The title compounds arose as unexpected by-products of an iodination reaction: in each case the fused-ring and isoxazole planes are almost perpendicular to each other.
Keywords: crystal structure, isoxazole, anthracenyl isoxazole, oxidation product
Abstract
The syntheses and structures of an unexpected by-product from an iodination reaction, namely, ethyl 5-methyl-3-(10-nitroanthracen-9-yl)isoxazole-4-carboxylate, C21H16N2O5, (I), and its oxidation product, ethyl 3-(9-hydroxy-10-oxo-9,10-dihydroanthracen-9-yl)-5-methylisoxazole-4-carboxylate, C21H17NO5 (V) are described. Compound (I) crystallizes with two molecules in the asymmetric unit in which the dihedral angles between the anthracene fused-ring systems and isoxazole ring mean planes are 88.67 (16) and 85.64 (16)°; both molecules feature a disordered nitro group. In (V), which crystallizes with one molecule in the asymmetric unit, the equivalent dihedral angle between the almost planar anthrone ring system (r.m.s. deviation = 0.029 Å) and the pendant isoxazole ring is 89.65 (5)°. In the crystal of (I), the molecules are linked by weak C—H⋯O interactions into a three-dimensional network and in the extended structure of (V), inversion dimers linked by pairwise O—H⋯O hydrogen bonds generate R 2 2(14) loops.
1. Chemical context
In the course of our study of aryl-isoxazole amide (AIM) anti-tumor agents, we have a standard operating procedure to identify by-products of the synthesis (Weaver, Campbell et al., 2020 ▸), and have used the mechanistic insights gained in order to optimize and improve subsequent syntheses.

During recent structure–activity relationship studies, we encountered complications in constructing sterically hindered examples, which we desired for their calculated pharmacokinetic properties. After obtaining mediocre results with bromine as a leaving group in Suzuki couplings, we pursued a fairly routine alternative of moving to the next halogen down in the periodic table. We have encountered more complications in this study than in the previous twenty papers we have published in this area (e.g. Weaver, Stump et al., 2020 ▸ and Weaver et al., 2015 ▸), and herein report the crystal structures of two compounds observed.
Using conditions usually reported for iodination, the main product observed for reaction of (II) was the nitro ester (I) rather than the expected iodo product (III), which was obtained in small amounts (Fig. 1 ▸). The nitro product so obtained exhibits most of the stereoelectronic properties of previously studied analogues that we have considered to be essential for their biological activity (Han et al., 2009 ▸). The nitro group is disordered and found in two distinct conformations in the unit cell. We attribute this to an extreme peri-effect, which substantially raises the energy of the co-planar conformer.
Figure 1.
Preparation and molecular structures of the title compounds.
In order to improve on the accuracy of the crystal structure of (I) we attempted numerous recrystallizations; however, what was observed was the addition of oxygen to compound (I), which we attribute to cycloaddition of dioxygen to an endo-peroxide (IV) (Klaper et al., 2016 ▸), and ring opening with loss of a leaving group to the oxidation product anthraquinone (V). Usually, anthracenes are oxidized in vivo predominantly by cytochrome P450, leading to a potentially toxic arene oxide (Silverman et al., 2014 ▸). The rationale for the isoxazole series is that the C-5 isoxazole methyl group represents an opportunity for safer metabolism (Natale et al., 2010 ▸). The observation in this manuscript suggests that intramolecular dioxygenation, which would likely be mediated in vivo by mono amine oxidase (MAO), is another plausible route (Silverman, 2002 ▸). The observation of a possible endo-peroxide pathway in this study suggests that the metabolism of these 10-substituted anthracenyl isoxazole analogues could go through dioxygenation catalysed by COX (cyclooxygenase) and other prostaglandin synthases in vivo (Silverman, 2002 ▸).
2. Structural commentary
The first title compound (I), C21H16N2O5, crystallizes in the monoclinic Cc space group with two independent molecules in the asymmetric unit (Fig. 2 ▸). The dihedral angle between the anthracene ring mean plane and the isoxazole ring mean plane indicate near orthogonality: 88.67 (16) and 85.64 (16)° for molecules A (containing C1) and B (containing C22), respectively. Each independent anthryl ring contains a 10-nitro group with the O atoms disordered over two orientations. The isoxazole group and its attached ethyl ester moiety are virtually co-planar, with the twist angles found to be 3.1 (2)° between the C15–C17/O1/N1 and O2/C19/O3/C20 planes in molecule A, and 4.2 (2)° between the C36–C38/O6/N3 and O7/C40/O8/C41 planes in molecule B. The ester ethyl group is exo- with respect to the anthryl ring in the solid state but this conformation is not completely retained in solution as the proton NMR indicates significant anisotropy at the methyl group of the ethyl ester (δ = 0.41), which indicates at the very least a significant population of the endo- orientation. In addition, many of our other reported anthracenyl isoxazole esters have shown the ester ethyl group in an endo- orientation (Weaver, Stump et al., 2020 ▸; Weaver et al., 2015 ▸; Li et al., 2013 ▸; Li et al., 2006 ▸; Han et al., 2003 ▸; Mosher et al., 1996 ▸).
Figure 2.
The asymmetric unit of compound (I) showing displacement ellipsoids drawn at the 50% probability level. The structure on the left is molecule A and that on the right is molecule B.
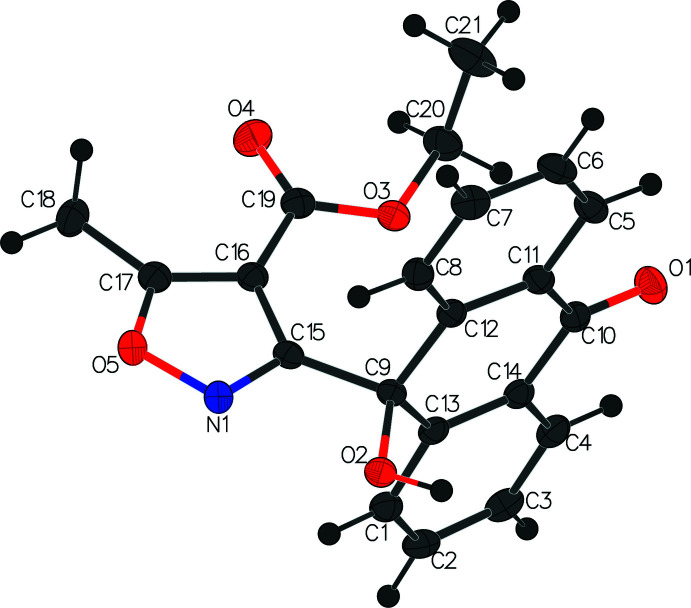
The second title compound (V), C21H17NO5, crystallizes in the monoclinic P21/c space group with one independent molecule in the asymmetric unit (Fig. 3 ▸). The anthrone ring system is virtually planar with an r.m.s. deviation of 0.029 Å. Like the other anthracenyl isoxazole structures we have reported (vide supra), the isoxazole ring is orthogonal to the anthracene ring, with a dihedral angle of 89.65 (5)°. The ester ethyl group is in endo- orientation and the C19—O3—C20—C21 grouping is twisted [torsion angle = 86.7 (2)°].
Figure 3.
The asymmetric unit of compound (V) with displacement ellipsoids drawn at the 50% probability level.
3. Supramolecular features
In compound (I), weak C—H⋯O hydrogen bonds between adjacent A molecules (C7—H7⋯O4 and C1—H1⋯O5) form a column running perpendicular to the [101] direction. Molecule B lies between the columns and its O7 atom accepts a hydrogen bond from H3 of molecule A (Table 1 ▸, Fig. 4 ▸). There is an aromatic π–π stacking interaction with a centroid–centroid separation of 3.537 (5) Å between the planes of the C22–C25/C32/C33 and C1–C4/C11/C12 rings. A σ–π interaction is observed at a distance of 3.774 Å from atom C42 to the plane centroid.
Table 1. Hydrogen-bond geometry (Å, °) for (I) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C1—H1⋯O5i | 0.95 | 2.46 | 3.366 (12) | 159 |
| C3—H3⋯O7ii | 0.95 | 2.44 | 3.339 (6) | 158 |
| C7—H7⋯O4iii | 0.95 | 2.40 | 3.24 (4) | 147 |
| C7—H7⋯O4A iii | 0.95 | 2.46 | 3.34 (6) | 154 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 .
.
Figure 4.
The partial packing of compound (I). For clarity, only hydrogen bonds C1—H1⋯O5i and C3—H3⋯O7ii are shown as dashed lines, and H atoms not involved in these hydrogen bonds are removed.
In the crystal of compound (V), inversion dimers linked by pairwise O2—H2⋯O1 hydrogen bonds occur (Table 2 ▸, Fig. 5 ▸). A short contact distance between the isoxazole ring of one molecule (ring mean plane C15–C17/O5N1) and the carbonyl oxygen (O4) of another molecule [3.1486 (16) Å] may contribute to the head-to-head, tail-to-tail arrangement in the crystal structure, also shown in Fig. 8 ▸ b.
Table 2. Hydrogen-bond geometry (Å, °) for (V) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2⋯O1i | 0.91 (3) | 1.93 (3) | 2.8359 (19) | 176 (2) |
Symmetry code: (i)
 .
.
Figure 5.
The packing of compound (V). Inversion dimers linked by pairwise O2—H2⋯O1 hydrogen bonds are shown in dashed lines.
Figure 8.
(a) The Hirshfeld surface of (V) mapped over d norm. Short and long contacts are indicated as red and blue spots, respectively. Contacts with distances approximately equal to the sum of the van der Waals radii are colored white. Hydroxyl and carbonyl groups on the anthrone ring contributed major short contacts. (b) π–π interactions (anthrone to anthrone and carbonyl to isoxazole ring) and σ–π interaction (C—H bond to carbonyl) are shown as orange–red spots with green dashed lines in the shape-index map.
4. Hirshfeld surface analysis
Hirshfeld surface analysis (Spackman & Jayatilaka, 2009 ▸) was performed, and the associated two-dimensional fingerprint plots (McKinnon et al., 2007 ▸) were generated to quantify the intermolecular interactions using Crystal Explorer 21.5 (Spackman et al., 2021 ▸). The Hirshfeld surface of (I) is mapped over d norm in a fixed color scale of −0.31 (red) to 1.26 (blue) arbitrary units (Fig. 6 ▸). The delineated two-dimensional fingerprint plots shown in Fig. 7 ▸ indicate that two main contributions to the overall Hirshfeld surface area arise from H⋯H contacts (35.3%) and O⋯H/H⋯O contacts (29.0%) with C⋯H/H⋯C interactions contributing 17.5% of the Hirshfeld surface.
Figure 6.
(a) The Hirshfeld surface of (I) mapped over d norm. Short and long contacts are indicated as red and blue spots, respectively. Contacts with distances approximately equal to the sum of the van der Waals radii are colored white. (b) Weak π–π interactions are shown as green dashed lines on a surface mapped over curvedness. The π–π stacking is indicated by the green flat regions surrounded by dark blue edges.
Figure 7.
The two-dimensional fingerprint plots for (I) delineated into (a) H⋯H contacts, (b) O⋯H/H⋯O contacts, (c) C⋯H/H⋯C contacts, and (d) N⋯H/H⋯N contacts. Other contact contributions less than 5% are omitted.
The Hirshfeld surface of compound V is mapped over d norm in a fixed color scale of −0.58 (red) to 1.31 (blue) arbitrary units (Fig. 8 ▸ a), showing two short contacts from O⋯H hydrogen bonds in red spots. The delineated two-dimensional fingerprint plots (Fig. 9 ▸) indicate that H⋯H contacts contribute 47.7% of the Hirshfeld surface. Aromatic π–π stacking is also identifiable from the Hirshfeld surface mapped over the shape-index property (Fig. 8 ▸ b).
Figure 9.
The two-dimensional fingerprint plots for (V) delineated into (a) H⋯H contacts, (b) O⋯H/H⋯O contacts, (c) C⋯H/H⋯C contacts, and (d) N⋯H/H⋯N contacts. Other contact contributions less than 5% are omitted.
5. Database survey
A search for the 9-nitroanthracenyl moiety in the Cambridge Structural Database (CSD version 5.43, November 2021 update; Groom et al., 2016 ▸) resulted in 14 hits, of which two crystal structures of 9-nitroanthracene itself were reported, namely refcodes NTRANT (Trotter, 1959 ▸) and NTRANT01 (Glagovich et al., 2004 ▸). The reported angles between the NO2 plane and the anthracene plane are 84.78 and 69.40°, respectively, which agree with our observation of the disordered NO2 group in (I).
A search in the same database for the 10-hydroxy anthrone fragment resulted in 59 hits, of which 10 structures had an aromatic ring at the 10-position, namely refcodes COBWEY (Barker et al., 2019 ▸), DULVUB (Skrzat & Roszak, 1986 ▸), ELULII (Stepovik et al., 2015 ▸), EVETIL (Mao et al., 2021 ▸), JAYPAA (Roszak et al., 1990 ▸), MOTJIQ (Chen et al., 2015 ▸), MOTKEN (Chen et al., 2015 ▸), QAJPUQ (Forensi et al., 2020 ▸), SAMNEC (Hoffend et al., 2013 ▸) and WOKYIH (Pullella et al., 2019 ▸). The anthrone unit in these 10 structures are either essentially planar or in a shallow boat conformation. The aromatic rings at the 10-position in these compounds are all at a vertical orientation relative to the anthrone ring. It may be noted that an anthrone isoxazole ester we reported in 2014, refcode TIYZEI, also shares similar structural features (Duncan et al., 2014 ▸).
6. Synthesis and crystallization
Iodination of aromatic hydrocarbons with molecular iodine has been accomplished by several methods, typically using an oxidizing agent to generate the iodonium cation electrophile. Among the conditions we surveyed, fuming nitric acid in particular (Bansal et al., 1987 ▸) with the anthracene isoxazole (II), appears to consistently produce the nitrated anthryl (I) rather than the desired iodo product (III). The anthryl isoxazole ester (II) was prepared as previously described (Mosher et al., 1996 ▸), and recrystallized before use. The ester (II) (67 mg, 0.19 mmol) was dissolved in acetic acid (1 ml), and iodine (24.1 mg) was added. To this solution was added concentrated sulfuric acid (1 ml) and sodium nitrite (13.1 mg, 0.19 mmol). The resulting solution was warmed to reflux for 30 minutes, after which it was poured over ice (3 g) and the precipitate collected by filtration. Silica gel chromatography using ethyl acetate in hexane provided the product, which was recrystallized from solutions in methylene chloride, ethyl acetate and hexane by slow evaporation, whereby the product was obtained as dull dark-yellow prisms (15 mg, 21%). 1H NMR: (CDCl3) δppm 7.95 (d, 2H, J = 8Hz); 7.69 (m, 4H); 7.6 (m, 2H); 3.735 (q, 2H, J = 4Hz); 2.94 (s, 3H); 0.41 (t, 3H, J = 4Hz). 13C NMR: (CDCl3) δppm 176.66, 161.03, 159.45, 145.97, 133.59, 130.34, 128.68, 127.11, 125.67, 121.81, 121.57, 111.45, 60.41, 13.47, 12.94. HPLC–MS: calculated for [C21H16N2O5+H]+ 377.1137, observed m/z 377 ([M + 1]+, 100% rel. intensity).
During the re-crystallization of compound (I), different solvent combinations of hexane, methanol, dichloromethane, and ethyl acetate were used. Instead of better crystals of compound (I), compound (V) was formed as translucent light-yellow prisms from the slow evaporation of the solvent mixture composed of hexane and methanol at room temperature over a period of two months. 1H NMR: (CDCl3) δppm 8.29 (dd, 2H, J = 1.37 and 7.79 Hz); 7.67 (d, 2H, J = 7.79 Hz); 7.60 (ddd, 2H, J = 1.37, 7.33, and 7.79 Hz); 7.50 (ddd, 2H, J = 1.37, 7.33, and 7.79 Hz); 4.06 (q, 2H, J = 6.87 Hz); 2.60 (s, 3H); 1.06 (t, 3H, J = 6.87 Hz). 13C NMR: (CDCl3) δppm 183.86, 177.58, 167.05, 162.74, 143.92, 133.68, 130.96, 128.86, 127.29, 126.72, 71.26, 61.71, 14.16, 13.96.
7. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. In compound (I), the nitro group is disordered in each of the two independent molecules in the asymmetric unit. The occupancies of each disordered part were refined, converging to 0.572 (13) and 0.428 (13) for molecule A, and 0.64 (3) and 0.36 (3) for molecule B. EADP constraints were applied (Sheldrick, 2015 ▸) to each nitro group. The C-bound hydrogen atoms on both compounds were fixed geometrically and treated as riding with C—H = 0.95–0.98 Å and refined with U
iso(H) = 1.2U
eq(CH, CH2) or 1.5U
eq(CH3). The O-bound H atom in (V) was found in a difference-Fourier map and refined freely. Four reflections (
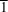 10, 110,
10, 110,
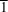 11 and 11
11 and 11
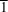 ) in compound (I) and four reflections (100,
) in compound (I) and four reflections (100,
 4 5, 110 and 011) in compound (V) affected by the beam stop were omitted from the final cycles of refinement because of poor agreement between the observed and calculated intensities. The absolute structure of (I) was indeterminate in the present refinement.
4 5, 110 and 011) in compound (V) affected by the beam stop were omitted from the final cycles of refinement because of poor agreement between the observed and calculated intensities. The absolute structure of (I) was indeterminate in the present refinement.
Table 3. Experimental details.
| (I) | (V) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C21H16N2O5 | C21H17NO5 |
| M r | 376.36 | 363.36 |
| Crystal system, space group | Monoclinic, C c | Monoclinic, P21/c |
| Temperature (K) | 100 | 100 |
| a, b, c (Å) | 16.4968 (10), 14.8697 (9), 16.1836 (9) | 8.2862 (4), 23.5895 (11), 8.6219 (4) |
| β (°) | 114.879 (3) | 97.728 (2) |
| V (Å3) | 3601.5 (4) | 1669.99 (14) |
| Z | 8 | 4 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 0.10 | 0.10 |
| Crystal size (mm) | 0.29 × 0.24 × 0.22 | 0.28 × 0.20 × 0.19 |
| Data collection | ||
| Diffractometer | Bruker SMART Breeze CCD | Bruker SMART Breeze CCD |
| Absorption correction | – | Numerical (SADABS; Krause et al., 2015 ▸) |
| T min, T max | – | 0.945, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 45790, 7615, 5596 | 44252, 4112, 3252 |
| R int | 0.054 | 0.051 |
| (sin θ/λ)max (Å−1) | 0.633 | 0.668 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.059, 0.158, 1.02 | 0.051, 0.114, 1.13 |
| No. of reflections | 7615 | 4112 |
| No. of parameters | 546 | 250 |
| No. of restraints | 2 | 0 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.55, −0.19 | 0.37, −0.21 |
| Absolute structure | Flack x determined using 2257 quotients [(I +)−(I −)]/[(I +)+(I −)] (Parsons et al., 2013 ▸) | – |
| Absolute structure parameter | 0.5 (4) | – |
Supplementary Material
Crystal structure: contains datablock(s) I, V, global. DOI: 10.1107/S2056989022005710/hb8020sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022005710/hb8020Isup2.hkl
Structure factors: contains datablock(s) V. DOI: 10.1107/S2056989022005710/hb8020Vsup3.hkl
Supporting information file. DOI: 10.1107/S2056989022005710/hb8020Isup4.cml
Supporting information file. DOI: 10.1107/S2056989022005710/hb8020Vsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Ethyl 5-methyl-3-(10-nitroanthracen-9-yl)isoxazole-4-carboxylate (I). Crystal data
| C21H16N2O5 | F(000) = 1568 |
| Mr = 376.36 | Dx = 1.388 Mg m−3 |
| Monoclinic, Cc | Mo Kα radiation, λ = 0.71073 Å |
| a = 16.4968 (10) Å | Cell parameters from 7597 reflections |
| b = 14.8697 (9) Å | θ = 2.7–21.0° |
| c = 16.1836 (9) Å | µ = 0.10 mm−1 |
| β = 114.879 (3)° | T = 100 K |
| V = 3601.5 (4) Å3 | Prism, yellow |
| Z = 8 | 0.29 × 0.24 × 0.22 mm |
Ethyl 5-methyl-3-(10-nitroanthracen-9-yl)isoxazole-4-carboxylate (I). Data collection
| Bruker SMART Breeze CCD diffractometer | Rint = 0.054 |
| Radiation source: 2 kW sealed X-ray tube | θmax = 26.7°, θmin = 2.7° |
| φ and ω scans | h = −20→20 |
| 45790 measured reflections | k = −18→18 |
| 7615 independent reflections | l = −20→20 |
| 5596 reflections with I > 2σ(I) |
Ethyl 5-methyl-3-(10-nitroanthracen-9-yl)isoxazole-4-carboxylate (I). Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.059 | w = 1/[σ2(Fo2) + (0.0864P)2 + 2.331P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.158 | (Δ/σ)max < 0.001 |
| S = 1.02 | Δρmax = 0.55 e Å−3 |
| 7615 reflections | Δρmin = −0.19 e Å−3 |
| 546 parameters | Absolute structure: Flack x determined using 2257 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
| 2 restraints | Absolute structure parameter: 0.5 (4) |
| Primary atom site location: structure-invariant direct methods |
Ethyl 5-methyl-3-(10-nitroanthracen-9-yl)isoxazole-4-carboxylate (I). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Ethyl 5-methyl-3-(10-nitroanthracen-9-yl)isoxazole-4-carboxylate (I). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O1 | 0.3319 (3) | 0.2201 (3) | 0.5466 (2) | 0.0487 (9) | |
| O2 | 0.3808 (2) | 0.0465 (3) | 0.3397 (2) | 0.0459 (9) | |
| O3 | 0.4880 (2) | 0.0385 (3) | 0.4815 (2) | 0.0471 (9) | |
| N1 | 0.2636 (3) | 0.2332 (3) | 0.4569 (3) | 0.0440 (10) | |
| C1 | 0.3269 (3) | 0.2892 (3) | 0.2833 (4) | 0.0392 (11) | |
| H1 | 0.363669 | 0.290494 | 0.346904 | 0.047* | |
| C2 | 0.3469 (4) | 0.3427 (4) | 0.2259 (4) | 0.0519 (14) | |
| H2 | 0.397851 | 0.380760 | 0.250073 | 0.062* | |
| C3 | 0.2934 (4) | 0.3424 (4) | 0.1320 (4) | 0.0548 (15) | |
| H3 | 0.308016 | 0.380624 | 0.093210 | 0.066* | |
| C4 | 0.2203 (4) | 0.2876 (4) | 0.0955 (4) | 0.0471 (13) | |
| H4 | 0.184644 | 0.288156 | 0.031651 | 0.056* | |
| C5 | 0.0334 (3) | 0.0476 (3) | 0.1397 (3) | 0.0375 (11) | |
| H5 | 0.001 (4) | 0.040 (3) | 0.083 (4) | 0.045 (15)* | |
| C6 | 0.0166 (4) | −0.0086 (4) | 0.1956 (4) | 0.0447 (13) | |
| H6 | −0.030 (4) | −0.042 (4) | 0.171 (4) | 0.042 (15)* | |
| C7 | 0.0677 (3) | −0.0057 (4) | 0.2910 (3) | 0.0433 (12) | |
| H7 | 0.054540 | −0.045527 | 0.329570 | 0.052* | |
| C8 | 0.1361 (3) | 0.0548 (3) | 0.3273 (3) | 0.0363 (11) | |
| H8 | 0.170089 | 0.056581 | 0.391448 | 0.044* | |
| C9 | 0.2294 (3) | 0.1746 (3) | 0.3054 (3) | 0.0335 (10) | |
| C10 | 0.1272 (3) | 0.1697 (3) | 0.1202 (3) | 0.0359 (11) | |
| C11 | 0.1972 (3) | 0.2301 (3) | 0.1519 (3) | 0.0354 (11) | |
| C12 | 0.2511 (3) | 0.2315 (3) | 0.2482 (3) | 0.0326 (10) | |
| C13 | 0.1573 (3) | 0.1148 (3) | 0.2714 (3) | 0.0295 (10) | |
| C14 | 0.1042 (3) | 0.1105 (3) | 0.1749 (3) | 0.0325 (10) | |
| C15 | 0.2857 (3) | 0.1785 (3) | 0.4056 (3) | 0.0330 (10) | |
| C16 | 0.3647 (3) | 0.1298 (3) | 0.4562 (3) | 0.0353 (10) | |
| C17 | 0.3904 (3) | 0.1590 (4) | 0.5441 (3) | 0.0404 (12) | |
| C18 | 0.4659 (4) | 0.1363 (4) | 0.6323 (3) | 0.0537 (15) | |
| H18A | 0.516186 | 0.176866 | 0.642774 | 0.081* | |
| H18B | 0.484587 | 0.074058 | 0.630503 | 0.081* | |
| H18C | 0.446894 | 0.143120 | 0.681695 | 0.081* | |
| C19 | 0.4107 (3) | 0.0671 (4) | 0.4186 (3) | 0.0402 (12) | |
| C20 | 0.5375 (4) | −0.0214 (4) | 0.4483 (4) | 0.0517 (14) | |
| H20A | 0.556563 | 0.011073 | 0.406087 | 0.062* | |
| H20B | 0.499576 | −0.072871 | 0.415356 | 0.062* | |
| C21 | 0.6175 (4) | −0.0541 (4) | 0.5297 (4) | 0.0585 (15) | |
| H21A | 0.651142 | −0.096711 | 0.509947 | 0.088* | |
| H21B | 0.597864 | −0.083994 | 0.572036 | 0.088* | |
| H21C | 0.655804 | −0.002855 | 0.560146 | 0.088* | |
| O4 | 0.1027 (18) | 0.111 (3) | −0.022 (3) | 0.072 (6) | 0.572 (13) |
| O5 | −0.0003 (7) | 0.1959 (10) | −0.0102 (7) | 0.068 (3) | 0.572 (13) |
| N2 | 0.076 (3) | 0.156 (2) | 0.025 (3) | 0.039 (4) | 0.572 (13) |
| O4A | 0.073 (3) | 0.105 (4) | −0.022 (4) | 0.072 (6) | 0.428 (13) |
| O5A | 0.0259 (10) | 0.2395 (14) | −0.0113 (10) | 0.068 (3) | 0.428 (13) |
| N2A | 0.066 (4) | 0.178 (3) | 0.017 (4) | 0.039 (4) | 0.428 (13) |
| O6 | 0.4350 (3) | 0.7239 (3) | 0.2788 (2) | 0.0553 (11) | |
| O7 | 0.3756 (2) | 0.5736 (2) | 0.4917 (2) | 0.0424 (8) | |
| O8 | 0.2920 (2) | 0.5316 (2) | 0.3474 (2) | 0.0422 (8) | |
| N3 | 0.4850 (3) | 0.7577 (3) | 0.3678 (3) | 0.0561 (13) | |
| N4 | 0.6061 (3) | 0.7796 (3) | 0.8043 (3) | 0.0495 (12) | |
| C22 | 0.3713 (4) | 0.8310 (4) | 0.4975 (4) | 0.0506 (14) | |
| H22 | 0.347573 | 0.821907 | 0.433552 | 0.061* | |
| C23 | 0.3270 (4) | 0.8841 (4) | 0.5326 (4) | 0.0600 (16) | |
| H23 | 0.272392 | 0.911450 | 0.492941 | 0.072* | |
| C24 | 0.3606 (4) | 0.8994 (4) | 0.6269 (4) | 0.0568 (15) | |
| H24 | 0.328476 | 0.936589 | 0.650609 | 0.068* | |
| C25 | 0.4393 (4) | 0.8609 (3) | 0.6845 (4) | 0.0506 (14) | |
| H25 | 0.461760 | 0.872032 | 0.748095 | 0.061* | |
| C26 | 0.6969 (3) | 0.6606 (4) | 0.7304 (3) | 0.0471 (13) | |
| H26 | 0.721891 | 0.669551 | 0.794480 | 0.057* | |
| C27 | 0.7377 (4) | 0.6055 (5) | 0.6938 (5) | 0.0650 (17) | |
| H27 | 0.792063 | 0.577059 | 0.732452 | 0.078* | |
| C28 | 0.7018 (4) | 0.5891 (5) | 0.6000 (4) | 0.0635 (17) | |
| H28 | 0.731967 | 0.549990 | 0.575834 | 0.076* | |
| C29 | 0.6250 (4) | 0.6284 (4) | 0.5438 (4) | 0.0481 (13) | |
| H29 | 0.600529 | 0.615429 | 0.480499 | 0.058* | |
| C30 | 0.4988 (4) | 0.7306 (4) | 0.5209 (3) | 0.0405 (12) | |
| C31 | 0.5696 (3) | 0.7629 (3) | 0.7055 (3) | 0.0364 (11) | |
| C32 | 0.4875 (3) | 0.8051 (3) | 0.6514 (3) | 0.0381 (11) | |
| C33 | 0.4527 (3) | 0.7886 (3) | 0.5551 (3) | 0.0400 (12) | |
| C34 | 0.5802 (3) | 0.6883 (3) | 0.5776 (3) | 0.0349 (11) | |
| C35 | 0.6165 (3) | 0.7057 (3) | 0.6735 (3) | 0.0369 (11) | |
| C36 | 0.4597 (4) | 0.7112 (3) | 0.4208 (3) | 0.0413 (12) | |
| C37 | 0.3956 (3) | 0.6454 (3) | 0.3713 (3) | 0.0370 (11) | |
| C38 | 0.3817 (3) | 0.6569 (4) | 0.2828 (3) | 0.0429 (12) | |
| C39 | 0.3211 (4) | 0.6178 (4) | 0.1946 (3) | 0.0478 (13) | |
| H39A | 0.316485 | 0.552762 | 0.201431 | 0.072* | |
| H39B | 0.344798 | 0.629473 | 0.149346 | 0.072* | |
| H39C | 0.261828 | 0.645184 | 0.174453 | 0.072* | |
| C40 | 0.3545 (3) | 0.5804 (3) | 0.4101 (3) | 0.0365 (11) | |
| C41 | 0.2435 (4) | 0.4683 (4) | 0.3801 (4) | 0.0487 (13) | |
| H41A | 0.226101 | 0.498471 | 0.424769 | 0.058* | |
| H41B | 0.282252 | 0.416477 | 0.410479 | 0.058* | |
| C42 | 0.1625 (4) | 0.4370 (4) | 0.3006 (4) | 0.0563 (15) | |
| H42A | 0.180119 | 0.410741 | 0.255014 | 0.084* | |
| H42B | 0.122459 | 0.488051 | 0.273756 | 0.084* | |
| H42C | 0.131623 | 0.391508 | 0.320559 | 0.084* | |
| O9 | 0.5612 (9) | 0.7525 (10) | 0.8442 (5) | 0.075 (5) | 0.64 (3) |
| O10 | 0.6744 (11) | 0.8219 (11) | 0.8389 (6) | 0.076 (4) | 0.64 (3) |
| O9A | 0.6187 (17) | 0.7124 (9) | 0.8578 (8) | 0.060 (6) | 0.36 (3) |
| O10A | 0.6276 (15) | 0.8540 (10) | 0.8356 (9) | 0.054 (5) | 0.36 (3) |
Ethyl 5-methyl-3-(10-nitroanthracen-9-yl)isoxazole-4-carboxylate (I). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.052 (2) | 0.056 (2) | 0.0267 (19) | 0.0025 (19) | 0.0058 (17) | −0.0110 (15) |
| O2 | 0.045 (2) | 0.058 (2) | 0.0280 (19) | 0.0043 (17) | 0.0083 (16) | −0.0063 (15) |
| O3 | 0.037 (2) | 0.066 (2) | 0.0312 (19) | 0.0086 (18) | 0.0079 (16) | −0.0030 (17) |
| N1 | 0.044 (2) | 0.049 (3) | 0.029 (2) | 0.006 (2) | 0.0054 (19) | 0.0002 (19) |
| C1 | 0.031 (3) | 0.041 (3) | 0.038 (3) | −0.006 (2) | 0.008 (2) | 0.003 (2) |
| C2 | 0.044 (3) | 0.057 (3) | 0.055 (4) | −0.015 (3) | 0.021 (3) | 0.001 (3) |
| C3 | 0.050 (3) | 0.069 (4) | 0.048 (3) | −0.011 (3) | 0.023 (3) | 0.018 (3) |
| C4 | 0.048 (3) | 0.060 (3) | 0.031 (3) | −0.003 (3) | 0.015 (2) | 0.006 (2) |
| C5 | 0.031 (3) | 0.050 (3) | 0.026 (2) | −0.009 (2) | 0.007 (2) | −0.008 (2) |
| C6 | 0.035 (3) | 0.053 (3) | 0.044 (3) | −0.019 (3) | 0.015 (2) | −0.009 (2) |
| C7 | 0.044 (3) | 0.050 (3) | 0.039 (3) | −0.012 (2) | 0.020 (2) | 0.001 (2) |
| C8 | 0.037 (3) | 0.046 (3) | 0.027 (2) | −0.004 (2) | 0.014 (2) | −0.003 (2) |
| C9 | 0.029 (2) | 0.040 (3) | 0.029 (2) | −0.001 (2) | 0.0097 (19) | −0.002 (2) |
| C10 | 0.032 (2) | 0.049 (3) | 0.022 (2) | 0.000 (2) | 0.0062 (19) | −0.001 (2) |
| C11 | 0.031 (2) | 0.042 (3) | 0.032 (2) | −0.002 (2) | 0.012 (2) | 0.006 (2) |
| C12 | 0.028 (2) | 0.038 (2) | 0.030 (2) | −0.0012 (19) | 0.0099 (19) | 0.0026 (19) |
| C13 | 0.029 (2) | 0.033 (2) | 0.025 (2) | 0.0011 (18) | 0.0104 (19) | −0.0022 (18) |
| C14 | 0.026 (2) | 0.043 (3) | 0.029 (2) | −0.001 (2) | 0.0112 (19) | −0.005 (2) |
| C15 | 0.037 (3) | 0.035 (3) | 0.024 (2) | −0.007 (2) | 0.011 (2) | −0.0030 (19) |
| C16 | 0.033 (2) | 0.043 (3) | 0.023 (2) | −0.006 (2) | 0.0047 (19) | −0.004 (2) |
| C17 | 0.038 (3) | 0.047 (3) | 0.026 (2) | −0.005 (2) | 0.004 (2) | −0.004 (2) |
| C18 | 0.053 (3) | 0.073 (4) | 0.019 (2) | 0.000 (3) | −0.001 (2) | −0.007 (2) |
| C19 | 0.041 (3) | 0.048 (3) | 0.028 (3) | −0.008 (2) | 0.011 (2) | −0.003 (2) |
| C20 | 0.051 (3) | 0.060 (4) | 0.045 (3) | 0.010 (3) | 0.021 (3) | 0.002 (3) |
| C21 | 0.049 (3) | 0.069 (4) | 0.050 (4) | 0.009 (3) | 0.014 (3) | 0.009 (3) |
| O4 | 0.100 (19) | 0.068 (6) | 0.035 (2) | 0.004 (14) | 0.015 (12) | −0.010 (3) |
| O5 | 0.031 (6) | 0.118 (10) | 0.044 (3) | 0.013 (5) | 0.007 (4) | 0.017 (5) |
| N2 | 0.037 (10) | 0.044 (15) | 0.025 (7) | 0.008 (10) | 0.003 (7) | 0.018 (9) |
| O4A | 0.100 (19) | 0.068 (6) | 0.035 (2) | 0.004 (14) | 0.015 (12) | −0.010 (3) |
| O5A | 0.031 (6) | 0.118 (10) | 0.044 (3) | 0.013 (5) | 0.007 (4) | 0.017 (5) |
| N2A | 0.037 (10) | 0.044 (15) | 0.025 (7) | 0.008 (10) | 0.003 (7) | 0.018 (9) |
| O6 | 0.065 (3) | 0.058 (2) | 0.031 (2) | −0.017 (2) | 0.0094 (19) | 0.0037 (16) |
| O7 | 0.047 (2) | 0.052 (2) | 0.0246 (17) | 0.0098 (17) | 0.0108 (15) | 0.0032 (14) |
| O8 | 0.050 (2) | 0.0442 (19) | 0.0293 (18) | −0.0018 (17) | 0.0132 (16) | −0.0044 (15) |
| N3 | 0.063 (3) | 0.058 (3) | 0.034 (2) | −0.016 (2) | 0.008 (2) | −0.003 (2) |
| N4 | 0.052 (3) | 0.050 (3) | 0.032 (2) | −0.015 (2) | 0.003 (2) | −0.006 (2) |
| C22 | 0.045 (3) | 0.056 (3) | 0.038 (3) | 0.005 (3) | 0.005 (2) | 0.002 (3) |
| C23 | 0.055 (4) | 0.053 (3) | 0.057 (4) | 0.017 (3) | 0.010 (3) | 0.003 (3) |
| C24 | 0.064 (4) | 0.045 (3) | 0.060 (4) | 0.014 (3) | 0.024 (3) | −0.002 (3) |
| C25 | 0.061 (4) | 0.039 (3) | 0.045 (3) | −0.005 (3) | 0.015 (3) | −0.008 (2) |
| C26 | 0.041 (3) | 0.059 (3) | 0.030 (3) | 0.002 (3) | 0.004 (2) | 0.014 (2) |
| C27 | 0.047 (3) | 0.085 (5) | 0.058 (4) | 0.024 (3) | 0.016 (3) | 0.016 (3) |
| C28 | 0.058 (4) | 0.085 (5) | 0.047 (3) | 0.026 (3) | 0.021 (3) | 0.008 (3) |
| C29 | 0.044 (3) | 0.067 (4) | 0.032 (3) | 0.007 (3) | 0.014 (2) | 0.007 (3) |
| C30 | 0.045 (3) | 0.041 (3) | 0.029 (3) | −0.004 (2) | 0.010 (2) | 0.000 (2) |
| C31 | 0.042 (3) | 0.037 (2) | 0.023 (2) | −0.008 (2) | 0.007 (2) | −0.0019 (19) |
| C32 | 0.042 (3) | 0.033 (2) | 0.034 (3) | −0.005 (2) | 0.012 (2) | −0.001 (2) |
| C33 | 0.041 (3) | 0.038 (3) | 0.030 (3) | −0.003 (2) | 0.004 (2) | −0.003 (2) |
| C34 | 0.035 (3) | 0.037 (2) | 0.027 (2) | −0.006 (2) | 0.007 (2) | 0.0038 (19) |
| C35 | 0.035 (3) | 0.040 (3) | 0.028 (2) | −0.009 (2) | 0.005 (2) | 0.005 (2) |
| C36 | 0.048 (3) | 0.041 (3) | 0.025 (2) | 0.004 (2) | 0.006 (2) | 0.006 (2) |
| C37 | 0.039 (3) | 0.039 (3) | 0.023 (2) | 0.005 (2) | 0.003 (2) | −0.0026 (19) |
| C38 | 0.043 (3) | 0.043 (3) | 0.032 (3) | −0.002 (2) | 0.006 (2) | 0.002 (2) |
| C39 | 0.052 (3) | 0.056 (3) | 0.028 (3) | −0.007 (3) | 0.009 (2) | −0.002 (2) |
| C40 | 0.038 (3) | 0.035 (3) | 0.034 (3) | 0.010 (2) | 0.012 (2) | −0.002 (2) |
| C41 | 0.051 (3) | 0.057 (3) | 0.042 (3) | −0.004 (3) | 0.024 (3) | −0.001 (2) |
| C42 | 0.050 (3) | 0.066 (4) | 0.054 (4) | −0.002 (3) | 0.023 (3) | −0.010 (3) |
| O9 | 0.093 (9) | 0.100 (9) | 0.038 (4) | −0.031 (8) | 0.032 (5) | −0.006 (4) |
| O10 | 0.065 (7) | 0.104 (8) | 0.044 (4) | −0.036 (7) | 0.008 (5) | −0.009 (5) |
| O9A | 0.100 (16) | 0.045 (7) | 0.032 (6) | 0.009 (7) | 0.023 (7) | 0.007 (5) |
| O10A | 0.048 (10) | 0.047 (8) | 0.048 (7) | −0.023 (7) | 0.003 (7) | −0.014 (5) |
Ethyl 5-methyl-3-(10-nitroanthracen-9-yl)isoxazole-4-carboxylate (I). Geometric parameters (Å, º)
| O1—N1 | 1.428 (5) | O6—N3 | 1.418 (6) |
| O1—C17 | 1.338 (7) | O6—C38 | 1.349 (7) |
| O2—C19 | 1.199 (6) | O7—C40 | 1.219 (6) |
| O3—C19 | 1.324 (6) | O8—C40 | 1.319 (6) |
| O3—C20 | 1.455 (7) | O8—C41 | 1.470 (6) |
| N1—C15 | 1.318 (6) | N3—C36 | 1.301 (7) |
| C1—H1 | 0.9500 | N4—C31 | 1.474 (6) |
| C1—C2 | 1.365 (7) | N4—O9 | 1.237 (9) |
| C1—C12 | 1.423 (7) | N4—O10 | 1.203 (11) |
| C2—H2 | 0.9500 | N4—O9A | 1.281 (13) |
| C2—C3 | 1.401 (8) | N4—O10A | 1.206 (14) |
| C3—H3 | 0.9500 | C22—H22 | 0.9500 |
| C3—C4 | 1.366 (8) | C22—C23 | 1.352 (8) |
| C4—H4 | 0.9500 | C22—C33 | 1.420 (8) |
| C4—C11 | 1.416 (7) | C23—H23 | 0.9500 |
| C5—H5 | 0.86 (6) | C23—C24 | 1.406 (9) |
| C5—C6 | 1.343 (7) | C24—H24 | 0.9500 |
| C5—C14 | 1.416 (7) | C24—C25 | 1.366 (9) |
| C6—H6 | 0.85 (6) | C25—H25 | 0.9500 |
| C6—C7 | 1.415 (7) | C25—C32 | 1.403 (8) |
| C7—H7 | 0.9500 | C26—H26 | 0.9500 |
| C7—C8 | 1.367 (7) | C26—C27 | 1.346 (9) |
| C8—H8 | 0.9500 | C26—C35 | 1.425 (7) |
| C8—C13 | 1.415 (6) | C27—H27 | 0.9500 |
| C9—C12 | 1.408 (7) | C27—C28 | 1.399 (9) |
| C9—C13 | 1.399 (6) | C28—H28 | 0.9500 |
| C9—C15 | 1.493 (6) | C28—C29 | 1.343 (8) |
| C10—C11 | 1.380 (7) | C29—H29 | 0.9500 |
| C10—C14 | 1.407 (7) | C29—C34 | 1.406 (7) |
| C10—N2 | 1.43 (5) | C30—C33 | 1.408 (8) |
| C10—N2A | 1.55 (6) | C30—C34 | 1.416 (7) |
| C11—C12 | 1.433 (6) | C30—C36 | 1.498 (7) |
| C13—C14 | 1.436 (6) | C31—C32 | 1.413 (7) |
| C15—C16 | 1.413 (7) | C31—C35 | 1.388 (7) |
| C16—C17 | 1.373 (7) | C32—C33 | 1.438 (7) |
| C16—C19 | 1.485 (7) | C34—C35 | 1.433 (7) |
| C17—C18 | 1.485 (7) | C36—C37 | 1.415 (7) |
| C18—H18A | 0.9800 | C37—C38 | 1.362 (7) |
| C18—H18B | 0.9800 | C37—C40 | 1.465 (7) |
| C18—H18C | 0.9800 | C38—C39 | 1.474 (7) |
| C20—H20A | 0.9900 | C39—H39A | 0.9800 |
| C20—H20B | 0.9900 | C39—H39B | 0.9800 |
| C20—C21 | 1.500 (8) | C39—H39C | 0.9800 |
| C21—H21A | 0.9800 | C41—H41A | 0.9900 |
| C21—H21B | 0.9800 | C41—H41B | 0.9900 |
| C21—H21C | 0.9800 | C41—C42 | 1.488 (8) |
| O4—N2 | 1.22 (6) | C42—H42A | 0.9800 |
| O5—N2 | 1.29 (4) | C42—H42B | 0.9800 |
| O4A—N2A | 1.27 (9) | C42—H42C | 0.9800 |
| O5A—N2A | 1.11 (4) | ||
| C17—O1—N1 | 109.5 (3) | O5A—N2A—O4A | 131 (6) |
| C19—O3—C20 | 114.9 (4) | C38—O6—N3 | 109.0 (4) |
| C15—N1—O1 | 104.3 (4) | C40—O8—C41 | 116.3 (4) |
| C2—C1—H1 | 119.9 | C36—N3—O6 | 105.7 (4) |
| C2—C1—C12 | 120.2 (5) | O9—N4—C31 | 116.7 (5) |
| C12—C1—H1 | 119.9 | O10—N4—C31 | 118.0 (6) |
| C1—C2—H2 | 119.5 | O10—N4—O9 | 125.2 (7) |
| C1—C2—C3 | 120.9 (5) | O9A—N4—C31 | 118.6 (6) |
| C3—C2—H2 | 119.5 | O10A—N4—C31 | 121.6 (8) |
| C2—C3—H3 | 119.7 | O10A—N4—O9A | 119.7 (9) |
| C4—C3—C2 | 120.6 (5) | C23—C22—H22 | 119.6 |
| C4—C3—H3 | 119.7 | C23—C22—C33 | 120.9 (5) |
| C3—C4—H4 | 119.7 | C33—C22—H22 | 119.6 |
| C3—C4—C11 | 120.7 (5) | C22—C23—H23 | 119.5 |
| C11—C4—H4 | 119.7 | C22—C23—C24 | 121.0 (5) |
| C6—C5—H5 | 115 (4) | C24—C23—H23 | 119.5 |
| C6—C5—C14 | 120.7 (5) | C23—C24—H24 | 120.0 |
| C14—C5—H5 | 124 (4) | C25—C24—C23 | 120.0 (6) |
| C5—C6—H6 | 116 (4) | C25—C24—H24 | 120.0 |
| C5—C6—C7 | 121.2 (5) | C24—C25—H25 | 119.4 |
| C7—C6—H6 | 122 (4) | C24—C25—C32 | 121.1 (5) |
| C6—C7—H7 | 120.2 | C32—C25—H25 | 119.4 |
| C8—C7—C6 | 119.6 (5) | C27—C26—H26 | 119.9 |
| C8—C7—H7 | 120.2 | C27—C26—C35 | 120.2 (5) |
| C7—C8—H8 | 119.3 | C35—C26—H26 | 119.9 |
| C7—C8—C13 | 121.4 (4) | C26—C27—H27 | 119.3 |
| C13—C8—H8 | 119.3 | C26—C27—C28 | 121.5 (5) |
| C12—C9—C15 | 118.5 (4) | C28—C27—H27 | 119.3 |
| C13—C9—C12 | 122.2 (4) | C27—C28—H28 | 119.8 |
| C13—C9—C15 | 119.3 (4) | C29—C28—C27 | 120.4 (6) |
| C11—C10—C14 | 125.3 (4) | C29—C28—H28 | 119.8 |
| C11—C10—N2 | 121.3 (18) | C28—C29—H29 | 119.6 |
| C11—C10—N2A | 115 (2) | C28—C29—C34 | 120.9 (5) |
| C14—C10—N2 | 113.2 (17) | C34—C29—H29 | 119.6 |
| C14—C10—N2A | 120 (2) | C33—C30—C34 | 122.7 (4) |
| C4—C11—C12 | 118.7 (4) | C33—C30—C36 | 119.0 (5) |
| C10—C11—C4 | 124.3 (4) | C34—C30—C36 | 118.3 (5) |
| C10—C11—C12 | 117.0 (4) | C32—C31—N4 | 116.4 (4) |
| C1—C12—C11 | 118.9 (4) | C35—C31—N4 | 118.1 (4) |
| C9—C12—C1 | 121.6 (4) | C35—C31—C32 | 125.5 (4) |
| C9—C12—C11 | 119.5 (4) | C25—C32—C31 | 125.2 (5) |
| C8—C13—C14 | 117.8 (4) | C25—C32—C33 | 118.9 (5) |
| C9—C13—C8 | 123.2 (4) | C31—C32—C33 | 115.9 (5) |
| C9—C13—C14 | 119.0 (4) | C22—C33—C32 | 118.1 (5) |
| C5—C14—C13 | 119.3 (4) | C30—C33—C22 | 122.2 (5) |
| C10—C14—C5 | 123.7 (4) | C30—C33—C32 | 119.7 (5) |
| C10—C14—C13 | 117.0 (4) | C29—C34—C30 | 122.7 (4) |
| N1—C15—C9 | 119.8 (4) | C29—C34—C35 | 119.1 (4) |
| N1—C15—C16 | 112.6 (4) | C30—C34—C35 | 118.2 (5) |
| C16—C15—C9 | 127.6 (4) | C26—C35—C34 | 117.8 (5) |
| C15—C16—C19 | 126.2 (4) | C31—C35—C26 | 124.1 (4) |
| C17—C16—C15 | 104.2 (4) | C31—C35—C34 | 118.0 (4) |
| C17—C16—C19 | 129.4 (4) | N3—C36—C30 | 119.9 (5) |
| O1—C17—C16 | 109.5 (4) | N3—C36—C37 | 111.5 (4) |
| O1—C17—C18 | 116.7 (4) | C37—C36—C30 | 128.6 (5) |
| C16—C17—C18 | 133.8 (5) | C36—C37—C40 | 125.7 (4) |
| C17—C18—H18A | 109.5 | C38—C37—C36 | 105.3 (5) |
| C17—C18—H18B | 109.5 | C38—C37—C40 | 129.0 (5) |
| C17—C18—H18C | 109.5 | O6—C38—C37 | 108.6 (4) |
| H18A—C18—H18B | 109.5 | O6—C38—C39 | 115.7 (4) |
| H18A—C18—H18C | 109.5 | C37—C38—C39 | 135.6 (5) |
| H18B—C18—H18C | 109.5 | C38—C39—H39A | 109.5 |
| O2—C19—O3 | 124.6 (5) | C38—C39—H39B | 109.5 |
| O2—C19—C16 | 123.0 (5) | C38—C39—H39C | 109.5 |
| O3—C19—C16 | 112.3 (4) | H39A—C39—H39B | 109.5 |
| O3—C20—H20A | 110.3 | H39A—C39—H39C | 109.5 |
| O3—C20—H20B | 110.3 | H39B—C39—H39C | 109.5 |
| O3—C20—C21 | 107.3 (5) | O7—C40—O8 | 124.2 (5) |
| H20A—C20—H20B | 108.5 | O7—C40—C37 | 123.0 (5) |
| C21—C20—H20A | 110.3 | O8—C40—C37 | 112.7 (4) |
| C21—C20—H20B | 110.3 | O8—C41—H41A | 110.0 |
| C20—C21—H21A | 109.5 | O8—C41—H41B | 110.0 |
| C20—C21—H21B | 109.5 | O8—C41—C42 | 108.4 (5) |
| C20—C21—H21C | 109.5 | H41A—C41—H41B | 108.4 |
| H21A—C21—H21B | 109.5 | C42—C41—H41A | 110.0 |
| H21A—C21—H21C | 109.5 | C42—C41—H41B | 110.0 |
| H21B—C21—H21C | 109.5 | C41—C42—H42A | 109.5 |
| O4—N2—C10 | 123 (3) | C41—C42—H42B | 109.5 |
| O4—N2—O5 | 122 (4) | C41—C42—H42C | 109.5 |
| O5—N2—C10 | 116 (3) | H42A—C42—H42B | 109.5 |
| O4A—N2A—C10 | 108 (3) | H42A—C42—H42C | 109.5 |
| O5A—N2A—C10 | 121 (5) | H42B—C42—H42C | 109.5 |
| O1—N1—C15—C9 | −179.2 (4) | N2A—C10—C14—C5 | 9 (3) |
| O1—N1—C15—C16 | 0.1 (5) | N2A—C10—C14—C13 | −172 (2) |
| N1—O1—C17—C16 | −0.1 (6) | O6—N3—C36—C30 | −179.1 (5) |
| N1—O1—C17—C18 | −179.4 (5) | O6—N3—C36—C37 | 1.3 (6) |
| N1—C15—C16—C17 | −0.1 (6) | N3—O6—C38—C37 | −0.5 (6) |
| N1—C15—C16—C19 | −176.0 (5) | N3—O6—C38—C39 | 176.6 (5) |
| C1—C2—C3—C4 | −0.7 (9) | N3—C36—C37—C38 | −1.6 (6) |
| C2—C1—C12—C9 | 179.1 (5) | N3—C36—C37—C40 | 178.6 (5) |
| C2—C1—C12—C11 | 0.6 (7) | N4—C31—C32—C25 | 0.1 (7) |
| C2—C3—C4—C11 | 0.0 (9) | N4—C31—C32—C33 | 179.7 (4) |
| C3—C4—C11—C10 | −176.7 (5) | N4—C31—C35—C26 | 1.3 (7) |
| C3—C4—C11—C12 | 1.0 (8) | N4—C31—C35—C34 | 179.0 (4) |
| C4—C11—C12—C1 | −1.2 (7) | C22—C23—C24—C25 | 0.4 (10) |
| C4—C11—C12—C9 | −179.8 (5) | C23—C22—C33—C30 | 177.8 (6) |
| C5—C6—C7—C8 | −0.6 (8) | C23—C22—C33—C32 | −1.1 (8) |
| C6—C5—C14—C10 | 178.2 (5) | C23—C24—C25—C32 | −0.6 (9) |
| C6—C5—C14—C13 | −1.0 (7) | C24—C25—C32—C31 | 179.5 (5) |
| C6—C7—C8—C13 | −0.2 (8) | C24—C25—C32—C33 | −0.1 (8) |
| C7—C8—C13—C9 | −176.8 (5) | C25—C32—C33—C22 | 0.9 (7) |
| C7—C8—C13—C14 | 0.3 (7) | C25—C32—C33—C30 | −178.0 (5) |
| C8—C13—C14—C5 | 0.2 (6) | C26—C27—C28—C29 | −0.1 (11) |
| C8—C13—C14—C10 | −179.0 (4) | C27—C26—C35—C31 | 179.3 (5) |
| C9—C13—C14—C5 | 177.5 (4) | C27—C26—C35—C34 | 1.6 (8) |
| C9—C13—C14—C10 | −1.7 (6) | C27—C28—C29—C34 | 1.6 (10) |
| C9—C15—C16—C17 | 179.1 (5) | C28—C29—C34—C30 | −179.5 (5) |
| C9—C15—C16—C19 | 3.2 (8) | C28—C29—C34—C35 | −1.5 (8) |
| C10—C11—C12—C1 | 176.6 (5) | C29—C34—C35—C26 | −0.1 (7) |
| C10—C11—C12—C9 | −2.0 (7) | C29—C34—C35—C31 | −177.9 (4) |
| C11—C10—C14—C5 | −178.2 (5) | C30—C34—C35—C26 | 178.0 (4) |
| C11—C10—C14—C13 | 1.0 (7) | C30—C34—C35—C31 | 0.1 (6) |
| C11—C10—N2—O4 | 77 (4) | C30—C36—C37—C38 | 178.9 (5) |
| C11—C10—N2—O5 | −101 (3) | C30—C36—C37—C40 | −0.9 (9) |
| C11—C10—N2A—O4A | 115 (4) | C31—C32—C33—C22 | −178.7 (5) |
| C11—C10—N2A—O5A | −64 (6) | C31—C32—C33—C30 | 2.3 (7) |
| C12—C1—C2—C3 | 0.4 (9) | C32—C31—C35—C26 | −176.7 (5) |
| C12—C9—C13—C8 | 177.7 (4) | C32—C31—C35—C34 | 0.9 (7) |
| C12—C9—C13—C14 | 0.7 (7) | C33—C22—C23—C24 | 0.5 (10) |
| C12—C9—C15—N1 | 91.6 (6) | C33—C30—C34—C29 | 178.1 (5) |
| C12—C9—C15—C16 | −87.6 (6) | C33—C30—C34—C35 | 0.1 (7) |
| C13—C9—C12—C1 | −177.3 (5) | C33—C30—C36—N3 | 95.1 (7) |
| C13—C9—C12—C11 | 1.3 (7) | C33—C30—C36—C37 | −85.4 (7) |
| C13—C9—C15—N1 | −88.6 (6) | C34—C30—C33—C22 | 179.7 (5) |
| C13—C9—C15—C16 | 92.3 (6) | C34—C30—C33—C32 | −1.4 (8) |
| C14—C5—C6—C7 | 1.2 (8) | C34—C30—C36—N3 | −85.9 (6) |
| C14—C10—C11—C4 | 178.5 (5) | C34—C30—C36—C37 | 93.6 (7) |
| C14—C10—C11—C12 | 0.8 (7) | C35—C26—C27—C28 | −1.5 (10) |
| C14—C10—N2—O4 | −97 (4) | C35—C31—C32—C25 | 178.2 (5) |
| C14—C10—N2—O5 | 85 (3) | C35—C31—C32—C33 | −2.2 (7) |
| C14—C10—N2A—O4A | −71 (5) | C36—C30—C33—C22 | −1.4 (8) |
| C14—C10—N2A—O5A | 110 (5) | C36—C30—C33—C32 | 177.5 (4) |
| C15—C9—C12—C1 | 2.6 (7) | C36—C30—C34—C29 | −0.8 (7) |
| C15—C9—C12—C11 | −178.9 (4) | C36—C30—C34—C35 | −178.8 (4) |
| C15—C9—C13—C8 | −2.1 (7) | C36—C37—C38—O6 | 1.2 (6) |
| C15—C9—C13—C14 | −179.2 (4) | C36—C37—C38—C39 | −175.0 (6) |
| C15—C16—C17—O1 | 0.1 (5) | C36—C37—C40—O7 | −3.9 (8) |
| C15—C16—C17—C18 | 179.3 (6) | C36—C37—C40—O8 | 175.1 (5) |
| C15—C16—C19—O2 | −5.4 (8) | C38—O6—N3—C36 | −0.5 (6) |
| C15—C16—C19—O3 | 173.5 (4) | C38—C37—C40—O7 | 176.3 (5) |
| C17—O1—N1—C15 | 0.0 (5) | C38—C37—C40—O8 | −4.6 (7) |
| C17—C16—C19—O2 | 179.8 (5) | C40—O8—C41—C42 | 166.0 (4) |
| C17—C16—C19—O3 | −1.2 (7) | C40—C37—C38—O6 | −179.0 (5) |
| C19—O3—C20—C21 | −174.3 (5) | C40—C37—C38—C39 | 4.7 (10) |
| C19—C16—C17—O1 | 175.8 (5) | C41—O8—C40—O7 | 2.6 (7) |
| C19—C16—C17—C18 | −5.1 (10) | C41—O8—C40—C37 | −176.5 (4) |
| C20—O3—C19—O2 | 0.5 (7) | O9—N4—C31—C32 | 63.2 (11) |
| C20—O3—C19—C16 | −178.4 (4) | O9—N4—C31—C35 | −115.0 (11) |
| N2—C10—C11—C4 | 4.9 (18) | O10—N4—C31—C32 | −113.3 (12) |
| N2—C10—C11—C12 | −172.8 (17) | O10—N4—C31—C35 | 68.5 (13) |
| N2—C10—C14—C5 | −4.1 (18) | O9A—N4—C31—C32 | 120.0 (14) |
| N2—C10—C14—C13 | 175.1 (17) | O9A—N4—C31—C35 | −58.2 (15) |
| N2A—C10—C11—C4 | −8 (3) | O10A—N4—C31—C32 | −63.1 (16) |
| N2A—C10—C11—C12 | 174 (2) | O10A—N4—C31—C35 | 118.7 (16) |
Ethyl 5-methyl-3-(10-nitroanthracen-9-yl)isoxazole-4-carboxylate (I). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1···O5i | 0.95 | 2.46 | 3.366 (12) | 159 |
| C3—H3···O7ii | 0.95 | 2.44 | 3.339 (6) | 158 |
| C7—H7···O4iii | 0.95 | 2.40 | 3.24 (4) | 147 |
| C7—H7···O4Aiii | 0.95 | 2.46 | 3.34 (6) | 154 |
Symmetry codes: (i) x+1/2, −y+1/2, z+1/2; (ii) x, −y+1, z−1/2; (iii) x, −y, z+1/2.
Ethyl 3-(9-hydroxy-10-oxo-9,10-dihydroanthracen-9-yl)-5-methylisoxazole-4-carboxylate (V). Crystal data
| C21H17NO5 | F(000) = 760 |
| Mr = 363.36 | Dx = 1.445 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.2862 (4) Å | Cell parameters from 9893 reflections |
| b = 23.5895 (11) Å | θ = 2.5–28.3° |
| c = 8.6219 (4) Å | µ = 0.10 mm−1 |
| β = 97.728 (2)° | T = 100 K |
| V = 1669.99 (14) Å3 | Prism, yellow |
| Z = 4 | 0.28 × 0.20 × 0.19 mm |
Ethyl 3-(9-hydroxy-10-oxo-9,10-dihydroanthracen-9-yl)-5-methylisoxazole-4-carboxylate (V). Data collection
| Bruker SMART Breeze CCD diffractometer | 3252 reflections with I > 2σ(I) |
| Radiation source: 2 kW sealed X-ray tube | Rint = 0.051 |
| φ and ω scans | θmax = 28.3°, θmin = 1.7° |
| Absorption correction: numerical (SADABS; Krause et al., 2015) | h = −10→11 |
| Tmin = 0.945, Tmax = 1.000 | k = −31→31 |
| 44252 measured reflections | l = −11→11 |
| 4112 independent reflections |
Ethyl 3-(9-hydroxy-10-oxo-9,10-dihydroanthracen-9-yl)-5-methylisoxazole-4-carboxylate (V). Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.051 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.114 | w = 1/[σ2(Fo2) + (0.0241P)2 + 1.7289P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.13 | (Δ/σ)max < 0.001 |
| 4112 reflections | Δρmax = 0.37 e Å−3 |
| 250 parameters | Δρmin = −0.21 e Å−3 |
| 0 restraints |
Ethyl 3-(9-hydroxy-10-oxo-9,10-dihydroanthracen-9-yl)-5-methylisoxazole-4-carboxylate (V). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Ethyl 3-(9-hydroxy-10-oxo-9,10-dihydroanthracen-9-yl)-5-methylisoxazole-4-carboxylate (V). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.65863 (16) | 0.55415 (6) | 0.82793 (16) | 0.0252 (3) | |
| O2 | 0.18140 (16) | 0.55228 (6) | 1.16490 (15) | 0.0203 (3) | |
| O3 | 0.44673 (16) | 0.68686 (5) | 0.90343 (16) | 0.0217 (3) | |
| O4 | 0.37570 (18) | 0.77723 (6) | 0.93929 (18) | 0.0306 (3) | |
| O5 | 0.03664 (16) | 0.70944 (5) | 1.19895 (16) | 0.0226 (3) | |
| N1 | 0.08101 (19) | 0.65146 (6) | 1.19429 (19) | 0.0212 (3) | |
| C1 | 0.0870 (2) | 0.57205 (7) | 0.8333 (2) | 0.0203 (4) | |
| H1 | −0.001664 | 0.578626 | 0.890399 | 0.024* | |
| C2 | 0.0569 (2) | 0.55858 (8) | 0.6759 (2) | 0.0228 (4) | |
| H2A | −0.052182 | 0.555788 | 0.625751 | 0.027* | |
| C3 | 0.1853 (2) | 0.54909 (8) | 0.5906 (2) | 0.0240 (4) | |
| H3 | 0.164699 | 0.541177 | 0.481704 | 0.029* | |
| C4 | 0.3430 (2) | 0.55131 (7) | 0.6659 (2) | 0.0218 (4) | |
| H4 | 0.431105 | 0.543796 | 0.608819 | 0.026* | |
| C5 | 0.7367 (2) | 0.57793 (8) | 1.1470 (2) | 0.0220 (4) | |
| H5 | 0.823547 | 0.568479 | 1.090262 | 0.026* | |
| C6 | 0.7694 (2) | 0.59175 (8) | 1.3033 (2) | 0.0247 (4) | |
| H6 | 0.878586 | 0.592110 | 1.353877 | 0.030* | |
| C7 | 0.6426 (2) | 0.60514 (8) | 1.3866 (2) | 0.0243 (4) | |
| H7 | 0.665219 | 0.614443 | 1.494505 | 0.029* | |
| C8 | 0.4833 (2) | 0.60507 (8) | 1.3136 (2) | 0.0205 (4) | |
| H8 | 0.397213 | 0.614388 | 1.371534 | 0.025* | |
| C9 | 0.2716 (2) | 0.59122 (7) | 1.0817 (2) | 0.0165 (3) | |
| C10 | 0.5456 (2) | 0.56479 (7) | 0.9026 (2) | 0.0189 (4) | |
| C11 | 0.5762 (2) | 0.57770 (7) | 1.0712 (2) | 0.0183 (4) | |
| C12 | 0.4490 (2) | 0.59136 (7) | 1.1556 (2) | 0.0172 (4) | |
| C13 | 0.2460 (2) | 0.57606 (7) | 0.9088 (2) | 0.0172 (4) | |
| C14 | 0.3747 (2) | 0.56448 (7) | 0.8250 (2) | 0.0182 (4) | |
| C15 | 0.1976 (2) | 0.64929 (7) | 1.1067 (2) | 0.0170 (4) | |
| C16 | 0.2349 (2) | 0.70473 (7) | 1.0516 (2) | 0.0181 (4) | |
| C17 | 0.1296 (2) | 0.73971 (8) | 1.1146 (2) | 0.0199 (4) | |
| C18 | 0.0996 (2) | 0.80170 (8) | 1.1084 (2) | 0.0253 (4) | |
| H18A | 0.008645 | 0.810745 | 1.165995 | 0.038* | |
| H18B | 0.072691 | 0.813653 | 0.999147 | 0.038* | |
| H18C | 0.197593 | 0.821674 | 1.156078 | 0.038* | |
| C19 | 0.3576 (2) | 0.72714 (8) | 0.9602 (2) | 0.0194 (4) | |
| C20 | 0.5726 (2) | 0.70681 (9) | 0.8136 (2) | 0.0263 (4) | |
| H20A | 0.534468 | 0.741549 | 0.755484 | 0.032* | |
| H20B | 0.593554 | 0.677531 | 0.736546 | 0.032* | |
| C21 | 0.7273 (3) | 0.71933 (11) | 0.9198 (3) | 0.0365 (5) | |
| H21A | 0.812088 | 0.730603 | 0.856913 | 0.055* | |
| H21B | 0.762503 | 0.685366 | 0.980518 | 0.055* | |
| H21C | 0.708331 | 0.750213 | 0.991184 | 0.055* | |
| H2 | 0.232 (3) | 0.5184 (12) | 1.162 (3) | 0.043 (7)* |
Ethyl 3-(9-hydroxy-10-oxo-9,10-dihydroanthracen-9-yl)-5-methylisoxazole-4-carboxylate (V). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0224 (7) | 0.0278 (7) | 0.0271 (7) | 0.0023 (6) | 0.0097 (6) | 0.0023 (6) |
| O2 | 0.0205 (7) | 0.0183 (6) | 0.0235 (7) | −0.0023 (5) | 0.0081 (5) | 0.0017 (5) |
| O3 | 0.0199 (7) | 0.0209 (6) | 0.0256 (7) | −0.0025 (5) | 0.0070 (5) | 0.0004 (5) |
| O4 | 0.0339 (8) | 0.0194 (7) | 0.0397 (9) | −0.0005 (6) | 0.0090 (7) | 0.0073 (6) |
| O5 | 0.0204 (7) | 0.0214 (7) | 0.0264 (7) | 0.0033 (5) | 0.0049 (5) | −0.0018 (5) |
| N1 | 0.0187 (8) | 0.0192 (8) | 0.0262 (8) | 0.0018 (6) | 0.0047 (6) | −0.0021 (6) |
| C1 | 0.0212 (9) | 0.0180 (8) | 0.0222 (9) | −0.0024 (7) | 0.0043 (7) | 0.0000 (7) |
| C2 | 0.0231 (10) | 0.0196 (9) | 0.0242 (10) | −0.0046 (7) | −0.0021 (8) | −0.0008 (7) |
| C3 | 0.0342 (11) | 0.0183 (9) | 0.0190 (9) | −0.0041 (8) | 0.0017 (8) | −0.0016 (7) |
| C4 | 0.0285 (10) | 0.0162 (9) | 0.0216 (9) | −0.0018 (7) | 0.0073 (8) | −0.0011 (7) |
| C5 | 0.0169 (9) | 0.0218 (9) | 0.0279 (10) | 0.0000 (7) | 0.0047 (7) | 0.0079 (7) |
| C6 | 0.0172 (9) | 0.0256 (10) | 0.0300 (11) | −0.0034 (7) | −0.0021 (8) | 0.0088 (8) |
| C7 | 0.0262 (10) | 0.0244 (9) | 0.0212 (9) | −0.0020 (8) | −0.0009 (8) | 0.0029 (7) |
| C8 | 0.0206 (9) | 0.0199 (9) | 0.0215 (9) | 0.0005 (7) | 0.0045 (7) | 0.0012 (7) |
| C9 | 0.0156 (8) | 0.0162 (8) | 0.0185 (9) | −0.0022 (6) | 0.0050 (7) | 0.0007 (6) |
| C10 | 0.0200 (9) | 0.0147 (8) | 0.0229 (9) | 0.0002 (7) | 0.0057 (7) | 0.0030 (7) |
| C11 | 0.0178 (9) | 0.0158 (8) | 0.0214 (9) | −0.0011 (7) | 0.0034 (7) | 0.0037 (7) |
| C12 | 0.0168 (9) | 0.0135 (8) | 0.0210 (9) | −0.0002 (6) | 0.0020 (7) | 0.0027 (6) |
| C13 | 0.0200 (9) | 0.0130 (8) | 0.0190 (9) | −0.0020 (6) | 0.0040 (7) | 0.0002 (6) |
| C14 | 0.0213 (9) | 0.0135 (8) | 0.0202 (9) | −0.0011 (7) | 0.0047 (7) | 0.0008 (6) |
| C15 | 0.0143 (8) | 0.0196 (8) | 0.0168 (8) | −0.0013 (7) | 0.0004 (7) | −0.0002 (7) |
| C16 | 0.0161 (9) | 0.0186 (8) | 0.0187 (9) | −0.0005 (7) | −0.0012 (7) | −0.0004 (7) |
| C17 | 0.0182 (9) | 0.0216 (9) | 0.0184 (9) | 0.0000 (7) | −0.0028 (7) | −0.0008 (7) |
| C18 | 0.0282 (10) | 0.0208 (9) | 0.0259 (10) | 0.0047 (8) | 0.0002 (8) | −0.0016 (8) |
| C19 | 0.0187 (9) | 0.0205 (9) | 0.0178 (9) | −0.0010 (7) | −0.0025 (7) | 0.0014 (7) |
| C20 | 0.0250 (10) | 0.0303 (10) | 0.0253 (10) | −0.0049 (8) | 0.0094 (8) | 0.0022 (8) |
| C21 | 0.0219 (11) | 0.0466 (13) | 0.0410 (13) | −0.0056 (9) | 0.0040 (9) | 0.0122 (10) |
Ethyl 3-(9-hydroxy-10-oxo-9,10-dihydroanthracen-9-yl)-5-methylisoxazole-4-carboxylate (V). Geometric parameters (Å, º)
| O1—C10 | 1.231 (2) | C7—C8 | 1.383 (3) |
| O2—C9 | 1.436 (2) | C8—H8 | 0.9500 |
| O2—H2 | 0.91 (3) | C8—C12 | 1.392 (3) |
| O3—C19 | 1.336 (2) | C9—C12 | 1.521 (2) |
| O3—C20 | 1.458 (2) | C9—C13 | 1.520 (2) |
| O4—C19 | 1.208 (2) | C9—C15 | 1.528 (2) |
| O5—N1 | 1.418 (2) | C10—C11 | 1.474 (3) |
| O5—C17 | 1.336 (2) | C10—C14 | 1.483 (3) |
| N1—C15 | 1.304 (2) | C11—C12 | 1.397 (3) |
| C1—H1 | 0.9500 | C13—C14 | 1.393 (3) |
| C1—C2 | 1.384 (3) | C15—C16 | 1.439 (2) |
| C1—C13 | 1.392 (3) | C16—C17 | 1.365 (3) |
| C2—H2A | 0.9500 | C16—C19 | 1.466 (3) |
| C2—C3 | 1.391 (3) | C17—C18 | 1.483 (3) |
| C3—H3 | 0.9500 | C18—H18A | 0.9800 |
| C3—C4 | 1.380 (3) | C18—H18B | 0.9800 |
| C4—H4 | 0.9500 | C18—H18C | 0.9800 |
| C4—C14 | 1.397 (3) | C20—H20A | 0.9900 |
| C5—H5 | 0.9500 | C20—H20B | 0.9900 |
| C5—C6 | 1.377 (3) | C20—C21 | 1.501 (3) |
| C5—C11 | 1.401 (3) | C21—H21A | 0.9800 |
| C6—H6 | 0.9500 | C21—H21B | 0.9800 |
| C6—C7 | 1.387 (3) | C21—H21C | 0.9800 |
| C7—H7 | 0.9500 | ||
| C9—O2—H2 | 106.1 (16) | C8—C12—C9 | 118.00 (16) |
| C19—O3—C20 | 115.78 (15) | C8—C12—C11 | 119.62 (17) |
| C17—O5—N1 | 109.24 (14) | C11—C12—C9 | 122.37 (16) |
| C15—N1—O5 | 105.61 (14) | C1—C13—C9 | 118.24 (16) |
| C2—C1—H1 | 119.7 | C1—C13—C14 | 119.11 (17) |
| C2—C1—C13 | 120.57 (18) | C14—C13—C9 | 122.62 (16) |
| C13—C1—H1 | 119.7 | C4—C14—C10 | 119.05 (17) |
| C1—C2—H2A | 119.8 | C13—C14—C4 | 119.83 (17) |
| C1—C2—C3 | 120.38 (18) | C13—C14—C10 | 121.11 (16) |
| C3—C2—H2A | 119.8 | N1—C15—C9 | 117.35 (15) |
| C2—C3—H3 | 120.4 | N1—C15—C16 | 111.38 (16) |
| C4—C3—C2 | 119.28 (18) | C16—C15—C9 | 131.27 (16) |
| C4—C3—H3 | 120.4 | C15—C16—C19 | 134.48 (17) |
| C3—C4—H4 | 119.6 | C17—C16—C15 | 103.91 (16) |
| C3—C4—C14 | 120.74 (18) | C17—C16—C19 | 121.48 (16) |
| C14—C4—H4 | 119.6 | O5—C17—C16 | 109.86 (16) |
| C6—C5—H5 | 119.8 | O5—C17—C18 | 116.12 (17) |
| C6—C5—C11 | 120.49 (18) | C16—C17—C18 | 134.02 (18) |
| C11—C5—H5 | 119.8 | C17—C18—H18A | 109.5 |
| C5—C6—H6 | 120.1 | C17—C18—H18B | 109.5 |
| C5—C6—C7 | 119.80 (18) | C17—C18—H18C | 109.5 |
| C7—C6—H6 | 120.1 | H18A—C18—H18B | 109.5 |
| C6—C7—H7 | 119.8 | H18A—C18—H18C | 109.5 |
| C8—C7—C6 | 120.49 (18) | H18B—C18—H18C | 109.5 |
| C8—C7—H7 | 119.8 | O3—C19—C16 | 113.44 (15) |
| C7—C8—H8 | 119.9 | O4—C19—O3 | 123.74 (18) |
| C7—C8—C12 | 120.15 (18) | O4—C19—C16 | 122.82 (18) |
| C12—C8—H8 | 119.9 | O3—C20—H20A | 109.5 |
| O2—C9—C12 | 109.31 (14) | O3—C20—H20B | 109.5 |
| O2—C9—C13 | 109.03 (14) | O3—C20—C21 | 110.68 (17) |
| O2—C9—C15 | 104.91 (14) | H20A—C20—H20B | 108.1 |
| C12—C9—C15 | 108.81 (14) | C21—C20—H20A | 109.5 |
| C13—C9—C12 | 114.27 (15) | C21—C20—H20B | 109.5 |
| C13—C9—C15 | 110.09 (14) | C20—C21—H21A | 109.5 |
| O1—C10—C11 | 121.06 (17) | C20—C21—H21B | 109.5 |
| O1—C10—C14 | 120.73 (17) | C20—C21—H21C | 109.5 |
| C11—C10—C14 | 118.21 (16) | H21A—C21—H21B | 109.5 |
| C5—C11—C10 | 119.19 (17) | H21A—C21—H21C | 109.5 |
| C12—C11—C5 | 119.44 (17) | H21B—C21—H21C | 109.5 |
| C12—C11—C10 | 121.35 (16) | ||
| O1—C10—C11—C5 | 0.1 (3) | C9—C15—C16—C17 | −179.04 (17) |
| O1—C10—C11—C12 | 178.18 (16) | C9—C15—C16—C19 | −3.3 (3) |
| O1—C10—C14—C4 | 0.7 (3) | C10—C11—C12—C8 | −177.95 (16) |
| O1—C10—C14—C13 | 179.53 (16) | C10—C11—C12—C9 | 2.8 (3) |
| O2—C9—C12—C8 | −57.8 (2) | C11—C5—C6—C7 | 0.5 (3) |
| O2—C9—C12—C11 | 121.43 (17) | C11—C10—C14—C4 | −178.84 (16) |
| O2—C9—C13—C1 | 54.2 (2) | C11—C10—C14—C13 | 0.0 (2) |
| O2—C9—C13—C14 | −123.89 (17) | C12—C9—C13—C1 | 176.78 (15) |
| O2—C9—C15—N1 | 1.7 (2) | C12—C9—C13—C14 | −1.3 (2) |
| O2—C9—C15—C16 | −179.40 (17) | C12—C9—C15—N1 | −115.21 (17) |
| O5—N1—C15—C9 | 179.43 (14) | C12—C9—C15—C16 | 63.7 (2) |
| O5—N1—C15—C16 | 0.30 (19) | C13—C1—C2—C3 | −0.3 (3) |
| N1—O5—C17—C16 | 0.41 (19) | C13—C9—C12—C8 | 179.71 (15) |
| N1—O5—C17—C18 | −179.49 (15) | C13—C9—C12—C11 | −1.0 (2) |
| N1—C15—C16—C17 | −0.1 (2) | C13—C9—C15—N1 | 118.84 (17) |
| N1—C15—C16—C19 | 175.66 (19) | C13—C9—C15—C16 | −62.2 (2) |
| C1—C2—C3—C4 | 2.3 (3) | C14—C10—C11—C5 | 179.66 (16) |
| C1—C13—C14—C4 | 2.6 (3) | C14—C10—C11—C12 | −2.3 (2) |
| C1—C13—C14—C10 | −176.28 (16) | C15—C9—C12—C8 | 56.2 (2) |
| C2—C1—C13—C9 | 179.77 (16) | C15—C9—C12—C11 | −124.52 (17) |
| C2—C1—C13—C14 | −2.1 (3) | C15—C9—C13—C1 | −60.4 (2) |
| C2—C3—C4—C14 | −1.8 (3) | C15—C9—C13—C14 | 121.53 (17) |
| C3—C4—C14—C10 | 178.25 (17) | C15—C16—C17—O5 | −0.22 (19) |
| C3—C4—C14—C13 | −0.6 (3) | C15—C16—C17—C18 | 179.7 (2) |
| C5—C6—C7—C8 | −0.4 (3) | C15—C16—C19—O3 | 6.9 (3) |
| C5—C11—C12—C8 | 0.1 (3) | C15—C16—C19—O4 | −172.96 (19) |
| C5—C11—C12—C9 | −179.15 (16) | C17—O5—N1—C15 | −0.44 (19) |
| C6—C5—C11—C10 | 177.76 (17) | C17—C16—C19—O3 | −178.00 (16) |
| C6—C5—C11—C12 | −0.3 (3) | C17—C16—C19—O4 | 2.2 (3) |
| C6—C7—C8—C12 | 0.2 (3) | C19—O3—C20—C21 | 86.7 (2) |
| C7—C8—C12—C9 | 179.26 (16) | C19—C16—C17—O5 | −176.64 (15) |
| C7—C8—C12—C11 | 0.0 (3) | C19—C16—C17—C18 | 3.2 (3) |
| C9—C13—C14—C4 | −179.41 (16) | C20—O3—C19—O4 | 0.8 (3) |
| C9—C13—C14—C10 | 1.7 (3) | C20—O3—C19—C16 | −179.03 (15) |
Ethyl 3-(9-hydroxy-10-oxo-9,10-dihydroanthracen-9-yl)-5-methylisoxazole-4-carboxylate (V). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···O1i | 0.91 (3) | 1.93 (3) | 2.8359 (19) | 176 (2) |
Symmetry code: (i) −x+1, −y+1, −z+2.
Funding Statement
The authors thank the University of Montana for grant No. 325490.
References
- Bansal, R. C., Eisenbraun, E. J. & Ryba, R. J. (1987). Org. Prep. Proced. Int. 19, 258–260.
- Barker, N. M., Krause, J. A. & Zhang, P. (2019). CSD Communication (refcode COBWEY). CCDC, Cambridge, England.
- Bruker (2012). APEX2. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2018). SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Chen, Y.-J., Yang, S.-C., Tsai, C.-C., Chang, K.-C., Chuang, W.-H., Chu, W.-L., Kovalev, V. & Chung, W.-S. (2015). Chem. Asian J. 10, 1025–1034. [DOI] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Duncan, N. S., Beall, H. D., Kearns, A. K., Li, C. & Natale, N. R. (2014). Acta Cryst. E70, o315–o316. [DOI] [PMC free article] [PubMed]
- Forensi, S., Stopin, A., de Leo, F., Wouters, J. & Bonifazi, D. (2020). Tetrahedron, 76, 131299.
- Glagovich, N. M., Foss, P. C. D., Michalewski, O., Reed, E. M., Strathearn, K. E., Weiner, Y. F., Crundwell, G., Updegraff, J. B., Zeller, M. & Hunter, A. D. (2004). Acta Cryst. E60, o2125–o2126.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Han, X., Li, C., Mosher, M. D., Rider, K. C., Zhou, P., Crawford, R. L., Fusco, W., Paszczynski, A. & Natale, N. R. (2009). Bioorg. Med. Chem. 17, 1671–1680. [DOI] [PMC free article] [PubMed]
- Han, X., Twamley, B. & Natale, N. R. (2003). J. Heterocycl. Chem. 40, 539–545.
- Hoffend, C., Schickedanz, K., Bolte, M., Lerner, H.-W. & Wagner, M. (2013). Tetrahedron, 69, 7073–7081.
- Klaper, M., Wessig, P. & Linker, T. (2016). Chem. Commun. 52, 1210–1213. [DOI] [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Li, C., Campbell, M. J., Weaver, M. J., Duncan, N. S., Hunting, J. L. & Natale, N. R. (2013). Acta Cryst. E69, o1804–o1805. [DOI] [PMC free article] [PubMed]
- Li, C., Twamley, B. & Natale, N. R. (2006). Acta Cryst. E62, o854–o856.
- Mao, X., Zhang, J., Wang, X., Zhang, H., Wei, P., Sung, H. H. Y., Williams, I. D., Feng, X., Ni, X.-L., Redshaw, C., Elsegood, M. R. J., Lam, J. W. Y. & Tang, B. Z. (2021). J. Org. Chem. 86, 7359–7369. [DOI] [PubMed]
- McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2007). Chem. Commun. pp. 3814–3816. [DOI] [PubMed]
- Mosher, M. D., Natale, N. R. & Vij, A. (1996). Acta Cryst. C52, 2513–2515.
- Natale, N. R., Rider, K. C., Burkhart, D. J., Li, C., McKenzie, A. R. & Nelson, J. K. (2010). ARKIVOC, part (viii), pp. 97–107.
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Pullella, G. A., Wdowiak, A. P., Sykes, M. L., Lucantoni, L., Sukhoverkov, K. V., Zulfiqar, B., Sobolev, A. N., West, N. P., Mylne, J. S., Avery, V. M. & Piggott, M. J. (2019). Org. Lett. 21, 5519–5523. [DOI] [PubMed]
- Roszak, A. & Engelen, B. (1990). Acta Cryst. C46, 240–243.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Silverman, R. B. (2002). Organic Chemistry of Enzyme-catalyzed Reactions, 2nd ed, pp. 227–239. San Diego: Academic Press.
- Silverman, R. B. & Holladay, M. W. (2014). Organic Chemistry of Drug Design and Drug Action, pp. 368–373. San Diego: Academic Press.
- Skrzat, Z. & Roszak, A. (1986). Acta Cryst. C42, 1194–1196.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Spackman, P. R., Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Jayatilaka, D. & Spackman, M. A. (2021). J. Appl. Cryst. 54, 1006–1011. [DOI] [PMC free article] [PubMed]
- Stepovik, L. P., Malysheva, Y. B. & Fukin, G. K. (2015). Russ. J. Gen. Chem. 85, 1401–1411.
- Trotter, J. (1959). Acta Cryst. 12, 237–242.
- Weaver, M. J., Campbell, M. J., Li, C. & Natale, N. R. (2020). Acta Cryst. E76, 1818–1822. [DOI] [PMC free article] [PubMed]
- Weaver, M. J., Kearns, A. K., Stump, S., Li, C., Gajewski, M. P., Rider, K. C., Backos, D. S., Reigan, P. R., Beall, H. D. & Natale, N. R. (2015). Bioorg. Med. Chem. Lett. 25, 1765–1770. [DOI] [PMC free article] [PubMed]
- Weaver, M. J., Stump, S., Campbell, M. J., Backos, D. S., Li, C., Reigan, P., Adams, E., Beall, H. D. & Natale, N. R. (2020). Bioorg. Med. Chem. 28, 115781. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, V, global. DOI: 10.1107/S2056989022005710/hb8020sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022005710/hb8020Isup2.hkl
Structure factors: contains datablock(s) V. DOI: 10.1107/S2056989022005710/hb8020Vsup3.hkl
Supporting information file. DOI: 10.1107/S2056989022005710/hb8020Isup4.cml
Supporting information file. DOI: 10.1107/S2056989022005710/hb8020Vsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report