Abstract
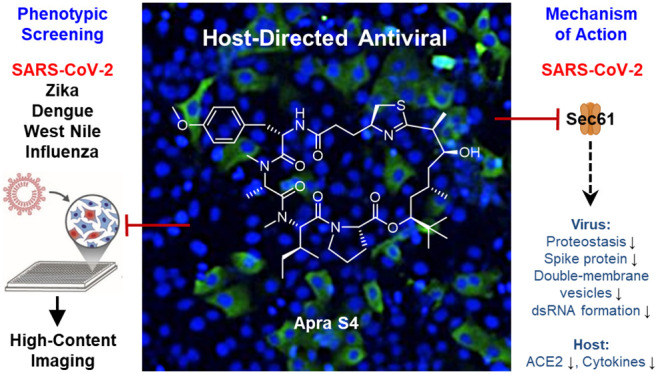
There is a pressing need for host-directed therapeutics that elicit broad-spectrum antiviral activities to potentially address current and future viral pandemics. Apratoxin S4 (Apra S4) is a potent Sec61 inhibitor that prevents cotranslational translocation of secretory proteins into the endoplasmic reticulum (ER), leading to anticancer and antiangiogenic activity both in vitro and in vivo. Since Sec61 has been shown to be an essential host factor for viral proteostasis, we tested Apra S4 in cellular models of viral infection, including SARS-CoV-2, influenza A virus, and flaviviruses (Zika, West Nile, and Dengue virus). Apra S4 inhibited viral replication in a concentration-dependent manner and had high potency particularly against SARS-CoV-2 and influenza A virus, with subnanomolar activity in human cells. Characterization studies focused on SARS-CoV-2 revealed that Apra S4 impacted a post-entry stage of the viral life-cycle. Transmission electron microscopy revealed that Apra S4 blocked formation of stacked double-membrane vesicles, the sites of viral replication. Apra S4 reduced dsRNA formation and prevented viral protein production and trafficking of secretory proteins, especially the spike protein. Given the potent and broad-spectrum activity of Apra S4, further preclinical evaluation of Apra S4 and other Sec61 inhibitors as antivirals is warranted.
Keywords: Sec61, COVID-19, host-directed therapeutics, broad-spectrum antivirals, double-membrane vesicles
COVID-19 has a broad clinical spectrum, ranging from asymptomatic to mild and severe symptoms. It is caused by the highly transmissible SARS-CoV-2, which belongs to the coronavirus (CoV) family, responsible for three major outbreaks in the 21st century (SARS-CoV-1, MERS-CoV, SARS-CoV-2). The SARS-CoV-2 genome encodes the Orf1ab polyprotein at the 5′ end, which is cleaved into 16 nonstructural proteins (Nsp), including the drug targets RNA polymerase (Nsp12) and main protease/3CL (Nsp5), as well as four major structural proteins encoded at the 3′ end (S, E, M, N) and accessory factors.1 Several vaccines are approved for preventing severe disease and hospitalization, and neutralizing antibody treatments have shown some success when administered at early stages of the disease.2,3 However, there are only few widely available specific small-molecule antiviral therapies for SARS-CoV-2. Remdesivir, targeting RNA-dependent RNA polymerase (RdRp), has pan-coronavirus inhibitory activity4 and has been FDA approved because of the reduced time to recovery;5 however, it requires injections and is not effective for patients with advanced disease, when the lung inflammation downstream of viral infection has already occurred. The RNA polymerase inhibitor molnupiravir, which received an emergency use authorization, has been shown to cut down the rate of hospitalizations and death by 30% (down from early clinical trial observation of 50%); however, administration near disease onset is important.6 The oral protease inhibitor PF-07321332, coadministered with low-dose ritonavir to enhance stability (Paxlovid), targeting the SARS-CoV-2 main protease (3CL) was also designed for patients exposed to or showing first signs of disease, and it showed an 89% reduction in risk of hospitalization and death and was consequently approved for emergency use.7 However, therapeutics with different mechanisms of action are needed. Drug repurposing has been at the forefront, and a large-scale repositioning survey for SARS-CoV-2 antivirals identified 21 candidates,8 including compounds that were subsequently validated in animal models.9 Because of the inflammatory component at later disease stages and the cytokine storm that correlates with the need for mechanical ventilation and mortality,10 anti-IL-6 treatments and corticosteroids have also been tested. Antibodies against the IL-6 receptor (tocilizumab or sarilumab) have shown some benefits to treat serious disease,11 and dexamethasone reduced mortality in patients already receiving respiratory support;12 however, delayed virus clearance is a concern if administered early.13,14
Looking forward, an ideal agent should also be active against other viruses of pandemic potential. For example, influenza A viruses cause respiratory distress and are responsible for four pandemics over the past hundred years in addition to annual epidemics. Changes in the influenza virus genome during replication and genetic reassortment between strains can result in novel strains, for which we lack immunity or that are resistant to current antivirals. Since many viruses rely on similar cellular pathways for their replication, drugs inhibiting these pathways could benefit from broad-spectrum antiviral activities.
Previous work revealed that Sec61 is essential for proteostasis of influenza A virus, HIV, and dengue virus.15−17 Sec61 is the translocon in the endoplasmic reticulum (ER) membrane, required for secretory protein transport and processing. Thus, it is also required for the processing and transport of viral glycoproteins and therefore essential for viral proteostasis.15−17 A SARS-CoV-2-human protein–protein interaction map revealed potential drug targets at the interface of host and viral proteins, including the interaction of Nsp8 with the signal recognition particle and Orf8 with the ER protein quality control, directly related to cotranslational translocation and processing.18 We have previously identified a marine-derived compound class that targets Sec61 and therefore pursued the hypothesis that it might exert broad-acting antiviral activities. Specifically, we discovered the apratoxin class of Sec61 inhibitors from a marine cyanobacterium19,20 synthesized and structurally modified the parent natural products,21−24 and identified and mechanistically characterized the effects on cotranslational translocation and downstream consequences.21−26 Apratoxins directly target Sec61α.27,28 Synthetic apratoxins were shown to be effective against pancreatic and colon cancer in animal models.21,22,29 We aimed to test if apratoxins could also combat SARS-CoV-2, and increase the therapeutic tool box for other viruses with pandemic potential.
There are several advantages of targeting host factors, such as Sec61, over virally encoded proteins: (i) targeting a host factor is expected to lead to reduced mutational resistance,30 and (ii) broad spectrum activity against other viruses. Apratoxins would be expected to inhibit viral proteostasis by interfering with endoplasmic reticulum trafficking of viral glycoproteins that require ER-mediated glycosylation and processing through interaction with Sec61,15 in addition to inhibitory effects on secretory host proteins, including the SARS-CoV-2 receptor ACE2 and cytokines.
The well-characterized anticancer agent apratoxin S4 (Apra S4, Figure 1), a synthetic analogue of marine cyanobacterial natural products, was previously repositioned and proved highly effective in ocular angiogenic disease models in vivo.31 Here, we report potent antiviral effects of Apra S4 against SARS-CoV-2 compared with RdRp inhibitor remdesivir (preventing viral RNA synthesis) and PIKfyve inhibitor apilimod (interfering with cell entry)32 (Figure 1). Apra S4 was also found to have activity against various influenza and flaviviruses. Characterization studies on the molecular and ultrastructural level in the context of SARS-CoV-2, employing electron microscopy and immunofluorescence, revealed that Apra S4 blocked the formation of stacked double-membrane vesicles and vesicles with multiple progeny viruses, and decreased levels of intracellular S protein in infected cells and viral protein trafficking to the cell surface, as the major mechanism of action.
Figure 1.
Structures and targets of apratoxin S4 (Apra S4), and of remdesivir and apilimod used as controls in our studies.
Results
Apra S4 Exhibits Potent Activity against SARS-CoV-2 in Vitro
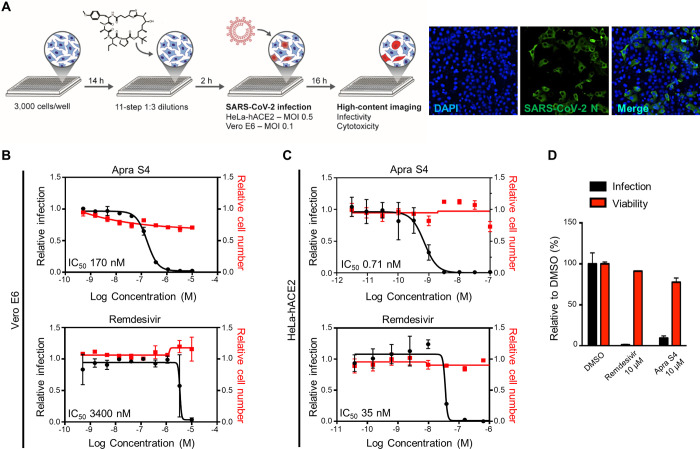
The antiviral activity of Apra S4 was characterized in multicycle virus growth assays in cell culture. Specifically, Vero E6 and HeLa-hACE2 (HeLa cells transduced with human ACE2) cells were preincubated with Apra S4 for 2 h, followed by infection with SARS-CoV-2. Sixteen hours postinfection, cells were fixed and immunostained for the SARS-CoV-2 nucleoprotein (N). Cellular nuclei were stained with DAPI, prior to automated imaging and analysis (Figure 2A). The percentage of infection for each well was calculated as the ratio of infected cells stained with N antibody, over the total number of cells. Apra S4 treatment reduced viral replication in a concentration-dependent manner, with IC50 values of 0.17 μM (Vero E6) and 0.71 nM (HeLa-hACE2), outperforming remdesivir by 20- to 50-fold in monkey (Vero) and human (HeLa-hACE2) cells, respectively. Cytotoxicity was not observed under these conditions, with nearly 100% inhibition of viral replication at 2 nM in human cells (Figure 2B,C).
Figure 2.
Screening approach and activities of Apra S4 against SARS-CoV-2 compared with remdesivir. (A) Three thousand cells per well were seeded in a 384-well black plate 16 h prior to infection. Two hours prior to infection, cells were treated with remdesivir or Apra S4 in 11-step 1:3 dilutions at indicated starting concentrations, in triplicate. SARS-CoV-2 USA-WA1/2020 was added to each well, at an MOI = 0.1 (Vero E6) or 1 (HeLa-hACE2). Sixteen hours postinfection, cells were fixed and analyzed by immunofluorescence imaging. For each condition, the percentage of infection was calculated as the ratio of the number of infected cells stained for coronavirus N to number of cells stained with DAPI. (B,C) Dose–response analysis for Vero E6 (B) and HeLa-hACE2 cells (C). (D) Antiviral activity of Apra S4 against SARS-CoV-2 infection in human stem cell derived pneumocyte-like cells. Human stem cell derived pneumocyte-like cells were treated with remdesivir or Apra S4 (10 μM) for 1 h at 37 °C followed by SARS-CoV-2 USA-WA1/2020 infection for 48 h at 37 °C. Then, the cells were dissociated using cell dissociation buffer and fixed in 4% methanol-free formaldehyde for the following FACS analysis. The fixed cells were incubated with antimouse SARS-CoV-2 N protein antibody for 1 h and the infected cells were counted by flow cytometry. The data were represented for each triplicate as the percentage of cells infected compared to the DMSO control. The duplicate set of cells were treated with same drug concentrations but left uninfected. After 48 h of incubation at 37 °C, the cells were analyzed for viability using an MTT assay.
Apra S4 Shows Antiviral Activity in a Human Primary Cell Model
Apra S4 was further evaluated for antiviral activity in human pneumocyte-like cells derived from human stem cells. These cells are considered a relevant model for SARS-CoV-2 replication in the lung. Cells were differentiated and then incubated with 10 μM Apra S4 or remdesivir 1 h prior to infection. Cells were infected with SARS-CoV-2 in the presence of the compound. After 48 h of infection, viral infection was quantified by flow-cytometry analysis of CoV N expression. Treatment with Apra S4 resulted in significantly decreased viral replication in the human pneumocyte-like cells. No cytotoxicity of the compounds was observed (Figure 2D).
Apra S4 Exhibits Broad-Spectrum Antiviral Effects in Vitro
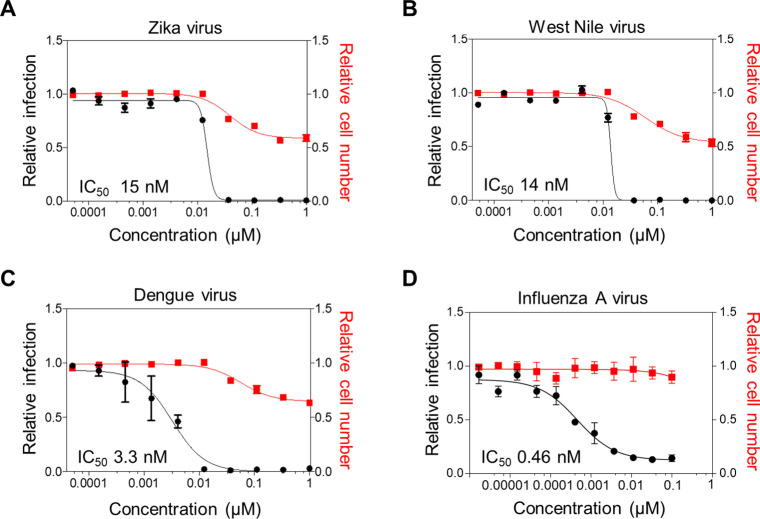
Apra S4 also showed potent antiviral activity in the low nM range against flaviviruses, including Zika virus, Dengue virus, and West Nile virus (Figure 3, Table 1), with no to negligible cytotoxicity (CC50) (Figure 3A–C). Apra S4-inhibited SARS-CoV-2 and influenza A virus very potently (Figure 3D), with subnanomolar IC50 values and high selectivity index (SI = CC50/IC50 at 16 h, Table 1), indicating that Apra S4-induced antiviral activity is not due to toxicity, which is usually a general concern with anticancer agents. It is known that apratoxins exert cytotoxic and/or growth inhibitory activity between 24 and 48 h, which is in the low to mid nM range for human cancer cells.23 Therefore, we only determined the CC50 up to 1 μM in human cells at 24 h (flaviviruses) or 36 h (influenza).
Figure 3.
Antiviral activity of Apra S4 against flaviviruses and influenza A virus. (A–C) Huh7.5 cells were seeded overnight and then treated with increasing concentrations of Apra S4 for 2 h prior to infection. Cells were then infected with (A) Zika virus/Ugandan MR766 at MOI = 0.25, (B) West Nile Virus/Kunjin isolate at MOI = 0.05, or (C) Dengue virus/16681 serotype 2 at MOI = 3 in the presence of Apra S4. (D) For influenza A virus/WSN infection, A549 cells were treated with Apra S4 for 2 h and infected at MOI = 0.1. At 24 h postinfection of flaviviruses (A–C) or 36 h postinfection of influenza virus (D), cells were fixed and infection was measured through immunofluorescence. The percentage of infection was calculated as the ratio between the number of infected cells stained for flavivirus envelope protein or influenza nucleoprotein, and the total amount of cells stained with DAPI. Dose–response curves for both infectivity (black) and cell number (red) are shown. Data represent the average ± standard deviation of infection and cell count relative to DMSO.
Table 1. Antiviral Activity of Apratoxin S4 against SARS-CoV-2, Flaviviruses, and Influenza.
| viruses | cell line | IC50 (nM) | CC50 (μM) | SI |
|---|---|---|---|---|
| SARS-CoV-2 USA-WA1/2020 | Vero E6 | 170 | >10 | >58 |
| SARS-CoV-2 USA-WA1/2020 | HeLa-hACE2 | 0.71 | >1 | >1400 |
| Zika virus/Ugandan MR766 | Huh-7.5 | 15 | >1 | >67 |
| West Nile virus/Kunjin isolate | Huh-7.5 | 14 | >1 | >71 |
| Dengue virus/16681 serotype 2 | Huh-7.5 | 3.3 | >1 | >300 |
| Influenza A virus/WSN | A549 | 0.46 | >1 | >2200 |
Apra S4 Reduces Multicycle Growth and Impacts on SARS-CoV-2 Post-Entry Steps
Apra S4 strongly reduced SARS-CoV-2 multicycle growth based on measurements at 24, 48, and 72 h post infection, while remdesivir was less potent at 10 μM under these experimental conditions (Figure 4A), underscoring the strong antiviral activity of Apra S4. To verify that these effects are Sec61-mediated, we treated with eeyarestatin I, which was suggested to also inhibit Sec61 (in addition to p97).33,34 Eeyarestatin I also inhibited SARS-CoV-2 in a dose-dependent manner, although less potently than Apra S4 (only at 10 μM, Supporting Information, Figure S1). Similarly, siRNA-mediated knockdown of Sec61A1 using validated siRNAs16 reduced SARS-CoV-2 growth by 10- to 100-fold (Supporting Information, Figure S2). Taken together, these results suggest that the antiviral activity of Apra S4 is an on-target (Sec61) effect. We next performed time-of-addition studies to evaluate if Apra S4 acts on entry or post-entry steps of the SARS-CoV-2 replication cycle. Specifically, Vero CCL81 cells were infected for 1 h with SARS-CoV-2 (MOI = 0.5) and incubated with Apra S4 at 1 μM at 2 h before and after, as well as concomitant with viral challenge (Figure 4B). Viral infection was determined after 16 h by virus titer determination. Apra S4 did not lose its antiviral activity even if added 2 h after infection, suggesting that it does not act on viral entry but targets post-entry steps (Figure 4B). To further validate these results, we evaluated the impact of Apra S4 on a vesicular stomatitis virus (VSV)-based virus-like particle pseudotyped with SARS-CoV-1 S protein, SARS-CoV-2 S protein, or VSV-G protein in Vero E6 cells. Apilimod, a PIKfyve inhibitor, is known to inhibit viral replication during the entry step.8 Apilimod inhibited viral entry of SARS-CoV-1 and SARS-CoV-2, consistent with previous observations (Figure 4C).32,35 On the other hand, Apra S4 treatment for 2 h prior to virus challenge for 1 h slightly enhanced infection compared to the DMSO control, suggesting that Apra S4 does not inhibit viral entry of SARS-CoV-1 or SARS-CoV-2 (Figure 4C). None of the compounds impacted VSV-G-mediated entry. No cytotoxic effect was observed under these conditions.
Figure 4.
Effect of Apra S4 on SARS-CoV-2 multicycle growth and entry. (A) SARS-CoV-2 growth curve. Left panel: Vero CCL81 cells were treated with Apra S4, Remdesivir, or DMSO (0.1%) at the indicated concentrations 2 h before, during, and after infection with SARS-CoV-2 BetaCoV/Germany/BavPat1/2020 (MOI = 0.01). Of note, compounds were not replenished during incubation. Cell supernatants were harvested 0, 16, 24, 48, and 72 h postinfection and virus titers were determined by plaque assay. Shown are mean virus titers of three independent experiments performed in duplicates. Error bars indicate standard deviation. LoD: limit of detection was at 100 PFU/mL. Of note, titers for 1 μM and 0.1 μM Apra S4 lie below the LoD. Right panel: Cell viability of Vero CCL81 after treatment similar to left panel, but left uninfected. At the indicated time points cell viability was determined using the CellTiter-Glo kit. (B) Time-of-Addition assay: Vero CCL81 cells were treated with Apra S4 (1 μM), Remdesivir (10 μM), or DMSO (0.1%) starting either 2 h before, during, or 2 h after infection with SARS-CoV-2 BetaCoV/Germany/BavPat1/2020 (MOI = 0.5). Cell supernatants were harvested 16 h postinfection and virus titers were determined by plaque assay. Shown are mean virus titers of three independent experiments performed in duplicates. Error bars indicate standard deviation. For both (A) and (B), an unpaired t test on log-transformed data was performed to test for statistical significance. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. LoD: limit of detection was at 100 PFU/mL. (C) VSV-based pseudotyped viral particles assay. Vero E6 cells were pretreated with Apra S4 for 2 h and then infected with SARS-CoV-1 S/SARS-CoV-2 S pseudotyped particles harboring firefly luciferase for additional 1 h. Luciferase signal was quantified at 24 h postinoculation (left). The cell viability was measured at 24 h postinfection using WST-8 assay and normalized to the DMSO-treated samples (right). Error bars represent standard error of the mean (SEM).
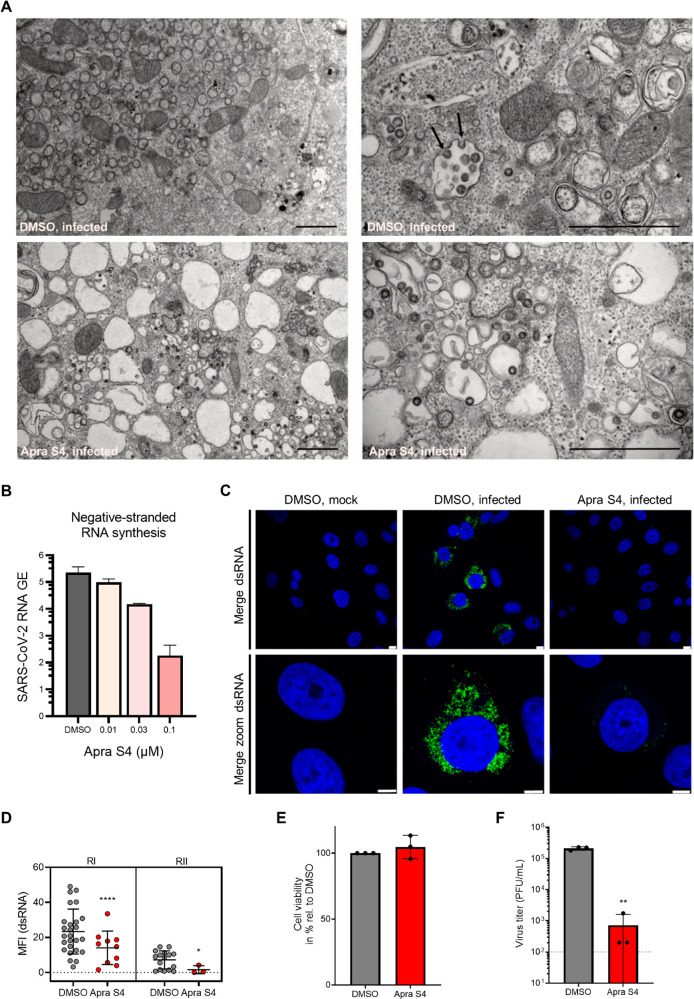
Apra S4 Inhibits Formation of Double-Membrane Vesicles and RNA Replication
To understand how Apra S4 inhibits viral replication in cells, we visualized viral replication factories by electron and confocal microscopy. First, we employed transmission electron microscopy on ultrathin sections of Vero CCL81 cells treated with Apra S4 (or solvent as control) and infected with SARS-CoV-2 (MOI = 2) (Figure 5A). In control-treated SARS-CoV-2 infected cells we detected large clusters of stacked double-membrane vesicles (Figure 5A, upper left) that represent the viral replication organelles as reported previously.36 In areas adjacent to the replication organelles, we observed larger vesicles containing multiple virus particles (Figure 5A, upper right). Progeny virions budding into these vesicles were also observed frequently (Figure 5A, upper right, arrows). In contrast, no stacked double-membrane vesicles (DMV) and vesicles carrying multiple virus particles could be detected in inhibitor-treated cells (Figure 5A, bottom panels). Instead, large empty vacuoles (Figure 5A, bottom left) and only a few vesicles with single virus particles were detected in Apra S4-treated samples (Figure 5A, bottom right). These data suggest that Apra S4 not only impacts viral proteostasis but also leads to a block of the formation of viral replication organelles, used as dedicated sites for viral RNA replication, and vesicles with multiple progeny viruses.37,38 As Apra S4 targets the translocon complex and affects primarily the expression of proteins within the secretory pathway, our observations suggest that viral RNA replication could be indirectly compromised by Apra S4 treatment. We therefore evaluated the impact of Apra S4 on viral RNA production by electroporating in vitro transcribed viral RNA into Vero E6 cells, and directly measured RNA synthesis (Figure 5B). At 12 h postelectroporation, Apra S4 reduced the negative-strand RNA at a concentration near the IC50 for antiviral activity (0.1 μM, Figure 5B). This inhibition of viral RNA production is presumably a consequence of the inhibition of DMV formation. To further dissect the mechanism of inhibition we stained for dsRNA, a marker for viral replication, 16 h after infection of Vero CCL81 cells with SARS-CoV-2. We found that treatment with 1 μM Apra S4 strongly reduced the presence of dsRNA intermediates in infected cells compared to DMSO treatment, indicating that viral genome replication was affected (Figure 5C,D). This inhibition of dsRNA formation was not due to cytotoxic effects of the inhibitor (Figure 5E) and correlated with a more than 100-fold reduction in infectious virus titers in the supernatant under these conditions at the same time point (Figure 5F). Apra S4 blocked the formation of stacked double-membrane vesicles, leading to the inhibition of viral replication, and prevented formation of vesicles with multiple progeny viruses.
Figure 5.
Apra S4 inhibits formation of double-membrane vesicles and SARS-CoV-2 replication. (A) Transmission electron microscopy demonstrates block of the formation of viral replication organelles. Vero CCL81 cells were pretreated with DMSO (top) or 1 μM Apra S4 (bottom) for 2 h before cells were infected with SARS-CoV-2 BetaCoV/Germany/BavPat1/2020 (MOI = 2) in the presence of DMSO or Apra S4. At 16 h postinfection cells were fixed, dehydrated, and embedded. Ultrathin sections were stained with lead citrate and analyzed by transmission electron microscopy. Arrows indicate budding viruses. Scale bars indicate 1 μm. (B) Negative-stranded viral RNA synthesis. Vero E6 cells were electroporated with in vitro transcribed viral RNA. Two hours after seeding, the cells were treated with Apra S4 at the indicated concentration. Negative-stranded RNA was then quantified at 12 h postelectroporation. Error bars represent SEM for n = 3 independent experiments. (C) dsRNA expression. Vero CCL81 cells were pretreated with DMSO or 1 μM Apra S4 for 2 h. Cells were infected with SARS-CoV-2 BetaCoV/Germany/BavPat1/2020 (MOI = 2) in the presence of DMSO or Apra S4. At 16 h postinfection, cells were fixed and stained for dsRNA (green). Nuclei are stained in blue. Scale bars indicate 8 and 5 μm for the merged and merged zoom images, respectively. (D) Quantification of dsRNA. The mean fluorescence intensity of dsRNA-expressing cells from (C) was quantified using ImageJ software. Shown is mean fluorescence intensity (MFI) of two independent experiments (RI/RII). Error bars indicate standard deviation. A Mann–Whitney U test was performed to test for statistical analyses. *P < 0.05; ****P < 0.0001 (E) Vero CCL81 cells were treated with DMSO or 1 μM Apra S4 for 18 h and cell viability was determined using the CellTiter-Glo kit. Mean values from three independent experiments are shown relative to the DMSO control. Error bars depict standard deviation. (F) Vero CCL81 cells were pretreated with DMSO or 1 μM Apra S4 for 2 h before cells were infected with SARS-CoV-2 BetaCoV/Germany/BavPat1/2020 (MOI = 2) in the presence of DMSO or Apra S4. At 16 h postinfection virus titers in the supernatants were determined. Mean values from three independent experiments are shown. Error bars depict the standard deviation. Dotted line represents the limit of detection. An unpaired t test on log-transformed data was performed to test for statistical significance. **P < 0.01.
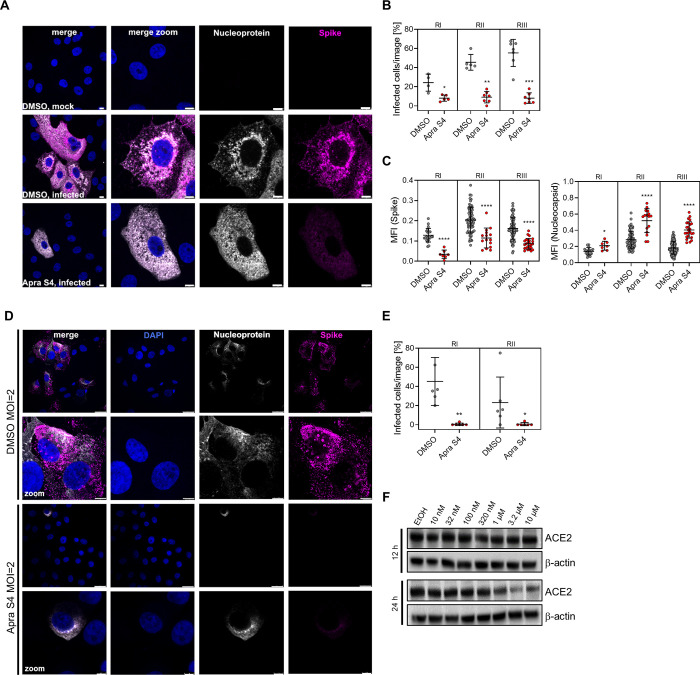
Apra S4 Reduces Production and Trafficking of Secretory Pathway Dependent Proteins
We then pretreated Vero CCL81 cells with Apra S4 or vehicle (DMSO) for 2 h, infected with SARS-CoV-2 for 16 h in the presence or absence of the inhibitor, and stained for the total viral nucleoprotein and the viral spike protein to assess viral protein production (Figure 6A). Quantification of infected cells revealed a significantly reduced number of infected cells in all three independent experiments (Figure 6B). In those few infected cells with detectable nucleoprotein and spike protein levels, the spike protein staining intensity was significantly reduced (Figure 6C). In contrast, the nucleoprotein signal intensity actually increased based on statistical analysis (Figure 6C), possibly due to accumulation of nucleoprotein as no particles are being formed and released. We then immunostained for nucleoprotein and spike protein levels on the cell surface, using nonpermeabilizing conditions (Figure 6D). Surprisingly, besides spike, also the nucleoprotein was detected in nonpermeabilized, infected cells. The inhibitory effect of Apra S4 was even more pronounced when monitoring viral protein expression at the cell surface and apparent for both nucleoprotein and spike protein (Figure 6E). In addition to viral glycoprotein expression, we also expected that ACE2 levels would be compromised by Apra S4 since ACE2 is a known substrate for Sec61-dependent cotranslational translocation. We thus monitored ACE2 protein expression in Vero E6 cells by immunoblot analysis over time (12 and 24 h), and observed decreased protein levels after 24 h starting at 1 μM, slightly above the IC50 for antiviral activity in this cell type (Figure 6F). We also tested human lung epithelial cells (Calu-3) with Apra S4 and observed decreased expression at 3.2 nM or higher concentrations (Supporting Information, Figure S3), consistent with human cells being more sensitive to Apra S4 than monkey cells. This additional on-target activity may provide additive or synergistic antiviral benefits at longer exposure times.
Figure 6.
Apra S4 reduces production and trafficking of secretory pathway dependent proteins. (A) Intracellular spike and nucleoprotein expression. Vero CCL81 cells were pretreated with DMSO or 1 μM Apra S4 for 2 h. Cells were infected with SARS-CoV-2 BetaCoV/Germany/BavPat1/2020 (MOI = 2) in the presence of DMSO or Apra S4. At 16 h postinfection, cells were fixed, permeabilized, and stained for SARS-CoV-2 nucleoprotein (N, gray) and spike protein (S, magenta). Nuclei are stained in blue. Scale bars indicate 8 μm. Of note, in the Apra S4-treated cells, infected cells were found only rarely. Shown are infected cells to compare localization of viral protein in the different experimental conditions. (B,C) Quantification of (A). The percentage infected cells (B) (cells positive for nucleoprotein or spike) per image and the intensity (C) of N (left panel) and S (right panel) was determined using CellProfiler software. Shown are quantification of three independent experiments (RI/RII/RIII). Error bars indicate standard deviation. A Mann–Whitney U test was performed to test for statistical significance. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (D) Cell surface nucleoprotein and spike protein expression. Experimental setup as in (A) but cells were left nonpermeabilized. Cell surface SARS-CoV-2 nucleoprotein (gray) and spike protein (magenta) were stained. Nuclei are stained in blue. Scale bars indicate 25 μm (upper rows) or 5 μm (lowest row). Of note, in the Apra S4-treated cells, infected cells were found only rarely. (E) Quantification of (D). The percentage infected cells was determined using CellProfiler software. Shown are quantifications of two independent experiments (RI/RII). Error bars indicate standard deviation. A Mann–Whitney U test was performed to test for statistical significance. *P < 0.05; **P < 0.01. (F) Immunoblot analysis in Vero E6 cells. Cells were treated with Apra S4 for 12 and 24 h, protein lysates were collected, and ACE2 immunoblot analysis was performed with β-actin serving as loading control.
Discussion
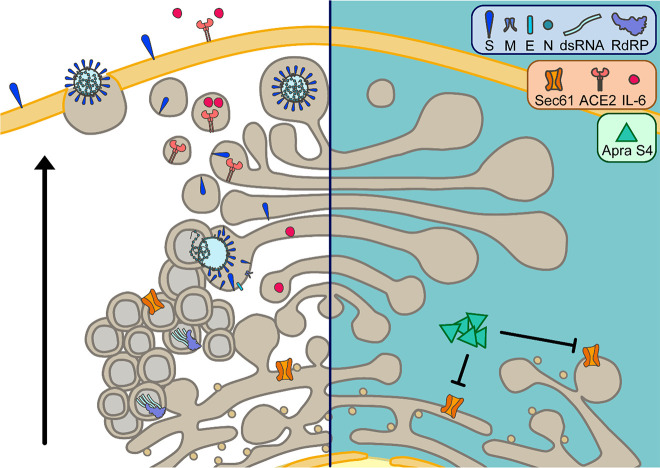
Apratoxin S4 (Apra S4) is a synthetic hybrid analogue of apratoxins A/E, which prevents protein translocation by directly targeting Sec61α, the central subunit of the protein translocation channel on the endoplasmic reticulum (ER) membrane.21,28 Among known Sec61 inhibitors, apratoxins possess unique resistant profiles in cancer cells, indicating a unique mode of target interaction.27,28 It prevents protein ER translocation at an earlier stage than cotransin and appears to impact ER import of Sec61 substrate proteins in a substrate-nonselective fashion based on biochemical data.27 However, the substrate selectivity for host proteins is tunable and highly context-dependent, as we recently reported.39 Since viral glycoproteins are dependent on Sec61-mediated secretion, we observed blockage of viral proteostasis. In cellular models of viral infection including SARS-CoV-2, Apra S4 inhibited viral replication in a concentration-dependent manner. The compound showed antiviral activity by impacting post-entry stages of the viral replication cycle. Immunofluorescence indicated a preference for inhibiting the spike protein as a major target, presumably since it requires ER-mediated glycosylation upon interaction with Sec61, in contrast to the N protein that is less sensitive to the inhibition of the secretory pathway. In addition to the expected inhibition of viral glycoprotein processing and transport, we also detected a block of DMV formation, dsRNA accumulation, and subsequent virion assembly. These findings suggest that Sec61 function is also required for DMV formation and consequently genome replication, in line with previous reports showing that other coronaviruses recruit the Sec61 complex to their replication sites.40,41 The proposed cellular mechanism indicating a post-entry inhibition of replication is depicted in Figure 7.
Figure 7.
Putative antiviral action and effects on host proteins induced by apratoxin treatment as a consequence of Sec61 inhibition (left: uninhibited state, right: inhibited state). Apratoxins, like Apra S4, potently inhibit Sec61α, a subunit of the Sec61-translocon in the ER membrane. Sec61 inhibition by Apra S4 reduces the SARS-CoV-2-induced formation of double-membrane vesicles, the site of viral genome replication. Consequently, replication intermediates such as double-stranded RNA are strongly reduced upon Apra S4 treatment. In addition, expression of viral proteins is negatively affected through Apra S4-mediated inhibition of Sec61. Especially expression of structural proteins that require cotranslational translocation into the ER, presumably in a Sec61-dependent fashion, to undergo N-glycosylation and other posttranslational modifications is prevented by Sec61 inhibition. As a consequence, virus assembly and budding in the ERGIC (ER-Golgi intermediate compartment), trafficking, and release from cells via exocytosis is abrogated in Apra S4-treated cells. Furthermore, apratoxin treatment affects expression of cellular proteins, which require Sec61-mediated cotranslational translocation. Among these proteins, downregulation of ACE2 expression and inhibition of cytokine secretion could further affect SARS-CoV-2 infection and disease progression.
A different type of Sec61 inhibitor, ipomoeassin F, was recently shown to inhibit the in vitro biogenesis of both spike protein and the SARS-CoV receptor ACE2; however, this observation remains to be confirmed in cell culture models.42 Consistent with the known effects of Apra S4 on host membrane proteins,39 Apra S4 additionally downregulates ACE2, although not completely. Further studies are required to determine if this indirect effect would additionally interfere with virus entry upon longer exposure to Apra S4 (Figure 7). By preventing cotranslational translocation, apratoxins also prevent secretion of cytokines, including IL-6. We have shown this in various cancer cells and in stromal cells in the context of tumor microenvironment.24,29 Apra S4 has potential anti-inflammatory activity and blocks LPS-induced cytokines in macrophage cells (Supporting Information, Figure S4). We speculate that apratoxins may also prevent the cytokine storm in COVID-19, which remains to be investigated. Given the clinical data with anti-inflammatory agents,13,14 viral clearance might be delayed if administered early during the disease; the antiviral effects and anti-inflammatory and potentially immunosuppressive effects would have to be balanced.
Apra S4 previously showed exquisite efficacy in ocular angiogenic disease models in vivo at the same concentration as for cancer (0.25 mg/kg) without toxicity or histological abnormalities.31 The eEF1A inhibitor plitidepsin, broadly inhibiting protein synthesis, showed preclinical efficacy in various SARS-CoV-2 animal models.43 We expect that selective inhibitors of subsets of secretory proteins15 (Sec61 inhibitors) could be even more effective and safe, especially given the short-term treatment regimen. Efficacy studies in animal models of SARS-CoV-2 and other viruses are warranted.
Conclusion
Sec61 inhibition by apratoxins provides a complementary mechanism of action to cell entry inhibitors and viral-targeting agents. Apra S4 possesses exquisite broad-spectrum antiviral activity, particularly against SARS-CoV-2, and the spike protein appeared to be one major target of Apra S4-mediated Sec61 inhibition. Apra S4 impaired DMV formation and RNA replication, transport, and processing, and trafficking of spike protein and thus progeny virus particle formation. There is an urgent need for host-directed therapeutics, and further evaluation of Apra S4 and other Sec61 inhibitors, alone or in combination with mechanistically complementary agents, is warranted to potentially address current and future pandemics of concern.
Methods
Synthesis
Apratoxin S4 was synthesized as previously described.22
Viruses and Cell Lines
SARS-CoV-2 USA-WA1/2020 strain, isolated from an oropharyngeal swab from a patient with a respiratory illness who developed clinical disease (COVID-19) in January 2020 in Washington, USA, was obtained from BEI Resources (NR-52281) and used for the Vero E6, HeLa-hACE2, and pneumocyte experiments. SARS-CoV-2 strain BetaCoV/Germany/BavPat1/2020 was obtained from the European Virus Archive GLOBAL (EVA-GLOBAL; ref-SKU: 026V-03883) and used for the Vero CCL81 experiments. SARS-CoV-2 BetaCoV/Germany/BavPat1/2020 virus stocks were generated as described previously.44
West Nile virus (Kunjin isolate) was obtained from Michael S. Diamond (Washington University School of Medicine at St. Louis), Dengue virus (16681 serotype 2) from Ana Fernandez-Sesma (Icahn School of Medicine at Mount Sinai), Zika virus (Ugandan MR766 strain) from ATCC, and influenza A/WSN/33 (H1N1) is a reverse genetics clone as described.45
Calu-3 and RAW264.7 cell lines were obtained from ATCC and used for ACE2 protein expression and cytokine profiling experiments.
SARS-CoV-2 was propagated in Vero E6 (ATCC CRL-1586) cells. A/WSN/33 (H1N1) was propagated in MDCK cells. Titer was determined by plaque assay on MDCK cells using agar overlay medium. Vero E6 cells were maintained in Dulbecco’s modified eagle medium (DMEM, Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco), 50 U/mL penicillin, 50 μg/mL streptomycin, 1 mM sodium pyruvate (Gibco), 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, Gibco), and 1× MEM nonessential amino acids solution (Gibco). Vero CCL81 cells (ATCC) were cultured at 37 °C and 5% CO2 in DMEM (Gibco), supplemented with 10% (v/v) fetal calf serum (FCS), 100 U/mL of penicillin and 100 μg/mL of streptomycin (#15140-122; Gibco). HeLa cells stably expressing ACE2 (HeLa-hACE2) were obtained from Thomas Rogers (University of California, San Diego), generated by transducing HeLa (ATCC) cells with ACE2-expressing lentivirus, followed by selection of resistant cells with puromycin (InvivoGen) at 2 μg/mL for 14 days. The resistant cells were maintained in DMEM supplemented with 10% FBS (Gibco), 50 U/mL penicillin, 50 μg/mL streptomycin, and 1 μg/mL puromycin. The expression of ACE2 in these ACE2 stable cell lines was determined by Western blot analysis.
Huh-7.5 cells were provided by Charles M. Rice (The Rockefeller University, NY)46 and maintained in DMEM supplemented with 10% FBS (Gibco), 50 U/mL penicillin, 50 μg/mL streptomycin, 1 mM sodium pyruvate (Gibco), 10 mM HEPES (Gibco), and 1× MEM nonessential amino acids solution (Gibco). A549 cells (ATCC CCL-185) were cultured in DMEM (Fisher Scientific) supplemented with 10% FBS (Gibco), 1 mM sodium pyruvate (Life Technologies), 2 mM l-glutamine (Fisher Scientific), 10 mM HEPES (Fisher Scientific), and 100 U/mL of penicillin–100 μg/mL of streptomycin. (Fisher Scientific). All cells were tested and were confirmed to be free of mycoplasma contamination.
Calu-3 cells (ATCC) in Eagle’s Minimum Essential Medium (EMEM, Corning) and RAW264.7 cells (ATCC) in DMEM (Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS), and 1% Anti-Anti (Gibco; 10 000 units/mL of penicillin, 10 000 μg/mL of streptomycin, and 25 μg/mL of amphotericin B) were cultured at 37 °C and 5% CO2.
A549-ACE2 were a kind gift from Sam Wilson (University of Glasgow).47 Cells were cultured at 37 °C and 5% CO2 in DMEM (Gibco), supplemented with 10% (v/v) fetal calf serum (FCS), 100 U/mL of penicillin and 100 μg/mL of streptomycin (#15140-122; Gibco) and 200 μg/mL hygromycin (ALX-380-309-G001, Enzo).
Human Pneumocyte-like Cell Generation
Human embryonic stem cells (H9, WiCell, WA09) were cultured with mTeSR (STEMCELL Technologies, 85850) on Vitronectin XF (STEMCELL Technologies, 07180)-coated tissue culture plates and split in a ratio of 1:6 to 1:12 every 4–6 days with Gentle Cell Dissociation Reagent (STEMCELL Technologies, 07174). Alveolar differentiation was induced as previously described.43 Briefly, cells were collected at 70–80% confluency, and 2 million cells per 10 cm2 were plated on Vitronectin-coated tissue culture plates in mTeSR. The next day, definitive endoderm differentiation was induced following the protocol described by Jacob et al.48 for 4 days. Cells were split and further differentiated following an adapted alveolar differentiation protocol49 in Iscove Modified Dulbecco Media (IMDM, Life Technologies, 31980030) supplemented with 10% FBS (Sigma, F4135), 2 mM l-glutamine (Life Technologies 25030081), 0.5 μM all-trans-retinoic acid (Sigma, R2626), 10 ng/mL FGF-10 (R&D Systems, 345-FG-025), 10 ng/mL EGF (R&D Systems, 236-EG-01M), 100 ng/mL Wnt3a (R&D Systems, 5036-WN-010), 10 ng/mL KGF (R&D Systems, 251-KG-050) and 5 ng/mL BMP-4 (R&D Systems, 314-BP-010). Viral infections were performed on day nine after induction of differentiation, and the cells were analyzed 2 days postinfection.
Pneumocyte-like Cell MTT Assay
The duplicate set of cells were treated with same drug concentrations but left uninfected. After 48 h of incubation at 37 °C, the cells were analyzed for viability using MTT assay kit (Roche). The viability was calculated as a percentage relative to DMSO control in triplicates.
Dose–Response Curves and IC50 Calculations
Apra S4 was evaluated by immunofluorescence in a dose–response experiment to determine EC50 and CC50 through a cell-based high-content imaging assay, labeling the viral nucleoproteins within infected cells. Three thousand cells per well were seeded in a 384-well black plate 16 h prior to infection. Two hours prior to infection, cells were dosed with DMSO (control), remdesivir, apilimod, or Apra S4 in 11-step 1:3 dilutions at indicated starting concentrations, in triplicate. Ten μL of SARS-CoV-2 USA-WA1/2020 were added to each well, at an MOI of 0.1 (Vero E6) or 0.5 (HeLa-hACE2). Sixteen hours postinfection, cells were fixed with 5% paraformaldehyde for 4 h at room temperature, washed with PBS, and then permeabilized with 0.5% Triton X-100 for 5 min. Cells were blocked in 3% BSA/PBS for 1 h, then incubated overnight at 4 °C with mouse anti-SARS-CoV-2 N protein antibody (Thermo Fisher MA529981) 1:1000 dilution in 3% BSA/PBS. Cells were washed twice, with PBS, and then incubated for 1 h with goat antimouse secondary Antibody Alexa 488 (Thermo Fisher A-11001) 1:2000 dilution and DAPI 1:3000 dilution in 3% BSA/PBS. Cells were washed twice with PBS and then imaged using the Celigo Image Cytometer (Nexcelom). The assay results and data analysis enabled to determine infectivity and viability/cytotoxicity. On the basis of all infectivity and cytotoxicity values, a 4-parameter logistic nonlinear regression model was used to calculate IC50 and CC50 concentration values.
Flavivirus (Zika, Dengue, West Nile) Assays
For Zika, Dengue, West Nile virus assay, 2000 Huh7.5 cells were seeded overnight and then treated with Apra S4 for 2 h prior to infection. Cells were then infected with Zika virus (Ugandan MR766 strain) at MOI = 0.25, West Nile Virus (Kunjin isolate) at MOI = 0.05, or Dengue virus (16681 serotype 2) at MOI = 3. At 24 h postinfection, cells were fixed with 4% formaldehyde and then subjected to immunostaining for viral Envelope (E) protein, and stained with DAPI.
Influenza Assay
For influenza virus assay, 3500 A549 cells were seeded overnight and then treated with Apra S4 for 2 h prior to infection. Cells were then infected with Influenza A virus (WSN strain) at MOI = 0.1. At 36 h postinfection, cells were fixed with 4% formaldehyde and then subjected to immunostaining for viral nucleoprotein, and stained with DAPI.
VSV-Based Pseudotyped Viral Particles Assay
Vero E6 cells were pretreated with the indicated compounds for 2 h and then infected with SARS-CoV-1 S and SARS-CoV-2 S pseudotyped particles harboring firefly luciferase for additional 2 h. Luciferase signals were quantified at 24 h postinoculation. Error bars represent SEM for n = 6 independent experiments. The cell viability was measured at 24 h postinfection using Cell Counting Kit-8 (WST-8; Sigma, 96992) and normalized to the DMSO-treated samples.
SARS-CoV-2 BetaCoV/Germany/BavPat1/2020 Infections and Titrations
SARS-CoV-2 infection and titration assays were performed as described previously.44,50 Briefly, cells were infected for 1 h with SARS-CoV-2 at the indicated MOI in PBS supplemented with 0.3% BSA, 1 mM Ca2+/Mg2+, 100 U/mL penicillin, and 100 μg/mL streptomycin. After washing with PBS, medium was replaced with DMEM supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 0.3% BSA, 20 mM HEPES, 0.1% FCS, and 0.5 μg/mL TPCK-treated trypsin. Cell culture supernatants were collected at the indicated time points, and titrated by plaque assay. Of note, data points below the detection limit of 100 PFU/mL are plotted as 99 PFU/mL.
Cell Viability by CellTiter-Glo
Cell viability was determined using the CellTiter-Glo luminescent cell viability assay (Promega) according to the manufacturer’s instructions.
In Vitro Transcribed Viral RNA Based on Viral Replication Assay
The full-length SARS-CoV-2 viral RNA transcripts were in vitro synthesized from an infectious clone of SARS-CoV-2 (kindly provided by Pei-Yong Shi, University of Texas Medical Branch) according to a recently published protocol.51 10 μg of total RNA transcripts and 5 μg SARS-CoV-2 N gene transcript were mixed with Vero E6 cells stably expressing SARS-CoV-2 N protein and then added into a 0.2 cm cuvette for nucleofection with the 4D-Nucleofector Core Unit (Lonza) using pulse code V-001. Immediately after electroporation, 1000 μL of prewarmed media was added to the cuvette and cells were subsequently aliquoted into 384-well plates. Two hours postseeding, Apra S4 at different concentrations were added into each well. At 12 h postelectroporation, intracellular and viral RNA was purified from the treated cells with TurboCapture 384 mRNA Kit (Qiagen) in accordance with the manufacturer’s instructions. The purified RNA was subjected to first-strand cDNA synthesis using the high-capacity cDNA reverse transcription kit (Applied Biosystems, Inc.) with the following primer (TagRdRp-F: 5′-CGGTCATGGTGGCGAATAACCCTGTGGGTTTTACACTTAA-3′). Real-time PCR analysis was performed using TaqPath 1-step RT-qPCR Master Mix (Applied Biosystems, Inc.). The following primers and probe were used for negative-stranded RNA detection: Tag-F: 5′-CGGTCATGGTGGCGAATAACCCTGT-3′, ORF1ab-R: 5′-ACGATTGTGCATCAGCTGA-3′, ORF1ab-P: 5′-6FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1-3′.
siRNA Transfection
A549 cells stably expressing human ACE2 were transfected in suspension with 30 nM siRNA, targeting Sec61A1 s26722 and s26723 (Thermo Fisher); nontargeting control: scrambled1777 (custom siRNA, 5′-AAGCGTTCGTCCTATGATCGA-3′, Qiagen); targeting RPS27A (custom siRNA, 5′-AAGCTGGAAGATGGACGTACT-3′, Qiagen) using lipofectamine RNAimax (Invitrogen) according to the manufacturer’s instructions. Infection and cell viability assays were performed 48 h post-transfection.
Immunofluorescence
Vero CCL81 cells were seeded on coverslips and incubated overnight at 37 °C and 5% CO2 in DMEM supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, and 10% FCS. The next day, medium was removed and cells were washed with PBS followed by an incubation of 1 μM Apra S4 or DMSO diluted in OptiMem for 2 h at 37 °C and 5% CO2. Then, cells were infected with BetaCoV/Germany/BavPat1/2020 at a multiplicity of infection (MOI) of 2 in PBS supplemented with 1 μM Apra S4 or DMSO and 0.3% BSA, 1 mM Ca2+/Mg2+, 100 U/mL penicillin, and 100 μg/mL streptomycin. After 1 h of incubation, the inoculum was removed, cells were washed with PBS, and then incubated in DMEM supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 0.3% BSA, 20 mM HEPES, 0.1% FCS, and 0.5 μg/mL TPCK-treated trypsin. At 16 h postinfection, medium was removed, cells were washed with PBS, and then fixed with 4% paraformaldehyde (50-980-487, Fisher Scientific) in PBS. For permeabilization of cells PBS supplemented with 50 mM ammonium chloride (#254134; Sigma-Aldrich), 0.1% saponin (#47036, Sigma-Aldrich), and 2% BSA was used. SARS-CoV-2 viral proteins were stained with a mouse monoclonal antibody against the nucleoprotein (MA529981, Thermo Fisher) and a human monoclonal antibody against the spike protein (cov2rbdc1-mab1, InvivoGen). Double-stranded RNA was stained with a mouse anti-dsRNA antibody (#9D5 Ab00458-1.1 Absolute Antibodies). Nuclei were stained with DAPI (#10236276001; Sigma-Aldrich). Coverslips were mounted using ProLong Gold Antifade Mountant (#P36930; Thermo Fisher Scientific) and images were acquired on a Leica SP8 confocal microscope.
Electron Microscopy
Vero CCL81 cells were seeded in DMEM, supplemented with 10% (v/v) fetal calf serum, 100 U/mL of penicillin, and 100 μg/mL of streptomycin on 12 mm cover glasses coated with 0.1% poly-l-lysine (Sigma-Aldrich) and incubated overnight at 37 °C and 5% CO2. The next day, cells were washed with PBS and incubated for 2 h with 1 μM Apra S4 or DMSO (final concentration of DMSO: 0.1%) diluted in OptiMEM. Next, cells were infected with SARS-CoV-2 at an MOI of 2 PFU/cell in PBS supplemented with 0.3% BSA, 1 mM Ca2+/Mg2+, 100 U/mL penicillin, and 100 μg/mL streptomycin for 1 h in the presence of either 1 μM Apra S4 or DMSO. After infection, cells were washed twice with PBS. Then, cells were incubated for another 15 h in DMEM supplemented with either 1 μM Apra S4 or DMSO and 100 U/mL penicillin, 100 μg/mL streptomycin, 0.3% BSA, 20 mM HEPES, 0.1% FCS, and 0.5 μg/mL TPCK-treated trypsin. Following incubation, medium was removed and cells were fixed in prewarmed 2.5% glutaraldehyde in PBS for 15 min followed by washing with PBS. Next, cells were sequentially fixed with prewarmed (to 37 °C) 2.5% glutaraldehyde in PBS for 1 h, with 1% osmium tetroxide in 0.1 M cacodylate buffer (pH 7.35) for 1 h at 0 °C, and 2% aqueous uranyl acetate for 1 h at 4 °C. Samples then were dehydrated in an ethanol series followed by propylene oxide and embedded in Epon/Araldite (Sigma-Aldrich). Ultrathin (70 nm) sections were poststained with lead citrate and examined with a Talos 120 transmission electron microscope at an acceleration voltage of 120 kV using a Ceta digital camera and the MAPS software package (Thermo Fisher Scientific).
Immunoblot Analysis for ACE2 Expression
Vero E6 or Calu-3 cells (2.5 × 105 cells/well) were seeded in 6-well clear bottom plates and allowed to attach over 24 h. Media was replaced next day prior to treatment with apratoxin S4 or solvent control (0.5% EtOH). Whole cell lysates were collected using PhosphoSafe buffer (EMD Chemicals, Inc., Gibbstown, NJ) after 12 and 24 h incubation with the compound. Protein concentrations were measured with the BCA Protein Assay kit (Thermo Fisher Scientific, Rockford, IL). Lysates containing equal amounts of protein were separated by SDS polyacrylamide gel electrophoresis (4–12%), transferred to polyvinylidene difluoride membranes, probed with primary and secondary antibodies, and detected with the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). Anti-ACE2 (AF933) and secondary antigoat (HAF109) antibodies were obtained from R&D Systems, Inc. (Minneapolis, MN), and β-actin (4970S) and secondary antirabbit antibodies were obtained from Cell Signaling Technology, Inc. (Danvers, MA).
All experiments involving live SARS-CoV-2 and other viruses followed the approved standard operating procedures of the Biosafety Level 3 facilities at Sanford Burnham Prebys Medical Discovery Institute, the University of Zurich, and Icahn School of Medicine at Mount Sinai.
Acknowledgments
This research was in part supported by the NIH, National Cancer Institute Grant R01CA172310 (to H.L.), and the Debbie and Sylvia DeSantis Chair professorship (H.L.). Work in the Chanda lab was supported in part by the DoD, Grant W81XWH-20-1-0270, and the NIH/NIAID, Grant Fluomics/NOSI U19AI135972 and DHIPC Grant U19AI118610; and NCI Grant P30CA030199-40; as well as generous philanthropic donations from D. Ruch, and S. and J. Blair (to S.K.C.). This research was also partly supported by CRIPT (Center for Research for Influenza Pathogenesis and Transmission), a NIAID supported Center of Excellence for Influenza Research and Response (CEIRR, Contract # 75N93021C00014), by DARPA Grant HR0011-19-2-0020, by supplements to NIAID Grants U19AI142733, U19AI135972 and DoD Grant W81XWH-20-1-0270, and by the generous support of the JPB Foundation, the Open Philanthropy Project (Research Grant 2020-215611 (5384)), and anonymous donors to A.G-S. Work in the Stertz and Hale groups was supported by the Swiss National Science Foundation (Grant Numbers 31003A_176170 (to S.St.) and 31003A_182464 (to B.G.H.)) and imaging performed with support of the Center for Microscopy and Image Analysis, University of Zurich. We thank Sonja Fernbach for her support with the immunofluorescence quantifications using CellProfiler. The authors would like to thank Charles M. Rice (The Rockefeller University, NY) for sharing Huh-7.5 cells, Michael S. Diamond (Washington University School of Medicine in St. Louis) for the West Nile virus, Ana Fernandez-Sesma (Icahn School of Medicine at Mount Sinai) for the Dengue virus, Thomas Rogers (University of California, San Diego) for providing HeLa-hACE2 cells, Sam Wilson (University of Glasgow) for providing the A659-ACE2 cells, and Pei-Yong Shi (University of Texas Medical Branch, Galveston) for the SARS-CoV-2 clone. We thank R. Albrecht for support with the BSL-3 facility and procedures at the Icahn School of Medicine at Mount Sinai, New York.
Glossary
Abbreviations
- ACE2
angiotensin-converting enzyme 2
- ATCC
American Type Culture Collection
- BMP-4
bone morphogenetic protein 4
- BSA
bovine serum albumin
- CC50
cytotoxic concentration (50%)
- CoV
coronavirus
- DAPI
4′,6-diamidino-2-phenylindole
- DMSO
dimethyl sulfoxide
- DMV
double-membrane vesicle
- EC50
half maximal effective concentration
- eEF1A
eukaryotic translation elongation factor 1 alpha
- ER
endoplasmic reticulum
- FDA
Food and Drug Administration
- EGF
epidermal growth factor
- FGF
fibroblast growth factor
- IC50
50% inhibitory concentration
- IL-6
interleukin 6
- KGF
keratinocyte growth factor
- MERS
Middle East respiratory syndrome
- MOI
multiplicity of infection
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- Nsp
nonstructural protein
- Orf8
open reading frame 8
- PBS
phosphate buffered saline
- PIKfyve
phosphoinositide kinase, FYVE-type zinc containing
- PFU
plaque forming units
- RdRp
RNA-dependent RNA polymerase
- RNA
ribonucleic acid
- SI
selectivity index
- SARS
severe acute respiratory syndrome
- SARS-CoV-2
severe acute respiratory syndrome-Corona virus 2
- Wnt3a
Wnt family member 3A.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.2c00008.
SARS-CoV-2 inhibition by eeyarestatin I (Figure S1) and Sec61A1 knockdown (Figure S2), ACE2 immunoblot analysis in Calu-3 cells (Figure S3), and anti-inflammatory activity in RAW264.7 cells (Figure S4 and Methods) (PDF)
Author Present Address
● Department of Immunology and Microbiology, Scripps Research, La Jolla, California, 92037, United States
Author Present Address
▲ Calibr, a division of Scripps Research, La Jolla, California, 92037, United States
Author Present Address
□ State Key Laboratory of Veterinary Biotechnology, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin 150069, China
Author Contributions
M.O.P., L.M.-S., R.Ra., K.M.W., L.R., Q.-Y.C., G.L., I.B., X.Y., S.L., Y.P., R.Ro., M.D., Y.C., A.B., S.Y., performed research. N.M., S.St., S.K.C., H.L., designed research. M.O.P, L.M.-S., R.Ra., K.M.W., L.R., L.P., S.St., H.L., data analysis. Q.-Y.C., chemical synthesis. P.D.D.J., assay coordination. B.G.H., T.P.Z., S.St., A.G-S., S.K.C., H.L., supervised research. B.G.H., A.G.-S., S.St., S.K.C., H.L., funding acquisition. M.O.P., L.M.-S., R.Ra., N.M., S.St., S.K.C., manuscript preparation. H.L., project conception.
The authors declare the following competing financial interest(s): H. Luesch is co-founder of Oceanyx Pharmaceuticals, Inc., which has licensed patents and patent applications related to apratoxins. The A.G.-S. laboratory has received research support from Pfizer, Senhwa Biosciences, Kenall Manufacturing, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, Model Medicines, Atea Pharma and Merck, outside of the reported work. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, Pharmamar, Paratus, CureLab Oncology, CureLab Veterinary and Pfizer, outside of the reported work. A.G.-S. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York, outside of the reported work.
Supplementary Material
References
- O’Leary V. B.; Dolly O. J.; Höschl C.; Černa M.; Ovsepian S. V. Unpacking Pandora From Its Box: Deciphering the molecular basis of the SARS-CoV-2 coronavirus. Int. J. Mol. Sci. 2021, 22 (1), 1–14. 10.3390/ijms22010386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary N.; Weissman D.; Whitehead K. A. MRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug Discovery 2021, 20 (11), 817–838. 10.1038/s41573-021-00283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien M. P.; Forleo-Neto E.; Musser B. J.; Isa F.; Chan K.-C.; Sarkar N.; Bar K. J.; Barnabas R. v.; Barouch D. H.; Cohen M. S.; Hurt C. B.; Burwen D. R.; Marovich M. A.; Hou P.; Heirman I.; Davis J. D.; Turner K. C.; Ramesh D.; Mahmood A.; Hooper A. T.; Hamilton J. D.; Kim Y.; Purcell L. A.; Baum A.; Kyratsous C. A.; Krainson J.; Perez-Perez R.; Mohseni R.; Kowal B.; DiCioccio A. T.; Stahl N.; Lipsich L.; Braunstein N.; Herman G.; Yancopoulos G. D.; Weinreich D. M. Subcutaneous REGEN-COV antibody combination to prevent Covid-19. N. Engl. J. Med. 2021, 385 (13), 1184–1195. 10.1056/NEJMoa2109682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T. P.; Sims A. C.; Graham R. L.; Menachery V. D.; Gralinski L. E.; Case J. B.; Leist S. R.; Pyrc K.; Feng J. Y.; Trantcheva I.; Bannister R.; Park Y.; Babusis D.; Clarke M. O.; MacKman R. L.; Spahn J. E.; Palmiotti C. A.; Siegel D.; Ray A. S.; Cihlar T.; Jordan R.; Denison M. R.; Baric R. S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017, 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J. H.; Tomashek K. M.; Dodd L. E.; Mehta A. K.; Zingman B. S.; Kalil A. C.; Hohmann E.; Chu H. Y.; Luetkemeyer A.; Kline S.; Lopez de Castilla D.; Finberg R. W.; Dierberg K.; Tapson V.; Hsieh L.; Patterson T. F.; Paredes R.; Sweeney D. A.; Short W. R.; Touloumi G.; Lye D. C.; Ohmagari N.; Oh M.; Ruiz-Palacios G. M.; Benfield T.; Fätkenheuer G.; Kortepeter M. G.; Atmar R. L.; Creech C. B.; Lundgren J.; Babiker A. G.; Pett S.; Neaton J. D.; Burgess T. H.; Bonnett T.; Green M.; Makowski M.; Osinusi A.; Nayak S.; Lane H. C. Remdesivir for the treatment of Covid-19 — Final Report. N. Engl. J. Med. 2020, 383 (19), 1813–1826. 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov M. Merck’s COVID pill loses its lustre: what that means for the pandemic. Nature 2021, 10.1038/d41586-021-03667-0. [DOI] [PubMed] [Google Scholar]
- Owen D. R.; Allerton C. M. N.; Anderson A. S.; Aschenbrenner L.; Avery M.; Berritt S.; Boras B.; Cardin R. D.; Carlo A.; Coffman K. J.; Dantonio A.; Di L.; Eng H.; Ferre R.; Gajiwala K. S.; Gibson S. A.; Greasley S. E.; Hurst B. L.; Kadar E.P.; Kalgutkar A. S.; Lee J. C.; Lee J.; Liu W.; Mason S. W.; Noell S.; Novak J. J.; Obach R. S.; Ogilvie K.; Patel N. C.; Pettersson M.; Rai D. K.; Reese M. R.; Sammons M. F.; Sathish J. G.; Singh R. S. P.; Steppan C. M.; Stewart A. E.; Tuttle J. B.; Updyke L.; Verhoest P. R.; Wei L.; Yang Q.; Zhu Y. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374 (6575), 1586–1593. 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- Riva L.; Yuan S.; Yin X.; Martin-Sancho L.; Matsunaga N.; Pache L.; Burgstaller-Muehlbacher S.; de Jesus P. D.; Teriete P.; Hull M. v.; Chang M. W.; Chan J. F.-W.; Cao J.; Poon V. K.-M.; Herbert K. M.; Cheng K.; Nguyen T.-T. H.; Rubanov A.; Pu Y.; Nguyen C.; Choi A.; Rathnasinghe R.; Schotsaert M.; Miorin L.; Dejosez M.; Zwaka T. P.; Sit K.-Y.; Martinez-Sobrido L.; Liu W.-C.; White K. M.; Chapman M. E.; Lendy E. K.; Glynne R. J.; Albrecht R.; Ruppin E.; Mesecar A. D.; Johnson J. R.; Benner C.; Sun R.; Schultz P. G.; Su A. I.; García-Sastre A.; Chatterjee A. K.; Yuen K.-Y.; Chanda S. K. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 2020, 586 (7827), 113–119. 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S.; Yin X.; Meng X.; Chan J. F. W.; Ye Z. W.; Riva L.; Pache L.; Chan C. C. Y.; Lai P. M.; Chan C. C. S.; Poon V. K. M.; Lee A. C. Y.; Matsunaga N.; Pu Y.; Yuen C. K.; Cao J.; Liang R.; Tang K.; Sheng L.; Du Y.; Xu W.; Lau C. Y.; Sit K. Y.; Au W. K.; Wang R.; Zhang Y. Y.; Tang Y. D.; Clausen T. M.; Pihl J.; Oh J.; Sze K. H.; Zhang A. J.; Chu H.; Kok K. H.; Wang D.; Cai X. H.; Esko J. D.; Hung I. F. N.; Li R. A.; Chen H.; Sun H.; Jin D. Y.; Sun R.; Chanda S. K.; Yuen K. Y. Clofazimine broadly inhibits coronaviruses including SARS-CoV-2. Nature 2021, 593 (7859), 418–423. 10.1038/s41586-021-03431-4. [DOI] [PubMed] [Google Scholar]
- Herold T.; Jurinovic V.; Arnreich C.; Lipworth B. J.; Hellmuth J. C.; von Bergwelt-Baildon M.; Klein M.; Weinberger T. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J. Allergy Clin. Immunol. 2020, 146 (1), 128–136. 10.1016/j.jaci.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin E. J.; Baden L. R.; Morrissey S. Audio Interview: A New mRNA Vaccine. N. Engl. J. Med. 2021, 384 (25), e109 10.1056/NEJMe2110531. [DOI] [PubMed] [Google Scholar]
- Horby P.; Lim W. S.; Emberson J. R.; Mafham M.; Bell J. L.; Linsell L.; Staplin N.; Brightling C.; Ustianowski A.; Elmahi E.; Prudon B.; Green C.; Felton T.; Chadwick D.; Rege K.; Fegan C.; Chappell L. C.; Faust S. N.; Jaki T.; Jeffery K.; Montgomery A.; Rowan K.; Juszczak E.; Bailie J. K.; Haynes R.; Landray M. J. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 2021, 384 (8), 693–704. 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y. M.; Mandourah Y.; Al-Hameed F.; Sindi A. A.; Almekhlafi G. A.; Hussein M. A.; Jose J.; Pinto R.; Al-Omari A.; Kharaba A.; Almotairi A.; al Khatib K.; Alraddadi B.; Shalhoub S.; Abdulmomen A.; Qushmaq I.; Mady A.; Mady O.; Al-Aithan A. M.; Al-Raddadi R.; Ragab A.; Balkhy H. H.; Balkhy A.; Deeb A. M.; al Mutairi H.; Al-Dawood A.; Merson L.; Hayden F. G.; Fowler R. A. Corticosteroid therapy for critically ill patients with middle east respiratory syndrome. Am. J. Respir. Crit. Care Med. 2018, 197 (6), 757–767. 10.1164/rccm.201706-1172OC. [DOI] [PubMed] [Google Scholar]
- Stockman L. J.; Bellamy R.; Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006, 3 (9), 1525–1531. 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicari D.; Chatziioannou A.; Koutsandreas T.; Sitia R.; Chevet E. Role of the early secretory pathway in SARS-CoV-2 Infection. J. Cell Biol. 2020, 219 (9), e202006005 10.1083/jcb.202006005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton N. S.; Moshkina N.; Fenouil R.; Gardner T. J.; Aguirre S.; Shah P. S.; Zhao N.; Manganaro L.; Hultquist J. F.; Noel J.; Sachs D. H.; Hamilton J.; Leon P. E.; Chawdury A.; Tripathi S.; Melegari C.; Campisi L.; Hai R.; Metreveli G.; Gamarnik A. V.; García-Sastre A.; Greenbaum B.; Simon V.; Fernandez-Sesma A.; Krogan N. J.; Mulder L. C. F.; van Bakel H.; Tortorella D.; Taunton J.; Palese P.; Marazzi I. Targeting viral proteostasis limits influenza virus, HIV, and dengue virus infection. Immunity 2016, 44 (1), 46–58. 10.1016/j.immuni.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P. S.; Link N.; Jang G. M.; Sharp P. P.; Zhu T.; Swaney D. L.; Johnson J. R.; Von Dollen J.; Ramage H. R.; Satkamp L.; Newton B.; Hüttenhain R.; Petit M. J.; Baum T.; Everitt A.; Laufman O.; Tassetto M.; Shales M.; Stevenson E.; Iglesias G. N.; Shokat L.; Tripathi S.; Balasubramaniam V.; Webb L. G.; Aguirre S.; Willsey A. J.; Garcia-Sastre A.; Pollard K. S.; Cherry S.; Gamarnik A. V.; Marazzi I.; Taunton J.; Fernandez-Sesma A.; Bellen H. J.; Andino R.; Krogan N. J. Comparative flavivirus-host protein interaction mapping reveals mechanisms of dengue and zika virus pathogenesis. Cell 2018, 175 (7), 1931–1945. 10.1016/j.cell.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. E.; Jang G. M.; Bouhaddou M.; Xu J.; Obernier K.; White K. M.; O’Meara M. J.; Rezelj V. v; Guo J. Z.; Swaney D. L.; Tummino T. A.; Hüttenhain R.; Kaake R. M.; Richards A. L.; Tutuncuoglu B.; Foussard H.; Batra J.; Haas K.; Modak M.; Kim M.; Haas P.; Polacco B. J.; Braberg H.; Fabius J. M.; Eckhardt M.; Soucheray M.; Bennett M. J.; Cakir M.; McGregor M. J.; Li Q.; Meyer B.; Roesch F.; Vallet T.; mac Kain A.; Miorin L.; Moreno E.; Naing Z. Z. C.; Zhou Y.; Peng S.; Shi Y.; Zhang Z.; Shen W.; Kirby I. T.; Melnyk J. E.; Chorba J. S.; Lou K.; Dai S. A.; Barrio-Hernandez I.; Memon D.; Hernandez-Armenta C.; Lyu J.; Mathy C. J. P.; Perica T.; Pilla K. B.; Ganesan S. J.; Saltzberg D. J.; Rakesh R.; Liu X.; Rosenthal S. B.; Calviello L.; Venkataramanan S.; Liboy-Lugo J.; Lin Y.; Huang X. P.; Liu Y. F.; Wankowicz S. A.; Bohn M.; Safari M.; Ugur F. S.; Koh C.; Savar N. S.; Tran Q. D.; Shengjuler D.; Fletcher S. J.; O’Neal M. C.; Cai Y.; Chang J. C. J.; Broadhurst D. J.; Klippsten S.; Sharp P. P.; Wenzell N. A.; Kuzuoglu-Ozturk D.; Wang H. Y.; Trenker R.; Young J. M.; Cavero D. A.; Hiatt J.; Roth T. L.; Rathore U.; Subramanian A.; Noack J.; Hubert M.; Stroud R. M.; Frankel A. D.; Rosenberg O. S.; Verba K. A.; Agard D. A.; Ott M.; Emerman M.; Jura N.; von Zastrow M.; Verdin E.; Ashworth A.; Schwartz O.; d’Enfert C.; Mukherjee S.; Jacobson M.; Malik H. S.; Fujimori D. G.; Ideker T.; Craik C. S.; Floor S. N.; Fraser J. S.; Gross J. D.; Sali A.; Roth B. L.; Ruggero D.; Taunton J.; Kortemme T.; Beltrao P.; Vignuzzi M.; García-Sastre A.; Shokat K. M.; Shoichet B. K.; Krogan N. J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583 (7816), 459. 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesch H.; Yoshida W. Y.; Moore R. E.; Paul V. J.; Corbett T. H. Total structure determination of apratoxin A, a potent novel cytotoxin from the marine cyanobacterium Lyngbya majuscula. J. Am. Chem. Soc. 2001, 123 (23), 5418–5423. 10.1021/ja010453j. [DOI] [PubMed] [Google Scholar]
- Matthew S.; Schupp P. J.; Luesch H. Apratoxin E, a cytotoxic peptolide from a Guamanian collection of the marine cyanobacterium Lyngbya bouillonii. J. Nat. Prod. 2008, 71 (6), 1113–1116. 10.1021/np700717s. [DOI] [PubMed] [Google Scholar]
- Chen Q. Y.; Liu Y.; Luesch H. Systematic chemical mutagenesis identifies a potent novel apratoxin A/E hybrid with improved in vivo antitumor activity. ACS Med. Chem. Lett. 2011, 2 (11), 861–865. 10.1021/ml200176m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q. Y.; Liu Y.; Cai W.; Luesch H. Improved total synthesis and biological evaluation of potent apratoxin S4 based anticancer agents with differential stability and further enhanced activity. J. Med. Chem. 2014, 57 (7), 3011–3029. 10.1021/jm4019965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P.; Cai W.; Chen Q.-Y.; Xu S.; Yin R.; Li Y.; Zhang W.; Luesch H. Total Synthesis and Biological Evaluation of Apratoxin E and Its C30 epimer: configurational reassignment of the natural product. Org. Lett. 2016, 18 (20), 5400–5403. 10.1021/acs.orglett.6b02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W.; Chen Q.-Y.; Dang L. H.; Luesch H. Apratoxin S10, a dual inhibitor of angiogenesis and cancer cell growth to treat highly vascularized tumors. ACS Med. Chem. Lett. 2017, 8 (10), 1007–1012. 10.1021/acsmedchemlett.7b00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesch H.; Chanda S. K.; Raya R. M.; DeJesus P. D.; Orth A. P.; Walker J. R.; Belmonte J. C. I.; Schultz P. G. A functional genomics approach to the mode of action of apratoxin A. Nat. Chem. Biol. 2006, 2 (3), 158–167. 10.1038/nchembio769. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Law B. K.; Luesch H. Apratoxin A reversibly inhibits the secretory pathway by preventing cotranslational translocation. Mol. Pharmacol. 2009, 76 (1), 91–104. 10.1124/mol.109.056085. [DOI] [PubMed] [Google Scholar]
- Paatero A. O.; Kellosalo J.; Dunyak B. M.; Almaliti J.; Gestwicki J. E.; Gerwick W. H.; Taunton J.; Paavilainen V. O. Apratoxin kills cells by direct blockade of the Sec61 protein translocation channel. Cell Chem. Biol. 2016, 23 (5), 561–566. 10.1016/j.chembiol.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Luesch H.; Paavilainen V. O. Natural products as modulators of eukaryotic protein secretion. Nat. Prod. Rep 2020, 37, 717–736. 10.1039/C9NP00066F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W.; Ratnayake R.; Gerber M. H.; Chen Q. Y.; Yu Y.; Derendorf H.; Trevino J. G.; Luesch H. Development of apratoxin S10 (Apra S10) as an anti-pancreatic cancer agent and its preliminary evaluation in an orthotopic patient-derived xenograft (PDX) model. Invest. New Drugs 2019, 37 (2), 364–374. 10.1007/s10637-018-0647-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann S. H. E.; Dorhoi A.; Hotchkiss R. S.; Bartenschlager R. Host-directed therapies for bacterial and viral infections. Nat. Rev. Drug Discovery 2018, 17 (1), 35–56. 10.1038/nrd.2017.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu B.; Tan A.; Veluchamy A. B.; Li Y.; Murray H.; Cheng W.; Liu C.; Busoy J. M.; Chen Q. Y.; Sistla S.; Hunziker W.; Cheung C. M. G.; Wong T. Y.; Hong W.; Luesch H.; Wang X. Apratoxin S4 inspired by a marine natural product, a new treatment option for ocular angiogenic diseases. Invest. Ophthalmol. Visual Sci. 2019, 60 (8), 3254–3263. 10.1167/iovs.19-26936. [DOI] [PubMed] [Google Scholar]
- Ou X.; Liu Y.; Lei X.; Li P.; Mi D.; Ren L.; Guo L.; Guo R.; Chen T.; Hu J.; Xiang Z.; Mu Z.; Chen X.; Chen J.; Hu K.; Jin Q.; Wang J.; Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11 (1), 1620. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamayun I.; O’Keefe S.; Pick T.; Klein M.-C.; Nguyen D.; McKibbin C.; Piacenti M.; Williams H. M.; Flitsch S. L.; Whitehead R. C.; Swanton E.; Helms V.; High S.; Zimmermann R.; Cavalié A. Eeyarestatin compounds selectively enhance Sec61-mediated Ca2+ leakage from the endoplasmic reticulum. Cell Chem. Biol. 2019, 26 (4), 571–583. 10.1016/j.chembiol.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Shinkre B. A.; Lee J.-G.; Weniger M. A.; Liu Y.; Chen W.; Wiestner A.; Trenkle W. C.; Ye Y. The ERAD inhibitor Eeyarestatin I is a bifunctional compound with a membrane-binding domain and a p97/VCP inhibitory group. PLoS One 2010, 5, e15479 10.1371/journal.pone.0015479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y. L.; Chou Y. Y.; Rothlauf P. W.; Liu Z.; Soh T. K.; Cureton D.; Case J. B.; Chen R. E.; Diamond M. S.; Whelan S. P. J.; Kirchhausen T. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (34), 20803–20813. 10.1073/pnas.2007837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese M.; Lee J. Y.; Cerikan B.; Neufeldt C. J.; Oorschot V. M. J.; Köhrer S.; Hennies J.; Schieber N. L.; Ronchi P.; Mizzon G.; Romero-Brey I.; Santarella-Mellwig R.; Schorb M.; Boermel M.; Mocaer K.; Beckwith M. S.; Templin R. M.; Gross V.; Pape C.; Tischer C.; Frankish J.; Horvat N. K.; Laketa V.; Stanifer M.; Boulant S.; Ruggieri A.; Chatel-Chaix L.; Schwab Y.; Bartenschlager R. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe. 2020, 28 (6), 853–866. 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S.; Cortese M.; Winter S. L.; Wachsmuth-Melm M.; Neufeldt C. J.; Cerikan B.; Stanifer M. L.; Boulant S.; Bartenschlager R.; Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020, 11 (1), 5885. 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E. J.; Limpens R. W. A. L.; de Wilde A. H.; de Jong A. W. M.; Zevenhoven-Dobbe J. C.; Maier H. J.; Faas F. F. G. A.; Koster A. J.; Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020, 18 (6), e3000715 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W.; Ratnayake R.; Wang M.; Chen Q.-Y.; Raisch K. P.; Dang L. H.; Law B. K.; Luesch H. Inhibition of cotranslational translocation by apratoxin S4: Effects on oncogenic receptor tyrosine kinases and the fate of transmembrane proteins produced in the cytoplasm. Curr. Res. Pharmacol. Drug Discovery 2021, 2, 100053. 10.1016/j.crphar.2021.100053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K.; Swett-Tapia C.; van den Worm S. H. E.; te Velthuis A. J. W.; Koster A. J.; Mommaas A. M.; Snijder E. J.; Kikkert M. Integrity of the early secretory pathway promotes, but is not required for, severe acute respiratory syndrome coronavirus RNA synthesis and virus-induced remodeling of endoplasmic reticulum membranes. J. Virol. 2010, 84 (2), 833–846. 10.1128/JVI.01826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P.; Gerber M.; Kelly J.; Pfaender S.; Ebert N.; Lagache S. B.; Simillion C.; Portmann J.; Stalder H.; Gaschen V.; Bruggmann R.; Stoffel M. H.; Heller M.; Dijkman R.; Thiel V. Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. eLife 2019, 8, e42037 10.7554/eLife.42037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe S.; Roboti P.; Duah K. B.; Zong G.; Schneider H.; Shi W. Q.; High S. Ipomoeassin-F inhibits the in vitro biogenesis of the SARS-CoV-2 spike protein and its host cell membrane receptor. J. Cell Sci. 2021, 134 (4), jcs257758. 10.1242/jcs.257758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K. M.; Rosales R.; Yildiz S.; Kehrer T.; Miorin L.; Moreno E.; Jangra S.; Uccellini M. B.; Rathnasinghe R.; Coughlan L.; Martinez-Romero C.; Batra J.; Rojc A.; Bouhaddou M.; Fabius J. M.; Obernier K.; Dejosez M.; Guillén M. J.; Losada A.; Avilés P.; Schotsaert M.; Zwaka T.; Vignuzzi M.; Shokat K. M.; Krogan N. J.; García-Sastre A. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science 2021, 371 (6532), 926–931. 10.1126/science.abf4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl M. O.; Busnadiego I.; Kufner V.; Glas I.; Karakus U.; Schmutz S.; Zaheri M.; Abela I.; Trkola A.; Huber M.; Stertz S. G.; Hale B. G. SARS-CoV-2 variants reveal features critical for replication in primary human cells. PLoS Biol. 2021, 19 (3), e3001006 10.1371/journal.pbio.3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor E.; Devenish L.; Engelhardt O. G.; Palese P.; Brownlee G. G.; García-Sastre A. Rescue of influenza A virus from recombinant DNA. J. Virol. 1999, 73 (11), 9679–9682. 10.1128/JVI.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight K. J.; McKeating J. A.; Rice C. M. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 2002, 76 (24), 13001–13014. 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihn S. J.; Merits A.; Bakshi S.; Turnbull M. L.; Wickenhagen A.; Alexander A. J. T.; Baillie C.; Brennan B.; Brown F.; Brunker K.; Bryden S. R.; Burness K. A.; Carmichael S.; Cole S. J.; Cowton V. M.; Davies P.; Davis C.; De Lorenzo G.; Donald C. L.; Dorward M.; Dunlop J. I.; Elliott M.; Fares M.; da Silva Filipe A.; Freitas J. R.; Furnon W.; Gestuveo R. J.; Geyer A.; Giesel D.; Goldfarb D. M.; Goodman N.; Gunson R. C.; Hastie J.; Herder V.; Hughes J.; Johnson C.; Johnson N.; Kohl A.; Kerr K.; Leech H.; Lello L. S.; Li K.; Lieber G.; Liu X.; Lingala R.; Loney C.; Mair D.; McElwee M. J.; McFarlane S.; Nichols J.; Nomikou K.; Orr A.; Orton R. J.; Palmarini M.; Parr Y. A.; Pinto R. M.; Raggett S.; Reid E.; Robertson D. L.; Royle J.; Cameron-Ruiz N.; Shepherd J. G.; Smollett K.; Stewart D. G.; Stewart M.; Sugrue E.; Szemiel A. M.; Taggart A.; Thomson E. C.; Tong L.; Torrie L. S.; Toth R.; Varjak M.; Wang S.; Wilkinson S. G.; Wyatt P. G.; Zusinaite E.; Alessi D. R.; Patel A. H.; Zaid A.; Wilson S. J.; Mahalingam S. A plasmid DNA-launched SARS-CoV-2 reverse genetics system and coronavirus toolkit for COVID-19 research. PLoS Biol. 2021, 19 (2), e3001091 10.1371/journal.pbio.3001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A.; Vedaie M.; Roberts D. A.; Thomas D. C.; Villacorta-Martin C.; Alysandratos K. D.; Hawkins F.; Kotton D. N. Derivation of self-renewing lung alveolar epithelial type II cells from human pluripotent stem cells. Nat. Protoc. 2019, 14 (12), 3303–3332. 10.1038/s41596-019-0220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaedi M.; Calle E. A.; Mendez J. J.; Gard A. L.; Balestrini J.; Booth A.; Bove P. F.; Gui L.; White E. S.; Niklason L. E. Human iPS cell-derived alveolar epithelium repopulates lung extracellular matrix. J. Clin. Invest. 2013, 123 (11), 4950–4962. 10.1172/JCI68793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busnadiego I.; Fernbach S.; Pohl M. O.; Karakus U.; Huber M.; Trkola A.; Stertz S.; Hale B. G. Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2. mBio 2020, 11 (5), e01928-20 10.1128/mBio.01928-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X.; Lokugamage K. G.; Zhang X.; Vu M. N.; Muruato A. E.; Menachery V. D.; Shi P.-Y. Engineering SARS-CoV-2 using a reverse genetic system. Nat. Prot. 2021, 16, 1761–1784. 10.1038/s41596-021-00491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.