Abstract
Sotos syndrome (SS), the most common overgrowth with intellectual disability (OGID) disorder, is caused by inactivating germline mutations of NSD1, which encodes a histone H3 lysine 36 methyltransferase. To understand how NSD1 inactivation deregulates transcription and DNA methylation (DNAm), and to explore how these abnormalities affect human development, we profiled transcription and DNAm in SS patients and healthy control individuals. We identified a transcriptional signature that distinguishes individuals with SS from controls and was also deregulated in NSD1-mutated cancers. Most abnormally expressed genes displayed reduced expression in SS; these downregulated genes consisted mostly of bivalent genes and were enriched for regulators of development and neural synapse function. DNA hypomethylation was strongly enriched within promoters of transcriptionally deregulated genes: overexpressed genes displayed hypomethylation at their transcription start sites while underexpressed genes featured hypomethylation at polycomb binding sites within their promoter CpG island shores. SS patients featured accelerated molecular aging at the levels of both transcription and DNAm. Overall, these findings indicate that NSD1-deposited H3K36 methylation regulates transcription by directing promoter DNA methylation, partially by repressing polycomb repressive complex 2 (PRC2) activity. These findings could explain the phenotypic similarity of SS to OGID disorders that are caused by mutations in PRC2 complex-encoding genes.
Introduction
Over the last decade, high-throughput sequencing studies have identified the genes that are causally mutated in many congenital disorders. This research has revealed that many congenital growth and neurodevelopmental disorders are caused by germline mutations in genes that encode epigenetic modifying enzymes, i.e. enzymes that ‘read,’ ‘write’ and ‘erase’ epigenetic modifications (1–4). These epigenetic modifications include DNA methylation (DNAm) (methyl groups added to cytosines followed by guanines; cytosine–phosphate–guanine, CpGs) and histone modifications. Epigenetic modifications function interactively to regulate processes such as transcription, primarily by regulating the transcriptional machinery’s access to DNA. Mutations in epigenetic modifying enzymes are presumed to cause congenital disorders by altering the deposition of their target modifications, resulting in altered transcription of genes that regulate growth and other affected phenotypes. However, the mechanisms that are altered by genetic mutations and affect disease phenotypes are not fully understood for any Mendelian disorder of the epigenetic machinery.
Sotos syndrome (SS) is an autosomal dominant neurodevelopmental and growth disorder that is caused by intragenic mutations or whole-gene deletions of NSD1 (5,6). Clinically, SS is primarily characterized by generalized overgrowth including tall stature and macrocephaly, global developmental delay often culminating in intellectual disability, distinct facial features, as well supranuclear hypotonia (7). SS is diagnosed in approximately one in 10 000 births and is the most common of the ‘overgrowth with intellectual disability (OGID) disorders,’ a class of congenital disorders that are defined by concurrent developmental overgrowth and intellectual disability (8). NSD1 appears to play a general role in the regulation of physiological growth because while inactivating NSD1 mutations cause overgrowth, rare germline NSD1 amplifications cause a dwarfism syndrome known as ‘reverse SS’ (9,10). NSD1 is deregulated in multiple cancer types (11–14) suggesting that its inactivation may drive cancer-related growth.
NSD1 encodes a histone lysine methyltransferase, i.e. an enzyme that catalyzes histone methylation (15). NSD1 deposits mono and di-methylation of histone 3 at lysine 36 (H3K36me1/2) (12,15,16). NSD1 mutations are understood to cause SS due to impairments of NSD1-catalyzed histone methylation, as mutations often occur within the gene region encoding the methyltransferase (SET) protein domain (5). While the function of H3K36me1 remains poorly understood, H3K36me2 is generally associated with active transcription and has been shown to prevent transcriptional silencing by polycomb repressive complex 2 (PRC2) in experimental systems (17,18).
H3K36me2 also regulates DNAm, an epigenetic modification that regulates transcription (19,20). Consequently, SS is associated with a genome-wide signature of abnormal DNAm that is primarily characterized by loss of DNAm (21). Moreover, we and others have previously reported that in squamous cell carcinomas of the head and neck (HNSCC) and of the lung (LUSCC), somatic NSD1 mutations are associated with signatures of abnormal DNAm that are similar to that observed in SS, and are associated with widespread transcriptional deregulation (12,14). These observations suggest that NSD1 regulates transcription by directing DNAm; however, the mechanism through which this occurs is unknown.
Here, we have investigated transcriptional and DNAm deregulation in SS, in order to characterize patterns of transcriptional deregulation that are caused by NSD1 inactivation and to gain insights into the mechanism through which NSD1 regulates transcription.
Results
Identification of transcriptionally deregulated genes
We profiled transcriptional deregulation in SS by applying RNA-sequencing (RNA-Seq) to primary whole blood samples of SS patients (i.e. probands, n = 10), and matched healthy control subjects (n = 15) (Table 1).
Table 1.
Study participants who were profiled using RNA-Seq and DNA methylation arrays
| Barcode | Case control | Participant type | Proband that control is matched to | Family | Study phase | Age | NSD1 lesion | Clinical significancea | Sex | Race | DNA methylation array performed |
|---|---|---|---|---|---|---|---|---|---|---|
| 3425-F | Control | Father | 3425-P | 3425 | Discovery | 57.3 | _ | M | Asian | Pooled (parental control) sample |
| 3425-P | Case | Proband | _ | 3425 | Discovery | 20.9 | NM_022455.5(NSD1):c.6049C > T (p.Arg2017Trp) | Pathogenic | M | Asian | Individual sample |
| M-19-23 | Control | Age- and sex-matched donor | 3425-P | NA | Discovery | 21 | _ | M | NA | Individual sample |
| 4681-F | Control | Father | 4681-P | 4681 | Discovery | _ | _ | M | White | _ |
| 4681-M | Control | Mother | 4681-P | 4681 | Discovery | _ | _ | F | White | _ |
| 4681-P | Case | Proband | _ | 4681 | Discovery | 3.3 | _ | M | White | _ |
| 4834-F | Control | Father | 4834-P | 4834 | Discovery | 39.6 | _ | M | White | _ |
| 4834-M | Control | Mother | 4834-P | 4834 | Discovery | 35.1 | _ | F | White | _ |
| 4834-P | Case | Proband | _ | 4834 | Discovery | 1.9 | Whole gene deletion involving exon 1-exon 24, at least 474.6 kb deletion | M | White | _ |
| 5767-F | Control | Father | 5767-P | 5767 | Discovery | 53.9 | _ | M | Asian | Pooled (parental control) sample |
| 5767-P | Case | Proband | _ | 5767 | Discovery | 17.8 | NM_022455.5(NSD1):c.6049C > T (p.Arg2017Trp) | Pathogenic | F | Asian | Individual sample |
| F-16-20 | Control | Age- and sex-matched donor | 5767-P | NA | Discovery | 18.5 | _ | F | NA | Individual sample |
| 8658-F | Control | Father | 8658-P | 8658 | Discovery | 65.7 | _ | M | White | Pooled (parental control) sample |
| z | Case | Proband | _ | 8658 | Discovery | 27 | _ | M | White | Individual sample |
| M-24-28 | Control | Age- and sex-matched donor | 8658-P | NA | Discovery | 25 | _ | M | NA | Individual sample |
| 3077-P | Case | Proband | _ | 3077 | Validation | 1.5 | NM_022455.5(NSD1):c.3549dup (p.Glu1184Ter) | Pathogenic | F | White | _ |
| 3077-S | Control | Age- and sex-matched sibling | 3077-P | 3077 | Validation | 4.4 | _ | F | White | _ |
| 5530-P | Case | Proband | _ | 5530 | Validation | 28.1 | NM_022455.5(NSD1):c.6014G > A (p.Arg2005Gln) | Pathogenic | F | White | _ |
| 3077-M | Control | Mother and age- and sex-matched control | 3077-P (Mother) and 5530-P (Age/sex/race matched unrelated control) | 3077 | Validation | 32.5 | _ | F | White | _ |
| 5996-P | Case | Proband | _ | 5996 | Validation | 4.7 | NM_022455.5(NSD1):c.6014G > A (p.Arg2005Gln)| Pathogenic | M | White | _ |
| 5996-S | Control | Age- and sex-matched sibling | 5996-P | 5996 | Validation | 7.3 | _ | M | White | _ |
| 6424-P | Case | Proband | _ | 6424 | Validation | 16.8 | _ | F | White | _ |
| 6424-S | Control | Age- and sex-matched sibling | 6424-S | 6424 | Validation | 23.5 | _ | F | White | _ |
| 9049-P | Case | Proband | _ | 9049 | Validation | 37.8 | Intragenic deletion involving exon 5 (Identified by real-time quantitative PCR amplification of exon 5) | F | White | _ |
| 7634-M | Control | Age/sex/race-matched control | 9049-P | 7634 | Validation | 42.7 | _ | F | White | _ |
Clinical signficance, as indicated by ClinVar and the Genetic Testing Registry, is shown for point mutations.
Choufani et al. (21) reported that SS displays a ‘genome-wide’ signature of abnormal DNAm, which can be used to diagnose SS with perfect accuracy (21,22). To confirm the diagnosis of SS in probands and to confirm that they had abnormal DNAm, we measured DNAm of cg07600533, a CpG that distinguished SS patients from healthy control subjects, in all subject DNA samples using pyrosequencing (Supplementary Material, Table S1). We selected cg07600533 as a diagnostic CpG because cg07600533 methylation perfectly separated SS patients (N = 38) from healthy control subjects (N = 53) in data from the Choufani study (21) (Supplementary Material, Fig. S1A). Consistently, pyrosequencing-based methylation of cg07600533 clearly distinguished all SS cases from controls within samples that were collected as part of the current study (Supplementary Material, Fig. S1B). All SS cases in the study were considered to be ‘de novo’ cases, since neither parent nor any family member of the SS patient was diagnosed with SS. Cg07600533 pyrosequencing was applied to both parents of SS probands in cases where blood samples of both parents could be collected (n = 6 cases). ‘Normal’ cg07600533 methylation levels were observed in all parents, confirming that the SS patient’s NSD1 lesion was not inherited from either parent.
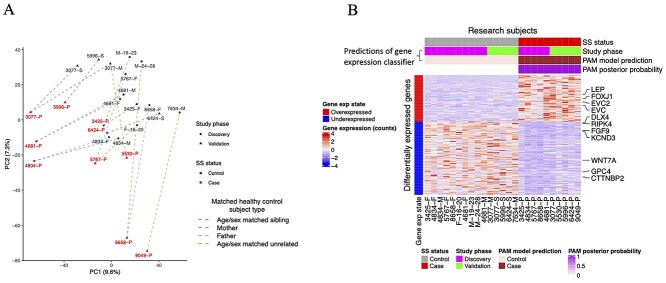
High RNA-Seq data quality was confirmed by the observation that all gene expression profiles were consistent with whole blood RNA-Seq data from the GTEx study (23) and displayed expression of sex chromosome marker genes that were consistent with the sex of the subject (Supplementary Material, Fig. S2). Principal component analysis (PCA) was applied to the top 10% of genes with the highest variation (mean absolute deviation) (N = 12 100 genes). This indicated clustering of SS cases and controls (Fig. 1A).
Figure 1.
Identification of abnormally expressed genes in SS. (A) PCA of transcriptome data of SS and healthy control subjects. PCA was applied to the top 10% of the most variable genes based on the mean absolute deviation. Lines connect SS patients to their matched healthy control subjects, with line colors indicating the relationship of the healthy control subjects to the SS patient. (B) Heatmap showing expression of all genes that were overexpressed (N = 72) and underexpressed (N = 113) in SS patients (case) relative to healthy control subjects (control), based on differentially expressed gene analysis applied the combined (discovery and validation) sample sets. Labels indicate genes with essential roles in developmental growth and neurodevelopment. Horizontal sidebars indicated by the bracket show gene expression classifier model predictions for classification of research subjects as either SS patients or healthy control subjects. This gene expression model was developed using PAM (24) applied to transcriptional data for genes shown in the heatmap. The PAM posterior probability indicates the probability of the subject being a case, as predicted by the classifier. The final predictions of the model are indicated by the ‘PAM model prediction’ horizontal sidebar. Abbreviations: SS = Sotos syndrome, exp = expression, PAM = Predictions Analysis of Microarrays.
We next applied differentially expressed gene (DEG) analysis to identify genes that are abnormally expressed in SS, defined as genes with an absolute log2-fold count difference of 1 or greater between SS cases and healthy control subjects (i.e. an expression fold change of 2 or greater), and with an false discovery rate (FDR)-corrected P-value of less than 0.05. We first identified genes that were differentially expressed between cases and controls in the discovery set (n = 77) and then investigated the differential expression of these genes within the validation set. The validation set represents an independent RNA-Seq experiment from the discovery set, as the validation set samples were collected and analyzed subsequent to the analysis of RNA-Seq data from the discovery set; however, the protocols for sample collection, processing, and analysis were consistent between the two study phases such that the validation set experiment represents a biological replicate of the discovery set experiment. 41/77 (57%) of these genes were significantly differentially expressed in the validation sample set after adjustment for multiple correction testing, all of which had directions of differential expressions that were consistent between the discovery and validation sample sets (23 overexpressed, 18 underexpressed) (Supplementary Material, Table S2). Log2-fold changes for expression differences between cases and controls were highly correlated between the discovery and validation sets, indicating reproducibility of differential expression between study phases (Supplementary Material, Fig. S3). In order to improve statistical power to identify DEGs, we next applied DEG analysis to the combined discovery and validation experiment datasets, including the study phase as a covariate within DEG models. This revealed 72 genes that were overexpressed, and 113 genes that were underexpressed in SS (Fig. 1B) (Supplementary Material, Figs S4 and S5) (Supplementary Material, Table S3). Notably, NSD1 was not differentially expressed between SS cases controls (Supplementary Material, Fig. S4C). This is not surprising since many SS cases (including four of seven cases for which NSD1 lesion data were available with the current study) are caused by missense variants that are predicted to perturb NSD1 methyltransferase activity rather than NSD1 expression. NSD1 was expressed at appreciable levels in all probands including 4834-P, an individual with a whole gene NSD1 deletion. This indicates that in patients with truncating mutations, NSD1 is expressed from the wild-type allele, which could partially compensate for NSD1 haploinsufficiency. Importantly, differential expression could not be accounted for by abnormalities of blood cell fractions in SS, as fractions of blood cell types were similar between cases and controls (Supplementary Material, Fig. S6) (see Supplementary results).
Gene expression-based diagnostic classification of SS
Multidimensional scaling was applied to transcriptional data of all (N = 185) deregulated genes, which indicated the separation of cases from controls (Supplementary Material, Fig. S7). To determine if the transcriptional abnormalities identified are sufficient to diagnose SS, we trained a gene expression classifier to distinguish SS cases from control subjects based on the expression of deregulated genes, using prediction analysis of microarrays (PAM) (24). We tested the accuracy of this model by applying it in combination with 10-fold cross-validation. The gene expression classifier accurately predicted the SS status of all subjects with high confidence (Fig. 1B). This indicates that SS could be diagnosed using the transcription signature, as can be done using DNAm data (21).
Downregulation of bivalent developmental genes
We next sought to functionally characterize the genes that were transcriptionally deregulated in SS. Downregulated genes included genes whose deregulation could account for phenotypes of SS, including genes that control skeletal development (WNT7A (25,26), FGF9 (27) and GPC4 (28)), neurodevelopment (CTTNBP2 (29), GPC4 (28), and KCND3 (30)) and craniofacial development (DLX4 (31), GPC4 (28), and RIPK4 (32)). Other underexpressed genes are causally implicated in phenotypes less frequently observed in SS (6), including autism spectrum disorder (CTTNBP2 (29), NEO1 (33)), congenital cardiac defects (HEY1 (34), DSC2 (35)), muscle hypotonia or weakness (COL6A1 (36)), ocular defects (NECTIN3 (37), GPC4 (28)) and hypodontia (GREM2 (38), TSPEAR (39)). Gene set enrichment analysis (GSEA) was applied to underexpressed genes and identified many enriched gene sets, of which the most significantly enriched was ‘ZHAN_MULTIPLE_MYELOMA_MS_UP,’ representing genes that were upregulated in a subtype of multiple myeloma that is defined by overexpression of NSD2 (also known as WHSC1) (40) (Supplementary Material, Table S6). This indicates overlap between genes that are positively regulated by NSD1 and its paralogue NSD2, presumably due to their shared H3K36me1/2 methyltransferase activity (41,42).
The second most significantly enriched gene set, as well as three additional enriched gene sets, consisted of genes that are regulated by PRC2, as determined by chromatin immunoprecipitation-sequencing (ChIP-Seq) (43,44). These gene sets include genes that were bound by PRC2 core component proteins SUZ12 and EED (43), as well as a set of bivalent genes. Bivalent genes represent a subset of PRC2 target genes that feature H3K27me3 (43,44), the PRC2-deposited repressive histone modification, in combination with the activating histone modification H3K4me3 at the same location within their promoters. Bivalent genes generally represent tissue-specific development genes that regulate cellular differentiation and include many neurodevelopmental genes (45). Network-based analysis of the enriched gene sets revealed that many of the enriched gene sets consisted of genes that encode components of the synaptic membrane as well as genes that are involved in embryogenesis (Supplementary Material, Fig. S8). Also enriched were genes that encode members of developmental signaling pathways, notably netrin signaling, which controls axon guidance (46), KRAS signaling, deregulation of which causes congenital growth disorders (47,48) and WNT signaling, which controls cellular differentiation (49).
Overexpressed genes included developmental genes whose overexpression could contribute to SS phenotypes, particularly EVC and EVC2, mutations within which cause short limb dwarfism and congenital heart defects (50), LEP, which regulates bone metabolism (51) and FOXJ1, which regulates nervous system development (52). Despite this, GSEA did not identify any gene set that significantly overlapped with overexpressed genes, most likely due to the relatively small number of overexpressed genes compared with underexpressed genes.
Confirming deregulation of bivalent genes
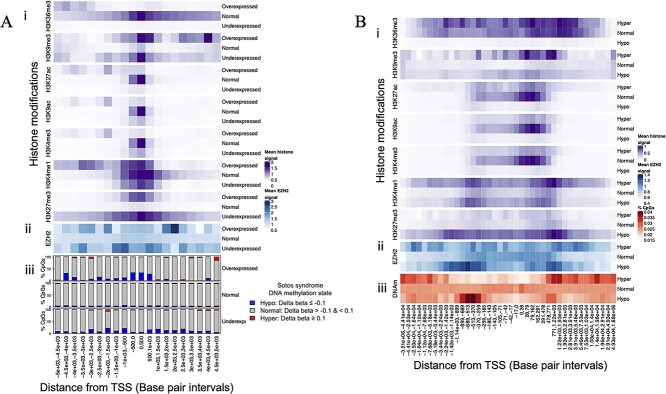
Underexpression of PRC2 target genes in SS could account for many of the phenotypes of SS, since germline mutations in genes encoding PRC2 core component enzymes cause OGID disorders that are highly similar to SS (53). To confirm that genes that are deregulated in SS contain an overrepresentation of PRC2 target genes, we analyzed reference epigenomes (54) to assess baseline chromatin profiles (Chromatin profiles in healthy NSD1 wild-type tissues) of genes that were abnormally expressed in SS. These included histone modification ‘tracks,’ which map genome-wide levels of histone modifications and were measured using ChIP-Seq applied to peripheral blood mononuclear cells (PBMCs) of healthy individuals. We also analyzed a consolidated ‘18 chromatin state’ map, which assigns each 500 bp genomic interval to one of the 18 distinct chromatin states based on its combination of histone modifications.
Relative to genes that were normally expressed in SS, genes that were underexpressed and overexpressed in SS featured higher levels of H3K27me3 as well as the repressive modification H3K9me3 within their promoter regions, in reference epigenomes (i.e. in blood of healthy individuals) (Fig. 2Ai). This indicates that genes that are transcriptionally deregulated in SS feature H3K27me3 bound promoters under normal physiological conditions, confirming that they represent PRC2-regulated genes as suggested by GSEA. Promoters of abnormally expressed genes featured lower levels of all other histone modifications in reference epigenomes; however, underexpressed genes featured appreciable H3K4me3 levels at their transcription start sites (TSSs), indicating that they have bivalent promoters. Indeed, analysis of consolidated chromatin states confirmed that bivalent chromatin was strongly enriched within the promoters of underexpressed genes relative to normally expressed genes (Supplementary Material, Fig. S9Ai). Sixty-two percent of underexpressed genes featured bivalent chromatin within their promoters, compared with 9% of normally expressed genes (Supplementary Material, Fig. S9Aii). Half of the remaining non-bivalent underexpressed genes featured non-bivalent polycomb-associated chromatin states within their promoters, indicating that NSD1 primarily promotes transcription of PRC2 target genes. Moreover, underexpressed genes were overrepresented for protein-coding genes and had a higher density of CpG sites within their promoters relative to genes that were normally expressed in SS; characteristics that are associated with bivalent genes (55) (Supplementary Material, Tables S4 and S5). Genes that were overexpressed in SS were also overrepresented for bivalent genes, albeit to a lesser extent than underexpressed genes, indicating that NSD1 regulates bivalent genes both positively and negatively.
Figure 2.
Profiles of baseline histone modifications and EZH2 occupancy at genomic regions that feature promoters of abnormally expressed genes and regions of abnormal DNA methylation in SS. Heatmaps show mean baseline levels (i.e. ChIP-Seq signal) of histone modifications and EZH2 occupancy within genomic regions that feature: (A) promoters of genes that display different gene expression states in SS; and (B) CpGs that display different methylation states in SS. ‘Baseline’ EZH2 occupancy and histone modifications levels refer to the levels of these variables in peripheral blood mononuclear cells of healthy individuals, as indicated by reference ChIP-Seq datasets that were generated as part of the ENCODE (106) and Epigenomics Roadmap (109) projects, respectively. (A) The heatmap shows (i) mean histone modification and (ii) EZH2 occupancy levels within 500 bp intervals (i.e. genomic windows) surrounding the TSSs of genes that were overexpressed, normally expressed (normal) and underexpressed in SS. EZH2 and histone modification levels are shown for TSS distance intervals within regions that span from 5 kb upstream to 5 kb downstream of TSSs. (iii) Shown for reference are bar plots illustrating the proportion of CpGs that were hypomethylated, hypermethylated and hypermethylated in SS, within each TSS distance interval. These bar plots are equivalent to those shown in Figure 4 (ii), where they are described in additional detail. In (B), the heatmap shows levels of baseline (i) histone modifications and (ii) EZH2 occupancy at the loci of CpGs that were hypomethylated, hypermethylated and normally methylated in SS. CpGs are split into groups (TSS distance intervals) along the horizontal axis based on their distance from the closest TSS (of any gene). This illustrates levels of EZH2 occupancy and histone modifications within TSS distance intervals that feature enrichment of abnormal DNA methylation (i.e. overrepresentation of hypermethylated and hypomethylated CpGs). (iii) Shown for reference is a heatmap that indicates the percentages of hypermethylated, hypomethylated and normally methylated CpGs that occurred within each TSS distance interval. Darker red regions indicate distances from TSSs at which CpGs were disproportionately hypermethylated or hypomethylated in SS. TSS distance intervals were generated by calculating the distance of each CpG to its closest TSS and then splitting CpGs into forty groups based on this distance. Interval boundaries were defined by frequency, such that each interval includes an approximately equal number of CpGs. This heatmap and the distribution of abnormal DNAm in SS are described in additional detail in Figure 3.
To determine if abnormally expressed genes are normally occupied by PRC2, we examined the levels of baseline EZH2 ChIP-Seq signal (i.e. EZH2 occupancy or binding) within the region surrounding the promoters of DEGs, using an EZH2 binding reference dataset that was generated by ENCODE (56). This indicated that relative to genes that were normally expressed in SS, genes that were underexpressed featured higher levels of EZH2 occupancy within their promoters, particularly within regions flanking their TSS, in the blood of healthy individuals (Fig. 2Aii). Taken together, these findings indicate that NSD1 inactivation primarily deregulates PRC2 target genes, particularly downregulating bivalent genes.
DNA hypomethylation of promoter CpG island shores and hypermethylation of intergenic regions
Choufani et al. reported that SS displays a ‘genome-wide’ signature of abnormal DNAm (21). Here, we profiled abnormal DNAm using the Illumina 850 K DNAm array (57) in order to investigate the role of DNAm in mediating transcriptional regulation by NSD1. We profiled genome-wide DNAm in the blood of three SS case subjects (n = 3), and age- and sex-matched healthy control subjects (n = 3). We also profiled a pooled blood DNA sample of the parents of the three probands (n = 5). We generated this dataset (hereafter referred to as the ‘Stanford’ dataset) in order to investigate the genomic distribution of abnormal DNAm in SS, since the 850 K array yields greater and more balanced genome coverage than the 450 k array that was used to generate the preexisting Choufani dataset (21). Delta beta values (i.e. the mean methylation beta value difference between cases and controls) of CpGs were highly consistent between the Stanford dataset and the larger Choufani dataset for CpGs that were included in both datasets (Supplementary Material, Fig. S10). This confirmed that despite its small sample size, the Stanford dataset provides a reliable estimate of abnormal DNAm in SS.
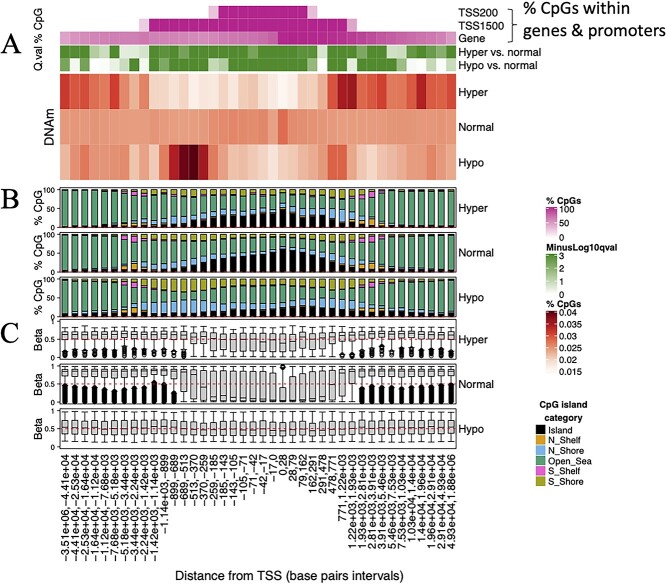
Differential methylation analysis identified many abnormally methylated CpGs, defined as CpGs with an absolute delta beta of 0.1 or greater; this approximately equates to a 10% difference in methylation between cases and controls. There was a strong bias toward hypomethylation; overall, 5% of CpGs (n = 41 660) were hypomethylated, whereas 0.016% of CpGs (n = 12 128) were hypermethylated. We next analyzed the distribution of abnormally methylated CpGs with respect to TSSs. Hypomethylated CpGs were strongly overrepresented within the regions flanking TSSs, particularly at 5’ TSS flanking regions where there was a clear spike of DNA hypomethylation (Fig. 3A) (Supplementary Material, Table S8). Hypomethylated CpGs within these regions were overrepresented for CpGs that are within promoter CGI shores, regions of intermediate CpG density at the borders of CGIs that exhibit high DNAm variability and are often associated with transcription (20) (Fig. 3B).
Figure 3.
Characterizing the genomic distribution of abnormal DNA methylation in SS. (A) Heatmap representation of the genomic distribution of abnormal DNAm in SS. The heatmap shows the distribution of CpGs in relation to the TSSs, with CpGs stratified into those that were hypermethylated (Hyper), hypomethylated (Hypo) and normally methylated (Normal) in the blood of SS patients, relative to healthy control subjects. The heatmap indicates the percentages of hypermethylated, hypomethylated and normally methylated CpGs that occur within genomic intervals (i.e. ‘windows’) of distance to the closest TSS. Dark red cells indicate intervals of distances from TSSs (TSS distance intervals) at which abnormally methylated CpGs states disproportionately occurred. TSS distance intervals were generated by calculating the distance of each CpG to its closest TSS and then splitting CpGs into 40 groups based on this distance. Interval boundaries were defined by frequency, such that each interval includes an approximately equal number of CpGs. Purple horizontal sidebars indicate the percentages of CpGs within each interval that occur within genes and promoter regions (TSS200 and TSS1500). TSS200 and TSS1500 regions encompass CpGs within 200 and 1500 bp of a TSS, respectively. For each TSS distance interval, green horizontal sidebars indicate the negative log 10 FDR-adjusted P-values (−log10 P-value) for differences in the frequencies of hypermethylated and hypomethylated CpGs relative to normally methylated CpGs (Fisher’s exact test). Negative log10 P-values greater than 3 (i.e. P < 0.001) were assigned a value of 3 to improve visualization of the P-value range. (B) Stacked bar plots indicating the percentages of CpGs that were within CpG islands (CGIs), CGI shores and CGI shelves, for hypermethylated, normally methylated and hypomethylated CpGs within each TSS distance interval. North ‘N’ and south ‘S’ CGI shores and shelves (Those that occur upstream and downstream of CGIs) are indicated, as defined within the Illumina 850 k array annotation. (C) Boxplots indicate the distribution of baseline DNA methylation for CpGs that were hypermethylated, normally methylated and hypomethylated in SS, within each TSS distance interval. Baseline DNA methylation refers to the level of DNA methylation in the whole blood of healthy control subjects (N = 15). Horizontal red dashed reference lines indicate the Y-axis position that indicates 50% (i.e. intermediate) baseline DNAm. Abbreviations: TSS = ‘Transcription start site,’ Island = ‘CpG island,’ Beta = ‘Beta value,’ Q.val = ‘Q value,’ Hyper = ‘Hypermethylated,’ ‘Hypo’ = ‘Hypomethylated,’ ‘Normal’ = ‘Normally methylated.’
A recent study (58) reported that NSD1-deposited H3K36me recruits DNAm to intergenic regions; this conclusion was based partially on the authors’ reanalysis of the Choufani study 450 k array data, which showed that DNA hypomethylation disproportionally occurred at intergenic regions. Our findings partially concur with this analysis, as hypomethylated CpGs were overrepresented within intergenic regions overall. Despite this, analysis of the distribution of hypomethylated CpGs in relation to TSSs indicated that hypomethylation was enriched only at intergenic CpGs that occurred at the borders of promoters, i.e., promoter-flanking regions, which represent cis-regulatory elements of genes. Hypomethylation was not enriched at intergenic CpGs that were more distant from genes, indicating that NSD1 recruits DNAm to promoter–proximal regions rather than intergenic regions per se. In contrast, hypermethylated CpGs were enriched within intergenic regions and introns as well as 3’ TSS flanking regions but were depleted at TSSs and 5’ TSS flanking regions (Fig. 3A). The distinct genomic distributions of hypomethylated and hypermethylated CpGs therefore indicate that the direction in which NSD1 regulates DNAm depends on the underlying genomic context.
DNAm is bimodally distributed such that in healthy tissues, DNAm levels of the vast majority of CpGs are either very low (where CpGs occur within gene promoters) or high (where CpGs occur within intergenic regions). A distinctive feature of CpGs that were abnormally methylated in SS was that they were generally restricted to CpGs that display intermediate baseline DNAm, i.e. approximately 50% DNAm levels in blood samples of healthy control subjects, across all genomic regions (Fig. 3C). This observation persisted despite the exclusion of CpGs within sex chromosomes and known imprinted regions, indicating that NSD1 maintains intermediate DNAm at non-imprinted autosomal regions.
To determine if NSD1 regulates DNAm within similar genomic contexts across tissues, we analyzed the distribution of abnormal DNAm in dermal fibroblasts of SS patients. We identified CpGs that were abnormally methylated in dermal fibroblasts of SS patients (n = 3) relative to those of healthy control subjects (n = 4) using a previously published Illumina 450 k array data (21) (Supplementary Material, Fig. S11A). This revealed a similar distribution of abnormal DNAm in SS fibroblasts as was observed in blood, characterized by strong enrichment of hypomethylation at 5’ TSS flanking CGI shores, enrichment of hypermethylation within intergenic regions, and intermediate baseline methylation (in healthy control fibroblasts) of CpGs that were abnormally methylated in SS. This indicates that NSD1 regulates DNAm within similar contexts across tissues.
Promoter DNA hypomethylation of transcriptionally deregulated genes
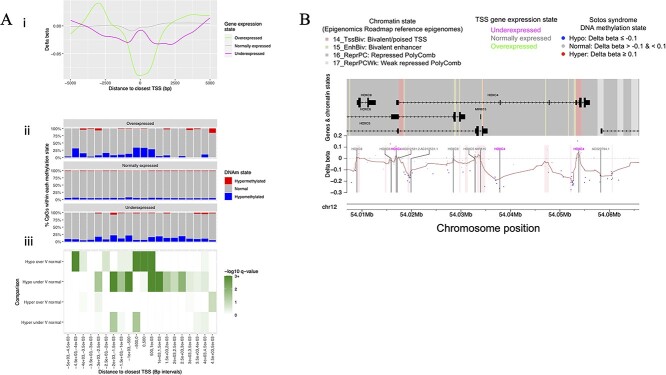
We next analyzed the distribution of abnormal DNAm within regions surrounding transcriptionally deregulated genes, to determine if DNAm deregulation could play a role in transcriptional deregulation. Indeed, DNA hypomethylation was strongly enriched at TSSs of overexpressed genes (Figs 2Aiii and 4A) (Supplementary Material, Tables S4 and S8). This suggests that transcription of these genes is normally silenced by (NSD1-directed) DNAm, as TSS DNAm is known to prevent transcriptional initiation (18,19). In contrast with overexpressed genes, unexpressed genes displayed DNA hypomethylation within their TSS flanking regions, precisely at the intervals of distance from TSSs at which we had observed spikes of DNA hypomethylation within promoter CGI shores. Moreover, genes that were normally expressed in SS displayed very little promoter hypomethylation, indicating that NSD1 primarily directs DNAm to promoters of genes that are transcriptionally regulated by NSD1. Underexpressed genes displayed significantly higher proportions of hypermethylated CpGs within their overall regions compared with normally expressed genes (P < 0.001), suggesting that hypermethylation plays a role in transcriptionally silencing these genes. For example, the underexpressed gene HOXC4 featured hypermethylation of CpGs within its bivalent TSS regions, consistent with epigenetic silencing; however, the most extreme alterations of DNAm within HOXC4 were within its TSS flanking regions, where most CpGs displayed strong DNA hypomethylation (Fig. 4B).
Figure 4.
Abnormal DNA methylated CpGs within promoters of abnormally expressed genes. (A i) Smoothed lines illustrating the distribution of abnormally methylated CpGs within promoters of genes that were abnormally expressed in SS. For CpGs that are within 5 kb of TSSs of genes that were overexpressed (Green), normally expressed (Gray) and underexpressed (Magenta) in SS, smoothed lines plot the delta beta (i.e. the mean DNA methylation beta value difference between SS patients and healthy control subjects) (Y-axis) against the distance of the CpG to its closest TSS (of a gene of the relevant expression state). (A ii) Bar plots illustrate the percentages of CpGs that were hypomethylated (Blue), normally methylated (Gray) or hypermethylated (Red) in SS, within 500 bp intervals (i.e. ‘windows’) surrounding the transcription start sites of genes that were overexpressed, normally expressed or underexpressed in SS. Each CpG is assigned to a ‘Distance to closest TSS’ interval based of its base pair (bp) distance to the closest TSS of any gene within the relevant gene expression state. Each bar plot represents all CpGs that are within 5 kb of a TSS of any gene of the relevant expression state. (A iii) Heatmap of P-values indicating the significance of differential representation of hypomethylated and hypermethylated CpGs within the promoters of abnormally expressed genes relative to normally expressed genes. For TSS distance intervals that are shown in ‘ii,’ the heatmap indicates –log10 P-values (Fisher’s exact test) for differences in the proportions of hypomethylated and hypermethylated CpGs within intervals surrounding the TSSs of overexpressed genes compared with normally expressed genes (over V normal) and underexpressed genes compared with normally expressed genes (under V normal). Negative log10 P-values greater than 3 (P < 0.001) were assigned a value of 3 to improve visualization of the P-value range. (B) Karyoplot illustrating levels of abnormal DNA methylation and baseline chromatin states within HOXC4, the most significantly underexpressed gene in SS. The upper panel (Labeled ‘Genes & chromatin states’) illustrates the structures of HOXC4 and surrounding genes, with background color blocks illustrating the baseline chromatin states (i.e. the Roadmap Epigenomics (109) blood chromatin states) within the region. The lower panel illustrates the genomic location and degree of abnormal DNA methylation in SS (Delta beta) of all CpGs within the region. Points indicate the delta beta value for each CpG, while the brown connecting line indicates the rolling mean of CpG delta beta values. A horizontal gray dashed marker line is shown at the ‘zero’ delta beta position, representing the Y-axis position of CpGs that display normal DNA methylation (i.e. no difference between cases and controls). Vertical black bars indicate positions of transcription states sites, with labels indicating the associated gene symbol and label colors indicating the expression state of the associated gene in SS. Transparent pink blocks indicate the regions covered by CpG islands. The Karyoplot was generated using KaryoplotR (108). Abbreviations: TSS=‘Transcription start site,’ DNAm = ‘DNA methylation,’ Bp = ‘Base pairs.’
Colocalization of DNA hypomethylation with PRC2 binding sites
We next investigated the levels of baseline histone modifications and EZH2 occupancy at the loci of abnormally methylated CpGs. This revealed that across all genomic regions, CpGs that were hypomethylated in SS featured much higher levels of EZH2 occupancy and H3K27me3 in reference datasets (in the blood of healthy individuals) than CpGs that were normally methylated in SS (Fig. 2B). This indicates that hypomethylation in SS disproportionally occurs at regions of PRC2 activity. Moreover, regions of DNA hypomethylation in SS dermal fibroblasts also featured enrichment of EZH2 occupancy and H3K27me3, as indicated by analysis of reference epigenomes that were derived from dermal fibroblast cultures (Supplementary Material, Fig. S11B). This indicates that the spikes of DNA hypomethylated within the TSS flanking regions of underexpressed genes correspond to sites of PRC2 binding and methyltransferase activity. Taken together with our earlier observation that underexpressed genes featured enrichment of EZH2 occupancy and H3K27me3 within their TSS flanking regions, this suggests that NSD1 preferentially directs DNAm to PRC2 binding sites within the promoters of NSD1-regulated genes. Consistent with the enrichment of DNA hypomethylation at bivalent gene promoters, hypomethylated CpGs within TSS-flanking regions displayed elevated levels of H3K4me1, a modification that marks bivalent promoters and enhancers (59), while hypomethylated CpGs displayed low levels of all activating histone modification. Hypermethylated CpGs displayed high levels of H3K4me1 as well as H3K9me3, a modification that marks heterochromatin, and H3K36me3, a modification that marks bodies of actively transcribed genes. Analysis of consolidated chromatin states at the loci of abnormally methylated CpGs confirmed that hypomethylated CpGs disproportionately occurred at regions that featured PRC2-associated chromatin states (Supplementary Material, Fig. S9B). Interestingly, hypermethylated CpGs were associated with active enhancer states. Taken together with our earlier observation that hypermethylation was enriched at underexpressed genes, this suggests that hypermethylation of enhancers could silence bivalent genes in SS.
Accelerated molecular aging in SS
DNAm is continuously altered with age, such that age can be predicted from DNAm data using algorithms known as epigenetic clocks (60,61). By applying epigenetic clocks to the Choufani dataset, two recent studies (62,63) reported that SS is associated with ‘accelerated epigenetic aging,’ i.e. with DNAm-predicted age values that exceed chronological age. To validate this finding, we calculated DNAm age for SS case and control samples using an updated version of the ‘Horvath clock,’ which improves the accuracy of age estimates by incorporating CpGs that are represented on the 850 K array (62,63). Consistent with previous findings, all three SS patients, but not healthy control subjects (n = 3) were predicted to be approximately 20 years older than their chronological age (Supplementary Material, Fig. S12A and Table S9). We next investigated whether SS patients display accelerated molecular aging at the level of transcription, by calculating the transcriptional age of all samples using two ‘transcriptional aging clocks,’ namely RNAAgeCalc (64) and TRAP (65). Indeed, SS patients displayed accelerated transcriptional age relative to controls based on estimates for both transcriptional age calculators (Supplementary Material, Fig. S12B). This postulates that deregulation of DNAm in SS results in age-associated transcription deregulation, indicating that NSD1 inactivation-associated molecular aging could affect phenotype.
Transcriptional deregulation of Sotos syndrome-deregulated genes in NSD1-mutated cancers
It has previously been reported that somatic NSD1 mutations are associated with ‘SS-like’ signatures of abnormal DNAm in squamous cell carcinomas (HNSCS and LUSCC) (14). To determine if NSD1 inactivation causes similar patterns of transcriptional deregulation between SS and cancer, we investigated differential expression of genes that were abnormally expressed in SS between SCCs that harbor somatic NSD1 mutations and those that are NSD1 wild type. Indeed, the mean expression of genes that were overexpressed and underexpressed in SS was higher and lower, respectively, in NSD1-mutated SCCs relative to NSD1 wild-type SCCs, in both HNSCC and LUSCC (Supplementary Material, Fig. S13). This indicates that the patterns of transcriptional and epigenetic deregulation that are caused by NSD1 inactivation are partially consistent across tissues and disease states.
Discussion
Here we characterized abnormal transcription and DNAm in SS, identifying genes that are deregulated as a result of NSD1 mutations as well as mechanisms through which NSD1 could regulate transcription. A key insight was that NSD1 preferentially regulates bivalent genes; this observation is consistent with experimental evidence that NSD1 represses PRC2 activity, as experimental NSD1 inactivation in embryonic stem cells (17) and spermatozoa (18) of mice caused accumulation of H3K27me3 and downregulation of PRC2 target genes. Despite this, our findings indicate that NSD1 specifically promotes transcription of (a subset of) bivalent genes, rather than PRC2 target genes overall, representing a much more specific regulatory role than previously appreciated. Moreover, almost 40% of transcriptionally deregulated genes were overexpressed, revealing that NSD1 regulates developmental genes either positively or negatively depending on context.
Our findings indicate that NSD1 regulates transcription by directing promoter DNAm, since abnormal DNAm in SS was enriched at promoters of transcriptionally deregulated genes. We found clear evidence that NSD1 represses transcription of a subset of genes by directing promoter DNAm, since overexpressed genes displayed DNA hypomethylation directly at their TSSs, regions at which DNAm prevents transcriptional initiation (19,66).
Intriguingly, we also found evidence that NSD1 promotes transcription of bivalent genes by recruiting promoter DNAm, as hypomethylation was strongly enriched within the TSS-flanking regions of underexpressed genes, particularly at CpG islands shores that normally feature PRC2 binding and H3K27me3. These observations lead us to hypothesize that NSD1-deposited H3K36me protects bivalent genes from PRC2-mediated silencing by recruiting DNAm to PRC2 binding sites within their promoters, thereby antagonizing PRC2 activity. In support of this model, DNAm of bivalent gene CGI shores has been implicated in preventing PRC2-mediated silencing of these genes (67). In evidence of a third mechanism, we found that DNA hypermethylation was enriched at underexpressed genes and enhancers, suggesting that NSD1-deposited H3K36me promotes transcription of bivalent genes by precluding DNAm at nearby enhancers.
NSD1 mutations could cause many of the phenotypes of SS by disrupting normal PRC2 activity, as SS is phenotypically very similar to OGID disorders that are caused by germline mutations in genes that encode the three core subunits of PRC2; these include weaver syndrome (MIM: 277590, caused by EZH2 mutations), Cohen–Gibson syndrome (MIM: 617561, caused by EED mutations) and Imagawa–Matsumoto syndrome (MIM: 618786, caused by SUZ12 mutations). The observation that SS closely mimics the phenotypes of disorders of PRC2 disruption, coupled with the collective evidence that NSD1 regulates PRC2 activity, suggests that NSD1 mutations cause SS at least partially by deregulating PRC2, as has been speculated previously (53). Moreover, this hypothesis suggests that disruption of PRC2 could represent a general causal mechanism underlying OGID, and could play a role in other OGID disorders that are phenotypically similar to SS, such as the conditions that are caused by germline mutations in NFIX (68), HIST1H1E (69) and APC2 (70).
Aberrant silencing of bivalent genes could account for neurodevelopmental phenotypes of SS, since neurodevelopmental genes often feature bivalent promoters (42). Moreover, underexpressed genes included a strong enrichment of synaptic membrane-expressed genes. This suggests that cognitive impairment in SS could be caused by the silencing of bivalent genes that orchestrate synaptic assembly, since disrupted synaptic assembly is a feature of many neurodevelopmental disorders (71). We identified other transcriptionally and epigenetically deregulated genes that are known to regulate skeletal overgrowth and craniofacial anomalies, suggesting that perturbation of these genes contributes to the SS phenotypes. Collectively, these findings suggest that NSD1 mutations cause SS by impairing epigenetic regulation of developmental genes.
Genes that were transcriptionally deregulated in SS were also altered in NSD1-mutated cancers, suggesting that NSD1 regulates similar transcriptional pathways through a consistent mechanism across tissues and disease states. Since bivalent genes are frequently deregulated in cancer (72), NSD1 mutations might promote tumorigenesis and oncogenic growth by perturbing PRC2 activity. Since H3K27me3 at bivalent genes is understood to maintain the identity of stem cells (45), silencing of bivalent genes in NSD1-mutated cancers could promote stem cell-like cellular states and could give rise to cancer stem-like cells that have increased metastatic and tumor-initiating potential (73). This hypothesis is supported by previous observations that NSD1-mutated cancers display stem cell-like transcriptional and DNAm profiles (14,74).
Another intriguing finding was that SS patients featured accelerated molecular aging at the levels of both DNAm (as previously reported (62,63)) and transcription. Deregulation at bivalent genes in SS could explain this phenomenon since aging-associated molecular deregulation occurs predominantly at bivalent domains (75–79). A validation study to confirm accelerated transcriptional aging in SS is warranted, as this would identify NSD1 as a master regulator of human molecular aging.
Functional studies are needed to elucidate the mechanisms through which NSD1 regulates transcription and to confirm that NSD1-directed DNAm precludes PRC2-mediated silencing of bivalent genes. Understanding this mechanism could enable the development of targeted therapies, such as epigenetic drugs or gene therapies (80,81), for the treatment of SS and related disorders.
Materials and methods
Research subject recruitment and study design
This study was conducted under an approved IRB protocol. Written informed consent was obtained from all participants. Ten individuals with clinical features suggestive of classic SS whose diagnoses were confirmed molecularly (i.e. with an NSD1 mutation or deletion) were enrolled in the study. Also enrolled were one or both (if available) parents of each SS patient (parent controls, n = 16), as well as siblings of patients that were of the same sex and close to the same age of the proband (n = 3).
Research subjects were recruited during two separate phases, which we refer to as the ‘discovery’ and ‘validation’ phases, each including five sets of patients and matched controls. The discovery phase sample set included five SS patients that were recruited at a local child health institute. For each proband, the discovery study included one or both parents whose samples were collected and processed at the same time. For three of these probands, anonymous sex and age-matched control subjects (blood donors) were included. The validation set included five SS probands as well as a healthy control subject that was matched to the case subject by sex, ancestry (self-declared) and age (within six years).
Given the challenge of collecting both RNA and DNA from fresh primary tissues of patients with a rare condition, this study was designed to achieve maximum statistical power from a limited number of samples. We achieved this by selecting healthy control subjects that were matched to probands by potential confounding factors that could affect gene expression, including age, sex and ancestry, as well as technical factors. This allowed us to perform DEG analysis without the need to adjust for these factors within the model, which would have reduced statistical power. Control subjects were especially well matched to probands for all of the aforementioned demographic and technical factors within the validation study, such that we could exclude these potential confounding factors for genes that were differentially expressed within the validation study. For each proband, at least one healthy control subject was included whose sample was collected and underwent all steps of processing and analysis at the same time as the probands sample, thereby serving as technical controls. Where available, siblings were included to act as controls that were matched for demographic factors (sex, age and ancestry), as well as family and technical factors. Where matched siblings were not available, blood samples were collected from unrelated individuals that were matched to the patient by age, sex and self-declared ancestry (Healthy volunteer controls) (N = 2). Healthy control volunteers’ blood samples were collected on the same date and in the same location as the matched patient’s sample. One of these healthy volunteer control subjects (ID: 5530-P) is the mother of a proband that is included in this study, whose unaffected sibling is also included. The other healthy volunteer control subject (ID: 7634-M) is the mother of a patient whose sample was not analyzed as part of this study. Where siblings or volunteer subjects were not available, blood samples of sex/age-matched healthy blood donors were accessed from a local blood collection facility. Where blood donor control samples were used, a sample of a parent of the proband was also included to control for technical variables and ancestry, representing the sex-matched parent where possible (for two out of three cases). For two infant subjects, where age/sex-matched siblings were not available, nor could age-matched blood donor samples be accessed (due to the patient’s age), samples of both parents were included in all analysis. See Table 1 for details of the study design.
NSD1 mutations were identified through clinical laboratory testing for all participants. When primary records of testing were not available DNAm pyrosequencing of cg07600533 was used to confirm that the patient had the characteristic DNAm signature of SS.
Blood collection and nucleic acid extraction and processing
Blood samples (EDTA tubes) were collected from all research subjects by venipuncture. DNA and RNA were extracted simultaneously from 1 mL fresh whole blood, using the AllPrep DNA/RNA Mini kit (Qiagen). In order to analyze intact RNA from living cells, RNA was extracted from fresh blood samples promptly after blood collection (within 5 h). Extracted RNA and DNA were stored at −80 and −20°C, respectively.
Pyrosequencing
Pyrosequencing was used to measure DNAm of cg07600533 using a custom-designed pyrosequencing assay. Assay design and pyrosequencing were performed using the PyroMark Q24 system (Qiagen). Pyrosequencing assay design was attempted for the top five CpGs with the largest difference in DNAm between SS case and control subjects within the Choufani et al. dataset. The cg07600533 assay was selected to confirm the diagnosis of all subjects because cg07600533 represented the CpG site with the second-largest difference in methylation between SS cases and controls and because the cg07600533 assay measured cg07600533 methylation with high accuracy, as indicated by the methylation levels observed in universally methylated and unmethylated control DNA samples (Supplementary Material, Fig. S1). Pyrosequencing was applied to PCR amplicons of bisulfite-converted DNA. PCR was performed using FASTSTART TAQ DNA polymerase. Bisulfite conversion was performed using the EZ DNAm kit (Zymo Research). Universally methylated and unmethylated DNA controls were purchased from EpiTect. PCR and pyrosequencing primer sequences are supplied in Supplementary Material, Table S10.
RNA sequencing
RNA sequencing (RNA-Seq) was applied to whole blood RNA samples of ten SS patients and 15 healthy control subjects. RNA-Seq was performed separately for the discovery and validation phase sample sets on separate dates approximately six months apart, such that the validation set experiment represents a biological replicate of the discovery set experiment.
RNA samples were quality controlled to ensure adequate concentration, integrity and purity of mRNA, by applying electrophoresis using the 2100 Bioanalyzer (Agilent). Globin mRNAs were depleted from RNA samples using the GLOBINclear kit (Invitrogen). Stranded library preparation and eukaryotic transcriptome resequencing were performed using the BGISeq-500 next-generation sequencing platform with paired-end 100 bp reads. RNA quality control, globin depletion, library preparation and sequencing were performed by BGI Group (BGI Group, Shenzhen, Guangdong, China).
RNA-Seq data preprocessing
Trim-Galore! was used to perform adaptor trimming and filtering of raw reads. Kallisto (82) was used to align reads to the GENCODE 32 human transcriptome (Genome build hg38). MultiQC was used to perform quality control of RNA-Seq samples based on the output of Trim-Galore! and Kallisto, including the percentages of reads that passed quality controls and that were aligned to the transcriptome. The percentages of aligned reads were between 83.3 and 91 for all samples. Transcript-level counts were summarized to gene level using tximport (83).
Principal component analysis
PCA was applied to the top 10% of most variable genes based on the mean absolute deviation. PCA was performed by applying the counts2PCA algorithm to normalized counts data that was batch corrected (Correcting for study phase) using COMBAT (84).
Differentially expressed gene analysis
Limma-Voom (85,86) was applied to identify DEGs between SS and controls. DEGs were defined as those with FDR-adjusted P-values less than 0.05 and absolute log2-fold changes of 1 or greater (corresponding to an absolute fold change of 2 or greater). To assess the reproducibility of differential expression, we first identified DEGs within the discovery set and the confirmed differential expression of these genes within the validation set. Having confirmed the reproducibility of differential expression, we then performed DEG analysis within the combined discovery and validation sets, including the study phase (discovery or validation) as a covariate in the Limma model matrix.
Development of a diagnostic gene expression classifier
PAM analysis (24) was used to develop a diagnostic gene expression classifier model to distinguish SS patients from control subjects based on gene expression. The ability of this model to distinguish cases from controls was assessed by performing PAM analysis in combination with 10-fold cross-validation. For each fold of cross-validation, a diagnostic model was trained on 90% of patient samples and then used to classify the remaining ‘held out’ 10%. The accuracy of PAM predictions across the ten folds of the outer cross-validation was evaluated based on the area under the ROC curve. The posterior probabilities of classifications were also assessed as a measure of model confidence, as posterior probabilities that are strongly weighted toward one class indicate high certainty.
TCGA data access and processing
TCGA gene RNA-Seq data were processed from raw bam files as part of our previously reported study (87). RNA-Seq reads were aligned to the GENCODE version 32 using STAR and counted using RSEM (88), ensuring optimal matching of gene identifiers between TCGA data and SS RNA Seq. STAR + RSEM counts were analyzed for consistency with our previously reported research into NSD1 using TCGA data, and with TCGA project studies in general. Transcript-level expression was summarized to gene level using tximport (83). Gene-level counts were then normalized using upper quartile normalization (89), the method that was reported for TCGA projects: Raw RSEM counts were normalized by the 75th percentile of the column (patient sample) after excluding zeroes, and multiplied by 1000.
TCGA gene-level copy number (gistic2_thresholded) data and somatic mutation (gene-level non-silent mutation) data were processed as part of the TCGA Pan-Cancer project (90). Somatic mutations and copy number alterations were called using MutSig2 (91) and Gistic2 (92), respectively. Fully processed genetic datasets were downloaded from the Xena browser (93) and analyzed without any additional processing.
Functional gene set enrichment analysis
Functional GSEA was carried out using MSigDB (94), selecting H, C1, C2, C5, C6, C7 and C8 gene set collections for comparison with the input gene set (genes deregulated in SS). C3 (regulatory gene sets) and C4 (computational) libraries were excluded from the analysis due to the difficulty in interpreting enrichments of these computationally derived and sparsely annotated gene sets. The top 100 most enriched gene sets were visualized as a network map generated using enrichment map (95).
Estimating blood cell fractions
CIBERSORTx was applied to transcriptional data in order to infer the fractions of major cell types within blood samples. CIBERSORTx was applied using two signature matrices that were previously shown to accurately impute the relative fractions of cell types from whole blood RNA-Seq data (96). The signature matrices consist of sets of genes (gene signatures) whose relative expression levels indicate the proportions of particular cell types. These included the LM22 signature matrix, which comprises of gene expression signatures for 22 immune cell types that were derived from bulk RNA-Seq data of isolated cell types (97), and the ‘non-small cell lung cancer (NSCLC) PBMC’ signature matrix, comprised of gene expression signatures for six blood cell types that were derived from single-cell RNA-Seq data of PBMCs of a patient with NSCLC (96). The relative expression levels of these signature matrix genes indicate the fractions of cell types and are used to train the CIBERSORTx algorithm. TPM data were generated using Kallisto, and transcript-level TPM data were summarized to gene-level data using tximport (83).
Processing DNAm array data
DNAm was profiled using the Illumina Infinium EPIC Beadchip array (i.e. the ‘850 K array’) (98). Raw 850 K array data were extracted using GenomeStudio. Methylation data preprocessing was performed using the minfi R package (99). Minfi quality controls were applied to ensure that each sample had high median intensities in the methylated and unmethylated channels, had mean P-values access all probes that was less than 0.01 and that the sex of the patient sample was accurately predicted from methylation data. WateRmelon (100) was used to ensure that all samples had high (>94%) bisulfite conversion rates. CpGs were discarded from the analysis that had single nucleotide polymorphisms within either the interrogated CpG or at the single nucleotide extension, that represented cross-reactive probes that are listed within the maxprobes R package (101), or that had detection P-values of greater than 0.01 in more than 10% of samples. CpGs within sex chromosomes were excluded from all analyses in order to exclude biases associated with the analysis of sex chromosome DNAm in samples of mixed sexes.
Processing Choufani et al. SS DNAm array data
Raw DNAm array data that were generated as part of the Choufani study were accessed from Gene Expression Omnibus (Accession number: GSE74432). This dataset included DNAm data that were generated from primary whole blood samples of SS patients (n = 38) and age-matched healthy controls (n = 53). Also included were DNAm array data that were generated from dermal fibroblasts of SS patients (n = 3) and age-matched healthy control subjects (n = 4). These data were generated using the Illumina Infinium HumanMethylation450 Beadchip array (i.e. the ‘450 k array’). Raw Choufani study data were preprocessed using the minfi package with a pipeline that were consistent with that used for 850 K array data.
Analysis of baseline DNAm
Baseline DNAm (i.e. DNAm in NSD1 wild-type tissues) was analyzed in healthy control subjects of the Stanford study (n = 15) for blood, and healthy control dermal fibroblasts from the Choufani study (n = 4) for dermal fibroblasts. Imprinted CpGs, which were accessed from Hernadez Mora et al. (102), were excluded from all analyses of baseline DNAm.
Transcriptional and DNAm aging clocks
DNAm age was calculated using the online DNAm age calculator (http://dnamage.genetics.ucla.edu/) (60). Transcriptional age was calculated using RNAAgeCalc (64), which was developed to calculate transcriptional age from RNA-Seq data, and TRAP (65), which was developed to calculate transcriptional age from peripheral blood gene expression microarrays. RNAAgeCalc was applied to FPKM values using the RNAAgeCalc R package according to the reference manual. Model parameters were set such that the model was trained using blood gene expression data from the GTEx study, including samples of individuals from all races. FPKM values were calculated from raw counts using the ‘count2FPKM’ function. Gene lengths (needed to calculate FPKM values) were calculated from the GENCODE gene transfer format (GTF) file using GenomicFeatures (103). TRAP was applied to raw counts using the online transcriptomic age predictor at https://trap.erasmusmc.nl/.
Accessing and processing reference epigenomes
Reference epigenomes were downloaded from the NIH Epigenomics Roadmap web portal. These included imputed hg38 signal tracks for seven histone modifications, each of which indicates the levels (ChIP-Seq signal) of a histone modification, as well as consolidated chromatin state maps, which consolidate data for the seven histone modifications, indicating the overall chromatin state within each 200 bp genomic interval. Consolidated chromatin states thereby indicate the locations of functional chromatin states including polycomb-associated chromatin regions and enhancers.
Consolidated, imputed hg38 signal tracks were converted to BedGraph files using the UCSC bigWigToBedGraph tool prior to analysis. LiftOver tool was used to convert chromatin state bed files from genome build hg19 to hg38. Reference epigenomes were selected to match the tissue types under investigation, including blood and dermal fibroblasts. These reference epigenomes were established from primary PBMCs (epigenome identifier: E062) and adult dermal fibroblasts (epigenome identifier: E126).
Mapping colocalization of genes, CpGs and genomic features
Bedtools (104) was used to perform operations that entailed mapping the spatial relationship between genomic features, such as genes and CpGs. Bed files were generated for each of the following genomic features:
Genes, TSSs and CpGs: the chromosome locations of all GENCODE genes and their TSSs were accessed using BioMart, selecting the ensemble BioMart database and the ‘hsapiens_gene_ensembl’ dataset. Hg38 genomic locations of all 850 K array CpGs were accessed from an annotation file that was generated by Zhou et al. (105). Bed files were generated that indicated the locations of all genes, TSSs and CpGs. Metadata columns were included within each bed file, indicating the expression and DNAm states of genes and CpGs, respectively. Additionally, separate bed files were generated for genes and TSSs that were associated with each gene expression state (overexpressed, underexpressed and normally expressed) in SS, as well as CpGs that were associated with each DNAm state (hypomethylated, hypermethylated and normally methylated) in SS.
Reference genomes: The Ensembl hg38 DNA primary genome assembly fasta file was accessed from the Ensembl FTP website (ftp.ensembl.org). Bed files indicating the positions of genes, exons and introns, were accessed from the UCSC Table Browser. The positions of intergenic regions were identified by subtracting genic regions from the full UCSC hg38 genome using the Bedtools ‘subtract.’ An hg38 bed file of the ‘cpgIslandExt’ track, indicating the positions of CpG islands, was also accessed from the UCSC Table Browser.
Reference epigenomes: Reference epigenomes were downloaded from the NIH Epigenomics Roadmap web portal. These included consolidated and imputed hg38 signal tracks, which indicate the levels of individual histone modifications, as well as chromatin state maps that indicate the overall chromatin state within each genomic interval. Consolidated, imputed hg38 signal tracks were converted to BedGraph files using the UCSC bigWigToBedGraph tool prior to analysis. The UCSC Genome Browser LiftOver tool was used to convert chromatin states from genome build hg19 to hg38.
EZH2 binding sites: ChIP-Seq broadPeak file were accessed from GEO (accession numbers GSM1003498 and GSM1003550). These datasets were generated as part of the ENCODE (106) project and included EZH2 ChIP-Seq datasets that were derived from the lymphoblastoid cell line GM12878, and the primary dermal fibroblast culture NHDF-Ad. LiftOver was used to convert the broadPeak files from genome build hg19 to hg38.
The Bedtools operations that were used to perform analysis outlined in the Results section are as follows:
Calculating the mean histone modification levels within gene promoter-proximal regions (Fig. 2Ai) (Supplementary Material, Table S7): Bedtools ‘map’ was used to calculate mean histone modification signal across all genomic windows that overlapped with promoters of each gene. The promoter of each gene was defined as the combined regions that are within 1500 base pairs of all TSSs of the gene. Prior to mapping of histone modifications to genes, Bedtools ‘slop’ was used to add 1500 bp to the start and end genomic coordinates of each TSS, yielding the ‘TSS1500’ region of the TSS. Since each gene can have multiple TSSs, histone modification levels were averaged across all TSS1500 regions that were associated with each gene, yielding the average promoter histone modification level of the gene.
Calculating mean levels of EZH2 occupancy within promoter-proximal regions (Fig. 2Aii): Bedtools ‘map’ was used to calculate mean EZH2 ChIP-Seq signal (from EZH2 ChIP-Seq broadPeak files) within consolidated chromatin state 200 bp windows (from chromatin state BedGraph files). This yielded 200 bp windows of average EZH2 occupancy (EZH2 occupancy windows). Values of zero were assigned to chromatin state windows that did not overlap with EZH2 broadPeak files, indicating the absence of EZH2 occupancy. Using this approach, we confirmed that EZH2 signal was elevated within regions that feature polycomb-associated chromatin states, providing proof of principle for our approach (data not shown). Bedtools ‘window’ was used to identify EZH2 occupancy windows that overlapped with regions spanning from 5 bp upstream to 5 kb downstream of TSSs of genes that were overexpressed, underexpressed and normally expressed in SS. This yielded three sets of EZH2 occupancy windows, occurring within 5 kb of TSSs of overexpressed, underexpressed and normally expressed genes. For each of these sets of EZH2 occupancy windows, Bedtools closest was then used to calculate the distance of each EZH2 occupancy window to its closest TSS of the relevant gene expression state. Bedtools closest was applied using the ‘D -b’ parameter (where TSSs are designated as ‘b’), such that the strand orientation of TSSs was considered when calculating the distance of EZH2 occupancy windows (which do not possess stranded information) to them. This stipulates that where EZH2 occupancy windows are upstream of TSSs, a negative distance is calculated. This parameter also specifies that where TSSs are on the antisense strand, EZH2 occupancy windows that have higher start and stop coordinates are considered to be upstream of TSSs. To calculate percentages of EZH2 occupancy within TSS distance intervals (Fig. 2Ai), EZH2 occupancy windows were split into 1 kb windows based on their distance from the closest TSS, using the arules R package (107). Mean EZH2 occupancy was then calculated across EZH2 occupancy windows that were within each TSS interval, for TSSs that were associated with each gene expression state.
Calculating levels of histone modifications and EZH2 occupancy within the loci of CpGs (Fig. 2B) (Supplementary Material, Fig. S11B): Bedtools ‘intersect’ was used to identify regions within histone modification tracks that overlapped with each CpG. Similarly, Bedtools ‘intersect’ was used to identify regions within EZH2 ChIP-Seq broadPeak files that overlapped with each CpG. CpGs that did not overlap with EZH2 broadPeak files were assigned values of zero, indicating that these loci did not feature EZH2 binding. Mean histone modification and EZH2 signal were then calculated across CpGs that were associated with each DNAm state, for CpGs within each TSS distance interval.
Calculating the percentages of hypermethylated, hypomethylated and normally methylated CpGs (in SS) that occur within intervals of distance from the closest TSS (Figs 2B and 3A) (Supplementary Material, Figs S9B and S11A): Bedtools ‘closest’ was used to calculate the distance of each CpG to its closest TSS (of any gene), using the ‘D -b’ parameter (where TSSs are designated as ‘b’), such that the strand orientation of TSSs was considered when calculating the distance of CpGs to TSS. This stipulates that where CpGs are upstream of TSSs, Bedtools closest yields a negative distance value, and that where the TSSs are on the antisense strand, CpGs that have higher genomic coordinates are considered to be upstream of the TSS. To calculate TSS distance intervals, CpGs were split into forty equally sized groups (i.e. with approximately equal numbers of CpGs) based on their distance from the closest TSS, using the arules R package (107).
Calculating distances of CpGs to TSSs of genes that were overexpressed, underexpressed, or normally expressed in SS (Figs 2Aiii and 4Aii): Separate bed files were generated that indicated the positions of TSSs of overexpressed, underexpressed, and normally expressed genes. For TSSs associated with each gene expression state, Bedtools ‘window’ was used to identify all CpGs that occur within 5 kb of any TSS of that state. This yielded three sets of CpGs, occurring within 5 kb of TSSs of overexpressed, underexpressed and normally expressed genes. For each of these sets of CpGs, Bedtools closest was then used to calculate the distance of each CpG to its closest TSS of the relevant gene expression state. Bedtools closest was applied using the ‘D-b’ parameter (where TSSs are designates as ‘b’), such that the strand orientation of TSSs was considered when calculating the distance of CpGs (which do not possess stranded information) to them. This stipulates that where CpGs are upstream of TSSs, a negative distance is calculated. This parameter also specifies that where TSSs are on the antisense strand, CpGs that have higher start and stop coordinates are considered to be upstream of TSSs.
Calculating percentages of CpGs that were hypomethylated and hypermethylated within 5 kb genomic windows (Supplementary Material, Fig. S5): Bedtools ‘makewindows’ was used to split the hg38 reference genome into 5 kb windows. Bedtools map was then used to map each CpGs to its 5 kb window. The percentage of CpGs within each 5 kb window that were hypomethylated and hypermethylated were then calculated, and rolling means of these percentages were used to plot the genomic distribution of abnormal DNAm using karyoploteR (108).
Calculating percentages of chromatin states within regions surrounding gene promoters (Supplementary Material, Fig. S9): Separate bed files were generated for TSSs of overexpressed, underexpressed and normally expressed genes. For TSSs associated with each gene expression state, Bedtools ‘window’ was used to identify consolidated chromatin state 200 bp windows that overlapped with regions spanning from 5 bp upstream to 5 kb downstream of any TSS that was associated with that gene expression state. This yielded three sets of chromatin state windows, occurring within 5 kb of TSSs of overexpressed, underexpressed and normally expressed genes. For each of these sets of chromatin state windows, Bedtools closest was then used to calculate the distance of each chromatin state window to its closest TSS of the relevant gene expression state. Bedtools closest was applied using the ‘D-b’ parameter (where TSSs are designated as ‘b’), such that the strand orientation of TSSs was considered when calculating the distance of chromatin state windows (which do not possess stranded information) to them. This stipulates that where chromatin state windows are upstream of TSSs, a negative distance is calculated. This parameter also specifies that where TSSs are on the antisense strand, chromatin state windows that have higher start and stop coordinates are considered to be upstream of TSSs. To calculate percentages of chromatin states within TSS distance intervals (Supplementary Material, Fig. S9Ai), chromatin state windows were split into 1 kb windows based on their distance from the closest TSS, using the arules R package (107). To identify chromatin state regions that overlapped with gene promoters (Supplementary Material, Fig. S9Aii), Bedtools window was used to identify all consolidated chromatin state regions that overlapped within TSS1500 regions of genes, i.e. regions within 1500 bp of TSSs of genes. Prior to the mapping of chromatin state regions to promoters, Bedtools slop was used to extend the genomic coordinates of TSSs by 1500 bp, yielding the TSS1500 genomic coordinates for each gene. Since each gene can have multiple TSSs, consolidated chromatin state regions that overlap the promoter of a gene were defined as those that overlap any TSS1500 region of that gene.
Calculating percentages of consolidated chromatin states within regions that overlap CpGs (Supplementary Material, Fig. S9B): Bedtools intersect was used to identify regions within chromatin state maps that overlap with each CpG. For CpGs that were associated with each DNAm state, the percentage of CpGs that overlapped with each chromatin state was calculated.
Calculating overlap of CpGs with genomic region types and genomic features (Supplementary Material, Table S8): Bed files were generated for each of the following types of genomic region categories: whole genes, introns, exons, 5’UTRs, 3’UTRs, intergenic regions, CpG islands and EZH2 ChIP-Seq peaks (i.e. EZH2 occupancy/binding sites). Bedtools intersect was used to identify the CpGs that overlap with genomic regions of each category. Bedtools intersect was also used to identify the CpGs that overlap with genes and TSS regions. Bedtools slop was used to calculate the genomic coordinates of TSS200 and TSS1500 regions by adding 200 or 1500 base pairs to the start and end coordinates of TSSs, respectively. Bedtools slop was also used to add 1 kb or 5 kb to the starts and ends of GENCODE genes.
Calculating levels of abnormal DNAm within genes (Supplementary Material, Tables S3 and S4): Bedtools map was used to calculate mean beta values and delta beta values across all CpGs that occur within each gene. Bedtools map was also used to count the number of hypomethylated, hypermethylated and normally methylated CpGs within each gene. Prior to mapping of CpGs to genes, Bedtools slop was used to add a specified number of base pairs to the start and end of the genomic coordinates of each gene, such that operations were applied to all CpGs that occur within the specified genomic region. This approach was used to calculate mean DNAm within genes and their surrounding regions, adding either one kilobase (1 kb) or five (5 kb) to each gene. Similarly, to analyze CpGs that occur within 200 or 1500 bp of each TSSs, Bedtools slop was used to add either 200 or 500 bp upstream and downstream to the genomic coordinates of each TSS, and Bedtools map was then used to calculate statistics across all CpGs that are within these regions. The regions within 200 and 500 bp of TSSs are referred to as the ‘TSS200’ and ‘TSS1500’ regions, respectively. TS200 and TS1500 regions were analyzed for consistency with the official annotation of Illumina DNAm arrays (98). Each gene can have multiple TSSs; therefore, to analyze the TSS200 and TSS1500 regions of each gene, statistics were averaged across all TS200 or TS1500 regions associated with the gene.
Data Analysis Software
Data analysis was performed using R version 3.6.0. Other programs and tools are indicated within the relevant methods and result sections.
Supplementary Material
Acknowledgements
We would like to thank the Sotos Syndrome Support Association for their advice and assistance with the recruitment of research participants. K.B. Acknowledges support from the Stanford Maternal and Child Health Research Institute (Pilot Grant Award) and the Human Growth Foundation (Small Grant Award). J.A.F. acknowledges support from The Hartwell Foundation (Individual Biomedical Research Award) and the National Institutes of Health (K08HD086250).
Conflict of Interest statement. The authors declare that they do not have any conflict of interest.
Contributor Information
Kevin Brennan, Stanford Center for Biomedical Informatics Research, Department of Medicine, Stanford University, Stanford, CA 94305, USA.
Hong Zheng, Stanford Center for Biomedical Informatics Research, Department of Medicine, Stanford University, Stanford, CA 94305, USA.
Jill A Fahrner, Department of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA; Department of Pediatrics, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
June Ho Shin, Department of Otolaryngology – Head and Neck Surgery, Stanford University School of Medicine, Palo Alto, CA 94305, USA.
Andrew J Gentles, Stanford Center for Biomedical Informatics Research, Department of Medicine, Stanford University, Stanford, CA 94305, USA.
Bradley Schaefer, Department of Pediatrics, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
John B Sunwoo, Department of Otolaryngology – Head and Neck Surgery, Stanford University School of Medicine, Palo Alto, CA 94305, USA.
Jonathan A Bernstein, Department of Pediatrics, Stanford University School of Medicine, Stanford, CA 94305, USA.
Olivier Gevaert, Stanford Center for Biomedical Informatics Research, Department of Medicine, Stanford University, Stanford, CA 94305, USA.
References
- 1. Millan, M.J. (2013) An epigenetic framework for neurodevelopmental disorders: from pathogenesis to potential therapy. An epigenetic framework for neurodevelopmental disorders: from pathogenesis to potential therapy. Neuropharmacology, 68, 2–82. [DOI] [PubMed] [Google Scholar]
- 2. Berdasco, M. and Esteller, M. (2013) Genetic syndromes caused by mutations in epigenetic genes. Genetic syndromes caused by mutations in epigenetic genes. Hum. Genet., 132, 359–383. [DOI] [PubMed] [Google Scholar]
- 3. Kingwell, K. (2017) Neurodevelopmental disorders: epigenetic targets on the table. Neurodevelopmental disorders: epigenetic targets on the table. Nat. Rev. Drug Discov., 16, 677. [DOI] [PubMed] [Google Scholar]
- 4. Fahrner, J.A. and Bjornsson, H.T. (2019) Mendelian disorders of the epigenetic machinery: postnatal malleability and therapeutic prospects. Mendelian disorders of the epigenetic machinery: postnatal malleability and therapeutic prospects. Hum. Mol. Genet., 28, 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Douglas, J., Hanks, S., Temple, I.K., Davies, S., Murray, A., Upadhyaya, M., Tomkins, S., Hughes, H.E., Cole, T.R.P. and Rahman, N. (2003) NSD1 mutations are the major cause of Sotos syndrome and occur in some cases of weaver syndrome but are rare in other overgrowth phenotypes. Am. J. Hum. Genet., 72, 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tatton-Brown, K. and Rahman, N. (2006) Sotos syndrome. Eur. J. Hum. Genet., 15, 264–271. [DOI] [PubMed] [Google Scholar]
- 7. Kurotaki, N. and Matsumoto, N. (2006) Sotos syndrome. In: Lupski J.R., Stankiewicz P. (eds) Genomic Disorders, Humana Press. 10.1007/978-1-59745-039-3_16. [DOI]
- 8. Tatton-Brown, K., Loveday, C., Yost, S., Clarke, M., Ramsay, E., Zachariou, A., Elliott, A., Wylie, H., Ardissone, A., Rittinger, O.et al. (2017) Mutations in epigenetic regulation genes are a major cause of overgrowth with intellectual disability. Am. J. Hum. Genet., 100, 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang, H., Lu, X., Beasley, J., Mulvihill, J.J., Liu, R., Li, S. and Lee, J.Y. (2011) Reversed clinical phenotype due to a microduplication of Sotos syndrome region detected by array CGH: microcephaly, developmental delay and delayed bone age. Am. J. Med. Genet. Part A, 155, 1374–1378. [DOI] [PubMed] [Google Scholar]
- 10. Quintero-Rivera, F., Eno, C.C., Sutanto, C., Jones, K.L., Nowaczyk, M.J.M., Wong, D., Earl, D., Mirzaa, G., Beck, A. and Martinez-Agosto, J.A. (2021) 5q35 duplication presents with psychiatric and undergrowth phenotypes mediated by NSD1 overexpression and mTOR signaling downregulation. Hum. Genet., 140, 681–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shiba, N., Ichikawa, H., Taki, T., Park, M., Jo, A., Mitani, S., Kobayashi, T., Shimada, A., Sotomatsu, M., Arakawa, H.et al. (2013) NUP98-NSD1 gene fusion and its related gene expression signature are strongly associated with a poor prognosis in pediatric acute myeloid leukemia. Genes Chromosomes Cancer, 52, 683–693. [DOI] [PubMed] [Google Scholar]
- 12. Papillon-Cavanagh, S., Lu, C., Gayden, T., Mikael, L.G., Bechet, D., Karamboulas, C., Ailles, L., Karamchandani, J., Marchione, D.M., Garcia, B.A.et al. (2017) Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat. Genet., 49, 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morishita, M. and Di Luccio, E. (2011) Cancers and the NSD family of histone lysine methyltransferases. Cancers and the NSD family of histone lysine methyltransferases. Biochim. Biophys. Acta - Rev. Cancer, 1816, 158–163. [DOI] [PubMed] [Google Scholar]
- 14. Brennan, K., Shin, J.H., Tay, J.K., Prunello, M., Gentles, A.J., Sunwoo, J.B. and Gevaert, O. (2017) NSD1 inactivation defines an immune cold, DNA hypomethylated subtype in squamous cell carcinoma. Sci. Rep., 7, 17064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qiao, Q., Li, Y., Chen, Z., Wang, M., Reinberg, D. and Xu, R.M. (2011) The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation. J. Biol. Chem., 286, 8361–8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tatton-Brown, K. and Rahman, N. (2013) The NSD1 and EZH2 overgrowth genes, similarities and differences. Am. J. Med. Genet. Part C Semin. Med. Genet., 163, 86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Streubel, G., Watson, A., Jammula, S.G., Scelfo, A., Fitzpatrick, D.J., Oliviero, G., McCole, R., Conway, E., Glancy, E., Negri, G.L.et al. (2018) The H3K36me2 methyltransferase Nsd1 demarcates PRC2-mediated H3K27me2 and H3K27me3 domains in embryonic stem cells. Mol. Cell. [DOI] [PubMed] [Google Scholar]
- 18. Shirane, K., Miura, F., Ito, T. and Lorincz, M.C. (2020) NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts polycomb-associated silencing. Nat. Genet., 52, 1088–1098. [DOI] [PubMed] [Google Scholar]
- 19. Greenberg, M.V.C. and Bourc’his, D. (2019) The diverse roles of DNA methylation in mammalian development and disease. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol., 20, 590–607. [DOI] [PubMed] [Google Scholar]
- 20. Jones, P.A. (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet., 13, 484–492. [DOI] [PubMed] [Google Scholar]
- 21. Choufani, S., Cytrynbaum, C., Chung, B.H.Y., Turinsky, A.L., Grafodatskaya, D., Chen, Y.A., Cohen, A.S.A., Dupuis, L., Butcher, D.T., Siu, M.T.et al. (2015) NSD1 mutations generate a genome-wide DNA methylation signature. Nat. Commun., 6, 10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aref-Eshghi, E., Rodenhiser, D.I., Schenkel, L.C., Lin, H., Skinner, C., Ainsworth, P., Paré, G., Hood, R.L., Bulman, D.E., Kernohan, K.D.et al. (2018) Genomic DNA methylation signatures enable concurrent diagnosis and clinical genetic variant classification in neurodevelopmental syndromes. Am. J. Hum. Genet., 102, 156–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carithers, L.J., Ardlie, K., Barcus, M., Branton, P.A., Britton, A., Buia, S.A., Compton, C.C., Deluca, D.S., Peter-Demchok, J., Gelfand, E.T.et al. (2015) A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv. Biobank., 13, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tibshirani, R., Hastie, T., Narasimhan, B. and Chu, G. (2002) Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. U. S. A., 99, 6567–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qu, Q., Sun, G., Murai, K., Ye, P., Li, W., Asuelime, G., Cheung, Y.-T. and Shi, Y. (2013) Wnt7a regulates multiple steps of neurogenesis. Mol. Cell. Biol., 33, 2551–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lan, L., Wang, W., Huang, Y., Bu, X. and Zhao, C. (2019) Roles of Wnt7a in embryo development, tissue homeostasis, and human diseases. J. Cell. Biochem., 120, 18588–18598. [DOI] [PubMed] [Google Scholar]
- 27. Hung, I.H., Schoenwolf, G.C., Lewandoski, M. and Ornitz, D.M. (2016) A combined series of Fgf9 and Fgf18 mutant alleles identifies unique and redundant roles in skeletal development. Dev. Biol., 411, 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amor, D.J., Stephenson, S.E.M., Mustapha, M., Mensah, M.A., Ockeloen, C.W., Lee, W.S., Tankard, R.M., Phelan, D.G., Shinawi, M., deBrouwer, A.P.M.et al. (2019) Pathogenic variants in GPC4 cause Keipert syndrome. Am. J. Hum. Genet., 104, 914–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shih, P.Y., Hsieh, B.Y., Lin, M.H., Huang, T.N., Tsai, C.Y., Pong, W.L., Lee, S.P. and Hsueh, Y.P. (2020) CTTNBP2 controls synaptic expression of zinc-related autism-associated proteins and regulates synapse formation and autism-like behaviors. Cell Rep. 31, 107700. [DOI] [PubMed] [Google Scholar]
- 30. Pollini, L., Galosi, S., Tolve, M., Caputi, C., Carducci, C., Angeloni, A. and Leuzzi, V. (2020) KCND3-related neurological disorders: from old to emerging clinical phenotypes. KCND3-related neurological disorders: from old to emerging clinical phenotypes. Int. J. Mol. Sci. 21, 5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu, D., Mandal, S., Choi, A., Anderson, A., Prochazkova, M., Perry, H., Gil-Da-Silva-Lopes, V.L., Lao, R., Wan, E., Tang, P.L.F.et al. (2015) DLX4 is associated with orofacial clefting and abnormal jaw development. Hum. Mol. Genet., 24, 4340–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kalay, E., Sezgin, O., Chellappa, V., Mutlu, M., Morsy, H., Kayserili, H., Kreiger, E., Cansu, A., Toraman, B., Abdalla, E.M.et al. (2012) Mutations in RIPK4 cause the autosomal-recessive form of popliteal pterygium syndrome. Am. J. Hum. Genet., 90, 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Siu, W.K., Lam, C.W., Gao, W.W., Vincent Tang, H.M., Jin, D.Y. and Mak, C.M. (2016) Unmasking a novel disease gene NEO1 associated with autism spectrum disorders by a hemizygous deletion on chromosome 15 and a functional polymorphism. Behav. Brain Res., 300, 135–142. [DOI] [PubMed] [Google Scholar]
- 34. Fischer, A., Steidl, C., Wagner, T.U., Lang, E., Jakob, P.M., Friedl, P., Knobeloch, K.P. and Gessler, M. (2007) Combined loss of Hey1 and HeyL causes congenital heart defects because of impaired epithelial to mesenchymal transition. Circ. Res., 100, 856–863. [DOI] [PubMed] [Google Scholar]
- 35. Heuser, A., Plovie, E.R., Ellinor, P.T., Grossmann, K.S., Shin, J.T., Wichter, T., Basson, C.T., Lerman, B.B., Sasse-Klaassen, S., Thierfelder, L.et al. (2006) Mutant desmocollin-2 causes arrhythmogenic right ventricular cardiomyopathy. Am. J. Hum. Genet., 79, 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pepe, G., Lucarini, L., Zhang, R.Z., Pan, T.C., Giusti, B., Quijano-Roy, S., Gartioux, C., Bushby, K.M.D., Guicheney, P. and Chu, M.L. (2006) COL6A1 genomic deletions in bethlem myopathy and ullrich muscular dystrophy. Ann. Neurol., 59, 190–195. [DOI] [PubMed] [Google Scholar]
- 37. Lachke, S.A., Higgins, A.W., Inagaki, M., Saadi, I., Xi, Q., Long, M., Quade, B.J., Talkowski, M.E., Gusella, J.F., Fujimoto, A.et al. (2012) The cell adhesion gene PVRL3 is associated with congenital ocular defects. Hum. Genet., 131, 235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Magruder, S., Carter, E., Williams, M.A., English, J., Akyalcin, S. and Letra, A. (2018) Further evidence for the role of WNT10A, WNT10B and GREM2 as candidate genes for isolated tooth agenesis. Orthod. Craniofac. Res., 21, 258–263. [DOI] [PubMed] [Google Scholar]
- 39. Peled, A., Sarig, O., Samuelov, L., Bertolini, M., Ziv, L., Weissglas-Volkov, D., Eskin-Schwartz, M., Adase, C.A., Malchin, N., Bochner, R.et al. (2016) Mutations in TSPEAR, encoding a regulator of notch signaling, affect tooth and hair follicle morphogenesis. PLoS Genet., 12, e1006369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhan, F., Huang, Y., Colla, S., Stewart, J.P., Hanamura, I., Gupta, S., Epstein, J., Yaccoby, S., Sawyer, J., Burington, B.et al. (2006) The molecular classification of multiple myeloma. Blood., 108, 2020–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lucio-Eterovic, A.K., Singh, M.M., Gardner, J.E., Veerappan, C.S., Rice, J.C. and Carpenter, P.B. (2010) Role for the nuclear receptor-binding SET domain protein 1 (NSD1) methyltransferase in coordinating lysine 36 methylation at histone 3 with RNA polymerase II function. Proc. Natl. Acad. Sci. U. S. A., 107, 16952–16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hoffmann, A., Zimmermann, C.A. and Spengler, D. (2015) Molecular epigenetic switches in neurodevelopment in health and disease. Front. Behav. Neurosci., 9, 1662–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ben-Porath, I., Thomson, M.W., Carey, V.J., Ge, R., Bell, G.W., Regev, A. and Weinberg, R.A. (2008) An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet., 40, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meissner, A., Mikkelsen, T.S., Gu, H., Wernig, M., Hanna, J., Sivachenko, A., Zhang, X., Bernstein, B.E., Nusbaum, C., Jaffe, D.B.et al. (2008) Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature, 454, 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blanco, E., González-Ramírez, M., Alcaine-Colet, A., Aranda, S. and Di Croce, L. (2020) The bivalent genome: characterization, structure, and regulation. Trends Genet., 36, 118–131. [DOI] [PubMed] [Google Scholar]
- 46. Round, J. and Stein, E. (2007) Netrin signaling leading to directed growth cone steering. Netrin signaling leading to directed growth cone steering. Curr. Opin. Neurobiol., 17, 15–21. [DOI] [PubMed] [Google Scholar]
- 47. Schubbert, S., Zenker, M., Rowe, S.L., Böll, S., Klein, C., Bollag, G., Van Der Burgt, I., Musante, L., Kalscheuer, V., Wehner, L.E.et al. (2006) Germline KRAS mutations cause Noonan syndrome. Nat. Genet., 38, 331–336. [DOI] [PubMed] [Google Scholar]
- 48. Aoki, Y., Niihori, T., Inoue, S.I. and Matsubara, Y. (2016) Recent advances in RASopathies. Recent advances in RASopathies. J. Hum. Genet., 61, 33–39. [DOI] [PubMed] [Google Scholar]
- 49. Steinhart, Z. and Angers, S. (2018) Wnt signaling in development and tissue homeostasis. Wnt signaling in development and tissue homeostasis. Development, 145, dev146589. [DOI] [PubMed] [Google Scholar]
- 50. Ruiz-Perez, V.L., Tompson, S.W.J., Blair, H.J., Espinoza-Valdez, C., Lapunzina, P., Silva, E.O., Hamel, B., Gibbs, J.L., Young, I.D., Wright, M.J.et al. (2003) Mutations in two nonhomologous genes in a head-to-head configuration cause Ellis-van Creveld syndrome. Am. J. Hum. Genet., 72, 728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reid, I.R., Baldock, P.A. and Cornish, J. (2018) Effects of leptin on the skeleton. Effects of leptin on the skeleton. Endocr. Rev., 39, 938–959. [DOI] [PubMed] [Google Scholar]
- 52. Li, X., Floriddia, E.M., Toskas, K., Chalfouh, C., Honore, A., Aumont, A., Vallières, N., Lacroix, S., Fernandes, K.J.L., Guérout, N.et al. (2018) FoxJ1 regulates spinal cord development and is required for the maintenance of spinal cord stem cell potential. Exp. Cell Res., 368, 84–100. [DOI] [PubMed] [Google Scholar]
- 53. Deevy, O. and Bracken, A.P. (2019) PRC2 functions in development and congenital disorders. PRC2 functions in development and congenital disorders. Development, 146, dev.181354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chadwick, L.H. (2012) The NIH roadmap Epigenomics program data resource. The NIH Roadmap Epigenomics Program data resource. Epigenomics, 4, 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Orlando, D.A., Guenther, M.G., Frampton, G.M. and Young, R.A. (2012) CpG island structure and trithorax/polycomb chromatin domains in human cells. Genomics, 100, 320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Consortium, E.P., Dunham, I., Kundaje, A., Aldred, S.F., Collins, P.J., Davis, C.A., Doyle, F., Epstein, C.B., Frietze, S., Harrow, J.et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dhingra, R., Kwee, L.C., Diaz-Sanchez, D., Devlin, R.B., Cascio, W., Hauser, E.R., Gregory, S., Shah, S., Kraus, W.E., Olden, K.et al. (2019) Evaluating DNA methylation age on the Illumina MethylationEPIC Bead Chip. PLoS One, 14, e0207834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Weinberg, D.N., Papillon-Cavanagh, S., Chen, H., Yue, Y., Chen, X., Rajagopalan, K.N., Horth, C., McGuire, J.T., Xu, X., Nikbakht, H.et al. (2019) The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature, 573, 281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bae, S. and Lesch, B.J. (2020) H3K4me1 distribution predicts transcription state and poising at promoters. Front. Cell Dev. Biol., 8, 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Horvath, S. (2013) DNA methylation age of human tissues and cell types. Genome Biol., 14, R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bell, C.G., Lowe, R., Adams, P.D., Baccarelli, A.A., Beck, S., Bell, J.T., Christensen, B.C., Gladyshev, V.N., Heijmans, B.T., Horvath, S.et al. (2019) DNA methylation aging clocks: challenges and recommendations. DNA methylation aging clocks: challenges and recommendations. Genome Biol., 20, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jeffries, A.R., Maroofian, R., Salter, C.G., Chioza, B.A., Cross, H.E., Patton, M.A., Dempster, E., Karen Temple, I., Mackay, D.J.G., Rezwan, F.I.et al. (2019) Growth disrupting mutations in epigenetic regulatory molecules are associated with abnormalities of epigenetic aging. Genome Res., 29, 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Martin-Herranz, D.E., Aref-Eshghi, E., Bonder, M.J., Stubbs, T.M., Choufani, S., Weksberg, R., Stegle, O., Sadikovic, B., Reik, W. and Thornton, J.M. (2019) Screening for genes that accelerate the epigenetic aging clock in humans reveals a role for the H3K36 methyltransferase NSD1. Genome Biol., 20, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ren, X. and Kuan, P.F. (2020) RNAAgeCalc: a multi-tissue transcriptional age calculator. PLoS One, 15, e0237006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Peters, M.J., Joehanes, R., Pilling, L.C., Schurmann, C., Conneely, K.N., Powell, J., Reinmaa, E., Sutphin, G.L., Zhernakova, A., Schramm, K.et al. (2015) The transcriptional landscape of age in human peripheral blood. Nat. Commun., 6, 8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. O’Neill, K.M., Irwin, R.E., Mackin, S.J., Thursby, S.J., Thakur, A., Bertens, C., Masala, L., Loughery, J.E.P., McArt, D.G. and Walsh, C.P. (2018) Depletion of DNMT1 in differentiated human cells highlights key classes of sensitive genes and an interplay with polycomb repression. Epigenet. Chromatin, 11, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Manzo, M., Wirz, J., Ambrosi, C., Villaseñor, R., Roschitzki, B. and Baubec, T. (2017) Isoform-specific localization of DNMT3A regulates DNA methylation fidelity at bivalent CpG islands. EMBO J., 36, 3421–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Malan, V., Rajan, D., Thomas, S., Shaw, A.C., Louis Dit Picard, H., Layet, V., Till, M., Van Haeringen, A., Mortier, G., Nampoothiri, S.et al. (2010) Distinct effects of allelic NFIX mutations on nonsense-mediated mRNA decay engender either a sotos-like or a Marshall-smith syndrome. Am. J. Hum. Genet., 87, 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Takenouchi, T., Uehara, T., Kosaki, K. and Mizuno, S. (2018) Growth pattern of Rahman syndrome. Am. J. Med. Genet. Part A, 176, 712–714. [DOI] [PubMed] [Google Scholar]
- 70. Almuriekhi, M., Shintani, T., Fahiminiya, S., Fujikawa, A., Kuboyama, K., Takeuchi, Y., Nawaz, Z., Nadaf, J., Kamel, H., Kitam, A.K.et al. (2015) Loss-of-function mutation in APC2 causes sotos syndrome features. Cell Rep., 10, 1585–1598. [DOI] [PubMed] [Google Scholar]
- 71. Washbourne, P. (2014) Synapse assembly and neurodevelopmental disorders. Neuropsychopharmacology, 401, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gan, L., Yang, Y., Li, Q., Feng, Y., Liu, T. and Guo, W. (2018) Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential. Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential. Biomark. Res., 6, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sauvageau, M. and Sauvageau, G. (2010) Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell, 7, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Malta, T.M., Sokolov, A., Gentles, A.J., Burzykowski, T., Poisson, L., Weinstein, J.N., Kamińska, B., Huelsken, J., Omberg, L., Gevaert, O.et al. (2018) Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell, 173, 338–354.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rakyan, V.K., Down, T.A., Maslau, S., Andrew, T., Yang, T.P., Beyan, H., Whittaker, P., McCann, O.T., Finer, S., Valdes, A.M.et al. (2010) Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res., 20, 434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mozhui, K. and Pandey, A.K. (2017) Conserved effect of aging on DNA methylation and association with EZH2 polycomb protein in mice and humans. Mech. Ageing Dev., 162, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cheung, P., Vallania, F., Warsinske, H.C., Donato, M., Schaffert, S., Chang, S.E., Dvorak, M., Dekker, C.L., Davis, M.M., Utz, P.J.et al. (2018) Single-cell chromatin modification profiling reveals increased epigenetic variations with aging. Cell, 173, 1385–1397.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ni, Z., Ebata, A., Alipanahiramandi, E. and Lee, S.S. (2012) Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell, 11, 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sen, P., Shah, P.P., Nativio, R. and Berger, S.L. (2016) Epigenetic mechanisms of longevity and aging. Epigenetic mechanisms of longevity and aging. Cell, 166, 822–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gadalla, K.K.E., Vudhironarit, T., Hector, R.D., Sinnett, S., Bahey, N.G., Bailey, M.E.S., Gray, S.J. and Cobb, S.R. (2017) Development of a novel AAV gene therapy cassette with improved safety features and efficacy in a mouse model of Rett syndrome. Mol. Ther. Methods Clin. Dev., 5, 180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tărlungeanu, D.C. and Novarino, G. (2018) Genomics in neurodevelopmental disorders: an avenue to personalized medicine. Exp. Mol. Med., 508, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bray, N.L., Pimentel, H., Melsted, P. and Pachter, L. (2016) Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol., 34, 525–527. [DOI] [PubMed] [Google Scholar]
- 83. Soneson, C., Love, M.I. and Robinson, M.D. (2016) Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences [version 2; referees: 2 approved]. F1000Res., 10.12688/f1000research.7563.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Johnson, W.E., Li, C. and Rabinovic, A. (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 8, 118–127. [DOI] [PubMed] [Google Scholar]
- 85. Law, C.W., Chen, Y., Shi, W. and Smyth, G.K. (2014) Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol., 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ritchie, M.E., Phipson, B., Wu, D., Hu, Y., Law, C.W., Shi, W. and Smyth, G.K. (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res., 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zheng, H., Brennan, K., Hernaez, M. and Gevaert, O. (2019) Benchmark of long non-coding RNA quantification for RNA sequencing of cancer samples. Gigascience., 8, giz145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Li, B. and Dewey, C.N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics., 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bullard, J.H., Purdom, E., Hansen, K.D. and Dudoit, S. (2010) Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics., 11, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Weinstein, J.N., Collisson, E.A., Mills, G.B., Shaw, K.R.M., Ozenberger, B.A., Ellrott, K., Sander, C., Stuart, J.M., Chang, K., Creighton, C.J.et al. (2013) The cancer genome atlas pan-cancer analysis project. The cancer genome atlas pan-cancer analysis project. Nat. Genet., 45, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lawrence, M.S., Stojanov, P., Polak, P., Kryukov, G.V., Cibulskis, K., Sivachenko, A., Carter, S.L., Stewart, C., Mermel, C.H., Roberts, S.A.et al. (2013) Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature, 499, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mermel, C.H., Schumacher, S.E., Hill, B., Meyerson, M.L., Beroukhim, R. and Getz, G. (2011) GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol., 12, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Goldman, M.J., Craft, B., Hastie, M., Repečka, K., McDade, F., Kamath, A., Banerjee, A., Luo, Y., Rogers, D., Brooks, A.N.et al. (2020) Visualizing and interpreting cancer genomics data via the Xena platform. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol., 38, 675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Subramanian, A., Subramanian, A., Tamayo, P., Tamayo, P., Mootha, V.K., Mootha, V.K., Mukherjee, S., Mukherjee, S., Ebert, B.L., Ebert, B.L.et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A., 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Isserlin, R., Merico, D., Voisin, V. and Bader, G.D. (2014) Enrichment map - a Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Res, 3, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Newman, A.M., Steen, C.B., Liu, C.L., Gentles, A.J., Chaudhuri, A.A., Scherer, F., Khodadoust, M.S., Esfahani, M.S., Luca, B.A., Steiner, D.et al. (2019) Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol., 37, 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Newman, A.M., Liu, C.L., Green, M.R., Gentles, A.J., Feng, W., Xu, Y., Hoang, C.D., Diehn, M. and Alizadeh, A.A. (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Moran, S., Arribas, C. and Esteller, M. (2016) Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics., 8, 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Aryee, M.J., Jaffe, A.E., Corrada-Bravo, H., Ladd-Acosta, C., Feinberg, A.P., Hansen, K.D. and Irizarry, R.A. (2014) Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics, 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Pidsley, R., Wong, C.C.Y., Volta, M., Lunnon, K., Mill, J. and Schalkwyk, L.C. (2013) A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics, 14, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. McCartney, D.L., Walker, R.M., Morris, S.W., McIntosh, A.M., Porteous, D.J. and Evans, K.L. (2016) Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genomics Data., 9, 22–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hernandez Mora, J.R., Tayama, C., Sánchez-Delgado, M., Monteagudo-Sánchez, A., Hata, K., Ogata, T., Medrano, J., Poo-Llanillo, M.E., Simón, C., Moran, S.et al. (2018) Characterization of parent-of-origin methylation using the Illumina Infinium MethylationEPIC array platform. Epigenomics., 10, 941–954. [DOI] [PubMed] [Google Scholar]
- 103. Lawrence, M., Huber, W., Pagès, H., Aboyoun, P., Carlson, M., Gentleman, R., Morgan, M.T. and Carey, V.J. (2013) Software for computing and annotating genomic ranges. PLoS Comput. Biol., 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Quinlan, A.R. and Hall, I.M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zhou, W., Laird, P.W. and Shen, H. (2017) Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res., 45, gkw967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Dunham, I., Kundaje, A., Aldred, S.F., Collins, P.J., Davis, C.A., Doyle, F., Epstein, C.B., Frietze, S., Harrow, J., Kaul, R.et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature., 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hahsler, M., Chelluboina, S., Hornik, K. and Buchta, C. (2011) The arules R-package ecosystem: analyzing interesting patterns from large transaction data sets. J. Mach. Learn. Res., 12, 2021–2025. [Google Scholar]
- 108. Gel, B. and Serra, E. (2017) KaryoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics, 33, 3088–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Roadmap Epigenomics Consortium, Kundaje, A., Meuleman, W., Ernst, J., Bilenky, M., Yen, A., Heravi-Moussavi, A., Kheradpour, P., Zhang, Z., Wang, J.et al. (2015) Integrative analysis of 111 reference human epigenomes. Nature., 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.