Abstract
We previously demonstrated a genetic basis in tomato for support of the growth of a biological control agent, Bacillus cereus UW85, in the spermosphere after seed inoculation (K. P. Smith, J. Handelsman, and R. M. Goodman, Proc. Natl. Acad. Sci. USA 96:4786–4790, 1999). Here we report results of studies examining the host effect on the support of growth of Bacillus and Pseudomonas strains, both inoculated on seeds and recruited from soil, using selected inbred tomato lines from the recombinant inbred line (RIL) population used in our previous study. Two tomato lines, one previously found to support high and the other low growth of B. cereus UW85 in the spermosphere, had similar effects on growth of each of a diverse, worldwide collection of 24 B. cereus strains that were inoculated on seeds and planted in sterilized vermiculite. In contrast, among RILs that differed for support of B. cereus UW85 growth in the spermosphere, we found no difference for support of growth of the biocontrol strains Pseudomonas fluorescens 2-79 or Pseudomonas aureofaciens AB254. Thus, while the host effect on growth extended to all strains of B. cereus examined, it was not exerted on other bacterial species tested. When seeds were inoculated with a marked mutant of B. cereus UW85 and planted in soil, RIL-dependent high and low support of bacterial growth was observed that was similar to results from experiments conducted in sterilized vermiculite. When uninoculated seeds from two of these RILs were planted in soil, changes in population levels of indigenous Bacillus and fluorescent Pseudomonas bacteria differed, as measured over time by culturing and direct microscopy, from growth patterns observed in the inoculation experiments. Neither RIL supported detectable levels of growth of indigenous Bacillus soil bacteria, while the line that supported growth of inoculated B. cereus UW85 supported higher growth of indigenous fluorescent pseudomonads and total bacteria. The vermiculite system used in these experiments was predictive for growth of B. cereus UW85 inoculated on seeds and grown in soil, but the patterns of growth of inoculated strains—both Bacillus and Pseudomonas spp.—did not reflect host genotype effects on indigenous microflora recruited from soil to the spermosphere.
When seeds are planted in soil, they are rapidly colonized by indigenous microorganisms that have a profound effect on plant disease, nutrition, and development. Consequently, strategies to enhance plant health by modifying the microflora on seeds have concentrated on inoculating seeds with high doses of specific microorganisms. This approach delivers high populations of the desired microorganisms on the seed, thereby enhancing events that are driven by population size.
For example, strategies to suppress seed and seedling diseases using biocontrol agents have concentrated on inoculation techniques that deliver high populations of the biocontrol agent on the seed at the time of planting (for reviews, see references 8 and 32). Several studies have shown that the level of disease suppression obtained with a biocontrol agent is correlated to the initial population size of the agent (6, 28). Colonization of the seed by the biocontrol agent is critical to controlling pathogens such as the oomycete Pythium, which is chemotactically attracted to seeds and infects them within hours of planting (21, 23). Left unchecked, Pythium, which has a wide host range, can cause serious economic losses on a large number of crops (13).
Approaches to improve plant vigor have also benefited from strategies that give an introduced microorganism the advantage over indigenous microflora for colonization. For example, the ability of a symbiotic, nitrogen-fixing Rhizobium etli strain to colonize the seed and rhizosphere of common bean (Phaseolus vulgaris) in the first 96 h after planting was correlated to its ability to occupy nodules of that plant (E. A. Robleto and J. Handelsman, unpublished data). Accordingly, inoculating seeds with strains capable of high levels of nitrogen fixation is an approach that has been used to increase nitrogen fixation in bean. This enhances the introduced strain's chances of competing successfully for nodulation against indigenous nitrogen-fixing bacteria, which can be poor nitrogen fixers (11).
The genotype of the host plant can also influence the numbers and types of microorganisms, both inoculated and recruited, that colonize the spermosphere. In tomato, we identified several quantitative trait loci that are associated with population growth of the biocontrol agent Bacillus cereus UW85 (27). One of these quantitative trait loci was also associated with disease suppression by UW85. In bean, certain genotypes preferentially nodulate with specific Rhizobium strains (9, 16, 25). Genetic crosses between such genotypes produce progeny that segregate for preferential nodulation in a manner that suggests simple inheritance of this trait (10, 19, 29). Taken together, these studies suggest that microbial communities of plants are affected by the host genotype during germination and seedling development and the composition of the communities that develop can have a profound impact on the health of the plant.
Here, we build upon previous work demonstrating a host genotype effect on support of growth of B. cereus UW85 on seeds. First, we evaluated other B. cereus and non-Bacillus biocontrol strains for growth on tomato seeds. Second, we used two independent approaches, one based on culturing and one on direct microscopy, to evaluate whether the bacterial growth phenotypes of tomato lines inoculated with bacteria and grown in sterilized vermiculite predict growth of bacteria on inoculated and uninoculated seeds planted in soil.
MATERIALS AND METHODS
Tomato seeds.
We obtained from D. Zamir seed for 87 lines from a recombinant inbred line (RIL) population (22) derived from a cross between Lycopersicon esculentum cv. UC204 and L. cheesmanii LA 483 (24). To produce seed for experiments, we grew these lines in the greenhouse in 1994 and in the field in Madison, Wis., in 1995 to 1997. Seeds were removed from the fruit using an acid extraction procedure (28).
Bacterial strains and seed treatment.
We evaluated a collection of strains of B. cereus and Pseudomonas spp. for their ability to grow in the tomato spermosphere. The B. cereus strains are representative, in terms of origin, antibiotic production, and biocontrol activity, of a worldwide collection studied previously (30). The B. cereus strain UW200 was constructed by transforming strain UW85 with plasmid pBC16 (5), which encodes resistance to tetracycline. B. cereus strains were grown on a rotary shaker at 200 rpm for 4 days in half-strength tryptic soy broth (Difco, Detroit, Mich.) at 29°C.
The Pseudomonas strains used, P. fluorescens 2-79 (31) and P. aureofaciens AB254 (7), are published strains with biocontrol activity against soil-borne pathogens. They were grown in King's B medium (15) under the same conditions used for B. cereus strains but for 2 instead of 4 days.
We treated tomato seeds with suspensions of bacteria as described previously (28). Briefly, broth cultures of bacteria, diluted in a volume of sterile water determined to yield the appropriate desired initial seed population levels, were applied to seeds that were subsequently vacuum dried. Seed inoculation with B. cereus strains was done using a partially sporulated culture. Seeds treated with B. cereus strains were sometimes stored dry for up to several days before being used in an experiment, while seeds treated with the other strains were used within hours of treatment.
Bacterial growth experiments in sterilized vermiculite.
To determine the effect of the tomato lines on population sizes of inoculated bacteria, we designed an experimental system to facilitate sampling individual seeds and to approximate the environment used in biocontrol assays conducted previously (28). We planted individual treated seeds in 1-ml plastic pipette tips (Research Products International Corp., Mount Prospect, Ill.) that had been filled with sterilized, medium-grade vermiculite (Strong-Lite, Pine Bluff, Ak.). After planting the seeds, we added enough vermiculite to cover the seeds and 1 ml of sterile distilled water to each tip. A box containing the tips was placed in a plastic tray (28 cm by 56 cm; Hummert, Int., Earth City, Mo.) filled with 4 liters of deionized water. Additional water was added to the tray at 12-h intervals to maintain a constant water level throughout the experiment. The experiments were conducted in a growth chamber at 24°C and 40% relative humidity, with 12 h of continuous light (415 μE m−2 s−1 provided by 40-W cool white fluorescent bulbs) in each 24-h period. To measure the spermosphere population, we removed the tip containing the seed, placed it in a test tube filled with a defined volume of sterile distilled water, sonicated (Branson Ultrasonics Corporation, Danbury, Conn.) the tube for 30 s, and plated dilutions with a mechanized spiral plater (Autoplate model 3000; Spiral Biotech, Bethesda, Md.) on 1/10 strength tryptic soy agar (TSA). After 2 days of incubation at 24°C, we counted the number of CFU.
Growth of bacteria on seeds planted in soil.
We evaluated selected tomato lines to determine the effect of host genotype on the bacterial spermosphere populations of seeds planted in soil. We placed individual, untreated seeds in polypropylene microcentrifuge tubes filled with 60 mg of sieved soil obtained from West Madison Research Station. We tapped the tubes to submerge the seed in the soil, added 20 μl of sterile distilled water to each, and placed them in a dark, 24°C incubator. We sampled the spermosphere population by removing the seed, rinsing it briefly in sterile water, placing it in a 1.5-ml microcentrifuge tube containing 1 ml of sterile distilled water, and sonicating the sample for 30 s. Total Bacillus and bacterial population sizes were determined by plating as described above, while those of fluorescent pseudomonads were determined by plating on King's B medium. The bacteria were also quantified using epifluorescence microscopy (see below).
We also evaluated the population levels of a tetracycline-resistant strain of UW85 (UW200) inoculated on seeds and planted in soil. These seeds were harvested and plated, as described above, except that they were also plated on TSA amended with tetracycline (10 mg/liter).
Oligonucleotide probes and labeling.
The following probes were used: S-D-Bact-0338-a-A-18 (Bact338) (designation in accordance with the Oligonucleotide Probe Database 1), a bacterial domain-specific small-subunit (SSU) rRNA-targeted DNA oligonucleotide probe with the sequence 5′-GCT-GCC-TCC-CGT-AGG-AGT-3′ (2); Bc440, a B. cereus (and closely related Bacillus thuringiensis and Bacillus mycoides) species-specific SSU rRNA-targeted DNA oligonucleotide probe designed by us with the sequence 5′-GTG-CCA-GCT-TAT-TCA-ACT-AGC-3′; and Ps, a large-subunit (LSU) rRNA-targeted DNA oligonucleotide probe specific for a variety of Pseudomonas species, including fluorescent pseudomonads, with the sequence 5′-GCT-GGC-CTA-GCC-TTC-3′ (26). All probes were synthesized and labeled with fluorescein phosporamidite (Glen Research, Sterling, Va.) or the cyanine dye CY3 (Glen Research) by the University of Wisconsin Biotechnology Center (Madison, Wis.).
Cell fixation and controls.
Exponentially growing B. cereus UW85 (12) and P. fluorescens 2-79 (31) cells were harvested after 12 h, fixed in 3% paraformaldehyde as described (17), and stored in 70% ethanol at −20°C for use as controls for the Bc440, Ps, and Bact338 oligonucleotide probes (see below). Other strains used for controls included a number of organisms from the Eucarya, Bacteria, and Archaea domains. These were received as gifts of cultures or grown to saturation using standard growth medium as described (4) or as indicated and harvested for fixation. Strains from the Department of Bacteriology strain collection (DBSC), University of Wisconsin-Madison, were grown in Difco nutrient broth (Becton Dickinson and Co., Franklin Lakes, N.J.). The control strains included Aureobasidium pullulans; Bacillus megaterium 7A1 (Bacillus Genetic Stock Center, Ohio State University, Columbus, Ohio); Erwinia herbicola 005; Escherichia coli 8009 (DBSC); Leptospirillum ferrooxidans; Methanobacterium formicicum (medium II 14); Methylosinus trichosporium; Rhizobium etli CE3 (20); Rhodobacter sphaeroides 9502 (DBSC); Rhodospirillum rubrum 9405 (DBSC); Salmonella enterica serovar Typhimurium LT2 (18); Staphylococcus aureus 3001 (DBSC); Sulfolobus shibatae; Sulfolobus solfataricus; Thermococcus litoralis; Thiobacillus ferrooxidans; Vibrio cholerae F115A (DBSC); and Yersinia enterocolitica WA.
Whole-cell hybridization.
Fixed cells from ethanol storage were placed in 10-μl aliquots for hybridization as described (17) on Teflon-printed slides (Electron Microscopy Sciences, Fort Washington, Pa.) that were coated with a mixture of 0.25% Bacto-gelatin (Difco Laboratories) and 0.01% chromium potassium sulfate (Sigma, St. Louis, Mo.). Briefly, samples were incubated with a hybridization solution (Sigma) that was 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–1× Denhardt's solution–100 μg of sheared DNA ml−1–deionized formamide to 20% for 1 h at 37°C for prehybridization. After 1 h, the prehybridization solution was replaced by hybridization solution, which is prehybridization solution to which 10% dextran sulfate and 5 ng of each appropriate probe per μl was added, and the samples were incubated for 12 to 16 h in the dark at 37°C. Following hybridizations, samples were washed three times (10 min) at 24°C in 0.1× SSC and then air dried. Samples were mounted in Fluorguard antifade medium (Bio-Rad, Hercules, Calif.) containing 2 μg of 4′,6-diamidino-2-phenylindole (DAPI) per ml and observed by microscopy.
Microscopy.
Four fields from each of 10 seeds from each RIL were examined. Specimens were examined with a BX-60 microscope (Olympus America Inc., Melville, N.Y.) equipped for epifluorescence with an HBO 100-W mercury arc lamp. Filters used were a Chroma narrow-excitation DAPI (82360X) and fluorescein isothiocyanate (FITC) (82485X) set, and a CY3/CY5 (51007) dual-filter set (Chroma Technology Corp., Brattleboro, Vt.). The specifications for the DAPI set were exciter (Ex) filter 357 ± 25.5 nm, emission (Em) filter 462 ± 7 nm; for the FITC set, they were Ex 484 ± 8.5 and Em 522 ± 9.5; for the CY3/CY5 dual set, they were Ex 543 ± 11 nm and Em 587 ± 18 nm for CY3 and Ex 634 ± 12 nm and Em 693.5 ± 30.5 nm for CY5. Images were recorded with a cooled charge-coupled device video camera (DEI-750: Optronics Engineering, Goleta, Calif.) and converted from an analog to a digital format (digitized) with a NuVista+ Videographics Card (Truevision Inc., Indianapolis, Ind.) controlled by IP Lab Spectrum 3.0 Software (Signal Analytics Corp., Vienna, Va.).
Data analysis.
Analysis of variance was conducted using Proc GLM from the Statistical Analysis System (SAS Institute, Cary, N.C.). Unless otherwise stated, significance was determined at α = 0.05. We repeated all experiments at least once, and present results that are representative of multiple experiments.
RESULTS
Effect of seed source on B. cereus UW85 growth.
To develop a system to study the effect of the host on bacterial growth and colonization of the spermosphere, we first examined whether host effects on B. cereus UW85 population sizes were consistent among different seed lots of the same tomato genotype. Our goal was to test and eliminate seed lots demonstrating significant variability. We evaluated the growth of UW85 on three tomato lines that supported growth and three lines that did not in a previous study (28). For these six lines, we evaluated three seed lots for each, produced in different years. In addition, we evaluated two more 1996 seed lots resulting from different harvests from the same plant for RIL 52. Population sizes of UW85 for the three lines that did not support growth did not change from 0 to 48 h for any of the seed lots (Table 1). Population sizes for the three lines that did support growth increased for all of the seed lots except the 1994 lot for RIL 14 and RIL 32, which were not used in subsequent experiments.
TABLE 1.
Effect of seed source on growth of B. cereus UW85 on seeds planted in autoclaved vermiculite
| RILa | Lotb | Mean log CFU/seed ± SEM (n = 6)
|
|
|---|---|---|---|
| 0 h | 48 h | ||
| 24 | 1994 | 4.84 ± 0.09 | 4.87 ± 0.10 |
| 1995 | 4.68 ± 0.15 | 4.92 ± 0.09 | |
| 1996 | 4.41 ± 0.06 | 4.42 ± 0.24 | |
| 25 | 1994 | 4.68 ± 0.08 | 4.40 ± 0.12 |
| 1995 | 4.25 ± 0.22 | 4.43 ± 0.04 | |
| 1996 | 4.45 ± 0.11 | 4.37 ± 0.30 | |
| 83 | 1994 | 4.69 ± 0.06 | 4.86 ± 0.18 |
| 1995 | 4.68 ± 0.07 | 4.57 ± 0.05 | |
| 1996 | 4.57 ± 0.04 | 4.65 ± 0.08 | |
| 14 | 1994 | 4.69 ± 0.12 | 4.72 ± 0.08 |
| 1995 | 4.54 ± 0.12 | 5.98 ± 0.32 | |
| 1996 | 4.89 ± 0.12 | 5.85 ± 0.17 | |
| 32 | 1994 | 5.15 ± 0.07 | 5.35 ± 0.10 |
| 1995 | 4.69 ± 0.05 | 5.66 ± 0.20 | |
| 1996 | 4.76 ± 0.07 | 5.90 ± 0.15 | |
| 52 | 1994 | 4.99 ± 0.03 | 5.66 ± 0.05 |
| 1995 | 4.74 ± 0.13 | 5.29 ± 0.05 | |
| 1996 | 4.66 ± 0.04 | 5.65 ± 0.11 | |
| 1996 | 4.95 ± 0.06 | 5.54 ± 0.09 | |
| 1996 | 4.95 ± 0.06 | 5.92 ± 0.10 | |
B. cereus UW85 growth-supportive RILs are 14, 32, and 52, and nonsupportive RILs are 24, 25, and 83.
The 1994 seeds were produced in a greenhouse. The 1995 and 1996 seeds were produced in two different field locations in Madison, Wis.
Variation among B. cereus strains for host effect on growth.
To determine if the host effect on growth of B. cereus UW85 was specific to UW85, we evaluated 24 additional B. cereus strains from a worldwide collection. With few exceptions, RIL 83, the line supporting low growth of B. cereus UW85, similarly supported low growth of the other strains, while RIL 52, the line supporting high growth of B. cereus UW85, similarly supported high growth of the other strains (Table 2). The average increase per seed in population size for RIL 83 and 52 over 48 h was 0.24 and 1.31 log CFU, respectively. Analysis of variance indicated there was no effect of RIL on initial UW85 population sizes (P = 0.97), but there were significant differences in initial population sizes among the B. cereus strains (P < 0.001). There was a significant difference between RIL 52 and RIL 83 in the change in population size (48 to 0 h) across all B. cereus strains (P < 0.001), but no significant differences within each RIL among strains (P = 0.42), and no significant strain-by-RIL interaction (P = 0.16).
TABLE 2.
Effect of plant genotype on population sizes of a diverse collection of B. cereus strains after planting in sterilized vermiculitea
| Strain | Mean log CFU/seed of RIL 83
|
Δ Mean log CFU/seed | Mean log CFU/seed of RIL 52
|
Δ Mean log CFU/seed | ||
|---|---|---|---|---|---|---|
| 0 h | 48 h | 0 h | 48 h | |||
| UW85 | 3.90 ± 0.15 | 4.39 ± 0.21 | 0.49 | 3.94 ± 0.24 | 4.75 ± 0.19 | 0.81 |
| Soy 130 | 3.50 ± 0.20 | 4.13 ± 0.23 | 0.63 | 3.75 ± 0.25 | 4.22 ± 0.47 | 0.47 |
| Alf 23 | 3.79 ± 0.33 | 4.32 ± 0.36 | 0.53 | 4.07 ± 0.16 | 4.77 ± 0.28 | 0.70 |
| Alf 83 | 3.50 ± 0.39 | 3.11 ± 1.00 | −0.39 | 3.37 ± 0.85 | 4.88 ± 0.42 | 1.51 |
| Alf 137 | 4.36 ± 0.54 | 4.95 ± 0.26 | 0.59 | 3.84 ± 0.15 | 4.96 ± 0.32 | 1.12 |
| SM 32 | 3.97 ± 0.23 | 3.53 ± 0.82 | 0.56 | 3.92 ± 0.22 | 5.13 ± 0.36 | 1.21 |
| VGA 137 | 3.46 ± 0.78 | 3.78 ± 0.92 | 0.32 | 3.57 ± 0.17 | 5.18 ± 0.29 | 1.61 |
| VGA 562 | 4.00 ± 0.12 | 4.32 ± 0.43 | 0.32 | 3.81 ± 0.13 | 5.31 ± 0.44 | 1.50 |
| Mor 28 | 3.72 ± 0.17 | 4.04 ± 0.19 | 0.32 | 3.93 ± 0.17 | 4.97 ± 0.29 | 1.04 |
| Mor 37 | 3.73 ± 0.31 | 3.68 ± 0.23 | −0.05 | 3.71 ± 0.16 | 5.43 ± 0.36 | 1.72 |
| BAR 177 | 3.37 ± 0.78 | 4.49 ± 0.25 | 1.12 | 3.43 ± 0.18 | 4.72 ± 0.18 | 1.29 |
| SNY 44 | 3.76 ± 0.26 | 3.83 ± 0.90 | 0.07 | 3.72 ± 0.20 | 5.53 ± 0.50 | 1.81 |
| SNY 73 | 2.59 ± 0.87 | 2.74 ± 1.00 | 0.25 | 3.87 ± 0.11 | 4.89 ± 0.23 | 1.02 |
| LUTZ 128 | 3.92 ± 0.19 | 3.72 ± 0.83 | −0.20 | 3.68 ± 0.27 | 5.13 ± 0.39 | 1.45 |
| LUTZ 58 | 2.56 ± 0.90 | 3.05 ± 1.06 | 0.49 | 2.18 ± 0.77 | 3.90 ± 1.02 | 1.72 |
| AS 8-4 | 3.74 ± 0.18 | 4.41 ± 0.14 | 0.67 | 3.35 ± 0.23 | 4.95 ± 0.28 | 1.60 |
| HS 23-11 | 4.20 ± 0.16 | 4.46 ± 0.23 | 0.26 | 4.05 ± 0.26 | 5.21 ± 0.39 | 1.16 |
| MS 1-9 | 3.86 ± 0.06 | 4.13 ± 0.23 | 0.27 | 3.83 ± 0.16 | 5.45 ± 0.13 | 1.62 |
| LS 2-2 | 3.87 ± 0.22 | 2.73 ± 0.97 | −1.14 | 3.68 ± 0.22 | 5.09 ± 0.20 | 1.41 |
| WS 4-12 | 4.06 ± 0.28 | 4.28 ± 0.22 | 0.22 | 3.83 ± 0.23 | 5.44 ± 0.20 | 1.61 |
| LN 100 | 4.28 ± 0.08 | 4.23 ± 0.27 | −0.05 | 3.93 ± 0.12 | 4.96 ± 0.17 | 1.06 |
| DGA 34 | 3.77 ± 0.16 | 3.98 ± 0.27 | 0.21 | 3.46 ± 0.20 | 5.02 ± 0.53 | 1.56 |
| DGA 94 | 3.45 ± 0.19 | 4.34 ± 0.12 | 0.89 | 2.94 ± 0.23 | 4.77 ± 0.57 | 1.83 |
| TNM 243 | 2.78 ± 0.69 | 3.10 ± 0.72 | 0.32 | 3.66 ± 0.30 | 4.46 ± 0.38 | 0.80 |
| TG 38 | 3.69 ± 0.28 | 3.97 ± 0.33 | 0.28 | 4.07 ± 0.18 | 5.20 ± 0.09 | 1.13 |
The seeds were inoculated with a suspension of each B. cereus strain, and population sizes were determined (see Materials and Methods) at 0 and 48 h after planting in autoclaved vermiculite. Data are means for six replicates ± standard error of the mean.
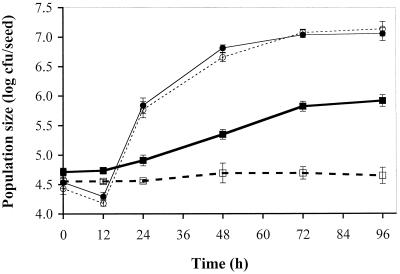
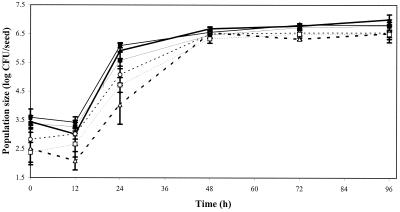
We evaluated growth of P. fluorescens 2-79 on the seeds of RIL 52 and RIL 83 to determine whether the host effect on B. cereus extended to other species of bacteria. The growth of P. fluorescens 2-79 did not differ significantly between the two lines, while the RILs differed in support of growth of B. cereus, as observed in previous experiments (Fig. 1). We next examined growth of P. fluorescens 2-79 and P. aureofaciens AB254 on RILs 52, 17, and 25, chosen because they support high, intermediate, and low growth of UW85, respectively. These lines did not differ in support of the growth of either strain between 48 and 96 h after planting in sterilized vermiculite (Fig. 2).
FIG. 1.
Effect of plant genotype on population sizes of B. cereus UW85 and P. fluorescens 2-79 on the seed over 96 h after planting in sterilized vermiculite. The seeds were treated with a suspension of the individual strains, and population sizes were determined at the indicated times after planting in sterilized vermiculite. RIL 52, P. fluorescens (●, solid line); RIL 83, P. fluorescens (○, dashed line); RIL 52, B. cereus UW85 (■, bold solid line); RIL 83, B. cereus UW85 (□, bold dashed line). RIL 52 and 83 are two inbred tomato lines shown to be supportive and nonsupportive, respectively, of growth of B. cereus UW85 in the spermosphere. Each bar represents the mean for eight seeds. The error bars indicate the standard error of the mean.
FIG. 2.
Effect of plant genotype on population sizes of P. aureofaciens AB254 and P. fluorescens 2-79 on the seed over 96 h after planting in sterilized vermiculite. RIL 52, P. aureofaciens AB254 (▴, bold solid line); RIL 52, P. fluorescens 2-79 (▵, bold dashed line); RIL 17, P. aureofaciens AB254 (●, solid line); RIL 17, P. fluorescens 2-79 (○, dashed line); RIL 25, P. aureofaciens AB254 (■, solid line); RIL 25, P. fluorescens 2-79 (□, dashed line). RILs 52, 17, and 25 supported high, intermediate, and low growth of B. cereus UW85, respectively, in previous experiments. Each data point represents the mean of six seeds. The error bars indicate the standard error of the mean.
Evaluation of host effect on Bacillus growth on Bacillus-inoculated seeds in soil quantified by culturing.
We used dilution plating on culture medium to evaluate the host genotype effect on B. cereus UW200 (UW85 with plasmid pBC16 conferring tetracycline resistance) population sizes of inoculated seeds that were planted in soil. In seven of eight experiments, the population size of B. cereus UW200 on RIL 52 increased between 0 and 48 h after inoculation, while on RIL 83 it did not change (Table 3). The total number of culturable bacteria also increased to a higher level on RIL 52 than on RIL 83, as did the number of fluorescent pseudomonads.
TABLE 3.
Effect of plant genotype on bacterial population size on seeds inoculated with B. cereus UW200 and planted in 60-mg of soil per sample
| Bacteria | Sample | Mean log CFU/seed or soil sample ± SEM (n = 10)
|
|
|---|---|---|---|
| 0 h | 48 h | ||
| Bacillus | RIL 52 | 4.36 ± 0.07 | 4.86 ± 0.23 |
| RIL 83 | 4.05 ± 0.07 | 4.10 ± 0.10 | |
| Soil | 4.24 ± 0.09 | 4.27 ± 0.04 | |
| TetrBacillus | RIL 52 | 4.35 ± 0.11 | 4.87 ± 0.22 |
| RIL 83 | 3.96 ± 0.11 | 4.03 ± 0.08 | |
| Soil | 2.78 ± 0.04 | 1.97 ± 0.14 | |
| Pseudomonadsa | RIL 52 | 0.18 ± 0.20 | 4.56 ± 0.14 |
| RIL 83 | 0.18 ± 0.20 | 2.91 ± 0.84 | |
| Soil | 3.33 ± 0.12 | 3.67 ± 0.07 | |
| Total Bacteria | RIL 52 | 4.42 ± 0.07 | 6.53 ± 0.11 |
| RIL 83 | 4.15 ± 0.09 | 5.69 ± 0.10 | |
| Soil | 5.39 ± 0.09 | 5.64 ± 0.04 | |
Fluorescent pseudomonads were detected.
Evaluation of host effect on Bacillus growth on Bacillus-inoculated seeds in soil quantified by FISH assays.
The population size of B. cereus relative to total bacterial population size was also evaluated for the RILs using a molecular phylogenetic approach. Spermosphere populations were simultaneously hybridized with Bc440, an oligonucleotide designed to be specific for the SSU rRNA of B. cereus (which is also predicted to hybridize to the SSU rRNA of B. thuringiensis and B. mycoides), together with Bact338, an oligonucleotide specific for the SSU rRNA of Bacteria (2). The probes were labeled with different fluorochromes, allowing positives with each to be distinguished.
While B. cereus UW200 grown in culture and harvested in exponential phase was positive with both Bc440 and Bact338, we were not able to detect B. cereus UW200 on seeds by fluorescence in situ hybridization (FISH). We hypothesized that the majority of these cells were present as spores at the 48-h time point and therefore would not be detected. Results from experiments in which the loss of FISH signal from cultures of B. cereus UW200 grown in the laboratory was correlated with the onset of sporulation (data not shown) supported the hypothesis. To test for the presence of spores in the spermosphere, we used culturing to compare the growth of B. cereus UW200 recovered from seeds at 48 h after inoculation, with and without heat treatment for 10 min at 80°C to kill vegetative cells. Results from four experiments showed that <1% of Bacillus cells survived the heat treatment at time zero, while >70% survived at the 48-h time point. In one experiment, we also examined B. cereus UW200 from non-heat-treated seeds using FISH. Samples taken at 8, 12, 24, 36, and 48 h after inoculation showed a distinct decrease in the number of Bacillus vegetative cells. This was accompanied by an appearance at 36 h of cells containing endospore-like structures and almost no detectable Bacillus-specific FISH signal at 48 h (data not shown).
Evaluation of host effect on growth of indigenous soil bacteria on the seed quantified by culturing.
To determine if the host genotype effect on inoculated B. cereus and Pseudomonas strains was predictive of growth of indigenous soil bacteria on the seed, we quantified bacterial populations of RIL 52 and 83 on uninoculated seeds planted in soil by culturing. In two experiments, the population size of indigenous Bacillus-like bacteria decreased for both RILs between 0 and 48 h after inoculation, whereas the population size of fluorescent pseudomonads increased for both RILs (Table 4). The decrease observed in Bacillus population size was in contrast to results from experiments using Bacillus-inoculated seeds. However, as also observed from the Bacillus-inoculated seeds, the increase in population size for both fluorescent pseudomonads and total cultivable bacteria was larger for RIL 52 than for RIL 83. Expressing these numbers as a percentage of the total bacteria indicates that RIL 52 preferentially supports higher populations of pseudomonads than does RIL 83 (Table 5).
TABLE 4.
Effect of plant genotype on bacterial population size on uninoculated seeds planted in soil
| Bacteria | Sample | Mean log CFU/seed or soil sample ± SEM (n = 10)
|
|
|---|---|---|---|
| 0 h | 48 h | ||
| Bacillus | RIL 52 | 2.13 ± 0.10 | NDa |
| RIL 83 | 2.03 ± 0.07 | 1.15 ± 0.31 | |
| Soil | 4.05 ± 0.07 | 4.10 ± 0.03 | |
| Pseudomonadsb | RIL 52 | 1.71 ± 0.13 | 4.94 ± 0.17 |
| RIL 83 | 1.57 ± 0.14 | 4.11 ± 0.20 | |
| Soil | 3.54 ± 0.10 | 3.99 ± 0.04 | |
| Total Bacteria | RIL 52 | 3.52 ± 0.08 | 6.81 ± 0.06 |
| RIL 83 | 3.47 ± 0.06 | 6.18 ± 0.11 | |
| Soil | 5.39 ± 0.07 | 5.60 ± 0.05 | |
ND, none detected—the population size of Bacillus bacteria on RIL 52 at 48 h was below our detection level of 20 bacteria/seed.
Fluorescent pseudomonads were detected.
TABLE 5.
Mean percentage of fluorescent pseudomonads relative to total bacteria in the spermosphere of uninoculated seedsa
| RIL | % Fluorescent pseudomonads
|
|||
|---|---|---|---|---|
| Expt I
|
Expt II
|
|||
| Culturing | Microscopy | Culturing | Microscopy | |
| 52 | 0.47 ± 0.32 | 1.46 ± 0.11 | 3.00 ± 1.94 | 2.05 ± 0.24 |
| 83 | 0.16 ± 0.11 | 0.93 ± 0.23 | 1.58 ± 1.00 | 1.46 ± 0.24 |
Samples were evaluated 48 h after planting in soil. Values represent means ± standard error of the mean (n = 10).
Evaluation of host effect on growth of indigenous Pseudomonas species on the seed quantified by FISH.
Population sizes of indigenous Pseudomonas species relative to total bacterial populations were also evaluated for the RILs by FISH. Spermosphere populations were simultaneously hybridized with Ps, an oligonucleotide specific for the LSU rRNA of a variety of different Pseudomonas species (26), together with Bact338 (2). Bacillus populations were not examined by FISH because results from the culturing experiments indicated that proportions of Bacillus to total bacteria were too low for it to be practical to examine in this manner (and because our results from the inoculation experiments suggested that at least some portion of the Bacillus population would be present as spores and therefore would not be detected).
Results from two experiments were consistent with RIL 52's supporting a higher proportion of indigenous Pseudomonas species to total bacteria than RIL 83 when sampled after 48 h of growth in soil (Table 5). Based on the values in Table 5, the ratio of RIL 52 to RIL 83 for the percentage of Pseudomonas species relative to total bacteria was 1.57 and 1.40, based on microscopy, and 2.94 and 1.90, based on culturing, for experiments 1 and 2, respectively. To determine the proportion of the cultured isolates that were positive with the LSU rRNA probe, 20 cultured Pseudomonas isolates that fluoresced on King's B medium (15) were chosen randomly and examined by FISH. Eighteen of 20 of those isolates hybridized with the probe Ps, and all hybridized with Bact338.
DISCUSSION
Our previous work described a genetic basis in the tomato host for support of growth of B. cereus UW85 and its correlation to disease suppression (27). Results from the present study indicate that this host effect on growth of B. cereus UW85 on the seed in sterilized vermiculite is a more general effect on B. cereus but does not extend to inoculated Pseudomonas bacteria. While there were no differences between RILs for population size of Pseudomonas bacteria after 48 h on the seed, it is worth noting that the Pseudomonas strains tested consistently reached higher population sizes at earlier times than did the Bacillus strains. This indicates that the Bacillus strains are limited for growth relative to Pseudomonas strains and that specific host factors are required for growth of B. cereus in the tomato spermosphere. This is also supported by the fact that sporulation occurred in the spermosphere over the course of our experiments, as evidenced through the observation of spores by microscopy and the fact that the majority of Bacillus cells on the seed were heat resistant 48 h after inoculation. Sporulation may be a critical factor in determining the efficacy of B. cereus UW85 as a biocontrol strain, for example, by allowing it to survive in high numbers in the putative nutrient-limiting environment of the pregerminated seed.
We were interested in whether the tomato genotype that supported growth of the inoculated biocontrol strains would be successful in recruiting potentially beneficial indigenous Bacillus bacteria to the spermosphere. While the ability of RIL 52 to support higher populations of bacteria on the seed extended to both total indigenous bacteria and fluorescent pseudomonads, neither RIL supported growth of detectable levels of indigenous Bacillus strains. These results differed from those observed when Bacillus and Pseudomonas strains were inoculated on seeds and incubated in sterilized vermiculite, in which RIL 52 and 83 differentially supported high and low growth of Bacillus strains, respectively, but did not differ for Pseudomonas strains. The differences may be associated with the timing of colonization or specific niches of indigenous bacteria, competition from other indigenous bacteria present in the soil system, or the fact that growth of the strains used for the inoculation experiments may not be indicative of the growth of indigenous strains. Even though the vermiculite system was less predictive of growth of particular indigenous bacteria on seeds in soil, RIL 52 was preferentially supportive for bacterial growth in general.
Exploring the role of host genotype in structuring microbial communities in a soil environment is inherently more complex than doing so in a sterilized vermiculite system. Since it is known that only a small proportion (<1%) of microorganisms can typically be cultured from environmental samples using standard techniques (3), use of both culturing and FISH on microbial SSU rRNA together could potentially provide a more comprehensive view of changes in total spermosphere microflora. The latter approach takes advantage of the ubiquitous and phylogenetically informative nature and signal amplification of rRNA molecules to detect single cells of different groups of microorganisms in situ. For these experiments, we did not use probes that allowed us to extend our conclusions concerning changes in groups of spermosphere-colonizing microorganisms other than those of particular interest to this study, but experiments in this study form the basis for future studies to address such changes. It is notable that in our hands FISH was a more precise method than culturing for quantifying spermosphere bacteria, as indicated by greater consistency of the values within and between different experiments (Table 5). The variability observed with culturing might be due to machine or human errors made in dilution plating, or, alternatively, it is possible that populations of culturing pseudomonads differed significantly between experiments while the number of total pseudomonads did not.
While culturing studies indicated that RIL 52 was supportive for growth of B. cereus inoculated on seeds and incubated in soil, we were not able to detect B. cereus on the seeds 48 h after inoculation using FISH. Our data indicate that this is because the bacterium is present in spore form at that time, which is not detected with rRNA-specific probes. Thus, one limitation of FISH is that the method cannot be used to quantify bacterial spores, which could be a drawback for working with the gram-positive spore-forming bacteria.
The use of microbial biocontrol methods to improve plant health must take into account the complexity of the interactions among the plant host, introduced biocontrol agent, and other members of the soil microbial community. An important step toward this goal is identifying the genetic determinants in the host and biocontrol agent that govern disease suppression. For example, previous research revealed that diverse B. cereus strains from around the world are capable of producing the antibiotic zwittermicin A, an important determinant for disease suppression (30). Exploiting host genes that enhance the growth and colonization of zwittermicin A-producing B. cereus strains on tomato seeds could result in improved disease suppression.
It is also important to understand how these strategies affect indigenous soil microbial communities. Introduction of B. cereus on seeds did not interfere with the supportiveness of RIL 52 for growth of either indigenous fluorescent pseudomonads or total bacteria. This indicates that the inoculation of B. cereus on seeds does not lead to a gross quantitative disturbance of indigenous spermosphere bacteria. Further development of biocontrol strategies will likely benefit from the use of both vermiculite and soil screening systems, as well as culturing and direct microscopy methods, for monitoring changes in microbial communities.
ACKNOWLEDGMENTS
This research was supported by a grant from the Consortium for Plant Biotechnology Research; a grant from Cargill Hybrid Seeds, Ciba Geigy Corporation, and DowElanco; the McKnight Foundation; the Lawrence University Individual Development and Enrichment Award (IDEA); and the Arthur Kelman Undergraduate Research Internship.
We are especially grateful to John Helgeson for the generous use of his Olympus BX-60 microscope and to Russ Spear for help with FISH. We thank Russ Spear, Matt Schrenk, Jorge Escalante-Semerena, Jonathon Trent, Paul Weimer, Dan Noguera, Lee Hood, Susan Straley, Jon Roll, John Lindquist, the Department of Bacteriology at the University of Wisconsin—Madison, and the Bacillus Genetic Stock Center at the University of Ohio for their generous gifts of paraformaldehyde-fixed microorganisms or cell cultures.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raski L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atlas R M. Microbiological media. Boca Raton, Fla: CRC Press; 1993. [Google Scholar]
- 5.Bernhard K, Schrempf H, Goebel W. Bacteriocin and antibiotic resistance plasmids in Bacillus cereus and Bacillus subtilis. J Bacteriol. 1978;133:897–903. doi: 10.1128/jb.133.2.897-903.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bull C T, Weller D M, Thomashow L S. Relationship between root colonization and suppression of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens strain 2-79. Phytopathology. 1991;81:954–959. [Google Scholar]
- 7.Callan N W. Bio-priming seed treatment for biological control of Pythium ultimum preemergence damping-off in sh2 sweet corn. Plant Dis. 1990;74:368–372. [Google Scholar]
- 8.Cook R J, Baker K F. The nature and practice of biological control of plant pathogens. St. Paul, Minn: APS Press; 1983. [Google Scholar]
- 9.Cregan P B, Kayser H H, Sadowsky M J. Host plant effects on nodulation and competitiveness of the Bradyrhizobium japonicum serotype strains constituting serocluster 123. Appl Environ Microbiol. 1989;55:2532–2536. doi: 10.1128/aem.55.10.2532-2536.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodman R M, Bintrim S B, Handelsman J, Quirino B F, Rosas J C, Simon H M, Smith K P. A dirty look: soil microflora and rhizosphere microbiology. In: Flores H E, Lynch J P, Eissenstat D, editors. Radical biology: advances and perspectives on the function of plant roots. Rockville, Md: American Society of Plant Physiologists; 1998. pp. 219–231. [Google Scholar]
- 11.Graham P H. Some problems of nodulation and symbiotic nitrogen fixation in Phaseolus vulgaris L: a review. Field Crops Res. 1981;4:93–111. [Google Scholar]
- 12.Handelsman J, Raffel S, Mester E H, Wunderlich L, Grau C R. Biological control of damping-off of alfalfa seedlings with Bacillus cereus UW85. Appl Environ Microbiol. 1990;56:713–718. doi: 10.1128/aem.56.3.713-718.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrix F F, Campbell W A. Pythiums as plant pathogens. Annu Rev Phytopathology. 1973;11:77–99. [Google Scholar]
- 14.Kim B-K, Conway de Macario E, Nölling J, Daniels L. Isolation and characterization of a copper-resistant methanogen from a copper-mining soil sample. Appl Environ Microbiol. 1996;62:2629–2635. doi: 10.1128/aem.62.7.2629-2635.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 16.Kipe-Nolt J A, Montealegre C M, Tohme J. Restriction of nodulation by the broad host range Rhizobium tropici strain CIAT899 in wild accessions of Phaseolus vulgaris L. New Phytol. 1992;120:484–494. [Google Scholar]
- 17.Li S, Spear R N, Andrews J H. Quantitative fluorescence in situ hybridization of Aureobasidium pullulans on microscope slides and leaf surfaces. Appl Environ Microbiol. 1997;63:3261–3267. doi: 10.1128/aem.63.8.3261-3267.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lilleengen K. Typing Salmonella typhimurium by means of bacteriophage. Acta Pathol Microbiol Scand Suppl. 1948;77:33–105. [Google Scholar]
- 19.Lohrke S M, Orf J H, Sadowsky M J. Inheritance of host-controlled restriction of nodulation by Bradyrhizobium japonicum strain USDA110. Crop Sci. 1996;36:1271–1276. doi: 10.1128/aem.61.6.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noel K D, Sanchez A, Fernandez L, Leemans J, Cevallos M A J. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J Bacteriol. 1984;158:148–155. doi: 10.1128/jb.158.1.148-155.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osburn R M, Schroth M N, Hancock J G, Hendson M. Dynamics of sugar beet seed colonization by Pythium ultimum and Pseudomonas species: effect on seed rot and damping-off. Phytopathology. 1989;79:709–716. [Google Scholar]
- 22.Paran I, Goldman I, Tanksley S D, Zamir D. Recombinant inbred lines for genetic mapping in tomato. Theor Appl Genet. 1995;90:542–548. doi: 10.1007/BF00222001. [DOI] [PubMed] [Google Scholar]
- 23.Parke J L. Population dynamics of Pseudomonas cepacia in the pea spermosphere in relation to biocontrol of Pythium. Phytopathology. 1990;80:1307–1311. [Google Scholar]
- 24.Paterson A H, Damon S, Hewitt J D, Zamir D, Rabinowitch H D, Lincoln S E, Lander E S, Tanksley S D. Mendelian factors underlying quantitative traits in tomato: comparisons across species, generations and environments. Genetics. 1991;127:181–197. doi: 10.1093/genetics/127.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosas J C, Castro J A, Robleto E A, Handelsman J. A method for screening Phaseolus vulgaris L. germplasm for preferential nodulation with a selected Rhizobium etli strain. Plant Soil. 1998;203:71–78. [Google Scholar]
- 26.Schleifer K H, Ludwig W, Amann R I. Nucleic acid. In: Goodfellow M, O'Donnell A G, editors. Handbook of new bacterial systematics. London, U.K: Academic Press Limited; 1993. pp. 463–510. [Google Scholar]
- 27.Smith K P, Handelsman J, Goodman R M. Genetic basis in plants for interactions with disease-suppressive bacteria. Proc Natl Acad Sci USA. 1999;96:4786–4790. doi: 10.1073/pnas.96.9.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith K P, Handelsman J, Goodman R M. Modeling dose-response relationships in biocontrol: partitioning host responses to the pathogen and biocontrol agent. Phytopathology. 1997;87:720–729. doi: 10.1094/PHYTO.1997.87.7.720. [DOI] [PubMed] [Google Scholar]
- 29.Smith K P, Goodman R M. Host variation for interactions with beneficial plant-associated microbes. Annu Rev Phytopathol. 1999;37:473–491. doi: 10.1146/annurev.phyto.37.1.473. [DOI] [PubMed] [Google Scholar]
- 30.Stabb E V, Jacobson L M, Handelsman J. Zwittermicin A-producing strains of Bacillus cereus from diverse soils. Appl Environ Microbiol. 1994;60:4404–4412. doi: 10.1128/aem.60.12.4404-4412.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weller D M, Cook R J. Suppression of take-all of wheat by seed treatments with fluorescent pseudomonads. Phytopathology. 1983;73:463–469. [Google Scholar]
- 32.Whipps J M. Developments in the biological control of soil-borne plant pathogens. Adv Bot Res. 1997;26:1–84. [Google Scholar]