Abstract
Background
Lack of easy-to-interpret disease activity prediction methods in early MS can lead to worse patient prognosis.
Objectives
Using machine learning (multiple kernel learning – MKL) models, we assessed the prognostic value of various clinical and MRI measures for disease activity.
Methods
Early MS patients (n = 148) with at least two associated clinical and MRI visits were investigated. T2-weighted MRIs were cropped to contain mainly the lateral ventricles (LV). High disease activity was defined as surpassing NEDA-3 Criteria more than once per year. Clinical demographic, MRI-extracted image-derived phenotypes (IDP), and MRI data were used as inputs for separate kernels to predict future disease activity with MKL. Model performance was compared using bootstrapped effect size analysis of mean differences.
Results
A total of 681 visits were included, where 81 (55%) patients had high disease activity in a combined end point measure using all follow-up visits. MKL model discrimination performance was moderate (AUC ≥ 0.62); however, modelling with combined clinical and cropped LV kernels gave the highest prediction performance (AUC = 0.70).
Conclusions
MRIs contain valuable information on future disease activity, especially in and around the LV. MKL techniques for combining different data types can be used for the prediction of disease activity in a relatively small MS cohort.
Keywords: Relapsing/remitting, MRI, disease activity prediction, multiple kernel learning, lateral ventricles, machine learning
Introduction
Currently, the lack of machine learning (ML) models that can be used in MS for predicting disease activity or prognosis could be due to the heterogeneity of disease cohorts and the variability of extracted measures from clinical questionnaires and MRI.1–3 Multiple kernel learning (MKL) offers the possibility to combine different data types represented by separate kernels4,5 to fit the prediction analyses.6,7 ML kernels are well-suited for analysing complex patterns in raw data. 8
With better prediction models for early MS patients using routine MRI data, it may become possible to make more personalised treatment decisions and prevent further relapses or future disease disability. It has been shown that the lateral ventricles (LV) contract and expand over the course of early MS, whereas patients with higher ventricular CSF volume change had lower disability scores. 9 Also, areas around the ventricles were found to be most predictive in discriminating between MS patients and healthy participants using a deep learning algorithm. 10 Recently, the choroid plexus (CP) within the LV was shown to have significantly higher gadolinium-enhancing (Gd) intensities in MS patients versus healthy participants, thus this region could be related to disease activity. 11 Rather than only extracting measures such as lesion load or brain volumetrics that are reliant on many intensive preprocessing and partially manual steps, using raw-MRIs centred and cropped at the LV-CP region could result in better modelling by ML algorithms. In this study, we aim to predict the level of disease activity in early relapsing-remitting MS (RRMS) patients using MKL, allowing for the combination of clinical, image-derived, and raw-MRI data. We hypothesise that clinical data, MRI-extracted features, and raw-MRIs cropped at the LV-CP region will yield clinically relevant models for the prediction of future disease activity using MKL models.
Methods
Study population
Patient data were acquired from our ongoing, longitudinal observational early MS cohort (Berlin of Clinically Isolated Syndrome (CIS) cohort: ClinicalTrials.gov Identifier: NCT01371071, start date: January 2011). The study includes patients over the age of 18, with a diagnosis CIS or RRMS according to the McDonald 2017 diagnostic criteria. 12 We screened 168 patients for the availability of at least two MRI scans performed within three months of clinical assessments. Criteria were met for 148 patients with CIS (N = 39) or early RRMS (N = 109). This study was approved by the local ethics committee (EA1/182/10) and conducted in accordance with the current applicable version of the Declaration of Helsinki and German law. All participants provided written informed consent. All relevant patient baseline clinical and MRI-derived information are shown in Table 1.
Table 1.
CIS and early RRMS cohort baseline characteristics.
| High disease activity (n = 81) | Low disease activity (n = 67) | All patients (n = 148) | Statistics | |
|---|---|---|---|---|
| Age (years) [mean ± SD] | 32.8 ± 8.2 | 34.4 ± 9.4 | 33.5 ± 8.8 | NA |
| Sex [F:M (%)] | 55:26 (68:32) | 42:25 (63:37) | 97:51 (66:34) | χ2=14.287, p=1.56e-04 |
| McDonald 2017 Diagnosis [CIS:RRMS (%)] | 19:62 (23:77) | 20:47 (30:70) | 39:109 (26:74) | χ2=33.108, p=8.717e-09 |
| Disease Duration at Baseline (months) [median (range)] | 4.7 (0–28) | 5 (1–35) | 4 (0–35) | t = −1.802, p =0.962 |
| EDSS Score at Baseline [median (range)] | 1.5 (0–4) | 1.5 (0–4) | 1.5 (0–4) | t = −1.174, p =0.879 |
| Follow-up visits (N) [median (range)] | 4 (2–9) | 5 (2–10) | 4 (2–10) | t = −2.313, p =0.989 |
| MS Therapies (N) [%] | Dimethylfumerate: 9 (11) Glatirameracetat: 15 (19)
Interferon Beta: 10 (12) NA: 47 (58) |
Dimethylfumerate: 3 (5) Glatirameracetat: 10 (15) Interferon
Beta: 4 (6) Natalizumab: 1 (1) NA: 49 (73) |
Dimethylfumerate: 12 (8) Glatirameracetat: 25 (17)
Interferon Beta: 14 (9.4) Natalizumab: 1 (0.6) NA: 96
(65) |
NA |
| Disease Activity based on NEDA-3 Status at each time point (N) [high:low (%)] | NA | NA | 81:67 (55:45) | χ2 = 1.324, p =0.249 |
| Brain T2-hyperintense lesion count (N) [median (range)] | 11 (1–83) | 9 (0–217) | 10 (0–217) | t = 0.115, p =0.454 |
Chi-squared test statistics were calculated only using data from the all patients column, not within each disease activity group. Welch two-sample t-tests were performed only between high and low disease activity groups. Bolded text indicates statistical significance (p < 0.05). It should be noted that for MS Therapies, NA denotes no treatment, since most patients at baseline were originally included into the observational study as CIS patients.
CIS: clinically isolated syndrome; RRMS: relapsing-remitting MS; EDSS: expanded disability status scale; NEDA-3: no evidence of disease activity; SD: standard deviation.
MRI acquisition
This study utilised longitudinally acquired MRIs with a protocol that included a 3D magnetization prepared rapid acquisition gradient echo (MPRAGE) sequence (1 mm isotropic resolution, TR = 1900 ms, TE = 3.03 ms, TI = 900 ms), and a 3D fluid-attenuated inversion recovery (FLAIR) sequence (1 mm isotropic resolution, TR = 6000 ms, TE = 388 ms, TI = 2100 ms) using a 3 Tesla Tim Trio MRI (Siemens Medical Systems, Erlangen, Germany).
MRI preprocessing
Preprocessing included white and grey matter brain masking 13 for brain extraction, N4-bias field correction 14 and linear, rigid body registration of FLAIR images to MPRAGE images. 15 MPRAGE and FLAIR scans were longitudinally co-registered as described in Cooper et al. 16 Cerebral Gd- and T2-hyperintense lesions were segmented manually and saved as binary masks using ITK-SNAP (www.itksnap.org) by two expert MRI technicians (greater than 10 years of experience). Longitudinally co-registered MPRAGEs were lesion-filled (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/lesion_filling) and used for whole brain, white and grey matter and CSF normalised volume extraction. 17 Each lesion from lesion masks were thresholded to contain at least 15 voxels to reduce the chance of including very small non-specific lesions and/or hyperintense-artefacts for lesion count and volume extraction (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/Fslutils). Brain-extracted FLAIRs were centred at the LV-CP and cropped by reducing the dimensions to 60 × 54 × 2 mm, using an in-house Python script. This size was chosen, as it effectively reduces the dimensionality of the image for ML processing and contains the most MRI slices with the LV and CP in view.
NEDA-3 criteria
We used a combined endpoint measure to define no evidence of disease activity (NEDA). NEDA-3 can be defined as a lack of clinical relapses, a lack of disease progression measured by the expanded disability status scale (EDSS) and an absence of new brain lesions (new/enlarged T2-hyperintense and/or Gd-enhancing lesion) in an MRI after the baseline visit. In the current study, NEDA-3 status is calculated between consecutive visits and not from the baseline MRI visit. Thus, patients may fail NEDA-3 at each follow-up session, indicating an increase in disease activity.
To maintain NEDA-3 status at each time point, a patient must have:
No new occurrence of relapses
No disability progression in terms of increases in EDSS scores. A binary variable for disability progression is introduced based on different thresholds 18 depending on the EDSS score at the previous time point.
No increases in the counts or volumes of T2-lesions and Gd-enhancing lesions from brain MRIs.
The above criteria were used to determine the NEDA-3 status at each follow-up visit. Since Gd MRIs were not collected for all patients at every time point, this information is only used for provided cases and ignored otherwise.
Future high and low disease activity
A binary measure of low (0) or high (1) disease activity for each patient was calculated. First, NEDA-3 status was evaluated per patient session, then all instances where NEDA-3 was surpassed were summed for each patient. The number of instances was divided by the total time in years from baseline to last follow-up visit using this formula:
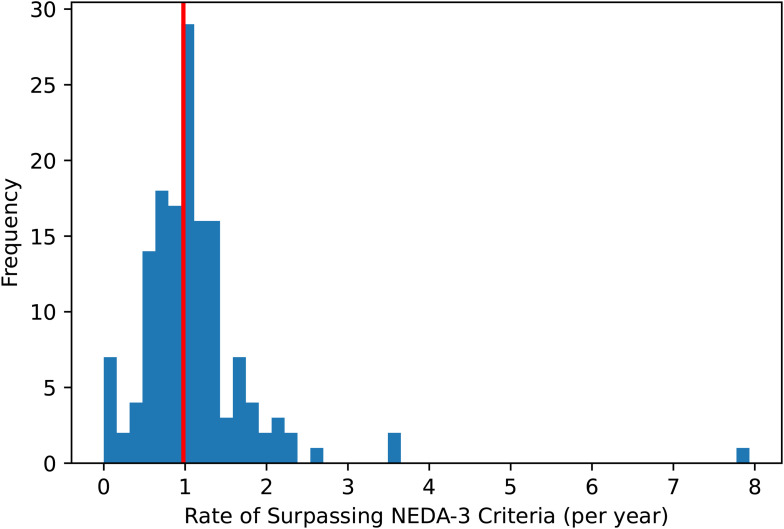
Values greater than 1 may occur, depending on the frequency of follow-up visits. For example, one patient had two visits 0.126 years apart, where NEDA-3 was surpassed in the second visit. This would give an annualised NEDA-3 of 1/0.126 = 7.93. Thus, this calculation is able to account for the temporal aspect of the prediction, as well as adjusting for differences in follow-up times and visits for each patient. The annualised NEDA-3 distribution of all patients showed centrality around 1 surpassing event per year (Figure 1). High disease activity was counted as ≥1 surpassing event per year, and low disease activity was designated as <1 surpassing event per year.
Figure 1.
Rate of surpassing NEDA-3 criteria per year (annualised NEDA-3) calculated in the cohort of patients with at least 2 clinical visits. The red solid line denotes the threshold for calculating high (≥1) and low (<1) disease activity.
Input features for the ML model
The prediction of low and high disease activity was based on the following sets of baseline features:
Clinical-related (Clin) – four variables: age, disease duration from first manifestation in months, EDSS score, sex
Image-derived phenotypes (IDP) – five variables: T2-hyperintense brain lesion count, total brain, brain grey, white matter and CSF normalised volumes
Raw-MRIs (MRI) – 6480 variables (intensity value per voxel): flattened one-dimensional vector from FLAIR images centred on the CP and cropped around the LV
Each set of features (Clin, IDP, and MRI) were standardised using sklearn StandardScaler 19 and input into separate radial basis function (RBF) kernels for fitting to the ML algorithm.
Mklaren model
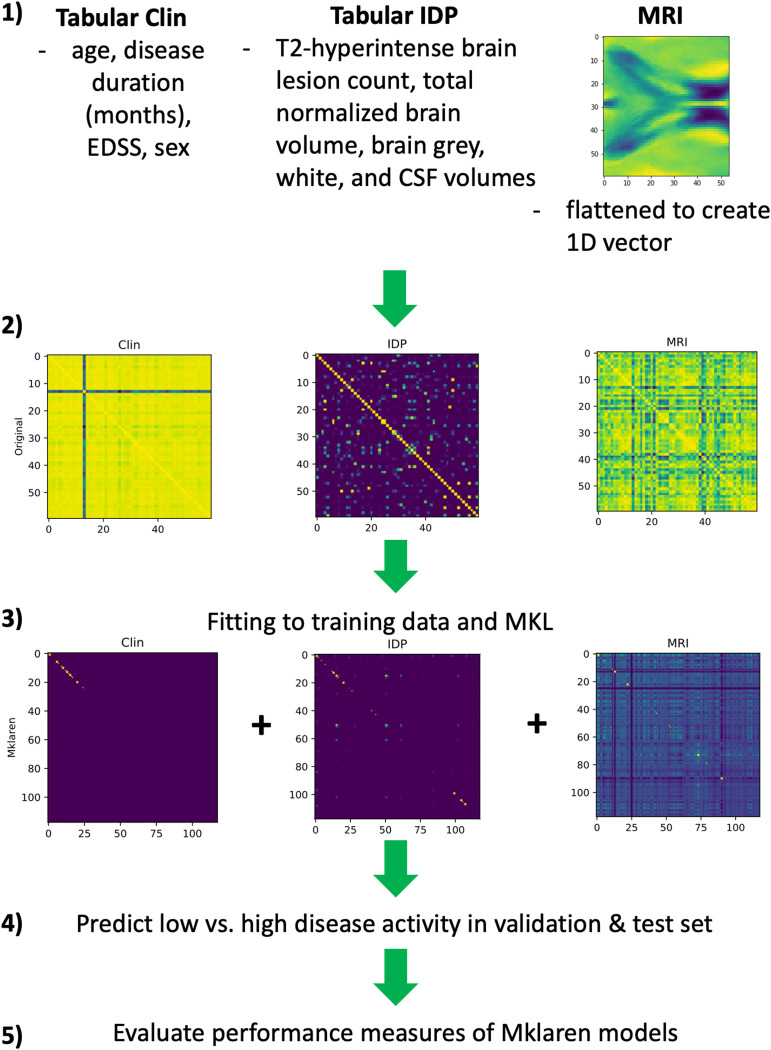
We used the Mklaren (Multiple kernel learning with least-angle regression) algorithm, which performs simultaneous low-rank approximation of MKL with least-angle regression to find the best combination of RBF kernel matrix columns to use for training and prediction (6). We optimised all available hyperparameters in this algorithm (Supplemental Table 1). Different combinations of RBF kernels were used to create the following Mklaren models (Figure 2):
Only Clin
Only IDP
Only MRI
Clin and IDP
Clin and MRI
IDP and MRI
Clin, IDP and MRI
Figure 2.
Outline of experimental design for MKL. (1) Types of different data used as inputs for (2) RBF kernels, which were then used for (3) fitting to training data and MKL. Kernels that are highly related to target outcomes are approximated with better accuracies. Combined approximations are used to (4) predict low vs. high disease activity in the validation and test set, and (5) calculate and evaluate performance measures for the Mklaren models.
EDSS: expanded disability status scale; CSF: cerebral spinal fluid; 1D: 1-dimensional; AUC: area under the curve; Clin: clinical data; IDP: image-derived phenotypes; MRI: cropped MRIs at the lateral ventricles; RBF: radial basis function; MKL: multiple kernel learning.
Interchangeably using IDP and MRI features allowed for an indirect comparison of whether non-extracted MRI measures (MRI) could be used in predictive models as well as preprocessed and MRI-extracted measures.
Statistical analysis
Chi-squared and Welch two-sample t-tests were performed to test for significant differences between cohort demographics (Table 1). Ten repetitions of five-fold nested cross-validation were performed for each Mklaren model. Outer and inner loops were split using stratified K-folds, 19 to give balanced sex and outcome measure grouping in training and validation/test sets. The inner loops were used to select models with hyperparameter sets that gave the Mklaren model with the highest validation receiver operating characteristic area under the curve (AUC) and lowest root mean square error. Since the Mklaren algorithm is a regression model, and our task is a classification, we used the validation AUC from each inner loop selected model to calculate an optimal value (using an in-house python script) that was applied to the regression output for binarising into high and low disease activity classes. In addition, we thresholded the selection of the top AUCs from each inner loop to be above 0.55, to reduce the possibility of selecting models which learned the classification based on chance (Supplemental Statistical Analysis Text). The identified sets of hyperparameters per inner loop were then used on the outer loop to obtain performance measures (test AUC, balanced accuracy, sensitivity, specificity). The mean performance measures per outer loop were then calculated (Figure 2). Performance measures were calculated using standard sklearn metrics tools. 19 To compare the AUC, balanced accuracy, sensitivity, and specificity of each different Mklaren model, mean differences between each model (Mklaren models ii – vii) against the model using only clinical data (Mklaren model i) were calculated using 5000 bootstrap resamples in R. 20 Kaplan–Meier estimation analysis 21 was used to evaluate whether baseline low (< mean of all mean intensity measures in the cohort) versus high mean intensities in the cropped MRIs have different risk of failing NEDA-3 Criteria. To investigate how mean intensity measures in the cropped MRIs compare between high and low disease activity patients, a Welch's two-sample t-test was used. Statistical significance was set to p < 0.05 for all tests. All statistics on demographics, Kaplan–Meier, performance measures, and t-tests were performed using R version 4.1.0 (https://www.r-project.org/).
Data availability
Persons interested in obtaining access to the data should contact Friedemann Paul (friedemann.paul@charite.de).
Results
Cohort demographics
As expected, the cohort included larger proportions of females and RRMS patients compared to CIS (Table 1). To account for sex, we stratified our training, validation and test sets for sex and outcome measure to reduce bias in model fitting and identification of sex as a predictor. In terms of diagnosis, we did not subset the group of CIS patients since only 18 patients (12%) in their last visit remained CIS, and these patients represent normal variation in an early MS cohort.
High versus low disease activity patients
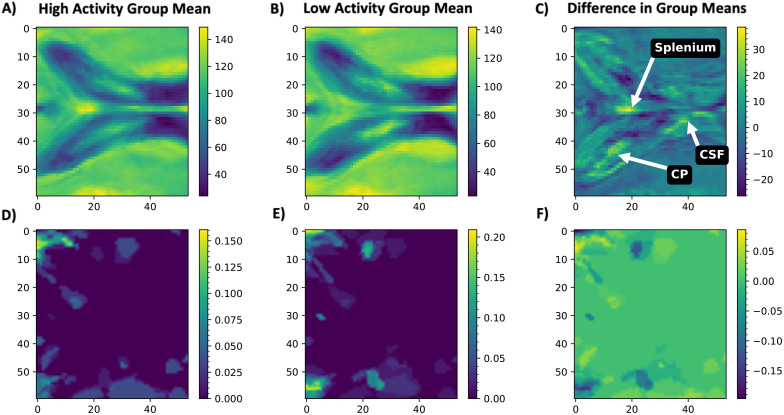
We visually inspected the 3D cropped raw FLAIR LV region, which was imported as arrays of intensity values, where the mean was calculated using all imported image arrays of high and low disease activity patients at baseline, respectively (Figure 3). An average increase in image intensity was observed in the patients with high disease activity (Figure 3C), confirming that this region could be of interest for MKL. Mean lesion masks in the same cropped regions from each group were assessed by importing as arrays, giving Figure 3D to F.
Figure 3.
Mean raw T2-weighted 3D FLAIR images cropped to contain only the lateral ventricles and centred on the choroid plexus. Shown are the centre slice of the mean cropped MRI for: (A) high disease activity patients, (B) low disease activity patients, and (C) the subtracted mean image (low subtracted from high disease activity). Mean lesion masks in the same cropped regions were created for (D) high disease activity and (E) low disease activity patients, and (F) the subtracted mean image (low subtracted from high disease activity). This mean difference in lesion masks visually indicates similar mean lesion load and lesion locations between the two groups. However, it can be seen in (C) that it seems there may be a higher incidence of T2-hyperintense lesions in the splenium of the corpus callosum, along with higher intensities in the choroid plexus and lateral ventricular CSF, in patients with high disease activity. Colour bars indicate intensity measures in first row of mean raw cropped-FLAIR images (A-C) and indicate an average incidence of lesions in specific voxels in the second row of mean lesion masks (D-F).
CP: corpus callosum; CSF: cerebrospinal fluid.
MKL experiments
Table 2 shows the performance measures of each model used to predict high versus low disease activity in this early RRMS cohort. The effect sizes of the mean differences between performance measures of each model (ii – vii) from model i (only clinical tabular data) are included.
Table 2.
Performance measures and effect sizes of mean differences of Mklaren models used for prediction of high versus low disease activity.
| Performance measures | Tabular clinical data kernel | Tabular IDP kernel | MRI kernel | Clinical + IDP kernels | Clinical + MRI kernels | IDP + MRI kernels | Clinical + IDP + MRI (all kernels) |
|---|---|---|---|---|---|---|---|
| Mean AUC ± SD (Effect-size [95% CI]) | 0.62 ± 0.06 (NA) | 0.62 ± 0.05 (−0.005 [−0.028 to 0.016]) | 0.65 ± 0.06 (0.028 [0.004 to 0.050]) | 0.63 ± 0.06 (0.011 [−0.013 to 0.033]) | 0.70 ± 0.07 (0.081 [0.056 to 0.11]) | 0.66 ± 0.04 (0.039 [0.017 to 0.059]) | 0.70 ± 0.05 (0.073 [0.052 to 0.094]) |
| Mean Balanced Accuracy ±SD (Effect-size [95% CI]) | 0.55 ± 0.06 (NA) | 0.51 ± 0.04 (−0.036 [−0.054 to 0.018]) | 0.58 ± 0.07 (0.038 [0.013 to 0.06]) | 0.55 ± 0.07 (0.004 [−0.020 to 0.028]) | 0.63 ± 0.07 (0.084 [0.061 to 0.11]) | 0.58 ± 0.07 (0.032 [0.007 to 0.055]) | 0.61 ± 0.06 (0.065 [0.043 to 0.089]) |
| Mean Sensitivity ±SD (Effect-size [95% CI]) | 0.45 ± 0.38 (NA) | 0.62 ± 0.42 (0.173 [0.017 to 0.33]) | 0.61 ± 0.17 (0.163 [0.045 to 0.27]) | 0.57 ± 0.31 (0.132 [−0.008 to 0.258]) | 0.52 ± 0.17 (0.076 [−0.039 to 0.186]) | 0.60 ± 0.29 (0.153 [0.015 to 0.283]) | 0.57 ± 0.23 (0.122 [−0.006 to 0.238]) |
| Mean Specificity ±SD (Effect-size [95% CI]) | 0.64 ± 0.41 (NA) | 0.40 ± 0.43 (−0.245 [−0.41 to −0.08]) | 0.56 ± 0.20 (−0.087 [−0.207 to 0.044]) | 0.52 ± 0.37 (−0.124 [−0.27 to 0.030]) | 0.73 ± 0.17 (0.092 [−0.022 to 0.214]) | 0.55 ± 0.33 (−0.089 [−0.229 to 0.057]) | 0.65 ± 0.21 (0.009 [−0.11 to 0.143]) |
Bolded text indicates highest effect sizes of mean differences in performance measures in comparison to using kernels input with only tabular clinical data.
AUC: area under the curve; SD: standard deviation; CI: confidence interval; IDP: image-derived phenotypes; MRI: cropped MRIs at the lateral ventricles.
It can be seen from the effect size analysis, the model fit with both tabular clinical and cropped raw-MRI of the LV (Clin + MRI) gave the highest mean difference against models using only tabular clinical data (Supplemental Figure 1). Using these kernels in the Mklaren model gave the highest mean performance measures for predicting high disease activity in CIS and RRMS patients (AUC = 0.70, balanced accuracy = 0.63, and specificity = 0.73). This model did not have the highest sensitivity in the tested models. This would suggest that the model with clinical and raw MRI data is not able to predict true positives of high disease activity as well as models using IDP data combinations, however, was better at distinguishing true negatives.
Cropped T2-weighted raw-MRIs around the lateral ventricles
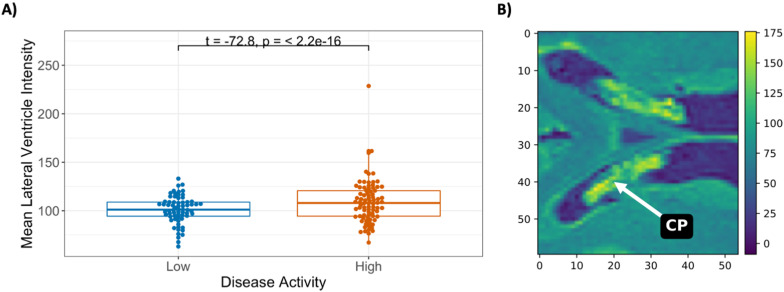
Baseline mean intensities of the cropped MRI region, stratified by low (patient baseline mean intensity < mean of all baseline mean intensities) and high values, were evaluated to see whether intensity groupings could distinguish between a higher risk of failing the NEDA-3 Criteria from baseline (Supplemental Figure 2). There was no significant difference in risk between the LV MRI intensity groups. However, it can be seen that in our cohort 50% of patients risk failing NEDA-3 Criteria around six months after baseline. We investigated further if the baseline mean intensities from the cropped MRIs around the LV showed differences between low and high disease activity patient groups using a Welch's two-sample t-test (Figure 4).
Figure 4.
(A) Mean intensity values from cropped MRIs of the lateral ventricle regions between low and high disease activity patient groups at baseline MRI. (B) Image of the highest mean intensity cropped raw FLAIR (from the high disease activity group), showing low-intensity values in the lateral ventricle CSF and surrounding white and grey matter, however, the choroid plexus has high intensities. The colour bar in part (B) indicates the intensity measures in the raw cropped-FLAIR image.
CSF: cerebrospinal fluid; CP: choroid plexus.
Figure 4B shows the hyperintense CP of a high disease activity patient with the highest mean image intensity shown in Figure 4A. Although N4-bias correction (14) of the FLAIR MRIs was performed during the MRI preprocessing stage, there are higher intensities from this LV region in patients with high disease activity (t = −64.1, p ≤ 2.2 × 10−16).
Discussion
Our study showed that the MKL Mklaren method can aid in the prediction of future disease activity in CIS and early RRMS patients. In addition, we trained MKL models using raw, cropped T2-FLAIR MRIs, allowing for the use of minimally preprocessed, clinically relevant MRIs in an ML classification task. RBF kernels combining baseline clinical (age, sex, disease duration, EDSS score) with cropped MRIs at the LV region gave the highest predictive performance measures for high vs. low disease activity. We also evaluated the mean intensity measures of the cropped LV regions, centred on the CP, which showed higher mean baseline MRI intensities in patients classified with high disease activity, than patients classified with low disease activity.
We did not see the utility of tabular clinical data alone in our Mklaren models for predicting future disease activity; however, it did give models with the highest specificity compared to others using single data types. The Mklaren models including IDP had slightly higher sensitivity, suggesting true positive (or high disease activity) are better identified with the inclusion of MRI-extracted information, which is in-line with other MS ML studies.22–24 Some other large studies were able to discern the probability of disease progression to secondary progressive MS using ML methods solely with baseline EDSS and age.25,26 Brain age in MS has become an important line of research, and age is associated with brain atrophy rates, thus it is important to continue to include age in MRI studies.27,28 Using an MKL approach with clinical tabular and IDP data may be useful in classification tasks within smaller cohorts.
Typical IDP data for ML prediction tasks involve brain-extracted and advanced MRI-extracted measures, such as connectivity measures based on lesions in the white matter.23,29 This is not only time-consuming to pre-process and post-process the MRI data, there is a need to reduce dimensionality for ML modelling. 30 In our study, the Mklaren MKL algorithm automatically combines the data by fitting RBF kernels to the task using simultaneous low-rank approximation with least angle regression. This reduces the computational complexity (O(n2)) of the model allowing for large amounts of observations to be analysed, such as that from raw-MRIs, where the removal of kernel columns that do not aid in the prediction task is also performed. 31 Since kernels are based on each different type of data, this approach is flexible for use in different cohorts with different types of observations. As discussed in Tozlu et al., 29 advanced MRI techniques until now are mainly for research purposes and thus, we need to shift towards a more clinically applicable tool, such as one able to read raw MRI data, for patient evaluation.
The LV intensities from FLAIR MRIs are a region of high interest in evaluating future disease activity in MS. We observed in patients classified as having high disease activity, based on the annualised NEDA-3 score, significantly higher mean intensities in the LV-CP compared to patients with low disease activity. Our visual evaluation of the LV region also showed that there may be a higher incidence of lesions/T2-hyperintensities in the splenium of the corpus callosum in high disease activity patients, consistent with a previous study that found higher disability scores in patients with white matter damage in the splenium. 32 Baseline mean intensity measures of the LV region did not show a different risk of failing NEDA-3 Criteria in a Kaplan–Meier analysis. However, 50% of the entire patient cohort risked evidence of disease activity six months after baseline, highlighting the need for early treatment to avoid further MS-related disease activity. We posit that for better prediction of future disease activity using MKL and information from the LV, patients may decide for disease-modifying therapy prior to their next relapse or MRI finding. The CP in the LV is pertinent to investigate since increased CP volume has been shown to be associated with inflammation in MS patients33,34 although, studies have found LV and CP enlargement from normal aging.35,36 In our study, we included LV intensities as a feature because a recent study showed increased Gd-enhancement of the LV-CP in MS patients. 11 Meanwhile, another study found increased intensity in FLAIR MRI from higher protein concentration detection usually suppressed in the CSF at normal concentrations. 37 Importantly, it has been found that the pattern of neurodegeneration in MS is associated with the load of inflammatory infiltrates in the CSF, such as chemokines that are produced by the CP and then affecting the LV regions. 38 Thus, along with previous findings that the ventricular volume in early RRMS patients transiently fluctuate, 9 our findings using raw-MRI data even without contrast enhancement, support observations that disease activity may be related to changes in the CP and LV. Our results could lead to a more easily accessible clinical tool, whereby patient raw MRIs (i.e. T2-FLAIR) are input directly from the scanner into an automated pipeline that evaluates the risk of a patient having high disease activity in the future.
One limitation of our study was the small cohort size for an ML prediction task. For small patient cohorts, not only could the dataset have low statistical power, the variability in the data splitting for ML training, as well as the choice of features, can lead to unstable performance results. 39 We decreased bias in the training, validation, and test sets of the data by stratifying splitting based on both the outcome variable (low and high disease activity groupings) and sex, which is often an important confounder of MRI brain data. 40 Since the Mklaren algorithm allows for flexibility in using large numbers of observations for prediction, it was also a well-suited choice for our small dataset. Although other types of ML methods such as ensemble learning could also be of value, as they are not limited to just one type of algorithm or model per class of data, this advanced modelling would require much more post-processing of the models and data. 41 Therefore, we believe using our stratification strategy with MKL for the prediction of disease activity in MS was a sound choice.
Another limitation of our study includes the difference in the number of follow-up visits between patients. To account for this, we calculated an annualised NEDA-3 score, which allowed us to evaluate at each visit, whether a patient surpassed the NEDA-3 Criteria. This was further binarised into patients with high activity (annualised NEDA-3 score ≥1) and low activity based on the overall distribution of the score in our cohort. Thus, we could still test for future disease activity using all the available follow-up visit data, which was successfully predicted with moderate performance using tabular clinical and raw-MRI data.
Conclusion
MKL can be applied to clinical, MRI-extracted, and raw T2-weighted MRI data for the prediction of future disease activity in CIS and early RRMS patients. MKL with individual kernels for different types of data may allow for a better and more personalised assessment of disease course. Automatically analysing raw-MRI data, especially in the LV/CP region could enable clinicians and patients to make treatment decisions based on probable future disease activity. These methods, including evaluation of feature importance, and brain regions warrant further investigation in larger MS studies.
Supplemental Material
Supplemental material, sj-docx-1-mso-10.1177_20552173221109770 for Prediction of high and low disease activity in early MS patients using multiple kernel learning identifies importance of lateral ventricle intensity by Claudia Chien, Moritz Seiler, Fabian Eitel, Tanja Schmitz-Hübsch, Friedemann Paul and Kerstin Ritter in Multiple Sclerosis Journal – Experimental, Translational and Clinical
Supplemental material, sj-tiff-2-mso-10.1177_20552173221109770 for Prediction of high and low disease activity in early MS patients using multiple kernel learning identifies importance of lateral ventricle intensity by Claudia Chien, Moritz Seiler, Fabian Eitel, Tanja Schmitz-Hübsch, Friedemann Paul and Kerstin Ritter in Multiple Sclerosis Journal – Experimental, Translational and Clinical
Supplemental material, sj-tiff-3-mso-10.1177_20552173221109770 for Prediction of high and low disease activity in early MS patients using multiple kernel learning identifies importance of lateral ventricle intensity by Claudia Chien, Moritz Seiler, Fabian Eitel, Tanja Schmitz-Hübsch, Friedemann Paul and Kerstin Ritter in Multiple Sclerosis Journal – Experimental, Translational and Clinical
Supplemental material, sj-docx-4-mso-10.1177_20552173221109770 for Prediction of high and low disease activity in early MS patients using multiple kernel learning identifies importance of lateral ventricle intensity by Claudia Chien, Moritz Seiler, Fabian Eitel, Tanja Schmitz-Hübsch, Friedemann Paul and Kerstin Ritter in Multiple Sclerosis Journal – Experimental, Translational and Clinical
Supplemental material, sj-docx-5-mso-10.1177_20552173221109770 for Prediction of high and low disease activity in early MS patients using multiple kernel learning identifies importance of lateral ventricle intensity by Claudia Chien, Moritz Seiler, Fabian Eitel, Tanja Schmitz-Hübsch, Friedemann Paul and Kerstin Ritter in Multiple Sclerosis Journal – Experimental, Translational and Clinical
Supplemental material, sj-pdf-6-mso-10.1177_20552173221109770 for Prediction of high and low disease activity in early MS patients using multiple kernel learning identifies importance of lateral ventricle intensity by Claudia Chien, Moritz Seiler, Fabian Eitel, Tanja Schmitz-Hübsch, Friedemann Paul and Kerstin Ritter in Multiple Sclerosis Journal – Experimental, Translational and Clinical
Acknowledgements
The authors would like to thank all patients who volunteered for this observational study and the study nurses, MRI technicians (C. Kraut and S. Pikol), the Berlin Center for Advanced Neuroimaging, and residents who aided in collecting the clinical data and the NeuroCure Clinical Research Center for infrastructure and regulatory support.
Footnotes
Declaration of conflicting interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: C.C. has received speaking honoraria from Bayer and funding for research by Novartis, unrelated to this study. T.S.H has received speaker honoraria from Bayer, travel grant from Celgene, and funding for research by Roche and Celgene, unrelated to this study. F.P. receives honoraria for lecturing, and travel expenses for attending meetings from Guthy Jackson Foundation, Bayer, Biogen, Merck Serono, Sanofi Genzyme, Novartis, Alexion, Viela Bio, Roche, UCB, Mitsubishi Tanabe and Celgene. His research is funded by the German Ministry for Education and Research (BMBF), Deutsche Forschungsgemeinschaft (DFG), Einstein Foundation, Guthy Jackson Charitable Foundation, EU FP7 Framework Program, Biogen, Genzyme, Merck Serono, Novartis, Bayer, Alexion, Roche, Parexel and Almirall.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Deutsche Multiple Sklerose Gesellschaft (DMSG), Deutsche Forschungsgemeinschaft (DFG grant number 389563835), Fondation Eugène Devic EDMUS contre la Sclérose en Plaques & Observatoire Français de la Sclérose en Plaques (grant name DEEP MS), and the German Research Foundation. We acknowledge financial support from the Open Access Publication Fund of Charité – Universitätsmedizin Berlin and the German Research Foundation (DFG).
ORCID iD: Claudia Chien https://orcid.org/0000-0001-8280-9513
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Claudia Chien, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, ECRC Experimental and Clinical Research Center, Berlin, Germany; Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, NeuroCure Clinical Research Center, Berlin, Germany; Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Department of Psychiatry and Neurosciences, Berlin, Germany.
Fabian Eitel, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Department of Psychiatry and Neurosciences, Berlin, Germany; Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Bernstein Center for Computational Neuroscience, Berlin Center for Advanced Neuroimaging, Berlin, Germany.
Tanja Schmitz-Hübsch, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, ECRC Experimental and Clinical Research Center, Berlin, Germany; Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, NeuroCure Clinical Research Center, Berlin, Germany.
Friedemann Paul, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, ECRC Experimental and Clinical Research Center, Berlin, Germany; Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, NeuroCure Clinical Research Center, Berlin, Germany; Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Department of Neurology, Berlin, Germany.
Kerstin Ritter, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Department of Psychiatry and Neurosciences, Berlin, Germany; Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Bernstein Center for Computational Neuroscience, Berlin Center for Advanced Neuroimaging, Berlin, Germany.
References
- 1.Pellegrini F, Copetti M, Sormani MP, et al. Predicting disability progression in multiple sclerosis: insights from advanced statistical modeling. Mult Scler 2020; 26: 1828–1836. [DOI] [PubMed] [Google Scholar]
- 2.Moazami F, Lefevre-Utile A, Papaloukas C, et al. Machine learning approaches in study of multiple sclerosis disease through magnetic resonance images. Front Immunol 2021; 12: 3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seccia R, Romano S, Salvetti Met al. et al. Machine learning use for prognostic purposes in multiple sclerosis. Life (Basel ) 2021; 11: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu S, Falck T, Daemen A, et al. L2-norm multiple kernel learning and its application to biomedical data fusion. BMC Bioinformatics 2010; 11: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen T, Zeng D, Wang Y. Multiple kernel learning with random effects for predicting longitudinal outcomes and data integration. Biometrics 2015; 71: 918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stražar M, Curk T. Approximate multiple kernel learning with least-angle regression. Neurocomputing 2019; 340: 245–258. [Google Scholar]
- 7.Lauriola I, Polato M, Aiolli F. The minimum effort maximum output principle applied to multiple kernel learning. Comput Intell 2018; 6: 183–188. [Google Scholar]
- 8.Schölkopf B, Schölkopf D of the MPI for I in TGP for MLB, Schölkopf Bernhard, Smola AJ, Bach F, Scholkopf MD of the MPI for BC in TGPB. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond. MIT Press; 2002. 658.
- 9.Millward JM, Delgado PR, Smorodchenko A, et al. Transient enlargement of brain ventricles during relapsing-remitting multiple sclerosis and experimental autoimmune encephalomyelitis. JCI Insight [Internet ] 2020; 5(21): 1–21. Available from: https://insight.jci.org/articles/view/140040 (accessed 6 November 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eitel F, Soehler E, Bellmann-Strobl J, et al. Uncovering convolutional neural network decisions for diagnosing multiple sclerosis on conventional MRI using layer-wise relevance propagation. Neuroimage Clin 2019; 24: 102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H, Lim Y-M, Kim G, et al. Choroid plexus changes on magnetic resonance imaging in multiple sclerosis and neuromyelitis optica spectrum disorder. J Neurol Sci 2020. [cited 2021 Feb 27]; 415: 116904.. Available from: https://www.jns-journal.com/article/S0022-510X(20)30241-0/abstract [DOI] [PubMed] [Google Scholar]
- 12.Thompson AJ, Banwell BL, Barkhof F, et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol 2018; 17: 162–173. [DOI] [PubMed] [Google Scholar]
- 13.Tohka J, Zijdenbos A, Evans A. Fast and robust parameter estimation for statistical partial volume models in brain MRI. Neuroimage 2004; 23: 84–97. [DOI] [PubMed] [Google Scholar]
- 14.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 2010; 29: 1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004; 23: S208–S219. [DOI] [PubMed] [Google Scholar]
- 16.Cooper G, Chien C, Zimmermann H, et al. Longitudinal analysis of T1w/T2w ratio in patients with multiple sclerosis from first clinical presentation. Mult Scler 2021; 27: 2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith SM, Zhang Y, Jenkinson M, et al. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. Neuroimage 2002; 17: 479–489. [DOI] [PubMed] [Google Scholar]
- 18.Zimmermann HG, Knier B, Oberwahrenbrock T, et al. Association of retinal ganglion cell layer thickness with future disease activity in patients with clinically isolated syndrome. JAMA Neurol 2018; 75: 1071–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in python. Machine Learning in Python; 6: 2825–2830. [Google Scholar]
- 20.Ho J, Tumkaya T, Aryal Set al. et al. Moving beyond P values: data analysis with estimation graphics. Nat Methods 2019; 16: 565–566. [DOI] [PubMed] [Google Scholar]
- 21.Kassambara A, Kosinski M, Biecek P. Survminer: drawing survival curves using ‘ggplot2’. R package version 0.4.9. [Internet]. 2021. Available from: https://rpkgs.datanovia.com/survminer/index.html
- 22.Wottschel V, Alexander DC, Kwok PP, et al. Predicting outcome in clinically isolated syndrome using machine learning. Neuroimage Clin 2015; 7: 281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tozlu C, Jamison K, Nguyen T, et al. Structural disconnectivity from paramagnetic rim lesions is related to disability in multiple sclerosis. Brain Behav 2021; 11: e2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bendfeldt K, Taschler B, Gaetano L, et al. MRI-based prediction of conversion from clinically isolated syndrome to clinically definite multiple sclerosis using SVM and lesion geometry. Brain Imaging Behav 2018; 13: 1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramanujam R, Zhu F, Fink K, et al. Accurate classification of secondary progression in multiple sclerosis using a decision tree. Mult Scler 2020; 27: 1240–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinto MF, Oliveira H, Batista S, et al. Prediction of disease progression and outcomes in multiple sclerosis with machine learning. Sci Rep 2020; 10: 21038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Høgestøl EA, Kaufmann T, Nygaard GO, et al. Cross-sectional and longitudinal MRI brain scans reveal accelerated brain aging in multiple sclerosis. Front Neurol 2019; 10: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Opfer R, Ostwaldt A-C, Sormani MP, et al. Estimates of age-dependent cutoffs for pathological brain volume loss using SIENA/FSL – a longitudinal brain volumetry study in healthy adults. Neurobiol Aging 2018; 65: 1–6. [DOI] [PubMed] [Google Scholar]
- 29.Tozlu C, Jamison K, Gu Zet al. et al. Estimated connectivity networks outperform observed connectivity networks when classifying people with multiple sclerosis into disability groups. Neuroimage Clin 2021; 32: 102827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wottschel V, Chard DT, Enzinger C, et al. SVM recursive feature elimination analyses of structural brain MRI predicts near-term relapses in patients with clinically isolated syndromes suggestive of multiple sclerosis. Neuroimage Clin [Internet ] 2019[cited 2020 Oct 5];24: 102011.. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6861587/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stražar M, Curk T. Learning the kernel matrix via predictive low-rank approximations. Neurocomputing 2019; 340: 245–258. [Google Scholar]
- 32.Ozturk A, Smith SA, Gordon-Lipkin EM, et al. MRI of the corpus callosum in multiple sclerosis: association with disability. Mult Scler 2010; 16: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fleischer V, Gonzalez-Escamilla G, Ciolac D, et al. Translational value of choroid plexus imaging for tracking neuroinflammation in mice and humans. Proc Natl Acad Sci U S A [Internet ] 2021; 118: 1–12. Available from: https://www.pnas.org/content/118/36/e2025000118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manouchehri N, Stüve O. Choroid plexus volumetrics and brain inflammation in multiple sclerosis. Proc Natl Acad Sci [Internet ] 2021; 118: 1–3. Available from: https://www.pnas.org/content/118/40/e2115221118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Currà A, Pierelli F, Gasbarrone R, et al. The ventricular system enlarges abnormally in the seventies, earlier in men, and first in the frontal horn: a study based on more than 3,000 scans. Front Aging Neurosci 2019; 11: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alisch JSR, Kiely M, Triebswetter C, et al. Characterization of age-related differences in the human choroid plexus volume, microstructural integrity, and blood perfusion using multiparameter magnetic resonance imaging. Front Aging Neurosci. 2021; 13: 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albayram MS, Smith G, Tufan F, et al. Non-invasive MR imaging of human brain lymphatic networks with connections to cervical lymph nodes. Nat Commun 2022; 13: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monaco S, Nicholas R, Reynolds R, et al. Intrathecal inflammation in progressive multiple sclerosis. Int J Mol Sci 2020; 21: E8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouthillier X, Delaunay P, Bronzi M, et al. Accounting for variance in machine learning benchmarks. Proc Mach Learn Syst [Internet]. 2021; 3: 747–769. Available from: https://proceedings.mlsys.org/paper/2021/hash/cfecdb276f634854f3ef915e2e980c31-Abstract.html [Google Scholar]
- 40.Ritchie SJ, Cox SR, Shen X, et al. Sex differences in the adult human brain: evidence from 5216 UK biobank participants. Cereb Cortex 2018; 28: 2959–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barile B, Marzullo A, Stamile Cet al. et al. Ensemble learning for multiple sclerosis disability estimation using brain structural connectivity. Brain Connect 2021; 12: 476–488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-mso-10.1177_20552173221109770 for Prediction of high and low disease activity in early MS patients using multiple kernel learning identifies importance of lateral ventricle intensity by Claudia Chien, Moritz Seiler, Fabian Eitel, Tanja Schmitz-Hübsch, Friedemann Paul and Kerstin Ritter in Multiple Sclerosis Journal – Experimental, Translational and Clinical
Supplemental material, sj-tiff-2-mso-10.1177_20552173221109770 for Prediction of high and low disease activity in early MS patients using multiple kernel learning identifies importance of lateral ventricle intensity by Claudia Chien, Moritz Seiler, Fabian Eitel, Tanja Schmitz-Hübsch, Friedemann Paul and Kerstin Ritter in Multiple Sclerosis Journal – Experimental, Translational and Clinical
Supplemental material, sj-tiff-3-mso-10.1177_20552173221109770 for Prediction of high and low disease activity in early MS patients using multiple kernel learning identifies importance of lateral ventricle intensity by Claudia Chien, Moritz Seiler, Fabian Eitel, Tanja Schmitz-Hübsch, Friedemann Paul and Kerstin Ritter in Multiple Sclerosis Journal – Experimental, Translational and Clinical
Supplemental material, sj-docx-4-mso-10.1177_20552173221109770 for Prediction of high and low disease activity in early MS patients using multiple kernel learning identifies importance of lateral ventricle intensity by Claudia Chien, Moritz Seiler, Fabian Eitel, Tanja Schmitz-Hübsch, Friedemann Paul and Kerstin Ritter in Multiple Sclerosis Journal – Experimental, Translational and Clinical
Supplemental material, sj-docx-5-mso-10.1177_20552173221109770 for Prediction of high and low disease activity in early MS patients using multiple kernel learning identifies importance of lateral ventricle intensity by Claudia Chien, Moritz Seiler, Fabian Eitel, Tanja Schmitz-Hübsch, Friedemann Paul and Kerstin Ritter in Multiple Sclerosis Journal – Experimental, Translational and Clinical
Supplemental material, sj-pdf-6-mso-10.1177_20552173221109770 for Prediction of high and low disease activity in early MS patients using multiple kernel learning identifies importance of lateral ventricle intensity by Claudia Chien, Moritz Seiler, Fabian Eitel, Tanja Schmitz-Hübsch, Friedemann Paul and Kerstin Ritter in Multiple Sclerosis Journal – Experimental, Translational and Clinical
Data Availability Statement
Persons interested in obtaining access to the data should contact Friedemann Paul (friedemann.paul@charite.de).