Abstract
Spermatogenesis is precisely controlled by sophisticated gene expression programs and is driven by epigenetic reprogramming, including histone modification alterations and histone-to-protamine transition. Nuclear receptor binding SET domain protein 2 (Nsd2) is the predominant histone methyltransferase catalyzing H3K36me2 and its role in male germ cell development remains elusive. Here, we report that NSD2 protein is abundant in spermatogenic cells. Conditional loss of Nsd2 in postnatal germ cells impaired fertility owing to apoptosis of spermatocytes and aberrant spermiogenesis. Nsd2 deficiency results in dysregulation of thousands of genes and remarkable reduction of both H3K36me2 and H3K36me3 in spermatogenic cells, with H3K36me2 occupancy correlating positively with expression of germline genes. Nsd2 deficiency leads to H4K16ac elevation in spermatogenic cells, probably through interaction between NSD2 and PSMA8, which regulates acetylated histone degradation. We further reveal that Nsd2 deficiency impairs EP300-induced H4K5/8ac, recognized by BRDT to mediate the eviction of histones. Accordingly, histones are largely retained in Nsd2-deficient spermatozoa. In addition, Nsd2 deficiency enhances expression of protamine genes, leading to increased protamine proteins in Nsd2-deficient spermatozoa. Our findings thus reveal a previously unappreciated role of the Nsd2-dependent chromatin remodeling during spermatogenesis and provide clues to the molecular mechanisms in epigenetic abnormalities impacting male reproductive health.
INTRODUCTION
Methylation of lysine 36 in histone H3 is the widespread modification at chromatin and is well known for its involvement in transcription (1,2). In mammalian cells, mono-, di- and trimethylation of H3K36 (H3K36me, H3K36me2 and H3K36me3, respectively) are catalyzed by distinct histone methyltransferases (HMTases). Nuclear SET domain (NSD)-containing methyltransferases, including NSD1, NSD2 and NSD3, induce H3K36me and H3K36me2, whereas SET domain-containing 2 (SETD2) is the major enzyme for catalyzing H3K36me3 (3). Setd2 is required for expression of acrosin-binding protein 1 and protamines and is essential for spermiogenesis in mice (4). Nsd1 plays a critical role in de novo DNA methylation in prospermatogonia and counteracts Polycomb-associated silencing (5). Nsd2, also known as Wolf–Hirschhorn syndrome candidate 1 (Whsc1), is responsible for deposition and spread of H3K36me2 through recognizing and catalyzing H3K36me2. In humans, heterozygous deletions of Nsd2 are implicated in the developmental disorder Wolf–Hirschhorn syndrome, characterized by cognitive and developmental defects (6). However, the functions of Nsd2 and its associated H3K36me2 during spermatogenesis remain elusive.
Acetylation of lysine 16 in histone H4 (H4K16ac) is another important histone modification that is essential for fly development (7). In Drosophila, reduction of both di- and trimethylation of H3K36 led to decreased acetylation of H4K16, which directly influenced the packaging of higher order chromatin (8). However, this process is currently understudied and still unclear in mammals, mainly due to the intricacy of the process itself and insufficiency of in vitro experimental systems for studying it. Chromatin remodelers, like HMTases, are believed to be essential for facilitating dynamic changes in chromatin compaction, but their roles in spermatogenesis have not been clearly elucidated. In this study, we show that Nsd2 orchestrates H4K16ac and H3K36me2 in spermatogenesis.
Spermatogenesis is a highly organized process that transforms diploid spermatogonial stem cells into haploid spermatozoa within seminiferous tubules of testis (9). It can be divided into three major phases: mitosis, meiosis and spermiogenesis. Spermatogonial stem cells first multiply by a series of mitotic divisions and differentiate into primary spermatocytes, which undergo one round of replication followed by meiotic homologous recombination and two rounds of chromosome segregation, resulting in the formation of haploid round spermatids (RS) (10,11). RS then mature into highly differentiated spermatozoa through morphological and molecular changes, including chromatin condensation, acrosome formation, flagellum development and cytoplasm elimination (12). Mouse seminiferous epithelial cycle can be subdivided into stages (stages I–XII), each of which represents a distinct repertoire of cell types (13). Mouse spermiogenesis is further divided into 16 steps according to morphological characteristics of acrosome and nucleus (14).
In this study, we demonstrate that Nsd2 is highly expressed in the pachytene spermatocytes (PS) and RS and plays important roles during sperm development. Deficiency of Nsd2 in mouse spermatogenic cells leads to sperm chromatin compaction defects and damaged male fertility. We reveal that Nsd2 both mediates a cascade of molecular events for histone removal and modulates the homeostasis of protamine, identifying Nsd2 as a critical regulator of the chromatin remodeling process during spermatogenesis.
MATERIALS AND METHODS
Ethics statement
All the animal procedures were approved by the Institutional Animal Care and Use Committee of Tongji Medical College, Huazhong University of Science and Technology, and the mice were housed in the specific pathogen-free facility of Huazhong University of Science and Technology. All experiments with mice were conducted ethically according to the Guide for the Care and Use of Laboratory Animals.
Animals
Floxed Nsd2 mice (Nsd2flox/flox) were generated by targeting mouse embryonic stem cells (ESCs) and blastocyst injection at Shanghai Research Center for Model Organisms. In brief, ESCs were targeted by carrying two loxP sites flanked in exons 14 and 16, and a neomycin selection cassette flanked by FRT sites in introns 15 and 16 of Nsd2 gene. The Nsd2+/flox mice were obtained by chimera formation and germline transmission. Mice were then crossed with FLP transgenic mice to remove the neomycin cassette and maintained on a C57BL/6J background. Stra8-Cre in the C57BL/6J background was purchased from the Jackson Laboratory. Stra8-Cre males were first crossed with Nsd2flox/flox females to generate the Stra8-Cre;Nsd2+/flox males, and then the Stra8-Cre;Nsd2+/flox male mice were bred with Nsd2flox/flox female mice to obtain the Stra8-Cre;Nsd2flox/flox (designated as Nsd2 cKO) males.
Antibodies
Antibodies used for western blot (WB), immunofluorescence (IF) and chromatin immunoprecipitation (ChIP) were as follows: anti-H3K36me3 (Solarbio no. K003337P, 1:400 for IF), anti-H3K36me2 (Solarbio no. K003336P, 1:400 for IF), anti-H3 (Solarbio no. K107283P, 1:800 for WB), anti-γ-H2A.X (Solarbio no. K006207P, 1:200 for IF, 1:1000 for WB), anti-mCherry (Solarbio no. K200015M, 1:1000 for WB), anti-H4K16ac (Abcam no. ab109463, 1:200 for IF), anti-NSD2 (Abcam no. ab-75359, 1:1000 for WB, 1:200 for IF), anti-PSMA8 (Proteintech no. 4022-1-AP, 1:1000 for WB), anti-GAPDH (Proteintech no. 60004-1-Ig, 1:1000 for WB), anti-PRM2 (Briar Patch Biosciences, Hup2B, 1:200 for IF, 1:800 for WB), anti-WT1 (Abcam no. ab89901, 1:200 for IF), anti-H4K5ac (ABclonal no. A19525, 1:1000 for WB, 1:200 for IF), anti-H4K8ac (ABclonal no. A7258, 1:1000 for WB, 1:200 for IF), anti-EP300 (Abcam no. ab-10485, 1:1000 for WB), anti-H4 (ABclonal no. A1131, 1:1000 for WB) and anti-α-Tubulin (Proteintech no. 66031-1-Ig, 1:1000 for WB).
Histological analysis
Mouse testes and epididymis were collected and fixed in Bouin’s solution (Sigma, HT10132) at 4°C overnight and then washed with 75% alcohol five times, 30 min each time. Samples were then embedded in paraffin; 5 μm sections were cut and stained with periodic acid–Schiff (PAS) after being dewaxed and rehydrated.
Electron microscopy
For transmission electron microscopy, mature sperm were fixed in 4% paraformaldehyde containing 0.05% glutaraldehyde in 0.1 M PBS, and then post-fixed in 1% osmium tetroxide. Dehydration was carried out in ethanol and the samples were embedded in Epon 812. Ultrathin sections were counterstained with uranyl acetate and lead citrate, and examined with a transmission electron microscope (Hitachi HT7700, Japan). For scanning electron microscopy, the samples were fixed in 2.5% glutaraldehyde solution in 0.1 M PBS, collected on poly-l-lysine-coated glass cover slips, post-fixed in osmium tetroxide, dehydrated in a graded ethanol series, subjected to critical point drying and then coated with gold/palladium. Samples were then examined with a scanning electron microscope (AZteclive Ultim Max 100, UK).
Immunofluorescence
Testes were fixed in 4% PFA in PBS overnight at 4°C and then were sequentially soaked in 5%, 10%, 12.5%, 15% and 20% sucrose in PBS and embedded in Tissue-Tek O.C.T. compound (Sakura Finetek, 4583) on dry ice. Embedded samples were stored at −80°C. Five-micrometer-thick cryosections were cut and washed with PBS three times. To perform antigen retrieval, cryo-sections were microwaved in 0.01 M sodium citrate buffer (pH 6.0) and then cooled down to room temperature. After washing with PBS three times, the sections were blocked in blocking solution (containing 3% normal goat serum and 3% fetal bovine serum in 1% bovine serum albumin) for 1 h. Later, tissue sections were incubated with primary antibodies in blocking solution overnight at 4°C. Slides were then incubated with secondary antibody for 1 h at room temperature after washing with PBS and stained with DAPI. Laser confocal scanning images were captured using a FluoView 1000 microscope (Olympus, Japan) with a digital camera (MSX2, Micro-shot Technology Limited, China).
TUNEL staining
Testes were fixed in 4% PFA, embedded in Tissue-Tek O.C.T. compound and sectioned (5 μm). TUNEL staining was performed using the TUNEL ApoGreen Detection Kit (YEASEN, 40307ES20). Images were obtained with a FluoView 1000 microscope (Olympus, Japan).
Western blot
Testis tissues were collected and proteins were extracted by using RIPA buffer (Beyotime, P0013J). In total, 40 μg of protein lysates were separated on a 10% SDS-PAGE gel, proteins were transferred to PVDF membranes (Bio-Rad) and the membranes were blocked in 5% non-fat milk (blocking solution) for 1 h. Primary antibodies were incubated overnight at 4°C after blocking. The membranes were washed with TBST three times and then incubated with a secondary antibody for 1 h before using luminol/enhancer solution and peroxide solution (Clarity™ Western ECL Substrate, Bio-Rad).
Immunoprecipitation
Fresh mouse adult testes from indicated genotypes were dissected and lysed in immunoprecipitation (IP) buffer (Beyotime, P0013J), clarified by centrifugation at 12 000 × g and then precleared with protein A beads (Bio-Rad, 161-4013). The lysate was incubated with primary antibodies overnight at 4°C on a rotator and conjugated with protein A beads. The beads were washed with IP buffer and then boiled in 2× SDS loading buffer for WB analysis.
Pulldown assay
Nsd2 cDNA fragments were cloned into the pmCherry-N1 vector encoding mCherry tag. HEK293T cells (obtained from Stem Cell Bank of Chinese Academy of Sciences, cat# GNHu43) were transfected with indicated plasmids using Lipofectamine 2000 (Life Technologies). After 48 h, IP was performed. Twenty-five microliters of mCherry-Trap bead 50% slurry (AlpaLife, ktsm1331) was used and all wash steps were performed with washing buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA). mCherry-Trap beads were washed with dilution buffer prior to addition to cell lysate. Beads were incubated with cell lysate at 4°C for 2 h following another wash step. To elute the proteins off the beads, 40 μl sample buffer (120 mM Tris–HCl, pH 6.8, 20% glycerol, 4% SDS, 0.04% bromophenol blue, 10% β-mercaptoethanol) was added and samples were boiled at 95°C for 5 min. WB was used for analysis. mCherry-tagged proteins were detected by WB using anti-mCherry antibody.
Isolation of PS and RS
Spermatogenic cells were isolated from the whole mouse testis using rigorous double enzymatic digestion with fraction enrichment on a discontinuous BSA density gradient as described previously (15). In brief, testes were collected from 8-week-old mice. Leydig cells were separated from collagenase-treated decapsulated testes by gravity sedimentation through a BSA cushion. The dispersed seminiferous tubules were then digested with trypsin and DNase I to single-cell suspensions. Next, Sertoli cells were separated from germ cells by filtration through a 40-mm cell strainer and by adhesion to lectin-coated culture plates. Different populations of germ cells were separated by using a manually prepared 0.5–5% discontinuous BSA density gradient for velocity sedimentation sediment. After sedimentation, enriched fractions of the three germ cell types (PS, RS) were manually collected.
RNA isolation and quantitative RT-PCR
Total RNAs were extracted from purified germ cell fractions using TRIzol reagent (Invitrogen) following the manufacturer’s procedure. The purity and concentration of RNA samples were determined by using a NanoDrop ND-2000 spectrophotometer (Thermo Scientific). Reverse transcriptional reactions contained 500 ng of purified total RNA using a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa) to remove the DNA contamination. RT-qPCR was performed with SYBR green master mix (TaKaRa) on the ABI Step One System (Applied Biosystems) according to the manufacturer’s instructions. The primers are listed in Supplementary Table S1. The relative gene expression was quantified using the comparative cycle threshold method, with the Gapdh expression used for normalization.
RNA-seq analysis
Total RNA was isolated from the isolated spermatogenic cells (two biological repeats for control and Nsd2 cKO, respectively) using TRIzol reagents (Invitrogen). The RNA concentration was verified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). One microgram of total RNA was used from each sample to prepare the mRNA libraries using TruSeq Stranded mRNA Library Preparation Kit Set A (cat. no. RS-122-2101, Illumina) according to the manufacturer’s instructions. All libraries were sequenced using the Illumina HiSeq 4000 platform. The FASTX-Toolkit was used to remove adaptor sequences and low-quality reads from the sequencing data. To identify all the transcripts, we used Tophat2 and Cufflinks to assemble the sequencing reads based on the UCSC mm10 mouse genome. The differential expression analysis was performed by Cuffdiff. The differentially expressed genes were set with the threshold of FDR < 0.05 and fold change >2.
Chromatin immunoprecipitation
ChIP was performed using SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling) with the indicated antibodies following the manufacturer’s procedure.
Statistical analysis
All data are presented as mean ± SD, unless otherwise noted in the figure legends. Statistical differences between datasets were assessed by one-way ANOVA or Student’s t-test using the GraphPad software. P-values are denoted in figures by *P < 0.05, **P < 0.01 and ***P < 0.001.
RESULTS
Nsd2 is robustly expressed in multiple stages of mouse spermatogenic cells
To explore the biological function of Nsd2 in mouse spermatogenesis, we first characterized expression pattern of Nsd2 in mouse spermatogenic cells. Reported RNA-seq result shows that Nsd2 transcript is enriched in all stages of spermatogenic cells, especially spermatocytes and spermatids (Figure 1A). Analysis of development-specific expression of NSD2 protein by WB indicated that NSD2 is continually expressed from postnatal day 0 (P0) testes to adult testes (Figure 1B). NSD2 protein was highly expressed in P0 and P8 (enriched for spermatogonia), with gradually decreased expression by P14 when meiosis I appeared (16). In adult mouse testes, the immunostaining signal of NSD2 was detected in the nuclei of PS of stages V–X and diplotene spermatocytes of stage XI, and RS (steps 1–8), with no NSD2 signal observed in elongated spermatids (steps 9–16) (Figure 1C). Moreover, Wilms’ tumor 1 (WT1) protein marks Sertoli cells and IF results revealed that NSD2 was also observed in WT1-positive cells (Figure 1D). Collectively, these data indicate that NSD2 is a nuclear protein that is expressed beginning in spermatogonia, early and late spermatocytes, and RS, but becomes downregulated at later stages of elongated spermatids (Figure 1E).
Figure 1.
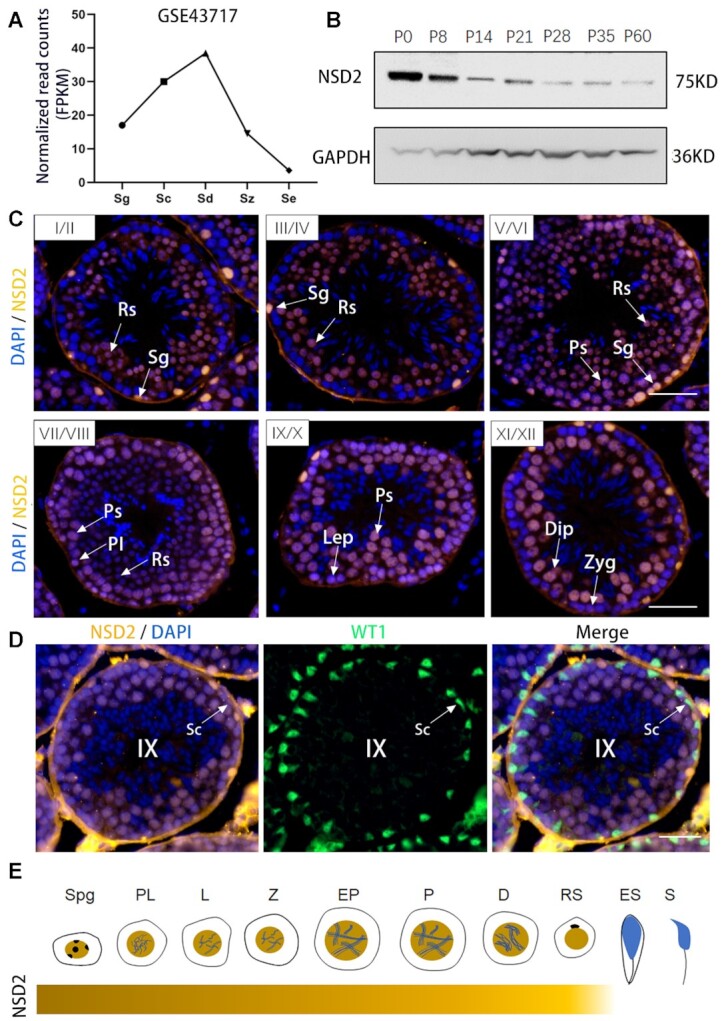
Nsd2 displays a dynamic expression during mouse spermatogenesis. (A) RNA-seq analysis of Nsd2 in mouse testicular cell types (GSE43717). Sg, spermatogonia; Sc, spermatocytes; Sd, spermatids; Sz, spermatozoa; Se, Sertoli cells. (B) WB analysis of NSD2 protein in lysates from mouse testis tissue collected at different time points during postnatal development. (C) Immunostaining of testis sections from 8-week-old WT male mice. DNA was counterstained with DAPI. Sg, spermatogonia; PI, pre-leptotene; Lep, leptotene; Zyg, zygotene; Ps, pachytene spermatocytes; Dip, diplotene; Rs, round spermatids. Scale bars, 50 μm. (D) Co-IF staining for NSD2 and WT1 on 8-week-old WT testis sections. Nuclei were stained with DAPI. Scale bar, 50 μm. (E) Graphic representation of NSD2 protein level during spermatogenesis.
Nsd2 deficiency led to defective spermatogenesis in mouse
To elucidate how Nsd2 regulates spermatogenesis, we generated germline-specific knockout mice by using Stra8-Cre transgenic mice in which Cre [beginning at postnatal day 3 in differentiating spermatogonia (17)] to delete exons 14–16 of Nsd2 gene (Stra8-Cre;Nsd2flox/flox, herein called Nsd2 cKO) (Supplementary Figure S1A and B). While Nsd2 cKO male mice were viable and appeared to be grossly normal, they displayed impaired fertility (Figure 2A). Six-month continuous breeding assessment showed that pup accumulation from Nsd2 cKO males (∼2 pubs/female) was significantly lower than that of WT controls (∼7 pubs/female). Consistent with the physiological defects, testis of Nsd2 cKO mice was significantly smaller than their controls (Figure 2B and C). Immunostaining assay verified that nuclear signal of NSD2 was absent in most spermatogenic cells in Nsd2 cKO mice (Figure 2D). Consistently, total protein levels of NSD2 in cKO adult testes were significantly decreased compared with controls by WB (Figure 2E), indicating that Nsd2 was inactivated specifically in testes with high efficiency (Supplementary Figure S1C). Further histological analyses showed that Nsd2 cKO mice had abnormal seminiferous tubules that were severely atrophic and contained fewer germ cells and vacuoles, and less spermatozoa in epididymis (Figure 2F and Supplementary Figure S1D). Moreover, TUNEL staining revealed increased numbers of apoptotic cells in seminiferous tubules of Nsd2 cKO testes (Figure 2G and H).
Figure 2.
Conditional inactivation of Nsd2 in postnatal male germ cells results in defective spermatogenesis and significantly reduced male fertility in mice. (A) Continuous breeding assessment shows decreased cumulative pubs per litter. Data are presented as mean ± SD (n = 3 for each genotype). (B) Gross morphology of the testis from control and Nsd2 cKO mice at 6, 9 and 12 weeks, respectively. (C) Comparison of testis weight from controls and Nsd2 cKO mice at 6, 9 and 12 weeks, respectively. Data are presented as mean ± SD (n = 3 for each genotype). (D) Representative IF images showing NSD2 was undetectable in adult Nsd2 cKO spermatogenic cells. (E) WB analysis of NSD2 protein in lysates from mouse testis tissue proved deficiency of NSD2 protein. (F) PAS staining showing the histology of testis sections from 6-week-old control and Nsd2 cKO mice (scale bar, 50 μm). Large vacuoles were seen in the seminiferous tubules of the Nsd2 cKO testes. (G) TUNEL staining of representative testicular sections from control Nsd2 cKO mice (scale bar, 50 μm). Apoptotic cells were labeled by TUNEL staining. (H) Comparison of TUNEL-positive seminiferous tubules from control and Nsd2 cKO mice. Data are presented as mean ± SD (n = 3 for each genotype). **P < 0.01; ***P < 0.001.
Nsd2 deficiency caused gene expression changes in PS and RS
To investigate molecular events underlying sperm developmental arrest in Nsd2-deficient males, we purified PS and RS from adult WT and Nsd2 cKO males (Supplementary Figure S2A) and performed RNA-seq (Figure 3A). Comparison of mRNA expression profiles revealed that 3528 and 640 genes were significantly up- and downregulated, respectively, in Nsd2 cKO versus WT PS. In Nsd2 cKO RS, expression of a larger number of genes was significantly different from control, including 4483 upregulated and 4017 downregulated genes. GO analysis of downregulated genes in Nsd2 cKO PS showed that these genes are mainly involved in sperm structure and chromosome condensation (Figure 3B). In Nsd2 cKO RS, GO analysis of downregulated genes was related to mRNA processing, histone modification and chromatin remodeling (Figure 3B). It is of note that the expression of many critical regulators for the meiotic cell cycle process (such as Hsf1, Brdt, Atrx, Cntd1, Bag6 and Ddx4), spermatic nucleus differentiation (such as Parp11, Hmgb2, Sycp1, Sun5, Kdm3a and Tssk6) and acrosome assembly (such as Gopc, Ggnbp2, Sox30, Nectin2, Nectin3 and Acrbp) was significantly impaired upon Nsd2 loss (Figure 3C). Therefore, Nsd2 is critical for expression of haploid RS-specific genes involved in spermiogenesis. In addition, we found that the most upregulated genes in Nsd2 cKO PS were mainly involved in regulation of protein folding, regulation of mitotic cell cycle, positive regulation of deacetylase activity, etc. (Supplementary Figure S2B). In Nsd2 cKO RS, the top enrichment for the upregulated genes was involved in regulation of mitochondrial electron transport, rRNA 3′-end processing, reciprocal meiotic recombination, etc. (Supplementary Figure S2B).
Figure 3.
RNA-seq reveals global gene expression change in Nsd2-deficient PS and RS. (A) Scatter plot of differentially expressed transcripts in Nsd2 cKO PS and RS compared with WT cells. Genes that were significantly changed (fold change >2, FDR < 0.05) were indicated. (B) GO term enrichment analysis for downregulated transcripts in PS and RS of Nsd2 cKO mice. (C) Expression analysis of the indicate genes in RS cells upon Nsd2 loss by RNA-seq analysis.
Nsd2 deficiency caused germline gene suppression by altering H3K36me2 in PS and RS
Notably, in Nsd2 cKO mouse testis, H3K36me2 signals were remarkably decreased in RS (Figure 4A and Supplementary Figure S3A). WB further confirmed decreased level of H3K36me2 in the mutant testis by analyzing the ratio of H3K36me2 to total H3 (Supplementary Figure S4A and B). Interestingly, we also found that the level of H3K36me3 significantly dropped in the mutants, which were more severe (Figure 4B and Supplementary Figure S3B). These results indicate that Nsd2 loss causes changes of both H3K36me2 and H3K36me3, consistent with another study that H3K36me3 is lost in ESCs derived from Nsd2-defective mice (18). To better gain mechanistic insights into transcriptional changes of male germline by Nsd2 loss, we set out to determine genome-wide distribution of H3K36me2 by ChIP-seq in PS and RS. In both PS and RS cells, we found that H3K36me2 is mainly enriched in intergenic regions (Figure 4C and Supplementary Figure S3C), in agreement with the canonical understanding of this histone mark in the somatic cells (19). In Nsd2-defective PS and RS cells, a lower level of H3K36me2 was measured throughout the intergenic regions (Figure 4C). Consistent with our hypothesis of Nsd2 being the major H3K36me2 methyltransferase in spermatogenesis, we found that germline genes (such as Spag17, Cdh18 and Inpp4b) in the mutant cells showed a drastic loss of H3K36me2 (Figure 4D). Notably, a previous study showed that knockout of Spag17 led to abnormal acrosome development and morphological defects in the head (20), which is similar to that seen in Nsd2 mutants. Further analyses showed that the downregulated genes are generally marked with higher levels of H3K36me2 than the upregulated genes in PS and RS (Figure 4E), suggesting that the downregulated expression is likely the direct effect of Nsd2 loss. We also noticed that the downregulated genes were expressed at higher levels than the upregulated genes in PS or RS (Figure 4E), indicating positive roles of H3K36me2 in transcriptional activation.
Figure 4.
Nsd2 deficiency alters normal H3K36me2 distribution in PS and RS. (A) Representative IF images showing H3K36me2 was significantly decreased in RS of adult Nsd2 cKO testis (scale bar, 50 μm). (B) H3K36me3 was largely decreased in PS and RS of adult Nsd2 cKO testis (scale bar, 50 μm). (C) Genomic distribution of H3K36me2 ChIP-seq peaks in PS and RS cells compared with genome background. (D) Genome browser view of H3K36me2 ChIP-seq reads on representative gene loci in isolated cells from control and Nsd2 cKO adult mice. (E) Signal plot analysis showing the H3K36me2 distribution over the up- and downregulated genes in PS and RS cells upon Nsd2 cKO. Box plot analyses of the expression levels of the up- and downregulated genes in PS and RS cells upon Nsd2 loss. ***P < 0.001.
Nsd2 deficiency resulted in increased H4K16ac through impacting PSMA8 protein level
Then, we examined the morphology of spermatogenic cells in Nsd2 cKO testis by co-staining with γ-H2AX. In mouse spermatogenesis, γ-H2AX is indicative of DNA double-strand breaks, which was detectable in leptotene, and zygotene spermatocytes, as well as elongating spermatid steps (21). Surprisingly, we found that Nsd2 deficiency in spermatogenic cells led to elevated level of H4K16ac implicated in chromatin decondensation and increased nuclear volume of spermatogenic cells (Figure 5A and B). H4K16ac is prevalent in elongating spermatids in WT mice, whereas higher levels of H4K16ac were detected in all stages of spermatocytes, and round and elongating spermatids in Nsd2 cKO mice (Figure 5B).
Figure 5.

Nsd2 deficiency leads to increased H4K16ac during spermiogenesis. (A) IF staining for H4K16ac in sections of adult WT and Nsd2 cKO testes. (B) Colocalization of γ-H2AX with H4K16ac was analyzed in the WT and Nsd2 cKO testes by immunostaining. (C) Co-IP of HEK293T cells transfected with empty mCherry or mCherry-Nsd2 vector. Anti-mCherry beads were used for IP. Antibodies used are indicated on the left. (D) Reciprocal co-IP of endogenous NSD2 and PSMA8 of the testis lysate from WT and Nsd2 cKO mice. (E) Expression of Psma8 in WT and Nsd2 cKO testes by qRT-PCR (n = 6 for each group); ns, not significant. (F) WB analysis of NSD2 and PSMA8 in WT and Nsd2 cKO testes.
To elucidate the molecular mechanism how Nsd2 regulates H4K16ac, we sought to determine other regulators with which NSD2 interacts. Interestingly, we identified PSMA8 in NSD2-immunoprecipitated protein complexes in mouse testis lysate (Supplementary Table S2). Since PSMA8, a testis-specific proteasome, is involved in acetylated histone degradation (22) and is also essential for spermatogenesis (23), we thus propose that NSD2 may cooperate with PSMA8 to recognize H4K16ac for histone cleaning in male germline. To confirm the association between NSD2 and PSMA8, we ectopically expressed tagged NSD2 in HEK293T cells, in which PSMA8 was detected in tagged NSD2 immunoprecipitates (Figure 5C). We then performed reciprocal IP assays in testes using antibodies specific for NSD2 and PSMA8. NSD2 was clearly detected in anti-PSMA8 antibody immunoprecipitants, and vice versa (Figure 5D). These results confirmed that NSD2 and PSMA8 are bona fide interacting partners in the testes. In addition, we found that protein level but not mRNA level of PSMA8 was dramatically decreased in Nsd2 cKO testes (Figure 5E and F). This indicates that NSD2–PSMA8 interaction ensures stability of PSMA8, which is unstable upon lack of NSD2 in testes, thus leading to increased levels of acetylated histones.
Nsd2 deficiency disturbed H4K5 and H4K8 acetylation during spermatogenesis
Previous studies reported that BRDT’s first bromodomain specifically recognized histone H4 bearing the simultaneous acetylation of K5 and K8 and mediated the eviction of histones (24,25). Defective histone retention may occur in Nsd2 cKO testis because Brdt expression was impaired in our RNA-seq data (Figure 3C). As expected, a lower protein level of BRDT and a higher protein level of H4 were observed in Nsd2 cKO testes compared with controls (Figure 6A). Moreover, we also found that the protein level of EP300 was significantly decreased in Nsd2 cKO testes (Figure 6A), and nuclear protein in testis (NUT) uses EP300 and/or CREB-binding protein to enhance acetylation of H4 at both K5 and K8 (26). We therefore performed WB analysis on testis extracts and identified decreased levels of H4K5ac and H4K8ac in the absence of NSD2 (Figure 6B). This result was in accordance with our observation by IF (Figure 6C). Specifically, in elongating spermatids of WT testis, H4K5ac and H4K8ac were widely distributed, but their levels were significantly reduced in Nsd2 cKO testes. To determine the basis for drastically reduced fertility in Nsd2 cKO mice, sperm were obtained from cauda epididymis and analyzed by electron microscopy. Scanning electron microscopy demonstrated that the sperm from Nsd2 cKO mice displayed abnormal head morphology (Figure 6D), lacking a typical hook-shaped appearance, a feature often related to nuclear shaping defects. To further characterize the inner structure of the abnormal sperm, we also carried out transmission electron microscope analysis (Figure 6D). As expected, deformed nuclei were observed in Nsd2 cKO sperm. We also found that Nsd2 cKO sperm showed detached acrosome and expanded perinuclear space, indicating that Nsd2 is crucial for structural organization of the male germ cell genome.
Figure 6.
Nsd2 is essential for sustaining H4K5 and H4K8 acetylation in testis and sperm. (A) Protein levels of EP300, BRDT and H4 in testis extracts from WT and Nsd2 cKO mice. Quantification of protein levels by immunoblotting is shown on the right. (B) Immunoblot analysis of H4K5ac and H4K8ac in WT and Nsd2 cKO testes with H4 protein amounts adjusted for equal. Quantification of histone modification levels by immunoblotting is shown on the right. (C) H4K5ac and H4K8ac (red) were significantly decreased in elongating spermatids of adult Nsd2 cKO testis (scale bar, 50 μm). Nuclei were stained with DAPI. The tubule sections shown are in stage X or XI of spermatogenesis. (D) Scanning electron micrographs (left panel) show abnormal head shaping (white arrow) in sperm of mutant mice. Transmission electron micrographs (right panel) display deformed nuclei (asterisk) and detached acrosome (black arrow) of sperm from mutant mice. Acr, acrosome; Nu, nucleus.
Nsd2 deficiency disrupted histone-to-protamine replacement
Chromatin reorganization during spermiogenesis begins concurrently with acrosome formation. In Nsd2 cKO mice, discernible changes were observed in the sperm, exhibiting abnormal shaping of the sperm head and acrosome defects via PNA staining (Figure 7A and B). These sperm morphological abnormalities seen in Nsd2 cKO mice were similar to those in Spag17 knockout mice. Nsd2 cKO mice had significantly lower sperm counts, and the sperm that were produced had compromised motility and a higher proportion of morphological abnormalities (Figure 7C). We also evaluated blastocyst rate of mutant sperm-fertilized WT oocytes collected from superovulated WT females. Compared with WT sperm, the rate of blastocyst formation was significantly decreased when Nsd2 cKO sperm were used (Supplementary Figure S4C and D). It is well known that, during late stages of sperm differentiation, the majority of the core histones are acetylated, degraded and replaced by transition nuclear proteins and protamines, subsequently forming highly condensed chromatin in elongated spermatids (27). Consistent with the spermatid developmental defects of the Nsd2 cKO mice, we identified significant upregulation of Tnps and Prms (Figure 7D), which was also confirmed by RT-qPCR (Figure 7E). Additionally, expressions of many critical regulators for spermiogenesis, such as Acrbp1, Pick1, Odf1 and Spem1, were also significantly impaired upon Nsd2 loss (Figure 7E). Several histone variant genes, such as H1fnt (H1-7), H2afb1 (H2A.L.2) and H3f3a (H3.3), were also downregulated (Figure 7E). Malformation of the acrosome caused by Nsd2 loss was probably due to misregulation of above genes. Furthermore, IF staining showed increased level of protamine proteins in Nsd2 cKO sperm (Figure 7F). Consistent with the IF observation in sperm, WB of lysates from sperm also revealed elevated protamine level and enhanced retention of histone in Nsd2 cKO sperm (Figure 7G). Collectively, histone retention in sperm can be interpreted by reduced BRDT and EP300 expression and dampened histone degradation due to decreased PSMA8 level, while increased protamine level for competition may compromise deleterious impact of histone retention. These findings indicate that the transition from histone-to-protamine replacement was abnormal upon Nsd2 loss, thereby contributing to defective spermiogenesis.
Figure 7.

Nsd2 modulates histone removal and homeostasis of transition proteins and protamines. (A) Representative abnormal head morphology of Nsd2 cKO spermatozoa. (B) Fluorescently tagged PNA staining of epididymal sperm from control and Nsd2 cKO adult mice. (C) Comparison of epididymal sperm counts, abnormal sperm and motility of control and Nsd2 cKO adult mice. Data are presented as mean ± SD (n = 3 for each genotype). (D) Genome browser view of RNA-seq reads on representative gene loci in isolated cells from WT and Nsd2 cKO mice. (E) qRT-PCR validation of RNA-seq revealed gene expression changes in the testis of WT and Nsd2 cKO mice. Results are normalized to Gapdh expression and data are presented as mean ± SD (n = 3 for each genotype). (F) IF colocalization of sperm genome with protamines in decondensed and permeabilized sperm nuclei. DNA was stained with DAPI. Scale bar, 50 μm. (G) WB analysis of histone and protamine in spermatozoa from the epididymis of WT and Nsd2 cKO mice. Quantification of protein levels of immunoblotting is shown on the right. *P < 0.05; **P < 0.01; ***P < 0.001.
NSD2 expression in spermatozoa from patients with oligoasthenoteratozoospermia
We also analyzed spermatozoa RNA profiles from patients with oligoasthenoteratozoospermia (OAT) (28) who presented with severe and consistent teratozoospermia, and ≤3% spermatozoa displayed the ideal form. According to Kruger’s guideline, spermatozoa with 4% percent ideal form already exhibited very poor prognosis of fertility (29,30). The patients included in our analysis had no other severe pathologies to exclude confounding conditions, and their sperm motility and concentration were normal. We found that NSD2 expression was significantly lower than that of normal subjects (Figure 8A). Therefore, NSD2 is a promising molecular maker to distinguish OAT from normal populations (Figure 8B).
Figure 8.

Correlation of NSD2 levels with OAT. (A) Relative expression level of the NSD2 in normal and OAT human sperm samples from the GSE6872 dataset. (B) Hierarchical clustering of normal and OAT human sperm samples. NSD2 was used to cluster the class assignment of the set of the normal and OAT samples. ***P < 0.001.
Taken together, our results indicate that Nsd2 is essential for normal spermatogenesis in mice, and that deletion of Nsd2 postnatally resulted in germ cell decrease, abnormal histone-to-protamine transition and sperm deficiency (Figure 9).
Figure 9.
A schematic model on the role of Nsd2 in spermatogenesis. We propose that in mouse spermatogenic cells, methylation of H3K36 by NSD2 regulates germline gene expression for normal spermatogenesis; NSD2 plays important roles in orchestrating H4 acetylation through regulating PSMA8, EP300 and BRDT expression to ensure histone removal in male germ cells.
DISCUSSION
NSD2-mediated H3K36me2 is a transcriptionally coupled histone methylation mark, enriched in intergenic regions undergoing active transcription in somatic cells (31). Our results showed that H3K36me2 patterns in PS and RS are also enriched in intergenic regions. Nsd2 loss leads to expression changes of hundreds to thousands of genes in PS and RS cells. Further analyses found that the downregulated genes are generally marked by higher levels of H3K36me2 in the WT germ cells and are expressed at higher levels. These observations not only show that the downregulated expression is likely the direct effect of Nsd2 loss but also indicate that Nsd2 and H3K36me2 are required for transcriptional activities of more highly expressed genes, which may play important roles in spermatogenesis. Indeed, we observed significant impaired expression of critical sperm regulators, including Spag17, Inpp4b, Cdh18 and spermiogenesis genes, which normally undergo transcriptional silencing at these stages. For example, loss of Spag17 resulted in abnormal acrosome similar to that observed in Nsd2 mutants. The high level of both Nsd2 and H3K36me2 was observed in PS and RS cells in the adult mouse testes. In addition, we found that the H3K36me3 level was largely decreased in PS and RS of the Nsd2 cKO mice when compared with the H3K36m2 level. Therefore, relationship between Nsd2 and H3K36me3 may need further investigation.
NSD2 is a key methyltransferase of histone H3K36me2, a mark associated with active transcription. In Drosophila, H3K36 methylation has crosstalk with H4K16ac (8). Our results show that Nsd2 deficiency leads to significant increase in H4K16ac and subsequent defects in histone-to-protamine replacement, supporting the notion that H4K16ac is indeed critical during spermiogenesis. Remarkably, our study identifies that testis-specific proteasome subunit PSMA8 interacts with NSD2 and PSMA8 expression is decreased in Nsd2 cKO mice, providing a new promising target for future study on the protein–protein interaction network. PSMA8 stimulated the degradation of the acetylated core histones, instead of nonacetylated histones in vitro (22). In mice, the loss of Psma8 led to the accumulation of H4K16ac in RS (23). These results suggest that Nsd2 is involved in the histone acetylation regulation. In addition, a previous study demonstrated that Psma8 is expressed specifically in spermatocytes from the pachytene stage and is required for the assembly/stabilization of proteasomes in testes. Psma8-deficient spermatocytes showed delayed onset of metaphase phase and were arrested at metaphase phase, while the processes of meiotic recombination and synapsis were less affected by Psma8 deletion (32). Therefore, we propose that the reduction of Psma8 mediated reduced RS counts in Nsd2 cKO mice.
NSD2 is a multidomain protein containing one catalytic SET (suppressor of variegation, enhancer of zeste and trithorax) domain, two PWWP (proline–tryptophan–tryptophan–proline motif) domains, one HMG (high-mobility group) box, five PHD (plant-homeodomain) zinc finger motifs and one Cys–His-rich C5HCH domain, which may enable its diverse functions during spermiogenesis (33). The PWWP, HMG and PHD domains have been shown to mediate chromatin interaction and recognition of histone marks (19). The SET domain confers catalytic function on this enzyme for H3K36me2 (34). The interaction between PWWP domain and H3K36me2 plays a key role in stabilizing NSD2 at chromatin (35). The HMG domain interacts with the DNA-binding domain of androgen receptor (36). It is known that the PHD of Chd5 plays important roles in regulating gene expression in spermatids (37). Chd5 deficiency results in elevated levels of both transition proteins and protamines, which is similar to that seen in Nsd2 mutants. We observed that the inactivation of NSD2 impaired male fertility with spermatogenesis abnormalities at the histone removal stage. Our results demonstrated that disturbed histone retention in spermatozoa can be interpreted by the downregulation of EP300 and BRDT. Here, we showed that expression of NSD2 exists in post-meiotic spermatogenic cells, where its presence can couple histone H4 acetylation, BRDT binding or the exchange of histone variants, to control protamine invasion of nucleosomes and histone replacement. The multiple functional domains of NSD2 may work independently or in concert, enabling the diverse functions of NSD2 observed during spermatogenesis.
Although the enzymology and molecular function of NSD2 have been documented, the biological consequences of NSD2 inactivation in testis development at the organism level have not been explored. Here, we show that NSD2 is the key H3K36me2 methyltransferase in PS and RS and is essential for normal spermatogenesis. Loss of Nsd2 led to significant decrease of both H3K36me2 and H3K36me3 in PS and RS cells. Furthermore, we find that Nsd2-dependent H3K36me2 plays a critical role in regulating germline gene expression in these germ cells. In contrast, Nsd2 deficiency stimulates H4K16ac in spermatocytes and spermatids. There was significant increase in expression of histones in mature sperm of Nsd2 cKO mice, suggesting a defect in histone-to-protamine transition. It was reported that NSD2 associated with HIRA, the H3.3-specific histone chaperone, and facilitated prolonged H3.3 deposition (38). We also identified H3.3 in NSD2-immunoprecipitated protein complexes from our mass spectrometry data. Therefore, NSD2 may interact with H3.3 to regulate chromatin packaging during spermatogenesis. Notably, H3f3a knockout mice had subfertility that was related to head defects of azoospermia (39), exhibiting similar phenotype with Nsd2 cKO sperm. Correspondingly, we revealed that Nsd2 modulates the homeostasis of transition proteins and protamines by increasing expression of Tnp1, Tnp2, Prm1 and Prm2 transcriptionally. These findings unravel pleiotropic functions of Nsd2 and highlight its multifaceted role in orchestrating the histone modification and histone-to-protamine remodeling that occurs during male germ cell development.
Using the germ cell-specific Nsd2 knockout mouse model, we found that targeted Nsd2 knockout in germ cells causes largely aberrant sperm with acrosomal malformation and dampens fertility. Our findings reveal a previously unappreciated role of the Nsd2-dependent H3K36me2 and H4K16ac modification in spermatogenesis and provide clues to the molecular mechanisms in epigenetic disorders underlying male infertility.
DATA AVAILABILITY
The RNA-seq data in this study have been deposited in the Gene Expression Omnibus website: PRJNA773637. The ChIP-seq data in this study have been deposited in the Gene Expression Omnibus website: PRJNA773601.
Supplementary Material
Contributor Information
Zhiming Li, Institute of Reproductive Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, China.
Xinzong Zhang, NHC Key Laboratory of Male Reproduction and Genetics, Guangdong Provincial Reproductive Science Institute, Guangzhou 510600, China.
Shiming Xie, Institute of Reproductive Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, China.
Xingping Liu, Institute of Reproductive Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, China.
Caifeng Fei, Institute of Reproductive Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, China.
Xunbin Huang, Institute of Reproductive Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, China.
Yunge Tang, NHC Key Laboratory of Male Reproduction and Genetics, Guangdong Provincial Reproductive Science Institute, Guangzhou 510600, China.
Li-quan Zhou, Institute of Reproductive Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2018YFC1004502]; NHC Key Laboratory of Male Reproduction and Genetics, Guangdong Provincial Reproductive Science Institute (Guangdong Provincial Fertility Hospital) [KF201902]; Health Commission of Hubei Province Scientific Research Project [WJ2021Q023]; National Natural Science Foundation of China [NSFC 82001620]; Program for HUST Academic Frontier Youth Team. Funding for open access charge: Program for HUST Academic Frontier Youth Team.
Conflict of interest statement. None declared.
REFERENCES
- 1. Suganuma T., Workman J.L.. Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 2011; 80:473–499. [DOI] [PubMed] [Google Scholar]
- 2. Wagner E.J., Carpenter P.B.. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 2012; 13:115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang C., Zhu B.. Roles of H3K36-specific histone methyltransferases in transcription: antagonizing silencing and safeguarding transcription fidelity. Biophys. Rep. 2018; 4:170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zuo X., Rong B., Li L., Lv R., Lan F., Tong M.H.. The histone methyltransferase SETD2 is required for expression of acrosin-binding protein 1 and protamines and essential for spermiogenesis in mice. J. Biol. Chem. 2018; 293:9188–9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shirane K., Miura F., Ito T., Lorincz M.C.. NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nat. Genet. 2020; 52:1088–1098. [DOI] [PubMed] [Google Scholar]
- 6. Stec I., Wright T.J., van Ommen G.J., de Boer P.A., van Haeringen A., Moorman A.F., Altherr M.R., den Dunnen J.T.. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf–Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum. Mol. Genet. 1998; 7:1071–1082. [DOI] [PubMed] [Google Scholar]
- 7. Krishnan V., Chow M.Z., Wang Z., Zhang L., Liu B., Liu X., Zhou Z.. Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc. Natl Acad. Sci. U.S.A. 2011; 108:12325–12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bell O., Wirbelauer C., Hild M., Scharf A.N., Schwaiger M., MacAlpine D.M., Zilbermann F., van Leeuwen F., Bell S.P., Imhof A.et al.. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J. 2007; 26:4974–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clermont Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol. Rev. 1972; 52:198–236. [DOI] [PubMed] [Google Scholar]
- 10. de Rooij D.G. Proliferation and differentiation of spermatogonial stem cells. Reproduction. 2001; 121:347–354. [DOI] [PubMed] [Google Scholar]
- 11. Zickler D., Kleckner N.. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 2015; 7:a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O’Donnell L. Mechanisms of spermiogenesis and spermiation and how they are disturbed. Spermatogenesis. 2014; 4:e979623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahmed E.A., de Rooij D.G.. Staging of mouse seminiferous tubule cross-sections. Methods Mol. Biol. 2009; 558:263–277. [DOI] [PubMed] [Google Scholar]
- 14. Meistrich M.L., Hess R.A.. Assessment of spermatogenesis through staging of seminiferous tubules. Methods Mol. Biol. 2013; 927:299–307. [DOI] [PubMed] [Google Scholar]
- 15. Da Ros M., Lehtiniemi T., Olotu O., Meikar O., Kotaja N.. Enrichment of pachytene spermatocytes and spermatids from mouse testes using standard laboratory equipment. J. Vis. Exp. 2019; 10.3791/60271. [DOI] [PubMed] [Google Scholar]
- 16. Zindy F., den Besten W., Chen B., Rehg J.E., Latres E., Barbacid M., Pollard J.W., Sherr C.J., Cohen P.E., Roussel M.F.. Control of spermatogenesis in mice by the cyclin D-dependent kinase inhibitors p18(Ink4c) and p19(Ink4d). Mol. Cell. Biol. 2001; 21:3244–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sadate-Ngatchou P.I., Payne C.J., Dearth A.T., Braun R.E.. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008; 46:738–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nimura K., Ura K., Shiratori H., Ikawa M., Okabe M., Schwartz R.J., Kaneda Y.. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf–Hirschhorn syndrome. Nature. 2009; 460:287–291. [DOI] [PubMed] [Google Scholar]
- 19. Popovic R., Martinez-Garcia E., Giannopoulou E.G., Zhang Q., Zhang Q., Ezponda T., Shah M.Y., Zheng Y., Will C.M., Small E.C.et al.. Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS Genet. 2014; 10:e1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kazarian E., Son H., Sapao P., Li W., Zhang Z., Strauss J.F., Teves M.E.. SPAG17 is required for male germ cell differentiation and fertility. Int. J. Mol. Sci. 2018; 19:1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ketchum C.C., Larsen C.D., McNeil A., Meyer-Ficca M.L., Meyer R.G.. Early histone H4 acetylation during chromatin remodeling in equine spermatogenesis. Biol. Reprod. 2018; 98:115–129. [DOI] [PubMed] [Google Scholar]
- 22. Zhang Z.H., Jiang T.X., Chen L.B., Zhou W., Liu Y., Gao F., Qiu X.B.. Proteasome subunit α4s is essential for formation of spermatoproteasomes and histone degradation during meiotic DNA repair in spermatocytes. J. Biol. Chem. 2021; 296:100130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gomez H.L., Felipe-Medina N., Condezo Y.B., Garcia-Valiente R., Ramos I., Suja J.A., Barbero J.L., Roig I., Sanchez-Martin M., de Rooij D.G.et al.. The PSMA8 subunit of the spermatoproteasome is essential for proper meiotic exit and mouse fertility. PLoS Genet. 2019; 15:e1008316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goudarzi A., Zhang D., Huang H., Barral S., Kwon O.K., Qi S., Tang Z., Buchou T., Vitte A.L., He T.et al.. Dynamic competing histone H4 K5K8 acetylation and butyrylation are hallmarks of highly active gene promoters. Mol. Cell. 2016; 62:169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gaucher J., Boussouar F., Montellier E., Curtet S., Buchou T., Bertrand S., Hery P., Jounier S., Depaux A., Vitte A.L.et al.. Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J. 2012; 31:3809–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shiota H., Barral S., Buchou T., Tan M., Coute Y., Charbonnier G., Reynoird N., Boussouar F., Gerard M., Zhu M.et al.. Nut directs p300-dependent, genome-wide H4 hyperacetylation in male germ cells. Cell Rep. 2018; 24:3477–3487. [DOI] [PubMed] [Google Scholar]
- 27. Wang T., Gao H., Li W., Liu C.. Essential role of histone replacement and modifications in male fertility. Front. Genet. 2019; 10:962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Platts A.E., Dix D.J., Chemes H.E., Thompson K.E., Goodrich R., Rockett J.C., Rawe V.Y., Quintana S., Diamond M.P., Strader L.F.et al.. Success and failure in human spermatogenesis as revealed by teratozoospermic RNAs. Hum. Mol. Genet. 2007; 16:763–773. [DOI] [PubMed] [Google Scholar]
- 29. Szczygiel M., Kurpisz M.. Teratozoospermia and its effect on male fertility potential. Andrologia. 1999; 31:63–75. [PubMed] [Google Scholar]
- 30. Van Waart J., Kruger T.F., Lombard C.J., Ombelet W.. Predictive value of normal sperm morphology in intrauterine insemination (IUI): a structured literature review. Hum. Reprod. Update. 2001; 7:495–500. [DOI] [PubMed] [Google Scholar]
- 31. Weinberg D.N., Papillon-Cavanagh S., Chen H., Yue Y., Chen X., Rajagopalan K.N., Horth C., McGuire J.T., Xu X., Nikbakht H.et al.. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019; 573:281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Q., Ji S.Y., Busayavalasa K., Shao J., Yu C.. Meiosis I progression in spermatogenesis requires a type of testis-specific 20S core proteasome. Nat. Commun. 2019; 10:3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang Z., Wu H., Chuai S., Xu F., Yan F., Englund N., Wang Z., Zhang H., Fang M., Wang Y.et al.. NSD2 is recruited through its PHD domain to oncogenic gene loci to drive multiple myeloma. Cancer Res. 2013; 73:6277–6288. [DOI] [PubMed] [Google Scholar]
- 34. Jaffe J.D., Wang Y., Chan H.M., Zhang J., Huether R., Kryukov G.V., Bhang H.E., Taylor J.E., Hu M., Englund N.P.et al.. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat. Genet. 2013; 45:1386–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sankaran S.M., Wilkinson A.W., Elias J.E., Gozani O.. A PWWP domain of histone-lysine N-methyltransferase NSD2 binds to dimethylated Lys-36 of histone H3 and regulates NSD2 function at chromatin. J. Biol. Chem. 2016; 291:8465–8474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kang H.B., Choi Y., Lee J.M., Choi K.C., Kim H.C., Yoo J.Y., Lee Y.H., Yoon H.G.. The histone methyltransferase, NSD2, enhances androgen receptor-mediated transcription. FEBS Lett. 2009; 583:1880–1886. [DOI] [PubMed] [Google Scholar]
- 37. Li W., Wu J., Kim S.Y., Zhao M., Hearn S.A., Zhang M.Q., Meistrich M.L., Mills A.A.. Chd5 orchestrates chromatin remodelling during sperm development. Nat. Commun. 2014; 5:3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sarai N., Nimura K., Tamura T., Kanno T., Patel M.C., Heightman T.D., Ura K., Ozato K.. WHSC1 links transcription elongation to HIRA-mediated histone H3.3 deposition. EMBO J. 2013; 32:2392–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang M.C., Jacobs S.A., Mattiske D.M., Soh Y.M., Graham A.N., Tran A., Lim S.L., Hudson D.F., Kalitsis P., O’Bryan M.K.et al.. Contribution of the two genes encoding histone variant H3.3 to viability and fertility in mice. PLoS Genet. 2015; 11:e1004964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data in this study have been deposited in the Gene Expression Omnibus website: PRJNA773637. The ChIP-seq data in this study have been deposited in the Gene Expression Omnibus website: PRJNA773601.







