Abstract
Prostate cancer (PCa) is the most common worldwide diagnosed malignancy in male population. The diagnosis, the identification of aggressive disease, and the post-treatment follow-up needs a more comprehensive and holistic approach. Radiomics is the extraction and interpretation of images phenotypes in a quantitative manner. Radiomics may give an advantage through advancements in imaging modalities and through the potential power of artificial intelligence techniques by translating those features into clinical outcome prediction. This article gives an overview on the current evidence of methodology and reviews the available literature on radiomics in PCa patients, highlighting its potential for personalized treatment and future applications.
Keywords: artificial intelligence, MRI, PET-CT, prostate cancer, radiomics
Introduction
Prostate cancer (PCa) represents a global healthcare burden, with a 13.5% incidence and a 6.7% mortality rate accounting as the fifth leading cause of death in the world in male population.1,2 Recent developments in imaging methods hold great promise in diagnosis and detection of PCa, especially clinically significant PCa (csPCa), screening and staging of the disease, treatment response, and personalized prediction of oncological outcomes.3–5 Through advances in artificial intelligence (AI) methods of evaluating radiological features, a recent area of interest, radiomics, is developing, moving the interest from qualitative evaluation of imaging modalities to measuring quantitative data from these images. 6 Radiomics is the extraction and analysis of quantitative imaging features from radiographic images7,8 that can be used to develop descriptive and predictive models, combining image features and phenotypes with gene and protein signatures.9,10 PCa is diagnosed from biopsy samples by evaluating its grade; 11 according to this grade, patients are stratified using International Society of Urologic Pathologists (ISUP) grade, serum prostate-specific antigen (PSA) levels, and clinical staging.12–14 The upstaging of the disease after radical prostatectomy (RP) is observed in many cases, in part due to PCa heterogeneity, therefore, is essential to better stratify PCa patients from diagnosis to early biochemical recurrence (BCR). Efforts are made to validate the use of multi-parametric magnetic resonance imaging (mpMRI) parameters for diagnosis and BCR prediction in patients with localized disease. 15 Prostate biopsies miss an important percentage of csPCa, which could be evaluated to be up to 38–46% in systematic biopsies. 16
By combining the recent advances in imaging techniques, such as mpMRI prior to the biopsy, 12 performing targeted biopsy (TBx) plus the systematic biopsy of the contralateral lobe can detect approximately 96% of csPCa.17–20 Still, there are undiagnosed csPCa that will threaten the life of patients, in part due to heterogeneity of PCa tumors, 20 in part due to spatial 21 alignment of heterogeneous tumor growth and, finally, due to clonal genomic diversity.4,22,23 To limit the inter-observer variability and reader’s experience, the Prostate Imaging—Reporting and Data System version 2.1 (PIRADS v2.1) was proposed. 24 Even if this standardization in image interpretation cannot limit the differences between expert healthcare sites 25 and centers outside highly specialized clinics, 26 the possibility to distinguish between low- and high-grade cancers due to a numerical score, is moderately reliable27–29 and permit to avoid unnecessary biopsies. 30 Up to date, there is no evidence that mpMRI can overcome the heterogeneity of PCa.27,31,32
Prostate-specific membrane antigen positron emission tomography (PSMA-PET) is being used for staging of PCa and for follow-up of recurrent disease.33–36 Recent evidence are reported for detection of PCa lesions (especially PIRADS 3 lesions), 37 tumor delineation, 38 tumor localization, 39 and segmentation of prostate volume and lesions using PSMA-PET.40,41 As current risk stratification models, which could predict oncological outcomes, are unable to accurately delineate the prognosis for each patient and for each stage of disease, there is an ongoing need for the detection of personalized and precise detections tools and treatment. 12 Radiomics features (RFs) analysis can give information for detection, risk stratification, and treatment. Images obtained from existing follow-up imaging tools do not require additional investigations and the whole tumor is characterized compared with biopsy.40,42 The AI techniques and the continuous research to clinical translations of different computational models in medicine and PCa43,44 will enhance the detection, and possible grade of PCa tumors and its classification.45,46 Radiomics and its combination with machine-learning (ML) techniques are assessing the possibility of differentiation between low- and high-grade PCas,32,47–53 tumor description,54–61 risk assessment,48,62–65 and treatment planning.66–71
This review gives an overview of the methodological aspects of radiomics, and current and future possible applications of radiomics in the management of PCa. This includes our literature search and a qualitative analysis of radiomics in PCa from different imaging techniques and its clinical significance. We focused our review on enclosing especially the latest studies that used different imaging modalities [most used imaging modality analyzed in reviews was magnetic resonance imagining (MRI)], following a predefined methodology, aiming to find studies with validation protocols and also to introduce a review of how AI can improve radiomics and translation of these results into clinical practice and a mini-review on strengths and limitations of different algorithms used in PCa radiomics.
Workflow of RFs extraction
Radiological images are usually qualitatively analyzed by human readers. Radiomics, instead, aim to quantitative mapping images. Such process is based on several image features extraction, analysis and modeling in relation to specific targets that could be either anatomical or functional, based on different imaging techniques used as a starting point. 72 A radiomics study is composed by different and consecutive phases, namely, data selection, medical imaging, feature extraction, exploratory analysis, and modeling. To assess the quality of all processes, Lambin et al. 72 reported the radiomics quality score, which is an item-based score. The radiomics quality score is composed of 16 different items and explore all the aspects of the radiomics process through the five phases described above. In particular, the radiomics quality score considered the image protocol quality, multiple segmentations, the phantom study, imaging time points, adjustment for multiple testing, the use of multivariable analyses, the detection and discussion of biological correlates, cut-off and accuracy analyses (calibration and accuracy statistics), trial registration and image availability, cost-effectiveness analyses, and comparisons with the current gold standard. The following paragraphs resume the main aspects of radiomics pipeline.
Radiomics pipeline
Image acquisition and pre-processing
The radiomics pipeline start with the acquisition of a robust data set of medical images and patients’ outcomes. Radiomics is based on the hypothesis that quantitative data extracted from radiological images are related to underlying genomic, pathologic, and clinical features. Unfortunately, the high variability in the image acquisition process reduces the reproducibility of the quantitative features extracted from each image and affects the external validity of any produced model. In this scenario, one of the most challenging aspects of radiomics is image acquisition and pre-processing to reduce the influence of imaging protocols on the extracted radiomics features. Several strategies have been developed in this regard. First, some imaging modalities are inherently endowed with lower variability. Computed tomography (CT) scans and anatomical T2 MRI sequences are more robust and reproducible images.73,74 The creation of AI models that evaluate only these sequences, however, would be very limiting and would result in the loss of large amounts of information that are instead essential for radiologists who qualitatively evaluate each sequence and take clinical decisions based on the combination of each sequence. 75 The way forward is the standardization of the imaging protocols and image pre-processing: the attempt to homogenize images from which RFs will be extracted with respect to pixel spacing, gray-level intensities, bins of the gray-level histogram, and so forth. 76 Preliminary results have shown that the test–retest robustness of RFs extracted largely depends on the image pre-processing settings used. 77 Given the difficulties encountered in the image standardization process, a radiomics model must first be tested internally and then externally validated in multiple validation cohorts. The availability of open-source data sets that include medical images will be of great help to the scientific community and will speed up the process of discovery and validation of radiomics models. 78
High-throughput feature extraction
Digital Imaging and Communications in Medicine (DICOM) is the standard file format for the management of medical imaging information and related data. A single DICOM file coming from X-rays, CT scans, or MRI, comprehend discrete information of each pixel, tridimensional coordinates and every pieces needed to digitally visualize the image, regrouping those information into data sets. Radiomics uses software-implemented mathematical algorithms to extract quantitative features from medical images. Feature extraction is performed using two main methods: handcrafted radiomics and deep learning (DL). For both radiomics approaches, delineation of the region of interest (ROI) in two and three dimensions is the crucial first step in the pipeline.
The handcrafted radiomics approach involves radiologist’s manual segmentation of the ROI on medical imaging, and the subsequent extraction of thousands of human-defined quantitative features, which describe tumor shape, spatial relationships, and texture among other characteristics. ML methods may then be applied to identify the imaging features that are associated with a given clinical outcome. The introduction of a source of human bias into the process raises, however, concerns on the reproducibility of handcrafted RF extraction due to the intra- and inter-reader variability that results from the reliance on manual segmentation of the tumor, and due to variation in imaging and pre-processing techniques for feature extraction. 79
The advent of DL methods has enabled a larger learning capacity, boosting generalizability and accuracy while reducing potential bias. 80
Most of the DL radiomics applications rely on the definition of the ROI based on a single-point placement within the tumor volume, replacing full human segmentations with approximate localization. Recently, DL methods have allowed also automated segmentation 81 and automated extraction, and learning of relevant radiographic features without the need for previous definition by researchers. 82
Data integration and data analysis
Once RFs have been extracted and normalized from an ROI, statistical methods are used to assess their association with clinical and molecular outcomes.
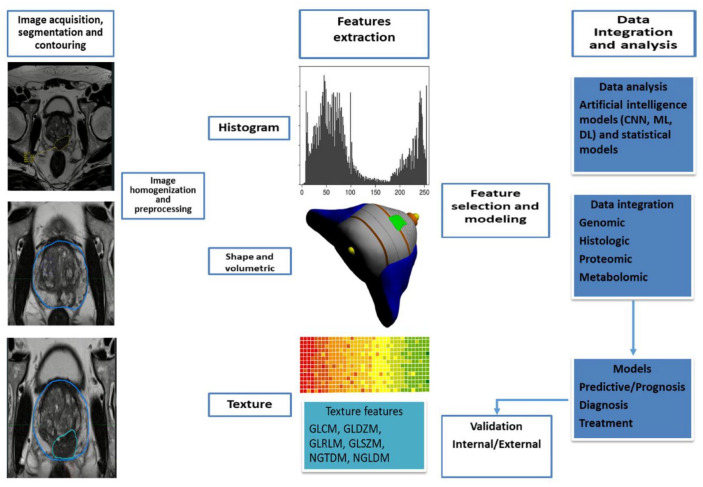
Data integration with information coming from histologic analysis, genomics, proteomics, and metabolomics can improve the predictive accuracy of a radiomics model and allow for a deeper understanding of the biological meaning of quantitative features extracted from medical images. 83 The aim is the prediction of patients’ outcomes, such as the presence of disease, disease recurrence, treatment response, and cancer-specific mortality. AI is used not only for the feature calculation but also for the selection of features associated with a specific outcome. 84 In this step, from thousands of features extracted and tested, only a small number are selected to be included in the final model. To make a model generalizable, it is important to avoid finding spurious correlations in the data that do not apply to other similar data sets, an occurrence termed overfitting. 85 Overfitting may be avoided by increasing the sample size and the variability of the training and development set of images and reducing the number of features ultimately included in the model. Figure 1 integrates the workflow of RFs.
Figure 1.
A typical radiomics workflow, including the extraction of features, the data integration and analysis, and the production of predictive model.
CNN, convolutional neural network; DL, deep learning; GLCM, gray-level co-occurrence matrix; GLDZM, gray-level distance zone matrix; GLRLM, gray-level run length matrix; GLSZM, gray-level size-zone matrix; ML, machine-learning.
Methodology
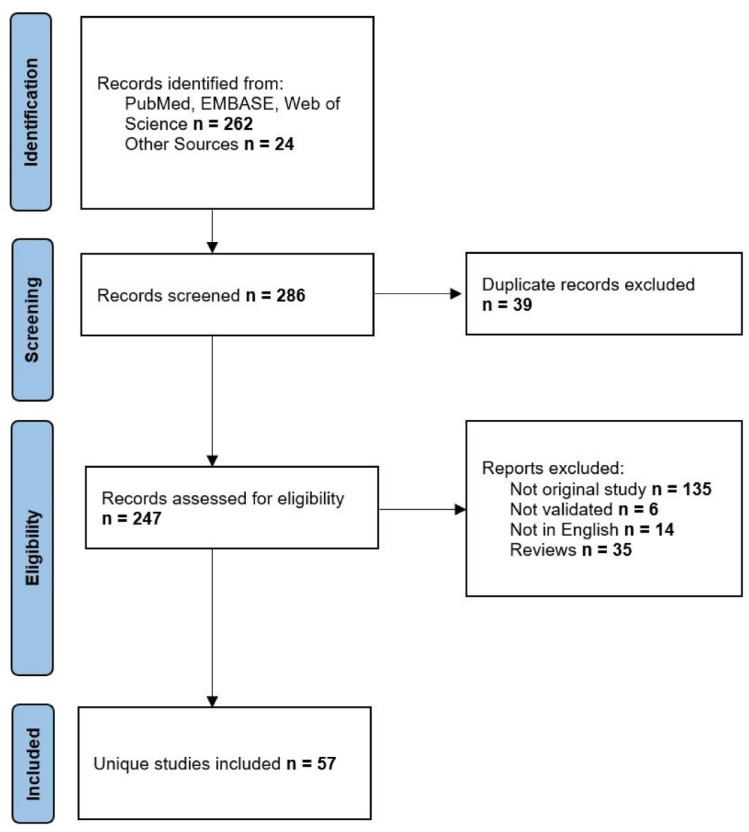
We had performed an extensive search through online databases (PubMed/Medline, EMBASE, clinicaltrials.gov) with the use of the following terms: prostate cancer AND radiomics AND [(MRI) OR (PSMA-PET) OR (CT) OR (TRUS)]. Inclusion criteria were as follows: articles with research on PCa radiomics with features extracted from choline/PSMA-PET, CT, MRI, and transrectal ultrasonography (TRUS) that were subjected to internal or external validation. Two authors performed independently the search of databases and one author assessed the eligibility of studies from the last 5 years with inclusion of early leading studies. A number of 262 studies were found with and 24 articles from other registers plus five ongoing clinical trials. At the end, a number of 57 studies were included in this analysis. Figure 2 depicts the description of methodology applied in accordance with Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidance. 86
Figure 2.
PRISMA flowchart of included studies.
Radiomics
Prostate-specific membrane antigen positron emission tomography
Current status of PSMA-PET in evaluation of PCa
PSMA-PET is the most promising imaging modality in PCa. Several studies have proved that PSMA-PET/CT is more accurate in N (node) and M (metastasis) staging as compared with MRI, abdominal contrast-enhanced CT or choline PET/CT. Results from randomized control trials (RCTs) evaluating the management and outcome of patients with (and without) metastases detected by PSMA-PET/CT are awaited before a decision can be made to use this test in primary staging. Guidelines, however, currently recommend PSMA-PET for M staging in patients with suspicious disease recurrence after primary treatment. Few studies have tested radiomics on this imaging modality that combine information coming with a PET and CT scans.
PSMA-PET radiomics studies in PCa
Erle et al. 87 recently confirmed the accuracy of radiomics decision tree (DT)-based classifiers in a larger population (N = 87) of patients undergoing 68Ga-PSMA-PET/CT for M and N staging of PCa. In the validation cohort, the extra trees classifier trained with the data of 30 patients resulted in an area under the curve (AUC) of 0.95, a sensitivity of 0.95, and a specificity of 0.80. The authors, however, pointed out suboptimal performances of the algorithm in liver, kidneys, genito-urinary (GU) tract, and lacrimal and salivary glands with a high rate of false positives. Peeken et al. 67 developed a CT-based radiomics model to predict lymph node (LN) metastasis status using a PSMA radio-guided surgery cohort with histological confirmation of all suspected LN. The best radiomics model (AUC = 0.95) outperformed all conventional CT parameters, including LN short diameter (AUC = 0.84), LN volume (AUC = 0.80), and an expert rating (AUC = 0.67) for detection of LN metastases. In the setting of primary staging of PCa, Zamboglou et al. evaluated a PSMA-PET/CT radiomics model for the prediction of any PCa and csPCa foci in radical RP specimens. Twenty and 52 patients were included in the training and validation cohorts. Visual PET image interpretation missed up to 60% of tumor lesions. Conversely, authors showed that two RFs [local binary pattern (LBP) size-zone non-uniformality normalized and LBP small-area emphasis] perform excellently in visually unknown PCa detection (p < 0.01, AUC ⩾ 0.93). The sensitivities of both random forests (RFs) in the validation cohort were ⩾ 0.8. 57 In a similar setting, Domachevsky et al. 88 also showed the accuracy of PSMA-PET/MRI for the diagnosis of intra-prostatic cancer nodules. Quantitative features extracted from PSMA-PET/MRI images, such as standardized uptake value (SUV) max and apparent diffusion coefficient (ADC) values are promising for differentiating between important intra-prostatic lesions and benign tissue in patients with naïve PCa. Ongoing studies will prove the utility of this multi-parametric approach to guide biopsy or focal treatment and even prevent unnecessary biopsies in certain scenarios. 88 In Table 1, we have listed the articles on the potential of radiomics and CT scans in risk stratification of PCa patients.
Table 1.
Radiomics and PSMA-PET/CT scan studies.
| Author | Clinical outcomes | Imaging modality | Results | No. patients/prospective or retrospective | Segmentation |
|---|---|---|---|---|---|
| Radiomics in diagnosis and detection of prostate cancer | |||||
| Moazemi et al. 89 | Comparing results from human readers to those of ML-based analyses | 68Ga-PSMA-PET/CT scan | DT-based classifiers showed the best performance with up to 0.98 AUC, 0.94 sensitivity, and 0.89 specificity | 2419 hotspots from 72 patients/retrospective | Manual |
| Erle et al. 87 | Comparing results from human readers to those of ML-based analyses | 68Ga-PSMA-PET/CT scan | Extra trees classifier: AUC of 0.95, a sensitivity of 0.95, and a specificity of 0.80 | 2452 hotspots from 87 patients/retrospective | Manual |
| Moazemi et al. 90 | Overall survival after treatment with (177) Lu-PSMA | 68Ga-PSMA-PET/CT scan | A radiomics signature based on SUVmin and kurtosis achieved p values less than 0.05 | 2070 pathological hotspots from 83 subjects/retrospective | Manual |
| Peeken et al. 67 | Detection of LN metastases in PSMA radio-guided surgery patients | 68Ga-PSMA-PET/CT scan | A CT-based radiomics model (AUC = 0.95) outperformed all conventional CT parameters for detection of LN metastases | 108 patients with recurrent PCa who received radio-guided surgery of 68Ga-PSMA-PET/CT-positive PCa recurrences/retrospective | Manual |
| Zamboglou et al. 57 | Detection of PCa areas in whole-gland RP specimens | 68Ga-PSMA-PET/CT scan | Local binary pattern size-zone non-uniformity normalized and LBP small-area emphasis AUC ⩾ 0.93; sensitivity >> 0.8 | 72 patients with PCa undergoing RP/retrospective | Manual |
| Domachevsky et al. 88 | Detection of PCa areas in whole-gland RP specimens | 68Ga-PSMA-PET/MRI scan | PET/MR SUVmax, ADCmin and ADC mean differentiate between normal and tumor prostatic tissue (all p < 0.001) | 22 Patients, 44 PCa areas/prospective | Manual |
ADC, apparent diffusion coefficient; AUC, area under the curve; CT, computed tomography; DT, decision tree; ECOG, Eastern cooperative oncology group; LBP, local binary pattern; LN, lymph node; ML, machine-learning; MRI, magnetic resonance imaging; Pca, prostate cancer; PET/MR, positron emission tomography/magnetic resonance; PSA, prostate-specific antigen; PSMA-PET, prostate-specific membrane antigen positron emission tomography; RP, radical prostatectomy; SUV, standardized uptake value.
CT scans
Most research on Pca radiomics mainly focuses on mpMRI, however, other imaging modalities—which are regularly used for Pca work-up and bear the potential to carry information that can be used to improve current Pca management at different clinical steps—have been evaluated. To date, specificity and sensitivity of CT scan in the detection of tumor lesions inside the prostate are lacking. Especially for patients with contraindications to mpMRI (e.g. pacemaker, metallic implants, claustrophobia) and to address some problems inherent to mpMRI (e.g. variability and challenges in imaging acquisition and interpretation), radiomics performance of CT imaging might add further clinical value in a variety of PCa scenarios.
CT scans and radiomics studies in Pca
Osman et al. 62 evaluated the value of CT-based RFs for Pca risk stratification. Authors retrospectively extracted RFs from CT scans of 342 patients with localized Pca treated with RT, and found that their model—based on 1618 quantitative imaging features—classified low- versus intermediate- and low- versus high-risk groups (AUC = 1.00 and 0.96, respectively), as well as Gleason score (GS) 6 versus 7 and 7 (3 + 4) versus 7 (4 + 3), with high accuracy (AUC = 0.90 and 0.98, respectively). Similarly, Tanadini-Lang et al. 91 performed a radiomics retrospective analysis of the entire prostate using CT perfusion imaging for predicting Pca classification. In their case series of 41 Pca patients referred for RP, 1701 RFs were determined in three perfusion maps (i.e. blood volume, blood flow, and mean transit time), and grouped into 10 groups—represented by the single RF with the best correlation with the group. They found that one RF (i.e. blood flow HHL joint average) was able to predict post-surgical GS (R 2 = 0.21, p = 0.01), and two radiomics parameters (i.e. blood flow HHH fractal dimension and blood volume HLH root mean square) to distinguish between intermediate- versus high-risk patients, and GS 7 (3 + 4) versus 7 (4 + 3) PCa (AUC = 0.81 and 0.77, respectively). Bosetti et al. 66 explored the role of cone-beam CT-based radiomics for risk assessment and prognosis. Cone-beam CT is routinely performed prior to RT sessions for treatment set-up (i.e. to verify daily treatment position). Authors evaluated 31 patients with PCa, using 300 RFs. According to their analysis, the best-performing models, which included both specific histogram and shape-based features identified low- or intermediate- versus high-risk cases with an AUC of 0.83 (95% CI = 0.73–0.92), and GS 6 versus ⩾ 7 cases with an AUC of 0.80 (95% CI = 0.74–0.85), suggesting a good performance in classifying Pca cases. With regards to the prediction of BCR, especially histogram-based features were found to have predictive value (Precision-recall curve [PRC] = 1.00)—yet their case series included only three patients with BCR. Mostafaei et al. 92 successfully used CT RFs alone and combined with clinical and dosimetric parameters to develop a prediction model for RT-induced toxicity (refer to ‘Radiomics and treatment response and toxicity’ paragraph for more detailed data). Peeken et al. 67 sought to develop a CT-based radiomics model to predict LN metastases status in a retrospective cohort of 80 men with recurrent Pca who underwent PSMA radio-guided surgery (i.e. a novel surgical approach that allows intra-operative detection and resection of PSMA-PET/CT- positive LN). Among the 156 RFs extracted, texture and shape features were found to correlate with LN volume. Among the radiomics models, the LBP model outperformed the intensity, shape, and texture models (AUC of 0.90 versus 0.73, 0.83 and 0.78, respectively), as well as conventional CT parameters, such as LN short diameter (AUC = 0.84); interestingly, the combined radiomics model—based on all RFs—achieved the best performance (AUC = 0.95) in predicting LN status, with LBP features providing the highest contribution to model performance. Moreover, the combined radiomics model showed a significant correlation with LN status on multivariate analysis [odds ratio (OR) = 15.5], and a clinical net benefit above the LN short diameter according to decision curve analysis. Acar et al. 93 aimed to use CT texture features and ML methods to distinguish bone lesions, imaged via 68Ga-PSMA-PET/CT, as metastatic from completely responded in Pca patients, and demonstrated a good accuracy of CT in detecting treatment response in bone metastasis (BM) (refer to ‘Radiomics and bone metastasis’ paragraph for more detailed data). In Table 2, we summarize the results of articles on radiomics and CT studies.
Table 2.
Radiomics and CT studies.
| Author | Clinical outcomes | Type of image acquisition | Results | Study design No. of patients |
Image segmentation |
|---|---|---|---|---|---|
| Radiomics and CT scans | |||||
| Osman et al. 62 | GS, risk-group classification | CT | Radiomics classifier: • Low- versus intermediate-risk groups, AUC = 1.00 • Low- versus high-risk groups, AUC = 0.96 • GS 6 versus ⩾7, AUC = 0.90 • GS 7 (3 + 4) versus 7(4 + 3), AUC = 0.98 |
Retrospective n = 342 |
All DICOM images and structure sets were reviewed by an experienced clinical oncologist |
| Tanadini-Lang et al. 91 | GS, risk-group classification | CT perfusion | • Intermediate- versus high-risk groups, AUC = 0.81 • GS 7 (3 + 4) versus 7 (4 + 3), AUC = 0.77 |
Retrospective n = 41 |
The prostate was delineated on one of the CT image batches of the perfusion series; perfusion parameters were only calculated inside these contours |
| Bosetti et al. 66 | Stage, GS, PSA level, risk-group classification, BCR | Cone-beam CT | • Low- or intermediate- versus high-risk groups, AUC = 0.83 • GS 6 versus ⩾7, AUC = 0.80 |
Retrospective n = 31 |
Manually segmentation the prostate gland on each CBCT scan and contouring the lesions |
| Mostafaei et al. 92 | RT toxicity (i.e. cystitis, proctitis) | CT | Cystitis: • Clinical–radiomics model, AUC = 0.77 • Radiomics model alone, AUC = 0.71 Proctitis: • Clinical–radiomics model, AUC = 0.65 • Radiomics model alone, AUC = 0.71 |
Prospective n = 64 |
ROIs were manually drawn on each slice, including the rectal and bladder walls and excluding the rectal lumen and bladder entity |
| Peeken et al. 67 | LN metastases | Contrast-enhanced CT (from PSMA-PET/CT) | • Combined radiomics model, AUC = 0.95 • Local binary pattern model, AUC = 0.90 • conventional CT parameters (i.e. LN short diameter), AUC = 0.84 |
Retrospective n = 80 |
LN segmentation was performed manually using Eclipse 13.0 on the contrast-enhanced diagnostic CT data sets |
| Acar et al. 93 | Detection of BM that responded after treatment | CT (from PSMA-PET/CT) | weighted k-nearest neighborhood algorithm, AUC = 0.76 | Retrospective n = 75 |
The VOI was drawn to entire sclerotic lesion via CT images with manually by a nuclear medicine physician |
AUC, area under the curve; BCR, biochemical recurrence; BM, bone metastasis; CBCT, 3D cone-beam computed tomography; CT, computed tomography; DICOM, Digital Imaging and Communications in Medicine; GS, Gleason score; LN, lymph node; PSA, prostate-specific antigen; PSMA-PET, prostate-specific membrane antigen positron emission tomography; ROI, region of interest; RT, radiotherapy; VOI, volume of interest.
TRUS imaging
Similarly, radiomics performance applied to ultrasound (US) imaging has been recently investigated. Historically, US has been widely used for prostate gland examination and Pca diagnosis—yet, to date, standard TRUS is no more suitable for Pca detection. 12 It is an appealing imaging method from a radiomics perspective, however, especially because of its several advantages (i.e. it is non-invasive, radiation-free, cost-effective, widely available, and practical).
TRUS imaging and radiomics studies in PCa
Wildeboer et al. 58 evaluated the role of multi-parametric US for the localization of Pca in 48 men with Pca referred for RP. In particular, authors combined the information from three different US modalities [i.e. B-mode US, shear-wave elastography (SWE), and dynamic contrast-enhanced US (DCE-US)] and their radiomics through ML technology, to develop a multi-parametric radiomics-based classification. According to their analysis, the multi-parametric classifier outperformed the best-performing single parameter (i.e. contrast velocity) in the detection of Pca and csPCa (defined as GS > 3 + 4) (AUC = 0.75 and 0.90 versus AUC = 0.69 and 0.76, respectively). Of note, the improvement was partly due to radiomics extraction and partly to the multi-parametric combination. Zhang et al. 94 aimed to use a computer-aided diagnosis (CAD) system on multimodal US images to diagnose PCa. Authors analyzed 47 men with Pca and 56 with BPH, using a combination of two US modalities [i.e. B-mode US and real-time elastography (RTE)] and a DL network (i.e. PGBM-RBM2) to fuse multimodal features by a multi-layer architecture. This proposed model yielded the best performance in the discrimination between benign disease and PCa (AUC = 0.85), compared with the competing methods (both baseline methods and DL methods based on B-mode US or RTE features only). Huang et al. 95 used a computer-assisted analysis of US through image processing to improve PCa detection. In this case series, texture features were extracted from TRUS images of 48 patients (22 negative and 22 PCa cases), fused and then put into support vector machine (SVM) classifier for classification purpose. Such a method achieved an accuracy of 70.9% in differentiating PCa from benign tissue. Wu et al. 96 proposed a novel automatic 2D prostate segmentation framework. To develop such a model, authors used a novel feature extraction and classification pipeline based on speckle features (i.e. size, orientation, and spatial coherence), the dark-to-light intensity transition near prostate boundary, and the prostate shape prior. According to results validated on two data sets of US images, their framework demonstrated a good classification performance, outperforming other proposed methods (error rate of 11.0% versus 46.6%, 18.8%, 48.3%, and 24.5% for Traditional Gabor, rotation-invariant, moments-invariant, and LBP features, respectively). Table 3 depicts results of radiomics and TRUS imaging studies.
Table 3.
Radiomics and TRUS imaging studies.
| Author | Clinical outcomes | Type of image acquisition | Results | Study design No. of patients |
Image segmentation |
|---|---|---|---|---|---|
| Radiomics and TRUS imaging | |||||
| Wildeboer et al. 58 | PCa detection, GS | TRUS: B-mode, contrast-enhanced US |
detection of PCa • Multi-parametric classifier, AUC = 0.75 • Best-performing single parameter (i.e. contrast velocity), AUC = 0.69 detection of GS > 3 + 4 PCa • Multi-parametric classifier, AUC = 0.90 • Best-performing single parameter (i.e. contrast velocity), AUC = 0.76 |
Retrospective n = 48 |
The prostate was located and delineated by an automated DL-based TRUS segmentation algorithm on the side-view fundamental B-mode images of both SWE and DCE-US acquisition; a detection algorithm was designed to outline calcifications in the B-mode images |
| Zhang et al. 94 | PCa detection | TRUS: B-mode, RTE |
• Multimodal method + deep learning network, AUC = 0.85 • B-mode features + deep learning network, AUC = 0.80 • RTE features + deep learning network, AUC = 0.79 |
Retrospective n = 103 |
The boundaries of the prostate peripheral gland were manually drawn on B-mode US images and then mapped to the retrieved elastograms to specify ROI |
| Huang et al. 95 | PCa detection | TRUS | • Proposed method, AUC = 0.70 | Retrospective n = 48 |
An optical density conversion technology was used of each pixel to dry the ROI and enhanced the contrast and to make the details of the image clearer for subsequent analysis |
| Wu et al. 96 | Prostate segmentation | TRUS: B-mode |
Segmentation framework using speckle-induced features, error rate = 11% | Retrospective n = 132 + 5 videos |
A 2D prostate segmentation framework utilizing speckle-induced texture features |
AUC, area under the curve; DCE-US, dynamic contrast-enhanced ultrasound; DL, deep learning; GS, Gleason score; PCa, prostate cancer; ROI, region of interest; RTE, real-time elastography; SWE, shear-wave elastography; TRUS, transrectal ultrasound; US, ultrasound.
Radiomics from MRI images
MRI has a high sensitivity and specificity as an imaging method in detecting csPCa and for lesion targeted biopsy of the prostate, and it gives high quantitative and qualitative images for the use in radiomics research. 97 The prediction of PCa characteristics (GS, PIRADS v2, risk class) and also the automated prostate segmentation in a fully automated quality control system makes MRI-based radiomics a valuable tool in PCa diagnosis.49,98 Another use of ML models was to develop new detection features, such as advanced zoomed diffusion-weighted imaging (DWI) and conventional full-field-of-view DWI, 99 performing texture analysis of prostate MRI in PIRADS v2 3 lesions, 100 delivering frameworks for automated PCa localization and detection,101,102 and managing RT treatment69,70,92,103,104 and predicting BCR and RT toxicity.66,71,105–108 In PCa, the use of radiomics comes with aid in prostate volume selection and segmentation,58,59,98,109,110 PCa screening,61,111 detection and classification,61,68,112–114 risk stratification,59,63,115 and treatment.66,116–118 The in-depth analysis of these studies can be found in the following sections.
Prostate volume selection and segmentation
Using mpMRI and an auto-fixed volume of interest (VOI)-based radiomics signature was studied by Bleker et al., 54 identifying a model with features from T2-weighted (T2w) and DWI through an auto-fixed volume of interest, AUC = 0.870 (95% CI = 0.980–0.754), enhancing the actual assessment of the peripheral zone lesions. Orzcyk et al., 109 compared the entropy score and visual score in the assessment of prostate lesions on mpMRI and found that the entropy score (numerical cut-off of 16.61) natural information unit (NAT) better predicted the csPCa lesions than the visual score (p < 0.001). Bernatz et al., 119 compared several ML algorithms used to predict csPCa with the help of mpMRI (T2w, dynamic contrast-enhanced (DCE) images and quantitative analysis on ADC map, and PIRADS v2. ADC-derived RFs and the PIRADS v2 categories in relation to the Gleason grade score were applied and results found that lower mean ADC values were revealed for malignant lesions (p < 0.001) which is in concordance with PIRADS v2. Lately, an effort by Sunoqrot et al., 98 to provide an automated quality control system for prostate segmentation from mpMRI images was created to evaluate the quality of prostate segmentation. Krauss et al. 110 used radiomics analysis to segment the prostate in a way that the peripheral zone, the transition zone and the entire prostate are available as analyzable volumes in 3D Slicer and PyRadiomics software. To facilitate detection of PCa, a combination of mpMRI, B-mode, SWE, and contrast-enhanced US radiomics fully automated techniques are developed at the moment. Wildeboer et al., 58 used radiomics to integrate information from all of the above-described modalities. ML was applied through an RF classification algorithm and found that combinations between perfusion, dispersion, and elasticity-related features were favored.
One of the challenges in spatial delimitation of tumor burden is related to tumor delineation and segmentation of ROI which in the conventional way is reader dependable and time-consuming. The standardization of MRI image acquisition across institutions should be encouraged. 69 More data are required from clinical trials to assess the robustness of radiomics-based predictive models and to further assess the automatic segmentation of gross tumor volume automatic.69,120–122
PCa screening
For screening purposes, non-invasive techniques like radiomics tools can be of great interest in identifying csPCa. McGarry et al., 123 used mpMRI included field of view (FOV)—optimized and constrained undistorted single-shot (FOCUS) DWI, DCE imaging, and T2w imaging. The authors introduced a new concept, the Gleason probability map, explained by the fact that each profile is replaced by its respective percentage value, creating four maps describing low grade, high grade, benign, and the presence of cancer, obtaining an AUC of 0.79 in distinguishing cancer from atrophy tissue, but having a low performance in the successful characterization of Gleason 3 from Gleason 4. Qi et al., 61 evaluated a combined model that involves RFs and clinical and radiological risk factors. The model involving radiomics, PSA density and PIRADS v2 score had AUC of 0.956 and 0.933 on trained cohorts and validation models, respectively, and performed better than just the clinical and radiological model (p < 0.05), therefore, identifying more negative lesions on mpMRI and potentially lowering the number of patients needing a prostate biopsy by 18.4%.
There are few studies that looked into the potential of screening the patients for PCa and this is due to the fact that the technology behind mpMRI, lack of standardization as reported for the inter-reader differences in the assessment of PIRADS score lesions.
Radiomics in diagnosis and detection of PCa
Initial research of radiomics in PCa An array of studies looking into the potential and future benefits in PCa detection and diagnosis through radiomics-based models using different features were studied. A few studies focused on the detection of PCa, on evaluating PIRADS performance for diagnosis of suspicious lesions. Cameron et al. 122 first experienced a model based on clinical use of imaging features by radiologists. The morphology, asymmetry, physiology, and size grouped together outperformed each one of them taken separately, in terms of accuracy, sensitivity and specificity compared with conventional mpMRI, with an average of 87% and 65%, respectively. Hang et al., 68 aimed to develop and validate a radiomics model based on mpMRI to predict PCa upgrading after RP. The combination model (radiomics signature, clinical stage, and time from biopsy to RP performed better than the clinical model and radiomics model alone (AUC: combined model, 0.910; clinical model, 0.646; and radiomics model, 0.868).
Clinical studies for detection of PCa involving radiomics One of the first studies that analyzed imaging features for PCa was performed by Khalvati et al., 124 with the goal of creating a radiomics-based auto-detection method utilizing an mpMRI feature model that combined computed high-b DWI and correlated diffusion imaging and was evaluated with an SVM classifier, reporting good results for specificity evaluated with AUC as a target [sensitivity = 0.84 (95% CI = 0.76–0.91), specificity = 0.86(95% CI = 0.82–0.91), AUC = 0.90 (95% CI = (0.88–0.93)]. Dulhanty et al., 113 investigated ML models using several radiomics-driven zone-level PCa sensing strategies and a discovery radiomics approach based on zone level. The best-performing model was a zone discovery radiomics model with an AUC of 0.86 performing better than the clinical model (AUC = 0.79). Bagher-Ebadian et al. 112 build an adaptive model with RFs from MRI images to characterize dominant intra-prostatic lesions using AI technique, the artificial neural network (ANN). The AUC between the ANN model and the conventional model is somehow comparable (95% and 94%, respectively). Qi et al., 61 evaluated a combined model that involves RFs, and clinical and radiological risk factors, and to detect PCa in patients with PSA range 4–10 ng/ml. The model involving radiomics, PSA density, and PIRADS v2 score had AUC of 0.956 and 0.933 on trained cohorts and validation models, respectively, and performed better than just the clinical and radiological model (p < 0.05), therefore, identifying more negative lesions on mpMRI and potentially lowering the number of patients needing a prostate biopsy by 18.4%. Chen et al., 114 developed and a radiomics-based model for diagnosis of intermediate-/high-grade tumors and assessing its aggressiveness compared with PIRADS v2 scores. Radiomics-based model outperformed a PIRADS v2 model in detecting PCa versus without PCa (AUC = 0.999, validation as well as in differentiating high- from low-grade PCa, AUC = 0.777, validation). Hu et al., 99 compared the efficiency of PCa detection using a radiomics signature based on advanced zoomed diffusion-weighted imaging (z-DWI) and conventional full-field-of-view (f-DWI). Radiomics signatures based on the z-DWI technology had better diagnostic accuracy for PCa than f-DWI technology (p z-DWIb1500 versus f-DWIb1500: p = 0.048; z-calDWIb2000 versus f-DWIb1500: p = 0.014). The mixed model (mpMRI and clinically independent risk factors and radiomics) was better at diagnosing PCa compared with mpMRI signatures and clinically independent risk factors alone (AUC = 0.81, 0.93, and 0.94 in training sets, and 0.74, 0.92, and 0.93 in validation sets, respectively). Brunese et al., 125 developed a deep architecture, based on several convolutional layers, with the purpose to automatically assign the GS to MRI. The researchers obtained an accuracy for GS prediction equal to 0.98473, 0.96667, 0.98780, and 0.97561 with regard to GS 3 + 3, GS 3 + 4, GS 4 + 3 and GS 4 + 4 prediction, respectively, from 71 RFs derived directly from MRI images.
The reproducibility and replication of results is the basis for standardized reporting protocols, at the moment, researchers have no consensus between studies to identify the most favorable model, along with external validation of those models to apply to populations outside the studied patients’ cohorts. To accurately distinguish cancerous versus benign tissue, radiomics has to benefit from technological improvement in segmentation, feature extraction, statistical analysis, multi-center, prospective RCTs to be integrated in clinical practice and in decision-making protocols.
Radiomics and detection of csPCa
Wang et al., 126 aimed to investigate whether ML-based analysis of MRI radiomics can help improve the performance PIRADS v2 in csPCa. A predictive model was developed using a novel SVM trained with radiomics, PIRADS scores and a combination of radiomics and PIRADS v2 scores. Performance of PIRADS was improved for PCa versus peripheral zone (AUC: 0.983) and PCa versus transitional zone (AUC = 0.968). Kwon et al., 127 aimed to classify csPCa in mpMRI and RFs. Best performance with AUC of 0.82 for RFs, 0.76 for CART, and 0.76 for adaptive LASSO (least absolute shrinkage and selection operator). Parra et al., 128 classifier models based on DCE mpMRI were built to find the best discriminating characteristics between clinically significant and insignificant prostate lesions defined by GS. The trained models had an AUC of 0.82 and an AUC of 0.82 on the validation cohort. Penzias et al., 129 analyzed the translation of radiomics to better understand the underlying morphologic tissue characteristics and found that Gabor texture features were identified as being most predictive of Gleason grade on MRI (AUC = 0.69) Giambelluca et al., 100 analyzed the diagnostic performance of texture analysis of prostate mpMRI for PIRADS 3 images. Texture analysis extraction was performed using radiomics software from T2w and ADC images. For the detection of csPCa, the predictive models for the diagnosis of PCa yielded an AUC of 0.769 and 0.817 on T2w or 0.749 and 0.744 on ADC maps images. Min et al., 130 evaluated the performance of an mpMRI-based radiomics signature for discriminating between csPCa and insignificant PCa. Logistic regression modeling yielded AUC of 0.872 in the training cohort and 0.823 in the test cohort. Brancato et al., 131 aimed to investigate the potential use of radiomics for detection of PCa with GS ⩾ 6 in PIRADS 3 images and in peripheral PIRADS 3 upgraded to PIRADS 4 images. RFs were extracted from T2w, ADC map, and DCE-MRI images using specific software. For PIRADS 3 images, second-order model was chosen due to a slightly higher mean sensitivity, specificity, and accuracy (80%, 51%, and 71%, respectively) with AUC = 0.76. For upgraded PIRADS 4 classification task, first-order model showed higher performances with AUC, 89%; sensitivity, 87%; specificity, 62%; and accuracy, 82%. Li et al., 132 a model with biparametric MRI-RFs and a clinical–radiomics combined model for predicting csPCa. Both the radiomics model (AUC = 0.98) and the clinical–radiomics combined model (AUC = 0.98) achieved greater predictive efficacy than the clinical model (AUC = 0.79). In 2018, Bonekamp et al., 133 compared biparametric contrast-free radiomics ML, mean ADC, and radiologist characterization of prostate images. Quantitative measurement of the mean ADC reduced the number of patients with false-positive lesions (specificity = 51%), but radiomics ML did not perform better than mean ADC assessment. Using different ML techniques, Hou et al., 55 look at the PIRADS 3 lesions to screen out csPCa. The AUC of radiomics ML model 1 is 0.89 and is significantly (p = 0.003) higher than that of radiomics ML model 2 with AUC of 0.87. The decision analysis curve demonstrated that the models 1 and 2 significantly improved risk prediction at threshold probabilities csPCa at 20–80% compared with doing-none or doing-all. Zhang et al., 134 in a retrospective study performed on 159 PCa extracted RFs from T2w, DWI, and ADC images to differentiate csPCa from insignificant one. The radiomics signature was significantly associated with csPCa (p < 0.001). AUC values were 0.95 (training group), 0.86 (internal validation group), and 0.81 (external validation group). The combination nomogram including the radiomics signature and ADC value demonstrated a favorable classification capability with the AUC of 0.95 (training group), 0.93 (internal validation group), and 0.84 (external validation group). Gong et al. 135 used logistic regression to evaluate detection of csPCa. The combined clinical and radiomics model (the T2w/DWI) acquired an AUC of 0.788. Woźnicki et al., 59 developed and evaluated ML models using quantitative RFs extracted from mpMRI to detect and classify PCa. The model combining radiomics, PIRADS, PSA density, and digital rectal examination (DRE) showed a significantly better performance compared with ADC for csPCa prediction (AUC = 0.571, p = 0.022). Bernatz et al., 119 analyzed radiomics of ADC maps using ML algorithms to discriminate csPCa versus indolent disease. The authors used mpMRI images determining VOIs and processed by radiomics analysis. Three classification models were trained and a subset of shape features improved the diagnostic accuracy of the clinical assessment categories (maximum increase in diagnostic accuracy ΔAUC = +0.05, p < 0.001). Gugliandolo et al., 49 tested the associations of a large number of quantitative imaging features with clinical characteristics. Texture features were the most predictive for GS, PIRADS v2 score, and risk class; AUC = 0.74–0.94. Krauss et al., 110 used radiomics MRI phenotype of the transition zone to explain PSA level in patients with low suspicion for csPCa. Five RFs are significantly correlated with PSA level (r = 0.53–0.69, p < 0.05). The regression model significantly improves the explanatory value for PSA level (p < 0.05). Song et al., 136 looked a designing open-source software to extract image features, preprocess the feature matrix, develop different models in an automatically fashion, and evaluate them with common clinical statistics. The model was used to classify the csPCa and indolent disease using the PROSTATEx data set. The AUC on the training, validation, and test data set achieved results of 0.838, 0.814, and 0.824, respectively. Lately, Castillo et al., 137 in a multi-center study, the authors evaluate the possibility of generalizing radiomics models for PCa classification and to compare the performance of these models to the performance of radiologists. Radiomics models with images from mpMRI were used to differentiate high- versus low-grade lesions which were generated by ML software. The three single-center models obtained a mean AUC of 0.75, outperforming the expert radiologist.
The combined radiological and clinical radiomics models were among the best methods to predict the csPCa in patients with PIRADS score 3 and more. The development of different models in an automatically fashion using ML and AI techniques and the construction of nomograms 136 can improve the radiomics potential to discriminate csPCa from indolent disease. Existing data are promising with radiomics outperforming PIRADS v2 in some aspects 137 and future studies will have to focus on the performance of radiomics compared with conventional radiologic analysis.
Radiomics and detection of extracapsular extension
Research has been analyzing radiomics models in the detection of prostate extracapsular extension (ECE). Losnegård et al., 138 investigated MRI-RFs combined with ML to predict ECE in non-favorable intermediate- and high-risk PCa patients. The authors obtained AUC for ECE prediction models individually and in different combinations [Radiomics (12 features and PSA density), 0.750; Radiology, 0.740; Memorial Sloan Kettering Cancer Center (MSKCC), 0.670; combination 1: MSKCC + radiomics, 0.770; combination 2: MSKCC + radiology, 0.770; combination 3: radiomics + radiology, 0.780; and combination 4: all (MSKCC + radiology + radiomics), 0.80]. Ma et al., 139 studied the probability of radiomics to identify PCa ECE. T2w and DCE images from mpMRI were extracted and further evaluated by experienced radiologist. The radiomics model obtained an AUC of 0.902 and 0.883 in the training and validation cohort, respectively. The model outperformed and was more sensitive than the radiologists results (AUC range = 0.600–0.697), (75.00% versus 46.88–50.00%, all p < 0.05), respectively. The same leading author, 140 published yet another research and performed a similar study (LASSO model with decision curve analysis). Obtaining an AUC of 0.906 and 0.821 for the training and validation data sets, respectively, and had a good calibration performance.
To obtain a predictive model for the pre-treatment detection of ECE, studies focusing on combined model between clinical, conventional radiology and radiomics achieved the best results and probably the future research models should rely on combined models.
In Table 4, we incorporated the research on the potential of radiomics to detect PCa, differentiate between aggressive and indolent disease, and detect ECE.
Table 4.
Clinical results of radiomics studies using MRI techniques.
| Author | Clinical outcomes | MRI modality | Results | No. patients/prospective or retrospective | Segmentation |
|---|---|---|---|---|---|
| Radiomics in diagnosis and detection of PCa | |||||
| Zhang et al. 68 | Prediction of PCa upgrading from biopsy to RP | MRI | AUC: combined clinical and radiomics model, 0.910; clinical model, 0.646; and radiomics model, 0.868 | 166/retrospective | Manually |
| Dulhanty et al. 113 | Identification of PCa | MRI | Zone-discovery radiomics model better than clinical heuristics model for positive or negative zones | 101/retrospective | Manually |
| Bagher-Ebadian et al. 112 | Detection intra-prostatic lesions and normal tissue | MRI | Comparison between conventional and AAN models | 117/retrospective | Manually |
| Qi et al. 61 | PCa detection for PSA range 4–10 ng/ml | MRI | Combination model, including RFs and clinical or radiological risk factors, p < 0.05 | 199/retrospective | Manually |
| Chen et al. 114 | Detection of PCa tumors with GS ⩾ 7 | MRI | Radiomics-based model better than PIRADS v2 model in detecting PCa versus non prostate cancer lesions and for differentiating high- from low-grade PCa | 381/retrospective | Manually |
| Khalvati et al. 124 | Detection of PCa (GS ⩾ 7) | MRI | Improved PCa detection with the use of SVM classifier | 20/retrospective | Manually |
| Hu et al. 99 | Detection of prostate cancer | MRI | Combined model had better performance compared with mpMRI signatures and clinically independent risk factors alone | 136/prospective | Manually |
| Radiomics and detection of csPCa | |||||
| Wang et al. 126 | Detection of clinically significant PCa lesions with a volume > 0.5 cm3 on histopathology | mpMRI | Performance of PIRADS was improved for prostate cancer versus peripheral zone (AUC = 0.983) and prostate cancer versus transitional zone (AUC = 0.968) | 176/retrospective | Manually |
| Kwon et al. 127 | Detection of csPCa | MRI | RFs highest AUC = 0.82 | 344/retrospective | Manually |
| Parra et al. 128 | Detection of csPCa | mpMRI | DCE-based classifier models best AUC = 0.82 | 52/retrospective | Manually |
| Penzias et al. 129 | Detection of high-grade PCa | MRI | Gabor texture features were identified as being most predictive of Gleason grade on MRI (AUC of 0.69) | 36/retrospective | Manually |
| Giambelluca et al. 100 | Presence of clinically significant prostate cancer in PIRADS 3 images | MRI | Texture analysis of PIRADS 3 lesions on T2-weighted and ADC maps images helps identifying prostate cancer AUC of 0.769 and 0.817 on T2w or 0.749 and 0.744 on ADC maps images |
43/retrospective | Manually |
| Min X et al. 130 | Detection of csPCa | mpMRI | Logistic regression modeling yielded AUC, 0.872 in the training cohort and 0.823 in the test cohort for GS 3 + 4 or lower | 280/retrospective | Manually |
| Brancato et al. 131 | Gleason score ⩾ 6 in PIRADS 3 images and in peripheral PIRADS 3 upgraded to PIRADS 4 images | MRI | Radiomics models showed high diagnostic efficacy in classify PIRADS 3 and up PIRADS 4 lesions, outperforming PIRADS v2.1 performance. | 116/retrospective | Manually |
| Hou et al. 55 | Detection of csPCa in PIRADS 3 lesions | mpMRI | Radiomics model can predict csPCa [AUC model one is 0.89 and higher than that of model two with AUC of 0.87 ( p = 0.003)] | 263/retrospective | Manually |
| Zhang et al. 134 | Differentiation between csPCa from insignificant PCa | MRI | The radiomics signature was significantly associated with clinically significant prostate cancer (p < 0.001) | 159/retrospective | Manually |
| Gong et al. 135 | Detection of csPCa | bpMRI | The combined clinical and radiomics model (the T2w/DWI) acquired an AUC of 0.788 | 489/retrospective | Manually |
| Woźnicki et al. 59 | Prediction of clinically significant prostate cancer | mpMRI | The model combining radiomics, PIRADS, PSA density and digital rectal examination showed a significantly better performance compared with ADC for csPCa prediction (p = 0.022) | 191/retrospective | Manually |
| Bernatz et al. 119 | Discriminating csPCa against indolent disease | mpMRI | Using RF, the additional application of max 3D outperformed PIRADS alone (p < 0.001, ΔAUC = +0.05) establishing the best working mode | 73/retrospective | Semi-automatic |
| Gugliandolo et al. 49 | Predictive of Gleason score, PIRADS v2 score, and risk class | mpMRI | Gleason score, PIRADS v2 score and risk class; AUC = 0.74–0.94 | 65/retrospective | Manually |
| Krauss et al. 110 | PSA level in patients with low suspicion for csPCa | MRI | Five RFs are significantly correlated with PSA level (r = 0.53–0.69, p < 0.05). The regression model significantly improves the explanatory value for PSA level (p < 0.05) | 36/retrospective | Manually |
| Song et al. 136 | Differentiate csPCa from indolent disease | mpMRI | AUC on the training, validation, and test data set achieved results of 0.838, 0.814, and 0.824, respectively | 185/retrospective | Manually |
| Castillo et al. 137 | Differentiate high- versus low-grade lesions | mpMRI | Radiomics models obtained a mean AUC of 0.75, outperforming the expert radiologist | 107/retrospective | Manually |
| Li et al. 132 | Prediction of csPCa | bpMRI | Both the radiomics model (AUC = 0.98) and the clinical–radiomics combined model (AUC = 0.98) achieved greater predictive efficacy than the clinical model (AUC = 0.79) | 381/retrospective | Manually |
| Li et al. 141 | SVM classification on classification of the GS of PCa in the central gland | mpMRI | The SVM classification based on mpMRI-derived image features obtains consistently accurate classification of the GS of PCa in the central gland | 63/retrospective | Manually |
| Bonekamp et al. 133 | Compare radiomics and mean ADC for characterization of prostate lesions (ISUP ⩾ 2) | MRI | Comparison of the area under the AUC for the mean ADC (AUCglobal = 0.84; AUCzone-specific ⩽ 0.87) versus the RML (AUCglobal = 0.88, p = 0.176; AUCzone-specific ⩽ 0.89, p ⩾ 0.493) | 316/retrospective | Manually |
| Bleker et al. 54 | Identification of clinically significant peripheral zone PCa | mpMRI | Combined model T2w and DWI images through an auto fixed VOI with AUC = 0.870 (95% CI = 0.980–0.754) | 206/prospective | Semi-automatic |
| Radiomics and detection of ECE | |||||
| Losnegård et al. 138 | Prediction of extraprostatic extension in non-favorable intermediate- and high-risk PCa patients | mpMRI | AUC ECE prediction models used in combination (MSKCC + radiology + radiomics) AUC = 0.80 | 228/retrospective | Manually |
| Ma et al. 139 | Identification of PCa ECE | mpMRI | Outperforming the radiologists results (AUC range = 0.600–0.697), (75.00% versus 46.88%-50.00%, all p < 0.05) | 285/retrospective | Manually |
| Ma et al. 140 | Identification of PCa ECE | mpMRI | AUC of 0.906 and 0.821 for the training and validation data sets | 165/retrospective | Manually |
ANN, artificial neural network; ADC, apparent diffusion coefficient; AUC, area under the curve; bpMRI, biparametric magnetic resonance imaging; CI, confidence interval; csPCa, clinically significant prostate cancer; DA, discriminant analysis; DCE, dynamic contrast enhanced; DNA, deoxyribonucleic acid; DRE, digital rectal examination; DWI, diffusion-weighted imaging; ECE, extracapsular extension; GLM, generalized linear model regression; GS, Gleason score; ISUP, International Society of Urological Pathology; LASSO, least absolute shrinkage and selection operator; mpMRI, multi-parametric magnetic resonance imaging; MRI, magnetic resonance imaging; MSKCC, Memorial Sloan Kettering Cancer Center; PCa, prostate cancer; PIRADS v2, prostate imaging reporting and data system version 2; PSA, prostate-specific antigen; PZ, peripheral zone; RFs, radiomics features; RML, radiomics machine-learning; RP, radical prostatectomy; SAVR, surface area-to-volume ratio; SVM, support vector machine; T2w, T2-weighted; TZ, transitional zone; VOI, volume of interest.
Radiomics and BM
Detection of BM in PCa
Although clinical outcomes are excellent for patients with localized PCa, patients with metastatic PCa have poor prognosis; indeed, median survival of patients with newly diagnosed metastases—although it is highly variable due to the heterogeneity of the M1 population—is approximately 42 months.12,142–144 PCa has the tendency to metastasize to bone sites (organ-specific homing), usually leading to osteoblastic BM—although it might also occur with osteolytic or mixed-type lesions.145–147 99mTc-Bone scan has been the most widely used method for M staging (i.e. evaluating BM); however, the field of non-invasive M staging of PCa patients is evolving rapidly and radiomics might offer a high potential. Yet, to date, most studies on PCa radiomics are focused on PCa detection, and research with specific regard to the detection of BM and the assessment of treatment response is still scarce.
Identification of BM using radiomics studies in PCa
Wang et al. 148 aimed to identify mpMRI-based texture features for pre-treatment prediction of BM in 176 PCa patients. Of the 976 extracted features, 15 were significantly associated with BM (p < 0.01). With regards to the prediction of BM, the model using MRI features derived from both T2w and DCE T1w showed better performance (AUC = 0.89) than those derived from a single sequence (AUC = 0.87 for T2w and AUC = 0.87 for DCE T1w) or GS (AUC = 0.73), yet the integration of the clinical risk factors (i.e. free PSA, age, and GS) into the MRI model achieved the highest performance (AUC = 0.92). Zhang et al. 149 sought to develop and validate a radiomics nomogram for predicting BM in PCa patients, combining mpMRI-based RFs and clinical risk factors. In their case series of 116 men with newly diagnosed PCa, of the 204 obtained features, 12 were significantly correlated with BM (p < 0.001). The proposed combined model—which included RFs from T2w, DWI, and DCE T1w images, and total PSA—had a higher discrimination efficiency (AUC = 0.92) than either the clinical or radiomics models alone (AUC = 0.85 and 0.84, respectively). Cysouw et al. 48 assessed an ML-based analysis of quantitative [18 F]DCFPyL PET metrics to predict high-risk pathological tumor features or metastatic PCa, analyzing 76 men with intermediate- to high-risk PCa scheduled for undergo RP + LND. This radiomics-based ML model—based on the extraction of 480 RFs—demonstrated a prediction of nodal or distant metastasis, with an AUC of 0.86, which was higher than that using standard PET metrics only (AUC = 0.81). Reischauer et al. 150 aimed to explore the potential role of whole-lesion volumetric texture analysis of ADC maps for treatment monitoring in PCa BM. A texture analysis—based on four first-order and 19 second-order statistical texture features—was performed in 12 treatment-naive men with BM before undergoing androgen deprivation therapy (ADT) and at 1, 2, and 3 months after starting hormone therapy. According to their analyses, almost all texture features showed moderate to high precision; moreover, both first- and second-order features changed significantly in response to ADT (Friedman’s tests). Acar et al. 93 explored the value of CT in evaluating treatment response in bone lesions. In particular, authors assessed a CT texture analysis in association with ML methods to differentiate bone lesions identified by 68Ga-PSMA-PET/CT as metastatic and completely responded sclerotic areas in a retrospective cohort of 75 PCa patients with BM who were previously treated. Texture features were obtained from lesions with and without PSMA expression and then compared. Results revealed a statistically significant difference in 28 of the obtained 35 features between the two groups. Furthermore, the weighted k-nearest neighborhood algorithm showed a good accuracy in distinguishing between the two types of lesions (AUC = 0.76). Currently, there is an ongoing trial (NCT04343885) designed to evaluate the prognostic and predictive role of radiomics using PET, CT, or bone scan after treatment with Lu-PSMA therapy and/or chemotherapy in a population of newly diagnosed high-volume metastatic hormone-naive PCa men.
Radiomics and BCR
General considerations of BCR in PCa
BCR is defined as a rise in PSA to 0.2 ng/ml after RP or a rise of 2 ng/ml (or more) above the nadir PSA after radiation therapy. 12 Following radical treatment, 27–53% of patients experience BCR within 10 years, with a particularly increased risk in patients who report locally advanced disease (> cT2c or pT3), positive margins or high GS > 8.151,152 Nevertheless, the risk of BCR among PCa patients is not uniform and, more importantly, not all patients with BCR develop disease progression or distant metastases. 153 Adjuvant therapy for RP or local RT, although could have a role in high-risk patients (as positive margins and pT3 disease) and improve BCR-free survival, has not been definitively established and still remains controversial, due to adverse effects and potential overtreatment. 154
The current role of radiomics and BCR in PCa
In this background, the use of radiomics in predicting BCR could address clinical decisions, permitting to tailor a proper treatment for Pca patients and, contextually, avoiding the risks of overtreatment. Although few studies are reported in the literature, growing evidence supporting the use of pre-treatment mpMRI radiomics in the prediction of BCR occurrence has been produced since 2018. In an exploratory retrospective study by Shiradkar et al., performed on 120 PCa patients of two different institutions, which performed a 3T mpMRI pre-treatment, a total of 150 RFs derived from T2w and ADC maps were evaluated, reporting, in 10 top-discriminating RFs, a significant difference between BCR + and BCR lesions (p < 0.05) ranked according to C-index for an AUC of 0.84 in the training data set, and an AUC of 0.73 on validation data set. The combination of clinical parameters and RFs resulted in an improved prediction in the training data set (AUC = 0.91) while a marginal improvement was reported in the validation data set (AUC = 0.74). Among the top-discriminating RFs, skewness and kurtosis statistics of Haralick features obtained from T2wI and CoLlaGe features obtained from ADC maps, reported the best results [hazard ratio (HR) = 2.09 and 2.28, respectively]. 155 A similar study performed by Dinis Fernandes et al., investigated the potential of whole-prostate mpMRI features for 5-year BCR prediction after RT of local PCa in a cohort of 120 high-risk patients. Using PyRadiomics 1.2.0 toolbox, a total of 254 region-level features were obtained per patient and, successively, two independent models were created using clinical or RFs. Overall, an AUC of 0.63 was obtained for whole-prostate pre-treatment mpMRI-RFs compared with an AUC of 0.51 for the clinical model. As stated by the authors, despite those AUC values were relatively low to permit to extrapolate significant clinical decisions, results obtained highlighted the potential of RFs in the prediction and evaluation of BCR. 107 In 2019, Bourbonne et al. validated an mpMRI radiomics-based model predicting BCR and BCR-free survival on 107 high-risk PCa patients (pT3–pT4, positive margins, GS 8–10) treated with RP and followed for a median of 52 months. After semi-automatic prostate tumor delineation, a total of 172 RFs were extracted for the successive analysis. Among those RFs, 10 non-redundant and uncorrelated features were significantly associated, both at uni- and multivariate analyses, with BCR, reporting, in particular a strong correlation for SZEGLSZM, SZLGEGLSZM, and HGREGLRLM with, respectively, OR of 16.6 (p = 0.026), 8.8 (p = 0.025), and 15.2 (p = 0.011). The radiomics model reached an AUC of 0.799, which outperformed the clinical model (AUC = 0.57), while the combination of both models resulted in a high prediction of BCR with an AUC of 0.849 (p < 0.0001) and a prediction of biochemical recurrence-free survival (bRFS) with a HR of 23.1 (p < 0.0001). In addition, one RF extracted from ADC maps, small-zone emphasis (SZE), reported the highest predictive ability regarding BCR alone, with an AUC of 0.79 and an HR of 17.9 (p = 0.0001). Overall, with only 17 patients who reported BCR, thus yielding a negative predictive value of 96%, the authors suggested their model as a helpful method to identify patients at very low risk of recurrence, permitting a better stratification for those eligible for adjuvant RT. 156 The findings reported, however, could have been relevantly influenced by the presence of 1.5 T mpMRI only in the cohort, permitting to hypothesize potentially better results using 3T coils only.156,157 A subsequent external validation was performed by the same authors on 88 PCa patients who underwent RP and were followed for a median of 49.2 months. The previously identified RF (ADC SZEGLSZM), confirmed the independent and significant correlation with BCR, yielding an AUC of 0.82 for the radiomics model, while the clinical prediction model reached an AUC of 0.68. Moreover, when a cut-off of 0.53 was set for ADC SZEGLSZM value, patients >0.53 reported 37 months of bRFS compared 19.2 months of patients <0.53 (p = 0.0013). Similarly, at the follow-up, 83.3% of patients <0.53 exhibited BCR compared with the 22% of patients >0.53. Moreover, the combined radiomics/clinical model did not outperform the radiomics-only model, reporting an accuracy of 67% and an HR of 5.7 for bRFS (p < 0.0001). Differently from the previous study, the validation cohort included patients with 3T mpMRI, which could partially explain the better results. 63 An analogous study by Zhong et al. evaluated the relationship between pre-treatment 3 T mpMRI-RFs and BCR in a retrospective analysis of 91 patients with localized PCa after RT. 1536 quantitative RFs were extracted; among them, only 45 RFs were used as input of neural network-based ML, reporting 32% of patients classified as BCR and 68% as non-BCR. As result, the overall classification accuracy was 74.1%, with a correct classification rate for positive samples of 50% and a correct classification rate for negative samples of 86.1%, for a mean AUC of 0.99 for the training cohort and 0.73 for the test cohort. 158 A further effort in the identification of patients at risk of developing BCR was made by Li et al. which sought to develop and evaluate a radiomics risk score (RadS), and creating, in addition, an integrated radiomics-clinicopathologic nomogram called RadClip. A total of 198 patients, who had undergone a pre-operative 3T mpMRI and followed for a mean of 35 months, were included in the study. Two hundred RFs were extracted from T2wI and ADC maps, including first- and second-order statistics (Gabor, Laws, Haralick, and CoLlaGe). RadS, which was built with five RFs, resulted in an HR of 7.01 (p < 0.05) and was significantly associated with bRFS while RadClip reported an HR of 1.9 (p < 0.05) and a C-index of 0.77, with an AUC of 0.71–0.66 (p < 0.05). These results outperformed Decipher prostate test and prostate cancer risk assessment (CAPRA), two risk assessments based on clinical and genomic features. 159 A short summary of this research is inserted in Table 5.
Table 5.
Radiomics and biochemical recurrence studies.
| Author | Clinical outcomes | Type of image acquisition | Results | No. patients/prospective or retrospective | Segmentation |
|---|---|---|---|---|---|
| Radiomics in detection and prediction of PCa biochemical recurrence | |||||
| Shiradkar et al. 155 | Prediction of BCR | MRI | AUC: 0.84 in training data set; AUC: 0.73 in validation data set. Mixed model AUC: 0.91 in training data set and AUC: 0.74 in validation data set | 120 /retrospective | Semi-automatic |
| Dinis Fernandes et al. 107 | Prediction of BCR | MRI | AUC: 0.63 for radiomics model compared with AUC: 0.51 of clinical model | 120/retrospective | Semi-automatic |
| Bourbonne et al. 156 | Prognostic value of BCR in high-risk PCa patients | MRI | Radiomics model AUC: 0.799; clinical model AUC: 0.57; Mixed model AUC: 0.849 | 107/retrospective | Semi-automatic |
| Bourbonne et al. 63 | External validation of radiomics model in prediction of BCR | MRI | Radiomics model AUC: 0.82; clinical model AUC: 0.68 | 88/retrospective | Semi-automatic |
| Zhong et al. 158 | Prediction of prognosis of localized PCa after RT | MRI | AUC: 0.99 in training data set; AUC: 0.73 validation data set | 91/retrospective | Semi-automatic |
| Li et al. 159 | Prediction of biochemical recurrence-free survival via a radiomics risk score (RadS) | MRI | AUC: 0.71, C-index 0.77 | 198/retrospective | Semi-automatic |
AUC, area under the curve; BCR, biochemical recurrence; MRI, magnetic resonance imaging; PCa, prostate cancer; RT, radiotherapy.
Radiomics, and treatment response and toxicity
Brief considerations of treatment response and toxicity in PCa
The management of PCa has widely changed with the availability of novel treatments. In particular, the imaging, which was previously limited for the initial diagnosis, has become a potential marker of treatment response and evaluation of RT toxicity in PCa. To minimize damage to surrounding structures and to deliver the best treatment, a radiomics-based RT planning has been suggested by several authors, emphasizing the performance of mpMRI in diagnosis, staging, grading, and therapy assessment. 160 One of the first studies toward this direction was performed by Lee et al., which reported a radiomics-based approach in the evaluation and assessment of RT protocols.
Treatment response and radiomics in PCa
As reported by Abdollahi et al., who evaluated a total of 33 PCa patients treated with nine coplanar intensity-modulated radiation therapy (IMRT) fields, the response evaluation utilizing changes in ADC values pre and post IMRT. After a first features extraction, which accounted for a total of 4540 features, 40 RFs (20 from T2w and 20 from ADC) were selected. Overall, an AUC of 0.61 was reached for T2w radiomics, while an AUC of 0.63 was reported for ADC radiomics. In particular, 2 and 15 RFs, of, respectively, T2 and ADC reported a statistical significance (p < 0.05). Although several limitations, as the limited number of patients involved and the retrospective nature of the study, the study by Abdollahi et al. represented the first step in the personalized PCa diagnosis and therapy due to an advanced ML and radiomics approaches. 161 The role of radiomics in treatment response was also assessed in novel RT techniques, as carbon ion radiotherapy (CIRT), which allows for improved tumor control while sparing adjacent tissues. In a study on 60 PCa patients, Wu et al. reported indeed, on a total of 26,601 features, a higher intra correlation coefficient (ICC) of ADC RFs (0.71) compared with T2w RFs (0.6) (p < 0.01), indicating higher robustness of the first against delineation uncertainty. By combining both features, the model yielded an AUC of 0.88. 103 Another use of radiomics in treatment response is reported by Yu et al. which investigated the utility of this new technology in ADT on a cohort of 43 patients (23 ADT non-responders and 20 ADT responders), extracting 396 RFs from T2wI maps performed after treatment. Compared with the clinical model, which reported an AUC of 0.774, the radiomics model reported an AUC of 0.807–0.855, based on texture features with an ICC > 0.8. Interestingly, the combined model reached a promising AUC of 0.811–0.997, yielding a sensitivity of 0.87 and a specificity of 0.9. 162 The importance of these findings is related to the possibility to properly evaluate, in single patients, the efficacy of treatment with cost and time-effective methods, confirming the role of mpMRI radiomics in monitoring ADT and IMRT responses. Finally, in an mpMRI dominated radiomics, Moazemi et al. reported an interestingly PSMA-PET/CT radiomics model evaluated before Lu-PSMA treatment: on a total of 2070 pathological hotspots (73 features per hotspot) in 83 PCa patients, a mixed PET/CT RFs reported an AUC of 0.8, for a sensitivity of 0.75 and an analogous specificity. 163
Toxicity and radiomics in PCa
mpMRI could also be utilized in the assessment of PCa therapies toxicities which regard in particular RT with radiation-induced bone changes, proctitis and cystitis. To predict early complications, a radiomics approach has been tried for femoral head changes in patients treated with IMRT, confirming early changes in ADC, T1- and T2-derived features. 164 Similarly, on 274 RFs extracted from T2w images in 33 patients who, analogously, underwent IMRT, it has been possible to predict urinary toxicity with an AUC ranging from 0.62 to 0.75. It is clear how, with the available technology related to radiomics, the possibility to use non-invasive, non-ionizing imaging in the prediction of complications and toxicities related to therapies, represent a promising step forward in the tailoring of treatment and reducing, in this case, bladder complications induced by radiation. 108 Pre-IMRT mpMRI radiomics could also help in the prediction of rectal toxicity, where, with RFs extracted from T2w and ADC images, the radiomics models reported a mean AUC of 0.68 and 0.61 for, respectively, pre and post IMRT mpMRI. 106 In a prospective study, Mostafaei et al. predicted, via a CT-based radiomics, RT-induced toxicities, reporting, on 64 PCa patients, an AUC of 0.66, 0.71 and 0.65 for, respectively, clinical, radiomics, and mixed models, regarding gastrointestinal toxicity (⩾grade 1 proctitis); similarly, AUC was, 071, 0.67, and 0.77 for urinary toxicity (⩾grade 1 cystitis). 92 Finally, an exploratory study by Lorenz et al. evaluated the role of delta-radiomics, i.e, the variation in quantitative image metrics over time, in the analysis of IMRT-induced toxicity. Although the small cohort of patients involved (4 PCa patients) and the non-conclusivity of data obtained, authors confirmed the feasibility of this approach in acquiring longitudinal radiomics data, potentially opening a wider opportunity in this future research. 165 The conclusions of these trials are shown in Table 6.
Table 6.
Radiomics and PCa treatment toxicities studies.
| Author | Clinical outcomes | Type of image acquisition | Results | No. patients/prospective or retrospective | Segmentation |
|---|---|---|---|---|---|
| Radiomics in the evaluation of treatment toxicity | |||||
| Abdollahi et al. 164 | Assessment of early changes in femoral heads in PCa patients treated with IMRT | MRI | All RFs underwent changes pre and post IMRT | 30/retrospective | Manual |
| Abdollahi et al. 108 | Assessment of urinary toxicity in PCa patients treated with IMRT | MRI | Radiomics model AUC: 0.62–0.75 | 33/retrospective | Manual |
| Abdollahi et al. 106 | Assessment of rectal toxicity in PCa patients treated with IMRT | MRI | Radiomics model AUC: 0.68 (pre-IMRT) and 0.61 (post-IMRT) | 33/retrospective | Manual |
| Mostafaei et al. 92 | Evaluation of urinary and GI toxicity | CT | GI toxicity Radiomics model AUC: 0.71; clinical model AUC: 0.66; mixed model AUC: 0.65 Urinary toxicity Radiomics model AUC: 0.67; clinical model AUC: 0.71; mixed model AUC: 0.77 |
64/prospective | Semi-automatic |
| Lorenz et al. 165 | Evaluation of delta radiomics in the analysis of IMRT toxicities | MRI | Feasibility of delta radiomics in the evaluation of IMRT toxicities | 4/retrospective | Manual |
AUC, area under the curve; CT, computed tomography; GI, gastrointestinal; IMRT, intensity-modulated radiation therapy; MRI, magnetic resonance imaging; PCa, prostate cancer; RFs, radiomics features.
Radiomics and AI in PCa
AI and radiomics for the management of PCa
Radiomics in the diagnostic assessment and urological aid for decision-making treatment choice for PCa is the ‘omic’ discipline focused on the withdrawal of the analysis related to the textures and features, 109 that provides an assessment of data sets from functional imaging modalities (e.g. transrectal US and/or mpMRI) through semi-automatic or automatic software. 47 The main goals of the discipline are to pinpoint the localization of the suspected PCa lesions and to predict csPCa versus indolent PCa behavior.111,125 Similar methodology has been previously investigated for different other malignancies,166,167 but recently with the advent of AI and ML algorithms, novel risk-grouping has been introduced regarding PCa, focusing on the differentiation of indolent from csPCa, 43 on the modeled prediction for the relative tumor GS percentage at final pathology both at prostate biopsy52,168 and on the BCR-free survival outcome after a curative intervention, such as RP and/or RT. 70 Moreover, the radiomics sciences have contributed developing novel potential detection features for PCa diagnosis, such as advanced z-DWI and conventional full-field-of-view DWI. 99 The new insights are also currently focused on the texture analysis of prostate MRI in the PIRADS for PIRADS 3 score lesions, 100 the creation of frameworks for automated PCa localization and detection;43,101 and, finally, the management of RT treatment and toxicity. 69 In addition, radiomics and AI together could eventually aid in limiting the inter-agreement variability among different GU radiologists with different levels of expertise. Indeed, mpMRI of the prostate is based on both qualitative parameters reflecting the ability of an expert radiologist to provide an accurate scoring for the lesion images in the prostate gland and prostate-specific quantitative measurements, such as tumor size, prostate volumes, and RFs previously mentioned which directly derives for the acquired images 156 that could be considered reader-independent thus potentially being converged in AI algorithms reducing the radiological discrepancy among non-tertiary centers.
The current trends and evidence of using AI and radiomics in PCa
Specifically, with regard of early detection of csPCa lesions biopsied after a positive mpMRI, Strom et al. 46 identified n = 10 MRI-based shape features which were extracted both from axial T2w and ADC map images, after lesion tridimensional segmentation on mpMRI. At their multivariable analysis, the authors revealed that the parameter defined as surface area to volume ratio (SAVR), especially when extracted from ADC maps was the strongest independent predictor of csPCa among tested shape features. As demonstrated in the article, quantitative radiomics was able indeed to allow for the extraction of more advanced shape features than previously possibly imaginable if we would consider that lesion maximum diameter and volume of the index areas are currently considered those with highest diagnostic accuracy. Santone et al. 52 described a novel radiomics-based algorithm that automatically inserts images from the Gleason grade group (GG) in mpMRI images. The authors proposed a methodology that originated as input from MRI and generated a formal model from the MRI slice itself. Thus, the model was verified with a set of properties (one property for each GG) with the formal verification tool. This RFs through formal verification reached a sensitivity ranging between 0.95 (for the GG1 and GG3 detection) and 1 (for the GG4 and GG5 detection) while specificity was equal to 1 for all the GGs. Nevertheless, the main limitations of the proposed methodology were the manual definition of the formulae as well as the time performances which were considered significantly high and poorly reproducible in clinical environment.
Focusing on the implications of the radiomics sciences on treatment influences, Gugliandolo et al. 49 just recently published a sub-analysis from a phase II trial on ultra-hypofractionated radiotherapy (AIRC IG-13218). In their article, the authors analyzed the MRI-based radiomics signature for localized PCa as a new potential tool for cancer aggressiveness prediction. Specifically, n = 65 patients undergone RT for cs-localized PCa, were reviewed and the T2w MRI images were normalized with the histogram intensity scale standardization method. The association of each RF with risk class, pT-stage, GS, ECE score, and PIRADS v2 score was assessed by logistic regression. The results demonstrated that those texture features were the most predictive for GS, PIRADS v2 score, and risk class, while different intensity features were associated with T-stage, ECE score, and risk class, with AUC ranging from 0.74 to 0.94.
Another field of application deserving particular attention for future implications of radiomic-clinicopathologic assessment within PCa patients has been represented by the study of Li et al. 159 who developed a nomogram so-called ‘RadClip’ for predicting post-surgical BCR and adverse pathology of PCa from pre-operative bpMRI. This comparative experience confronted the nomogram based on approximately 200 RF, including first- and second orders of statistics, together with novel specific RFs previously described, which were extracted on a per-voxel basis within PCa lesion regions on both T2w images and ADC maps. These features analyzed with the aim to characterize the underlying texture, and to quantify heterogeneity on bpMRI, were melted into the nomogram for to characterize the PCa lesion regions. The score was then compared with other predicting tools, such as the CAPRA score, post-surgery prostate cancer risk assessment (CAPRA-S) score, and Decipher genomic test, which are already available tools to identify patients at risk of developing BCR or adverse pathology at RP. Surprisingly, ‘RadClip’ resulted in a higher AUC (0.71, 95% CI = 0.62–0.81) for predicting AP compared with Decipher (0.66, 95% CI = 0.56–0.77) and CAPRA (0.69, 95% CI = 0.59–0.79) even if several limitations of the study have to be acknowledged like the retrospective design itself allowing for covariate and selection imbalances.
Diving deeply into treatment-related possible applications, Lee et al. 169 investigated early changes in the ADC RFs for patients undergoing RT with dose escalation to the gross tumor volume. Overall, n=101 RFs were tested to verify for diminished image contrast post-external beam radiation therapy and possible differences in prostate gland volume, shape, and orientation. The findings revealed significant changes in numerous gross tumor volume and prostate features assessed from ADC and T2w images during RT. The authors therefore concluded that the findings could be potential biomarkers of early radiation response. In addition, Mostafaei et al. 92 recently presented prediction models of RT-induced toxicities in PCa patients through the use of CT radiomics. The authors described two models (radiomics and clinical) for predicting RT-induced urinary and gastrointestinal toxicities. The AUC for clinical, radiomics, and clinical–radiomics models was 0.66, 0.71, and 0.65, respectively, for gastrointestinal toxicity, while for urinary events, the AUC for radiomics, clinical, and clinical radiomics models was 0.71, 0.67, and 0.77, respectively. For cystitis, the authors observed that radiomics with clinical and dosimetric features were able to amplify the predictive performance to an AUC of 0.77. Differently, for proctitis modeling, outcomes were non-comparable and the association of radiomics and clinic presented together lower performance than the radiomics model alone.
Finally, regardless of the improvements in the diagnostic accuracy and prediction strategy which clearly appears promising yet still far from daily clinical practice, the novel RFs associated with AI developments in particular DL techniques might alleviate the problems regarding the inter- and intra-observer variability as well as long learning curves associated with multi-parametric scoring systems, such as PIRADS criteria thus potentially reducing discrepancy and implementing the wide spread for mpMRI diagnostic modalities.
Notes on the advantages and limitations of classifications algorithms
The CAD tools commonly used to classify resampled instances to analyze medical images from pictures databases are RF, SVM, 170 and ML algorithms.171,172 Other classifiers used are naive Bayesian classifier (NBC) and k-nearest neighbor (KNN). 44 Lately, most of the AI methods used are convolutional neural networks(CNNs), because they have the potential to extract complex variables. 173 Other researches focused on ML methods, like SVM, DT for classification, and LASSO and Ridge regression methods, for features extraction, are heavily implemented in recent studies.
To ease the work of researchers, a comparative analysis between the proposed and used classifiers that could enhance the performance, costs and training of image data sets is mandatory. 174 Usually, a team of researchers are using computer science or statistics they are familiarized with. Findings revealed that the best-performing method as a radiomic classifier is boosted quadratic discriminant analysis (QDA) that has a relatively short execution time and it seems that all variants of the DT classifiers have overestimated performance. All result should be independently validated, as it is a less generalizable model. The SVM classifier has a reasonable performance.174,175 The limitation for these classifiers resides in a longer time to train and test and without convergence in a validation setting. 174 Bayesian and QDA classifiers have low variance, subjected to bias, and hard to generalize results.
The classifiers taken into account are arranged as different sets of families. Some are linear discriminant analysis and generalized linear models and rule-based classifiers or DT. First have roots in statistics and the second family is related to AI methods. The generalizability and implementation of this classifiers could represent a breakthrough in using automated algorithms, but this can be limited by the use of simple statistical correlation analysis, like Waikato Environment for Knowledge Analysis (WEKA 3) 176 and neural network algorithms, like Matlab.177,178 The R collection of algorithms 179 is easy to use for automatic tuning of features and it is available for researchers that are just familiarized with the methodology of statistics. All classifiers in a family have strong differences between data sets and the mean accuracy can have a limited significance if the data sets used are at low volume. 180 The assessment of how well different ML methods perform is using benchmark experiments. These experiments try to compare the performance of different algorithms, then cross-validation and resampling models analyze the best algorithm for the given task.181,182 The AI models have been often used for diagnosing, mapping and intimate description of prostate tumors. 183 From the clinical point of view, latest evidence is that CAD systems can improve the reading and interpretation of imaging images by the radiologists.184,185
Research that searched to distinguish between classifiers revealed that RF has the best performance when compared with other methods. 186 RF compared with SVM leads to reduced training and testing costs and in the same time performance is comparable. 187 Following the same line, another study, 188 showed that RFs have the best performance with the highest sensitivity (92.6%) and negative predictive value (92.6%). RF method compiles a high number of regression trees (RTs) to build a forest, to address the instability of the classification, and RT. 127 RFs’ limitations are also related to the time and the training of individually random trees on random subsets of data. 189 The researcher has to have very good understanding and knowledge of how to use the appropriate algorithm for the task at hand. An RF advantage is that has generalizability and robustness in overall performance and does not result in overfitting problems. 190
KNN is a performing model used for data mining, 191 and a nonparametric model for classification and regression. 192 In PCa, KNN is a basic algorithm that classifies data based on a similarity measure and pattern recognition. 191 One advantage is that is fast, and can learn from small data volumes and have good results in PCa diagnosis. 193
SVM had the best accuracy when compared with RF and KNN in image augmentation to differentiate Gleason groups. 194 In different radiomics studies, pulmonary195,196 and colorectal 197 achieved good performance in MRI sequences (T2w and DWI). Lately, Xuefi et al., 198 identified that LASSO has advantages in radiomics studies and in combination with clinical data can improve the standardization of the model used. In the classification of suspicious lesions on prostate, MRI-RFs have better AUCs, compared with LASSO. 127
When we use AI to help the reader in boosting the generated results, we have to involve maintaining, preserving and adding value to digital research data throughout its lifecycle by trained professionals. To develop the automated classifier leads to a rise in expenditure and time to finish the process. 199 Advantages of unsupervised and self-supervised methods derived from deep neural networks (DNNs) decrease the time, costs and balances the method used. 200 A drawback in using AI methods is the lack of consensus on data sets to improve performance, generalizability, and reproducibility, benchmarking being mandatory for the process.199,201 Limitations of AI are linked to the interpretability and ability to interrogate these methods and if the method will lead to the expected results. Therefore, institutions bound to watch over the acceptance of such methods in detecting, staging and treatment of diseases are challenging the ability of AI over human experts. In present time, CAD systems can be considered unethical due to confidentiality of patient’s medical data. 202 There are some studies that compared the feasibility of different algorithms in head and neck cancers subjected to radiotherapy and assessing cochlear damage, LASSO versus MLs methods, 203 with LASSO preventing overfitting due to collinearity of the covariates and high-dimensionality, which is a clear advantage of the method. 204 The LASSO shrinkage methods have good performance in correlated high-dimensional data. 127 To overcome the drawback of not-important variables chosen by standard algorithm, the adaptive LASSO work better to deliver consistent variable selection. 205 In PCa, MRI-derived delta-radiomics is linked to similar performance to lead radiologists for predicting histopathological progression of PCa, which used three classifiers (LASSO, RF, and SVM). All signatures had good performances in differentiating between benign and cancerous lesions. 198 And advantage of LASSO is that it can extract features and regularize simultaneously data from radiomics. The PRECISE scoring system studied in PCa, with medical images from MRI, compared the performance with parenclitic networks, LASSO regression, and RFs ML methods, and had better results. 188 A recent meta-analysis showed that there are no differences in heterogeneity, performance, sensitivity, and specificity among different ML algorithms used in PCa radiomics studies. 206 In DL studies, heterogeneity was higher than among the studies employing other ML approaches to detect PCa. DL performs complex tasks and they use large amount of data. For this reason, it seems that they are better used to analyze vast amount of medical images to automatically identify patterns and achieve high performance. This is also available for unbalanced and small data sets. Among the advantages of CNNs are consistent diagnoses, cost-effectiveness, and improved efficiency. 171 Ishioka et al., 207 developed a CNN algorithm to pinpoint a specific zone for the detection of PCa through biopsy. Other studies have shown no additional performance for differentiating PCa from benign tissue using radiomic ML. 132
Direct comparison between different ML algorithms is scarce in terms of available research. There is also an ongoing debate on which of the proposed, ‘newest’, ‘most used’, ‘familiarized’, ‘at hand’ algorithms are better to study the computer-aided radiomics to detect, characterize, treat, and follow PCa patients. Advantages come with limitations and therefore further algorithm development and research is mandatory. Existing research did not compare AI with conventional statistical analysis. Conventional statistics can analyze a small number of training features and AI can process vast amounts of data, and for future comparison studies, it will be challenging. Almost every of the above-mentioned research did not compare the classifiers between them and a conclusion regarding the best algorithm classifier to be used for the study of radiomics in PCa cannot be used at this time.
Discussion
Radiomics is the next-level research field that could have the potential to offer personalized medicine for PCa patients. The included papers in this review are from all areas with an impact in PCa management. Our search found 60 studies that used mostly imaging provided by MRI as by recommendation of the guidelines 12 as imaging option for PCa patients. Nonetheless, CT, PSMA-PET, and TRUS are imaging options not commonly used, but there are several works researching their potential in that field in the last years. MRI is mostly used in the detection of PCa, 69 identifying and differentiate between csPCa and favorable PCa, 208 treatment with RT and monitoring toxicity, the prediction of ECE, BCR, and GS.
Radiomics represent a promising diagnostic tool that relies on a proper extraction and analysis of quantitative radiological features mediated through proper contouring of ROI, effected via semi-automatic or automatic methods. 76 As result, a certain degree of contouring variability has to be taken into account. 209 In particular, even for contour created by a trained radiologist, inter- and intra-observer contouring variability could significantly impact the successive radiomics extracted features, while in automatically generated contouring, despite promising results, current findings are still unripe.210,211 Despite this could potentially lead to important limitations related to reproducibility, novel approaches assessing the sensitivity of RFs in relation to inter-observer variability (via the normalization and quantization of images before extraction or the use of morphological features) could limit this issue.212,213 Different combination models between clinical features, imaging phenotypes, and radiomics gave better results in diagnosing PCa, but their use is still limited to clinical research. Most of the research papers used manually interpretation and segmentation of the images, with few articles that used semi-automatic or automatic processing of RFs. The well-known inter-observer variability, 214 different acquisition protocols, different machines used for these images are biases for the standardization of the techniques. The rise of AI and ML, and the automatization of acquisition and segmentation of these intra-prostatic lesions could potentially limit this bias. 43 One important researched aspect was the radiomics assessment of GS. MRI, CT, and TRUS looked at the RFs that can predict the known intra-tumoral heterogeneity of PCa.
CT evaluated GS in three studies explored the post-surgical GS and post-RT prognosis and risk stratification of different RFs to predict the GS. CT articles used in research the 68Ga-PSMA-PET/CT scan and radiomics assessed overall survival after treatment, detection of LN metastasis, metastasis, and detection of PCa areas in whole gland after RP. Recently, Chu et al. 215 also investigated the possibility of PSMA-PET RFs and the examination of PSMA expression and found a correlation between PSMA expression and GS and Zamboglou et al. showed that PSMA-PET RFs can excellently perform in visualizing unknown PCa, thus paving the road to more studies using PSMA-PET and radiomics for risk stratification of PCa patients. PSMA-PET studies are of valuable use in the setting of detecting LN metastasis, guiding treatment, evaluating the radio-toxicity and assessing distant organ metastasis. Only one study is indicating the accuracy of PSMA-PET/MRI for the diagnosis of intra-prostatic cancer nodules. 88
A few studies addressed PCa detection and GS with the use of TRUS RFs. It draws attention because it has several advantages: is non-invasive, radiation-free, cost-effective, widely available, and practical. Even if the AUCs are high for the proposed models, but we know that radiomics is a computer-based order of operations that can be modified and there are a number of proposed models that are modified, approximately by each research team and not standardized, therefore the results to be validated further, the researchers need to lower the variability, enhance robustness and reproducibility for all imaging technologies used. 216
Using MRI-RFs for screening purposes is low on clinical evidence. We only found two studies that looked into this field with moderate to good AUCs, but with the same biases as depicted in the upper paragraph. Chen et al., 114 for instance, found that radiomics-based model better detected PCa than PIRADS v2 model versus non-PCa lesions and for differentiating high- from low-grade PCa, and Khalvati et al. 124 found that radiomics improved PCa detection with the use of SVM classifier, both using MRI images. Assigning GS to lesions obtained through MRI-RFs seems feasible to be investigated as also Brunese et al. have shown earlier. But the researchers have no consensus between studies to identify the most favorable model, along with external validation of those models to apply to populations outside the studied patients’ cohorts. As heterogeneity of PCa tumors pose management challenges on how to distinguish between csPCa and indolent disease, several studies, using also AI and ML techniques in a very well-designed environment of clinical studies, managed to outperform just PIRADS or clinical expertise of radiologists. These findings were pointed out by Zhang et al. from T2w, DWI, and ADC images, Gugliandolo et al. in complex combined models, and also studies like Bonekamp et al. where radiomics ML did not perform better than mean ADC assessment. For ECE assessment just combined predictive models between clinical, conventional radiology and radiomics achieved the best results.
Several articles investigated the potential of using radiomics in BCR achieving moderate results in terms of radiomics models AUCs (range = 0.63–0.99) and clinical models (range = 0.51–0.82) with the best results in mixed models (0.74–0.84). The potential is there but standardization is the goal for future studies.
Radio-toxicity has been assessed by radiomics studies, but there are only few studies with modest AUCs reported so far. By combining RFs with AI and ML methods, these applications may hold promise for future studies and clinical implementation from diagnosis to outcome and toxicity prediction.
AI methods are an attractive research field in PCa. In clinical settings, there are several limitations, such as the variability in study design, algorithms used, and training methodologies. The validation of results is often within the data set and not externally. There is also a need to compare AI studies with conventional imaging and interpretation. But the predictive precision of AI and ML has the ability to provide personalized medicine by including prospective, large data sets to obtain more information for the molecular and biological aspects of PCa.
Future perspectives for clinical translation
Radiomics represents an emerging field, and it will play an important role in the future of personalized medicine. Its fundamental element is the extraction of quantitative features from radiological images that cannot be processed by radiologists, although today there are still limitations on the complete automation of the model selection, analysis, and creation process. 44 The most recent studies analyze mostly images from MRI, and this is an important limit of radiomics in PCa. For this reason, future studies should use multimodality imaging, such as PET, MRI, CT, and others. In this way, the study of Papp et al. 50 showed the power to integrate risk classification in PCa through PET/MRI, radiomics and ML in patients without a previous prostate biopsy. The advantage of combining the PSMA-PET information with the mpMRI overcomes the limitations of each of the methods in PCa detection, and is needed to provide a deep identification of the entire intra-prostatic tumor amount. 44 Kesch et al. 217 also showed a strong correspondence between the multi-imaging features and genomic index lesion which was based on the close correlation between mpMRI, 68Ga-PSMA-PET imaging, and chromosomal copy number alteration at the biopsy. These studies pave the way of the clinical translation of radiomics in PCa. Moreover, most of the existing studies in literature are retrospective and carried out in single institutions. The prospective studies, instead, are in a limited number, as shown in Table 6, and many of these are not yet in the recruitment phase. The results of these studies, however, could be groundbreaking in the next future, opening a new era in the diagnostics and tailored treatment of patients. For this reason, more prospective studies should be performed in the future. Another important field in which radiomics could play a fundamental role is theranostic, which combines specified therapeutics and specified diagnostics. In this way, PSMA-PET could have in the near future good results not only in diagnosis but also in treatment of patients with hormone-refractory PCa. 218 PET radiomics could also become fundamental in immunotherapy, given the close correlation that has been highlighted between tumor metabolism and the immune presence in the tumor microenvironment. 219 Furthermore, radiomics represents a milestone in radio-genomics, integrating tumor morphology with genetic characteristics of the tumor, thus allowing deriving useful radio-phenotypes to integrate the already existing validated genomic risk stratification biomarkers.220–222 A very important limitation, however, is the lack of a robust methodological radiomics framework. For this reason, one of the goals in the near future should be a good standardization in imaging and reconstruction settings to create a harmonization of images of different qualities for more accurate and robust results. 223 Furthermore, research needs to be directed toward a deeper analysis of the inter-tumor and intra-tumor heterogeneity. 219 Despite these important limitations, radiomics has enormous potential to become a cornerstone of personalized medicine, radio-genomics and theranostics, thanks to the possibility of providing important information useful for the prognosis, treatment, and response of the tumor. All these trials are summarized in Table 7.
Table 7.
Radiomics ongoing trials.
| Trial | Date | Status | Interventions | Characteristics | Outcomes |
|---|---|---|---|---|---|
| Prospective evaluation of mpMRI, MR-guided biopsy, and molecular markers for AS of prostate cancer (PROMM-AS) 224 | Start: October 2017 | Unknown, no results available | Diagnostic test: *mpMRI *MRI-guided *Biopsy *Radiomics *Molecular markers |
Type: interventional Phase: not applicable |
Reduction of the discontinuation of AS • Value of MRI (ADC) regarding aggressiveness • Detection rates of targeted FUS-GB versus systematic TRUS-GB biopsies • Correlation of clinical parameters with Gleason score progression or MRI quantified progression • Patient compliance to recommended MRI-based observation • Evaluation of resolve DWI |
| Can MRI of the prostate combined with a radiomics evaluation determine the invasive capacity of a tumor (MRI-PREDICT) 225 | Start: September 2021 |
Not yet recruiting | Diagnostic test: MRI accuracy and stability |
Type: interventional Phase: not applicable |
Outcome measures: • MRI classification change • MRI classification: baseline • MRI Classification: Week 8 • Model optimization with novel RFs and clinical covariates |
| PSMA-PET: deep radiomics biomarkers of progression and response prediction in prostate cancer 226 | Start: December 2018 | Recruiting, no results available. | Diagnostic test: 18FDC FPyL IV injection |
Type: Interventional Phase III |
Outcome measures: • Overall survival • Progression-free survival |
| MR radiomics features in prostate cancer 227 | Start: September 2019 |
Recruiting, no results available | Type: observational | Outcome measures: • Number of surrogate bioimaging markers of histological characteristics in MR images • Extract RFs from MRI |
|
| ProsTIC registry of men treated With PSMA theranostics 228 | Start: May 2021 |
Recruiting, no results available | Diagnostic test: 177 Lu-PSMA |
Type: observational | Outcome measures: • PSA-RR • Number of participants with AE and Serious adverse events • Radiographic progression free survival • PSA progression-free survival • Overall survival • EORTC QLQ-C30 |
18FDC FPyL, 18F-fluorodeoxyglucose; 177LU-PSMA, 177Lutentium-prostate-specific membrane antigen; ADC, apparent diffusion coefficient; AE, adverse events; AS, active surveillance; DWI, diffusion-weighted imaging; EORTC, European Organization for Research and Treatment of Cancer; FUS-BG, fusion ultrasound-guided biopsies; mpMRI, multi-parametric magnetic resonance imaging; MRI, magnetic resonance imaging; PSA, prostate-specific antigen; PSMA-PET, prostate-specific membrane antigen positron emission tomography; RF, radiomic features; TRUS-BG, transrectal ultrasound-guided biopsies.
Conclusion
Radiomics research mainly uses MRI as an imaging modality and is focused on PCa diagnosis and risk stratification by identifying the GS. For tumor delineation and segmentation of ROIs, standardized reporting protocols are a key component. An improvement in image acquisition and AI DL protocols represents the future of radiomic research. For replication of results and ECE, complex models (clinical and conventional radiology) are probably the basis for future studies. These models show the possibility of prediction of csPCa in susceptible to misinterpretation lesions, such as PIRADS 3 lesions. High-resolution images from MRI detect many low volume PCa tumors with high sensitivity and low specificity. Although this is encouraging remains the issue of inter-reader agreement that has low specificity. PSMA-PET is the latest researched and the current evidence points out that it will have a potential as an imaging tool of PCa patients in the near future. The PSMA-PET/CT RFs achieved good results, higher than the visually detection of PCa lesions, in available studies looking at PCa detection as well as for metastatic involvement of LN. The results taken as a whole are good, the models performed well, but at this point the variability has to be taken into consideration before translating it to clinical practice. MRI and PSMA-PET will be the focus of research for the future after standardizing the RF and by the combination of the PSMA-PET and mpMRI overcomes the limitations of each of the methods in PCa detection. AI developments, in particular, DL techniques, already show that some of the issues regarding the inter- and intra-observer variability are being improved. The learning curves associated with PIRADS scoring systems are also overcome by the use of AI. The results taken as a whole are good, the models performed well, but at this point, the variability has to be taken into consideration before translating it to clinical practice. MRI and PSMA-PET will be the focus of research for the future after standardizing the RF.
Footnotes
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Author contributions: Matteo Ferro: Conceptualization; Formal analysis; Methodology; Supervision; Validation; Visualization; Writing—original draft; Writing—review & editing.
Ottavio de Cobelli: Supervision; Validation; Writing—review & editing.
Gennaro Musi: Supervision; Validation; Writing—review & editing.
Francesco del Giudice: Methodology; Validation; Writing—original draft; Writing—review & editing.
Giuseppe Carrieri: Supervision; Validation; Writing—review & editing.
Gian Maria Busetto: Supervision; Validation; Writing—review & editing.
Ugo Giovanni Falagario: Methodology; Validation; Visualization; Writing—original draft; Writing—review & editing.
Alessandro Sciarra: Supervision; Validation; Writing—review & editing.
Martina Maggi: Methodology; Validation; Visualization; Writing—original draft; Writing—review & editing.
Felice Crocetto: Validation; Writing—original draft; Writing—review & editing.
Biagio Barone: Methodology; Supervision; Validation; Visualization; Writing—original draft; Writing—review & editing.
Vincenzo Francesco Caputo: Writing—original draft; Writing—review & editing.
Michele Marchioni: Methodology; Validation; Visualization; Writing—original draft; Writing—review & editing.
Giuseppe Lucarelli: Supervision; Validation; Visualization; Writing—review & editing.
Ciro Imbimbo: Supervision; Validation; Writing—review & editing.
Francesco Alessandro Mistretta: Supervision; Validation; Writing—review & editing.
Stefano Luzzago: Supervision; Validation; Writing—review & editing.
Mihai Dorin Vartolomei: Supervision; Validation; Writing—review & editing.
Luigi Cormio: Supervision; Validation; Writing—review & editing.
Riccardo Autorino: Methodology; Supervision; Validation; Writing—review & editing.
Octavian Sabin Tătaru: Conceptualization; Formal analysis; Methodology; Supervision; Validation; Visualization; Writing—original draft; Writing—review & editing.
ORCID iDs: Matteo Ferro  https://orcid.org/0000-0002-9250-7858
https://orcid.org/0000-0002-9250-7858
Gian Maria Busetto  https://orcid.org/0000-0002-7291-0316
https://orcid.org/0000-0002-7291-0316
Biagio Barone  https://orcid.org/0000-0003-4884-132X
https://orcid.org/0000-0003-4884-132X
Vincenzo Francesco Caputo  https://orcid.org/0000-0002-9462-1427
https://orcid.org/0000-0002-9462-1427
Riccardo Autorino  https://orcid.org/0000-0001-7045-7725
https://orcid.org/0000-0001-7045-7725
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
Conflict of interest statement: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Availability of data and materials: Not applicable.
Contributor Information
Matteo Ferro, Department of Urology, European Institute of Oncology, IRCCS, Milan, Italy, via Ripamonti 435 Milano, Italy.
Ottavio de Cobelli, Department of Urology, European Institute of Oncology, IRCCS, Milan, Italy; Department of Oncology and Hematology-Oncology, Università degli Studi di Milano, Milan, Italy.
Gennaro Musi, Department of Urology, European Institute of Oncology, IRCCS, Milan, Italy; Department of Oncology and Hematology-Oncology, Università degli Studi di Milano, Milan, Italy.
Francesco del Giudice, Department of Urology, Policlinico Umberto I, Sapienza University of Rome, Rome, Italy.
Giuseppe Carrieri, Department of Urology and Organ Transplantation, University of Foggia, Foggia, Italy.
Gian Maria Busetto, Department of Urology and Organ Transplantation, University of Foggia, Foggia, Italy.
Ugo Giovanni Falagario, Department of Urology and Organ Transplantation, University of Foggia, Foggia, Italy.
Alessandro Sciarra, Department of Urology, Policlinico Umberto I, Sapienza University of Rome, Rome, Italy.
Martina Maggi, Department of Urology, Policlinico Umberto I, Sapienza University of Rome, Rome, Italy.
Felice Crocetto, Department of Neurosciences, Reproductive Sciences and Odontostomatology, University of Naples ‘Federico II’, Naples, Italy.
Biagio Barone, Department of Neurosciences, Reproductive Sciences and Odontostomatology, University of Naples ‘Federico II’, Naples, Italy.
Vincenzo Francesco Caputo, Department of Neurosciences, Reproductive Sciences and Odontostomatology, University of Naples ‘Federico II’, Naples, Italy.
Michele Marchioni, Department of Medical, Oral and Biotechnological Sciences, G. d’Annunzio, University of Chieti, Chieti, Italy; Urology Unit, ‘SS. Annunziata’ Hospital, Chieti, Italy; Department of Urology, ASL Abruzzo 2, Chieti, Italy.
Giuseppe Lucarelli, Department of Emergency and Organ Transplantation, Urology, Andrology and Kidney Transplantation Unit, University of Bari, Bari, Italy.
Ciro Imbimbo, Department of Neurosciences, Reproductive Sciences and Odontostomatology, University of Naples ‘Federico II’, Naples, Italy.
Francesco Alessandro Mistretta, Department of Urology, European Institute of Oncology, IRCCS, Milan, Italy; Università degli Studi di Milano, Milan, Italy.
Stefano Luzzago, Department of Urology, European Institute of Oncology, IRCCS, Milan, Italy; Università degli Studi di Milano, Milan, Italy.
Mihai Dorin Vartolomei, Department of Cell and Molecular Biology, George Emil Palade University of Medicine, Pharmacy, Science, and Technology of Târgu Mures, Târgu Mures, Romania; Department of Urology, Medical University of Vienna, Vienna, Austria.
Luigi Cormio, Urology and Renal Transplantation Unit, Department of Medical and Surgical Sciences, University of Foggia, Foggia, Italy; Urology Unit, Bonomo Teaching Hospital, Foggia, Italy.
Riccardo Autorino, Division of Urology, VCU Health, Richmond, VA, USA.
Octavian Sabin Tătaru, Institution Organizing University Doctoral Studies, I.O.S.U.D., George Emil Palade University of Medicine, Pharmacy, Science, and Technology of Târgu Mures, Târgu Mures, Romania.
References
- 1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. [DOI] [PubMed] [Google Scholar]
- 2. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71: 209–249. [DOI] [PubMed] [Google Scholar]
- 3. Bruno SM, Falagario UG, d’Altilia N, et al. PSA density help to identify patients with elevated PSA due to prostate cancer rather than intraprostatic inflammation: a prospective single center study. Front Oncol 2021; 11: 693684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maggi M, Gentilucci A, Salciccia S, et al. Psychological impact of different primary treatments for prostate cancer: a critical analysis. Andrologia 2019; 51: e13157. [DOI] [PubMed] [Google Scholar]
- 5. Logozzi M, Angelini DF, Giuliani A, et al. Increased plasmatic levels of PSA-expressing exosomes distinguish prostate cancer patients from benign prostatic hyperplasia: a prospective study. Cancers 2019; 11: E1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith CP, Czarniecki M, Mehralivand S, et al. Radiomics and radiogenomics of prostate cancer. Abdom Radiol 2019; 44: 2021–2029. [DOI] [PubMed] [Google Scholar]
- 7. Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012; 30: 1234–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 1990; 201248: 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tătaru OS, Martha O, Crocetto F, et al. Fascin-1 and its role as a serological marker in prostate cancer: a prospective case-control study. Future Sci OA 2021; 7: FSO745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Busetto GM, Del Giudice F, Maggi M, et al. Prospective assessment of two-gene urinary test with multiparametric magnetic resonance imaging of the prostate for men undergoing primary prostate biopsy. World J Urol 2021; 39: 1869–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Epstein JI. Prostate cancer grading: a decade after the 2005 modified system. Mod Pathol 2018; 31: S47–S63. [DOI] [PubMed] [Google Scholar]
- 12. Mottet N, van den Bergh RCN, Briers E, et al. EAU-EANM-ESTRO-ESUR-SIOG guidelines on prostate cancer-2020 update. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol 2021; 79: 243–262. [DOI] [PubMed] [Google Scholar]
- 13. Sciarra A, Maggi M, Del Proposto A, et al. Impact of uni- or multifocal perineural invasion in prostate cancer at radical prostatectomy. Transl Androl Urol 2021; 10: 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tarantino G, Crocetto F, Vito CD, et al. Clinical factors affecting prostate-specific antigen levels in prostate cancer patients undergoing radical prostatectomy: a retrospective study. Future Sci OA 2021; 7: FSO643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mazzone E, Gandaglia G, Ploussard G, et al. Risk stratification of patients candidate to radical prostatectomy based on clinical and multiparametric magnetic resonance imaging parameters: development and external validation of novel risk groups. Eur Urol 2022; 81: 193–203. [DOI] [PubMed] [Google Scholar]
- 16. Kvåle R, Møller B, Wahlqvist R, et al. Concordance between Gleason scores of needle biopsies and radical prostatectomy specimens: a population-based study. BJU Int 2009; 103: 1647–1654. [DOI] [PubMed] [Google Scholar]
- 17. Bryk DJ, Llukani E, Taneja SS, et al. The role of ipsilateral and contralateral transrectal ultrasound-guided systematic prostate biopsy in men with unilateral magnetic resonance imaging lesion undergoing magnetic resonance imaging-ultrasound fusion-targeted prostate biopsy. Urology 2017; 102: 178–182. [DOI] [PubMed] [Google Scholar]
- 18. Freifeld Y, Xi Y, Passoni N, et al. Optimal sampling scheme in men with abnormal multiparametric MRI undergoing MRI-TRUS fusion prostate biopsy. Urol Oncol 2019; 37: 57–62. [DOI] [PubMed] [Google Scholar]
- 19. Fasulo V, Cowan JE, Maggi M, et al. Characteristics of cancer progression on serial biopsy in men on active surveillance for early-stage prostate cancer: implications for focal therapy. Eur Urol Oncol 2022; 5: 61–69. [DOI] [PubMed] [Google Scholar]
- 20. Haffner MC, Zwart W, Roudier MP, et al. Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev Urol 2021; 18: 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brastianos HC, Murgic J, Salcedo A, et al. Determining the impact of spatial heterogeneity on genomic prognostic biomarkers for localized prostate cancer. Eur Urol Oncol 2022; 5: 362–365. [DOI] [PubMed] [Google Scholar]
- 22. Salciccia S, Capriotti AL, Laganà A, et al. Biomarkers in prostate cancer diagnosis: from current knowledge to the role of metabolomics and exosomes. Int J Mol Sci 2021; 22: 4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sciarra A, Maggi M, Salciccia S, et al. Tissue expression of androgen receptor splice variant 7 at radical prostatectomy predicts risk of progression in untreated nonmetastatic prostate cancer. Oncology 2021; 99: 251–255. [DOI] [PubMed] [Google Scholar]
- 24. Turkbey B, Rosenkrantz AB, Haider MA, et al. Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. Eur Urol 2019; 76: 340–351. [DOI] [PubMed] [Google Scholar]
- 25. Greer MD, Shih JH, Lay N, et al. Interreader variability of prostate imaging reporting and data system version 2 in detecting and assessing prostate cancer lesions at prostate MRI. Am J Roentgenol. Epub ahead of print 27 March 2019. DOI: 10.2214/AJR.18.20536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kohestani K, Wallström J, Dehlfors N, et al. Performance and inter-observer variability of prostate MRI (PI-RADS version 2) outside high-volume centres. Scand J Urol 2019; 53: 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mazzone E, Stabile A, Pellegrino F, et al. Positive predictive value of prostate imaging reporting and data system version 2 for the detection of clinically significant prostate cancer: a systematic review and meta-analysis. Eur Urol Oncol 2021; 4: 697–713. [DOI] [PubMed] [Google Scholar]
- 28. Wadera A, Alabousi M, Pozdnyakov A, et al. Impact of PI-RADS category 3 lesions on the diagnostic accuracy of MRI for detecting prostate cancer and the prevalence of prostate cancer within each PI-RADS category: a systematic review and meta-analysis. Br J Radiol 2021; 94: 20191050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rapisarda S, Bada M, Crocetto F, et al. The role of multiparametric resonance and biopsy in prostate cancer detection: comparison with definitive histological report after laparoscopic/robotic radical prostatectomy. Abdom Radiol 2020; 45: 4178–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Flammia S, Frisenda M, Maggi M, et al. Cribriform pattern does not have a significant impact in Gleason Score ⩾7/ISUP Grade ⩾2 prostate cancers submitted to radical prostatectomy. Medicine 2020; 99: e22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stewart GD, Van Neste L, Delvenne P, et al. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: results of the MATLOC study. J Urol 2013; 189: 1110–1116. [DOI] [PubMed] [Google Scholar]
- 32. Chaddad A, Kucharczyk MJ, Niazi T. Multimodal radiomic features for the predicting Gleason score of prostate cancer. Cancers 2018; 10: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fendler WP, Eiber M, Beheshti M, et al. 68Ga-PSMA PET/CT: joint EANM and SNMMI procedure guideline for prostate cancer imaging: version 1.0. Eur J Nucl Med Mol Imaging 2017; 44: 1014–1024. [DOI] [PubMed] [Google Scholar]
- 34. Hofman MS, Murphy DG, Williams SG, et al. A prospective randomized multicentre study of the impact of gallium-68 prostate-specific membrane antigen (PSMA) PET/CT imaging for staging high-risk prostate cancer prior to curative-intent surgery or radiotherapy (proPSMA study): clinical trial protocol. BJU Int 2018; 122: 783–793. [DOI] [PubMed] [Google Scholar]
- 35. Hofman MS, Lawrentschuk N, Francis RJ, et al. Prostate-specific membrane antigen PET-CT in patients with high-risk prostate cancer before curative-intent surgery or radiotherapy (proPSMA): a prospective, randomised, multicentre study. Lancet 2020; 395: 1208–1216. [DOI] [PubMed] [Google Scholar]
- 36. Gillessen S, Attard G, Beer TM, et al. Management of patients with advanced prostate cancer: report of the advanced prostate cancer consensus conference 2019. Eur Urol 2020; 77: 508–547. [DOI] [PubMed] [Google Scholar]
- 37. Chen M, Zhang Q, Zhang C, et al. Combination of 68Ga-PSMA PET/CT and multiparametric MRI improves the detection of clinically significant prostate cancer: a lesion-by-lesion analysis. J Nucl Med 2019; 60: 944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spohn SKB, Kramer M, Kiefer S, et al. Comparison of manual and semi-automatic [18F]PSMA-1007 PET based contouring techniques for intraprostatic tumor delineation in patients with primary prostate cancer and validation with histopathology as standard of reference. Front Oncol 2020; 10: 600690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zamboglou C, Kramer M, Kiefer S, et al. The impact of the co-registration technique and analysis methodology in comparison studies between advanced imaging modalities and whole-mount-histology reference in primary prostate cancer. Sci Rep 2021; 11: 5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spohn SKB, Bettermann AS, Bamberg F, et al. Radiomics in prostate cancer imaging for a personalized treatment approach – current aspects of methodology and a systematic review on validated studies. Theranostics 2021; 11: 8027–8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Massanova M, Robertson S, Barone B, et al. The comparison of imaging and clinical methods to estimate prostate volume: a single-centre retrospective study. Urol Int 2021; 105: 804–810. [DOI] [PubMed] [Google Scholar]
- 42. Porreca A, Bianchi FM, Salvaggio A, et al. Prognostic performance of magnetic resonance imaging-guided biopsy in defining prostate cancer anterior lesions. World J Urol 2021; 39: 1473–1479. [DOI] [PubMed] [Google Scholar]
- 43. Tătaru OS, Vartolomei MD, Rassweiler JJ, et al. Artificial Intelligence and Machine Learning in Prostate Cancer Patient Management-Current Trends and Future Perspectives. Diagnostics 2021; 11: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Checcucci E, Autorino R, Cacciamani GE, et al. Artificial intelligence and neural networks in urology: current clinical applications. Minerva Urol Nefrol 2020; 72: 49–57. [DOI] [PubMed] [Google Scholar]
- 45. Mortensen MA, Borrelli P, Poulsen MH, et al. Artificial intelligence-based versus manual assessment of prostate cancer in the prostate gland: a method comparison study. Clin Physiol Funct Imaging 2019; 39: 399–406. [DOI] [PubMed] [Google Scholar]
- 46. Ström P, Kartasalo K, Olsson H, et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol 2020; 21: 222–232. [DOI] [PubMed] [Google Scholar]
- 47. Chaddad A, Niazi T, Probst S, et al. Predicting Gleason score of prostate cancer patients using radiomic analysis. Front Oncol 2018; 8: 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cysouw MCF, Jansen BHE, van de Brug T, et al. Machine learning-based analysis of [18F]DCFPyL PET radiomics for risk stratification in primary prostate cancer. Eur J Nucl Med Mol Imaging 2021; 48: 340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gugliandolo SG, Pepa M, Isaksson LJ, et al. MRI-based radiomics signature for localized prostate cancer: a new clinical tool for cancer aggressiveness prediction? Sub-study of prospective phase II trial on ultra-hypofractionated radiotherapy (AIRC IG-13218). Eur Radiol 2021; 31: 716–728. [DOI] [PubMed] [Google Scholar]
- 50. Papp L, Spielvogel CP, Grubmüller B, et al. Supervised machine learning enables non-invasive lesion characterization in primary prostate cancer with [68Ga]Ga-PSMA-11 PET/MRI. Eur J Nucl Med Mol Imaging 2021; 48: 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bian Y, Li J, Cao K, et al. Magnetic resonance imaging radiomic analysis can preoperatively predict G1 and G2/3 grades in patients with NF-pNETs. Abdom Radiol 2021; 46: 667–680. [DOI] [PubMed] [Google Scholar]
- 52. Santone A, Brunese MC, Donnarumma F, et al. Radiomic features for prostate cancer grade detection through formal verification. Radiol Med 2021; 126: 688–697. [DOI] [PubMed] [Google Scholar]
- 53. Han C, Ma S, Liu X, et al. Radiomics models based on apparent diffusion coefficient maps for the prediction of high-grade prostate cancer at radical prostatectomy: comparison with preoperative biopsy. J Magn Reson Imaging 2021; 54: 1892–1901. [DOI] [PubMed] [Google Scholar]
- 54. Bleker J, Kwee TC, Dierckx RAJO, et al. Multiparametric MRI and auto-fixed volume of interest-based radiomics signature for clinically significant peripheral zone prostate cancer. Eur Radiol 2020; 30: 1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hou Y, Bao ML, Wu CJ, et al. A radiomics machine learning-based redefining score robustly identifies clinically significant prostate cancer in equivocal PI-RADS score 3 lesions. Abdom Radiol 2020; 45: 4223–4234. [DOI] [PubMed] [Google Scholar]
- 56. Alongi P, Laudicella R, Stefano A, et al. Choline PET/CT features to predict survival outcome in high risk prostate cancer restaging: a preliminary machine-learning radiomics study. Q J Nucl Med Mol Imaging. Epub ahead of print 15 June 2020. DOI: 10.23736/S1824-4785.20.03227-6. [DOI] [PubMed] [Google Scholar]
- 57. Zamboglou C, Bettermann AS, Gratzke C, et al. Uncovering the invisible-prevalence, characteristics, and radiomics feature-based detection of visually undetectable intraprostatic tumor lesions in 68GaPSMA-11 PET images of patients with primary prostate cancer. Eur J Nucl Med Mol Imaging 2021; 48: 1987–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wildeboer RR, Mannaerts CK, van Sloun RJG, et al. Automated multiparametric localization of prostate cancer based on B-mode, shear-wave elastography, and contrast-enhanced ultrasound radiomics. Eur Radiol 2020; 30: 806–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Woźnicki P, Westhoff N, Huber T, et al. Multiparametric MRI for prostate cancer characterization: combined use of radiomics model with PI-RADS and clinical parameters. Cancers 2020; 12: 1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xu L, Zhang G, Zhao L, et al. Radiomics based on multiparametric magnetic resonance imaging to predict extraprostatic extension of prostate cancer. Front Oncol 2020; 10: 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Qi Y, Zhang S, Wei J, et al. Multiparametric MRI-based radiomics for prostate cancer screening with PSA in 4-10 ng/mL to reduce unnecessary biopsies. J Magn Reson Imaging 2020; 51: 1890–1899. [DOI] [PubMed] [Google Scholar]
- 62. Osman SOS, Leijenaar RTH, Cole AJ, et al. Computed tomography-based radiomics for risk stratification in prostate cancer. Int J Radiat Oncol Biol Phys 2019; 105: 448–456. [DOI] [PubMed] [Google Scholar]
- 63. Bourbonne V, Fournier G, Vallières M, et al. External validation of an MRI-derived radiomics model to predict biochemical recurrence after surgery for high-risk prostate cancer. Cancers 2020; 12: 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sushentsev N, Rundo L, Blyuss O, et al. MRI-derived radiomics model for baseline prediction of prostate cancer progression on active surveillance. Sci Rep 2021; 11: 12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Varghese B, Chen F, Hwang D, et al. Objective risk stratification of prostate cancer using machine learning and radiomics applied to multiparametric magnetic resonance images. Sci Rep 2019; 9: 1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bosetti DG, Ruinelli L, Piliero MA, et al. Cone-beam computed tomography-based radiomics in prostate cancer: a mono-institutional study. Strahlenther Onkol 2020; 196: 943–951. [DOI] [PubMed] [Google Scholar]
- 67. Peeken JC, Shouman MA, Kroenke M, et al. A CT-based radiomics model to detect prostate cancer lymph node metastases in PSMA radioguided surgery patients. Eur J Nucl Med Mol Imaging 2020; 47: 2968–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang GM, Han YQ, Wei JW, et al. Radiomics based on MRI as a biomarker to guide therapy by predicting upgrading of prostate cancer from biopsy to radical prostatectomy. J Magn Reson Imaging 2020; 52: 1239–1248. [DOI] [PubMed] [Google Scholar]
- 69. Shiradkar R, Podder TK, Algohary A, et al. Radiomics based targeted radiotherapy planning (Rad-TRaP): a computational framework for prostate cancer treatment planning with MRI. Radiat Oncol 2016; 11: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Macomber MW, Phillips M, Tarapov I, et al. Autosegmentation of prostate anatomy for radiation treatment planning using deep decision forests of radiomic features. Phys Med Biol 2018; 63: 235002. [DOI] [PubMed] [Google Scholar]
- 71. Wong J, Fong A, McVicar N, et al. Comparing deep learning-based auto-segmentation of organs at risk and clinical target volumes to expert inter-observer variability in radiotherapy planning. Radiother Oncol 2020; 144: 152–158. [DOI] [PubMed] [Google Scholar]
- 72. Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017; 14: 749–762. [DOI] [PubMed] [Google Scholar]
- 73. Bernatz S, Zhdanovich Y, Ackermann J, et al. Impact of rescanning and repositioning on radiomic features employing a multi-object phantom in magnetic resonance imaging. Sci Rep 2021; 11: 14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bianchini L, Santinha J, Loução N, et al. A multicenter study on radiomic features from T2-weighted images of a customized MR pelvic phantom setting the basis for robust radiomic models in clinics. Magn Reson Med 2021; 85: 1713–1726. [DOI] [PubMed] [Google Scholar]
- 75. Perez IM, Jambor I, Kauko T, et al. Qualitative and quantitative reporting of a unique biparametric MRI: towards biparametric MRI-based nomograms for prediction of prostate biopsy outcome in men with a clinical suspicion of prostate cancer (IMPROD and MULTI-IMPROD trials). J Magn Reson Imaging 2020; 51: 1556–1567. [DOI] [PubMed] [Google Scholar]
- 76. van Timmeren JE, Cester D, Tanadini-Lang S, et al. Radiomics in medical imaging- ‘how-to’ guide and critical reflection. Insights Imaging 2020; 11: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bailly C, Bodet-Milin C, Couespel S, et al. Revisiting the robustness of PET-based textural features in the context of multi-centric trials. PLoS ONE 2016; 11: e0159984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Prior F, Almeida J, Kathiravelu P, et al. Open access image repositories: high-quality data to enable machine learning research. Clin Radiol 2020; 75: 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chalkidou A, O’Doherty MJ, Marsden PK. False discovery rates in PET and CT studies with texture features: a systematic review. PLoS ONE 2015; 10: e0124165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Suzuki K. Overview of deep learning in medical imaging. Radiol Phys Technol 2017; 10: 257–273. [DOI] [PubMed] [Google Scholar]
- 81. Zhou Z, Siddiquee MMR, Tajbakhsh N, et al. UNet++: a nested U-net architecture for medical image segmentation. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support 2018; 11045: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521: 436–444. [DOI] [PubMed] [Google Scholar]
- 83. Beksac AT, Cumarasamy S, Falagario U, et al. Multiparametric magnetic resonance imaging features identify aggressive prostate cancer at the phenotypic and transcriptomic level. J Urol 2018; 200: 1241–1249. [DOI] [PubMed] [Google Scholar]
- 84. Mayerhoefer ME, Materka A, Langs G, et al. Introduction to radiomics. J Nucl Med 2020; 61: 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rizzo S, Botta F, Raimondi S, et al. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp 2018; 2: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 2009; 151: 264–269, W64. [DOI] [PubMed] [Google Scholar]
- 87. Erle A, Moazemi S, Lütje S, et al. Evaluating a machine learning tool for the classification of pathological uptake in whole-body PSMA-PET-CT scans. Tomography 2021; 7: 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Domachevsky L, Goldberg N, Bernstine H, et al. Quantitative characterisation of clinically significant intra-prostatic cancer by prostate-specific membrane antigen (PSMA) expression and cell density on PSMA-11. Eur Radiol 2018; 28: 5275–5283. [DOI] [PubMed] [Google Scholar]
- 89. Moazemi S, Khurshid Z, Erle A, et al. Machine learning facilitates hotspot classification in PSMA-PET/CT with nuclear medicine specialist accuracy. Diagnostics 2020; 10: E622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Moazemi S, Erle A, Lütje S, et al. Estimating the potential of radiomics features and radiomics signature from pretherapeutic PSMA-PET-CT scans and clinical data for prediction of overall survival when treated with 177Lu-PSMA. Diagnostics 2021; 11: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tanadini-Lang S, Bogowicz M, Veit-Haibach P, et al. Exploratory radiomics in computed tomography perfusion of prostate cancer. Anticancer Res 2018; 38: 685–690. [DOI] [PubMed] [Google Scholar]
- 92. Mostafaei S, Abdollahi H, Kazempour Dehkordi S, et al. CT imaging markers to improve radiation toxicity prediction in prostate cancer radiotherapy by stacking regression algorithm. Radiol Med 2020; 125: 87–97. [DOI] [PubMed] [Google Scholar]
- 93. Acar E, Leblebici A, Ellidokuz BE, et al. Machine learning for differentiating metastatic and completely responded sclerotic bone lesion in prostate cancer: a retrospective radiomics study. Br J Radiol 2019; 92: 20190286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhang Q, Xiong J, Cai Y, et al. Multimodal feature learning and fusion on B-mode ultrasonography and sonoelastography using point-wise gated deep networks for prostate cancer diagnosis. Biomed Tech 2020; 65: 87–98. [DOI] [PubMed] [Google Scholar]
- 95. Huang X, Chen M, Liu P, et al. Texture feature-based classification on transrectal ultrasound image for prostatic cancer detection. Comput Math Methods Med 2020; 2020: 7359375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wu P, Liu Y, Li Y, et al. Robust prostate segmentation using intrinsic properties of TRUS images. IEEE Trans Med Imaging 2015; 34: 1321–1335. [DOI] [PubMed] [Google Scholar]
- 97. Bevilacqua A, Mottola M, Ferroni F, et al. The primacy of high B-value 3T-DWI radiomics in the prediction of clinically significant prostate cancer. Diagnostics 2021; 11: 739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sunoqrot MRS, Selnæs KM, Sandsmark E, et al. A quality control system for automated prostate segmentation on T2-weighted MRI. Diagnostics 2020; 10: 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hu L, Zhou DW, Fu CX, et al. Advanced zoomed diffusion-weighted imaging vs. full-field-of-view diffusion-weighted imaging in prostate cancer detection: a radiomic features study. Eur Radiol 2021; 31: 1760–1769. [DOI] [PubMed] [Google Scholar]
- 100. Giambelluca D, Cannella R, Vernuccio F, et al. PI-RADS 3 lesions: role of prostate MRI texture analysis in the identification of prostate cancer. Curr Probl Diagn Radiol 2021; 50: 175–185. [DOI] [PubMed] [Google Scholar]
- 101. Khalvati F, Zhang J, Chung AG, et al. MPCaD: a multi-scale radiomics-driven framework for automated prostate cancer localization and detection. BMC Med Imaging 2018; 18: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Maggi M, Panebianco V, Mosca A, et al. Prostate imaging reporting and data system 3 category cases at multiparametric magnetic resonance for prostate cancer: a systematic review and meta-analysis. Eur Urol Focus 2020; 6: 463–478. [DOI] [PubMed] [Google Scholar]
- 103. Wu S, Jiao Y, Zhang Y, et al. Imaging-based individualized response prediction of carbon ion radiotherapy for prostate cancer patients. Cancer Manag Res 2019; 11: 9121–9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kairemo K, Kappadath SC, Joensuu T, et al. A retrospective comparative study of sodium fluoride (NaF-18)-PET/CT and fluorocholine (F-18-CH) PET/CT in the evaluation of skeletal metastases in metastatic prostate cancer using a volumetric 3-D radiomics analysis. Diagnostics 2020; 11: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Isaksson LJ, Pepa M, Zaffaroni M, et al. Machine learning-based models for prediction of toxicity outcomes in radiotherapy. Front Oncol 2020; 10: 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Abdollahi H, Mahdavi SR, Mofid B, et al. Rectal wall MRI radiomics in prostate cancer patients: prediction of and correlation with early rectal toxicity. Int J Radiat Biol 2018; 94: 829–837. [DOI] [PubMed] [Google Scholar]
- 107. Dinis Fernandes C, Dinh CV, Walraven I, et al. Biochemical recurrence prediction after radiotherapy for prostate cancer with T2w magnetic resonance imaging radiomic features. Phys Imaging Radiat Oncol 2018; 7: 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Abdollahi H, Tanha K, Mofid B, et al. MRI radiomic analysis of IMRT-induced bladder wall changes in prostate cancer patients: a relationship with radiation dose and toxicity. J Med Imaging Radiat Sci 2019; 50: 252–260. [DOI] [PubMed] [Google Scholar]
- 109. Orczyk C, Villers A, Rusinek H, et al. Prostate cancer heterogeneity: texture analysis score based on multiple magnetic resonance imaging sequences for detection, stratification and selection of lesions at time of biopsy. BJU Int 2019; 124: 76–86. [DOI] [PubMed] [Google Scholar]
- 110. Krauss T, Engel H, Jilg CA, et al. MRI phenotype of the prostate: transition zone radiomics analysis improves explanation of prostate-specific antigen (PSA) serum level compared to volume measurement alone. Eur J Radiol 2020; 129: 109063. [DOI] [PubMed] [Google Scholar]
- 111. Gillies RJ, Schabath MB. Radiomics improves cancer screening and early detection. Cancer Epidemiol Biomarkers Prev 2020; 29: 2556–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Bagher-Ebadian H, Janic B, Liu C, et al. Detection of dominant intra-prostatic lesions in patients with prostate cancer using an artificial neural network and MR multi-modal radiomics analysis. Front Oncol 2019; 9: 1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Dulhanty C, Wang L, Cheng M, et al. Radiomics driven diffusion weighted imaging sensing strategies for zone-level prostate cancer sensing. Sensors 2020; 20: 1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Chen T, Li M, Gu Y, et al. Prostate cancer differentiation and aggressiveness: assessment with a radiomic-based model vs. PI-RADS v2. J Magn Reson Imaging 2019; 49: 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Algohary A, Shiradkar R, Pahwa S, et al. Combination of peri-tumoral and intra-tumoral radiomic features on bi-parametric MRI accurately stratifies prostate cancer risk: a multi-site study. Cancers 2020; 12: 2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Brunese L, Mercaldo F, Reginelli A, et al. Formal methods for prostate cancer Gleason score and treatment prediction using radiomic biomarkers. Magn Reson Imaging 2020; 66: 165–175. [DOI] [PubMed] [Google Scholar]
- 117. Schick U, Lucia F, Dissaux G, et al. MRI-derived radiomics: methodology and clinical applications in the field of pelvic oncology. Br J Radiol 2019; 92: 20190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Delgadillo R, Ford JC, Abramowitz MC, et al. The role of radiomics in prostate cancer radiotherapy. Strahlenther Onkol 2020; 196: 900–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Bernatz S, Ackermann J, Mandel P, et al. Comparison of machine learning algorithms to predict clinically significant prostate cancer of the peripheral zone with multiparametric MRI using clinical assessment categories and radiomic features. Eur Radiol 2020; 30: 6757–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Madabhushi A, Feldman MD, Metaxas DN, et al. Automated detection of prostatic adenocarcinoma from high-resolution ex vivo MRI. IEEE Trans Med Imaging 2005; 24: 1611–1625. [DOI] [PubMed] [Google Scholar]
- 121. Lopes R, Ayache A, Makni N, et al. Prostate cancer characterization on MR images using fractal features. Med Phys 2011; 38: 83–95. [DOI] [PubMed] [Google Scholar]
- 122. Cameron A, Khalvati F, Haider MA, et al. MAPS: a quantitative radiomics approach for prostate cancer detection. IEEE Trans Biomed Eng 2016; 63: 1145–1156. [DOI] [PubMed] [Google Scholar]
- 123. McGarry SD, Bukowy JD, Iczkowski KA, et al. Gleason probability maps: a radiomics tool for mapping prostate cancer likelihood in MRI space. Tomography 2019; 5: 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Khalvati F, Wong A, Haider MA. Automated prostate cancer detection via comprehensive multi-parametric magnetic resonance imaging texture feature models. BMC Med Imaging 2015; 15: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Brunese L, Mercaldo F, Reginelli A, et al. Radiomics for Gleason score detection through deep learning. Sensors 2020; 20: 5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wang J, Wu CJ, Bao ML, et al. Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. Eur Radiol 2017; 27: 4082–4090. [DOI] [PubMed] [Google Scholar]
- 127. Kwon D, Reis IM, Breto AL, et al. Classification of suspicious lesions on prostate multiparametric MRI using machine learning. J Med Imaging 2018; 5: 034502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Parra NA, Lu H, Li Q, et al. Predicting clinically significant prostate cancer using DCE-MRI habitat descriptors. Oncotarget 2018; 9: 37125–37136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Penzias G, Singanamalli A, Elliott R, et al. Identifying the morphologic basis for radiomic features in distinguishing different Gleason grades of prostate cancer on MRI: preliminary findings. PLoS ONE 2018; 13: e0200730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Min X, Li M, Dong D, et al. Multi-parametric MRI-based radiomics signature for discriminating between clinically significant and insignificant prostate cancer: cross-validation of a machine learning method. Eur J Radiol 2019; 115: 16–21. [DOI] [PubMed] [Google Scholar]
- 131. Brancato V, Aiello M, Basso L, et al. Evaluation of a multiparametric MRI radiomic-based approach for stratification of equivocal PI-RADS 3 and upgraded PI-RADS 4 prostatic lesions. Sci Rep 2021; 11: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Li M, Chen T, Zhao W, et al. Radiomics prediction model for the improved diagnosis of clinically significant prostate cancer on biparametric MRI. Quant Imaging Med Surg 2020; 10: 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Bonekamp D, Kohl S, Wiesenfarth M, et al. Radiomic machine learning for characterization of prostate lesions with MRI: comparison to ADC values. Radiology 2018; 289: 128–137. [DOI] [PubMed] [Google Scholar]
- 134. Zhang Y, Chen W, Yue X, et al. Development of a novel, multi-parametric, MRI-based radiomic nomogram for differentiating between clinically significant and insignificant prostate cancer. Front Oncol 2020; 10: 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Gong L, Xu M, Fang M, et al. Noninvasive prediction of high-grade prostate cancer via biparametric MRI radiomics. J Magn Reson Imaging 2020; 52: 1102–1109. [DOI] [PubMed] [Google Scholar]
- 136. Song Y, Zhang J, Zhang YD, et al. FeAture Explorer (FAE): a tool for developing and comparing radiomics models. PLoS ONE 2020; 15: e0237587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Castillo TJM, Starmans MPA, Arif M, et al. A multi-center, multi-vendor study to evaluate the generalizability of a radiomics model for classifying prostate cancer: high grade vs. low grade. Diagnostics 2021; 11: 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Losnegård A, Reisæter LAR, Halvorsen OJ, et al. Magnetic resonance radiomics for prediction of extraprostatic extension in non-favorable intermediate- and high-risk prostate cancer patients. Acta Radiol 2020; 61: 1570–1579. [DOI] [PubMed] [Google Scholar]
- 139. Ma S, Xie H, Wang H, et al. MRI-based radiomics signature for the preoperative prediction of extracapsular extension of prostate cancer. J Magn Reson Imaging 2019; 50: 1914–1925. [DOI] [PubMed] [Google Scholar]
- 140. Ma S, Xie H, Wang H, et al. Preoperative prediction of extracapsular extension: radiomics signature based on magnetic resonance imaging to stage prostate cancer. Mol Imaging Biol 2020; 22: 711–721. [DOI] [PubMed] [Google Scholar]
- 141. Li J, Weng Z, Xu H, et al. Support vector machines (SVM) classification of prostate cancer Gleason score in central gland using multiparametric magnetic resonance images: a cross-validated study. Eur J Radiol 2018; 98: 61–67. [DOI] [PubMed] [Google Scholar]
- 142. James ND, Spears MR, Clarke NW, et al. Survival with newly diagnosed metastatic prostate cancer in the ‘docetaxel era’: data from 917 patients in the control arm of the STAMPEDE trial (MRC PR08, CRUK/06/019). Eur Urol 2015; 67: 1028–1038. [DOI] [PubMed] [Google Scholar]
- 143. Maggi M, Cowan JE, Fasulo V, et al. The long-term risks of metastases in men on active surveillance for early stage prostate cancer. J Urol 2020; 204: 1222–1228. [DOI] [PubMed] [Google Scholar]
- 144. Sciarra A, Fasulo A, Ciardi A, et al. A meta-analysis and systematic review of randomized controlled trials with degarelix versus gonadotropin-releasing hormone agonists for advanced prostate cancer. Medicine 2016; 95: e3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Maggi M, Salciccia S, Del Giudice F, et al. A systematic review and meta-analysis of randomized controlled trials with novel hormonal therapies for non-metastatic castration-resistant prostate cancer: an update from mature overall survival data. Front Oncol 2021; 11: 700258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Bauckneht M, Rebuzzi SE, Signori A, et al. The prognostic power of inflammatory indices and clinical factors in metastatic castration-resistant prostate cancer patients treated with radium-223 (BIO-Ra study). Eur J Nucl Med Mol Imaging 2022; 49: 1063–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Sciarra A, Busetto GM, Salciccia S, et al. Does exist a differential impact of degarelix versus LHRH agonists on cardiovascular safety? Evidences from randomized and real-world studies. Front Endocrinol 2021; 12: 695170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Wang Y, Yu B, Zhong F, et al. MRI-based texture analysis of the primary tumor for pre-treatment prediction of bone metastases in prostate cancer. Magn Reson Imaging 2019; 60: 76–84. [DOI] [PubMed] [Google Scholar]
- 149. Zhang W, Mao N, Wang Y, et al. A radiomics nomogram for predicting bone metastasis in newly diagnosed prostate cancer patients. Eur J Radiol 2020; 128: 109020. [DOI] [PubMed] [Google Scholar]
- 150. Reischauer C, Patzwahl R, Koh D-M, et al. Texture analysis of apparent diffusion coefficient maps for treatment response assessment in prostate cancer bone metastases – a pilot study. Eur J Radiol 2018; 101: 184–190. [DOI] [PubMed] [Google Scholar]
- 151. Van den Broeck T, van den Bergh RCN, Briers E, et al. Biochemical recurrence in prostate cancer: the European Association of Urology Prostate Cancer Guidelines Panel Recommendations. Eur Urol Focus 2020; 6: 231–234. [DOI] [PubMed] [Google Scholar]
- 152. Nkengurutse G, Tian F, Jiang S, et al. Preoperative predictors of biochemical recurrence-free survival in high-risk prostate cancer following radical prostatectomy. Front Oncol 2020; 10: 1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Kurbegovic S, Berg KD, Thomsen FB, et al. The risk of biochemical recurrence for intermediate-risk prostate cancer after radical prostatectomy. Scand J Urol 2017; 51: 450–456. [DOI] [PubMed] [Google Scholar]
- 154. Artibani W, Porcaro AB, De Marco V, et al. Management of biochemical recurrence after primary curative treatment for prostate cancer: a review. Urol Int 2018; 100: 251–262. [DOI] [PubMed] [Google Scholar]
- 155. Shiradkar R, Ghose S, Jambor I, et al. Radiomic features from pretreatment biparametric MRI predict prostate cancer biochemical recurrence: preliminary findings. J Magn Reson Imaging 2018; 48: 1626–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Bourbonne V, Vallières M, Lucia F, et al. MRI-derived radiomics to guide post-operative management for high-risk prostate cancer. Front Oncol 2019; 9: 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Ullrich T, Quentin M, Oelers C, et al. Magnetic resonance imaging of the prostate at 1.5 versus 3.0T: a prospective comparison study of image quality. Eur J Radiol 2017; 90: 192–197. [DOI] [PubMed] [Google Scholar]
- 158. Zhong QZ, Long LH, Liu A, et al. Radiomics of multiparametric MRI to predict biochemical recurrence of localized prostate cancer after radiation therapy. Front Oncol 2020; 10: 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Li L, Shiradkar R, Leo P, et al. A novel imaging based nomogram for predicting post-surgical biochemical recurrence and adverse pathology of prostate cancer from pre-operative bi-parametric MRI. EBioMedicine 2021; 63: 103163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Yao S, Jiang H, Song B. Radiomics in prostate cancer: basic concepts and current state-of-the-art. Chin J Acad Radiol 2020; 2: 47–55. [Google Scholar]
- 161. Abdollahi H, Mofid B, Shiri I, et al. Machine learning-based radiomic models to predict intensity-modulated radiation therapy response, Gleason score and stage in prostate cancer. Radiol Med 2019; 124: 555–567. [DOI] [PubMed] [Google Scholar]
- 162. Yu N, Wang B, Ren J, et al. The clinical guiding value of a radiomics model for androgen deprivation therapy for prostate cancer. J Int Med Res 2021; 49: 3000605211014301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Moazemi S, Erle A, Khurshid Z, et al. Decision-support for treatment with 177Lu-PSMA: machine learning predicts response with high accuracy based on PSMA-PET/CT and clinical parameters. Ann Transl Med 2021; 9: 818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Abdollahi H, Mahdavi SR, Shiri I, et al. Magnetic resonance imaging radiomic feature analysis of radiation-induced femoral head changes in prostate cancer radiotherapy. J Cancer Res Ther 2019; 15(Suppl.): S11–S19. [DOI] [PubMed] [Google Scholar]
- 165. Lorenz JW, Schott D, Rein L, et al. Serial T2-weighted magnetic resonance images acquired on a 1.5 Tesla magnetic resonance linear accelerator reveal radiomic feature variation in organs at risk: an exploratory analysis of novel metrics of tissue response in prostate cancer. Cureus 2019; 11: e4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Bardis MD, Houshyar R, Chang PD, et al. Applications of artificial intelligence to prostate multiparametric MRI (mpMRI): current and emerging trends. Cancers 2020; 12: 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Sun Y, Reynolds HM, Parameswaran B, et al. Multiparametric MRI and radiomics in prostate cancer: a review. Australas Phys Eng Sci Med 2019; 42: 3–25. [DOI] [PubMed] [Google Scholar]
- 168. Ferro M, de Cobelli O, Vartolomei MD, et al. Prostate cancer radiogenomics-from imaging to molecular characterization. Int J Mol Sci 2021; 22: 9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Lee SL, Lee J, Craig T, et al. Changes in apparent diffusion coefficient radiomics features during dose-painted radiotherapy and high dose rate brachytherapy for prostate cancer. Phys Imaging Radiat Oncol 2019; 9: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Yoo S, Gujrathi I, Haider MA, et al. Prostate cancer detection using deep convolutional neural networks. Sci Rep 2019; 9: 19518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Suarez-Ibarrola R, Hein S, Reis G, et al. Current and future applications of machine and deep learning in urology: a review of the literature on urolithiasis, renal cell carcinoma, and bladder and prostate cancer. World J Urol 2020; 38: 2329–2347. [DOI] [PubMed] [Google Scholar]
- 172. Li H, Lee CH, Chia D, et al. Machine learning in prostate MRI for prostate cancer: current status and future opportunities. Diagnostics 2022; 12: 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Shah U, Biswas MR, Alzubaidi MS, et al. Recent developments in artificial intelligence-based techniques for prostate cancer detection: a scoping review. Stud Health Technol Inform 2022; 289: 268–271. [DOI] [PubMed] [Google Scholar]
- 174. Viswanath SE, Chirra PV, Yim MC, et al. Comparing radiomic classifiers and classifier ensembles for detection of peripheral zone prostate tumors on T2-weighted MRI: a multi-site study. BMC Med Imaging 2019; 19: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Lemaître G, Martí R, Freixenet J, et al. Computer-Aided Detection and diagnosis for prostate cancer based on mono and multi-parametric MRI: a review. Comput Biol Med 2015; 60: 8–31. [DOI] [PubMed] [Google Scholar]
- 176. Mah SA, Avci R, Du P, et al. Antral variation of murine gastric pacemaker cells informed by confocal imaging and machine learning methods. Annu Int Conf IEEE Eng Med Biol Soc 2021; 2021: 3105–3108. [DOI] [PubMed] [Google Scholar]
- 177. Alshehri AHD, Osman SOS, Prise KM, et al. A novel tool for improving the interpretation of isotope bone scans in metastatic prostate cancer. Br J Radiol 2020; 93: 20200775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Bianconi F, Antonini C, Tomassoni L, et al. CRA toolbox: software package for conditional robustness analysis of cancer systems biology models in MATLAB. BMC Bioinformatics 2019; 20: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Kuhn M. Building predictive models in R using the caret package. J Stat Softw 2008; 28: 1–26.27774042 [Google Scholar]
- 180. Macia N, Bernadó-Mansilla E. Towards UCI+: a mindful repository design. Inf Sci 2014; 261: 237–262. [Google Scholar]
- 181. Hothorn T, Leisch F, Zeileis A, et al. The design and analysis of benchmark experiments. J Comput Graph Stat 2005; 14: 675–699. [Google Scholar]
- 182. Eugster MJ, Hothorn T, Leisch F. Domain-based benchmark experiments: exploratory and inferential analysis. Austrian J Stat 2012; 41: 5–26. [Google Scholar]
- 183. Liu L, Tian Z, Zhang Z, et al. Computer-aided detection of prostate cancer with MRI: technology and applications. Acad Radiol 2016; 23: 1024–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Giannini V, Mazzetti S, Armando E, et al. Multiparametric magnetic resonance imaging of the prostate with computer-aided detection: experienced observer performance study. Eur Radiol 2017; 27: 4200–4208. [DOI] [PubMed] [Google Scholar]
- 185. Hambrock T, Vos PC, Hulsbergen-van de Kaa CA, et al. Prostate cancer: computer-aided diagnosis with multiparametric 3-T MR imaging – effect on observer performance. Radiology 2013; 266: 521–530. [DOI] [PubMed] [Google Scholar]
- 186. Ham J, Chen Y, Crawford MM, et al. Investigation of the random forest framework for classification of hyperspectral data. IEEE Trans Geosci Remote Sens 2005; 43: 492–501. [Google Scholar]
- 187. Bosch A, Zisserman A, Munoz X. Image classification using random forests and ferns. In: 2007 IEEE 11Th international conference on computer vision, Rio de Janeiro, 14–21 October 2007, pp. 1–8. New York: IEEE. [Google Scholar]
- 188. Sushentsev N, Rundo L, Blyuss O, et al. Comparative performance of MRI-derived PRECISE scores and delta-radiomics models for the prediction of prostate cancer progression in patients on active surveillance. Eur Radiol 2022; 32: 680–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Lay N, Tsehay Y, Greer MD, et al. Detection of prostate cancer in multiparametric MRI using random forest with instance weighting. J Med Imaging 2017; 4: 024506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Yang X, Wu W, Yan B, et al. Infrared image super-resolution with parallel random forest. Int J Parallel Program 2018; 46: 838–858. [Google Scholar]
- 191. Bouazza SH, Hamdi N, Zeroual A, et al. Gene-expression-based cancer classification through feature selection with KNN and SVM classifiers. In: 2015 intelligent systems and computer vision (ISCV), Fez, Morocco, 25–26 March 2015, pp. 1–6. New York: IEEE. [Google Scholar]
- 192. Maillo J, Ramírez S, Triguero I, et al. kNN-IS: an iterative spark-based design of the k-nearest neighbors classifier for big data. Knowl-Based Syst 2017; 117: 3–15. [Google Scholar]
- 193. Jensen C, Carl J, Boesen L, et al. Assessment of prostate cancer prognostic Gleason grade group using zonal-specific features extracted from biparametric MRI using a KNN classifier. J Appl Clin Med Phys 2019; 20: 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Makowski MR, Bressem KK, Franz L, et al. De novo radiomics approach using image augmentation and features from T1 mapping to predict Gleason scores in prostate cancer. Invest Radiol 2021; 56: 661–668. [DOI] [PubMed] [Google Scholar]
- 195. Zhao W, Xu Y, Yang Z, et al. Development and validation of a radiomics nomogram for identifying invasiveness of pulmonary adenocarcinomas appearing as subcentimeter ground-glass opacity nodules. Eur J Radiol 2019; 112: 161–168. [DOI] [PubMed] [Google Scholar]
- 196. Huang Y, Liu Z, He L, et al. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology 2016; 281: 947–957. [DOI] [PubMed] [Google Scholar]
- 197. Liang C, Huang Y, He L, et al. The development and validation of a CT-based radiomics signature for the preoperative discrimination of stage I-II and stage III-IV colorectal cancer. Oncotarget 2016; 7: 31401–31412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Ji X, Zhang J, Shi W, et al. Bi-parametric magnetic resonance imaging based radiomics for the identification of benign and malignant prostate lesions: cross-vendor validation. Phys Eng Sci Med 2021; 44: 745–754. [DOI] [PubMed] [Google Scholar]
- 199. Bi WL, Hosny A, Schabath MB, et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J Clin 2019; 69: 127–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Jamaludin A, Kadir T, Zisserman A. Self-supervised learning for spinal MRIs. In: Jorge Cardoso M, Arbel T, Carneiro G, et al. (eds) Deep learning in medical image analysis and multimodal learning for clinical decision support. Cham: Springer, 2017, pp. 294–302. [Google Scholar]
- 201. Purushotham S, Meng C, Che Z, et al. Benchmarking deep learning models on large healthcare datasets. J Biomed Inform 2018; 83: 112–134. [DOI] [PubMed] [Google Scholar]
- 202. Song SE, Seo BK, Cho KR, et al. Computer-aided detection (CAD) system for breast MRI in assessment of local tumor extent, nodal status, and multifocality of invasive breast cancers: preliminary study. Cancer Imaging 2015; 15: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Abdollahi H, Mostafaei S, Cheraghi S, et al. Cochlea CT radiomics predicts chemoradiotherapy induced sensorineural hearing loss in head and neck cancer patients: a machine learning and multi-variable modelling study. Phys Med 2018; 45: 192–197. [DOI] [PubMed] [Google Scholar]
- 204. Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med 1997; 16: 385–395. [DOI] [PubMed] [Google Scholar]
- 205. Zou H. The adaptive lasso and its oracle properties. J Am Stat Assoc 2006; 101: 1418–1429. [Google Scholar]
- 206. Castaldo R, Cavaliere C, Soricelli A, et al. Radiomic and genomic machine learning method performance for prostate cancer diagnosis: systematic literature review. J Med Internet Res 2021; 23: e22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Ishioka J, Matsuoka Y, Uehara S, et al. Computer-aided diagnosis of prostate cancer on magnetic resonance imaging using a convolutional neural network algorithm. BJU Int 2018; 122: 411–417. [DOI] [PubMed] [Google Scholar]
- 208. Zhang L, Jiang D, Chen C, et al. Development and validation of a multiparametric MRI-based radiomics signature for distinguishing between indolent and aggressive prostate cancer. Br J Radiol 2022; 95: 20210191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Zhao B. Understanding sources of variation to improve the reproducibility of radiomics. Front Oncol 2021; 11: 633176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Schmidt RM, Delgadillo R, Ford JC, et al. Assessment of CT to CBCT contour mapping for radiomic feature analysis in prostate cancer. Sci Rep 2021; 11: 22737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Delgadillo R, Spieler BO, Ford JC, et al. Repeatability of CBCT radiomic features and their correlation with CT radiomic features for prostate cancer. Med Phys 2021; 48: 2386–2399. [DOI] [PubMed] [Google Scholar]
- 212. Gudmundsson S, Runarsson TP, Sigurdsson S. Test–retest reliability and feature selection in physiological time series classification. Comput Methods Programs Biomed 2012; 105: 50–60. [DOI] [PubMed] [Google Scholar]
- 213. Traverso A, Kazmierski M, Welch ML, et al. Sensitivity of radiomic features to inter-observer variability and image pre-processing in apparent diffusion coefficient (ADC) maps of cervix cancer patients. Radiother Oncol 2020; 143: 88–94. [DOI] [PubMed] [Google Scholar]
- 214. Steenbergen P, Haustermans K, Lerut E, et al. Prostate tumor delineation using multiparametric magnetic resonance imaging: inter-observer variability and pathology validation. Radiother Oncol 2015; 115: 186–190. [DOI] [PubMed] [Google Scholar]
- 215. Chu CE, Alshalalfa M, Sjöström M, et al. Prostate-specific membrane antigen and fluciclovine transporter genes are associated with variable clinical features and molecular subtypes of primary prostate cancer. Eur Urol 2021; 79: 717–721. [DOI] [PubMed] [Google Scholar]
- 216. Schwier M, van Griethuysen J, Vangel MG, et al. Repeatability of multiparametric prostate MRI radiomics features. Sci Rep 2019; 9: 9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Kesch C, Radtke J-P, Wintsche A, et al. Correlation between genomic index lesions and mpMRI and 68Ga-PSMA-PET/CT imaging features in primary prostate cancer. Sci Rep 2018; 8: 16708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Lütje S, Heskamp S, Cornelissen AS, et al. PSMA ligands for radionuclide imaging and therapy of prostate cancer: clinical status. Theranostics 2015; 5: 1388–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Ha S. Perspectives in radiomics for personalized medicine and theranostics. Nucl Med Mol Imaging 2019; 53: 164–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Stoyanova R, Takhar M, Tschudi Y, et al. Prostate cancer radiomics and the promise of radiogenomics. Transl Cancer Res 2016; 5: 432–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Ferro M, Crocetto F, Bruzzese D, et al. Prostate Health Index and multiparametric MRI: partners in crime fighting overdiagnosis and overtreatment in prostate cancer. Cancers 2021; 13: 4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Maggi M, Del Giudice F, Falagario UG, et al. SelectMDx and multiparametric magnetic resonance imaging of the prostate for men undergoing primary prostate biopsy: a prospective assessment in a multi-institutional study. Cancers 2021; 13: 2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Galavis PE, Hollensen C, Jallow N, et al. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol 2010; 49: 1012–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Heinrich-Heine University, Duesseldorf. Prospective phase II trial evaluating multiparametric MRI, radiomics, MR-guided biopsy, and molecular markers for active surveillance of patients with low- and intermediate-risk prostate cancer. Clinical Trial Registration NCT03979573, clinicaltrials.gov, https://clinicaltrials.gov/ct2/show/NCT03979573 (2019, accessed 5 October 2021).
- 225. Kucharczyk M. Can magnetic resonance imaging of the prostate combined with a radiomics evaluation determine the invasive capacity of a tumour (can MRI-PREDICT). Clinical Trial Registration NCT05024162, clinicaltrials.gov, https://clinicaltrials.gov/ct2/show/NCT05024162 (2021, accessed 5 October 2021).
- 226. Centre hospitalier de l’Université de Montréal (CHUM). PSMA-PET: deep radiomic biomarkers of progression and response prediction in prostate cancer. Clinical Trial Registration NCT03594760, clinicaltrials.gov, https://clinicaltrials.gov/ct2/show/NCT03594760 (2021, accessed 5 October 2021).
- 227. Cobelli FD. MR radiomic features in prostate cancer. Clinical Trial Registration NCT04219059, clinicaltrials.gov, https://clinicaltrials.gov/ct2/show/NCT04219059 (2021, accessed 5 October 2021).
- 228. Hofman M. Prostate cancer theranostics and imaging centre of excellence (ProsTIC) prospective patient registry of men treated with PSMA theranostics. Clinical Trial Registration NCT04769817, clinicaltrials.gov, https://clinicaltrials.gov/ct2/show/NCT04769817 (2021, accessed 5 October 2021).