Abstract
Efficient host-vector systems have been developed for the versatile, strictly anaerobic, halo- and fumarate-respiring gram-positive bacterium Desulfitobacterium dehalogenans. An electroporation-based transformation procedure resulting in approximately 103 to 104 transformants per μg of the cloning vector pIL253 was developed and validated. The broad-host-range vector pG+host9 was shown to replicate at a permissive temperature of 30°C, whereas the replicon was not functional at 40°C. The D. dehalogenans frdCAB operon, predicted to encode a fumarate reductase, was cloned, characterized, and targeted for insertional inactivation by pG+host9 carrying a 0.6-kb internal frdA fragment. Single-crossover integration at the frdA locus occurred at a frequency of 3.3 × 10−4 per cell and resulted in partially impaired fumarate reductase activity. The gene cloning and inactivation systems described here provide a solid basis for the further elucidation of the halorespiratory network in D. dehalogenans and allow for its further exploitation as a dedicated degrader.
It has been shown for a wide range of haloorganic compounds that reductive dechlorination is the first crucial step in the degradation of such pollutants (15, 25). Halorespiring bacteria have received increasing attention during the past decade due to a significant contribution to reductive dehalogenation processes occurring in anoxic polluted environments such as soils, aquifers, and sediments (14, 24). In contrast to the cometabolic reductive dehalogenation catalyzed by various metal-containing tetrapyrrol cofactors in a variety of anaerobic bacteria, this reaction is catalyzed at much higher rates by specific enzymes in halorespiring microbes, where it is coupled to energy conservation by electron transport-coupled phosphorylation (14, 18, 31). One of these strains is the versatile, low-G+C, gram-positive bacterium Desulfitobacterium dehalogenans, which is able to link the oxidation of several electron donors such as hydrogen, formate, lactate, and pyruvate to the reduction of various organic and inorganic acceptors, including ortho-chlorinated phenols (o-CP), fumarate, and nitrate (37). Recently, the o-CP-reductive dehalogenase (CPR) from D. dehalogenans has been purified and characterized at the biochemical and genetic levels (33, 39). Comparison with other chloroalkene- and haloaromate-reductive dehalogenases isolated and characterized from various phylogenetically distinct halorespiring bacteria indicated that these enzymes share significant similarities in both structural and functional properties, suggesting that they constitute a novel class of corrinoid-containing reductases (for recent reviews, see references 18 and 31).
The detailed molecular analysis of the cpr gene cluster in D. dehalogenans led to the identification of genes encoding putative regulatory proteins and protein-folding catalysts, the transcription of which was specifically induced under halorespiring conditions. From these results, their potential involvement in regulation and maturation of the reductive dehalogenase complex has been suggested (33). Additional genomic loci that appear essential for functional o-CP respiration of D. dehalogenans have been identified by means of random chromosomal integration of the conjugational transposon Tn916 (32). Nevertheless, detailed structural and functional analysis of these proteins has been hampered by the absence of genetic techniques for D. dehalogenans, including transformation, gene cloning, and specific gene disruption and insertion. Moreover, the development of such genetic modification tools would also enable the design of strains with improved performance in the bioremediation of polluted environments (19, 35).
Host-vector systems that allow for the genetic, metabolic, and protein engineering of low-G+C gram-positive bacteria (LGB) have been developed and optimized mainly for industrially applied strains of lactic acid bacteria, for bacilli, and, to a lesser extent, for clostridia (for reviews see references (10 to 12 and 41). It has been shown that vectors based on the theta replicon of the broad-host-range conjugative plasmid pAMβ1 (6), among which are the cloning vectors pIL252 and pIL253, are functional in all genera of LGB studied, indicating their potential use for halorespiring genera of LGB, such as Desulfitobacterium and Dehalobacter (12, 30). Similarly, vectors of the pG+host series of thermosensitive derivatives of yet another broad-host-range plasmid, pWV01, have been proven to be instrumental for high-efficiency gene inactivation, replacement, and insertional mutagenesis, especially in poorly transformable LGB (2, 12, 22, 23).
The main objectives of this study were (i) to develop an efficient protocol for the transformation of D. dehalogenans, (ii) to investigate the suitability of gene transfer systems previously developed for other LGB, (iii) to confirm temperature-sensitive replication of pG+host9 in D. dehalogenans, and (iv) to demonstrate its applicability for specific gene disruption using the putative fumarate reductase-encoding frdA gene as a model target.
MATERIALS AND METHODS
Materials.
All gases were obtained from Hoek Loos (Schiedam, The Netherlands). When appropriate, experiments were carried out in an anaerobic glove box (Coy Laboratory Products, Grass Lake, Mich.) under an atmosphere of 96% N2 and 4% H2. The oxygen concentration was kept low with the palladium catalyst RO-20, provided by BASF (Arnhem, The Netherlands).
Bacterial strains, plasmids, and culture conditions.
D. dehalogenans strain JW/IU-DC1 (DSM 9161) (37) was routinely grown under anaerobic conditions (gas phase, 100% N2) at 37°C in basal mineral medium as described by Neumann et al. (26), supplemented with 0.1% peptone, 30 mM NaHCO3, and trace elements and vitamin solution as recommended by the German Collection of Microorganisms and Cell Cultures (Brunswick, Germany). An electron donor and an electron acceptor were added to the appropriate concentrations from sterile anaerobic stock solutions.
Strains of Escherichia coli were grown in Luria-Bertani medium at 37°C (28). E. coli XL1-Blue (Stratagene, La Jolla, Calif.) was generally used as a host for cloning vectors. As a host for the rolling-circle pG+host vectors, E. coli MC1061 (8) was used. Lactococcus lactis MG1614 (16) was grown at 30°C in M17 broth (Difco, Detroit, Mich.) supplemented with 0.5% glucose (GM17). Where appropriate, media were amended with ampicillin (100 μg/ml) or erythromycin (150 μg/ml for E. coli, 10 μg/ml for L. lactis, and 5 μg/ml for D. dehalogenans). The MIC of erythromycin for D. dehalogenans was determined on plates containing 0 to 5 μg of erythromycin/ml and 20 mM lactate and fumarate as the electron donor and electron acceptor, respectively.
The cloning vectors pUC18 and pUC19 were purchased from Amersham Pharmacia Biotech (Uppsala, Sweden), and the PCR product cloning vectors pGEM-T and pMON38201 (3) were obtained from Promega (Madison, Wis.) and Monsanto (St. Louis, Mo.), respectively. Plasmids pIL253 (30) and pG+host9 (23) were kindly provided by Richard van Kranenburg (NIZO Food Research, Ede, The Netherlands) and Emmanuelle Maguin (Laboratoire de Génetique Microbienne, Institut National de la Recherche Agronomique, Jouy en Josas Cedex, France).
DNA isolation and manipulation.
Total DNA of D. dehalogenans was isolated as described previously (39). Plasmid DNA was isolated from E. coli by using the alkaline lysis method, and standard DNA manipulations were performed according to established procedures (28) and manufacturers' instructions. Isolation of plasmid DNA from L. lactis was performed as described previously (13). L. lactis was transformed according to the method of Wells et al. (40). Large-scale preparations of plasmid DNA (pIL253, pG+host9) were purified by CsCl density gradient centrifugation (28).
Enzymes were purchased from Life Technologies B.V. (Breda, The Netherlands), Roche Molecular Biochemicals (Mannheim, Germany), or New England Biolabs (Beverly, Mass.). Oligonucleotides were obtained from Eurogentec (Seraing, Belgium), Life Technologies Inc., and MWG Biotech (Ebersberg, Germany). PCR products were purified prior to subsequent manipulation using the QIAquick PCR purification kit (Qiagen GmbH, Hilden, Germany).
Transformation procedure and competence of D. dehalogenans.
For the transformation of D. dehalogenans, the electroporation-based method described by Wells et al. (40) was modified and optimized for use with anaerobic bacteria. Cells of D. dehalogenans were grown in the presence of 40 mM glycine with 20 mM lactate as the electron donor and 20 mM fumarate as the electron acceptor. Unless otherwise indicated, all subsequent steps were carried out in the anaerobic glove box. Exponentially growing cells were harvested at an A600 of approximately 0.2 by centrifugation at 2,600 × g for 10 min at 4°C and then resuspended in 0.15 volume of ice-cold anaerobic washing buffer (0.5 M sucrose–10% glycerol). Cells were recovered by centrifugation at 4,000 × g for 10 min at 4°C, washed with 0.05 volume of washing buffer, recentrifuged, and finally resuspended in 0.001 volume of washing buffer. For electroporation, DNA was added in 0.5 to 1.0 μl of deionized water to 40 μl of concentrated cell suspension and transferred to precooled 0.2-cm electroporation cuvettes. A single pulse was applied outside the glove box at different settings (field strength, 12.5 kV · cm−1; capacitance, 25 μF; resistance, 200 to 800 Ω) using a Gene Pulser (Bio-Rad, Hercules, Calif.). Immediately after electroporation, cells were moved back into the anaerobic glove box, mixed with 0.96 ml of recovery medium (growth medium containing 20 mM lactate and fumarate and 0.5 M sucrose), and incubated at 37°C for 5 h. To determine the influence of the transformation procedure on the viability of D. dehalogenans, appropriate dilutions were inoculated onto plates without erythromycin containing 20 mM lactate and fumarate as described previously (32). Transformants were selected on plates containing 20 mM lactate and fumarate and 5 μg of erythromycin/ml. Further subcultivation of single colonies in liquid medium was performed as described previously (32). Plasmid DNA was isolated from D. dehalogenans using a protocol modified from reference 13. Briefly, protoplasts were prepared from 12 ml of early-stationary-phase culture in 250 μl of THMS buffer (30 mM Tris-HCl [pH 8.0] and 3 mM MgCl2 in 25% sucrose) containing 20 mg of lysozyme/ml (38). Subsequently, plasmid DNA was purified by alkaline lysis and recovered by isopropanol precipitation. Agarose gel electrophoresis and Southern blot analysis were used to check for the presence of plasmids.
Thermosensitivity of pG+host9 in D. dehalogenans.
To determine the segregational stability of the thermosensitive vector pG+host9 in D. dehalogenans, an early-stationary-phase culture of plasmid-carrying D. dehalogenans that was grown in the presence of 40 mM pyruvate and 5 μg of erythromycin/ml at 30°C was diluted 40-fold into medium without antibiotics and incubated at 30°C to stationary phase (0 h). This culture was then diluted 100-fold into fresh medium without antibiotics and incubated at 30, 37, and 40°C. Appropriate dilutions were inoculated onto plates with or without erythromycin (5 μg/ml) at 0.1, 16, and 40 h after dilution and were incubated at 30°C. After 40 h of growth, all cultures had reached stationary phase. Cultures were again diluted 10-fold and kept for an additional 24 h at the respective temperatures until stationary phase was reached (68 h). Total DNA was isolated from samples taken at 0, 40, and 68 h, digested with EcoRI, and analyzed by Southern blot analysis. Linearized pG+host9 was used as a plasmid-specific probe for hybridization. Hybridization with a probe specific for the D. dehalogenans frdAC genes was used as an internal standard. This probe was a PCR product obtained with primers BG355 (positions 942 to 974 of the frd gene cluster) and IK04 [5′-(A/G)TG NGC NCC NC(G/T) NS(A/T) (C/T)TC-3′; positions 3157 to 3140 of the frd gene cluster] (see below and Fig. 3). A Hybond-N+ nylon transfer membrane (Amersham Life Science, Little Chalfont, United Kingdom) was used for Southern blot analysis, and probes for hybridization experiments were labeled by nick translation in the presence of [α-32P]dATP (Amersham Pharmacia Biotech).
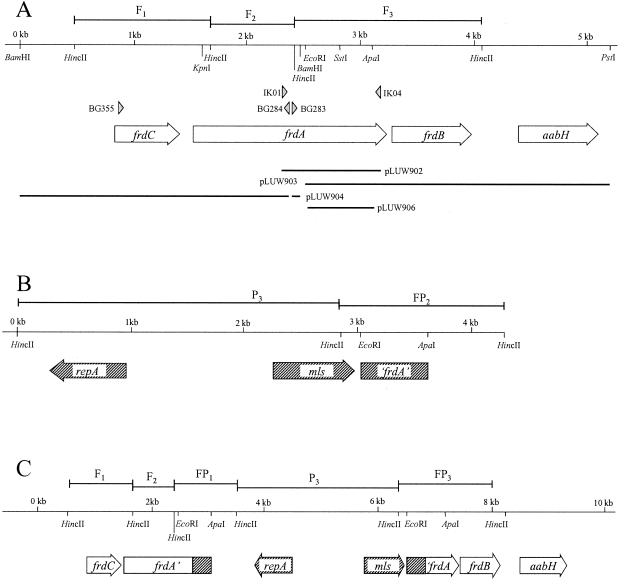
FIG. 3.
Physical maps of the D. halogenans frd gene locus (A), pLUW906 (linearized) (B), and the frd gene locus after recombination (C). Horizontal arrows, open reading frames; triangles, oligonucleotides used in this study; horizontal lines, clones, constructs, and hybridizing fragments (F1 to F3, fragments hybridizing solely with the frdAC-specific probe; P1 to P3 fragments hybridizing solely with the pG+host9-specific probe: FP1 to FP3, fragments hybridizing with both probes). DNA restriction sites which were relevant for the construction of clones and constructs are indicated.
Cloning of a putative fumarate reductase-encoding operon.
The degenerated primers IK01 [5′-GA(A/G) (A/G/T)(G/C)N (G/T)(G/C)N A/C)GN GGN GAN GGN GG-3′; positions 2312 to 2334] and IK04, which were designed based on an amino acid sequence alignment of known bacterial fumarate reductases, were used to PCR amplify a fragment of a putative fumarate reductase-encoding operon from the chromosomal DNA of D. dehalogenans. The resulting 0.85-kb PCR product was cloned in E. coli using XcmI-digested pMON38201, yielding pLUW902. Subsequently, Southern blot analysis of PstI-EcoRI-digestedchromosomal DNA of D. dehalogenans revealed a 3-kb fragment that strongly hybridized with the radiolabeled 0.85-kb PCR product. The 3-kb fragment was cloned in E. coli using PstI-EcoRI-digested pUC18, resulting in pLUW903. pLUW904 was obtained by inverse PCR (36) that was performed as described previously (39) with BamHI-digested and self-ligated chromosomal DNA of D. dehalogenans by using the divergent primer pair BG283 and BG284 (positions 2456 to 2477 and positions 2356 to 2335, respectively).
Plasmid constructions and single-crossover integration into the D. dehalogenans chromosome.
A 578-bp ApaI-EcoRI internal fragment of the D. dehalogenans frdA gene was cloned in E. coli MC1061 using ApaI-EcoRI-digested pG+host9, yielding pLUW906. Subsequently, electrocompetent cells of D. dehalogenans were transformed with plasmid DNA isolated from E. coli MC1061 using a QAprep Spin Miniprep kit (Qiagen GmbH). Recovery after electroporation and cultivation on selective plates were performed at 30°C. Erythromycin-resistant colonies that appeared within 5 days were transferred to liquid selective medium containing 40 mM pyruvate and were incubated at 30°C. Cultures were diluted 20-fold in the same medium, grown at 30°C for 8 h to reach log phase, and then shifted to 40°C for 16 h (3 to 5 generations). Appropriate dilutions were incubated on plates in the presence of 20 mM pyruvate and erythromycin at 40°C in order to detect integration events and on nonselective plates at 40°C for the determination of viable cell counts. The ratio of the two counts was used to determine the frequency of integration per cell as described by Biswas et al. (2). Integrants that were isolated at 40°C were subsequently routinely maintained in selective medium containing 20 to 40 mM pyruvate. Southern blot analysis of HincII-digested chromosomal and plasmid DNA and preparation of pG+host9- and D. dehalogenans frdAC-specific probes were performed as described above.
DNA sequencing and sequence analysis.
DNA sequencing was performed using a LiCor (Lincoln, Nebr.) DNA sequencer 4000L. Plasmid DNA used for sequencing reactions was purified with the QIAprep Spin Miniprep kit (Qiagen GmbH). Reactions were performed using the Thermo Sequenase fluorescent-labeled primer cycle sequencing kit (Amersham Pharmacia Biotech). Fluorescently (IRD 800) labeled universal sequencing primers were purchased from MWG Biotech. Sequence similarity searches and alignments were performed using the BLAST 2.0 program (1) (National Center for Biotechnology Information, Bethesda, Md.) and the Clustal X (34) and GeneDoc (K. B. Nicholas and H. B. J. Nicholas, GeneDoc: a tool for editing multiple sequence alignments, 1997) programs and DNAstar package (DNASTAR Inc., Madison, Wis.), respectively.
Enzyme and protein assays.
Harvesting of cells and preparation of cell extracts by sonication under anoxic conditions were performed as described previously (39). Fumarate reductase activities were determined spectrophotometrically at 30°C in 1 ml of 100 mM Tris-HCl (pH 8.0) as described previously (32). One unit of enzyme activity corresponds to the amount of enzyme catalyzing the conversion of 1 μmol of substrate or 2 μmol of benzyl viologen per min. Succinate dehydrogenase activity was measured with 2,6-dichlorophenolindophenol and phenazine methosulfate as an artificial electron acceptor as described by Schirawski and Unden (29). Protein was determined according to the method of Bradford, with bovine serum albumin as the standard (5).
Nucleotide sequence accession number.
The nucleotide sequence of the putative fumarate reductase-encoding operon has been deposited in GenBank under accession no. AF299117.
RESULTS
Development of an electroporation-based transformation protocol for D. dehalogenans.
To allow for the application of plasmid vector systems for genetic manipulation of the strict anaerobe D. dehalogenans, an electroporation-based transformation protocol for this bacterium was designed and optimized using the promiscuous plasmid pIL253 (30). Because this cloning vector, which is a derivative of the broad-host-range thetareplicating plasmid pAMβ1, carries an erythromycin resistance marker, we checked D. dehalogenans for its sensitivity to this antibiotic. On plates that contained 0.1 μg of erythromycin/ml, 6.5 × 106 CFU/ml was obtained, compared to 4 × 107 CFU/ml on plates without any antibiotic. At erythromycin concentrations of 0.25 and 0.5 μg/ml, microcolonies colonies appeared after 4 days, whereas no colonies developed at concentrations of ≥1 μg of erythromycin/ml, indicating that the frequency of spontaneous resistance to erythromycin is below 2.5 × 10−7 per CFU. Subsequently, a concentration of 5 μg of erythromycin/ml was used in solid and liquid media for the selection of strains of D. dehalogenans carrying the erythromycin resistance marker.
Electrocompetent cells of D. dehalogenans were prepared from exponential-phase cells that had grown in the presence of the cell wall-weakening agent glycine as described by Wells et al. (40). Cells were washed and finally concentrated approximately 1,000-fold in ice-cold anaerobic washing buffer. On average, approximately 70% of the cells could be recovered as viable CFU on nonselective plates after the cell collection and washing procedure. Electroporation of 40-μl aliquots of concentrated cell suspension in the presence or absence of different amounts of plasmid DNA was performed outside the anaerobic chamber at a field strength of 12.5 kV · cm−1, a capacitance of 25 μF, and a resistance of 200 to 800 Ω. After a subsequent incubation of 5 h in the presence of 0.5 M sucrose, cells were inoculated onto plates with or without 5 μg of erythromycin/ml. After 3 days of incubation at 37°C, colonies were counted to determine survival and transformation efficiency. The pulse resulted in a 40 to 65% decrease in CFU on nonselective plates with decreasing resistance compared to the survival of an aliquot of concentrated cells that was kept inside the anaerobic chamber and was not subjected to a pulse (Fig. 1). The highest numbers of transformants were obtained at a resistance of 400 Ω, resulting in a pulse time constant of approximately 7.5 ms. Both shorter and longer pulse times (200 Ω, 4.7 ms; 600 Ω, 12.8 ms; 800 Ω, 16.4 ms) resulted in significantly lower numbers of transformants (Fig. 1). Routinely, 3,000 ± 1,900 transformants (maximal value, 6.6 × 103) were obtained per μg of CsCl-purified pIL253, independent of the amount of plasmid DNA (ranging from 50 to 800 ng) used in the electrotransformation. In order to check the transformants for the presence and concentration of plasmid, erythromycin-resistant colonies were transferred to liquid selective medium containing 20 mM lactate and fumarate, and plasmid DNA was isolated from early-stationary-phase cultures (A600 = 0.25). Plasmid DNA was detectable by agarose gel electrophoresis, and quantification indicated a concentration of 5 ng of pIL253/ml of culture, corresponding to approximately 10 copies per cell (data not shown).
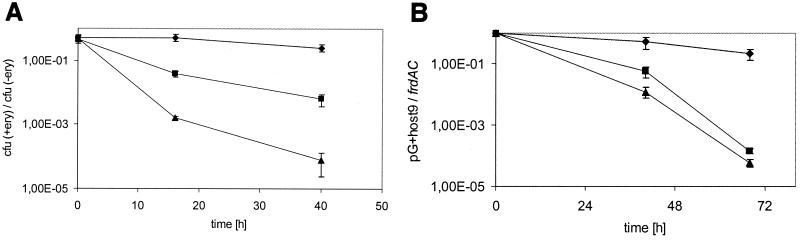
FIG. 1.
Survival and transformation efficiency of cells of D. dehalogenans after electroporation. Survival was defined as CFU per milliliter of cell suspension on nonselective plates (⧫). Efficiency of transformation of D. dehalogenans was determined on plates containing 5 μg of erythromycin/ml and calculated as CFU per microgram of CsCl-purified pIL253 (■).
Segregational stability and thermosensitivity of pG+host9 in D. dehalogenans.
The thermosensitive broad-host-range pG+host vector family has been shown to be instrumental for high-efficiency gene inactivation and replacement in gram-positive bacteria (2). In order to study the applicability of this system in the halorespiring bacterium D. dehalogenans, electrocompetent cells were transformed with CsCl-purified pG+host9. To ensure functional replication, posttransformation incubation and cultivation on selective media were performed at 30°C. Transformation yielded, on average, 600 transformants per μg of plasmid DNA. Colonies that appeared on selective plates were transferred to liquid medium. Plasmid DNA was isolated from early-stationary-phase cultures and could be detected by agarose gel electrophoresis (data not shown). In order to determine the permissive and nonpermissive temperatures for the replication of pG+host9 in D. dehalogenans, the segregational stability of the plasmid at nonselective concentrations of erythromycin was analyzed at different temperatures. A culture of D. dehalogenans containing the plasmid was diluted into fresh medium without any antibiotic and incubated at 30, 37, and 40°C. The ratio of the CFU on selective plates to the CFU on nonselective plates at 30°C was determined at 0, 16, and 40 h after dilution. Whereas this ratio decreased only 50% for the culture that was incubated at 30°C (0.51 at 0 h and 0.26 at 40 h), it dropped 72- and 48,000-fold at 37 and 40°C, respectively, within 7 generations (Fig. 2A). No influence of the incubation temperature on segregational stability was observed in the case of the nonthermosensitive plasmid pIL253 (data not shown). Similar results were obtained by Southern blot analysis of total DNA that was isolated before and 7 (40 h) and 10 generations (68 h) after the shift to nonselective conditions, respectively (Fig. 2B). The amount of plasmid-derived sequences detected following growth for 10 generations at 37 or 40°C was found to be more than 1,000-fold lower than that detected in cells grown at 30°C.
FIG. 2.
Segregational stability and thermosensitivity of pG+host9 in D. dehalogenans under nonselective culture conditions at 30°C (⧫), 37°C (■), and 40°C (▴). (A) Ratio of CFU on selective and nonselective plates. (B) Normalized ratio of hybridization signal intensities obtained with probes specific for pG+host9 and frdAC.
Cloning and sequence analysis of a putative fumarate reductase-encoding frdBAC gene cluster.
The versatile gram-positive anaerobe D. dehalogenans has the ability to utilize fumarate as the terminal electron acceptor for anaerobic respiration with H2, formate, lactate, or pyruvate as the electron donor. High fumarate reductase activity is readily detectable in cell extracts of D. dehalogenans grown in the presence of fumarate or yeast extract (32, 37). In order to provide an easy-to-screen target gene for the development of genetic modification approaches, we amplified a 0.85-kb fragment from the chromosome of D. dehalogenans using degenerated primers that were designed based on a primary sequence alignment of known succinate:quinone oxidoreductases (17). Sequence analysis indicated significant similarity with the flavoproteins of fumarate reductases and succinate dehydrogenases present in the databases. The subsequent isolation and analysis of a 5.3-kb PstI-BamHI chromosomal fragment from D. dehalogenans revealed the presence of three closely linked genes, frdCAB, and a fourth open reading frame, aabH, potentially encoding a polypeptide with significant similarity to ATP-binding cassette transporter binding proteins. The three genes frdC, frdA, and frdB potentially code for polypeptides of 208, 578, and 251 amino acids with calculated molecular masses of 23,728, 64,441, and 28,043 Da, respectively (Fig. 3A). The predicted gene products exhibit significant similarities with the type B membrane anchor, flavoprotein, and iron-sulfur-protein subunits of known succinate:quinone oxidoreductases, respectively (17). The highest similarities were found with the succinate dehydrogenases of Bacillus subtilis and Paenibacillus macerans (identities on the amino acid level, 72 and 74% for FrdA, 58 and 59% for FrdB, and 42 and 29% for FrdC, respectively). Upstream of each of the genes, potential Shine Dalgarno sequences that are complementary to the 3′ end of the D. dehalogenans 16S rRNA (33) could be identified (data not shown).
Gene-specific single-crossover integration in the D. dehalogenans chromosome.
An internal 0.6-kb fragment of the D. dehalogenans frdA gene was cloned into pG+host9 in E. coli MC1061. The resulting plasmid, pLUW906 (Fig. 3B), was introduced by transformation into D. dehalogenans, where it was stably maintained at 30°C. Subsequently, cultures of D. dehalogenans containing either pLUW906 or pG+host9 were shifted to 40°C to induce single-crossover or spontaneous chromosomal integration, respectively. Single-crossover integration at the frdA locus would result in the generation of two, chromosomal copies of the frdA gene, truncated at either the 3′ or the 5′ end, and interrupted by the vector (Fig. 3C). Integrants were selected as erythromycin-resistant colonies appearing at 40°C, and integration of pLUW906 occurred at a frequency of 3.3 × 10−4 ± 6.6 × 10−5 per cell compared to 4.8 × 10−6 ± 6.9 × 10−6 per cell, for pG+host9.
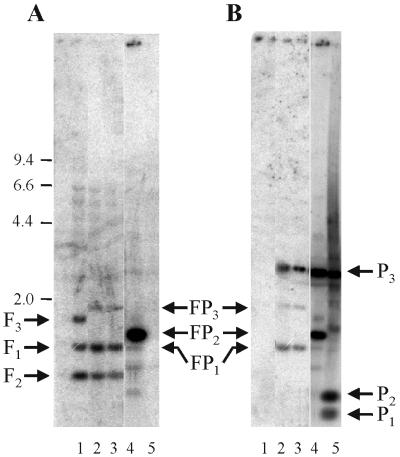
In order to investigate whether the significantly higher number of integration events was due to specific chromosomal integration into the frdA gene, pLUW906 integrants were further analyzed at the physiological, biochemical, and genetic level, Southern blot analysis of HincII-digested total DNA from pLUW906 integrants with radiolabeled frdAC- and pG+host9-specific probes revealed the loss of a 1.7-kb wild-type genomic frdBA fragment (F3), whereas two fragments (FP1 and FP3) appeared in the integrant DNA, which also hybridized with the pG+host9 probe, as would be expected in the case of specific integration of pLUW906 into the frdA gene of D. dehalogenans (Fig. 3C and 4). Furthermore, the 1.4-kb pLUW906 fragment (FP2) hybridizing with both probes was absent from integrant DNA, indicating the lack of free plasmid (Fig. 4). Similar results were obtained by PCR analysis with primers IK04 and BG355, as the 2-kb wild-type amplification product shifted to a distinct integrant-specific 6-kb fragment (data not shown).
FIG. 4.
Hybridization of HincII-digested total DNA from wild-type D. dehalogenans and pLUW906 integrants of D. dehalogenans with probes specific for frdAC (A) and pG+host9 (B). pG+host9 and pLUW906 plasmid DNAs were used as controls. The autoradiograph was digitally corrected for differences in signal intensities between lanes containing total DNA and lanes containing plasmid DNA. Lanes: 1, wild type; 2 and 3, pLUW906 integrants; 4, pLUW906; 5, pG+host9. DNA size markers are in kilobase pairs. F1 to F3 fragments hybridizing solely with the frdAC-specific probe; P1 to P3, fragments hybridizing solely with the pG+host9-specific probe (P1 and P2 originate from an additional HincII site between the EcoRI and ApaI sites in pG+host9); FP1 to FP3, fragments hybridizing with both probes.
Whereas fumarate-dependent growth was not significantly impaired in the pLUW906 integrants grown with lactate as the electron donor and fumarate as the electron acceptor, fumarate reductase activity was reduced, although not completely diminished (0.17 ± 0.01 and 0.09 ± 0.06 U/mg in two independently obtained pLUW906 integrants, compared to 0.36 ± 0.11 U/mg in a pG+host9 integrant control strain).
DISCUSSION
The recent detailed molecular analysis of the halorespiratory system in the o-CP-respiring gram-positive bacterium D. dehalogenans has brought us to a deeper understanding of structure, function, and control of this novel respiratory pathway (32, 33, 39). Previously, we described the development of an efficient plating, delivery, and screening system that has been useful for the isolation of halorespiration-deficient mutants following the chromosomal integration of the conjugative transposon Tn916 (32). These mutants have been instrumental in the identification of genes potentially encoding polypeptides which might be involved as structural components of the halorespiratory network or might play a role in their control and functional assembly. However, the instability of some of these mutants and the occurrence of preferential integration has to some extent hampered their further physiological and biochemical characterization. Here we report on the development and validation of host-vector systems for the genetic modification of the environmentally important, strictly anaerobic, low-G+C, gram-positive bacterium D. dehalogenans.
An efficient electroporation-based transformation procedure was designed using a protocol that had previously been optimized for the high-frequency electrotransformation of L. lactis (40). Routinely, we obtained 1.0 × 103 to 6.6 × 103 erythromycin-resistant transformants per μg of plasmid DNA from the 4.8-kb theta-replicating pAMβ1 derivative pIL253. These values observed for D. dehalogenans are in the same range as or higher than transformation frequencies obtained for several other LGB, such as Clostridium spp., but are lower than those obtained in genetic model strains of L. lactis (7, 9, 21, 41). Although pIL253 was maintained in D. dehalogenans at only moderate copy numbers of approximately 10 copies per cell, compared with 45 to 85 copies for L. lactis (12, 30), the stable replication of the vector indicates its potential use as a cloning vector in D. dehalogenans.
Plasmids based on the thermosensitive replicon pG+host were previously shown to conditionally replicate in various LGB as well as in E. coli (22, 23). One of these, the 3.8-kb rolling-circle-replicating, thermosensitive pWV01 derivative pG+host9, was used for the development of a system for specific gene disruption in D. dehalogenans. Transformation efficiencies for pG+host9 were on average 1 order of magnitude lower (6 × 102) than those for pIL253. These differences in frequency of transformation might be due to the difference in the mode of replication, as was previously reported for various other strains of LGB, but could also be caused by differences in marker gene expression (12, 20, 27). We were able to confirm thermosensitive replication, which was essentially absent at 40°C in D. dehalogenans. Although moderate segregational instability was also observed at the permissive temperature of 30°C, the relative number of viable cells able to grow on selective plates was reduced to 2 × 10−5 at 40°C within 7 generations. The nonpermissive temperature that we found for D. dehalogenans is somewhat higher than that reported for L. lactis (22). However, as D. dehalogenans is still growing at almost maximum growth rates at 40°C, this does not affect the applicability of the pG+host system (37).
In order to provide a model target to test the thermosensitive vector pG+host9 for its applicability for specific gene disruption, we cloned and sequenced the putative fumarate reductase-encoding frdCAB operon from D. dehalogenans. A pG+host9 derivative containing a 0.6-kb internal frdA fragment was successfully introduced and maintained in D. dehalogenans under permissive conditions. Chromosomal integration at nonpermissive temperatures was significantly more efficient in the case of pLUW906 compared to the empty vector, and the observed integration frequencies were similar to those found for L. lactis (2). However, although stable site-specific chromosomal integration of pLUW906 into the frdA gene could be unambiguously demonstrated by Southern blot analysis and PCR analysis, the fumarate reductase activity was only partly reduced and no changes in growth with fumarate were observed compared to the growth of D. dehalogenans containing pG+host9. One possible explanation could be that at least one of the truncated frdA genes present in the pLUW906 integrant is still coding for a (partially) active fumarate reductase enzyme due to a polar effect from the inserted vector sequences. This, however, is rather unlikely, since both the 3′- and 5′-truncated frdA copies lack several conserved residues that are probably essential for fumarate reductase activity (4, 17). Another possibility could be that the frdCAB operon actually encodes a succinate dehydrogenase. Nevertheless, no significant succinate dehydrogenase activity could be detected in cell extracts of D. dehalogenans. Northern analysis of total RNA isolated from cultures of D. dehalogenans grown with different electron donors and 3-chloro-4-hydroxy-phenylacetic acid, nitrate, or fumarate as the electron acceptor indicated that transcription of the frdCAB operon is constitutive rather than being induced in the presence of fumarate. This, however, is not in agreement with the highly induced fumarate reductase activity that has been measured in fumarate-grown cells of D. dehalogenans (H. Smidt et al., unpublished data). This suggests that the frdCAB operon only partially codes for the fumarate reductase activity, which is measured with benzyl viologen as an artificial electron donor. If so, this strongly supports the presence of at least one additional fumarate reductase-encoding gene cluster.
The development of the various gene transfer systems reported here is the first example of a genetic system for a halorespiring microbe. It has significantly improved our possibilities for studying the function and regulation of chromosomal genes in D. dehalogenans, including those relevant for the novel halorespiratory pathway this organism possesses. Moreover, the present set of genetic tools will enable the further exploitation of D. dehalogenans and related strains as dedicated degraders of recalcitrant environmental pollutants.
ACKNOWLEDGMENTS
We thank Richard van Kranenburg and Emmanuelle Maguin for providing plasmids pIL253 and pG+host9.
This work was partly supported by a grant from the Studienstiftung des Deutschen Volkes and contract BIO4-98-0303 of the European Union.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas I, Gruss A, Ehrlich S D, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 1993;175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borovkov A Y, Rivkin M I. XcmI-containing vector for direct cloning of PCR products. BioTechniques. 1997;22:812–814. doi: 10.2144/97225bm04. [DOI] [PubMed] [Google Scholar]
- 4.Bourgeron T, Rustin P, Chretien D, Birch Machin M, Bourgeois M, Viegas Pequignot E, Munnich A, Rotig A. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat Genet. 1995;11:144–149. doi: 10.1038/ng1095-144. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 6.Bruand C, Ehrlich S D, Janniere L. Unidirectional theta replication of the structurally stable Enterococcus faecalis plasmid pAMβ1. EMBO J. 1991;10:2171–2177. doi: 10.1002/j.1460-2075.1991.tb07752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckley N D, Vadeboncoeur C, LeBlanc D J, Lee L N, Frenette M. An effective strategy, applicable to Streptococcus salivarius and related bacteria, to enhance or confer electroporation competence. Appl Environ Microbiol. 1999;65:3800–3804. doi: 10.1128/aem.65.9.3800-3804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 9.Chen C K, Boucle C M, Blaschek H P. Factors involved in the transformation of previously nontransformable Clostridium perfringens type B. FEMS Microbiol Lett. 1996;140:185–191. doi: 10.1111/j.1574-6968.1996.tb08334.x. [DOI] [PubMed] [Google Scholar]
- 10.de Vos W M. Gene expression systems for lactic acid bacteria. Curr Opin Biotechnol. 1999;2:289–295. doi: 10.1016/S1369-5274(99)80050-2. [DOI] [PubMed] [Google Scholar]
- 11.de Vos W M, Kleerebezem M, Kuipers O P. Expression systems for industrial gram-positive bacteria with low guanine and cytosine content. Curr Opin Biotechnol. 1997;8:547–553. doi: 10.1016/s0958-1669(97)80027-4. [DOI] [PubMed] [Google Scholar]
- 12.de Vos W M, Simons G F M. Gene cloning and expression systems in Lactococci. In: Gasson M J, de Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. Glasgow, United Kingdom: Blackie Academic & Professional; 1994. pp. 52–105. [Google Scholar]
- 13.de Vos W M, Vos P, de Haard H, Boerrigter I. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine protease. Gene. 1989;85:169–176. doi: 10.1016/0378-1119(89)90477-0. [DOI] [PubMed] [Google Scholar]
- 14.El Fantroussi S, Naveau H, Agathos S N. Anaerobic dechlorinating bacteria. Biotechnol Prog. 1998;14:167–188. doi: 10.1021/bp980011k. [DOI] [PubMed] [Google Scholar]
- 15.Fetzner S. Bacterial dehalogenation. Appl Microbiol Biotechnol. 1998;50:633–657. doi: 10.1007/s002530051346. [DOI] [PubMed] [Google Scholar]
- 16.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hägerhäll C. Succinate:quinone oxidoreductases: variations on a conserved theme. Bioenergetics. 1997;1320:107–141. doi: 10.1016/s0005-2728(97)00019-4. [DOI] [PubMed] [Google Scholar]
- 18.Holliger C, Wohlfarth G, Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev. 1999;22:383–398. [Google Scholar]
- 19.Keasling J D, Bang S-D. Recombinant DNA techniques for bioremediation and environmentally friendly synthesis. Curr Opin Biotechnol. 1998;9:135–140. doi: 10.1016/s0958-1669(98)80105-5. [DOI] [PubMed] [Google Scholar]
- 20.Kiewit R, Kok J, Seegers J F M L, Venluna G, Bron S. The mode of replication is a major factor in segregational plasmid instability in Lactococcus lactis. Appl Environ Microbiol. 1993;59:358–364. doi: 10.1128/aem.59.2.358-364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klapatch T R, Guerinot M L, Lynd L R. Electrotransformation of Clostridium thermosaccharolyticum. J Ind Microbiol. 1996;16:342–347. doi: 10.1007/BF01570112. [DOI] [PubMed] [Google Scholar]
- 22.Maguin E, Duwat P, Hege T, Ehrlich S D, Gruss A. New thermosensitive plasmid for gram-positive bacteria. J Bacteriol. 1992;174:5633–5638. doi: 10.1128/jb.174.17.5633-5638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maguin E, Prevost H, Ehrlich S D, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Middeldorp P J M, Luijten M L G C, van de Pas B A, van Eekert M H A, Kengen S W M, Schraa G, Stams A J M. Anaerobic microbial reductive dehalogenation of chlorinated ethenes. Bioremediation J. 1999;3:151–169. [Google Scholar]
- 25.Mohn W W, Tiedje J M. Microbial reductive dehalogenation. Microbiol Rev. 1992;56:482–507. doi: 10.1128/mr.56.3.482-507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumann A, Scholz-Muramatsu H, Diekert G. Tetrachloroethene metabolism of Dehalospirillum multivorans. Arch Microbiol. 1994;162:295–301. doi: 10.1007/BF00301854. [DOI] [PubMed] [Google Scholar]
- 27.O'Sullivan T F, Fitzgerald G F. Electrotransformation of industrial strains of Streptococcus thermophilus. J Appl Microbiol. 1999;86:275–283. doi: 10.1046/j.1365-2672.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schirawski J, Unden G. Anaerobic respiration of Bacillus macerans with fumarate, TMAO, nitrate and nitrite and regulation of the pathways by oxygen and nitrate. Arch Microbiol. 1995;163:148–154. [Google Scholar]
- 30.Simon D, Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988;70:559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- 31.Smidt, H., A. D. L. Akkermans, J. van der Oost, and W. M. de Vos. Molecular characterisation of key enzymes in halorespiration. In S. N. Agathos and W. Reineke (ed.), Biotechnology for the environment. Focus on biotechnology, vol. 3, in press. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 32.Smidt H, Song D L, van der Oost J, de Vos W M. Random transposition by Tn916 in Desulfitobacterium dehalogenans allows for isolation and characterization of halorespiration-deficient mutants. J Bacteriol. 1999;181:6882–6888. doi: 10.1128/jb.181.22.6882-6888.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smidt H, van Leest M, van der Oost J, de Vos W M. Transcriptional regulation of the cpr gene cluster in ortho-chlorophenol-respiring Desulfitobacterium dehalogenans. J Bacteriol. 2000;182:5683–5691. doi: 10.1128/jb.182.20.5683-5691.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL-X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timmis K N, Pieper D H. Bacteria designed for bioremediation. Trends Biotechnol. 1999;17:200–204. doi: 10.1016/s0167-7799(98)01295-5. [DOI] [PubMed] [Google Scholar]
- 36.Triglia T, Peterson M G, Kemp D J. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Utkin I, Woese C, Wiegel J. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int J Syst Bacteriol. 1994;44:612–619. doi: 10.1099/00207713-44-4-612. [DOI] [PubMed] [Google Scholar]
- 38.van Asseldonk M, de Vos W M, Simons G. Functional analysis of the Lactococcus lactis usp45 secretion signal in the secretion of a homologous proteinase and a heterologous alpha-amylase. Mol Gen Genet. 1993;240:428–434. doi: 10.1007/BF00280397. [DOI] [PubMed] [Google Scholar]
- 39.van de Pas B A, Smidt H, Hagen W R, van der Oost J, Schraa G, Stams A J M, de Vos W M. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J Biol Chem. 1999;274:20287–20292. doi: 10.1074/jbc.274.29.20287. [DOI] [PubMed] [Google Scholar]
- 40.Wells J M, Wilson P W, Le Page R W. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol. 1993;74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 41.Young D I, Evans V J, Jefferies J R, Jennert K C B, Phillips Z E V, Ravagnani A, Young M. Genetic methods in clostridia. Methods Microbiol. 1999;29:191–207. [Google Scholar]