Abstract
Transfer of antibiotic resistance genes by conjugation is thought to play an important role in the spread of resistance. Yet virtually no information is available about the extent to which such horizontal transfers occur in natural settings. In this paper, we show that conjugal gene transfer has made a major contribution to increased antibiotic resistance in Bacteroides species, a numerically predominant group of human colonic bacteria. Over the past 3 decades, carriage of the tetracycline resistance gene, tetQ, has increased from about 30% to more than 80% of strains. Alleles of tetQ in different Bacteroides species, with one exception, were 96 to 100% identical at the DNA sequence level, as expected if horizontal gene transfer was responsible for their spread. Southern blot analyses showed further that transfer of tetQ was mediated by a conjugative transposon (CTn) of the CTnDOT type. Carriage of two erythromycin resistance genes, ermF and ermG, rose from <2 to 23% and accounted for about 70% of the total erythromycin resistances observed. Carriage of tetQ and the erm genes was the same in isolates taken from healthy people with no recent history of antibiotic use as in isolates obtained from patients with Bacteroides infections. This finding indicates that resistance transfer is occurring in the community and not just in clinical environments. The high percentage of strains that are carrying these resistance genes in people who are not taking antibiotics is consistent with the hypothesis that once acquired, these resistance genes are stably maintained in the absence of antibiotic selection. Six recently isolated strains carried ermB genes. Two were identical to erm(B)-P from Clostridium perfringens, and the other four had only one to three mismatches. The nine strains with ermG genes had DNA sequences that were more than 99% identical to the ermG of Bacillus sphaericus. Evidently, there is a genetic conduit open between gram-positive bacteria, including bacteria that only pass through the human colon, and the gram-negative Bacteroides species. Our results support the hypothesis that extensive gene transfer occurs among bacteria in the human colon, both within the genus Bacteroides and among Bacteroides species and gram-positive bacteria.
Concern over the safety implications of antibiotic-resistant bacteria in foods has centered around the question of how likely such bacteria are to transfer resistance genes to human intestinal bacteria during their passage through the intestinal tract and what might happen to the transferred genes once they enter colonic bacteria. This question is part of a larger question about the amount of horizontal gene transfer that actually occurs in nature. Few attempts have been made to determine how much gene transfer occurs among bacteria in the colon or in other environments. Some studies have been done to assess the extent of horizontal gene transfer among microorganisms in soil and water (9, 13, 16), the intestines of laboratory mice (26), and experimental abscesses (4). These studies found evidence that horizontal gene transfer events do occur in these settings at frequencies similar to or higher than those observed in the laboratory.
In an earlier paper, Nikolich et al. examined a small number of tetracycline-resistant Bacteroides and Prevotella species from the human colon and the colons of farm animals (27). The results of that study suggested that horizontal gene transfer had occurred between members of these two genera. Transfer was also demonstrated between the black-pigmented oral Prevotella and Bacteroides species in the laboratory (11). Results of recent studies of vancomycin-resistant enterococci isolated from the intestines of animals and humans also support the hypothesis that horizontal gene transfer events, occur in the intestinal tract (15, 48; L. B. Jensen, A. M. Hammerum, R. L. Poulsen, and H. Westh, Letter, Antimicrob. Agents Chemother. 43:724–725, 1999). In this paper, we report the results of the first systematic investigation of horizontal gene transfer events involving a major population of human colonic bacteria, Bacteroides species. Although this study focuses on transfer of antibiotic resistance genes, the conclusions could presumably be applied to the transfer of other genes that perform accessory functions.
The human colon is an environment that should be very conducive to horizontal gene transfer events. Nutrients are abundant, the concentration of bacteria is high (1012 per g [wet weight]), and there are many surfaces such as plant particles to which bacteria can adhere. Colonic bacteria have been shown to carry a variety of plasmids and integrated elements that can be transferred by conjugation. Yet, under optimized laboratory conditions, transfer of these elements occurs at a relatively low frequency, 10−5 to 10−7 per recipient or lower. This raised the question of how effectively such conjugal elements could spread in the colonic environment. Moreover, if a transfer event took place, how likely would the recipient be to maintain the newly acquired element?
A way to assess the degree of horizontal transfer among different strains of bacteria in a natural setting is to determine whether identical or virtually identical copies of the same gene are found in different species. This approach does not provide transfer rates in vivo, but it can answer questions about the extent to which horizontal transfer has occurred and what types of elements are most often responsible for such transfers. For this study, we chose to focus on Bacteroides species. Bacteroides species comprise a major part of the human colonic microbiota, accounting for about 25% of all colonic isolates (36, 52). Bacteroides species, especially Bacteroides fragilis and Bacteroides thetaiotaomicron, are opportunistic pathogens, which can cause life-threatening infections if they escape from the colon as a result of abdominal trauma or surgery. Over the past several decades, Bacteroides clinical isolates have become increasingly resistant to antibiotics. Resistance to tetracycline has become so common that strains are often not tested for susceptibility to this antibiotic. In recent years, resistance to drugs used to treat Bacteroides infections, such as clindamycin, has also been increasing. Conjugal elements such as plasmids and conjugative transposons (CTn's) have been found in Bacteroides clinical isolates (34). Thus, horizontal gene transfer could conceivably have played a role in the rising incidence of resistance in this bacterial group. In this study, we assess the role of CTn's and plasmids in the transfer of antibiotic resistance genes.
MATERIALS AND METHODS
Bacteroides strains.
The 88 VPI strains are from the Anaerobe Laboratory at the Virginia Polytechnical Institute in Blacksburg. Some of these strains had been isolated from healthy volunteers (community isolates); others had been isolated from infected patients (clinical isolates). All of them were isolated before 1980, many before 1960 (17). The clinical isolates from 1980 to the present were obtained from various medical centers and hospitals: the majority were from the Wadsworth Anaerobe Laboratory (WAL) in Los Angeles, Calif. (n = 34) and Loyola Strich School of Medicine, in Maywood, Ill. (n = 65; designations begin with DH). The 1996-to-1997 community isolates were from rectal swabs taken from volunteers attending the Microbial Diversity Course at Woods Hole, Mass., in 1996 and 1997 (n = 86; designations begin with WH). The isolation of Bacteroides strains from these volunteers took advantage of the aerotolerance of Bacteroides species and their resistance to high levels of gentamicin (200 μg/ml). Samples were diluted and streaked directly onto supplemented BHI medium (7) containing 200 μg of gentamicin/ml under aerobic conditions and were then incubated anaerobically in BBL GasPak jars for 48 h. Fifty or more isolates were first checked by colony hybridization using tetQ and CTnDOT probes (Table 1), and four or five isolates from each source were selected at random, stocked, and saved for further analysis. The further verification of the identity of the isolates included hybridization to Bacteroides-specific probes (19), Gram staining, and some selective 16S ribosomal DNA (rDNA) sequencing using a universal prokaryotic forward primer and a Bacteroides group-specific reverse primer (30). This mode of isolation could have had a slight bias in favor of the species that are somewhat more aerotolerant and less fastidious than other species, such as B. thetaiotaomicron and B. fragilis. This bias could not have been too strong, however, because we isolated no B. fragilis using this procedure, and whereas many of the isolates were B. thetaiotaomicron, several were also Bacteroides uniformis and Bacteroides eggerthii. These ratios reflected the ratios of Bacteroides species in the colon, where the concentration of B. fragilis is more than 10-fold lower than that of B. thetaiotaomicron or B. uniformis.
TABLE 1.
Sources of the DNA fragments used to probe the Bacteroides isolates
| Target gene(s) | Source of DNA for the probea |
|---|---|
| Antibiotic resistance genes | |
| tet(Q) | The tetQ gene of CTnDOT cloned onto pNFD13-2 (28). A 1.55-kbp EcoRI-PvuII internal fragment was used as a probe. Primers Q for (GGC TTCT ACG ACA TCT ATT A) and Q rev (CAT CAA CAT TTA TCT CTC TG) were used to PCR amplify a 758-bp internal fragment (21). |
| tet(C) | HindIII- NruI fragment of pBR328 (tetC) |
| tet(L) | pMV158 (entire) (tetL) (20) |
| tet(M) | BamHI-KpnI tetM (Tn916) fragment from pFD310 (45) |
| tetB(P) | PstI-EcoRI internal fragment of tetB(P) from pJIR667 (43) |
| ermB | A 639-bp internal fragment of pTV1-OK (Tn 917) was used for sequencing and as a probe. Primers were B1 (GAA AAG GTA CTC AAC CAA ATA) and B2 (AGT AAC GGT ACT TAA ATT GTT TAC). The accession no. is X58285) (47). |
| ermF | A 0.85-kbp EcoRI fragment containing ermFS (accession no. M37699) of Tn4551 cloned in pFD214 (44) |
| ermG | PCR primers G for1 (ACA TTT CCT AGC CAC AAT C) and G rev1 (CGC TAT GTT TAA CAA GC) were used to obtain a 442-bp internal product (bp 465 to bp 907) (accession no. L42817) (8). |
| ermA, ermC, ermQ | PCR products or fragments of cloned genes. The primers and targets for ermA and ermC are described by Sutcliffe et al. (47). A 380-bp internal fragment of ermQ was isolated from pJIR745 (2). |
| Element probes | |
| CTnDOT joined ends | 1.1-kbp PCR product containing the CTnDOT joined ends; includes att site and part of the integrase gene (5) |
| NBU1–NBU2 shared region | prmN1 oriT mobN1 region of NBU1 (41) |
| IS4351 | 1.2-kbp EcoRI-HindIII fragment (accession no. M17124) of Tn4400 cloned onto pEG920 (31, 42) |
| tetQ-rteA-rteB and rteC region of CTnDOT | 7.7-kbp EcoRI fragment of CTnDOT isolated from a cosmid clone of CTnDOT (40, 46) |
| rteC alone | 1.1-kbp subclone from CTnDOT region on pLYL52 (23) (accession no. L02419) |
| rteB alone | 500-bp EcoRV-BsaI fragment of rteB from CTnDOT (accession no. M81439) |
Fragments of tet(C), tet(L), tet(M), and tetB(P) were isolated from plasmids and used as probes. See reference 22 for accession numbers.
The species of the VPI community and clinical isolates was first identified by biochemical analysis and later confirmed by DNA-DNA or rRNA hybridization studies (17, 18). The species of the isolates from the Loyola and Wadsworth Veterans Affairs (VA) hospitals and the other clinical sources were determined by biochemical analyses carried out at the institutions from which they were obtained. We checked some of these strains by partial sequencing of their 16S rRNA genes. In the case of the clinical isolates, there was probably a slight bias in favor of resistant over susceptible strains because patients infected with resistant strains are more likely to experience treatment failure and are thus more likely to have specimens of the infecting bacterium sent to the clinical laboratory. The two sets of strains from the Wadsworth VA hospital were obtained because of their resistance to chloramphenicol or tetracycline and therefore are not representative of all clinical isolates. Data for these are reported separately.
Antibiotic resistance phenotypes.
Fresh overnight cultures were streaked onto supplemented BHI (7) agar plates containing the antibiotic. The strains were considered resistant if they grew on plates containing erythromycin (3 μg/ml), clindamycin (3 μg/ml), or tetracycline (1 μg/ml). B. thetaiotaomicron 5482 and B. fragilis 638 did not grow at these concentrations. No attempt was made to determine the MICs for each strain because the purpose of these phenotypic tests was to correlate the resistance phenotype with the presence of the resistance genes responsible and not to determine the precise level of resistance. Many of the clinical strains came with MIC results, and the resistances we observed correlated with the information provided.
DNA hybridizations.
DNA dot blots using total DNA prepared from 2-ml overnight cultures of each strain (33, 50) were probed using fluorescein-dUTP-labeled DNA fragments or purified PCR products. Labeling was done as described in the Renaissance kit protocols (NEN Life Sciences). The blots were developed using a chemiluminescent substrate. Southern blotting was done on the DNA of 60 strains that hybridized to tetQ to determine how related they were to the known Bacteroides CTn's, especially CTnDOT. The DNA from each strain was digested with EcoRI and EcoRV, and the blots were probed with a series of fluorescein-labeled probes. The first probe used was a 7,700-bp EcoRI fragment of CTnDOT which contained tetQ, rteA, rteB, and rteC (46). Some of the blots were probed sequentially with an rteC probe, an rteB probe, and then a probe containing tetQ and rteA in order to identify restriction fragments containing each sequence. This was necessary because of the restriction site polymorphisms observed for some of the strains. See Table 1 for descriptions of primers for PCR-generated products and the sources of the isolated fragments used to make the probes.
PCR amplification of 16S rRNA sequences and tetQ genes.
The PCR primers used to amplify 16S rDNA and the internal fragments for tetQ, ermG, and ermB genes are described in Table 1. The amplification was done using Taq I polymerase in 100-μl reaction mixtures containing 10 to 100 ng of DNA and 200 ng of each primer in a solution containing 1× Gibco-BRL PCR buffer, 1.5 mM MgCl2, and 0.2 mM deoxyribonucleoside triphosphate mixture. The amplification cycles were as follows: 95°C for 5 min; 30 cycles of 95°C for 1 min, 50 to 52°C for 1 min, and 72°C for 2 min; and a final elongation step of 72°C for 5 min. The PCR products were extracted directly from the reaction mix using a Wizard PCR Clean up kit (Promega). The PCR products were sequenced using the PCR primers by the University of Illinois Biotechnology Center.
RESULTS
Hybridization analyses of community and clinical isolates.
For an initial assessment of the extent to which horizontal gene transfer has occurred between colonic Bacteroides strains over the past 3 decades, we compared tetracycline and erythromycin resistance profiles and the carriage of possible resistance genes of two sets of isolates. One set (88 isolates) had been collected before 1980, many of the strains before 1960. The second set (211 isolates) had been collected after 1980, most during the 1990s. Both sets contained strains isolated from patients with Bacteroides infections (clinical isolates) and strains isolated from the intestines of healthy humans (community isolates). By comparing clinical and community isolates, it was possible to determine whether patterns seen in clinical isolates mirrored those seen in community isolates. If the spread of resistance was occurring in the community isolates, later causing clinical infections, the patterns should be similar. If resistance was arising in hospitals, the incidence of phenotype resistance and carriage of resistance genes should be higher in the clinical isolates than in the community isolates.
Results of the initial survey are shown in Table 2. As expected from studies of clinical isolates, the incidence of phenotypic resistance to tetracycline and erythromycin had risen dramatically since the pre-1970 period. All of the tetracycline-resistant strains tested contained a single tetracycline resistance gene, tetQ. tetQ encodes a ribosome protection type of tetracycline resistance (28). No hybridization was detected when probes representing other tetracycline resistance genes, tetC, tetL, tetM, or tetB(P), were used. It is clear from Table 2 that the incidence of strains carrying tetQ has risen steadily from the pre-1970 period to the 1990s. tetQ was also found in strains obtained from a sewage treatment plant, indicating that carriage of the gene was maintained in this environment.
TABLE 2.
Distribution of tetQ and of MLS- and CTnDOT-specific genes found in community and clinical Bacteroides sp. isolates
| Isolate groupc (n) | % of isolates with resistance or hybridizing to the indicated probe
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tcr | CTnDOT element probesa
|
Emr | MLS and IS probesb
|
|||||||
| tetQ | CTn ends | rteB | rteC | ermF | ermG | ermB | IS4351d | |||
| Community | ||||||||||
| VPI, pre-1970 (32) | 28 | 28 | 34 | 47 | 31 | 0 | 0 | 0 | 0 | 0 |
| VPI, 1970–1980 (33) | 42 | 42 | 48 | 48 | 42 | 0 | 0 | 0 | 0 | 0 |
| WH, 1996–1997 (86) | 80 | 83 | 81 | 83 | 76 | 29 | 15 | 0 | 2 | 5 |
| Sewage, 1997 (16) | 62 | 62 | 88 | 81 | 75 | 38 | 12 | 6 | 6 | 0 |
| Clinical | ||||||||||
| VPI, pre-1970 (23) | 22 | 22 | 30 | 43 | 26 | 9 | 9 | 0 | 0 | 0 |
| Loyola VA (65) | 83 | 83 | 85 | 83 | 80 | 31 | 17 | 8 | 2 | 3 |
| WAL Cm (20) | NDe | 60 | 60 | 90 | 60 | 5 | 5 | 0 | 0 | 0 |
| WAL Tc (14) | 100 | 100 | 86 | 93 | 79 | 64 | 50 | 14 | 14 | 0 |
| Other (10) | 80 | 80 | 90 | 90 | 80 | 70 | 60 | 10 | 0 | 20 |
Genes found on a family of Bacteroides 50- to 70-kbp CTn's called the CTnDOT family (38). The sources of the DNAs used for probes are described in Table 1.
MLS genes or erm genes found by hybridization. Active genes confer resistance to all three groups of antibiotics. Several strains hybridized to two different erm probes.
Strains are described in Materials and Methods. Cmr, all strains in this group were chloramphenicol resistant. Only two other strains, one community isolate and one clinical isolate, were found to be chloramphenicol resistant. Tcr, all the strains in this group were tetracycline resistant.
IS4351 has usually been found associated with Bacteroides regular compound transposons that also contain ermF. All of the IS4351-containing strains also hybridized to the ermF probe.
ND, not done.
Carriage of erythromycin resistance genes (erm genes) has also increased. Seventy-two percent of the erythromycin-resistant strains (51 of 71) contained either ermF or ermG. ermF and ermG are members of the macrolide-lincosamide-streptogramin B (MLS) family of resistance genes (32, 51). These genes also confer resistance to clindamycin, a drug that has been used to treat Bacteroides infections in the past. The fact that so many resistant strains harbored these two erm genes suggests that they too are being transferred horizontally. Although ermF has been associated primarily with Bacteroides and related genera (10, 12), ermG was first found in a gram-positive soil bacterium, Bacillus sphaericus (25). None of the strains in this survey hybridized to the ermA, ermC, or ermQ probes (Table 1); however, six of the strains surveyed contained DNA that cross-hybridized with the ermB probe. ermB, like ermG, has been found mainly in the gram-positive bacteria and is prevalent in clinical isolates of Clostridium, Streptococcus, and Enterococcus spp. (32). All of the Bacteroides strains carrying ermB or ermG were recent isolates, whereas some ermF-containing strains were found among the pre-1970 clinical isolates. Thus, it appears that ermB and ermG have entered the Bacteroides species more recently than ermF.
The strains included in the survey represented 10 different Bacteroides species in both the clinical and community isolates. These species are only distantly related to each other and share DNA-DNA hybridization values ranging from 5 to 45% (17). Finding the same resistance genes detectable by Southern hybridization, which indicates >80% nucleotide identity in such distantly related strains, suggested that horizontal gene transfer, and not dissemination of one or a few resistant strains, was responsible for the high incidence of carriage of tetQ, ermB, ermF, and ermG.
Sequence analysis of tetQ genes from different strains.
If recent horizontal gene transfer was responsible for the widespread carriage of tetQ and the erm genes, the genes in different strains should be virtually identical at the DNA sequence level. Of the erm genes, we were particularly interested in the ermB and ermG genes because of their known gram-positive origin. The Bacteroides ermF genes found on transposons and CTn's are >99% identical, and the original source of this gene is not known (10, 12). tetQ PCR products from 33 isolates representing 10 species (13 pre-1970 strains and 20 post-1980 strains) were sequenced and compared. The results are summarized in Table 3. The sequences were compared to the tetQ sequences deposited in GenBank. The sequences of the 33 strains had 96 to 100% identity to the tetQ-3 gene found on the conjugative transposon CTnDOT (Table 3) (accession no. X58717). Genes from 20 of the 33 strains were 98 to 100% identical to tetQ-1 from B. fragilis strain BF2 (accession no. Y08615), including 9 of the 13 pre-1970 strains. The tetQ on CTnV479 is still the most divergent, with only 89 to 90% identity to the other tetQ genes in either Bacteroides or Prevotella strains (27). The high sequence identity of the tetQ alleles supports the hypothesis that tetQ has been spread by horizontal gene transfer. This result also rules out convergent evolution, the independent evolution of two versions of the same gene. The amino acid sequences of such genes can be very similar if there is strong selection for a particular sequence. Even two proteins with the same amino acid sequence, however, can be encoded by genes whose sequences differ by as much as 20% due to 3rd-base wobble.
TABLE 3.
Summary of the Bacteroides tetQ sequence data
| Group (no. of strains) | % Identity (no. of strains) to the following tetQ allele, source, and RFLP patterna:
|
||
|---|---|---|---|
| tetQ-1, CTnDOT, A | tetQ-2, BF1126, C or D | tet-3, BF-2, B | |
| VPI, 1960–1970s (13)b | 100 (1) | 100 (1) | 100 (3) |
| >96 (12) | >98 (6) | ||
| Woods Hole community isolates (10) | >96 (10) | 99–100 (2) | 99–100 (6) |
| Clinical isolates, 1980–presentc (10) | >96 (3) | 99–100 (2) | 100 (5) |
The accession numbers for the three Bacteroides tetQ alleles are as follows: for tetQ-1, X58717; for tetQ-2, Z21523; and for tetQ-3, Y08615. They are all >96% identical to each other, and they all were isolated from Bacteroides strains containing a CTn element. The RFLP pattern for the tetQ rteA rteB rteC region is shown in Fig. 1 and 2. The highest correlation (there were exceptions) of the tetQ allele identity with the Southern blot RFLP pattern observed with the tetQ, rteA, rteB, and rteC probes is also indicated.
Ten different species are represented by these strains.
The known CTnDOT element sequences (e.g., CTnDOT, CTnERL, CTn12256) described by Nikolich et al. (27) are not included. CTnV479 is still the only tetQ sequence with <96% identity to any of the three tetQ alleles shown.
Sequence analysis of the six ermB-containing isolates revealed that two contained genes that were identical to the erm(B)-P gene found previously in Clostridium perfringens and Streptococcus pneumoniae (3, 32), and the remaining four differed only by 1 to 3 nucleotides (Table 4). A similarly high level of sequence identity was seen when the 442-bp internal sequences of nine ermG genes were compared. These had 1 to 5 nucleotide differences within this region compared to the ermG in Bacillus sphaericus (25) and could be grouped into four groups by sequence (Table 5). The ermG genes from two of the strains were identical to the ermG found on CTn7853 and fell into group II, with 3 nucleotide differences. The finding that there was more sequence diversity among alleles of tetQ than among alleles of ermG and ermB is consistent with our results in Table 2, which indicate that tetQ has been in Bacteroides species longer than either ermG or ermB.
TABLE 4.
Bacteroides ermB sequence comparisons to C. perfringens erm(B)-P
| Strain | Source | Change at nucleotide positiona:
|
||
|---|---|---|---|---|
| A433 | C450 | C522 | ||
| Bov7991b | Clinical, WAL | |||
| WH202c | Community, Woods Hole, 1997 | |||
| WH207 | Community, Woods Hole, 1997 | G | ||
| BF8371 | Clinical, WAL | G | ||
| DH3760 | Clinical, Loyola | C | G | |
| WH714 | Community (sewage), Woods Hole, 1997 | C | G | T |
The 639-bp PCR-amplified ermB product from each of the Bacteroides strains was sequenced and compared to the ermB [erm(B)-P] of C. perfringens (accession no. X58285). Only the differences are indicated.
A Bacteroides ovatus strain that contains both ermB and ermG sequences.
A 1997 Woods Hole isolate that is phenotypically sensitive to erythromycin.
TABLE 5.
Differences between the Bacteroides ermG sequences and Bacillus sphaericus ermG
| Strain | Source | Groupa | Change at nucleotide positionb:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| C509 | G582 | G602 | T678 | G715 | T810 | C853 | |||
| Bov7991 | WAL | I | T | ||||||
| BT7853 | WAL | II | T | A | C | ||||
| DH4083 | Loyola | II | T | A | C | ||||
| DH4072 | Loyola | II | T | A | C | ||||
| DH3716 | Loyola | III | A | T | C | T | |||
| DH3717 | Loyola | III | A | T | C | T | |||
| DH4140 | Loyola | III | A | T | C | T | |||
| WH713 | Sewage, 1997 | III | A | T | C | T | |||
| BF6436-5 | Clinical, Yale | IV | T | A | T | C | T | ||
The strains are grouped according to sequence identities. BT7853, group II, contains CTn7853. The other two strains in group II also hybridized to the CTn7853-specific probe.
The sequences of the 442-bp Bacteroides ermG PCR products were compared to the sequence of ermG from Bacillus sphaericus (accession no. M15332). The G602-to-T602 difference shared by all of the Bacteroides sequences is indicated by boldface.
Type of gene transfer elements associated with the horizontal gene transfer.
Previous studies, which were limited to a small number of clinical isolates, had shown that tetQ in Bacteroides spp. was carried on two different types of CTn's, one represented by CTnDOT and one represented by CTn7853 (29, 39). ermF was found previously on several CTn's of the CTnDOT group and on three transmissible plasmids (24). ermG, however, has been found only on CTn7853 (8). Virtually all of the strains that harbored tetQ also harbored DNA that hybridized with a probe from the ends of CTnDOT (5). Whereas more than 80% of CTnDOT has now been sequenced, making it easier to design probes that identify elements of this family of CTn's, only a small amount of sequence outside the tetQ region of CTn7853 is available. Using a 1-kbp fragment located 6 kbp upstream of tetQ as a probe for CTn7853 type CTn's, we found only two additional strains that cross-hybridized to the probe; they both contained ermG and were the other two strains in Group II (Table 5), with an ermG sequence identical to that of CTn7853 in B. thetaiotaomicron 7853. Thus, it appears that CTn7853 type CTn's are not widespread in Bacteroides and that the CTnDOT type elements are the predominant type of CTn. There were strains that hybridized to the CTnDOT end probe that did not contain tetQ, for example, the VPI strain B. uniformis 0061. B. uniformis 0061 has been shown previously to carry a cryptic CTn, CTnXBU4422, which is related to CTnDOT (42).
Bacteroides strains commonly have one or more plasmids, and some of these plasmids are either self-transmissible or mobilizable (24, 35, 37). tetQ has not been found on plasmids in Bacteroides spp., but it has been found on plasmids, e.g., pRR14, from Prevotella strains (27). ermF-carrying Bacteroides plasmids have been described, and in all of these plasmids the ermF gene was linked to an insertion sequence, IS4351, which provided a promoter for the resistance gene (24). IS4351 is rare in Bacteroides; it was found in only 8 of 299 strains tested and was detected only in association with ermF sequences from the strains isolated after 1980 in this study (Table 2). IS4351 has not been found on any CTn's (36). Only 1/5 of the strains that contained ermF also contained IS4351. In the other 4/5, ermF may well be carried on CTn's such as CTnDOT.
Finding tetQ in a strain containing DNA that also hybridizes to the CTnDOT probe does not necessarily prove that tetQ is carried on a CTnDOT type element. Downstream of tetQ on CTnDOT are three regulatory genes, rteA, rteB, and rteC (Fig. 1). The CTnDOT family of CTn's represents the only elements so far found to contain this entire region (37). CTn7853 and tetQ from some Prevotella strains have a small fragment of rteA adjacent to tetQ but none of the other genes (29). If tetQ is carried on a CTnDOT type element, the rteA rteB rteC region should be intact and adjacent to tetQ. Results of the survey shown in Table 1 indicated that strains carrying the CTnDOT end sequences also carried DNA that hybridized with rteB and rteC (which are missing on CTn7853). To confirm that there was genetic linkage between tetQ and the rte genes in such strains, 60 strains were analyzed by Southern blotting. The majority of the strains tested (79%) had restriction patterns indicating that the tetQ rteA rteB rteC region was present, although there were some restriction fragment length polymorphisms (RFLP) (Fig. 1 and 2). The CTnDOT pattern A or A′ (EcoRV site missing or not cutting) with the two small fragments containing rteC sequences was not the predominant one. Instead, elements with pattern B, with the rteC sequences all contained on a 2.6-kbp fragment, as shown in Fig. 1 and 2, predominated in all but one of the groups of strains tested (Fig. 3). This pattern correlated fairly well with the strains whose tetQ sequence had 98 to 100% identity to tetQ-3 from B. fragilis BF-2 (accession no. Y08615; G. Reysset, unpublished data). Several of the strains tested contained more than one element (four instead of two end junction fragments) and had a mixed pattern such as A∗. For example, B. fragilis ERL, a clinical isolate, had two CTnDOT type elements: one (CTnERL) had pattern A and the other (CTnERL2) had the A′ pattern (42).
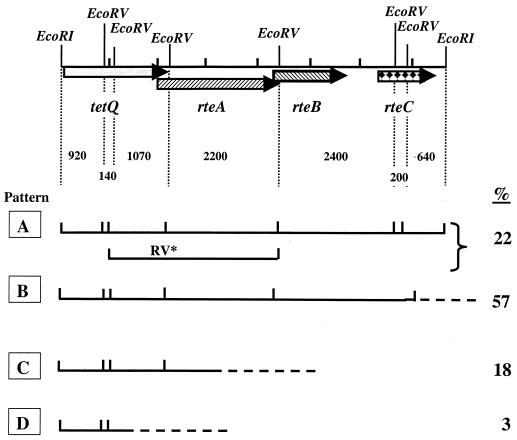
FIG. 1.
Diagrammatic representation of the restriction patterns seen on Southern blots of DNA from different strains, which was hybridized with a probe that detects the tetQ rteA rteB rteC region of the CTnDOT type CTn's. The fragment sizes and the locations of the genes within the 7.7-kbp EcoRI fragment used as the probe are shown at the top. Pattern A has the restriction fragment profile of CTnDOT and closely related elements. Some of the CTnDOT family of elements (CTnERL2) lack an EcoRV site and have the fragment labeled RV∗. This pattern is referred to as A′. Occasionally this EcoRV site does not cut completely, and a mixed pattern is observed (indicated as A∗ in the Southern blot in Fig. 2). Strains exhibiting pattern B are missing the two small rteC fragments (C0.2 and C0.64 in Fig. 2), and all of the rteC homology is located in the 2.6-kbp fragment labeled BC2.6 in Fig. 2. Strains that exhibit the C pattern are heterogeneous. A few lack rteC completely (CTn7853; C3 in Fig. 2), and others just have a very different pattern, but all of the genes on the probe are present (CTnV479; C2 in Fig. 2). Pattern D is rare; these strains have tetQ and usually neither rteB nor rteC sequences. The percentage of the 60 strains exhibiting each of the patterns is shown at the right.
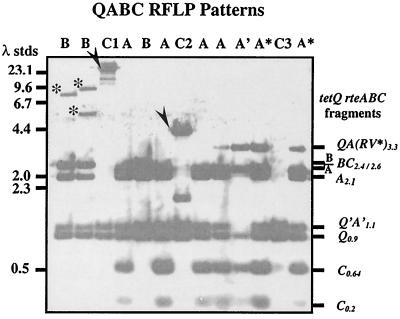
FIG. 2.
Southern blot of the EcoRI- and EcoRV-digested cellular DNA from tetQ-containing Bacteroides isolates. The blot was first probed with an rteC probe and then reprobed with the tetQ-rteA-rteB-rteC-containing probe. The blot is overexposed so that the small rteC-containing bands (C0.2 and C0.64) can be observed for pattern A. These sequences appear in the BC2.6 band of pattern B. The sizes of the HindIII lambda DNA size standards (stds) are given on the left. The sizes and contents of the major bands hybridizing to the probes are shown on the right. A schematic of the region being probed and the expected sizes is shown in Fig. 1. The patterns for each lane are labeled according to the scheme described in Fig. 1. The rteC-containing fragments for patterns C1 and C2 are indicated by arrows. These fragments also contain rteB, and C2 is for CTnV479. The B patterns in lanes 1 and 2 also contain extra hybridizing bands (indicated by asterisks) that hybridize to rteB but not rteC or tetQ probes. The A* patterns may be due to a strain containing both an A and an A′ CTn as is seen for B. fragilis ERL or the pattern may be due to partial digestion of the EcoRV site between tetQ and rteA (Fig. 1), as is sometimes observed for CTn12256 (data not shown).
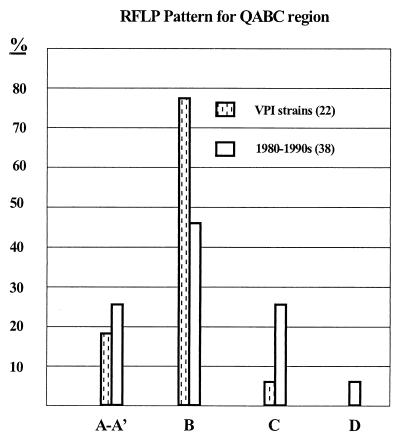
FIG. 3.
Differences in restriction patterns (depicted in Fig. 1 and 2) between older isolates and modern isolates. Both groups of strains include both community and clinical isolates. The 22 VPI strains were isolated before 1970. The RFLP patterns are shown in Fig. 1, and Southern blot patterns A, B, and C are shown in Fig. 2.
An interesting trend was seen in CTn's contained in more recently isolated strains (Fig. 3). Whereas virtually all the older isolates carried CTn's with the A–A′ or B restriction pattern, the pattern in many of the newer isolates indicated that parts of the rteA rteB rteC region had been lost or rearranged, as seen in the patterns of C1, C2, and C3 in the Southern blot shown in Fig. 2 and summarized in Fig. 3. C3 is the rare pattern observed for CTn7853 that lacks both rteB and rteC, whereas patterns C1 and C2 contained both rteB and rteC sequences (Fig. 3). C1 and C2 type isolates also hybridized to the CTnDOT end probe, whereas C3 isolates did not. Since the rte genes are essential for transfer of CTnDOT type elements, this may indicate that these C pattern strains that hybridized to the CTnDOT ends contain CTn's that are no longer transmissible.
DISCUSSION
Taken together, our results show that the substantial increase in tetracycline resistance among Bacteroides isolates that has occurred over the past few decades was due to the horizontal spread of a single gene, tetQ, on CTn's of the CTnDOT type. ermF and ermG also appear to be spreading by horizontal gene transfer, possibly also on CTn's such as CTnDOT and CTn7853. It is likely that these gene transfer events took place in the human colon, because Bacteroides species are found primarily in the human colon and are present, if at all, in low numbers in the intestines of other animals or in the environment. The only environmental site outside the human colon to harbor significant numbers of Bacteroides strains would be a sewage treatment plant. Transfer of resistant Bacteroides strains from sewage treatment plants to humans, however, would be unlikely to have produced the widespread colonization seen in our studies. Also, as is evident from the 16 sewage plant isolates tested (Table 2), carriage seems to be lower in strains obtained from this setting than in strains isolated from the human colon.
A feature of the CTnDOT type elements allows us to speculate about what may have caused this extensive spread of resistance genes. Most of the CTnDOT type elements exhibit regulated transfer (38). That is, transfer occurs only if the donors are first stimulated with low levels of the antibiotic tetracycline. After tetracycline induction, transfer frequencies rise 1,000- to 10,000-fold. No other class of antibiotics has this effect. Thus, tetracycline use in the community probably played a role not only in selecting for maintenance of tetQ but also in causing it to be transferred in the first place.
Our results show clearly that once a resistance gene enters Bacteroides species, it can be spread widely among these species if it becomes part of a CTn or some other transmissible element. Our survey also provided evidence that Bacteroides species may share DNA with members of genera outside the Bacteroides-Prevotella group. ermF was first found in Bacteroides species (12) but clearly originated in a low-G+C organism (33%). tetQ, which appears to have originated in Bacteroides spp., has 40% G+C, which is the average observed for this genus (17). The origin of ermG (27% G+C) is also unknown, but this gene has been found in Bacillus sphaericus, a gram-positive soil bacterium, as well as in B. thetaiotaomicron 7853 (8). Our survey also turned up six Bacteroides strains that had acquired an ermB gene virtually identical to an ermB from C. perfringens, Streptococcus pneumoniae, and Enterococcus faecalis. This finding suggests that even bacteria that do not normally reside in the colon (S. pneumoniae) or that reside there in low numbers (C. perfringens and E. faecalis) can donate DNA to members of numerically predominant groups of colonic bacteria. Our results do not prove that DNA was transferred directly from these species to Bacteroides species, but the presence of ermG and ermB in multiple isolates from different geographical locations indicates that some genetic connection, however indirect, is open between the gram-positive bacteria where the genes appear to have originated and the gram-negative Bacteroides species.
Given that some gram-positive resistance genes appear to have moved into Bacteroides, it is surprising that tetM, a resistance gene that is distantly related to tetQ and confers the same type of resistance, was not found in any of these Bacteroides isolates. The tetM gene is carried on several gram-positive CTn's that appear to have a broad host range, at least under laboratory conditions. The failure to find tetM could indicate that there might be barriers to transfer of some types of elements in vivo or that there is a lack of selective pressure for tetracycline resistance determinants other than tetQ, since 80% of the Bacteroides strains are already tetracycline resistant due to tetQ.
Once in Bacteroides species, conjugal transfer mediated primarily by CTn's can spread these genes widely among different Bacteroides species. This study is the first to demonstrate what scientists have long suspected, that the human colon is a site that is highly conducive to horizontal gene transfer and to stable maintenance of transferred resistance genes. It is also the first study to associate an increase in antibiotic resistance with carriage of a particular type of conjugal element, in this case CTn's of the CTnDOT class. CTn's have also been found in gram-positive bacteria (1, 6) and in the Escherichia coli phylogenetic group of gram-negative bacteria (14, 49). The results of our study suggest that CTn's are making a major contribution to the transmission of antibiotic resistance genes.
ACKNOWLEDGMENTS
We are grateful to the laboratories of S. Feingold and D. Hecht for providing clinical isolates and to the students in the Microbial Diversity Course at Woods Hole, Mass., and Caroline Plugge for the community and sewage isolates. Andy Cooper provided the clone used to identify CTn7853-related elements. We thank Anamika Gupta for furnishing the sequences of some of the ermB alleles.
This work was supported by grant AI22383 from the National Institutes of Health.
REFERENCES
- 1.Ayoubi P, Kilic A O, Vijaykumar M N. Tn5253, the pneumococcal omega (cat tet) BM6001 element, is a composite structure of two conjugative transposons, Tn5251 and Tn5252. J Bacteriol. 1991;173:1617–1622. doi: 10.1128/jb.173.5.1617-1622.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berryman D I, Lyristis M, Rood J I. Cloning and sequence analysis of ermQ, the predominant macrolide-lincosamide-streptogramin B resistance gene in Clostridium perfringens. Antimicrob Agents Chemother. 1994;38:1041–1046. doi: 10.1128/aac.38.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berryman D I, Rood J I. The closely related ermB-ermAM genes from Clostridium perfringens, Enterococcus faecalis (pAMβ1), and Streptococcus agalactiae (pIP501) are flanked by variants of a directly repeated sequence. Antimicrob Agents Chemother. 1995;39:1830–1834. doi: 10.1128/aac.39.8.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler E, Joiner K A, Malamy M, Barlett J C, Tally F P. Transfer of tetracycline or clindamycin resistance among strains of Bacteroides fragilis in experimental abscesses. J Infect Dis. 1984;150:20–24. doi: 10.1093/infdis/150.1.20. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Q, Paszkiet B J, Shoemaker N B, Gardner J F, Salyers A A. Integration and excision of a Bacteroides conjugative transposon, CTnDOT. J Bacteriol. 2000;182:4035–4043. doi: 10.1128/jb.182.14.4035-4043.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clewell D B, Flannagan S E. The conjugative transposons of gram-positive bacteria. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 369–393. [Google Scholar]
- 7.Cooper A J, Kalinowski A P, Shoemaker N B, Salyers A A. Construction and characterization of a Bacteroides thetaiotaomicron recA mutant: transfer of Bacteroides integrated conjugative elements is RecA independent. J Bacteriol. 1997;179:6221–6227. doi: 10.1128/jb.179.20.6221-6227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper A J, Shoemaker N B, Salyers A A. The erythromycin resistance gene from the Bacteroides conjugal transposon Tcr Emr 7853 is nearly identical to ermG from Bacillus sphaericus. Antimicrob Agents Chemother. 1996;40:506–508. doi: 10.1128/aac.40.2.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson J. Genetic exchange between bacteria in the environment. Plasmid. 1999;42:73–91. doi: 10.1006/plas.1999.1421. [DOI] [PubMed] [Google Scholar]
- 10.Fletcher H M, Macrina F L. Molecular survey of clindamycin and tetracycline resistance determinants in Bacteroides species. Antimicrob Agents Chemother. 1991;35:2415–2418. doi: 10.1128/aac.35.11.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guiney D G, Hasegawa P. Transfer of conjugal elements in oral black-pigmented Bacteroides (Prevotella) spp. involves DNA rearrangements. J Bacteriol. 1992;174:4853–4855. doi: 10.1128/jb.174.14.4853-4855.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halula M C, Manning S, Macrina F L. Nucleotide sequence of ermFU, a macrolide-lincosamide-streptogramin (MLS) resistance gene encoding an RNA methylase from the conjugal element of Bacteroides fragilis V503 (BF12256) Nucleic Acids Res. 1991;19:3453. doi: 10.1093/nar/19.12.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrick J B, Stuart-Keil K G, Ghiorse W C, Madsen E L. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl Environ Microbiol. 1997;63:2330–2337. doi: 10.1128/aem.63.6.2330-2337.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochhut B, Jahreis K, Lengeler J W, Schmid K. CTnscr94, a conjugative transposon found in enterobacteria. J Bacteriol. 1997;179:2097–2101. doi: 10.1128/jb.179.7.2097-2102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen L B, Ahrens P, Dons L, Jones R N, Hammerum A M, Aarestrup F M. Molecular analysis of Tn1546 from Enterococcus faecium isolated from animals and humans. J Clin Microbiol. 1998;36:437–442. doi: 10.1128/jcm.36.2.437-442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang S C, Paul J H. Gene transfer by transduction in the marine environment. Appl Environ Microbiol. 1998;64:2780–2787. doi: 10.1128/aem.64.8.2780-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson J L. Taxonomy of the Bacteroides: deoxyribonucleic acid homologies among Bacteroides fragilis and other saccharolytic Bacteroides species. Int J Syst Bacteriol. 1978;28:245–256. [Google Scholar]
- 18.Johnson J L, Harich B. Ribosomal ribonucleic acid homology among species of the genus Bacteroides. Int J Syst Bacteriol. 1986;36:71–79. [Google Scholar]
- 19.Kuritza A P, Getty C E, Shaughnessy P, Hesse R, Salyers A A. DNA probes for identification of clinically important Bacteroides species. J Clin Microbiol. 1986;23:343–349. doi: 10.1128/jcm.23.2.343-349.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lacks S A, Lopez P, Greenberg B, Espinosa M. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J Mol Biol. 1986;192:753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- 21.Lacroix J M, Walker C B. Detection and prevalence of the tetracycline resistance determinant Tet Q in the microbiota associated with adult periodontitis. Oral Microbiol Immunol. 1996;11:282–288. doi: 10.1111/j.1399-302x.1996.tb00182.x. [DOI] [PubMed] [Google Scholar]
- 22.Levy S B, McMurry L M, Barbosa T M, Burdett V, Courvalin P, Hillen W, Roberts M C, Rood J I, Taylor D E. Nomenclature for new tetracycline resistance determinants. Antimicrob Agents Chemother. 1999;43:1523–1524. doi: 10.1128/aac.43.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L Y, Shoemaker N B, Salyers A A. Location and characteristics of the transfer region of a Bacteroides conjugative transposon and regulation of transfer genes. J Bacteriol. 1995;177:4992–4999. doi: 10.1128/jb.177.17.4992-4999.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macrina F L, Smith C J. Gene transmission, MLS, and tetracycline resistance in Bacteroides. In: Sebald M, editor. Genetics and molecular biology of anaerobic bacteria. New York, N.Y: Springer-Verlag Inc.; 1993. pp. 474–489. [Google Scholar]
- 25.Monod M, Mohan S, Dubnau D. Cloning and analysis of ermG, a new macrolide-lincosamide-streptogramin B resistance element from Bacillus sphaericus. J Bacteriol. 1987;169:340–350. doi: 10.1128/jb.169.1.340-350.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Netherwood T, Bowden R, Harrison P, O'Donnell A G, Parker D S, Gilbert H J. Gene transfer in the gastrointestinal tract. Appl Environ Microbiol. 1999;65:5139–5141. doi: 10.1128/aem.65.11.5139-5141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikolich M P, Hong G, Shoemaker N B, Salyers A A. Evidence for natural horizontal transfer of tetQ between bacteria that normally colonize humans and bacteria that normally colonize livestock. Appl Environ Microbiol. 1994;60:3255–3260. doi: 10.1128/aem.60.9.3255-3260.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikolich M P, Shoemaker N B, Salyers A A. A Bacteroides tetracycline resistance gene represents a new class of ribosome protection tetracycline resistance. Antimicrob Agents Chemother. 1992;36:1005–1012. doi: 10.1128/aac.36.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikolich M P, Shoemaker N B, Wang G R, Salyers A A. Characterization of a new type of Bacteroides conjugative transposon, Tcr Emr 7853. J Bacteriol. 1994;176:6606–6612. doi: 10.1128/jb.176.21.6606-6612.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paster B J, Dewhirst F E, Olsen I, Fraser G J. Phylogeny of Bacteroides, Prevotella, and Porphyromonas spp. and related bacteria. J Bacteriol. 1994;176:725–732. doi: 10.1128/jb.176.3.725-732.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasmussen J L, Odelson D A, Macrina F L. Complete nucleotide sequence of insertion element IS4351 from Bacteroides fragilis. J Bacteriol. 1987;169:3573–3580. doi: 10.1128/jb.169.8.3573-3580.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts M C, Sutcliffe J, Courvalin P, Jensen L B, Rood J, Seppala H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother. 1999;43:2823–2830. doi: 10.1128/aac.43.12.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saito H, Miura K I. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- 34.Salyers A A, Amabile-Cuevas C F. Why are antibiotic resistance genes so resistant to elimination? Antimicrob Agents Chemother. 1997;41:2321–2325. doi: 10.1128/aac.41.11.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salyers A A, Shoemaker N B. Conjugative transposons: the force behind the spread of antibiotic resistance genes among Bacteroides clinical isolates. Anaerobe. 1995;1:143–150. doi: 10.1006/anae.1995.1011. [DOI] [PubMed] [Google Scholar]
- 36.Salyers A A, Shoemaker N B. Resistance gene transfer in anaerobes: new insights, new problems. Clin Infect Dis. 1996;23(Suppl. 1):S36–S43. doi: 10.1093/clinids/23.supplement_1.s36. [DOI] [PubMed] [Google Scholar]
- 37.Salyers A A, Shoemaker N B, Li L Y. In the driver's seat: the Bacteroides conjugative transposons and the elements they mobilize. J Bacteriol. 1995;177:5727–5731. doi: 10.1128/jb.177.20.5727-5731.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salyers A A, Shoemaker N B, Stevens A M. Tetracycline regulation of conjugal transfer genes. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 393–400. [Google Scholar]
- 39.Salyers A A, Shoemaker N B, Stevens A M, Li L Y. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shoemaker N B, Barber R D, Salyers A A. Cloning and characterization of a Bacteroides conjugal tetracycline-erythromycin resistance element by using a shuttle cosmid vector. J Bacteriol. 1989;171:1294–1302. doi: 10.1128/jb.171.3.1294-1302.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shoemaker N B, Wang G-R, Salyers A A. Multiple gene products and sequences required for the excision of the mobilizable integrated Bacteroides element NBU1. J Bacteriol. 2000;182:928–936. doi: 10.1128/jb.182.4.928-936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoemaker N B, Salyers A A. A cryptic 65-kilobase-pair transposonlike element isolated from Bacteroides uniformis has homology with Bacteroides conjugal tetracycline resistance elements. J Bacteriol. 1990;172:1694–1702. doi: 10.1128/jb.172.4.1694-1702.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sloan J, McMurry L M, Lyras D, Levy S B, Rood J I. The Clostridium perfringens Tet P determinant comprises two overlapping genes: tetA(P), which mediates active tetracycline efflux, and tetB(P), which is related to the ribosomal protection family of tetracycline-resistance determinants. Mol Microbiol. 1994;11:403–415. doi: 10.1111/j.1365-2958.1994.tb00320.x. [DOI] [PubMed] [Google Scholar]
- 44.Smith C J. Nucleotide sequence analysis of Tn4551: use of ermFS operon fusions to detect promoter activity in Bacteroides fragilis. J Bacteriol. 1987;169:4589–4596. doi: 10.1128/jb.169.10.4589-4596.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith C J, Rogers M B, McKee M L. Heterologous gene expression in Bacteroides fragilis. Plasmid. 1992;27:141–154. doi: 10.1016/0147-619x(92)90014-2. [DOI] [PubMed] [Google Scholar]
- 46.Stevens A M, Shoemaker N B, Li L Y, Salyers A A. Tetracycline regulation of genes on Bacteroides conjugative transposons. J Bacteriol. 1993;175:6134–6141. doi: 10.1128/jb.175.19.6134-6141.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Bogaard A E, Stobberingh E E. Epidemiology of resistance to antibiotic. Links between animals and humans. Int J Antimicrob Agents. 2000;14:327–335. doi: 10.1016/s0924-8579(00)00145-x. [DOI] [PubMed] [Google Scholar]
- 49.Waldor M K, Tschape H, Mekalanos J J. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 1996;178:4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Shoemaker N, Wang G-R, Salyers A A. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J Bacteriol. 2000;182:3559–3571. doi: 10.1128/jb.182.12.3559-3571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson K H, Ikeda J S, Blitchington R B. Phylogenic placement of community members of human colonic biota. Clin Infect Dis. 1997;25(Suppl. 2):S114–S116. doi: 10.1086/516230. [DOI] [PubMed] [Google Scholar]