Abstract
We describe the development and validation of a method for the qualitative analysis of complex bifidobacterial communities based on PCR and denaturing gradient gel electrophoresis (DGGE). Bifidobacterium genus-specific primers were used to amplify an approximately 520-bp fragment from the 16S ribosomal DNA (rDNA), and the fragments were separated in a sequence-specific manner in DGGE. PCR products of the same length from different bifidobacterial species showed good separation upon DGGE. DGGE of fecal 16S rDNA amplicons from five adult individuals showed host-specific populations of bifidobacteria that were stable over a period of 4 weeks. Sequencing of fecal amplicons resulted in Bifidobacterium-like sequences, confirming that the profiles indeed represent the bifidobacterial population of feces. Bifidobacterium adolescentis was found to be the most common species in feces of the human adult subjects in this study. The methodological approach revealed intragenomic 16S rDNA heterogeneity in the type strain of B. adolescentis, E-981074. The strain was found to harbor five copies of 16S rDNA, two of which were sequenced. The two 16S rDNA sequences of B. adolescentis E-981074T exhibited microheterogeneity differing in eight positions over almost the total length of the gene.
The human gastrointestinal (GI) tract hosts a rich and complex microbiota. Bifidobacteria are part of the normal microbiota of the human intestine, and they are considered to be important in maintaining well-balanced intestinal microbiota (4, 31). It has been postulated that Bifidobacterium spp. have several health-promoting effects, including the prevention of diarrhea and intestinal infections, alleviation of constipation, production of antimicrobials against harmful intestinal bacteria, and immunostimulation (4, 31). Therefore, many attempts have been made to increase the number of bifidobacteria in the intestine by administration of certain bifidobacterial strains (probiotics) or oligo- and polysaccharides that stimulate the growth of bifidobacteria (prebiotics) (2, 7, 10, 13). For the enumeration and isolation of bifidobacteria, several selective plating techniques have been developed (5, 14, 32). So far, 12 species have been associated with the human host: Bifidobacterium adolescentis, Bifidobacterium infantis, Bifidobacterium longum, Bifidobacterium bifidum, Bifidobacterium breve, Bifidobacterium catenulatum, Bifidobacterium pseudocatenulatum, Bifidobacterium angulatum, Bifidobacterium gallicum, Bifidobacterium inopinatum, Bifidobacterium dentium, and Bifidobacterium denticolens, the last three being found primarily in the oral cavity (6, 11, 20, 23, 25).
Our present knowledge of the GI tract microbiota is largely based on cultivation studies, but according to recent estimates up to 85% of the entire microbial population in the human intestine might be uncultured (19, 36). Consequently, our picture of the intestinal microbiota has been biased in favor of the more easily cultured members of the community. Moreover, cultivation techniques are laborious and time-consuming, especially if bacterial isolates are to be identified. In order to overcome the limitations associated with culturing techniques, molecular biological methods are increasingly being applied to study the GI tract ecology (38). One of the most widely used approaches in ecological studies has been the use of rRNA and its encoding genes as target molecules (3). Specific PCR primers and probes can be designed based on the variable regions of this molecule to detect certain species or groups of bacteria. Numerous genus- and species-specific PCR primers and probes have been developed also for bifidobacteria (15, 19, 23, 24, 40). Species-specific primers and probes are excellent tools for targeting certain Bifidobacterium species in mixed populations, providing valuable help in identification, which is laborious and sometimes unreliable by phenotypic characterization. However, the use of specific primers and probes in ecological studies rules out the possibility of finding other than the target Bifidobacterium species possibly also present in the sample. On the other hand, genus-specific primers or probes can give a good overall picture of the bifidobacterial population, but no information is obtained about the species or strain composition.
Another way of utilizing the rRNA sequence heterogeneity in microbial ecology is to use universal bacterial PCR primers to amplify a fragment of rRNA or ribosomal DNA (rDNA) and then separate the obtained PCR products in a sequence-specific manner in temperature gradient gel electrophoresis (TGGE) or denaturing gradient gel electrophoresis (DGGE) (27, 28). The TGGE or DGGE profile thus obtained represents the prominent bacteria in the community. This technique has already been successfully applied to monitor the most predominant bacterial populations in human fecal samples (43).
In this study we describe the development and validation of a method that combines Bifidobacterium genus-specific PCR with DGGE that allowed us to analyze complex bifidobacterial communities. This approach was applied to study the bifidobacterial communities in the feces of adult subjects. The newly developed method also revealed intragenomic 16S rDNA heterogeneity in B. adolescentis E-981074T that was demonstrated to contain at least two distinct copies of 16S rDNA.
MATERIALS AND METHODS
Strains and growth conditions.
The 19 strains of bifidobacteria belonging to 13 different species used in this study are presented in Table 1. Bifidobacterium lactis and Bifidobacterium animalis species are often utilized in probiotic preparations for human and animal use whereas the other species listed in Table 1 are associated with the human host. Bacteria were obtained from the VTT Culture Collection (VTT Biotechnology, Espoo, Finland), Chr. Hansen A/S (Hørsholm, Denmark), and CSIRO Starter Culture Collection (CSCC) (Melbourne, Australia). The strains were grown in Man-Rogosa-Sharpe medium supplemented with 0.5 g of cysteine liter−1 in anaerobic jars with Anaerocult A-strips (Merck, Darmstadt, Germany) at 37°C.
TABLE 1.
Bifidobacterium strains used in the present study
| Species | Strain | Source | Code in other collection |
|---|---|---|---|
| B. adolescentis | E-981074T | VTT Culture Collection | ATCC 15703 |
| E-991436 | VTT Culture Collection | ATCC 15705 | |
| B. angulatum | CSCC 1925T | CSCC | ATCC 27535 |
| B. animalis | E-96663T | VTT Culture Collection | ATCC 25527 |
| B. bifidum | E-97795T | VTT Culture Collection | ATCC 29521 |
| Bb-11 | Chr. Hansen A/S | ||
| B. breve | E-981075T | VTT Culture Collection | ATCC 15700 |
| B. catenulatum | CSCC 1967T | CSCC | ATCC 27539 |
| B. denticolens | E-991434T | VTT Culture Collection | DSM 10105 |
| B. dentium | E-991438T | VTT Culture Collection | ATCC 27534 |
| B. gallicum | CSCC 5492T | CSCC | ATCC 49850 |
| B. infantis | E-97796T | VTT Culture Collection | ATCC 15697 |
| Bb-02 | Chr. Hansen A/S | ||
| B. lactis | E-97847T | VTT Culture Collection | DSM 10140 |
| E-94508 | VTT Culture Collection | Chr. Hansen A/S Bb-12 | |
| B. longum | E-96664T | VTT Culture Collection | ATCC 15707 |
| E-94505 | VTT Culture Collection | Chr. Hansen A/S Bb-46 | |
| E-96702 | VTT Culture Collection | ||
| B. pseudocatenulatum | E-991439T | VTT Culture Collection | ATCC 27919 |
Fecal samples.
Fecal samples were collected from six Finnish individuals (subjects I to VI) of different ages (21 to 55 years) and sex (three women and three men). Samples were frozen at −70°C immediately after defecation. Bifidobacterial counts of fecal samples were determined by selective plating on Beerens agar (5) under anaerobic conditions in a Whitley Anaerobic Cabinet (model MK II; Don Whitley Scientific Ltd., Shipley, United Kingdom) with an atmosphere of N2 (80%), CO2 (10%), and H2 (10%). The plates were incubated at 37°C for 4 days in anaerobic jars filled with mixed gas (85% N2, 5% CO2, and 10% H2) by evacuation-replacement method (Anoxomat; Hart, Lichtenvoorde, The Netherlands).
Nucleic acid isolation.
Isolation of chromosomal DNA from pure cultures was performed as described elsewhere (1). When necessary, the method was slightly modified by prolonging the time for enzymatic lysis from 1 h to 2 or 3 h. Methods previously described (43) were used to extract RNA from pure cultures and DNA from fecal samples.
Primers.
All primers used in the study are listed in Table 2. Bifidobacterium genus-specific PCR was performed using 16S rDNA-targeted primers Bif164-f and Bif662-r or Im26-f and Im3-r, which produce approximately 520- or 1,420-bp PCR amplicons, respectively. For DGGE analysis of PCR products a 40-bp GC clamp was attached to the 5′ end of either Bif164-f or Bif662-r (Table 2). Complete 16S rDNA was amplified using primers 7-f and 1510-r. For reverse transcription of 16S rRNA to cDNA, primer 1401-r was used. Primers T7, Sp6, 338-r, 515-r, 1100-r, 338-f, and 968-f labeled with IRD800 were used for sequencing. All primers were purchased from MWG-Biotech (Ebersberg, Germany).
TABLE 2.
Primers used in the present study
| Primer | Sequence (5′ to 3′) | Use | Specificity or target | Reference |
|---|---|---|---|---|
| Bif164-f | GGGTGGTAATGCCGGATG | PCR | Bifidobacterium 16S | 17, 19 |
| Bif662-r | CCACCGTTACACCGGGAA | PCR | Bifidobacterium 16S | 17, 19 |
| Bif164-GC-f | CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGG-GGGTGGTAATGCCGGATG | PCR | Bifidobacterium 16S | This study |
| Bif662-GC-r | CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG-CCACCGTTACACCGGGAA | PCR | Bifidobacterium 16S | This study |
| Im26-f | GATTCTGGCTCAGGATGAACG | PCR | Bifidobacterium 16S | 15 |
| Im3-r | CGGGTGCTICCCACTTTCATG | PCR | Bifidobacterium 16S | 15 |
| 7-f | AGAGTTTGAT(C/T)(A/C)TGGCTCAG | PCR | Eubacterial 16S | 18 |
| 1510-r | ACGG(C/T)TACCTTGTTACGACTT | PCR | Eubacterial 16S | 18 |
| 1401-r | CGGTGTGTACAAGACCC | RT-PCR | Eubacterial 16S | 30 |
| 338-r | CTGCTGCCTCCCGTAGGAGT | Sequencing | Eubacterial 16S | 18 |
| 338-f | CTCCTACGGGAGGCAGCAG | Sequencing | Eubacterial 16S | 18 |
| 515-r | ATCGTATTACCGCGGCTGCTGGCAC | Sequencing | Eubacterial 16S | 18 |
| 968-f | AACGCGAAGAACCTTA | Sequencing | Eubacterial 16S | 18 |
| 1100-r | GGGTTGCGCTCGTTG | Sequencing | Eubacterial 16S | 18 |
| T7 | TAATACGACTCACTATAGGG | Sequencing | pGEMT | Promega |
| Sp6 | GATTTAGGTGACACTATAG | Sequencing | pGEMT | Promega |
Reverse transcriptase PCR (RT-PCR) and PCR amplification.
PCRs were performed using a Taq DNA polymerase kit from Life Technologies (Gaithersburg, Md.). The reaction mixture consisted of 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 3 mM MgCl2, 0.2 mM deoxynucleoside triphosphate (dNTP), a 0.2 μM concentration of each primer, 1.25 U of Taq polymerase, and 1 μl of appropriately diluted template DNA in a final volume of 50 μl. In PCR with primers 7-f and 1510-r, the dNTP concentration was increased to 0.3 mM and the amount of Taq polymerase was increased to 1.5 U. The PCR thermocycling program with Bif164-f and Bif662-r primers was the following: 94°C for 5 min; 35 cycles of 94°C for 30 s, 62°C for 20 s, and 68°C 40 s; 62°C for 20 s; and 68°C for 7 min. The reactions were subsequently cooled to 4°C. For the amplification with primers 7-f and 1510-r the denaturation and elongation times were prolonged to 1 min 30 s and the annealing step was performed at 52°C for 30 s. The thermocycling program with primers Im26-f and Im3-r was: 94°C for 5 min; 30 cycles of 94°C for 30 s, 57°C for 30 s, and 68°C for 1 min 30 s; 57°C for 30 s; and 68°C for 7 min.
RT-PCR was performed with the Geneamp Thermostable rTth Reverse Transcriptase RNA PCR kit (Perkin-Elmer, Norwalk, Conn.). Reverse transcription reaction mixtures (10 μl) consisted of 10 mM Tris-HCl (pH 8.3), 90 mM KCl, 1 mM MnCl2, 0.25 mM dNTP, 0.75 μM primer 1401-r, 1.25 U of recombinant Tth DNA polymerase, and 1 μl of appropriately diluted RNA. The RT reaction was performed at 68°C for 30 min and followed by the addition of 40 μl of PCR mixture consisting of 5% glycerol, 10 mM Tris-HCl (pH 8.3), 100 mM KCl, 0.05% Tween 20, 0.75 mM EGTA, 3.75 mM MgCl2, 0.2 mM dNTP, and a 0.25 μM concentration of each of the primers Bif164-f and Bif662-GC-r. The PCR thermocycling program was the same as described above for these primers.
The size and amounts of PCR products were estimated by analyzing 5-μl samples by 1.2% agarose gel (wt/vol) electrophoresis and ethidium bromide staining.
DGGE analysis of PCR products.
DGGE analysis of PCR amplicons was performed essentially as described previously (27, 29) using the DCode or D GENE System apparatus (Bio-Rad, Hercules, Calif.). Polyacrylamide gels (8% [wt/vol] acrylamide–bisacrylamide [37.5:1]) in 0.5× Tris-acetic acid-EDTA buffer with a denaturing gradient were prepared with a gradient mixer and Econo-pump (Bio-Rad) using solutions containing 45 and 55% denaturant. A 100% denaturant corresponds to 7 M urea and 40% (vol/vol) formamide. PCR amplicons were separated by electrophoresis at a constant voltage of 85 V and a temperature of 60°C for 16 h. The DNA fragments were visualized by AgNO3 staining and developing basically as described previously (35).
Cloning of the PCR products.
The PCR products were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions and cloned in E. coli JM109 by using the pGEM-T vector system (Promega, Madison, Wis.). Colonies were picked and transferred into 20 μl of 10 mM Tris-HCl (pH 8.0)–1 mM EDTA and boiled for 15 min to lyse the cells, and the cell lysates were used to screen the transformants by PCR with Bif164-f and Bif662-GC-r primers followed by DGGE analysis. Plasmid DNA of selected transformants was isolated using a QIAprep spin miniprep kit (Qiagen).
Sequence analysis.
Sequence analysis was carried out using purified plasmid DNA and sequencing primers T7 and Sp6 complementary to the adjacent sequences of the pGEMT cloning site and other primers complementary to the 16S rDNA sequences (see Table 2). Sequencing was performed with the Sequenase sequencing kit (Amersham, Slough, United Kingdom) according to manufacturer's instructions. The sequences were analyzed with an automatic LI-COR (Lincoln, Nebr.) DNA sequencer 4000L and corrected manually. Pairwise and multiple sequence alignments and similarity comparisons between individual sequences were carried out using BCM services available on the Internet (http://www.hgsc.bcm.tmc.edu/SearchLaunher/) or from the DNASTAR (Madison, Wis.) program. Homology searches of 16S rDNA sequences derived from fecal clones and the DNA databases were carried out by using the BCM Nucleic acid sequence search service. In addition to the comparison with sequences in the databases, sequences of fecal clones were compared to the B. adolescentis E-981074T sequences determined in this study, because the B. adolescentis 16S rDNA sequence deposited in the GenBank appears to contain many ambiguous bases.
Southern hybridization.
Chromosomal DNA (2μg) was digested with EcoRI, EcoRV, or NruI restriction enzymes (GibcoBRL, Paisley, United Kingdom) and the DNA fragments were separated by electrophoresis in 1% agarose. Fragments larger than 500 bp were transferred to Hybond-N+ membrane (Amersham, Aylesbury, United Kingdom) by vacuum blotting with a VacuGene XL vacuum blotting system (Pharmacia, Uppsala, Sweden), and hybridizations were carried out according to established protocols (34). The Bif164-f–to–Bif662-r PCR amplicon from strain B. adolescentis E-981074T was labeled with [α-32P]ATP by nick translation and used as a probe.
Extraction of chromosomal NruI fragments from agarose gel.
Chromosomal DNA (10 μg) was digested with NruI restriction enzyme, and the DNA fragments were separated by agarose gel electrophoresis as mentioned above. DNA fragments of a size between 3 and 23 kb were recovered using Concert matrix gel extraction system (GibcoBRL) and checked for the presence of 16S rDNA by PCR with primers Bif164-f and Bif662-GC-r.
Nucleotide sequence accession numbers.
The sequences of the two different 16S rDNA copies of B. adolescentis E-981074T were deposited in the GenBank database and have been assigned accession numbers AF275881 (nru-1) and AF275882 (nru-5). The accession numbers of the fecal clones in GenBank are the following (clone code in parenthesis): AF275890 (7B), AF275891 (7G), AF275892 (9A), AF275893 (9B), AF275894 (9C), AF275884 (13D), AF275885 (15A), AF275883 (15B), AF275886 (15D), AF275887 (16B), AF275888 (16C), and AF275889 (16F).
RESULTS
Development of the DGGE method for separation of bifidobacteria.
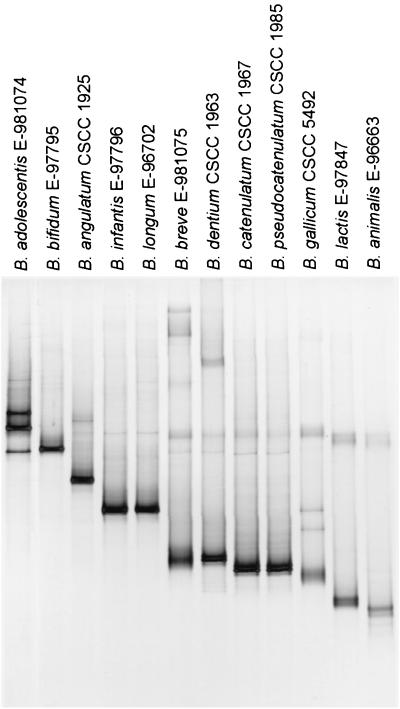
In order to set up the method based on genus-specific PCR and DGGE, primers that amplify a fragment that is separable by DGGE were selected and tested. The specificity of the previously described Bifidobacterium genus-specific primers Bif164-f and Bif662-r (17, 19) was confirmed using numerous bacterial species occurring in feces as the reference material. The primers showed good specificity for the genus Bifidobacterium, and the approximately 520-bp product was amplified exclusively from bifidobacteria (data not shown). In order to separate the bifidobacterial sequences by DGGE, a GC clamp was attached to either of the primers (Table 2). The use of primers Bif164-GC-f and Bif662-r to amplify bifidobacterial 16S rDNA resulted in PCR fragments from different species that showed very limited separation upon 45 to 55% DGGE (data not shown). In contrast, when the GC clamp was attached to the reverse primer (Bif662-GC-r) instead of the forward (Bif164-f), a good separation of different species in DGGE was obtained (Fig. 1). However, some closely related species could not be separated from each other by this approach. B. longum and B. infantis gave fragments in the same position in the gel as well as B. catenulatum and B. pseudocatenulatum. Also B. breve and B. dentium PCR fragments migrated to the same position. Different strains of the same species (Table 1) gave fragments in the same position upon DGGE. Two species, B. breve and B. gallicum gave diffuse fragments. An individual fragment from one strain was frequently observed as a doublet with two fragments very close to each other. This is very likely due to abortion of the elongation reaction during PCR caused by the GC clamp (hairpin formation), resulting in DNA molecules with slightly different migration behavior (30). However, B. adolescentis produced three strong fragments relatively far apart from each other (Fig. 1), and the cause for this was further examined in detail (see below).
FIG. 1.
Separation of PCR products from different Bifidobacterium species with genus-specific primers in 45 to 55% DGGE (increasing gradient of denaturant from top to bottom).
Host-specific and stable DGGE patterns of bifidobacteria from human feces.
The applicability of the DGGE method to monitoring complex bifidobacterial communities was first tested by using DNA from several Bifidibacterium species as the template in a competitive PCR. Fragments from all species were found in the DGGE profile, but in addition some extra fragments appeared above the single-stranded DNA in the DGGE profile (data not shown). These fragments are presumably heteroduplexes, which are more unstable and therefore, remain in the upper part of the DGGE gel.
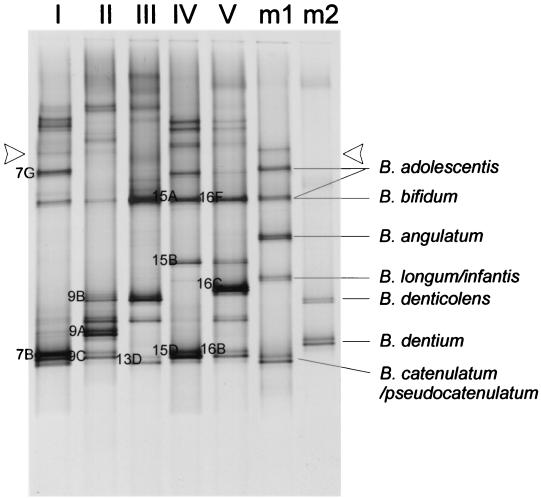
The bifidobacterial composition in fecal samples from six adult subjects (subjects I to VI) was studied. PCR products were obtained from samples I to V that had bifidobacterial counts reaching approximately 108 to 1010 CFU/g (wet weight) (Table 3), but no PCR product was obtained from subject VI, whose sample gave a low bifidobacterial count (approximately 104 CFU/g [wet weight]). The cultivation was performed from frozen samples, which is likely to have introduced a bias to the bifidobacterial counts, but allowed us to monitor fluctuations in the counts over time. DGGE analysis of bifidobacterial PCR products of fecal samples revealed complex host-specific patterns of bifidobacteria (Fig. 2, lanes I to V). In order to identify the Bifidobacterium species present in the feces, two samples consisting of a mixture of PCR products from identified species were run alongside the fecal samples (Fig. 2). However, most fragments of the fecal samples migrated to a different position than those of the culture collection strains and could not be identified in this way.
TABLE 3.
Bifidobacterial counts of the fecal samples
| Subject | Bifidobacterial count (CFU g [wet wt]−1) at wk:
|
||
|---|---|---|---|
| 0 | 3 | 4 | |
| I | 9.0 × 108 | 2.2 × 109 | 1.3 × 109 |
| II | 1.8 × 108 | 3.4 × 1010 | 2.5 × 109 |
| III | 2.2 × 109 | 2.9 × 109 | 7.3 × 109 |
| IV | 2.8 × 108 | 4.3 × 107 | 1.8 × 109 |
| V | 5.7 × 107 | 2.3 × 108 | 1.1 × 108 |
| VI | 4.1 × 104 | <104 | 3.9 × 104 |
FIG. 2.
DGGE of bifidobacterial PCR products of fecal samples from adult individuals (lanes I to V) and mixed PCR products from pure cultures (lanes m1 and m2). Single-stranded DNA and presumed heteroduplexes are above the line indicated with arrowheads. Indications 7B to 16F refer to the corresponding clones in Table 4.
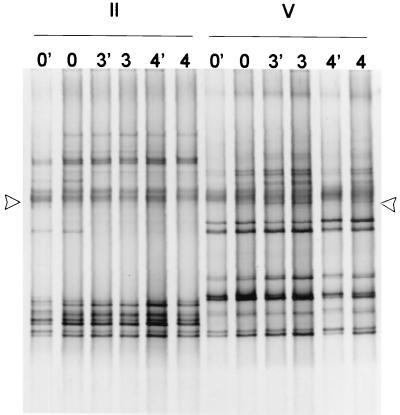
The stability of the bifidobacterial community over a period of 4 weeks was studied (Fig. 3). Analysis of three samples taken within this period showed stable bifidobacterial profiles, indicating that the composition of bifidobacterial community did not alter over this period despite some slight fluctuation in the bifidobacterial numbers (Table 3). Only subject II had a minor change in profile, where a faint fragment present in the first sample disappeared in the following samples (Fig. 3). DGGE profiles from undiluted and 10-fold-diluted fecal DNA samples were similar (Fig. 3), indicating that the template DNA from fecal samples can be diluted at least 10 times to avoid possible inhibition of the PCR without affecting the DGGE profile.
FIG. 3.
DGGE of bifidobacterial PCR products of fecal samples from two adult individuals (II and V) from a 4-week period (samples from weeks 0, 3, and 4). In samples 0′, 3′, and 4′ 10-fold-diluted DNA was used for PCR. Single-stranded DNA and presumed heteroduplexes are above the line indicated with arrowheads.
Validation of bifidobacterial profiles from feces.
In order to identify some of the fragments in fecal profiles, a longer fragment of approximately 1,400 bp was amplified from the fecal samples with another set of Bifidobacterium-specific primers, lm26-f and lm3-r. The amplified fragments were cloned into E. coli JM109 by using pGEM-T, and transformants were amplified with primers Bif164-f and Bif662-GC-r. The mobility of these PCR products in DGGE was compared to the PCR pattern of the fecal sample obtained with the same primer set in order to determine which fragment they corresponded to. The plasmids from selected clones were purified, and the 16S rDNA insert was sequenced from both ends. The sequencing results confirmed that the DGGE profiles obtained with the primers Bif164-f and Bif662-GC-r indeed represent the bifidobacterial population in feces, since all sequenced fragments were derived from Bifidobacterium species. Clone sequences showed very high similarity to many Bifidobacterium species, and therefore could not be unambiguously identified to the species level (Table 4). All sequenced clones had the highest similarity to the sequence of B. adolescentis or its close phylogenetic relative species (B. ruminantium and B. dentium) or to B. pseudocatenulatum.
TABLE 4.
Sequencing of bifidobacterial clones from fecal samples
| Subject no. | Clone | Sequence length (bp) | Ambiguity
|
Closest relatives (% sequence similarityb) | |
|---|---|---|---|---|---|
| bp | % | ||||
| I | 7B | 1,267 | 16 | 1.3 | B. adolescentis (98)a, B. ruminantium (97), B. dentium (95) |
| I | 7G | 1,365 | 15 | 1.1 | B. adolescentis (98)a, B. ruminantium (97), B. dentium (96) |
| II | 9A | 1,346 | 11 | 0.8 | B. adolescentis (97)a, B. ruminantium (96), B. dentium (95) |
| II | 9B | 1,360 | 5 | 0.4 | B. pseudocatenulatum (98), B. angulatum (97), B. dentium (96) |
| II | 9C | 1,267 | 8 | 0.6 | B. adolescentis (98)a, B. ruminantium (97), B. dentium (96) |
| III | 13D | 1,297 | 12 | 0.9 | B. pseudocatenulatum (98), B. ruminantium (97), B. adolescentis (97)a |
| IV | 15A | 1,363 | 5 | 0.4 | B. adolescentis (99)a, B. ruminantium (98), B. dentium (97) |
| IV | 15B | 1,357 | 6 | 0.4 | B. adolescentis (99)a, B. ruminantium (98), B. dentium (96) |
| IV | 15D | 1,365 | 10 | 0.7 | B. adolescentis (98)a, B. ruminantium (97), B. dentium (96) |
| V | 16B | 1,268 | 18 | 1.4 | B. adolescentis (97)a, B. ruminantium (96), B. dentium (96) |
| V | 16C | 1,287 | 15 | 1.2 | B. ruminantium (97), B. adolescentis (97)a, B. pseudocatenulatum (96) |
| V | 16F | 1,362 | 17 | 1.3 | B. adolescentis (98)a, B. ruminantium (97), B. dentium (96) |
Compared to the 16S rDNA sequences determined in this study.
Percent similarity values from pairwise sequence alignments.
B. adolescentis shows 16S rDNA heterogeneity.
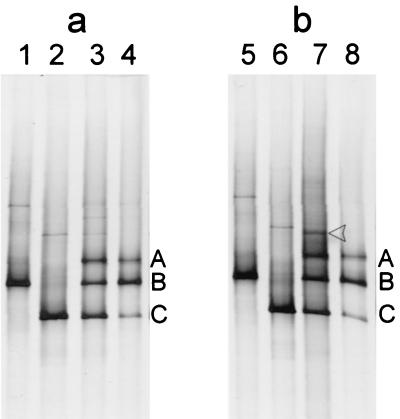
The B. adolescentis E-981074T PCR product obtained with the primers Bif164-f and Bif662-GC-r appeared in the DGGE gel as three distinct fragments (bands A to C in Fig. 4). In order to study the possible differences in sequences of the PCR fragments migrating to different positions, the PCR product was purified and cloned into E. coli JM109 using pGEM-T vector. The rDNA inserts of 40 clones were amplified by PCR with the primers Bif164-f and Bif662-GC-r, and analysis of their migration in DGGE showed that the majority of them produced 16S rRNA PCR amplicons that corresponded to either the middle fragment (B) or the lowest fragment (C) (clones b and c, respectively) but none that corresponded to the upper fragment (A). However, one clone (a1) was obtained that produced all three fragments, A, B, and C (data not shown). In order to obtain a clone corresponding to the single fragment A, we repeatedly colony purified the clone but only obtained colonies that upon PCR produced fragments corresponding to either B or C or all three fragments. Fragment A had the uppermost migration position in DGGE, indicating that it melts under weaker denaturing conditions than fragments B and C and thus is the most unstable fragment. We therefore came to the assumption that fragment A was a heteroduplex of fragments B and C formed during melting and reannealing of sequences in the PCR thermocycling. Presumably, clone al contained at least two plasmids carrying either fragment B or C and thereby produced all three fragments during PCR. Further evidence that fragment A was a heteroduplex of fragments B and C was obtained with the following experiments (Fig. 4). Firstly, purified plasmids from clones b1 and c1 were used separately and together as templates in PCR with the primers Bif164-f and Bif662-GC-r. Plasmid from clone b1 produced fragment B in DGGE, and similarly, clone c1 produced fragment C. When plasmids of b1 and c1 were both present in the PCR the three fragments (A, B, and C) could be observed after DGGE (lanes 1 to 3 in Fig. 4a). The same result was obtained following a nested-PCR amplification using fragments B and C separately and in combination as template in a subsequent PCR with the same primers (data not shown). Secondly, when PCR products B and C were mixed, heat denatured, and cooled to room temperature, analysis by DGGE showed the presence of all three fragments again (lanes 5 to 7 in Fig. 4b). Moreover, the inserts of four clones (b1, b2, c1, and c2) were sequenced. Sequence comparison revealed a minor difference in the sequences between clones b and c, i.e., a T deletion at position 219 (numbering begins at the 5′ end of the 16S rDNA) in clones c1 and c2 (Fig. 5).
FIG. 4.
DGGE profiles of B. adolescentis E-981074T 16S PCR product and its derivative clones. (a) Heteroduplex formation experiment by PCR. Templates used in PCR are as follows: lane 1, plasmid from clone b1; lane 2, plasmid from clone c1; lane 3, plasmids from clones b1 and c2; lane 4, DNA from E-981074T. (b) Heteroduplex formation experiment by melting PCR products. PCR products are as follows: lane 5, clone b1; lane 6, clone c1; lane 7, clones b1 and c1 heat denaturated together and cooled slowly to allow reannealing of complementary strands; lane 8, E-981074T. Single-stranded DNA is indicated with an arrowhead.
FIG. 5.
Sequence alignment of B. adolescentis E-981074T clones with the B. adolescentis sequence from GenBank (M58729), showing sequence differences that were found (in boldface type). b1, b2, c1, nru-1, and nru-5 are double-stranded sequences, and c2 is a single-stranded sequence.
Heterogeneity, complete sequence analysis, and expression of the 16S rRNA gene of B. adolescentis E-981074T.
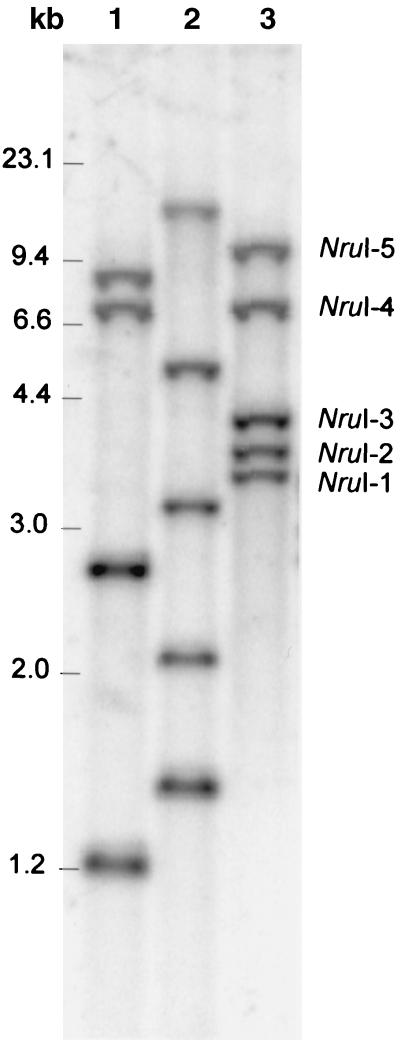
The above observations prompted us to determine whether the microheterogeneity found in the PCR products resulted from sequence heterogeneity in the V2 region of different copies of 16S rDNA in the chromosome, thus ruling out the possibility of a PCR bias. Genomic DNA of B. adolescentis was prepared from a culture grown from a single colony and cleaved with restriction enzymes EcoRI, EcoRV, and NruI. According to the GenBank sequence data (accession number M58729) (D. Yang and C. R. Woese, unpublished data) the first two enzymes cut the B. adolescentis 16S rDNA sequence only once at positions 660 and 691, respectively, while NruI has no cleavage site within the 16S rDNA. Subsequent Southern hybridization was performed with a fragment homologous to the 16S rDNA sequence between bp 164 and 662 (Fig. 6). Hence, the number of fragments in EcoRI and EcoRV digests that hybridize with the probe correspond to the copy number of rrn operons in the chromosome. Four fragments were visible in the EcoRI digest, and five were visible in the EcoRV digest. The approximately 2.7-kb EcoRI fragment (lane 1 in Fig. 6) was relatively more intense than the other fragments and probably contains two fragments containing 16S rDNA sequences. From these results we concluded that B. adolescentis E-981074T harbors five copies of rrn operon, a conclusion which is also supported by the observation that five NruI fragments (NruI-1 to NruI-5) (Fig. 6), supposedly containing intact copies of the 16S rDNA, hybridized with the probe. The five NruI fragments were isolated, and parts of the 16S rDNA sequences were amplified using primers Bif164-f and Bif662-GC-r. The resultant PCR products were analyzed by DGGE, and this showed that fragment B was produced from four of the 16S rDNA copies (NruI-1 to NruI-4 fragments) and that fragment C was produced from one copy (NruI-5 fragment) (Fig. 6). Fragment A was not produced from any of the copies. Next, primers 7-f and 1510-r were used to amplify the full-length 16S rDNA from fragments NruI-1 and NruI-5, and the PCR products were cloned into E. coli JM109 using pGEM-T, generating clones nru-1 and nru-5. The heteroduplex formation experiments were repeated with the plasmids of these clones, and results analogous to those described above were obtained; i.e., clone nru-1 produced fragment B and nru-5 produced fragment C, but together they produced the additional fragment A.
FIG. 6.
Southern blot analysis of rrn operons of B. adolescentis E-981074T. The genomic DNA cleaved with EcoRI (lane 1), EcoRV (lane 2), and NruI (lane 3) and hybridized with 16S rDNA probe.
The full-length 16S rDNA fragments from clones nru-1 and nru-5 were sequenced and compared. The two copies of 16S rDNA had a similarity of 99.4%, showing differences in eight positions. The difference previously described for the b and c clones was confirmed (Fig. 5); i.e., the T deletion in clones c1 and c2 at position 219 was also found in clone nru-5. In addition, clone nru-1 had a substitution of T to C at position 467. Thus, the deletion of one T at position 219 in clones c1, c2, and nru-5 changed the migration of the PCR fragment in DGGE, but the T-to-C substitution in clone nru-1 was not sufficient to alter its migration. The most prominent heterogeneity between the two chromosomal copies of 16S rDNA was, however, found outside the region that was amplified with primers Bif164-f and Bif662-r. Clones nru-1 and nru-5 differed in several base pairs between positions 77 to 80 and 93 to 96 (Fig. 5) in the V1 region of the 16S rDNA. The numerous ambiguous nucleotides present in the B. adolescentis 16S rDNA GenBank sequence (M58729) were determined in this study. Both sequences nru-1 and nru-5 are different from M58729 at positions 96, 391, 459, 998, and 1401, while nru-5 shows additional differences in the aforementioned positions in the V1 region.
In order to get a picture of the expression of the different 16S rDNA copies in B. adolescentis, an RT-PCR experiment was performed. First the 16S rRNA was transcribed to cDNA, which was then amplified in PCR with the primers Bif164-f and Bif662-GC-r. When the PCR product was analyzed by DGGE, the same pattern of three fragments, A, B, and C, was obtained. This showed that the 16S rDNA copy producing fragment C is transcribed together with the copies producing fragment B. As fragment B appeared significantly more intense than fragment C in DGGE, we estimated that more than one copy and possibly all four corresponding to fragment B are transcribed. Consistently, the intensity of the heteroduplex fragment A corresponded to that of fragment C.
DISCUSSION
In this study we describe the development and validation of a sensitive method for the qualitative analysis of complex bifidobacterial communities based on genus-specific PCR and DGGE. During the optimization of the method it was noticed that the location of the GC clamp in the DNA fragment greatly influenced the melting behavior and subsequently the migration of the fragment in the DGGE gel. Due to the use of different sequences in the GC clamps, direct comparison of the results obtained with the two primer pairs, Bif164-GC-f–Bif662-r and Bif164-f–Bif662-GC-r, was not possible, but it is more than likely that the location of the GC clamp has more effect on the separation than its sequence. When the GC clamp was attached to the forward primer (Bif164-GC-f) the separation of different Bifidobacterium species by DGGE was not good, whereas an efficient separation was obtained when the GC clamp was attached to the reverse primer (Bif662-GC-r). Alignment of bifidobacterial sequences from GenBank showed considerable sequence heterogeneity close to the Bif164-f primer end (data not shown), which is apparently critical for the sequence-specific separation of the PCR products from different Bifidobacterium species.
The developed method based on genus-specific PCR and DGGE allows us to monitor the qualitative composition of the whole bifidobacterial population with merely a single PCR. In DGGE the PCR products from all culture collection strains of the same species migrated to the same position, but it was not possible to identify species in fecal samples by comparing the position of the fragment to those of the identified culture collection strains. Molecular typing methods such as ribotyping and pulsed-field gel electrophoresis have shown considerable genomic heterogeneity in strains of the same Bifidobacterium species (8, 22, 33). This heterogeneity is also present in the 16S-to-23S internally transcribed spacer sequences, but 16S rDNA sequences are very conserved among bifidobacteria and show 93% similarity between most of the species of the genus Bifidobacterium (21, 26). Even minor differences in the 16S rDNA sequence may, however, alter the migration behavior of a PCR fragment in DGGE, as shown in the case of B. adolescentis E-981074T. This allows us to rapidly monitor changes occurring in the predominant members of the bifidobacterial community. The method may provide a valuable alternative to molecular typing techniques (22, 25) in rapidly monitoring qualitative changes in the bifidobacterial populations, although it does not allow definite discrimination or quantification of different strains. The DGGE method has an advantage of being independent of prior time-consuming culturing of the isolates on selective medium, which may favor the growth of some strains, thereby biasing the results. The PCR approach can also, however, lead to some distortions, because some sequences may amplify better than others, and heteroduplexes can be formed during PCR (39), as also observed in this study and further discussed below. In the PCR-DGGE approach identification of fragments can be done by subsequent cloning and sequencing of the PCR products, but it is hampered by the high similarity of 16S rDNA sequences between different Bifidobacterium species and the inadequate sequence data quality for many of the sequences in GenBank. We contributed to the construction of a more comprehensive database by depositing to the GenBank two accurate 16S sequences of B. adolescentis E-981074T, which can be used for identification of new strains and phylogenetic studies.
The DGGE profiles of 16S PCR amplicons of bifidobacteria were found to be unique for each individual. This supports the results of previous studies that intestinal Bifidobacterium communities, like the dominant microbial populations, are host specific (16, 22, 25). The bifidobacterial populations were also found to be stable in composition during the 4-week study period. In general, the bifidobacterial population in the adult gut seems to be relatively stable for strain composition over several months or even a year, although some individual variations have also been detected (22, 25). In contrast, in the developing gut microbiota of infants bifidobacterial species change in time (C. Favier, E. E. Vaughan, W. M. de Vos, and A. D. L. Akkermans, unpublished data). Further studies with larger test groups are needed to make conclusions about development and the long-term stability of bifidobacterial communities.
Matsuki et al. (23) applied species- and group-specific PCR directly to fecal samples and found B. catenulatum group species (B. catenulatum and B. pseudocatenulatum) in 92% of adult fecal samples and B. longum, B. adolescentis, and B. bifidum in 65, 60, and 38% of the samples, respectively. Comparison of the species-specific PCR method with the classical culture method revealed that some species, most frequently B. adolescentis, were detected by the direct PCR method but not by culturing followed by specific PCR of the isolates (23). In these individuals B. adolescentis either was not among the most numerous bifidobacteria or it failed to grow on the selective media used. Our results indicate that B. adolescentis or closely related species are numerically the most prevailing bifidobacteria in some individuals, as it was most frequently found in the clone library. B. adolescentis was also the most widely distributed Bifidobacterium species among the subjects of the test group. Taking into account the possible heteroduplex formation and the fact that some species or strains may give more than one fragment in DGGE, it is difficult to give accurate estimates on the diversity of bifidobacteria in the fecal samples. The DGGE patterns show that the bifidobacterial diversity in individual samples is quite restricted, and according to sequence data some of these strains may belong to the same species. This result is in good agreement with previous studies showing that in most adults the bifidobacterial community is a combination of one to four species and that several distinct strains of the same species can coexist in one community (22, 23).
Our results show that B. adolescentis E-981074T carries five rRNA gene clusters and exhibits intragenomic 16S rDNA sequence heterogeneity. Previously, B. breve has been found to have at least three rrn operons, and B. bifidum has been found to have two (8, 42). The two 16S rDNA copies of B. bifidum were sequenced, but no differences were found between the sequences (42). It is anticipated that the greater the number of rRNA operons is the higher the possibility of heterogeneity among rRNA genes within an organism is. Indeed, a Paenibacillus polymyxa strain that harbors at least twelve rrn operons was found to display a high degree of sequence diversity among its 16S rRNA genes (30). Intragenomic 16S rDNA heterogeneity, both microheterogeneities and larger changes, has been found also in other bacteria (9, 12, 37, 41). Microheterogeneities of a few base changes most likely result from mutations during DNA replication, whereas higher levels of sequence variation are considered to be a result of horizontal gene transfer (37, 41). The two 16S rDNA sequences of B. adolescentis E-981074T differed in only eight bases over the almost total length of the gene and showed the highest similarity (99.4%) to each other over all the sequences available in GenBank. Therefore, we consider that the variability has resulted from mutations in one copy of the 16S rRNA gene. Intragenomic sequence variation has effects on the phylogeny of organisms and biodiversity estimates. The heterogeneities may also interfere with analysis of denaturing gel patterns, and therefore some caution must be exercised when interpreting the results, especially when estimating strain and species numbers and diversity.
Probiotic and prebiotic research aims at developing functional food products that are able to modify the gut microbiota to a potentially more healthy one. A particular interest in these studies is to follow the marker organisms of well-balanced gut microbiota or the probiotic strains, often bifidobacteria and lactobacilli, and the changes of their proportions in the intestinal microbiota. Recently, genus-specific primers were designed for Lactobacillus spp. and also used successfully in combination with DGGE to analyze communities of lactobacilli (G. H. J. Heilig, E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. L. Akkermans, and W. M. de Vos, unpublished data). In conclusion, this study demonstrates that the combination of genus- or group-specific PCR with DGGE is a powerful tool to study targeted microbial populations in the complex GI tract ecosystem. The approach opens new possibilities to follow the qualitative changes in the bifidobacterial and lactobacilli populations in response to probiotic or prebiotic administration as well as to study the effect of age, genetic background and other factors on the composition and diversity of these bacterial groups.
ACKNOWLEDGMENTS
We are indebted to Christine Favier for valuable technical advice and to Ineke Heikamp-de Jong for her excellent technical assistance in sequencing. We thank Ross Crittenden for providing the CSCC strains and Benedikte Grenov for the Chr. Hansen A/S strains. We also thank the volunteers for their cooperation.
The financial support from EU project FAIR-CT96-1028, the Technology Development Centre Of Finland (TEKES) project 40302/98, and VTT Biotechnology, is gratefully acknowledged.
REFERENCES
- 1.Alander M, Satokari R, Korpela R, Saxelin M, Vilpponen-Salmela T, Mattila-Sandholm T, von Wright A. Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl Environ Microbiol. 1999;65:351–354. doi: 10.1128/aem.65.1.351-354.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alles M S, Hartemink R, Meyboom S, Harryvan J L, Van Laere K M, Nagengast F M, Hautvast J G. Effect of transgalactooligosaccharides on the composition of the human intestinal microflora and on putative risk markers for colon cancer. Am J Clin Nutr. 1999;69:980–991. doi: 10.1093/ajcn/69.5.980. [DOI] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballongue J. Bifidobacteria and probiotic action. In: Salminen S, von Wright A, editors. Lactic acid bacteria, microbiology and functional aspects. 2nd ed. New York, N.Y: Marcel Dekkers, Inc; 1998. pp. 519–587. [Google Scholar]
- 5.Beerens H. An elective and selective isolation medium for Bifidobacterium spp. Lett Appl Microbiol. 1990;11:155–175. [Google Scholar]
- 6.Biavati B, Sgorbati B, Scardovi V. The genus Bifidobacterium. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1991. pp. 816–833. [Google Scholar]
- 7.Bouhnik Y, Pochart P, Marteau P, Arlet G, Goderel I, Rambaud J C. Fecal recovery in humans of viable Bifidobacterium sp ingested in fermented milk. Gastroenterology. 1992;102:875–878. doi: 10.1016/0016-5085(92)90172-u. [DOI] [PubMed] [Google Scholar]
- 8.Bourget N, Simonet J M, Decaris B. Analysis of the genome of the five Bifidobacterium breve strains: plasmid content, pulsed-field gel electrophoresis genome size estimation and rrn loci number. FEMS Microbiol Lett. 1993;110:11–20. doi: 10.1111/j.1574-6968.1993.tb06288.x. [DOI] [PubMed] [Google Scholar]
- 9.Clayton R A, Sutton G, Hinkle P S J, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–599. doi: 10.1099/00207713-45-3-595. [DOI] [PubMed] [Google Scholar]
- 10.Collins M D, Gibson G R. Probiotics, prebiotics, and synbiotics: approaches for modulating the microbial ecology of the gut. Am J Clin Nutr. 1999;69:1052s–1057s. doi: 10.1093/ajcn/69.5.1052s. [DOI] [PubMed] [Google Scholar]
- 11.Crociani F, Biavati B, Alessandrini A, Chiarini C, Scardovi V. Bifidobacterium inopinatum sp. nov. and Bifidobacterium denticolens sp. nov., two new species isolated from human dental caries. Int J Syst Bacteriol. 1996;46:564–571. doi: 10.1099/00207713-46-2-564. [DOI] [PubMed] [Google Scholar]
- 12.Ebersbach H, Breitenstein A, Lechner U. All species of Desulfitobacterium contain two types of 16S rRNA genes. In: Böck A, Heesemann J, Schleifer K H, Wagner H, editors. Biospektrum. Proceedings of the Microbiology 2000 Congress, Munich, Germany. Heidelberg, Germany: Spektrum Akademischer Verlag GmbH; 2000. p. 157. [Google Scholar]
- 13.Gibson G R, Beatty E R, Wang X, Cummings J H. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology. 1995;108:975–982. doi: 10.1016/0016-5085(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 14.Hartemink R, Kok B J, Weenk G H, Rombouts F M. Raffinose-bifidobacterium (RB) agar, a new selective medium for bifidobacteria. J Microbiol Methods. 1996;27:33–43. [Google Scholar]
- 15.Kaufmann P, Pfefferkorn A, Teuber M, Meile L. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl Environ Microbiol. 1997;63:1268–1273. doi: 10.1128/aem.63.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura K, McCartney A L, McConnell M A, Tannock G W. Analysis of populations of bifidobacteria and lactobacilli and investigation of the immunological responses of their human hosts to the predominant strains. Appl Environ Microbiol. 1997;63:3394–3398. doi: 10.1128/aem.63.9.3394-3398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kok R G, de Waal A, Schut F, Welling G W, Weenk G, Hellingwerf K J. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl Environ Microbiol. 1996;62:3668–3672. doi: 10.1128/aem.62.10.3668-3672.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E R, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley and Sons Ltd.; 1991. pp. 115–175. [Google Scholar]
- 19.Langendijk P S, Schut F, Jansen G J, Raangs G C, Kamphuis G R, Wilkinson M H, Welling G W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;61:3069–3075. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauer E. Bifidobacterium gallicum sp. nov. isolated from human feces. Int J Syst Bacteriol. 1990;40:100–102. doi: 10.1099/00207713-40-1-100. [DOI] [PubMed] [Google Scholar]
- 21.Leblond-Bourget N, Philippe H, Mangin I, Decaris B. 16S rRNA and 16S to 23S internal transcribed spacer sequence analyses reveal inter- and intraspecific Bifidobacterium phylogeny. Int J Syst Bacteriol. 1996;46:102–111. doi: 10.1099/00207713-46-1-102. [DOI] [PubMed] [Google Scholar]
- 22.Mangin I, Bouhnik Y, Bisetti N, Decaris B. Molecular monitoring of human intestinal Bifidobacterium strain diversity. Res Microbiol. 1999;150:343–350. doi: 10.1016/s0923-2508(99)80060-6. [DOI] [PubMed] [Google Scholar]
- 23.Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol. 1999;65:4506–4512. doi: 10.1128/aem.65.10.4506-4512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuki T, Watanabe K, Tanaka R, Oyaizu H. Rapid identification of human intestinal bifidobacteria by 16S rRNA-targeted species-and group-specific primers. FEMS Microbiol Lett. 1998;167:113–121. doi: 10.1111/j.1574-6968.1998.tb13216.x. [DOI] [PubMed] [Google Scholar]
- 25.McCartney A L, Wang W, Tannock G W. Molecular analysis of the composition of the bifidobacterial and lactobacillus microflora of humans. Appl Environ Microbiol. 1996;62:4608–4613. doi: 10.1128/aem.62.12.4608-4613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyake T, Watanabe K, Watanabe T, Oyaizu H. Phylogenetic analysis of the genus Bifidobacterium and related genera based on 16S rDNA sequences. Microbiol Immunol. 1998;42:661–667. doi: 10.1111/j.1348-0421.1998.tb02337.x. [DOI] [PubMed] [Google Scholar]
- 27.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- 29.Myers R M, Maniatis T, Lerman L S. Detection and localization of single base changes by denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:501–527. doi: 10.1016/0076-6879(87)55033-9. [DOI] [PubMed] [Google Scholar]
- 30.Nubel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R I, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Sullivan D, Kullen M J. Tracking of probiotic bifidobacteria in the intestine. Int Dairy J. 1998;8:513–525. [Google Scholar]
- 32.Rowland I R, Tanaka R. The effects of transgalactosylated oligosaccharides on gut flora metabolism in rats associated with a human faecal microflora. J Appl Bacteriol. 1993;74:667–674. doi: 10.1111/j.1365-2672.1993.tb05201.x. [DOI] [PubMed] [Google Scholar]
- 33.Roy D, Ward P, Champagne G. Differentiation of bifidobacteria by use of pulsed-field gel electrophoresis and polymerase chain reaction. Int J Food Microbiol. 1996;29:11–29. doi: 10.1016/0168-1605(95)00013-5. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sanguinetti C J, Dias Neto E, Simpson A J. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. BioTechniques. 1994;17:914–921. [PubMed] [Google Scholar]
- 36.Suau A, Bonnet R, Sutren M, Godon J J, Gibson G R, Collins M D, Dore J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueda K, Seki T, Kudo T, Yoshida T, Kataoka M. Two distinct mechanisms cause heterogeneity of 16S rRNA. J Bacteriol. 1999;181:78–82. doi: 10.1128/jb.181.1.78-82.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaughan E E, Schut F, Heilig H G H J, Zoetendaal E G, de Vos W M, Akkermans A D L. A molecular view of the intestinal ecosystem. Curr Issues Intest Microbiol. 2000;1:1–12. [PubMed] [Google Scholar]
- 39.Wintzingerode F V, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto T, Morotomi M, Tanaka R. Species-specific oligonucleotide probes for five Bifidobacterium species detected in human intestinal microflora. Appl Environ Microbiol. 1992;58:4076–4079. doi: 10.1128/aem.58.12.4076-4079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yap W H, Zhang Z, Wang Y. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J Bacteriol. 1999;181:5201–5209. doi: 10.1128/jb.181.17.5201-5209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon J-H, Yim D K, Goodfellow M, Park Y-H. Sequence analysis of 16S rRNA genes amplified from two ribosomal RNA gene clusters of Bifidobacterium bifidum. Antonie Leeuwenhoek. 1999;75:329–333. doi: 10.1023/a:1002025100444. [DOI] [PubMed] [Google Scholar]
- 43.Zoetendal E G, Akkermans A D L, De Vos W M. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]