Abstract
Hepatocellular carcinoma (HCC) is the most common malignancy of hepatocytes accounting for 75-85% of primary hepatic carcinoma cases. Small extracellular vesicles (sEVs), previously known as exosomes with a diameter of 30-200 nm, can transport a variety of biological molecules between cells, and have been proposed to function in physiological and pathological processes. Recent studies have indicated that the cargos of sEVs are implicated in intercellular crosstalk among HCC cells, paratumor cells and the tumor microenvironment. sEV-encapsulated substances (including DNA, RNA, proteins and lipids) regulate signal transduction pathways in recipient cells and contribute to cancer initiation and progression in HCC. In addition, the differential expression of sEV cargos between patients facilitates the potential utility of sEVs in the diagnosis and prognosis of patients with HCC. Furthermore, the intrinsic properties of low immunogenicity and high stability render sEVs ideal vehicles for targeted drug delivery in the treatment of HCC. The present review article summarizes the carcinogenic and anti-neoplastic capacities of sEVs and discusses the potential and prospective diagnostic and therapeutic applications of sEVs in HCC.
Keywords: hepatocellular carcinoma, small extracellular vesicles, exosomal RNAs, biomarker
1. Introduction
Liver cancer is ranked as the sixth most prevalent malignancy worldwide, and was the third highest cause of cancer-associated mortality worldwide in 2020 with ~905,677 new cases and 830,180 cancer-associated mortalities annually (1). Hepatocellular carcinoma (HCC) is the predominant subtype of hepatic carcinoma, and accounts for 75-85% of all primary liver cancer cases (2). Infection with hepatitis B or C viruses (HBV or HCV, respectively) causes chronic liver injury and has recently been reported to play a pivotal place in the carcinogenesis and development of HCC (3). Due to the vaccination against HBV, the prevalence of HBV and the incidence rate of HCC have markedly decreased in numerous high-risk regions, such as China (4). However, the current situation is far from satisfactory in numerous low- and middle-income countries, due to the shortage of HBV vaccines, and the lack of improved sanitation and regular screening (5). Thus, the 5-year overall survival rate of patients with HCC remains low (<25%), and large-scale efforts are urgently required to elucidate the mechanisms underlying the development of neoplasia and to improve the preliminary diagnostic rate of HCC (6,7).
Small extracellular vesicles (sEVs), which were previously known as exosomes, have a diameter of 30-200 nm and are a subset of EVs that were first described by Johnstone et al (8) in the 1980s. Following several decades of research, it was observed that sEVs not only function in cellular waste disposal, but also serve as an excellent vehicle for cell-cell communications. sEVs contain complex and diverse materials, including DNAs, RNAs, proteins, lipids and metabolites, and they shuttle these bioactive molecules between cells (9). The cargos of sEVs can be internalized by recipient cells, thus mediating the metabolic activities of recipient cells and consequently participating in both normal physiology and acquired abnormalities, such as immune responses, mammalian reproduction and development, central nervous system-related diseases and cancers (10). In HCC, accumulated evidence has indicated that sEVs play an essential role in carcinogenesis and in the remodeling of the tumor microenvironment (TME), as well as in proliferation, metastasis, angiogenesis and drug resistance (11). The profiles of sEV cargos are origin-specific, and the distinct expression of sEV cargos between patients with HCC and healthy subjects renders sEVs a potential diagnostic biomarker for HCC (12). Furthermore, certain sEV RNAs may serve as molecular markers for the early detection, TNM staging, prognostic evaluation and recurrence monitoring in HCC, which may contribute more effective to diagnosis and treatment options (13). Considering the intrinsic property of sEVs of transferring information and altering the biological response of recipient cells, recent studies have highlighted their potential utility values in the therapeutic fields of several diseases, including cardiovascular diseases and cancers (14,15).
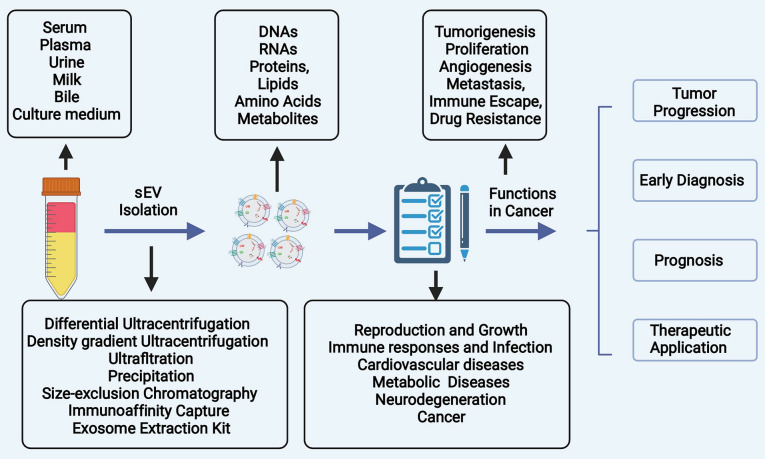
The present review article summarizes the biogenesis of sEVs, as well as the role of sEVs in the tumorigenesis and progression of HCC. In addition, the potential and emerging clinical applications of sEVs in the diagnosis and treatment of HCC are discussed (Fig. 1).
Figure 1.
The fundamental purpose of the present review was to introduce the biogenesis of sEVs, generalize the role of sEVs payloads in the initiation and development of HCC, and dialectically discuss the clinical applications of sEVs in the diagnosis and possible treatment applications of HCC. sEVs, small extracellular vesicles; HCC, hepatocellular carcinoma.
2. Biology of sEVs
'EV' is a heterogeneous collective term for phospholipid bilayer membrane-encapsulated nano or microvesicles. Traditionally, EVs were broadly categorized into cytoplasmic membrane-derived ectosomes and exosomes of endosome-origin (16). However, without optimal isolation methods and real-time imaging technologies to visualize the process of release or specific markers of different subtypes of EVs, the differentiation between exosomes and small ectosomes is unlikely due to their analogous intrinsic properties and the overlapping size. Thus, the latest guideline of Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018) proposed the use of standard terminologies for EV subtypes followed by physical characteristics, biochemical composition and the condition of progenitor cells (17). In the present review article, the term 'EV' encompasses a heterogeneous population of both exosomes and nano-scaled ectosomes with a diameter <200 nm.
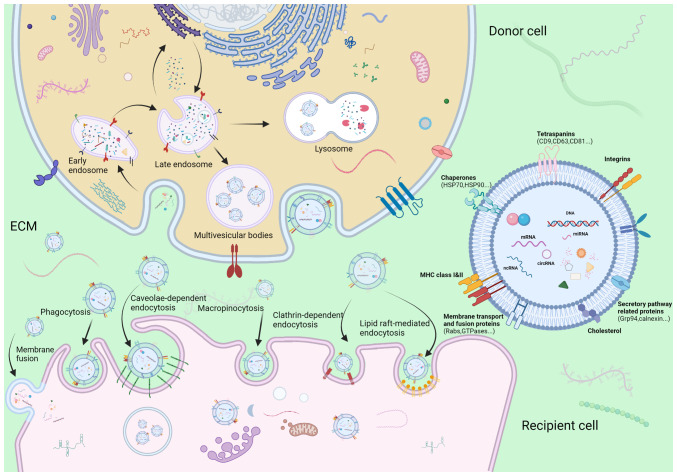
Ectosomes, which are microvesicles and microparticles with a diameter ranging from 50 to 1,000 nm, are vesicles produced directly by the outward budding of the plasma membrane (18). By contrast, the process of the synthesis and release of exosomes is a more complicated and intricate sequence of multiple fusion events, budding of the plasma membrane and releasing of specific payloads (Fig. 2). The first inward invagination of a lipid bilayers contributes to the generation of early-sorting endosomes (ESEs) (19). ESEs can mature towards late-sorting endosomes (LSEs), which is followed by the formation of multivesicular bodies (MVBs) through the second intraluminal budding of the endosomal membrane, during which, specific bioactive compounds such as nucleic acids, proteins, and lipids are gradually enriched in intraluminal vesicles (ILVs) (20). Although the mechanisms underlying the formation of ILVs and specific bioactive compound sorting system have not yet been well elucidated, the majority of oncologists hypothesize that endosomal sorting complex required for transport (ESCRT) facilitates exosome budding. Of note, an ESCRT-independent mechanism may play a role in the biogenesis of exosomes, since no notable decrease in the release of exosomes was observed following the inhibition of ESCRT family activity (21). These two pathways may not be completely separated, although they function synergistically in the synthesis of exosomes (22). MVBs mainly have two endings: i) These mature MVBs may be incorporated into autophagosomes or lysosomes for hydrolysis of vesicular contents; or ii) they can be incorporated into the cellular plasma membrane and be subsequently expelled into the extracellular space as exosomes (23). When arriving to their recipient cells, sEVs are recognized and assimilated into cells via the following mechanisms: donor-acceptor interaction, membrane fusion, phagocytosis, and clathrin-independent and clathrin-dependent endocytosis, depending on their physical and biological properties (24,25). For example, angiopoietin-2 (ANGPT2)-bearing sEVs derived from HCC cells are transferred into human umbilical vein endothelial cells (HUVECs) via endocytosis (26). The processes of formation, secretion and uptake of exosomes are depicted in Fig. 2, as reported in a previous study by the authors (27).
Figure 2.
Schematic diagram illustrating the process of formation, secretion and uptake of sEVs. Inward budding of the cellular plasma membrane forms the early-sorting endosomes. Subsequently, intraluminal budding of endosomes generates MVBs encapsulating intraluminal vesicles. sEVs are ultimately liberated by incorporating of MVBs to plasma membrane and the exocytosis of intraluminal vesicles. The mechanism of sEV uptake includes donor-acceptor interaction, membrane fusion, phagocytosis, and clathrin-independent and -dependent endocytosis. sEVs, small extracellular vesicles; MVBs, multivesicular bodies; ECM, extracellular matrix.
sEVs can be excreted by almost all cells types and are abundant in the human body, existing in biological fluids, such as plasma, urine, tears, plasma and breast milk (28). sEVs can be isolated from cell culture conditioned media, multiple biofluids, or tissue using several methods. The separation of sEVs principally involves five approaches, including differential ultracentrifugation, sucrose and iodixanol density ultracentrifugation, polyethylene glycol precipitation, size exclusion chromatography (SEC) and immunoaffinity capture (29). However, it is extremely difficult to identify a single separation strategy with both a high recovery rate and high specificity. The present study aimed to systematically review the recent, cutting-edge research on sEVs and HCC, focusing on high-quality studies using differential ultracentrifugation or density gradient centrifugation as the separation methods of sEVs, with an intermediate recovery rate and purity according to MISEV2018 (17). Notably, all these aforementioned approaches have their own advantages and disadvantages; thus, a combined method, such as differential ultracentrifugation followed by SEC is scalable for future sEVs-based studies (30). Other articles (31-68) discussing methods of isolation of sEVs involving only ultracentrifugation or commercial kits are cited Table SI. Further investigations on a more effective and reproducible approach for separating sEVs are urgently required.
3. Roles of sEVs in HCC tumor formation and progression
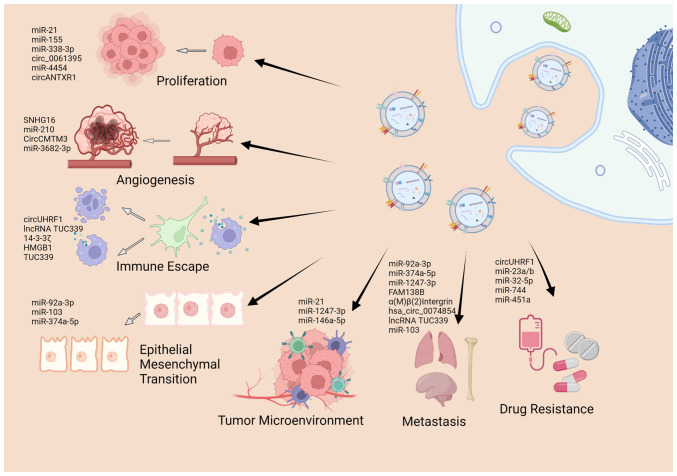
As aforementioned, sEVs encapsulate a series of cargos, including nucleic acids, proteins and lipids, and sEV-related research has mainly focused on the ability of sEVs to exchange of these cargos between cells (69,70). Previous studies on the roles of sEV cargos in cancer have demonstrated that sEVs are involved in almost all hallmarks of cancers, including tumor initiation and formation (71-75), in the remodeling of the TME (76,77), apoptosis (50), angiogenesis (78,79), metastasis (80-83), immune escape (52), and drug resistance (84). The present review article summarizes the literature that highlights the significance of sEV cargos in the carcinogenesis and development of HCC (Fig. 3), as presented in Table I.
Figure 3.
Biological function of sEV cargos in development of HCC. sEVs cargos are involved in numerous hallmarks of HCC, including proliferation, angiogenesis, epithelial-mesenchymal transition, metastasis, immune escape, drug resistance as well as remolding of the tumor microenvironment. The figure was created using Biorender (https://biorender.com/). sEVs, small extracellular vesicles; HCC, hepatocellular carcinoma.
Table I.
Roles of sEVs in the initiation and development of hepatocellular carcinoma.
| Type | Molecule | Function | Signaling/target | Year of publication | (Refs.) |
|---|---|---|---|---|---|
| miRNA | miR-21↑ | Promotes proliferation, migration, angiogenesis and invasion; inhibits apoptosis | PTEN/E-cadherin | 2019, 2018 | (71,80,91) |
| miR-10b↑ | Promotes proliferation, migration and invasion | PTEN/E-cadherin | 2019 | (80) | |
| miR-23a/b↑ | Promotes proliferation, migration and chemoresistance to 5-Fu | VHL-HIF-1α | 2019 | (115) | |
| miR-15a↓ | Inhibits proliferation, migration and invasion | SALL4 | 2021 | (76) | |
| miR-25↑ | Promotes proliferation and migration | Wnt/β-catenin | 2021 | (72) | |
| miR-155↑ | Promotes proliferation | PTEN | 2019 | (96) | |
| miR-451a↑ | Inhibits resistance to paclitaxel, proliferation, migration and invasion | ADAM10 | 2021 | (117) | |
| miR-3682-3p↑ | Inhibits angiogenesis | RAS-MEK1/2-ERK1/2 | 2021 | (101) | |
| miR-374a-5p↑ | Promotes EMT, migration and invasion | GADD45A | 2020 | (105) | |
| miR-210↑ | Promotes angiogenesis | SMAD4 and STAT6 | 2018 | (100) | |
| miR-92a-3p↑ | Promotes EMT and migration | PTEN/Akt/Snail | 2020 | (103) | |
| miR-92a-2-5p↑ | Promotes invasion | AR/PHLPP/p-AKT/β-catenin | 2020 | (82) | |
| miR-146a-5p↑ | Remodels the TME | STAT3/SALL4 | 2019 | (77) | |
| miR-30a-3p↑ | Inhibit migration and invasion | SNAP23 | 2021 | (83) | |
| miR-1247-3p↑ | Promotes migration | β1-integrin-NF-κB | 2018 | (92) | |
| miR-25-5p↑ | Promotes migration and invasion | LRRC7 | 2018 | (81) | |
| miR-4454↑ | Promotes proliferation, migration, invasion, and angiogenesis, and inhibits cycle arrest, apoptosis | Vps4A, Rab27A | 2021 | (95) | |
| miR-338-3p↑ | Inhibits proliferation, invasion and migration, and induce apoptosis, | ETS1 | 2021 | (97) | |
| miR-744↓ | Promotes proliferation and inhibits chemosensitivity to sorafenib | PAX2 | 2019 | (118) | |
| lncRNA | lncRNA H19↑ | Promotes angiogenesis and adhesion of endothelial cell | VEGF, ICAM1 | 2015 | (78) |
| FAM138B↑ | Inhibits proliferation, migration and invasion | miRNA-765 | 2020 | (106) | |
| SNHG16↑ | Promotes angiogenesis | PI3K/Akt/mTOR | 2021 | (99) | |
| lncRNA 85↑ | Promotes proliferation and migration and polarization | miRNA-324-5p | 2020 | (73) | |
| TUC339↑ | Regulation of macrophage activation | IL-1β and TNF-α | 2018 | (109) | |
| circRNA | hsa_circ_0074854↑ | Promotes migration and invasion | HuR | 2021 | (108) |
| circFBLIM1↑ | Promote progression and glycolysis | miRNA-338/LRP | 2020 | (75) | |
| circRNA-100338↑ | Promotes angiogenesis and invasion | mTOR | 2020 | (102) | |
| Protein | 14-3-3ζ↑ | Impairs the function, proliferation and activation of TILs | AXL or TGF-β/ERK | 2018 | (112) |
| NSMase1↑ | Inhibits proliferation and induce apoptosis | JNK | 2018 | (94) | |
| ENO1↑ | Promotes proliferation and metastasis | FAK/Src-p38MAPK | 2020 | (74) | |
| LOXL4↑ | Promotes migration and angiogenesis | FAK/Src | 2019 | (79) | |
| α(M) β(2) Integrin↑ | Promotes metastasis | MMP9 | 2021 | (107) | |
| HMGB1 | Promotes immune evasion | TLR 2/4, MAPK | 2018 | (113) | |
| VEGF↑ | Induces acquired resistance to AATs | - | 2019 | (84) |
Upward arrows (↑) indicate upregulation and downward arrows (↓) indicate downregulation. EMT, epithelial-mesenchymal transition; TME, tumor microenvironment; TILs, tumor-infiltrating T-lymphocytes; AATs, anti-angiogenic therapies; PTEN, phosphatase and tensin homolog; VHL-HIF-1α, von Hippel-Lindau/hypoxia-inducible factor; SALL4, spalt-like transcription factor 4; ADAM10, a disintegrin and metalloprotease 10; MEK1/2, mitogen-activated proteinkinase kinase 1/2; ERK1/2, extracellular regulated protein kinases 1/2; GADD45A, growth arrest and DNA damage 45-alpha; SMAD4, mothers against decapentaplegic homolog 4; STAT, signal transducer and activator of transcription; AR, androgen receptor; PHLPP, PH domain leucine-rich repeat protein phosphatase; SALL4, transcription factor Sal-like protein-4; SNAP23, synaptosome-associated protein 23; LRRC7, leucine-rich repeat-containing 7; Vps4A, vacuolar protein sorting 4 homolog A; ETS1, E26 transformation specific-1; PAX2, paired box gene 2; VEGF, vascular endothelial growth factor; ICAM1, intercellular adhesion molecule 1; FAM138B, family with sequence similarity 138 member B; SNHG16, small nucleolar RNA host gene 16; PI3K, phosphatidylin-ositol-3-kinase; mTOR, mechanistic target of rapamycin; IL-1β, interleukin-1β; TNF-α, tumor necrosis factor-α; HuR, human antigen; circFBLIM1, circRNA filamin binding LIM protein 1; LRP6, lipoprotein receptor-related protein 6; AXL, activity of the receptor tyrosine kinase; NSMase1, neutral sphingomyelinase 1; TGF-β, transforming growth factor-β; JNK, c-Jun N-terminal kinase; ENO1, alpha-enolase; FAK, focal adhesion kinase; LOXL4, lysyl oxidase-like 4; MAPK, mitogen-activated protein kinase; MMP9, matrix metallopeptidase 9; HMGB1, high mobility group box 1; TLR, Toll like receptor.
TME
Tumorigenesis is not a single-step event, but a consequence of long-term alteration of mutations of genes and functional changes in the TME (85). Emerging evidence suggests that sEVs participate in the initiation, formation and remodeling of the TME in HCC (11). Chronic hepatitis B (CHB) remains a main factor responsible for HCC development, and sEVs are implicated in the spread, immune regulation and antiviral response of HBV infections (86). For example, exosomes from macrophages can deliver IFN-α-related microRNAs (miRNAs or miRs) to HBV-infected hepatocytes, and activate the antiviral response to suppress HBV replication and expression (87). The exosomal long non-coding RNA (lncRNA) HOTTIP has been shown to play a role in mediating the antiviral effect of tenofovir alafenamide following HBV infection (88). It has been demonstrated that sEVs from CD4+ T-cells can enhance B cell responses and potentiate the efficacy of the hepatitis B surface antigen vaccine (89). These studies indicate that sEVs can mediate immune regulation and antiviral response in HBV infection. A previous study also indicated that sEVs may exert negative immune regulatory effects, and that they are indispensable in the transformation from liver cirrhosis (LC) to liver cancer (90). The interplay between cancer cells and the TME is an essential activity that supports or prevents tumor development and progression. In HCC, tumor cells co-exist with other non-cancerous cells that constitute the TME and enhance tumor growth via various mechanisms. sEVs can exert an effect on the information and remodeling of the TME. For instance, exosomal miR-21 derived from HCC has been found to promote the conversion of hepatic stellate cells into cancer-associated fibroblasts (CAFs), and to facilitate the formation of the TME (91). Exosomal-miR-1247-3p from HCC cells has been shown to reduce the expression of B4GALT3 in CAFs and stabilize β1-integrin, leading to the activation of fibroblasts via the NF-κB signaling pathway (92). To summarize, sEVs play an essential role in the pathogenesis of HBV-related hepatic diseases, transformation from precancerous diseases to HCC and in the formation of the TME to confer tumorigenesis in HCC.
Proliferation and apoptosis
The development of HCC can be partly attributed to the rapid proliferation and uncontrolled expansion of tumor cells, which also accounts for tumor progression and resistance to therapy. sEVs mediate tumor growth and expansion by affecting the cell cycle, proliferation rate and apoptosis of HCC cells (93-95). Cao et al (71) suggested that exosomal miR-21 can influence HCC by altering the expression of the tumor suppressor genes, PTEN and PTEN pseudogene 1. Sun et al (96) indicated that exosome-specific miR-155 targeted PTEN and consequently stimulated the proliferation of HCC cells. On the contrary, certain sEV-encapsulated cargos, such as miR-338-3p can inhibit cell proliferation, induce cell apoptosis and consequently repress the progression of HCC (97). In addition, another study demonstrated that the proliferative and migratory abilities of HCC cell lines were potentiated, while their apoptosis was counteracted via the enforced expression of the exosomal lncRNA H19 (98). Furthermore, sEV constituents may intervene in the cell cycle to regulate the progression of HCC. It has been corroborated that circ_0061395 silencing can trigger cell cycle arrest and apoptosis, and suppress the proliferation of HCC in vitro, as well as inhibit tumor growth (56). Similarly, miR-4454 inhibitor-mediated exosomes can substantially exacerbate cycle arrest, apoptosis and the formation of reactive oxygen species in HCC (95). Of note, the progression of HCC is a result of the accumulation of several time-intersecting steps, including invasion, migration, angiogenesis, immune escape and metastasis, and sEV cargos may also function via several mechanisms. Huang et al (62) suggested that the silencing of circANTXR1 can suppress HCC progression, not only by inhibiting the proliferative ability of HCC, but also by hampering the migration, invasion and metastasis of tumor cells. The roles of sEVs in other hallmarks of tumor progression will be further discussed in the following section.
Angiogenesis
Angiogenesis refers to the formation of new blood vessels from pre-existing ones. It is a complex, multistep process involving extracellular matrix remodeling, endothelial cell migration and ultimately generation of microvessels. Angiogenesis not only provides sufficient oxygen and nutrition for cancer cells, but is also essential for HCC proliferation, local invasion and distant metastasis. The significance of sEVs in cancer angiogenesis has been widely explored and documented recently (98). In HCC, exosomal SNHG16 can sponge miR-4500 and activate angiogenesis in HUVECs by regulating polypeptide N-acetylgalactosaminyltransferase 1 via the PI3K/Akt/mTOR pathway (99). Lin et al (100) reported that tumor-derived exosomes (TDEs) containing miR-210 could target SMAD4 and STAT6 in endothelial cells, and thereby promote the angiogenesis of HCC. The functions of these sEV cargos are multifaceted, and they alter the gene expression of the recipient cells, which become more aggressive and exhibit malignant characteristics. Apart from regulating angiogenesis, they may also control phenotypic changes, such as the proliferative or migratory abilities of cancer cells. A previous study demonstrated that circCMTM3-bearing sEVs can drive the angiogenesis of HUVECs, as well as their viability, migration and invasion (58). Certain studies have found that serval sEV cargos may play the opposite role and suppress angiogenesis in HCC (34,101). For instance, HCC-derived exosomes containing miR-3682-3p have been shown to attenuate angiogenesis via targeting ANGPT1, which is dependent on the RAS-MEK1/2-ERK1/2 pathway (101). Taken together, these results indicate that angiogenesis is a complex process that is orchestrated by multiple biological factors, and the treatment of angiogenesis may provide a novel prospective therapeutic approach for HCC.
Epithelial-mesenchymal transition (EMT) and metastasis
Widespread metastasis in patients with HCC remains a major challenge for treatment, and a main reason for treatment failure, as well as one of the leading-causes of cancer-associated mortality (102). Metastasis is a multistep process involving EMT, invasion into vessels, intravascular transport and organ-specific seeding. The most common mode of metastasis in HCC is intrahepatic metastasis, followed by lymphatic metastasis and distant metastasis to the lungs. sEVs are involved in multiple steps of HCC metastasis, and the importance of sEVs in HCC metastasis has recently been widely reported. Firstly, sEVs contribute to the EMT of HCC cells. Yang et al (103) demonstrated that exosomal miR-92a-3p from high-metastatic HCC cell lines can potentiate EMT and metastasis by inactivating PTEN and activating Akt/Snail signaling. Similarly, Chen et al (104) suggested that TDEs from HCC cells can accelerate EMT, and induce HCC progression and recurrence by activating the MAPK/ERK signaling pathway; however, those studies did not clarify the specific sEVs-carrying cargo that is involved in this process. sEVs exacerbate the migratory and invasive abilities of HCC, which may promote the metastasis of HCC. It has been observed that miR-374a-5p in exosomes potentiates the migration and invasion of HCC by regulating growth arrest and DNA damage inducible alpha (105). In addition, sEVs can orchestrate the organotropic metastasis of HCC by converting the pre-metastatic microenvironment into a tumor cell-friendly site. A previous study demonstrated that Exo-miR-1247-3p derived from HCC can trigger β1-integrin-NF-κB signaling in fibroblasts in the lungs, and is positively associated with several pro-inflammatory cytokines, such as IL-8 and IL-6, which promote the lung metastasis of HCC (92). Recent studies have identified numerous sEV cargos that are involved in the metastasis of HCC, including FAM138B (106), α(M) β(2) integrin (107), hsa_circ_0074854 (108) and lncRNA TUC339 (109). It should be noted that the aforementioned sEV cargos may not only participate in one step of metastasis, but may play multifaceted roles in the whole process of metastasis. For example, Fang et al (42) indicated that, apart from promoting EMT and enhancing tumor motility in vitro, HCC cells can secrete sEV-encapsulated miR-103, which also potentiates vascular permeability and lung metastasis in mouse models.
Immune response and therapeutic resistance
The immune system plays a paramount role in recognizing and eliminating malignant cells and foreign invaders. In the processes of tumor initiation and progression, aberrant proliferation and gene alteration in cancer cells can generate abnormally expressed antigens, which should be adequately presented, recognized and eliminated by the immune system (110). During their fight against the immune system, cancer cells also evolve, and may acquire the ability to evade immunosurveillance via various mechanisms. Among the immune escape effects, that contribute to cancer progression and drug resistance, engagement of the attenuation or abrogation of immunocytes is worth mentioning, and sEVs play a pivotal role in this process (111). Recent studies have suggested that HCC-derived sEVs can impair the function of natural killer (NK) and T-cells, as well as activate immuno-suppressive cells such as M2 macrophages. For instance, exosomal circUHRF1 from HCC triggers the exhaustion of NK cells and subsequently induces resistance to therapy (52). Exo-lncRNA TUC339 has also been shown to be internalized by macrophages, and to modulate M1/M2 polarization and suppress the antitumor immune response in HCC (109). Apart from diminishing the activity of the innate immune system, previous studies have indicated that TDEs from HCC can also impede the activation and function of specific immunocytes such as T- and B-cells (32,46,52). Wang et al (112) found that exosomal 14-3-3ζ released by HCC cells suppressed the antineoplastic characteristics of tumor-infiltrating T-lymphocytes. Tumor-derived exosomal HMGB1 can enhance the expansion the T-cell Ig and mucin domain (TIM)-1 (+) regulatory B-cells and facilitate HCC immune evasion (113). Collectively, sEVs play multiple roles in the communication of HCC and immune cells, and are critical for the immune escape of HCC cells and tumor progression. Thus, sEVs may serve as ideal therapeutic targets for HCC, although further investigations into this matter are warranted.
HCC is one of the most aggressive cancer types, and hepatic resection remains the gold standard of treatment for HCC if the patients can withstand surgery. For patients who experience HCC recurrence or cannot tolerate surgery, targeted therapies involving the use of sorafenib, a multi-kinase inhibitor compound, and chemotherapy including paclitaxel and 5-fluorouracil (5-FU) are first-line treatments. Resistance to drugs remains a main obstacle to the effective treatment of these patients. The mechanisms underlying drug resistance remain complex and elusive; however, but the roles of sEVs in this process is emerging and have captured the interest of researchers. For instance, a previous study found that transfection with GRP78 small interfering RNA into bone marrow-derived mesenchymal stem cells could yield sEVs containing siGRP78, thus mediating targeted RNA silencing, which increased the sensitivity of drug-resistant cancer cells to sorafenib and improved the drug resistance reversion (114). Furthermore, as previously demonstrated, sEV-encapsulated miR-23a/b derived from adipocytes was transferred to neighboring HCC cells, which enhanced their chemoresistance to 5-FU by targeting the VHL/HIF axis (115). The upregulation of miR-32-5p-bearing sEVs has been shown to induce multidrug resistance by potentiating EMT and angiogenesis via targeting the PI3K/PTEN/Akt signaling pathway (34). Another study demonstrated that sEVs secreted from cancer stem cells induced regorafenib insensitivity by upregulating Nanog expression (116). These findings highlight the significance of sEVs in the drug resistance of HCC, which results from sEVs directly suppressing drug efficacy against tumor cells, or from sEVs regulating the gene expression of recipient cells to facilitate cancer survival. Since sEVs can alter the drug sensitivity of HCC cells, it is tempting to engineer a sEV-derived vehicle to deliver specific agents for HCC treatment (117). Studies on this topic are currently underway, and the preliminary results are promising. Wang et al (118) indicated that the downregulation of miR-744 in HCC tissue and cell lines was implicated in the chemoresistance to sorafenib, and HCC cell lines treated with miR-744-upregulating sEVs were more sensitive to sorafenib, which provides a potential approach to reduce the occurrence of drug resistance.
4. Clinical applications of sEVs in HCC
sEVs as diagnostic and prognostic biomarkers in HCC
Despite the advances in the diagnosis of HCC, the number of new cases and cancer mortalities associated with HCC remain high (1). The diagnosis of HCC relies heavily on imaging analyses, such as magnetic resonance imaging or computed tomography; however, the diagnosis of the majority of patients is confirmed at an advanced stage, and thus these patients miss the optimal treatment period (119). Immense efforts have been made as regards the early diagnosis of HCC, although success has been limited. Alpha-fetoprotein (AFP) is a traditional HCC marker with a low specificity, which has a limited value in the differential diagnosis between HCC and other liver diseases (120,121). As regards other biomarkers, such as golgi glycoprotein 73, AFP-L3, phosphatidylinositol proteoglycan 3 and decarboxylated prothrombin, they do not provide any obvious advantage in the early diagnosis of HCC compared with AFP (122,123). Therefore, a non-invasive method with a high diagnostic sensitivity and specificity is urgently required. Recently, liquid biopsies and particularly circulating tumor cells, have attracted extensive interest for the diagnosis and monitoring of HCC (124). Studies on other tumor-derived components, such as circulating tumor DNA, sEVs and serum miRNAs are also increasing (125-127).
sEVs have the following advantages: i) Due to being protected by the sEV membrane, sEV cargos have a high stability and cannot easily degraded by lysosomes; ii) since the secretion of sEVs is a normal physiological event for tumor cells, sEVs can be detected in the majority of fluids, and their extraction is relatively non-invasive; iii) compared with plasma biomarkers, bioactive molecules from sEVs contain less interference of plasma; and iv) markedly, cargos of sEVs have extensive homology with recipient cells, which can confer sEVs superior sensitivity and specificity than traditional methods (128-130). Differential ultracentrifugation is the most common method used to separate sEVs from the cell culture medium. However, each biological fluid presents specific biophysical and chemical characteristics that render it different from culture conditioned medium. Despite current mainstream commercial kits are based on precipitation, which may result in EV populations bound to or mixed with introduced components, such as antibodies, beads or polymers; the majority of studies on the potential clinical applications of sEVs use this method as it is user-friendly, cost-effective and has potential for scale-up production (17). Furthermore, the efficiency and repeatability of sEVs separated using the ExoQuick™ kit have been demonstrated to be comparable with those of differential ultracentrifugation (131). Therefore, the articles cited in the current section include those that using commercial kits to isolate sEVs.
Numerous studies on the role of sEVs as HCC promising biomarkers have been conducted (132-136). The importance of sEVs as HCC biomarkers is reflected in numerous aspects, including the fact that sEV cargos may serve as biomarkers for the early detection of HCC; among these sEV cargos, miRNAs are the most extensively investigated ones. For instance, the expression of miR-21 and miR-10b in sEVs is markedly increased in patients with HCC compared with that of healthy individuals and patients with CHB, indicating that sEVs-carrying miR-21 and miR-10b may be used as early diagnostic biomarkers for HCC (80). Similarly, by comparison with that of patients with LC, the expression of miR-221, miR-192 and miR-146a in exosomes was increased in patients with HCC, and Fründt et al (137) indicated that sEVs carrying miR-146a could distinguish patients with HCC from patients with LC with an area under the curve value of 0.80±0.14 in a logistic regression model, and miR-96, miR-122, miR-200a had similar effect (138-140). Other sEV cargos, such as proteins and other non-coding RNAs (ncRNAs), including lncRNAs and circRNAs, may also play a role in the preliminary diagnosis of HCC, and it has been observed that LINC00161, circRNA 0006602, LDHC, sphingosines, dilysocardiolipins, lysophosphatidylserines, and (O-acyl)-1-hydroxy fatty acids are early diagnostic biomarker candidates (48,121,141-143).
Apart from their early diagnostic value, sEV cargos may be involved in the prediction of tumor staging and metastasis (144-147). Exo-miR-1307-5p expression in plasma has been found to be positively associated with tumor stage and progression, while sEVs carrying miR-125b have been shown to possess anti-metastatic features and are indicators of early metastasis in HCC (148-150). Other sEV cargos can play a similar role in tumor staging or metastasis prediction, and the function of lncRNA ATB, hnRNPH1 and ASMTL-AS1 in this regard has been reported (151-153). In addition, certain sEV cargos may serve as prognostic indicators and may predict the prognosis of patients with HCC. It has been corroborated that miR-638, miR-150-3p, lncRNA CRNDE and circAKT3 in the sEVs of patients with HCC are implicated in overall survival and disease-free survival and may serve as independent indicators of a poor prognosis (31,154-157). It should be noted that the abnormal expression of certain sEV cargos, such as miR-718, miR-125b and miR-92b is not only an effective tool to evaluate survival, but is also a potential marker to predict the recurrence of HCC (32,149,158). Notably, sEVs-carrying miR-122, hsa-circRNA-G004213 and DANCR are also potential markers to evaluate the efficacy of HCC surgical and interventional treatment (153,159-161). In addition to the above, a panel of tumor specific biochemical indicators has been proposed for a higher sensitivity and specificity compared with single one (162-166). For example, Sorop et al (167) established exosomal miR HCC Score including serum AFP and the level of plasma sEVs-carrying miR-21-5p and miR-92a-3p with a great diagnostic ability of HCC (AUC=0.85). Taken together, previous studies have demonstrated that certain sEV cargos, including ncRNAs, mRNAs, lipids and proteins, may serve as potential HCC diagnostic and prognostic biomarkers. The potential HCC biomarkers are summarized in Table II and the efficacy of these candidates warrants further validation.
Table II.
sEVs carrying cargos as biomarkers for HCC.
| Type of sEVs contents | Molecules | Origin | Potential functions | Year of publication | (Refs.) | |
|---|---|---|---|---|---|---|
| miRNA | miR-21 | Serum | Differentiates HCC patients from healthy control, and predict survival | 2019 | (80,126) | |
| miR-10b | Serum | Differentiates E-HCC patients from healthy control, and predict DFS | 2019 | (80) | ||
| miR-638 | Serum | Is negatively associated with survival and predicts recurrence | 2018, 2021 | (154,157) | ||
| miR-122 | Serum | The ratio of miR-122 predicts DSS for HCC patients with liver cirrhosis treated with TACE, and miR-122 differentiate HCC patients from healthy controls | 2018 | (140,159) | ||
| miR-212 | Serum | Differentiates HBV-infection HCC patients from non-HBV-infection HCC | 2019 | (120) | ||
| patients, and predict survival | ||||||
| miR-455-5p, miR-30c-5p | Plasma | Predicts survival | 2020 | (127) | ||
| miR-150-3p | Plasma | Is negatively associated with survival | 2021 | (31) | ||
| miR-1307-5p | Serum/Plasma | Predicts metastasis | 2020 | (148) | ||
| miR-92b | Serum | Predicts recurrence for HCC patients after LDLT | 2019 | (32) | ||
| miR-125b | Serum | Predicts early metastasis, survival and recurrence | 2017, 2021 | (149,150) | ||
| miR-146a | Plasma | Differentiate HCC patients from cirrhosis patients | 2021 | (137) | ||
| miR-192 | Plasma | Predicts survival | 2021 | (137) | ||
| miR-320d | Serum | Differentiates HCC patients from healthy controls, and predicts TNM stage, lymph node metastasis, and survival and survival | 2020 | (144) | ||
| miR-4661-5p | Serum | Differentiates HCC patients at early stage and predict prognosis | 2020 | (135) | ||
| miR-93 | Serum | Differentiates HCC patients from healthy controls, predict TNM stage, and survival | 2018 | (36) | ||
| miR-665 | Serum | Differentiates HCC patients from healthy controls, predicts TNM stage, local metastasis, and survival | 2017 | (145) | ||
| miR-718 | Serum | Predicts recurrence for HCC patients after liver transplantation | 2015 | (158) | ||
| miR-224 | Serum | Differentiates HCC from healthy controls, and predict survival | 2019 | (136) | ||
| lncRNA | lncRNA ATB | Serum | Predicts TNM stage and survival | 2019 | (151) | |
| DANCR | Serum | Predicts recurrence for HCC patients with HCV after curative HCC resection | 2021 | (160) | ||
| ASMTL-AS1 | Serum | Predicts recurrence and metastasis for HCC patients after insufficient RFA | 2020 | (153) | ||
| LINC00161 | Serum | Differentiates HCC patients from healthy controls | 2018 | (48) | ||
| LINC00853 | Serum | Differentiates E-HCC from non-HCC | 2020 | (142) | ||
| lncRNA-HEIH | Serum | Differentiates HCC from HCV patients | 2018 | (133) | ||
| CRNDE | Serum | Predicts TNM stage, OS and DFS | 2021 | (155) | ||
| circRNA | circRNA 0006602 | Plasma | Differentiates HCC from non-HCC | 2021 | (121) | |
| hsa_circ_0070396 | Plasma | Differentiates HCC patients from healthy controls | 2021 | (134) | ||
| circAKT3 | Serum | Predict recurrence and OS | 2020 | (156) | ||
| hsa-circRNA-G004213 | Plasma | Predict efficacy of TACE | 2021 | (161) | ||
| mRNA | hnRNPH1 | Serum | Differentiates HCC patients from healthy controls and CHB patients, and predicts lymph node metastasis, TNM stage and OS | 2018 | (152) | |
| LDHC | Serum | Differentiates E-HCC patients from healthy controls, and predicts efficacy, recurrence | 2020 | (141) | ||
| Protein | ENO1 | Serum | Predicts TNM stage, and metastasis | 2020 | (74) | |
| Lipid | Sphingosines, | Plasma | Differentiates E-HCC patients from non-HCC | 2021 | (143) | |
| dilysocardiolipins, lysophosphatidylserines, and (O-acyl)-1-hydroxy fatty acids | ||||||
| Panel | miR-10b, miR-21, miR-122 and miR-200a | Serum | Differentiates E-HCC patients from healthy controls and cirrhosis patients | 2015 | (139) | |
| miR-21-5p, and | Plasma | Together with serum AFP establishing Exosomal miR HCC score, and | 2020 | (167) | ||
| miR-92a-3p | differentiates HCC patients from healthy controls and cirrhosis patients | |||||
| miR-18a, miR-20b, and miR-221 | Plasma | Predicts metastasis | 2021 | (165) | ||
| miR-122, miR-21, miR-96 | Serum | Differentiates HCC patients from cirrhosis patients | 2020 | (138) | ||
| miR-10b-5p, miR-221-3p, miR-223-3p, and miR-21-5p | Plasma | Differentiates HCC patients from non-HCC | 2020 | (162) | ||
| miR-122, and miR-148a | Serum | Together with serum AFP, differentiates E-HCC patients from cirrhosis patients | 2018 | (140) | ||
| miR-4661-5p, and miR-4746-5p | Serum | Differentiate E-HCC patients from non-HCC | 2020 | (135) | ||
| miR-4718, miR-642a-5p, miR-6826-3p, and miR-762 | Serum | Differentiates HCC patients from HCV patients, and predicts recurrence for SVR-HCC with DAA | 2019 | (166) | ||
| hsa_circ_0004001, hsa_circ_0004123, hsa_circ_0075792 | Serum | Differentiates HCC patients from healthy controls | 2020 | (163) | ||
| MALAT1 and SNHG1, DLEU2 and AFP | Serum | Differentiates E-HCC patients from non-HCC | 2021 | (164) | ||
| ENSG00000258332.1 LINC00635 | Serum | Predicts TNM stage, lymph node metastasis, and OS. Together with AFP, differentiates HCC patients from non-HCC | 2018 | (146) | ||
| Cofilin-1 and CCT8 | Serum | Predict vascular invasion, TNM stage, survival. Together with AFP,differentiates HCC patients from non-HCC | 2021 | (147) | ||
HCC, hepatocellular carcinoma; E-HCC, early-stage hepatocellular carcinoma; HCV, hepatitis C viruses; CHB, chronic hepatitis B; TACE, transarterial chemoembolization; LDLT, living donor liver transplantation; DAA, direct-acting antiviral therapy; SVR, sustained viral response; DFS, disease-free survival; DSS, disease-specific survival; OS, overall survival; AFP, alpha-fetoprotein; TNM, tumor, node, metastasis.
Therapeutic potential of sEVs in the treatment of HCC
Liver cancer is ranked as the third leading cause of cancer-associated mortality worldwide due to resistance to traditional drugs, as well as diagnosis in the late stage (1,168). The identification of novel drugs for targeted therapy is imperative for patients with HCC (169). As aforementioned, the distinctive property of sEVs in delivering functional molecules and altering the biological behavior of recipient cells highlights their potential application as ideal therapeutic vehicles in cancer therapy, both at the theoretical and practical level. The development of engineered sEVs, with a purpose of acting as alternatives to chemotherapeutic and targeted agents, is currently ongoing. sEVs have several advantages compared with previous drug carriers, such as liposomes: First, sEVs achieve highly efficient drug delivery due to their facility of penetrating biological barriers. In addition, the cellular origin of sEVs makes them well tolerated, and they can easily escape immune clearance, which also reduces drug dose and toxicity (111,170,171); Furthermore, the heterogeneity of proteins on the sEVs membrane facilitates the targeting abilities of sEVs. In addition, sEVs are more biocompatible, safe and stable than liposomes (172,173). Taken together, sEVs have a great potential to serve as nano-carriers in the treatment field. The present section mainly focuses on the advancements made in sEV research as regards their application in therapy.
To achieve a better understanding of sEVs as vehicles for therapeutic agents, methods of sEV preparation, the engineering of sEVs and the selection of cargos are under investigation, and the preliminary results are promising. Multiple therapeutic agents, including chemotherapeutic drugs and nucleic acids or their inhibitors, can be loaded. For example, in a previous study, the subcutaneous injection of sEVs containing miR-let-7a into a breast cancer mouse model exhibited an antitumor ability by targeting EGFR (174). In HCC, recent research has validated the importance of sEVs in the delivery of EV-packaged drugs (175). A previous study provided a prospective approach for generating sEV-associated adeno-associated virus containing inducible caspase 9 (iCasp9) suicide gene (Vexo-AAV6-iCasp9). The engineered sEVs possessed a low immunogenicity and toxicity, and were readily absorbed by HCC cells, consequently increasing HCC regression in an in vivo xenograft model (176). Another study encapsulated erastin (a typical ferroptosis inducer) and rose bengal (RB, a well-known photosensitizer) into sEVs and engineered CD47 on the surface of sEVs to protect the designed sEVs from phagocytosis by macrophages. The sEVs induced obvious ferroptosis in HCC, with minimized toxicity in the liver and kidneys (177). Apart from packing antitumor payloads into sEVs, previous studies have developed nanoparticles targeting specific adhesion or receptor proteins on the surface of sEVs membranes for targeted delivery. Tian et al (80) designed a nano-drug based on the PDCM system by targeting sEVs shuttling miR-21 and miR-10b, which markedly decreased HCC growth and the numbers of metastatic lung nodules. TDEs not only assist in exacerbating tumor progression, but also increase the resistance of cancer cells to antitumor treatments (178). Sorafenib and transarterial chemoembolization have been considered optional treatments for terminal-stage HCC for numerous years. Due to acquired drug resistance to commonly used chemotherapeutic agents, the clinical outcome and overall survival of patients with HCC remain unsatisfactory (179). The expression of programmed cell death protein 1 (PD-1) in HCC tissues from patients with HCC who accepted sorafenib treatment was upregulated and induced T-cell apoptosis (180). Therefore, immune checkpoint inhibitors, such as commonly used PD-1 antibodies and PD-L1 anti-bodies have been introduced into medical practice as part of the HCC regimen; however, the efficacy of the combination of sorafenib and immunotherapy has not yet been fully elucidated. Shi et al (180) treated mouse models of HCC with the triple treatment of sorafenib, PD-1 antibodies and DC-derived exosomes (DEXs), which markedly prolonged the survival time mice with HCC by comparison with the mice treated with sorafenib alone, DEXs alone, or the combination of DEXs and sorafenib. Taken together, these preclinical studies offer encouragement for the application of sEVs as vehicles for HCC treatment.
Despite the rapid development of advanced techniques, major limitations remain in the current understanding of sEVs vs. ideal treatment scenarios: i) For sEV technology to play a role as a drug delivery vector, it is necessary to ensure high purity and adequate production. There are still several obstacles which hamper the efficacy of current methods, such as being time consuming, having a high cost and generating polluting by-products. Oncologists increase the total yield of sEVs by intracellular calcium production, external stress, cytoskeletal blocking, drug stimulation and the induction of gene expression factors. Furthermore, sEVs usually represent heterogeneous populations from different cell sources, and no standard separation process has been established to date to achieve product consistency (181); ii) the efficient incorporation of external antitumor agents and molecules is another demanding challenge that needs to be optimized. The high drug-loading content in sEVs must be sufficient to obtain a therapeutic response. Several approaches, including transfection, electroporation and sonication (182,183), can be applied to upload the desired biomolecules into sEVs. However, it is difficult to ensure the integrity and biostability of the plasma membrane and the function of sEVs; iii) the current evidence for the ability of sEVs to deliver specific messages derives from cell culture studies. The biodistribution and tissue or cellular tropism in vivo will determine the application of therapeutic sEVs in clinical practice. At present, there is insufficient evidence for in vivo and clinical applications, which is a critical topic for future research in this area. Due to being subjected to elimination by the mononuclear phagocyte system, the half-life of sEVs in the systemic circulation is relatively short (184). Thus, further studies on the balance between prolonged circulation time and increased risk of toxicity on major organs are warranted; and iv) currently, sEVs need to be stored under-20 and -80°C in phosphate buffer saline (185). Therefore, identifying a suitable storage method is one of the barriers to be overcome.
In summary, while the application of sEVs as a therapeutic drug delivery system remains in its infancy, the deeper understanding of the aforementioned obstacles will provide a new orientation for cancer nanomedicine and immunotherapy.
5. Conclusions and future perspectives
sEVs can trigger the alteration of gene expression and induce aggressive behaviors in HCC cells; however, whether such observations can be replicated in vivo needs to be further investigated, since the precise isolation and high concentration of cell culture-derived sEVs could not be achieved in the majority of in vivo studies published thus far. Paradigm-shifting findings in the field of HCC diagnosis have resulted in new avenues for research on HCC biomarkers. Their easy availability, vesicle-tethering stability and high donor-homology confer sEVs an unparalleled advantage as HCC biomarkers compared with traditional biomarkers. However, the clinical value of sEVs as HCC biomarkers is still limited due to the absence of clinical research with large sample sizes. Extensive efforts are currently being made to identify sEVs biomarkers with high specificity and sensitivity, and apply them to clinical practice. The role of sEVs in cancer therapy has been studied extensively in recent years; however, research on sEVs for HCC remains limited. Before applying them in clinical practice, it is important to validate the purity, safety and effectiveness of sEVs-encapsulated agents. Further research is warranted to guarantee the homogeneity of sEVs, improve the efficiency of their isolation methods and reduce the associated side-effects. The targeting of sEVs is another issue that needs to be resolved. Surface modification is a typical approach to harvest targeted sEVs by modifying the proteins or peptides that specifically expressed on the cell membrane through gene transfection. Engineered sEVs can be selectively delivered to target cells and reach the standard in terms of yield and targeted therapy. However, the safety, mutagenesis and time-consuming limit their clinical applications. Currently, aptamers, also known as chemical antibodies, have attracted the attention of oncologists. The majority of aptamers have been utilized to guide nanoparticles, therapeutic and imaging agents to target locations in several promising anticancer preclinical studies, whereby they are able to modulate tumor retention and biodistribution. However, all these issues cannot be solved in a short period of time, and as the number of clinical studies increases, more patients will gain clinical benefit from research in sEVs.
Supplementary Data
Acknowledgments
Not applicable.
Funding Statement
The present review article was funded by the National Natural Science Foundation of China (grant no. 82070575), the Digestive Medical Coordinated Development Center of Beijing Municipal Administration of Hospitals (grant no. XXZ0205) and the Beijing Municipal Science and Technology Commission (grant no. Z191100006619080).
Availability of data and materials
Not applicable.
Authors' contributions
GZ and PL conceptualized the study. SY and JW were involved in the writing and preparation of the original draft. SW and AZ were involved in the writing, reviewing and editing of the manuscript. All authors have read and approved the final manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Petrick JL, Florio AA, Znaor A, Ruggieri D, Laversanne M, Alvarez CS, Ferlay J, Valery PC, Bray F, McGlynn KA. International trends in hepatocellular carcinoma incidence, 1978-2012. Int J Cancer. 2020;147:317–330. doi: 10.1002/ijc.32723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ringelhan M, McKeating JA, Protzer U. Viral hepatitis and liver cancer. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160274. doi: 10.1098/rstb.2016.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen SQ, Li J, Wang D, Fung H, Wong LY, Zhao L. The hepatitis B epidemic in China should receive more attention. Lancet. 2018;391:1572. doi: 10.1016/S0140-6736(18)30499-9. [DOI] [PubMed] [Google Scholar]
- 5.Sarin SK, Kumar M, Eslam M, George J, Al Mahtab M, Akbar SMF, Jia J, Tian Q, Aggarwal R, Muljono DH, et al. Liver diseases in the Asia-pacific region: A lancet gastroenterology & hepatology commission. Lancet Gastroenterol Hepatol. 2020;5:167–228. doi: 10.1016/S2468-1253(19)30342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao C, Xing F, Yeo YH, Jin M, Le R, Le M, Jin M, Henry L, Cheung R, Nguyen MH. Only one-third of hepatocellular carcinoma cases are diagnosed via screening or surveillance: A systematic review and meta-analysis. Eur J Gastroenterol Hepatol. 2020;32:406–419. doi: 10.1097/MEG.0000000000001523. [DOI] [PubMed] [Google Scholar]
- 7.Wu G, Wu J, Wang B, Zhu X, Shi X, Ding Y. Importance of tumor size at diagnosis as a prognostic factor for hepatocellular carcinoma survival: A population-based study. Cancer Manag Res. 2018;10:4401–4410. doi: 10.2147/CMAR.S177663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–9420. doi: 10.1016/S0021-9258(18)48095-7. [DOI] [PubMed] [Google Scholar]
- 9.Chen W, Mao Y, Liu C, Wu H, Chen S. Exosome in hepatocellular carcinoma: An update. J Cancer. 2021;12:2526–2536. doi: 10.7150/jca.54566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang W, Xiao D, Fang X, Wang J. Phospholipids in small extracellular vesicles: Emerging regulators of neurodegenerative diseases and cancer. Cytotherapy. 2022;24:93–100. doi: 10.1016/j.jcyt.2021.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Sun F, Wang JZ, Luo JJ, Wang YQ, Pan Q. Exosomes in the oncobiology, diagnosis, and therapy of hepatic carcinoma: A new player of an old game. Biomed Res Int. 2018;2018:2747461. doi: 10.1155/2018/2747461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen SW, Lima LG, Lobb RJ, Norris EL, Hastie ML, Krumeich S, Möller A. Breast cancer-derived exosomes reflect the cell-of-origin phenotype. Proteomics. 2019;19:e1800180. doi: 10.1002/pmic.201800180. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Xu M, Li X, Su X, Xiao X, Keating A, Zhao RC. Exosomes released by hepatocarcinoma cells endow adipocytes with tumor-promoting properties. J Hematol Oncol. 2018;11:82. doi: 10.1186/s13045-018-0625-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng X, Hermann DM, Bähr M, Doeppner TR. The role of small extracellular vesicles in cerebral and myocardial ischemia-molecular signals, treatment targets, and future clinical translation. Stem Cells. 2021;39:403–413. doi: 10.1002/stem.3329. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, He F, Gao L, Cong M, Sun J, Xu J, Wang Y, Hu Y, Asghar S, Hu L, Qiao H. Engineering exosome-like nanovesicles derived from asparagus cochinchinensis can inhibit the proliferation of hepatocellular carcinoma cells with better safety profile. Int J Nanomedicine. 2021;16:1575–1586. doi: 10.2147/IJN.S293067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim OY, Lee J, Gho YS. Extracellular vesicle mimetics: Novel alternatives to extracellular vesicle-based theranostics, drug delivery, and vaccines. Semin Cell Dev Biol. 2017;67:74–82. doi: 10.1016/j.semcdb.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy DE, de Jong OG, Brouwer M, Wood MJ, Lavieu G, Schiffelers RM, Vader P. Extracellular vesicle-based therapeutics: Natural versus engineered targeting and trafficking. Exp Mol Med. 2019;51:1–12. doi: 10.1038/s12276-019-0223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aslan C, Maralbashi S, Salari F, Kahroba H, Sigaroodi F, Kazemi T, Kharaziha P. Tumor-derived exosomes: Implication in angiogenesis and antiangiogenesis cancer therapy. J Cell Physiol. 2019;234:16885–16903. doi: 10.1002/jcp.28374. [DOI] [PubMed] [Google Scholar]
- 20.Antimisiaris SG, Mourtas S, Marazioti A. Exosomes and exosome-inspired vesicles for targeted drug delivery. Pharmaceutics. 2018;10:218. doi: 10.3390/pharmaceutics10040218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian X, Shen H, Li Z, Wang T, Wang S. Tumor-derived exosomes, myeloid-derived suppressor cells, and tumor microenvironment. J Hematol Oncol. 2019;12:84. doi: 10.1186/s13045-019-0772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle LM, Wang MZ. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. 2019;8:727. doi: 10.3390/cells8070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farooqi AA, Desai NN, Qureshi MZ, Librelotto DRN, Gasparri ML, Bishayee A, Nabavi SM, Curti V, Daglia M. Exosome biogenesis, bioactivities and functions as new delivery systems of natural compounds. Biotechnol Adv. 2018;36:328–334. doi: 10.1016/j.biotechadv.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Wang Y, Wang Q, Liu Y, Bao W, Wu S. Exosomes in cancer: Small transporters with big functions. Cancer Lett. 2018;435:55–65. doi: 10.1016/j.canlet.2018.07.037. [DOI] [PubMed] [Google Scholar]
- 26.Xie JY, Wei JX, Lv LH, Han QF, Yang WB, Li GL, Wang PX, Wu SB, Duan JX, Zhuo WF, et al. Angiopoietin-2 induces angiogenesis via exosomes in human hepatocellular carcinoma. Cell Commun Signal. 2020;18:46. doi: 10.1186/s12964-020-00535-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Z, Yang S, Zhou A, Li X, Fang R, Zhang S, Zhao G, Li P. Small extracellular vesicles in the development, diagnosis, and possible therapeutic application of esophageal squamous cell carcinoma. Front Oncol. 2021;11:732702. doi: 10.3389/fonc.2021.732702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu S, Cao H, Shen B, Feng J. Tumor-derived exosomes in cancer progression and treatment failure. Oncotarget. 2015;6:37151–37168. doi: 10.18632/oncotarget.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greening DW, Xu R, Ji H, Tauro BJ, Simpson RJ. A protocol for exosome isolation and characterization: Evaluation of ultra-centrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol. 2015;1295:179–209. doi: 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- 30.Vaswani K, Koh YQ, Almughlliq FB, Peiris HN, Mitchell MD. A method for the isolation and enrichment of purified bovine milk exosomes. Reprod Biol. 2017;17:341–348. doi: 10.1016/j.repbio.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Yugawa K, Yoshizumi T, Mano Y, Itoh S, Harada N, Ikegami T, Kohashi K, Oda Y, Mori M. Cancer-associated fibroblasts promote hepatocellular carcinoma progression through downregulation of exosomal miR-150-3p. Eur J Surg Oncol. 2021;47:384–393. doi: 10.1016/j.ejso.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Nakano T, Chen IH, Wang CC, Chen PJ, Tseng HP, Huang KT, Hu TH, Li LC, Goto S, Cheng YF, et al. Circulating exosomal miR-92b: Its role for cancer immunoediting and clinical value for prediction of posttransplant hepatocellular carcinoma recurrence. Am J Transplant. 2019;19:3250–3262. doi: 10.1111/ajt.15490. [DOI] [PubMed] [Google Scholar]
- 33.Moh-Moh-Aung A, Fujisawa M, Ito S, Katayama H, Ohara T, Ota Y, Yoshimura T, Matsukawa A. Decreased miR-200b-3p in cancer cells leads to angiogenesis in HCC by enhancing endothelial ERG expression. Sci Rep. 2020;10:10418. doi: 10.1038/s41598-020-67425-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu X, Liu M, Qu S, Ma J, Zhang Y, Shi T, Wen H, Yang Y, Wang S, Wang J, et al. Exosomal microRNA-32-5p induces multidrug resistance in hepatocellular carcinoma via the PI3K/Akt pathway. J Exp Clin Cancer Res. 2018;37:52. doi: 10.1186/s13046-018-0677-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Tang J, Li Y, Liu K, Zhu Q, Yang WH, Xiong LK, Guo DL. Exosomal miR-9-3p suppresses HBGF-5 expression and is a functional biomarker in hepatocellular carcinoma. Minerva Med. 2018;109:15–23. doi: 10.23736/S0026-4806.17.05167-9. [DOI] [PubMed] [Google Scholar]
- 36.Xue X, Wang X, Zhao Y, Hu R, Qin L. Exosomal miR-93 promotes proliferation and invasion in hepatocellular carcinoma by directly inhibiting TIMP2/TP53INP1/CDKN1A. Biochem Biophys Res Commun. 2018;502:515–521. doi: 10.1016/j.bbrc.2018.05.208. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Wang G, Niu L, Zhao S, Li J, Zhang Z, Jiang H, Zhang Q, Wang H, Sun P, et al. Exosomal MiR-1290 promotes angiogenesis of hepatocellular carcinoma via targeting SMEK1. J Oncol. 2021;2021:6617700. doi: 10.1155/2021/6617700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ouyang Y, Tang Y, Fu L, Peng S, Wu W, Tan D, Fu X. Exosomes secreted by chronic hepatitis B patients with PNALT and liver inflammation grade ≥ A2 promoted the progression of liver cancer by transferring miR-25-3p to inhibit the co-expression of TCF21 and HHIP. Cell Prolif. 2020;53:e12833. doi: 10.1111/cpr.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Y, Yang X, Xue X, Sun D, Cai P, Song Q, Zhang B, Qin L. HANR promotes lymphangiogenesis of hepatocellular carcinoma via secreting miR-296 exosome and regulating EAG1/VEGFA signaling in HDLEC cells. J Cell Biochem. 2019;120:17699–17708. doi: 10.1002/jcb.29036. [DOI] [PubMed] [Google Scholar]
- 40.Xiong L, Zhen S, Yu Q, Gong Z. HCV-E2 inhibits hepatocellular carcinoma metastasis by stimulating mast cells to secrete exosomal shuttle microRNAs. Oncol Lett. 2017;14:2141–2146. doi: 10.3892/ol.2017.6433. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Chen W, Huang L, Liang J, Ye Y, He S, Niu J. Hepatocellular carcinoma cells-derived exosomal microRNA-378b enhances hepatocellular carcinoma angiogenesis. Life Sci. 2021;273:119184. doi: 10.1016/j.lfs.2021.119184. [DOI] [PubMed] [Google Scholar]
- 42.Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin YF, Yuan Y, Zhuang SM. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology. 2018;68:1459–1475. doi: 10.1002/hep.29920. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z, Li X, Sun W, Yue S, Yang J, Li J, Ma B, Wang J, Yang X, Pu M, et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017;397:33–42. doi: 10.1016/j.canlet.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Bai ZZ, Li HY, Li CH, Sheng CL, Zhao XN. M1 macrophage-derived exosomal MicroRNA-326 suppresses hepatocellular carcinoma cell progression via mediating NF-κB signaling pathway. Nanoscale Res Lett. 2020;15:221. doi: 10.1186/s11671-020-03432-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Chen QL, Xie CF, Feng KL, Cui DY, Sun SL, Zhang JC, Xiong CM, Huang JH, Chong Z. microRNAs carried by exosomes promote epithelial-mesenchymal transition and metastasis of liver cancer cells. Am J Transl Res. 2020;12:6811–6826. [PMC free article] [PubMed] [Google Scholar]
- 46.Fan F, Chen K, Lu X, Li A, Liu C, Wu B. Dual targeting of PD-L1 and PD-L2 by PCED1B-AS1 via sponging hsa-miR-194-5p induces immunosuppression in hepatocellular carcinoma. Hepatol Int. 2021;15:444–458. doi: 10.1007/s12072-020-10101-6. [DOI] [PubMed] [Google Scholar]
- 47.Wang LP, Lin J, Ma XQ, Xu DY, Shi CF, Wang W, Jiang XJ. Exosomal DLX6-AS1 from hepatocellular carcinoma cells induces M2 macrophage polarization to promote migration and invasion in hepatocellular carcinoma through microRNA-15a-5p/CXCL17 axis. J Exp Clin Cancer Res. 2021;40:177. doi: 10.1186/s13046-021-01973-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Sun L, Su Y, Liu X, Xu M, Chen X, Zhu Y, Guo Z, Bai T, Dong L, Wei C, et al. Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular carcinoma. J Cancer. 2018;9:2631–2639. doi: 10.7150/jca.24978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J, Pu J, Zhang Y, Yao T, Luo Z, Li W, Xu G, Liu J, Wei W, Deng Y. Exosome-transmitted long non-coding RNA SENP3-EIF4A1 suppresses the progression of hepatocellular carcinoma. Aging (Albany NY) 2020;12:11550–11567. doi: 10.18632/aging.103302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Li N. LINC ROR from hepatocarcinoma cell-derived exosomes modulates inflammation in human macrophages. Sichuan Da Xue Xue Bao Yi Xue Ban. 2019;50:177–181. In Chinese. [PubMed] [Google Scholar]
- 51.Li B, Mao R, Liu C, Zhang W, Tang Y, Guo Z. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018;197:122–129. doi: 10.1016/j.lfs.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 52.Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, Cai JB, Ke AW. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19:110. doi: 10.1186/s12943-020-01222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A, Qi F. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY) 2019;11:8183–8203. doi: 10.18632/aging.102312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song X, Gorshkov K, Sun Q, Lin H, Zheng X, et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Target Ther. 2020;5:298. doi: 10.1038/s41392-020-00375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu C, Su Y, Liu L, Wang S, Liu Y, Wu J. Circular RNA hsa_circ_0004277 stimulates malignant phenotype of hepatocellular carcinoma and epithelial-mesenchymal transition of peripheral cells. Front Cell Dev Biol. 2021;8:585565. doi: 10.3389/fcell.2020.585565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu Y, Bian L, Liu R, Wang Y, Xiao X. Circular RNA hsa_circ_0061395 accelerates hepatocellular carcinoma progression via regulation of the miR-877-5p/PIK3R3 axis. Cancer Cell Int. 2021;21:10. doi: 10.1186/s12935-020-01695-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, Zang H, Zhang X, Huang G. Exosomal circ-ZNF652 promotes cell proliferation, migration, invasion and glycolysis in hepatocellular carcinoma via miR-29a-3p/GUCD1 axis. Cancer Manag Res. 2020;12:7739–7751. doi: 10.2147/CMAR.S259424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu K, Li NF, Li JR, Chen ZG, Wang JH, Sheng LQ. Exosome circCMTM3 promotes angiogenesis and tumorigenesis of hepatocellular carcinoma through miR-3619-5p/SOX9. Hepatol Res. 2021;51:1139–1152. doi: 10.1111/hepr.13692. [DOI] [PubMed] [Google Scholar]
- 59.Liu D, Kang H, Gao M, Jin L, Zhang F, Chen D, Li M, Xiao L. Exosome-transmitted circ_MMP2 promotes hepatocellular carcinoma metastasis by upregulating MMP2. Mol Oncol. 2020;14:1365–1380. doi: 10.1002/1878-0261.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, Zhang Y, Li H, Zhang Q, Yang Y, Chen G. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. 2019;40:432–445. doi: 10.1016/j.ebiom.2018.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, Chen L, Liu Q, He P, Ye Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128. doi: 10.1016/j.canlet.2020.01.022. [DOI] [PubMed] [Google Scholar]
- 62.Huang C, Yu W, Wang Q, Huang T, Ding Y. CircANTXR1 contributes to the malignant progression of hepatocellular carcinoma by promoting proliferation and metastasis. J Hepatocell Carcinoma. 2021;8:1339–1353. doi: 10.2147/JHC.S317256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dai W, Wang Y, Yang T, Wang J, Wu W, Gu J. Downregulation of exosomal CLEC3B in hepatocellular carcinoma promotes metastasis and angiogenesis via AMPK and VEGF signals. Cell Commun Signal. 2019;17:113. doi: 10.1186/s12964-019-0423-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang A, Dong J, Li S, Wang C, Ding H, Li H, Su X, Ge X, Sun L, Bai C, et al. Exosomal transfer of vasorin expressed in hepatocellular carcinoma cells promotes migration of human umbilical vein endothelial cells. Int J Biol Sci. 2015;11:961–969. doi: 10.7150/ijbs.11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng Z, Lei Z, Yang P, Si A, Xiang D, Tang X, Guo G, Zhou J, Hüser N. Exosome-transmitted p120-catenin suppresses hepatocellular carcinoma progression via STAT3 pathways. Mol Carcinog. 2019;58:1389–1399. doi: 10.1002/mc.23022. [DOI] [PubMed] [Google Scholar]
- 66.Li M, Lu Y, Xu Y, Wang J, Zhang C, Du Y, Wang L, Li L, Wang B, Shen J, et al. Horizontal transfer of exosomal CXCR4 promotes murine hepatocarcinoma cell migration, invasion and lymphangiogenesis. Gene. 2018;676:101–109. doi: 10.1016/j.gene.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 67.Xia Y, Zhen L, Li H, Wang S, Chen S, Wang C, Yang X. MIRLET7BHG promotes hepatocellular carcinoma progression by activating hepatic stellate cells through exosomal SMO to trigger Hedgehog pathway. Cell Death Dis. 2021;12:326. doi: 10.1038/s41419-021-03494-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu Q, Zhang Q, Lou Y, Yang J, Nie G, Chen Q, Chen Y, Zhang J, Wang J, Wei T, et al. Primary tumor-derived exosomes facilitate metastasis by regulating adhesion of circulating tumor cells via SMAD3 in liver cancer. Oncogene. 2018;37:6105–6118. doi: 10.1038/s41388-018-0391-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou Y, Zhang Y, Gong H, Luo S, Cui Y. The role of exosomes and their applications in cancer. Int J Mol Sci. 2021;22:12204. doi: 10.3390/ijms222212204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang H, Lu Z, Zhao X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol. 2019;12:133. doi: 10.1186/s13045-019-0806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao LQ, Yang XW, Chen YB, Zhang DW, Jiang XF, Xue P. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol Cancer. 2019;18:148. doi: 10.1186/s12943-019-1075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fu X, Tang Y, Wu W, Ouyang Y, Tan D, Huang Y. Exosomal microRNA-25 released from cancer cells targets SIK1 to promote hepatocellular carcinoma tumorigenesis. Dig Liver Dis. 2021;S1590-8658(21):00385–6. doi: 10.1016/j.dld.2021.07.017. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 73.Huang X, Sun L, Wen S, Deng D, Wan F, He X, Tian L, Liang L, Wei C, Gao K, et al. RNA sequencing of plasma exosomes revealed novel functional long noncoding RNAs in hepatocellular carcinoma. Cancer Sci. 2020;111:3338–3349. doi: 10.1111/cas.14516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang K, Dong C, Yin Z, Li R, Mao J, Wang C, Zhang J, Gao Z, Liang R, Wang Q, Wang L. Exosome-derived ENO1 regulates integrin α6β4 expression and promotes hepatocellular carcinoma growth and metastasis. Cell Death Dis. 2020;11:972. doi: 10.1038/s41419-020-03179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lai Z, Wei T, Li Q, Wang X, Zhang Y, Zhang S. Exosomal circFBLIM1 promotes hepatocellular carcinoma progression and glycolysis by regulating the miR-338/LRP6 axis. Cancer Biother Radiopharm. 2020 Sep 9; doi: 10.1089/cbr.2020.3564. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 76.Ma YS, Liu JB, Lin L, Zhang H, Wu JJ, Shi Y, Jia CY, Zhang DD, Yu F, Wang HM, et al. Exosomal microRNA-15a from mesenchymal stem cells impedes hepatocellular carcinoma progression via downregulation of SALL4. Cell Death Discov. 2021;7:224. doi: 10.1038/s41420-021-00611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin C, Han Q, Xu D, Zheng B, Zhao X, Zhang J. SALL4-mediated upregulation of exosomal miR-146a-5p drives T-cell exhaustion by M2 tumor-associated macrophages in HCC. Oncoimmunology. 2019;8:1601479. doi: 10.1080/2162402X.2019.1601479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F, Manno M, Raccosta S, Mancone C, Tripodi M, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li R, Wang Y, Zhang X, Feng M, Ma J, Li J, Yang X, Fang F, Xia Q, Zhang Z, et al. Exosome-mediated secretion of LOXL4 promotes hepatocellular carcinoma cell invasion and metastasis. Mol Cancer. 2019;18:18. doi: 10.1186/s12943-019-0948-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tian XP, Wang CY, Jin XH, Li M, Wang FW, Huang WJ, Yun JP, Xu RH, Cai QQ, Xie D. Acidic microenvironment up-regulates exosomal miR-21 and miR-10b in early-stage hepatocellular carcinoma to promote cancer cell proliferation and metastasis. Theranostics. 2019;9:1965–1979. doi: 10.7150/thno.30958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu H, Chen W, Zhi X, Chen EJ, Wei T, Zhang J, Shen J, Hu LQ, Zhao B, Feng XH, et al. Tumor-derived exosomes promote tumor self-seeding in hepatocellular carcinoma by transferring miRNA-25-5p to enhance cell motility. Oncogene. 2018;37:4964–4978. doi: 10.1038/s41388-018-0309-x. [DOI] [PubMed] [Google Scholar]
- 82.Liu G, Ouyang X, Sun Y, Xiao Y, You B, Gao Y, Yeh S, Li Y, Chang C. The miR-92a-2-5p in exosomes from macrophages increases liver cancer cells invasion via altering the AR/PHLPP/p-AKT/β-catenin signaling. Cell Death Differ. 2020;27:3258–3272. doi: 10.1038/s41418-020-0575-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu C, Zhou X, Long Q, Zeng H, Sun Q, Chen Y, Wu D, Liu L. Small extracellular vesicles containing miR-30a-3p attenuate the migration and invasion of hepatocellular carcinoma by targeting SNAP23 gene. Oncogene. 2021;40:233–245. doi: 10.1038/s41388-020-01521-7. [DOI] [PubMed] [Google Scholar]
- 84.Zeng Y, Yao X, Liu X, He X, Li L, Liu X, Yan Z, Wu J, Fu BM. Anti-angiogenesis triggers exosomes release from endothelial cells to promote tumor vasculogenesis. J Extracell Vesicles. 2019;8:1629865. doi: 10.1080/20013078.2019.1629865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu Q, Zhou L, Lv D, Zhu X, Tang H. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J Hematol Oncol. 2019;12:53. doi: 10.1186/s13045-019-0739-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang Y, Han Q, Hou Z, Zhang C, Tian Z, Zhang J. Exosomes mediate hepatitis B virus (HBV) transmission and NK-cell dysfunction. Cell Mol Immunol. 2017;14:465–475. doi: 10.1038/cmi.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu W, Wu D, Yan W, Wang Y, You J, Wan X, Xi D, Luo X, Han M, Ning Q. Interferon-induced macrophage-derived exosomes mediate antiviral activity against hepatitis B virus through miR-574-5p. J Infect Dis. 2021;223:686–698. doi: 10.1093/infdis/jiaa399. [DOI] [PubMed] [Google Scholar]
- 88.Liu QM, He YY, Liu LL, Wang LK. Exosomal lncRNA HOTTIP mediates antiviral effect of tenofovir alafenamide (TAF) on HBV infection. J Inflamm Res. 2021;14:5489–5500. doi: 10.2147/JIR.S315716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu J, Wu J, Xie F, Tian J, Tang X, Guo H, Ma J, Xu P, Mao L, Xu H, Wang S. CD4+ T cell-released extracellular vesicles potentiate the efficacy of the HBsAg vaccine by enhancing B cell responses. Adv Sci (Weinh) 2019;6:1802219. doi: 10.1002/advs.201802219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wei Y, Tang X, Ren Y, Yang Y, Song F, Fu J, Liu S, Yu M, Chen L, Wang S, et al. An RNA-RNA crosstalk network involving HMGB1 and RICTOR facilitates hepatocellular carcinoma tumorigenesis by promoting glutamine metabolism and impedes immunotherapy by PD-L1+ exosomes activity. Signal Transduct Target Ther. 2021;6:421. doi: 10.1038/s41392-021-00801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, Shi X. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res. 2018;37:324. doi: 10.1186/s13046-018-0965-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fang T, Lv H, Lv G, Li T, Wang C, Han Q, Yu L, Su B, Guo L, Huang S, et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191. doi: 10.1038/s41467-017-02583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lou G, Song X, Yang F, Wu S, Wang J, Chen Z, Liu Y. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J Hematol Oncol. 2015;8:122. doi: 10.1186/s13045-015-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lin M, Liao W, Dong M, Zhu R, Xiao J, Sun T, Chen Z, Wu B, Jin J. Exosomal neutral sphingomyelinase 1 suppresses hepatocellular carcinoma via decreasing the ratio of sphingomyelin/ceramide. FEBS J. 2018;285:3835–3848. doi: 10.1111/febs.14635. [DOI] [PubMed] [Google Scholar]
- 95.Lin H, Zhang R, Wu W, Lei L. miR-4454 promotes hepatic carcinoma progression by targeting Vps4A and Rab27A. Oxid Med Cell Longev. 2021;2021:9230435. doi: 10.1155/2021/9230435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sun JF, Zhang D, Gao CJ, Zhang YW, Dai QS. Exosome-mediated MiR-155 transfer contributes to hepatocellular carcinoma cell proliferation by targeting PTEN. Med Sci Monit Basic Res. 2019;25:218–228. doi: 10.12659/MSMBR.918134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li YH, Lv MF, Lu MS, Bi JP. Bone marrow mesenchymal stem cell-derived exosomal MiR-338-3p represses progression of hepatocellular carcinoma by targeting ETS1. J Biol Regul Homeost Agents. 2021;35:617–627. doi: 10.23812/20-638-A. [DOI] [PubMed] [Google Scholar]
- 98.Wang D, Xing N, Yang T, Liu J, Zhao H, He J, Ai Y, Yang J. Exosomal lncRNA H19 promotes the progression of hepatocellular carcinoma treated with Propofol via miR-520a-3p/LIMK1 axis. Cancer Med. 2020;9:7218–7230. doi: 10.1002/cam4.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li S, Qi Y, Huang Y, Guo Y, Huang T, Jia L. Exosome-derived SNHG16 sponging miR-4500 activates HUVEC angiogenesis by targeting GALNT1 via PI3K/Akt/mTOR pathway in hepatocellular carcinoma. J Physiol Biochem. 2021;77:667–682. doi: 10.1007/s13105-021-00833-w. [DOI] [PubMed] [Google Scholar]
- 100.Lin XJ, Fang JH, Yang XJ, Zhang C, Yuan Y, Zheng L, Zhuang SM. Hepatocellular carcinoma cell-secreted exosomal MicroRNA-210-promotes angiogenesis in vitro and in vivo. Mol Ther Nucleic Acids. 2018;11:243–252. doi: 10.1016/j.omtn.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dong SS, Dong DD, Yang ZF, Zhu GQ, Gao DM, Chen J, Zhao Y, Liu BB. Exosomal miR-3682-3p suppresses angiogenesis by targeting ANGPT1 via the RAS-MEK1/2ERK1/2 pathway in hepatocellular carcinoma. Front Cell Dev Biol. 2021;9:633358. doi: 10.3389/fcell.2021.633358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, Zhou J, Tang ZY. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39:20. doi: 10.1186/s13046-020-1529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang B, Feng X, Liu H, Tong R, Wu J, Li C, Yu H, Chen Y, Cheng Q, Chen J, et al. High-metastatic cancer cells derived exosomal miR92a-3p promotes epithelial-mesenchymal transition and metastasis of low-metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene. 2020;39:6529–6543. doi: 10.1038/s41388-020-01450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen L, Guo P, He Y, Chen Z, Chen L, Luo Y, Qi L, Liu Y, Wu Q, Cui Y, et al. HCC-derived exosomes elicit HCC progression and recurrence by epithelial-mesenchymal transition through MAPK/ERK signalling pathway. Cell Death Dis. 2018;9:513. doi: 10.1038/s41419-018-0534-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lin Q, Zhou CR, Bai MJ, Zhu D, Chen JW, Wang HF, Li MA, Wu C, Li ZR, Huang MS. Exosome-mediated miRNA delivery promotes liver cancer EMT and metastasis. Am J Transl Res. 2020;12:1080–1095. [PMC free article] [PubMed] [Google Scholar]
- 106.Zhuo C, Yi T, Pu J, Cen X, Zhou Y, Feng S, Wei C, Chen P, Wang W, Bao C, et al. Exosomal linc-FAM138B from cancer cells alleviates hepatocellular carcinoma progression via regulating miR-765. Aging (Albany NY) 2020;12:26236–26247. doi: 10.18632/aging.202430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu J, Gao W, Tang Q, Yu Y, You W, Wu Z, Fan Y, Zhang L, Wu C, Han G, et al. M2 macrophage-derived exosomes facilitate HCC metastasis by transferring αM β2 integrin to tumor cells. Hepatology. 2021;73:1365–1380. doi: 10.1002/hep.31432. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 108.Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, Li S. Downregulation of hsa_circ_0074854 suppresses the migration and invasion in hepatocellular carcinoma via interacting with HuR and via suppressing exosomes-mediated macrophage M2 polarization. Int J Nanomedicine. 2021;16:2803–2818. doi: 10.2147/IJN.S284560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li X, Lei Y, Wu M, Li N. Regulation of macrophage activation and polarization by HCC-derived exosomal lncRNA TUC339. Int J Mol Sci. 2018;19:2958. doi: 10.3390/ijms19102958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ozga AJ, Chow MT, Luster AD. Chemokines and the immune response to cancer. Immunity. 2021;54:859–874. doi: 10.1016/j.immuni.2021.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Syn NL, Wang L, Chow EK, Lim CT, Goh BC. Exosomes in cancer nanomedicine and immunotherapy: Prospects and challenges. Trends Biotechnol. 2017;35:665–676. doi: 10.1016/j.tibtech.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 112.Wang X, Shen H, Zhangyuan G, Huang R, Zhang W, He Q, Jin K, Zhuo H, Zhang Z, Wang J, et al. 14-3-3ζ delivered by hepatocellular carcinoma-derived exosomes impaired anti-tumor function of tumor-infiltrating T lymphocytes. Cell Death Dis. 2018;9:159. doi: 10.1038/s41419-017-0180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ye L, Zhang Q, Cheng Y, Chen X, Wang G, Shi M, Zhang T, Cao Y, Pan H, Zhang L, et al. Tumor-derived exosomal HMGB1 fosters hepatocellular carcinoma immune evasion by promoting TIM-1+ regulatory B cell expansion. J Immunother Cancer. 2018;6:145. doi: 10.1186/s40425-018-0451-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li H, Yang C, Shi Y, Zhao L. Exosomes derived from siRNA against GRP78 modified bone-marrow-derived mesenchymal stem cells suppress Sorafenib resistance in hepatocellular carcinoma. J Nanobiotechnology. 2018;16:103. doi: 10.1186/s12951-018-0429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu Y, Tan J, Ou S, Chen J, Chen L. Adipose-derived exosomes deliver miR-23a/b to regulate tumor growth in hepatocellular cancer by targeting the VHL/HIF axis. J Physiol Biochem. 2019;75:391–401. doi: 10.1007/s13105-019-00692-6. [DOI] [PubMed] [Google Scholar]
- 116.Huang H, Hou J, Liu K, Liu Q, Shen L, Liu B, Lu Q, Zhang N, Che L, Li J, et al. RAB27A-dependent release of exosomes by liver cancer stem cells induces Nanog expression in their differentiated progenies and confers regorafenib resistance. J Gastroenterol Hepatol. 2021;36:3429–3437. doi: 10.1111/jgh.15619. [DOI] [PubMed] [Google Scholar]
- 117.Xu Y, Lai Y, Cao L, Li Y, Chen G, Chen L, Weng H, Chen T, Wang L, Ye Y. Human umbilical cord mesenchymal stem cells-derived exosomal microRNA-451a represses epithelial-mesenchymal transition of hepatocellular carcinoma cells by inhibiting ADAM10. RNA Biol. 2021;18:1423. doi: 10.1080/15476286.2020.1851540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang G, Zhao W, Wang H, Qiu G, Jiang Z, Wei G, Li X. Exosomal MiR-744 inhibits proliferation and sorafenib chemoresistance in hepatocellular carcinoma by targeting PAX2. Med Sci Monit. 2019;25:7209–7217. doi: 10.12659/MSM.919219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pinyol R, Montal R, Bassaganyas L, Sia D, Takayama T, Chau GY, Mazzaferro V, Roayaie S, Lee HC, Kokudo N, et al. Molecular predictors of prevention of recurrence in HCC with sorafenib as adjuvant treatment and prognostic factors in the phase 3 STORM trial. Gut. 2019;68:1065–1075. doi: 10.1136/gutjnl-2018-316408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Y, Xi H, Nie X, Zhang P, Lan N, Lu Y, Liu J, Yuan W. Assessment of miR-212 and other biomarkers in the diagnosis and treatment of HBV-infection-related liver diseases. Curr Drug Metab. 2019;20:785–798. doi: 10.2174/1389200220666191011120434. [DOI] [PubMed] [Google Scholar]
- 121.Guo S, Hu C, Zhai X, Sun D. Circular RNA 0006602 in plasma exosomes: A new potential diagnostic biomarker for hepatocellular carcinoma. Am J Transl Res. 2021;13:6001–6015. [PMC free article] [PubMed] [Google Scholar]
- 122.De Stefano F, Chacon E, Turcios L, Marti F, Gedaly R. Novel biomarkers in hepatocellular carcinoma. Dig Liver Dis. 2018;50:1115–1123. doi: 10.1016/j.dld.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 123.Galle PR, Foerster F, Kudo M, Chan SL, Llovet JM, Qin S, Schelman WR, Chintharlapalli S, Abada PB, Sherman M, Zhu AX. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int. 2019;39:2214–2229. doi: 10.1111/liv.14223. [DOI] [PubMed] [Google Scholar]
- 124.Ahn JC, Teng PC, Chen PJ, Posadas E, Tseng HR, Lu SC, Yang JD. Detection of circulating tumor cells and their implications as a biomarker for diagnosis, prognostication, and therapeutic monitoring in hepatocellular carcinoma. Hepatology. 2021;73:422–436. doi: 10.1002/hep.31165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.von Felden J, Garcia-Lezana T, Schulze K, Losic B, Villanueva A. Liquid biopsy in the clinical management of hepatocellular carcinoma. Gut. 2020;69:2025–2034. doi: 10.1136/gutjnl-2019-320282. [DOI] [PubMed] [Google Scholar]
- 126.Mjelle R, Dima SO, Bacalbasa N, Chawla K, Sorop A, Cucu D, Herlea V, Sætrom P, Popescu I. Comprehensive transcriptomic analyses of tissue, serum, and serum exosomes from hepatocellular carcinoma patients. BMC Cancer. 2019;19:1007. doi: 10.1186/s12885-019-6249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sheng LQ, Li JR, Qin H, Liu L, Zhang DD, Zhang Q, Huang ML, Li XL, Xu XY, Wei YN, et al. Blood exosomal micro ribonucleic acid profiling reveals the complexity of hepatocellular carcinoma and identifies potential biomarkers for differential diagnosis. World J Gastrointest Oncol. 2020;12:1195–1208. doi: 10.4251/wjgo.v12.i10.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016;5:31292. doi: 10.3402/jev.v5.31292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Salehi M, Sharifi M. Exosomal miRNAs as novel cancer biomarkers: Challenges and opportunities. J Cell Physiol. 2018;233:6370–6380. doi: 10.1002/jcp.26481. [DOI] [PubMed] [Google Scholar]
- 130.Fitts CA, Ji N, Li Y, Tan C. Exploiting exosomes in cancer liquid biopsies and drug delivery. Adv Healthc Mater. 2019;8:e1801268. doi: 10.1002/adhm.201801268. [DOI] [PubMed] [Google Scholar]
- 131.Caradec J, Kharmate G, Hosseini-Beheshti E, Adomat H, Gleave M, Guns E. Reproducibility and efficiency of serum-derived exosome extraction methods. Clin Biochem. 2014;47:1286–1292. doi: 10.1016/j.clinbiochem.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 132.Ge Y, Mu W, Ba Q, Li J, Jiang Y, Xia Q, Wang H. Hepatocellular carcinoma-derived exosomes in organotropic metastasis, recurrence and early diagnosis application. Cancer Lett. 2020;477:41–48. doi: 10.1016/j.canlet.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 133.Zhang C, Yang X, Qi Q, Gao Y, Wei Q, Han S. lncRNA-HEIH in serum and exosomes as a potential biomarker in the HCV-related hepatocellular carcinoma. Cancer Biomark. 2018;21:651–659. doi: 10.3233/CBM-170727. [DOI] [PubMed] [Google Scholar]
- 134.Lyu L, Yang W, Yao J, Wang H, Zhu J, Jin A, Liu T, Wang B, Zhou J, Fan J, et al. The diagnostic value of plasma exosomal hsa_circ_0070396 for hepatocellular carcinoma. Biomark Med. 2021;15:359–371. doi: 10.2217/bmm-2020-0476. [DOI] [PubMed] [Google Scholar]
- 135.Cho HJ, Baek GO, Seo CW, Ahn HR, Sung S, Son JA, Kim SS, Cho SW, Jang JW, Nam SW, et al. Exosomal microRNA-4661-5p-based serum panel as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. Cancer Med. 2020;9:5459–5472. doi: 10.1002/cam4.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cui Y, Xu HF, Liu MY, Xu YJ, He JC, Zhou Y, Cang SD. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J Gastroenterol. 2019;25:1890–1898. doi: 10.3748/wjg.v25.i15.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fründt T, Krause L, Hussey E, Steinbach B, Köhler D, von Felden J, Schulze K, Lohse AW, Wege H, Schwarzenbach H. Diagnostic and prognostic value of miR-16, miR-146a, miR-192 and miR-221 in exosomes of hepatocellular carcinoma and liver cirrhosis patients. Cancers (Basel) 2021;13:2484. doi: 10.3390/cancers13102484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang S, Yang Y, Sun L, Qiao G, Song Y, Liu B. Exosomal MicroRNAs as liquid biopsy biomarkers in hepatocellular carcinoma. Onco Targets Ther. 2020;13:2021–2030. doi: 10.2147/OTT.S232453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu WH, Ren LN, Wang X, Wang T, Zhang N, Gao Y, Luo H, Navarro-Alvarez N, Tang LJ. Combination of exosomes and circulating microRNAs may serve as a promising tumor marker complementary to alpha-fetoprotein for early-stage hepatocellular carcinoma diagnosis in rats. J Cancer Res Clin Oncol. 2015;141:1767–1778. doi: 10.1007/s00432-015-1943-0. [DOI] [PubMed] [Google Scholar]
- 140.Wang Y, Zhang C, Zhang P, Guo G, Jiang T, Zhao X, Jiang J, Huang X, Tong H, Tian Y. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018;7:1670–1679. doi: 10.1002/cam4.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cui Z, Li Y, Gao Y, Kong L, Lin Y, Chen Y. Cancer-testis antigen lactate dehydrogenase C4 in hepatocellular carcinoma: A promising biomarker for early diagnosis, efficacy evaluation and prognosis prediction. Aging (Albany NY) 2020;12:19455–19467. doi: 10.18632/aging.103879. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim SS, Baek GO, Ahn HR, Sung S, Seo CW, Cho HJ, Nam SW, Cheong JY, Eun JW. Serum small extracellular vesicle-derived LINC00853 as a novel diagnostic marker for early hepatocellular carcinoma. Mol Oncol. 2020;14:2646–2659. doi: 10.1002/1878-0261.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sanchez JI, Jiao J, Kwan SY, Veillon L, Warmoes MO, Tan L, Odewole M, Rich NE, Wei P, Lorenzi PL, et al. Lipidomic profiles of plasma exosomes identify candidate biomarkers for early detection of hepatocellular carcinoma in patients with cirrhosis. Cancer Prev Res (Phila) 2021;14:955–962. doi: 10.1158/1940-6207.CAPR-20-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li W, Ding X, Wang S, Xu L, Yin T, Han S, Geng J, Sun W. Downregulation of serum exosomal miR-320d predicts poor prognosis in hepatocellular carcinoma. J Clin Lab Anal. 2020;34:e23239. doi: 10.1002/jcla.23239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Qu Z, Wu J, Wu J, Ji A, Qiang G, Jiang Y, Jiang C, Ding Y. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8:80666–80678. doi: 10.18632/oncotarget.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xu H, Chen Y, Dong X, Wang X. Serum exosomal long noncoding RNAs ENSG00000258332.1 and LINC00635 for the diagnosis and prognosis of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2018;27:710–716. doi: 10.1158/1055-9965.EPI-17-0770. [DOI] [PubMed] [Google Scholar]
- 147.Cho HJ, Baek GO, Yoon MG, Ahn HR, Son JA, Kim SS, Cheong JY, Eun JW. Overexpressed proteins in HCC cell-derived exosomes, CCT8, and cofilin-1 are potential biomarkers for patients with HCC. Diagnostics (Basel) 2021;11:1221. doi: 10.3390/diagnostics11071221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Eun JW, Seo CW, Baek GO, Yoon MG, Ahn HR, Son JA, Sung S, Kim DW, Kim SS, Cho HJ, Cheong JY. Circulating exosomal MicroRNA-1307-5p as a predictor for metastasis in patients with hepatocellular carcinoma. Cancers (Basel) 2020;12:3819. doi: 10.3390/cancers12123819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao B, Dai Z, Cao Y, Fan J, Zhou J. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther. 2017;10:3843–3851. doi: 10.2147/OTT.S140062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kim HS, Kim JS, Park NR, Nam H, Sung PS, Bae SH, Choi JY, Yoon SK, Hur W, Jang JW. Exosomal miR-125b exerts anti-metastatic properties and predicts early metastasis of hepatocellular carcinoma. Front Oncol. 2021;11:637247. doi: 10.3389/fonc.2021.637247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee YR, Kim G, Tak WY, Jang SY, Kweon YO, Park JG, Lee HW, Han YS, Chun JM, Park SY, Hur K. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer. 2019;144:1444–1452. doi: 10.1002/ijc.31931. [DOI] [PubMed] [Google Scholar]
- 152.Xu H, Dong X, Chen Y, Wang X. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin Chem Lab Med. 2018;56:479–484. doi: 10.1515/cclm-2017-0327. [DOI] [PubMed] [Google Scholar]
- 153.Ma D, Gao X, Liu Z, Lu X, Ju H, Zhang N. Exosome-transferred long non-coding RNA ASMTL-AS1 contributes to malignant phenotypes in residual hepatocellular carcinoma after insufficient radiofrequency ablation. Cell Prolif. 2020;53:e12795. doi: 10.1111/cpr.12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shi M, Jiang Y, Yang L, Yan S, Wang YG, Lu XJ. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J Cell Biochem. 2018;119:4711–4716. doi: 10.1002/jcb.26650. [DOI] [PubMed] [Google Scholar]
- 155.Wang T, Zhu H, Xiao M, Zhou S. Serum exosomal long noncoding RNA CRNDE as a prognostic biomarker for hepatocellular carcinoma. J Clin Lab Anal. 2021;35:e23959. doi: 10.1002/jcla.23959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Luo Y, Liu F, Gui R. High expression of circulating exosomal circAKT3 is associated with higher recurrence in HCC patients undergoing surgical treatment. Surg Oncol. 2020;33:276–281. doi: 10.1016/j.suronc.2020.04.021. [DOI] [PubMed] [Google Scholar]
- 157.Yokota Y, Noda T, Okumura Y, Kobayashi S, Iwagami Y, Yamada D, Tomimaru Y, Akita H, Gotoh K, Takeda Y, et al. Serum exosomal miR-638 is a prognostic marker of HCC via downregulation of VE-cadherin and ZO-1 of endothelial cells. Cancer Sci. 2021;112:1275–1288. doi: 10.1111/cas.14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, Shinden Y, Iguchi T, Eguchi H, Shirabe K, et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112:532–538. doi: 10.1038/bjc.2014.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Suehiro T, Miyaaki H, Kanda Y, Shibata H, Honda T, Ozawa E, Miuma S, Taura N, Nakao K. Serum exosomal microRNA-122 and microRNA-21 as predictive biomarkers in transarterial chemoembolization-treated hepatocellular carcinoma patients. Oncol Lett. 2018;16:3267–3273. doi: 10.3892/ol.2018.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang SC, Li CY, Chang WT, Cheng WC, Yen CH, Tu WY, Lin ZY, Lin CC, Yeh ML, Huang CF, et al. Exosome-derived differentiation antagonizing non-protein coding RNA with risk of hepatitis C virus-related hepatocellular carcinoma recurrence. Liver Int. 2021;41:956–968. doi: 10.1111/liv.14772. [DOI] [PubMed] [Google Scholar]
- 161.Qin L, Zhan Z, Wei C, Li X, Zhang T, Li J. Has-circRNA-G004213 promotes cisplatin sensitivity by regulating miR-513b-5p/PRPF39 in liver cancer. Mol Med Rep. 2021;23:421. doi: 10.3892/mmr.2021.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ghosh S, Bhowmik S, Majumdar S, Goswami A, Chakraborty J, Gupta S, Aggarwal S, Ray S, Chatterjee R, Bhattacharyya S, et al. The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low alpha-fetoprotein. Int J Cancer. 2020;147:2934–2947. doi: 10.1002/ijc.33111. [DOI] [PubMed] [Google Scholar]
- 163.Sun XH, Wang YT, Li GF, Zhang N, Fan L. Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int. 2020;20:226. doi: 10.1186/s12935-020-01302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kim SS, Baek GO, Son JA, Ahn HR, Yoon MK, Cho HJ, Yoon JH, Nam SW, Cheong JY, Eun JW. Early detection of hepatocellular carcinoma via liquid biopsy: Panel of small extracellular vesicle-derived long noncoding RNAs identified as markers. Mol Oncol. 2021;15:2715–2731. doi: 10.1002/1878-0261.13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Huang C, Tang S, Shen D, Li X, Liang L, Ding Y, Xu B. Circulating plasma exosomal miRNA profiles serve as potential metastasis-related biomarkers for hepatocellular carcinoma. Oncol Lett. 2021;21:168. doi: 10.3892/ol.2021.12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Itami-Matsumoto S, Hayakawa M, Uchida-Kobayashi S, Enomoto M, Tamori A, Mizuno K, Toyoda H, Tamura T, Akutsu T, Ochiya T, et al. Circulating exosomal miRNA profiles predict the occurrence and recurrence of hepatocellular carcinoma in patients with direct-acting antiviral-induced sustained viral response. Biomedicines. 2019;7:87. doi: 10.3390/biomedicines7040087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sorop A, Iacob R, Iacob S, Constantinescu D, Chitoiu L, Fertig TE, Dinischiotu A, Chivu-Economescu M, Bacalbasa N, Savu L, et al. Plasma small extracellular vesicles derived miR-21-5p and miR-92a-3p as potential biomarkers for hepatocellular carcinoma screening. Front Genet. 2020;11:712. doi: 10.3389/fgene.2020.00712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xu LX, He MH, Dai ZH, Yu J, Wang JG, Li XC, Jiang BB, Ke ZF, Su TH, Peng ZW, et al. Genomic and transcriptional heterogeneity of multifocal hepatocellular carcinoma. Ann Oncol. 2019;30:990–997. doi: 10.1093/annonc/mdz103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Huang A, Yang XR, Chung WY, Dennison AR, Zhou J. Targeted therapy for hepatocellular carcinoma. Signal Transduct Target Ther. 2020;5:146. doi: 10.1038/s41392-020-00264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yang Y, Hong Y, Cho E, Kim GB, Kim IS. Extracellular vesicles as a platform for membrane-associated therapeutic protein delivery. J Extracell Vesicles. 2018;7:1440131. doi: 10.1080/20013078.2018.1440131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm Sin B. 2016;6:287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ding J, Wang J, Chen J. Exosomes as therapeutic vehicles in liver diseases. Ann Transl Med. 2021;9:735. doi: 10.21037/atm-20-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tai YL, Chen KC, Hsieh JT, Shen TL. Exosomes in cancer development and clinical applications. Cancer Sci. 2018;109:2364–2374. doi: 10.1111/cas.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lowry MC, Gallagher WM, O'Driscoll L. The role of exosomes in breast cancer. Clin Chem. 2015;61:1457–1465. doi: 10.1373/clinchem.2015.240028. [DOI] [PubMed] [Google Scholar]
- 175.Xue D, Han J, Liu Y, Tuo H, Peng Y. Current perspectives on exosomes in the diagnosis and treatment of hepatocellular carcinoma (review) Cancer Biol Ther. 2021;22:279–290. doi: 10.1080/15384047.2021.1898728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Khan N, Maurya S, Bammidi S, Jayandharan GR. AAV6 vexosomes mediate robust suicide gene delivery in a murine model of hepatocellular carcinoma. Mol Ther Methods Clin Dev. 2020;17:497–504. doi: 10.1016/j.omtm.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Du J, Wan Z, Wang C, Lu F, Wei M, Wang D, Hao Q. Designer exosomes for targeted and efficient ferroptosis induction in cancer via chemo-photodynamic therapy. Theranostics. 2021;11:8185–8196. doi: 10.7150/thno.59121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Li I, Nabet BY. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol Cancer. 2019;18:32. doi: 10.1186/s12943-019-0975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Le Grazie M, Biagini MR, Tarocchi M, Polvani S, Galli A. Chemotherapy for hepatocellular carcinoma: The present and the future. World J Hepatol. 2017;9:907–920. doi: 10.4254/wjh.v9.i21.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shi S, Rao Q, Zhang C, Zhang X, Qin Y, Niu Z. Dendritic cells pulsed with exosomes in combination with PD-1 antibody increase the efficacy of sorafenib in hepatocellular carcinoma model. Transl Oncol. 2018;11:250–258. doi: 10.1016/j.tranon.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Loch-Neckel G, Matos AT, Vaz AR, Brites D. Challenges in the development of drug delivery systems based on small extracellular vesicles for therapy of brain diseases. Front Pharmacol. 2022;13:839790. doi: 10.3389/fphar.2022.839790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pospichalova V, Svoboda J, Dave Z, Kotrbova A, Kaiser K, Klemova D, Ilkovics L, Hampl A, Crha I, Jandakova E, et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J Extracell Vesicles. 2015;4:25530. doi: 10.3402/jev.v4.25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zarovni N, Corrado A, Guazzi P, Zocco D, Lari E, Radano G, Muhhina J, Fondelli C, Gavrilova J, Chiesi A. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods. 2015;87:46–58. doi: 10.1016/j.ymeth.2015.05.028. [DOI] [PubMed] [Google Scholar]
- 184.Bala S, Csak T, Momen-Heravi F, Lippai D, Kodys K, Catalano D, Satishchandran A, Ambros V, Szabo G. Biodistribution and function of extracellular miRNA-155 in mice. Sci Rep. 2015;5:10721. doi: 10.1038/srep10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhou H, Yuen PS, Pisitkun T, Gonzales PA, Yasuda H, Dear JW, Gross P, Knepper MA, Star RA. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 2006;69:1471–1476. doi: 10.1038/sj.ki.5000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.