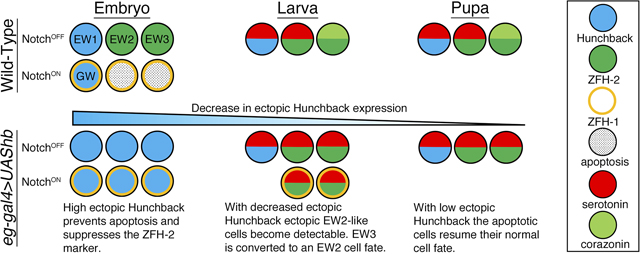
Abstract
The serotonergic lineage (NB7–3) in the Drosophila ventral nerve cord produces six cells during neurogenesis. Four of the cells differentiate into neurons: EW1, EW2, EW3 and GW. The other two cells undergo apoptosis. This simple lineage provides an opportunity to examine genes that are required to induce or repress apoptosis during cell specification. Previous studies have shown that Notch signaling induces apoptosis within the NB7–3 lineage. The three EW neurons are protected from Notch-induced apoptosis by asymmetric distribution of Numb protein, an inhibitor of Notch signaling. In a numb1 mutant EW2 and EW3 undergo apoptosis. The EW1 and GW neurons survive even in a numb1 mutant background suggesting that these cells are protected from Notch-induced apoptosis by some factor other than Numb. The EW1 and GW neurons are mitotic sister cells, and uniquely express the transcription factor Hunchback. We present evidence that Hunchback prevents apoptosis in NB7–3 lineage during normal CNS development and can rescue the two apoptotic cells in the lineage when it is ectopically expressed. We show that hunchback overexpression produces ectopic cells that express markers similar to the EW2 neuron and changes the expression pattern of the EW3 neuron to a EW2 neuron. In addition we show that hunchback overexpression can override apoptosis that is genetically induced by the pro-apoptotic genes grim and hid.
Keywords: Hunchback, Apoptosis, NB7–3, Serotonin, Corazonin
Graphical Abstract
INTRODUCTION
Apoptosis is an essential process in the development and maintenance of an organism. It eliminates excess cells during development, it plays a role in tissue homeostasis, and it removes cells that have undergone DNA damage (Ambrosini et al., 2017; Fuchs and Steller, 2011; Pinto-Teixeira et al., 2016; Suzanne and Steller, 2013; Teng and Toyama, 2011). Defects in apoptosis can lead to various pathologies including cancer (Fadeel and Orrenius, 2005; Favaloro et al., 2012; Fogarty and Bergmann, 2017; Hanahan and Weinberg, 2000; Perez-Garijo, 2018; Perez-Garijo and Steller, 2015; Singh et al., 2019). During neurogenesis all cell types in the CNS including neural stem cells, neurons and glial cells undergo apoptosis. In the ventral nerve cord (VNC) of Drosophila almost all of the cell lineages have been shown to have a specific number of progeny cells that undergo apoptosis (Pinto-Teixeira et al., 2016; Rogulja-Ortmann et al., 2007; Schmid et al., 1999; Yamaguchi and Miura, 2015). Here we examine the role of Hunchback (Hb) as an inhibitor of Notch-induced apoptosis in the Drosophila serotonergic NB7–3 lineage.
In the segmented VNC of the Drosophila CNS each hemisegment repeats a stereotypic array of approximately 30 neuroblasts (NB) (Bossing et al., 1996; Broadus et al., 1995; Doe, 1992; Goodman and Doe, 1993; Hartenstein and Campos-Ortega, 1984; Schmid et al., 1999; Schmidt et al., 1997). The NBs give rise to smaller precursor cells called ganglion mother cells (GMCs). Each GMC divides once to generate two progeny cells. The NB7–3 lineage generates three GMCs designated, GMC1, GMC2, and GMC3 (Fig. 1A) (Bossing et al., 1996; Dittrich et al., 1997; Higashijima et al., 1996; Isshiki et al., 2001; Lundell and Hirsh, 1998; Schmid et al., 1999). GMC1 gives rise to two cells, EW1 and GW, which differentiate into the medial serotonin neuron and a motor neuron respectively. GMC2 gives rise to EW2, which differentiates into the lateral serotonin neuron, and EW2-sib that undergoes Notch-induced apoptosis. GMC3 gives rise to EW3, which differentiates into a corazonin producing neuron, and EW3-sib that undergoes Notch-induced apoptosis (Isshiki et al., 2001; Karcavich and Doe, 2005; Lee and Lundell, 2007; Lundell et al., 2003; Novotny et al., 2002). The EW neurons are protected from Notch-induced apoptosis by Numb protein that asymmetrically distributed into these cells during cell division (Karcavich and Doe, 2005) and is an inhibitor of Notch-signaling (Guo et al., 1996). In a numb1 mutant EW2 and EW3 undergo apoptosis but EW1 survives 90% of the time (Lundell et al., 2003). The GW motor neuron which does not receive Numb protein during cell division has active Notch signaling, but is protected from apoptosis unlike the EW2-sib and EW3-sib cells (Lundell et al., 2003). These observations suggest that the EW1 serotonin neuron and the GW motor neuron have some factor other than Numb that can inhibit Notch-induced apoptosis. One possible candidate is the transcription factor, Hunchback (Hb), which is uniquely expressed in the EW1 and GW neurons (Isshiki et al., 2001; Novotny et al., 2002).
Figure 1. A delta7 loss-of-function mutation duplicates the NB7–3 EW neurons.
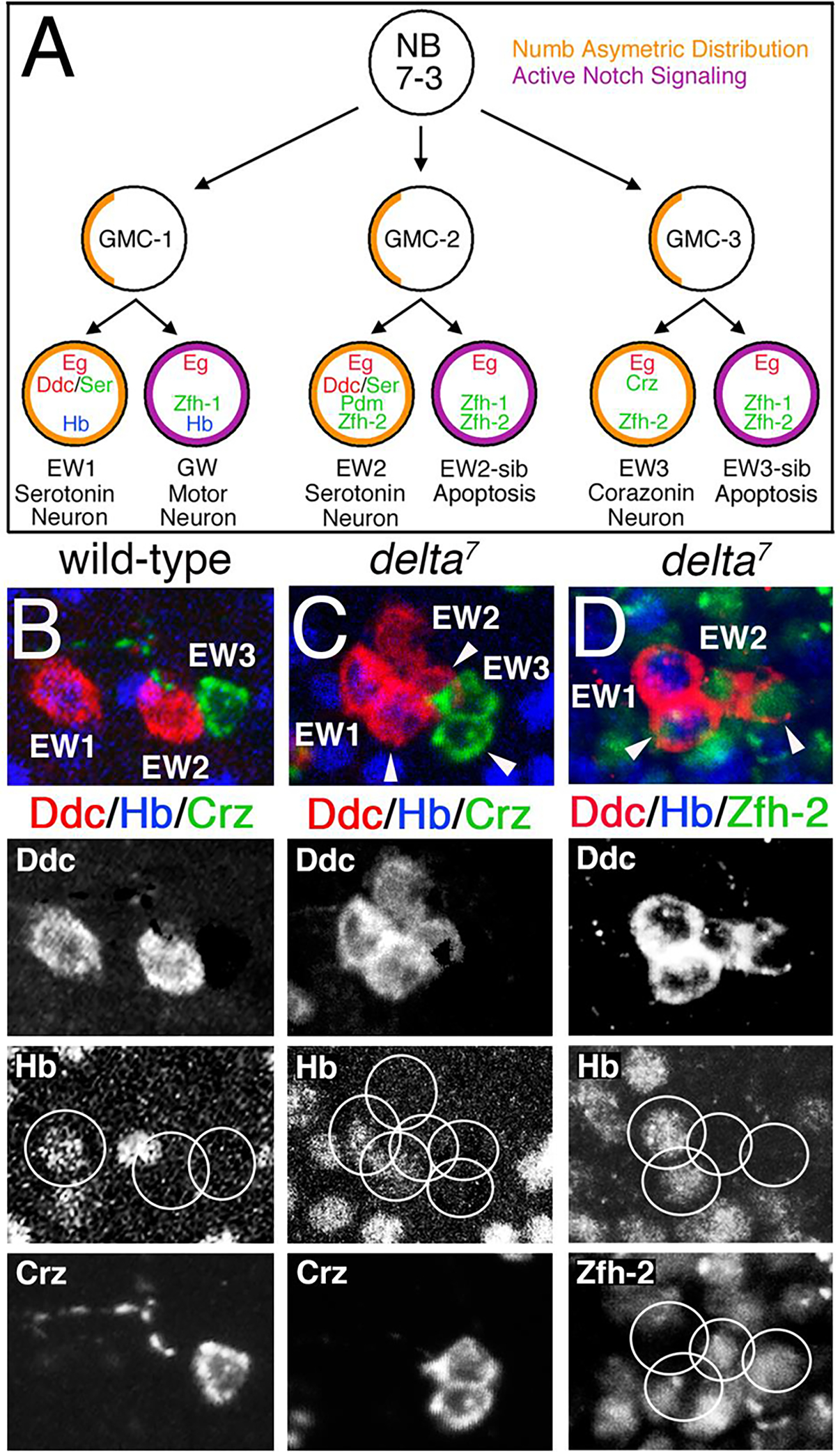
(A) Depiction of NB7–3 lineage and relevant cell markers. The NB7–3 lineage produces three GMCs and six progeny cells, two of which undergo apoptosis. Numb asymmetrically partitions into the EW neurons blocking Notch signaling. (B-D) A comparison of stage 17 VNC for wild-type and the delta7 mutation. (B-C) Immunostaining for Ddc (red), Corazonin (green) and Hb (blue) shows duplication in the number of detectible cells in the delta7 mutation. (D) Immunostaining for Ddc (red), Hb (blue) and Zfh-2 (green) demonstrates that two of the four Ddc cells are GMC1 progeny (Hb positive) and two are GMC2 progeny (Zfh-2 positive). Ectopic cells are marked with arrowheads. Separate fluorochromes are shown below the merged images for the same hemisegments. The midline is to the left in all panels. (Panel A is adapted from Dev Biol 475, 65–79, Guntur, A.R., Venkatanarayan, A., Gangula, S., Lundell, M.J., Zfh-2 facilitates Notch-induced apoptosis in the CNS and appendages of Drosophila melanogaster. (2021), with permission from Elsevier.)
During neurogenesis neuronal identity is specified by transcription factors inherited from the neuroblasts that are established by both spatial and temporal cues, reviewed in (Doe, 2017; Kohwi and Doe, 2013; Li et al., 2013; Sen et al., 2019). Hb is the first temporal transcription factor expressed in most VNC NBs, and its expression is both necessary and sufficient to specify the first-born GMC and respective neuronal progeny (Cleary and Doe, 2006; Isshiki et al., 2001; Novotny et al., 2002; Pearson and Doe, 2003; Tran and Doe, 2008). The overexpression of hb in multiple lineages, including NB7–3, produces ectopic cells. The interpretation of these results is that the overexpression of hb maintains the neuroblast in a temporal “young” state such that it can undergo supernumerary divisions and produces extra GMC1-like progeny (Cleary and Doe, 2006; Grosskortenhaus et al., 2005; Isshiki et al., 2001; Novotny et al., 2002; Pearson and Doe, 2003; Tran and Doe, 2008). Here we re-examine the identity of the ectopic cells in the NB7–3 lineage at the larval stage, and present evidence for an alternative hypothesis where the overexpression of hb produces ectopic GMC2-like cells by inhibiting Notch-induced apoptosis of the EW2-sib and EW3-sib cells, and by converting the EW3 cell to an EW2 cell. We also show that overexpression of hb can limit apoptosis that is genetically induced in the NB7–3 lineage.
MATERIALS AND METHODS
Drosophila stocks
The following fly lines were used in this study: eg-Gal4 (egmz360) (from G. Technau); hkb-Gal4, UAS-hb, UAS-Partner of Numb-GFP (UAS-PON-GFP) (from C. Doe); UAS-notchACT (from Y. N. Jan); Canton S, delta7, hb9, elav-Gal-4, UAS-hid, UAS-grim, and UAS-lac Z (from the Bloomington Stock Center).
Isolation of Drosophila CNS
Embryonic CNS were prepared from eggs that were washed in ddH2O to eliminate yeast contamination, and then dechlorinated in Clorox for approximately 5 minutes. Samples were washed briefly in ddH2O followed by one wash in 0.4% NaCl/0.3% TrintonX-100 in Phosphate Buffered Saline (PBT). Embryos were homogenized in a 1.5 ml centrifuge tube using a pestle. The homogenate was fixed in 4% paraformaldehyde for one hour. During this time CNS were sorted from the debris. Isolated CNS were washed four times for five minutes in PBT plus 3% Normal Goat Serum prior to immunohistochemistry.
Larvae and white pupae CNS were individually dissected under a microscope in 4% paraformaldehyde. After approximately 40 minutes the collected CNS were washed four times for five minutes in PBT plus 3% Normal Goat Serum prior to immunohistochemistry.
Immunohistochemistry
Prepared CNS were incubated with primary and secondary antisera as previously described (Lundell and Hirsh, 1994). Primary antibodies used were: rabbit anti-eagle (1:2000, G. Technau), guinea pig anti-Hb (1:600, East Asia Distribution Center for Segmentation Antibodies; (Kosman et al., 1998)), rabbit anti-Ddc (1:50, M. Lundell), rat anti-Ddc (1:200, M. Lundell), rabbit anti-corazonin (1:200, C. Doe), rat anti-serotonin (1:100, Millipore Corporation), rabbit anti-Zfh-1 (1:400, R. Lehman), rabbit anti-pdm1 (1:1000, T. Dick), and rat anti-zfh-2 (1:200, A. Tomlinson). All fluorescent secondary antibodies were from Jackson Laboratories and used at 1:100 to1:400 dilution.
Microscopy
Confocal images were obtained using a BioRad 1024 laser-scanning microscope and a Zeiss LSM 510 confocal microscope. Images were processed with Imaris and edited with Adobe Photoshop.
TUNEL Assay
Apoptotic cells were detected by TUNEL assay using a Trevigen TACS 2 TDT-Fluor In Situ Kit.
RESULTS
Delta facilitates Notch-induced apoptosis in the NB7–3 lineage.
Previous work has demonstrated that Notch signaling is required for apoptosis of the EW2-sib and EW3-sib cells (Lee and Lundell, 2007; Lundell et al., 2003). Here we confirm this observation by showing a loss-of-function mutation for the Notch ligand, Delta, can rescue the EW2-sib and EW3-sib cells from apoptosis and alter the fate of the GW motor neuron to an EW1-like cell. Serotonin cells can be detected with an antibody to Dopa-decarboxylase (Ddc), which is required for the biosynthesis of serotonin. A wild-type ventral cord has two Ddc expressing neurons, EW1 and EW2, and one corazonin neuron EW3 (Fig.1B). The delta7 mutation (Bridges and Brehme, 1944) can duplicate the number of cells showing four Ddc (two of which are Hb positive) and two corazonin expressing neurons (Fig. 1C). Fig. 1D shows that two of the four Ddc neurons express Hb which is expressed only in GMC1 progeny (Isshiki et al., 2001; Lundell et al., 2003; Novotny et al., 2002) and that the other two Ddc neurons express Zfh-2 which is expressed only in GMC2 and GMC3 progeny (Isshiki et al., 2001; Lundell and Hirsh, 1998; Lundell et al., 2003; Novotny et al., 2002). This phenotype of duplicated cells has low penetrance, only 21% hemisegments show ectopic Ddc or corazonin neurons (n=48). This could be due to the hypomorphic nature of the delta7 mutation and/or redundant function of the other Notch ligand, Serrate. This result confirms the role of Delta/Notch signaling in specifying cell fates in the NB7–3 lineage.
Hunchback is necessary to prevent Notch-induced apoptosis of the GW motor neuron in the NB7–3 lineage.
To investigate whether Hb protects the GW motor neuron from apoptosis we examined the hypomorphic allele hb9 (Lehmann and Nusslein-Volhard, 1987) and assayed for apoptosis using the TUNEL assay. In a stage 15 wild-type VNC all four NB7–3 progeny (GW, EW1, EW2, and EW3) are detected with Eagle (Eg) immunoreactivity (Fig. 2A). Hb is detected only in the GMC1 progeny, GW and EW1 neurons. TUNEL immunoreactivity is never detected in the four wild-type NB7–3 neurons (n=10). In a stage 15 hb9 mutant VNC the GW motor neuron is positive for TUNEL 70% of the time (n=10) (Fig. 2B). In some hemisegments apoptosis has progressed far enough that the GW can no longer be accurately identified. These results suggest that Hb is necessary to prevent Notch-induced apoptosis in the GW motor neuron. Although the hb9 allele retains some Hb expression it is apparently insufficient to prevent apoptosis of the GW neuron. We did not observe a significant loss of EW1 or TUNEL immunoreactivity in EW1. EW1 is protected from apoptosis by the asymmetric distribution of Numb which inhibits Notch signaling inhibits Notch-signaling. Previous reports have shown the absence of EW1 with hb null alleles (Isshiki et al., 2001; Novotny et al., 2002). It was suggested in these reports that the inability to detect EW1was most likely due to a change in cell fate rather than apoptosis.
Figure 2. Hunchback is necessary to prevent Notch-induced apoptosis in the GW motor neuron.
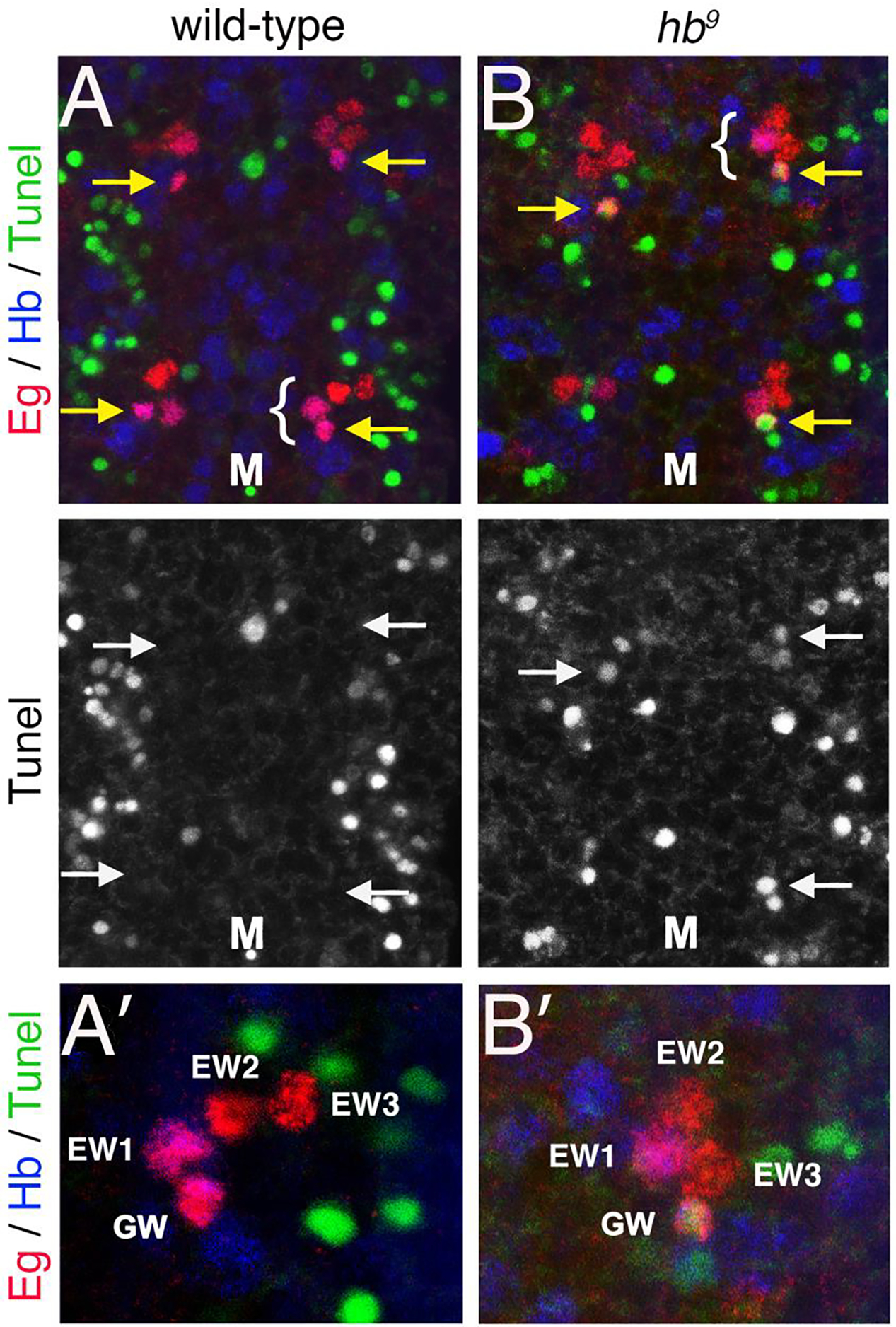
Stage 15 VNC from wild-type and hb9 embryos were immunostained for TUNEL (green), Eg (red) and Hb (blue). Yellow arrows indicate the GW motor neurons. The brackets indicate hemisegments that are shown at a higher magnification in A’ and B’. Tunel immunoreactivity is shown separately for panels A and B. (A) In a wild-type VNC all four NB7–3 neurons in a hemisegment are positive for Eg and the EW1 and GW neurons co-express Hb. None of the cells are positive for TUNEL. (B) In a hb9 mutant VNC identifiable GW motor neurons are positive for TUNEL. M denotes the midline.
Temporal expression pattern of the eg-gal4 and hkb-gal4 drivers in the NB7–3 lineage.
To test whether Hb is sufficient to prevent Notch-induced apoptosis in the NB7–3 lineage, we asked whether overexpression of hb can prevent apoptosis of the EW2-sib and EW3-sib cells. Previous studies have shown that ectopic expression of hb within the NB7–3 lineage generates extra Eg positive cells at stage 15 and extra serotonin positive cells in the larval VNC (Isshiki et al., 2001; Novotny et al. 2002). To further investigate the origin and identity of these ectopic cells we used two Gal4 drivers, eagle-gal4 (eg-gal4) and huckebein-gal4 (hkb-gal4), to induce UAS-hb. Both eg and hkb are expressed early in NB7–3 (Broadus et al., 1995; Higashijima et al., 1996).
We first examined the expression pattern of the two Gal4 drivers by crossing each to a UAS-lacZ reporter line (Fig. 3). Both Eg and β-gal immunoreactivity were analyzed during stages 12 and 14, which is the period when the NB7–3 lineage decreases from six cells to four cells due to apoptosis of the EW2-sib and EW3-sib cells. With eg-gal4>UAS-lacZ, β-gal immunoreactivity is strong at stage 12 (Fig. 3A) but there is variable β-gal immunoreactivity in cells a stage 14 (arrows; Fig. 3B). This suggests that eg-gal4 expression decays over this time period. Although, β-gal immunoreactivity can persist past the point when Gal4 expression stops, this result demonstrates both the Gal4 and β-gal are decaying during this time frame. In comparison, hkb-gal4>UAS-lacZ β-gal immunoreactivity is strong at both stage 12 (Fig. 3C) and stage 14 (Fig. 3D) indicating consistency in the expression of hkb-gal4 during this time frame. Thus in the NB7–3 lineage, both eg-gal4 and hkb-gal4 have strong expression at stage 12, but eg-gal4 expression begins to fade by stage 14. This decay of eg expression during embryogenesis has been suggested in previous publications (Dittrich et al., 1997; Guntur et al., 2021; Higashijima et al., 1996; Lundell et al., 2003).
Figure 3. LacZ and GFP reporters demonstrate the temporal expression of two Gal4 drivers in the NB7–3 lineage.
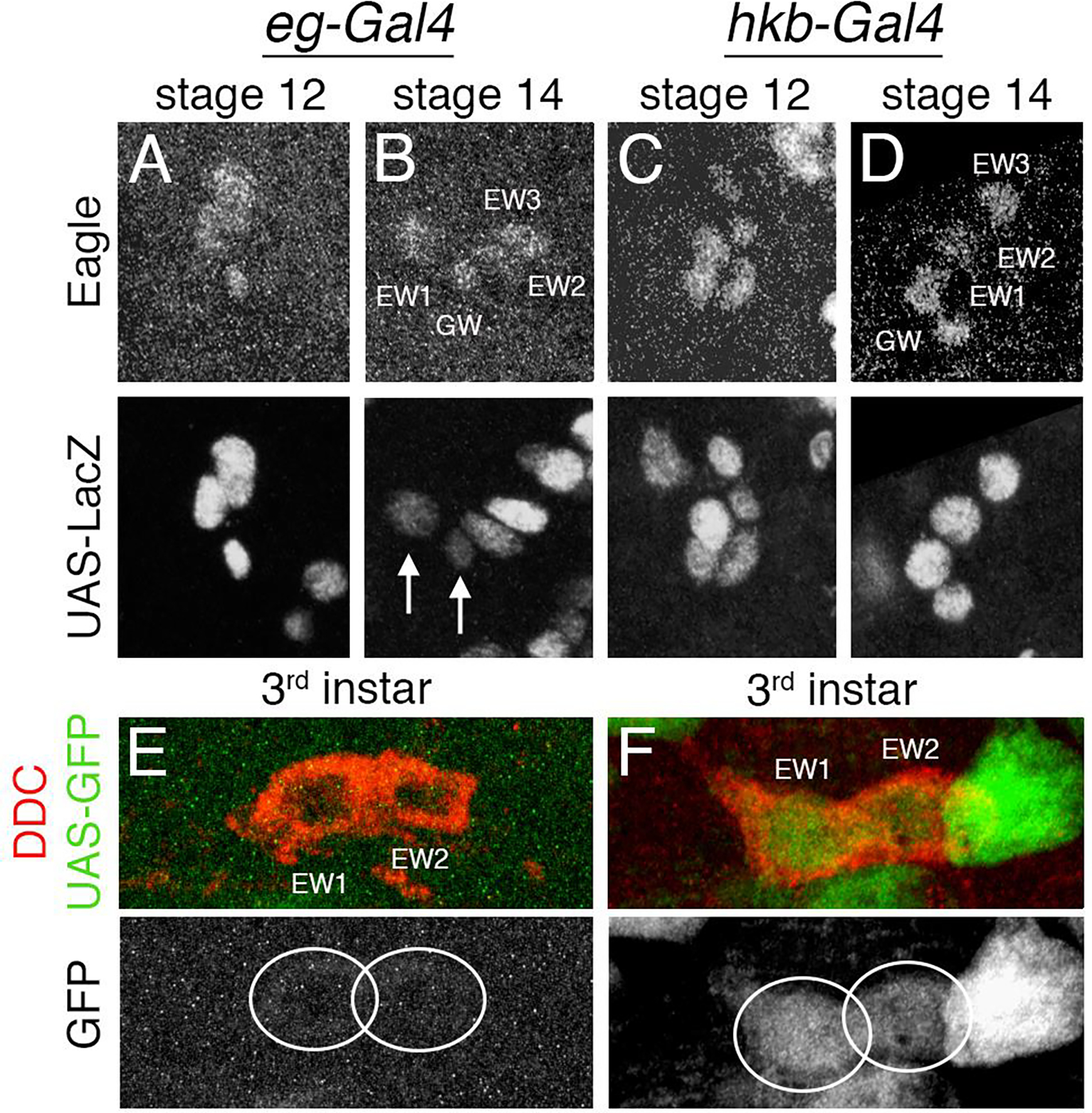
(A-D) The top panels show Eg immunostaining, the bottom panels show β-gal immunostaining for the same hemisegments. The midline is to the left in all panels. (A) An eg-gal4>UAS-lacZ stage 12 CNS shows Eg and β-gal expression in all cells of the NB7–3 lineage. (B) An eg-gal4>UAS-lacZ stage 14 CNS shows a decay of β-gal expression (arrows). (C) A hkb-gal4>UAS-lacZ stage 12 CNS shows strong expression of β-gal in all the cells in the lineage. (D) A hkb-gal4>UAS-lacZ stage 14 CNS shows β-gal expression remains strong. (E-F) 3rd instar CNS immunostained for Ddc (red) and GFP (green). GFP immunoreactivity of the same hemisegments is shown separately below. (E) An eg-gal4>UAS-gfp 3rd instar larval CNS shows a lack of GFP expression in EW1 and EW2. (F) A hkb-gal4>UAS-gfp CNS shows GFP expression in EW1 and EW2.
The expression pattern of the two Gal4 drivers was also examined in 3rd instar larvae. Each Gal4 line was crossed to the reporter UAS-gfp (Fig. 3E–F). The results show while eg-gal4 (Fig. 3E) shows no GFP expression in EW1 and EW2, hkb-gal4 (Fig. 3F) has continued GFP expression in EW1 and EW2. Thus, the larval expression of GFP reflects the embryonic expression of lacZ in that eg-gal4 expression declines earlier than hkb-gal4 expression.
Ectopic expression of hb in the NB7–3 lineage produces ectopic serotonin neurons and reduces the number of corazonin neurons.
We next examined the effects of hb overexpression in the NB7–3 lineage by using eg-gal4 and hkb-gal4 to drive expression of UAS-hb. At stage 15, a wild-type VNC has four eagle positive cells in each hemisegment and only the GW and EW1 neurons show Hb expression (Fig. 4A and Table 1A). In eg-gal4>UAS-hb 80% of the hemisegments have five or six Eg immunoreactive cells (Fig. 4B and Table 1A). In hkb-gal4>UAS-hb 71% of the hemisegments have five or six Eg immunoreactive cells (Fig. 4C and Table 1A). All the cells show expression of hb since this gene is being ectopically expressed although the level of expression can be variable between cells. Only 1–2% of the hemisegments show more than six Eg immunoreactive cells (Table 1A). In previous reports as many as eight (Novotny et al., 2002) or eleven (Isshiki et al., 2001) Eg positive cells per hemisegment were observed, but it was not reported what percentage of hemisegments had more than six cells.
Figure 4. Ectopic expression of hb in the NB7–3 lineage generates ectopic Ddc/serotonin cells and a loss of detectable corazonin cells.
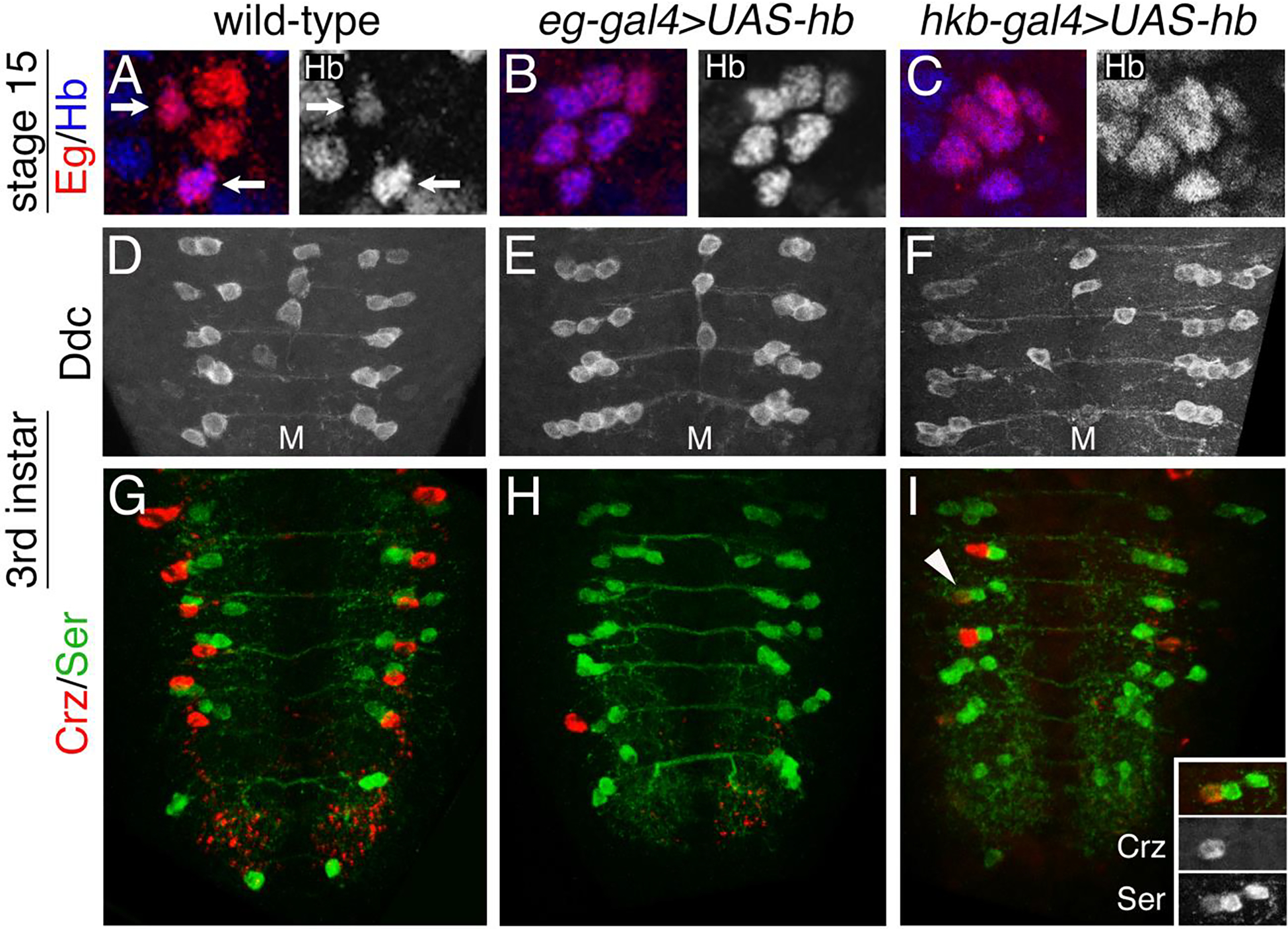
(A-C) Stage 15 CNS were immunostained for Eg (red) and Hb (blue). Hb immunoreactivity of the same hemisegments is shown separately to the right. The midline is to the left. (A) A wild-type hemisegment shows four cells immunoreactive for Eg. GW and EW1 cells are positive for Hb (arrows). (B) An eg-Gal4>UAS-hb hemisegment shows six cells immunoreactive for Eg. All cells are positive for Hb. (C) A hkb-Gal4>UAS-hb hemisegment shows six cells immunoreactive for Eg. All cells are positive for Hb. (D-F) 3rd instar larval CNS were immunostained for Ddc. (D) In a wild-type VNC two Ddc cells are detected in each hemisegment. (E) In an eg-Gal4>UAS-hb VNC three to six Ddc cells are detected in each hemisegment. (F) In a hkb-Gal4>UAS-hb VNC three to six Ddc cells are detected in each hemisegment. (G-I) 3rd instar larval CNS were immunostained for Corazonin (red) and Serotonin (green). (G) In a wild-type VNC two Ser neurons and one Crz neuron are detected in hemisegments A1-A6. (H) In an eg-Gal4>UAS-hb VNC ectopic Ser neurons and very few Crz neurons are detected. (I) In a hkb-Gal4>UAS-hb VNC ectopic Ser neurons and a few Crz neurons are detected. The hemisegment marked with an arrowhead in panel I is shown at higher magnification in the inset and shows an EW3 Crz neuron that also expresses Ser. Quantification of these results is presented in Table 1.
Table 1.
Statistical Analysis of Eg, Ddc, Hb, Serotonin and Corazonin expression in the NB7-3 lineage when UAS-hb is ectopically expressed with eg-gal4 and hkb-gal4
|
A. genotype |
stage | Number of Eg positive cells / hemisegment | n | X ± SEM | ||||
| 3 | 4 | 5 | 6 | 7 or 8 | ||||
| wild-type | 15 | 17.2% | 80.5% | 1.9% | 0.4% | 0 | 478 | 3.86 ±_0.02 |
| eg-gal4>UAS-hb | 15 | 1.6% | 16.3% | 47.9% | 31.9% | 2.3% | 430 | 5.17 ± 0.04 |
| hkb-gal4>UAS-hb | 15 | 3.3% | 24.4% | 50.5% | 20.7% | 1.1% | 368 | 4.92 ± 0.04 |
|
B. genotype |
stage | Number of Ddc positive cells / hemisegment | n | X ± SEM | ||||
| 2 | 3 | 4 | 5 | 6 | ||||
| wild-type | 1st
instar 3rd instar pupal |
100% | 0 | 0 | 0 | 0 | 280 | 2.0 ± 0.0 |
| 100% | 0 | 0 | 0 | 0 | 140 | 2.0 ± 0.0 | ||
| 100% | 0 | 0 | 0 | 0 | 84 | 2.0 ± 0.0 | ||
| eg-gal4>UAS-hb | 1st
instar 3rd instar pupal |
4.9% | 21.6% | 43.0% | 26.5% | 3.9% | 509 | 4.03 ± 0.04 |
| 5.1% | 44.2% | 34.0% | 14.0% | 2.6% | 588 | 3.65 ± 0.04 | ||
| 10.6% | 64.2% | 17.7% | 7.6% | 0 | 198 | 3.22 ± 0.05 | ||
| hkb-gal4>UAS-hb | 1st
instar 3rd instar |
5.0% | 16.3% | 56.6% | 22.0% | 0 | 159 | 3.96 ± 0.06 |
| 3.8% | 23.3% | 57.1% | 15.0% | 0.8% | 385 | 3.84 ± 0.04 | ||
|
C. genotype |
stage | Number of Hb positive Ddc cells / hemisegment | n | X ± SEM | ||||
| 0 | 1 | 2 | 3 | 4 | ||||
| wild-type | 1st
instar 3rd instar pupal |
0 | 100% | 0 | 0 | 0 | 35 | 1.0 ± 0.0 |
| 0 | 100% | 0 | 0 | 0 | 44 | 1.0 ± 0.0 | ||
| 0 | 100% | 0 | 0 | 0 | 35 | 1.0 ± 0.0 | ||
| eg-gal4>UAS-hb | 1st
instar 3rd instar pupal |
2.5% | 2.5% | 17.5% | 70.0% | 7.5% | 40 | 2.78 ± 0.09 |
| 1.5% | 0 | 51.5% | 39.4% | 7.5% | 66 | 2.52 ± 0.07 | ||
| 2.2% | 42.2% | 55.6% | 0 | 0 | 45 | 1.53 ± 0.08 | ||
| hkb-gal4>UAS-hb | 1st
instar 3rd instar |
0 | 0 | 14.3% | 78.6% | 7.1% | 14 | 2.92 ± 0.13 |
| 0 | 0 | 25.0% | 65.0% | 10.0% | 20 | 2.75 ± 0.13 | ||
|
D. genotype |
stage | Number of Serotonin positive cells / hemisegment | n | X ± SEM | ||||
| 1 | 2 | 3 | 4 | 5 | ||||
| wild-type | 3rd instar | 5.6% | 94.4% | 0 | 0 | 0 | 54 | 1.94 ± 0.03 |
| eg-gal4>UAS-hb | 3rd instar | 5.1% | 7.7% | 53.8% | 33.3% | 0 | 39 | 3.05 ± 0.11 |
| hkb-gal4>UAS-hb | 3rd instar | 7.3% | 20.0% | 49.1% | 21.8% | 1.8% | 55 | 2.91 ± 0.12 |
|
E. genotype |
stage | Number of Corazonin positive cells / hemisegment | n | X ± SEM | ||||
| 0 | 1 | 2 | 3 | 4 | ||||
| wild-type | 3rd instar | 100% | -- | -- | -- | 264 | 1.0 ± 0.0 | |
| eg-gal4>UAS-hb | 3rd instar | 99.0% | 1.0% | -- | -- | -- | 300 | 0.01 ± 0.01 |
| hkb-gal4>UAS-hb | 3rd instar | 70.8% | 29.2% | -- | -- | -- | 504 | 0.29 ± 0.02 |
Table 1. For each genotype, hemisegments were counted for the number of NB7-3 cells that expressed the specific antigen. The results are presented as a percentage of total number of hemisegments counted and as an average number of cells per hemisegments. Abdominal segments 1–6 were counted. A. NB7-3 lineage assayed for Eg at embryonic stage 15. B. NB7-3 lineage assayed for Ddc at three stages. C. NB7-3 lineage assayed for Hb at three stages. D. NB7-3 lineage assayed for serotonin in third instar VNC. E. NB7-3 lineage assayed for corazonin in third instar VNC.
We then examined whether the ectopic Eg cells observed at stage 15 result in ectopic serotonin cells in the larval VNC. In a wild-type third instar larval VNC there are normally two Ddc cells in each hemisegment (Fig. 4D and Table 1B). In third instar VNCs from either eg-gal4>UAS-hb (Fig. 4E) or hkb-gal4>UAS-hb (Fig. 4F) 95% of the hemisegments show ectopic Ddc cells. Most hemisegments have 3–5 Ddc cells with a small number of hemisegments showing six cells (Table 1B). Ectopic cells expressing Ddc results in ectopic serotonin (Ser) producing cells (Figs. 4H–4I). 87% of eg-gal4>UAS-hb hemisegments, and 73% of hkb-gal4>UAS-hb hemisegments show ectopic Ser cells (Table 1D). This result is similar to a previous report which showed 90% of hemisegments had three to four Ser cells (Novotny et al., 2002). Our results show a slight reduction in the number of hemisegments that show ectopic serotonin cells relative to the number of hemisegments that show ectopic Ddc cells, suggesting that not all Ddc expressing cells may be able to make detectable levels of serotonin.
We also examined whether the ectopic Eg cells observed at stage 15 result in ectopic EW3 corazonin cells in the larval VNC. In wild-type larval VNC there is normally one corazonin cell in hemisegment A1-A6 (Fig. 4G and Table 1E). Ectopic expression of hb results in very few detectable corazonin cells with both Gal4 drivers. For eg-gal4>UAS-hb 99% of the hemisegments lacked corazonin cells and for hkb-Gal4>UAS-hb 71% of the hemisegments lacked corazonin cells (Figs. 4H and I, and Table 1E). Thus, ectopic expression of hb in the NB7–3 lineage inhibits the EW3 cell fate. This result is similar to a previous report which showed no detectable corazonin cells in the larval VNC (Novotny et al., 2002). The inset in Fig. 3I shows a hemisegment where the EW3 cell is simultaneously expressing corazonin and serotonin, which is not observed in wild-type VNC. This suggests that the overexpression of Hb may convert the EW3 corazonin cell fate to an EW2 serotonin cell fate.
The number of ectopic NB7–3 cells induced with eg-gal4>UAS-hb decreases with time to a terminal state of three Ddc expressing cells.
The segmental variation in the number of Ddc cells that is observed with the overexpression of hb might be due in part to temporal changes in the expression of the Gal4 drivers. To investigate this possibility, an analysis of the number of Ddc positive cells and the fraction of those cells that are Hb positive at different developmental time points was examined for both Gal4 drivers.
Wild-type VNC consistently show two Ddc cells per hemisegment with one being Hb positive (EW1) throughout development. In eg-gal4>UAS-hb the average number of Ddc cells/hemisegment decreases from 1st instar to 3rd instar larval stages and decreases further in the pupal stage. At the pupal stage the most common number of Ddc cells/hemisegment is three cells (64%; Table 1B), which is one additional cell compared to wild-type. Thus, although an eg-gal4>UAS-hb VNC can show as many as six Ddc cells/hemisegment at embryonic stage 15, with time the number of cells detected decreases towards a terminal state of three Ddc cells at the pupal stage (Fig. 5 and Table 1B). Hb expression in these cells also decreases toward a terminal state of one Hb cell (Fig. 5 and Table 1C). This single Hb positive cell is the EW1 cell that normally expresses Hb (Figs. 7E–F and 8D–G). In comparison, hkb-gal4>UAS-hb which maintains high Gal4 expression through 3rd instar (Fig. 3F), shows very little change in the number of Ddc and Hb immunoreactive cells between the 1st and 3rd instar larval stages (Fig. 5 and Table 1). Overexpression of hb with hkb-gal4 results in lethality at the pupal stage. The conclusion of these results is that as Hb expression decays in eg- gal4>UAS-hb VNC the number of detectable Ddc cells in the lineage decreases to three, which are most likely EW1, EW2 and the converted EW3 corazonin neuron, with only the EW1 cell retaining Hb immunoreactivity. The variability of Hb decay from cell to cell will contribute to the variation in the number of Ddc observed between hemisegments. Thus, although the overexpression of hb can transiently produce as many as six Ddc cells, with time the number of Ddc cells decreases to three.
Figure 5. Decrease in the number of ectopic Ddc and Hb positive cells with time.
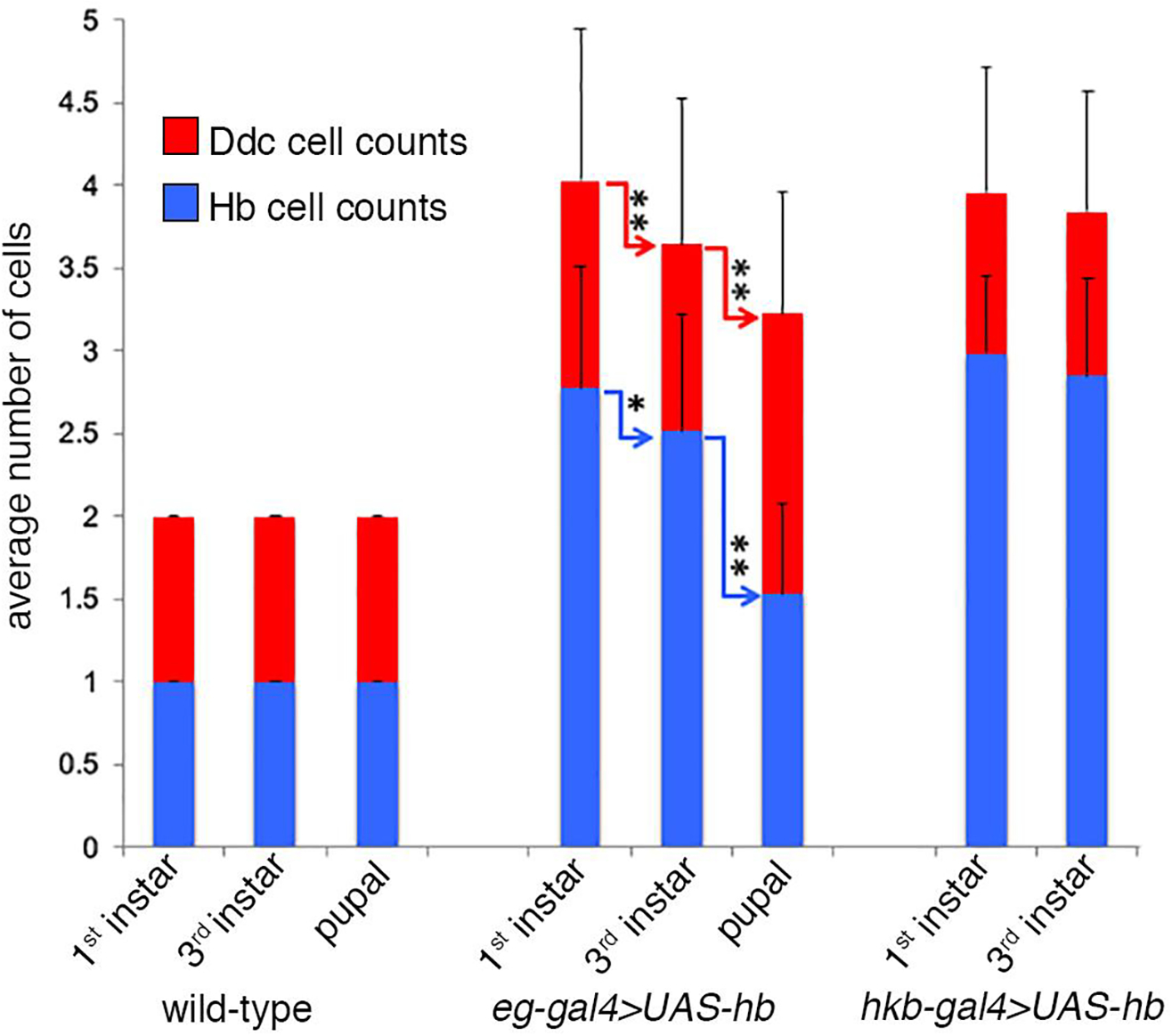
This is a graphical representation of the data presented in Table 1B–C. The average number of Ddc cells (red) detected per hemisegment and the average number that are simultaneously Hb positive (blue) are shown at different developmental stages for the overexpression of hb using the eg-gal4 and hkb-gal4 drivers. Overexpression of hb with hkb-gal4 produces lethality at the pupal stage. The eg-gal4 driver shows a decrease in the number of Ddc cells per hemisegment over time, as well as decrease in the number that are Hb positive. Connecting arrows indicate a significant difference in the number of labeled cells between different developmental stages (** indicates a p value <0.0001 and * indicates a p value <0.04). Statistical analysis was done using nonparametric comparisons with either the Wilcoxon-Mann-Whitney or Kruskal-Wallis statistical tests (Sprent, 1988).
Ectopic expression of hb in NB7–3 generates ectopic EW2-like Ddc cells
To determine the identity of the ectopic Ddc cells produced with the overexpression of hb we examined several molecular markers. Hb is used to identify EW1 cell fate. Pdm is expressed specifically in EW2 cells at the larval stage (Lundell and Hirsh, 1998; Lundell et al., 2003), and Zfh-2 is expressed in both EW2 and EW3 (Isshiki et al., 2001; Lundell and Hirsh, 1998; Lundell et al., 2003; Novotny et al., 2002). We first checked whether overexpression of hb affects the expression of pdm and zfh-2. When the general neuronal driver elav-Gal4 is used to drive expression of UAS-hb many more Hb positive cells are detected compared to a wild-type VNC (Figs. 6A and 6B). In a wild-type VNC both Zfh-2 and Pdm are detected in many neurons (Figs 6C and E). In an elav-Gal4>UAS-hb VNC, cells that express zfh-2 and pdm are no longer detectable (Figs. 6D and F). The suppression of zfh-2 and pdm by Hb has been previously reported (Guntur et al., 2021; Hirono et al., 2017; Isshiki et al., 2001; Kambadur et al., 1998; Novotny et al., 2002; Tran et al., 2010). Thus, relying on the lack of Pdm and Zfh-2 expression as markers of cell identity is problematic when hb is ectopically expressed. However, given the temporal decay of eg-Gal4>UAS-hb we reasoned it might be possible to detect these markers in some hemisegments at the larval stage.
Figure 6. Hb represses expression of zfh-2 and pdm.
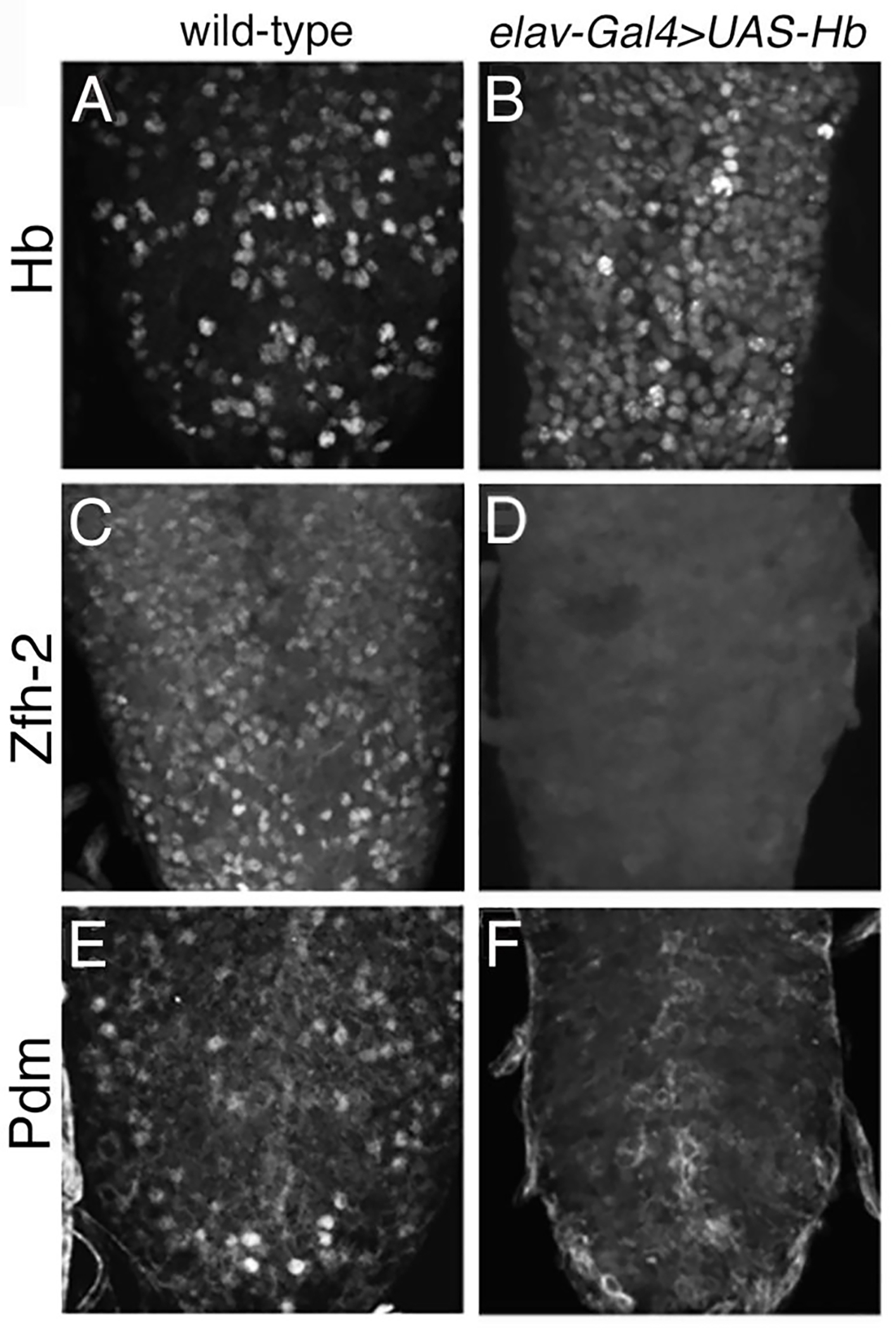
All panels show 3rd instar larval VNC immunostained for Hb, Zfh-2, or Pdm. Compared to wild-type, elav-Gal4>UAS-hb shows an increase in cells positive for Hb (A-B) and a dramatic decrease in cells positive for Zfh-2 (C-D) and Pdm (E-F).
In a wild-type stage 15 VNC all four NB7–3 progeny, GW, EW1, EW2, and EW3, are detected by Eg immunoreactivity. The GW and EW1 neurons express Hb, and the EW2 and EW3 neurons express Zfh-2 (Fig. 7A). In eg-Gal4>UAS-hb, 4–6 Eg immunoreactive cells are detected in each hemisegment (Fig. 7B and Table 1A). All of the cells show continued expression of hb since this gene is being ectopically expressed, most of the cells do not show expression of Zfh-2, which is to be expected since we have shown that Hb can suppress zfh-2 expression (Fig. 6E). However, occasionally an Eg cell with Zfh-2 expression is observed if that cell has a reduced level of Hb (Fig. 7B; arrow).
Figure 7. Ectopic expression of hb in NB7–3 generates ectopic EW2-like Ddc cells.
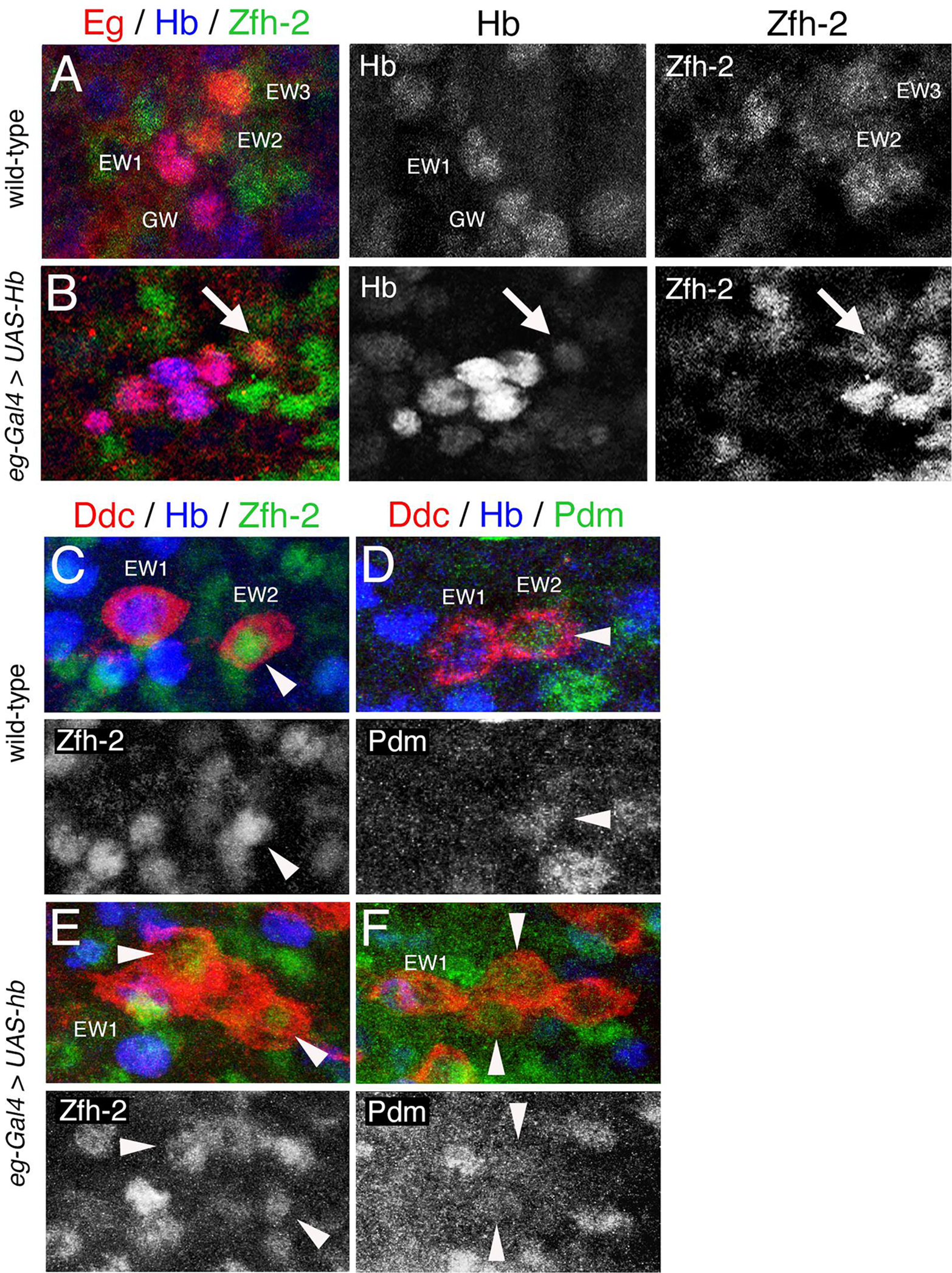
(A-B) Stage 15 VNC were immunostained for Eg (red), Hb (blue), and Zfh-2 (green). Hb and Zfh-2 immunoreactivity of the same hemisegments is shown separately to the right. (A) A wild-type hemisegment showing four Eg cells. GW and EW1 cells express Hb. EW2 and EW3 cells express Zfh-2. (B) An eg-Gal4>UAS-hb hemisegment showing six Eg cells. All cells express Hb. One cell that expresses Hb at low levels (arrow) also expresses Zfh-2. (C-F) show 3rd instar VNC immunostained for Ddc (red), Hb (blue) and either Zfh-2 or Pdm (green). Zfh-2 and Pdm immunoreactivity of the same hemisegments is shown separately below. Arrowheads indicate co-localization between Ddc and either Zfh-2 or Pdm. (C-D) Two wild-type hemisegments, showing EW1 expresses Hb and EW2 expresses Zfh2 (C) and Pdm (D). (E-F) Two eg-gal4>UAS-hb hemisegments each showing four Ddc cells where two cells express Zfh-2 (E) or Pdm (F). This suggest that the ectopic Ddc cells have an EW2-like cell fate. The midline is to the left in all panels.
When Hb is overexpressed many of the ectopic Ddc cells even at third instar stage still express Hb (Fig. 5 and Table 1C). However, Fig.7E–F shows select hemisegments where Hb has decreased sufficiently to relieve the suppression on zfh-2 and pdm expression such that these markers become detectable. In the wild-type VNC both Zfh-2 (Fig. 7C) and Pdm (Fig. 7D) are expressed in EW2. Fig. 7E shows an eg-gal4>UAS-hb hemisegment with four Ddc cells. The EW1 is distinguishable by Hb expression and of the remaining three cells: two show Zfh-2 expression. Similarly, Fig. 7F, shows an eg-gal4>UAS-hb hemisegment with four Ddc cells, one expresses Hb and two express Pdm. The detection of Pdm and Zfh-2 in more than one Ddc cell indicates that overexpression of Hb in the NB7–3 lineage generates ectopic EW2-like Ddc cells rather than EW1 cells. Not all the cells that lack Hb expression show expression of Zfh-2 or Pdm. It is possible in these cells that Hb expression has decayed beyond detection, but not enough time has elapsed to allow for detectable expression of Zfh-2 or Pdm. Alternatively, some of the ectopic Ddc cells may not be capable of expressing markers that resemble either EW1 or EW2. We propose that the ectopic EW2-like Ddc cells produced but the overexpression of hb are either the rescued EW2-sib and EW3-sib apoptotic cells, or EW3 cells converted from a corazonin cell fate to an EW2 Ddc cell fate.
Ectopic expression of hb in the NB7–3 lineage produces ectopic Ddc cells that express Zfh-1.
Zfh-1 is normally expressed in the GW neuron and in the apoptotic EW2-sib and EW3-sib cells, but not in the EW neurons (Lee and Lundell, 2007). We examined whether ectopic Ddc cells observed with eg-gal4>UAS-hb are immunoreactive for Zfh-1. At stage 15, a hemisegment with four eg-gal4>UAS-GFP positive cells shows a wild-type pattern in that only the GW neuron expresses Zfh-1(Fig. 8A). A hemisegment with six eg-gal4>UAS-GFP positive cells shows three Zfh-1 positive cells (Fig. 8B). This pattern is identical to the wild-type pattern of Zfh-1 expression at stage 12 before apoptosis or when apoptosis is blocked with the apoptosis inhibitor p35 (Lee and Lundell, 2007).
Figure 8. Ectopic expression of hb in the NB7–3 lineage produces ectopic Ddc cells that express Zfh-1.
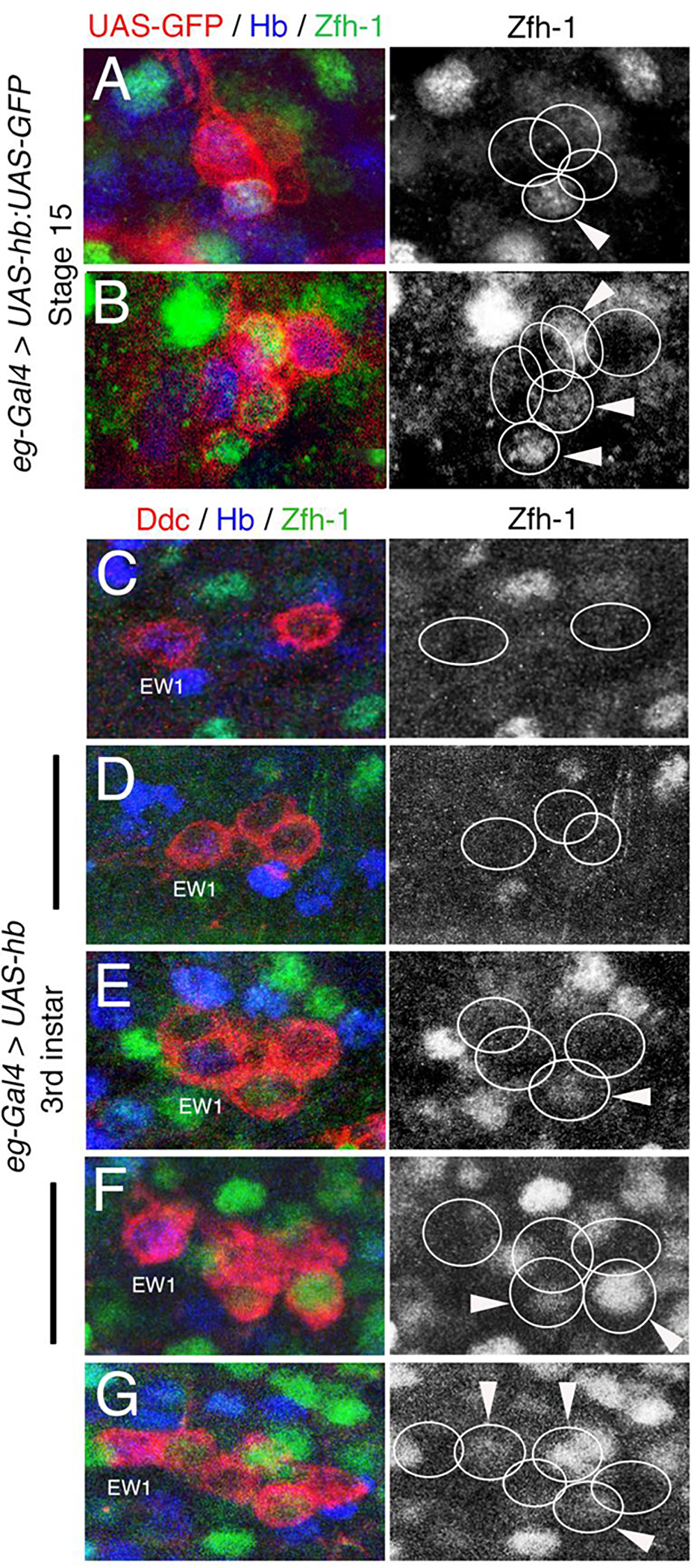
(A-B) Stage 15 embryonic eg-Gal4>UAS-Hb:UAS-PON-GFP VNC labeled for Eg-GFP (red), and Zfh-1 (green). Red and green colors were reversed electronically to consistently label all lineage cells in red. Zfh-1 immunoreactivity of the same hemisegments is shown separately to the right. (A) A hemisegment showing a wild-type pattern of four Eg-GFP and one cell (GW) that expresses Zfh-1. (B) A hemisegment showing six Eg-GFP cells and three that express Zfh-1. (C-G) Third instar larval eg-gal4>UAS-hb VNC immunostained for Ddc (red), Hb (blue) and Zfh-1 (green). Zfh-1 immunoreactivity of the same hemisegments is shown separately to the right. (C) A hemisegment showing a wild-type patten of two Ddc cells, neither cell shows Zfh-1 expression and the EW1 is positive for Hb. (D) A hemisegment showing three Ddc cells, none of which express Zfh1. (E) A hemisegment showing four Ddc cells, one of which expresses Zfh-1. (F) A hemisegment showing five Ddc cells, two of which express Zfh-1. (G) A hemisegment showing six Ddc cells, three of which express Zfh-1. The midline is to the left in all panels.
In eg-gal4>UAS-hb larval hemisegments that have two (Fig. 8C) or three (Fig. 8D) Ddc cells, none of the cells show Zfh-1 immunoreactivity (n=17). These segments presumably contain only the EW neurons, including the conversion of EW3 to a Ddc neuron. In larval hemisegments with four-six Ddc cells, 51% (n=35) have one (Fig. 8E), two (Fig. 8F), or three (Fig. 8G) Zfh-1 positive Ddc cells. The novel co-expression of Ddc and Zfh-1 in hemisegments with four or more Ddc cells, supports our hypothesis that the apoptotic cells in the NB7–3 lineage are rescued by the overexpression of hb.
Ectopic expression of hb is sufficient to prevent apoptosis induced by the ectopic expression of pro-apoptotic genes.
Since our results suggest that overexpression of hb can prevent apoptosis in the EW2-sib and EW3-sib cells, we asked whether overexpression of hb can prevent apoptosis induced by the misexpression of pro-apoptotic genes within the NB7–3 lineage. We have previously shown that ectopic expression of UAS-notchAct in the NB7–3 lineage with eg-gal4 induces apoptosis of the EW neurons (Fig. 9C) (Lundell et al., 2003). Here we also show that ectopic expression of the pro-apoptotic genes grim (Fig. 9E) and hid (Fig. 9G) with eg-gal4 can also promote apoptosis of the EW neurons but with a less severe phenotype than UAS-notchAct.
Figure 9. Ectopic expression of hb is sufficient to prevent apoptosis induced by ectopic expression of grim or hid but not Notch.
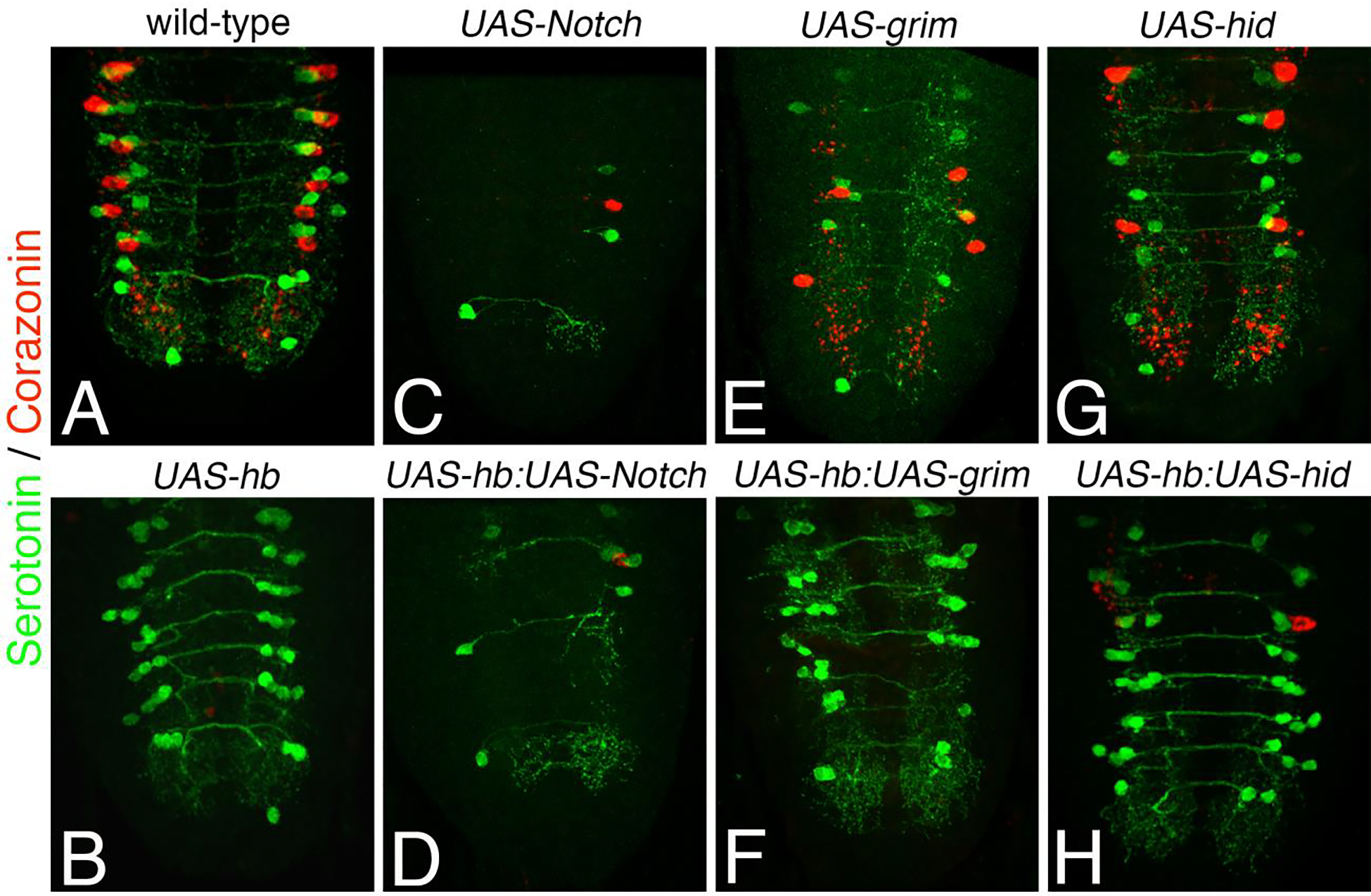
All UAS constructs were activated with eg-Gal4. 3rd instar larval VNC were immunostained for serotonin (green) and corazonin (red). (A) A wild-type VNC shows the EW1 and EW2 Ser neurons and the EW3 Crz neurons. (B) An eg-Gal4>UAShb VNC shows ectopic Ser neurons and no Crz neurons. (C, E and G) Ectopic expression of UAS-notchACT (C), UAS-grim (E) and UAS-hid (G) all lead to reduced numbers of detectable EW neurons, and panels are ordered in decreasing severity of the phenotype. (D) Simultaneous expression of UAS-hb and UAS-notchACT does not change the UAS-notchACT phenotype. (F and H) Simultaneous expression of UAS-hb with either UAS-grim (F) or UAS-hid (H) can reduce apoptosis in the NB7–3 lineage.
When UAS-notchAct is expressed with eg-gal4 there is a 97% reduction in the number of detectable serotonin neurons compared to wild-type (Fig. 9C and Table 2). When both UAS-notchAct and UAS-hb are simultaneously expressed with eg-gal4 (Fig. 9D) the UAS-NotchAct phenotype persists and there is no detectable rescue of serotonin cells (Table 2). When UAS-grim is expressed with eg-gal4 there is a 57% reduction in the number of detectable serotonin neurons compared to wild-type (Fig. 9E and Table 2). When UAS-hid is expressed with eg-gal4 there is a 39% reduction in the number of detectable serotonin neurons compared to wild-type (Fig. 9G and Table 2). When either UAS-grim or UAS-hid are simultaneously expressed with UAS-hb there is an increase in the number of detectable serotonin cells to within 75% of the number of cells detected in wild-type (Fig. 9F, 9H and Table 2). Therefore, the simultaneous expression of UAS-hb can only partially rescue apoptotic cells induced by the expression of UAS-grim or UAS-hid. It is not possible to determine from these experiments if there is any preference as to which cells in the lineage are blocked from apoptosis by Hb, but some of the hemisegments have 4–5 serotonin neurons (8.0% in UAS-Hb:UAS-grim and 9.5% in UASHb:UAS-hid) which suggests that both the EW neurons and the sibling apoptotic cells can be recovered. These results demonstrate that overexpression of hb in NB7–3 can prevent apoptosis that is genetically induced by UAS-grim or UAS-hid, but not apoptosis genetically induced with UAS-Notch. This may be because Notch signaling may activate multiple components of the apoptotic pathway whereas Hb may only block one component, or it could be that the strength of UAS-NotchAct out-competes the effects of UAS-Hb.
Table 2.
Comparison of the number of seratonin and corazonin cells in the NB7-3 lineage when hb is ectopically expressed simultaneously with three pro-apoptotic genes.
| genotype | # of Ser cells/hemisegment X ± SEM | # of Crz cells/hemisegment X ± SEM | Total cells/hemisegment | % change from wild-type | Ser n value | Crz n value |
|---|---|---|---|---|---|---|
| wild-type | 1.98 ± .01 | 1.00 ± 0 | 2.98 | --- | 168 | 168 |
| UAS-hb | 3.38 ± .02 | 0.00 ± 0 | 3.38 | +13% | 123 | 123 |
| UAS-Notch | 0.09 ± .02 | 0.01 ± .01 | 0.10 | −97% | 378 | 324 |
| UAS-Notch:hb | 0.10 ± .02 | 0.01 ± .01 | 0.11 | −97% | 448 | 384 |
| UAS-grim | 0.95 ± .06 | 0.32 ± .03 | 1.27 | −57% | 161 | 312 |
| UAS-grim:hb | 2.19 ± .09 | 0.04 ± .02 | 2.23 | −25% | 138 | 144 |
| UAS-hid | 1.39 ± .05 | 0.43 ± .03 | 1.82 | −39% | 159 | 240 |
| UAS-hid:hb | 2.16 ± .07 | 0.10 ± .02 | 2.20 | −26% | 241 | 276 |
Table 2. For each genotype, hemisegments were counted for the number of NB7-3 cells that expressed serotonin and corazonin. The results are presented as an average of the number serotonin and corazonin of cells per hemisegments, the total number of cells per hemisegment and the percent change in the number of cells per hemisegement compared to wild-type. Abdominal segments 1–6 were counted.
Interestingly we have found that, unlike UAS-hid and UAS-grim, expression of UAS-rpr in the NB7–3 lineage does not significantly induce apoptosis of the EW neurons (data not shown). It could be that the NotchOFF EW cells are not sensitive to the induced expression of UAS-rpr because there is a robust mechanism to inhibit Rpr activity since it may be the primary inducer of apoptosis in the lineage. This would agree with previous studies that suggested rpr is primarily responsible for apoptosis during neurogenesis (Lee et al., 2013; Peterson et al., 2002; Robinow et al., 1997; Tan et al., 2011; White et al., 1994) and that NotchON cells specifically use rpr to induce apoptosis in optic lobe neuroblasts (Bertet et al., 2014).
DISCUSSION
Apoptosis in the NB7–3 lineage is positively regulated by both Notch (Lundell et al., 2003) and Zfh-2 (Guntur et al., 2021). In this manuscript we show that apoptosis in the NB7–3 lineage is negatively regulated by Hb. Our results demonstrate, that in wild-type embryos hb expression in the GMC1 lineage of NB7–3 prevents early apoptosis of the GW neuron, and that hb expression must be suppressed in the GMC2 and GMC3 lineages to allow for apoptosis of the EW2-sib and EW3-sib cells.
With a hb loss-of-function allele we have shown that the GW neuron undergoes early apoptosis (Fig.2). During normal development the GW motor neurons eventually undergo Ubx-mediated apoptosis at late stage 16 (Rogulja-Ortmann et al., 2007; Rogulja-Ortmann et al., 2008). Hb is a known repressor of Ubx during embryonic segmentation (Irish et al., 1989; Qian et al., 1991; White and Lehmann, 1986; Zhang and Bienz, 1992). It may be that Hb temporarily represses Ubx-mediated apoptosis in the GW neuron, but a subsequent decrease in hb expression at stage 16 may allow the GW neuron to undergo Notch-induced apoptosis similar to the apoptotic EW2-sib and EW3-sib cells. Hb has also been shown to prevent apoptosis of bridge cell precursors during development of the Drosophila tracheal system (Wolf and Schuh, 2000).
Overexpression of hb in several neuronal lineages has been shown to retain the neuroblast in a “young state” such that it undergoes supernumerary divisions and produces ectopic neurons with first-born identity, reviewed in (Doe, 2017; Kohwi and Doe, 2013; Li et al., 2013; Sen et al., 2019). Subsequent downregulation of hb expression allows the neuroblast to resume normal gene expression patterns and normal lineage divisions (Grosskortenhaus et al., 2005). There is a window of competency for the supernumerary divisions that closes at the end of stage 12 (Cleary and Doe, 2006; Kohwi et al., 2013; Pearson and Doe, 2003; Touma et al., 2012; Tran and Doe, 2008). For example in the well-studied NB7–1 lineage, the neuroblast can produce three extra cells of first-born identity before the competency window closes (Pearson and Doe, 2003). NB7–1 is one of the earliest NBs to form (late stage 8), whereas NB7–3 is one of the last (late stage 11) (Doe, 1992). Therefore, the window of competency for NB7–3 to generate extra GMC-1 type progeny would be much shorter.
In our studies examining the overexpression of hb we find that most hemisegments do not appear to make extra NB7–3 divisions. At stage 15, the majority of hemisegements (80%) show five to six Eg positive cells (Table 1A), and by the larval stage we mostly observe three to five Ddc cells per hemisegment (Table 1B). All of these cells can be accounted for by the three normal NB7–3 lineage divisions. Hemisegements with five Ddc cells would most likely contain, EW1, EW2, the EW3 corazonin neuron that has been converted to a Ddc neuron, and the rescue of the two sibling apoptotic cells. Hemisegments with six Ddc cells are observed at a very low frequency (<2.6%) (Table 1B). The sixth cell could be the conversion of the GW into a Ddc cell which occurs under with other genetic mutations such as the loss of Delta (Fig. 1), loss of Sanpodo (Lundell et al., 2003) and loss of Zfh-1 (Lee and Lundell, 2007). Alternatively, if the GW undergoes normal apoptosis at late stage 16 (Rogulja-Ortmann et al., 2007; Rogulja-Ortmann et al., 2008), despite elevated levels of Hb, then hemisegments with six Ddc cells could be due to retention of NB7–3 or an extra NB7–3 division.
We have also demonstrated that overexpression of hb produces ectopic Ddc cells that express identity markers similar to GMC2 progeny (Zfh-2 and Pdm) rather than GMC1 progeny (Hb) (Fig. 7E and F). The visualization of these markers is only possible at the larval stage when the decline in eg-Gal4>UAShb expression removes the repressive effect that Hb has on zfh-2 and pdm expression (Fig. 6). In addition, we have shown that the ectopic Ddc cells express Zfh-1 (Fig. 8), which is a feature of all the NotchON progeny in the lineage, including the apoptotic cells (Lee and Lundell, 2007). These ectopic Ddc/Zfh-2/Pdm/Zfh-1 positive cells are most likely the rescue of the two apoptotic cells in the lineage. The cell context that allows the overexpression of Hb to maintain the neuroblast in a “young state” may also maintain the neuronal progeny in a “young state” such that the EW-sib cells do not immediately undergo apoptosis and are initially maintained like the GW neuron. However, this alteration in developmental history is transient because once the expression of eg-Gal4>UAShb declines the cells resume their original fate, undergoing apoptosis and producing a terminal state of three Ddc cells (Fig. 5 and Table 1B) which are most likely EW1, EW2 and the converted EW3.
In comparison, the conversion of the EW3 corazonin cell to a Ddc cell appears to be a permanent change in cell fate. Corazonin expression does not reappear as the expression of eg-Gal4>UAShb declines. When krüpple (kr) is overexpressed in the NB7–3 lineage there is also a reduction in the number of corazonin neurons (Isshiki et al., 2001). The hierarchy of temporal transcription factors shows that Hb activates the expression of kr, and that Kr activates the expression of pdm (Brody and Odenwald, 2000, 2002; Doe, 2017; Isshiki et al., 2001; Kambadur et al., 1998; Novotny et al., 2002). Thus, the overexpression of hb could indirectly convert receptive EW3 cells to an EW2 Pdm/Ddc expressing cell fate.
Our results suggest that overexpression of hb in the NB7–3 lineage has heterogeneous effects on neuronal phenotypes. Using single cell labeling to resolve the EW1 and EW2 innervation patterns, it was shown that eg-Gal4>UAShb generates EW1-type projections in the NB7–3 lineage (Chen and Condron, 2008). Our interpretation is that these neurons are the terminal Ddc cells that survive once Hb levels decline. These results along with our results suggest that overexpression of hb is sufficient to disrupt terminal phenotypes of the EW neurons, but is insufficient to alter the programed apoptotic fate of the EW sibling cells.
Similar heterogeneity in response to ectopic hb expression has been observed in the NB7–1 lineage. Although as many as 10–15 Hb expressing motor neurons are generated by the overexpression of hb in NB7–1 (Isshiki et al., 2001; Kohwi et al., 2013; Meng et al., 2019; Pearson and Doe, 2003), only the earliest cells within the competency window are fully transformed to first-born U1 motor neurons, expressing definitive first-born molecular markers (Cleary and Doe, 2006; Kohwi et al., 2013; Pearson and Doe, 2003) and having the appropriate muscle and neuropil targeting (Seroka and Doe, 2019). Many of the later motor neurons lose Hb expression by late larval stages and have muscle targets that are inconsistent with first-born identity, suggesting that these cells are not fully transformed (Meng et al., 2019). Interestingly the end of the competency window that can generate first-born identity in NB7–1 has been correlated with an inability of Hb to repress zfh-2 expression (Seroka and Doe, 2019). These studies reflect what we have presented for the NB7–3 lineage in that Hb can transform the apoptotic EW sibling cells such that they initially express Ddc similar to the EW neurons, but once Hb levels decline and zfh-2 and pdm expression resumes the cells undergo apoptosis in late larval and pupal stages.
Determining how Hb transiently inhibits apoptosis will require further investigation. Hb is a bifunctional transcription factor that can activate or repress gene expression by binding to cis-regulatory elements (Papatsenko and Levine, 2008; Schulz and Tautz, 1994; Staller et al., 2015; Struhl et al., 1992; Vincent et al., 2018; Zuo et al., 1991). Hb has also been shown to have chromatin remodeling activity (Hirono et al., 2017; Kehle et al., 1998; Zhang and Bienz, 1992). Given these diverse regulatory capabilities, Hb could impose developmental decisions by regulating various genetic components of the apoptosis mechanism. Hb could work directly by repressing pro-apoptotic genes (e.g. rpr, hid, grim, caspases) or by activating inhibitors of apoptosis (IAPS). Alternatively, Hb could work indirectly by repressing or activating transcriptional regulators of apoptosis.
In this manuscript we have shown when apoptosis is genetically induced in the EW neurons by the expression of UAS-grim or UAS-hid, that simultaneous expression of UAS-hb can partially rescue these phenotypes (Figs. 9F and 9H). Since the UAS promoters are unlikely to be influenced by Hb, this suggests that the ability of Hb to override apoptosis in these experiments is due to activation of a component that can inhibit Hid and Grim function, possibly an IAP. Alternatively, Hb could repress Zfh-2, a positive regulator of apoptosis. In a previous manuscript we have shown that Zfh-2 is necessary to induce apoptosis in the NB7–3 lineage, and that Hb and Zfh-2 reciprocally repress each other to help define the GMC1 lineage from the GMC2 and GMC3 lineages (Guntur et al., 2021). This could explain how in the numb1 mutant, where Notch is activated in all three EW neurons, that the hb expressing EW1 neurons survive but the zfh-2 expressing EW2 and EW3 neurons undergo apoptosis (Lundell et al., 2003).
The experiments we have presented in this manuscript do not rule out the possibility that overexpression of hb in the NB7–3 lineage could, depending on the cellular context of each hemisegment, either rescue the apoptotic cells or induce extra neuroblast divisions. Such behavior might account for the variability in the number of Ddc cells seen between hemisegments. The rescue of NotchON apoptotic cells by the overexpression of hb may likely apply to other neuronal lineages and may complicate the interpretation of specific genetic phenotypes. Further investigation on how hb expression can transiently inhibit apoptosis will be important to elucidating the molecular regulation of apoptosis in the Drosophila CNS lineages.
Highlights:
Hunchback is necessary to prevent apoptosis of the GW neuron during specification of the Drosophila NB 7–3 lineage.
Ectopic Hunchback expression rescues apoptotic cells in the Drosophila NB 7–3 lineage.
Ectopic Hunchback produces ectopic serotonin neurons that resemble the EW2 neuron in the Drosophila NB 7–3 lineage.
Ectopic Hunchback can prevent apoptosis induced by the misexpression of pro-apoptotic genes in the Drosophila NB 7–3 lineage.
ACKNOWLEDGEMENTS
This work was supported by NIH/MBRS/SCORE grant GM 08184 to M. J. L. and a NIH MBRS Rise Fellowship GM 60655 and SLOAN Fellowship to E. P. The authors acknowledge the Cell Analysis Core Facility at the University of Texas at San Antonio for support during this work. Confocal imaging for this project was supported by grants from the National Center for Research Resources (5 G12RR013646–12) and the National Institute on Minority Health and Health Disparities (G12MD007591) from the National Institutes of Health. We thank G. Technau, C. Doe, A. Whitworth, Y. N. Jan and the Bloomington Stock Center for provision of stocks. We thank G. Technau, C. Doe, R. Lehman, T. Dick and A. Tomlinson for provision of antisera. We thank Colleen Witt, Robert Cook, Sandra Cordona and Judy Haschenbruger for technical assistance. We thank Claire Cronmiller for comments on the manuscript.
Footnotes
Declarations of Interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ambrosini A, Gracia M, Proag A, Rayer M, Monier B, Suzanne M, 2017. Apoptotic forces in tissue morphogenesis. Mech Dev 144, 33–42. 10.1016/j.mod.2016.10.001 [DOI] [PubMed] [Google Scholar]
- Bertet C, Li X, Erclik T, Cavey M, Wells B, Desplan C, 2014. Temporal patterning of neuroblasts controls Notch-mediated cell survival through regulation of Hid or Reaper. Cell 158, 1173–1186. 10.1016/j.cell.2014.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM, 1996. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol 179, 41–64. 10.1006/dbio.1996.0240 [DOI] [PubMed] [Google Scholar]
- Bridges CB, Brehme KS, 1944. The mutants of Drosophila melanogaster, Washington, D.C.,.
- Broadus J, Skeath JB, Spana EP, Bossing T, Technau G, Doe CQ, 1995. New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mech Dev 53, 393–402. 10.1016/0925-4773(95)00454-8 [DOI] [PubMed] [Google Scholar]
- Brody T, Odenwald WF, 2000. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Dev Biol 226, 34–44. 10.1006/dbio.2000.9829 [DOI] [PubMed] [Google Scholar]
- Brody T, Odenwald WF, 2002. Cellular diversity in the developing nervous system: a temporal view from Drosophila. Development 129, 3763–3770 [DOI] [PubMed] [Google Scholar]
- Chen J, Condron BG, 2008. Branch architecture of the fly larval abdominal serotonergic neurons. Dev Biol 320, 30–38. 10.1016/j.ydbio.2008.03.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary MD, Doe CQ, 2006. Regulation of neuroblast competence: multiple temporal identity factors specify distinct neuronal fates within a single early competence window. Genes Dev 20, 429–434. 10.1101/gad.1382206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich R, Bossing T, Gould A, Technau G, Urban J, 1997. The differentiation of the serotonergic neurons in the Drosophila ventral nerve cord depends on the combined function of the zinc finger proteins Eagle and Huckebein. Development 124, 2515–2525 [DOI] [PubMed] [Google Scholar]
- Doe C, 1992. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development 116, 855–863 [DOI] [PubMed] [Google Scholar]
- Doe CQ, 2017. Temporal Patterning in the Drosophila CNS. Annu Rev Cell Dev Biol 33, 219–240. 10.1146/annurev-cellbio-111315-125210 [DOI] [PubMed] [Google Scholar]
- Fadeel B, Orrenius S, 2005. Apoptosis: a basic biological phenomenon with wide-ranging implications in human disease. J Intern Med 258, 479–517. 10.1111/j.1365-2796.2005.01570.x [DOI] [PubMed] [Google Scholar]
- Favaloro B, Allocati N, Graziano V, Di Ilio C, De Laurenzi V, 2012. Role of apoptosis in disease. Aging (Albany NY) 4, 330–349. 10.18632/aging.100459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty CE, Bergmann A, 2017. Killers creating new life: caspases drive apoptosis-induced proliferation in tissue repair and disease. Cell Death Differ 24, 13901400. 10.1038/cdd.2017.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y, Steller H, 2011. Programmed cell death in animal development and disease. Cell 147, 742–758. 10.1016/j.cell.2011.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman C, Doe C, 1993. Embryonic development in the Drosophila central nervous system in: Bate M, Martinez-Aris A (Eds.), The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press, Plainview, NY, pp. 1131–1206. [Google Scholar]
- Grosskortenhaus R, Pearson BJ, Marusich A, Doe CQ, 2005. Regulation of temporal identity transitions in Drosophila neuroblasts. Dev Cell 8, 193–202. 10.1016/j.devcel.2004.11.019 [DOI] [PubMed] [Google Scholar]
- Guntur AR, Venkatanarayan A, Gangula S, Lundell MJ, 2021. Zfh-2 facilitates Notch-induced apoptosis in the CNS and appendages of Drosophila melanogaster. Dev Biol 475, 65–79. 10.1016/j.ydbio.2021.02.009 [DOI] [PubMed] [Google Scholar]
- Guo M, Jan LY, Jan YN, 1996. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17, 27–41. 10.1016/s0896-6273(00)80278-0 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA, 2000. The hallmarks of cancer. Cell 100, 57–70. 10.1016/s0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Campos-Ortega JA, 1984. Early neurogenesis in wild-typeDrosophila melanogaster. Development Genes and Evolution 193, 308–325. 10.1007/BF00848159 [DOI] [PubMed] [Google Scholar]
- Higashijima S, Shishido E, Matsuzaki M, Saigo K, 1996. eagle, a member of the steroid receptor gene superfamily, is expressed in a subset of neuroblasts and regulates the fate of their putative progeny in the Drosophila CNS. Development 122, 527–536 [DOI] [PubMed] [Google Scholar]
- Hirono K, Kohwi M, Clark MQ, Heckscher ES, Doe CQ, 2017. The Hunchback temporal transcription factor establishes, but is not required to maintain, early-born neuronal identity. Neural Dev 12, 1. 10.1186/s13064-017-0078-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish VF, Martinez-Arias A, Akam M, 1989. Spatial regulation of the Antennapedia and Ultrabithorax homeotic genes during Drosophila early development. EMBO J 8, 1527–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ, 2001. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511–521. 10.1016/s0092-8674(01)00465-2 [DOI] [PubMed] [Google Scholar]
- Kambadur R, Koizumi K, Stivers C, Nagle J, Poole SJ, Odenwald WF, 1998. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev 12, 246–260. 10.1101/gad.12.2.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcavich R, Doe CQ, 2005. Drosophila neuroblast 7–3 cell lineage: a model system for studying programmed cell death, Notch/Numb signaling, and sequential specification of ganglion mother cell identity. J Comp Neurol 481, 240–251. 10.1002/cne.20371 [DOI] [PubMed] [Google Scholar]
- Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, Muller J, 1998. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282, 1897–1900. 10.1126/science.282.5395.1897 [DOI] [PubMed] [Google Scholar]
- Kohwi M, Doe CQ, 2013. Temporal fate specification and neural progenitor competence during development. Nat Rev Neurosci 14, 823–838. 10.1038/nrn3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Lupton JR, Lai SL, Miller MR, Doe CQ, 2013. Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell 152, 97–108. 10.1016/j.cell.2012.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Small S, Reinitz J, 1998. Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev Genes Evol 208, 290–294. 10.1007/s004270050184 [DOI] [PubMed] [Google Scholar]
- Lee G, Sehgal R, Wang Z, Nair S, Kikuno K, Chen CH, Hay B, Park JH, 2013. Essential role of grim-led programmed cell death for the establishment of corazonin-producing peptidergic nervous system during embryogenesis and metamorphosis in Drosophila melanogaster. Biol Open 2, 283–294. 10.1242/bio.20133384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-K, Lundell MJ, 2007. Differentiation of the Drosophila serotonergic lineage depends on the regulation of Zfh-1 by Notch and Eagle. Molecular and Cellular Neuroscience 36, 47–58. 10.1016/j.mcn.2007.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C, 1987. hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev Biol 119, 402–417. 10.1016/0012-1606(87)90045-5 [DOI] [PubMed] [Google Scholar]
- Li X, Chen Z, Desplan C, 2013. Temporal patterning of neural progenitors in Drosophila. Curr Top Dev Biol 105, 69–96. 10.1016/B978-0-12-396968-2.00003-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundell M, Hirsh J, 1998. eagle is required for the specification of serotonin neurons and other neuroblast 7–3 progeny in the Drosophila CNS. Development 125, 463–472 [DOI] [PubMed] [Google Scholar]
- Lundell MJ, Hirsh J, 1994. Temporal and Spatial Development of Serotonin and Dopamine Neurons in the Drosophila CNS. Developmental Biology 165, 385–396. 10.1006/dbio.1994.1261 [DOI] [PubMed] [Google Scholar]
- Lundell MJ, Lee H-K, Perez E, Chadwell L, 2003. The regulation of apoptosis by Numb/Notch signaling in the serotonin lineage of Drosophila. Development 130, 4109–4121. 10.1242/dev.00593 [DOI] [PubMed] [Google Scholar]
- Meng JL, Marshall ZD, Lobb-Rabe M, Heckscher ES, 2019. How prolonged expression of Hunchback, a temporal transcription factor, re-wires locomotor circuits. Elife 8. 10.7554/eLife.46089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny T, Eiselt R, Urban J, 2002. Hunchback is required for the specification of the early sublineage of neuroblast 7–3 in the Drosophila central nervous system. Development 129, 1027–1036 [DOI] [PubMed] [Google Scholar]
- Papatsenko D, Levine MS, 2008. Dual regulation by the Hunchback gradient in the Drosophila embryo. Proc Natl Acad Sci U S A 105, 2901–2906. 10.1073/pnas.0711941105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ, 2003. Regulation of neuroblast competence in Drosophila. Nature 425, 624–628. 10.1038/nature01910 [DOI] [PubMed] [Google Scholar]
- Perez-Garijo A, 2018. When dying is not the end: Apoptotic caspases as drivers of proliferation. Semin Cell Dev Biol 82, 86–95. 10.1016/j.semcdb.2017.11.036 [DOI] [PubMed] [Google Scholar]
- Perez-Garijo A, Steller H, 2015. Spreading the word: non-autonomous effects of apoptosis during development, regeneration and disease. Development 142, 3253–3262. 10.1242/dev.127878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C, Carney GE, Taylor BJ, White K, 2002. reaper is required for neuroblast apoptosis during Drosophila development. Development 129, 1467–1476 [DOI] [PubMed] [Google Scholar]
- Pinto-Teixeira F, Konstantinides N, Desplan C, 2016. Programmed cell death acts at different stages of Drosophila neurodevelopment to shape the central nervous system. FEBS Lett 590, 2435–2453. 10.1002/1873-3468.12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S, Capovilla M, Pirrotta V, 1991. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J 10, 1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow S, Draizen TA, Truman JW, 1997. Genes that induce apoptosis: transcriptional regulation in identified, doomed neurons of the Drosophila CNS. Dev Biol 190, 206–213. 10.1006/dbio.1997.8696 [DOI] [PubMed] [Google Scholar]
- Rogulja-Ortmann A, Luer K, Seibert J, Rickert C, Technau GM, 2007. Programmed cell death in the embryonic central nervous system of Drosophila melanogaster. Development 134, 105–116. 10.1242/dev.02707 [DOI] [PubMed] [Google Scholar]
- Rogulja-Ortmann A, Renner S, Technau GM, 2008. Antagonistic roles for Ultrabithorax and Antennapedia in regulating segment-specific apoptosis of differentiated motoneurons in the Drosophila embryonic central nervous system. Development 135, 3435–3445. 10.1242/dev.023986 [DOI] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ, 1999. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development 126, 4653–4689 [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM, 1997. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol 189, 186–204. 10.1006/dbio.1997.8660 [DOI] [PubMed] [Google Scholar]
- Schulz C, Tautz D, 1994. Autonomous concentration-dependent activation and repression of Kruppel by hunchback in the Drosophila embryo. Development 120, 3043–3049 [DOI] [PubMed] [Google Scholar]
- Sen SQ, Chanchani S, Southall TD, Doe CQ, 2019. Neuroblast-specific open chromatin allows the temporal transcription factor, Hunchback, to bind neuroblast-specific loci. Elife 8. 10.7554/eLife.44036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seroka AQ, Doe CQ, 2019. The Hunchback temporal transcription factor determines motor neuron axon and dendrite targeting in Drosophila. Development 146, dev175570. 10.1242/dev.175570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Letai A, Sarosiek K, 2019. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol 20, 175–193. 10.1038/s41580-018-0089-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent P, 1988. Applied Nonparametric Statistical Methods. Springer Netherlands. [Google Scholar]
- Staller MV, Vincent BJ, Bragdon MD, Lydiard-Martin T, Wunderlich Z, Estrada J, DePace AH, 2015. Shadow enhancers enable Hunchback bifunctionality in the Drosophila embryo. Proc Natl Acad Sci U S A 112, 785–790. 10.1073/pnas.1413877112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Johnston P, Lawrence PA, 1992. Control of Drosophila body pattern by the hunchback morphogen gradient. Cell 69, 237–249. 10.1016/0092-8674(92)90405-2 [DOI] [PubMed] [Google Scholar]
- Suzanne M, Steller H, 2013. Shaping organisms with apoptosis. Cell Death Differ 20, 669–675. 10.1038/cdd.2013.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Yamada-Mabuchi M, Arya R, St Pierre S, Tang W, Tosa M, Brachmann C, White K, 2011. Coordinated expression of cell death genes regulates neuroblast apoptosis. Development 138, 2197–2206. 10.1242/dev.058826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng X, Toyama Y, 2011. Apoptotic force: active mechanical function of cell death during morphogenesis. Dev Growth Differ 53, 269–276. 10.1111/j.1440-169X.2011.01251.x [DOI] [PubMed] [Google Scholar]
- Touma JJ, Weckerle FF, Cleary MD, 2012. Drosophila Polycomb complexes restrict neuroblast competence to generate motoneurons. Development 139, 657–666. 10.1242/dev.071589 [DOI] [PubMed] [Google Scholar]
- Tran KD, Doe CQ, 2008. Pdm and Castor close successive temporal identity windows in the NB3–1 lineage. Development 135, 3491–3499. 10.1242/dev.024349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran KD, Miller MR, Doe CQ, 2010. Recombineering Hunchback identifies two conserved domains required to maintain neuroblast competence and specify early-born neuronal identity. Development 137, 1421–1430. 10.1242/dev.048678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent BJ, Staller MV, Lopez-Rivera F, Bragdon MDJ, Pym ECG, Biette KM, Wunderlich Z, Harden TT, Estrada J, DePace AH, 2018. Hunchback is counter-repressed to regulate even-skipped stripe 2 expression in Drosophila embryos. PLoS Genet 14, e1007644. 10.1371/journal.pgen.1007644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H, 1994. Genetic control of programmed cell death in Drosophila. Science 264, 677–683. 10.1126/science.8171319 [DOI] [PubMed] [Google Scholar]
- White RA, Lehmann R, 1986. A gap gene, hunchback, regulates the spatial expression of Ultrabithorax. Cell 47, 311–321. 10.1016/0092-8674(86)90453-8 [DOI] [PubMed] [Google Scholar]
- Wolf C, Schuh R, 2000. Single mesodermal cells guide outgrowth of ectodermal tubular structures in Drosophila. Genes Dev 14, 2140–2145. 10.1101/gad.180900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Miura M, 2015. Programmed cell death in neurodevelopment. Dev Cell 32, 478–490. 10.1016/j.devcel.2015.01.019 [DOI] [PubMed] [Google Scholar]
- Zhang CC, Bienz M, 1992. Segmental determination in Drosophila conferred by hunchback (hb), a repressor of the homeotic gene Ultrabithorax (Ubx). Proc Natl Acad Sci U S A 89, 7511–7515. 10.1073/pnas.89.16.7511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo P, Stanojevic D, Colgan J, Han K, Levine M, Manley JL, 1991. Activation and repression of transcription by the gap proteins hunchback and Kruppel in cultured Drosophila cells. Genes Dev 5, 254–264. 10.1101/gad.5.2.254 [DOI] [PubMed] [Google Scholar]


