Abstract
Strong rationale and a growing number of preclinical and clinical studies support combining radiotherapy and immunotherapy to improve patient outcomes. However, several critical questions remain, such as the identification of patients who will benefit from immunotherapy and the identification of the best modalities of treatment to optimize patient response. Imaging biomarkers and radiomics have recently emerged as promising tools for the non-invasive assessment of the whole disease of the patient, allowing comprehensive analysis of the tumor microenvironment, the spatial heterogeneity of the disease and its temporal changes. This review presents the potential applications of medical imaging and the challenges to address, in order to help clinicians choose the optimal modalities of both radiotherapy and immunotherapy, to predict patient’s outcomes and to assess response to these promising combinations.
Keywords: radioimmunotherapy, radiotherapy, immunotherapy, tumor biomarkers
Introduction
Immunotherapy has revolutionized cancer management in recent years. Most advanced patients receiving frontline immune checkpoint inhibitors (ICIs) will, however, develop disease progression. In order to improve the response rates of immunotherapy, a growing number of studies are evaluating different treatment combinations or strategies, one of which is the combination of systemic immunotherapy and local radiotherapy.1–7 Indeed, there is substantial evidence that radiotherapy induces immune stimulation with the release of tumor antigens and proinflammatory cytokines at the local and systemic levels and thus, may act synergistically with immunotherapy in the systemic control of advanced metastatic cancers.1 8–12 Radiotherapy activates immune response by triggering immunogenic cell death (with extracellular release of release of high-mobility group box 1 protein and adenosine-5′-triphosphate, and cell surface translocation of calreticulin) and production of type I interferon (IFN) via the activation of the cGAS-STING pathway, leading to dendritic cells and cytotoxic T cells activation, which may eventually lead to an out-of-field ‘abscopal’ response.13 14 Inversely, it has been shown that radiotherapy also promotes immunosuppressive effects, with notably a PD-L1 upregulation via different pathway such as IFN-dependent pathway and IL-6/JAK/STAT pathway,15 or DNA damages through ATM/ATR/Chk1 kinase activation,16 along with the attraction of immunosuppressive cells (ie, tumor-associated macrophages, myeloid-derived suppressor cells, and regulatory T cells) and the release of immunosuppressive cytokines such as TGF-β and IL-10.13 These results support the potential of immunotherapy as an adjunct to radiotherapy to counteract its immunosuppressive effects, as illustrated by the phase III PACIFIC trial showing a benefit of a consolidation therapy with the anti-PD-L1 drug durvalumab in unresectable locally advanced non-small cell lung cancer (NSCLC) after chemoradiotherapy.17
However, only a small number of controlled randomized trials evaluating the benefit of adding radiotherapy to immunotherapy exist with mostly negative results.3 4 7 18–20 For instance, in advanced NSCLC patients, a pooled analysis of two clinical trials evaluating pembrolizumab (anti-PD1)±radiotherapy showed that adding radiotherapy to pembrolizumab improved outcomes. However, a randomized phase 2 trial was not able to identify a benefit of low-dose or high-dose radiation therapy with durvalumab (anti-PD-L1)+tremelimumab (anti-CTLA4) in advanced immunotherapy-pretreated NSCLC patients.4
Irradiation modalities to optimize the achievement of a systemic antitumor immune response are then subject to numerous studies.10 21–23
Fine tuning delivered dose and fractionation (dose per session) may be crucial for achieving an out-of-field abscopal response.24–27 For example, an abscopal response was obtained in immunocompetent mice bearing syngeneic mammary and colorectal tumor cell transplants after hypofractionated radiotherapy (three sessions of 8 Gy delivered 3 days in a row) but was not obtained with a single fraction radiotherapy of 20 Gy.24 Doses above 12 Gy increase the activity of 3′ repair exonuclease 1, a DNA exonuclease that degrades DNA accumulating in the cytosol of irradiated cells, decreasing STING activation and Batf3-dependent dendritic cell recruitment.28 These studies would suggest that the optimal dose in radioimmunotherapy may be lower than the maximum tolerated doses used in radiotherapy alone.22 However, low dose radiotherapy (1 Gy/fraction) has also been shown to potentially stimulate immune responses by activating and stimulating helper T cells, promoting a generation of immune memory and modulating the tumor stroma with the reduction of TGF-β induced Tregs in the tumor microenvironment.29 30
Open questions such as the number and choice of lesions to irradiate remain unanswered.31 32 Indeed, the tumor heterogeneity of extensive disease could limit the likelihood of a systemic (abscopal) antitumor immune response induced by radiotherapy, especially in the case of single-site irradiation.21 Thus, a wide range of possibilities in the choice of doses and volumes to irradiate makes possible combinations more complex.
In this context where both disease and treatment are characterized by heterogeneity, medical imaging may express its full potential as it allow analysis of the entire tumor mass and individual lesions, unlike traditional biopsies.33 Indeed, each lesion may have a different microenvironment and may receive different local and/or systemic treatment effects, leading to heterogeneous response profiles.34 Imaging-derived biomarkers especially may help to better understand the relationship between radiotherapy and immunotherapy and to eventually guide the clinician in the choice of treatment and its modalities.
Here, we will present the potential applications and challenges of imaging to guide immunotherapeutic combinations with a particular focus on radiomics. We will also discuss the particularities to be taken into account in imaging for the evaluation of response in these new combinations.
Imaging to increase knowledge, to predict responses, and to guide therapeutics
A brief introduction to computational medical imaging principles
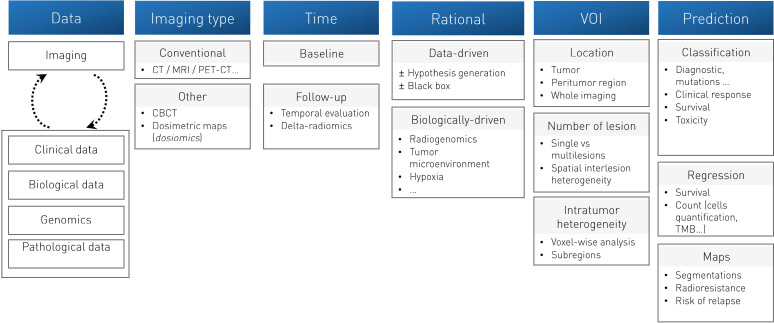
Artificial intelligence approaches applied to medical imaging have been particularly developed in recent decades with great results for detection tasks, image reconstruction, and generation of imaging biomarkers for diagnostic support, prognostic assessment, or treatment response prediction.33 35 36 The term ‘radiomics’ was introduced by Gillies et al 37 in 2010 to define the process of computational translation of medical imaging into high-dimensional quantitative data (similar to other ‘omics’) and the use of machine learning for the development of imaging biomarkers. A typical pipeline of analysis is usually constituted by the following steps: (1) definition of the volumes of interest (VOIs) to be analyzed, (2) extraction of the quantitative information represented by the radiomics features, and (3) application of machine learning approaches and evaluation of the validity and generalizability of the model.35 38 While machine learning methods may seem complex at first glance to the clinician, we will outline some general and relatively simple concepts that allow for a better understanding of this discipline.
Computational translation of the image into numbers relies either on the calculation of ‘classical’ radiomic features (in particular from the histogram of the image intensities, texture matrices, or the shape of the VOI, defined a priori and which can be extracted by different software such as LIFEx39 or Pyradiomics40), or on deep learning approaches that allow ‘discovering’ the relevant information directly from the raw image (‘deep features’) using neural networks (especially conventional neural networks).41 While classical radiomic features are mostly used for biomarker development, neural networks can also be used for image detection, segmentation and reconstruction tasks. This data extraction can be performed in one or more volume of interest to be analyzed (eg, a tumor lesion), or on the whole image (especially when using deep learning techniques). It is important to note that the different acquisition protocols and reconstruction parameters of the imaging process (CT, MRI, or positron emission tomography scan (PET-CT)) directly affect the image and may be a confounding factor. This can particularly impact the performance of algorithms in the case of heterogeneous cohorts. Thus, image preprocessing and normalization steps are performed to make the images of different patients with different acquisition protocols comparable.42 43
Once the quantitative data have been extracted from the image, machine learning algorithms aim to train a model for a particular task. Classically, three distinct data sets are used in machine learning approaches: a training set to learn a specific task, a validation set to optimize the algorithm and an independent test set for the final validation and evaluation of the generalizability of the algorithm. Assessment of the model performance in an independent data set is essential to ensure the external validity of an algorithm. Indeed, overfitting may happen during the training, especially when the number of features extracted is higher than the number of patients, leading to overoptimistic performance estimation and subpar performance when used in real clinical applications.
A growing number of initiatives and recommendations are being issued to ensure the quality, the reproducibility and the methodological validity of artificial intelligence (AI) approaches applied on medical images.44–46 Harmonization of imaging data and/or radiomic features a posteriori is essential to enable robust development of generalisable imaging biomarkers in a multicenter setting.47 Although standardization of image acquisition protocols is a simple way to obtain consistent images across different centers,48 it can be difficult to implement on a large scale and does not take into account the potential batch effect of acquisitions from multiple devices from different manufacturers. Image processing methods, including at least isotropic spatial resampling of voxels and, for example, skull stripping for image intensity normalization (using, eg, the Z-score method), and bias field correction for brain MRI images, have been shown to be effective for medical imaging normalization.49 50 Adjusting radiomic features after extraction to correct for batch effect using methods such as the ComBat approach has also shown interesting results in reducing discrepancies between different imaging protocols and manufacturers.51 Standardization of radiomic feature definitions have also been proposed by the image biomarker standardization initiative to increase the reproducibility and comparability of software used for feature extraction.52 Regarding deep learning studies, some recommendations to ensure their proper conduct have also recently been published.53 The Radiomic Quality Score calculated from a 16-point checklist allows the assessment of the various technical and statistical aspects and the clinical validity of the radiomic analysis method.54
Radiomics to predict response to immunotherapy and radiotherapy combinations: toward a selective destruction of predicted immune refractory lesions and immunogenic low-dose radiotherapy for abscopal effect friendly lesions?
Since the first studies evaluating radiomics to predict the immune microenvironment in 2017,55 an increasing number of radiomic studies have confirmed the potential of imaging biomarkers for use in immuno-oncology (table 1). Promising results have been obtained using a wide variety of approaches on many different tumors types, such as lung cancers,56–61 head and neck cancers,62 melanoma,56 63 64 urothelial65–67 and renal cancers,68 with either CT scans,56–58 69 PET-CTs,60 70 or MRIs71 72 (figure 1).
Table 1.
Summary of radiomics study evaluating response to radioimmunotherapy and immunotherapy
| Reference | Cancer type | Patients | Multilesion analysis | Modality | Endpoints | Features | Machine learning | Findings |
| Prediction of response to radiotherapy and immunotherapy | ||||||||
| Korpics et al 202074 | Solid tumor | 68 pts with solid tumors SBRT (30–50 Gy, 3–5 fractions)+pembrolizumab combination. 139 irradiated lesions |
Y | CT | RT+ICI lesion and patient response | Published radiomic signature of CD8 cells (Sun et al, Lancet Oncol 201869) – average score. Cut-off=1st and 3rd quartiles. No machine learning. |
Association with tumor response: OR=10.2; 95% CI 1.76 to 59.17; p=0.012. PFS (HR 0.47, 95% CI 0.26 to 0.85; p=0.013) and OS (HR 0.39, 95% CI 0.20 to 0.75; p=0.005). |
|
| Sun et al 202073 | Solid tumor | 94 pts with RT (8 Gy × 3 mostly)+immunotherapy. 100 irradiated+189 non-irradiated lesions. |
Y | CT | RT+ICI lesion and patient response | Published radiomic signature of CD8 cells (Sun et al, Lancet Oncol 201869). No machine learning. |
Association with lesion response AUC=0.63, p=0.0020. Spatial heterogeneity assessment using MinCD8RS and entropy was associated with OS and PFS. |
|
| Biologically driven radiomic biomarker for immunotherapy response prediction | ||||||||
| Sun et al 201869 | Solid tumors | Training: n=135 for CD8 cells prediction. Validation: n=119 (CD8 cells validation), n=100 (immunophenotype) and n=137 (ICI response). |
N | CT |
|
78 radiomic features from tumor+border, five location variables, and one technical variable. | Elastic net regularized regression. Eight variables retained |
Validation of CD8 cells prediction: AUC=0.67; 95% CI 0.57 to 0.77; p=0.0019. Association with immune inflamed tumors: AUC=0.76; 95% CI 0.66 to 0.86; p<0.0001. Association with IO response and OS (HR 0.58, 95% CI 0.39 to 0.87; p=0.0081). |
| He et al 202091 | NSCLC | n=327 pts with complete resection of lung ADK for TMBRB (TMB radiomic biomarker) development (Tr/V/Te : 236/26/65 pts). n=123 NSCLC: ICI response. |
N | CT | TMB ICI response |
1020 deep learning features | Feature extraction: 3D-densenet. Classification: fully connected network. |
TMB prediction: AUC=0.81, 95% CI 0.77 to 0.85 in test cohort. TMBRB was associated with ICI-treated patients. OS: HR=0.54, 95% CI 0.31 to 0.95; p=0.030, and PFS=HR: 1.78, 95% CI 1.07 to 2.95; p=0.023. |
| Mu et al 202199 | NSCLC | Tr=284, V: 116, test: 85. | N | PET | PD-L1 | Deep learning | PD-L1: AUC ≥0.82 in all the cohorts PFS, OS |
|
| Immunotherapy response prediction | ||||||||
| Tunali et al 201957 | NSLCC | n=228 NSCLC patients treated with single agent or double agent immunotherapy. No validation set. |
N | CT | Rapid progression phenotypes | 600 features from the largest tumor+border. Logistic regression. No validation |
AUC 0.804 to 0.865 to predict rapid disease progression phenotypes (TTP <2 months or hyperprogressive disease). | |
| Trebeschi et al 201956 | NSCLC and melanoma | n=203 patients with advanced melanoma and NSCLC undergoing anti-PD-1 therapy. Accounting for 1055 target lesions. Training, tuning and test sets. |
Y | CT | Lesion progression | Features extracted from original CT and image transformations, with different scales. | Comparisons of different feature selection methods and eight trained classifiers. | Prediction of NSCLC lesions progression (AUC up to 0.83; p<0.001) and melanoma lymph nodes progression (0.64 AUC, p=0.05). Patient response prediction based on lesion progression probability: AUC of up to 0.76 for both cancer types (p<0.001). |
| Alessandrino et al 201965 | Urothelial | n=31 pts with metastatic urothelial cancer treated with anti-PD-1/PD-L1. 65 lesions ≥1 cm analyzed at baseline, 72 at the first evaluation. |
Y | CT 2D |
PFS <12 months | Histogram features from single slice of each lesion at different spatial scale filters (TexRad). Aggregation by mean value. Logistic stepwise regression – no validation. |
Entropy and mean were associated with patients with PFS <12 months. | |
| Khorrami et al 2019(58 | NSCLC | n=139 patients with NSCLC treated with ICI. Discovery set (D1=50) and two validation sets (D2=62, D3=27). 36 pts for TILs evaluation. |
N | CT |
|
495 delta texture features 2D+49 shape features (DelRADx) (intranodular and perinodular). | Linear discriminant analysis (LDA) classifier was trained with eight DelRADx features. | Responders AUC of 0.88, 0.85 and 0.81 in D1, D2 and D3 OS: HR=1.64; 95% CI 1.22 to 2.21; p=0.0011; deltaradiomics. Peritumoral Gabor features were associated with the density of TILs on diagnostic biopsy samples. |
| Mu et al 202059 | NSCLC | n=194 stage IIIB–IV NSCLC pts treated with ICI. Tr: 99 retrospective patients. V: retrospective (n=47) and prospective test cohorts (n=48). |
N | PET | Durable clinical benefit (DCB) (6 months PFS) |
790 features from PET, CT and PET+CT fusion images. | Feature selection Pearson LASSO with 100 times fivefold CV eight features retained. |
Prediction of DCB=AUC 0.86 (95% CI 0.79 to 0.94), 0.83 (95% CI 0.71 to 0.94), and 0.81 (95% CI 0.68 to 0.92). Association with OS and PFS. |
| Khatua et al 2020104 | Medulloblastoma and ependymoma. | n=12 pediatric pts treated with intraventricular infusions of ex vivo expanded autologous NK cells (7 pts for the radiomic study). |
N | MRI | Responders | Features not detailed LASSO. No validation. |
Exploratory results: accuracy and specificity 100% but not significant (only five patients were analyzed). |
|
| Polverari et al 202060 | NSCLC | n=57 NSLSC pts (stage IIIb/c or IV). Treated with ICI. |
N | PET | Progression | PET parameters and radiomic features (shape, histogram, texture). Univariate analysis (Fisher, Wilcoxon). No validation. |
Metabolic tumor volume (MTV) (p=0.028) and total lesion glycolysis (TLG) (p=0.035) were associated with progression. High tumor volume, TLG and heterogeneity (‘skewness’ and ‘kurtosis’) had a higher probability of failing immunotherapy. | |
| Park et al 202066 | Urothelial carcinoma | n=62 pts with metastatic urothelial carcinoma treated with ICI. Tr: n = 41/V: n=21. 224 lesions analyzed. |
Y | CT | Objective response and disease control | 49 RFs (histogram, GLCM, GLRLM): 26 RFs were reliable. Feature selection by LASSO (progressive lesions). |
Five features and the presence of visceral organ involved. | A radiomics signature for each lesion was built to predict patient response (objective response and disease control). The median signature of each lesion was used at the patient level for patients with multiple lesions. Optimum cut-off (Youden index) for disease control. Objective response: AUC 0.87 (95% CI 0.65 to 0.97) disease control: AUC 0.88 (95% CI 0.67 to 0.98). Association with OS and PFS. |
| Khene et al 202068 | mRCC | n=48 mRCC pts treated with nivolumab. 1–5 lesions per pt (aggregation method not described). Random split for Tr and V. |
Y | CT 2D |
PD versus SD/PR/CR | 279 RFs histogram, GLCM, GLRLM, autoregressive model features, Haar wavelet. | Feature selection: LASSO: 5 RFs. Four models tested. |
Prediction of PD: accuracy of 0.82, 0.71, 0.91 and 0.81 (KNN, random forest tree, logistic regression and SVM, respectively) AUC of 0.79, 0.67, 0.92 and 0.71, respectively. |
| Valentinuzzi et al 202061 | NSCLC | n=30 pts with NSCLC treated with pembrolizumab. | N | PET | Responders (OS>median) | Five preselected features at baseline, months 1 and 4. Logistic regression analyses and fivefold cross-validation. No test set. | Association between features and OS. | |
| Colen et al 2021134 | Advanced rare cancers | n=57 pts in pembrolizumab phase II trials. | N | CT | Controlled disease versus progression | 610 features | Feature selection: LASSO. ML: XGBoost+LOOCV. |
Progressive disease (RECIST): accuracy, SE, and Sp of 94.7%, 97.3%, and 90%, respectively; p<0.001. |
| Tunali et al 202192 | NSCLC | Advanced NSCLC treated with IO. Tr=180, V1=90, V2=62. |
N | CT | OS | 213 Intra+peritumoral features, reduced to 67 stability and reproducibility (segm. algorithms, image parameters, RIDER). | Univariate analysis of RF and OS, then ML: CART 1RF+2 clinical variables (dependency ?). |
Radioclinical model: OS (four risk groups). The RF (GLCM inverse): association with CAIX (hypoxia) using retrospective radiogenomics cohort of 103 surgically resected adenocarcinomas. Validation by IHC on 16 patients. |
| Del Re et al 2021135 | NSCLC | Advanced NSCLC treated with anti-PD1 n=32;. Radmioc analysis for 11 pts. |
N | CT | PFS | 25 RFs, exosomal mRNA expression of PD-L1 and IFN-γ, PD-L1 polymorphisms, TML. |
LASSO. 11-fold CV. |
Association with PD-L1. |
| Granata et al 2021136 | NSCLC | n=38 IO and 50 with chemo- or targeted therapy. No validation set. |
N | CT | OS, PFS | 573 RFs | LASSO, SVM, Tree-based methods. | OS (AUC 0.89, accuracy 81%). RFs to predict OS or PFS time were different between the control group and the IO group |
| Yang et al 2021137 | NSCLC | n=92. Tr=64, V=28. |
N | CT | DCB, PFS | 88 RFs | Random forest. | DCB (model 1): AUC 0.848 in Tr and 0.795 in V. PFS (model 2): AUC 0.717 in Tr and 0.760 in V. |
| Rundo et al 202167 | Urothelial | n=42 metastatic urothelial cancer. Tr 70%, V 30%. |
N | CT | OS | 3D deep radiomics. | 3D deep radiomics. | Acuracy 82.5%, SE 96%, Sp 60%. |
| Liu et al 202182 | NSCLC | n=197. 322 RECIST target lesions. Tr=137, V=60. |
Y | CT | Responders at 6 months. | Largest lesion (LL) model. Target lesion (TL) model: average RF of all lesions. |
mRMR (feature selection) and LASSO (model). | LL model and TL models performance where comparable. Baseline signatures performance were not significant Best model: TL-delta radiomics with clinical factor of distant metastasis, AUC=0.81 (95% CI 0.68 to 0.95). |
| Trebeschi et al 2021102 | NSCLC | 152 stage IV patients treated with nivolumab. 73 discovery, 79 test, 903 CTs. |
N | CT | 1 year OS from the last acquisition. | Chest CT morphological changes. | Deep learning. | Using CTs from the first 3–5 months of treatment: AUC of 0.69–0.75. Independent of clinical, radiological, PDL1, and histopathological factors. |
| Shen et al 2021138 | NSCLC | 63 patients. 72 lesions. No validation set. |
Y | CT | Lesion Progression | Texture features 3 Feature selection methods (Fisher, MI, POE+ACC) |
three classifiers evaluated (PCA, LDA, NDA) | Lesion-wise model of lesion progression Best model performance: AUC=0.812) |
| Yang et al 2021139 | NSCLC | n=200 patients. 1633 CTs. No independent validation set (cross-validation). |
Y | CT | 90-day responders. | Deep radiomics±clinical and biological features. | Deep learning model with simple temporal attention. | AUC for response prediction=0.80. The model was associated with OS and PFS. |
| Aoude et al 64 | Melanoma | 52 III/IV treated with BRAF inhibitors and/or immunotherapy. WES+RNAseq+immune signature. No validation set. |
N | PET | OS and PFS | Histogram features+MTV, SUV, TLG, extracted from largest lesion (node or metastasis). | Univariate analysis+optimal cut-offs analyses for survival. | High SD or high mean of MPP associated with PFS (p=0.00047 and p=0.0014) OS (0.0223, p=0.0389) CD8 expression p=0.0028. |
| Liu et al 2021140 | NSCLC | 46 IIB/IV NSCLC treated with nivolumab. No validation set (performance estimated by LOOCV). |
N | CT | OS and PFS | 1106 RFs from the largest tumor. | SVM, logistic regression, Gaussian Naïve Bayes. | AUC of the model 0.73 and 0.61 for PFS and OS. |
| Zerunian et al 141 | NSCLC | 21 pts treated with pembrolizumab. No validation. |
Y | CT | OS and PFS | TexRad features extracted from aggregation of VOIs. | Univariate analysis AUC and log-rank tests. |
Association of MPP and OS (HR=0.89). |
| Corino et al 202162 | HNSCC | 85 recurrent or metastatic pts treated with nivolumab. Tr=68, V=17 pts. |
N | CT | 10-month OS | 536 RFs from the largest tumor. | LASSO+SVM | AUC in validation set=0.67. Performance of radiomic score was higher than the one obtainable with clinical variables. |
| Chen et al 202163 | Melanoma | 50 patients. | N | CT | PD | Automated multi-objective delta-radiomics (Auto-MODR) – 2D largest lesion. |
497 RFs × 3 (pre+post +deltaRFs) from largest lesion. | AUC 86 in cross-validation and 0.73 in independent study. |
| Brendlin et al 2021142 | Melanoma | 140 stage IV pts. 776 lesions. Tr=70 pts V=70 pts. 1291 follow-up examinations (6533 lesions). |
Y | DECT. SECT. |
Lesion reponse. Patient response (PD vs CRPRSD). |
Pyradiomics features. Aggregation of lesion features. |
Feature selection. Multiple logistic regression. Random Forest. |
Patient response: AUC SECT=0.5, DECT=0.75; lesion response AUROC SECT=0.61, DECT=0.85; p<0.001. |
| Barabino et al 2022132 | NSCLC | 33 patients. 43 lesions delineated. No test – aggregation not described. |
Y | CT | PD, PR and SD. | 93 features extracted at baseline and first evaluation. | ANOVA | 27 delta radiomics features were associated with response (univariate). Nine features correlated with pseudoprogression. |
| Dercle et al 202281 | Melanoma | 575 patients. Tr=252, V=287. |
Y | CT | OS at the month 6 post-treatment. | Features extracted from the aggregation of lesions volumes into tumor burden. | 50 best features at baseline and 50 month 3 delta features. Random forest. |
Radiomics signature performed better than RECIST 1.1 with AUC for estimation of OS of 0.92 (95% CI 0.89 to 0.95) versus AUC=0.80 (95% CI 0.75 to 0.84). |
| Preclinical studies | ||||||||
| Mihaylov et al 202176 | Mice | 15 mice treated with RT (8 Gy × 3)+IO, 4 for control. Tr=6 mice, V=9 mice. 1 irradiated lesion and 1 non-irradiated. |
Y | CT MRI |
Response of a non-irradiated lesion (occurred in four mice). | 92 CT and 92 MRI radiomics features from both lesion. Lesion-level analysis to predict abscopal response occurence. |
ANOVA for feature selection, logistic regression for training. | Imaging model (either CT or MRI) combined with NLR achieved good performance to predict abscopal response (AUC close to 1, to be interpreted with caution due to the limited sample size). |
| Eresen et al 2021143 | Mice pancreatic cancer | 8 mice with dendritic cell vaccine+8 mice for control. | N | MRI | OS | 264 delta features. Feature selection using SVM (LOOCV) for identification of the treatment relative changes. |
Regression for OS prediction | Association of RFs with OS and histological tumor markers (fibrosis percentage, CK19+area, Ki67+cells). |
| Devkota et al 202098 | Mice | Xenograft tumors with or without MDSC and some mice treated with MDSC-targeting immunotherapy. | N | Nanoparticle contrast-enhanced CT, CT angiograms and T2w-MR. | Immunotherapy-treated group. | 107 RFs. | Univariate analysis (Kruskal-Wallis test) and Bonferroni correction. | Nano-radiomics revealed texture-based features capable of differentiating immune-treated tumors and untreated tumors. |
ANOVA, analysis of variance; AUC, area under the curve; CART, classification and regression trees; CI, confidence interval; CK19, cytokeratin 19; CR, complete response; DCB, durable clinical benefit; DECT, dual energy CT; GLCM, gray-level co-occurrence matrix; GLRLM, gray-level run-length matrix; HR, hazard ratio; ICI, immune checkpoint inhibitors; IFN, interferon; IHC, immunohistochemistry; IO, immuno-oncology; KNN, k nearest neighbors; LASSO, least absolute shrinkage and selection operator logistic regression model; LDA, linear discriminant analysis; LOOCV, leave-one-out cross validation; MDSC, myeloid-derived suppressor cells; MI, mutual information; MinCD8RS, Minimal value of the CD8 radiomic score; ML, machine learning; MPP, mean value of positive pixels; mRMR, minimum redundancy maximum relevance; mRNA, messenger ribonucleic acid; N, no; NDA, non-linear discriminant analysis; NK, natural killer cells; NLR, neutrophil-to-lymphocyte ratio; NSCLC, non-small cell lung cancer; OR, odds ratio; OS, overall survival; PCA, principal component analysis; PD-1, programmed death 1; PD, progressive disease; PD-L1, programmed death ligand 1; PFS, progression-free survival; POE+ACC, minimization of classification error probability combined average correlation coefficients; PR, partial response; RECIST, response evaluation criteria in solid tumours; RFs, radiomic features; RT, radiotherapy; SBRT, stereotactic body radiation therapy; SD, stable disease; SECT, single energy CT; SUVmax, maximum standardized uptake value; SVM, support vector machine; Te, test set; TIL, tumor-infiltrating lymphocyte; TLG, total lesion glycolysis; TMB, tumor mutational burden; TML, tumor mutational load; Tr, training set; TTP, time-to-progression; V, validation set; WES, whole exome sequencing; Y, yes.
Figure 1.
Overview of the different methodologies and strategies applicable for developing imaging biomarkers. Adapted from Sun et al Artificial intelligence, radiomics and pathomics to predict response and survival of patients treated with radiations. Cancer/Radiothérapie 2021; Volume 25 (Issues 6–7): 630–637. Copyright 2021 Elsevier Masson SAS. All rights reserved.
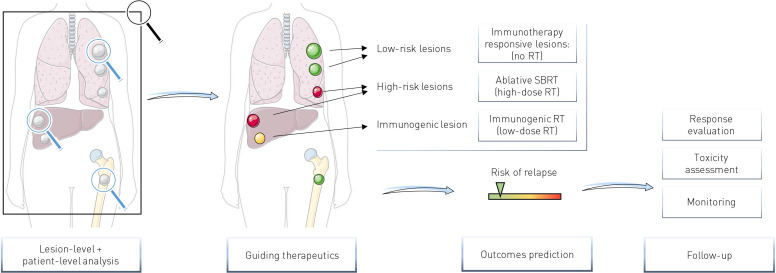
One of the major considerations for implementation of imaging biomarkers in radiotherapy and immunotherapy combinations in a metastatic context is whether they can be used to assess the spatial heterogeneity of the disease and more specifically the heterogeneity between lesions of a same patient to potentially guide radiotherapy on specific lesions (figure 2).
Figure 2.
Potential clinical interest of imaging biomarkers for radiotherapy-immunotherapy combinations. Lesion-level analyses may help to identify potential immune-refractory lesions (high-risk lesions) needing focal destruction with ablative SBRT (high dose RT) or immunogenic lesions for which low dose RT may improve systemic response. Patient-level analyses allow overall response prediction. RT, radiotherapy; SBRT, stereotactic body radiation therapy.
To the best of our knowledge, only two studies have evaluated radiomics to predict response of metastatic cancer patients treated with immunotherapy combined with radiotherapy.73 74 Interestingly, each of these independently evaluated the previously published radiomics signature predicting CD8-T cells (CD8-Rscore) developed by our team.69 This CD8-Rscore was initially developed using contrast enhanced CT scans of 135 patients for whom RNA sequencing data of a biopsied lesion were available and allowed estimation of the abundance of CD8 T cells. This signature, based on eight variables including five radiomics features extracted from the lesion analyzed and a peripheral ring around the lesion, two variables of VOI location and one technical variable (the kiloVoltage Peak), was validated using a Cancer Genome Atlas dataset of 119 patients (area under the curve (AUC)=0.67; 95% CI 0.57 to 0.77; p=0.0019)69 and was also associated with clinical response and overall survival of 137 patients treated with immunotherapy (HR=0.58, 95% CI 0.39 to 0.87; p=0.0081). However, in the primary study, only one lesion per patient was analyzed for predicting outcomes.69 In ref 73, we applied the CD8-Rscore on multiple lesions on the baseline CT scan of 94 solid cancer patients treated with immunotherapy and radiotherapy. Almost all patients underwent hypofractionated radiotherapy (n=91 (96.9%)). The main radiotherapy dose-fractionation schedule was 3×8 Gy delivered on a single lesion. Seven patients (7.4%) had more than one irradiated lesion. The CD8-Rscore was applied on a total of 100 irradiated lesions and 189 non-irradiated lesions. At a lesion level, the CD8-Rscore at baseline of responding lesions was higher than those of stable or progressive lesions (AUC=0.63, 95% CI 0.56 to 0.71), p=0.0020). To assess whether the CD8-Rscore of multiple lesions may be aggregated to predict outcomes at a patient level, several metrics consisting of the mean value of the CD8-Rscores, its maximal and the minimal values, the SD, and the entropy of the distribution of the CD8-Rscores have been evaluated, using median value to dichotomize two groups of patients. In this study, the entropy of the distribution of the CD8-Rscores was associated with patient progression-free survival (PFS) (HR=1.67, p=0.040), out-of-field abscopal response (AUC=0.70, p=0.014) and overall survival (HR=2.08, p=0.023). The minimal value of the CD8-Rscores, representing the lesion considered to be the least infiltrated by CD8 T cells by the radiomics signature, was also interesting with a statistical trend identified with PFS (p=0.07). Significance was reached when looking at the extreme values (first vs third quartile, HR=0.34,95% CI 0.17 to 0.68, p=0.0021). This last metric has the advantage to allow direct designation of the most pejorative lesion in a non-invasive manner, which are more likely to be non-responsive to immunotherapy and eventually may benefit from ablative targeted radiotherapy (high dose radiotherapy).
Korpics et al 74 also evaluated the CD8-Rscore in a cohort of 68 patients with advanced solid cancer patients treated with multisite stereotactic body radiation therapy (SBRT) and pembrolizumab. Two to four metastases per patient were irradiated, with a prescription dose of 30–50 Gy in 3–5 fractions. A total of 139 metastases were treated with SBRT and evaluated using the CD8-Rscore. Low pretreatment CD8-Rscore was defined as a CD8-Rscore lower than the 25% percentile in their study. The average of the CD8-Rscores of irradiated lesions of each patient was used to evaluate the clinical outcomes at a patient-level. High CD8-Rscore was associated with improved lesion control (HR=0.18; 95% CI 0.04 to 0.74; p=0.018). At a patient-level, high scores were correlated with better PFS (HR 0.47; 95% CI 0.26 to 0.85; p=0.013) and OS (HR=0.39; 95% CI 0.20 to 0.75; p=0.005). As a continuous variable, higher scores were also associated with improved PFS (HR=0.12; 95% CI 0.03 to 0.51; p=0.004) but did not reach significance for overall survival and local control.74 Overall, although these two studies evaluating the CD8-Rscore were retrospective and need prospective validation, they illustrate the potential of radiomics for lesion response and patient outcomes prediction in the context of radiotherapy and immunotherapy combinations. However, it remains unclear whether this type of signature, developed in pan-cancer cohorts, can be as effective in the case of homogeneous cohort of patients with specific tumor type. Indeed, tumor type might be a confounding factor since tumor immune infiltrate may vary according to tumor types.75 The development of tumor-specific signatures could therefore improve the performance of these models.
Regarding the prediction of out-of-field ‘abscopal’ response, a preclinical study evaluated CT and MRI radiomics on Lewis Lung Carcinoma in a syngeneic, subcutaneous murine model.76 Tumors were implanted on both flanks of 19 mice. Fifteen mice received radiotherapy on the right flank (3×8 Gy) followed by immunotherapy (anti PD-1), and four mice presented abscopal response on the left flank. A total of 92 CT and 92 MRI pretreatment radiomics features were extracted from each lesion (right and left flanks). Using six mice (12 tumors) for training and nine mice (18 tumors for validation), the authors identified some pretreatment CT and MRI features and a biological variable, the neutrophil-to-lymphocyte ratio, that were associated with abscopal response. This proof-of-concept study, although very promising, must be interpreted with caution due to the low number of mice and events (occurrence of abscopal response).76 However, it suggests that radiomics may help to identify lesions favorable to the occurrence of an abscopal response. Such biomarkers would be of particular interest to guide potential low-dose immunogenic radiotherapy to improve systemic responses.29 30
It is interesting to note that, vice versa, these imaging tools may help to indicate lesions—and patients—with a high chance of response under immunotherapy and not requiring radiotherapy. This is of particular interest in order to optimize and limit radiotherapy as parsimoniously as possible. Indeed, side effects of radiotherapy have to be taken into account, in particular lymphopenia, which may decrease the response to immunotherapy.77 In the same spirit, analyses of intratumor heterogeneity could also be of interest for the partial irradiation of tumors, which is also an approach being explored in the combination of radiotherapy and immunotherapy. Moreover, computational imaging approaches have also shown promising results for intratumor heterogeneity assessment.78 79 This could be key for guiding partial tumor irradiation, which is also an approach being explored in radioimmunotherapy with the concept of ‘adscopal effect’, the immune mediated effect of radiotherapy close to the irradiated field.80
Spatial heterogeneity assessment: the key to allow characterization and targeting of lesions
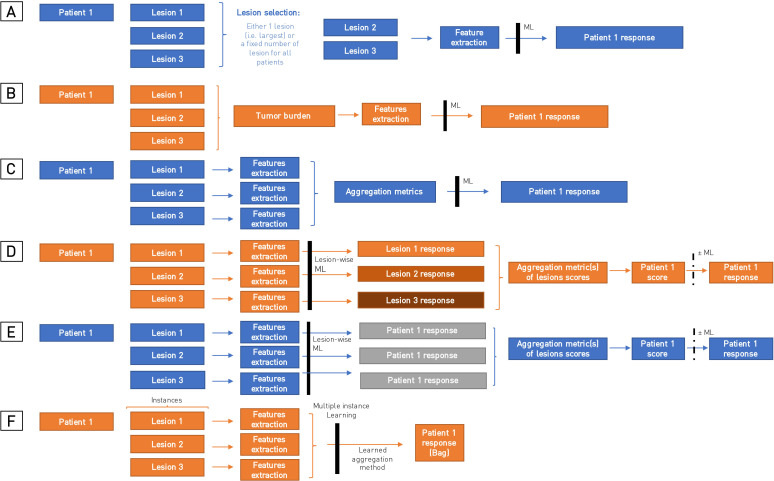
Aggregating spatial heterogeneity from predictions made at a lesion level to perform predictions at a patient level is particularly challenging, and no consensual method is clearly defined (figure 3). Indeed, such models need to take into account the possibility for each patient to have different numbers of lesions and to be robust to variation in lesion selection if exhaustive delineation is not performed. This may explain why the majority of radiomics studies only analyzed a single lesion to predict outcomes of patients (table 2). Some relatively simple ways to analyze several lesions are the aggregation of either all the lesions delineated into one volume corresponding to the tumor burden before the extraction of features,81 or the aggregation of the features extracted from several lesions using metrics such as the average value, the minimal or the maximal value of the features, irrespective of the lesion from which the value was extracted.65 82 However, although they have shown very interesting results to predict response or survival of patients treated with immunotherapy, these methods do not allow evaluation of the aggressiveness or the immunogenicity of a particular lesion, which is determinant in order to identify a possible target for destruction in radiotherapy (cooperative effect of radiotherapy) or to promote an abscopal response (synergistic effect of radiotherapy). Similarly, some authors have trained a lesion-wise radiomics model to predict patient response by assigning the same patient response to each different lesions.66 This, however, does not take into account the possibility of dissociated response, with presence of both responding and progressive lesions.
Figure 3.
Summary of different aggregating methods for multiple lesion analyses. (A) Defining a fixed number of a lesion to analyze for each patient. (B) Aggregating lesions into one volume (tumor burden). (C) Aggregating features extracted from several lesions (ie, using metrics such as the average value or the minimal value). (D) Predicting lesion-level response then aggregating the predictions to assess patient outcomes. (E and F) Assigning for each lesion the patient outcome to predict then using predefined aggregation metrics (E) or learned aggregation methods (attention) in multiple-instance learning approaches (F).
Trebeschi et al 56 have applied a very interesting method to address these challenges. In a study analyzing 1055 primary and metastatic lesions from 203 patients with advanced melanoma and NSCLC treated with anti-PD1, they trained a lesion-wide model predicting lesion progression. Best performances were obtained with NSCLC pulmonary and nodal metastases (0.83 AUC, Mann-Whitney U test p<0.001 and 0.78 AUC, p<0.001, respectively). By combining predictions made on individual lesions, this method allowed identification of patients with at least one predicted progressive lesion. The patients experiencing either a uniform progressive disease or a dissociated response had lower overall survival than patients with a uniform response (0.76 AUC for all patients).56 This study highlighted the potential of imaging to identify high-risk lesions for predicting survival of patient treated with immunotherapy.
Only few studies have compared the performance of different aggregation methods for multiple lesions.73 82 83 Liu et al compared the predictive value of radiomic features extracted from the largest lesion (LL model) with the average of features extracted from several target lesions defined according to RECIST V.1.184 (TL model) with a maximum of five lesions analyzed, at baseline (pretreatment), as well as the relative change of these features on the follow-up CT (delta-radiomics), in a cohort of 197 NSCLC patients (and 322 target lesions) treated with anti-PD1 immunotherapy. LL and TL models seemed comparable in their study, with good performance when using delta-radiomics to predict responders at 6 months; however, none of the methods showed significant predictive value when using only the baseline features.82 Chang et al evaluated six different aggregating methods of radiomics features extracted from brain metastases: ‘Unweighted average’ of radiomic features (except for the size and shape features which were summed), ‘Weighted average’ of radiomic features according to the total volume of all metastases, ‘Weighted average of largest three metastases’, ‘Largest+number of metastases’, and ‘Largest metastasis and Smallest metastasis’.83 The ‘volume-weighted average of the largest three tumors’ model had the best performance for predicting patient survival with C-index of 0.627 (0.595–0.661), 0.628 (0.591–0.666), and 0.652 (0.565–0.727) for Cox proportional hazards model, LASSO and random forest models, respectively. In our study evaluating the CD8-Rscores in patients treated with radiotherapy and immunotherapy, we have shown that entropy of the distribution of the score and the least score were metrics to assess patient outcomes, while average or maximal score were not.73 Interestingly, Van den Eynde et al have shown similar results with an immunohistochemistry and digital pathology-based scoring system of lymphocyte populations (CD3 and CD8), the Immunoscore, in metastatic colorectal cancer patients.85 86 They evaluated the Immunoscore in 603 resected metastases from 222 patients. The value of the least infiltrated lesion had a stronger association with patient outcomes than the most infiltrated lesion or the average value of the different lesions of a patient.
Evaluating spatial heterogeneity also raises questions of the effect of the number of lesions chosen for analysis, as well as the minimal number of lesions to analyze, in order to obtain maximum information regarding disease heterogeneity, and whether a comprehensive analysis of all lesions is mandatory. These questions have been studied particularly for the number of lesions to be measured in radiology87 but remains pending for radiomic analyses.
Finally, advanced machine learning approaches such as deep learning and multiple instance learning (MIL) may be key for spatial heterogeneity assessment.88–90 Unlike the methods described previously where one class label is assigned to each observation or ‘instance’, in MIL, class labels are assigned to a ‘bag of instance’, which is a collection of observations. Here, the bag of instance could be the patient global response, where the instances would be the different lesions. The challenge of the MIL approach is to determine which instance(s) influence the overall bag label. This approach is particularly interesting since obtaining the annotation and labels of each lesion is difficult and time-consuming, while patient response label is easier to obtain.
Other promising immunotherapy radiomics biomarker
Radiomics biomarkers have shown promising results to predict several biological and histological features that can be used as a rationale for characterizing the patient’s disease and developing radiomic scores predictive of response to high or low doses of radiotherapy. A study used a deep learning method in a cohort of 327 patients with NSCLC to predict tumor mutational burden (TMB) (AUC=0.81, 95% CI 0.77 to 0.85) in the test cohort).91 This radiomic signature of TMB was associated with overall survival, in an independent cohort of NSCLC patients treated with immunotherapy (OS: HR=0.54; 95% CI 0.31 to 0.95; p=0.030, and PFS: HR=1.78; 95% CI 1.07 to 2.95; p=0.023).91
Tunali et al 92 have developed a radiomic signature predicting overall survival of NSCLC patients treated with immunotherapy using a large cohort of 180 patients for training, and two cohorts of 90 and 62 patients for validation. Interestingly, the signature they identified, allowing discrimination of four risk groups was based on one radiomic feature: gray level co-occurrence matrix inverse difference, which has been associated with the expression of CAIX, an hypoxia-related gene in their analyses.92
Other radiomic studies, although conducted without immunotherapy, have also shown interesting results in large cohorts for the prediction of lymphocyte infiltration according to the Immunoscore on MRI images,71 93 PD-L1 on either CT94 95 or PET images,70 or of microsatellite instability on CT or MRI images96 97 and may also show promise for the prediction of response to immunotherapy. The diversity of biological mechanisms that can be assessed by radiomics is of definite interest for the development of new immunotherapy combinations such as myeloid-derived suppressor cells (MDSC)-targeting immunotherapy.98
While both biologically driven radiomic signatures (ie, predicting TILs,69 PD-L199 or TMB91) and radiomic signatures directly designed to predict patient outcomes from images seem to achieve good performance to predict response to immunotherapy (table 1), both strategies have their advantages and disadvantages. Using radiomics to estimate a biological intermediate biomarker allows to bring a rational to the interpretation of the patient’s outcomes predictions, which makes the signature appearing less like a ‘black box’. However, the final performance on the output of clinical interest, for example, OS or PFS, may be limited by this two-step process and the associated accumulation of errors. Radiomic signatures trained without any biological assumptions are less interpretable but may help to discover new mechanisms and to generate new hypotheses. Thus, some radiomic signatures have shown to be independent from known biological biomarkers, which paves the way for potential multimodal signatures integrating genomics and TMB, pathomics, biology and/or metabolomics.95 100 101 Trebeschi et al 102 have developed an AI-derived score predicting 1-year survival of advanced NSCLC patients treated with nivolumab from morphological changes on chest CT acquired during patient’s follow-up (73 patients in the training set, 79 in the test set). This showed that their tool outperformed PD-L1 expression (>1%) and volumetric changes of tumor burden in a multivariate analysis; however, it was only evaluable for 22 patients. Khorrami et al 58 have shown in a similar population that a delta-radiomic feature combined with PD-L1 was better than PD-L1 alone to predict patient overall survival. Finally, Luo et al have shown in a cohort of 247 NSCLC patients treated with immunotherapy that a multimodal signature integrating radiology, pathology and genomic data was able to predict response to anti-PD-1 (AUC=0.80, 95% CI 0.74 to 0.86) and outperformed unimodal radiomic model (0.65, 95% CI [0.57 to 0.73), PD-L1 (AUC=0.73, 95% CI 0.65 to 0.81), and TMB (AUC=0.61, 95% CI 0.52 to 0.70).103
Despite this, a low level of evidence for radiomic studies remains, with many studies often performed in small numbers and some without independent testing cohorts.60 61 72 104 To date, only one study has a prospective testing cohort,59 and only the CD8-Rscore has had testing by an external team.74 Thus, further research still remains, prior to implementing strategies based on radiomics into clinical practice. Standardization of imaging protocols and prospective clinical trials will be key.
Contribution of functional imaging
Many functional and molecular imaging techniques have been developed in recent years with promising applications for radiotherapy and immunotherapy combinations.105 106
Functional images have the advantage of directly evaluating cellular functions or metabolic pathways, while machine learning approaches can only seek statistical associations of these processes with imaging patterns.
Immuno-PET is the combination of ultrasensitive functional imaging, PET imaging and the high affinity and specificity of monoclonal antibodies or ligands based on monoclonal antibodies. This approach allows detection, quantification and longitudinal monitoring of specific immune receptors or cells. Several studies have shown promising PET-imaging tracers allowing detection of PD1 expression such as the 64Cu-DOTA-PD-1,107 or PD-L1 expression such as the 89Zr-C4.108 Combined with radiotherapy, these approaches have helped increase knowledge by allowing evaluation of changes induced after radiation.109–112 For instance, several immune-PET studies have illustrated radiotherapy-induced PD-L1 upregulation on tumor cells.110–112 Christensen et al evaluated an 89Zr radiolabeled anti-PD-L1 immuno-PET tracer, the 89Zr-DFO-6E11, in tumor-bearing mice treated with radiotherapy with and without antimouse PD-L1 immunotherapy or with antimouse PD-L1 immunotherapy alone. The PET-CT imaging was performed in all mice and 72 hours after the selective irradiation of the tumor (2 Gy × 3) for irradiated mice. In their study, radiotherapy+anti-PD-L1 and ant-PD-L1 alone resulted in significant tumor control compared with the control group and RT alone. They reported that the 89Zr-DFO-6E11 allowed objective quantification of PD-L1 in tumors and spleens of irradiated mice. Moreover, the maximum 89Zr-DFO-6E11 tumor-to-muscle ratio was associated with treatment response (p=0.025). Several early-phase clinical trials have confirmed the potential of immuno-PET targeting PD-L1 approaches for immunotherapy with different promising tracers113–115: the 89Zr-atezolizumab,98 the 89Zr-CX-072, which is a PD-L1 targeting probody therapeutic, which is engineered to be activated in the tumor microenvironment by tumor-associated proteases,114 and the 89Zr-durvalumab.115
Immuno-PET approaches have also shown interesting results to monitor tumor-infiltrating CD8 T cells.116 117 Farwell et al 117 evaluated an anti-CD8 radiolabeled minibody, 89Zr-Df-IAB22M2C, in a phase 1 trial including 15 patients with metastatic solid tumors treated with immunotherapy (n=8), targeted therapy (n=2) and 5 treatment-naïve patients. Radiotracer uptake in tumors occurred in 10 patients, seven of whom were receiving immunotherapy. A post-treatment increase of the 89Zr-Df-IAB22M2C uptake in tumor lesions occurred in three patients treated with immunotherapy and was correlated with response, which is very promising for immunotherapy early response prediction. Whether the 89Zr-Df-IAB22M2C may characterize the impact of radiotherapy on CD8 T cell infiltration within irradiated and non-irradiated tumor lesions in patients with polymetastatic solid cancers is evaluated in an ongoing clinical trial (ABSCOTEP trial, EudraCT: 2016-000665-23).
Overall, these functional imaging approaches are very promising for immunotherapy-radiotherapy combinations and will surely help to better understand determinant of abscopal response and to identify the best targets for radiotherapy. However, these approaches do have limitations such as the difficult access to the tracers that may impact the repeatability of these exams, the need for standardization to allow reproducibility, and the need to take into account the tracer biological distribution, especially in certain normal lymphoid tissues such as spleen and nodes.110 112
Imaging to assess response to radioimmunotherapy
Addition of radiotherapy to immunotherapy has raised specific questions for response assessment.
The term ‘abscopal effect’ has been introduced to describe an immune-mediated response of lesions occurring after radiotherapy outside the radiation field.118 While rare abscopal responses have been described with radiotherapy alone, immunotherapy has been used to try to enhance the systemic effect of radiotherapy. However, the proper systemic benefit of radiotherapy added to the direct effect of immunotherapy is difficult to evaluate outside of a controlled trial. Moreover, definition of abscopal effect varies between studies. Some have considered as abscopal response, a reduction in the size of one non-irradiated metastasis by ≥30%,119–121 while others have considered out-of-field response using aggregate diameter of non-irradiated target lesions, in accordance with RECIST 1.1 criteria (‘best out-of-field (abscopal) response rate’).6 122 123 To the best of our knowledge, only few data comparing both definitions are published. In our cohort of 94 patients treated with immunotherapy and radiotherapy, 15 patients (16.0%) presented an out-of-field response according to RECIST 1.1 criteria, while nine patients had at least one non-irradiated responding lesion but the aggregated change in diameter of all the non-irradiated lesions did not reach the 30% threshold. Overall survival of patients with ‘RECIST-negative’ out-of-field response was not significantly different than survival of patients with no responding out-of-field lesion, while ‘RECIST-compliant’ out-of-field patients had significantly better overall survival (p=0.039).73 This result therefore encourages staying with the RECIST 1.1 criterion for out-of-field response assessment, which considers the evaluation of patient response on non-irradiated targets.124 To note, immunotherapy has also brought new patterns of response, such as pseudoprogression, dissociated response, and hyperprogression,125 126 which led to the introduction of additional response assessment criteria such as Immune related Response Criteria,127 immune RECIST,128 immune modified RECIST,129 and immune-related RECIST.130
Interestingly, the concern of evaluating treated and untreated lesions has also been raised with intratumoral immunotherapy injections. The itRECIST criteria proposed to assess injected and non-injected lesions response separately, with the possible definition of target and non-target lesions in both injected and non-injected lesions.131 Sum of diameters (SOD) is performed on target-injected lesions and target non-injected lesions separately, then a combined SOD includes all target lesions (injected and non-injected).131 The relevance of such strategies for radiotherapy and immunotherapy combination does not seem clear yet might be worth investigating.
Use of machine learning and functional imaging for early response assessment might also be useful to predict patient outcomes and discrimination of progression and pseudoprogression.81 82 117 132 Delta-radiomics, which considers the relative changes of radiomic features between two time points, might help to improve the assessment of treatment response. Dercle et al 81 have shown in a study analyzing 575 melanoma patients from the KEYNOTE-002 and KEYNOTE-006 studies that a radiomics signature taking into account changes in tumor imaging phenotype at month 3, performed better than RECIST 1.1 criteria for discriminating patients overall survival (AUC=0.92, 95% CI 0.89 to 0.95 vs 0.80, 95% CI 0.75 to 0.84).81
Radiomics analysis of follow-up images have also shown interesting results for the assessment of radiotherapy-immunotherapy toxicity.133 Cheng et al 113 have developed a radiomics signature to differentiate immune checkpoint inhibitor-related pneumonitis (CIP) and radiation pneumonitis (RP) on CT scan images. They trained their model using CT images of patients with pneumonitis while treated with ICI (n=28) or radiotherapy only (n=31) and tested their model in a cohort of 14 patients treated with ICI+radiotherapy. Performance of the best-trained model in the test set was promising with an AUC=0.896, although further prospective validation is needed.
Finally, another challenge will be to assess whether imaging biomarkers may help to monitor patients undergoing treatment. Indeed, the development of imaging biomarkers has particularly focused on the prediction of response using pretreatment scans, with or without the use of first evaluation images (ie, with delta-radiomics). However, no study has assessed whether these tools are still effective in predicting patient responses or variations in the tumor microenvironment during treatment over time. Development of automatic segmentation tools and deep learning will probably be key to automate such analyses.
Conclusion
While radioimmunotherapy combinations seem very promising for improving patients’ outcomes, novel performant biomarkers are needed to identify patients who are most likely to benefit from these treatments and to help clinicians choose the optimal modalities of both radiotherapy and immunotherapy. Imaging biomarkers have shown promising results to characterize the whole disease with its spatial heterogeneity, which eventually may guide different radioimmunotherapy strategies such as a synergistic immunogenic low-dose radiotherapy to improve systemic effects, or a complementary preemptive ablative high-dose radiotherapy to destroy selective lesions at high risk of progression under immunotherapy. Further efforts and randomized prospective studies are needed to validate such strategies, but imaging biomarkers will surely be key for development of precision medicine in immunotherapy and radioimmunotherapy combinations and monitoring of patients.
Footnotes
Twitter: @rgrsun, @lescientifiik
Contributors: Conception and drafting of the manuscript: RS and ED; critical revision of the manuscript: all authors. Final approval of manuscript: all authors.
Funding: Fondation pour la Recherche Médicale (grant DIC20161236437), Fondation ARC pour la recherche contre le cancer (grant SIGN'IT20181007805), SIRIC-SOCRATE 2.0 (grant INCa-DGOS-INSERM_12551) and Amazon AWS grant provided funding to INSERM U1030 Radiomics team - Gustave Roussy. Fondation ARC pour la recherche contre le cancer and Paris-Saclay University provided support to RS with a grant for international mobility. RS received support from Fondation BETTENCOURT-SCHUELLER and Ecole INSERM.
Competing interests: ED has declared consulting fees and support from Roche, BMS, Boehringer, Astrazeneca, Lilly Amgen, and Merck-Serono. The remaining authors have no conflicts of interest to declare that are relevant to the content of this article.
Provenance and peer review: Not commissioned; externally peer reviewed.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1. Twyman-Saint Victor C, Rech AJ, Maity A, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 2015;520:373–7. 10.1038/nature14292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Theurich S, Rothschild SI, Hoffmann M, et al. Local tumor treatment in combination with systemic ipilimumab immunotherapy prolongs overall survival in patients with advanced malignant melanoma. Cancer Immunol Res 2016;4:744–54. 10.1158/2326-6066.CIR-15-0156 [DOI] [PubMed] [Google Scholar]
- 3. McBride S, Sherman E, Tsai CJ, et al. Randomized phase II trial of nivolumab with stereotactic body radiotherapy versus nivolumab alone in metastatic head and neck squamous cell carcinoma. J Clin Oncol 2021;39:30–7. 10.1200/JCO.20.00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schoenfeld JD, Giobbie-Hurder A, Ranasinghe S, et al. Durvalumab plus tremelimumab alone or in combination with low-dose or hypofractionated radiotherapy in metastatic non-small-cell lung cancer refractory to previous PD(L)-1 therapy: an open-label, multicentre, randomised, phase 2 trial. Lancet Oncol 2022;23:279–91. 10.1016/S1470-2045(21)00658-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Theelen WSME, Peulen HMU, Lalezari F, et al. Effect of pembrolizumab after stereotactic body radiotherapy vs pembrolizumab alone on tumor response in patients with advanced non-small cell lung cancer: results of the PEMBRO-RT phase 2 randomized clinical trial. JAMA Oncol 2019;5:1276–82. 10.1001/jamaoncol.2019.1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Welsh J, Menon H, Chen D, et al. Pembrolizumab with or without radiation therapy for metastatic non-small cell lung cancer: a randomized phase I/II trial. J Immunother Cancer 2020;8:e001001. 10.1136/jitc-2020-001001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Theelen WSME, Chen D, Verma V, et al. Pembrolizumab with or without radiotherapy for metastatic non-small-cell lung cancer: a pooled analysis of two randomised trials. Lancet Respir Med 2021;9:467–75. 10.1016/S2213-2600(20)30391-X [DOI] [PubMed] [Google Scholar]
- 8. Demaria S, Ng B, Devitt ML, et al. Ionizing radiation inhibition of distant untreated tumors (abscopal effect) is immune mediated. Int J Radiat Oncol Biol Phys 2004;58:862–70. 10.1016/j.ijrobp.2003.09.012 [DOI] [PubMed] [Google Scholar]
- 9. Bockel S, Antoni D, Deutsch É, et al. [Immunotherapy and radiotherapy]. Cancer Radiother 2017;21:244–55. 10.1016/j.canrad.2016.12.005 [DOI] [PubMed] [Google Scholar]
- 10. Weichselbaum RR, Liang H, Deng L, et al. Radiotherapy and immunotherapy: a beneficial liaison? Nat Rev Clin Oncol 2017;14:365–79. 10.1038/nrclinonc.2016.211 [DOI] [PubMed] [Google Scholar]
- 11. Herrera FG, Irving M, Kandalaft LE, et al. Rational combinations of immunotherapy with radiotherapy in ovarian cancer. Lancet Oncol 2019;20:e417–33. 10.1016/S1470-2045(19)30401-2 [DOI] [PubMed] [Google Scholar]
- 12. Mondini M, Levy A, Meziani L, et al. Radiotherapy-immunotherapy combinations - perspectives and challenges. Mol Oncol 2020;14:1529–37. 10.1002/1878-0261.12658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sato H, Okonogi N, Nakano T. Rationale of combination of anti-PD-1/PD-L1 antibody therapy and radiotherapy for cancer treatment. Int J Clin Oncol 2020;25:801–9. 10.1007/s10147-020-01666-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reynders K, Illidge T, Siva S, et al. The abscopal effect of local radiotherapy: using immunotherapy to make a rare event clinically relevant. Cancer Treat Rev 2015;41:503–10. 10.1016/j.ctrv.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen M-F, Chen P-T, Chen W-C, et al. The role of PD-L1 in the radiation response and prognosis for esophageal squamous cell carcinoma related to IL-6 and T-cell immunosuppression. Oncotarget 2016;7:7913–24. 10.18632/oncotarget.6861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sato H, Niimi A, Yasuhara T, et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat Commun 2017;8:8. 10.1038/s41467-017-01883-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Antonia SJ, Villegas A, Daniel D, et al. Overall survival with durvalumab after chemoradiotherapy in stage III NSCLC. N Engl J Med 2018;379:2342–50. 10.1056/NEJMoa1809697 [DOI] [PubMed] [Google Scholar]
- 18. Kwon ED, Drake CG, Scher HI, et al. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol 2014;15:700–12. 10.1016/S1470-2045(14)70189-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhu X, Cao Y, Liu W, et al. Stereotactic body radiotherapy plus pembrolizumab and trametinib versus stereotactic body radiotherapy plus gemcitabine for locally recurrent pancreatic cancer after surgical resection: an open-label, randomised, controlled, phase 2 trial. Lancet Oncol 2022;23:e105–15. 10.1016/S1470-2045(22)00066-3 [DOI] [PubMed] [Google Scholar]
- 20. Lee NY, Ferris RL, Psyrri A, et al. Avelumab plus standard-of-care chemoradiotherapy versus chemoradiotherapy alone in patients with locally advanced squamous cell carcinoma of the head and neck: a randomised, double-blind, placebo-controlled, multicentre, phase 3 trial. Lancet Oncol 2021;22:450–62. 10.1016/S1470-2045(20)30737-3 [DOI] [PubMed] [Google Scholar]
- 21. Brooks ED, Chang JY. Time to abandon single-site irradiation for inducing abscopal effects. Nat Rev Clin Oncol 2019;16:123–35. 10.1038/s41571-018-0119-7 [DOI] [PubMed] [Google Scholar]
- 22. Deutsch E, Chargari C, Galluzzi L, et al. Optimising efficacy and reducing toxicity of anticancer radioimmunotherapy. Lancet Oncol 2019;20:e452–63. 10.1016/S1470-2045(19)30171-8 [DOI] [PubMed] [Google Scholar]
- 23. Wei J, Montalvo-Ortiz W, Yu L, et al. Sequence of αPD-1 relative to local tumor irradiation determines the induction of abscopal antitumor immune responses. Sci Immunol 2021;6:eabg0117. 10.1126/sciimmunol.abg0117 [DOI] [PubMed] [Google Scholar]
- 24. Dewan MZ, Galloway AE, Kawashima N, et al. Fractionated but not single-dose radiotherapy induces an immune-mediated abscopal effect when combined with anti-CTLA-4 antibody. Clin Cancer Res 2009;15:5379–88. 10.1158/1078-0432.CCR-09-0265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verbrugge I, Hagekyriakou J, Sharp LL, et al. Radiotherapy increases the permissiveness of established mammary tumors to rejection by immunomodulatory antibodies. Cancer Res 2012;72:3163–74. 10.1158/0008-5472.CAN-12-0210 [DOI] [PubMed] [Google Scholar]
- 26. Filatenkov A, Baker J, Mueller AMS, et al. Ablative tumor radiation can change the tumor immune cell microenvironment to induce durable complete remissions. Clin Cancer Res 2015;21:3727–39. 10.1158/1078-0432.CCR-14-2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee Y, Auh SL, Wang Y, et al. Therapeutic effects of ablative radiation on local tumor require CD8+ T cells: changing strategies for cancer treatment. Blood 2009;114:589–95. 10.1182/blood-2009-02-206870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vanpouille-Box C, Alard A, Aryankalayil MJ, et al. DNA exonuclease TREX1 regulates radiotherapy-induced tumour immunogenicity. Nat Commun 2017;8:15618. 10.1038/ncomms15618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patel RR, Verma V, Barsoumian H. Use of multi-site radiation therapy as systemic therapy: a new treatment approach personalized by patient immune status. Int J Radiat Oncol Biol Phys 2020:S0360301620341146. [Google Scholar]
- 30. Herrera FG, Ronet C, Ochoa de Olza M, et al. Low-Dose radiotherapy reverses tumor immune desertification and resistance to immunotherapy. Cancer Discov 2022;12:108–33. 10.1158/2159-8290.CD-21-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Frey B, Rückert M, Deloch L, et al. Immunomodulation by ionizing radiation-impact for design of radio-immunotherapies and for treatment of inflammatory diseases. Immunol Rev 2017;280:231–48. 10.1111/imr.12572 [DOI] [PubMed] [Google Scholar]
- 32. Rückert M, Deloch L, Fietkau R, et al. Immune modulatory effects of radiotherapy as basis for well-reasoned radioimmunotherapies. Strahlenther Onkol 2018;194:509–19. 10.1007/s00066-018-1287-1 [DOI] [PubMed] [Google Scholar]
- 33. Limkin EJ, Sun R, Dercle L, et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol 2017;28:1191–206. 10.1093/annonc/mdx034 [DOI] [PubMed] [Google Scholar]
- 34. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–92. 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun R, Limkin EJ, Dercle L, et al. Imagerie médicale computationnelle (radiomique) et potentiel en immuno-oncologie. Cancer/Radiothérapie 2017;21:648–54. 10.1016/j.canrad.2017.07.035 [DOI] [PubMed] [Google Scholar]
- 36. Tang X. The role of artificial intelligence in medical imaging research. BJR|Open 2020;2:20190031. 10.1259/bjro.20190031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gillies RJ, Anderson AR, Gatenby RA, et al. The biology underlying molecular imaging in oncology: from genome to anatome and back again. Clin Radiol 2010;65:517–21. 10.1016/j.crad.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dercle L, Henry T, Carré A, et al. Reinventing radiation therapy with machine learning and imaging bio-markers (radiomics): state-of-the-art, challenges and perspectives. Methods 2021;188:44–60. 10.1016/j.ymeth.2020.07.003 [DOI] [PubMed] [Google Scholar]
- 39. Nioche C, Orlhac F, Boughdad S. A freeware for tumor heterogeneity characterization in PET, SPECT, CT, MRI and US to accelerate advances in radiomics. J Nucl Med 2017;58:1316. [Google Scholar]
- 40. van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017;77:e104–7. 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. LeCun Y, Bengio Y. Convolutional networks for images, speech, and time-series. In: The handbook of brain theory and neural networks. MIT Press, 1995. [Google Scholar]
- 42. Reuzé S, Schernberg A, Orlhac F, et al. Radiomics in nuclear medicine applied to radiation therapy: methods, pitfalls, and challenges. Int J Radiat Oncol Biol Phys 2018;102:1117–42. 10.1016/j.ijrobp.2018.05.022 [DOI] [PubMed] [Google Scholar]
- 43. Orlhac F, Frouin F, Nioche C, et al. Validation of a method to compensate multicenter effects affecting CT Radiomics. Radiology 2019;291:53–9. 10.1148/radiol.2019182023 [DOI] [PubMed] [Google Scholar]
- 44. Welch ML, McIntosh C, Haibe-Kains B, et al. Vulnerabilities of radiomic signature development: the need for safeguards. Radiother Oncol 2019;130:2–9. 10.1016/j.radonc.2018.10.027 [DOI] [PubMed] [Google Scholar]
- 45. Fornacon-Wood I, Faivre-Finn C, O’Connor JPB, et al. Radiomics as a personalized medicine tool in lung cancer: separating the hope from the hype. Lung Cancer 2020;146:197–208. 10.1016/j.lungcan.2020.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fournier L, Costaridou L, Bidaut L, et al. Incorporating radiomics into clinical trials: expert consensus endorsed by the European Society of Radiology on considerations for data-driven compared to biologically driven quantitative biomarkers. Eur Radiol 2021;31:6001–12. 10.1007/s00330-020-07598-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yip SSF, Aerts HJWL. Applications and limitations of radiomics. Phys Med Biol 2016;61:R150–66. 10.1088/0031-9155/61/13/R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ellingson BM, Bendszus M, Boxerman J, et al. Consensus recommendations for a standardized brain tumor imaging protocol in clinical trials. Neuro Oncol 2015;17:1188–98. 10.1093/neuonc/nov095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dewey BE, Zhao C, Reinhold JC, et al. DeepHarmony: a deep learning approach to contrast harmonization across scanner changes. Magn Reson Imaging 2019;64:160–70. 10.1016/j.mri.2019.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carré A, Klausner G, Edjlali M, et al. Standardization of brain MR images across machines and protocols: bridging the gap for MRI-based radiomics. Sci Rep 2020;10:12340. 10.1038/s41598-020-69298-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fortin J-P, Parker D, Tunç B, et al. Harmonization of multi-site diffusion tensor imaging data. Neuroimage 2017;161:149–70. 10.1016/j.neuroimage.2017.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zwanenburg A, Vallières M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020;295:328–38. 10.1148/radiol.2020191145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bluemke DA, Moy L, Bredella MA, et al. Assessing radiology research on artificial intelligence: a brief guide for authors, reviewers, and readers from the radiology editorial board. Radiology 2020;294:487–9. 10.1148/radiol.2019192515 [DOI] [PubMed] [Google Scholar]
- 54. Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749–62. 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 55. Grossmann P, Stringfield O, El-Hachem N, et al. Defining the biological basis of radiomic phenotypes in lung cancer. eLife 2017;6. 10.7554/eLife.23421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Trebeschi S, Drago SG, Birkbak NJ, et al. Predicting response to cancer immunotherapy using non-invasive radiomic biomarkers. Ann Oncol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tunali I, Gray JE, Qi J, et al. Novel clinical and radiomic predictors of rapid disease progression phenotypes among lung cancer patients treated with immunotherapy: an early report. Lung Cancer 2019;129:75–9. 10.1016/j.lungcan.2019.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Khorrami M, Prasanna P, Gupta A, et al. Changes in CT radiomic features associated with lymphocyte distribution predict overall survival and response to immunotherapy in non-small cell lung cancer. Cancer Immunol Res 2020;8:108–19. 10.1158/2326-6066.CIR-19-0476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mu W, Tunali I, Gray JE, et al. Radiomics of 18F-FDG PET/CT images predicts clinical benefit of advanced NSCLC patients to checkpoint blockade immunotherapy. Eur J Nucl Med Mol Imaging 2020;47:1168–82. 10.1007/s00259-019-04625-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Polverari G, Ceci F, Bertaglia V, et al. 18F-FDG PET parameters and radiomics features analysis in advanced NSCLC treated with immunotherapy as predictors of therapy response and survival. Cancers 2020;12:1163. 10.3390/cancers12051163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Valentinuzzi D, Vrankar M, Boc N, et al. [ 18 F]FDG PET immunotherapy radiomics signature (iRADIOMICS) predicts response of non-small-cell lung cancer patients treated with pembrolizumab. Radiol Oncol 2020;54:285–94. 10.2478/raon-2020-0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Corino VDA, Bologna M, Calareso G, et al. A CT-based radiomic signature can be prognostic for 10-Months overall survival in metastatic tumors treated with nivolumab: an exploratory study. Diagnostics 2021;11:979. 10.3390/diagnostics11060979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen X, Zhou M, Wang Z, et al. Immunotherapy treatment outcome prediction in metastatic melanoma through an automated multi-objective delta-radiomics model. Comput Biol Med 2021;138:104916. 10.1016/j.compbiomed.2021.104916 [DOI] [PubMed] [Google Scholar]
- 64. Aoude LG, Wong BZY, Bonazzi VF, et al. Radiomics biomarkers correlate with CD8 expression and predict immune signatures in melanoma patients. Mol Cancer Res 2021;19:950–6. 10.1158/1541-7786.MCR-20-1038 [DOI] [PubMed] [Google Scholar]
- 65. Alessandrino F, Gujrathi R, Nassar AH, et al. Predictive role of computed tomography texture analysis in patients with metastatic urothelial cancer treated with programmed death-1 and programmed death-ligand 1 inhibitors. Eur Urol Oncol 2020;3:680–6. 10.1016/j.euo.2019.02.002 [DOI] [PubMed] [Google Scholar]
- 66. Park KJ, Lee J-L, Yoon S-K, et al. Radiomics-based prediction model for outcomes of PD-1/PD-L1 immunotherapy in metastatic urothelial carcinoma. Eur Radiol 2020;30:5392–403. 10.1007/s00330-020-06847-0 [DOI] [PubMed] [Google Scholar]
- 67. Rundo F, Bersanelli M, Urzia V, et al. Three-dimensional deep noninvasive radiomics for the prediction of disease control in patients with metastatic urothelial carcinoma treated with immunotherapy. Clin Genitourin Cancer 2021;19:396–404. 10.1016/j.clgc.2021.03.012 [DOI] [PubMed] [Google Scholar]
- 68. Khene Z-E, Mathieu R, Peyronnet B, et al. Radiomics can predict tumour response in patients treated with nivolumab for a metastatic renal cell carcinoma: an artificial intelligence concept. World J Urol 2021;39:3707–9. 10.1007/s00345-020-03334-5 [DOI] [PubMed] [Google Scholar]
- 69. Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 2018;19:1180–91. 10.1016/S1470-2045(18)30413-3 [DOI] [PubMed] [Google Scholar]
- 70. Jiang M, Sun D, Guo Y, et al. Assessing PD-L1 expression level by radiomic features from PET/CT in nonsmall cell lung cancer patients: an initial result. Acad Radiol 2020;27:171–9. 10.1016/j.acra.2019.04.016 [DOI] [PubMed] [Google Scholar]
- 71. Chen S, Feng S, Wei J, et al. Pretreatment prediction of immunoscore in hepatocellular cancer: a radiomics-based clinical model based on Gd-EOB-DTPA-enhanced MRI imaging. Eur Radiol 2019;29:4177–87. 10.1007/s00330-018-5986-x [DOI] [PubMed] [Google Scholar]
- 72. Hectors SJ, Lewis S, Besa C, et al. MRI radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur Radiol 2020;30:3759–69. 10.1007/s00330-020-06675-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sun R, Sundahl N, Hecht M, et al. Radiomics to predict outcomes and abscopal response of patients with cancer treated with immunotherapy combined with radiotherapy using a validated signature of CD8 cells. J Immunother Cancer 2020;8:e001429. 10.1136/jitc-2020-001429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Korpics MC, Polley M-Y, Bhave SR, et al. A validated T cell Radiomics score is associated with clinical outcomes following multisite SBRT and pembrolizumab. Int J Radiat Oncol Biol Phys 2020;108:189–95. 10.1016/j.ijrobp.2020.06.026 [DOI] [PubMed] [Google Scholar]
- 75. Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity 2018;48:812–30. 10.1016/j.immuni.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mihaylov IB, Totiger TM, Giret TM, et al. Toward prediction of abscopal effect in radioimmunotherapy: pre-clinical investigation. PLoS One 2021;16:e0255923. 10.1371/journal.pone.0255923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cho Y, Park S, Byun HK, et al. Impact of treatment-related lymphopenia on immunotherapy for advanced non-small cell lung cancer. Int J Radiat Oncol Biol Phys 2019;105:1065–73. 10.1016/j.ijrobp.2019.08.047 [DOI] [PubMed] [Google Scholar]
- 78. Jiang Y, Liang X, Han Z, et al. Radiographical assessment of tumour stroma and treatment outcomes using deep learning: a retrospective, multicohort study. Lancet Digit Health 2021;3:e371–82. 10.1016/S2589-7500(21)00065-0 [DOI] [PubMed] [Google Scholar]
- 79. Sun R, Orlhac F, Robert C, et al. In Regard to Mattonen et al. Int J Radiat Oncol Biol Phys 2016;95:1544–5. 10.1016/j.ijrobp.2016.03.038 [DOI] [PubMed] [Google Scholar]
- 80. Lemons JM, Luke JJ, Janisch L, et al. The ADscopal effect? Control of partially irradiated versus completely irradiated tumors on a prospective trial of pembrolizumab and SBRT per NRG-BR001. Int J Radiat Oncol Biol Phys 2017;99:S87. 10.1016/j.ijrobp.2017.06.209 [DOI] [Google Scholar]
- 81. Dercle L, Zhao B, Gönen M, et al. Early readout on overall survival of patients with melanoma treated with immunotherapy using a novel imaging analysis. JAMA Oncol 2022;8:6818 10.1001/jamaoncol.2021.6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu Y, Wu M, Zhang Y, et al. Imaging biomarkers to predict and evaluate the effectiveness of immunotherapy in advanced non-small-cell lung cancer. Front Oncol 2021;11:657615. 10.3389/fonc.2021.657615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chang E, Joel MZ, Chang HY, et al. Comparison of radiomic feature aggregation methods for patients with multiple tumors. Sci Rep 2021;11:9758. 10.1038/s41598-021-89114-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Schwartz LH, Litière S, de Vries E, et al. RECIST 1.1-Update and clarification: from the RECIST Committee. Eur J Cancer 2016;62:132–7. 10.1016/j.ejca.2016.03.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Van den Eynde M, Mlecnik B, Bindea G, et al. The link between the multiverse of immune microenvironments in metastases and the survival of colorectal cancer patients. Cancer Cell 2018;34:1012–26. 10.1016/j.ccell.2018.11.003 [DOI] [PubMed] [Google Scholar]
- 86. Van den Eynde M, Mlecnik B, Bindea G, et al. Multiverse of immune microenvironment in metastatic colorectal cancer. Oncoimmunology 2020;9:1824316. 10.1080/2162402X.2020.1824316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schwartz LH, Mazumdar M, Brown W, et al. Variability in response assessment in solid tumors: effect of number of lesions chosen for measurement. Clin Cancer Res 2003;9:4318–23. [PubMed] [Google Scholar]
- 88. Quellec G, Cazuguel G, Cochener B, et al. Multiple-instance learning for medical image and video analysis. IEEE Rev Biomed Eng 2017;10:213–34. 10.1109/RBME.2017.2651164 [DOI] [PubMed] [Google Scholar]
- 89. Zhang X, Lu D, Gao P, et al. Survival-relevant high-risk subregion identification for glioblastoma patients: the MRI-based multiple instance learning approach. Eur Radiol 2020;30:5602–10. 10.1007/s00330-020-06912-8 [DOI] [PubMed] [Google Scholar]
- 90. Cheplygina V, de Bruijne M, Pluim JPW. Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med Image Anal 2019;54:280–96. 10.1016/j.media.2019.03.009 [DOI] [PubMed] [Google Scholar]
- 91. He B, Dong D, She Y, et al. Predicting response to immunotherapy in advanced non-small-cell lung cancer using tumor mutational burden radiomic biomarker. J Immunother Cancer 2020;8:e000550. 10.1136/jitc-2020-000550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Tunali I, Tan Y, Gray JE, et al. Hypoxia-Related Radiomics and immunotherapy response: a Multicohort study of non-small cell lung cancer. JNCI Cancer Spectr 2021;5:pkab048. 10.1093/jncics/pkab048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Galon J, Mlecnik B, Bindea G, et al. Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumours. J Pathol 2014;232:199–209. 10.1002/path.4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yoon J, Suh YJ, Han K, et al. Utility of CT radiomics for prediction of PD-L1 expression in advanced lung adenocarcinomas. Thorac Cancer 2020;11:993–1004. 10.1111/1759-7714.13352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wen Q, Yang Z, Dai H, et al. Radiomics study for predicting the expression of PD-L1 and tumor mutation burden in non-small cell lung cancer based on CT images and clinicopathological features. Front Oncol 2021;11:620246. 10.3389/fonc.2021.620246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Golia Pernicka JS, Gagniere J, Chakraborty J, et al. Radiomics-based prediction of microsatellite instability in colorectal cancer at initial computed tomography evaluation. Abdom Radiol 2019;44:3755–63. 10.1007/s00261-019-02117-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Li Z, Dai H, Liu Y, et al. Radiomics analysis of multi-sequence MR images for predicting microsatellite instability status preoperatively in rectal cancer. Front Oncol 2021;11:697497. 10.3389/fonc.2021.697497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Devkota L, Starosolski Z, Rivas CH, et al. Detection of response to tumor microenvironment-targeted cellular immunotherapy using nano-radiomics. Sci Adv 2020;6:eaba6156. 10.1126/sciadv.aba6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mu W, Jiang L, Shi Y, et al. Non-invasive measurement of PD-L1 status and prediction of immunotherapy response using deep learning of PET/CT images. J Immunother Cancer 2021;9:e002118. 10.1136/jitc-2020-002118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhou J-G, Donaubauer A-J, Frey B, et al. Prospective development and validation of a liquid immune profile-based signature (LIPS) to predict response of patients with recurrent/metastatic cancer to immune checkpoint inhibitors. J Immunother Cancer 2021;9:e001845. 10.1136/jitc-2020-001845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Pan Y, Lei X, Zhang Y. Association predictions of genomics, proteinomics, transcriptomics, microbiome, metabolomics, pathomics, radiomics, drug, symptoms, environment factor, and disease networks: a comprehensive approach. Med Res Rev 2022;42:441–61. 10.1002/med.21847 [DOI] [PubMed] [Google Scholar]
- 102. Trebeschi S, Bodalal Z, Boellaard TN, et al. Prognostic value of deep learning-mediated treatment monitoring in lung cancer patients receiving immunotherapy. Front Oncol 2021;11:609054. 10.3389/fonc.2021.609054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Luo J, Vanguri RS, Aukerman AT, et al. Multimodal integration of radiology, pathology, and genomics for prediction of response to PD-1 blockade in patients with non–small cell lung cancer. JCO 2022;40:9064. 10.1200/JCO.2022.40.16_suppl.9064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Khatua S, Cooper LJN, Sandberg DI, et al. Phase I study of intraventricular infusions of autologous ex vivo expanded NK cells in children with recurrent medulloblastoma and ependymoma. Neuro Oncol 2020;22:1214–25. 10.1093/neuonc/noaa047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang W, Gao Z, Wang L, et al. Application and prospects of molecular imaging in immunotherapy. Cancer Manag Res 2020;12:9389–403. 10.2147/CMAR.S269773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Truillet C, Oh HLJ, Yeo SP, et al. Imaging PD-L1 expression with ImmunoPET. Bioconjug Chem 2018;29:96–103. 10.1021/acs.bioconjchem.7b00631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Natarajan A, Mayer AT, Xu L, et al. Novel radiotracer for ImmunoPET imaging of PD-1 checkpoint expression on tumor infiltrating lymphocytes. Bioconjug Chem 2015;26:2062–9. 10.1021/acs.bioconjchem.5b00318 [DOI] [PubMed] [Google Scholar]
- 108. Maute RL, Gordon SR, Mayer AT, et al. Engineering high-affinity PD-1 variants for optimized immunotherapy and immuno-PET imaging. Proc Natl Acad Sci U S A 2015;112:E6506–14. 10.1073/pnas.1519623112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hettich M, Braun F, Bartholomä MD, et al. High-Resolution PET imaging with therapeutic antibody-based PD-1/PD-L1 checkpoint tracers. Theranostics 2016;6:1629–40. 10.7150/thno.15253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kikuchi M, Clump DA, Srivastava RM, et al. Preclinical immunoPET/CT imaging using Zr-89-labeled anti-PD-L1 monoclonal antibody for assessing radiation-induced PD-L1 upregulation in head and neck cancer and melanoma. Oncoimmunology 2017;6:e1329071. 10.1080/2162402X.2017.1329071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ehlerding EB, Lee HJ, Barnhart TE, et al. Noninvasive Imaging and Quantification of Radiotherapy-Induced PD-L1 Upregulation with 89Zr-Df-Atezolizumab. Bioconjug Chem 2019;30:1434–41. 10.1021/acs.bioconjchem.9b00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Christensen C, Kristensen LK, Alfsen MZ, et al. Quantitative PET imaging of PD-L1 expression in xenograft and syngeneic tumour models using a site-specifically labelled PD-L1 antibody. Eur J Nucl Med Mol Imaging 2020;47:1302–13. 10.1007/s00259-019-04646-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bensch F, van der Veen EL, Lub-de Hooge MN, et al. 89Zr-atezolizumab imaging as a non-invasive approach to assess clinical response to PD-L1 blockade in cancer. Nat Med 2018;24:1852–8. 10.1038/s41591-018-0255-8 [DOI] [PubMed] [Google Scholar]
- 114. Kist de Ruijter L, Hooiveld-Noeken JS, Giesen D, et al. First-in-human study of the biodistribution and pharmacokinetics of 89Zr-CX-072, a novel immunopet tracer based on an anti-PD-L1 probody. Clin Cancer Res 2021;27:5325–33. 10.1158/1078-0432.CCR-21-0453 [DOI] [PubMed] [Google Scholar]
- 115. Verhoeff S, van de Donk PP, Aarntzen EHJG, et al. 89 Zr-durvalumab PD-L1 PET in recurrent or metastatic (R/M) squamous cell carcinoma of the head and neck. JCO 2020;38:3573. 10.1200/JCO.2020.38.15_suppl.3573 [DOI] [Google Scholar]
- 116. Tavaré R, Escuin-Ordinas H, Mok S, et al. An effective immuno-PET imaging method to monitor CD8-dependent responses to immunotherapy. Cancer Res 2016;76:73–82. 10.1158/0008-5472.CAN-15-1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Farwell MD, Gamache RF, Babazada H, et al. CD8-targeted PET imaging of tumor infiltrating T cells in patients with cancer: A phase I first-in-human study of 89 Zr-Df-IAB22M2C, a radiolabeled anti-CD8 minibody. J Nucl Med 2021:jnumed.121.262485. 10.2967/jnumed.121.262485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Mole RH. Whole body irradiation; radiobiology or medicine? Br J Radiol 1953;26:234–41. 10.1259/0007-1285-26-305-234 [DOI] [PubMed] [Google Scholar]
- 119. Golden EB, Chhabra A, Chachoua A, et al. Local radiotherapy and granulocyte-macrophage colony-stimulating factor to generate abscopal responses in patients with metastatic solid tumours: a proof-of-principle trial. Lancet Oncol 2015;16:795–803. 10.1016/S1470-2045(15)00054-6 [DOI] [PubMed] [Google Scholar]
- 120. Formenti SC, Demaria S. Systemic effects of local radiotherapy. Lancet Oncol 2009;10:718–26. 10.1016/S1470-2045(09)70082-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Formenti SC, Demaria S. Combining radiotherapy and cancer immunotherapy: a paradigm shift. J Natl Cancer Inst 2013;105:256–65. 10.1093/jnci/djs629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Luke JJ, Lemons JM, Karrison TG, et al. Safety and clinical activity of pembrolizumab and multisite stereotactic body radiotherapy in patients with advanced solid tumors. J Clin Oncol 2018;36:1611–8. 10.1200/JCO.2017.76.2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Sundahl N, Vandekerkhove G, Decaestecker K, et al. Randomized phase 1 trial of pembrolizumab with sequential versus concomitant stereotactic body radiotherapy in metastatic urothelial carcinoma. Eur Urol 2019;75:707–11. 10.1016/j.eururo.2019.01.009 [DOI] [PubMed] [Google Scholar]
- 124. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 125. Borcoman E, Kanjanapan Y, Champiat S, et al. Novel patterns of response under immunotherapy. Ann Oncol 2019;30:385–96. 10.1093/annonc/mdz003 [DOI] [PubMed] [Google Scholar]
- 126. Champiat S, Dercle L, Ammari S, et al. Hyperprogressive disease is a new pattern of progression in cancer patients treated by anti-PD-1/PD-L1. Clin Cancer Res 2017;23:1920–8. 10.1158/1078-0432.CCR-16-1741 [DOI] [PubMed] [Google Scholar]
- 127. Hodi FS, Hwu W-J, Kefford R, et al. Evaluation of immune-related response criteria and RECIST v1.1 in patients with advanced melanoma treated with pembrolizumab. J Clin Oncol 2016;34:1510–7. 10.1200/JCO.2015.64.0391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Seymour L, Bogaerts J, Perrone A, et al. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol 2017;18:e143–52. 10.1016/S1470-2045(17)30074-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Hodi FS, Ballinger M, Lyons B, et al. Immune-modified response evaluation criteria in solid tumors (imRECIST): refining guidelines to assess the clinical benefit of cancer immunotherapy. J Clin Oncol 2018;36:850–8. 10.1200/JCO.2017.75.1644 [DOI] [PubMed] [Google Scholar]
- 130. Wolchok JD, Hoos A, O'Day S, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res 2009;15:7412–20. 10.1158/1078-0432.CCR-09-1624 [DOI] [PubMed] [Google Scholar]
- 131. Goldmacher GV, Khilnani AD, Andtbacka RHI, et al. Response criteria for intratumoral immunotherapy in solid tumors: itRECIST. J Clin Oncol 2020;38:2667–76. 10.1200/JCO.19.02985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Barabino E, Rossi G, Pamparino S, et al. Exploring response to immunotherapy in non-small cell lung cancer using delta-radiomics. Cancers 2022;14:350. 10.3390/cancers14020350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Cheng J, Pan Y, Huang W, et al. Differentiation between immune checkpoint inhibitor-related and radiation pneumonitis in lung cancer by CT radiomics and machine learning. Med Phys 2022;49:1547–58. 10.1002/mp.15451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Colen RR, Rolfo C, Ak M, et al. Radiomics analysis for predicting pembrolizumab response in patients with advanced rare cancers. J Immunother Cancer 2021;9:e001752. 10.1136/jitc-2020-001752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Del Re M, Cucchiara F, Rofi E, et al. A multiparametric approach to improve the prediction of response to immunotherapy in patients with metastatic NSCLC. Cancer Immunol Immunother 2021;70:1667–78. 10.1007/s00262-020-02810-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Granata V, Fusco R, Costa M, et al. Preliminary report on computed tomography Radiomics features as biomarkers to immunotherapy selection in lung adenocarcinoma patients. Cancers 2021;13:3992. 10.3390/cancers13163992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Yang B, Zhou L, Zhong J, et al. Combination of computed tomography imaging-based radiomics and clinicopathological characteristics for predicting the clinical benefits of immune checkpoint inhibitors in lung cancer. Respir Res 2021;22:189. 10.1186/s12931-021-01780-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Shen L, Fu H, Tao G, et al. Pre-immunotherapy contrast-enhanced CT texture-based classification: a useful approach to non-small cell lung cancer immunotherapy efficacy prediction. Front Oncol 2021;11:591106. 10.3389/fonc.2021.591106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Yang Y, Yang J, Shen L, et al. A multi-omics-based serial deep learning approach to predict clinical outcomes of single-agent anti-PD-1/PD-L1 immunotherapy in advanced stage non-small-cell lung cancer. Am J Transl Res 2021;13:743–56. [PMC free article] [PubMed] [Google Scholar]
- 140. Liu C, Gong J, Yu H, et al. A CT-based radiomics approach to predict nivolumab response in advanced non-small-cell lung cancer. Front Oncol 2021;11:544339. 10.3389/fonc.2021.544339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Zerunian M, Caruso D, Zucchelli A, et al. CT based radiomic approach on first line pembrolizumab in lung cancer. Sci Rep 2021;11:6633. 10.1038/s41598-021-86113-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Brendlin AS, Peisen F, Almansour H, et al. A machine learning model trained on dual-energy CT radiomics significantly improves immunotherapy response prediction for patients with stage IV melanoma. J Immunother Cancer 2021;9:e003261. 10.1136/jitc-2021-003261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Eresen A, Yang J, Shangguan J, et al. Detection of immunotherapeutic response in a transgenic mouse model of pancreatic ductal adenocarcinoma using multiparametric MRI radiomics: a preliminary investigation. Acad Radiol 2021;28:e147–54. 10.1016/j.acra.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]