Abstract
The cancer-preventive activities of quercetin have been extensively studied in invasive breast cancer; however, the role of quercetin on triple-negative breast cancer (TNBC) with overexpression of insulin-like growth factor-1 receptor (IGF1R) has not been resolved. In this article, we demonstrated that quercetin inhibited the activation of IGF1R and its downstream kinases Akt and Erk1/2 in a dose-dependent manner in human MDA-MB-231 breast cancer cells (TNBC cell line). Related to this, quercetin markedly suppressed the metastatic phenotype and epithelial–mesenchymal transition (EMT) of MDA-MB-231 cells by inhibiting the expression of EMT transcription factors Snail and Slug. Quercetin also increased the secretion of IGF-binding protein-3 in the conditioned medium of MDA-MB-231 cells while reducing the secretion of IGF1; thus, quercetin interrupted the autocrine or paracrine loop of IGF1 signaling. In xenograft mouse models, the administration of quercetin blocked the growth of MDA-MB-231 tumor xenografts and their lung metastasis, accompanied by the inactivation of IGF1R and the downregulation of the expression of Snail, Slug, and mesenchymal markers fibronectin and vimentin. These results suggest that quercetin has cancer-preventive value for TNBC by inhibiting IGF1/IGF1R signaling and preventing the consequent EMT and metastasis of TNBC.
Keywords: Quercetin, MDA-MB-231 cells, IGF1R, IGF1, EMT
1. Introduction
About 15% to 20% of patients with breast cancer are diagnosed as triple-negative breast cancer (TNBC) phenotype, which is characterized by the lack of the expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). TNBC tends to be aggressive and could be predicted for a substantial association with relapse and unfavorable outcome [1,2]. Given the absence of target receptors, TNBC is resistant to current hormone therapy and HER2-targeted therapy and has been considered a clinical challenge [3]. Therefore, developing new effective chemotherapeutics or targeted therapies for the treatment of advanced TNBC is urgently needed.
Recent epidemiological evidences suggest that elevated levels of circulating insulin-like growth factor-1 (IGF1) are highly linked to the increased recurrence and risk of all-cause mortality in female breast cancer [4–6]. The overexpression or high activation of IGF1 or IGF1 receptor (IGF1R) increases cancer incidence and progression with reduced response to treatment and survival rate because the IGF1 signaling pathway is involved in cell proliferation, adhesion, and anchorage-independent growth, which eventually result in cancer cell invasion and metastatic spreading [7]. About 22% to 46% of TNBCs overexpress IGF1R protein [8], and the IGF1/IGF1R signaling pathway, which increases cell proliferation and promotes cell survival, may be active in TNBC cell lines [9]. TNBC cell lines and xenografts with high IGF1R and IGF1R-induced gene expression are more sensitive to anti-IGF1R therapy in combination with docetaxel [10]. These findings indicate that IGF1/IGF1R may act as a prospective anticancer candidate for targeted therapy in patients suffering from TNBC [11].
IGF1R is a typical receptor of tyrosine kinase and can transmit signals from insulin receptor substrate-1 (IRS1) and Src homology 2-containing protein (SHC). Activated IRS1 recruits phosphoinositide 3-kinase (PI3K) to activate the PI3K–Akt signaling pathway whereas SHC initiates the mitogen-activated protein kinase (MAPK) signaling cascade to promote cell proliferation and survival [12]. Furthermore, IGF1R signaling also plays a critical role in the initiation of invasion and metastasis and consequent cancer mortality through the induction of epithelial–mesenchymal transition (EMT) program [13]. EMT is a process of trans-differentiation that endows cancer cells with mobile and cancer cell stem-like phenotypes during tumor dissemination [14–16]. EMT-relevant changes in cell polarity and motility are accompanied by the loss of expression or mislocalization of cell junction proteins, such as E-cadherin, zonula occludens (ZO-1), keratin 18, and keratin 19, and the acquisition of the expression of mesenchymal-like proteins, such as fibronectin and vimentin [17]. IGF1 triggers the EMT program, which results in metastasis in breast, lung, gastric, and prostate cancers [18–24]. The overexpression of IGF1R leads to the depolarization and mesenchymal-like transition of breast carcinoma cells in response to IGF1 [19]. The PI3K–Akt and MAPK pathways promote IGF1-stimulated EMT progression by upregulating EMT transcription factors, such as zinc finger E-box binding homeobox (ZEB)1 and ZEB2 [22,24], or stabilizing EMT transcription factor Snail by the inactivation of glycogen synthase kinase-3β (GSK-3β) [25]. These findings provide molecular basis for new drug development by targeting IGF1R signaling pathways.
Quercetin (3,3′,4′,5,7-pentahydroxy-flavone), a naturally occurring flavonoid, is a dietary antioxidant with substantial anti-biofilm, anti-tumor, antiallergic, and anti-inflammatory activities [26,27]. Quercetin has cancer chemopreventive activity in colon, prostate, and lung cancers and exerts anti-metastatic effect on breast cancer [28,29]. Quercetin can inhibit a broad range of protein kinases, including PI3K, and is the basis for the structural design of the synthetic PI3K inhibitor LY294002 [30,31]. In H-Ras-transformed MCF10A cells, quercetin potently blocks H-Ras-induced invasion and migration by directly inhibiting PI3K activity and thereby suppressing the activity of matrix metalloproteinase (MMP)2 and MMP9 [28]. Quercetin can inhibit cell proliferation and EMT or induce apoptosis in different cancer cells by modulating the cell signaling pathways elicited by IGF1, hepatocyte growth factor, and epidermal growth factor (EGF) [32,33]. In addition, Srinivasan et al. reported that quercetin reverses EMT and suppresses the migratory ability of TNBC by preventing the nuclear localization of β-catenin [34]. However, whether quercetin regulates the IGF1R signaling pathway to inhibit EMT and the metastasis of TNBC remains unclear. Thus, we used a TNBC cell line, human MDA-MB-231 cells, to determine whether quercetin suppresses the invasive phenotypes inMDA-MB-231 cells by mediating the IGF1/IGF1R signaling pathway and also validated the underlying mechanisms by cell-based and xenograft animal models.
2. Materials and methods
2.1. Materials
Recombinant human IGF1 and IGF-binding protein-3 (IGFBP3) were obtained from PeproTech, Inc. (Rocky Hill, NJ). Quercetin, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), dimethyl sulfoxide (DMSO) and Bouin’s solution were obtained from Sigma Chemical Co. (St. Louis, MO). Snail and Slug siRNAs were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). 4′,6-Diamidino-2-phenylindole (DAPI) was purchased from Southern Biotech (Birmingham, AL). Picropodophyllin (PPP) was available from MedChem Express (Monmouth Junction, NJ).
2.2. Cell culture and treatment
Two human breast cancer cell lines MCF-7 (a luminal A breast cancer cell line with ER and PR expression) and MDA-MB-231 (a TNBC cell line) characterized as low and high metastatic phenotypes, respectively, were purchased from American Type Culture Collection. Both breast cancer cell lines were grown in Dulbecco’s minimal essential medium (DMEM) containing 10% fetal bovine serum (FBS; Gibco BRL, Grand Island, NY), 100 units/mL of penicillin, and 100 μg/mL of streptomycin (Gibco BRL, Grand Island, NY) and incubated in a humidified atmosphere at 37 °C with 5% CO2. For experiments, cells were harvested from the culture monolayers at 80%–90% confluence and then rinsed with cold phosphate-buffered saline (PBS). Viable cells were isolated using 0.25% trypsin containing 0.1% EDTA, centrifuged at 1000 rpm for 5 min, and then resuspended in growth medium. Finally, the cells were subcultured and then processed as described in the following sections.
2.3. Western blot
After treatment with various concentrations of quercetin as indicated in the figure legends, cell lysates were prepared by incubating the harvested cells with a homobuffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.5 mM phenylmethanesulfonylfluoride, 0.5 mM dithiothreitol, and 0.5% NP-40) for 30 min at 4 °C. Then 50 μg of the total cellular proteins of each sample were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Immobilon P, Millipore, Billerica, MA). Western blot was performed by incubation with primary antibodies followed by horseradish peroxidase (HRP)-conjugated secondary antibodies. The immune complexes were stained with enhanced chemiluminescence kits (Amersham, UK), and band density was calculated using a Fujifilm LAS-4000 luminescent image analyzer (Fujifilm UK Ltd., Bedford, UK). Antibodies against IGF1, IGF1R, phospho-IRS1 (Ser307), IRS-1, phospho-Akt (Ser 473), Akt, phospho-Erk1/2 (Thr 202/Tyr 204), Erk1/2, vascular endothelial growth factor (VEGF), ZO-1, Oct4, Sox2, and Nanog were purchased from Cell Signaling Technology (New England Biolabs, Ipswich, MA). Antibodies against E-cadherin and vimentin were purchased from Millipore (Billerica, MA). IGFBP3, Snail, Slug, and phospho-IGF1R (Tyr 1161) antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). β-Actin antibody was sourced from Sigma Chemical (Sigma, St. Louis, MO). In order to determine the secreted protein in the condition medium, MDA-MB-231 cells (5 × 105 cells/mL) were treated with the indicated quercetin for 24 h, and the protein concentration in the condition medium was measured by the Bradford method to ensure that the load in the Western blot was equal.
2.4. Wound healing assay
A linear wound was created on the cultured cell monolayer with approximately 90% confluence using a sterile 200 μL micropipette tip. Then, the cells were rinsed with cold PBS to remove the detached cells and re-cultured in 1% FBS–DMEM. The cells were treated with a variety of concentrations of quercetin for 24 h, and the closure of wound edge was measured with a microscope.
2.5. In vitro invasion assay
MDA-MB-231 cells (2.5 × 104) were plated in the upper chamber of a Corning Transwell (cat. no. 3422, Corning Incorporated, New York, NY) with polycarbonate film (8-μm pores) covered with Matrigel (BD Biosciences, San Jose, NJ) in DMEM containing 0.5% FBS at 37 °C. After treatment with quercetin, the cells remaining in the upper side of the membrane were scraped off with a cotton swab. The number of cells that invaded the lower surface of the membrane was visualized by propidium iodide staining and counted in at least five random fields using a fluorescence microscope (Carl Zeiss MicroImaging GmbH, Jena, Germany).
2.6. Cell adhesion assay
Cell adhesion assay was performed based on a colorimetric method, in which extracellular matrix (ECM)-adherent cells are stained with crystal violet dye. Briefly, a 6-well plate was coatedwith 10 μg/mL of collagen or 20 μg/mL of fibronectin for 1 h at 4 °C and then blocked with 1% bovine serum albumin in DMEM for 1 h before seeding. Then, MDA-MB-231 cells pretreated with quercetin for 24 h at a density of 1 × 105 cells were plated in fibronectin-coated plate by incubation with 10% FBS-containing DMEM at 37 °C for 1 h. After washing with PBS to remove nonattached cells, the adherent cells were fixed using 70% ethanol, stained with 0.1% crystal violet, and then visualized using a microscope (magnification 200 times). The stain of the adherent cells was extracted in 0.2% Triton X-100 and further quantified colorimetrically by measuring the absorbance at 550 nm.
2.7. Immunofluorescence staining
MDA-MB-231 cells were seeded on coverslips to 60% confluence and then treated with various concentrations of quercetin for 24 h. For F-actin staining, the cells were washed with cold PBS, fixed with 4% paraformaldehyde for 20 min at room temperature, and labeled with Alexa Fluor 488-conjugated Phalloidin (Molecular Probes, Eugene, OR) (1:10,000). For immunofluorescence staining, the cells were fixed with 4% paraformaldehyde; immunolabeled with primary antibody, such as mouse monoclonal anti-vimentin, anti-fibronectin (Santa Cruz Biotechnology, CA), and anti-keratin 18 or anti-keratin 19 (Cell Signaling Technology, Danvers, MA), at 4 °C overnight in the dark; incubated with secondary antibodies, including rabbit anti-mouse secondary antibody conjugated with fluorescein isothiocyanate (FITC; Sigma, St. Louis, MO) for 30 min at room temperature. The cells were then mounted with DAPI–Fluoromount-G (Southern Biotech, Birmingham, AL), and their fluorescent images were observed using a fluorescence microscope (Carl Zeiss MicroImaging GmbH, Jena, Germany).
2.8. Cell viability assay
Breast cancer cells with a density of 5 × 103 cells/mL were grown overnight in a 96-well plate and then treated with various concentrations of quercetin for 24 h. After incubation, the cell viability was determined by the following MTT assay method. 20 μL of MTT solution (5 mg/mL) was added to each well and incubated for 30 min at 37 °C. Then the supernatant was discarded, the MTT-formazan crystals formed by the metabolism of living cells were dissolved in 200 μL DMSO and the absorbance of the dissolved crystals at a wavelength of 570 nm was measured by the colorimetric method.
2.9. Quantitative reverse transcription-polymerase chain reaction (Q-PCR)
Total RNA was extracted using TRIzol reagent (Sigma, St. Louis, MO) according to the manufacturer’s instructions. Briefly, total RNA (5 μg) was reverse-transcribed into cDNA by incubating with the reaction mixture (25 μL) for 90 min at 40 °C using Moloney murine leukemia virus (MMLV) reverse transcriptase with oligo-dT18 primer. The sequences of the forward and reverse primers used are listed in Table 1. Q-PCR was performed in a final volume of 20 μL using an optimized concentration of each primer pair with the Fastart Universal SYBR Green Master Mix (Roche, Indianapolis, IN), and the products were analyzed with an ABI 7000 sequence detection system.
Table 1.
Primers for quantitative RT-PCR and cloning snail and slug promoters.
| Gene symbol | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| Quantitative qPCR primers | ||
| Snail | GGCGTGTGCTCG GACCTTCT | AGGCAGGGGCAGGTATGGAG |
| Slug | CCTTCTCCAGAATGTCTCTC | ATTTGGTTGGTCAGCACAGGAGA |
| Zeb1 | GCACAACCAAGTGCAGAAGA | CATTTGCAGATTGAGGCTGA |
| GAPDH | GAGTCAACGGATTTGGTCGT | GACAAGCTTCCCGTTCTCAG |
| Primers for promoters cloning | ||
| Snail | AAGCGCTCAGACCACCGGGC | GGGAAGAGACTGAAGTAGAGGAGA |
| Slug | TTGTGCAAGGCAAACCTCTC | GTATGACAGGCATGGAGTAACTCTC |
2.10. Luciferase assay
The promoter regions of Snail (−350 to +1126) and Slug (−450 to +1035) were cloned into a yT&A vector (Yeastern Biotech, Taiwan) according to the procedure described by Chen Y. et. al. [35]. The upstream regions of Snail (−350 to +84) and Slug (−450 to +164) were subsequently cloned into a firefly luciferase expression vector (pGL3; Promega, Madison, WI). Then, 30 ng of pGL3-basic, pGL3-Snail promoter, or pGL3-Slug promoter vectors were co-transfected with 30 ng of βgalactosidase expression vector (pCH110, internal control vectors; Addgene, Watertown, MA) into MDA-MB-231 cells (at a density of 5 × 104 cells per well in 6-well culture plate) using a Turbofect kit (Fermentas, Carlsbad, CA) following the manufacturer’s instructions. After 16 h of transfection, the cells were exposed to quercetin (0, 5, 10, and 20 μM) for an additional 24 h. Cell lysates were then collected and assayed for normalized luciferase activity using a luciferase assay kit (Promega, Madison, WI).
2.11. Animal care and xenograft study
All procedures in the animal experiments were approved by Taiwan University’s Institutional Animal Care and Use Committee. Briefly, 4-week-old female SCID mice were purchased from BioLASCO Experimental Animal Center (Taiwan Co., Ltd.). After 1 week of adaptation, the abdominal hair of each mouse was shaved the day before the start of the experiment. 5-Week-old mice were injected with MDA-MB-231 cells (2 × 106 cells/0.1 mL Hanks’ balanced salt solution) into the bottom left mammary fat pad. After 3 days of transplantation, the mice were randomly divided into two groups: control group and experimental group. Each group had six mice. The mice were injected intraperitoneally (i.p.) with an aliquot of corn oil (control group) or with 20 or 50 mg/kg quercetin (experimental group) five times per week for 42 days. Then, the mice were sacrificed for further histological and immunohistochemical studies as described below.
2.12. Histological analysis
For histological studies, the lung tissues were preserved in 10% neutral buffered formalin and embedded in paraffin. Then, the paraffin-embedded tissues were cut into 4 μm-thick sections and stained with hematoxylin and eosin. Histological patterns were observed using a microscope.
2.13. Immunohistochemistry (IHC)
The 3-μm tumor sections were deparaffinized and rehydrated by treatment with 3% hydrogen peroxide for 15 min to block endogenous peroxidase. The sections were cooked under pressure in 10 mM citrate buffer at pH 6.0 (ImmunoDNA retriever citrate, BIO SB, Santa Barbara, CA) for antigen retrieval. The sections were then incubated with primary antibody against phospho-IGF1R (pIGF1R), IGF1R, Snail, vimentin, keratin 18 and keratin 19 (diluted 1:200 in PBS) overnight and incubated with biotinylated secondary antibody for 30 min. The tissue sections were further reacted with streptavidin–HRP for 30 min, then colored with 0.05% 3,3′-diaminobenzidine tetrahydrochloride, and observed under a light microscope.
2.14. Statistical analysis
Experimental data were obtained via three or more repeated experiments and expressed as mean ± standard error of the mean (SEM). One-way ANOVA post hoc Tukey’s b test was used to evaluate multiple comparison, and the test was considered significantly different when p < 0.05.
3. Results
3.1. Quercetin inhibits the activation of IGF1R and its downstream signaling pathways in MDA-MB-231 cells
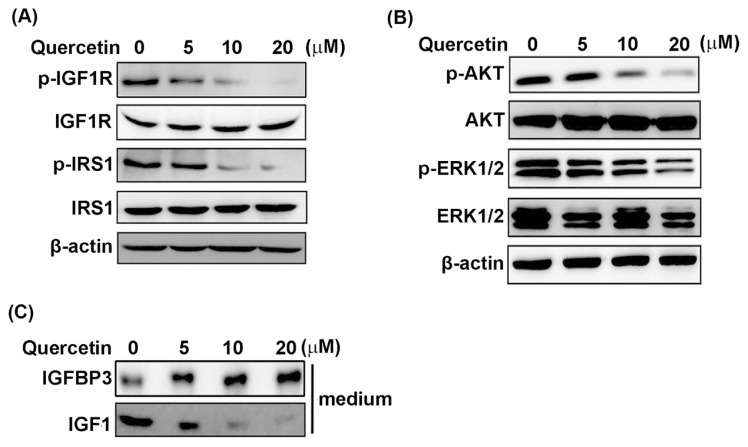
The expression and phosphorylation status (activation) of IGF1R and its related downstream signaling molecules, including IRS1, Akt, and Erk1/2, were examined by Western blot to elucidate whether quercetin affects the IGF1R-mediated signaling in MDA-MB-231 cells. As shown in Fig. 1A, quercetin reduced the phosphorylation statuses of IGF1R and IRS1 in a dose-dependent manner while downregulating the phosphorylation statuses of Akt and Erk1/2 (Fig. 1B). Quercetin also reduced IGF1 secretion in conditioned medium in a concentration-dependent manner but increased the accumulation of IGFBP3 (Fig. 1C). These results indicate that quercetin can downregulate IGF1R-mediated signaling, which is correlated with the reduced autocrine/paracrine action of IGF1 in MDA-MB-231 cells.
Fig. 1.
Downregulation of IGF1R phosphorylation and its downstream signaling by quercetin in MDA-MB-231 cells. MDA-MB-231 cells were treated with the indicated concentrations of quercetin for 24 h. (A) Effect of quercetin on the phosphorylation status of IGF1R and IRS1 was determined by Western blot. (B) The phosphorylation status of two main IGF1R-downstream signaling kinases, AKT and ERK, was also analyzed by Western blot. (C) The secretion of IGFBP3 and IGF1 proteins in conditioned medium were monitored by Western blot in quercetin-treated MDA-MB-231 cells. β-Actin was used as internal control in Western blot, and all data are the mean of at least three measurements.
3.2. Quercetin suppresses the invasive phenotypes of MDA-MB-231 Cells
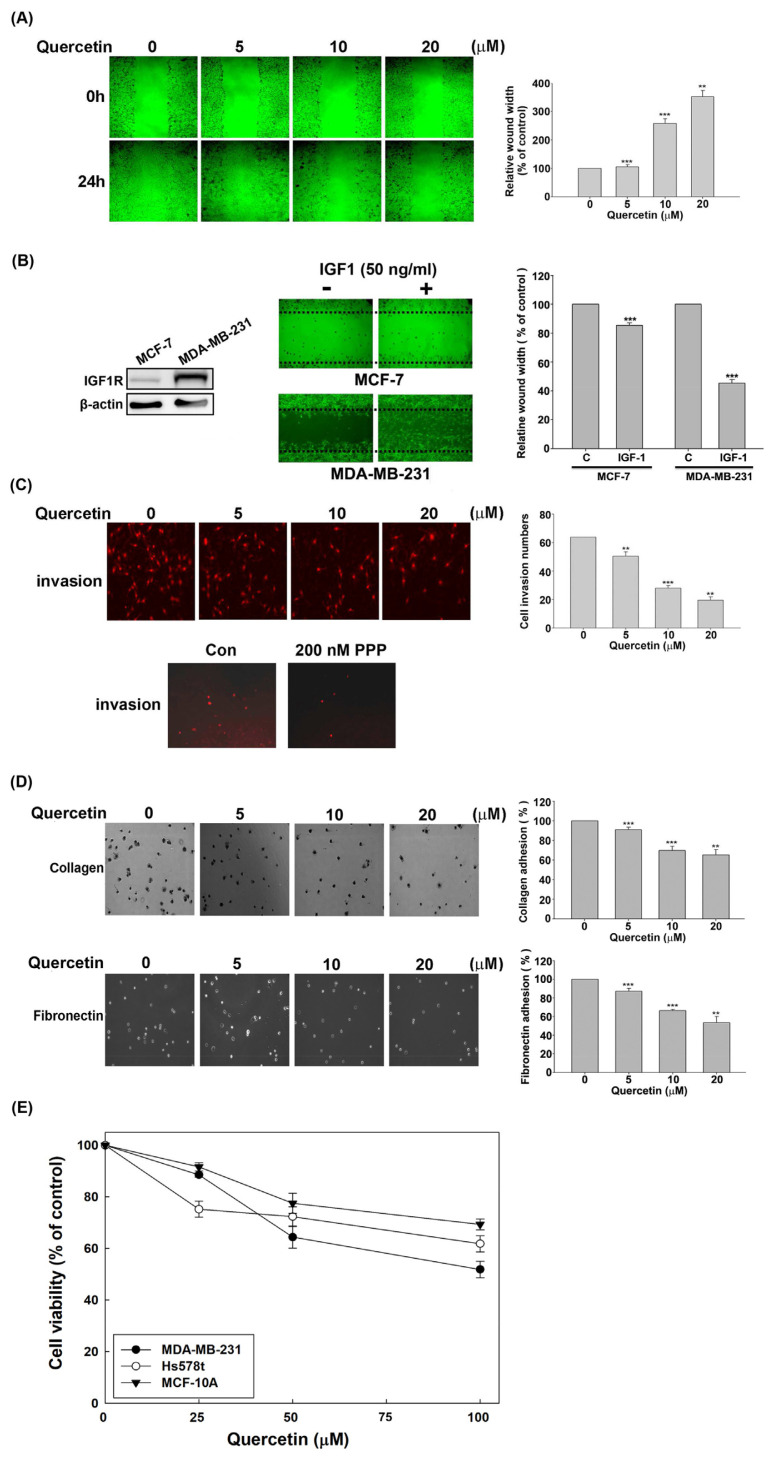
A scratch wound healing assay was performed to decipher the effect of quercetin on cell motility to assess whether the suppression of IGF1R signaling by quercetin is tightly correlated with the downregulation of the invasive phenotypes of MDA-MB-231 cells. Fig. 2A displays that quercetin remarkably exhibited a dose-related delay in wound healing after 24 h of incubation with 1% FBS in MDA-MB-231 cells. Fig. 2B shows that IGF1R was overexpressed in MDA-MB-231 cells compared with MCF-7 cells and IGF1 has the ability to drive the wound healing of MDA-MB-231 cells but has no substantial effect on the wound healing of MCF-7 cells. This result demonstrated that IGF1/IGF1R signaling is essential for the wound closure of MDA-MB-231 cells. Then, invasion assay using Matrigel-coated Transwell was performed to evaluate the effect of quercetin on the invasive ability of MDA-MB-231 cells. The result showed that quercetin caused a concentration-dependent decrease in Transwell invasion cells with a reduction from ~20% to ~64% in response to quercetin treatment (Fig. 2C). Fig. 2C (lower panel) displayed that picropodophyllin (PPP), a selective IGF1R inhibitor, suppressed the Transwell invasion of MDA-MB-231 cells, indicating that IGF1R signaling can drive the invasion of MDA-MB-231 cells. In addition, the culture dishes were pre-coated with type I collagen or fibronectin, and the degree of cell adhesion was evaluated by colorimetry to verify whether quercetin affects cell adhesion. The data show that quercetin remarkably reduced the MDA-MB-231 cells that adhered to the tested ECM in a dose-dependent manner (Fig. 2D).
Fig. 2.
Inhibition of the aggressive invasion phenotypes of MDA-MB-231 cells by quercetin. (A) Wound healing assay was performed by treatment with the indicated doses of quercetin for 24 h in MDA-MB-231 cells after scraping a 90% confluent culture into a wound. (B) After 90% confluent cells were scraped with a pipette tip to produce a wound, the cells were incubated with IGF1 (50 ng/mL) for 24 h, and the images of the wound were further examined using a microscope. The quantified data represent the relative distance of the wound width, which is expressed as the measured wound width divided by the width at the beginning of the wound. The left panel represents the expression of IGF1R in MCF-7 and MDA-MB-231 cells determined by Western blot, and β-actin was used as internal control. (C) Invasion assay was performed using a diluted Matrigel-coated Transwell. After 24 h of cell culture in the upper chamber of the Transwell, the MDA-MB-231 cells were exposed to the indicated concentrations of quercetin or 200 nM PPP (IGF1R inhibitor) for an additional 24 h. The gel-invading cells on the lower surface of the filter were stained and quantified by microscopy with 10 random fields per well. (D) MDA-MB-231 cells pretreated with quercetin for 24 h were cultured in 24-well plates coated with 10 μg/mL of collagen or 20 μg/mL of fibronectin for 1 h. Adherent cells were counted by colorimetry using a microscope. (E) The indicated cell lines were treated with 0, 25, 50, or 100 μM quercetin for 24 h and cell viability was measured using a MTT-based method. All quantified data was derived from the statistics of at least three repeated experiments and expressed as mean ± SEM. Significant difference compared with the control group was signed as asterisks (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
In order to rule out that the anti-invasive effect of quercetin in MDA-MB-231 cells was due to the cytotoxicity of quercetin, the cell viability in MDA-MB-231 cells treated with quercetin was determined by MTT method. Fig. 2E shows that quercetin slightly inhibited the proliferation of MDA-MB-231 cells at low concentrations (<25 μM), but the degree of inhibition was remarkably lower than that of quercetin on motility and invasiveness (Fig. 2A and C, respectively).Meanwhile, we also determined the cytotoxic effect of quercetin on MCF-10A cell line (non-tumorigenic breast epithelial cells) and Hs578t cell line (TBNC) by MTT assay to evaluate the biological safety of quercetin. We found that quercetin has a low growth inhibitory effect on MDA-MB-231, Hs578t and MCA-10A cells, and the IC50 of all cells exceeds 100 μM. In particular, non-carcinogenic MCF-10A cells are more resistant to quercetin than the two TNBC cell lines MDA-MB-231 and Hs578t cells, which indicates that quercetin is selective against TNBC and suggested to be less toxic to normal cells. Taken together, the results confirmed that the inhibition of IGF1R signaling by quercetin is closely related to the downregulation of the invasiveness and migration of MDA-MB-231 cells.
3.3. Quercetin reverses the EMT program by the acquisition of cancer stem cell-like (CSCL) phenotype in MDA-MB-231 cells
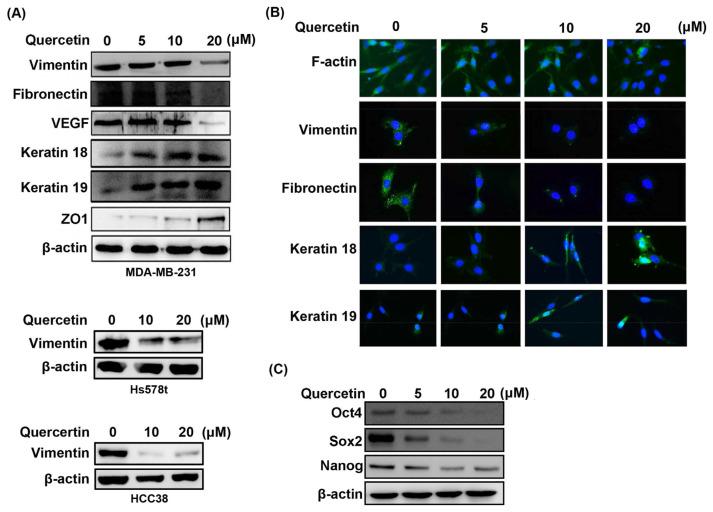
EMT is considered a prerequisite for IGF1/IGF1R-induced cancer cell migration and metastatic potential [36]. The expression levels of EMT markers were monitored by Western blot in quercetin-treated MDA-MB-231 cells to explore whether the downregulation of IGF1R signaling pathway and the invasive phenotype of MDA-MB-231 cells by quercetin are accompanied by EMT reversal. Fig. 3A shows that the protein levels of mesenchymal markers, including vimentin, fibronectin, and VEGF, decreased in a dose-dependent manner in quercetin-treated cells. By contrast, the protein levels of epithelial-like markers, such as keratin 18, keratin 19, and ZO1, increased in a dose-dependent manner after quercetin treatment. Quercetin also dose-dependently inhibited the protein expression of vimentin in the other two TNBC cell lines Hs578t and HCC38, demonstrating that the inhibition of quercetin on EMT of TNBC cells is universal, not only for MDA-MB-231 cells (Fig. 3A). Moreover, immunofluorescence staining revealed that the high concentration of quercetin reduced the formation of stress fibers assembled by F-actin and induced the localization of F-actin at the cell junction (Fig. 3B). Consistent with Fig. 3A, immunofluorescence images show that quercetin decreased the protein levels of vimentin and fibronectin and increased the protein levels of keratin 18 and keratin 19 in MDA-MB-231 cells (Fig. 3B). The EMT program confers metastasis to cancer cells and promotes the development of cancer cells into an undifferentiated state with drug resistance, which are possessed by stem cells [37]. We further investigated whether the decrease in mesenchymal-like phenotype caused by quercetin is correlated with a possible decrease in the stemness of the phenotype of MDA-MB-231 cells. We assessed the expression of embryonic stem cell transcription factors Oct4, Sox2, and Nanog by Western blot and found that quercetin diminished the expression of Oct4, Sox2, and Nanog in a concentration-dependent manner (Fig. 3C), which was accompanied by the reversal of EMT and the suppression of the invasion phenotype in MDA-MB-231 cells. These findings implicated that quercetin leads to the reversal of EMT in MDA-MB-231 cells.
Fig. 3.
Reversal of EMT and inhibition of expression of CSCL markers by quercetin in TNBC cell lines. (A) MDA-MB-231, Hs578t and HCC38 cells were treated with quercetin in the quantified dose for 24 h, and the protein expression of EMT markers was examined by Western blot. (B) Immunofluorescence by probing F-actin with Alexa Fluor 488 phalloidin (green) or by labeling vimentin, fibronectin, keratin18, or keratin19 with secondary antibody conjugated with FITC (green). Nuclei were stained with DAPI (blue). (C) The expression of Oct4, Sox2, and Nanog in MDA-MB-231 cells were determined by Western blot after 24 h of quercetin treatment. β-Actin was used as internal control. All of the measurements were performed at least three times.
3.4. Quercetin downregulates the expression of Snail and Slug transcription factors required to maintain the mesenchymal phenotype of MDA-MD-231 cells
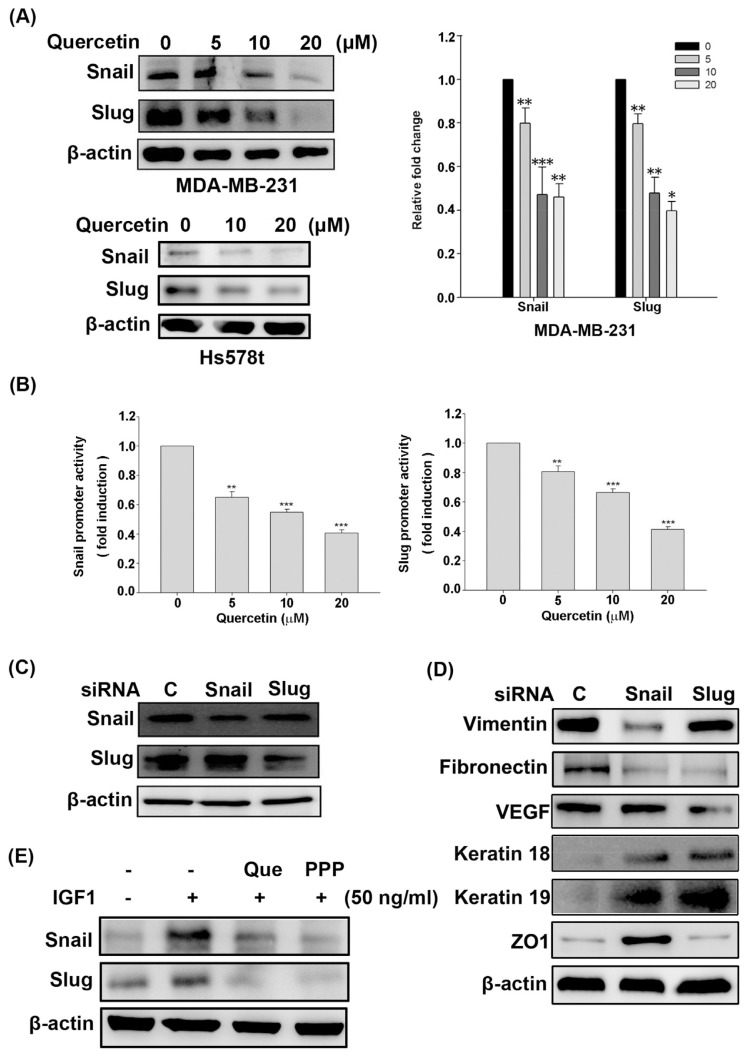
EMT-driven transcription factors, such as Snail and Slug, orchestrate EMT and induce breast cancer entrance into the tumor-initiating and metastatic states [38]. The expression of Snail and Slug was verified in quercetin-treated MDA-MB-231 cells to further demonstrate whether quercetin affects the EMT profile of MDA-MB-231 cells through a Snailor Slug-dependent mechanism. As shown in Fig. 4A, quercetin dose-dependently inhibited the protein and mRNA expression of Snail and Slug in MDA-MB-231 cells. Quercetin also inhibited the protein expression of Snail and Slug in Hs578t cells in Fig. 4A (bottom left). Consistently, quercetin markedly inhibited Snail and Slug promoter activities in MDA-MB-231 cells in a concentration-dependent fashion (Fig. 4B). Relative siRNA was applied to silence the expression of Snail and Slug in MDA-MB-231 cells to explore whether Snail and Slug are essential for the EMT of MDA-MB-231 cells. Fig. 4C demonstrates that specific siRNA transfection efficiently inhibited the protein expression of Snail and Slug. In parallel, the expression of epithelial markers keratin 18, keratin 19, and ZO1 was upregulated, whereas the expression of mesenchymal markers vimentin, fibronectin, and VEGF was downregulated in Snail or Slug siRNA-transfected MDA-MB-231 cells (Fig. 4D). The protein levels of these two EMT transcription factors were determined by Western blot in IGF1-stimulated cells to further verify whether IGF1/IGF1R signaling is responsible for Snail and Slug expression in MDA-MB-231 cells. Fig. 4E also shows that IGF1 dramatically triggered the Snail and Slug expression, and that quercetin and PPP can inhibit IGF1-stimulated Snail and Slug expression in MDA-MB-231 cells. Overall, these results suggest that quercetin inhibited the EMT and aggressive phenotype of MDA-MB-231 cells by blocking IGF1/IGF1R-mediated Snail and Slug expression.
Fig. 4.
Effects of quercetin on the expression of Snail and Slug required for EMT in MDA-MB-231 cells. (A) After 24 h of exposure to quercetin, the protein and mRNA expression levels of Snail and Slug in the indicated breast cancer cells were monitored by Western blot (left panel) and Q-PCR (right panel), respectively. The relative mRNA level of Snail or Slug is expressed as the measured transcript level divided by glyceraldehyde 3-phosphate dehydrogenase’s transcript level. (B) The pGL3-Snail or pGL3-Slug promoter vector was co-transfected into MDA-MB-231 cells for 16 h with the β-galactosidase reporter vector. Then, the transfected cells were treated with the indicated concentration of quercetin for 24 h. Snail or Slug promoter activity was determined by luciferase reporter assay and normalized to β-galactosidase activity. (C) MDA-MB-231 cells were transfected with siRNA against Snail or Slug for 24 h. The expression of Snail and Slug was analyzed by Western blot. (D) The effect of Snail or Slug siRNAs on the expression of epithelial markers (keratin18, keratin 19, and ZO-1), and mesenchymal markers (vimentin, fibronectin, and VEGF) was monitored by Western blot. (E) Fifty-percent confluent MDA-MB-231 cells were incubated for 12 h under serum-free conditions. The cells were pretreated with 20 μM quercetin (Que) or 200 nM PPP for 30 min, then stimulated with IGF1 (50 ng/mL) for 18 h and subjected to Western blot to analyze the expression of Snail and Slug. All data measurement were performed at least three times and considered significantly different compared with the control group (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
3.5. Quercetin suppresses xenograft tumor growth and metastasis by suppressing IGF1R signaling pathway and inducing EMT reversal in vivo
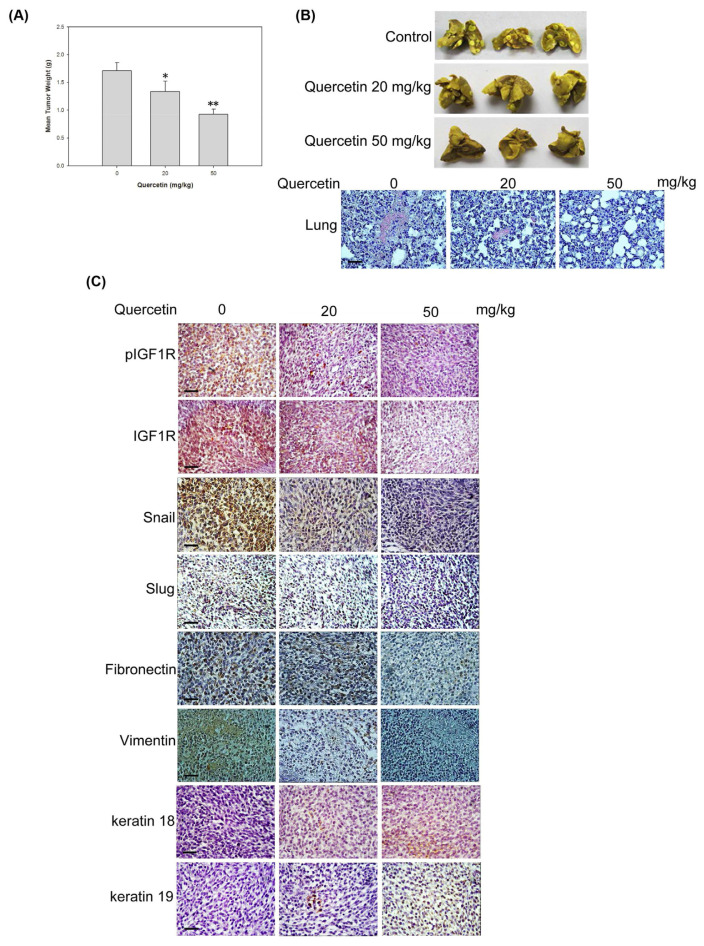
MDA-MB-231 cells were implanted into the mammary fat pad of female SCID mice with an i.p. injection of 20 or 50 mg/kg quercetin to elucidate whether quercetin blocks IGF1-induced invasion and metastasis in vivo. No detectable toxicity and weight loss were observed in mice after the administration of quercetin (data not shown). In addition, the transplanted mice showed no signs of adverse health effects and pain. After 42 days of quercetin administration, the mice were sacrificed to assess tumor weight and the extent of breast cancer metastasis in the lungs. Fig. 5A exhibits that quercetin treatment at the dosages of 0, 20, and 50 mg/kg gradually reduced xenograft tumor weights to 1.696 ± 0.184, 1.358 ± 0.398, and 0.934 ± 0.257 g, respectively. The histological examination of the overall appearance of the pulmonary nodules after fixation and hematoxylin and eosin staining revealed that quercetin markedly decreased the degree of pulmonary nodules and lung metastases in xenograft mice (Fig. 5B). As expected, quercetin downregulated the expression of pIGF1R; IGF1R; and mesenchymal-like markers, such as Snail, Slug, fibronectin, and vimentin, and conversely upregulated the expression of epithelial-like markers, such as keratin 18 and keratin 19, in xenograft tumor tissues (Fig. 5C). The results suggest that quercetin prevented the xenograft tumors from undergoing EMT by inhibiting IGF1R signaling and reversing EMT program; therefore, metastasis was inhibited in vivo (see Fig. 6).
Fig. 5.
Antitumor and anti-metastatic activity of quercetin in MDA-MB-231 xenograft mouse model. Female SCID mice were transplanted with MDA-MB-231 cells (2 × 106 cells) in the mammary fat pad and treated with quercetin (20 or 50 mg/kg, i.p.) for 42 days. (A) Tumor weight was measured after 42 days of quercetin treatment. Data are expressed as mean ± SEM (n = 6). *p < 0.05 and **p < 0.01 present that quercetin administration is significantly different from the vehicle control. (B) The lungs were removed at 42 days and fixed with 10% neutral buffered formalin for 24 h. The overall appearance of the pulmonary nodules was observed under a light microscope after hematoxylin and eosin staining (Scale bar = 100 μm). (C) Xenograft tumor tissues were removed and fixed with 10% formalin for 24 h. The expression of indicated proteins was determined in triplicate by IHC in xenograft tumor specimens (Scale bar = 100 μm).
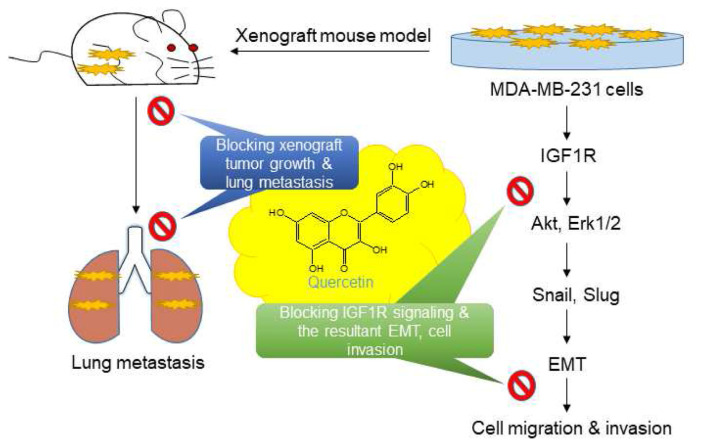
Fig. 6.
Inhibitory effects of quercetin on IGF1R signaling and the consequent EMT and invasion in MDA-MB-231 cells and xenograft mouse model.
4. Discussion
Despite our understanding of the highly aggressive and metastatic nature of the triple-negative subtype of breast cancer, we have yet to develop truly effective therapies to target this subset of disease. The development of many new therapies is focused on targeting primary breast tumor [39]; however, the major deaths from TNBC are due to distant metastasis. Targeting IGF1/IGF1R signaling has been considered a promising strategy for treating breast cancers, including TNBC, because breast cancer cells that overexpress IGF-IR exhibit loss of cell polarity and EMT after IGF1 stimulation [19], and at least ~22% to 46% of patients with TNBC have high IGF1R protein expression. In this study, we demonstrated that quercetin had a negative effect on the invasion and metastasis of MDA-MB-231 cells (Fig. 2) and inhibited IGF1R and its downstream Akt and Erk1/2 activities (Fig. 1A and B). Additionally, quercetin inhibited xenograft tumor growth and reduced lung nodules and metastasis in MDA-MB-231 xenograft mouse model (Fig. 5A and B). These findings suggest that such inhibition is due to the modulation of IGF1/IGF1R signaling by quercetin to prevent TNBC cells from expanding their metastatic subpopulation.
A growing number of reports indicate that IGF1-mediated PI3K–Akt and MAPK pathways enhance cancer cell proliferation, invasion, and metastasis by initiating EMT and modulating MMP activities [40,41]. The PI3K–Akt or MAPK pathway enhances the expression of EMT-resultant transcription factors, such as Snail, Slug, ZEB1, and ZEB2, to promote the induction of EMT and finally increases the motility of cancer cells [42,43]. Slug transfectant enhanced IGF1/IGF1R signaling in an autocrine manner by increasing the sensitivity of IGF1R to IGFs [20]. In view of these findings, our results showed that IGF1 increased the protein expression of Snail, and Slug (Fig. 4E), and the knockdown of Snail and Slug was sufficient to lead to the reversal of EMT in MDA-MB-231 cells (Fig. 4D). In addition, our study provides strong evidence that quercetin has the ability to inhibit the expression of Snail and Slug (Fig. 4A and B), which is related to the decreased expression of mesenchymal-related proteins fibronectin and vimentin in MDA-MB-231 cells (Fig. 3A and B) and xenograft tumors (Fig. 5C). In fact, some reports have shown that quercetin has an inhibitory effect on EMT-related diseases. For example, quercetin inhibits renal tubular EMT and renal fibrosis induced by mTORC1-mTOR-p70S6K signaling pathway in diabetic nephropathy [44]. Quercetin inhibits the EGF-induced transcription activities of Snail, Slug, and Twist by modulating the PI3K–Akt and Erk1/2 pathways and therefore prevents EMT in PC-3 prostate cancer cells [45]. Quercetin also downregulates the EMT and migration of TNBC by restraining the nuclear localization of β-catenin [34]. Based on these results, we suggest that quercetin may prevent the EMT and aggressive invasion of TNBC cells by blocking IGF1/IGF1R-mediated Snail and Slug expression.
In Fig. 1C, we found that quercetin induced the secretion level of IGFBP3 in the conditioned medium of MDA-MB-231 cells in a dose-dependent manner but remarkably reduced the secretion level of IGF1. Similar results were found in quercetin-treated androgen-independent prostate cancer PC-3 cells. Quercetin expands the accumulation of IGFBP-3 and reduces the level of IGFs in the conditioned medium of PC-3 cells; these changes are accompanied by caspase3-dependent apoptosis [46,47]. Quercetin increases the mRNA expression of IGFBP3, downregulates IGF1R-mediated signaling pathways, and therefore induces apoptosis in PC-3 cells [48]. IGFBP3 has multiple effects on tumor development; it negatively regulates autocrine or paracrine growth and cancer metastasis by binding to IGF in the circulation and enhances tumor progression by an IGF1-binding-independent manner [41]. Dependent of IGF1 binding, exogenous IGFBP3 inhibits the binding of IGFs to receptors by sequestrating IGFs and therefore retards the growth of breast cancer and several other cancer cells [7]. The knockdown of IGFBP3 expression enhances cancer progression and metastasis at the early stage of tumor process in mice [49]. Herein, we found that quercetin reduces the secretion ratio of IGF1 to IGFBP3 and suppressed the activation of IGF1R, IRS1, Akt, and Erk1/2 (Fig. 1A and B). These findings suggest that quercetin may prevent the autocrine or paracrine loop of IGF1 signaling and the malignancy of TNBC by increasing the expression and secretion of IGFBP3, but the detailed mechanism needs further investigation.
IGF1R and focal adhesion kinase maintain the mesenchymal features, CSCL characteristics, and the invasive phenotypes of TNBCs [50]. 6-Bromoindirubin-3′-oxime, a GSK-3β inhibitor, was able to reduce the EMT program, stem cell-like propensity, and metastatic behaviors of TNBCs [51]. These findings suggest that TNBC can acquire stem cell-like and invasive features by IGF1R signaling-dependent EMT; thus, its resistance to drug treatment is increased. Here, we observed that the protein expression of Sox2, Oct4, and Nanog, the well-identified transcription factors that maintain the pluripotency of CSCL cells, was dose-dependently downregulated by quercetin in MDA-MB-231 cells (Fig. 3C), similar to the inhibition of mesenchymal-like and invasive phenotypes. Thus, we suggest that EMT reversion by quercetin could make MDA-MB-231 cells well-differentiated, but this issue requires further study.
In summary, quercetin was able to reduce the activation of IGF1R and its downstream kinases Akt and Erk1/2 and simultaneously decrease Snail and Slug expression levels in MDA-MB-231 cells, reverse EMT program, and suppress the invasive phenotypes of MDA-MD-231 cells. Quercetin also inhibited IGF1R activation, reduced the expression levels of mesenchymal markers (Snail, Slug, fibronectin, and vimentin), and upregulated the expression levels of epithelial markers (keratins 18 and 19), which suppressed the growth of xenograft tumors and lung metastasis in xenograft mouse experiment. These results suggest that the anti-EMT and metastatic activity of quercetin on TNBC may be partly caused by the modulation of IGF1/IGF1R signaling pathway and point out the application value of quercetin in the treatment of TNBC.
Supplementary Information
Acknowledgments
This study was supported by grants from the Ministry of Science and Technology, Republic of China (MOST 106-2320-B-040-006- and MOST 107-2320-B-040-002-MY3). We are grateful to our collaborator, Dr. Min-Hsiung Pan, for the technical support in the animal experiments in this study. We are also thankful for the Instrument Center of Chung Shan Medical University, which provides fluorescence microscope supported by National Science Council, Ministry of Education and Chung Shan Medical University.
Funding Statement
This study was supported by grants from the Ministry of Science and Technology, Republic of China (MOST 106-2320-B-040-006- and MOST 107-2320-B-040-002-MY3).
Footnotes
Conflict of interest
The authors declare no competing financial interest.
References
- 1. Reis-Filho JS, Tutt ANJ. Triple negative tumours: a critical review. Histopathology. 2008;52:108–18. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- 2. Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA : the journal of the American Medical Association. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 3. Hudis CA, Gianni L. Triple-negative breast cancer: an unmet medical need. The oncologist. 2011;16(Suppl 1):1–11. doi: 10.1634/theoncologist.2011-S1-01. [DOI] [PubMed] [Google Scholar]
- 4. Sachdev D, Yee D. Inhibitors of insulin-like growth factor signaling: a therapeutic approach for breast cancer. J Mammary Gland Biol Neoplasia. 2006;11:27–39. doi: 10.1007/s10911-006-9010-8. [DOI] [PubMed] [Google Scholar]
- 5. Lann D, LeRoith D. The role of endocrine insulin-like growth factor-I and insulin in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:371–9. doi: 10.1007/s10911-008-9100-x. [DOI] [PubMed] [Google Scholar]
- 6. Duggan C, Wang CY, Neuhouser ML, Xiao L, Smith AW, Reding KW, et al. Associations of insulin-like growth factor and insulin-like growth factor binding protein-3 with mortality in women with breast cancer. International journal of cancer Journal international du cancer. 2013;132:1191–200. doi: 10.1002/ijc.27753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rota LM, Albanito L, Shin ME, Goyeneche CL, Shushanov S, Gallagher EJ, et al. IGF1R inhibition in mammary epithelia promotes canonical Wnt signaling and Wnt1-driven tumors. Cancer Res. 2014;74:5668–79. doi: 10.1158/0008-5472.CAN-14-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shin SJ, Gong G, Lee HJ, Kang J, Bae YK, Lee A, et al. Positive expression of insulin-like growth factor-1 receptor is associated with a positive hormone receptor status and a favorable prognosis in breast cancer. J Breast Cancer. 2014;17:113–20. doi: 10.4048/jbc.2014.17.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davison Z, de Blacquiere GE, Westley BR, May FE. Insulinlike growth factor-dependent proliferation and survival of triple-negative breast cancer cells: implications for therapy. Neoplasia. 2011;13:504–15. doi: 10.1593/neo.101590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Litzenburger BC, Creighton CJ, Tsimelzon A, Chan BT, Hilsenbeck SG, Wang T, et al. High IGF-IR activity in triple-negative breast cancer cell lines and tumorgrafts correlates with sensitivity to anti-IGF-IR therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:2314–27. doi: 10.1158/1078-0432.CCR-10-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bahhnassy A, Mohanad M, Shaarawy S, Ismail MF, El-Bastawisy A, Ashmawy AM, et al. Transforming growth factorbeta, insulin-like growth factor I/insulin-like growth factor I receptor and vascular endothelial growth factor-A: prognostic and predictive markers in triple-negative and nontriple-negative breast cancer. Mol Med Rep. 2015;12:851–64. doi: 10.3892/mmr.2015.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin CW, Yang LY, Shen SC, Chen YC. IGF-I plus E2 induces proliferation via activation of ROS-dependent ERKs and JNKs in human breast carcinoma cells. J Cell Physiol. 2007;212:666–74. doi: 10.1002/jcp.21061. [DOI] [PubMed] [Google Scholar]
- 13. Kim HJ, Litzenburger BC, Cui X, Delgado DA, Grabiner BC, Lin X, et al. Constitutively active type I insulin-like growth factor receptor causes transformation and xenograft growth of immortalized mammary epithelial cells and is accompanied by an epithelial-to-mesenchymal transition mediated by NF-kappaB and snail. Mol Cell Biol. 2007;27:3165–75. doi: 10.1128/MCB.01315-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. The Journal of clinical investigation. 2009;119:1438–49. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. The Journal of clinical investigation. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guan X. Cancer metastases: challenges and opportunities. Acta Pharm Sin B. 2015;5:402–18. doi: 10.1016/j.apsb.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lorenzatti G, Huang W, Pal A, Cabanillas AM, Kleer CG. CCN6 (WISP3) decreases ZEB1-mediated EMT and invasion by attenuation of IGF-1 receptor signaling in breast cancer. Journal of cell science. 2011;124:1752–8. doi: 10.1242/jcs.084194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jones RA, Campbell CI, Wood GA, Petrik JJ, Moorehead RA. Reversibility and recurrence of IGF-IR-induced mammary tumors. Oncogene. 2009;28:2152–62. doi: 10.1038/onc.2009.79. [DOI] [PubMed] [Google Scholar]
- 20. Sivakumar R, Koga H, Selvendiran K, Maeyama M, Ueno T, Sata M. Autocrine loop for IGF-I receptor signaling in SLUG-mediated epithelial-mesenchymal transition. Int J Oncol. 2009;34:329–38. [PubMed] [Google Scholar]
- 21. Zhou J, Wang J, Zeng Y, Zhang X, Hu Q, Zheng J, et al. Implication of epithelial-mesenchymal transition in IGF1R-induced resistance to EGFR-TKIs in advanced non-small cell lung cancer. Oncotarget. 2015;6:44332–45. doi: 10.18632/oncotarget.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H, Xu L, Li C, Zhao L, Ma Y, Zheng H, et al. Ubiquitin ligase Cbl-b represses IGF-I-induced epithelial mesenchymal transition via ZEB2 and microRNA-200c regulation in gastric cancer cells. Molecular cancer. 2014;13:136. doi: 10.1186/1476-4598-13-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H, Xu L, Zhao L, Ma Y, Zhu Z, Liu Y, et al. Insulin-like growth factor-I induces epithelial to mesenchymal transition via GSK-3beta and ZEB2 in the BGC-823 gastric cancer cell line. Oncology letters. 2015;9:143–8. doi: 10.3892/ol.2014.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Graham TR, Zhau HE, Odero-Marah VA, Osunkoya AO, Kimbro KS, Tighiouart M, et al. Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2008;68:2479–88. doi: 10.1158/0008-5472.CAN-07-2559. [DOI] [PubMed] [Google Scholar]
- 25. Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nature cell biology. 2004;6:931–40. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 26. Pradhan SJ, Mishra R, Sharma P, Kundu GC. Quercetin and sulforaphane in combination suppress the progression of melanoma through the down-regulation of matrix metalloproteinase-9. Exp Ther Med. 2010;1:915–20. doi: 10.3892/etm.2010.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pejin B, Ciric A, Markovic JD, Glamoclija J, Nikolic M, Stanimirovic B, et al. Quercetin Potently Reduces Biofilm Formation of the Strain Pseudomonas aeruginosa PAO1 in vitro. Current pharmaceutical biotechnology. 2015;16:733–7. doi: 10.2174/1389201016666150505121951. [DOI] [PubMed] [Google Scholar]
- 28. Song NR, Chung MY, Kang NJ, Seo SG, Jang TS, Lee HJ, et al. Quercetin suppresses invasion and migration of H-Ras-transformed MCF10A human epithelial cells by inhibiting phosphatidylinositol 3-kinase. Food Chem. 2014;142:66–71. doi: 10.1016/j.foodchem.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 29. Bishayee K, Ghosh S, Mukherjee A, Sadhukhan R, Mondal J, Khuda-Bukhsh AR. Quercetin induces cytochrome-c release and ROS accumulation to promote apoptosis and arrest the cell cycle in G2/M, in cervical carcinoma: signal cascade and drug-DNA interaction. Cell Prolif. 2013;46:153–63. doi: 10.1111/cpr.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, et al. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Molecular cell. 2000;6:909–19. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 31. Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–8. [PubMed] [Google Scholar]
- 32. Jung M, Bu SY, Tak KH, Park JE, Kim E. Anticarcinogenic effect of quercetin by inhibition of insulin-like growth factor (IGF)-1 signaling in mouse skin cancer. Nutr Res Pract. 2013;7:439–45. doi: 10.4162/nrp.2013.7.6.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sharmila G, Bhat FA, Arunkumar R, Elumalai P, Raja Singh P, Senthilkumar K, et al. Chemopreventive effect of quercetin, a natural dietary flavonoid on prostate cancer in in vivo model. Clin Nutr. 2014;33:718–26. doi: 10.1016/j.clnu.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 34. Srinivasan A, Thangavel C, Liu Y, Shoyele S, Den RB, Selvakumar P, et al. Quercetin regulates β-catenin signaling and reduces the migration of triple negative breast cancer. Molecular carcinogenesis. 2016;55:743–56. doi: 10.1002/mc.22318. [DOI] [PubMed] [Google Scholar]
- 35. Chen Y, Wang K, Qian CN, Leach R. DNA methylation is associated with transcription of Snail and Slug genes. Biochem Biophys Res Commun. 2013;430:1083–90. doi: 10.1016/j.bbrc.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cevenini A, Orru S, Mancini A, Alfieri A, Buono P, Imperlini E. Molecular Signatures of the Insulin-like Growth Factor 1-mediated Epithelial-Mesenchymal Transition in Breast, Lung and Gastric Cancers. International journal of molecular sciences. 2018:19. doi: 10.3390/ijms19082411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Turdo A, Veschi V, Gaggianesi M, Chinnici A, Bianca P, Todaro M, et al. Meeting the Challenge of Targeting Cancer Stem Cells. Front Cell Dev Biol. 2019;7:16. doi: 10.3389/fcell.2019.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–60. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weigelt B, Peterse JL, van ‘t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 40. Miyazono K. Transforming growth factor-beta signaling in epithelial-mesenchymal transition and progression of cancer. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:314–23. doi: 10.2183/pjab.85.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zielinska HA, Bahl A, Holly JM, Perks CM. Breast Cancer. Vol. 7. Dove Med Press; 2015. Epithelial-to-mesenchymal transition in breast cancer: a role for insulin-like growth factor I and insulin-like growth factor-binding protein 3? pp. 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 43. Mallini P, Lennard T, Kirby J, Meeson A. Epithelial-to-mesenchymal transition: what is the impact on breast cancer stem cells and drug resistance. Cancer Treat Rev. 2014;40:341–8. doi: 10.1016/j.ctrv.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 44. Lu Q, Ji XJ, Zhou YX, Yao XQ, Liu YQ, Zhang F, et al. Quercetin inhibits the mTORC1/p70S6K signaling-mediated renal tubular epithelial-mesenchymal transition and renal fibrosis in diabetic nephropathy. Pharmacol Res. 2015;99:237–47. doi: 10.1016/j.phrs.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 45. Bhat FA, Sharmila G, Balakrishnan S, Arunkumar R, Elumalai P, Suganya S, et al. Quercetin reverses EGF-induced epithelial to mesenchymal transition and invasiveness in prostate cancer (PC-3) cell line via EGFR/PI3K/Akt pathway. J Nutr Biochem. 2014;25:1132–9. doi: 10.1016/j.jnutbio.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 46. Vijayababu MR, Arunkumar A, Kanagaraj P, Arunakaran J. Effects of quercetin on insulin-like growth factors (IGFs) and their binding protein-3 (IGFBP-3) secretion and induction of apoptosis in human prostate cancer cells. Journal of carcinogenesis. 2006;5:10. doi: 10.1186/1477-3163-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vijayababu MR, Kanagaraj P, Arunkumar A, Ilangovan R, Dharmarajan A, Arunakaran J. Quercetin induces p53-independent apoptosis in human prostate cancer cells by modulating Bcl-2-related proteins: a possible mediation by IGFBP-3. Oncology research. 2006;16:67–74. doi: 10.3727/000000006783981224. [DOI] [PubMed] [Google Scholar]
- 48. Senthilkumar K, Elumalai P, Arunkumar R, Banudevi S, Gunadharini ND, Sharmila G, et al. Quercetin regulates insulin like growth factor signaling and induces intrinsic and extrinsic pathway mediated apoptosis in androgen independent prostate cancer cells (PC-3) Molecular and cellular biochemistry. 2010;344:173–84. doi: 10.1007/s11010-010-0540-4. [DOI] [PubMed] [Google Scholar]
- 49. Blouin MJ, Bazile M, Birman E, Zakikhani M, Florianova L, Aleynikova O, et al. Germ line knockout of IGFBP-3 reveals influences of the gene on mammary gland neoplasia. Breast Cancer Res Treat. 2015;149:577–85. doi: 10.1007/s10549-015-3268-8. [DOI] [PubMed] [Google Scholar]
- 50. Taliaferro-Smith L, Oberlick E, Liu T, McGlothen T, Alcaide T, Tobin R, et al. FAK activation is required for IGF1R-mediated regulation of EMT, migration, and invasion in mesenchymal triple negative breast cancer cells. Oncotarget. 2015;6:4757–72. doi: 10.18632/oncotarget.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vijay GV, Zhao N, Den Hollander P, Toneff MJ, Joseph R, Pietila M, et al. GSK3β regulates epithelial-mesenchymal transition and cancer stem cell properties in triple-negative breast cancer. Breast cancer research : BCR. 2019;21:37. doi: 10.1186/s13058-019-1125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.