Abstract
SARS-CoV-2 neutralizing antibodies play a critical role in COVID-19 prevention and treatment but are challenged by viral evolution and the emergence of novel escape variants. Importantly, the recently identified Omicron sublineages BA.2.12.1 and BA.4/5 are rapidly becoming predominant in various countries. By determining polyclonal serum activity of 50 convalescent or vaccinated individuals against BA.1, BA.1.1, BA.2, BA.2.12.1, and BA.4/5, we reveal a further reduction in BA.4/5 susceptibility to vaccinee sera. Most notably, delineation of sensitivity to an extended 163-antibody panel demonstrates pronounced antigenic differences with distinct escape patterns among Omicron sublineages. Antigenic distance and/or higher resistance may therefore favor immune-escape-mediated BA.4/5 expansion after the first Omicron wave. Finally, while most clinical-stage monoclonal antibodies are inactive against Omicron sublineages, we identify promising antibodies with high pan-SARS-CoV-2 neutralizing potency. Our study provides a detailed understanding of Omicron-sublineage antibody escape that can inform on effective strategies against COVID-19.
Keywords: SARS-CoV-2, Omicron, sublineages, BA.4, BA.5, BA.2.12.1, neutralizing antibodies, immune escape, resistance, COVID-19
Graphical abstract
Gruell and Vanshylla et al. study the immune escape properties of emerging Omicron sublineages including BA.2.12.1 and BA.4/5. Antigenic profiling using a large panel of monoclonal antibodies revealed distinct viral escape patterns. While most clinical antibodies lose activity against Omicron, highly potent SARS-CoV-2 antibodies with retained pan-Omicron activity were identified.
Introduction
Despite increasing levels of immunity against SARS-CoV-2 induced by vaccination and infection, the Omicron variant resulted in a global surge of infections that reflects its high transmissibility and immune evasion mediated by a highly mutated spike protein (Altarawneh et al., 2022; Andrews et al., 2022; Carreño et al., 2022; Cele et al., 2022; Garcia-Beltran et al., 2022; Gruell et al., 2022a; Hoffmann et al., 2022; Liu et al., 2022; Madhi et al., 2022; Planas et al., 2022; Schmidt et al., 2022; Tseng et al., 2022; Viana et al., 2022). Although prolonged vaccine dosing intervals and booster immunizations based on the ancestral Wu01 strain elicit Omicron-neutralizing serum activity, titers against Omicron are considerably lower compared with those against other variants (Garcia-Beltran et al., 2022; Gruell et al., 2022a; Pérez-Then et al., 2022; Schmidt et al., 2022; Wratil et al., 2022; Zhao et al., 2022). Moreover, the spike mutations of Omicron have rendered several therapeutic monoclonal antibodies ineffective (Gruell et al., 2022a; Liu et al., 2022; VanBlargan et al., 2022). Most experimental evidence on the resistance of Omicron to antibody-mediated neutralization is limited to analyses of the initial BA.1 strain. However, novel sublineages of Omicron are rapidly emerging and associated with increasing numbers of infections (Tegally et al., 2022; Yamasoba et al., 2022). Determining their antibody escape properties is therefore of critical importance to effectively guide preventive and therapeutic measures. To this end, we analyzed in detail antibody-mediated neutralization of prevalent and emerging Omicron sublineages (BA.1, BA.1.1, BA.2, BA.2.12.1, and BA.4/5) both on a polyclonal and monoclonal level.
Results
Rapid spread of Omicron sublineages
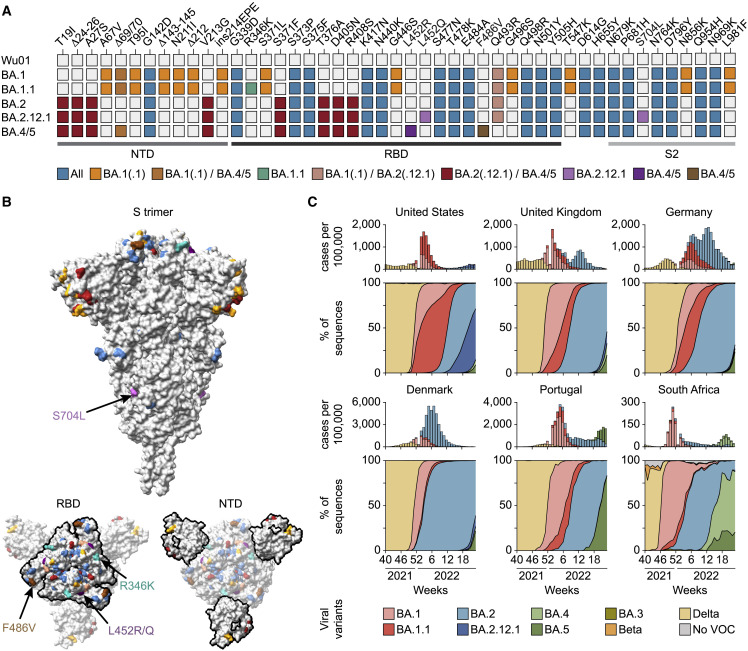
The spike protein of BA.1 differs in 39 amino acid residues from that of the ancestral Wu01 strain of SARS-CoV-2 (Figure 1A). While other Omicron sublineages share several mutations with BA.1, they diverge at various amino acid positions including in critical antibody epitopes (Figures 1A and 1B). For example, the BA.1.1 spike protein contains an R346K substitution in the receptor-binding domain (RBD) that was also observed in the SARS-CoV-2 Mu variant and associated with escape from neutralizing antibodies (Figure 1A) (Greaney et al., 2021). While BA.1 and BA.1.1 dominated the initial Omicron surge, they were rapidly outcompeted by BA.2 (Figure 1C). BA.2 shares 21 of its 31 spike protein changes with BA.1 but differs considerably in both the N-terminal domain (NTD) and the RBD, regions targeted by the most potent SARS-CoV-2 neutralizing antibodies (Figures 1A and 1B).
Figure 1.
Omicron sublineage differences
(A) Spike amino acid changes in Omicron sublineages relative to Wu01.
(B) Locations of Omicron-sublineage amino acid changes on the SARS-CoV-2 spike (PDB: 6XR8) with colors as in (A), and RBD and NTD outlined in the bottom models. Residues with mutations exclusive to individual sublineages are indicated by arrows.
(C) Top panels indicate weekly reported SARS-CoV-2 infections as aggregated by the Johns Hopkins University CSSE COVID-19 data repository and Our World in Data, with variant proportions extrapolated from weekly GISAID SARS-CoV-2 database variant sequences (accessed on June 20, 2022) shown in bottom panels.
NTD, N-terminal domain; RBD, receptor-binding domain; PDB, Protein Data Bank.
Following a decline in cases from the BA.1-/BA.2-related first Omicron wave, new sublineages are rapidly emerging (Figure 1C). For example, BA.2.12.1 is outcompeting other BA.2 sublineages in the United States resulting in an increase of infections. Moreover, within weeks of their identification, the BA.4 and BA.5 sublineages with an identical spike protein have become dominant in South Africa and Portugal and driven rises in case numbers (Figure 1C) (Tegally et al., 2022). BA.2.12.1 and BA.4/5 share the majority of their spike mutations with BA.2 (94% and 90%, respectively) but contain additional changes at sites associated with antibody escape. These include substitutions at residue 452 (L452Q in BA.2.12.1 and L452R in BA.4/5, respectively) that were also recorded for the Lambda and Delta variants (Figure 1A) (Kimura et al., 2022; Planas et al., 2021). In addition, the BA.4/5 spike protein harbors the F486V RBD mutation that can reduce monoclonal antibody sensitivity but had not yet been observed in other variants of interest or concern (Baum et al., 2020). Thus, newly emerged Omicron sublineages differ from BA.1 in key residues of the spike protein. Given their apparent growth advantages compared with BA.1 and the prevalent BA.2, the sublineages BA.2.12.1 and BA.4/5 are likely to be among the dominant variants of the SARS-CoV-2 pandemic in the near future.
Reduced serum-neutralizing activity against Omicron sublineages
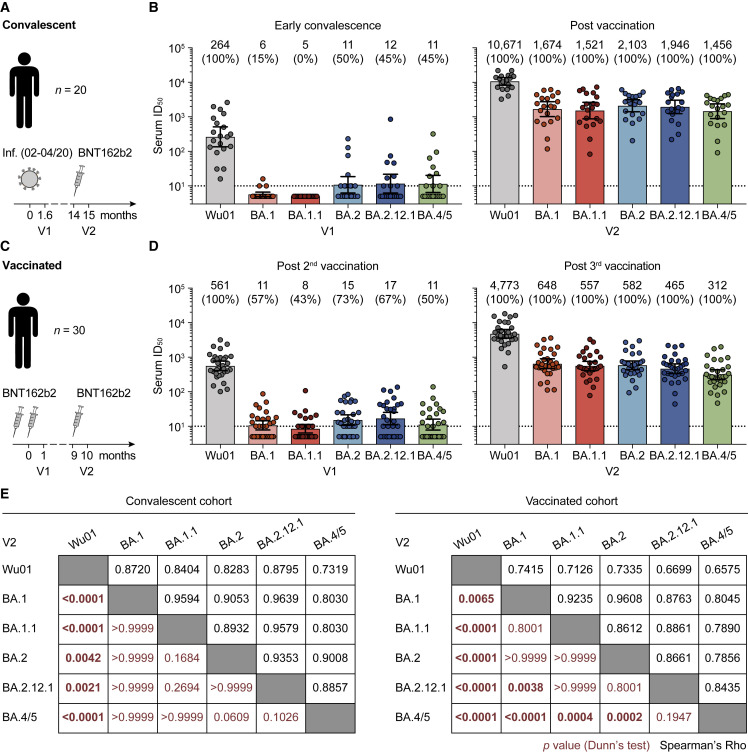
To determine Omicron-sublineage escape profiles, we measured neutralizing activity against the ancestral Wu01 strain as well as BA.1, BA.1.1, BA.2, BA.2.12.1, and BA.4/5 using lentivirus-based pseudovirus assays (Crawford et al., 2020; Vanshylla et al., 2021). In order to assess escape from polyclonal serum activity, we collected samples from two longitudinal cohorts of (1) SARS-CoV-2-convalescent individuals (n = 20) and (2) vaccinated health care workers (n = 30) (Table S1) (Hillus et al., 2021; Vanshylla et al., 2021). In both cohorts, individuals received an mRNA vaccine booster immunization (BNT162b2) after a median of 14 and 9 months following infection or two doses of BNT162b2, respectively.
Convalescent individuals had a median age of 51 years (interquartile range [IQR] 35–60) and were diagnosed with mild or asymptomatic SARS-CoV-2 infection. All individuals were infected during the initial phase of the pandemic between February and April 2020, prior to the emergence of designated variants of concern. Early post-infection samples (V1) were collected at a median of 48 days (IQR 34–58) after disease onset or diagnosis and neutralizing activity was assessed by determining the 50% inhibitory serum dilutions (ID50s) (Figure 2A). Neutralization of the Wu01 strain was detected in all samples (100%) obtained early after infection, with individual ID50 values ranging from 16 to 2,607 (geometric mean ID50 [GeoMeanID50] of 264) (Figure 2B). In contrast, serum activity against Omicron sublineages was strongly reduced and only detectable in 0%–15% for BA.1 and BA1.1, and 45%–50% for BA.2, BA.2.12.1, and BA.4/5 (Figure 2B). However, analysis of sera obtained at a median of 33 days (IQR 27–54) after a single BNT162b2 booster immunization revealed neutralizing activity against all tested Omicron sublineages (Figures 2B and S1A), with GeoMeanID50s ranging from 1,456 against BA.4/5 to 2,103 against BA.2 (Figures 2B and S1B).
Figure 2.
Omicron-sublineage-neutralizing serum activity in vaccinated and convalescent individuals
(A) Study scheme in COVID-19-convalescent individuals infected between February and April 2020.
(B) Pseudovirus neutralization assay 50% inhibitory serum dilutions (ID50s) in convalescent individuals. Bars indicate geometric mean ID50s (GeoMeanID50s) with 95% confidence intervals (CIs). Numbers indicate GeoMeanID50s and fraction of individuals with detectable neutralizing activity (ID50 > 10; in parentheses).
(C) Study scheme in vaccinated individuals.
(D) Pseudovirus neutralization assay serum ID50s in vaccinated individuals. Bars indicate GeoMeanID50s with 95% CIs. Numbers indicate GeoMeanID50s and fraction of individuals with detectable neutralizing activity (ID50 > 10; in parentheses).
(E) Black numbers indicate Spearman’s rank correlation coefficients (rho) at V2. Red numbers indicate p values determined after the Friedman test by Dunn’s multiple comparison tests at V2 (statistically significant differences with p < 0.05 in bold).
In (B) and (D), ID50s below the lower limit of quantification (LLOQ, ID50 = 10; indicated by black dotted lines) were imputed to ½ × LLOQ (ID50 = 5), and ID50s above the upper limit of quantification (21,870) were imputed to 21,871. See also Figure S1 and Table S1.
In addition, we determined Omicron-sublineage-neutralizing activity induced solely by vaccination (Figure 2C). At a median of 28 days (IQR 27–32) after completion of the initial two-dose course of BNT162b2 (V1), Wu01 neutralizing serum activity was detected in all 30 individuals with a GeoMeanID50 of 561 (Figure 2D). Although Omicron-sublineage neutralization was detectable in 43%–73% of vaccinated individuals at V1, GeoMeanID50s were very low (ranging from 8 to 17) (Figure 2D). Follow-up samples obtained at a median of 29 days (IQR 26–35) after booster immunization showed 8-fold higher activity against Wu01 (GeoMeanID50 of 4,773) and strongly increased activity against all Omicron sublineages (27- to 70-fold GeoMeanID50 increases) (Figures 2D, S1C, and S1D). Similar to the convalescent cohort, the lowest GeoMeanID50 after booster immunization was observed against the BA.4/5 sublineages (GeoMeanID50 of 312, Figure 2D). Of note, among vaccinated individuals, the neutralizing activity against BA.4/5 was statistically significantly lower compared with that against BA.1, BA.1.1, and BA.2 (GeoMeanID50s of 648, 557, and 582, respectively) (Figure 2E).
We conclude that booster immunizations are critical to elicit neutralizing serum activity against all prevalent Omicron sublineages in vaccinated as well as convalescent individuals. Notably, BA.4/5 consistently demonstrated the lowest sensitivity of all sublineages against polyclonal serum, suggesting increased antigenic escape despite the small number of spike protein differences relative to BA.2.
Dissecting escape of Omicron sublineages
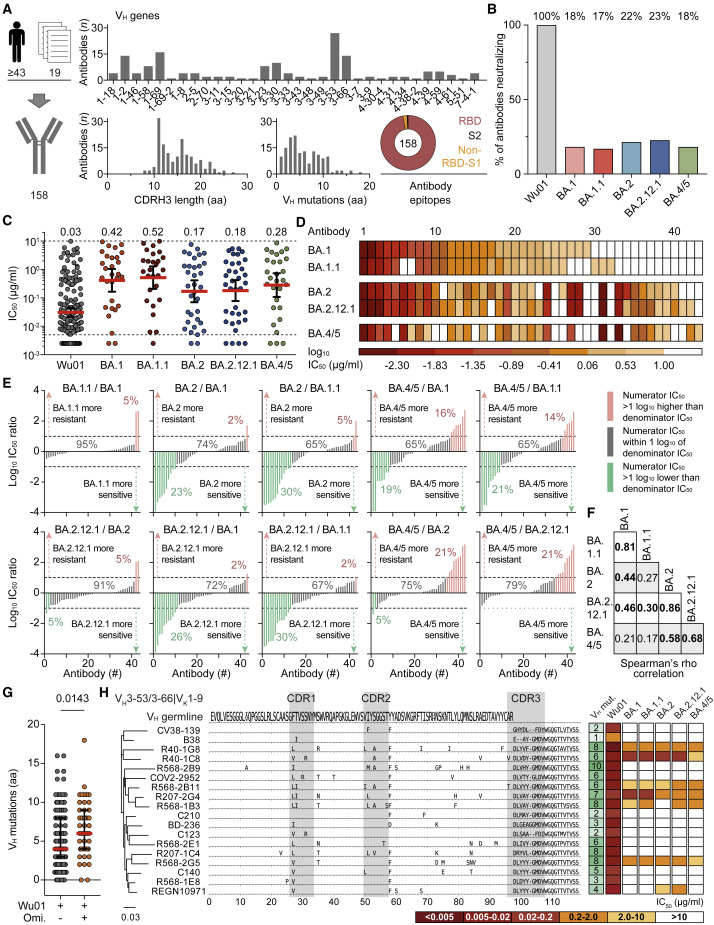
To decode Omicron escape from neutralizing antibodies on a molecular level, we produced and tested 158 monoclonal antibodies isolated from SARS-CoV-2-convalescent individuals against all sublineages. These included 67 randomly selected antibodies from the Coronavirus Antibody Database (CoV-AbDab) (Raybould et al., 2021), 79 antibodies isolated in our previous work (Kreer et al., 2020; Vanshylla et al., 2022), as well as 12 antibodies that have been clinically tested. In total, this antibody panel originated from at least 43 different individuals out of 19 independent studies (Figure 3A; Table S2). All antibodies were identified in unvaccinated individuals infected prior to the emergence of Omicron. The selection encompassed a broad spectrum of SARS-CoV-2-neutralizing antibodies with 92 VH/VL combinations and a diverse range of CDR3 lengths and V gene mutations (Figure 3A; Table S2). Most antibodies targeted the RBD (97%), and the panel included previously described public clonotypes such as the VH3-53/3-66 subgroup (Figure 3A; Table S2).
Figure 3.
Determining Omicron-sublineage immune escape using monoclonal antibodies
(A) SARS-CoV-2-neutralizing monoclonal antibodies (n = 158) were derived from 19 studies and ≥43 convalescent individuals. Bar and pie charts indicate numbers of antibodies per heavy-chain variable (VH) gene segment, amino acid (aa) length of the heavy-chain complementarity-determining region 3 (CDRH3), number of VH aa mutations relative to VH germline-encoded domain, and epitope. RBD, receptor-binding domain.
(B) Fractions of antibodies neutralizing Wu01 and Omicron sublineages (IC50 < 10 μg/mL).
(C) Neutralizing antibody IC50s against sublineages (Wu01, n = 158; BA.1, n = 29; BA.1.1, n = 27; BA.2, n = 34; BA.2.12.1, n = 36; BA.4/5, n = 29). Solid lines indicate geometric mean IC50s and 95% confidence intervals, and dashed lines indicate lower (LLOQ, 0.005 μg/mL) and upper limits of quantification (ULOQ, 10 μg/mL). IC50s < LLOQ were imputed to ½ × LLOQ (IC50 = 0.0025).
(D) IC50 heatmap of the subset of antibodies (n = 43) with neutralizing activity (IC50 < 10 μg/mL) against ≥1 Omicron sublineage. Antibodies are sorted based on their potency against BA.1.
(E) Log10 IC50 ratios of antibodies neutralizing any Omicron sublineage for indicated sublineage comparisons. Within each panel, antibodies are sorted by increasing IC50 ratios. Numbers indicate fractions of antibodies with higher, similar, or lower sublineage activity. IC50s < LLOQ were imputed to ½ × LLOQ (IC50 = 0.0025) and IC50s > ULOQ were imputed to 2 × ULOQ (IC50 = 20).
(F) Spearman’s rank correlation coefficients for ≥1 Omicron-sublineage-neutralizing antibodies (n = 43) as shown in Figure S2C. Numbers in bold indicate values with p < 0.05.
(G) VH aa mutations of antibodies neutralizing Wu01 only or both Wu01 and ≥1 Omicron sublineage. Lines indicate medians and interquartile ranges. Groups were compared using a two-tailed Mann-Whitney U test.
(H) Phylogenetic tree and heavy-chain sequence alignment of VH3-53/3-66|VK1-9 public clonotype antibodies. Letters indicate aa mutations relative to VH germline-encoded residues. Number of aa mutations and neutralizing activity are indicated. Germline VH represents consensus of identified antibody germline alleles (VH3-53∗01, VH3-53∗04, and VH3-66∗01).
While all 158 antibodies neutralized the Wu01 strain, only 18%, 17%, 22%, 23%, and 18% remained active against BA.1, BA.1.1, BA.2, BA.2.12.1, and BA.4/5, respectively (Figures 3B and S2A). Moreover, among antibodies with retained neutralizing activity, the overall potency against Omicron sublineages was 6- to 14-fold lower compared with Wu01 with GeoMeanIC50s of 0.42 (BA.1), 0.52 (BA.1.1), 0.17 (BA.2), 0.18 (BA.2.12.1), and 0.28 μg/mL (BA.4/5) (Figure 3C).
The neutralization profiles revealed both commonalities and diversity in antibody sensitivity of the different Omicron sublineages (Figures 3D–3F, S2B, and S2C). Between the most closely related sublineages (BA.1 and BA.1.1; BA.2 and BA.2.12.1), only small differences in antibody activity were observed (rs of 0.81 and 0.86, respectively) (Figures 3E, 3F, and S2C). In contrast, comparisons between the main sublineages BA.1, BA.2, and BA.4/5 demonstrated higher degrees of variation in antibody susceptibility (Figures 3E and 3F). For example, BA.2 was more resistant (>1 log10 IC50 difference) than BA.1 to only a single antibody (2%), but more sensitive to ten antibodies (23%) (Figure 3E). Moreover, higher fractions of antibodies showed >10-fold higher resistance against BA.4/5 relative to other variants (Figure 3E). While the comparison of BA.4/5 to BA.1 and BA.1.1 revealed heterogeneity in sensitivity to the antibody panel with no correlation, the sensitivity of BA.4/5 and BA.2 or BA.2.12.1 more strongly correlated (rs of 0.58 and 0.68, respectively) (Figures 3E and 3F). Importantly, however, whereas BA.4/5 was more sensitive to only 0%–5% of antibodies compared with BA.2 and BA.2.12.1, BA.4/5 showed higher resistance to 21% of the Omicron-neutralizing antibodies (Figure 3E).
Based on the analysis of the sublineage neutralization profiles, three prevalent classes of Omicron-neutralizing antibodies became apparent: (1) antibodies with broadly comparable activity against Omicron sublineages (21/43, 49%), (2) antibodies with strongly reduced activity against BA.1 and BA.1.1 compared with BA.2, BA.2.12.1, and BA.4/5 (7/43, 16%), and (3) antibodies with strongly reduced activity against BA.4/5 compared with BA.1, BA.1.1, as well as BA.2 and/or BA.2.12.1 (6/43, 14%).
We conclude that in contrast to the more subtle differences in polyclonal serum sensitivity, Omicron sublineages can considerably differ in their sensitivity to monoclonal antibodies. Importantly, BA.4/5 demonstrated a strong bias to higher resistance compared with the prevalent BA.2 sublineages.
Minor sequence variations critically affect antibody activity
Antibodies with Omicron-neutralizing activity carried a modestly higher number of amino acid mutations in the heavy- and light-chain variable genes compared with antibodies neutralizing Wu01 only (median of 6 versus 4 and 4 versus 3 mutations; p = 0.0143 and p = 0.0170, respectively) (Figures 3G and S2D). This suggests that higher sequence diversification might be overall favorable for Omicron neutralization.
High convergence in antibody responses against SARS-CoV-2 is demonstrated by public clonotypes with conserved sequence characteristics and neutralization mechanisms (Nielsen et al., 2020; Robbiani et al., 2020; Yuan et al., 2020). Among the analyzed antibody panel, 18 sequences from 11 individuals could be assigned to the prominent VH3-53/3-66|VK1-9 clonotype (Cao et al., 2020; Vanshylla et al., 2022; Zhang et al., 2021). Although these antibodies are highly conserved on a sequence level, they substantially differed in their Omicron-neutralizing capacity (Figures 3H and S2E). For example, antibodies R207-1C4 and R568-2G5 harbor eight amino acid mutations in their VH gene-encoded segment (without CDRH3) of which five are at the same residue, and three are identical. Both antibodies had similar Wu01 neutralizing activity, but R207-1C4 did not neutralize any Omicron sublineage, whereas R568-2G5 showed neutralizing activity against all variants. Notably, antibody C140, another member of this clonotype, which has the identical CDRH3 as R568-2G5, did not neutralize any Omicron variant.
We conclude that minimal variation in antibody sequences can tip the scale between Omicron neutralization and resistance. Thus, antibody sequence- or class-based predictions of Omicron neutralization can be difficult and experimental assessment of individual monoclonal antibody activity remains essential.
Impact on clinical and novel monoclonal antibodies
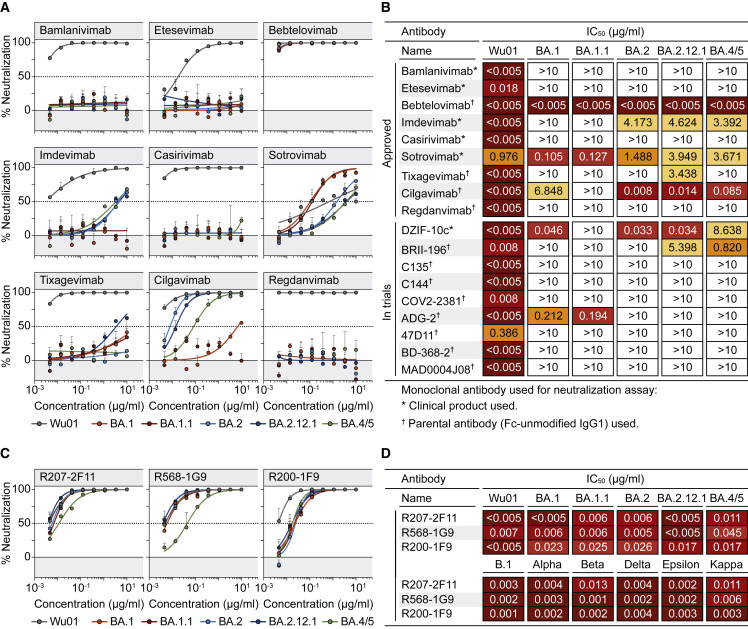
SARS-CoV-2-neutralizing monoclonal antibodies can reduce morbidity and mortality in infected individuals and are critical for passive immunization to protect individuals that do not mount an adequate immune response upon vaccination (Cohen et al., 2021; Corti et al., 2021; RECOVERY Collaborative Group, 2022; Gruell et al., 2022b; Gupta et al., 2021; Levin et al., 2022; Weinreich et al., 2021). To determine how antibodies in clinical use are affected by Omicron sublineages, we analyzed 9 monoclonal antibodies that received authorization for clinical use (Figures 4A and 4B) and 9 that are advanced in clinical development (Figures 4B and S3). All antibodies target the SARS-CoV-2 spike RBD and were tested for neutralizing activity against Wu01 and the Omicron sublineages.
Figure 4.
Omicron-sublineage-neutralizing activity of monoclonal antibodies in clinical use
(A) Neutralization dose response curves in pseudovirus-based assay. Circles show averages and error bars indicate standard deviation. Dotted lines indicate 50% neutralization (IC50).
(B) IC50s of antibodies in current or previous clinical use or development. Symbols indicate whether clinical products or parental antibodies produced as human IgG1 were used for neutralization testing.
(C) Neutralization dose response curves in pseudovirus-based assay. Circles show averages and error bars indicate standard deviation. Dotted lines indicate 50% neutralization (IC50).
(D) IC50s of antibodies in (C) against Wu01 and Omicron sublineages (upper rows) and previously circulating variants (lower rows, as previously determined; Vanshylla et al., 2022).
See also Figure S3.
Most antibodies showed highly potent neutralizing activity against Wu01 with IC50s below 0.005 μg/mL (Figure 4B). Less potent and incomplete Wu01 neutralizing activity was observed for sotrovimab, consistent with reduced activity against pseudoviruses lacking the dominant D614G spike mutation (Figure 4A) (Liu et al., 2021; Weissman et al., 2021). Only five (28%) out of the 18 tested antibodies neutralized BA.1 with IC50s < 10 μg/mL, and of these, three had strongly reduced potency (Figure 4B). While the antibody neutralization profile of BA.1.1 was generally similar to that of BA.1, there were some differences. For example, DZIF-10c neutralized BA.1 with an IC50 of 0.046 μg/mL but completely lost activity against BA.1.1 (Figures 4B and S3).
Although the number of antibodies in clinical use with neutralizing activity against BA.2 (5/18, 28%), BA.2.12.1 (7/18, 39%), and BA.4/5 (6/18, 33%) was similarly small, the neutralization profiles did differ (Figure 4B). For example, cilgavimab neutralized BA.2 and BA.2.12.1 with high potency (IC50s of 0.008 and 0.014 μg/mL, respectively) but was >480-fold less potent against BA.1 and BA.1.1 (Figures 4A and 4B). While cilgavimab also remained potent against BA.4/5 (IC50 of 0.085 μg/mL), the activity of antibody DZIF-10c against BA.4/5 was strongly reduced when compared with BA.2 and BA.2.12.1 (8.64 versus 0.03 μg/mL) (Figure 4B). In contrast, whereas antibody imdevimab showed no appreciable activity against BA.1 and BA.1.1, neutralization of BA.2, BA.2.12.1, and BA.4/5 was detectable at low levels (Figures 4A and 4B).
Out of all clinical antibodies tested, only bebtelovimab demonstrated potent neutralization against all Omicron sublineages (IC50 < 0.005 μg/mL; Figures 4A and 4B). To identify additional promising candidates, we examined antibodies previously isolated from individuals with “elite” IgG neutralizing activity (Vanshylla et al., 2022). Remarkably, antibodies R207-2F11, R568-1G9, and R200-1F9 demonstrated highly potent neutralizing activity against previously circulating variants (Wu01, B.1, Alpha, Beta, Delta, Epsilon, and Kappa), as well as against BA.1, BA.1.1, BA.2, BA.2.12.1, and BA.4/5 with IC50s < 0.05 μg/mL (Figure 4D; Table S2). These antibodies were identified from three different convalescent individuals within weeks of infection during the early phase of the pandemic (Vanshylla et al., 2022). Thus, the results highlight the capability of the immune system to generate potent antibodies against the highly divergent Omicron variants in response to contact with the ancestral SARS-CoV-2 spike protein.
We conclude that the prevalent and newly emerged Omicron sublineages, including BA.2.12.1 and BA.4/5, escape from most monoclonal antibodies in clinical use. Sensitivity to these antibodies can, however, strongly differ on the sublineage level.
Discussion
Antibody-mediated immunity is a critical component of prophylactic and therapeutic measures against SARS-CoV-2 infection (Corti et al., 2021; Feng et al., 2021; Gruell et al., 2022b; Khoury et al., 2021). Throughout the COVID-19 pandemic, it has however been confronted by the periodic emergence of antibody escape variants developing in the context of increasing population immunity (Harvey et al., 2021). Omicron sublineages with diverse spike proteins gaining prevalence suggests a further increase in immune escape and/or transmissibility (Lyngse et al., 2022; Tegally et al., 2022; Yamasoba et al., 2022). Thus, establishing the impact of the recent BA.2.12.1 and BA.4/5 sublineages on polyclonal and monoclonal immunity is critical to guide antibody-mediated strategies for prevention and treatment.
Our results demonstrate that Wu01-based mRNA vaccine boosters are effective in eliciting serum-neutralizing activity against the diverse Omicron sublineages. However, this activity is strongly limited compared with Wu01 neutralization and lower BA.4/5-neutralizing titers indicate even more pronounced immune evasion relative to previous Omicron sublineages. Interestingly, the relative reduction in activity against BA.4/5 after booster immunization was higher in infection-naive individuals than in convalescent individuals. This might indicate differences in the breadth and potency of the neutralizing antibody response elicited by vaccination only compared with those induced by “hybrid immunity.”
While the differences in polyclonal serum sensitivity between the Omicron sublineages were overall modest, our analysis of an extended antibody panel revealed distinct patterns in sublineage sensitivity on a monoclonal level. Comparisons of Omicron sublineages indicate a large antigenic distance between BA.4/5 and BA.1/BA.1.1 by the lack of correlation in antibody sensitivity and notable fractions of antibodies showing higher sensitivity to one or the other sublineage. Whereas BA.4/5 and BA.2/BA.2.12.1 showed more similar antibody susceptibility profiles, almost all differences manifested as higher resistance of BA.4/5.
Notably, minor antibody sequence differences strongly affected Omicron activity of potent Wu01-neutralizing antibodies, highlighting the relative difficulty of inducing antibodies with optimized Omicron neutralization after a single Wu01-related spike contact. Accordingly and consistent with previous analyses on BA.1 and BA.2 (Bruel et al., 2022; Iketani et al., 2022), almost all antibodies in clinical investigation showed strongly reduced or abolished activity against all Omicron sublineages. However, we identified novel antibodies isolated after ancestral strain infection with remarkable activity against all tested variants that may provide options for immune-mediated treatment and prevention in the Omicron era of the pandemic.
Our results suggest that outcompetition of BA.1(.1) by BA.2 may have been driven mostly by higher transmissibility rather than by immune evasion. In contrast, higher antigenic distance and antibody resistance of BA.4/5 may support its expansion after the initial Omicron wave. The rapid emergence of Omicron sublineages with reduced antibody susceptibility highlights the challenges posed by constant viral evolution and the critical need for genomic surveillance and sensitivity assessments to inform on antibody-based prophylactic and therapeutic measures.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 2-7, 2-15, 2-30, 2-36, 2-38, 2-43, 4-20 | Liu et al, 2020 | N/A |
| 47D11 | Wang et al., 2020 | N/A |
| ADG-2 | Rappazzo et al., 2021 | N/A |
| B-38 | Wu et al., 2020 | N/A |
| Bamlanivimab | Eli Lilly; Primary paper: Jones et al., 2021 | N/A |
| BD23, BD-236, BD-368-2 | Cao et al., 2020 | N/A |
| C002, C022, C101, C102, C104, C105, C123, C125, C128, C135, C140, C144, C155, C165, C210 | Robbiani et al., 2020 | N/A |
| CC6.31, CC6.33, CC12.4 | Rogers et al., 2020 | N/A |
| CnC2t1p1_B4, CnC2t1p1_D6, CnC2t1p1_E8, CnC2t1p1_E12, CnC2t1p1_G6, FnC1t2p1_D4, FnC1t2p1_G5, HbnC2t1p2_D9, HbnC3t1p1_C6, HbnC3t1p1_G4, HbnC3t1p2_B10, HbnC3t1p2_C6, MnC1t3p1_G9, MnC2t1p1_A3, MnC2t1p1_C5, MnC2t2p1_C11, MnC4t2p1_B3, MnC4t2p1_D10, MnC4t2p1_E6, MnC4t2p2_A4, MnC5t2p1_G1 | Kreer et al., 2020 | N/A |
| COV2-2050, COV2-2064, COV2-2068, COV2-2098, COV2-2130, COV2-2196, COV2-2268, COV2-2308, COV2-2354, COV2-2381, COV2-2479, COV2-2499, COV2-2531, COV2-2539, COV2-2562, COV2-2677, COV2-2678, COV2-2752, COV2-2841, COV2-2919, COV2-2952, COV2-2955 | Zost et al., 2020a; Zost et al., 2020b | N/A |
| COVA2-29 | Brouwer et al., 2020 | N/A |
| CV-X2-106, CV07-262, CV07-270, CV38-139, CV38-142 | Kreye et al., 2020 | N/A |
| CT-P59 (Regdanvimab) | Kim et al., 2021 | N/A |
| DH1042, DH1128, DH1138, DH1184, DH1210 | Li et al., 2021 | N/A |
| DZIF-10c | Boehringer Ingelheim; Primary paper: Kreer et al., 2020 | N/A |
| Etesevimab | Eli Lilly; Primary paper: Shi et al., 2020 | N/A |
| GW01 | Wang et al., 2022 | N/A |
| Casirivimab, Imdevimab | Regeneron; Primary paper: Hansen et al., 2020 | N/A |
| LY-CoV1404 (Bebtelovimab) | Westendorf et al., 2022 | N/A |
| MAD0004J08 | Andreano et al., 2021 | N/A |
| P2B-2F6, P2C-1F11 | Ju et al., 2020 | N/A |
| R40-1A1, R40-1A8, R40-1B4, R40-1B9, R40-1C8, R40-1D3, R40-1E1, R40-1E4, R40-1G6, R40-1G8, R40-1G12, R40-1H4, R121-1F1, R121-3F7, R121-3F11, R121-3G2, R200-1B8, R200-1B9, R200-1F9, R200-1G11, R200-4F4, R207-1C1, R207-1C4, R207-1G1, R207-2A6, R207-2A10, R207-2C2, R207-2F11, R207-2G4, R207-2H1, R259-1B9, R339-1B11, R339-3B5, R339-3C6, R410-1A8, R568-1A9, R568-1B3, R568-1C6, R568-1E8, R568-1G9, R568-2A1, R568-2A3, R568-2B9, R568-2B11, R568-2E1, R568-2E7, R568-2F1, R568-2G5, R568-2G11, R616-1A11, R616-1D6, R616-1F10, R616-1G4, R849-1C11, R849-1G7, R849-1H1R849-3H2 | Vanshylla et al., 2022 | N/A |
| REGN10954, REGN10955, REGN10964, REGN10970, REGN10971, REGN10977 REGN10986, REGN10989 | Hansen et al., 2020 | N/A |
| S2X35 | Piccoli et al., 2020 | N/A |
| Sotrovimab | GSK; Primary paper: Pinto et al., 2020 | N/A |
| Bacterial and virus strains | ||
| SARS-CoV-2 Wu01 pseudovirus | Vanshylla et al., 2021 | N/A |
| SARS-CoV-2 BA.1 pseudovirus | Gruell et al, 2022a | N/A |
| SARS-CoV-2 BA.1.1 pseudovirus | This paper | N/A |
| SARS-CoV-2 BA.2 pseudovirus | This paper | N/A |
| SARS-CoV-2 BA.2.12.1 pseudovirus | This paper | N/A |
| SARS-CoV-2 BA.4/5 pseudovirus | This paper | N/A |
| Biological samples | ||
| COVID-19 convalescent cohort serum samples | This paper; Vanshylla et al., 2021 | N/A |
| Vaccinated cohort serum samples | This paper; Hillus et al., 2021 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| ATP | Sigma-Aldrich | Cat#A2383; CAS: 34369-07-8 |
| Branched polyethylenimine, 25 kDa | Sigma-Aldrich | Cat#408727; CAS: 9002-98-6 |
| Coenzyme A sodium salt hydrate | Sigma-Aldrich | Cat#C3144; CAS: 55672-92-9 |
| D-Luciferin, sodium salt | GoldBio | Cat#LUCNA-1G; CAS: 103404-75-7 |
| Dulbecco’s Modified Eagle Medium (DMEM) | Thermo Fisher | Cat#11960044 |
| Fetal bovine serum (FBS) | Sigma-Aldrich | Cat#F9665 |
| FreeStyle™ 293 Expression Medium | Thermo Fisher | Cat#12338001 |
| FuGENE® 6 Transfection Reagent | Promega | Cat#E2691 |
| IGEPAL | Sigma-Aldrich | Cat#I8896; CAS: 9002-93-1 |
| L-Glutamine | Thermo Fisher | Cat#25030024 |
| MgCl2 | Sigma-Aldrich | Cat#M8266; CAS: 7786-30-3 |
| NEBuilder® HiFi DNA Assembly Cloning Kit | New England Biolabs | Cat#E5520S |
| Penicillin-Streptomycin | Thermo Fisher | Cat#15140122 |
| Protein G Sepharose 4 Fast Flow | Cytiva | Cat#17061805 |
| Q5® Site-Directed Mutagenesis Kit | New England Biolabs | Cat#E0554 |
| Sodium pyruvate | Thermo Fisher | Cat#11360070 |
| Experimental models: Cell lines | ||
| 293T-ACE2 cells |
Jesse Bloom Lab; Crawford et al., 2020; BEI Resources | Cat#NR-52511 |
| 293-6E cells | National Research Council of Canada | NRC file 11565 |
| HEK293T cells | ATCC | Cat#CRL-11268 |
| Recombinant DNA | ||
| Antibody heavy and light chain cloning fragments | This paper; Integrated DNA Technologies | N/A |
| Human antibody expression vectors (IgG1, Igκ, Igλ) | Tiller et al., 2008 | N/A |
| pcDNA™3.1/V5-His TOPO™ TA Expression Kit | Thermo Fisher | Cat#K480001 |
| pHAGE-CMV-Luc2-IRES-ZsGreen-W | Jesse Bloom Lab; Crawford et al., 2020 | RRID: Addgene_164432 |
| pHDM-Hgpm2 | Jesse Bloom Lab; Crawford et al., 2020 | RRID: Addgene_164441 |
| pHDM-tat1b | Jesse Bloom Lab; Crawford et al., 2020 | RRID: Addgene_164442 |
| pRC-CMV-Rev1b | Jesse Bloom Lab; Crawford et al., 2020 | RRID: Addgene_164443 |
| SARS-CoV-2 spike gene fragments | This paper; Thermo Fisher | N/A |
| Software and algorithms | ||
| ChimeraX-1.3 | Goddard et al., 2018; Pettersen et al., 2021 | RRID: SCR_015872 |
| Codon Usage Database | Nakamura et al., 2000 | N/A |
| Codon Optimization Tool | Integrated DNA Technologies | N/A |
| EMBL-EBI search and sequence analysis tools API | Madeira et al., 2019 | N/A |
| Geneious Prime | Biomatters | RRID: SCR_010519 |
| IgBlast | National Library of Medicine; Ye et al., 2013 | RRID: SCR_002873 |
| MAFFT algorithm | Katoh et al., 2002 | N/A |
| Matplotlib | http://matplotlib.sourceforge.net/ | RRID: SCR_008624 |
| NumPy | https://numpy.org | RRID: SCR_008633 |
| Pandas | https://pandas.pydata.org | RRID: SCR_018214 |
| Prism | GraphPad | RRID: SCR_002798 |
| Python | http://www.python.org/ | RRID: SCR_008394 |
| SciPy | http://www.scipy.org/ | RRID: SCR_008058 |
| Sequence Manipulation Suite | Stothard,2000 | N/A |
| Other | ||
| COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (weekly SARS-CoV-2 infections) | Dong et al.,2020 | https://coronavirus.jhu.edu |
| CoV-AbDab (The Coronavirus Antibody Database) | Raybould et al.,2021 | http://opig.stats.ox.ac.uk/webapps/covabdab/ |
| GISAID (deposited SARS-CoV-2 sequences by variant) | Elbe and Buckland-Merrett, 2017; Khare et al., 2021; Shu and McCauley, 2017 | https://www.gisaid.org; RRID: SCR_018279 |
| IMGT Database | Lefranc,2011 |
https://www.imgt.org; RRID: SCR_012780 |
| Our World in Data (aggregated weekly SARS-CoV-2 infections) | Global Change Data Lab | https://ourworldindata.org |
Resource availability
Lead contact
Requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Florian Klein (florian.klein@uk-koeln.de).
Materials availability
Raw data and/or materials will be made available by the Lead contact upon reasonable request but may require completion of a Material Transfer Agreement. Clinical samples may be subject to restrictions due to data and privacy protection regulations.
Experimental model and subject details
Study cohort and sample collection
Serum samples from COVID-19-convalescent individuals (n=20) were collected at the University Hospital Cologne under study protocols approved by the ethics committee (EC) of the Medical Faculty of the University of Cologne (16-054 and 20-1187). Between April and May, 2020, individuals with a history of SARS-CoV-2 infection confirmed by polymerase chain reaction (documented through a test certificate or as reported to study investigators by the participants) were enrolled within eight weeks of symptom onset and/or diagnosis. As all participants were enrolled early during the pandemic (i.e., prior to the emergence of variants of concern as designated by the World Health Organization), most individuals were likely to have been infected with an early viral strain similar to Wu01. Participants were followed longitudinally to analyze long-term immunity to SARS-CoV-2. Follow-up samples after booster immunization were collected between May and August, 2021. No additional SARS-CoV-2 infections between the sampling time points were reported by any of the participants.
Serum samples from vaccinated individuals (n=30) were collected under protocols approved by the EC of Charité - Universitätsmedizin Berlin (EICOV, EA4/245/20) as well as the EC of the Federal State of Berlin and the Paul Ehrlich Institute (COVIM, EudraCT-No. 2021-001512-28). Study participation irrespective of medical conditions was offered to health-care workers vaccinated at Charité – Universitätsmedizin (Berlin, Germany). All serum samples were tested for antibodies targeting the SARS-CoV-2 nucleocapsid using the SeraSpot Anti-SARS-CoV-2 IgG microarray-based immunoassay (Seramun Diagnostica). Samples from individuals with a history of SARS-CoV-2 infection, a positive SARS-CoV-2 nucleic acid amplification test (performed at sampling), or detectable anti-nucleocapsid antibodies were not included in this analysis. Serum samples after two vaccinations were collected in February and March 2021 (for 29/30 participants; from one participant, the early serum sample was obtained in July 2021); samples after booster immunization were collected in December 2021 and January 2022.
All study participants provided written informed consent. Vaccinations in both cohorts were performed as part of routine care outside of the observational studies. Selection of participants and samples for analysis was based on receipt of identical vaccines and comparable sampling time points relative to vaccinations. Details on the individual study cohorts, including gender distribution and age, are provided in Table S1. Serum samples were collected after centrifugation and stored at -80°C until analysis.
Cell lines
HEK293T cells and 293T-ACE2 cells (Crawford et al., 2020) were maintained at 37°C and 5% CO2 in high glucose Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 2 mM L-glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate (all Thermo Fisher), and 10% fetal bovine serum (FBS; Sigma-Aldrich). 293-6E cells were maintained at 37°C and 6% CO2 under constant shaking in FreeStyle Expression Medium supplemented with penicillin (20 U/ml) and streptomycin (20 μg/ml) (all Thermo Fisher). The sex of all cell lines is female. Cell lines were not specifically authenticated.
Method details
SARS-CoV-2 pseudovirus constructs
All SARS-CoV-2 spike proteins were expressed using codon-optimized expression plasmids. Wu01 (EPI_ISL_406716), BA.2.12.1, and BA.4/5 pseudoviruses were produced using an expression plasmid that incorporated a C-terminal deletion of 21 cytoplasmic amino acids that results in increased pseudovirus titers. Expression plasmids for Omicron sublineage spike proteins were produced by assembling and cloning codon-optimized overlapping gene fragments (Thermo Fisher) into the pCDNA3.1/V5-HisTOPO vector (Thermo Fisher) using the NEBuilder HiFi DNA Assembly Kit (New England Biolabs) and/or by site-directed PCR mutagenesis (Q5 Site-Directed Mutagenesis Kit; New England Biolabs), and included the spike sequences with the following amino acid changes relative to Wu01:
BA.1: A67V, Δ69-70, T95I, G142D, Δ143-145, N211I, Δ212, ins215EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, and L981F.
BA.1.1: As for BA.1 with an additional R346K mutation.
BA.2: T19I, Δ24-26, A27S, A67V, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K.
BA.2.12.1: As for BA.2 with additional L452Q and S704L mutations.
BA.4/5: As for BA.2 with additional Δ69-70, L452R, and F486V mutations, but lacking the Q493R mutation.
All plasmid sequences were verified by sequencing.
Monoclonal antibodies
Monoclonal antibodies previously isolated in our lab had been obtained by single cell-sorting of SARS-CoV-2 spike-specific B cells followed by reverse transcription, PCR amplification and cloning of antibody variable regions (Kreer et al., 2020; Vanshylla et al., 2022). For monoclonal antibodies derived from the CoV-AbDab (Raybould et al., 2021), variable region amino acid sequences were reverse translated with the reverse translate tool from the Sequence Manipulation Suite (Stothard, 2000) using the Homo sapiens codon table obtained from the Codon Usage Database (Nakamura et al., 2000), and sequences were ordered as gene fragments from Integrated DNA Technologies (IDT) with 5' and 3' overhangs. The variable regions were inserted into heavy and light chain expression plasmids (Tiller et al., 2008) by sequence- and ligation-independent cloning (SLIC). For antibodies ADG-2, COV2-2130 (cilgavimab), COV2-2196 (tixagevimab), COV2-2381, MAD0004J08, and P2C-1F11 (BRII-196), gene fragments based on the nucleotide sequences published in GenBank were ordered at IDT and cloned as above. For antibodies C135, CT-P59 (regdanvimab), and LY-CoV1404 (bebtelovimab), gene fragments based on antibody structures deposited in the Protein Data Bank (accession nos. 7K8Z, 7CM4, and 7MMO) were ordered at IDT after codon optimization using the IDT Codon Optimization Tool and cloned as above. For antibodies 47D11, BD-368-2, C144, and P2B-2F6, amino acid sequences were derived from CoV-AbDaB, corresponding nucleotide sequences generated and codon-optimized using the IDT Codon Optimization Tool, and gene fragments cloned as above.
Monoclonal antibody production was performed using 293-6E cells (National Research Council of Canada) by co-transfection of heavy and light chain expression plasmids using 25 kDa branched polyethylenimine (Sigma-Aldrich). Culture supernatants were harvested after an incubation period of 6-7 days at 37°C and 6% CO2 under constant shaking in FreeStyle Expression Medium supplemented with penicillin (20 U/ml) and streptomycin (20 μg/ml) (all Thermo Fisher). Clarified cell supernatants were incubated with Protein G Sepharose 4 FastFlow (Cytiva) overnight at 4°C. After centrifugation, antibodies bound to Protein G beads were eluted in chromatography columns (Bio-Rad) using 0.1 M glycine (pH=3.0) and buffered in 1 M Tris (pH=8.0). Buffer exchange to PBS was performed using centrifugal filter units (Millipore). For antibodies bamlanivimab, casirivimab, DZIF-10c, etesevimab, imdevimab, and sotrovimab, aliquots from clinical stocks were used.
Pseudovirus neutralization assays
Neutralization assays were performed using lentivirus-based pseudoviruses and ACE2-expressing 293T cells (Crawford et al., 2020; Vanshylla et al., 2021). Pseudovirus particle production was performed in HEK293T cells by co-transfection of individual expression plasmids encoding for the SARS-CoV-2 spike protein, HIV-1 Tat, HIV-1 Gag/Pol, HIV-1 Rev, and luciferase-IRES-ZsGreen using FuGENE 6 Transfection Reagent (Promega). Culture supernatants were exchanged with fresh medium (high glucose DMEM supplemented with 2 mM L-glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate (all Thermo Fisher), and 10% FBS (Sigma-Aldrich)) at 24 h post transfection. Pseudovirus-containing supernatants were harvested between 48 h and 72 h after transfection, centrifuged, clarified using a 0.45 μm filter, and stored at -80°C. Pseudoviruses were titrated by infection of 293T-ACE2 cells and luciferase activity was determined after a 48-hour incubation at 37°C and 5% CO2 by addition of luciferin/lysis buffer (10 mM MgCl2, 0.3 mM ATP, 0.5 mM coenzyme A, 17 mM IGEPAL CA-630 (all Sigma-Aldrich), and 1 mM D-Luciferin (GoldBio) in Tris-HCL) using a microplate reader (Berthold).
Serum samples were heat-inactivated at 56°C for 45 min before use. Three-fold serial dilutions of serum (starting at 1:10) and monoclonal antibodies (starting at 10 μg/ml) were prepared in culture medium and co-incubated with pseudovirus supernatants for one hour at 37°C and 5% CO2 prior to addition of 293T-ACE2 cells. Following a 48-hour incubation at 37°C and 5% CO2, luciferase activity was determined as described above. Average background relative light units (RLUs) of non-infected cells were subtracted, and serum ID50s and antibody IC50s were determined as the serum dilutions and antibody concentrations resulting in a 50% RLU reduction compared to the average of virus-infected untreated controls cells using a non-linear fit model plotting an agonist vs. normalized dose response curve with variable slope using the least squares fitting method in Prism 7.0 (GraphPad).
All serum and monoclonal antibody samples were tested in duplicates or triplicates. For further details and imputation rules for samples with values outside the limits of quantification see below (section on Quantification and Statistical Analysis).
Neutralizing antibody panel and sequence analysis
The panel of 158 SARS-CoV-2 neutralizing monoclonal antibodies isolated from SARS-CoV-2 convalescent individuals included in the analysis in Figure 3 is based on 79 antibodies obtained in our previous work (Kreer et al., 2020; Vanshylla et al., 2022), 67 randomly selected (retrieved on January 1, 2021) human SARS-CoV-2 neutralizing antibodies deposited at CoV-AbDab (Raybould et al., 2021), and 12 antibodies in clinical use or development. We did not include five of the antibodies in clinical development shown in Figure 4 into this analysis, as they were obtained from individuals infected with SARS-CoV (ADG-2, sotrovimab), from immunized mice harboring human immunglobulin gene repertoires (47D11, casirivimab), or using phage display technology that does not ensure native pairing of antibody heavy and light chains (regdanvimab).
Antibody amino acid sequences were annotated with IgBLASTp (Ye et al., 2013) based on the IMGT database (Lefranc, 2011). For sequence statistics, top V gene calls were counted without individual alleles, CDR3 lengths are reported according to the IMGT numbering system, and numbers of V mutations refer to the top V gene call from IgBLASTp. Phylogenetic analysis of antibodies belonging to the VH3-53/3-66|VK1-9 public clonotype was performed by alignment of amino acid sequences with the MAFFT algorithm (Katoh et al., 2002) via the EMBL-EBI search and sequence analysis tools API (Madeira et al., 2019) and the Tree Builder tool from Geneious Prime 2020.0.4 (Biomatters) using the Jukes-Cantor distance model for tree building with the neighbour-joining method without resampling. Data aggregation and visualization was performed with the Python libraries pandas (v1.1.5), NumPy (v1.19.2), SciPy (v1.5.2), and Matplotlib (v3.3.4) with Python (v3.6.8), and GraphPad Prism (v7).
Visualization of spike amino acid changes
Amino acid changes relative to the Wu01 spike protein were visualized on a cryo-electron microscopy 3D-reconstruction of the SARS-CoV-2 spike protein (PDB ID: 6XR8 (Cai et al., 2020)) using ChimeraX (v. 1.3) (Goddard et al., 2018; Pettersen et al., 2021).
SARS-CoV-2 variant distribution
Numbers of weekly cases as curated by the COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (Dong et al., 2020) were retrieved from http://ourworldindata.org (accessed on June 20, 2022). GISAID-curated clade and lineage statistics of sequences submitted to the GISIAD database (Elbe and Buckland-Merrett, 2017; Khare et al., 2021; Shu and McCauley, 2017) were retrieved from GISAID (accessed on June 20, 2022) and frequency of individual variants was plotted as fraction of all submitted sequences per week and variant. For visualization, all BA.1 sublineages except for BA.1.1 and its sublineages were classified as BA.1, all BA.2 sublineages except for BA.2.12.1 were classified as BA.2, and all BA.3 sublineages were classified as BA.3.
Quantification and statistical analysis
Serum samples and antibodies were tested in duplicates or triplicates. For serum samples, the average IC50 of 2-3 single dilution series experiments was determined (with the exception of one sample tested in technical duplicates in a single experiment). In cases of low-level detectable serum neutralization (ID50 >10) in only a single run, an ID50 value equal to the limit of quantification (ID50=10) was assigned to the sample. Antibodies in clinical use or investigation were tested in technical duplicates within the same experiment. Additional antibodies included in the panel analysis were tested in 2-3 single dilution experiments and average IC50s were determined.
For graphical representation and statistical evaluation of serum samples in Figures 2 and S1, samples that did not achieve 50% inhibition at the lowest tested dilution of 10 (lower limit of quantification, LLOQ) were imputed to ½ × of the LLOQ (ID50=5) and serum samples with ID50s >21,870 (upper limit of quantification) were imputed to ID50=21,871. For graphical representation and statistical analysis of monoclonal neutralizing antibodies in Figures 3 and S2, IC50 values of antibodies with an IC50 <0.005 μg/ml (LLOQ) were imputed to ½ × LLOQ (IC50=0.0025), and IC50 values >10 μg/ml (ULOQ) were imputed to 2 × ULOQ (IC50=20 μg/ml).
Testing for statistical significance of differences in serum neutralization titers against different variants/sublinages was performed with the Friedman test with Dunn’s multiple comparison post-hoc test using Prism 7.0 (GraphPad). Spearman’s rank correlation coefficients (Rho) were determined using Prism 7.0 (GraphPad). Numbers of amino acid mutations relative to germline between antibodies neutralizing Wu01 only and those neutralizing any Omicron sublineage were compared with two-sided Mann-Whitney U tests using Prism 7.0 (GraphPad). Statistical significance was defined as p < 0.05. Details are additionally provided in the Figure legends. Sample sizes of the convalescent and vaccinated cohorts were based on sample availability and the criteria described in the section on experimental model and subject details.
Acknowledgments
We are grateful to all study participants for their dedication to our research. We thank F. Dewald, L. Gieselmann, B. Kurt, C. Lehmann, P. Mayer, N. Riet, S. Salomon, M. Schlotz, R. Schröder, R. Stumpf, and H. Wüstenberg, as well as the members of the EICOV/COVIM Study Group (Y. Ahlgrimm, L. Bardtke, K. Behn, N. Bethke, H. Bias, D. Briesemeister, C. Conrad, V.M. Corman, C. Dang-Heine, S. Dieckmann, C. Eroglu, D. Frey, J.-A. Gabelich, J. Gerdes, U. Gläser, A. Hetey, L. Hasler, E.T. Helbig, D. Hillus, W. Hirst, A. Horn, C. Hülso, S. Jentzsch, C. von Kalle, L. Kegel, A. Krannich, W. Koch, P. Kopankiewicz, P. Kroneberg, H. Le, L.J. Lippert, C. Lüttke, P. de Macedo Gomes, B. Maeß, C. Matthaiou, J. Michel, A. Nitsche, A.-M. Ollech, C. Peiser, A. Pioch, C. Pley, K. Pohl, C. Rubisch, L. Ruby, A. Sanchez Rezza, I. Schellenberger, V. Schenkel, J. Schlesinger, S. Schmidt, G. Schwanitz, T. Schwarz, J. Seybold, A. Solarek, A. Stege, S. Steinbrecher, P. Stubbemann, C. Thibeault, and D. Treue) for sample acquisition and processing; L. Buchholz and M. Wunsch for help with antibody production and preparation; T. Lange and J.J. Malin for providing clinical antibody stocks; and all laboratories providing SARS-CoV-2 sequencing information to GISAID for facilitating variant distribution analyses. This work was supported by grants from COVIM: NaFoUniMedCovid19 (FKZ: 01KX2021) (to L.E.S. and F. Klein), the Federal Institute for Drugs and Medical Devices (V-2021.3/1503_68403/2021–2022) (to L.E.S. and F. Kurth), the German Center for Infection Research (DZIF) (to F. Klein), and the Deutsche Forschungsgemeinschaft (DFG) CRC1310 (to C.K. and F. Klein).
Author contributions
Conceptualization, H.G., K.V., C.K., and F. Klein; methodology, H.G., K.V., L.E.S., F. Kurth, C.K., and F. Klein; investigation, H.G. and K.V.; resources, P.T-L., M.K., M.Z., F.M., H.J., M.A., P.S., and C.K.; formal analysis, H.G., K.V., and C.K.; writing—original draft, H.G., K.V., C.K., and F. Klein; writing—review & editing, M.K., P.T.-L., and F. Kurth; visualization, H.G., K.V., C.K., and F. Klein; supervision, L.E.S., F. Kurth, and F. Klein; funding acquisition, L.E.S., F. Kurth, and F. Klein.
Declaration of interests
H.G., K.V., M.Z., C.K., and F. Klein are listed as inventors on patent applications on SARS-CoV-2 neutralizing antibodies filed by the University of Cologne that encompass aspects of this work.
Published: July 7, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2022.07.002.
Supplemental information
Data and code availability
-
•
Raw data reported in this paper will be shared by the lead contact upon reasonable request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
References
- Altarawneh H.N., Chemaitelly H., Hasan M.R., Ayoub H.H., Qassim S., AlMukdad S., Coyle P., Yassine H.M., Al-Khatib H.A., Benslimane F.M., et al. Protection against the omicron variant from previous SARS-CoV-2 infection. N. Engl. J. Med. 2022;386:1288–1290. doi: 10.1056/NEJMc2200133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., Manenti A., Pantano E., Kabanova A., Troisi M., et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell. 2021;184:1821–1835.e16. doi: 10.1016/j.cell.2021.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O'Connell A.M., et al. Covid-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N. Engl. J. Med. 2022;386:1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruel T., Hadjadj J., Maes P., Planas D., Seve A., Staropoli I., Guivel-Benhassine F., Porrot F., Bolland W.H., Nguyen Y., et al. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat. Med. 2022;28:1297–1302. doi: 10.1038/s41591-022-01792-5. [DOI] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreño J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H., Sominsky L.A., Clark J.J., Adelsberg D.C., Bielak D.A., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M.S., Nirula A., Mulligan M.J., Novak R.M., Marovich M., Yen C., Stemer A., Mayer S.M., Wohl D., Brengle B., et al. Effect of bamlanivimab vs placebo on incidence of COVID-19 among residents and staff of skilled nursing and assisted living facilities: a randomized clinical trial. JAMA. 2021;326:46–55. doi: 10.1001/jama.2021.8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Purcell L.A., Snell G., Veesler D. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell. 2021;184:3086–3108. doi: 10.1016/j.cell.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–466.e4. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Barnes C.O., Weisblum Y., Schmidt F., Caskey M., Gaebler C., Cho A., Agudelo M., Finkin S., et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 2021;12:4196. doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Schommers P., Lehmann C., Kurth F., Sander L.E., Klein F. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat. Med. 2022;28:477–480. doi: 10.1038/s41591-021-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruell H., Vanshylla K., Weber T., Barnes C.O., Kreer C., Klein F. Antibody-mediated neutralization of SARS-CoV-2. Immunity. 2022;55:925–944. doi: 10.1016/j.immuni.2022.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Gonzalez-Rojas Y., Juarez E., Crespo Casal M., Moya J., Falci D.R., Sarkis E., Solis J., Zheng H., Scott N., et al. Early treatment for Covid-19 with SARS-CoV-2 neutralizing antibody sotrovimab. N. Engl. J. Med. 2021;385:1941–1950. doi: 10.1056/NEJMoa2107934. [DOI] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., et al. COVID-19 Genomics UK (COG-UK) Consortium SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillus D., Schwarz T., Tober-Lau P., Vanshylla K., Hastor H., Thibeault C., Jentzsch S., Helbig E.T., Lippert L.J., Tscheak P., et al. Safety, reactogenicity, and immunogenicity of homologous and heterologous prime-boost immunisation with ChAdOx1 nCoV-19 and BNT162b2: a prospective cohort study. Lancet Respir. Med. 2021;9:1255–1265. doi: 10.1016/S2213-2600(21)00357-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.S., Winkler M.S., et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2022;185:447–456.e11. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iketani S., Liu L., Guo Y., Liu L., Chan J.F., Huang Y., Wang M., Luo Y., Yu J., Chu H., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S., Gurry C., Freitas L., Schultz M.B., Bach G., Diallo A., Akite N., Ho J., Lee R.T., Yeo W., et al. GISAID’s role in pandemic response. China CDC Wkly. 2021;3:1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Kim C., Ryu D.K., Lee J., Kim Y.I., Seo J.M., Kim Y.G., Jeong J.H., Kim M., Kim J.I., Kim P., et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12:288. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura I., Kosugi Y., Wu J., Zahradnik J., Yamasoba D., Butlertanaka E.P., Tanaka Y.L., Uriu K., Liu Y., Morizako N., et al. The SARS-CoV-2 Lambda variant exhibits enhanced infectivity and immune resistance. Cell Rep. 2022;38:110218. doi: 10.1016/j.celrep.2021.110218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreer C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K., et al. Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell. 2020;182:843–854.e12. doi: 10.1016/j.cell.2020.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreye J., Reincke S.M., Kornau H.C., Sánchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.D., et al. A therapeutic non-self-reactive SARS-CoV-2 antibody protects from lung pathology in a COVID-19 hamster model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc M.P. IMGT, the international ImMunoGeneTics information system. Cold Spring Harb. Protoc. 2011;2011:595–603. doi: 10.1101/pdb.top115. [DOI] [PubMed] [Google Scholar]
- Levin M.J., Ustianowski A., De Wit S., Launay O., Avila M., Templeton A., Yuan Y., Seegobin S., Ellery A., Levinson D.J., et al. Intramuscular AZD7442 (tixagevimab-cilgavimab) for prevention of Covid-19. N. Engl. J. Med. 2022;386:2188–2200. doi: 10.1056/NEJMoa2116620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Edwards R.J., Manne K., Martinez D.R., Schäfer A., Alam S.M., Wiehe K., Lu X., Parks R., Sutherland L.L., et al. In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell. 2021;184:4203–4219.e32. doi: 10.1016/j.cell.2021.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236.e13. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Lyngse F.P., Kirkeby C.T., Denwood M., Christiansen L.E., Mølbak K., Møller C.H., Skov R.L., Krause T.G., Rasmussen M., Sieber R.N., et al. Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: evidence from Danish Households. medRxiv. 2022 doi: 10.1101/2022.01.28.22270044. Preprint at. [DOI] [Google Scholar]
- Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S.A., Kwatra G., Myers J.E., Jassat W., Dhar N., Mukendi C.K., Nana A.J., Blumberg L., Welch R., Ngorima-Mabhena N., Mutevedzi P.C. Population immunity and Covid-19 severity with omicron variant in South Africa. N. Engl. J. Med. 2022;386:1314–1326. doi: 10.1056/NEJMoa2119658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Gojobori T., Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S.C.A., Yang F., Jackson K.J.L., Hoh R.A., Röltgen K., Jean G.H., Stevens B.A., Lee J.Y., Rustagi A., Rogers A.J., et al. Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Cell Host Microbe. 2020;28:516–525.e5. doi: 10.1016/j.chom.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Then E., Lucas C., Monteiro V.S., Miric M., Brache V., Cochon L., Vogels C.B.F., Malik A.A., De la Cruz E., Jorge A., et al. Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat. Med. 2022;28:481–485. doi: 10.1038/s41591-022-01705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybould M.I.J., Kovaltsuk A., Marks C., Deane C.M. CoV-AbDab: the coronavirus antibody database. Bioinformatics. 2021;37:734–735. doi: 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RECOVERY Collaborative Group Casirivimab and imdevimab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2022;399:665–676. doi: 10.1016/S0140-6736(22)00163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Muecksch F., Weisblum Y., Da Silva J., Bednarski E., Cho A., Wang Z., Gaebler C., Caskey M., Nussenzweig M.C., et al. Plasma neutralization of the SARS-CoV-2 omicron variant. N. Engl. J. Med. 2022;386:599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques. 2000;28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- Tegally H., Moir M., Everatt J., Giovanetti M., Scheepers C., Wilkinson E., Subramoney K., Makatini Z., Moyo S., Amoako D.G., et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022 doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H.F., Ackerson B.K., Luo Y., Sy L.S., Talarico C.A., Tian Y., Bruxvoort K.J., Tubert J.E., Florea A., Ku J.H., et al. Effectiveness of mRNA-1273 against SARS-CoV-2 Omicron and Delta variants. Nat. Med. 2022;28:1063–1071. doi: 10.1038/s41591-022-01753-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Jr., Purcell L.A., Kawaoka Y., Corti D., Fremont D.H., Diamond M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022;28:490–495. doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanshylla K., Di Cristanziano V., Kleipass F., Dewald F., Schommers P., Gieselmann L., Gruell H., Schlotz M., Ercanoglu M.S., Stumpf R., et al. Kinetics and correlates of the neutralizing antibody response to SARS-CoV-2 infection in humans. Cell Host Microbe. 2021;29:917–929.e4. doi: 10.1016/j.chom.2021.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanshylla K., Fan C., Wunsch M., Poopalasingam N., Meijers M., Kreer C., Kleipass F., Ruchnewitz D., Ercanoglu M.S., Gruell H., et al. Discovery of ultrapotent broadly neutralizing antibodies from SARS-CoV-2 elite neutralizers. Cell Host Microbe. 2022;30:69–82.e10. doi: 10.1016/j.chom.2021.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu M., Shen Y., Ma Y., Li X., Zhang Y., Liu M., Yang X.L., Chen J., Yan R., et al. Novel sarbecovirus bispecific neutralizing antibodies with exceptional breadth and potency against currently circulating SARS-CoV-2 variants and sarbecoviruses. Cell Discov. 2022;8:36. doi: 10.1038/s41421-022-00401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Xiao J., Hooper A.T., Hamilton J.D., Musser B.J., et al. REGEN-COV antibody combination and outcomes in outpatients with Covid-19. N. Engl. J. Med. 2021;385:e81. doi: 10.1056/NEJMoa2108163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Alameh M.G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S., et al. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe. 2021;29:23–31.e4. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf K., Žentelis S., Wang L., Foster D., Vaillancourt P., Wiggin M., Lovett E., van der Lee R., Hendle J., Pustilnik A., et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39:110812. doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wratil P.R., Stern M., Priller A., Willmann A., Almanzar G., Vogel E., Feuerherd M., Cheng C.C., Yazici S., Christa C., et al. Three exposures to the spike protein of SARS-CoV-2 by either infection or vaccination elicit superior neutralizing immunity to all variants of concern. Nat. Med. 2022;28:496–503. doi: 10.1038/s41591-022-01715-4. [DOI] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasoba D., Kimura I., Nasser H., Morioka Y., Nao N., Ito J., Uriu K., Tsuda M., Zahradnik J., Shirakawa K., et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell. 2022;185:2103–2115.e19. doi: 10.1016/j.cell.2022.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Ma N., Madden T.L., Ostell J.M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Ju B., Ge J., Chan J.F., Cheng L., Wang R., Huang W., Fang M., Chen P., Zhou B., et al. Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2. Nat. Commun. 2021;12:4210. doi: 10.1038/s41467-021-24514-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Li D., Ruan W., Chen Z., Zhang R., Zheng A., Qiao S., Zheng X., Zhao Y., Dai L., et al. Effects of a prolonged booster interval on neutralization of omicron variant. N. Engl. J. Med. 2022;386:894–896. doi: 10.1056/NEJMc2119426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw data reported in this paper will be shared by the lead contact upon reasonable request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.