Abstract
In order to perform single-cell analysis and online studies of N-acyl homoserine lactone (AHL)-mediated communication among bacteria, components of the Vibrio fischeri quorum sensor encoded by luxR-PluxI have been fused to modified versions of gfpmut3∗ genes encoding unstable green fluorescent proteins. Bacterial strains harboring this green fluorescent sensor detected a broad spectrum of AHL molecules and were capable of sensing the presence of 5 nM N-3-oxohexanoyl-l-homoserine lactone in the surroundings. In combination with epifluorescent microscopy, the sensitivity of the sensor enabled AHL detection at the single-cell level and allowed for real-time measurements of fluctuations in AHL concentrations. This green fluorescent AHL sensor provides a state-of-the-art tool for studies of communication between the individuals present in mixed bacterial communities.
In recent years it has become apparent that bacteria coordinate their interaction and association with higher organisms by intercellular communication systems. In gram-negative bacteria, one type of communication system functions via small, diffusible N-acyl homoserine lactone (AHL) signal molecules. The signals are synthesized from precursors by a synthase protein, “I,” and once they have reached a certain threshold concentration, they interact with a transcriptional activating “R” protein to induce expression of different target genes (for reviews see references 11, 13, and 43). Such regulatory systems operate as a quorum-sensing mechanism that allows bacteria to sense and express target genes in relation to their cell density.
Several methods to detect the presence of AHL have been described. AHLs can be extracted from liquid cultures, purified to homogeneity by semipreparative high-performance liquid chromatography (HPLC), and identified by mass spectrometry and 1H nuclear magnetic resonance (NMR) spectroscopy (10). A number of bacterial sensor systems such as the pigment-developing Chromobacterium violaceum (30) and luxAB- and lacZ-based systems have been described (36, 50). Bioluminescent sensor systems have been conveniently used in Escherichia coli and have enabled the isolation and cloning of a number of I genes (31, 41, 42). A simple and convincing method for separation and tentative identification of AHL molecules in extracts of whole cultures has been developed; it consists of thin-layer chromatography (TLC) followed by detection of AHL molecules by means of agar overlay with sensor bacteria (36).
Although these methods are very useful and highly sensitive, they do not allow for detection at the single-cell level or at the local environment. Furthermore, investigation of AHL expression based on population level analysis does not give information about local concentrations. A live bacterial AHL sensor that signals the presence of AHL molecules by expressing a reporter such as green fluorescent protein (GFP) can fulfil these requirements. GFP, obtained from the jellyfish Aequorea victoria, requires only trace amounts of oxygen to mature, i.e., no external compounds need to be added to organisms expressing GFP in order to detect green fluorescence (3). Previous work has demonstrated that GFP works well as a reporter for monitoring gene expression at the single-cell level in biofilms (4, 32, 39).
Unfortunately, once formed, GFP is extremely stable (1, 47). Consequently, the GFP version used for monitoring fluctuations in AHL concentration should possess a species-independent instability. Previously, we have constructed a number of gfp genes (derived from gfpmut3 [6]) each of which encodes proteins with different half-lives ranging from 40 min to a few hours (1). Briefly, this was done by manipulating the gfp gene in such a way that the resultant protein carried C-terminal peptide tags which are recognized and to various extent rapidly degraded by housekeeping/intracellular tail-specific proteases (ClpP) (1, 17, 22, 23). In the present constructs the product of the luxR gene derived from Vibrio fischeri comprises the quorum sensor. In the presence of exogenous AHL molecules, LuxR positively affects the expression of the luxI promoter (PluxI), which then in turn controls expression of the gfp reporter. LuxR functions in a number of different bacterial strains (10, 19, 34, 41) and is responsive to a variety of AHL molecules (35, 50). Here we describe the construction and application of GFP-based AHL sensors.
MATERIALS AND METHODS
Bacterial strains.
E. coli, Burkholderia cepacia, Serratia liquefaciens, Serratia ficaria, Pseudomonas aureofaciens, and Pseudomonas aeruginosa strains used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| Burkholderia cepacia | ||
| DSM50180 | AHL+ | Deutsche Stammsammlung |
| ATCC 25416 | AHL+ | American Type Culture Collection |
| Escherichia coli | ||
| JM105 | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/thi rpsL (Strr) endA sbcB15 sbcC hsdR4 (rK− mK−) Δ(lac-proAB) | 52 |
| MC1000 | araD139 (ara-leu)7697 Δlac thi hsdR | 37 |
| MT102 | Restriction-negative mutant derived from MC1000 | M. T. Hansen, unpublished data |
| Pseudomonas aureofaciens ATCC 13985 | wt, AHL+ | American Type Culture Collection |
| Serratia ficaria ATCC 33105 | wt, AHL+ | American Type Culture Collection |
| Serratia liquefaciens | ||
| MG1 | wt, AHL+ | 14 |
| MG44 | swrI mutant derived from MG1 | 10 |
| Pseudomonas aeruginosa | ||
| PAO1 | wt, AHL+ | B. H. Iglewski |
| JP2 | lasI rhlI double mutant derived from PAO1 | B. H. Iglewski |
| Plasmids | ||
| pQE70 | Apr | Qiagen product guide |
| pSB403 | Tcr; donor of luxR-PluxI cassette | 50 |
| pJBA27 | Apr; pUC18Not-PA1/04/03-RBSII-gfpmut3∗-T0-T1 | 1 |
| pJBA113 | Apr; pUC18Not-PA1/04/03-RBSII-gfp(ASV)-T0-T1 | 1 |
| pJBA88 | Apr; pUC18Not-luxR-PluxI-RBSII-gfpmut3∗-T0-T1 | This study |
| pJBA89 | Apr; pUC18Not-luxR-PluxI-RBSII-gfp(ASV)-T0-T1 | This study |
| pME6010 | Tcr; pVS1-pACY177 shuttle vector carrying the plasmid-stabilizing region sta of pVS1 | 25 |
| pME6031 | Tcr; derivative of pME6010, where the kanamycin promoter of pME6010 has been replaced with the transcription terminator T4 from pHP45 | 20 |
| pJBA130 | Tcr; pME6031-luxR-PluxI-RBSII-gfpmut3∗-T0-T1 | This study |
| pJBA132 | Tcr; pME6031-luxR-PluxI-RBSII-gfp(ASV)-T0-T1 | This study |
wt, wild type.
Media.
The basic medium used was either modified Luria-Bertani (LB) medium (2) containing 4 g of NaCl/liter instead of the normal 10 g of NaCl/liter or ABT minimal medium (AB minimal medium [5] containing 2.5 mg of thiamine/liter).
Plasmids.
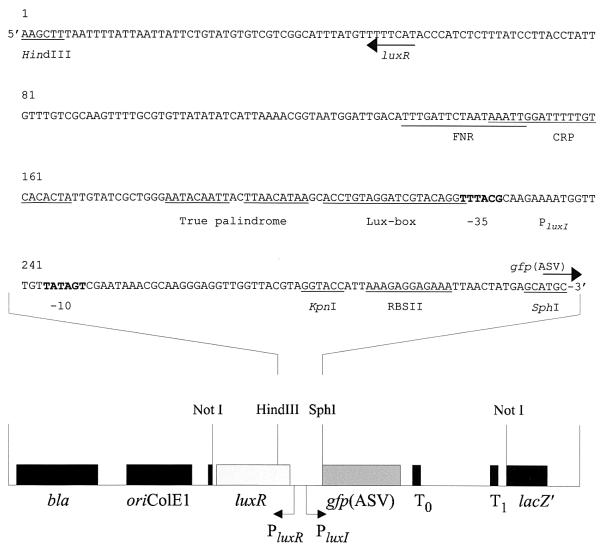
The plasmids used in this study are listed in Table 1. pJBA88 (Table 1) and pJBA89 (Table 1 and Fig. 1) were constructed as follows. PCR amplification with the primer set P1 (5′-CATTATTGCTTCTACAAGCTTTA-3′) and P2 (5′-ACACAGCATGCTCATAGTTAATTTCTCCTCTTTAATGGTACCTACGTAACCAACCTCCCTT-3′) and with pSB403 (50) as the template produced a 0.31-kb HindIII-SphI fragment encoding the N-terminal part of the V. fischeri luxR gene (9) including the regulatory region of the lux operon and an efficient synthetic ribosome binding site (RBSII from pQE70 [Qiagen product guide, 1997, Qiagen GmbH, Hilden, Germany]). To create pJBA88, this PCR fragment was combined with a 0.71-kb EcoRI-HindIII fragment (encoding the remaining C-terminal part of luxR) from pSB403 (50) and ligated to EcoRI-SphI-digested pJBA27 (a pUC18 NotI derivative carrying gfpmut3∗ followed by two transcriptional terminators, T0 [from phage lambda] and T1 [from the rrnB operon of E. coli]) (1). Subsequently, pJBA89 was constructed by inserting the 0.71-kb EcoRI-HindIII fragment of pSB403 (50) and the 0.31-kb HindIII-SphI fragment of pJBA88 into EcoRI-SphI-digested pJBA113 [a pUC18 NotI derivative carrying gfp(ASV) followed by two transcriptional terminators, T0 (from phage lambda) and T1 (from the rrnB operon of E. coli)] (1). pJBA130 and pJBA132 (Table 1) were constructed as follows. To allow insertion of the AHL sensor cassettes of pJBA88 and pJBA89 into the polylinker of pME6031 (a kind gift from Stephan Heeb) (20), the 5′ overhangs of the 2.8-kb NotI fragment of pJBA88 (containing the luxR-PluxI-gfpmut3∗-T0-T1 cassette), the 2.85-kb NotI fragment of pJBA89 [containing the luxR-PluxI-gfp(ASV)-T0-T1 cassette], and the 8.3-kb HindIII fragment of pME6031 were filled in using the Klenow fragment of DNA polymerase I. Finally, the 2.8-kb (NotI) fragment of pJBA88 and the 2.85-kb (NotI) fragment of pJBA89 were ligated to the 8.3-kb (HindIII) fragment of pME6031 to create pJBA130 and pJBA132, respectively.
FIG. 1.
Schematic drawing of pJBA89 and the DNA nucleotide sequence of the PCR-amplified luxR-PluxI-RBSII fragment used for the construction of pJBA88, pJBA89, pJBA130, and pJBA132. Arrows indicate directions of transcription of luxR and gfp(ASV). The positions of FNR, cAMP-CRP binding sites and the lux-box sequences are indicated, and the −35 and −10 regions of the luxI promoter (PluxI) are shown in bold letters (38). Important restriction sites are underlined.
DNA nucleotide sequencing.
Sequencing of the 0.31-kb HindIII-SphI fragment of pJBA88, was performed with a model 373A automatic DNA sequencer (Applied Biosystems, Foster City, Calif.) on double-stranded plasmid DNA in accordance with the manufacturer's recommendations.
Macroscopic detection of AHL-producing bacteria.
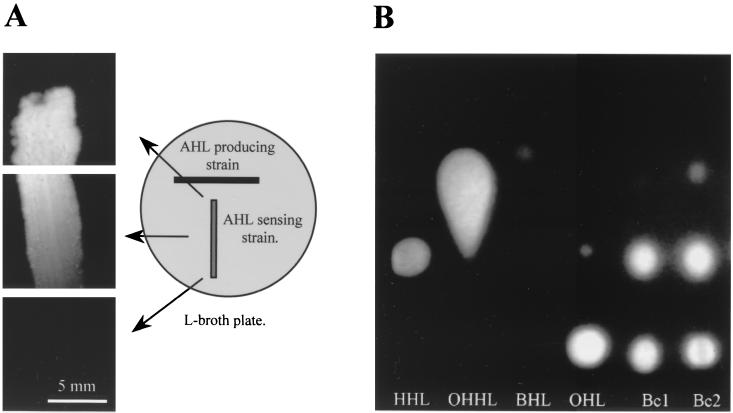
On an LB plate, the AHL producer S. liquefaciens MG1 (10) and the gfp(ASV)-based AHL sensor E. coli JM105 harboring pJBA89 were streaked close to each other to form a T (approximately 5 cm wide [MG1 streak] and 5 cm high [JB357 streak]). Following 20 h of incubation at 30°C, the green fluorescence phenotype of the AHL sensor streak was recorded with a charge-coupled device camera mounted on an epifluorescence microscope (Axioplan; Zeiss, Oberkochen, Germany) equipped with a 2.5× lens (see “Microscopy and image analysis” below for further details).
TLC.
A 250-ml volume of spent supernatants from B. cepacia cultures grown to an optical density at 600 nm (OD600) of 1.0 in ABT minimal medium supplemented with 0.2% glucose was extracted twice with dichloromethane. The combined extracts were dried over anhydrous magnesium sulfate, filtered, and evaporated to dryness. Residues were dissolved in 250 μl of ethyl acetate. Samples (10 μl) were then applied to C18 reversed-phase TLC plates (Merck no. 1.15389) and dried with a stream of cold air. As reference compounds, synthetic AHLs were included in the following concentrations: 5 ng of N-3-oxohexanoyl-l-homoserine lactone (OHHL), 50 ng of N-hexanoyl-l-homoserine lactone (HHL), 2.5 μg of N-octanoyl-l-homoserine lactone (OHL), and 5 μg of N-butyryl-l-homoserine lactone (BHL). Samples were separated by using methanol (60%, vol/vol) in water as the solvent, as described by Shaw and coworkers (36). For detection of AHLs, the TLC plate was overlaid with a thin film of a gfp-based AHL sensor (JM105 harboring pJBA89) seeded in LB medium solidified with 0.7% agar. Following incubation for 24 at 30°C, the TLC plate was illuminated with blue light using an HQ 480/40 filter (F44-001; AF Analysentechnik, Tübingen, Germany) in combination with a halogen lamp (Intralux 5000-1; Volpi, Zurich, Switzerland) as a light source. Illumination took place in a darkbox (Unit 1, Birkeroed, Denmark) that was equipped with a light-sensitive camera (C2400-47; Hamamatsu, Herrsching, Germany) with a Pentax CCTV camera lens and an HQ 535/50 filter (F44-001; AF Analysentechnik). The Argus 20 image analysis system (Hamamatsu) was used for detection and documentation of green fluorescent spots. The Rf values of the AHL molecules produced by B. cepacia were compared to those of reference AHL compounds for tentative identification of their structures.
AHL dose-response.
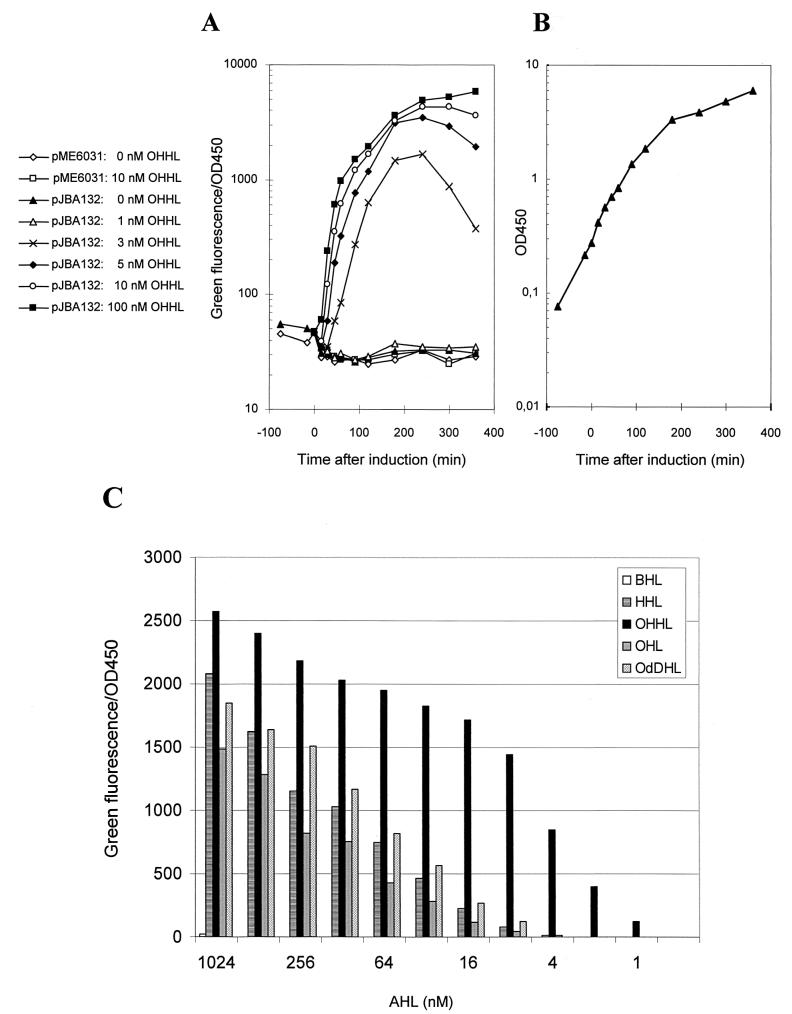
E. coli MT102 harboring either pJBA132 [expressing GFP(ASV) in response to OHHL] or pME6031 (nonfluorescent reference) (Table 1) was grown exponentially in LB medium at 30°C. At an OD450 of approximately 0.25, both cultures were divided into six subcultures. Next, OHHL was added to the subcultures, resulting in cultures of both strains containing 0, 1, 3, 5, 10, or 100 nM OHHL. The cultures were further incubated at 30°C, culture samples were withdrawn at various time intervals, and green fluorescence was measured with a fluorometer (model RF-1501; Shimadzu, Tokyo, Japan) set at an excitation wavelength of 475 nm and emission detection at 515 nm. For each sample, the measured value of green fluorescence was normalized to 1 ml of culture and then converted into specific green fluorescence by dividing normalized values by the OD450 of the bacterial culture. Subsequently, specific green fluorescence was plotted as a function of time in a semilogarithmic plot (see Fig. 3A).
FIG. 3.
(A) Dose-response of the AHL sensor MT102 harboring pJBA132 to OHHL. The strain was grown at 30°C in LB medium. In the early-mid-log phase (t = 0 min), the culture was divided into six subcultures and OHHL was added as indicated. The plot shows specific green fluorescence as a function of time. (B) Bacterial growth of a single representative culture. (C) Sensitivity to different AHL molecules (see Materials and Methods for details).
To compare the responsiveness of E. coli MT102 harboring pJBA132 towards BHL, HHL, OHHL, OHL, and N-3-oxododecanoyl-l-homoserine lactone (OdDHL) signal molecules, an overnight culture of this AHL monitor strain was initially diluted 4 times into fresh prewarmed (30°C) LB medium and then distributed as 200-μl aliquots into the wells of a microtiter plate. Next the AHL molecules mentioned above were added to the wells to obtain cultures containing either 1,024, 512, 256, 128, 64, 32, 16, 8, 4, 2, 1, or 0 nM AHL molecules. Following 2 h of incubation at 30°C, the microtiter plate was placed on ice and samples of each culture were withdrawn for measurement of OD450 and green fluorescence (see above). The measured values of green fluorescence were converted into specific green fluorescence as described earlier and plotted as a function of AHL concentration in a bar diagram (see Fig. 3C).
Response of the sensor to fluctuations in OHHL concentrations.
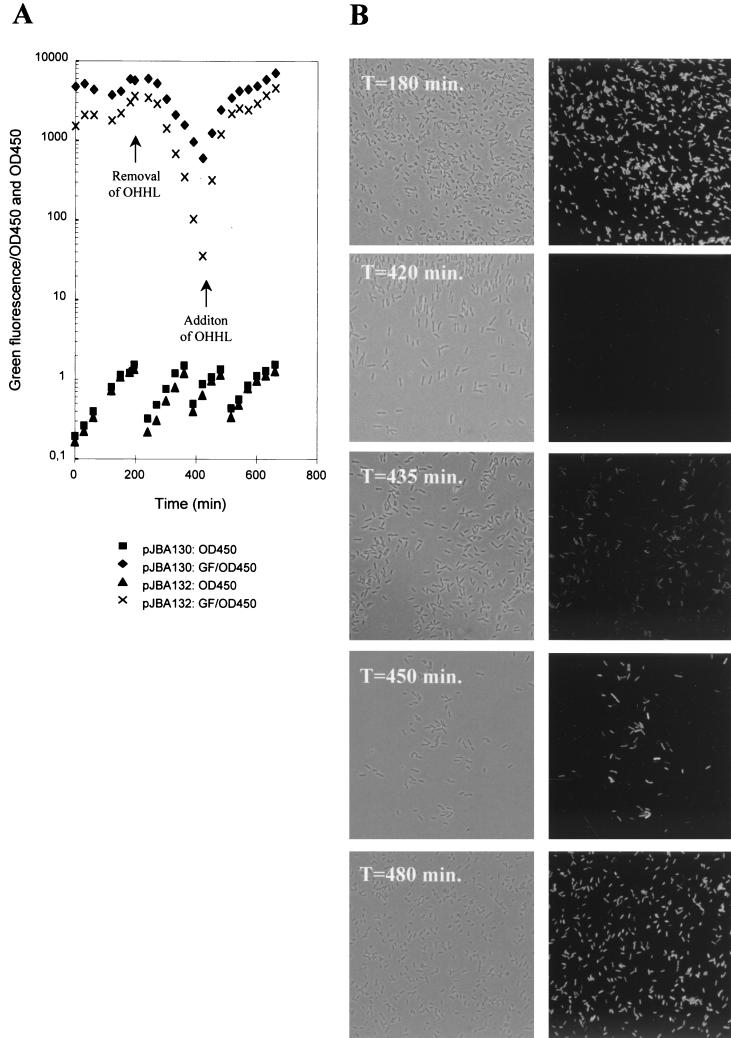
E. coli MT102 harboring either pJBA130 (expressing GFPmut3∗ in response to OHHL), pJBA132 [expressing GFP(ASV) in response to OHHL], or pME6031 (nonfluorescent control) was kept exponentially growing at 30°C throughout the entire experiment. This was achieved by diluting the cultures 5 times into fresh prewarmed medium whenever the cultures reached an OD450 of approximately 1.5. During the first 3 h of the experiment, the three strains were grown in LB medium containing 10 nM OHHL. When the cultures reached an OD450 of approximately 1.5 (t = 180 min), the three cultures were harvested by centrifugation (10,000 × g for 8 min at 4°C), washed once with ice-cold LB medium, and then resuspended into prewarmed LB medium lacking OHHL. Following 4 h of exponential growth in the absence of OHHL, OHHL was added back to the three cultures (t = 420 minutes) to give a final concentration of 10 nM and the cultures were grown for additional 4.5 h. During the experiment, culture samples were withdrawn at various time points and the green fluorescence was measured using a fluorometer (see the preceding section for details). Subsequently, the measured values of green fluorescence were converted into specific green fluorescence and plotted as a function of time in a semilogarithmic plot (see Fig. 4A).
FIG. 4.
OHHL down- and upshift experiment with two AHL sensors, MT102 harboring pJBA130 (encoding stable GFPmut3∗) and MT102 harboring pJBA132 [encoding the unstable GFP(ASV)]. The cultures were grown with 10 nM OHHL. At 180 min, OHHL was washed away, and the cultures were grown in fresh OHHL-free medium. At 420 min, 10 nM OHHL was added to the cultures. (A) Growth and specific green fluorescence. Arrows indicate the time of AHL removal (downshift) and the time of AHL readdition. (B) Phase-contrast (left) and epifluorescence (right), microscopy images of selected culture samples of MT102 harboring pJBA132 withdrawn during the AHL down- and upshift.
In order to visualize the GFP(ASV) sensor's ability to respond to fluctuations in AHL concentrations, culture samples were withdrawn following 180, 420, 435, 450, and 480 min of cultivation. The green fluorescent phenotype of single cells was recorded using an epifluorescence microscope equipped with a 63× lens (see “Microscopy and image analysis” below).
Swarming experiments.
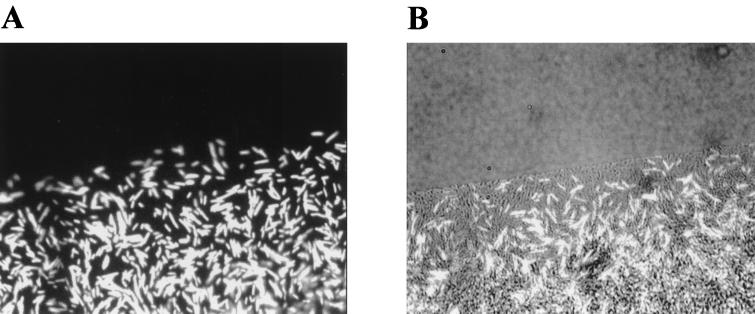
S. ficaria and S. liquefaciens MG44 harboring pJBA132 were inoculated either alone or mixed in a ratio of approximately 1:1 on semisolid agar plates containing ABT medium (see above) supplemented with 0.4% (wt/wt) glucose, 0.4% (wt/wt) Casamino Acids, and 0.7% Bacto Agar. Following 20 h of incubation at 30°C, the resulting colonies were examined using an epifluorescence microscope equipped with a 50× lens. Finally, the green fluorescent phenotypes of single cells in the respective colonies were captured as epifluorescent images (see Fig. 6) as described under “Microscopy and image analysis” below.
FIG. 6.
Binary swarming colony consisting of S. liquefaciens MG44 harboring pJBA132 and S. ficaria. Shown are an epifluorescence image (A) and a combined epifluorescence and phase-contrast image (B) of the moving colony.
Flow chamber experiments.
Surface-attached monospecies biofilms were cultivated in flow chambers (32) with channel dimensions of 1 by 4 by 40 mm. The substratum consisted of a microscope coverslip (24 mm by 50 mm; Knittel Gläser, Brunswick, Germany), and the flow chambers were supplied with a flow of 50-fold-diluted LB medium. Prior to each experiment, the flow system was assembled and prepared as described previously (32).
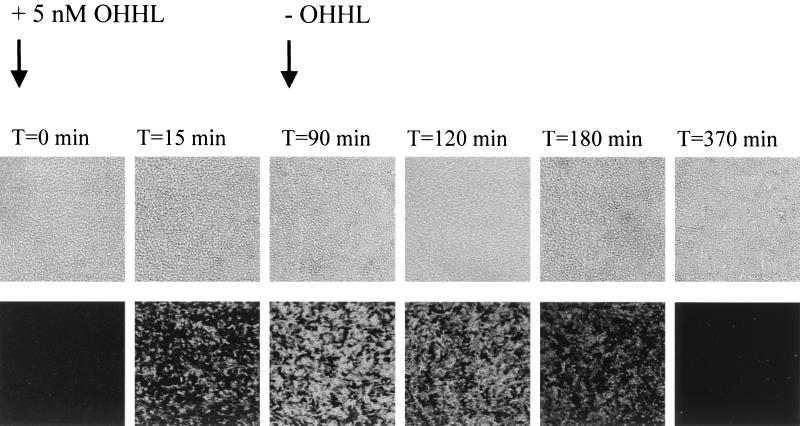
In order to evaluate the responsiveness of an AHL-sensing biofilm to fluctuations in the concentration of OHHL, a flow chamber was inoculated with 350 μl of an overnight culture of S. liquefaciens MG44 harboring pJBA132 [expressing GFP(ASV) in response to OHHL] diluted to an OD450 of 0.1 in 0.9% NaCl. After inoculation, the medium flow was arrested for 1 h to allow efficient colonization of the glass surface. The medium flow was then started, and the growth medium was pumped through the flow chamber at a constant rate of 0.2 mm/s using a peristaltic pump (Watson Marlow 205S). Following 24 h of incubation at 30°C, an OHHL up- and downshift was performed on the generated biofilm. At time zero, the biofilm was shifted to medium supplemented with 5 nM OHHL, and following 90 min of induction, the biofilm was shifted back to medium lacking OHHL. Throughout the OHHL up- and downshift, images of the biofilm were captured using a scanning confocal laser microscope (see the next section for details).
Microscopy and image analysis.
The green fluorescence phenotypes of colonies on semisolid surfaces, single cells in colonies on semisolid surfaces, or single cells from liquid cultures were recorded with a charge-coupled device camera mounted on an epifluorescence microscope (Zeiss Axioplan) equipped with a 2.5×, 50×, or 63× lens, respectively. For fluorescence microscopy we used Zeiss filter set no. 10 (excitation, 470 to 490 nm; emission, 515 to 565 nm; dichroic, 510 nm) and a Zeiss HBO-100 mercury lamp. A CH250 charge-coupled device equipped with a KAF 1400 liquid-cooled chip (Photometrics, Tucson, Ariz.) was used for imaging.
All microscopic observations and image acquisitions of biofilms were performed on a scanning confocal laser microscope (TCS4D; Leica Lasertechnik, GmbH, Heidelberg, Germany) equipped with a detector and a filter set for monitoring GFP. In addition, a reflection detector for acquiring bright-field images was installed. Images were obtained with a 63×/1.32 oil objective, and image scanning was carried out with the 488-nm laser line from an Ar/Kr laser. Images were processed for displaying using Photoshop software (Adobe, Mountain View, Calif.).
RESULTS
Construction of a GFP-based AHL sensor.
The luxR-PluxI quorum sensor derived from V. fischeri was chosen as the molecular component governing control over expression of the GFP reporters. Others have successfully exploited this AHL regulator in combination with a bioluminescent reporter to generate pSB403, an AHL-sensing system carried on a broad-host-range plasmid (50). Encouraged by the high sensitivity of AHL sensor strains harboring pSB403, we isolated from pSB403 a fragment containing luxR-PluxI and the adjacent region preceding the start codon of the luxCDABE operon, transcriptionally fused it to gfpmut3∗, and introduced it into a high-copy-number plasmid. Although it was responsive to OHHL, a major problem in the design of the first version of the gfpmut3∗-based sensor system was to match the output of the AHL quorum sensor with the accumulation of GFPmut3∗ in the cells. Colonies of E. coli strains harboring the first version of the gfpmut3∗-based sensor system were observed to develop only weak green fluorescence when cultivated on LB plates supplemented with 100 nM OHHL (data not shown). To improve the sensitivity, the region including the Shine-Dalgarno (SD) sequence and translation initiation site of gfpmut3∗ was modified. By PCR amplification, the SD sequence RBSII obtained from the pQE expression vectors (Qiagen product guide) was introduced downstream of PluxI to express the gfpmut3∗ reading frame and translational stop codons were introduced in the two other reading frames (see Fig. 1 and Materials and Methods for details). This modification increased GFPmut3∗ expression severalfold, and E. coli strains harboring this luxR-PluxI-RBSII-gfpmut3∗ cassette on a high-copy-number plasmid (pJBA88, of ColE1 origin) clearly responded to the presence of AHL molecules by expressing a bright GFP signal. In the absence of exogenous AHL, basic transcription from PluxI combined with the stability of the reporter resulted in some accumulation of GFPmut3∗, causing development of detectable green fluorescence. Consequently, discrimination between AHL-deficient strains and strains producing small amounts of AHL could not be accomplished with the pJBA88-based AHL sensor strain in AHL cross-feeding experiments (data not shown).
In order to obtain the lowest possible background signal, we constructed a transcriptional fusion between the luxR-PluxI-RBSII casette from pJBA88 and gfp(ASV), which encodes an unstable version of GFP exhibiting a half-life of approximately 45 min in exponentially growing E. coli cells (Fig. 1). Colonies of E. coli harboring this luxR-PluxI-RBSII-gfp(ASV) cassette on a high-copy-number plasmid (pJBA89, of ColE1 origin, see Table 1 and Fig. 1) appeared completely dark under the epifluorescence microscope following 20 h of incubation at 30°C on LB plates (data not shown). In striking contrast, the same strain produced bright-green fluorescent colonies when cultivated under identical conditions in the presence of 1 nM OHHL. Subsequent analysis revealed that the detection limits in liquid cultures of this gfp(ASV)-based sensor were in the range of 1 nM OHHL, 10 nM HHL, 10 nM OHL, 10 nM OdDHL, and 1000 nM BHL (51). When grown in close proximity to AHL-producing strains such as S. liquefaciens MG1 on LB plates, the sensor strain turned bright-green fluorescent, clearly demonstrating that physiological concentrations of AHLs are detectable by the sensor (Fig. 2A). In accordance with the existence of an AHL gradient arising from the MG1 streak, sensor cells located a few millimeters from MG1 were bright-green fluorescent, whereas sensor cells located approximately 35 mm from MG1 appeared completely nonfluorescent (Fig. 2A). To further evaluate its usefulness in macroscopic detection of AHL molecules, the gfp(ASV)-based AHL sensor was employed for visualization of AHLs separated by means of TLC (Fig. 2B). Samples extracted from culture supernatants of two B. cepacia strains and AHL references of known concentrations were separated by TLC. For detection of AHL, the TLC plate was subsequently overlaid with a thin film of LB agar seeded with the gfp(ASV)-based sensor. When illuminated with blue light, the TLC plate (Fig. 2B) showed that not only OHHL (the cognate autoinducer of LuxR) but also HHL and OHL could easily be detected, whereas the sensor was almost blind to BHL. By comparing the Rf values of the green fluorescent spots in the extracts from the B. cepacia strains with the Rf values of the known AHL references, the TLC analysis (Fig. 2B) strongly suggested that both B. cepacia strains produced HHL in addition to the OHL determined by Lewenza and coworkers (26).
FIG. 2.
(A) Epifluorescent image of the AHL sensor JM105 harboring pJBA89 cross-streaked with the AHL producer S. liquefaciens MG1. (B) Fluorescent image of a TLC plate used to separate extracts of B. cepacia (strains DM50180 [Bc1] and ATCC 25416 [Bc2]) and the reference compounds HHL, OHHL, BHL, and OHL overlaid with the AHL sensor (see Materials and Methods for details).
Broad-host-range sensors.
Although the sensors described above proved highly sensitive to AHLs, their applicability as reporters in vivo was limited. Due to the lack of plasmid-stabilizing functions, segregational stability could be obtained only in the presence of a selective pressure and only in the narrow host range dictated by the ColE1 replicon. To circumvent this and expand the application range of the gfp-based AHL monitor systems, the AHL-responsive cassettes of pJBA88 (luxR-PluxI-RBSII-gfpmut3∗) and pJBA89 [luxR-PluxI-RBSII-gfp(ASV)] were inserted into the polylinker of pME6031 (20) to give pJBA130 and pJBA132, respectively (Table 1). pME6031 is a shuttle vector based on the p15A replicon and the pVS1 replicon, making it capable of replicating in most gram-negative bacteria. Unfortunately, the pVS1 replicon does not appear to belong to any known incompatibility group; however, this replicon is compatible in P. aeruginosa with plasmids of incompatibility group IncP1 (equivalent to IncP in E. coli), IncP4 (equivalent to IncQ in E. coli), IncP8, IncP10, and IncP11 (20). In addition, pME6031 carries the stability region of pVS1, rendering the plasmid segregationally stable. To verify the host range and the stability of the resulting AHL-sensing plasmids, pJBA130 and pJBA132 were electroporated into competent cells of E. coli, S. liquefaciens, P. aureofaciens, and P. aeruginosa. As expected, both plasmids were capable of replicating in the species mentioned above, and they also appeared to be 100% segregationally stable. In fact, no plasmid-free bacteria were detected in batch cultures of the respective species following cultivation for 200 generations in the absence of selective pressure (data not shown). When introduced into AHL-producing strains like S. liquefaciens MG1, P. aeruginosa PAO1, and P. aureofaciens ATCC 13985, both plasmids gave rise to bright-green fluorescent colonies following cultivation for 24 h on LB plates (data not shown). Although minor variations in fluorescence intensity were observed, the plate assay clearly demonstrated that physiological concentrations of AHLs can be detected with these broad-host-range monitors. Since an equally important feature of AHL sensors is the capability of faithful detection of AHL signal molecules, the performances of pJBA130 and pJBA132 were also investigated in AHL-deficient strains. In an identical plate assay, colonies of E. coli MT102, S. liquefaciens MG44 (swrI), and P. aeruginosa JP2 (lasI rhlI), all harboring pJBA132, were observed to be completely nonfluorescent, whereas colonies of the same strains harboring pJBA130 acquired a weak green fluorescent phenotype. As observed in the previous section, reliable AHL detection can apparently be achieved only with fusions between the quorum sensor luxR-PluxI and the unstable GFP(ASV) variant. Consequently, further analysis was primarily performed using pJBA132-based monitor strains.
In order to estimate the response time of pJBA132-based sensor strains, we performed an AHL upshift experiment, in which different amounts of OHHL were added to early-mid-log-phase subcultures. Quantitative measurements of green fluorescence revealed that the detection limit for E. coli strain MT102 harboring pJBA132 was in the range of 1 to 3 nM OHHL (Fig. 3A). In the presence of 1 nM OHHL, accumulation of detectable amounts of GFP(ASV) could apparently not be achieved, as the measured green fluorescence never exceeded the background fluorescence. However, when the sensor strain was incubated in the presence of 3 nM OHHL a dramatic increase in measured green fluorescence was observed. The green fluorescence measured from the AHL sensor in response to either 10 nM OHHL or 100 nM OHHL was only slightly higher than the green fluorescence measured in response to 5 nM OHHL, indicating that maximal induction of the AHL sensor was achieved with approximately 10 nM OHHL. In a similar way, we found that the sensor was approximately 10-fold less sensitive to HHL, OHL, and OdDHL and at least 500-fold less sensitive to BHL (Fig. 3C). Epifluorescence microscopy revealed that cells cultivated in the absence of OHHL remained completely nonfluorescent throughout the entire experiment. Likewise, no green fluorescent single cells of the AHL sensor strain could be detected in the culture containing 1 nM OHHL. However, in response to the addition of 3 nM OHHL, single cells of the AHL sensor strain were observed to change from nonfluorescent to bright-green fluorescent within 15 to 30 min (data not shown).
Visual response time.
In nutrient downshift experiments, E. coli cells expressing GFP(ASV) were previously observed to lose green fluorescence within a few hours (1), indicating that sensors based on this GFP (ASV) protein should possess the potential of measuring fluctuations in AHL concentration. In order to determine the response time of the sensor to OHHL down- and upshifts, MT102 carrying either pJBA132 or pJBA130 was grown in LB in the presence of 10 nM OHHL. Following one wash and a shift to LB medium without OHHL, the specific GFP(ASV) signal expressed as fluorescence per OD450 (cell density) decreased rapidly, with a half-life of approximately 20 min (Fig. 4A). Decay of the fluorescent signal reflects protein turnover as well as dilution by growth. The GFPmut3∗ signal decreased with a half-life of approximately 40 min. Since the doubling time of the culture is approximately 40 min, this indicates that the sensor within a 30-min period after the shift becomes completely silent. This 30-min delay in response was also observed with the GFP(ASV) version and is likely a combination of turnover of activated LuxR, residual translation of mRNA encoding the GFP variants, and ongoing maturation of newly synthesized GFP polypeptides into a fluorescent conformation. Cellular OHHL is freely diffusible over the E. coli membranes and is expected to be washed out of the cells within a few minutes (21). Within 3 h after the downshift, the specific fluorescent signal intensity of the culture containing the GFP(ASV) version was reduced approximately 100-fold compared to 10-fold for the stable GFP version. This suggests that the activity of the ClpP protease during the 180 min of the downshift period accounted for a roughly 10-fold reduction and that another 10-fold reduction was due to dilution caused by bacterial growth. Upon addition of 10 nM OHHL, the signal doubled every 8 min and reached the signal intensity of the GFPmut3∗ sensor within 2 h.
In fact, green fluorescent cells could easily be detected by an epifluorescence microscope as early as 15 min after the addition of OHHL. Under the microscope, the cells appeared fully induced after 30 min, indicating that the “visual response time” of the AHL sensor strains is in the range of 15 to 30 min (Fig. 4B). This demonstrated that the sensor is a fast and highly sensitive upshift responder but a relative slow downshift responder. In nature bacteria predominantly grow attached to surfaces in biofilm structures. Recently, it has been demonstrated that the level of synthesis of curli in classical laboratory E. coli K-12 strains is insufficient to enable adherence to inert surfaces (49), rendering strains like MT102 unable to establish a biofilm on a glass surface. So in order to test the visual response time of a GFP(ASV)-based AHL sensor strain growing attached to a surface, we equipped S. liquefaciens MG44 with pJBA132. The swrI mutant MG44 is AHL negative because the signal-generating part of the quorum sensor has been knocked out (10, 11). We were able to grow this Serratia AHL sensor as a thin biofilm on the glass surfaces of a flow cell. Inspection of the cells by means of scanning confocal laser microscopy (SCLM) allowed us to follow the response to exogenous OHHL online at the single-cell level. As can be seen in Fig. 5, the single cells appeared completely dark at time zero; however, when OHHL was fed into the flow chamber via the medium supply, the single cells turned bright-green fluorescent within 15 min. After that time, only minor increases in green fluorescence were obtained during the following 75 min. At 90 min, the medium supply was shifted back to an OHHL-free stock. As seen in Fig. 5, the signal intensity dropped to the level before induction during the following 4 h. Since, no plasmid-free cells of MG44 were ever detected from the effluent of the flow cell, these observations clearly demonstrate that GFP(ASV) is also turned over in surface-attached cells. Like the E. coli sensor, the Serratia sensor was also a fast and highly sensitive upshift responder but a relatively slow downshift responder. Taken together, these up- and downshift experiments clearly demonstrated that AHL sensor strains based on pJBA132 are capable of responding to fluctuations in AHL concentrations and allow online studies of AHL-mediated communication at the single-cell level.
FIG. 5.
Glass surface-attached biofilm of the AHL sensor S. liquefaciens MG44 harboring pJBA132 in a flow cell. At 0 min, 5 nM OHHL was added to the medium flow. At 90 min, the chamber was shifted to medium without OHHL (−OHHL). (Upper panels) Phase-contrast images; (lower panels) epifluorescent images.
Interspecies communication.
AHL-mediated interspecies communication can be monitored online in a binary swarming colony. In S. liquefaciens the ability to move over semisoft surfaces by means of swarming motility is controlled by two major regulatory systems encoded by flhDC and swrI (16). The products of flhDC control swarm cell differentiation, whereas the putative swrI- and swrR-encoded quorum sensor controls production of the biosurfactant serrawettin W2 (27). These systems can be disconnected in individual cells by means of mutations in the key regulatory genes (11). In a swrI mutant, the signal generation has been knocked out and the strain is therefore unable to synthesize the surfactant W2 (27). The nonswarming S. ficaria does not produce a biosurfactant, but it produces OHHL signal molecules. A swarming culture can therefore be formed from a mixture of S. ficaria and S. liquefaciens MG44 (swrI). When the MG44 strain harbors pJBA132, signal molecules originating from S. ficaria could be monitored in situ by combined phase-contrast and epifluorescence microscopy (Fig. 6). The appearance of bright-green swrI cells harboring the AHL sensor system is indicative of interspecies communication (Fig. 6). This indicates that AHL signals originating from the AHL producers triggered surfactant synthesis in the population of swrI cells. The production of serrawettin creates a liquid interface layer in which the flow caused by the vigorous movement of the rafted swrI cells distributes both types of cells to the periphery of the expanding colony. Similarly, swarming colonies have been formed among more distantly related species such as P. aeruginosa PAO1 and the swrI mutant (11).
DISCUSSION
Communication systems based on AHLs have been described in a number of pathogenic gram-negative bacteria causing such diverse problems as food poisoning and infection of the human lungs. In most known cases these quorum-sensing systems control expression of virulence factors and hydrolytic enzymes (12, 44). In addition, there is growing evidence that the ability to form surface-associated, structured, and cooperative consortia (referred to as biofilms) may also be quorum sensing controlled (7, 8, 11). These properties all play a significant role in bacterial pathogenesis and are a common cause of persistent infections. In favor of this is the finding that communication-deficient strains are less pathogenic. This is true for Erwinia caratovora and Erwinia stewartii causing soft rot (34), as well as for the opportunistic human pathogen P. aeruginosa, which infects the lungs of cystic fibrosis patients (45). The latter communication-deficient variant has a decreased ability to bind host epithelial cells and is significantly impaired in virulence when tested in a mouse infection model. Furthermore, quorum-sensing systems have been identified in some of the most important fish-pathogenic bacteria, Vibrio anguillarum and Aeromonas salmonicida (31, 42). The ability to monitor bacterial communication at the single-cell level in situ enables investigation of cell-to-cell communication, intercellular signal transduction, and quorum sensing, and it gives us a tremendous potential for studying the efficiency of molecules that inhibit communication systems in biofilm systems and in other complex scenarios. The present highly sensitive GFP-based AHL monitor is a prototype of such a system. The sensitivity of the sensor was clearly influenced by plasmid copy number, promoter strength, translational signals, and turnover of the GFP reporter. All these factors have been optimized in the present constructs to match the sensitivity of current epifluorescent microscopes and SCLM, which then in turn allow for visual inspection of AHL-based cell-cell communication at the single-cell level.
A sensor in which the fluorescent signal intensity decays faster would have been preferred. Such constructs have been achieved by fusing more unstable versions of gfp to the quorum sensor, but unfortunately they turned out to be less sensitive. The reason is that a much higher synthesis rate is required to accumulate visualizable amounts of GFP, or, alternatively, detection would require more-sensitive microscopes. The present system [based on gfp(ASV)] fits the sensitivity of the state-of-the-art epimicroscopes and was designed to respond rapidly to increases in AHL concentrations in the lower nanomolar range. The use of broad-host-range, stabilized vectors expanded the system's application capability and proved to be useful for online and in vivo studies. The system was found to work well in P. aeruginosa PAO1 and its signal-generating null mutants, although the system was approximately 10-fold less sensitive to OHHL (data not shown). We have been able to detect communication among several gram-negative bacteria such as S. liquefaciens, S. ficaria, P. aeruginosa, P. aureofaciens, and E. coli. The system was recently used successfully in the lungs of animals (51). We were able to demonstrate that the infecting P. aeruginosa produced extracellular AHL molecules throughout a 3-day period. The broad-host-range, stabilized pJB130 and pJB132 constructs used in this study were perfectly stable without antibiotic selection. The utilization of the AHL sensor in biofilms is appealing because there are indications that AHL communication signals play a role in structuring and developing microbial communities in biofilms. For P. aeruginosa it has been demonstrated that the ability to form biofilms in flowthrough continuous-culture vessels is affected by the las but not the rhl quorum-sensing system (8), and it is speculated that the process of lung infection in cystic fibrosis patients may result in part from quorum-sensing control of virulence factors (33, 48). This supports the view of Tang et al. (45) that quorum sensing plays a critical role in the virulence of the organism. In addition, Telford et al. (46) demonstrated that the bacterial signal molecules not only function to regulate bacterial virulence gene expression through cell-cell communication but also, by virtue of immunomodulatory properties, may be a virulence determinant per se. It has also been demonstrated that P. aeruginosa biofilms on indwelling urethal catheters produce quorum-sensing signal molecules not only in vitro but also in situ (40). Food spoilage and expression of virulence factors by food-borne pathogens are microbial processes that are likely to be controlled by quorum-sensing mechanisms. Understanding of the role of quorum sensing-controlled gene expression in food spoilage bacteria is limited, and tools that enable in vivo studies of cell communication will expand our knowledge and understanding. It has been shown that AHL production is a common phenomenon in some types of gram-negative bacteria (Enterobacteriaceae) associated with food spoilage (18). Several gram-negative food-borne pathogenic bacteria (Aeromonas hydrophila, Vibrio parahaemolyticus) regulate virulence factors or other phenotypic behaviors by AHLs (29, 42, 44).
The multitude of bacteria that regulate expression of virulence and colonization-relevant traits by means of quorum-sensing systems suggests that efficient inhibitors of quorum sensors will have tremendous potential. Furanone compounds produced by the red algae Delisea pulchra are examples of naturally occurring inhibitors of quorum-sensing-controlled process (15, 24, 28). In the process of design and further modification of inhibitors, we might select compounds that function well in planktonic cells but less efficiently on the quorum sensors present in biofilm cells. For that reason analysis of the efficiency of quorum-sensing inhibitor compounds must be extended to include biofilm structures and in vivo studies. By means of the present sensor, the in vivo functionality of inhibitors can be addressed as expression of green fluorescence. Use of the GFP-based single-cell technology in combination with confocal microscopy will enable us to determine the penetration and half-life of inhibitory compounds in biofilm model systems and to assess the functionality of such compounds in animal model systems. Work on this is currently in progress in our laboratory.
ACKNOWLEDGMENTS
This work was supported in part by grants from the Danish Medical Research Council (to M.G.) and European Union (to M.G. and S.M.) and from the Deutche Forschungsgemeinschaft EB 2051/1-1 (to L.E.).
We thank Paul Williams and Gordon S. A. B. Stewart for supplying plasmid pSB403, Barbara H. Iglewski for supplying P. aeruginosa PAO1 and JP2, Rocky de Nys and Simon Swift for supplying different synthetic AHL molecules, and Linda Stabell for excellent technical assistance.
REFERENCES
- 1.Andersen J B, Sternberg C, Poulsen L K, Bjørn S P, Givskov M, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 4.Christensen B B, Sternberg C, Andersen J B, Eberl L, Møller S, Givskov M, Molin S. Establishment of new genetic traits in a microbial biofilm community. Appl Environ Microbiol. 1998;64:2247–2255. doi: 10.1128/aem.64.6.2247-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark D J, Maaløe O. DNA replication and the division cycle in Escherichia coli. J Mol Biol. 1967;23:99–112. [Google Scholar]
- 6.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 7.Costerton J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 8.Davies D, Parsek M, Pearson J, Iglewski B, Costerton J, Greenberg E. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 9.Devine J H, Countryman C, Baldwin T O. Nucleotide sequence of the luxR and luxI genes and structure of the primary regulatory region of the lux regulon of Vibrio fischeri ATCC 7744. J Biochem. 1988;27:837–842. [Google Scholar]
- 10.Eberl L, Winson M K, Sternberg C, Stewart G S, Christiansen G, Chhabra S R, Bycroft B, Williams P, Molin S, Givskov M. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol Microbiol. 1996;20:127–136. doi: 10.1111/j.1365-2958.1996.tb02495.x. [DOI] [PubMed] [Google Scholar]
- 11.Eberl L, Molin S, Givskov M. Surface motility of Serratia liquefaciens MG1. J Bacteriol. 1999;181:1703–1712. doi: 10.1128/jb.181.6.1703-1712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberl L. N-Acyl homoserinelactone-mediated gene regulation in Gram-negative bacteria. Syst Appl Microbiol. 1999;22:493–506. doi: 10.1016/S0723-2020(99)80001-0. [DOI] [PubMed] [Google Scholar]
- 13.Fuqua C, Winans S, Greenberg P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 14.Givskov M, Olsen L, Molin S. Cloning and expression in Escherichia coli of the gene for extracellular phospholipase A1 from Serratia liquefaciens. J Bacteriol. 1988;170:5855–5862. doi: 10.1128/jb.170.12.5855-5862.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Givskov M, de Nys R, Manefield M, Gram L, Maximilien R, Eberl L, Molin S, Steinberg P D, Kjelleberg S. Eukaryotic interference with homoserine lactone-mediated procaryotic signalling. J Bacteriol. 1996;178:6618–6622. doi: 10.1128/jb.178.22.6618-6622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Givskov M, Östling J, Lindum P W, Eberl L, Christiansen G, Molin S, Kjelleberg S. The participation of two separate regulatory systems in controlling swarming motility of Serratia liquefaciens. J Bacteriol. 1998;180:742–745. doi: 10.1128/jb.180.3.742-745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman S, Roche E, Zhou Y N, Sauer R T. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gram L, Christensen A B, Ravn L, Molin S, Givskov M. Production of acylated homoserine lactones by psychrotrophic members of the Enterobacteriaceae isolated from foods. Appl Environ Microbiol. 1999;65:3458–3463. doi: 10.1128/aem.65.8.3458-3463.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray K M, Passador L, Iglewski B H, Greenberg E P. Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa. J Bacteriol. 1994;176:3076–3080. doi: 10.1128/jb.176.10.3076-3080.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, Walsh U, O'Gara F, Haas D. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant-Microbe Interact. 2000;13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan H, Greenberg E. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol. 1985;163:1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keiler K C, Waller P R H, Sauer R T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 23.Keiler K C, Sauer R T. Sequence determinants of C-terminal substrate recognition by the Tsp protease. J Biol Chem. 1996;271:2589–2593. doi: 10.1074/jbc.271.5.2589. [DOI] [PubMed] [Google Scholar]
- 24.Kjelleberg S, Steinberg P D, Givskov M, Gram L, Manefield M, De Nys R. Do marine products interfere with procaryotic AHL regulatory systems? Rev Aquat Microbiol Ecol. 1997;13:85–93. [Google Scholar]
- 25.Laville J, Blumer C, von Schroetter C, Gaia V, Defago G, Keel C, Haas D. Characterization of the hncABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J Bacteriol. 1998;180:3187–3196. doi: 10.1128/jb.180.12.3187-3196.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewenza S, Conway B, Greenberg E P, Sokol P A. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J Bacteriol. 1999;181:748–756. doi: 10.1128/jb.181.3.748-756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindum P W, Anthoni U, Christophersen C, Eberl L, Molin S, Givskov M. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J Bacteriol. 1998;180:6384–6388. doi: 10.1128/jb.180.23.6384-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manefield M, de Nys R, Kumar N, Read R, Givskov M, Steinberg P, Kjelleberg S. Evidence that halogenated furanones from Delisea pulchra inhibit AHL-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology. 1999;145:283–291. doi: 10.1099/13500872-145-2-283. [DOI] [PubMed] [Google Scholar]
- 29.McCarter L L. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity in Vibrio parahaemolyticus. J Bacteriol. 1998;180:3166–3173. doi: 10.1128/jb.180.12.3166-3173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McClean K H, Winson M K, Fish L, Taylor A, Chhabra S R, Camara M, Daykin M, Lamb J H, Swift S, Bycroft B W, Stewart G S A B, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 31.Milton D L, Hardman A, Camara M, Chhabra S R, Bycroft B W, Stewart G S A B, Williams P. Quorum sensing in Vibrio anguillarum: characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-l-homoserine lactone. J Bacteriol. 1997;179:3004–3012. doi: 10.1128/jb.179.9.3004-3012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Møller S, Sternberg C, Andersen J B, Christensen B B, Ramos J L, Givskov M, Molin S. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl Environ Microbiol. 1998;64:721–732. doi: 10.1128/aem.64.2.721-732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 34.Pirhonen M, Flego D, Heikinheimo R, Palva E T. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaefer A L, Hanzelka B L, Eberhard A, Greenberg E P. Quorum sensing in Vibrio fischeri: probing autoinducer-LuxR interactions with autoinducer analogs. J Bacteriol. 1996;178:2897–2901. doi: 10.1128/jb.178.10.2897-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw P, Ping G, Daly S, Cha C, Cronan J J, Rinehart K, Farrand S. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silhavy T J, et al. Experiments with gene fusions, p. xi–xii. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 38.Sitnikov D M, Schineller J B, Baldwin T O. Transcriptional regulation of bioluminescence genes from Vibrio fischeri. Mol Microbiol. 1995;17:801–812. doi: 10.1111/j.1365-2958.1995.mmi_17050801.x. [DOI] [PubMed] [Google Scholar]
- 39.Sternberg C, Christensen B B, Johansen T, Nielsen A T, Andersen J B, Givskov M, Molin S. Distribution of bacterial growth activity in flow-chamber biofilms. Appl Environ Microbiol. 1999;65:4108–4117. doi: 10.1128/aem.65.9.4108-4117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stickler D J, Morris N S, McLean R J C, Fuqua C. Biofilms on indwelling urethral catheters produce quorum sensing signal molecules in situ and in vitro. Appl Environ Microbiol. 1998;64:3486–3490. doi: 10.1128/aem.64.9.3486-3490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swift S, Winson M K, Chan P F, Bainton N J, Birdsall M, Reeves P J, Rees C E D, Chhabra S R, Hill P H, Throup J P, Bycroft B W, Salmond G P C, Williams P, Stewart G S A B. A novel strategy for the isolation of luxI homologues: evidence for the widespread distribution of a LuxR:LuxI superfamily in enteric bacteria. Mol Microbiol. 1993;10:511–520. doi: 10.1111/j.1365-2958.1993.tb00923.x. [DOI] [PubMed] [Google Scholar]
- 42.Swift S, Karlyshev A, Fish L, Durant E, Winson M, Chhabra S, Williams P, Macintyre S, Stewart G. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J Bacteriol. 1997;179:5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swift S, Williams P, Stewart G S A B. N-Acylhomoserine lactones and quorum sensing in proteobacteria. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: American Society for Microbiology; 1999. pp. 297–313. [Google Scholar]
- 44.Swift S, Lynch M J, Fish L, Kirke D F, Tomas J M, Stewart G S A B, Williams P. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophilia. Infect Immun. 1999;67:5192–5199. doi: 10.1128/iai.67.10.5192-5199.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang H B, DiMango D, Bryan R, Gambello M, Iglewski B H, Goldberg J B, Prince A. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect Immun. 1996;64:37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Telford G, Wheeler D, Williams P, Tomkins P T, Appleby P, Sewell H, Stewart G S A B, Bycroft B W, Pritchard D I. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-l-homoserine lactone has immunomodulatory activity. Infect Immun. 1998;66:36–42. doi: 10.1128/iai.66.1.36-42.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tombolini R, Unge A, Davey M E, de Bruijn F J, Jansson J K. Flow cytometric and microscopic analysis of GFP-tagged Pseudomonas fluorescens bacteria. FEMS Microbiol Ecol. 1997;22:17–28. [Google Scholar]
- 48.Van Delden C, Iglewski B. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winson M K, Swift S, Fish L, Throup J P, Jørgensen F, Chhabra S R, Bycroft B W, Williams P, Stewart G S A B. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol Lett. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- 51.Wu H, Song Z, Hentzer M, Andersen J B, Heydorn A, Mathee K, Moser C, Eberl L, Molin S, Høiby N, Givskov M. Detection of N-acyl-homoserine lactones in lung tissue of mice infected with Pseudomonas aeruginosa. Microbiology. 2000;146:2481–2493. doi: 10.1099/00221287-146-10-2481. [DOI] [PubMed] [Google Scholar]
- 52.Yanisch-Perron C, Vieira J, Messing J. Improved M13 cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]