Abstract
Mechanical thrombectomy (MTB) is one of the two standard treatment options for Acute Ischemic Stroke (AIS) patients. Current clinical guidelines instruct the use of pretreatment imaging to characterize a patient’s cerebrovascular flow, as there are many factors that may underlie a patient’s successful response to treatment. There is a critical need to leverage pretreatment imaging, taken at admission, to guide potential treatment avenues in an automated fashion. The aim of this study is to develop and validate a fully automated machine learning algorithm to predict the final modified thrombolysis in cerebral infarction (mTICI) score following MTB. A total 321 radiomics features were computed from segmented pretreatment MRI scans for 141 patients. Successful recanalization was defined as mTICI score >= 2c. Different feature selection methods and classification models were examined in this study. Our best performance model achieved 74.42±2.52% AUC, 75.56±4.44% sensitivity, and 76.75±4.55% specificity, showing a good prediction of reperfusion quality using pretreatment MRI. Results suggest that MR images can be informative to predicting patient response to MTB, and further validation with a larger cohort can determine the clinical utility.
Keywords: Structural MRI, Radiomics, Machine Learning, Stroke Treatment
I. INTRODUCTION
A total of 795,000 strokes are diagnosed each year in the United States, which causes more than 140,000 deaths. Acute Ischemic Stroke (AIS) accounts for 87% of all strokes. [1] For AIS patients, thrombolysis and thrombectomy treatments can restore blood flow ischemic tissue. Successful mechanical thrombectomy (MTB), which physically removes the clot from the occluded artery, has many potential factors that could influence a patient’s response to treatment. [2] In practice, success is measured by restoration of blood flow to the stroke area, quantified by the modified treatment in cerebral infarction (mTICI) score. [3] During MTB, this score is assessed after every attempt to remove the clot, and a final mTICI score is assigned to the patient at the end of the procedure to signify how much blood flow has been restored. [4] Clinical trials and other studies have illustrated that patients who experience partial and/or full recanalization of the blood vessel typically experience better outcomes, particularly if recanalization is achieved in three attempts or less. [5]-[7] Thus, predicting a patient’s post-treatment mTICI can inform interventional neuroradiologists about the potential benefit of MTB.
Per current clinical guidelines, advanced imaging (i.e., perfusion-weighted imaging) may be taken at admission to determine a patient’s vascular status such as collateral flow. [8] Some imaging biomarkers have been statistically linked to successful recanalization, but they are not perfect predictors, and some are prone to inter-reader assessment. Previous machine learning methods applied to mTICI classification have used CT sequences as input, and the top performing models required manually segmented regions of interest (ROI) performed by an expert. [9]-[12] On the other hand, there is no reliable publicly available automated stroke lesion or thrombus segmentation algorithm, and none has been validated at a large scale . [13] Even with the help of segmentation by experts, which may not be feasible in a time-sensitive clinical setting, to our knowledge, there does not yet exist a method to automatically predict final mTICI from MR imaging. MR imaging has already proven to contain signal that can elucidate individual patient characteristics, informing how one might respond to thrombolytic, or the more invasive MTB treatment. [14] Developing an automatic classification of the mTICI score from admission MR imaging alone can help clinicians assess the most efficient course of treatment.
II. METHODS
A. Dataset
We retrospectively evaluated 141 patients from UCLA Ronald Reagan Medical Center who received MTB from 2014-2019. This work was performed under the approval of the UCLA Institutional Review Board (#18-000329). A patient was included if they were diagnosed with AIS, underwent a diffusion-weighted MR prior to treatment, and received MTB. Although perfusion imaging may provide more information about collateral flow status, we decided to only use diffusion-weighted image, as it included the broadest patient cohort within our study. Clinical characteristics are summarized in table 1. The study cohort had a median age of 73 (61-81) years, a median National Institutes of Health Stroke Scale (NIHSS) score of 15 (8-19), and were 55% female. The distribution of final mTICI score was 6.53% 0, 1.96% 1, 11.11% 2a, 45.75% 2b, 15.03% 2c, and 19.61% 3.
TABLE I.
Patient cohort demographics. Numbers are n (%) or median (interquartile ranges). NIHSS, National Institutes of Health Stroke Scale.
| Training Set (n = 112) |
Validation Set (n = 29) |
|
|---|---|---|
| Age (years) | 73 (62-81) | 73 (52-85) |
| Female | 63 (57%) | 22 (61%) |
| NIHSS | 15 (8 - 19) | 17 (10 - 21.75) |
| mTICI < 2c | 40 (36%) | 12 (41%) |
B. Image Preprocessing
All patients underwent MRI using a 1.5T or 3T echo-planar Siemens MR imaging scanner, performed with 12-channel head coils. DWI, FLAIR and ADC sequences were used in this study. The DWI images were acquired using a TR range of 4,000-9,000ms and a TE range of 78-122ms. The pixel dimension for DWI varied from 0.859x0.859x6.000mm to 1.850x1.850x6.500mm. FLAIR images were acquired using a TR range of 8,000-9,000ms and a TE range of 88-134ms. The corresponding pixel dimension varied from 0.688x0.688x6.000mm to 0.938x0.938x6.500mm. ADC images were calculated from DWI sequences. After image retrieval, the sequences were fed into our previously published automated preprocessing pipeline to ensure data consistency across individuals and reduce noisy information [15]. First, N4 bias field correction was applied to all sequences. Then, DWI image was skull-stripped and registered to a T2w MNI-152 atlas. FLAIR and ADC were co-registered afterwards. Finally, intensity normalization and histogram matching were performed using a reference case. Last, a vascular territory template was mapped on the registered images to extract ROI where the stroke lesion was located [16]. Instead of manually segmenting the stroke lesion or thrombus, or using unreliable algorithms, our detection method extracted the affected brain region.
C. Feature Extraction
Radiomic features were extracted from the ROI for DWI, FLAIR and ADC sequences separately using pyradiomics [17]. The features included a) 13 3D shape features and 19 first order features b) 24 texture features computed from the Gray Level Co-occurrence Matrix (GLCM) c) 14 features from Gray Level Dependence Matrix (GLDM) d) 16 features from Gray Level Size Zone Matrix (GLSZM) e) 16 features from Gray Level Run Length Matrix (GLRLM) and f) 5 features from Neighbouring Gray Tone Difference Matrix (NGTDM).
D. Feature Selection
Many extracted radiomic features have a high degree of correlation, and they may provide overlapping information to our machine learning model. To minimize information overlap, we implemented three feature selection methods and compared the performance of each using different classifiers. Two supervised methods, Least Absolute Shrinkage and Selection Operator (LASSO), Random Forest (RF), and one unsupervised method, Principle Component Analysis (PCA), were selected due to their popularity and efficiency in literature [18]. Briefly, LASSO applies a regularization process that penalizes the coefficient of regression features to minimize the prediction error and the features with non-zero coefficient after shrinking process are selected. For RF, features are selected by calculating each feature’s contribution to the decrease of the weighted impurity of a tree. Feature importance is calculated by averaging the decrease of impurity across trees and ranking the features according to this measure, only keeping the top 50th percentile of features in this rank. For PCA, the current features are transformed to a representation with fewer new features (principle components) by a dimensionality reduction process that involves orthogonal linear transformation while preserving the variance presented in the data.
E. Classification Modeling
To predict mTICI, the following classifiers were compared: Support Vector Machines (SVM), K-Nearest Neighbor (KNN), Logistic Regression (LR), and RF. Classifiers were trained using 5-fold cross-validation in the training set. The best model hyper-parameters were selected via grid search. For example, different kernel functions (radial basis, linear, sigmoid), gamma and C values were examined for SVM to generate best combination of hyper-parameters. The selected features were scaled using min-max normalization before being used in classification training. All feature selection and classification models were evaluated using area under the curve (AUC) of receiver operating characteristic (ROC) curve, as well as sensitivity and specificity, which were calculated using optimal Youden’s J statistic as defined below:
Performance ranges for a held-out validation set were calculated for each combination of feature selection method and classifier.
III. Results
A total of 321 features were extracted for each patient with DWI, FLAIR, and ADC sequences. 112 patients were assigned to the training set and 29 patients were assigned to the validation set following a 4-1 split, where the validation set was never seen by neither the feature selection, grid search nor classifier training processes. The demographic distribution for both training and validation sets remained the same for all experiments. In order to examine the stability of model performance, we trained and validated the results 100 times by changing the random seed to shuffle the training stage cross-validation. The results were reported as the mean +/− 95% confidence interval. All model training was performed using scikit-learn in Python 3. The best parameters for each model from grid search were: Random Forest with 80 max depth of the tree, max features 3, minimum number of samples required at a leaf node 3, minimum number of samples required to split an internal node 12, number of trees 200; SVM with radial basis kernel, penalty parameter C 10 and gamma 0.001; LR with L2 penalty and inverse regularization C 0.1; KNN with 10 neighbors.
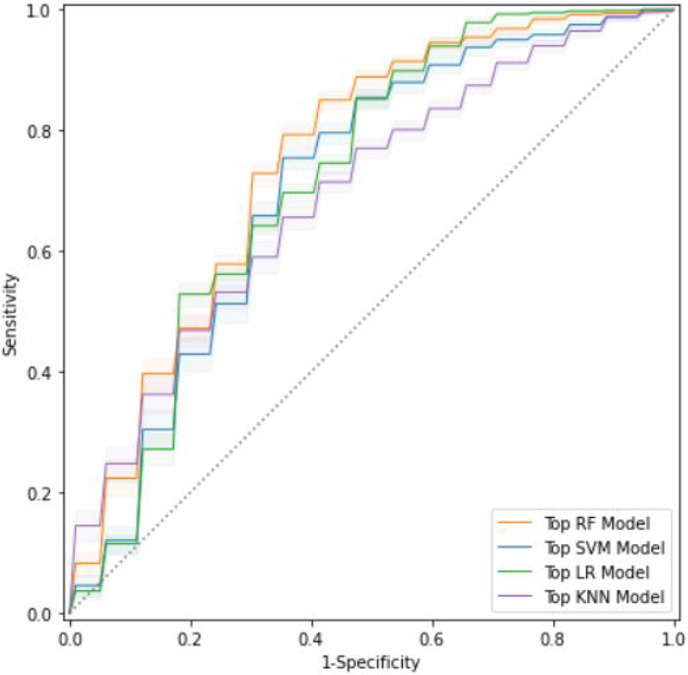
The RF feature selection determined 138 features were informative for mTICI classification. The LASSO feature selection kept 36 important features. We implemented PCA at 0.99 explained variance cutoff with 5 principle components for modeling. The average performance was reported in table 2. In addition, ROC curves on the validation set for each of the top models with best feature selection was reported in figure 2. RF feature selection and LASSO feature selection yielded comparable performance across different models where the combination of RF feature selection and RF classification model achieved best ROC-AUC 74.29%±0.68%. On the other hand, PCA feature selection yielded lower performance across models. Moreover, models with PCA features tended to be less stable with a larger variance except for LR model, showing that the PCA method may exclude much important information during dimension reduction step. Although the RF model achieved the highest performance, the LR model achieved 72.91%±0.84% using the RF feature selector and 72.29%±0.85% using LASSO feature selector. SVM achieved > 70% ROC-AUC for RF features but lower for LASSO and PCA features. In general, KNN model achieved lower ROC-AUC values for all three feature selectors with higher variance across experiments.
TABLE II.
Performance metrics across feature selection methods and classification models. Double lines separate models with different outputs. FS = Feature Selection, Sens = Sensitivity, Spec = Specificity, AUC = Receiver Operating Characteristic Area Under Curve
| Model | FS | AUC | Sens. | Spec. |
|---|---|---|---|---|
| RF | RF | 74.29±0.68% | 69.92±2.53% | 80.59±2.29% |
| LASSO | 70.94±0.75% | 77.67±2.27% | 68.18±2.22% | |
| PCA | 65.71±2.01% | 65.00±9.64% | 72.06±9.15% | |
| SVM | RF | 71.15±1.21% | 81.67±2.29% | 59.65±1.13% |
| LASSO | 66.33±3.32% | 83.50±1.66% | 64.18±5.27% | |
| PCA | 65.37±3.12% | 74.33±6.63% | 59.18±3.51% | |
| LR | RF | 72.91±0.84% | 57.67±1.84% | 90.59±1.47% |
| LASSO | 72.29±0.85% | 67.75±1.51% | 80.53±1.32% | |
| PCA | 67.16±0.02% | 66.67±0.63% | 82.35±0.05% | |
| KNN | RF | 67.03±2.91% | 80.42±6.56% | 57.65±7.72% |
| LASSO | 69.52±1.54% | 77.50±5.31% | 65.00±5.38% | |
| PCA | 65.34±1.77% | 67.92±7.51% | 67.65±7.08% |
Fig. 2.
ROC curves - Best feature selection method for each model. Top RF model = RF feature + RF model, top SVM model = RF feature + SVM model, top LR model = RF feature + LR model, top KNN model = LASSO feature + KNN model.
IV. DISCUSSION AND CONCLUSION
To our knowledge, this study presents the first algorithm to predict successful MTB recanalization from MRI taken from patients pre-treatment imaging. It is clear that there is important information from standard diffusion MR images before treatment that is directly related to MTB recanalization, leading to a potential new path of investigation in pre-treatment MR imaging and thrombectomy outcome. Our results produce a few findings. First, we demonstrated that the RF feature selector combined with the RF model achieved the best performance. Based on the 95% Confidence Interval for 100 times repeated experiments, RF and LR models both show stable performance, that is, minimal variation when shuffling the training data. Random forest models have been illustrated to be robust across many iterations, and moreover, have proven to achieve high classification performance for other radiomics tasks. Other tasks have found optimal performance using other classifiers, however, so more testing is needed to evaluate these models at a broader scale. A second finding is that our automated region extraction method yielded benchmark performance comparable to those generated from manually segmented regions on CT images. It could serve as a reference for future radiomics based MR image studies in the MTB recanalization field. Given the time-sensitive nature of stroke treatment decisions, this fully automatic method can quickly provide a model with the relevant stroke region without sacrificing performance.
There are a few areas of future research direction this study provides given the promising result of this preliminary study. Our analysis and modeling were conducted from a single center with images acquired retrospectively over eight years. A multi-center prospective study with a larger sample size is needed in the next step. In this study, we only examined several popular feature selection methods and machine learning algorithms. Many other feature selection methods and ML classification should be examined in future study. A reliable automated stroke lesion or thrombus segmentation algorithm is expected to provide more accurate radiomics features for use. By incorporating more accurate 3D image features, stratification by the type of MTB technique, and clinical factors as features into the machine learning model, the model performance can be improved.
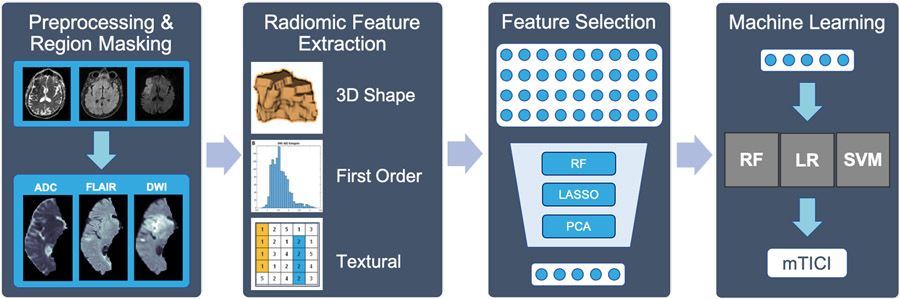
Fig. 1.
Our methodological pipeline. RF: Random Forest, LASSO: Least Absolute Shrinkage and Selection Operator, PCA: Principal Component Analysis, LR: Logistic Regression, SVM: Support Vector Machine.
Acknowledgments
This work was supported by the following grants: NIH T32EB016640-07, NIH R01NS100806-02, and NVIDIA Academic Hardware Grant.
Contributor Information
Haoyue Zhang, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA, USA..
Jennifer Polson, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA, USA..
Kambiz Nael, Department of Radiology, University of California, Los Angeles, Los Angeles, CA, USA..
Noriko Salamon, Department of Radiology, University of California, Los Angeles, Los Angeles, CA, USA..
Bryan Yoo, Department of Radiology, University of California, Los Angeles, Los Angeles, CA, USA..
William Speier, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA, USA..
Corey Arnold, Departments of Bioengineering, Radiology, and Pathology, University of California, Los Angeles, Los Angeles, CA, USA.
References
- [1].Benjamin EJ et al. , “Heart Disease and Stroke Statistics-2019 Update: A Report From the American Heart Association,” Circulation, vol. 139, no. 10, pp. e56–e528, 03 2019. [DOI] [PubMed] [Google Scholar]
- [2].Nogueira R, Jadhav A et al. , “Thrombectomy 6 to 24 hours after stroke with a mismatch between deficit and infarct,” The New England Journal of Medicine, vol. 378, p. 11–21, 2018. [DOI] [PubMed] [Google Scholar]
- [3].Tomsick T, “Timi, tibi, tici: I came, i saw, i got confused.” AJNR. American journal of neuroradiology, vol. 28 2, pp. 382–4, 2007. [PMC free article] [PubMed] [Google Scholar]
- [4].Liebeskind D, Bracard S et al. , “etici reperfusion: defining success in endovascular stroke therapy,” Journal of NeuroInterventional Surgery, vol. 11, pp. 433 – 438, 2018. [DOI] [PubMed] [Google Scholar]
- [5].Dargazanli C, Consoli A, Barral M, Labreuche J, Redjem H, Ciccio G, Smajda S, Desilles J, Taylor G, Preda C, Coskun O, Rodesch G, Piotin M, Blanc R, and Lapergue B, “Impact of modified tici 3 versus modified tici 2b reperfusion score to predict good outcome following endovascular therapy,” American Journal of Neuroradiology, vol. 38, pp. 90 – 96, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Goyal N, Tsivgoulis G, Frei D, Turk A, Baxter B, Froehler M, Mocco J, Ishfaq MF, Malhotra K, Chang J, Hoit D, Elijovich L, Loy D, Turner R, Mascitelli J, Espaillat K, Alexandrov A, and Arthur A, “Comparative safety and efficacy of modified tici 2b and tici 3 reperfusion in acute ischemic strokes treated with mechanical thrombectomy,” Neurosurgery, vol. 84, p. 680–686, 2019. [DOI] [PubMed] [Google Scholar]
- [7].Volny O, Cimflova P, and Szeder V, “Inter-rater reliability for thrombolysis in cerebral infarction with tici 2c category.” Journal of stroke and cerebrovascular diseases : the official journal of National Stroke Association, vol. 26 5, pp. 992–994, 2017. [DOI] [PubMed] [Google Scholar]
- [8].Powers WJ et al. , “Guidelines for the Early Management of Patients With Acute Ischemic Stroke: 2019 Update to the 2018 Guidelines for the Early Management of Acute Ischemic Stroke: A Guideline for Healthcare Professionals From the American Heart Association/American Stroke Association,” Stroke, vol. 50, no. 12, pp. e344–e418, Dec 2019. [DOI] [PubMed] [Google Scholar]
- [9].Qiu W, Kuang H, Nair J, Assis Z, Najm M, McDougall C, McDougall B, Chung K, Wilson A, Goyal M, Hill M, Demchuk A, and Menon B, “Radiomics-based intracranial thrombus features on ct and cta predict recanalization with intravenous alteplase in patients with acute ischemic stroke,” American Journal of Neuroradiology, vol. 40, pp. 39 – 44, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hofmeister J, Bernava G, Rosi A, Vargas M, Carrera E, Montet X, Burgermeister S, Poletti P, Platon A, Lovblad K, and Machi P, “Clot-based radiomics predict a mechanical thrombectomy strategy for successful recanalization in acute ischemic stroke,” Stroke, vol. 51, pp. 2488 – 2494, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].van Os HJA, Ramos LA, Hilbert A, van Leeuwen M, van Walderveen MV, Kruyt N, Dippel D, Steyerberg E, van der Schaaf I, Lingsma HF, Schonewille W, Majoie C, Olabarriaga S, Zwinderman K, Venema E, Marquering H, and Wermer M, “Predicting outcome of endovascular treatment for acute ischemic stroke: Potential value of machine learning algorithms,” Frontiers in Neurology, vol. 9, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hilbert A, Ramos LA, Os HJV, Olabarriaga S, Tolhuisen M, Wermer M, Barros RS, Schaaf IVD, Dippel D, Roos Y, Zwam WV, Yoo A, Emmer B, Nijeholt G, Zwinderman A, Strijkers G, Majoie C, and Marquering H, “Data-efficient deep learning of radiological image data for outcome prediction after endovascular treatment of patients with acute ischemic stroke,” Computers in biology and medicine, vol. 115, p. 103516, 2019. [DOI] [PubMed] [Google Scholar]
- [13].Winzeck S, Hakim A, McKinley R, Pinto JA, Alves V, Silva C, Pisov M, Krivov E, Belyaev M, Monteiro M et al. , “Isles 2016 and 2017-benchmarking ischemic stroke lesion outcome prediction based on multispectral mri,” Frontiers in neurology, vol. 9, p. 679, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].G.T., et al. , “Mri-guided thrombolysis for stroke with unknown time of onset,” The New England Journal of Medicine, vol. 379, p. 611–622, 2018. [DOI] [PubMed] [Google Scholar]
- [15].Zhang H, Polson JS, Nael K, Salamon N, Yoo B, El-Saden S, Scalzo F, Speier W, and Arnold C, “Intra-domain task-adaptive transfer learning to determine acute ischemic stroke onset time,” ArXiv, vol. abs/2011.03350, 2020. [DOI] [PubMed] [Google Scholar]
- [16].Fransen P, Beumer D, Berkhemer O, van den Berg LA, Lingsma HF, van der Lugt A, van Zwam WV, van Oostenbrugge RV, Roos Y, Majoie C, and Dippel D, “Mr clean, a multicenter randomized clinical trial of endovascular treatment for acute ischemic stroke in the netherlands: study protocol for a randomized controlled trial,” Trials, vol. 15, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].van Griethuysen J, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan R, Fillion-Robin J, Pieper S, and Aerts H, “Computational radiomics system to decode the radiographic phenotype.” Cancer research, vol. 77 21, pp. e104–e107, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Parmar C, Grossmann P, Bussink J, Lambin P, and Aerts HJWL, “Machine learning methods for quantitative radiomic biomarkers,” Scientific Reports, vol. 5, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]