Abstract
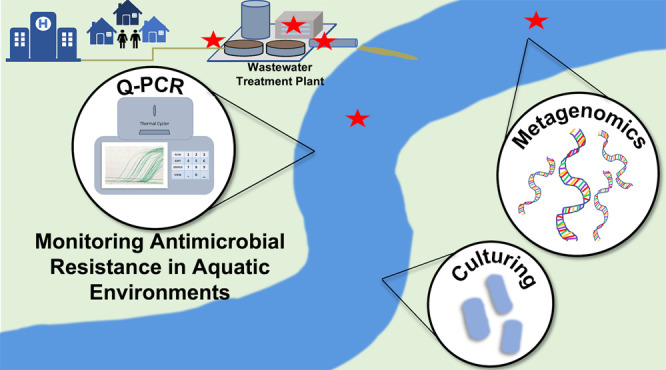
Antimicrobial resistance (AMR) is a grand societal challenge with important dimensions in the water environment that contribute to its evolution and spread. Environmental monitoring could provide vital information for mitigating the spread of AMR; this includes assessing antibiotic resistance genes (ARGs) circulating among human populations, identifying key hotspots for evolution and dissemination of resistance, informing epidemiological and human health risk assessment models, and quantifying removal efficiencies by domestic wastewater infrastructure. However, standardized methods for monitoring AMR in the water environment will be vital to producing the comparable data sets needed to address such questions. Here we sought to establish scientific consensus on a framework for such standardization, evaluating the state of the science and practice of AMR monitoring of wastewater, recycled water, and surface water, through a literature review, survey, and workshop leveraging the expertise of academic, governmental, consulting, and water utility professionals.
Keywords: antibiotic resistance, surveillance, standardization, wastewater, recycled water, surface water
Short abstract
Standardized methods are a crucial next step in creating internationally comparable databases for antibiotic resistance monitoring of water environments.
Standardized Environmental Monitoring Is Urgently Needed to Address the Global Public Health Threat of Antimicrobial Resistance
Antimicrobial resistance (AMR) is now recognized as among the top 10 threats to global health, with current trends in resistant infections in humans and livestock pointing toward a potential postantibiotic era.1 The societal and economic burden of AMR is still being realized, but some stark projections predict that resistant bacterial infections will become the leading cause of death worldwide by 2050.2
Global and national action plans have generally embraced a One Health (humans–animals–environment) framework for tackling AMR;3,4 however, it is increasingly recognized that more attention should be focused on environmental dimensions. The need to better understand the role of the environment in the spread of antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARGs) within and between humans, plants, and animals was emphasized in the U.S. National Action Plan for Combating Antibiotic-Resistant Bacteria,5 where a key objective in the 2020 update is to expand surveillance efforts. In the environmental sector, there are numerous avenues by which AMR can evolve and spread, but a large body of research has demonstrated that water environments are a unifying transmission pathway (Figure 1). Human exposure to environmental AMR can take place through water- or aerosol-borne exposures; this includes contamination of drinking water, food, or recreational water, which can result in skin, gastrointestinal, urogenital, and respiratory colonization and/or infection with resistant microbes.6−9 Despite evidence that water environments play a role, studies quantifying that role and the risks associated are lacking, further emphasizing the need to establish long-term comparable One Health monitoring data.
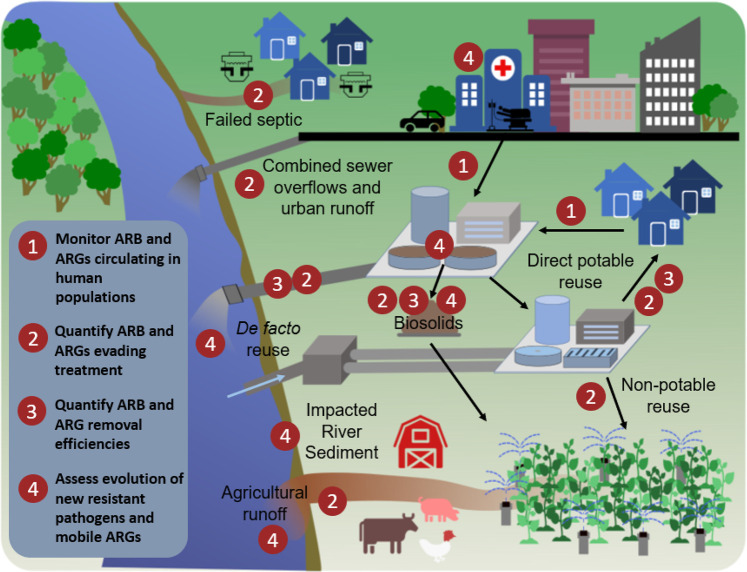
Figure 1.
Monitoring objectives and transmission pathways for antimicrobials, resistant microorganisms, mobile genetic elements (MGEs), and antibiotic resistance genes (ARGs) in humans and the environment.
A fundamental stumbling block to the advancement of AMR monitoring of water environments is a lack of agreed upon targets and standardized methods, including a lack of benchmarking and threshold data to inform evolutionary, epidemiological, and other risk modeling efforts. Here we synthesize key findings from a systematic literature review, expert survey, and expert workshop in working toward a framework for meeting this need. In the following sections we delve into critical environmental dimensions of AMR that should be taken into consideration in developing a monitoring program; expound upon the strengths and weaknesses of culture-, quantitative polymerase chain reaction (qPCR)-, and metagenomics-based methodologies; and summarize fundamental insights that have been gained through their application to the study of wastewater, surface water, and recycled water environments. Subsequently, we distill down the insights gained through engaging a diverse array of experts across sectors in an online survey and subsequent workshop and propose a corresponding framework for standardized monitoring of AMR in water environments that taps into the benefits of various targets and methods, depending on the monitoring objective. The ultimate goal was to develop a framework for engaging the water sector, particularly U.S. water utilities,10 in a manner that is supported by the current state of the science and also harmonized with ongoing international efforts to advance AMR monitoring of water environments. The framework is intended to support both local and global monitoring goals.
Water Environments Are an Interconnecting Thread in AMR Evolution and Spread
The environment is now recognized not as only a key avenue for AMR transmission, e.g., from fecal sources to potential points of exposure, but also as a source of acquisition of new ARGs by pathogens.11−14 A major challenge is that ARGs can be shared among bacteria via mobile genetic elements (MGEs), and therefore it is necessary to consider their movement along with ARB and ARGs in the environment. The current understanding is that most ARGs found in pathogens were originally recruited from the vast and diverse genetic reservoir embodied by environmental bacteria.15 Pollution is a direct source of ARB, ARGs, and MGEs into the environment. Pollution is also a direct source of antimicrobials, which can induce a selection pressure and can further maintain and amplify ARB and ARGs, contributing to the evolution of new forms of resistance. In fact, recent works have generally recognized the pollution and other inputs associated with industrialization over the past 150 years as a primary driver of AMR proliferation.16,17 ARB, ARGs, and MGEs can ultimately reach water environments via direct contamination from aquaculture,18 landfill leachates,19,20 improper treatment of pharmaceutical manufacturing waste streams,21,22 runoff from agricultural production,23 combined sewer overflow events,24 and generally from human and animal excreta25 (Figure 1). MGEs pose a special concern because they can carry several ARGs and represent a risk to pathogens acquiring resistance.
Sewage is known to contain a confluence of antibiotics, ARB, ARGs, MGEs, and human bacterial pathogens emanating from various domestic, industrial, and clinical sources,26,27 which are received by wastewater treatment plants (WWTPs). WWTPs have important potential to mitigate the spread of AMR to waterbodies, but they are not intentionally designed for this purpose. Concern has been raised that special measures may be warranted for pretreatment of hospital sewage before discharge,28 or otherwise for treating wastewaters enriched in clinically relevant forms of AMR. Recycled water, i.e., wherein WWTP effluent is further treated to a quality appropriate for the intended potable or nonpotable purpose (e.g., irrigation of fields or crops), is of special concern because there may be less opportunity for AMR to attenuate prior to human exposure.29 Research has also brought to light concerns about regrowth of resistant microorganisms in distribution systems, particularly in situations of nonpotable recycled water.30,31 Thus, wastewater, recycled water, and impacted receiving waters have been identified as key candidate monitoring points for AMR in the environment32−34 (Figure 1).
In order to address the rising threat of drug-resistant infections and disease, the field must move toward cooperative understanding of environmental occurrence, human exposures, effective mitigation strategies, and ultimately a dose–response assessment for human health risk assessments.35 The first step in achieving such goals is the development of standardized methods that can be used both locally and globally to establish baseline environmental occurrence data specific to all relevant water environments.36
A New Millennium, Evolving Methods, and Understanding: What Have we Learned?
Culture-Based Methods
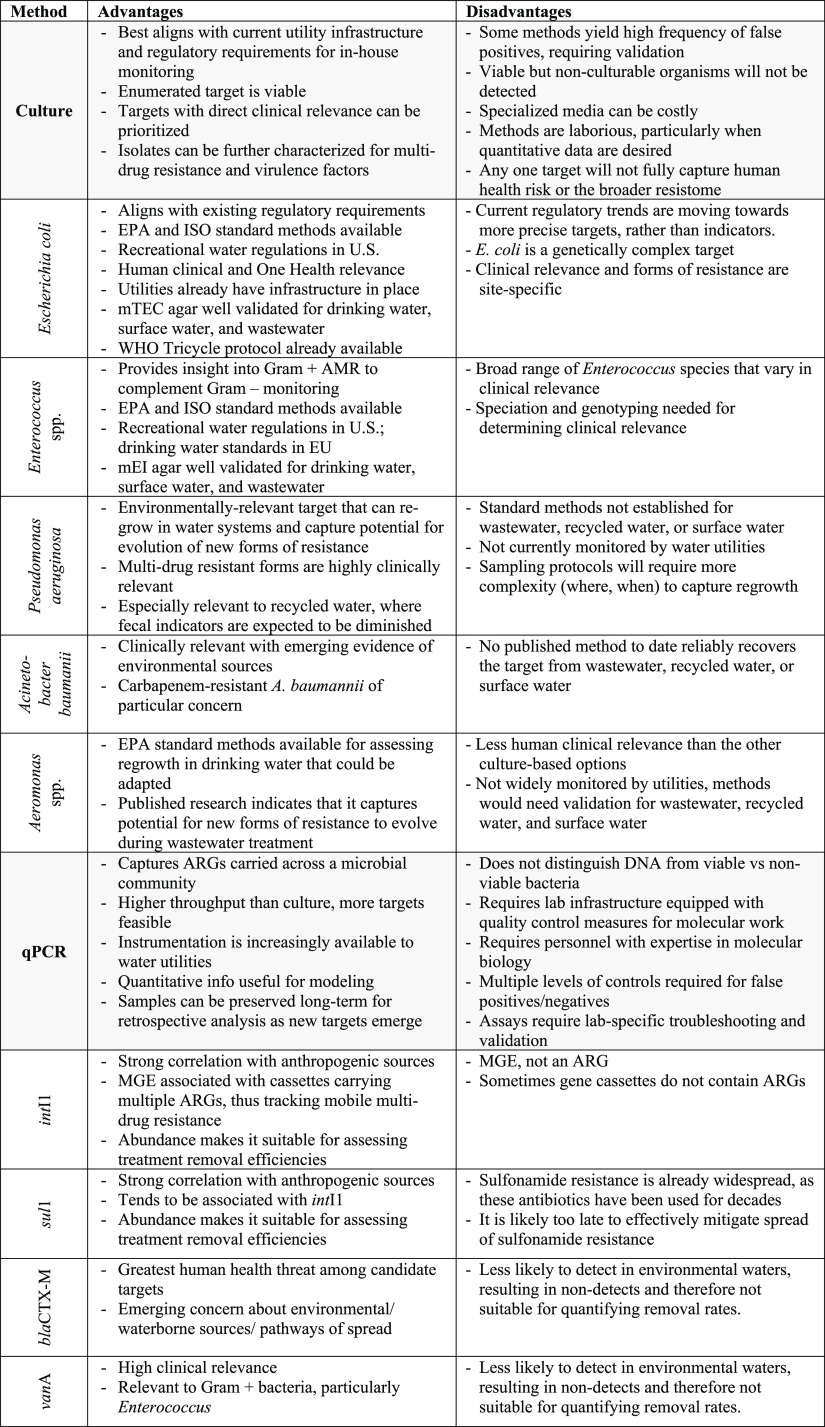
Culture-based methods are attractive for AMR monitoring because a target can be selected with known clinical relevance (e.g., taxonomic groups containing human pathogens), methods are fairly well standardized for defining clinical resistance levels (e.g., EUCAST,37 CLSI,38 Kirby-Bauer39), and, by definition, the recovered target is viable. Further, once isolates have been recovered, they can be subject to further analysis, such as multidrug-resistance testing, sequence-based typing, or whole genome sequencing, which can aid in identification of ARG-carrying MGEs (e.g., plasmids), delineating phylogenetic relationships among strains, and identifying sources of outbreak strains. Culture-based targets also are most amenable to informing human health risk assessments. A challenge to culture methods for AMR monitoring, however, is that while there are numerous genera/species found in wastewater and surface water environments that could be informative as targets, the typically high levels of background microorganisms in those environments can increase the likelihood of interference with methods for isolation of the target. In addition, the vast majority of environmental microbes cannot readily be cultured and therefore the broader pool of resistance in a given environmental reservoir will be overlooked by culture-based methods.34 No one target can comprehensively capture the AMR status of a given environment.
Fecal indicator bacteria, such as fecal coliforms, Escherichia coli, and Enterococcus spp., are obvious candidates due to the long history of regulatory monitoring in water and wastewater systems and, correspondingly, the high level of standardization for existing methods.40,41 The World Health Organization’s Global Antimicrobial Resistance Surveillance System (GLASS) utilizes a one-target integrated multisectoral approach that focuses on extended spectrum β-lactamase (ESBL) producing E. coli as an indicator due to its relevance in human health, food chain, and the environment.42Enterococcus spp. could potentially be a good complement as a Gram-positive organism, thus representing a distinct suite of antibiotics for which AMR is a concern.43 On the other hand, bacterial targets commonly present in sewage that also have broader niches in the environment than traditional fecal indicators, such as Pseudomonas aeruginosa and Aeromonas spp., are also important to consider.44 Because such targets survive and grow in the environment, they may serve as informative indicators of AMR that is acquired through horizontal gene transfer (HGT) of ARGs taking place through anthropogenic activities and maintained by ambient pollution sources in the receiving environments.45
Prior application of culture-based methods has proved useful for tracking the fate and persistence of vancomycin-resistant Enterococcus faecium in environmental waters due to sewage overflow.46 Culture-based monitoring has also been widely applied to examine changes in E. coli populations during wastewater treatment. One comprehensive study found no evidence of E. coli acquisition of ARGs during treatment, but this could be, in part, because E. coli tend to die off during treatment.47,48 Thus, others have examined resistance patterns in organisms, such as Aeromonas, which are capable of growth in WWTPs.49 Research on surface waters and aquaculture systems has exposed a high rate at which Aeromonas harbor ARGs, indicating Aeromonas may be an organism of particular interest when monitoring with the goal of mitigating human health exposures.45,50
qPCR-Based Methods
Molecular methods opened the door to assessing AMR in uncultured and unculturable bacteria, providing an integrated measure across the microbial community. In particular, qPCR began to be applied in the mid-2000s for the purpose of directly quantifying ARGs in various environmental samples, revealing widespread and striking patterns of elevated ARGs in anthropogenically impacted aquatic environments.51−53
qPCR can provide very sensitive detection and quantification over several logs, which is useful for identifying and characterizing patterns of anthropogenic influence, as well as assessing removal of ARGs during wastewater or other water treatments and informing modeling, such as environmental fate and human health risk assessment. For example, Pruden et al.54 found a near-perfect correlation between the number of sul1 ARG copies measured in the Poudre River, Colorado, sediments and the capacities of upstream WWTPs and animal feeding operations (total volume treated/day and number of livestock, respectively). Czekalski et al.55 demonstrated that subsurface discharge of WWTP effluent to Lake Geneva was a point source of sul, tet, and qnrA genes, whose fate could be modeled using the quantitative information yielded by qPCR. Similarly, class 1 integrons (typically tracked by targeting the corresponding intI1 integrase gene) have been found to be highly indicative of anthropogenic pollution and are now well-established as indicators of multiantibiotic resistance and potential for ARG mobility.56,57 Such patterns of sul1 and intI1 occurrence were readily apparent because (a) these genes have proven to be strongly associated with anthropogenic sources and (b) they are abundant in environments such as wastewater, facilitating efforts to measure their dispersal and attenuation. However, there is also value in evaluating less abundant, but more clinically relevant ARGs, such as those encoding resistance to last-resort antibiotics (e.g., on the CDC urgent, serious, or concerning threat lists58). vanA (vancomycin resistance) and blaCTX-M (extended spectrum β-lactamase (ESBL) resistance) have been proposed for this purpose, but sparse detection of such genes can undermine analytical potential. Mobility of ARGs, i.e., their association with MGEs and thus potential to move across species, is also an important consideration. Others have similarly sought to classify ARGs from omics data according to the tendency to be enriched in anthropogenic environments, mobility, and pathogenicity.59
A challenge with qPCR is that it requires the researcher to select ARGs or other relevant gene target(s) a priori, among thousands of choices, as is apparent from current ARG databases (CARD, DeepARG-db). Recently, high throughput qPCR arrays that include hundreds of ARG targets have been developed, reducing the need to choose the “right” suite of ARGs.60 Other markers, such as MGE and pathogen-specific genes, can also be included. However, the high throughput qPCR instrumentation is not widely available, quality assurance protocols are not well-established, and the addition of a preamplification step is required to achieve sensitive detection limits via specific target amplification, all of which are drawbacks for environmental monitoring.61 Also, guidance would still be needed on how to interpret high throughput qPCR data in terms of which occurrence patterns of ARGs are of concern. Digital droplet PCR (ddPCR) is gaining traction in the field, lauded as an improved version of qPCR that is more sensitive and less affected by inhibition. However, few published ARG studies have yet reported the use of ddPCR.
Metagenomic-Based Methods
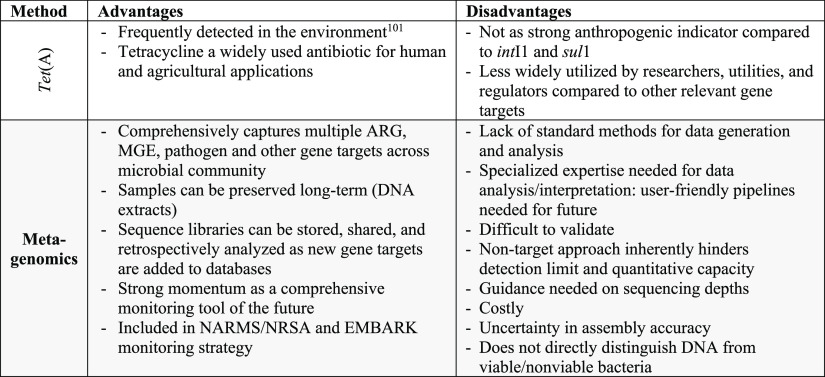
Next-generation DNA sequencing (NGS) applications became accessible in 2009, enabling for the first time the possibility of profiling “all” ARGs and other genes in an environmental sample without a priori selection of targets, i.e., metagenomic sequencing. Metagenomic sequencing is carried out through direct extraction of genomic DNA, fragmentation, and application of NGS to obtain millions of reads representing the bacterial community of environmental samples. Metagenomics was initially used to examine river sediments of highly impacted surface waters exposed to pharmaceutical (specifically antibiotic) wastewater discharges.62 Researchers found high levels of ARGs (sul2, aph-, tet-, qnr-, erm-), transposons, plasmids, and integrons, which are risk factors for HGT as a propagation mechanism for AMR in water. Metagenomics is now widely applied for examining shifts in the resistome (i.e., the collection of all ARGs across an environmental sample) through WWTPs and identifying MGEs in order to estimate the extent of HGT events.63−65 Metagenomics has also been used to characterize resistomes and treatment efficiencies in recycled water and drinking water treatment plants.31,66 Most recently, it has been recognized that metagenomics has potential as a powerful tool for wastewater-based epidemiology, i.e., monitoring raw sewage to estimate ARG carriage and shedding of corresponding human populations.67
A major challenge with comparing metagenomic data across studies is that there are numerous ways to generate, analyze, and interpret the data.68,69 For example, differences in sampling (composite versus grab sampling), sample storage conditions, DNA extraction methodologies, and sequencing depths and coverages have been shown to bias the generation and comparison of metagenomic libraries.70−72 Further, ARGs, MGEs, and other genes of interest can be annotated through comparison to numerous publicly available databases, including CARD,73,74 DeepARG,75 Resfinder,76 MEGARes,77 ResFams,78 resqu,79 SARG,80 ARG-ANNOT,81 BacMet,82 PlasFLow,83 ICEberg,84 and ACLAME,85 which vary in completeness, nomenclature, and degree/frequency of curation. The sequence libraries produced by NGS are semiquantitative; i.e., they must be normalized to a denominator in order to compare across studies. Such denominators include 16S rRNA genes, the β subunit of RNA polymerase (rpoB), single copy genes, or simply the size of the sequence library itself, resulting in relative abundance estimates of ARGs. Another major challenge is that the random nature of shotgun sequencing results in high representation of genes in the libraries that are not of interest, e.g., not ARGs, MGEs, or pathogen markers, and thus the limit of quantification is typically higher than that of qPCR86 and very rare genes will not likely be detected. NGS is already very costly, and attempts at deeper sequencing to capture more rare sequences further increase the cost.
Assembly, in which assemblers (e.g., IDBA-UD,87 MEGAHIT,88 and metaSpades89) are used to repiece together portions of the genomes of the organisms representing the original microbial community, is also a key aspect of metagenomic analysis that requires consideration for standardization.90 Assembly is often applied toward identifying putative linkages between ARGs, MGEs, and pathogens, a key frontier in the application of AMR risk assessment.91 A major challenge to assembly has also been that the dominant sequencing technology currently applied for environmental metagenomics, Illumina (Illumina, Inc., San Diego, CA), produces large sequence libraries, but with very short (∼150 bp) reads, which are computationally challenging to accurately piece together. Newly emerging methods such as long-read DNA sequencing, hybrid assembly, or proximity ligation prior to sequencing can help reduce such uncertainties.90,92
Critical Research Needs Call for Standardized Methods for AMR Monitoring of Water Environments
At this point it is clear that human activities profoundly shape the levels and types of AMR encountered in natural ecosystems, especially impacted water environments (Figure 1). There is also evidence that the environment is a source of resistant infections in clinical settings. However, there are still several critical questions that must be addressed in order to effectively tackle the spread of AMR, particularly from an environmental perspective. Some of these questions include the following:
What kinds and levels of ARGs in various water environments (wastewater, recycled water, surface water) result in elevated exposure and risk of acquiring a resistant infection?
What are the key hotspots for horizontal transfer of ARGs and the evolution of new forms of resistant pathogens, and how might such hotspots be prioritized for mitigation efforts?
Which environmental factors in aquatic environments, such as concentrations/mixtures of antibiotics/heavy metals/antimicrobials and physicochemical parameters (e.g., pH, temperature), substantially elevate selective pressure for resistant microbes and maintenance of ARGs?
What are the relative contributions of various environmental sources of AMR to resistant infections observed in humans?
What are the most concerning epidemiological linkages between AMR observed in the environment, plants, and animals and infections found in humans?
Coordinated environmental surveillance efforts employing standard methods, ideally at local, regional, national, and global scales, will be required to address these questions and to inform corresponding policy and practice to stem the spread of AMR.36
Local and Global Collaboration Required to Standardize Monitoring
There have been a number of recent calls for standardization of targets and methods for environmental AMR monitoring.32,93−96 Because AMR is complex, a multitarget, adaptable approach will be required, depending on the objective of the monitoring program. The complexity of AMR also mandates special attention to quality assurance/quality control in experimental design, execution, and reporting.97
Berendonk et al.93 remains a highly influential paper that set the stage for environmental surveillance of antibiotic resistance and a need for standardized methods. There it is argued that consistency and comparability are needed to assess the global antibiotic resistome, to inform future policy, science, and medicine.93 Pruden et al.32 outlined how the environmental link to AMR makes it a uniquely interdisciplinary challenge that will require engineering, environmental science, medicine, agriculture, public health, and policy to generate meaningful data and coordinate effective mitigation strategies. Overall, some of the authors’ recommendations included standardization of methods for surveillance, agreement on targets for monitoring, and coordinated global research representing varying water matrixes and contexts. Hujibers et al.94 outlined five main monitoring objectives within a One Health conceptual framework (paraphrased) addressing the following: the risk of transmission of already antibiotic-resistant bacteria to humans; the risk of accelerating the evolution of antibiotic resistance in pathogens through pollution with selective agents and bacteria of human or animal origin; the risks antibiotics pose to aquatic and terrestrial ecosystem health; the population-level prevalence of ARBs/ARGs; population-level antibiotic use.
The World Health Organization (WHO) recently proposed a standard method for ESBL-producing E. coli, under the Global Tricycle Surveillance program.42 The ESBL-EC Tricycle Protocol aims for a simple, feasible, and comparable method to enable global and multisectoral engagement in the surveillance of AMR that is relevant across One Health matrixes.98 Establishing a Monitoring Baseline for Antimicrobial Resistance in Key Environments (EMBARK) is an international program by the Joint Programming Initiative for Antimicrobial Resistance (JPIAMR), which aims to identify baseline levels of environmental AMR by region and types of resistance and thereafter standardize methods for surveillance and monitoring of those resistances worldwide.99 In the United States, a multiagency program was initiated in 1996 to encourage collaboration toward tackling AMR, the National Antimicrobial Resistance Monitoring System (NARMS), which has historically focused on culture-based monitoring of retail meat. Currently an effort is underway to extend monitoring efforts into the environment by utilizing existing EPA National Rivers and Streams Assessment (NRSA) infrastructure to detect AMR in surface waters, as an integrator of multiple environmental inputs (e.g., Figure 1).100
A Way Forward: Guidance from an Expert Survey and Workshop
An expert survey was conducted to provide insight into which methods are most commonly used for AMR monitoring, identify common barriers to implementation, and consider boundaries to application (e.g., cost, time, labor, expertise). The survey was conducted online and reached 105 experts, who resided mainly in North America, Europe, Asia, and Africa and spanned various relevant fields across academia, research institutions, state and federal governments, consulting, and water/wastewater utilities. The queries solicited input on recommended culture, qPCR, and metagenomic targets for monitoring AMR in water environments. The survey ultimately served to narrow down the list of commonly employed targets and protocols to a core list of methods whose efficacy had been validated. These methods were then discussed at the expert workshop in terms of appropriateness for standardization. A summary of the methods and findings of the survey is provided in the Supporting Information (section 1, Figures SI 1–SI 7).
The expert survey participants and/or their organizations were confirmed to be familiar with all three categories of methods, with 81% indicating use of qPCR, 75% culturing fecal coliforms, 66% engaging in some form of metagenomics (Table SI 2), and 81% claiming to be “familiar with next-generation sequencing” (Figure SI 4). In terms of the desired attributes of the targets, both “relevance to human health” and ability to inform “human health risk assessment” were the most frequently selected as the first or second most important among nine choices, using a sliding scale to order importance (Figure SI 7). A “quantifiable” target and one that captures “HGT potential” also were commonly selected within the top three most important attributes.
From a selection of seven culture targets, in which participants could choose two options, the majority selected E. coli (53%), followed by Enterococci (30%) (Figure SI 2). Enterobacteriaceae ranked a close third (29%), followed by Pseudomonas aeruginosa (15%), Acinetobacter baumannii (11.4%), Salmonella spp. (8.6%), and Aeromonas spp. (6.6%). On the basis of the survey, E. coli and Enterococci spp. were obvious choices to pursue further as complementary Gram-positive and Gram-negative fecal indicator organisms that already have widely applied standard methods for water samples. Enterobacteriaceae and Salmonella spp., which are also Gram-negative fecal-derived bacteria, were not considered further because of redundancy with the clearly popular E. coli. P. aeruginosa, A. baumannii, and Aeromonas spp. were considered further as candidate targets known for regrowth in water environments, which could contribute to the evolution of multiantibiotic resistant strains, and where further evaluation was warranted to assess suitability for standardization.
Similarly, survey participants were directed to select three targets from a list of 16 ARGs and one integron commonly targeted by qPCR for AMR monitoring of water environments (Figure SI 3). The five most frequently selected qPCR-based targets were ranked as follows: intI1 (36%), blaCTX-M (23%), sul1 (21%), vanA (15%), and tet(A) (12%). These five gene targets naturally captured key attributes of interest, such as strongly correlating with anthropogenic inputs (intI1, sul1), occurring at sufficient abundances in wastewater to quantify removal rates (intI1, sul1, tet(A)), encoding highly clinically relevant forms of resistance with suspected environmental linkages (blaCTX-M, vanA), high associations with multiantibiotic resistance (intI1, sul1), and relevance to HGT (intI1, sul1), while also representing resistance to four different antibiotic classes (sulfonamides, β-lactams, glycopeptides, and tetracyclines). Therefore, these five gene targets were selected for further consideration for potential standard methods.
A systematic Web of Science literature review was conducted to evaluate the application of the five culture targets, the five qPCR targets, and metagenomics for monitoring of AMR in wastewater, recycled water, and surface water environments. This literature review was provided to participants to review in advance of the expert workshop. The search terms and protocol for the literature review are provided in the Supporting Information (section 1a), with the portion devoted to qPCR recently being published.95 The workshop took place on four days over a two-week period (May 2021) and was attended by 49 (43 U.S., 6 international) experts representing academia (17), industry (8), federal governmental agencies (e.g., EPA, FDA, USDA) (13), state and local governmental organizations (2), United Nations agencies (WHO) (1), and water utilities (9). In addition to the literature review, the workshop participants were provided with a summary of the results of the online survey. The workshop itself was generally structured around leading with an expert presentation on the state of the science of one of four categories of methods (culture of fecal indicators, culture of targets with environmental niches, qPCR, and metagenomics) followed by discussion in breakout groups and a poll to rank the methods discussed within each category in terms of suitability for water utility monitoring of AMR. The workshop concluded with a panel discussion led by U.S. water utility representatives focused on the feasibility, benefits, challenges, and limitations of AMR monitoring of water environments and a final poll to assess opinions on these aspects and obtain a ranking of all of the methods for AMR monitoring of water environments that were discussed at the workshop. A summary of the overall assessment of the method categories and targets gathered from the workshop presentations, discussions, and polls is provided in Table 1 and the Supporting Information (section 3).
Table 1. Summary of Advantages and Disadvantages of the Methods and Targets for AMR Monitoring of Water Environments Identified in This Study through Literature Review, Expert Survey, and Expert Workshop.
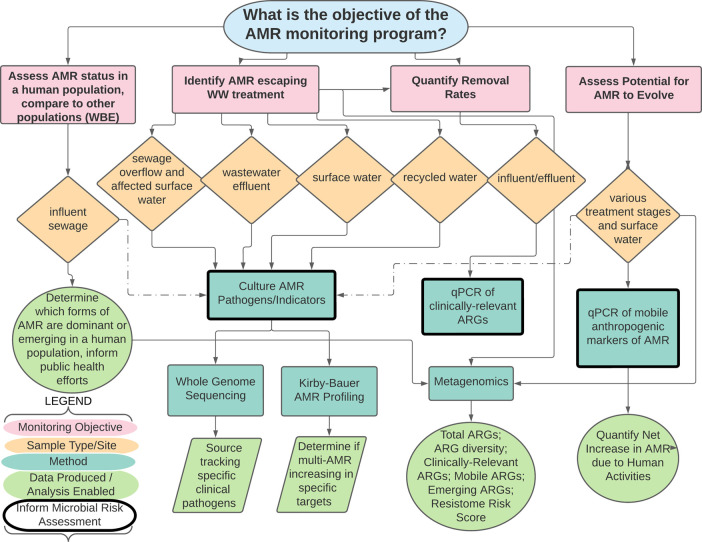
On the basis of the analysis of workshop discussion notes, recordings, and polls, a major outcome of the workshop that became well apparent was that the question “which is the best target for AMR monitoring of water environments?” is highly dependent on the objective or motivating research question (Figures SI 8–SI 10 and SI 15–SI 17). Together, the participants and moderators worked throughout the workshop to develop a decision tree based on four fundamental objectives that can subsequently guide the selection of culture-, qPCR-, and metagenomic-based methods, corresponding targets, and where to sample (Figure 2). Targets and methods that can also conceivably inform human health risk assessment are bolded in the tree, given that human health relevance and ability to inform risk assessment were prioritized attributes for both survey and workshop participants (Figures SI 7 and SI 12).
Figure 2.
Decision tree for selecting culture-, qPCR-, or metagenomics-derived monitoring methods for AMR monitoring of wastewater, recycled water, or surface water, depending on the monitoring objective. Dashed lines indicate the potential for enhanced realization of research objectives when molecular methods are coupled with culture. WBE, wastewater-based epidemiology.
In general, the workshop participants tended to be forward thinking, yet practical, in terms of recommending a framework for advancing standardized monitoring of AMR in water environments. Many highlighted the need to establish sample storage guidance to enable sharing of samples across partnering researchers and countries as well as archiving samples for future analysis. As new resistances evolve and our understanding of the role ARGs plays develops, studies on archived water samples can showcase temporal relationships and assess effectiveness of mitigation strategies, such as the research done by Knapp et al.101 with a 48 year archive of soils. Based on results from the expert survey (Figure SI 5), one aim was to narrow down the list of targets to what experts considered reasonable (three to five assays). In terms of culture-based methods, the top three selections based on the workshop polls were E. coli, Enterococcus spp., and P. aeruginosa, which conveniently captured a Gram-negative and Gram-positive organism with long regulatory histories and P. aeruginosa as an organism capable of regrowth in water environments and notoriously prone to multidrug resistance.
While recognizing the benefits of culture, participants were concerned that it would be a step backward not to move forward with inclusion of molecular measures in an AMR monitoring program. Such perspectives were reflected in the selection of qPCR-based methods as best balancing being “both informative and feasible” and the selection of metagenomics as being the “most informative” method for AMR monitoring (Figure SI 8). The ranking of qPCR targets based on the workshop polls were in strong accordance with the expert survey, with intI1 as the clear top choice, sul1 and vanA tending to be second choice, and blaCTX-M being split across first, second, and third choices (Figure SI 10).
There was notable enthusiasm for metagenomics, but with a recognition that it will be the most difficult to standardize. One challenge is the cost of conducting NGS; most experts surveyed thought $10–$50 per sample was a reasonable cost for monitoring, with only three participants selecting >$300 as a reasonable cost. Advice gathered at the workshop could help guide such an endeavor, considering aspects such as required sequencing depths, recommended annotation databases and parameters, denominators for normalization, accuracy of assembly, and positive and negative controls for validation (Figure SI 11, Table 1).
When workshop participants were polled regarding when they thought that the U.S. water sector would be ready to implement standard methods for AMR monitoring, 40.7% of respondents chose 10 years, followed by 5 years (33.3%), 3 years (14.8%), now (7.4%), and never (3.7%). It was clear throughout workshop discussions that regulatory requirements or other incentives are likely necessary to encourage broad-scale participation of water utilities in AMR monitoring. Emerging practice and regulation around water reuse was discussed as a potential example to follow, where molecular methods for pathogen detection have gained a strong footing. Larger WWTPs with more resources are also in a position to proactively set the standards of approaches applied. Taking action without regulatory requirement could earn favor with the general public, which is very concerned about the potential for future public health threats, given the current pandemic.
Interestingly, when polled at the end of the workshop about which AMR monitoring objectives would be of most interest to water utilities, it was “assess AMR status in a human population and compare to other populations (e.g., wastewater-based epidemiology)” that received the most votes (37%). This suggests that recent momentum in monitoring sewage for SARS-CoV-2 could spur adoption of monitoring of AMR-relevant targets and open the door to broader AMR monitoring to meet other objectives as well.36 For example, “quantifying removal of AMR through wastewater/recycled water” (29.6%) and “identifying types of AMR of clinical concern that might escape treatment” (25.9%) also ranked high. However, none of these monitoring objectives can be met without an agreed-upon framework and standard methods and approaches for monitoring AMR in aquatic environments.
Recommendations
The framework proposed in this feature article helps to better align target and method selection with monitoring objectives, while also laying out a path for standardizing approaches for monitoring AMR in water and wastewater in the United States and globally. On the basis of the feedback provided from the expert survey and workshop, it was possible to prioritize methods that are widely suitable and applicable for utilities, academics, government researchers, and industry scientists alike. The decision tree developed herein can provide a framework that is adaptable to specific monitoring locations and objectives and can support more detailed guidance, including standard operating procedures, to support data comparability. Such an effort is critically needed to break down current silos that limit data comparability and aid in supporting broader global AMR surveillance goals, including tracking and mitigating the spread of AMR via water environments and informing epidemiological studies and human health risk assessments.
Acknowledgments
We thank the participants in our expert survey and workshop. Workshop participants included the following: Alison Franklin, Amy Kirby, Andrea Ottesen, Anthea Lee, Ayella Maile-Moskowitz, Bina Nayak, Connor Brown, Daniel Quintanar, Daniel Gerrity, Ed Topp, Emily Garner, Erin Swanson, Gaya Ram Mohan, Jade Mitchell, Jay Garland, Jean McLain, Jeff Soller, Jennifer Jay, Johan Bengtsson-Palme, John Griffith, Jorge Matheu Alvarez, Kara Nelson, Kati Bell, Kerry Hamilton, Kim Cook, Lauren Stadler, Lenwood Heath, Liqing Zhang, Lisa Durso, Mark Borchardt, Mark Ibekwe, Mark LeChevallier, Mark Sobsey, Gertjan Medema, Nicholas Ashbolt, Peter Vikesland, Raul Gonzalez, Satoshi Ishii, Scott Keely, Sharon Nappier, Sunny Jiang, Suraj Gupta, Tanja Rauch-Williams, Thomas Berendonk, Walter Jakubowksi, and Zia Bukhari. Thanks also go to Anna Kurowski for assisting with workshop video transcripts. This contribution was funded by the Water Research Foundation Project 5052. Additional funding was provided by U.S. National Science Foundation Award Nos. OIE 1545756, OAC 2004751, CBET 1936319, and NRT 2125798.
Biography

Amy Pruden is a University Distinguished Professor at Virginia Tech. She received her B.S. in biological sciences in 1997 and her Ph.D. in environmental science in 2002 from the University of Cincinnati. Pruden’s research focuses on advancing means of monitoring and mitigating the spread of pathogens and antimicrobial resistance in the environment. She is the recipient of the Presidential Early Career Award in Science and Engineering, the Paul L. Busch Award, and the Reciparm International Environmental Award and is a fellow of the International Water Association.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.1c08918.
Methods, results, figures, and tables summarizing the approach and findings of the expert survey and workshop (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- World Health Organization (WHO); Food and Agriculture Organization of the United Nations (FAO); and World Organisation for Animal health (OIE) . Monitoring and Evaluation of The Global Action Plan on Antimicrobial Resistance: Framework and Recommended Indicators; World Health Organization: 2019. 10.20506/AMR.2969 [DOI]
- O’Neill J.The Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations, 2014. https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf.
- European Commission . A European One Health Action Plan against Antimicrobial Resistance (AMR); European Commission: 2017.
- Hernando-Amado S.; Coque T. M.; Baquero F.; Martínez J. L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nature microbiology 2019, 4 (9), 1432–1442. 10.1038/s41564-019-0503-9. [DOI] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services (DHHS), The Assistant Secretary for Planning and Evaluation (ASPE), Federal Task Force on Combating Antibiotic-Resistant Bacteria . National Action Plan for Combating Antibiotic-Resistant Bacteria: 2020–2025; U.S. Department of Health and Human Services: 2020.
- Price L. B.; Stegger M.; Hasman H.; Aziz M.; Larsen J.; Andersen P. S.; Aarestrup F. M.; et al. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 2012, 3 (1), e00305-11 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A. F.; Zhang L.; Balfour A. J.; Garside R.; Gaze W. H. Human recreational exposure to antibiotic resistant bacteria in coastal bathing waters. Environ. Int. 2015, 82, 92–100. 10.1016/j.envint.2015.02.013. [DOI] [PubMed] [Google Scholar]
- Xie J.; Jin L.; He T.; Chen B.; Luo X.; Feng B.; Huang W.; Li J.; Fu P.; Li X. Bacteria and Antibiotic Resistance Genes (ARGs) in PM 2.5 from China: Implications for Human Exposure. Environ. Sci. Technol. 2019, 53, 963–972. 10.1021/acs.est.8b04630. [DOI] [PubMed] [Google Scholar]
- Nordstrom L.; Liu C. M.; Price L. B. Foodborne urinary tract infections: a new paradigm for antimicrobial-resistant foodborne illness. Frontiers in Microbiology 2013, 4, 29. 10.3389/fmicb.2013.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Water Research Foundation (WRF) . Standardizing Methods with QA/QC Standards for Investigating the Occurrence and Removal of Antibiotic Resistant Bacteria/Antibiotic Resistance Genes (ARB/args) in Surface Water, Wastewater, and Recycled Water. 2020. https://www.waterrf.org/research/projects/standardizing-methods-qaqc-standards-investigating-occurrence-and-removal (accessed 2021-12-15).
- Larsson D. G. J.; Flach C. F. Antibiotic resistance in the environment. Nat. Rev. Microbiol 2022, 20, 257–269. 10.1038/s41579-021-00649-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassallo A.; Kett S.; Purchase D.; Marvasi M. Antibiotic-resistant genes and bacteria as evolving contaminants of emerging concerns (e-CEC): is it time to include evolution in risk assessment?. Antibiotics 2021, 10 (9), 1066. 10.3390/antibiotics10091066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg K. J.; Reyes A.; Wang B.; Selleck E. M.; Sommer M. O.; Dantas G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337 (6098), 1107–1111. 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund F.; Böhm M. E.; Martinsson A.; Ebmeyer S.; Österlund T.; Johnning A.; Kristiansson E.; Larsson D. G. J. Comprehensive screening of genomic and metagenomic data reveals a large diversity of tetracycline resistance genes. Microbial Genomics 2020, 6 (11), mgen000455. 10.1099/mgen.0.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez J. L. Ecology and evolution of chromosomal gene transfer between environmental microorganisms and pathogens. Microbiology Spectrum 2018, 6 (1), 6.1.06. 10.1128/microbiolspec.MTBP-0006-2016. [DOI] [PubMed] [Google Scholar]
- Baquero F.; Coque T. M.; Martínez J. L.; Aracil-Gisbert S.; Lanza V. F. Gene transmission in the one health microbiosphere and the channels of antimicrobial resistance. Frontiers in Microbiology 2019, 10, 2892. 10.3389/fmicb.2019.02892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando-Amado S.; Coque T. M.; Baquero F.; Martínez J. L. Antibiotic resistance: moving from individual health norms to social norms in one health and global health. Frontiers in Microbiology 2020, 11, 1914. 10.3389/fmicb.2020.01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello F. C.; Godfrey H. P.; Buschmann A. H.; Dölz H. J. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infectious Diseases 2016, 16 (7), e127 10.1016/S1473-3099(16)00100-6. [DOI] [PubMed] [Google Scholar]
- Anwar M.; Iqbal Q.; Saleem F. Improper disposal of unused antibiotics: an often overlooked driver of antimicrobial resistance. Expert review of anti-infective therapy 2020, 18 (8), 697–699. 10.1080/14787210.2020.1754797. [DOI] [PubMed] [Google Scholar]
- Yu X.; Sui Q.; Lyu S.; Zhao W.; Liu J.; Cai Z.; Yu G.; Barcelo D. Municipal solid waste landfills: An underestimated source of pharmaceutical and personal care products in the water environment. Environ. Sci. Technol. 2020, 54 (16), 9757–9768. 10.1021/acs.est.0c00565. [DOI] [PubMed] [Google Scholar]
- Nijsingh N.; Munthe C.; Larsson D. G. J. Managing pollution from antibiotics manufacturing: charting actors, incentives and disincentives. Environ. Health 2019, 18 (95), 1–17. 10.1186/s12940-019-0531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson D. J. Pollution from drug manufacturing: review and perspectives. Philosophical Transactions of the Royal Society B: Biological Sciences 2014, 369 (1656), 20130571. 10.1098/rstb.2013.0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almakki A.; Jumas-Bilak E.; Marchandin H.; Licznar-Fajardo P. Antibiotic resistance in urban runoff. Sci. Total Environ. 2019, 667, 64–76. 10.1016/j.scitotenv.2019.02.183. [DOI] [PubMed] [Google Scholar]
- Ahmed W.; Zhang Q.; Lobos A.; Senkbeil J.; Sadowsky M. J.; Harwood V. J.; Saeidi N.; Marinoni O.; Ishii S. Precipitation influences pathogenic bacteria and antibiotic resistance gene abundance in storm drain outfalls in coastal sub-tropical waters. Environ. Int. 2018, 116, 308–318. 10.1016/j.envint.2018.04.005. [DOI] [PubMed] [Google Scholar]
- Wang J.; Chu L.; Wojnárovits L.; Takács E. Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: an overview. Science of The Total Environment 2020, 744 (1–12), 140997. 10.1016/j.scitotenv.2020.140997. [DOI] [PubMed] [Google Scholar]
- Guo J.; Li J.; Chen H.; Bond P. L.; Yuan Z. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water research 2017, 123, 468–478. 10.1016/j.watres.2017.07.002. [DOI] [PubMed] [Google Scholar]
- Baquero F.; Martínez J. L.; Cantón R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19 (3), 260–265. 10.1016/j.copbio.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Novo A.; Manaia C. M. Factors influencing antibiotic resistance burden in municipal wastewater treatment plants. Appl. Microbiol. Biotechnol. 2010, 87 (3), 1157–1166. 10.1007/s00253-010-2583-6. [DOI] [PubMed] [Google Scholar]
- Pruden A. Balancing water sustainability and public health goals in the face of growing concerns about antibiotic resistance. Environ. Sci. Technol. 2014, 48 (1), 5–14. 10.1021/es403883p. [DOI] [PubMed] [Google Scholar]
- Zhu N. J.; Ghosh S.; Edwards M. A.; Pruden A. Interplay of Biologically Active Carbon Filtration and Chlorine-Based Disinfection in Mitigating the Dissemination of Antibiotic Resistance Genes in Water Reuse Distribution Systems. Environmental Science & Technology. Environ. Sci. Technol. 2021, 55 (12), 8329–8340. 10.1021/acs.est.1c01199. [DOI] [PubMed] [Google Scholar]
- Garner E.; Chen C.; Xia K.; Bowers J.; Engelthaler D. M.; McLain J.; Edwards M.; Pruden A. Metagenomic characterization of antibiotic resistance genes in full-scale reclaimed water distribution systems and corresponding potable systems. Environ. Sci. Technol. 2018, 52 (11), 6113–6125. 10.1021/acs.est.7b05419. [DOI] [PubMed] [Google Scholar]
- Pruden A.; Alcalde R. E.; Alvarez P. J. J.; Ashbolt N.; Bischel H.; Capiro N. L.; Crossette E.; Frigon D.; Grimes K.; Haas C. N.; Ikuma K.; Kappell A.; LaPara T.; Kimbell L.; Li M.; Li X.; McNamara P.; Seo Y.; Sobsey M. D.; Sozzi E.; Navab-Daneshmand T.; Raskin L.; Riquelme M. V.; Vikesland P.; Wigginton K.; Zhou Z. An environmental science and engineering framework for combating antimicrobial resistance. Environmental Engineering Science 2018, 35 (10), 1005–1011. 10.1089/ees.2017.0520. [DOI] [Google Scholar]
- Bürgmann H.; Frigon D.; Gaze W.; Manaia C.; Pruden A.; Singer A. C.; Smets B. F.; Zhang T. Water and sanitation: an essential battlefront in the war on antimicrobial resistance. FEMS Microbiology Ecology 2018, 94 (9), fiy101. 10.1093/femsec/fiy101. [DOI] [PubMed] [Google Scholar]
- Rizzo L.; Manaia C.; Merlin C.; Schwartz T.; Dagot C.; Ploy M. C.; Michael I.; Fatta-Kassinos D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Science of the total environment 2013, 447, 345–360. 10.1016/j.scitotenv.2013.01.032. [DOI] [PubMed] [Google Scholar]
- Amarasiri M.; Sano D.; Suzuki S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered. Critical Reviews in Environmental Science and Technology 2020, 50 (19), 2016–2059. 10.1080/10643389.2019.1692611. [DOI] [Google Scholar]
- Pruden A.; Vikesland P. J.; Davis B. C.; de Roda Husman A. M. Seizing the moment: now is the time for integrated global surveillance of antimicrobial resistance in wastewater environments. Curr. Opin. Microbiol. 2021, 64, 91–99. 10.1016/j.mib.2021.09.013. [DOI] [PubMed] [Google Scholar]
- European Society of Microbiology and Infectious Diseases . MIC and zone diameter distributions and ECOFFs. European Committee on Antimicrobial Susceptibility Testing (EUCAST), 2021. https://www.eucast.org/mic_distributions_and_ecoffs/ (accessed 2021-06-11).
- Clinical and Laboratory Standards Institute (CLSI). 2021. CLSI Susceptibility Testing Subcommittees and Resources. https://clsi.org/meetings/susceptibility-testing-subcommittees. (accessed 2021-06-11).
- Hudzicki J.Kirby-Bauer Disk Diffusion Susceptibility Test Protocol; American Society for Microbiology: 2009
- U.S. Environmental Protection Agency (U.S. EPA) . National Primary Drinking Water Regulations: Total Coliform Rule; Corrections; EPA-HQ-OW-2008-0878; U.S. U.S. EPA: 2014.
- U.S. Environmental Protection Agency (U.S. EPA) . 2012 Recreational Water Quality Criteria; EPA-HQ-OW-2011-0466; U.S. EPA: 2012.
- WHO Integrated Global Surveillance on ESBL-Producing E. coli Using a “One Health” Approach. World Health Organization (WHO), March 16, 2021. https://www.who.int/publications/i/item/who-integrated-global-surveillance-on-esbl-producing-e.-coli-using-a-one-health-approach (accessed 2021-11-16).
- Hamiwe T.; Kock M. M.; Magwira C. A.; Antiabong J. F.; Ehlers M. M. Occurrence of enterococci harbouring clinically important antibiotic resistance genes in the aquatic environment in Gauteng, South Africa. Environ. Pollut. 2019, 245, 1041–1049. 10.1016/j.envpol.2018.11.040. [DOI] [PubMed] [Google Scholar]
- Vaz-Moreira I.; Varela A. R.; Pereira T. V.; Fochat R. C.; Manaia C. M. Multidrug resistance in quinolone-resistant gram-negative bacteria isolated from hospital effluent and the municipal wastewater treatment plant. Microbial Drug Resistance 2016, 22 (2), 155–163. 10.1089/mdr.2015.0118. [DOI] [PubMed] [Google Scholar]
- Blasco M. D.; Esteve C.; Alcaide E. Multiresistant waterborne pathogens isolated from water reservoirs and cooling systems. Journal of applied microbiology 2008, 105 (2), 469–475. 10.1111/j.1365-2672.2008.03765.x. [DOI] [PubMed] [Google Scholar]
- Young S.; Nayak B.; Sun S.; Badgley B. D.; Rohr J. R.; Harwood V. J. Vancomycin-resistant enterococci and bacterial community structure following a sewage spill into an aquatic environment. Applied and environmental microbiology 2016, 82 (18), 5653–5660. 10.1128/AEM.01927-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach C. F.; Genheden M.; Fick J.; Joakim Larsson D. G. A comprehensive screening of Escherichia coli isolates from Scandinavia’s largest sewage treatment plant indicates no selection for antibiotic resistance. Environ. Sci. Technol. 2018, 52 (19), 11419–11428. 10.1021/acs.est.8b03354. [DOI] [PubMed] [Google Scholar]
- Mahfouz N.; Caucci S.; Achatz E.; Semmler T.; Guenther S.; Berendonk T. U.; Schroeder M. High genomic diversity of multi-drug resistant wastewater Escherichia coli. Sci. Rep. 2018, 8 (1), 8928. 10.1038/s41598-018-27292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueira V.; Vaz-Moreira I.; Silva M.; Manaia C. M. Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water research 2011, 45 (17), 5599–5611. 10.1016/j.watres.2011.08.021. [DOI] [PubMed] [Google Scholar]
- Jacobs L.; Chenia H. Y. Characterization of integrons and tetracycline resistance determinants in Aeromonas spp. isolated from South African aquaculture systems. International journal of food microbiology 2007, 114 (3), 295–306. 10.1016/j.ijfoodmicro.2006.09.030. [DOI] [PubMed] [Google Scholar]
- Schwartz T.; Kohnen W.; Jansen B.; Obst U. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS microbiology ecology 2003, 43 (3), 325–335. 10.1111/j.1574-6941.2003.tb01073.x. [DOI] [PubMed] [Google Scholar]
- Volkmann H.; Schwartz T.; Bischoff P.; Kirchen S.; Obst U. Detection of clinically relevant antibiotic-resistance genes in municipal wastewater using real-time PCR (TaqMan). J. Microbiol. Methods 2004, 56 (2), 277–286. 10.1016/j.mimet.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Pruden A.; Pei R.; Storteboom H.; Carlson K. H. Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ. Sci. Technol. 2006, 40 (23), 7445–7450. 10.1021/es060413l. [DOI] [PubMed] [Google Scholar]
- Pruden A.; Arabi M.; Storteboom H. N. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ. Sci. Technol. 2012, 46 (21), 11541–11549. 10.1021/es302657r. [DOI] [PubMed] [Google Scholar]
- Czekalski N.; Gascón Díez E.; Bürgmann H. Wastewater as a point source of antibiotic-resistance genes in the sediment of a freshwater lake. ISME J. 2014, 8, 1381–1390. 10.1038/ismej.2014.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings M. R.; Gaze W. H.; Pruden A.; Smalla K.; Tiedje J. M.; Zhu Y. G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME journal 2015, 9 (6), 1269–1279. 10.1038/ismej.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. C.; Riquelme M. V.; Ramirez-Toro G.; Bandaragoda C.; Garner E.; Rhoads W. J.; Vikesland P.; Pruden A. Demonstrating an Integrated Antibiotic Resistance Gene Surveillance Approach in Puerto Rican Watersheds Post-Hurricane Maria. Environ. Sci. Technol. 2020, 54 (23), 15108–15119. 10.1021/acs.est.0c05567. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) . Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services, CDC: Atlanta, GA, 2019. [Google Scholar]
- Zhang A. N.; Gaston J. M.; Dai C. L.; Zhao S.; Poyet M.; Groussin M.; Yin X.; Li L. G.; van Loosdrecht M. C. M.; Topp E.; Gillings M. R.; Hanage W. P.; Tiedje J. M.; Moniz K.; Alm E. J.; Zhang T. An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nat. Commun. 2021, 12 (1), 4765. 10.1038/s41467-021-25096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S. Quantification of antibiotic resistance genes for environmental monitoring: Current methods and future directions. Current Opinion in Environmental Science & Health 2020, 16, 47–53. 10.1016/j.coesh.2020.02.004. [DOI] [Google Scholar]
- Ishii S.; Segawa T.; Okabe S. Simultaneous quantification of multiple food-and waterborne pathogens by use of microfluidic quantitative PCR. Appl. Environ. Microbiol. 2013, 79 (9), 2891–2898. 10.1128/AEM.00205-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansson E.; Fick J.; Janzon A.; Grabic R.; Rutgersson C.; Weijdegård B.; Söderström H.; Larsson D. G. J. Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PloS one 2011, 6 (2), e17038 10.1371/journal.pone.0017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Li B.; Zou S.; Fang H. H.; Zhang T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water research 2014, 62, 97–106. 10.1016/j.watres.2014.05.019. [DOI] [PubMed] [Google Scholar]
- Ju F.; Beck K.; Yin X.; Maccagnan A.; McArdell C. S.; Singer H. P.; Johnson D. R.; Zhang T.; Bürgmann H. Wastewater treatment plant resistomes are shaped by bacterial composition, genetic exchange, and upregulated expression in the effluent microbiomes. ISME journal 2019, 13 (2), 346–360. 10.1038/s41396-018-0277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed H. J.; Riquelme M. V.; Davis B. C.; Gupta S.; Angeles L.; Aga D. S.; Garner E.; Pruden A.; Vikesland P. J. Evaluation of Metagenomic-Enabled Antibiotic Resistance Surveillance at a Conventional Wastewater Treatment Plant. Frontiers in Microbiology 2021, 12, 657954. 10.3389/fmicb.2021.657954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamps B. W.; Leddy M. B.; Plumlee M. H.; Hasan N. A.; Colwell R. R.; Spear J. R. Characterization of the microbiome at the world’s largest potable water reuse facility. Frontiers in Microbiology 2018, 9, 2435. 10.3389/fmicb.2018.02435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriksen R. S.; Munk P.; Njage P.; Van Bunnik B.; McNally L.; Lukjancenko O.; Röder T.; Nieuwenhuijse D.; Pedersen S. K.; Kjeldgaard J.; Kaas R. S.; Conradsen Clausen P. T. L.; Vogt J. K.; Leekitcharoenphon P.; van de Schans M. G. M.; Zuidema T.; Maria de Roda Husman A.; Rasmussen S.; Petersen B.; Amid C.; Cochrane G.; Sicheritz-Ponten T.; Schmitt H.; Matheu Alvarez J. R.; Aidara-Kane A.; Pamp S. J.; Lund O.; Hald T.; Woolhouse M.; Koopmans M. P.; Vigre H.; Petersen T.; Aarestrup F. M. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019, 10 (1), 1124. 10.1038/s41467-019-08853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta C. L.; Tiwari R. K.; Cytryn E. Platforms for elucidating antibiotic resistance in single genomes and complex metagenomes. Environ. Int. 2020, 138, 105667. 10.1016/j.envint.2020.105667. [DOI] [PubMed] [Google Scholar]
- Garner E.; Davis B. C.; Milligan E.; Blair M. F.; Keenum I.; Maile-Moskowitz A.; Pan J.; Gnegy M.; Liguori K.; Gupta S.; Prussin A. J.; Marr L. C.; Heath L. S.; Vikesland P. J.; Zhang L.; Pruden A. Next generation sequencing approaches to evaluate water and wastewater quality. Water Res. 2021, 194, 116907. 10.1016/j.watres.2021.116907. [DOI] [PubMed] [Google Scholar]
- Knudsen B. E.; Bergmark L.; Munk P.; Lukjancenko O.; Prieme A.; Aarestrup F. M.; Pamp S. J. Impact of sample type and DNA isolation procedure on genomic inference of microbiome composition. mSystems 2016, 1 (5), e00095-16 10.1128/mSystems.00095-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gweon H. S.; Shaw L. P.; Swann J.; De Maio N.; AbuOun M.; Niehus R.; Hubbard A.; Stoesser N.; et al. The impact of sequencing depth on the inferred taxonomic composition and AMR gene content of metagenomic samples. Environmental Microbiome 2019, 14 (1), 7. 10.1186/s40793-019-0347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen C. S.; Kaas R. S.; Aarestrup F. M.; Pamp S. J. Standard sample storage conditions have an impact on inferred microbiome composition and antimicrobial resistance patterns. Microbiology Spectrum 2021, 9 (2), e01387-21 10.1128/Spectrum.01387-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur A. G.; Waglechner N.; Nizam F.; Yan A.; Azad M. A.; Baylay A. J.; Bhullar K.; Canova M. J.; De Pascale G.; Ejim L.; Kalan L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B.; Raphenya A. R.; Alcock B.; Waglechner N.; Guo P.; Tsang K. K.; Lago B. A.; Dave B. M.; Pereira S.; Sharma A. N.; Doshi S.; Courtot M.; Lo R.; Williams L. E.; Frye J. G.; Elsayegh T.; Sardar D.; Westman E. L.; Pawlowski A. C.; Johnson T. A.; Brinkman F. S. L.; Wright G. D.; McArthur A. G. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Research 2017, 45, D566. 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango-Argoty G.; Garner E.; Pruden A.; Heath L. S.; Vikesland P.; Zhang L. DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 2018, 6 (1), 23. 10.1186/s40168-018-0401-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolaia V.; Kaas R. S.; Ruppe E.; Roberts M. C.; Schwarz S.; Cattoir V.; Philippon A.; Allesoe R. L.; Rebelo A. R.; Florensa A. F.; Fagelhauer L.; Chakraborty T.; Neumann B.; Werner G.; Bender J. K.; Stingl K.; Nguyen M.; Coppens J.; Xavier B. B.; Malhotra-Kumar S.; Westh H.; Pinholt M.; Anjum M. F.; Duggett N. A.; Kempf I.; Nykäsenoja S.; Olkkola S.; Wieczorek K.; Amaro A.; Clemente L.; Mossong J.; Losch S.; Ragimbeau C.; Lund O.; Aarestrup F. M. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75 (12), 3491–3500. 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doster E.; Lakin S. M.; Dean C. J.; Wolfe C.; Young J. G.; Boucher C.; Belk K. E.; Noyes N. R.; Morley P. S. MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic acids research 2020, 48 (D1), D561–D569. 10.1093/nar/gkz1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson M. K.; Forsberg K. J.; Dantas G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME journal 2015, 9 (1), 207–216. 10.1038/ismej.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1928 Diagnostics. resqu database, 2021. https://www.1928diagnostics.com/resdb/ (accessed 2021-12-28).
- Yang Y.; Jiang X.; Chai B.; Ma L.; Li B.; Zhang A.; Cole J. R.; Tiedje J. M.; Zhang T. ARGs-OAP: online analysis pipeline for antibiotic resistance genes detection from metagenomic data using an integrated structured ARG-database. Bioinformatics 2016, 32 (15), 2346–2351. 10.1093/bioinformatics/btw136. [DOI] [PubMed] [Google Scholar]
- Gupta S. K.; Padmanabhan B. R.; Diene S. M.; Lopez-Rojas R.; Kempf M.; Landraud L.; Rolain J. M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58 (1), 212–220. 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C.; Bengtsson-Palme J.; Rensing C.; Kristiansson E.; Larsson D. J. BacMet: antibacterial biocide and metal resistance genes database. Nucleic acids research 2014, 42 (D1), D737–D743. 10.1093/nar/gkt1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk P. S.; Lipinski L.; Dziembowski A. PlasFlow: predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acids Research 2018, 46 (6), e35 10.1093/nar/gkx1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.; Li X.; Xie Y.; Bi D.; Sun J.; Li J.; Tai C.; Deng Z.; Ou H. Y. ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic acids research 2019, 47 (D1), D660–D665. 10.1093/nar/gky1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R.; Lima-Mendez G.; Toussaint A. ACLAME: a CLAssification of Mobile genetic Elements, update 2010. Nucleic Acids Research 2010, 38, D57–D61. 10.1093/nar/gkp938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossette E.; Gumm J.; Langenfeld K.; Raskin L.; Duhaime M.; Wigginton K. Metagenomic Quantification of Genes with Internal Standards. mBio 2021, 12 (1), e03173-20 10.1128/mBio.03173-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y.; Leung H. C.; Yiu S. M.; Chin F. Y. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28 (11), 1420–1428. 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- Li D.; Liu C. M.; Luo R.; Sadakane K.; Lam T. W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31 (10), 1674–1676. 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Nurk S.; Meleshko D.; Korobeynikov A.; Pevzner P. A. metaSPAdes: a new versatile metagenomic assembler. Genome research 2017, 27 (5), 824–834. 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. L.; Keenum I. M.; Dai D.; Zhang L.; Vikesland P. J.; Pruden A. Critical evaluation of short, long, and hybrid assembly for contextual analysis of antibiotic resistance genes in complex environmental metagenomes. Sci. Rep 2021, 11 (1), 3753. 10.1038/s41598-021-83081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M.; Pruden A.; Chen C.; Heath L. S.; Xia K.; Zhang L. MetaCompare: a computation pipeline for prioritizing enivornmental resistome risk. FEMS Microbiology Ecology 2018, 94 (7), fiy079. 10.1093/femsec/fiy079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R.; Ley R. E.; Raes J.; Grice E. A. Expanding the scope and scale of microbiome research. Genome Biology 2019, 20 (1), 191. 10.1186/s13059-019-1804-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendonk T. U.; Manaia C. M.; Merlin C.; Fatta-Kassinos D.; Cytryn E.; Walsh F.; Bürgmann H.; Sørum H.; Norström M.; Pons M.-N.; Kreuzinger N.; Huovinen P.; Stefani S.; Schwartz T.; Kisand V.; Baquero F.; Martinez J. L. Tackling Antibiotic Resistance: The Environmental Framework. Nature Reviews Microbiology 2015, 13 (5), 310–317. 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- Huijbers P. M. C.; Flach C. F.; Larsson D. G. J. A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environ. Int. 2019, 130, 104880. 10.1016/j.envint.2019.05.074. [DOI] [PubMed] [Google Scholar]
- Keenum I.; Liguori K.; Calarco J.; Davis B. C.; Milligan E.; Harwood V. J.; Pruden A. A framework for standardized qPCR-targets and protocols for quantifying antibiotic resistance in surface water, recycled water and wastewater. Critical Reviews in Environmental Science and Technology 2022, 1–25. 10.1080/10643389.2021.2024739. [DOI] [Google Scholar]
- Hassoun-Kheir N.; Stabholz Y.; Kreft J. U.; de la Cruz R.; Dechesne A.; Smets B. F.; Romalde J. L.; Lema A.; Balboa S.; García-Riestra C.; Torres-Sangiao E.; Neuberger A.; Graham D.; Quintela-Baluja M.; Stekel D. J.; Graham J.; Pruden A.; Nesme J.; Sørensen S. J.; Hough R.; Paul M. EMBRACE-WATERS statement: Recommendations for reporting of studies on antimicrobial resistance in wastewater and related aquatic environments. One Health 2021, 13, 100339. 10.1016/j.onehlt.2021.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu J.WHO Model for Integrated Surveillance on AMR, The ESBL Ec Tricycle Protocol, Challenges and Lessons Learned. Presented at the WRF 5052 Expert Workshop on AMR Monitoring in Water, May 18, 2021, Virtual.
- JPIAMR . Establishing a Monitoring Baseline for Antimicrobial Resistance in Key Environments (EMBARK). Mission. https://antimicrobialresistance.eu/mission/ (accessed 2021-11-19).
- Garland J. L.; Brinkman N.; Jahne M.; Keely S.. Geospatial Distribution of Antimicrobial Resistance Genes in US Rivers and Streams; U.S. Environmental Protection Agency: 2019.
- Berglund B. Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics. Infection ecology & epidemiology 2015, 5 (1), 28564. 10.3402/iee.v5.28564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp C. W.; Dolfing J.; Ehlert P. A.; Graham D. W. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 2010, 44 (2), 580–587. 10.1021/es901221x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.