Abstract
The diagnostic performance of radiologist readers exhibits substantial variation that cannot be explained by CT acquisition protocol differences. Studying reader detectability from CT images may help identify why certain types of lesions are missed by multiple or specific readers. Ten subspecialized abdominal radiologists marked all suspected metastases in a multi-reader-multi-case study of 102 deidentified contrast-enhanced CT liver scans at multiple radiation dose levels. A reference reader marked ground truth metastatic and benign lesions with the aid of histopathology or tumor progression on later scans. Multi-slice image patches and 3D radiomic features were extracted from the CT images. We trained deep convolutional neural networks (CNN) to predict whether an average (generalized) or individual radiologist reader would detect or miss a specific metastasis from an image patch containing it. The individualized CNN showed higher performance with an area under the receiver operating characteristic curve (AUC) of 0.82 compared to a generalized one (AUC = 0.78) in predicting reader-specific detectability. Random forests were used to build the respective versions from radiomic features. Both the individualized (AUC = 0.64) and generalized (AUC = 0.59) predictors from radiomic features showed limited ability to differentiate detected from missed lesions. This shows that CNN can identify and learn automated features that are better predictors of reader detectability of lesions than radiomic features. Individualized prediction of difficult lesions may allow targeted training of idiosyncratic weaknesses but requires substantial training data for each reader.
Keywords: Convolutional Neural Network, Observer Performance, Liver metastasis detection, Low contrast detection
1. INTRODUCTION
In multi-reader, multi-case observer studies, the performance across radiologist readers exhibits wider variability than can be explained by variations due to image dose, quality, or reconstruction parameters. There is a critical need to study interobserver variability to devise targeted training strategies and achieve a consistent level of diagnostic performance. Quantifying case and reader-specific qualities affecting diagnostic performance is of prime importance before further optimization in terms of low-dose protocols can be realized. While there has been substantial investigation of artificial intelligence (AI)-based methods for the task of detecting metastatic cancer, there may be complementary value in using them for radiology education.
We aim to investigate the lesion-based characteristics of hepatic metastases from abdominal CT images that cause radiologists to miss detecting them. Using two AI-based approaches, i) radiomics analysis, and ii) deep convolutional networks (CNNs), we predict the radiologists’ performance for detecting malignant lesions from contrast-enhanced CT images. For each approach, we produce both a generalized and an individualized prediction. The generalized prediction is trained to identify the difficult lesions that are missed by many radiologists, whereas the individualized prediction is trained to identify lesions difficult for a specific radiologist, which may be affected by reader-specific fallacies. Accurate predictions of reader-specific detectability of lesions could enable targeted training of idiosyncratic weaknesses by optimizing the collection of cases that a resident learns from.
2. METHODS
2.1. Reader Performance Study
We used the data from a previous study [1] that collected abdominal CT images from 102 patients. A total of 124 hepatic metastatic lesions were identified from 51 patients through histopathology or progression on subsequent scans. Ten abdominal radiologists from our institution were recruited after IRB approval for the reader performance study.
2.2. Image processing
Raw CT images were reconstructed using five combinations of algorithms and quality reference mAs (QRM) levels. The five combinations were i) 200 QRM filtered back projection (FBP), ii) 160 QRM iterative reconstruction (IR), iii) 120 QRM IR, iv) 120 QRM FBP, and v) 100 QRM IR. We used the PyRadiomics package [2] to extract 110 3D radiomic features from all reconstructed images. 3D image patches comprising 3 slices of size 128×128 pixels, centered around the lesions, were extracted from the images to be used as the inputs to the CNNs.
2.3. Prediction of reader detectability
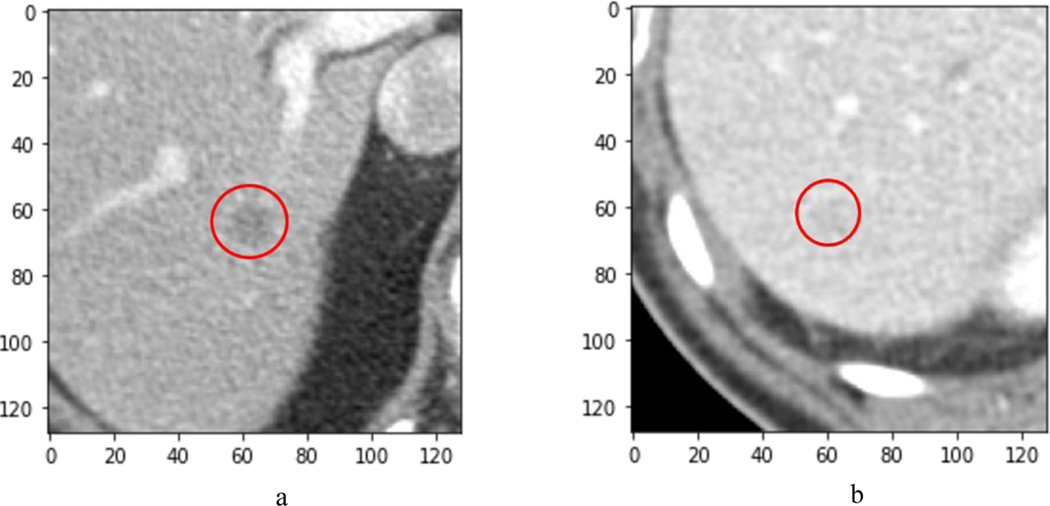
The ground truth was established by a reference reader according to predefined criteria. In particular, ground truth metastases were established on the basis of progression in subsequent imaging or on histopathology. The ten radiologists marked all suspected metastases from the reconstructed images with a confidence score between 1 and 100 (examples in Figures 1.a and 1.b).
Figure 1.
a) Example metastatic lesion detected by all 10 radiologists. b) Example metastatic lesion missed by all radiologists
We use the term learner to signify any AI mechanism that is automatically able to learn patterns in reader detectability of lesions. Two sets of prediction tasks were defined: i) Generalized: a single learner that learns simultaneously for all 10 radiologists, and ii) Individualized: separate learners for each radiologist. Each of the reconstructed lesions were assigned binary labels based on the prediction task. The generalized learner uses labels of “detected” if found by >75% of all readers or “missed” otherwise, while the individualized learner uses the labels of “detected” or “missed” by individual readers. The reader performance data was partitioned into 5 groups for five-fold cross-validation, without data leakage. Specifically, separation of lesions in the train and test sets were with respect to a unique patient identifier. This ensured the avoidance of same-patient bias and repetition of data at different reconstruction combinations. The training data was up-sampled using random sampling to overcome the class-imbalance issue with the minority class of missed lesions. The classification performances for the prediction tasks were tested using the portion of reserved data in each iteration. We used the following approaches to compare generalized and individualized prediction of missed lesions.
2.4. Radiomic data analysis
The 110 handcrafted features extracted by the PyRadiomics package [2] belong to 7 subgroups depicted in Table 1. PyRadiomics includes features such as first-order statistics that describe the distribution of voxel intensities within the image region, shape-based descriptors for the specifics of the 3D shape and sizes, gray level cooccurrence matrix (GLCM) for the second order joint probability of image levels, gray level size zone matrix (GLSZM) for the gray level zones in an image, gray level run length matrix (GLRLM) for length of pixels with the same gray level value, neighboring gray tone difference matrix (NGTDM) for the difference between gray levels with neighbors, and gray level dependency matrix for the adjacent voxels that are dependent (gray level difference is below a threshold) on a central voxel. Masking was not applied to the lesions because the tumors were not segmented.
Table 1.
Selected PyRadiomics features with P-values resulted from the ANOVA F-test
| Feature group | No. features | Generalized | Individualized | ||
|---|---|---|---|---|---|
| Selected feature | P-value | Selected feature | P-value | ||
| First Order Statistics | 19 | Range | <0.001 | Median | <0.001 |
| Shape-based (3D) | 16 | Imc1 | <0.001 | Inverse Variance | <0.001 |
| Gray Level Cooccurrence Matrix | 24 | Dependence Non-Uniformity | <0.001 | Dependence Non-Uniformity | <0.001 |
| Gray Level Run Length Matrix | 16 | Run Entropy | <0.001 | Gray Level Non-Uniformity | <0.001 |
| Gray Level Size Zone Matrix | 16 | Coarseness | <0.001 | Coarseness | <0.001 |
| Neighboring Gray Tone Difference Matrix | 5 | Gray Level Non-Uniformity | <0.001 | Gray Level Non-Uniformity | <0.001 |
| Gray Level Dependence Matrix | 14 | Least Axis Length | <0.001 | Flatness | <0.001 |
Removing zero-variance features that do not contribute to the classification resulted in 106 features. As the number of radiomic features outnumber the sample images, we employed feature selection as the first step to avoid overfitting. Our strategy was to choose one feature from each of these groups that could predict whether readers would miss/detect lesions with the highest accuracy. The features are selected based on the ANOVA F-test, where the F-statistic is the variation between sample means and within the samples, that are expected to vary in a similar manner under the null hypothesis. Higher F-values indicate higher variability among the samples. The most significant feature from each group is selected based on the highest F-value along with p-values lower than the significance level to reject the null hypothesis. As the train and test set changes per iteration, the selected features also change. For the final model fit, we considered the feature that appeared the greatest number of times from a group across the various iterations.
Next, we trained classifiers for the generalized and individualized tasks to predict the radiologists’ capability to detect lesions. A set of classifiers that included 3-nearest neighbors, linear and radial basis support vector machine, decision tree, random forest, multilayer perceptron, naive Bayes, and quadratic determinant analysis were used. We chose random forest as the final classification model due to better performance on the test set (generalized AUC = 0.59, individualized AUC = 0.64) and lower proneness to overfitting. Hyperparameter tuning of the random forest classifier in terms of maximum tree depth, number of samples per node, and number of trees was performed through grid search. The validation AUC for the other classifiers ranged between 0.52 and 0.57 for the generalized version and 0.54–0.59 for the individualized version.
2.5. Deep Convolutional Neural Network
Pretrained VGG-16 [3] CNNs with ImageNet classification task weights [4] were used to automatically extract and process image features from resized 3D image patches. Instead of RGB channels in color images which the network was initially trained on, we used the slice at the center, one slice before, and one slice after to mimic the three dimensionalities of data. The densely connected layers of these CNNs trained for ImageNet classification task were removed and two more convolution, batch normalization, pooling layers were added for fine tuning. A final dense layer with a SoftMax activation was used for the predictions. A pipeline of data augmentation to avoid class imbalance, dropout to reduce overfitting, and transfer learning with fine-tuning was used to train the CNNs on the reader performance data.
3. RESULTS
Reader sensitivity across the 10 radiologists ranged from 67.3% to 93.0% with an average sensitivity of 85.74%. Reader 4 missed the least number of lesions and reader 1 the most. Reader-specific eye-scanning patterns and image-specific characteristics would also contribute to the detectability of lesions.
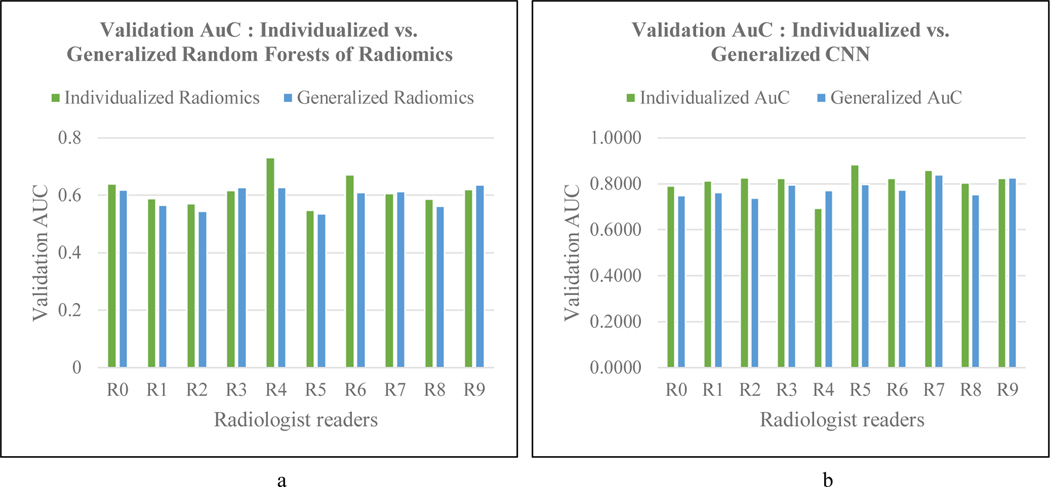
Table 2 lists the meaning of each of the selected features from PyRadiomics. Figure 2.a compares the individualized and generalized classification of radiomic features in predicting the detectability of lesions in the test set by each reader. The individualized version at an average area under the receiver operating characteristic curve (ROC-AUC) of 0.64 performed slightly better than the generalized version (AUC = 0.59) on the test set. The highest difference in AUC between the individualized and generalized random forests was for reader 4, with the former reporting a 16% increase. As discussed above reader 4 has the best diagnostic sensitivity. The generalized random forest performed better only for readers 8 and 9, which however, is only a 2% and 1% improvement over the individualized version.
Table 2.
Semantics of selected radiomic features from generalized and individualized predictions
| Generalized | Individualized | ||
|---|---|---|---|
| Feature | Meaning of Measure | Feature | Meaning of Measure |
| Range | Range of gray values in the region of interest (RoI) in an image | Median | Meadian gray level intensity within the RoI |
| Imc1 | Informational measure of correlation between pixels in the RoI | Inverse Variance | Inverse variance of the intensities for a length of pixels with the same gray level value |
| Dependence Non-Uniformity | Similarity of dependence in the image, with lower values denoting homogeneous dependences | Dependence Non-Uniformity | |
| Run Entropy | Uncertainty in the distribution of gray levels | Gray Level Non-Uniformity | Similarity of gray-level intensity values in the image |
| Coarseness | Spatial rate of change between the center voxel and it’s neighborhood | Coarseness | |
| Gray Level Non-Uniformity | Variability of gray-level intensities in the image | Gray Level Non-Uniformity | |
| Least Axis Length | Smallest axis length of the ellipsoid that encloses the RoI | Flatness | Relationship between the largest and smallest principal components in the Principal Component Analysis (PCA) performed using the voxel coordinates of the image |
Figure 2.
a) Comparing AUCs of individualized and generalized radiomics. b) Comparing AUCs of individualized CNN and generalized CNN for reader-specific lesion detectability
Individualized CNNs obtained an average area under the receiver operating characteristic curve (AUC) of 0.82 () in categorizing lesions as missed or detected by the 10 readers across the test sets for each fold. The generalized CNN obtained an average AUC of 0.78 (). The performance of the two CNN versions across the readers is depicted in Figure 2.b. As in the case with radiomic features, individualized CNN fared better than the generalized CNN for all readers except readers 4 and 9. Individualized CNN outdid the generalized one by 8.8% in the best case (reader 2), while the latter bettered the former at 7.8% for reader 4.
4. CONCLUSIONS
Image features extracted from deep CNNs outperformed handcrafted radiomic features in predicting radiologist detectability of metastatic liver lesions. An individualized CNN provides higher performance than a generalized CNN in predicting reader-specific lesion detectability. However, this requires substantial training data and effort for each reader. In situations where this training data cannot be collected, a generalized CNN may be an efficient method for identifying metastases that could be used for radiologist training.
REFERENCES
- [1].Fletcher JG, Yu L, Fidler JL, Levin DL, DeLone DR, Hough DM, Takahashi N, Venkatesh SK, Sykes AMG, White D. and Lindell RM, Estimation of observer performance for reduced radiation dose levels in CT: eliminating reduced dose levels that are too low is the first step. Academic radiology, 24(7), 876–890 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Van Griethuysen JJ, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RG, Fillion-Robin JC, Pieper S. and Aerts HJ, Computational radiomics system to decode the radiographic phenotype. Cancer research, 77(21), e104–e107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Simonyan K. and Zisserman A, Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014). [Google Scholar]
- [4].Krizhevsky A, Sutskever I. and Hinton GE, Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems, 25, 1097–1105 (2012). [Google Scholar]