Abstract
Long noncoding RNA (lncRNA) differentiation antagonizing noncoding RNA (DANCR) is a proven oncogenic lncRNA across multiple cancer types. Its effects on cancer cell migration and invasion position it as a potential target for therapy on multiple levels of gene regulation. DANCR is overexpressed in non-small cell lung cancer (NSCLC), the most common lung cancer subtype with poor patient survival. To effectively deliver small interfering RNA (siRNA) against DANCR for NSCLC therapy, we used arginine-glycine-aspartic acid (RGD)-poly(ethylene glycol) (PEG)-(1-aminoethyl)-iminobis[N-oleicylcysteinyl-1-aminoethyl)propionamide] (ECO)/small interfering RNA against DANCR (siDANCR) nanoparticles to transfect A549 and NCI-H1299 cells. Over 90% DANCR silencing was observed along with inhibition of cell migration, invasion, and spheroid formation relative to transfection with negative control siRNA in RGD-PEG-ECO nanoparticles. DANCR knockdown further showed efficacy in reducing migration and invasion of epidermal growth factor receptor (EGFR)-inhibitor resistant NSCLC along with resensitization to the inhibitor. RGD-PEG-ECO/siDANCR demonstrated silencing for up to 7 d following a single transfection. The results suggest nanoparticle-mediated RNA interference against DANCR as a potential approach for NSCLC treatment by regulating cell migration and invasion in addition to improving EGFR inhibitor response.
1. Introduction
In the United States, 228 820 new cases and 135 720 lung cancer deaths are anticipated in 2020.1 Lung cancer is the second leading cause of new cases and first overall in deaths for both men and women. While the incidence and mortality rates have declined due to improved screening and reduced cigarette smoking rates, the overall five-year relative survival rate is only 19%.1 Approximately 85% of cases are non-small cell lung cancer (NSCLC), which is normally treated with surgical resection of the tumor if diagnosed at an early stage where the cancer has not spread to other regions of the body.1 At an advanced stage, NSCLC treatment involves chemotherapy, radiation, immunotherapy, or a combination of multiple strategies. With these approaches there may be initial remission, but cancer recurs in 30–50% of patients in a form that is resistant to further treatment.2 Molecularly targeted therapies, which are designed to target proteins abundantly expressed on cancer cell surfaces used for cell growth and division, can also become ineffective as cellular reliance on these proteins shifts to new ones. There is no single cure for NSCLC that works for all patients, and treatments aim instead to prolong survival if the disease is at an advanced stage.
Chemotherapy requires sustained high doses to achieve therapeutically effective concentrations within the tumor.3 Repeated doses are required to treat cells in different cycles of growth and overcome barriers presented by solid tumor heterogeneity. Chemotherapy that kills rapidly dividing cancer cells also has significant toxicity to healthy cells.4 Obstacles associated with therapeutic efficacy and cancer recurrence during chemotherapy arise from the rapidly adapting biology of cancer cells. Lung cancer develops new compensatory mitogenic pathways to adapt to chemotherapy and conventional targeted therapies. Therapies against new molecular targets involved in unique NSCLC pathways are thus required. To overcome the limitations of conventional anticancer therapeutics, regulation of oncogenes is proposed as an alternative. Long noncoding RNAs (lncRNAs) regulate gene expression on multiple levels, including DNA, RNA, protein, and epigenome,5 and they have varied biological roles, but a subset is oncogenic. Because oncogenic lncRNAs are overexpressed in cancer, they are a suitable target for modulating multiple oncogenic pathways with few adverse effects on healthy tissue. lncRNAs cannot be downregulated with traditional pharmacological strategies and require the use of RNA interference. Targeting multiple oncogenic pathways via lncRNA can be achieved with small interfering RNA (siRNA).6
Differentiation antagonizing noncoding RNA (DANCR) was discovered and named for its function in suppressing epidermal progenitor cell differentiation.7 It is upregulated across multiple cancer types and plays a key role in regulating cancer cell proliferation, invasion, metastasis, and drug resistance.8 DANCR physically interacts with microRNAs, mRNAs, or proteins—depending on the specific cancer type—to exert oncogenic effects. Its targets normally suppress tumor progression but are inactivated upon interacting with DANCR. Silencing DANCR can overcome multiple oncogenic pathways with one type of treatment. In NSCLC, DANCR was reported to increase cell proliferation and migration and promote metastasis by sponging the microRNAs 214 and 138.9,10 These microRNAs normally regulate proteins that promote migration and metastasis, but DANCR contains complementary binding sites that “sponge” the microRNAs. Downregulating DANCR frees these microRNAs to once again inhibit proteins involved in pathways of proliferation and metastasis. Contrary to conventional chemotherapies, multiple oncogenic pathways can be simultaneously inhibited with RNA interference (RNAi) with siRNA. To achieve the same result with conventional treatments, multiple chemotherapeutic drugs with different mechanisms of action must be used.11 Such a combination of nonselective drugs leads to sides effects that extend recovery time and are associated with cancer relapse.11 RNAi presents a strategy that is more specific to the genetic composition of a patient’s cancer and achieves a comparable outcome as conventional therapy with less toxicity. Silencing overexpressed oncogenes, including oncogenic lncRNA, offers a targeted approach that chemotherapy or radiation fail to deliver.
DANCR has been successfully silenced across multiple cancer types using siRNA.12 Our group previously developed a multifunctional lipid carrier termed (1-aminoethyl)-iminobis[N-oleicylcysteinyl-1-aminoethyl)propionamide] (ECO)13 that forms stable nanoparticles with therapeutic nucleic acids. Nanoparticles form by electrostatic complexation of the ECO with negatively charged siRNA and hydrophobic condensation of the oleic acid tails. ECO nanoparticles protect siRNA from nuclease degradation and allow efficient siRNA delivery into the cytosol through pH-sensitive amphiphilic endosomal escape and reductive cytosolic delivery (PERC).14 The nanoparticles can be modified with poly(ethylene glycol) (PEG) to prolong circulation time and minimize immune response. PEG can be further conjugated with cyclic arginine-glycine-aspartic acid (RGD) peptide to improve uptake in lung cells.
We previously reported that DANCR was upregulated in triple negative breast cancer (TNBC) and found silencing with RGD-PEG-ECO/siDANCR reduced cell proliferation and invasion and inhibited tumor growth.15 Here, we evaluated the efficacy of silencing DANCR with RGD-PEG-ECO/siDANCR nanoparticles for inhibition of lung cancer cell migration and invasion. We assessed the effects of DANCR silencing in two NSCLC cell lines that overexpress DANCR. Transfection with tumor-targeted RGD-PEG-ECO/siDANCR nanoparticles showed significant and sustained DANCR knockdown that inhibited cell migration, invasion, and spheroid formation in vitro. Because DANCR is associated with promoting drug resistance in other cancer types,16 we further investigated the role of DANCR in epidermal growth factor receptor (EGFR)-inhibitor resistant NSCLC. Drug-resistant NSCLC expressed DANCR at higher levels than parental cells, but silencing DANCR resensitized the cells to treatment and similarly inhibited cell migration and invasion.
2. Results
2.1. Formation and Characterization of RGD-PEG-ECO/siDANCR Nanoparticles
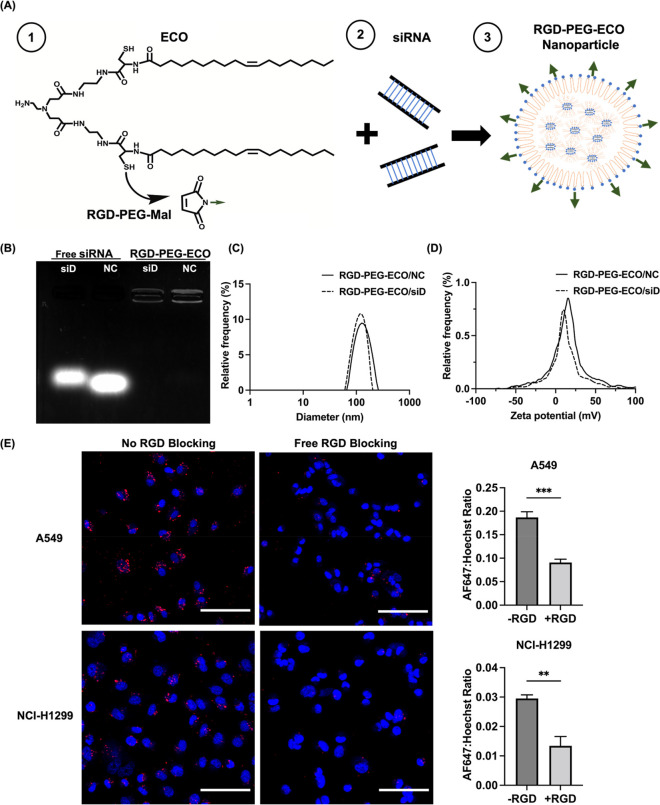
The RGD-PEG-ECO/siDANCR nanoparticles were formulated by the self-assembly of ECO and siDANCR at N/P = 8 after reaction of ECO with 2.5 mol % of RGD-PEG-MAL (MAL = maleimide). RGD-PEG-ECO/siNS (non-specific siRNA)nanoparticles were similarly formulated as a nonspecific control. Figure 1A shows the structure of ECO and a schematic representation of how it forms nanoparticles with siRNA. The nanoparticles were characterized using an agarose gel encapsulation assay, which revealed complete encapsulation of siRNA in nanoparticles and no free siRNA bands below the wells (Figure 1B). Free siRNA, in comparison, was not retained in the well and traveled down the agarose gel. Nanoparticle formulation was further characterized with dynamic light scattering. RGD-PEG-ECO/siDANCR nanoparticles were found to be 119 nm in size with zeta potential +14 mV, and RGD-PEG-ECO/siNS nanoparticles were 116 nm in size with zeta potential +13 mV (Figure 1C,D).
Figure 1.
Formation and characterization of RGD-PEG-ECO/siRNA nanoparticles. (A) Structure of ECO and scheme of RGD-PEG-ECO/siRNA formation. (B) Gel encapsulation assay shows strong entrapment of siDANCR and siNS in RGD-PEG-ECO nanoparticles with free siRNA as controls. RGD-PEG-ECO nanoparticles have uniform size (C) and zeta potential distributions (D) as measured with dynamic light scattering. The measured size is 119 nm for RGD-PEG-ECO/siDANCR nanoparticles and 116 nm for RGD-PEG-ECO/siNS nanoparticles; zeta potential is ca. +14 and +13 mV, respectively. (E) Confocal images (20× magnification) taken after 24 h blocking with excess free RGD peptide show reduced RGD-PEG-ECO nanoparticle uptake. Ratio of AF647 to Hoechst 33342 signal intensity is higher when no free RGD peptide is present in the transfection media (−RGD) compared to when excess RGD peptide is added (+RGD). Red: AF647; blue: Hoechst 33342 (n = 3). NC represents the negative control siRNA, siNS. siD represents siDANCR. Error bars denote standard error of measure; scale bars in all panels = 100 μm. ** p < 0.01, and *** p < 0.005 using unpaired t-test.
The role of RGD targeting was evaluated in blocking experiments using RGD-PEG-ECO nanoparticles containing AllStars Negative Control siRNA labeled with AF647 with and without the presence of an excess of free RGD peptide. AF647 fluorescence intensity was observed around the nuclei 24 h post-transfection in both cell lines, indicating RGD-PEG-ECO nanoparticle uptake and cytosolic siRNA delivery. The AF647 fluorescence intensity was compared to that of nuclei stained with Hoechst 33342, and it was revealed that blocking with excess free RGD peptide significantly reduced the uptake of RGD-targeted nanoparticles relative to the group without blocking (Figure 1E). This result supports the use of RGD targeting in NSCLC to improve siRNA delivery.
2.2. DANCR is Overexpressed in NSCLC and Can Be Downregulated by RGD-PEG-ECO Nanoparticles
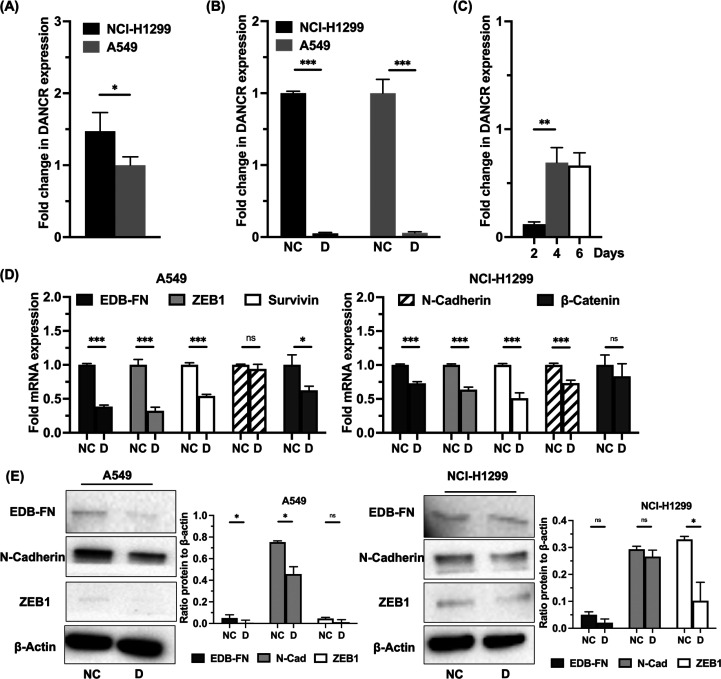
DANCR was previously shown to be overexpressed in NSCLC cell lines relative to normal human bronchial epithelial cells.17,18 As measured by qRT-PCR, NCI-H1299 cells express DANCR at a level 1.5 times greater than A549 cells (Figure 2A). A549 was chosen as a less aggressive model relative to the NCI-H1299 cells. When each was transfected with RGD-PEG-ECO/siDANCR nanoparticles for 48 h, over 90% silencing of DANCR expression was observed for both cell lines as measured by qRT-PCR relative to the corresponding cells treated with RGD-PEG-ECO/siNS nanoparticles (Figure 2B). After a single 48 h transfection, cells were collected at 2 d intervals, and RNA was extracted. DANCR silencing persisted over the course of 6 d (Figure 2C). This is expected, as cells divide and recover from transient knockdown. We hypothesize that, with repeated doses, cells will sustain DANCR silencing for longer periods of time. DANCR was previously shown to influence cell migration and invasion in triple negative breast cancer.15 Markers associated with lung cancer aggressiveness and epithelial-to-mesenchymal transition were examined following DANCR silencing without targeted nanoparticles. On the mRNA level, there was significant downregulation of EMT markers ZEB1 and EDB-fibronectin (EDB-FN), the antiapoptosis marker survivin, and the Wnt-signaling transcription factor β-catenin in A549 cells following siDANCR transfection as compared to the nonspecific control. NCI-H1299 cells showed similar downregulation in the same markers in addition to reduction of N-cadherin (Figure 2D). Western blot results confirmed some downregulation of the proteins in both cells lines following transfection with siDANCR relative to negative control transfection (Figure 2E).
Figure 2.
DANCR expression and downstream effects of silencing. (A) The NCI-H1299 cell line expresses DANCR mRNA at a 1.5× greater level relative to the A549 cell line as determined with a qRT-PCR analysis (n = 3). (B) Treatment with RGD-PEG-ECO/siDANCR nanoparticles silence DANCR expression over 90% relative to control (n = 3). (C) DANCR silencing in the NCI-H1299 cell line persists over the course of 6 d, with strongest silencing observed at 2 d post-transfection (n = 3). (D) Markers of epithelial-to-mesenchymal transition (EMT) are reduced on the mRNA level following DANCR silencing with ECO/siDANCR nanoparticles as shown with qRT-PCR (n = 3). (E) On the protein level, the same markers as in (D) similarly show reduced expression after silencing DANCR (n = 3). Quantification of western blot images shown as a ratio of the band of interest normalized to the loading control, β-actin. Error bars denote standard error of measure, * p < 0.05, ** p < 0.01, and *** p < 0.005 using unpaired t-test.
2.3. RGD-PEG-ECO/siDANCR Suppresses NSCLC Cell Migration and Invasion
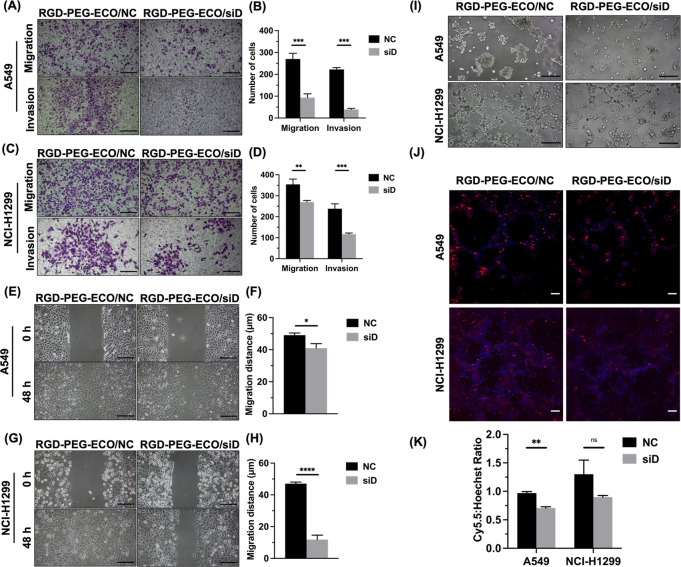
To assess the efficacy of downregulation of DANCR expression beyond gene and protein expression, we performed functional assays to investigate cell migration and invasion. Transwell migration and invasion assays were performed with each cell line transfected with RGD-PEG-ECO/siDANCR or RGD-PEG-ECO/siNS. Transfection with RGD-PEG-ECO/siDANCR significantly reduced migration and invasion for both A549 (Figure 3A) and NCI-H1299 cells (Figure 3E). Treatment with RGD-PEG-ECO/siDANCR reduced the number of migrated A549 cells by 63% and the number of invaded A549 cells by 78% relative to the control (Figure 3B). Treatment of NCI-H1299 with RGD-PEG-ECO/siDANCR reduced the number of migrated cells by 21% and the number of invaded cells by 60% (Figure 3F). The effect of DANCR silencing on cell migration was also assessed by a scratch wound healing assay. The wound healed completely for both cell lines treated with RGD-PEG-ECO/siNS transfection within 48 h but did not close for the cells treated with RGD-PEG-ECO/siDANCR transfection (Figure 3C,G). Quantifying the wound healing revealed significant reduction in the migration distance when either cell line was transfected with siDANCR (Figure 3D,H). Together with Transwell assay results, this evidence suggests treatment with siDANCR reduces NSCLC cell invasion and migration relative to transfection with the negative control siRNA.
Figure 3.
ECO/siDANCR nanoparticles inhibit lung cancer cell migration and invasion in Transwell assays. Across both A549 (A) and NCI-H1299 (C) cell lines (n = 3), cell migration (without Matrigel coating) and invasion (with Matrigel coating) are reduced upon silencing DANCR expression. Counting the number of stained cells migrating or invading across the Transwell membrane quantifies the reduction in migration and invasion for A549 (B) and NCI-H1299 (D) cells. Wound-healing assays revealed slower migration after DANCR knockdown in A549 (E) and NCI-H1299 (G) (n = 3). Quantification of wound-healing assay images as distance migrated relative to the original boundary positions over the course of the experiment for both A549 (F) and NCI-H1299 (H). (I) 3D growth on Matrigel after 5 d revealed smaller spheroid size after DANCR knockdown (n = 3). (J) Staining of the spheroids in (I) with ZD2-Cy5.5 (red) and Hoechst 33342 (blue) and subsequent confocal imaging showed the relative expression of EDB protein in the extracellular matrix. (K) Ratio of Cy5.5 maximum intensity to Hoechst maximum intensity in staining from (J) was reduced with DANCR silencing. Error bars denote the standard error of measure; scale bars in all panels = 100 μm. “NC” represents negative control RGD-PEG-ECO/siNS, “siD” represents RGD-PEG-ECO/siDANCR. * p < 0.05, ** p < 0.01, and *** p < 0.005 using unpaired t-test.
To assess tumor formation potential, we plated cells on a layer of Matrigel to observe three-dimensional (3D) growth. A549 cells formed spheroids with control RGD-PEG-ECO/siNS, while NCI-H1299 cells grew in networks. Following transfection with RGD-PEG-ECO/siDANCR, spheroids and networks were smaller in size for A549 and NCI-H1299, respectively (Figure 3I). Knockdown of siDANCR limited the tumor spheroid formation ability of NSCLC cells in vitro. Spheroids were stained with ZD2-Cy5.5, an EDB-FN specific fluorescence probe, to assess expression of EDB and marginally higher staining intensity in NCI-H1299 cells over A549 cells (Figure 3J). Quantification of this expression by comparing the ratio of Cy5.5 staining intensity to Hoechst staining intensity showed a reduction upon treatment with RGD-PEG-ECO/siDANCR relative to the negative control nanoparticles (Figure 3K).
2.4. DANCR is Upregulated in Drug-Resistant NSCLC
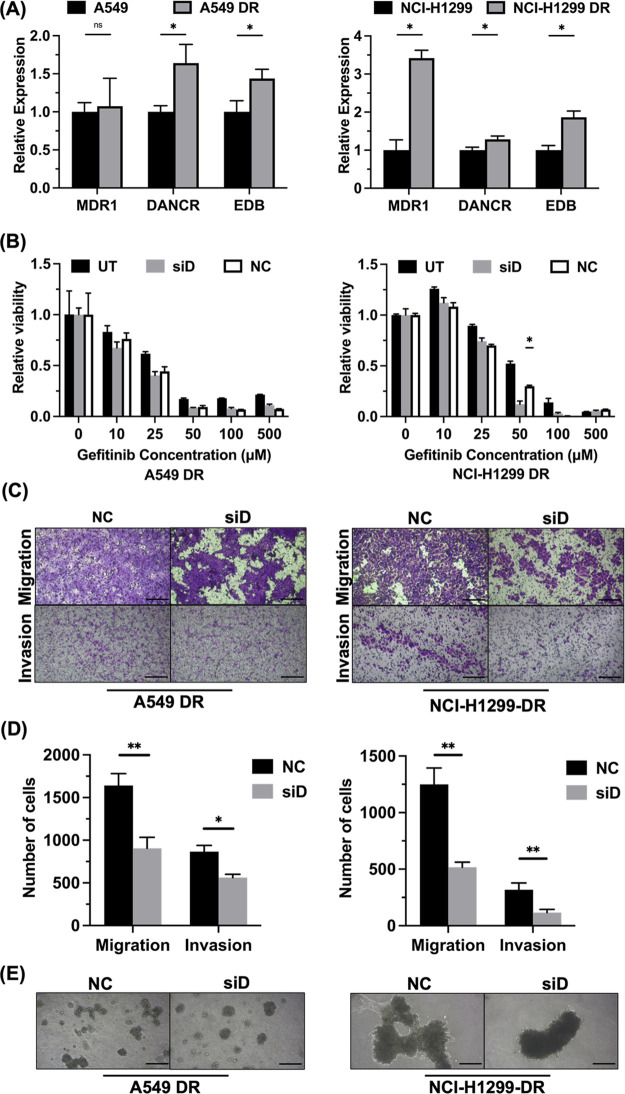
Because NSCLC is frequently detected at a late stage, it is prone to metastasis and drug resistance.19 To study whether DANCR is involved in NSCLC drug resistance, we developed gefitinib-resistant versions of the A549 and NCI-H1299 cell lines. EGFR inhibitors are commonly used in NSCLC treatment, and the cell lines express EGFR.20 EGFR inhibition is a common clinical treatment strategy. Gene expression of drug-resistant cells was compared to their parental lines using qRT-PCR. MDR1, DANCR, and EDB-FN are observed to be overexpressed in aggressive cancers.21 DANCR and EDB-FN were significantly overexpressed in A549-DR relative to parental A549, while MDR1 was slightly overexpressed in A549-DR but not significantly. The expression of MDR1, DANCR, and EDB-FN was significantly greater in NCI-H1299-DR (drug resistant) relative to parental NCI-H1299 (Figure 4A). Transfection with RGD-PEG-ECO/siDANCR significantly lowered the half-maximal inhibitory concentration (IC50) of NCI-H1299-DR relative to RGD-PEG-ECO/siNS and nontransfected groups, from an initial IC50 of 50 μM gefitinib down to 25 μM gefitinib (Figure 4B). The difference was not significant between treatment and control groups for the A549-DR cells, suggesting these cells did not develop the same level of gefitinib resistance or relied on different pathways for drug resistance.22 Functional effects of DANCR silencing were observed in drug-resistant cells in the same manner as they were in the parental cells. Both A549-DR and NCI-H1299-DR cells exhibited significant reduction in cell migration and invasion in Transwell assays (Figure 4C). Drug-resistant cells appeared to have increased levels of migration relative to their parental lines, consistent with the observation that drug-resistant cells are more likely to have metastasized and be more aggressive.23,24 Tumor spheroid formation was assessed, and smaller spheroids were observed with the RGD-PEG-ECO/siDANCR-treated cells relative to the control-treated cells (Figure 4D).
Figure 4.
Drug-resistant NSCLC cells respond to DANCR silencing. (A) mRNA expression of the drug transporter MDR1, DANCR, and ECM protein EDB fibronectin are elevated in drug-resistant cells relative to parental cells (n = 3). (B) Cell viability relative to untreated control shows significant reduction in gefitinib IC50 following transfection with RGD-PEG-ECO/siDANCR (n = 3). The IC50 was initially 50 μM gefitinib for both A549-DR and NCI-H1299-DR but was reduced to 25 μM gefitinib for NCI-H1299-DR with RGD-PEG-ECO/siDANCR transfection. (C) Transwell migration and invasion assays (n = 3) show significant reduction in the number of migrated and invaded cells, respectively, following DANCR knockdown relative to transfection with control siRNA in both cell lines. (D) Quantification of number of cells migrated or invaded in A549 DR or NCI-H1299 DR cells from (C). (E) Smaller 3D spheroid growth was observed with DANCR silencing relative to control siRNA transfection (n = 3). Error bars denote standard error of measure, scale bars in all panels = 100 μm. * p < 0.05 and ** p < 0.01 using unpaired t-test.
2.5. Kinetics of DANCR Silencing
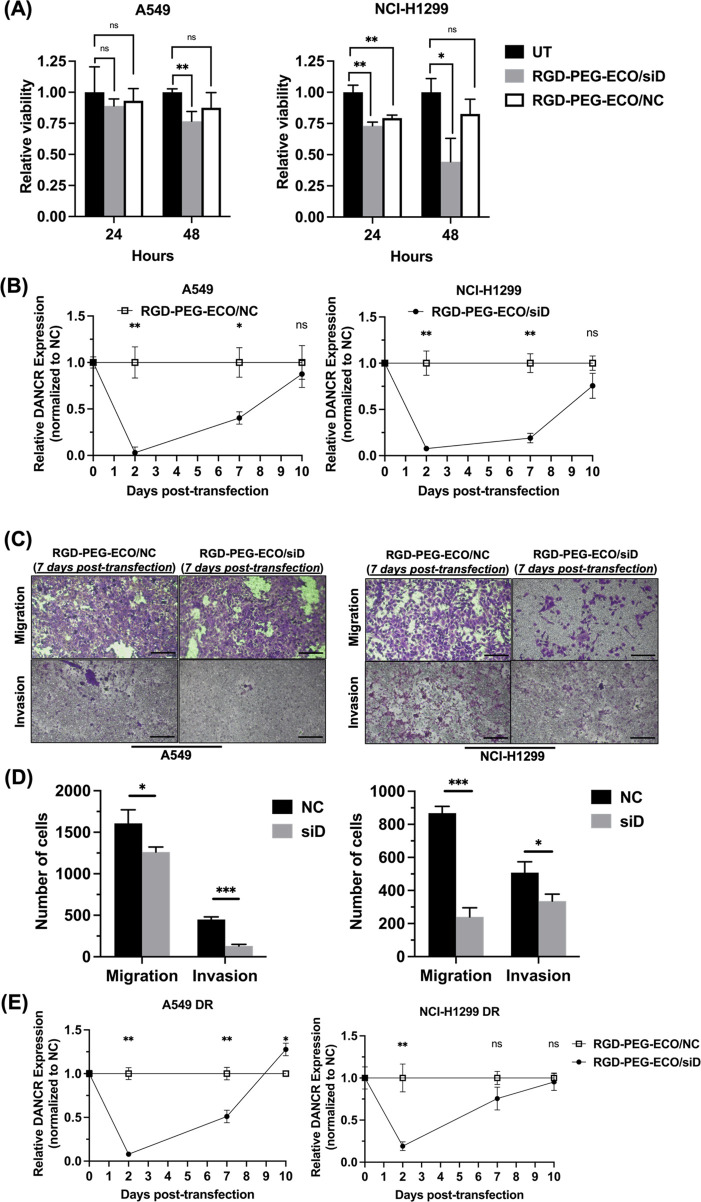
To further examine effects of RGD-PEG-ECO/siDANCR treatment, we studied the impact of silencing on DANCR expression over time. The viability of untreated cells was compared to those transfected with siDANCR or siNS negative control nanoparticles. The relative viability of cells was measured using the CCK-8 assay and normalized to the untreated cells. After 24 h post-transfection, there was no difference in viability of cells between siDANCR and negative control transfection. After 48 h, however, siDANCR-treated cells exhibited greater reduction in viability relative to the siNS-treated cells (Figure 5A). A subsequent analysis of DANCR expression over time following initial transfection revealed the greatest silencing after 2 d with partial recovery of expression by 7 d (Figure 5B). Additionally, functional effects of the single transfection were still observed after 7 d, Figure 5C. In NCI-H1299 cells, where DANCR silencing was maintained at 70% following the initial transfection, cell migration and invasion were both significantly reduced relative to the control. For A549 cells, where 55% silencing was maintained after the initial transfection, cell invasion, but not migration, was significantly different relative to the control. This suggests that a single transfection is sufficient to cause a reduction in cell migration and invasion for time beyond the initial transfection period depending on cell types.
Figure 5.
DANCR silencing with RGD-PEG-ECO/siDANCR is sustained over time. (A) RGD-PEG-ECO/siDANCR nanoparticles showed a functional effect in reducing cell viability after transfection relative to RGD-PEG-ECO/siNS nanoparticles or untreated control groups in both NCI-H1299 and A549 cells (n = 3). Viability was expressed relative to untransfected cells. (B) A single 48 h transfection with RGD-PEG-ECO/siDANCR was sufficient to reduce DANCR expression in A549 and NCI-H1299 cells for 7 d relative to a single 48 h transfection with RGD-PEG-ECO/siNS (n = 3). (C) Cell migration and invasion (n = 3) assessed 7 d following the initial transfection performed in (B) showed a greater reduction in NCI-H1299 cells than in A549 cells, where DANCR expression recovered to 20% and 50%, respectively. (D) Quantification of the number of cells migrated or invaded in A549 DR or NCI-H1299 DR cells. (E) In drug-resistant NSCLC cells, a single transfection of RGD-PEG-ECO/siDANCR showed a reduction in DANCR expression lasting for 7 d and fully recovering to pretransfection level at 10 d (n = 3). Error bars denote standard error of measure, scale bars = 100 μm. * p < 0.05, ** p < 0.01, and *** p < 0.005 using unpaired t-test.
DANCR silencing kinetics were also monitored in drug-resistant cells. With a single transfection, cells were cultured and collected at different time points to extract RNA for qRT-PCR. DANCR knockdown was lowest following the initial 2 d transfection and maintained at 50% in A549-DR and 25% in NCI-H1299-DR. Expression returned to pretransfection levels by 10 d (Figure 5D).
3. Discussion
We have shown here the potential of lncRNA DANCR as a target for non-small cell lung cancer therapy. Using RGD-PEG-ECO/siDANCR nanoparticles, we achieved over 90% reduction in DANCR expression in two different NSCLC cell lines (Figure 2B). Significant inhibition of cell migration and invasion was observed following DANCR silencing, along with a reduction in the oncogenic markers EDB-FN, ZEB1, survivin, and N-cadherin. Because DANCR has also been found to promote drug resistance, we further investigated its role in EGFR inhibitor-resistant NSCLC. DANCR expression was elevated in gefitinib-resistant versions of A549 and NCI-H1299 cells, and DANCR knockdown similarly reduced migration and invasion in these cells. More importantly, resensitization to gefitinib was observed in the NCI-H1299 cells following transfection with RGD-PEG-ECO/siDANCR nanoparticles.
Nanoparticles were formulated with RGD given the high expression of integrin αvβ3 in cancer cells resulting from neovascularization associated with NSCLC tumor growth.25 Human NSCLC tumors were reported to express greater levels of αv-family integrins relative to healthy lung tissue.26 The A549 and NCI-H1299 cell lines have been further shown to have elevated expression of these integrins,27 suggesting RGD-targeted nanoparticles are expected to show improved uptake over nontargeted nanoparticles. Previous studies using RGD-targeted PET imaging agents for NSCLC demonstrated higher accumulation in lung tumor tissue compared to scrambled peptide agents.28,29 Nanoparticles formed effectively and consistently with the RGD-PEG targeting ligand (Figure 1B). Characterization revealed efficient siRNA entrapment along with uniform size and zeta potential distributions (Figure 1C,D). Sizes below 200 nm and positive charge less than 50 mV provide optimal siRNA delivery with minimal toxicity.14,30 RGD peptide blocking during transfection with RGD-PEG-ECO nanoparticles limited uptake of the targeted nanoparticles, as seen with confocal imaging (Figure 1E). The free RGD peptide competitively binds to the same receptors as the targeted nanoparticles, thus reducing the ability for siRNA uptake into the cytosol of cells. Our previous studies demonstrated improved nanoparticle uptake using RGD-PEG-ECO in triple negative breast cancer and, further, showed nanoparticles formulated with nontargeted control RAD-PEG-ECO did not achieve the same level of uptake.31,32 We similarly evaluated siRNA entrapment and release from RGD-PEG-ECO nanoparticles33 and expect the same behavior using siDANCR in RGD-PEG-ECO nanoparticles.
NCI-H1299 cells exhibited higher expression of DANCR relative to A549 cells (Figure 2A). RGD-PEG-ECO showed superior silencing efficiency compared to other reported transfection agents, which typically reach only 50% silencing compared to over 90% observed here (Figure 2B),34 and silencing was maintained for up to one week (Figure 2C). The high level of DANCR silencing affected multiple downstream oncogenic pathways and markers of epithelial-to-mesenchymal transition (Figure 2D,E). DANCR knockdown presents an alternative to therapies targeting only one pathway that may lead to eventual tumor recurrence. Reductions in cell migration and invasion support the conclusion that DANCR knockdown has functional effects in preventing cancer spread. These effects persisted as long as DANCR expression was downregulated. Depending on the cell growth rate, downregulation by siDANCR lasted up to 7 d and still demonstrated reductions in migration and invasion. lncRNAs are a diverse group of molecules, and differences between cell lines are expected, as seen with variations in both functional behavior and expression analysis of the two cell lines. NCI-H1299 cells originated from a lymph node metastatic site of a patient receiving radiation therapy, while A549 cells were derived from a primary tumor.35 These inherent variations between the two cell lines may explain differences in DANCR expression and the responses associated with silencing it. DANCR expression is elevated in NCI-H1299 relative to A549, and DANCR potentially contributes more toward the oncogenic processes of this cell line. Beyond the molecular changes associated with DANCR silencing, functional effects reveal strong inhibition of cell migration and invasion (Figure 3). Inhibiting these functions is a critical step toward preventing the spread of metastatic cancer cells. Systemic delivery of various siRNAs against lncRNAs has successfully inhibited tumor progression in mouse models across multiple cancer types,36 and targeting DANCR will provide a novel treatment approach for non-small cell lung cancer in vivo.
DANCR has been previously implicated in the development of drug resistance across multiple cancers.37−39 In gastric cancer, DANCR was reported to promote multidrug resistance by upregulating expression of MDR1 and MRP1.40 We developed gefitinib-resistant NSCLC cell lines to determine the role of DANCR on drug resistance in these cells. EGFR inhibitors are initially successful in the treatment of NSCLC when EGFR is overexpressed, but most patients develop resistance to treatment.41 Once NSCLC cells became gefitinib-resistant, DANCR expression was significantly increased (Figure 4A). Expression of the drug transporter MDR1 was elevated in NCI-H1299-DR cells but not A549-DR, suggesting that NCI-H1299-DR could have developed greater resistance or that A549-DR relies on different pathways to maintain resistance. DANCR knockdown allowed resensitization to gefitinib from an IC50 concentration of 50 to 25 μM for NCI-H1299-DR but not A549-DR. At doses higher or lower than 50 μM the effect was not distinguishable between treatment and control groups (Figure 4B). High doses of inhibitor potentially overwhelm cellular drug elimination mechanisms and thus are not impacted by combination with transfection, while low doses are insufficient to impact cell viability. Drug-resistant cell migration, invasion, and tumor spheroid formation ability was reduced consistent with the trend observed in parental cell lines (Figure 4C–E).
To better characterize the response to RGD-PEG-ECO/siDANCR transfection, silencing effects were monitored over the course of 10 d. Maximum silencing was achieved 48 h following a single transfection, and the cytotoxic effects of siDANCR relative to siNS were similarly highest at 48 h (Figure 5A). Uptake of ECO/siRNA nanoparticles was previously reported as soon as 4 h but peaked over 72 h.42 Nanoparticles containing siDANCR are expected to have the same release and uptake behavior. While DANCR recovers to original levels after 7–10 d, functional effects are observed after 48 h. Because of varying cell growth rates, we noticed RGD-PEG-ECO/siDANCR transfection in NCI-H1299 cells maintained a 75% DANCR knockdown even at 7 d following initial transfection, whereas expression in A549 cells returned to 50% (Figure 5B,C). As a result, inhibition of cell migration and invasion was still noticed at 7 d in NCI-H1299 but only inhibiting invasion in A549. The effects of repeated RGD-PEG-ECO/siDANCR transfection on cell response remain to be explored but could more closely replicate the effects of an in vivo or clinical treatment regimen.
4. Conclusion
Tumor-targeted RGD-PEG-ECO/siDANCR nanoparticles achieve effective silencing that decreases migration, invasion, and spheroid formation in two NSCLC cell models. lncRNA DANCR is a potential target in the treatment of NSCLC and prevention of metastasis that overcomes limitations of traditional therapies targeting single oncogenic pathways. The in vitro results presented in this study create a foundation for a future in vivo evaluation of systemic NSCLC treatment through regulation of oncogenic lncRNA. Silencing oncogenic lncRNA DANCR is a promising strategy for treating and overcoming EGFR-inhibitor resistance in NSCLC.
5. Methods
5.1. Cell Lines and Reagents
Multifunctional pH-sensitive amino lipid ECO and targeting ligand RGD-PEG-MAL were synthesized as previously described (Cyclo(Arg-Gly-Asp-d-Phe-Lys), Vivitide, Gardner, MA, and Mal-PEG-NHS, 3.4k, Nanocs).33 A549 and NCI-H1299 cells were acquired from American Type Culture Collection (ATCC) and cultured according to manufacturer’s instructions in Dulbecco’s Modified Eagle’s Medium and Roswell Park Memorial Institute 1640 Medium, respectively, supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Life Technologies). Cells were maintained in a humidified incubator at 37 °C with 5% CO2. The siDANCR duplex [sense 5′-GGU CAU GAG AAA CGU GGA UUA CAdCdC-3′ and antisense 5′-GGU GUA AUC CAC GUU UCU CAU GAC CUC-3′] and negative control siLuciferase (referred to as siNS) duplex [sense 5′-UUA GCG UAG AUG UAA UGU GdTdT-3′and antisense 5′-CAC AUU ACA UCU ACG CUA A-3′] were purchased from IDT. The EDB-FN-specific fluorescent probe ZD2-Cy5.5 was synthesized as previously described.43
5.2. Nanoparticle Formulation and Transfection
RGD-PEG-ECO/siRNA nanoparticles were formulated as previously described.32 ECO at stock concentration of 50 mM in ethanol was diluted to 5 mM with nuclease-free water and mixed under gentle agitation with RGD-PEG-MAL (RGD-PEG/ECO = 2.5 mol %) in nuclease-free water at room temperature (RT) for 30 min. Subsequent complexation with siDANCR or siNS in nuclease-free water (25 μM) was achieved by mixing for an additional 30 min to obtain targeted ECO/siRNA nanoparticles at N/P = 8 and final siRNA concentration of 100 nM upon formulation. For transfection, cells were first washed three times with Dulbecco’s phosphate-buffered saline (DPBS). Nanoparticle formulations were mixed with cell culture media to a final volume of 1.5 mL and added to cells in six-well plates grown to 60% confluence. Nanoparticles were allowed to transfect for 48 h.
5.3. Nanoparticle Characterization
Agarose gels (1% w/v) were prepared in Tris/borate/ethylenediaminetetraacetic acid (EDTA) buffer with ethidium bromide added for a gel retardation assay. A nanoparticle formulation (10 μL) was added to 2 μL of DNA gel loading dye (6X, Life Technologies) and run at 100 V for 20 min. The gel was imaged with the ChemiDoc XRS system (BioRad). Nanoparticle size and zeta potential were measured with dynamic light scattering on the Anton Paar Litesizer.
5.4. RGD Peptide Competitive Binding and Nanoparticle Uptake
A549 or NCI-H1299 cells (50 000) were seeded in each well of an Ibidi four-well μ-Slide (Ibidi GmbH) and allowed to grow overnight. RGD-PEG-ECO nanoparticles were prepared as described above using AllStars Negative Control siRNA AF647 (Qiagen). Nanoparticles were added to media with or without a 10-fold molar excess of free RGD peptide and transfected onto cells for 24 h. The transfection medium was replaced with a medium containing Hoechst 33342 (1:2000 dilution, Invitrogen, Waltham, MA) for 25 min. Cells were washed three times with DPBS and then imaged using an Olympus FV1000 confocal microscope (Olympus Life Science).
5.5. RNA Extraction and qRT-PCR Analysis
Cells were transfected with nanoparticles mixed in culture medium and added to plated cells for 48 h. Total RNA was extracted from cells using the RNEasy Plus Mini Kit (Qiagen). Reverse transcription was performed using the miScript RT II kit (Qiagen), and qPCR was performed using the SyBr Green PCR Master Mix (Life Technologies). Gene expression was analyzed by the 2–ΔΔCt method with 18S expression as the control. Primer sequences were as follows: DANCR: Fwd 5′-GCGCCACTATGTAGCGGGTT-3′ and Rev 5′-TCAATGGCTTGTGCCTGTAGTT-3′; 18S: Fwd 5′-TCAAGAACGAAAGTCGGAGG-3′ and Rev 5′-GGACATCTAAGGGCATCACA-3′.
5.6. Western Blot
Total protein was extracted from cells using 1:1 complete protease inhibitor (Roche Diagnostics) in PBS and Laemmli buffer. Lysates were incubated at 100 °C for 10 min and centrifuged at 13 200 rpm at 4 °C for 15 min. Protein concentration was determined using the RC DC Protein Assay Kit (Bio-Rad). 40 μg of protein was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel at 100 V and then transferred to a nitrocellulose membrane at 60 V. The membrane was blocked with 5% milk in Tris-buffered saline with TWEEN 20 (TBST) for 1 h, then washed three times before the following antibodies were added at 1:1000 dilution with an overnight incubation at 4 °C: anti-ZEB1, anti-N-cadherin, and anti-β-actin monoclonal antibodies (Cell Signaling Technology) as well as anti-EDB G4 clone (Absolute Antibody). The membranes were incubated for 1 h with antirabbit IgG, HRP linked secondary antibody (Cell Signaling Technology) diluted 1:2000, then developed with Signal Fire Plus ECL Kit (Cell Signaling Technology) and imaged on the ChemiDoc XRS+ Imager (Bio-Rad). Images were quantified using FIJI (FIJI is just ImageJ) software by measuring protein band intensity corrected for background signal, then normalized to β-actin loading control.
5.7. Transwell Migration and Invasion Assay
Cells were transfected for 48 h with targeted ECO/siDANCR or ECO/siNS nanoparticles and cultured in serum-free medium. Starved cells (100 000) were seeded onto ThinCert Cell Culture Inserts (Greiner Bio-One) either uncoated or coated with 0.28 mg/mL Matrigel Membrane Matrix (Corning). The transwell invasion assay included a layer of Matrigel to simulate cell invasion through the basement membrane, while the migration assay did not. After 24 h the inserts were swabbed to remove unmigrated cells. The inserts were fixed with 10% formalin for 10 min and then stained with 0.05% crystal violet for 20 min. After they were dry, the inserts were imaged using a Moticam T2 camera (Motic Microscopes).
5.8. Scratch Wound Assay
NSCLC cells (1 × 106) were plated in a six-well plate and allowed to grow to confluence for 24 h. A 10 μL pipet tip was used to create a scratch along the center of the plate. Cells were washed with DPBS to remove unattached cells and transfected with RGD-PEG-ECO/siDANCR or RGD-PEG-ECO/siNS. The scratch wound was monitored for 48 h until closure, and images were taken with the Moticam T2 camera.
5.9. 3D Culture and Confocal Imaging
To an eight-well μ-Slide (Ibidi), 200 μL of Matrigel membrane matrix was added and allowed to solidify for 30 min at 37 °C. 100 000 cells were seeded in each well and were allowed to grow for 48 h before imaging. Spheroids were stained with ZD2-Cy5.5 (125 nM) and Hoechst 33342 (1:2000 dilution) for 25 min, then washed three times with DPBS. Images were obtained using an Olympus FV1000 confocal microscope. Images were analyzed in FIJI by calculating staining intensities. The average signal intensity for ZD2-Cy5.5 and Hoechst staining were calculated, and the ratio between average signals was found.
5.10. Development of Drug-Resistant Cells and Cytotoxicity Testing
Gefitinib-resistant cells were developed by culturing cells in media containing gefitinib (Sigma-Aldrich) dissolved in dimethyl sulfoxide (DMSO). Cells were maintained in media containing gefitinib for 72 h and allowed to recover in gefitinib-free media for 48 h. Drug exposure was repeated in this manner over time, and the shift in the IC50 concentration of gefitinib was used as a metric of confirming drug resistance.
To assess the level of drug resistance, 5000 cells were seeded in wells of a 96-well culture plate. After they were allowed to adhere for 24 h, these cells were treated with increasing concentrations of gefitinib for 48 h. Wells were washed with PBS, and a medium containing CCK-8 assay solution was added according to the manufacturer’s instructions (Dojindo Molecular Technologies). Plates were incubated at 37 °C for 2–4 h, and then the absorbance at 450 nm was measured with a microplate reader (Molecular Devices). Cell viability was determined by normalizing to the absorbance of control cells.
5.11. Statistical Analysis
All experiments were independently performed at least three times. A statistical analysis was done with GraphPad Prism 9, and p < 0.05 was considered statistically significant. Data between two groups was compared using the unpaired Student’s t-test. Data between three or more groups was compared using a one-way analysis of variance (ANOVA) test.
Acknowledgments
This research was funded by National Cancer Institute of the National Institutes of Health, Grant No. R01 CA235152. Z.R.L. is M. Frank Rudy and Margaret Domiter Rudy Professor of Biomedical Engineering.
The authors declare the following competing financial interest(s): The delivery platform has been licensed to Helios Biopharmaceuticals. Dr. Lu has ownership interest in the company.
References
- Siegel R. L.; Miller K. D.; Fuchs H. E.; Jemal A. Cancer Statistics, 2021. Ca-Cancer J. Clin. 2021, 71 (1), 7–33. 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- Uramoto H.; Tanaka F. Recurrence after surgery in patients with NSCLC. Transl. Lung Cancer Res. 2014, 3 (4), 242–9. 10.3978/j.issn.2218-6751.2013.12.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriraman S. K.; Aryasomayajula B.; Torchilin V. P. Barriers to drug delivery in solid tumors. Tissue Barriers 2014, 2, e29528. 10.4161/tisb.29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam K. M.; Anggondowati T.; Deviany P. E.; Ryan J. E.; Fetrick A.; Bagenda D.; Copur M. S.; Tolentino A.; Vaziri I.; McKean H. A.; et al. Patient preferences of chemotherapy treatment options and tolerance of chemotherapy side effects in advanced stage lung cancer. BMC Cancer 2019, 19 (1), 835. 10.1186/s12885-019-6054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G. D.; Lu X. Z.; Yuan L. J. LncRNA: A link between RNA and cancer. Biochim. Biophys. Acta, Gene Regul. Mech. 2014, 1839 (11), 1097–1109. 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- Charles Richard J. L.; Eichhorn P. J. A. Platforms for Investigating LncRNA Functions. SLAS Technol. 2018, 23 (6), 493–506. 10.1177/2472630318780639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.; Shi Q.; Yuan X.; Xue C.; Shen S.; He Y. DANCR: an emerging therapeutic target for cancer. Am. J. Transl. Res. 2020, 12 (7), 4031–4042. [PMC free article] [PubMed] [Google Scholar]
- Kretz M.; Webster D. E.; Flockhart R. J.; Lee C. S.; Zehnder A.; Lopez-Pajares V.; Qu K.; Zheng G. X.; Chow J.; Kim G. E.; et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012, 26 (4), 338–43. 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. R.; Wu Y. S.; Wang W. S.; Zhang J. S.; Wu Q. G. Upregulation of lncRNA DANCR functions as an oncogenic role in non-small lung cancer by regulating miR-214–5p/CIZ1 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24 (5), 2539–2547. 10.26355/eurrev_202003_20521. [DOI] [PubMed] [Google Scholar]
- Bai Y.; Zhang G.; Chu H.; Li P.; Li J. The positive feedback loop of lncRNA DANCR/miR-138/Sox4 facilitates malignancy in non-small cell lung cancer. Am. J. Cancer Res. 2019, 9 (2), 270–284. [PMC free article] [PubMed] [Google Scholar]
- Mahmoodi Chalbatani G.; Dana H.; Gharagouzloo E.; Grijalvo S.; Eritja R.; Logsdon C. D.; Memari F.; Miri S. R.; Rezvani Rad M.; Marmari V. Small interfering RNAs (siRNAs) in cancer therapy: a nano-based approach. Int. J. Nanomed. 2019, 14, 3111–3128. 10.2147/IJN.S200253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. J.; Jin M. Z.; Xia B. R.; Jin W. L.. Long Non-coding RNA DANCR as an Emerging Therapeutic Target in Human Cancers. Front. Oncol., 2019. 9. 10.3389/fonc.2019.01225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilb A. L.; Scheidt J. H.; Vaidya A. M.; Sun Z. H.; Sun D.; Lee S. J.; Lu Z. R. Optimization of Synthesis of the Amino Lipid ECO for Effective Delivery of Nucleic Acids. Pharmaceuticals 2021, 14 (10), 1016. 10.3390/ph14101016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. R.; Laney V. E. A.; Hall R.; Ayat N. Environment-Responsive Lipid/siRNA Nanoparticles for Cancer Therapy. Adv. Healthcare Mater. 2021, 10 (5), 2001294. 10.1002/adhm.202001294. [DOI] [PubMed] [Google Scholar]
- Vaidya A. M.; Sun D.; Ayat N.; Sun Z. H.; Jiang H. F.; Qian V.; Lu Z. R. Nanoparticle-Mediated Gene Therapy against DANCR Suppresses TNBC by Targeting Multiple Oncogenic Pathways. Mol. Ther. 2018, 26 (5), 61–61. [Google Scholar]
- Ma Y.; Fan B.; Ren Z.; Liu B.; Wang Y. Long noncoding RNA DANCR contributes to docetaxel resistance in prostate cancer through targeting the miR-34a-5p/JAG1 pathway. OncoTargets Ther. 2019, 12, 5485–5497. 10.2147/OTT.S197009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L. F.; Gu J. M.; Hou S. N.; Liu D. B.; Zhou M. J.; Hua T. J.; Zhang J. Y.; Ge Z. J.; Xu J. Long non-coding RNA DANCR promotes the progression of non-small-cell lung cancer by inhibiting p21 expression. OncoTargets Ther. 2019, 12, 135–146. 10.2147/OTT.S186607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Q.; Gao L. N.; Wang R. F.; Chu W. W.; Zhang Y. X.; Zhao X. J.; Lv B. L.; Liu J. B. LncRNA DANCR Promotes Lung Cancer by Sequestering miR-216a. Cancer Control 2018, 25 (1), 107327481876984. 10.1177/1073274818769849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst R. S.; Morgensztern D.; Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018, 553 (7689), 446–454. 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- Zhang F. N.; Wang S. P.; Yin L. L.; Yang Y. Z.; Guan Y.; Wang W.; Xu H.; Tao N. J. Quantification of Epidermal Growth Factor Receptor Expression Level and Binding Kinetics on Cell Surfaces by Surface Plasmon Resonance Imaging. Anal. Chem. 2015, 87 (19), 9960–9965. 10.1021/acs.analchem.5b02572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z.; Lu Z. R. Targeting fibronectin for cancer imaging and therapy. J. Mater. Chem. B 2017, 5 (4), 639–654. 10.1039/C6TB02008A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgillo F.; Della Corte C. M.; Fasano M.; Ciardiello F. Mechanisms of resistance to EGFR-targeted drugs: lung cancer. ESMO Open 2016, 1 (3), e000060. 10.1136/esmoopen-2016-000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoof S.; Mulford I. J.; Frisco-Cabanos H.; Nangia V.; Timonina D.; Labrot E.; Hafeez N.; Bilton S. J.; Drier Y.; Ji F.; et al. Targeting FGFR overcomes EMT-mediated resistance in EGFR mutant non-small cell lung cancer. Oncogene 2019, 38 (37), 6399–6413. 10.1038/s41388-019-0887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue T.; Weinberg R. A. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14 (10), 611–629. 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temming K.; Schiffelers R. M.; Molema G.; Kok R. J. RGD-based strategies for selective delivery of therapeutics and imaging agents to the tumour vasculature. Drug Resistance Updates 2005, 8 (6), 381–402. 10.1016/j.drup.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Elayadi A. N.; Samli K. N.; Prudkin L.; Liu Y. H.; Bian A.; Xie X. J.; Wistuba I. I.; Roth J. A.; McGuire M. J.; Brown K. C. A peptide selected by biopanning identifies the integrin alphavbeta6 as a prognostic biomarker for nonsmall cell lung cancer. Cancer Res. 2007, 67 (12), 5889–95. 10.1158/0008-5472.CAN-07-0245. [DOI] [PubMed] [Google Scholar]
- Irigoyen M.; Pajares M. J.; Agorreta J.; Ponz-Sarvise M.; Salvo E.; Lozano M. D.; Pio R.; Gil-Bazo I.; Rouzaut A.. TGFBI expression is associated with a better response to chemotherapy in NSCLC. Mol. Cancer, 2010. 9. 10.1186/1476-4598-9-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Y.; Sievers E.; Hou Y. P.; Park R.; Tohme M.; Bart R.; Bremner R.; Bading J. R.; Conti P. S. Integrin alpha(V)beta(3)-targeted imaging of lung cancer. Neoplasia 2005, 7 (3), 271–279. 10.1593/neo.04538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. S.; Park H. S.; Lee B. C.; Jung J. H.; Lee H. Y.; Kim S. E. Imaging of Integrin alpha(v)beta(3) Expression in Lung Cancers and Brain Tumors Using Single-Photon Emission Computed Tomography with a Novel Radiotracer Tc-99m-IDA-D-[c(RGDfK)](2). Cancer Biother. Radiopharm. 2017, 32 (8), 288–296. 10.1089/cbr.2017.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 2012, 7, 5577–5591. 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvani J. G.; Gujrati M. D.; Mack M. A.; Schiemann W. P.; Lu Z. R. Silencing beta3 Integrin by Targeted ECO/siRNA Nanoparticles Inhibits EMT and Metastasis of Triple-Negative Breast Cancer. Cancer Res. 2015, 75 (11), 2316–2325. 10.1158/0008-5472.CAN-14-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujrati M.; Vaidya A. M.; Mack M.; Snyder D.; Malamas A.; Lu Z. R. Targeted Dual pH-Sensitive Lipid ECO/siRNA Self-Assembly Nanoparticles Facilitate In Vivo Cytosolic sieIF4E Delivery and Overcome Paclitaxel Resistance in Breast Cancer Therapy. Adv. Healthcare Mater. 2016, 5 (22), 2882–2895. 10.1002/adhm.201600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayat N. R.; Sun Z. H.; Sun D.; Yin M.; Hall R. C.; Vaidya A. M.; Liu X. J.; Schilb A. L.; Scheidt J. H.; Lu Z. R. Formulation of Biocompatible Targeted ECO/siRNA Nanoparticles with Long-Term Stability for Clinical Translation of RNAi. Nucleic Acid Ther. 2019, 29 (4), 195–207. 10.1089/nat.2019.0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q. C.; Rui Z. H.; Guo Z. L.; Xie W.; Shan S.; Ren T. LncRNA-DANCR contributes to lung adenocarcinoma progression by sponging miR-496 to modulate mTOR expression. J. Cell. Mol. Med. 2018, 22 (3), 1527–1537. 10.1111/jcmm.13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. E.; Ju J. A.; Musacchio N.; Mathias T. J.; Vitolo M. I. Long Noncoding RNA DANCR Activates Wnt/beta-Catenin Signaling through MiR-216a Inhibition in Non-Small Cell Lung Cancer. Biomolecules 2020, 10 (12), 1646. 10.3390/biom10121646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Li Z.; Chen X.; Zhang S. Long non-coding RNAs: From disease code to drug role. Acta Pharm. Sin. B 2021, 11 (2), 340–354. 10.1016/j.apsb.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.; Zhou G.; Li M.; Hu D.; Zhang L.; Liu P.; Lin K. Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF-kappaB signaling pathway. Neurochem. Int. 2018, 118, 233–241. 10.1016/j.neuint.2018.03.011. [DOI] [PubMed] [Google Scholar]
- Zhao H. F.; Zhang Z. C.; Shi B. K.; Jiang X. Z. DANCR sponges miR-135a to regulate paclitaxel sensitivity in prostate cancer. Eur. Rev. Med. Pharmacol Sci. 2019, 23 (16), 6849–6857. [DOI] [PubMed] [Google Scholar]
- Xiong M.; Wu M.; Dan Peng; Huang W.; Chen Z.; Ke H.; Chen Z.; Song W.; Zhao Y.; Xiang A. P.; Zhong X.; et al. LncRNA DANCR represses Doxorubicin-induced apoptosis through stabilizing MALAT1 expression in colorectal cancer cells. Cell Death Dis. 2021, 12 (1), 24. 10.1038/s41419-020-03318-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. D.; Shang J.; Li M.; Zhang Y. Y. LncRNA DANCR accelerates the development of multidrug resistance of gastric cancer. Eur. Rev. Med. Pharmacol Sci. 2019, 23 (7), 2794–2802. [DOI] [PubMed] [Google Scholar]
- Stewart E. L.; Tan S. Z.; Liu G.; Tsao M. S. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations-a review. Transl. Lung Cancer Res. 2015, 4 (1), 67–81. 10.3978/j.issn.2218-6751.2014.11.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujrati M.; Malamas A.; Shin T.; Jin E. L.; Sun Y. L.; Lu Z. R. Multifunctional Cationic Lipid-Based Nanoparticles Facilitate Endosomal Escape and Reduction-Triggered Cytosolic siRNA Release. Mol. Pharmaceutics 2014, 11 (8), 2734–2744. 10.1021/mp400787s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z.; Zhou Z.; Shi X.; Wang J.; Wu X.; Sun D.; Chen Y.; Zhu H.; Magi-Galluzzi C.; Lu Z. R. EDB Fibronectin Specific Peptide for Prostate Cancer Targeting. Bioconjug Chem. 2015, 26 (5), 830–8. 10.1021/acs.bioconjchem.5b00178. [DOI] [PubMed] [Google Scholar]