1.2423 (5) | 0.5705 (4) | 0.9910 (3) | 0.0360 (13) | |
| H4A | 1.318132 | 0.607996 | 0.956861 | 0.043* | |
| H4B | 1.279137 | 0.517994 | 1.041693 | 0.043* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Ni1 | 0.0278 (5) | 0.0183 (4) | 0.0225 (4) | 0.0031 (4) | −0.0059 (3) | −0.0108 (4) |
| P2 | 0.0380 (8) | 0.0178 (6) | 0.0273 (7) | −0.0020 (6) | −0.0117 (6) | −0.0089 (5) |
| P1 | 0.0314 (7) | 0.0195 (6) | 0.0258 (6) | 0.0026 (6) | −0.0079 (5) | −0.0139 (5) |
| P3 | 0.0495 (9) | 0.0290 (7) | 0.0222 (7) | −0.0109 (7) | −0.0112 (6) | −0.0074 (6) |
| O5 | 0.056 (2) | 0.0220 (18) | 0.042 (2) | 0.0005 (18) | −0.0129 (18) | −0.0184 (17) |
| C29 | 0.031 (3) | 0.023 (2) | 0.019 (2) | 0.000 (2) | −0.0063 (19) | −0.010 (2) |
| O4 | 0.051 (2) | 0.0229 (18) | 0.0265 (18) | −0.0077 (17) | −0.0132 (16) | −0.0072 (15) |
| O3W | 0.0317 (19) | 0.0298 (19) | 0.0354 (19) | 0.0021 (16) | −0.0082 (15) | −0.0185 (16) |
| O4W | 0.064 (3) | 0.059 (3) | 0.060 (3) | −0.013 (2) | −0.008 (2) | −0.026 (2) |
| C22 | 0.017 (2) | 0.022 (2) | 0.020 (2) | −0.002 (2) | 0.0003 (18) | −0.012 (2) |
| C38 | 0.040 (3) | 0.022 (2) | 0.025 (3) | −0.007 (2) | −0.011 (2) | −0.010 (2) |
| C26 | 0.030 (3) | 0.019 (2) | 0.022 (2) | −0.002 (2) | −0.005 (2) | −0.011 (2) |
| N1 | 0.036 (2) | 0.021 (2) | 0.026 (2) | 0.0064 (19) | −0.0090 (18) | −0.0131 (18) |
| O3 | 0.049 (2) | 0.027 (2) | 0.054 (2) | 0.0082 (18) | −0.0240 (18) | −0.0272 (19) |
| C34 | 0.019 (2) | 0.025 (2) | 0.021 (2) | 0.006 (2) | −0.0042 (18) | −0.014 (2) |
| N2 | 0.039 (3) | 0.031 (2) | 0.030 (2) | −0.003 (2) | −0.0034 (19) | −0.016 (2) |
| C3 | 0.033 (3) | 0.052 (4) | 0.046 (3) | −0.004 (3) | 0.006 (2) | −0.032 (3) |
| O2 | 0.047 (2) | 0.031 (2) | 0.0312 (19) | −0.0057 (17) | 0.0045 (16) | −0.0216 (17) |
| O9 | 0.090 (3) | 0.034 (2) | 0.028 (2) | −0.021 (2) | −0.022 (2) | −0.0065 (17) |
| C35 | 0.028 (3) | 0.023 (2) | 0.024 (2) | −0.004 (2) | −0.004 (2) | −0.012 (2) |
| C41 | 0.023 (2) | 0.019 (2) | 0.027 (2) | 0.000 (2) | −0.0026 (19) | −0.012 (2) |
| C23 | 0.026 (3) | 0.022 (2) | 0.019 (2) | 0.004 (2) | −0.0033 (19) | −0.010 (2) |
| C30 | 0.028 (3) | 0.022 (3) | 0.046 (3) | 0.003 (2) | −0.006 (2) | −0.014 (2) |
| C25 | 0.021 (2) | 0.018 (2) | 0.025 (2) | 0.001 (2) | −0.0017 (19) | −0.010 (2) |
| C5 | 0.053 (4) | 0.030 (3) | 0.034 (3) | −0.007 (3) | −0.010 (3) | −0.014 (2) |
| C33 | 0.035 (3) | 0.022 (2) | 0.031 (3) | 0.000 (2) | −0.008 (2) | −0.016 (2) |
| C39 | 0.033 (3) | 0.040 (3) | 0.023 (3) | −0.007 (3) | −0.003 (2) | −0.013 (2) |
| C42 | 0.052 (3) | 0.021 (3) | 0.027 (3) | −0.004 (2) | −0.009 (2) | −0.010 (2) |
| O7 | 0.053 (2) | 0.038 (2) | 0.0282 (19) | −0.0107 (19) | −0.0119 (17) | −0.0096 (17) |
| C36 | 0.038 (3) | 0.025 (3) | 0.024 (3) | 0.001 (2) | −0.006 (2) | −0.014 (2) |
| C19 | 0.023 (2) | 0.018 (2) | 0.015 (2) | 0.001 (2) | −0.0015 (18) | −0.0077 (19) |
| C21 | 0.026 (3) | 0.032 (3) | 0.036 (3) | −0.002 (2) | −0.006 (2) | −0.023 (2) |
| O8 | 0.054 (2) | 0.053 (3) | 0.0258 (19) | −0.001 (2) | −0.0044 (17) | −0.0116 (19) |
| C20 | 0.023 (3) | 0.033 (3) | 0.036 (3) | 0.004 (2) | −0.001 (2) | −0.024 (2) |
| C2 | 0.036 (3) | 0.048 (4) | 0.041 (3) | 0.010 (3) | −0.005 (2) | −0.029 (3) |
| C37 | 0.038 (3) | 0.025 (3) | 0.030 (3) | 0.007 (2) | −0.015 (2) | −0.013 (2) |
| C32 | 0.020 (2) | 0.023 (2) | 0.022 (2) | 0.004 (2) | −0.0025 (18) | −0.013 (2) |
| C24 | 0.050 (3) | 0.021 (2) | 0.025 (3) | −0.003 (2) | −0.007 (2) | −0.012 (2) |
| Ni2 | 0.0292 (5) | 0.0196 (4) | 0.0220 (4) | 0.0031 (4) | −0.0055 (4) | −0.0117 (4) |
| O1W | 0.073 (3) | 0.036 (2) | 0.033 (2) | −0.014 (2) | −0.0077 (19) | −0.0169 (18) |
| N3 | 0.042 (3) | 0.027 (2) | 0.023 (2) | −0.007 (2) | −0.0027 (18) | −0.0130 (18) |
| C10 | 0.049 (3) | 0.044 (3) | 0.031 (3) | −0.007 (3) | −0.005 (2) | −0.024 (3) |
| N4 | 0.046 (3) | 0.031 (2) | 0.040 (3) | −0.002 (2) | −0.009 (2) | −0.019 (2) |
| C9 | 0.041 (3) | 0.047 (3) | 0.037 (3) | −0.002 (3) | 0.001 (2) | −0.025 (3) |
| C7 | 0.046 (3) | 0.041 (3) | 0.037 (3) | 0.001 (3) | −0.014 (3) | −0.016 (3) |
| C6 | 0.050 (3) | 0.029 (3) | 0.031 (3) | 0.001 (3) | −0.015 (2) | −0.016 (2) |
| C8 | 0.047 (4) | 0.039 (3) | 0.043 (3) | 0.002 (3) | −0.009 (3) | −0.020 (3) |
| Ni3 | 0.0357 (5) | 0.0258 (5) | 0.0275 (5) | −0.0032 (4) | −0.0080 (4) | −0.0148 (4) |
| O2W | 0.046 (2) | 0.043 (2) | 0.034 (2) | −0.0071 (19) | −0.0093 (17) | −0.0240 (18) |
| N5 | 0.058 (3) | 0.045 (3) | 0.039 (3) | −0.017 (3) | −0.008 (2) | −0.023 (2) |
| O7W | 0.037 (2) | 0.102 (4) | 0.104 (4) | 0.006 (2) | −0.008 (2) | −0.089 (3) |
| N6 | 0.050 (3) | 0.050 (3) | 0.056 (3) | 0.008 (3) | −0.023 (2) | −0.037 (3) |
| C14 | 0.042 (4) | 0.071 (5) | 0.079 (5) | 0.013 (3) | −0.014 (3) | −0.059 (4) |
| O6W | 0.052 (3) | 0.059 (3) | 0.042 (2) | 0.004 (2) | −0.0005 (19) | −0.023 (2) |
| O5W | 0.035 (2) | 0.041 (2) | 0.041 (2) | −0.0007 (18) | −0.0030 (16) | −0.0199 (18) |
| O1 | 0.045 (2) | 0.0221 (18) | 0.0296 (19) | 0.0041 (16) | −0.0121 (16) | −0.0115 (15) |
| O6 | 0.042 (2) | 0.026 (2) | 0.044 (2) | −0.0058 (18) | −0.0156 (17) | −0.0044 (17) |
| C17 | 0.023 (2) | 0.022 (2) | 0.021 (2) | 0.000 (2) | 0.0004 (19) | −0.010 (2) |
| C16 | 0.028 (3) | 0.020 (2) | 0.019 (2) | 0.000 (2) | −0.0066 (19) | −0.010 (2) |
| C18 | 0.025 (3) | 0.022 (2) | 0.028 (3) | −0.004 (2) | −0.005 (2) | −0.014 (2) |
| C40 | 0.025 (3) | 0.034 (3) | 0.031 (3) | 0.001 (2) | −0.007 (2) | −0.015 (2) |
| C1 | 0.048 (3) | 0.030 (3) | 0.043 (3) | 0.010 (3) | −0.016 (3) | −0.025 (3) |
| C28 | 0.036 (3) | 0.027 (3) | 0.032 (3) | −0.007 (2) | 0.009 (2) | −0.017 (2) |
| C31 | 0.021 (3) | 0.024 (3) | 0.041 (3) | 0.000 (2) | 0.004 (2) | −0.011 (2) |
| C13 | 0.090 (5) | 0.038 (4) | 0.060 (4) | 0.013 (4) | −0.044 (4) | −0.020 (3) |
| C15 | 0.041 (4) | 0.070 (5) | 0.058 (4) | −0.014 (3) | 0.006 (3) | −0.046 (4) |
| C11 | 0.087 (5) | 0.057 (4) | 0.042 (4) | −0.035 (4) | −0.005 (3) | −0.017 (3) |
| C27 | 0.035 (3) | 0.023 (3) | 0.031 (3) | 0.000 (2) | 0.006 (2) | −0.013 (2) |
| C12 | 0.106 (6) | 0.034 (4) | 0.048 (4) | −0.015 (4) | −0.034 (4) | −0.002 (3) |
| C4 | 0.039 (3) | 0.035 (3) | 0.034 (3) | −0.003 (3) | −0.011 (2) | −0.014 (3) |
Geometric parameters (Å, º)
| Ni1—N1i | 2.067 (4) | C24—H24A | 0.9800 |
| Ni1—N1 | 2.067 (4) | C24—H24B | 0.9800 |
| Ni1—N2 | 2.064 (4) | C24—H24C | 0.9800 |
| Ni1—N2i | 2.064 (4) | Ni2—N3ii | 2.072 (4) |
| Ni1—O1i | 2.134 (3) | Ni2—N3 | 2.072 (4) |
| Ni1—O1 | 2.134 (3) | Ni2—N4 | 2.076 (4) |
| P2—O5 | 1.495 (4) | Ni2—N4ii | 2.076 (4) |
| P2—C29 | 1.804 (5) | Ni2—O1Wii | 2.105 (4) |
| P2—O4 | 1.502 (3) | Ni2—O1W | 2.105 (4) |
| P2—O6 | 1.576 (4) | O1W—H1WA | 0.8701 |
| P1—O3 | 1.570 (4) | O1W—H1WB | 0.8691 |
| P1—O2 | 1.518 (3) | N3—H3 | 1.0000 |
| P1—O1 | 1.483 (3) | N3—C10 | 1.481 (6) |
| P1—C16 | 1.811 (5) | N3—C6 | 1.479 (6) |
| P3—C38 | 1.813 (5) | C10—H10A | 0.9900 |
| P3—O9 | 1.562 (4) | C10—H10B | 0.9900 |
| P3—O7 | 1.506 (4) | C10—C9ii | 1.496 (7) |
| P3—O8 | 1.499 (4) | N4—H4 | 1.0000 |
| C29—C30 | 1.391 (7) | N4—C9 | 1.486 (6) |
| C29—C28 | 1.381 (7) | N4—C8 | 1.457 (7) |
| O3W—H3WA | 0.8523 | C9—H9A | 0.9900 |
| O3W—H3WB | 0.8700 | C9—H9B | 0.9900 |
| O4W—H4WA | 0.8687 | C7—H7A | 0.9900 |
| O4W—H4WB | 0.8702 | C7—H7B | 0.9900 |
| C22—C41 | 1.400 (6) | C7—C6 | 1.504 (7) |
| C22—C23 | 1.402 (6) | C7—C8 | 1.513 (7) |
| C22—C19 | 1.497 (6) | C6—H6A | 0.9900 |
| C38—C39 | 1.380 (7) | C6—H6B | 0.9900 |
| C38—C37 | 1.390 (7) | C8—H8A | 0.9900 |
| C26—C25 | 1.488 (6) | C8—H8B | 0.9900 |
| C26—C31 | 1.388 (6) | Ni3—N5 | 2.070 (4) |
| C26—C27 | 1.392 (6) | Ni3—N5iii | 2.070 (5) |
| N1—H1 | 1.0000 | Ni3—N6 | 2.056 (5) |
| N1—C5 | 1.482 (6) | Ni3—N6iii | 2.056 (5) |
| N1—C1 | 1.474 (6) | Ni3—O2Wiii | 2.137 (3) |
| O3—H3C | 0.8400 | Ni3—O2W | 2.136 (3) |
| C34—C35 | 1.497 (6) | O2W—H2WA | 0.8638 |
| C34—C41 | 1.414 (6) | O2W—H2WB | 0.8553 |
| C34—C32 | 1.401 (6) | N5—H5 | 1.0000 |
| N2—H2 | 1.0000 | N5—C15 | 1.497 (7) |
| N2—C3 | 1.469 (6) | N5—C11 | 1.431 (8) |
| N2—C4 | 1.477 (6) | O7W—H7WA | 0.8613 |
| C3—H3A | 0.9900 | O7W—H7WB | 0.8521 |
| C3—H3B | 0.9900 | N6—H6 | 1.0000 |
| C3—C2 | 1.521 (7) | N6—C14 | 1.452 (7) |
| O9—H9C | 0.8400 | N6—C13 | 1.487 (8) |
| C35—C36 | 1.368 (7) | C14—H14A | 0.9900 |
| C35—C40 | 1.392 (6) | C14—H14B | 0.9900 |
| C41—C42 | 1.507 (6) | C14—C15iii | 1.507 (9) |
| C23—C25 | 1.396 (6) | O6W—H6WA | 0.8706 |
| C23—C24 | 1.510 (6) | O6W—H6WB | 0.8688 |
| C30—H30 | 0.9500 | O5W—H5WA | 0.8696 |
| C30—C31 | 1.382 (7) | O5W—H5WB | 0.8704 |
| C25—C32 | 1.409 (6) | O6—H6C | 0.8400 |
| C5—H5A | 0.9900 | C17—H17 | 0.9500 |
| C5—H5B | 0.9900 | C17—C16 | 1.387 (6) |
| C5—C4i | 1.513 (7) | C17—C18 | 1.396 (6) |
| C33—H33A | 0.9800 | C18—H18 | 0.9500 |
| C33—H33B | 0.9800 | C40—H40 | 0.9500 |
| C33—H33C | 0.9800 | C1—H1A | 0.9900 |
| C33—C32 | 1.506 (6) | C1—H1B | 0.9900 |
| C39—H39 | 0.9500 | C28—H28 | 0.9500 |
| C39—C40 | 1.399 (6) | C28—C27 | 1.375 (7) |
| C42—H42A | 0.9800 | C31—H31 | 0.9500 |
| C42—H42B | 0.9800 | C13—H13A | 0.9900 |
| C42—H42C | 0.9800 | C13—H13B | 0.9900 |
| C36—H36 | 0.9500 | C13—C12 | 1.506 (9) |
| C36—C37 | 1.385 (6) | C15—H15A | 0.9900 |
| C19—C20 | 1.391 (6) | C15—H15B | 0.9900 |
| C19—C18 | 1.393 (6) | C11—H11A | 0.9900 |
| C21—H21 | 0.9500 | C11—H11B | 0.9900 |
| C21—C20 | 1.389 (6) | C11—C12 | 1.514 (10) |
| C21—C16 | 1.398 (6) | C27—H27 | 0.9500 |
| C20—H20 | 0.9500 | C12—H12A | 0.9900 |
| C2—H2A | 0.9900 | C12—H12B | 0.9900 |
| C2—H2B | 0.9900 | C4—H4A | 0.9900 |
| C2—C1 | 1.513 (7) | C4—H4B | 0.9900 |
| C37—H37 | 0.9500 | ||
| N1—Ni1—N1i | 180.0 | N4—Ni2—O1W | 89.10 (17) |
| N1—Ni1—O1i | 91.79 (14) | N4ii—Ni2—O1W | 90.90 (17) |
| N1i—Ni1—O1i | 88.21 (14) | N4ii—Ni2—O1Wii | 89.10 (17) |
| N1i—Ni1—O1 | 91.79 (14) | N4—Ni2—N4ii | 180.00 (13) |
| N1—Ni1—O1 | 88.21 (14) | Ni2—O1W—H1WA | 106.8 |
| N1—Ni1—N2i | 85.31 (16) | Ni2—O1W—H1WB | 108.1 |
| N1i—Ni1—N2 | 85.31 (16) | H1WA—O1W—H1WB | 104.5 |
| N1i—Ni1—N2i | 94.69 (16) | Ni2—N3—H3 | 107.3 |
| N1—Ni1—N2 | 94.69 (16) | C10—N3—Ni2 | 105.7 (3) |
| N2i—Ni1—N2 | 180.0 | C10—N3—H3 | 107.3 |
| N2i—Ni1—O1 | 90.47 (15) | C6—N3—Ni2 | 114.6 (3) |
| N2—Ni1—O1 | 89.53 (15) | C6—N3—H3 | 107.3 |
| N2i—Ni1—O1i | 89.53 (15) | C6—N3—C10 | 114.3 (4) |
| N2—Ni1—O1i | 90.47 (15) | N3—C10—H10A | 109.9 |
| O1i—Ni1—O1 | 180.0 | N3—C10—H10B | 109.9 |
| O5—P2—C29 | 110.3 (2) | N3—C10—C9ii | 109.1 (4) |
| O5—P2—O4 | 115.4 (2) | H10A—C10—H10B | 108.3 |
| O5—P2—O6 | 111.3 (2) | C9ii—C10—H10A | 109.9 |
| O4—P2—C29 | 108.0 (2) | C9ii—C10—H10B | 109.9 |
| O4—P2—O6 | 110.5 (2) | Ni2—N4—H4 | 106.9 |
| O6—P2—C29 | 100.2 (2) | C9—N4—Ni2 | 106.4 (3) |
| O3—P1—C16 | 102.1 (2) | C9—N4—H4 | 106.9 |
| O2—P1—O3 | 110.2 (2) | C8—N4—Ni2 | 115.2 (3) |
| O2—P1—C16 | 107.1 (2) | C8—N4—H4 | 106.9 |
| O1—P1—O3 | 111.4 (2) | C8—N4—C9 | 114.0 (4) |
| O1—P1—O2 | 115.2 (2) | C10ii—C9—H9A | 110.1 |
| O1—P1—C16 | 110.06 (19) | C10ii—C9—H9B | 110.1 |
| O9—P3—C38 | 101.5 (2) | N4—C9—C10ii | 108.1 (4) |
| O7—P3—C38 | 107.6 (2) | N4—C9—H9A | 110.1 |
| O7—P3—O9 | 109.8 (2) | N4—C9—H9B | 110.1 |
| O8—P3—C38 | 109.4 (2) | H9A—C9—H9B | 108.4 |
| O8—P3—O9 | 111.8 (2) | H7A—C7—H7B | 107.3 |
| O8—P3—O7 | 115.7 (2) | C6—C7—H7A | 108.1 |
| C30—C29—P2 | 120.9 (4) | C6—C7—H7B | 108.1 |
| C28—C29—P2 | 121.0 (4) | C6—C7—C8 | 116.9 (5) |
| C28—C29—C30 | 118.0 (4) | C8—C7—H7A | 108.1 |
| H3WA—O3W—H3WB | 98.3 | C8—C7—H7B | 108.1 |
| H4WA—O4W—H4WB | 104.4 | N3—C6—C7 | 112.6 (4) |
| C41—C22—C23 | 120.1 (4) | N3—C6—H6A | 109.1 |
| C41—C22—C19 | 118.6 (4) | N3—C6—H6B | 109.1 |
| C23—C22—C19 | 120.9 (4) | C7—C6—H6A | 109.1 |
| C39—C38—P3 | 121.3 (4) | C7—C6—H6B | 109.1 |
| C39—C38—C37 | 118.6 (4) | H6A—C6—H6B | 107.8 |
| C37—C38—P3 | 120.1 (4) | N4—C8—C7 | 111.9 (5) |
| C31—C26—C25 | 123.6 (4) | N4—C8—H8A | 109.2 |
| C31—C26—C27 | 116.3 (4) | N4—C8—H8B | 109.2 |
| C27—C26—C25 | 120.1 (4) | C7—C8—H8A | 109.2 |
| Ni1—N1—H1 | 106.9 | C7—C8—H8B | 109.2 |
| C5—N1—Ni1 | 104.8 (3) | H8A—C8—H8B | 107.9 |
| C5—N1—H1 | 106.9 | O2W—Ni3—O2Wiii | 180.0 |
| C1—N1—Ni1 | 116.4 (3) | N5iii—Ni3—O2W | 91.54 (15) |
| C1—N1—H1 | 106.9 | N5—Ni3—O2W | 88.46 (15) |
| C1—N1—C5 | 114.2 (4) | N5iii—Ni3—O2Wiii | 88.46 (15) |
| P1—O3—H3C | 109.5 | N5—Ni3—O2Wiii | 91.54 (15) |
| C41—C34—C35 | 117.8 (4) | N5—Ni3—N5iii | 180.0 |
| C32—C34—C35 | 121.4 (4) | N6—Ni3—O2W | 89.19 (16) |
| C32—C34—C41 | 120.8 (4) | N6iii—Ni3—O2W | 90.81 (16) |
| Ni1—N2—H2 | 106.6 | N6iii—Ni3—O2Wiii | 89.19 (16) |
| C3—N2—Ni1 | 116.1 (3) | N6—Ni3—O2Wiii | 90.81 (16) |
| C3—N2—H2 | 106.6 | N5—Ni3—N6iii | 85.2 (2) |
| C3—N2—C4 | 114.4 (4) | N5—Ni3—N6 | 94.8 (2) |
| C4—N2—Ni1 | 106.0 (3) | N5iii—Ni3—N6 | 85.2 (2) |
| C4—N2—H2 | 106.6 | N5iii—Ni3—N6iii | 94.8 (2) |
| N2—C3—H3A | 109.1 | N6iii—Ni3—N6 | 180.0 |
| N2—C3—H3B | 109.1 | Ni3—O2W—H2WA | 110.2 |
| N2—C3—C2 | 112.3 (4) | Ni3—O2W—H2WB | 109.7 |
| H3A—C3—H3B | 107.9 | H2WA—O2W—H2WB | 103.1 |
| C2—C3—H3A | 109.1 | Ni3—N5—H5 | 106.1 |
| C2—C3—H3B | 109.1 | C15—N5—Ni3 | 105.8 (4) |
| P3—O9—H9C | 109.5 | C15—N5—H5 | 106.1 |
| C36—C35—C34 | 119.7 (4) | C11—N5—Ni3 | 117.4 (4) |
| C36—C35—C40 | 118.5 (4) | C11—N5—H5 | 106.1 |
| C40—C35—C34 | 121.7 (4) | C11—N5—C15 | 114.5 (5) |
| C22—C41—C34 | 119.4 (4) | H7WA—O7W—H7WB | 94.0 |
| C22—C41—C42 | 119.5 (4) | Ni3—N6—H6 | 106.1 |
| C34—C41—C42 | 121.1 (4) | C14—N6—Ni3 | 107.9 (4) |
| C22—C23—C24 | 118.7 (4) | C14—N6—H6 | 106.1 |
| C25—C23—C22 | 120.0 (4) | C14—N6—C13 | 114.0 (5) |
| C25—C23—C24 | 121.2 (4) | C13—N6—Ni3 | 115.9 (4) |
| C29—C30—H30 | 119.4 | C13—N6—H6 | 106.1 |
| C31—C30—C29 | 121.2 (5) | N6—C14—H14A | 109.8 |
| C31—C30—H30 | 119.4 | N6—C14—H14B | 109.8 |
| C23—C25—C26 | 118.9 (4) | N6—C14—C15iii | 109.4 (5) |
| C23—C25—C32 | 120.8 (4) | H14A—C14—H14B | 108.2 |
| C32—C25—C26 | 120.2 (4) | C15iii—C14—H14A | 109.8 |
| N1—C5—H5A | 110.0 | C15iii—C14—H14B | 109.8 |
| N1—C5—H5B | 110.0 | H6WA—O6W—H6WB | 104.5 |
| N1—C5—C4i | 108.4 (4) | H5WA—O5W—H5WB | 104.5 |
| H5A—C5—H5B | 108.4 | P1—O1—Ni1 | 167.3 (2) |
| C4i—C5—H5A | 110.0 | P2—O6—H6C | 109.5 |
| C4i—C5—H5B | 110.0 | C16—C17—H17 | 119.7 |
| H33A—C33—H33B | 109.5 | C16—C17—C18 | 120.5 (4) |
| H33A—C33—H33C | 109.5 | C18—C17—H17 | 119.7 |
| H33B—C33—H33C | 109.5 | C21—C16—P1 | 122.4 (4) |
| C32—C33—H33A | 109.5 | C17—C16—P1 | 118.7 (3) |
| C32—C33—H33B | 109.5 | C17—C16—C21 | 118.7 (4) |
| C32—C33—H33C | 109.5 | C19—C18—C17 | 121.1 (4) |
| C38—C39—H39 | 119.6 | C19—C18—H18 | 119.5 |
| C38—C39—C40 | 120.8 (5) | C17—C18—H18 | 119.5 |
| C40—C39—H39 | 119.6 | C35—C40—C39 | 120.0 (5) |
| C41—C42—H42A | 109.5 | C35—C40—H40 | 120.0 |
| C41—C42—H42B | 109.5 | C39—C40—H40 | 120.0 |
| C41—C42—H42C | 109.5 | N1—C1—C2 | 112.0 (4) |
| H42A—C42—H42B | 109.5 | N1—C1—H1A | 109.2 |
| H42A—C42—H42C | 109.5 | N1—C1—H1B | 109.2 |
| H42B—C42—H42C | 109.5 | C2—C1—H1A | 109.2 |
| C35—C36—H36 | 119.1 | C2—C1—H1B | 109.2 |
| C35—C36—C37 | 121.8 (5) | H1A—C1—H1B | 107.9 |
| C37—C36—H36 | 119.1 | C29—C28—H28 | 120.0 |
| C20—C19—C22 | 124.0 (4) | C27—C28—C29 | 120.1 (5) |
| C20—C19—C18 | 117.9 (4) | C27—C28—H28 | 120.0 |
| C18—C19—C22 | 118.0 (4) | C26—C31—H31 | 119.3 |
| C20—C21—H21 | 119.9 | C30—C31—C26 | 121.4 (4) |
| C20—C21—C16 | 120.2 (4) | C30—C31—H31 | 119.3 |
| C16—C21—H21 | 119.9 | N6—C13—H13A | 109.2 |
| C19—C20—H20 | 119.3 | N6—C13—H13B | 109.2 |
| C21—C20—C19 | 121.5 (4) | N6—C13—C12 | 112.1 (5) |
| C21—C20—H20 | 119.3 | H13A—C13—H13B | 107.9 |
| C3—C2—H2A | 108.4 | C12—C13—H13A | 109.2 |
| C3—C2—H2B | 108.4 | C12—C13—H13B | 109.2 |
| H2A—C2—H2B | 107.5 | N5—C15—C14iii | 110.1 (5) |
| C1—C2—C3 | 115.4 (4) | N5—C15—H15A | 109.6 |
| C1—C2—H2A | 108.4 | N5—C15—H15B | 109.6 |
| C1—C2—H2B | 108.4 | C14iii—C15—H15A | 109.6 |
| C38—C37—H37 | 120.0 | C14iii—C15—H15B | 109.6 |
| C36—C37—C38 | 120.1 (5) | H15A—C15—H15B | 108.2 |
| C36—C37—H37 | 120.0 | N5—C11—H11A | 109.3 |
| C34—C32—C25 | 118.8 (4) | N5—C11—H11B | 109.3 |
| C34—C32—C33 | 120.2 (4) | N5—C11—C12 | 111.5 (5) |
| C25—C32—C33 | 121.0 (4) | H11A—C11—H11B | 108.0 |
| C23—C24—H24A | 109.5 | C12—C11—H11A | 109.3 |
| C23—C24—H24B | 109.5 | C12—C11—H11B | 109.3 |
| C23—C24—H24C | 109.5 | C26—C27—H27 | 118.5 |
| H24A—C24—H24B | 109.5 | C28—C27—C26 | 123.0 (5) |
| H24A—C24—H24C | 109.5 | C28—C27—H27 | 118.5 |
| H24B—C24—H24C | 109.5 | C13—C12—C11 | 118.5 (6) |
| O1Wii—Ni2—O1W | 180.0 | C13—C12—H12A | 107.7 |
| N3—Ni2—O1W | 88.65 (15) | C13—C12—H12B | 107.7 |
| N3ii—Ni2—O1Wii | 88.65 (15) | C11—C12—H12A | 107.7 |
| N3—Ni2—O1Wii | 91.35 (15) | C11—C12—H12B | 107.7 |
| N3ii—Ni2—O1W | 91.35 (15) | H12A—C12—H12B | 107.1 |
| N3—Ni2—N3ii | 180.0 | N2—C4—C5i | 107.4 (4) |
| N3—Ni2—N4ii | 84.66 (16) | N2—C4—H4A | 110.2 |
| N3ii—Ni2—N4ii | 95.34 (16) | N2—C4—H4B | 110.2 |
| N3ii—Ni2—N4 | 84.66 (16) | C5i—C4—H4A | 110.2 |
| N3—Ni2—N4 | 95.34 (16) | C5i—C4—H4B | 110.2 |
| N4—Ni2—O1Wii | 90.90 (17) | H4A—C4—H4B | 108.5 |
| Ni1—N1—C5—C4i | 44.0 (4) | O7—P3—C38—C37 | 38.0 (5) |
| Ni1—N1—C1—C2 | −55.0 (5) | C36—C35—C40—C39 | −2.7 (7) |
| Ni1—N2—C3—C2 | 55.3 (5) | C19—C22—C41—C34 | 169.3 (4) |
| Ni1—N2—C4—C5i | −42.6 (4) | C19—C22—C41—C42 | −9.3 (6) |
| P2—C29—C30—C31 | −177.8 (4) | C19—C22—C23—C25 | −169.7 (4) |
| P2—C29—C28—C27 | 177.0 (4) | C19—C22—C23—C24 | 12.3 (6) |
| P3—C38—C39—C40 | −176.5 (4) | O8—P3—C38—C39 | −15.9 (5) |
| P3—C38—C37—C36 | 177.2 (4) | O8—P3—C38—C37 | 164.4 (4) |
| O5—P2—C29—C30 | 29.0 (5) | C20—C19—C18—C17 | −1.5 (7) |
| O5—P2—C29—C28 | −149.2 (4) | C20—C21—C16—P1 | 173.3 (4) |
| C29—C30—C31—C26 | 0.7 (8) | C20—C21—C16—C17 | −2.0 (7) |
| C29—C28—C27—C26 | 1.1 (8) | C37—C38—C39—C40 | 3.2 (7) |
| O4—P2—C29—C30 | −98.0 (4) | C32—C34—C35—C36 | −105.9 (5) |
| O4—P2—C29—C28 | 83.8 (4) | C32—C34—C35—C40 | 77.4 (6) |
| C22—C23—C25—C26 | 175.0 (4) | C32—C34—C41—C22 | 2.2 (7) |
| C22—C23—C25—C32 | −1.0 (7) | C32—C34—C41—C42 | −179.2 (4) |
| C22—C19—C20—C21 | −174.9 (4) | C24—C23—C25—C26 | −7.1 (7) |
| C22—C19—C18—C17 | 175.5 (4) | C24—C23—C25—C32 | 176.9 (4) |
| C38—C39—C40—C35 | −0.6 (8) | Ni2—N3—C10—C9ii | 43.0 (5) |
| C26—C25—C32—C34 | −177.1 (4) | Ni2—N3—C6—C7 | −54.7 (5) |
| C26—C25—C32—C33 | 2.1 (7) | Ni2—N4—C9—C10ii | −41.1 (5) |
| O3—P1—O1—Ni1 | 149.1 (9) | Ni2—N4—C8—C7 | 55.8 (5) |
| O3—P1—C16—C21 | 25.1 (4) | C10—N3—C6—C7 | −176.9 (4) |
| O3—P1—C16—C17 | −159.6 (3) | C9—N4—C8—C7 | 179.2 (5) |
| C34—C35—C36—C37 | −173.4 (4) | C6—N3—C10—C9ii | 170.0 (4) |
| C34—C35—C40—C39 | 174.0 (4) | C6—C7—C8—N4 | −71.7 (6) |
| N2—C3—C2—C1 | −71.6 (6) | C8—N4—C9—C10ii | −169.2 (4) |
| C3—N2—C4—C5i | −171.9 (4) | C8—C7—C6—N3 | 71.3 (6) |
| C3—C2—C1—N1 | 71.1 (6) | Ni3—N5—C15—C14iii | 37.7 (5) |
| O2—P1—O1—Ni1 | 22.6 (10) | Ni3—N5—C11—C12 | −53.9 (6) |
| O2—P1—C16—C21 | 141.0 (4) | Ni3—N6—C14—C15iii | −39.2 (5) |
| O2—P1—C16—C17 | −43.8 (4) | Ni3—N6—C13—C12 | 52.9 (6) |
| O9—P3—C38—C39 | 102.4 (4) | N5—C11—C12—C13 | 69.6 (7) |
| O9—P3—C38—C37 | −77.3 (4) | N6—C13—C12—C11 | −69.2 (7) |
| C35—C34—C41—C22 | −175.0 (4) | C14—N6—C13—C12 | 178.9 (5) |
| C35—C34—C41—C42 | 3.6 (7) | O1—P1—C16—C21 | −93.2 (4) |
| C35—C34—C32—C25 | 177.6 (4) | O1—P1—C16—C17 | 82.0 (4) |
| C35—C34—C32—C33 | −1.6 (7) | O6—P2—C29—C30 | 146.4 (4) |
| C35—C36—C37—C38 | −0.8 (8) | O6—P2—C29—C28 | −31.8 (4) |
| C41—C22—C23—C25 | 3.8 (7) | C16—P1—O1—Ni1 | −98.5 (10) |
| C41—C22—C23—C24 | −174.2 (4) | C16—C21—C20—C19 | −0.2 (7) |
| C41—C22—C19—C20 | 85.7 (6) | C16—C17—C18—C19 | −0.7 (7) |
| C41—C22—C19—C18 | −91.1 (5) | C18—C19—C20—C21 | 1.9 (7) |
| C41—C34—C35—C36 | 71.2 (6) | C18—C17—C16—P1 | −173.0 (3) |
| C41—C34—C35—C40 | −105.5 (5) | C18—C17—C16—C21 | 2.4 (7) |
| C41—C34—C32—C25 | 0.6 (7) | C40—C35—C36—C37 | 3.4 (7) |
| C41—C34—C32—C33 | −178.6 (4) | C1—N1—C5—C4i | 172.6 (4) |
| C23—C22—C41—C34 | −4.4 (7) | C28—C29—C30—C31 | 0.4 (8) |
| C23—C22—C41—C42 | 177.0 (4) | C31—C26—C25—C23 | 110.8 (5) |
| C23—C22—C19—C20 | −100.7 (5) | C31—C26—C25—C32 | −73.2 (6) |
| C23—C22—C19—C18 | 82.5 (5) | C31—C26—C27—C28 | 0.1 (7) |
| C23—C25—C32—C34 | −1.2 (7) | C13—N6—C14—C15iii | −169.4 (5) |
| C23—C25—C32—C33 | 178.0 (4) | C15—N5—C11—C12 | −179.0 (5) |
| C30—C29—C28—C27 | −1.3 (7) | C11—N5—C15—C14iii | 168.7 (5) |
| C25—C26—C31—C30 | 178.9 (5) | C27—C26—C25—C23 | −69.4 (6) |
| C25—C26—C27—C28 | −179.7 (5) | C27—C26—C25—C32 | 106.6 (5) |
| C5—N1—C1—C2 | −177.5 (4) | C27—C26—C31—C30 | −0.9 (7) |
| C39—C38—C37—C36 | −2.5 (7) | C4—N2—C3—C2 | 179.3 (5) |
| O7—P3—C38—C39 | −142.3 (4) |
Symmetry codes: (i) −x+2, −y+1, −z+2; (ii) −x+2, −y+2, −z+1; (iii) −x+1, −y+3, −z+1.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O6iv | 1.00 | 2.32 | 3.196 (5) | 146 |
| N2—H2···O6W | 1.00 | 2.18 | 3.039 (6) | 143 |
| N3—H3···07v | 1.00 | 2.13 | 3.102 (6) | 162 |
| N4—H4···O4W | 1.00 | 2.06 | 3.056 (6) | 173 |
| N5—H5···O9vi | 1.00 | 2.07 | 3.003 (6) | 155 |
| N6—H6···O7Wii | 1.00 | 1.98 | 2.956 (6) | 166 |
| O3—H3C···O5iv | 0.84 | 1.84 | 2.654 (5) | 162 |
| O6—H6C···O3Wvii | 0.84 | 1.75 | 2.550 (5) | 159 |
| O9—H9C···O4viii | 0.84 | 1.74 | 2.517 (5) | 154 |
| O1W—H1WB···O7v | 0.87 | 1.81 | 2.679 (5) | 173 |
| O1W—H1WA···O4W | 0.87 | 2.45 | 3.256 (6) | 155 |
| O2W—H2WB···O4 | 0.86 | 1.90 | 2.729 (5) | 164 |
| O2W—H2WA···O7Wix | 0.86 | 1.81 | 2.675 (6) | 174 |
| O3W—H3WB···O2 | 0.87 | 1.81 | 2.676 (4) | 177 |
| O3W—H3WA···O7v | 0.85 | 1.84 | 2.689 (5) | 174 |
| O4W—H4WB···O3 | 0.87 | 2.26 | 3.115 (6) | 167 |
| O4W—H4WA···O8v | 0.87 | 1.93 | 2.796 (6) | 172 |
| O5W—H5WB···O5x | 0.87 | 1.98 | 2.813 (5) | 159 |
| O5W—H5WA···O8xi | 0.87 | 1.87 | 2.725 (5) | 168 |
| O6W—H6WB···O2 | 0.87 | 2.02 | 2.799 (6) | 149 |
| O6W—H6WA···O5W | 0.87 | 2.00 | 2.842 (5) | 164 |
| O7W—H7WB···O3W | 0.85 | 2.02 | 2.731 (5) | 140 |
| O7W—H7WA···O5W | 0.86 | 1.83 | 2.688 (5) | 173 |
Symmetry codes: (ii) −x+2, −y+2, −z+1; (iv) x, y−1, z; (v) x, y, z−1; (vi) −x+1, −y+2, −z+2; (vii) x, y+1, z; (viii) x, y−1, z+1; (ix) x−1, y+1, z; (x) x+1, y−1, z; (xi) x+1, y, z−1.
References
- Alexandrov, E. V., Blatov, V. A. & Proserpio, D. M. (2017). CrystEngComm, 19, 1993–2006.
- Barefield, E. K., Bianchi, A., Billo, E. J., Connolly, P. J., Paoletti, P., Summers, J. S. & Van Derveer, D. G. (1986). Inorg. Chem. 25, 4197–4202.
- Beckmann, J., Rüttinger, R. & Schwich, T. (2008). Cryst. Growth Des. 8, 3271–3276.
- Bosnich, B., Poon, C. K. & Tobe, M. L. (1965a). Inorg. Chem. 4, 1102–1108.
- Bosnich, B., Tobe, M. L. & Webb, G. A. (1965b). Inorg. Chem. 4, 1109–1112.
- Firmino, A. D. G., Figueira, F., Tomé, J. P. C., Paz, F. A. A. & Rocha, J. (2018). Coord. Chem. Rev. 355, 133–149.
- Gagnon, K. J., Perry, H. P. & Clearfield, A. (2012). Chem. Rev. 112, 1034–1054. [DOI] [PubMed]
- Gong, Y.-N., Zhong, D.-C. & Lu, T.-B. (2016). CrystEngComm, 18, 2596–2606.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hermer, N., Reinsch, H., Mayer, P. & Stock, N. (2016). CrystEngComm, 18, 8147–8150.
- Kaskel, S. (2016). Editor. The Chemistry of Metal–Organic Frameworks: Synthesis, Characterization and Applications. Weinheim: Wiley-VCH.
- Lampeka, Ya. D. & Tsymbal, L. V. (2004). Theor. Exp. Chem. 40, 345–371.
- Lampeka, Ya. D., Tsymbal, L. V., Barna, A. V., Shuĺga, Y. L., Shova, S. & Arion, V. B. (2012). Dalton Trans. 41, 4118–4125. [DOI] [PubMed]
- MacGillivray, L. R. & Lukehart, C. M. (2014). Editors. Metal–Organic Framework Materials. Hoboken: John Wiley and Sons.
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst. 53, 226–235. [DOI] [PMC free article] [PubMed]
- Ouellette, W., Wang, G., Liu, H., Yee, G. T., O’Connor, C. J. & Zubieta, J. (2009). Inorg. Chem. 48, 953–963. [DOI] [PubMed]
- Pili, S., Argent, S. P., Morris, C. G., Rought, P., García-Sakai, V., Silverwood, I. P., Easun, T. L., Li, M., Warren, M. R., Murray, C. A., Tang, C. C., Yang, S. & Schröder, M. (2016). J. Am. Chem. Soc. 138, 6352–6355. [DOI] [PMC free article] [PubMed]
- Rao, C. N. R., Natarajan, S. & Vaidhyanathan, R. (2004). Angew. Chem. Int. Ed. 43, 1466–1496. [DOI] [PubMed]
- Rigaku OD (2020). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, England.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Stackhouse, C. A. & Ma, S. (2018). Polyhedron, 145, 154–165.
- Suh, M. P. & Moon, H. R. (2007). Advances in Inorganic Chemistry, Vol. 59, edited by R. van Eldik & K. Bowman-James, pp. 39–79. San Diego: Academic Press.
- Suh, M. P., Park, H. J., Prasad, T. K. & Lim, D.-W. (2012). Chem. Rev. 112, 782–835. [DOI] [PubMed]
- Taddei, M., Costantino, F., Vivani, R., Sabatini, S., Lim, S.-H. & Cohen, S. M. (2014). Chem. Commun. 50, 5737–5740. [DOI] [PubMed]
- Tsymbal, L. V., Andriichuk, I. L., Lozan, V., Shova, S. & Lampeka, Y. D. (2022). Acta Cryst. E78, 625–628. [DOI] [PMC free article] [PubMed]
- Vaidhyanathan, R., Mahmoudkhani, A. H. & Shimizu, G. K. H. (2009). Can. J. Chem. 87, 247–253.
- Vilela, S. M. F., Navarro, J. A. R., Barbosa, P., Mendes, R. F., Pérez-Sánchez, G., Nowell, H., Ananias, D., Figueiredo, F., Gomes, J. R. B., Tomé, J. P. C. & Paz, F. A. A. (2021). J. Am. Chem. Soc. 143, 1365–1376. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Yatsimirskii, K. B. & Lampeka, Ya. D. (1985). Physicochemistry of Metal Complexes with Macrocyclic Ligands. Kiev: Naukova Dumka (in Russian).
- Yücesan, G., Zorlu, Y., Stricker, M. & Beckmann, J. (2018). Coord. Chem. Rev. 369, 105–122.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989022006624/hb8026sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022006624/hb8026Isup2.hkl
CCDC reference: 2178456
Additional supporting information: crystallographic information; 3D view; checkCIF report