Abstract
Introduction.
NRG1 rearrangements produce chimeric ligands that subvert the ERBB pathway to drive tumorigenesis. A better understanding of the signaling networks that mediate transformation by NRG1 fusions is needed to inform effective therapeutic strategies. However, this has been hampered by a paucity of patient-derived disease models that faithfully recapitulate this molecularly defined cancer subset.
Methods.
Patient-derived xenograft (PDX) and cell line models were established from NRG1-rearranged lung adenocarcinoma samples. Transcriptomic, proteomic and biochemical analyses were performed to identify activated pathways. Efficacy studies were conducted to evaluate HER3- and MTOR-directed therapies.
Results.
We established a pair of PDX and cell line models of invasive mucinous lung adenocarcinoma (LUAD-0061AS3, SLC3A2-NRG1), representing the first reported paired in vitro/in vivo model of NRG1-driven tumors. Growth of LUAD-0061AS3 models was reduced by the anti-HER3 antibody GSK2849330. Transcriptomic profiling revealed activation of the MTOR pathway in lung tumor samples with NRG1 fusions. Phosphorylation of several MTOR effectors (S6 and 4EBP1) was higher in LUAD-0061AS3 cells compared to HBEC cells and the breast cancer cell line MDA-MB-175-VII (DOC4-NRG1 fusion). Accordingly, LUAD-0061AS3 cells were more sensitive to MTOR inhibitors than MDA-MB-175-VII cells and targeting the MTOR pathway with rapamycin blocked growth of LUAD-0061AS3 PDX tumors in vivo. In contrast, MDA-MB-175-VII breast cancer cells had higher MAPK pathway activation and were more sensitive to MEK inhibition.
Conclusions.
We identify the MTOR pathway as a candidate vulnerability in NRG1 fusion-positive lung adenocarcinoma that may warrant further pre-clinical evaluation, with the eventual goal of finding additional therapeutic options for patients in whom ERBB-directed therapy fails. Moreover, our results uncover heterogeneity in downstream oncogenic signaling among NRG1-rearranged cancers, possibly histology-dependent, the therapeutic significance of which requires additional investigation.
Keywords: NRG1 fusion, HER3 antibody, lung adenocarcinoma, GSK2849330
Introduction
NRG1 fusions are recurrent but uncommon clinically actionable somatic alterations identified in 0.1–0.2% of all tumors1, 2. In the largest published study that looked at the distribution of NRG1 fusions among different cancer types, 41/21,858 tumors had a fusion and the incidences varied by tumor type - 0.5% of gallbladder cancer, 0.5% of renal clear cell carcinoma, 0.5% of pancreatic cancer, 0.4% of ovarian cancer, 0.2% of sarcoma, and 0.2% of breast cancer1. NRG1 fusions have also been identified in uterine cancer and head and neck cancer2. However, in absolute numbers, non-small cell lung cancer accounts for the largest number of NRG1 fusion-positive cancers, representing 0.3% of NSCLC cases tested1.
Although NRG1 fusions were first described in the 1990s3, it was not until the recent emergence of the possibility of targeting NRG1 fusion-driven cancers2 that the biology of these fusions gained attention. Nearly all NRG1 gene fusions retain the EGF-like domain2, which is essential for transformation by this chimeric oncogene4. Binding of the EGF-like domain of NRG1 to ERBB3 in an autocrine or paracrine manner leads to formation and activation of an ERBB2-ERBB3 heterodimer, which then activates downstream signaling via the PI3K/AKT and RAS/MAPK pathways2, 5. Moreover, NRG1 has diverse physiological functions in different tissues6 and it is unknown if tissue-specific dependencies exist in cancers arising from NRG1 fusions. One significant limitation in studying NRG1 fusion-positive cancers is a paucity of patient-derived disease models that faithfully recapitulate the disease and are available to academic laboratories. To the best of our knowledge, there is only one NRG1 fusion-positive cell line that is widely available to researchers with a confirmed fusion (DOC4-NRG1 fusion in MDA-MB-175-VII)3.
All efforts made to date to identify and develop new therapies for NRG1-driven tumors have primarily focused on agents that block activation of the most upstream components of NRG1-activated pathways such as ERBB3 and ERBB2. Our group has shown that targeting ERBB3 with the anti-ERBB3 antibody GSK28493307 achieved a durable response in one patient with invasive mucinous adenocarcinoma of the lung harboring a CD7-NRG1 fusion2. The pan-ERBB tyrosine kinase inhibitor afatinib has been used to treat NRG1 fusion-positive cancers with mixed outcomes8,2, 9. To date, more NRG1 fusion-positive patients have received afatinib than any other ERBB-targeted agents, with at least four patients showing a partial response of >18 months9. Several anti-ERBB2 agents were shown to partially reduce growth of xenograft tumors developed from an artificial system in which an NRG1 fusion was expressed in transformed lung cells10. These studies suggest that there are multiple potential therapeutic targets for NRG1 fusion-driven lung cancer. However, examining these therapies in more appropriate preclinical patient-derived models would yield more appropriate data that can determine the translational potential of specific agents.
Here we report the generation and characterization of the first paired in vitro/in vivo model of NRG1-rearranged (SLC3A2-NRG1) lung cancer. We leveraged this model to study activated cell signaling pathways in a lung-specific context and report novel therapeutic vulnerabilities that can be explored for treatment of NRG1-driven malignancies in the context of resistance to current experimental anti-ERBB therapies.
Materials and Methods
Materials and cell lines.
Cell culture growth media, antibiotics and phosphate-buffered saline (PBS) were prepared by the MSK Media Preparation Core Facility. Fetal bovine serum (FBS) was procured from Atlanta Biologicals (Flowery Branch, GA). Primocin was purchased from InvivoGen (San Diego, CA). The immortalized, non-transformed breast epithelial cell line MCF10A11 and the breast cancer cell line MDA-MB-175-VII harboring a DOC4-NRG1 fusion12 were obtained from American Type Culture Collection (Manassas, VA). All studies were conducted within 6 months of thawing the purchased cells and cells were routinely tested for mycoplasma. Human bronchial epithelial cells were immortalized by overexpression of CDK4 and TERT (HBEC-3KT cell line)13 and were obtained from Dr. John Minna (UT South Western, TX). A p53 C-terminal mutant was introduced into HBEC-3KT (HBEC-DNp53) as described previously and these cells were used in this study14. Details of antibodies raised against total or phosphorylated proteins used for Western blotting and IHC are given in Supplementary Table S1. Small molecule inhibitors were obtained from Selleckchem (Houston, TX). Reagents for PCR, Promega’s ApoOne Homogenous Caspase 3/7 activity assay kit, AlamarBlue viability dye, keratinocyte serum-free medium (KSFM), tissue culture plastic wares and Western blotting reagents not listed elsewhere were obtained from ThermoFisher Scientific (Waltman, MA). Precision plus protein kaleidoscope pre-stained molecular weight markers used for Western blotting was obtained from Bio-Rad (Hercules, CA). Mammary epithelial cell growth medium (MEGM) and supplements were purchased from Lonza (Portsmouth, NH). Protease inhibitor cocktail, RIPA lysis buffer (10X) and all other chemicals not listed above were purchased from EMD-Millipore Sigma (St. Louis, MO). All oligonucleotides used for PCR assays were custom designed by the authors and obtained from Integrated DNA technologies (Coralville, IA). The monoclonal anti-ERBB3 antibody GSK2849330 was provided by GlaxoSmithKline.
Generation of patient-derived xenograft models and cell lines, and efficacy studies.
Tissue samples were collected under an institutional IRB-approved biospecimen collection protocol and informed consent was obtained. All animals were monitored daily and cared for in accordance with guidelines approved by the Memorial Sloan Kettering Cancer Center Institutional Animal Care and Use Committee and Research Animal Resource Center. Pleura effusion fluid samples were obtained from thoracentesis procedures and samples were collected in a sterile container in which heparin was added (10 USP units of heparin/mL fluid). Cells in the sample were collected by centrifugation (1,000 RPM for 5 min) in a table top centrifuge, washed twice with cold PBS, centrifuged again and resuspended in DMEM:F12 (1:1 ratio) growth medium supplemented with 10% FBS and 1% antibiotics (complete growth media). Forty million cells were pelleted, mixed with 50% matrigel (vol/vol) and then injected at 20 million cells per subcutaneous flank of female NOD/SCID gamma (NSG) mice (Jackson Laboratory, Bar Harbor, ME) to generate xenografts15. The LUAD-0061AS3 and LUAD-0061AS4 PDX models were generated from samples collected approximately six weeks apart and were considered established after three serial passages in NSG mice. The LUAD-0061AS3 cell line was generated from PDX tumor tissue obtained after seven serial passages. Briefly, fresh PDX tumors were cut into small pieces and then digested in a cocktail of tumor dissociation enzymes obtained from Miltenyl Biotech (130–095-929) in 5 mL serum-free DME:F12 media for one hour at 37 oC, with vortexing every 5–10 min, according to manufacturer’s instructions. Digested samples were resuspended in 45 mL complete growth media to inactivate the dissociation enzymes and then cells pelleted by centrifugation. Finally, cells were plated in complete growth media and allowed to propagate over multiple generations, trypsinized when necessary to subculture and eventually only single cells remained. The PDX and cell line models were generated and propagated in the absence of afatinib.
For efficacy studies, fresh PDX tumor samples were cleaned and then minced, mixed with matrigel and implanted into a subcutaneous flank of female NSG mice to generate xenografts. After randomizing into groups of 5, tumor-bearing animals were treated with vehicle, afatinib (5 mg/kg QD on a five day on, two days off schedule), IgG (25 mg/kg BIW), GSK2849330 (25 mg/kg BIW) or rapamycin (1–4 mg/kg QD, on a five day on, two days off schedule) when tumors reached approximately 100–150 mm3 volume. GSK2849330 and IgG were diluted in PBS and administered via intraperitoneal injection. Afatinib was resuspended in 0.5% methylcellulose and 0.1% Tween-80 and administered by oral gavage. Rapamycin was resuspended in 2% DMSO, 30% PEG 300 and 5% Tween-80, and administered via intraperitoneal injection. Tumor size and body weight were measured twice weekly and tumor volume was calculated using the modified ellipsoid formula V = length x width2 × 0.52 16
Genomic characterization of preclinical models.
Cell lines and PDX were profiled by the MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets) platform, which is a large panel next generation sequencing (NGS) assay designed to detect mutations, copy-number alterations and select fusions involving up to 505 cancer-associated genes 17. Paired analysis of PDX tissue or cell line and matched-normal sample was performed to unambiguously identify somatic mutations. MSK-IMPACT does not identify the SLC3A2-NRG1 fusion and therefore, RT-PCR was used to confirm the presence of the fusion mRNA.
Growth and propagation of cell lines.
All cell lines were maintained in a humidified incubator infused with 5% CO2 and subcultured when stock flasks reached approximately 75% confluence at a 1:3 dilution. The LUAD-0061AS3 cell line was maintained in DMEM:F12 growth medium supplemented with 10% FBS and 100 μg/mL primocin. MDA-MB-175-VII cells were maintained in DMEM:F12 growth medium supplemented with 20% FBS and 100 μg/mL primocin. MCF10A cells were maintained in MEGM containing 52 μg/mL bovine pituitary extract (BPE), 500 ng/mL hydrocortisone, 10 ng/mL EGF, 5 μg/mL insulin and 100 μg/mL primocin. HBEC-DNp53 cells were maintained in KSF medium supplemented with 50 μg/mL BPE, 5 ng/mL recombinant EGF and 100 μg/mL primocin. For determination of growth kinetics and how it was affected by treatment, cells were plated at a density of 10,000 cells per well in 12-well plates and 24 h after plating (day 0) cells were placed in standard growth media with or without therapeutic compounds. Cells were counted on day 0, and then every 24–48 h thereafter. Data points were fitted to an exponential growth equation using Graphpad Prism to establish a growth curve.
Histology and immunohistochemistry.
Hematoxylin- and eosin-stained slides from the original patient tumor and patient-derived xenograft were closely inspected and compared. Unstained tissue sections on slides were deparaffinized and rehydrated, and after antigens were retrieved, immunohistochemistry staining for the following antibodies was performed: TTF1, p-HER3, Napsin, Thyroglobulin, PAX8, WT1, PR, ER. The antibody clones and suppliers are listed in the Supplementary Table S1. A relevant positive control tissue was concurrently stained for each antibody.
Cytology.
For cytological examination of the cell suspension, cells were spread on a glass slide to form a thin and even monolayer and the suspension was left to air-dry for 2–5 min. The slide was then immersed in 100% methanol for 2 min at room temperature to fix the sample. Fixed slides were stained by immersion into Liu solution A (30 seconds) followed by Liu solution B (90 seconds). Stained slides were gently rinsed with water and air-dried.
Viability and caspase 3/7 assays.
For viability assays, cells were plated in clear-bottom, white 96-well plates at a density of 7,500 cells per well and incubated with compounds for 96 h. The relative number of viable cells was determined using alamarBlue viability dye and fluorescence was measured using a Molecular Devices SpectraMax M2 multimodal plate reader (Ex: 485 nm, Em: 530 nm)18. Data was analyzed by non-linear regression and curves fitted using Graphpad Prism software to generate IC50 values. For caspase 3/7 activity, cells were plated at a density of 20,000 cells/well directly into inhibitors in white, clear-bottom 96-well plates, grown for 48 h and then caspase 3/7 enzymatic activity determined using Apo-One Homogenous caspase 3/7 activity assay kit (Promega) following the manufacturer’s instructions. All data is expressed relative to control values and is an average of 2–5 independent experiments where each condition was assayed in triplicate determinations.
Preparation of whole-cell extracts and Western blotting.
Protein levels and phosphorylation state were detected by Western blotting. Cells were lysed in 1X RIPA lysis buffer containing phosphatase and protease inhibitors. Lysates were denatured in 2X sample buffer at 55°C for 15 min, resolved on 4–12% NuPAGE gels (Invitrogen) and transferred onto PVDF (polyvinylidene fluoride) membranes. Membranes were blocked in 3% bovine serum albumin in tris-buffered saline supplemented with 0.1% Tween-20 (vol/vol) for 1 h at room temperature and probed with primary antibodies with specificity as outlined in Supplementary Table S1. Bound antibodies were detected with peroxidase-labeled goat antibody raised to mouse or rabbit IgG (R&D Systems, Minneapolis, MN) and imaged with enhanced chemiluminescence (ECL) Western blotting detection reagent (GE Healthcare). Images were captured on x-ray films. Western blotting was conducted at least two times from independently prepared samples. Films with bands in the linear range of exposure were scanned and bands quantitated by densitometry using Image J software ((http://imagej.nih.gov/ij/) and all values are expressed relative to vehicle-treated controls.
Proteome profiling arrays.
We used a human proteome profiling array system (R&D Systems) that contain duplicate validated positive and negative controls, and capture antibodies that can simultaneously detect the phosphorylation state of 43 human kinases (Proteome Profiler Human Phospho-kinase Array kit). Five million cells were plated in 10-cm dishes and grown for 48 h. Cells were deprived of serum by culturing for 24 h in growth media supplemented with 0.05% FBS, then treated with 1 μM GSK2849330 or afatinib for 0.5 h, and detection of protein phosphorylation was carried out according to the manufacturer’s instructions. In brief, the array membranes were blocked, incubated with 350 μg total cellular protein per array overnight at 4 °C on a rocking platform, washed, and incubated with phospho-specific detection antibodies. Captured phosphorylated proteins were detected by ECL and imaged on x-ray films. The average pixel densities of duplicate spots were measured using ImageJ software (http://imagej.nih.gov/ij/), and are expressed relative to the positive control on each array.
RT-PCR and qPCR experiments.
For detection of the SLC3A2-NRG1 fusion transcript and RPTOR expression in the cell lines and PDX, RNA was extracted using a Qiagen RNA mini kit and cDNAs were synthesized using SuperScript IV VILO (ThermoFisher) according to the manufacturer’s instructions. The SLC3A2-NRG1 fusion was detected by RT-PCR using 5’-ATGCTTGCTGGTGCCGTGGTCA-3’ (forward, SLC3A2 exon 4) and 5’-GGTCTTTCACCATGAAGCACTCCCC-3’ (reverse, NRG1 exon 6) primers. For qPCR of RPTOR and GAPDH the Taqman assays Hs00375332_m1 and Hs02786624_g1 were used respectively. Three replicates were produced for each cell line and probe combination. RPTOR expression values were normalized to GAPDH expression level and compared to HBEC-DNP53. For detection of SLC3A2-NRG1 mRNA expression, RNA was isolated from rapamycin-treated cells (24 h), cDNAs synthesized as above and then qPCR performed using the primers listed above for SLC3A2 and NRG1 and SYBR Green PCR master mix (ThermoFisher).
Analysis of RNA expression data.
Gene expression data from The Cancer Genome Atlas were retrieved from a public genomics data repository Gene Expression Omnibus (accession GSE62944) 19. Data were normalized and analyzed using DESeq2 package in the R programming environment. Gene Set Enrichment Analysis (GSEA) was conducted and visualized using clusterProfileR R package. Additionally, ggplot2 R package was used for data visualization.
Statistical analysis.
Student’s t-test was used to compare caspase activity or protein phosphorylation. For animal studies, area under the curve (AUC) analysis was used to compare the average tumor volume between groups. Briefly, area under the curve values and their standard errors were computed as an estimation of the surface area between baseline values (mean value of the tumor volumes at the beginning of the treatment) and growth curves for vehicle and each treatment conditions. Regression of tumor volume below the baseline value is indicated by negative AUC values. Treatment response was compared to the vehicle group using One-way ANOVA test. All data was plotted and analyzed using Graphpad Prism 8 software. P < 0.05 was considered significant.
Results
Patient history
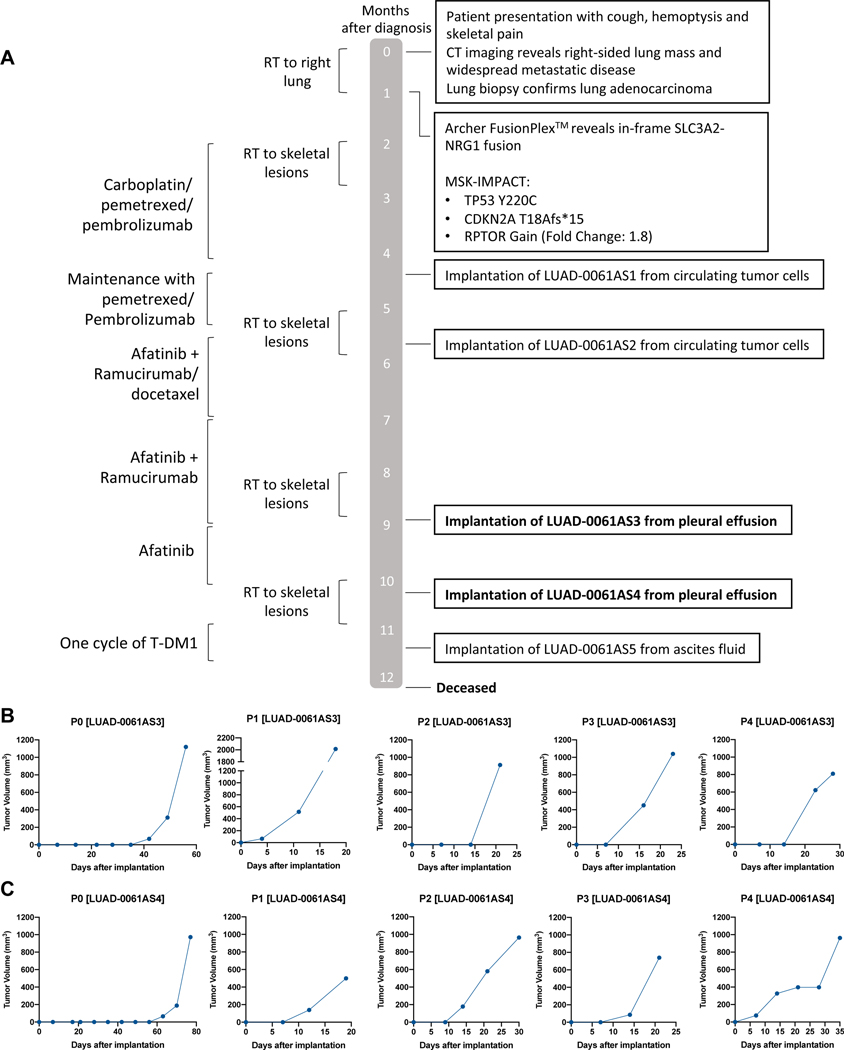
The treatment history of the patient from which the models were generated is depicted in Figure 1A. This 37-year old female never-smoker presented with hemoptysis and back pain. She was found to have a right upper lobe mass, extensive mediastinal and hilar adenopathy, right pleural effusion and distant disease including widespread osseous lesions, peritoneal carcinomatosis with ascites and a large cystic adnexal mass on cross-sectional imaging. Biopsies of the right upper lobe and the iliac bone were consistent with lung adenocarcinoma with acinar and cribriform patterns. Immunohistochemistry analysis revealed that the tumor was positive for TTF1 and napsin A, and negative for PAX8 and WT1, confirming the diagnosis. An SLC3A2-NRG1 fusion was detected via targeted RNAseq using MSK-Fusion Solid, a panel based on anchored multiplex PCR 20, 21. Additionally, a DNA-based exon capture assay (MSK-IMPACT) was performed to profile the tumor sample 22, 23. Results are summarized in Figure 1A. The patient underwent palliative intent radiation to painful skeletal lesions followed by systemic therapy with four cycles of carboplatin, pemetrexed, and pembrolizumab with a mixed response. She was continued on maintenance pemetrexed and pembrolizumab for four months. Therapy was then switched to afatinib with docetaxel and ramucirumab. Docetaxel was discontinued after two cycles due to an infusion reaction and ultimately, she continued on afatinib and ramucirumab for four months followed by widespread progression including multiple new brain metastases. The patient was started on ado-trastuzumab emtansine (T-DM1) but passed away after one cycle at 11.4 months after initial diagnosis, likely due to disease progression.
Figure 1. Generation of a lung cancer PDX model with NRG1 fusion.
A. Patient history showing major therapeutic interventions, results of molecular diagnostics and time points of PDX establishment. B-C. Growth curves of the first five serial passages demonstrated continuous growth of the xenograft models LUAD-0061AS3 (B) and LUAD-0061AS4 (C).
Establishment of patient-derived lung cancer models with NRG1 fusion
Five attempts were made to establish a patient-derived xenograft from the tissues obtained from this patient. While three attempts (two from circulating tumor cells and one from ascites fluid) failed, we successfully established two different PDX models from pleural effusion samples, LUAD-0061AS3pdx and LUAD-0061AS4pdx. These samples were collected approximately six weeks apart. Both models grew robustly, as shown in Figure 1B and C. The models took approximately 40 (LUAD-0061AS3pdx) and 60 (LUAD-0061AS4) days before palpable tumors were detected, respectively, following the first implantation. However, after the first transplant, the tumors grew much faster. To date, LUAD-0061AS3pdx has continuously grown for 14 passages, with tumors cryopreserved at every transplant, and this model was used in this study. LUAD-0061AS4pdx was cryopreserved after five serial passages and not used for any studies.
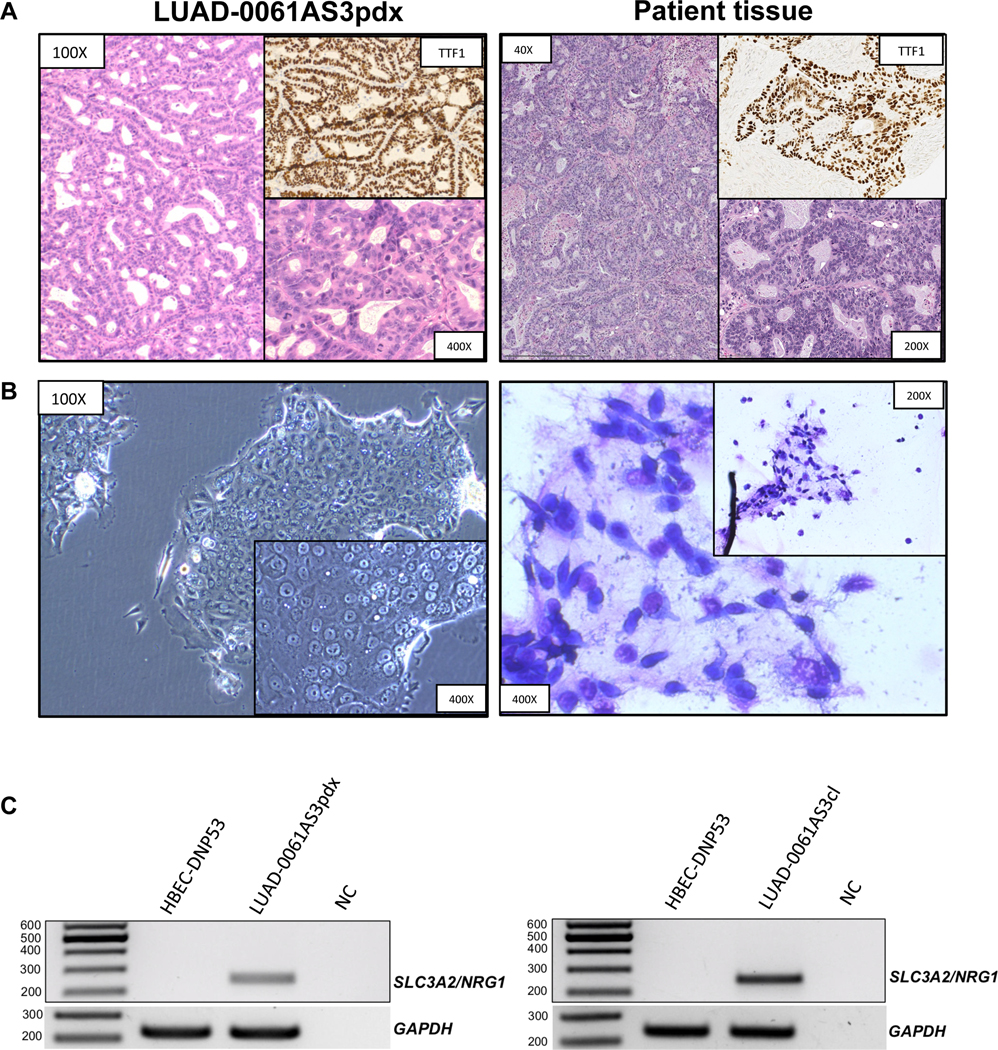
To validate the LUAD-0061AS3 PDX model we performed morphological characterization of the xenograft tissue. Hematoxylin and eosin-stained samples demonstrated a typical appearance of mucinous adenocarcinoma cells with abundant eosinophilic cytoplasm and eccentric nuclei forming gland-like structures filled with mucin (Figure 2A, left). This morphology closely recapitulated that of the original patient sample (Figure 2A, right). Further, we performed IHC staining for common differential diagnostic markers and as expected, thyroglobulin, WT1, PAX8, PR and ER were negative, whereas TTF1 displayed a strong positive signal (Figure 2A, Supplementary Figure S1). Staining for napsin-A, which was weakly positive in the original patient sample, was negative in the PDX tissue, likely due to suboptimal activity of the mouse anti-napsin-A antibody in murine xenograft tissue (Supplementary Figure S1). Phospho-ERBB3 staining revealed membranous positivity, consistent with previous reports of this immunostaining pattern in NRG1-rearranged invasive mucinous adenocarcinoma (Supplementary Figure S1) 24.
Figure 2. Histopathological characterization of the LUAD-0061AS3 PDX and sensitivity to ERBB therapy.
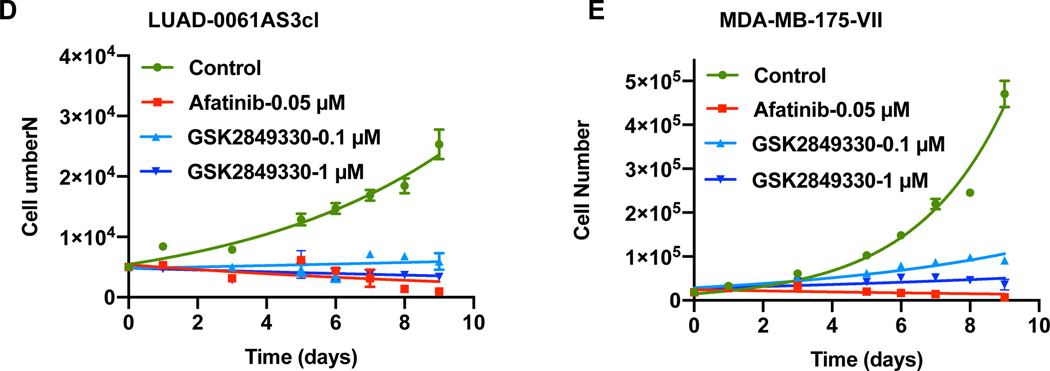
A. H&E stained samples demonstrated a typical appearance of mucinous adenocarcinoma cells with abundant eosinophilic cytoplasm and eccentric nuclei forming gland-like structures filled with mucin. IHC staining of TTF1 was performed to compare with the original patient sample. B. Cytopathological characterization was performed for an attached cell culture of LUAD-0061AS3cl (unstained, phase-contrast microscopy) and stained cell suspension (Liu stain). C. SLC3A2-NRG1 fusion-specific PCR was performed from cDNA isolated from PDX tissue (left) and cell line (right). cDNA from HBEC-DNP53 was used as a negative control to exclude non-specific amplification. NC: negative control. D-E. LUAD-0061AS3 (D) and MDA-MB-175-VII (E) cells were treated with afatinib (0.05 μM) or GSK2849330 (0.1 μM and 1 μM) and counted at 48 h intervals to establish growth curves. Results represent the mean ± SD of two independent measurements.
In addition to the two PDX models, we established a cell line from the LUAD-0061AS3 PDX tissue. The cell line has demonstrated continuous growth without any signs of senescence for the past 15 months. Inspection of unstained attached cell line culture by phase contrast microscopy showed highly atypical cells with foci of multilayer growth in spots of high cellular density, a sign of tumorigenicity. Atypical cells were also observed on the stained cytology slide (Figure 2B) 25. RT-PCR confirmed the presence of the SLC3A2-NRG1 fusion in the PDX tissue and the cell line (Figure 2C). Genomic profiling of the cell line and PDX models were performed using the latest version of MSK-IMPACT, which profiles 505 genes for somatic alterations. The three disease models retained the TP53 and CDKN2A mutations that were detected in the patient sample (Supplementary Table S2). The SLC3A2-NRG1 fusion is not detected by MSK-IMPACT but as noted above, was confirmed by RT-PCR (Figure 2C).
GSK2849330 efficiently reduces growth of the NRG1 fusion-positive models in vitro
Here we examined the effect of GSK2849330 on growth of LUAD-0061AS3cl and MDA-MB-175-VII cells 2 in time-course experiments. As a control for growth inhibition, we used the pan-ERBB TKI afatinib. Cells were treated with either GSK2849330 (0.1 or 1 μM) or afatinib (0.05 μM) for nine days and the number of cells counted at the time points indicated in Figure 2D and E. GSK2849330 treatment inhibited growth to a similar extent in both cell lines (Figure 2D and E). This inhibition of growth was equivalent to that observed following afatinib treatment (Figure 2D and E). The high sensitivity of the LUAD-0061AS3 cell line to higher dose afatinib in vitro contrasts with the poor response of the patient to the standard clinical dosing, representing the highest tolerable dose in humans, which corresponds to a dose of approximately 6.15 mg/kg in the mouse. Studies performed in vitro are not subject to systemic toxicity constraints, hence, higher dose afatinib appears as an effective agent in vitro. Alternatively, given that the PDX and cell line models were developed in the absence of afatinib over the course of several months, it is possible that afatinib-sensitive clones could have re-emerged to dominate the culture.
Characterization of activated intracellular signaling pathways in NRG1 fusion-positive cells
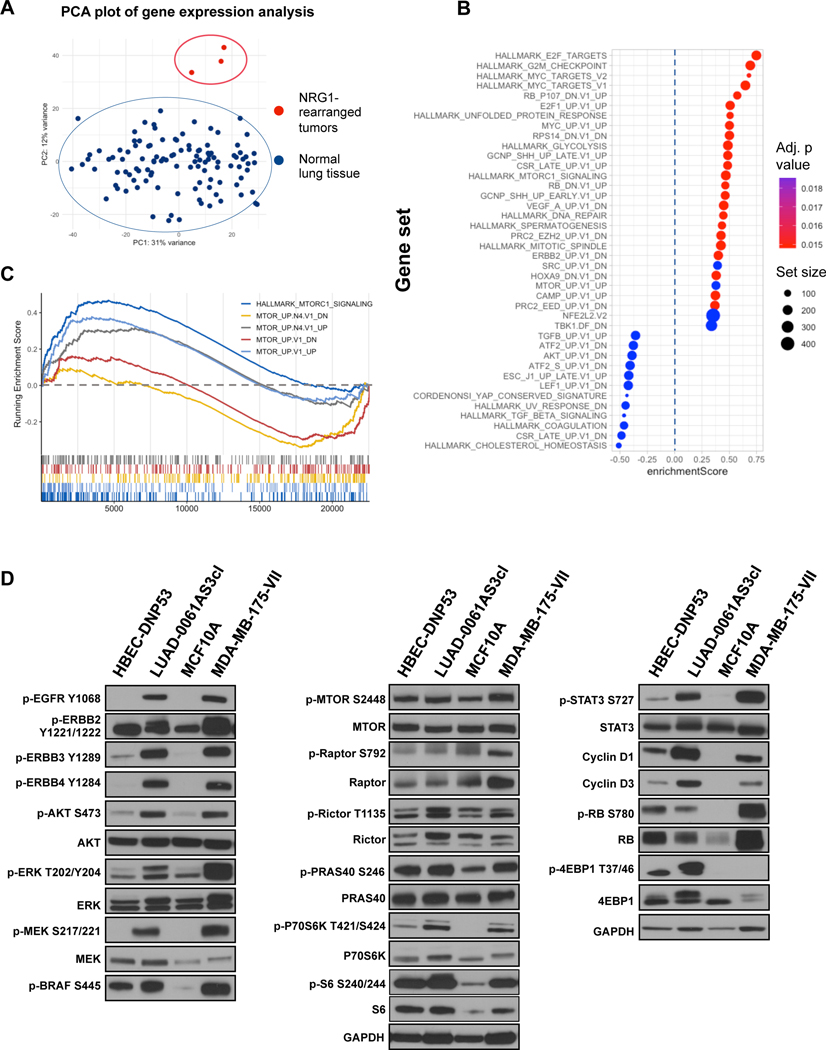
Little is known about the signaling pathways activated in NRG1-rearranged tumors. Although some studies have attempted to address this, all studies were performed in either murine cells or transformed lung cells, where NRG1 fusions were ectopically expressed, likely at supraphysiological levels5, 10, 26. As changes in intracellular signaling are reflected downstream in specific transcriptional signatures, we decided to analyze available gene expression data to define prominent gene expression signatures in NRG1-rearranged lung tumors. We compared three NRG1-rearranged lung carcinomas (two squamous carcinomas, one adenocarcinoma) with normal lung tissue RNA sequencing data from the TCGA dataset. Using principal component analysis, we found that NRG1-rearranged tumors grouped separately from healthy lung tissue samples as expected (Figure 3A). Differential gene expression analysis found 6,455 (29%) and 4,852 (22%) significantly up- and down-regulated transcripts, respectively, with NRG1 being in the top of the list of up-regulated genes (log2 fold change: 4.1, adjusted p value < 0.0001). To classify this output, we ranked the list of differentially expressed genes and performed Gene Set Enrichment Analysis (GSEA). Figure 3B illustrates the top 40 most significant expression signatures (ranked by p-value). This analysis showed activation of multiple facets of oncogenesis: G2-M progression and mitotic spindle activation, activation of E2F and MYC, glycolysis, etc. As for intracellular kinase signaling, the AKT/MTOR pathway was the most prominent expression signature present, with consistent up and down regulation of corresponding gene sets (Figure 3C and Supplementary Figure S3B).
Figure 3. Identification and characterization of activated signaling pathways in NRG1 fusion-driven lung cancers.
A. Principal component analysis demonstrated distinctive grouping of NRG1-rearranged lung tumor samples. B. GSEA analysis was performed to compare NRG1-rearragned lung cancer samples and healthy lung tissue gene expression (TCGA LUAD and LUSC datasets). The top 40 significantly enriched pathways from MSigDB (HALLMARK and Oncogenic signatures, combined) are demonstrated. C. Enrichment plot depicting significantly enriched AKT-MTOR related signatures. While corresponding upregulation were positively enriched, downregulation signatures demonstrated negative scores, consistent with a hypothesis of AKT-MTOR-associated transcriptomic changes. D. Whole-cell extracts were prepared from serum-starved cells and subjected to Western blotting analysis. Representative immunoblots are shown. Two independent experiments were conducted.
As noted above, most studies to date on NRG1 fusion-driven biology have used isogenic cancer cell lines that overexpress cDNAs encoding NRG1 fusions. Indeed, the only patient derived cell line in which NRG1 signaling has been investigated is the breast cancer cell line MDA-MB-175-VII that expresses a DOC4-NRG1 fusion 2. It is believed that the EGF-like domain of NRG1 directly interacts with ERBB3 and ERBB4 to activate ERBB-family receptors and trigger a downstream cascade of phosphorylation and activation of effector proteins27. Here we sought to validate the data obtained in Figure 3A and C, and to identify additional activated signaling pathways in NRG1 fusion-positive cell lines, using LUAD-0061AS3 and MDA-MB-175-VII cells. As comparisons, we used immortalized, untransformed human bronchial epithelial cells (HBEC-DNP53) and MCF10A breast epithelial cells. Western blotting analysis with phospho-specific antibodies showed that EGFR, ERBB2, ERBB3 and ERBB4 were all highly phosphorylated in the two NRG1 fusion-positive cell lines compared to the respective tissue-matched control lines (Figure 3D). Similarly, the NRG1 fusion-positive cell lines had higher level of phosphorylation of multiple effectors of the RAS-MAPK (ERK, MEK, BRAF), AKT-MTOR (AKT, p70S6K, 4EBP1) and STAT pathways (STAT3). The increased phosphorylation of AKT, p70S6K, 4EBP1 and RICTOR, a negative regulator of MTOR, in LUAD-0061AS3 cells compared to the HBEC-DNP53 cells confirmed the signaling changes inferred from the transcriptomic profiling data. The LUAD-0061AS3 cells had lower levels of phospho-ERK and phospho-BRAF than MDA-MB-175-VII 0061AS3 cells.
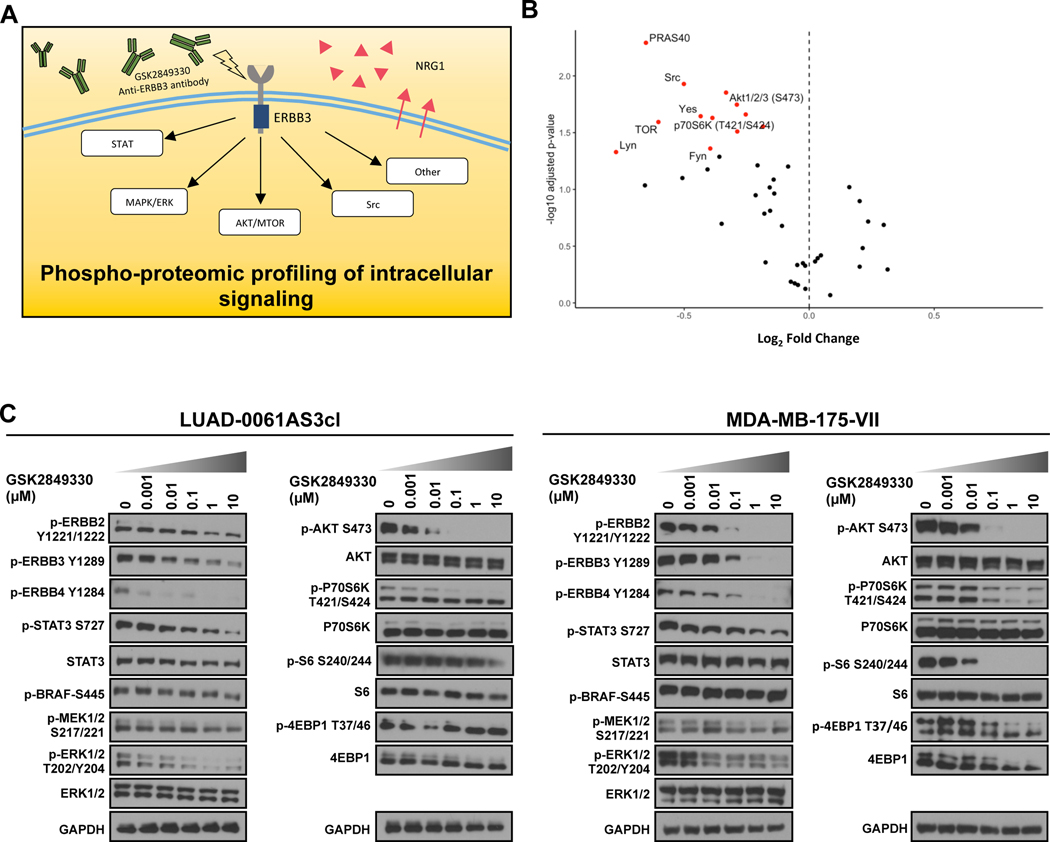
Phospho-proteomic profiling of NRG1 fusion-associated intracellular signaling downstream of ERBB3
Uncontrolled activation of the ERBB family receptors and downstream signaling presents a target for biology-oriented therapy of NRG1 fusion-driven tumors. Currently, multiple ERBB-family TKI or antibodies are available for the treatment of NRG1-rearranged tumors, with some being tested in clinical trials (NCT04100694, NCT02912949, NCT03805841). Despite this, little is known about how downstream signaling is affected by therapy in lung malignancies with NRG1 fusions. We have previously reported that the anti-ERBB3 antibody GSK2849330 was efficacious in a lung cancer patient with a CD74-NRG1 rearrangement. Here we used GSK2849330 to attempt to elucidate the signaling networks that are used by ERBB3 to drive growth in the SLC3A2-NRG1-expressing LUAD-0061AS3 cell line. To this end, we utilized proteomic arrays (a schematic outline of the experimental design is given in Figure 4A), as reported previously14. This uncovered significant inhibition of multiple effectors in the AKT-MTOR pathway (Figure 4B). In contrast, phosphorylation of ERK was inhibited less prominently (phospho-ERK, 0.01 μM, p = 0.035; 0.1 μM, p = 0.081). The complete data is shown in Supplementary Figure S2. To confirm the results obtained by phospho-proteomic profiling, we performed Western blotting analysis of cell extracts obtained from cells treated with multiple concentrations of GSK2849330. We observed dose-dependent inhibition of multiple kinases including ERBB2, ERBB3, ERBB4, AKT, p70 S6K, STAT3, and ERK (Figure 4C). Some MTOR effectors such as p70 S6 kinase, S6 and 4EBP1 were less readily inhibited in LUAD-0061AS3 cells compared to MDA-MB-175-VII, perhaps due to higher intrinsic activity of MTOR in these cells. Of note, the inhibition of ERK and STAT3 phosphorylation following GSK2849330 treatment was less prominent than inhibition of AKT in both cell lines.
Figure 4. Phospho-proteomic profiling of intracellular signaling pathways.
A. Schematic outline of the experiment. B. Volcano plot of phospho-kinase array data obtained in LUAD-0061AS3 cells treated with 1 μM GSK2849330 for 3 h. The points that reached significance (alpha = 0.05) are highlighted in red. AKT-MTOR and SRC pathway effectors are labeled. C. Western blot validation of protein kinase array results in LUAD-0061AS3 (left) and MDA-MB-175-VII (right) cells lines. Cells were treated for 3 h with GSK2849330 and then extracts prepared for Western blotting. Representative immune blots from two independent experiments are shown.
Inhibition of MTOR but not MEK decreases growth of lung adenocarcinoma cells with NRG1 fusion
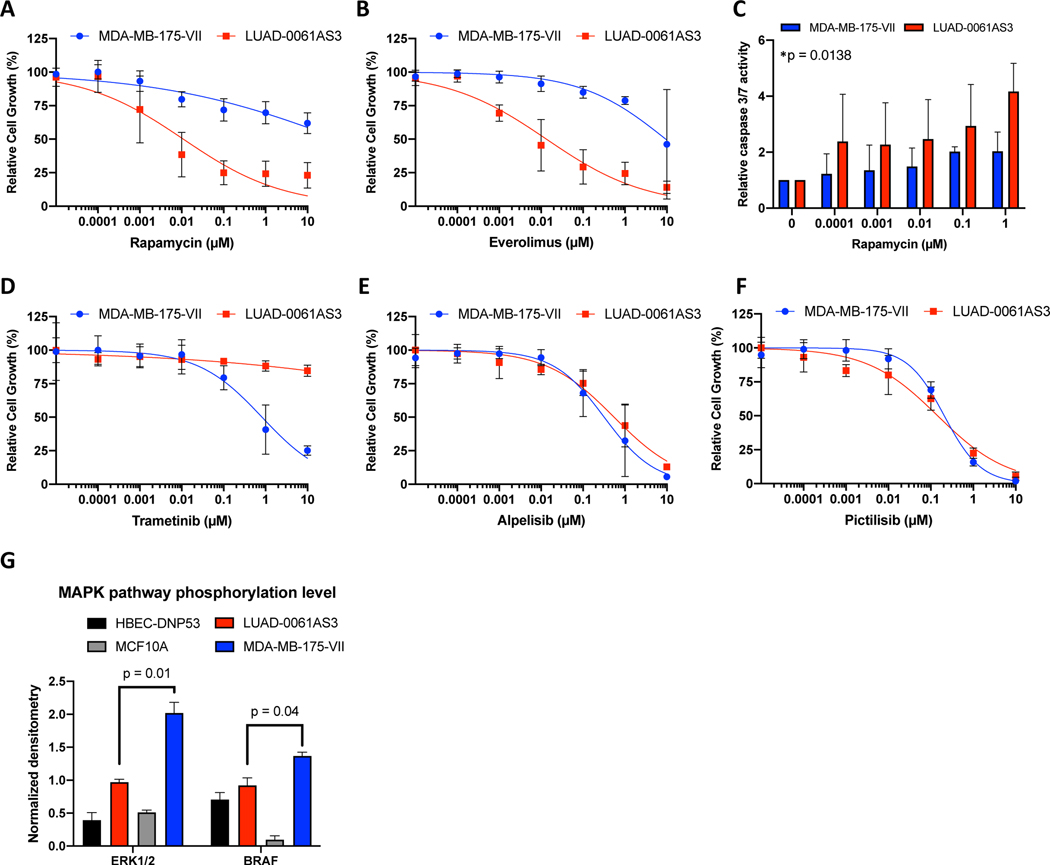
Based on the proteomic and transcriptomic evidence of activation of AKT/MTOR pathways described above, we hypothesized that MTOR inhibition can be effective in suppressing growth of tumor cells with NRG1 rearrangements. As demonstrated in Figure 5A and B, rapamycin and everolimus, two MTOR inhibitors, decreased viability of LUAD-0061AS3 cells at low nanomolar concentrations, achieving 75–90% reduction in growth. In contrast, rapamycin and everolimus only partially inhibited the growth of MDA-MB-175-VII cells. The two cell lines exhibited similar sensitivity to two PI 3-kinase inhibitors, alpelisib and pictilisib (Figure 5E and F). We next tested the effect of two pan AKT inhibitors on growth of the NRG1 fusion-positive cell lines. Growth of LUAD-0061AS3 and MDA-MB-175-VII cells were only affected at very high concentrations of AKT inhibitors (Supplementary Figure S4) beyond that which was shown to inhibit growth of AKT-dependent cells 28. Rapamycin treatment induced higher levels of caspase 3/7 activation in LUAD-0061AS3 cells than in the MDA-MB-175-VII cell line (Figure 5C). While a low level RPTOR copy number gain was identified in the patient sample by MSK-IMPACT analysis, we did not detect RPTOR overexpression at the transcriptomic or proteomic level (Supplementary Figure S3A and Figure 3D, respectively), so it is unlikely to explain the increased sensitivity to MTOR inhibitors. LUAD-0061AS3 cells had lower sensitivity to the MEK inhibitor trametinib (Figure 5D) compared to MDA-MB-175-VII and this correlated with the lower level of MAPK pathway activation (Figures 3D and 5G).
Figure 5. Efficacy of PI3K, MTOR and MEK inhibitors on growth of NRG1-rearranged cell lines.
Cells were treated for 96 h with the indicated concentrations of inhibitors and then growth determined using alamarBlue proliferation dye. Results represent the mean ± SD of 2–4 experiments in which each condition was assayed in triplicate determination. The MTOR inhibitors rapamycin (A) and everolimus (B) suppressed growth of LUAD-0061AS3cl at low nanomolar doses, while much higher doses were required to inhibit MDA-MB-175-VII cell line. (C) LUAD-0061AS3 and MDA-MB-175-VII cells were treated with rapamycin for 48 h and then caspase 3/7 activity determined. Results represent the mean ± SD of two independent experiments in which each condition was assayed in triplicate determination. *Treatment compared to untreated controls for LUAD-0061AS3, ANOVA. Values were not significantly different for MDA-MB-175-VII. D-F. MEK inhibitors demonstrated higher activity in MDA-MB-175-VII cells (D), while no difference in sensitivity to the PI3K inhibitors alpelisib and pictilisib between cell lines was observed (E, F). G. Comparison of MAPK pathway activation in the cell lines. Immunoblots for p-ERK and p-BRAF shown in Figure 3D was quantitated by densitometry.
Rapamycin effectively inhibits growth of LUAD-0061AS3 models in vivo.
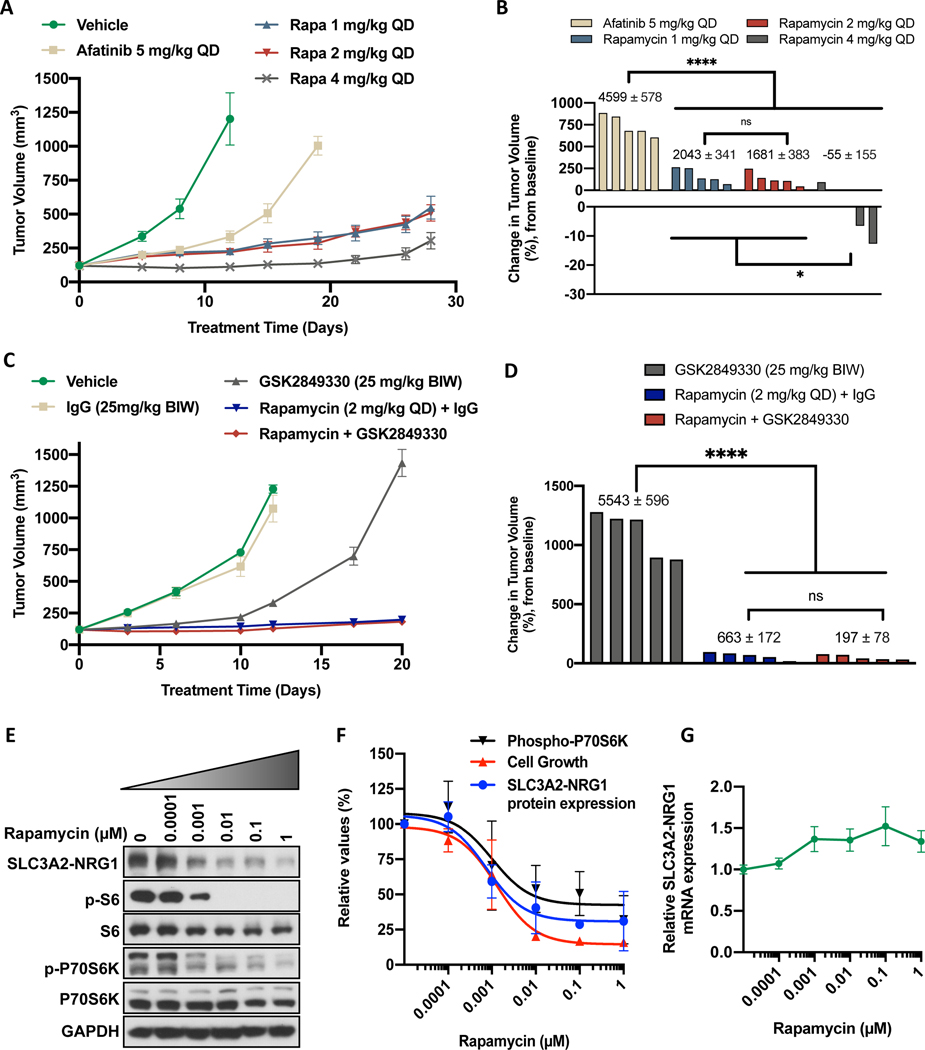
To begin to translate our in vitro findings showing increased sensitivity of LUAD-0061AS3 cells to MTOR inhibitors, we examined the efficacy of rapamycin in the LUAD-0061AS3 PDX model in comparison to afatinib. The data is presented in Figure 6 with tumor volumes over the course of treatment shown in Figure 6A and the percent change in volume of each tumor shown in Figure 6B for afatinib and rapamycin. All doses of rapamycin (1, 2 and 4 mg/kg, QD) were more effective at controlling tumor growth than afatinib when used at a dose in mice (5 mg/kg QD), which is similar to the clinical dose (Figure 6A). The highest dose of rapamycin tested resulted in regression of two of the five tumors (Figure 6B). We used area under the curve analysis (AUC) to compare the effect of treatment between the groups as this analysis considers the magnitude and duration of the treatment effect (Supplementary Figure S5A). As shown, all treatments caused a significant decrease in tumor growth (Supplementary Figure S5A) when compared to the vehicle (calculated on day 11 when all tumor bearing animals were alive). To better compare afatinib to rapamycin, the AUC values were calculated on day 19, which was the last treatment with afatinib (Figure 6B, numbers above the bars). This analysis showed that the three doses of rapamycin were significantly better than afatinib at blocking growth of LUAD-0061AS3 PDX tumors. Although afatinib caused a significant reduction in tumor volume compared to vehicle-treated tumors, all tumors continued to grow slowly in the afatinib-treated group (Figure 6A). A higher dose of afatinib (25 mg/kg QD) was more effective than the 5 mg/kg QD dose at inhibiting tumor growth (89.4 ± 0.7% tumor shrinkage, p < 0.0001), however, this dosage resulted in 12% loss in animal weight (p = 0.004) and cannot be achieved clinically. There was no reduction in animal weight and no toxicity was observed in any of the treatment groups (Supplementary Figure S5C).
Figure 6. Rapamycin treatment inhibits growth of LUAD-0061AS3 PDX tumors and reduce expression of the SLC3A2-NRG1 fusion.
LUAD-0061AS3 PDX tumors were implanted into a subcutaneous flank of NSG mice and treated as indicated on the graphs. Tumor volume and animal weight were measured twice weekly. Animal weight is given in Supplementary Figure S5. Tumor volume over time (A and C) and change in tumor volume (B and D) are shown. Each group consisted of five animals and data represent the mean ± SE of tumor volumes. Area under the curve (AUC) analysis was used to compare treatment effects and the AUC ± SE values are shown above the bars in B and D for days 19 and 20 respectively. Additional AUC data is provided in Supplementary Figure S5 for days 11 and 12 respectively. E-G. LUAD-0061AS3 cells were treated with the indicated concentrations of rapamycin for 24 h and then either whole-cell extracts were prepared for Western blotting (E). F. SLC3A2-NRG1 levels in comparison to cell viability and phosphorylation of p70S6K as a function of rapamycin concentration. Immunoblots were quantitated by densitometry and results represent the mean ± SD of two independent experiments. G. RNA was isolated from cells treated with rapamycin for 24 h and SLC3A2-NRG1 fusion mRNA level was then determined by qPCR using primers that target SLC3A2 and NRG1. Results are the mean ± SD of three replicate determinations in one experiment. * p < 0.05, **** p < 0.0001, ns: not significant (p > 0.05).
We next attempted to determine if the anti-tumor effect of rapamycin might be enhanced by combination with the anti-HER3 therapeutic antibody GSK2849330. The data is presented in Figure 6C–D with tumor volumes over the course of treatment shown in Figure 6C and the percent change in volume of each tumor is shown in Figure 6D for GSK2849330, rapamycin or the combination of the two agents. As above, we used area under the curve analysis (AUC) to compare the effect of treatment between the groups. Administration of a control IgG did not significantly alter growth of LUAD-0061AS3 PDX tumors compared to PBS vehicle treatment (Figure 6C and Supplementary Figure S5B, showing AUC for day 12). GSK2849330 (25 mg/kg BIW) treatment resulted in a significant reduction in tumor growth compared to IgG-treated tumors (Supplementary Figure S5B, p < 0.0001). However, rapamycin (2 mg/kg QD) was more effective at blocking growth of LUAD-0061AS3 PDX tumors than GSK2849330 (Figure 6D). A combination of rapamycin (2 mg/kg QD) and GSK2849330 (25 mg/kg BIW) was not more effective than rapamycin treatment alone (Figure 6D and Supplementary Figure S5B). There was no reduction in animal weight and no toxicity was observed in any of the treatment groups (Supplementary Figure S5D).
Expression of SLC3A2-NRG1 fusion is dependent on MTOR activity
Finally, to explore potential mechanisms by which rapamycin treatment inhibited growth of the LUAD-0061AS3 cell line and PDX tumors, we examined the effect of the drug on expression of the SLC3A2-NRG1 fusion. Cells were treated for 24 h with various concentrations of rapamycin and then the level of SLC3A2-NRG1 fusion protein (Western blotting) or SLC3A2-NRG1 mRNA (qPCR) was determined. Rapamycin treatment caused a significant reduction in expression of SLC3A2-NRG1 fusion protein at concentrations that correlated with inhibition of P70S6K and S6 phosphorylation (Figure 6E). Quantitation of the immunoblots by densitometry illustrated that down regulation of SLC3A2-NRG1 expression following rapamycin treatment also correlated with loss of cell viability (Figure 6F). Treatment of cells with rapamycin did not cause a reduction in SLC3A2-NRG1 fusion mRNA level (Figure 6G).
Discussion
Whereas an NRG1 fusion was first described in 1999 in a breast cancer cell line 3, it was not until 2014 that the first observation of lung cancers harboring this genetic alteration was reported 5. The subsequent discovery of numerous tumor types with NRG1 rearrangements using NGS-based techniques prompted the examination of targeted therapies for this subset of molecularly-defined tumors. As the ERBB family receptors represent direct binding partners of NRG1, multiple agents that antagonize the ERBB family are being evaluated, some with promising results. In the two largest analyses of data looking for cancer drivers, NRG1 fusions were identified in approximately 0.3% of NSCLC 1, 29. Based on an estimated diagnosis of 2.1 million lung cancer cases in 2018 (1.76 million NSCLC) 30, more than 5,000 NSCLC patients per year would benefit from therapy for this group of lung cancers. In addition, NRG1 fusions have been found to co-occur with ALK fusion 31, 32 and in one study, the NRG1 fusion was shown to arise after resistance to ALK inhibitor treatment 31, suggesting that there are additional patients who may benefit from therapeutically targeting NRG1 fusions in the setting of resistance to targeted therapy.
Investigation of the mechanisms of NRG1-induced tumorigenesis is lagging behind the therapeutic work, largely due to a paucity of preclinical models that faithfully represent this molecular subset of lung cancers. Fernandez-Cuesta et al. reported expression of CD74-NRG1 fusions in invasive mucinous lung adenocarcinomas 5. Ectopic expression of the NRG1 fusion in two lung cancer cell lines (H322 and H1568) showed increased levels of p-ERBB2, p-ERBB3, p-AKT, and p-P70S6K. H1568 cells ectopically expressing a CD74-NRG1 fusion exhibited enhanced colony formation in soft-agar assays 5. Co-culture of NIH-3T3 cells ectopically expressing CD74-NRG1 with Ba/F3 cells genetically engineered to overexpress human ERBB2 and ERBB3 also led to activation of AKT. Likewise, Murayama et al. demonstrated activation of PI3K/AKT/NF-κB signaling pathways and induction of an IGF2-IGF1R autocrine/paracrine circuit upon ectopic expression of a CD74-NRG1 transcript in lung (H322) and breast cancer (BT20) cell lines 26. However, expression of the CD74-NRG1 cDNA in H322 cells did not enhance tumor growth in mice. The limitation of interpreting these published results is that NRG1 fusions were introduced into systems that already had fully activated transforming genetic alterations, perhaps blunting the effect of an additional oncogene, thereby making it difficult to delineate NRG1 fusion-specific biology. While these isogenic comparisons in cancer cell lines have been somewhat informative, further confirmation of the findings has been hampered by a lack of patient-derived models with NRG1 rearrangements. To address this urgent need, we established and characterized a novel paired in vitro/in vivo model, LUAD-0061AS3, of NRG1 fusion-positive lung invasive mucinous adenocarcinoma harboring an SLC3A2-NRG1 fusion. We demonstrated efficacy of the therapeutic anti-ERBB3 antibody GSK2849330 in vitro and in vivo in the LUAD-0061AS3 models. The LUAD-0061AS3 cell line was sensitive to afatinib, in contrast to the poor response of the patient and the LUAD-0061AS3 PDX model, which showed no tumor regression when treated with 5 mg/kg QD afatinib. We estimate that this dose of afatinib in mice is equivalent to a human dose of 32 mg/day, derived by allometric scaling based on FDA draft guidelines 33, 34. A higher dose of afatinib (25 mg/kg) was able to cause tumor shrinkage in the PDX model but this was accompanied by toxicities, as reflected by substantial weight loss of tumor-bearing animals. This dose is not achievable clinically as most published cases of patients with NRG1 fusion-driven cancers were treated with 40 mg and several had dose reductions 9, 35. The discordant responses to afatinib of the cell line compared to the patient and the PDX model are a reminder that caution should be exercised in interpreting data obtained only in vitro.
As NRG1 fusions are identified across a variety of cancers, and numerous tissue-dependent functions of physiological NRG1 isoforms are described, the question of cross-organ differences of NRG1-induced tumorigenesis arises. Our study identified some differences between the breast cancer cell line MDA-MB-175-VII and our novel lung cancer model, LUAD-0061AS3. We observed higher phosphorylation of BRAF and ERK1/2 in the MDA-MB-175-VII cell line, concomitantly with increased sensitivity to MEK inhibitors. In contrast, MTOR (but not PIK3CA) inhibitors displayed relatively higher activity in LUAD-0061AS3 cells than in the breast cancer cell line. In support of these findings, transcriptomic profiling of lung tumors with NRG1 chimeric transcripts revealed an activated MTOR signature and two proteins that function downstream of MTOR, namely p70S6K and 4EBP1, showed higher phosphorylation in the LUAD-0061AS3 cell line compared to non-tumor HBEC cells and the MDA-MB-175-VII cell line. These findings prompted us to examine the therapeutic capacity of rapamycin for NRG1 fusion-positive lung cancers. Administration of rapamycin to mice bearing LUAD-0061AS3 PDX tumors resulted in a significant reduction in tumor growth compared to afatinib or GSK2849330 with the magnitude and duration of the anti-tumor effect of rapamycin being superior. The doses of rapamycin that we tested in mice are clinically achievable 36. Combination of GSK2849330 and rapamycin did not provide any additional benefit over rapamycin alone. These results support our hypothesis that MTOR is a significant effector of NRG1-dependent tumorigenesis in lung cancers with NRG1 fusions. Given that we utilized only one lung cancer PDX model to evaluate the efficacy of rapamycin, caution should be exercised in interpreting these encouraging data. Future studies should examine the efficacy of rapamycin and other MTOR inhibitors in additional in vivo preclinical disease models of NRG1 fusion-driven lung cancers.
Importantly, we found that rapamycin treatment down-regulated SLC3A2-NRG1 fusion protein levels in the LUAD-0061AS3 cell line. To our knowledge this is the first observation of a targeted agent causing downregulation of an oncogenic fusion protein and helps to explain the exquisite sensitivity of the LUAD-0061AS3 cell line to rapamycin. It remains unclear which additional pathways downstream of MTOR may also be co-opted by NRG1 fusions to drive tumorigenesis. Inhibition of AKT and PI3K were not sufficient to block cell growth, either because compensatory signaling pathways exist or that antagonizing these pathways are not very effective in cells where these effectors are not constitutively activated by genetic alterations.
Although we report some differences between the cell lines of different tissue origin, caution should be applied in interpreting these results as these models harbor different 5’ partners of NRG1. It is possible that different partners of NRG1 in the chimeric fusion protein may affect lipid raft localization, dimerization, extracellular cleavage, etc., events that can influence the mechanism and intensity of their oncogenicity. Nevertheless, anti-ERBB therapy was effective in both tumor types and therefore, we believe that it would be more practical to use such therapy for patients with NRG1 fusion-dependent cancers given the low prevalence of this oncogene. Our results suggest that the use of MTOR inhibitors in combination with HER3 antibodies (or other effective anti-ERBB therapy) may not provide additional benefit over inhibition of MTOR alone. Perhaps therapeutically targeting MTOR alone may be possible in unique situations where an anti-ERBB agent no longer offer substantial benefit, for e.g., if resistance to therapy occurs. However as noted above, further studies are required to clarify differences between tissues and fusion partners using additional patient-derived disease models with NRG1 fusions as they can substantially guide and transform therapy in this patient subpopulation.
Supplementary Material
Supplementary Figure S1. IHC characterization of LUAD-0061AS3 PDX. IHC staining of LUAD-0061AS3pdx sample was done to confirm the origin of the PDX tissue. PR, ER, WT1, PAX8 and thyroglobulin were consistent with the original patient sample. Phospho-ERBB3 stain demonstrated membranous positivity, as expected in NRG1 fusion-positive tumors. Napsin-A, although weakly positive in the original patient sample, was negative in the PDX tissue, likely due to suboptimal activity of anti-mouse napsin-A antibody in mouse xenograft tissue.
Supplementary Figure S2. Bar graphs of the results of protein kinase array performed on LUAD-0061AS3 cells treated with GSK2849330 0.01 μM (GSK330 0.01 μM) and GSK2849330 0.1 μM (GSK330 0.1 μM). *p < 0.05, **p < 0.01. Images were captured on x-ray films and then quantitated by densitometry. Results represent the mean ± SD of two measurements.
Supplementary Figure S3. A. qPCR analysis of RPTOR expression in HBEC-DNP53, LUAD-0061AS3cl, MCF10A and MDA-MB-175-VII. B. GSEA plot for MTOR gene sets in NRG1 fusion-positive cell lines as compared to normal lung tissue.
Supplementary Figure S4. Effect of AKT inhibitors on cell growth. Cells were treated with the pan-AKT inhibitors ipatasertib and MK2206 for 96 h and then viability determined. Data represent the mean ± SD of 3 replicate determinations in one experiment.
Supplementary Figure S5. Area under the curve analysis of tumor volume and animal weight A-B. The data shown in Figure 6A and C were analyzed by area under the curve analysis to compare treatment effects. AUC values were calculated for day 11 and 12 respectively. C-D. Animal weight over treatment course. Tumor bearing mice were weighed twice weekly. Data represent the mean ± SE. ****p < 0.0001. ns: not significant (p > 0.05).
Supplementary Table S1. Antibody source and specificity used in Western blotting and IHC. Excel File.
Supplementary Table S2. Genomic Characterization of PDX and Cell line models. Models were profiled by MSK-IMPACT. Excel file.
Acknowledgments
We acknowledge BioWa, Inc. (U.S. subsidiary of Kyowa Hakko Kirin Co., Ltd.) for their POTELLIGENT Technology and COMPLEGENT Technology in developing GSK2849330, an ADCC- and CDC-enhanced anti-ERBB3 mAb.
Financial support for this study was provided by a grant from Cycle for Survival and grant P01 CA129243 from the National Institutes of Health (NIH) to Marc Ladanyi, U54 OD020355 from NIH to Elisa de Stanchina, and a Memorial Sloan Kettering Cancer Center Support Grant (P30 CA008748).
Conflicts of Interest Statement
Igor Odintsov, Marissa S. Mattar, Allan J.W. Lui, Christopher Kurzatkowski, Lukas Delasos, Inna Khodos, Elisa de Stanchina, Marina Asher, Robert M. Daly and Natasha Rekhtman report no potential conflict of interest.
Michael Offin has received advisory board compensation from Novartis, PharmaMar and Targeted Oncology.
Gopinath Ganji is an employee and stockholder of GlaxoSmithKline.
Marc Ladanyi has received advisory board compensation from Merck, Bristol-Myers Squibb, Takeda, Bayer, Lilly Oncology, Janssen, and Paige.AI. In addition, he has received research support from LOXO Oncology, Helsinn Healthcare, Elevation Oncology Inc. and Merus.
Romel Somwar has received research grants from Merus, Helsinn Healthcare, LOXO Oncology, and Elevation Oncology Inc.
References
- 1.Jonna S, Feldman RA, Swensen J, et al. Detection of NRG1 Gene Fusions in Solid Tumors. Clin Cancer Res 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drilon A, Somwar R, Mangatt BP, et al. Response to ERBB3-Directed Targeted Therapy in NRG1-Rearranged Cancers. Cancer Discov 2018;8:686–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang XZ, Jolicoeur EM, Conte N, et al. gamma-heregulin is the product of a chromosomal translocation fusing the DOC4 and HGL/NRG1 genes in the MDA-MB-175 breast cancer cell line. Oncogene 1999;18:5718–5721. [DOI] [PubMed] [Google Scholar]
- 4.Shin DH, Lee D, Hong DW, et al. Oncogenic function and clinical implications of SLC3A2-NRG1 fusion in invasive mucinous adenocarcinoma of the lung. Oncotarget 2016;7:69450–69465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez-Cuesta L, Plenker D, Osada H, et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov 2014;4:415–422. [DOI] [PubMed] [Google Scholar]
- 6.Guma A, Martinez-Redondo V, Lopez-Soldado I, et al. Emerging role of neuregulin as a modulator of muscle metabolism. Am J Physiol Endocrinol Metab 2010;298:E742–750. [DOI] [PubMed] [Google Scholar]
- 7.Alsaid H, Skedzielewski T, Rambo MV, et al. Non invasive imaging assessment of the biodistribution of GSK2849330, an ADCC and CDC optimized anti HER3 mAb, and its role in tumor macrophage recruitment in human tumor-bearing mice. PLoS One 2017;12:e0176075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gay ND, Wang Y, Beadling C, et al. Durable Response to Afatinib in Lung Adenocarcinoma Harboring NRG1 Gene Fusions. J Thorac Oncol 2017;12:e107–e110. [DOI] [PubMed] [Google Scholar]
- 9.Cadranel J, Liu SV, Duruisseaux M, et al. Therapeutic Potential of Afatinib in NRG1 Fusion-Driven Solid Tumors: A Case Series. Oncologist 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin DH, Jo JY, Han JY. Dual Targeting of ERBB2/ERBB3 for the Treatment of SLC3A2-NRG1-Mediated Lung Cancer. Mol Cancer Ther 2018;17:2024–2033. [DOI] [PubMed] [Google Scholar]
- 11.Soule HD, Maloney TM, Wolman SR, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res 1990;50:6075–6086. [PubMed] [Google Scholar]
- 12.Schaefer G, Fitzpatrick VD, Sliwkowski MX. Gamma-heregulin: a novel heregulin isoform that is an autocrine growth factor for the human breast cancer cell line, MDA-MB-175. Oncogene 1997;15:1385–1394. [DOI] [PubMed] [Google Scholar]
- 13.Ramirez RD, Sheridan S, Girard L, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res 2004;64:9027–9034. [DOI] [PubMed] [Google Scholar]
- 14.Li GG, Somwar R, Joseph J, et al. Antitumor Activity of RXDX-105 in Multiple Cancer Types with RET Rearrangements or Mutations. Clin Cancer Res 2017;23:2981–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattar M, McCarthy CR, Kulick AR, et al. Establishing and Maintaining an Extensive Library of Patient-Derived Xenograft Models. Front Oncol 2018;8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Euhus DM, Hudd C, LaRegina MC, et al. Tumor measurement in the nude mouse. J Surg Oncol 1986;31:229–234. [DOI] [PubMed] [Google Scholar]
- 17.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Somwar R, Shum D, Djaballah H, et al. Identification and preliminary characterization of novel small molecules that inhibit growth of human lung adenocarcinoma cells. J Biomol Screen 2009;14:1176–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahman M, Jackson LK, Johnson WE, et al. Alternative preprocessing of RNA-Sequencing data in The Cancer Genome Atlas leads to improved analysis results. Bioinformatics 2015;31:3666–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benayed R, Offin M, Mullaney K, et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin Cancer Res 2019;25:4712–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014;20:1479–1484. [DOI] [PubMed] [Google Scholar]
- 22.Cheng DT, Prasad M, Chekaluk Y, et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med Genomics 2017;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trombetta D, Graziano P, Scarpa A, et al. Frequent NRG1 fusions in Caucasian pulmonary mucinous adenocarcinoma predicted by Phospho-ErbB3 expression. Oncotarget 2018;9:9661–9671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu CY, Chen TC, Chang CB, et al. Rehydration before wet fixation in conventional body fluid cytology - An 18-year experience. Cytopathology 2018;29:179–183. [DOI] [PubMed] [Google Scholar]
- 26.Murayama T, Nakaoku T, Enari M, et al. Oncogenic Fusion Gene CD74-NRG1 Confers Cancer Stem Cell-like Properties in Lung Cancer through a IGF2 Autocrine/Paracrine Circuit. Cancer Res 2016;76:974–983. [DOI] [PubMed] [Google Scholar]
- 27.Trombetta D, Rossi A, Fabrizio FP, et al. NRG1-ErbB Lost in Translation: A New Paradigm for Lung Cancer? Curr Med Chem 2017;24:4213–4228. [DOI] [PubMed] [Google Scholar]
- 28.Slotkin EK, Diolaiti D, Shukla NN, et al. Patient-Driven Discovery, Therapeutic Targeting, and Post-Clinical Validation of a Novel AKT1 Fusion-Driven Cancer. Cancer Discov 2019;9:605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jonna S, Feldman RA, Ou SH, et al. Characterization of NRG1 gene fusion events in solid tumors. Journal of Clinical Oncology 2020;38:3113. [Google Scholar]
- 30.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 31.Muscarella LA, Trombetta D, Fabrizio FP, et al. ALK and NRG1 Fusions Coexist in a Patient with Signet Ring Cell Lung Adenocarcinoma. J Thorac Oncol 2017;12:e161–e163. [DOI] [PubMed] [Google Scholar]
- 32.McCoach CE, Le AT, Gowan K, et al. Resistance Mechanisms to Targeted Therapies in ROS1(+) and ALK(+) Non-small Cell Lung Cancer. Clin Cancer Res 2018;24:3334–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nair AB, Jacob S. A simple practice guide for dose conversion between animals and human. J Basic Clin Pharm 2016;7:27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nair A, Morsy MA, Jacob S. Dose translation between laboratory animals and human in preclinical and clinical phases of drug development. Drug Dev Res 2018;79:373–382. [DOI] [PubMed] [Google Scholar]
- 35.Laskin J, Liu SV, Tolba K, et al. NRG1 fusion-driven tumors: biology, detection, and the therapeutic role of afatinib and other ErbB-targeting agents. Ann Oncol 2020;31:1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bishu K, Ogut O, Kushwaha S, et al. Anti-remodeling effects of rapamycin in experimental heart failure: dose response and interaction with angiotensin receptor blockade. PLoS One 2013;8:e81325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. IHC characterization of LUAD-0061AS3 PDX. IHC staining of LUAD-0061AS3pdx sample was done to confirm the origin of the PDX tissue. PR, ER, WT1, PAX8 and thyroglobulin were consistent with the original patient sample. Phospho-ERBB3 stain demonstrated membranous positivity, as expected in NRG1 fusion-positive tumors. Napsin-A, although weakly positive in the original patient sample, was negative in the PDX tissue, likely due to suboptimal activity of anti-mouse napsin-A antibody in mouse xenograft tissue.
Supplementary Figure S2. Bar graphs of the results of protein kinase array performed on LUAD-0061AS3 cells treated with GSK2849330 0.01 μM (GSK330 0.01 μM) and GSK2849330 0.1 μM (GSK330 0.1 μM). *p < 0.05, **p < 0.01. Images were captured on x-ray films and then quantitated by densitometry. Results represent the mean ± SD of two measurements.
Supplementary Figure S3. A. qPCR analysis of RPTOR expression in HBEC-DNP53, LUAD-0061AS3cl, MCF10A and MDA-MB-175-VII. B. GSEA plot for MTOR gene sets in NRG1 fusion-positive cell lines as compared to normal lung tissue.
Supplementary Figure S4. Effect of AKT inhibitors on cell growth. Cells were treated with the pan-AKT inhibitors ipatasertib and MK2206 for 96 h and then viability determined. Data represent the mean ± SD of 3 replicate determinations in one experiment.
Supplementary Figure S5. Area under the curve analysis of tumor volume and animal weight A-B. The data shown in Figure 6A and C were analyzed by area under the curve analysis to compare treatment effects. AUC values were calculated for day 11 and 12 respectively. C-D. Animal weight over treatment course. Tumor bearing mice were weighed twice weekly. Data represent the mean ± SE. ****p < 0.0001. ns: not significant (p > 0.05).
Supplementary Table S1. Antibody source and specificity used in Western blotting and IHC. Excel File.
Supplementary Table S2. Genomic Characterization of PDX and Cell line models. Models were profiled by MSK-IMPACT. Excel file.