Abstract
Purpose
We investigated the feasibility of measuring the hydronephrosis area to renal parenchyma (HARP) ratio from ultrasound images using a deep-learning network.
Materials and Methods
The coronal renal ultrasound images of 195 pediatric and adolescent patients who underwent pyeloplasty to repair ureteropelvic junction obstruction were retrospectively reviewed. After excluding cases without a representative longitudinal renal image, we used a dataset of 168 images for deep-learning segmentation. Ten novel networks, such as combinations of DeepLabV3+ and UNet++, were assessed for their ability to calculate hydronephrosis and kidney areas, and the ensemble method was applied for further improvement. By dividing the image set into four, cross-validation was conducted, and the segmentation performance of the deep-learning network was evaluated using sensitivity, specificity, and dice similarity coefficients by comparison with the manually traced area.
Results
All 10 networks and ensemble methods showed good visual correlation with the manually traced kidney and hydronephrosis areas. The dice similarity coefficient of the 10-model ensemble was 0.9108 on average, and the best 5-model ensemble had a dice similarity coefficient of 0.9113 on average. We included patients with severe hydronephrosis who underwent renal ultrasonography at a single institution; thus, external validation of our algorithm in a heterogeneous ultrasonography examination setup with a diverse set of instruments is recommended.
Conclusions
Deep-learning-based calculation of the HARP ratio is feasible and showed high accuracy for imaging of the severity of hydronephrosis using ultrasonography. This algorithm can help physicians make more accurate and reproducible diagnoses of hydronephrosis using ultrasonography.
Keywords: Congenital anomalies of kidney and urinary tract, Deep learning, Hydronephrosis, Ultrasonography
Graphical Abstract
INTRODUCTION
The most frequently used method for classifying hydronephrosis by ultrasonography is the Society for Fetal Urology (SFU) system or anteroposterior pelvic diameter (APD). However, the SFU system is well recognized for its shortcomings regarding its subjectivity and inconsistency in separating and reporting grades II and III hydronephrosis [1]. The APD varies according to hydration status, position, and bladder filling and sometimes misleads interpreters because of these dynamic variations [2]. As a more objective assessment tool for evaluating hydronephrosis, the combination of renal parenchymal area (RPA) and hydronephrosis area has been successfully implemented for predicting the necessity of surgical treatment or renal functional recovery after surgery [3]. In a recent study, the hydronephrosis area to renal parenchyma (HARP) ratio as a significant prognostic factor for renal functional deterioration after surgical treatment of ureteropelvic junction obstruction (UPJO) [4]. However, manually tracing the outline of the hydronephrotic kidney also creates the possibility for inter- and intra-operator variability [5].
Deep learning has demonstrated remarkable results in image data analysis, including kidney ultrasonography [6]. Recent advances in image segmentation, classification, and registration using deep-learning algorithms have shown the possibility of using deep-learning image interpretation as an adjunct or complementary to clinical interpretation of medical imaging by physicians [6]. The potential of deep-learning algorithms to overcome substantial inter- or intraobserver variability in kidney ultrasonography has been proposed in previous studies [6,7]. Lin et al. [8] reported the feasibility of deep-learning quantification of the HARP ratio in healthy subjects and pediatric patients with mild hydronephrosis without renal or urinary tract anomalies. However, deep-learning segmentation of the kidney and hydronephrosis areas on ultrasonography in pediatric patients with severe hydronephrosis due to UPJO has not been reported in the literature. Therefore, this study was designed to investigate the feasibility of measuring the HARP ratio from ultrasound images using a deep-learning network in pediatric cohorts with severe hydronephrosis due to UPJO.
MATERIALS AND METHODS
1. Study population and definition of variable
The study design was reviewed and approved by the Institutional Review Board of the Asan Medical Center (approval no. 2021-1612) in accordance with the Declaration of Helsinki. Informed consent was waived for this retrospective study by the institutional review board because all data were handled with anonymity. The medical records of 171 patients aged 0 to 18 years who underwent renal ultrasonography at our institution before and after pyeloplasty for UPJO between August 2002 and May 2016 were included in this study. Coronal renal ultrasound images with the greatest longitudinal dimension obtained before and after pyeloplasty were collected and analyzed for developing algorithms. We excluded images showing significant differences in image texture from deep-learning segmentation. Finally, 168 ultrasound images were included. The serum creatinine level within 2 days of pyeloplasty was collected. The estimated glomerular filtration rate (eGFR) was calculated using the following equation: eGFR=k×height (cm)/serum creatinine (mg/dL), where k=0.45 for infants, 0.70 for pubertal males, and 0.55 for all other children [9].
2. Sonography acquisition and label process
The manufacturer of the ultrasound systems was Philips (models: iU22, ATL HDI 5000, and EPIQ 5G; Bothell, WA, USA). All images were obtained in B-mode using abdominal convex probes with the patients in a supine position. The ultrasound images were cropped to fit in the ultrasonography field of view and then resized to a resolution of 512×512 pixels using zero padding. The boundaries of the dilated pelvicalyceal and renal regions on ultrasonography were manually annotated by an experienced urologist (SHS) using ImageJ software (National Institutes of Health, Bethesda, MD, USA, http://rsb.info.nih.gov/ij/). The RPA was estimated by subtracting the pelvicalyceal area from the total kidney area as pixel unit values. The HARP ratio was calculated by dividing the total hydronephrosis area by the RPA as previously demonstrated and designated as the ground truth HARP ratio [4]. Pixel information from the annotations was extracted using a custom Python script to build a labeled dataset for deep-learning segmentation models.
3. Networks
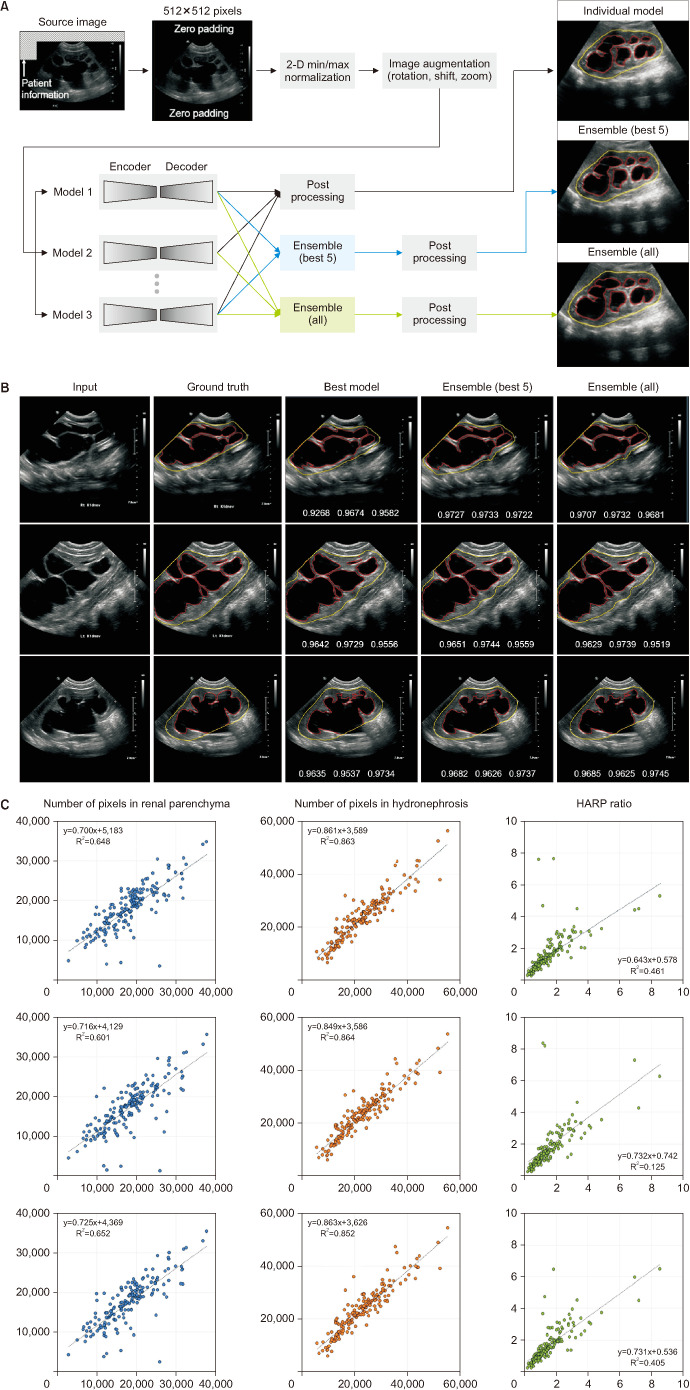
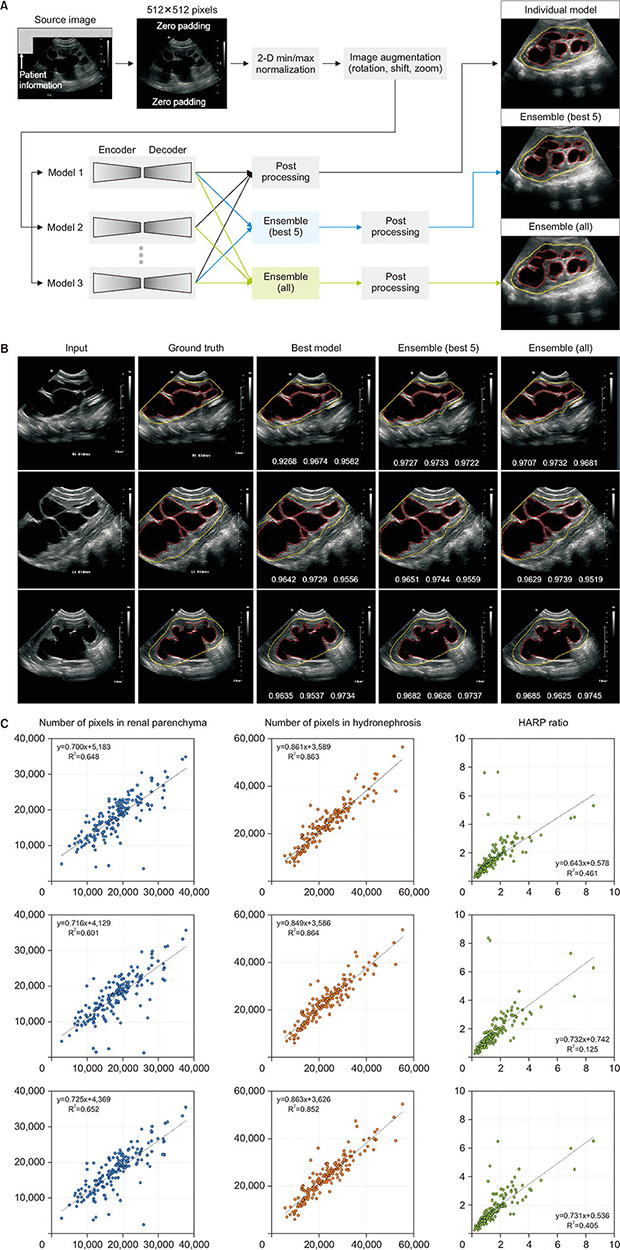
For the segmentation of the kidney contour and hydronephrosis area, 10 models were evaluated by combining novel networks, such as DeepLabV3+ [10] and UNet++ [11] (Fig. 1A). The combined outputs using a majority-voting ensemble of 10 networks [10,11,12,13,14,15,16,17,18,19,20] were also assessed. To determine the hydronephrosis and kidney areas, pixel-wise voting was performed using the outcomes from base models. With two sets of base models, ensemble models were constructed: all individual models and the five best-performing models.
Fig. 1. (A) Schematic diagram for deep-learning segmentation of ultrasonography. (B) Representative examples of hydronephrotic kidney segmentation. The boundaries of the kidney and hydronephrosis area are colored yellow and red, respectively. At the bottom of the images in the third to fifth columns, the dice similarity coefficients are presented in order of average, kidney boundary, and hydronephrosis area. The “best model” in the third column indicates the combination of DeepLabV3+ and EfficientNet-B4. (C) Scatter plots of deep-learning prediction (ensemble model) versus manually traced label: best model (combination of DeepLabV3+ and EfficientNet-B4 in the first row); ensemble of the five best models (second row); ensemble of all models (third row). HARP, hydronephrosis area to renal parenchyma.
Input images of 512×512 pixels were normalized using two-dimensional min/max normalization, and the initial weight was adopted from ImageNet for transfer learning, except for UNet++ [11]. The two output channels produced the probability maps for the kidney contour and hydronephrosis area, respectively. The nephrosis area predicted using deep-learning models was corrected to be bounded inside the kidney area. In the post-processing stage, the holes in the predicted masks were filled, and small blobs in the kidney area prediction were removed. Dice loss was applied for training the segmentation models, which was defined as 1 - dice similarity coefficient (DSC), where DSC=2×TP/(2×TP+FN+FP). TP, FN, and FP were the number of pixels corresponding to true positive, false negative, and false positive, respectively.
4. Training setup
Prediction models for the HARP ratio (predicted HARP) were trained for 400 epochs at maximum with a mini-batch size of 12. Data augmentation was performed using rotation (-20° to 20°), shift limit (0%–10% of image size in horizontal and vertical axes), and scale limit (0%–20%). For training, an Adam optimizer was applied with β1=0.9 and β2=0.999, and the learning rate, which was set to 10-3. The deep-learning networks were implemented in Python using PyTorch and trained on a workstation with Ryzen 5950X and two NVIDIA Geforce GTX 3090Ti graphics processing units. Four-fold cross-validation was performed to assess the sample bias. The first fold contained 42 images. In the cross-validation, the fold ratio of training, validation, and test sets was 2:1:1, and their composition was changed in the order of cyclic permutations.
5. Statistical analysis
Segmentation performance was measured using sensitivity, specificity, and DSC by comparing the manual annotations and the segmentations derived from deep-learning networks. Continuous data were reported as the mean±standard deviation and were compared using the Mann–Whitney U-test and Kruskal–Wallis test. Correlations were analyzed using Pearson’s correlation coefficient and demonstrated using scatter plots. Linear regression analysis was performed to estimate the regression equation. All data were calculated using PASW Statistics (version 18.0, SPSS Inc., Chicago, IL, USA).
RESULTS
1. Patient characteristics
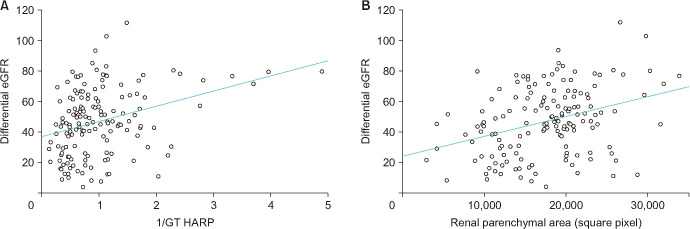
In this study, 168 patients with a mean age of 32.6 months were included. The mean APD was 31.6 mm (range, 7.0–73.0 mm), and the SFU grade was III in 19 patients (11.3%) and IV in 149 patients (88.7%) (Table 1). The mean APD and ground truth HARP ratio were significantly higher in patients with SFU grade IV hydronephrosis than in patients with SFU III (Fig. 2). The ground truth HARP ratio was inversely correlated with differential eGFR of the affected kidney with a Pearson coefficient of 0.327 (p<0.001), and the linear regression formula was as follows: y=9.97x+36.77. RPA was positively correlated with the differential eGFR with a Pearson correlation coefficient of 0.318 (p<0.001) and a linear regression prediction line of y=0.0013x+23.75 (Fig. 3).
Table 1. Patient characteristics (n=168).
| Variable | Value | |
|---|---|---|
| Age, mo | 32.6±54.6 | |
| Sex | ||
| Female | 42 (25.0) | |
| Male | 126 (75.0) | |
| Laterality | ||
| Right | 43 (25.6) | |
| Left | 125 (74.4) | |
| Society for Fetal Urology grade | ||
| III | 19 (11.3) | |
| IV | 149 (88.7) | |
| Anteroposterior pelvis diameter, mm | 31.6±12.5 | |
| Hydronephrosis area to renal parenchyma ratio | 1.35±0.89 | |
| Serum creatinine, mg/dL | 0.42±0.23 | |
| Estimated glomerular filtration rate, mL/min/1.73 m2 | 107.0±38.1 | |
| Differential renal function on renal scan, % | 46.6±22.1 | |
Values are presented as mean ± standard deviation or number (%).
Fig. 2. Bar graph of the mean anteroposterior pelvic diameter (APD) and hydronephrosis area to renal parenchyma (HARP) ratio in patients with Society for Fetal Urology (SFU) grade III and IV hydronephrosis. The mean APD, predicted HARP, and ground truth HARP were significantly different between patients with SFU grade III and IV. Asterisk indicates a statistical difference of the mean value in the Kruskal–Wallis test with a p-value of less than 0.01.
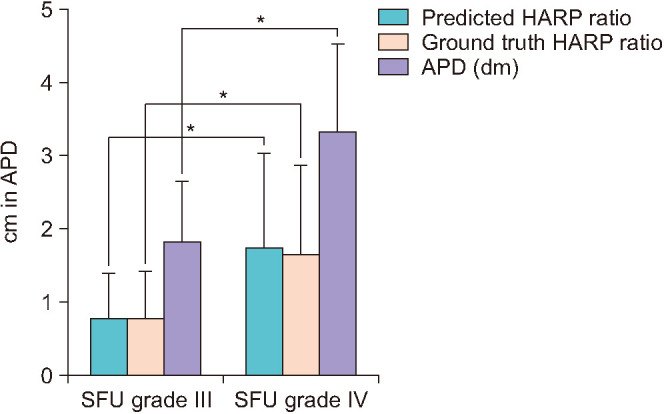
Fig. 3. Scatter plot of the ground truth (GT) hydronephrosis area to renal parenchyma (HARP) ratio and differential estimated glomerular filtration rate (eGFR) with a linear regression prediction line (y=9.97x+36.77) (A) and renal parenchymal area and the differential eGFR with a linear regression prediction line (y=0.0013x+23.75) (B). The Pearson correlation coefficients were 0.327 (p<0.001) (A) and 0.318 (p<0.001) (B).
2. Segmentation performance
Deep-learning network models and their prediction time are summarized in Table 2. The deep-learning models showed good visual correlation with the manually traced kidney and hydronephrosis areas (Fig. 1B). The relationship between the predicted and actual HARP ratios was visualized using a scatter plot (Fig. 1C). Three deep-learning models achieved DSCs higher than 0.9, and the remaining models scored higher than 0.87 (Table 3). DeepLabV3+ [10] with EfficientNet-B4 [12] scored the highest with an average DSC of 0.9087, of which the hydronephrosis segmentation and kidney outlines scored 0.8949 and 0.9226, respectively. The DSC of the 10-model ensemble was 0.9108 on average, and the best 5-model ensemble had a DSC of 0.9113 on average.
Table 2. Summary of deep-learning networks.
| No. | Encoder | Decoder/base architecture | Total parameter (million) | Prediction time per image (ms) |
|---|---|---|---|---|
| 1 | ResNet-34 [13] | PSPNet [14] | 21.44 | 46.7 |
| 2 | ResNet-34 [13] | LinkNet [15] | 21.77 | 52.8 |
| 3 | ResNet-34 [13] | DeepLabV3+ [10] | 22.43 | 51.6 |
| 4 | ResNet-34 [13] | UNet++ [11] | 26.07 | 55.9 |
| 5 | DenseNet-121 [16] | UNet [17] | 13.60 | 73.6 |
| 6 | Res2Net-50 [18] | UNet [17] | 31.63 | 62.1 |
| 7 | Xception [19] | DeepLabV3+ [10] | 37.77 | 67.1 |
| 8 | EfficientNet-B4 [12] | FPN [20] | 19.35 | 76.2 |
| 9 | EfficientNet-B4 [12] | DeepLabV3+ [10] | 18.62 | 68.1 |
| 10 | EfficientNet-B4 [12] | UNet++ [11] | 20.81 | 80.0 |
Table 3. Segmentation performance of deep-learning modelsa.
| Ranking | Network (encoder) | Average | Hydronephrosis in kidney | Kidney outline |
|---|---|---|---|---|
| 1 | DeepLabV3+ [10] (EfficientNet-B4 [12]) | 0.9087 | 0.8949 | 0.9226 |
| 2 | UNet [17] (Res2Net-50 [18]) | 0.9043 | 0.8915 | 0.9171 |
| 3 | FPN [20] (EfficientNet-B4 [12]) | 0.9027 | 0.8909 | 0.9146 |
| 4 | UNet [17] (DenseNet-121 [16]) | 0.89947 | 0.8855 | 0.9135 |
| 5 | UNet++ [11] (ResNet-34 [13]) | 0.89945 | 0.8839 | 0.9150 |
| 6 | LinkNet [15] (ResNet-34 [13]) | 0.8976 | 0.8815 | 0.9137 |
| 7 | DeepLabV3+ [10] (ResNet-34 [13]) | 0.8931 | 0.8853 | 0.901 |
| 8 | UNet++ [11] (EfficientNet-B4 [12]) | 0.8899 | 0.8864 | 0.8936 |
| 9 | PSPNet [14] (ResNet-34 [13]) | 0.8839 | 0.8717 | 0.896 |
| 10 | DeepLabV3+ [10] (Xception [19]) | 0.8714 | 0.8662 | 0.8767 |
| Ensemble (best 5) | 0.9113 | 0.8988 | 0.9239 | |
| Ensemble (all) | 0.9108 | 0.9000 | 0.9217 |
a:Models are ordered according to the ranking determined by the average dice similarity coefficient value.
DISCUSSION
To the best of our knowledge, this is the first study that has demonstrated the feasibility of using a deep-learning network for automatically calculating the HARP ratio by use of ultrasonography in children with UPJO. The deep-learning network model for automated segmentation of renal parenchymal and hydronephrosis areas showed excellent performance in terms of sensitivity, specificity, and accuracy in patients with severe hydronephrosis. Moreover, we achieved a DSC of 0.9 in the segmentation of hydronephrosis pelvicalyceal areas, which is the highest performance reported in the literature. Using this method, we may expect a more objective and accurate evaluation of hydronephrosis in patients with UPJO by minimizing the possibility of intra- and interobserver variability.
In studies on deep-learning algorithms to grade hydronephrosis severity, the performance was reported to be comparable to or less than that reported in this study. Smail et al. [7] explored the capability of deep convolutional neural networks to classify hydronephrosis according to the SFU system. They showed that the model discriminated low grades from high grades with an average DSC of 0.78. However, their model achieved an accuracy of 51% and an average DSC of 0.49 when it classified SFU grade in five scales (grade 0 to V). Lin et al. [8] used an attention-Unet to segment the kidney and pelvicalyceal system and showed a DSC of 0.92 and 0.83 for the kidney and pelvicalyceal system, respectively. However, they excluded patients with congenital kidney and urinary tract anomalies from their study. Therefore, their algorithm studied only minimal or mild hydronephrosis. In contrast, our study included patients who underwent pyeloplasty for UPJO, all of whom showed hydronephrosis with an average APD of 31.6 mm (range, 7.0–73 mm). Thus, we suggest that our deep-learning algorithm is more broadly applicable and predictive for the automated calculation of the HARP ratio in a wide range of hydronephrosis severity.
The prediction performance of the deep-learning models showed a larger deviation for the kidney area than for the hydronephrosis area. Because ultrasonography of high-grade hydronephrosis lacks the imaging features necessary to determine the renal boundary, the prediction errors were mostly found at the lower boundary of the kidney, affecting the predictability of the hydronephrosis area. The complex structures of hydronephrosis inside the kidney were accurately predicted, including small blobs. Among the individual models, the combination of DeepLabV3+ and EfficientNet-B4 exhibited the best performance in terms of DSC and demonstrated a high correlation with the manual traces in the HARP ratio (Fig. 1). From a practical perspective, the deep-learning model had advantages in terms of model size and prediction time (Table 2).
The ensemble method is a frequently used technique to ameliorate the segmentation performance by combining the prediction outcomes from multiple deep-learning models in various medical imaging modalities [21]. The ensemble application has been demonstrated to improve the accuracy of ultrasound images in diagnosing prostate [22] and breast cancers [23]. In this study, the highest DSCs for the kidney and pelvicalyceal system were achieved using ensemble models (i.e., 10 deep-learning models with different characteristics). The ensemble of the five best-performing models also showed the highest correlation with the manual trace in the prediction of hydronephrosis (Fig. 1). Adding a poor-performing model to the ensemble could degrade the prediction performance compared with that of a single model with the highest performance (kidney outline in Table 3) and increase the prediction time (Table 2).
The ultimate goal of monitoring patients with hydronephrosis and those who undergo surgical treatment for UPJO is to preserve renal function. The standard method for evaluating renal function, such as serum creatinine measurement and radioisotope renal scan, is invasive, which is a considerable hurdle, especially for children. Moreover, the eGFR may not be an accurate estimate for predicting recovery in patients with urinary tract obstruction [24]. The RPA on ultrasonography has been evaluated to detect reflux nephropathy, UPJO, and risk of end-stage renal disease in the posterior urethral valve and was suggested to be a surrogate predictor of renal function [25]. The HARP ratio is a parameter of the severity of hydronephrosis, in which RPA is a denominator in its calculation [4]. We demonstrated that the HARP ratio and RPA are similarly correlated with the differential eGFR (Fig. 2). However, the predictive ability of the HARP ratio for renal function estimation should be validated in a larger cohort.
This study indicated that the deep-learning calculation of the HARP ratio is feasible for pediatric patients with UPJO; however, there were several limitations. Inherent limitations of this study are its retrospective design and the limited sample size. Although we retrospectively collected longitudinal renal ultrasound images performed by multiple radiologists without standardizing the image acquisition protocol, we could minimize the selection bias by selecting specific images obtained to measure the maximal longitudinal diameter of the affected kidney. However, in some patients, images of the entire coronal kidney were not secured, and a small portion of the upper pole kidney was cropped off the ultrasonic window. Therefore, the HARP ratio may be exaggerated by omitting a small portion of the renal parenchyma. To overcome the sample size limitation, we used a four-fold cross-validation method. Nevertheless, a larger number of patients is necessary to validate the good predictive performance of deep-learning models. We included patients with severe hydronephrosis; therefore, the performance of the proposed algorithm should be validated in patients with lower-grade hydronephrosis. We manually selected coronal kidney images, which underwent preprocessing for training. For the further and wider application of this method for automatically calculating the HARP, an investigation of the feasibility for full automation of this process with real-time segmentation and calculation is warranted.
CONCLUSIONS
Deep-learning-based calculation of the HARP is feasible and showed high performance with high accuracy. This algorithm can help physicians make more accurate and reproducible diagnoses of the severity of hydronephrosis using ultrasonography.
ACKNOWLEDGMENTS
We thank Jung Eun Kim for her valuable contribution as an assistant research coordinator to this study.
Footnotes
CONFLICTS OF INTEREST: The authors have nothing to disclose.
FUNDING: This study was partially supported by a grant (no. 2017IT0536-1) from the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant no. HR21C0198).
- Research conception and design: Sang Hoon Song.
- Data acquisition: Sang Hoon Song, Jae Hyeon Han, Kun Suk Kim, Young Ah Cho, and Hye Jung Youn.
- Statistical analysis: Young In Kim and Jihoon Kweon.
- Data analysis and interpretation: Sang Hoon Song, Hye Jung Youn, Young In Kim, and Jihoon Kweon.
- Drafting of the manuscript: Sang Hoon Song and Jihoon Kweon.
- Critical revision of the manuscript: Sang Hoon Song, Young Ah Cho, Young In Kim, and Jihoon Kweon.
- Obtaining funding: Sang Hoon Song and Jihoon Kweon.
- Administrative, technical, or material support: Jae Hyeon Han, Kun Suk Kim, and Jihoon Kweon.
- Supervision: Jihoon Kweon.
- Approval of the final manuscript: all authors.
References
- 1.Nguyen HT, Benson CB, Bromley B, Campbell JB, Chow J, Coleman B, et al. Multidisciplinary consensus on the classification of prenatal and postnatal urinary tract dilation (UTD classification system) J Pediatr Urol. 2014;10:982–998. doi: 10.1016/j.jpurol.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Onen A. Grading of hydronephrosis: an ongoing challenge. Front Pediatr. 2020;8:458. doi: 10.3389/fped.2020.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rickard M, Lorenzo AJ, Braga LH. Renal parenchyma to hydronephrosis area ratio (PHAR) as a predictor of future surgical intervention for infants with high-grade prenatal hydronephrosis. Urology. 2017;101:85–89. doi: 10.1016/j.urology.2016.09.029. [DOI] [PubMed] [Google Scholar]
- 4.Han JH, Song SH, Lee JS, Nam W, Kim SJ, Park S, et al. Best ultrasound parameter for prediction of adverse renal function outcome after pyeloplasty. Int J Urol. 2020;27:775–782. doi: 10.1111/iju.14299. [DOI] [PubMed] [Google Scholar]
- 5.Wieczorek AP, Woźniak MM, Tyloch JF. Errors in the ultrasound diagnosis of the kidneys, ureters and urinary bladder. J Ultrason. 2013;13:308–318. doi: 10.15557/JoU.2013.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo CC, Chang CM, Liu KT, Lin WK, Chiang HY, Chung CW, et al. Automation of the kidney function prediction and classification through ultrasound-based kidney imaging using deep learning. NPJ Digit Med. 2019;2:29. doi: 10.1038/s41746-019-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smail LC, Dhindsa K, Braga LH, Becker S, Sonnadara RR. Using deep learning algorithms to grade hydronephrosis severity: toward a clinical adjunct. Front Pediatr. 2020;8:1. doi: 10.3389/fped.2020.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin Y, Khong PL, Zou Z, Cao P. Evaluation of pediatric hydronephrosis using deep learning quantification of fluid-to-kidney-area ratio by ultrasonography. Abdom Radiol (NY) 2021;46:5229–5239. doi: 10.1007/s00261-021-03201-w. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz GJ, Haycock GB, Edelmann CM, Jr, Spitzer A. A simple estimate of glomerular filtration rate in children derived from body length and plasma creatinine. Pediatrics. 1976;58:259–263. [PubMed] [Google Scholar]
- 10.Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H. Encoder-decoder with atrous separable convolution for semantic image segmentation [abstract]; 15th European Conference on Computer Vision (ECCV); 2018 Sept 8-14; Munich, Germany. Cham: Springer; 2018. pp. 833–851. [Google Scholar]
- 11.Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J. UNet++: a nested U-Net architecture for medical image segmentation. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support (2018) 2018;11045:3–11. doi: 10.1007/978-3-030-00889-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan M, Le Q. EfficientNet: rethinking model scaling for convolutional neural networks [abstract] Proc Mach Learn Res. 2019;97:6105–6114. [Google Scholar]
- 13.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition [abstract]; 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27-30; Las Vegas, USA. Piscataway: IEEE; 2016. pp. 770–778. [Google Scholar]
- 14.Zhao H, Shi J, Qi X, Wang X, Jia J. Pyramid scene parsing network [abstract]; 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21-26; Honolulu, USA. Piscataway: IEEE; 2017. pp. 6230–6239. [Google Scholar]
- 15.Chaurasia A, Culurciello E. LinkNet: exploiting encoder representations for efficient semantic segmentation [abstract]; 2017 IEEE Visual Communications and Image Processing (VCIP); 2017 Dec 10-13; St. Petersburg, USA. Piscataway: IEEE; 2017. pp. 1–4. [Google Scholar]
- 16.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks [abstract]; 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21-26; Honolulu, USA. Piscataway: IEEE; 2017. pp. 2261–2269. [Google Scholar]
- 17.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation [abstract]; 18th International Conference on Medical Image Computing and Computer-Assisted Intervention; 2015 Oct 5-9; Munich, Germany. Cham: Springer International Publishing; 2015. pp. 234–241. [Google Scholar]
- 18.Gao SH, Cheng MM, Zhao K, Zhang XY, Yang MH, Torr P. Res2Net: a new multi-scale backbone architecture. IEEE Trans Pattern Anal Mach Intell. 2021;43:652–662. doi: 10.1109/TPAMI.2019.2938758. [DOI] [PubMed] [Google Scholar]
- 19.Chollet F. Xception: deep learning with depthwise separable convolutions [abstract]; 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21-26; Honolulu, USA. Piscataway: IEEE; 2017. pp. 1800–1807. [Google Scholar]
- 20.Lin TY, Dollár P, Girshick R, He K, Hariharan B, Belongie S. Feature pyramid networks for object detection [abstract]; 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21-26; Honolulu, USA. Piscataway: IEEE; 2017. pp. 936–944. [Google Scholar]
- 21.Weisman AJ, Kieler MW, Perlman SB, Hutchings M, Jeraj R, Kostakoglu L, et al. Convolutional neural networks for automated PET/CT detection of diseased lymph node burden in patients with lymphoma. Radiol Artif Intell. 2020;2:e200016. doi: 10.1148/ryai.2020200016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karimi D, Zeng Q, Mathur P, Avinash A, Mahdavi S, Spadinger I, et al. Accurate and robust deep learning-based segmentation of the prostate clinical target volume in ultrasound images. Med Image Anal. 2019;57:186–196. doi: 10.1016/j.media.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Moon WK, Lee YW, Ke HH, Lee SH, Huang CS, Chang RF. Computer-aided diagnosis of breast ultrasound images using ensemble learning from convolutional neural networks. Comput Methods Programs Biomed. 2020;190:105361. doi: 10.1016/j.cmpb.2020.105361. [DOI] [PubMed] [Google Scholar]
- 24.Thompson A, Gough DC. The use of renal scintigraphy in assessing the potential for recovery in the obstructed renal tract in children. BJU Int. 2001;87:853–856. doi: 10.1046/j.1464-410x.2001.02213.x. [DOI] [PubMed] [Google Scholar]
- 25.Pulido JE, Furth SL, Zderic SA, Canning DA, Tasian GE. Renal parenchymal area and risk of ESRD in boys with posterior urethral valves. Clin J Am Soc Nephrol. 2014;9:499–505. doi: 10.2215/CJN.08700813. [DOI] [PMC free article] [PubMed] [Google Scholar]