Abstract
Homologous replication module genes were identified for four P335 type phages. DNA sequence analysis revealed that all four phages exhibited more than 90% DNA homology for at least two genes, designated rep2009 and orf17. One of these genes, rep2009, codes for a putative replisome organizer protein and contains an assumed origin of phage DNA replication (ori2009), which was identical for all four phages. DNA fragments representing the ori2009 sequence confer a phage-encoded resistance (Per) phenotype on lactococcal hosts when they are supplied on a high-copy-number vector. Furthermore, cloning multiple copies of the ori2009 sequence was found to increase the effectiveness of the Per phenotype conferred. A number of antisense plasmids targeting specific genes of the replication module were constructed. Two separate plasmids targeting rep2009 and orf17 were found to efficiently inhibit proliferation of all four phages by interfering with intracellular phage DNA replication. These results represent two highly effective strategies for inhibiting bacteriophage proliferation, and they also identify a novel gene, orf17, which appears to be important for phage DNA replication. Furthermore, these results indicate that although the actual mechanisms of DNA replication are very similar, if not identical, for all four phages, expression of the replication genes is significantly different in each case.
Lactococcus lactis strains are widely used as starter strains for the production of fermented dairy products, such as sour cream, buttermilk, and a variety of cheeses. These lactococcal starter strains are susceptible to attack by bacteriophages which are ubiquitous in the fermentation environment (24, 32). Such infection may lead to a number of problems ranging from slow fermentation to complete failure and may thus result in serious economic losses. The commercial and scientific impetus to develop modified starter strains with the desired fermentative qualities and improved resistance to bacteriophages is well documented (9, 10, 17, 19, 37, 41). Traditionally, this was accomplished by isolating bacteriophage-insensitive mutants which can be obtained following infection of a bacterial population with a specific phage at a high titer (20, 23). More recently, conjugative transfer of plasmids encoding natural resistance mechanisms from phage-resistant strains to phage-sensitive dairy strains with good fermentation or flavor properties has been described (9, 17, 37). Many of the naturally occurring resistance mechanisms have been identified, and they are divided into four main groups depending on the mode of action: inhibition of phage adsorption, blockage of phage DNA injection, restriction-modification, and abortive infection (2, 12, 18, 20, 28). Since the early 1990s members of the P335 bacteriophage species have been isolated with increasing frequency (1, 4, 34, 36). The emergence and subsequent characterization of this new species coincided with rapid advances in the molecular technologies available to researchers, such as automated DNA sequencing and bioinformatics, and consequently a wealth of information pertaining to P335 type phage biology has become available, which has enabled researchers to develop “intelligent” or “engineered” phage resistance systems.
One such system, termed “a triggered-suicide system,” employs a plasmid-located phage-specific promoter (φ31P) from bacteriophage φ31 placed upstream of the lethal LlaIR+ restriction gene of the LlaI restriction-modification system from lactococcal plasmid pTR2030 (14). Upon φ31 infection of a cell harboring this plasmid, the inducible promoter is activated; this causes LlaIR+ to produce its lethal product, which results in death of the host cell before infective phage particles are produced. Another example is phage-encoded resistance (Per), which was first used to confer phage resistance on L. lactis NCK203 against small isometric-headed phage φ50 (21). This so-called Per50 system consisted of a fragment of genomic φ50 DNA containing the putative origin of replication (ori50) cloned in a high-copy-number vector. Intracellular φ50 DNA replication was shown to be impeded in cells harboring ori50-containing plasmids, while the intracellular concentration of these plasmids was observed to increase following infection by φ50. These observations led to the conclusion that the plasmid-borne ori50 was competing with the incoming φ50 DNA for an essential and limiting phage replication function or functions. Per systems have also been described for other lactococcal phages (31, 38) and more recently for a number of phages infecting Streptococcus thermophilus (16, 42).
Since the early 1990s a number of reports have described utilization of antisense mRNA approaches to control phage multiplication (7, 25–27, 45). This approach involves cloning a target gene in the reverse orientation behind a promoter on a plasmid. The resulting antisense mRNA is assumed to bind to the target mRNA and form a nontranslatable double-stranded mRNA molecule which either prevents proper ribosome loading or makes the double-stranded molecule more susceptible to attack by RNA-degrading enzymes (22). Antisense strategies targeting structural proteins, a transcriptional activator, and genes of unknown function have been reported (8, 25–27, 46; K. M. Polzin, L. J. Collins, L. W. Lubbers, and A. W. Jarvis, 5th Symp. Lactic Acid Bacteria, abstr. F2); however, these strategies have yielded variable results. Recently, an ingenious combination of the Per and antisense strategies was described (46). In this system different φ31 genes (two middle expressed genes and four late expressed genes) are cloned between the strong Lactobacillus P6 promoter and the T7 terminator (TT7) in low-copy-number plasmid pTRK360, which contains the putative φ31 origin of replication (ori31). ori31 allows “explosive” amplification of pTRK360 following infection by φ31, thereby increasing the levels of antisense transcripts late in the lytic cycle. The presence of ori31 alone lowers the burst size of φ31 fourfold, which results in fewer sense target mRNAs being expressed from the phage genome.
It has been demonstrated that two previously described Per mechanisms, Per31 and Per50, inhibit proliferation of a number of different phages of the P335 species (35). Homologous replication module genes were identified for four P335 type phages (Q30, Q33, ul36, and Tuc2009), which indicated that these genes may be suitable targets for development of engineered phage resistance mechanisms.
In this report we describe an improved Per system and an optimized antisense strategy, both of which provide significant protection for L. lactis hosts against a number of phages belonging to the P335 group.
MATERIALS AND METHODS
Bacteria, bacteriophages, and plasmids.
The bacterial strains, bacteriophages, and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacteriophages, bacterial strains, and plasmids used in this study
| Phage, bacterial strain, or plasmid | Relevant feature(s) | Source or reference |
|---|---|---|
| Phages | ||
| Tuc2009 | P335 type lysogenic phage of L. lactis subsp. cremoris UC509 | 4 |
| Q30 | P335 type lytic phage of L. lactis subsp. lactis | 33 |
| Q33 | P335 type lytic phage of L. lactis subsp. lactis | 33 |
| ul36 | P335 type lytic phage of L. lactis subsp. cremoris | 34 |
| E. coli strains | ||
| JM101 | Transformation host | New England Biolabs |
| TOP10 | Transformation host | Invitrogen |
| L. lactis strains | ||
| UC509.9 | Prophage-cured derivative of L. lactis subsp. cremoris UC509, host for Tuc2009 | 11 |
| SMQ86 | Propagating host for phages Q30, Q33, and u136 | 15 |
| Plasmids | ||
| pCR 2.1-TOPO | Cloning vector, Apr | Invitrogen |
| pNZ8048 | E. coli-L. lactis high-copy-number shuttle vector, Cmr (Per−) | 13 |
| pNZRep-Cii | pNZ8048 containing one copy of ori2009(Per+) | 31 |
| pNZRep-3 | pNZ8048 containing rep2009 in antisense orientation (Per+) | 31 |
| pSMG-1 | pNZ8048 containing two copies of ori2009 (Per+) | This study |
| pSMG-2 | pNZ8048 containing three copies of ori2009 (Per+) | This study |
| pNZ44 | pNZ8048 containing constitutive P44 promoter from L. lactis chromosome | This study |
| pNZ44-rep2009 | pNZ44 containing rep2009 in sense orientation (Per+) | This study |
| pNZ44-orf14rev | pNZ44 containing orf14 in antisense orientation | This study |
| pNZ44-orf15rev | pNZ44 containing orf15 in antisense orientation | This study |
| pNZ44-rep2009rev | pNZ44 containing rep2009 in antisense orientation | This study |
| pNZ44-orf17rev | pNZ44 containing orf17 in antisense orientation | This study |
| pNZ44-orf18rev | pNZ44 containing orf18 in antisense orientation | This study |
| pNZ44-orf19rev | pNZ44 containing orf19 in antisense orientation | This study |
| pNZ44-msp1rev | pNZ44 containing msp1 in antisense orientation | This study |
| pNZ44-msp2rev | pNZ44 containing msp2 in antisense orientation | This study |
Media and growth conditions.
L. lactis strains were cultured at 30°C in M17 broth (Difco Laboratories, Detroit, Mich.) containing 0.5% glucose or in GSB, a modified version of LSB which contains glucose instead of lactose (4). Medium for plaque assays was prepared as described by Lillehaug (29). Briefly, double-layer agar plates containing M17 medium supplemented with glucose (5 g/liter), glycine (5 g/liter), and CaCl2 (10 mM) were prepared; 1% agar was used for the bottom layer, and 0.4% agar was used for the top layer. The medium was heat treated by boiling it for 5 min in a microwave. Escherichia coli strains were grown in Luria-Bertani medium at 37°C (40). Chloramphenicol (10 μg/ml) and (ampicillin (100 μg/ml) were added when appropriate.
Propagation and isolation of bacteriophages.
Tuc2009 was propagated on a prophage-free derivative of UC509 designated UC509.9. Bacteriophages Q30, Q33, and ul36 were propagated on SMQ86. Lactococcal strains were grown to the early log phase in GSB, and then CaCl2 (10 mM) and phage were added. Incubation was continued until lysis occurred. The lysates, containing approximately 109 PFU/ml, were filtered (pore size, 0.45 μm) and stored at 4°C until they were needed.
Plasmid and phage DNA isolation and molecular cloning.
Isolation of E. coli plasmid DNA and phage genomic DNA was accomplished by using a QIAprep spin plasmid miniprep kit (Qiagen, Inc., Chatsworth, Calif.) and a Qiagen lambda minikit kit, respectively, as recommended by the manufacturer. Restriction enzymes and T4 DNA ligase were purchased and used according to the instructions of the manufacturer (Boehringer GmbH, Mannheim, Germany). Genes homologous to rep2009 and orf17 from Q30, Q33, and ul36 were PCR amplified by using synthetic primers designed on the basis of the Tuc2009 DNA sequence (GenBank accession no. AF109874) and were cloned by using a TOPO TA Cloning kit according to the instructions of the manufacturer (Invitrogen Corporation, Carlsbad, Calif.).
DNA sequence analysis.
DNA sequence analysis was performed with a model 373A automated DNA sequencer (Applied Biosystems Inc., Foster City, Calif.) by using synthetic oligonucleotides (Oligo 1000M; Beckman Instruments) as primers. Sequences were assembled by using the seqman program of the DNASTAR software package. Database searches were performed by using the FASTA, BLASTN, and TBLASTN (3) programs with sequences present in the latest release of the nonredundant sequence databases (http://www.ncbi.nlm.nih.gov/). Sequences were aligned by using the Clustal method of the MEGALINE release 3.06 program of the DNASTAR 1996 release software package.
Electroporation procedure.
Electrotransformation of plasmid DNA into E. coli was performed essentially as described by Sambrook et al. (40). Electrotransformation of plasmid DNA into L. lactis was performed as described by Wells et al. (47).
Bacteriophage assays.
Plaque assays were performed as described by Lillehaug (29). Lysis-in-broth experiments were performed by growing strains in 10-ml portions of GSB to an optical density at 600 nm (OD600) of approximately 0.2, adding CaCl2 to a final concentration of 10 mM and 10 μl of a solution containing the desired concentration of phage particles, and measuring the OD600 over time.
Construction of Per and antisense constructs.
Construction of pNZRep-Cii has been described previously (31). pSMG-1 was constructed by cloning tandem copies of ori2009 in the NcoI-XbaI site of pNZ8048, while pSMG-2 was obtained by cloning a third copy of ori2009 in the XhoI site of pSMG-1. To create pNZ44, the nisin-inducible promoter (PnisA) of pNZ8048 was replaced with the constitutive P44 promoter from the L. lactis chromosome (43). Antisense constructs were generated by PCR amplifying open reading frames of interest, including potential Shine-Dalgarno sequences, with synthetic primers having suitable restriction sites at their 5′ ends and cloning in the reverse orientation behind the P44 promoter of pNZ44. Coordinates of the regions cloned and the oligonucleotides used are listed in Table 2.
TABLE 2.
Coordinates of the regions of the Tuc2009 chromosome amplified by PCR to construct antisense plasmidsa
| Antisense construct | Start
|
Stop
|
||
|---|---|---|---|---|
| Position | Oligonucleotide | Position | Oligonucleotide | |
| pNZ44-toporev | 5881 | AAATCTAGAAGTTTGAGGTAAATTATG | 6519 | AAACTGCAGTTATTTACTCCAATCTATATATG |
| pNZ44-ssbrev | 6495 | AAATCTAGAAGCATATATAGATTGGAG | 6971 | AAACTGCAGTTAAAATGGTAGGTCATC |
| pNZ44-rep2009rev | 7077b | AAATCTAGAATAGAAGAAAGGAGTATT | 7878b | AAACTGCAGTTAGATAAGACCCCCGAG |
| pNZ44-orf17rev | 7858b | AAATCTAGAACTCTCGGGGGTCTTATC | 8603b | AAACTGCAGTCATCTTTGTTGTCCTGTAC |
| pNZ44-methrev | 8586 | AAATCTAGAACAGGACAACAAAGATGAAG | 9358 | AAACTGCAGTTAAATTTCAAAGAGCGACAC |
| pNZ44-orf19rev | 9331 | AAATCTAGAATATGAGGTGTC | 9737 | AAACTGCAGTTATACCTCTGTAATTTC |
| pNZ44-msp1rev | 33453 | AAATCTAGACCAAATAATAGAAATAG | 34000 | AAACTGCAGTTTCTAATTCCGATAAAG |
| pNZ44-msp2rev | 23701 | AAATCTAGAATTTAGACTAAGATAGG | 24224 | AAACTGCAGGTCAGTAATATTTTCAGC |
GenBank accession no. AF109874.
Oligonucleotide also used to PCR amplify homologous genes from phages Q30, Q33, and u136.
Southern blot analysis.
Bacteriophage DNA was restricted with EcoRI and separated by 1.0% agarose gel electrophoresis (40). Following electrophoresis, the DNA was transferred to a Hybond N+ membrane (Amersham Corp., Amersham, United Kingdom) by capillary transfer by using 40 mM NaOH (40). An enhanced chemiluminescence kit (ECL; Amersham) was used to label DNA probes and to detect specific DNA fragments by using conditions specified by the supplier.
RNA isolation.
Bacterial cells were grown to the mid-log phase (OD600, 0.4 to 0.6), and then cells (4-ml aliquots) were harvested by centrifugation and the cell pellets were drop frozen in liquid nitrogen. Following sampling, the cell pellets were resuspended in 500 μl of buffer containing 20 mM sodium acetate, 1% sodium dodecyl sulfate, and 1 mM EDTA. Each cell suspension was immediately mixed with acid phenol-chloroform (1:1) (pH 4.7) that had been preheated to 65°C and 200 μl of glass beads (diameter, <100 μm). Samples were incubated for 10 min at 65°C with repeated vortexing, and this was followed by centrifugation at the maximum speed in a microcentrifuge for 10 min. The aqueous phase was extracted once with an equal amount of acid phenol-chloroform and transferred to a fresh Eppendorf tube containing 2 volumes of 96% ethanol. After incubation at −20°C for at least 1 h, RNA was collected by centrifugation at the maximum speed in a microcentrifuge for 20 min. RNA pellets were washed once in 70% ethanol and resuspended in TE buffer (10 mM Tris, 1 mM EDTA; pH 8.0) supplemented with 10 mM MgCl2. RNase-free DNase (Boehringer) and RNase inhibitor (Boehringer) were added to final concentrations of 10 and 5 U, respectively. Samples were incubated at room temperature for 30 min, after which they were treated with buffered phenol-chloroform (1:1) (pH 4.7) to remove the DNase and the RNase inhibitor. RNA was precipitated by adding 0.1 volume of 3 M sodium acetate (pH 5.2) and then 2.5 volumes of 96% ethanol. Samples were incubated overnight at −20°C to ensure the maximal yield. After centrifugation, pellets were washed with 70% ethanol and allowed to air dry for 15 min. The RNA was resuspended in 11 μl of diethyl pyrocarbonate-treated deionized water.
Northern analysis.
RNA samples were denatured with formamide and formaldehyde and were separated by 1.2% agarose gel electrophoresis (40). After electrophoresis, the RNA was transferred to a Hybond N+ membrane (Amersham) by capillary transfer by using 10 mM NaOH (40). An enhanced chemiluminescence kit (ECL; Amersham) was used to label DNA probes and detect transcripts by using conditions specified by the supplier.
Visualization of intracellular phage DNA replication.
The procedure described by Hill et al. (21) was used to isolate DNA at various times following infection. Briefly, UC509.9 cultures were grown to the early log phase (OD600, 0.3 to 0.4) and infected with Tuc2009 phage (multiplicity of infection, >1). At zero time, 2 ml of each culture was removed, and the cells were harvested by centrifugation for 2 min at the maximum speed with a bench top centrifuge. The pellets were then drop frozen in liquid nitrogen. The volume of culture from which cells was harvested was adjusted at subsequent times so that the same mass of cells was present in each sample. Total cellular DNA was isolated, and samples were restricted with the enzyme PstI, subjected to 1% agarose gel electrophoresis, and transferred to a Hybond N+ membrane (Amersham) (40). An ECL kit (Amersham) was used to label DNA probes and to detect phage DNA by using conditions specified by the supplier.
Nucleotide sequence accession numbers.
The rep2009 and orf17 homologues identified for Q30 have been deposited in the GenBank database under accession no. AF264632 and AF264633, respectively. Similarly, the rep2009 and orf17 homologues identified for Q33 have been deposited in the GenBank database under accession no. AF264634 and AF264635, respectively.
RESULTS
Tuc2009, Q30, Q33, and ul36 have similarly organized replication modules.
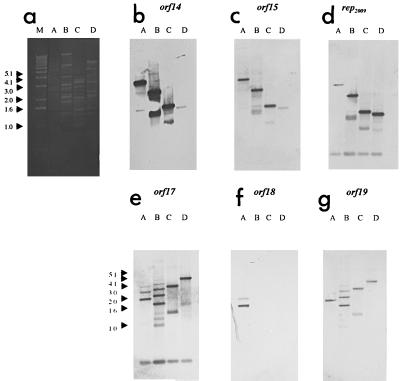
In order to determine if Per2009, the Tuc2009-derived Per system (31), was active against other phages and to examine if genes of the replication module are suitable targets for development of antisense strategies, Southern blot analysis was employed to compare genes of the Tuc2009 replication module with genes on the chromosomes of a number of P335 type phages (Fig. 1). DNA probes representing orf14 (encoding a putative topoisomerase), orf15 (encoding a putative single-stranded DNA binding protein), orf16 (rep2009 encoding the putative replisome organizer), orf17 (undetermined function), and orf19 (undetermined function) of Tuc2009 all hybridized with DNA of phages Q30, Q33, and ul36. Furthermore, synthetic oligonucleotide primers designed by using the Tuc2009 DNA sequence (Table 2) allowed amplification of genes homologous to rep2009 and orf17 of Tuc2009 from these three phages. The sequence of the replication module of ul36 has recently been made available (6) (GenBank accession no. AF212845) and contains two genes, orf255 and orf241, which are highly homologous to rep2009 and orf17 of Tuc2009, respectively. Sequence analysis revealed that these genes exhibit more than 90% DNA homology in all four phages. Significantly, the region previously designated ori2009 (31) is the same in all four phages. A DNA probe representing orf18 (putative methylase) of Tuc2009 did not hybridize to DNA from phages Q30, Q33, or ul36, indicating that this gene is apparently not present on any of these phages. Furthermore, PCR analysis verified that the organization of the replication module genes of both Q30 and Q33 is the same as the organization of orf14 to orf19 of Tuc2009 except for orf18, which is not present on either phage. This gene organization is consistent with that recently reported for ul36 (6).
FIG. 1.
Southern blot analysis showing hybridization of labeled Tuc2009 DNA probes to DNA fragments of bacteriophages Q30, Q33, and u136. (a) Ethidium bromide-stained agarose gel with EcoRI-restricted phage DNA. (b through g) Southern blot analysis. The Tuc2009 DNA probe used is indicated above each panel. Lane M contained DNA molecular weight marker X (Boehringer). Lanes A, Tuc2009; lanes B, Q30; lanes C, Q33; lanes D, ul36.
Increasing the number of origins of replication cloned increases the Per phenotype.
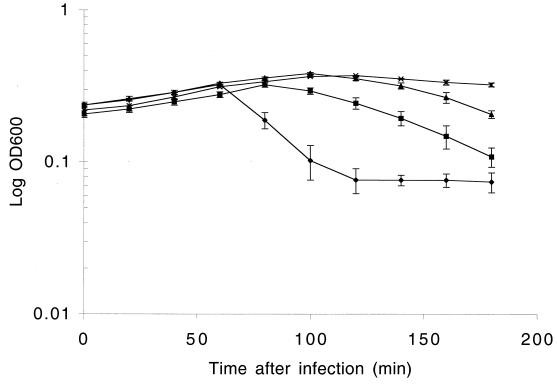
It has been reported that the degree of phage resistance conferred by Per plasmids is directly related to the plasmid copy number (38, 42). The Per2009 plasmid, pNZRep-Cii (31), was constructed by using vector pNZ8048, which has a reported copy number of approximately 50 copies per cell (42). In order to further increase the number of phage Tuc2009 origins (ori2009) supplied, pSMG-1 and pSMG-2 were constructed, which contained two and three copies of ori2009, respectively. Both of these plasmids were maintained in a stable condition in UC509.9 cells. As Table 3 shows, a direct relationship between the number of ori2009 fragments carried by the pNZ8048 derivatives and the phage resistance conferred was observed when UC509.9 harboring these plasmids was challenged with Tuc2009. Plaques on strains harboring pSMG-1 and pSMG-2 were difficult to enumerate because of their small size, and the difference in Per phenotype conferred was visualized better by lysis-in-broth experiments (Fig. 2).
TABLE 3.
Comparison of the effects of Per2009 and antisense plasmids on the EOP of the phages testeda
| Plasmid | Phage EOP
|
|||
|---|---|---|---|---|
| Tuc2009 | Q30 | Q33 | ul36 | |
| pNZ8048 | 1 | 1 | 1 | 1 |
| pNZrep-Cii | 1 × 10−1 (pp)b | 5 × 10−1 (sr) | 1 × 10−5 (I) | 1 × 10−5 (I) |
| pSMG-1 | 1 × 10−2 (pp) | 5 × 10−1 (sr) | 1 × 10−5 (I) | 1 × 10−5 (I) |
| pSMG-3 | 1 × 10−3 (pp) | 1 × 10−5 (I) | 1 × 10−6 (I) | 1 × 10−5 (I) |
| pNZRep-3 | 1 × 10−1 (pp) | NTc | NT | NT |
| pNZ44 | 1 | 1 | 1 | 1 |
| pNZ44-rep2009 | 5 × 10−1 (sr) | NT | NT | NT |
| pNZ44-toporev | 1 × 10−1 (sr) | 1 | 1 | 1 |
| pNZ44-ssbrev | 1 × 10−1 (sr) | 1 | 1 | 1 |
| pNZ44-rep2009rev | 1 × 10−6 (pp) | 1 × 10−3 (I) | 1 × 10−6 (I) | 1 × 10−5 (I) |
| pNZ44-orf17rev | 1 × 10−6 (pp) | 5 × 10−1 (sr) | 1 × 10−5 (I) | 1 × 10−4 (I) |
| pNZ44-methrev | 5 × 10−1 (sr) | 1 | 1 | 1 |
| pNZ44-orf19rev | 1 | 1 | 1 | 1 |
| pNZ44-msp1rev | 1 | NT | NT | NT |
| pNZ44-msp2rev | 1 (sr) | NT | NT | NT |
Tuc2009 was titrated on uc509.9. Q30, Q33, and ul36 were titrated on SMQ86.
pp, pinpoint plaques; sr, slight reduction in plaque size; I, irregularly shaped plaques (both small and large).
NT, not tested.
FIG. 2.
Lysis-in-broth experiment, demonstrating the increased levels of resistance conferred by increasing the numbers of copies of ori2009 cloned. Phage was added at zero time at a multiplicity of infection of approximately 1, and the OD600 was measured over time. Symbols: ⧫, UC509.9(pNZ8048); ■, UC509.9(pNZRep-Cii); ▴, UC509.9(pSMG-1); ×, UC509.9(pSMG-2). The data are averages based on three independent experiments.
The finding that phages Q30, Q33, and u136 contained sequences identical to the sequence of ori2009 indicated that ori2009-harboring plasmids (Per2009-conferring plasmids) would be effective at inhibiting proliferation of these phages. As indicated in Table 3, Per2009 plasmids confer resistance on SMQ86 against phages Q30, Q33, and u136, albeit to different degrees. The Per phenotype conferred by pNZRep-Cii is much more dramatic for Q33 and u136 than it is for Q30 or indeed Tuc2009, and increasing the copy number of ori2009 had little additional effect on the former two phages. In the case of Q30 there was little discernible phenotypic difference between the presence of one copy of ori2009 on a plasmid and the presence of two copies of ori2009 on a plasmid. However, when three copies of ori2009 were present, the resistance phenotype was greatly increased. These data demonstrate that the Per2009 system successfully inhibits proliferation of other phages belonging to the P335 group, which contain replication functions similar to those of Tuc2009.
Genes of the replication module are suitable targets for antisense strategies.
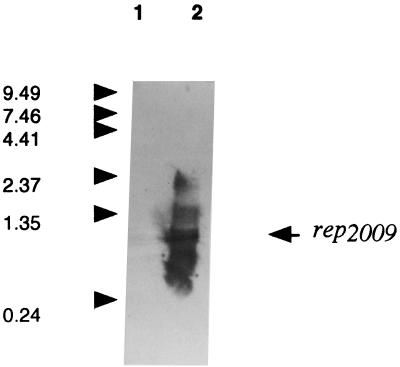
We have previously described a Tuc2009 genomic region containing genes assumed to be involved in DNA replication, including rep2009, which appears to be essential to the replication process (31). Results have also indicated that at least one additional phage-encoded protein other than Rep2009 is necessary for DNA replication initiation (31). In order to determine if antisense constructs directed against rep2009 and other genes from the assumed replication module of phages Tuc2009, Q30, Q33, and ul36 are effective in preventing phage proliferation, a number of antisense plasmids were constructed as described in Materials and Methods. Two plasmids targeting major structural proteins were also constructed in order to compare the efficiency when genes other than those of the replication module are targeted. As Table 3 shows, the levels of resistance conferred on UC509.9 by antisense constructs targeted against genes of the replication module were significantly greater than the level of resistance conferred by targeting the major structural proteins. The most dramatic resistance phenotypes were conferred by pNZ44-rep2009rev and pNZ44-orf17rev, and these two constructs also conferred significant protection on SMQ86 against Q33 and ul36 infections, indicating that the gene products of the rep2009 and orf17 homologues identified for these phages also play important roles in DNA replication. However, these plasmids conferred significantly less resistance against Q30 than against the other phages tested. This finding is in good agreement with the phenotypes conferred by Per2009 plasmids and indicates that expression of these proteins is significantly different for Q30 than it is for the other phages tested. Previous experiments have shown that the resistance conferred on UC509.9 against Tuc2009 is independent of the orientation in which the complete rep2009 gene is cloned and is identical to the resistance conferred by the minimum Per-conferring DNA fragment ori2009; i.e., there are pinpoint plaques and a 10-fold reduction in the efficiency of plating (EOP) (31). However, the resistance phenotype conferred by pNZ44-rep2009rev is dramatically increased; i.e., there are pinpoint plaques and a 6-log reduction in the EOP. The rep2009 gene cloned in the sense orientation in pNZ44 (pNZ44-rep2009) actually conferred a less efficient Per phenotype (EOP, 0.5) than pNZRep-Cii conferred (EOP, 0.1). This was probably due to transcription from the P44 promoter through ori2009, which interfered with Rep2009 binding. Northern blot analysis demonstrated that pNZ44-rep2009rev generates several RNA transcripts, the most prominent of which is approximately 800 bp long, whereas no such transcripts were detected with pNZRep-3 (Fig. 3). These results indicate that the observed phage resistance conferred by the antisense constructs was in fact due to transcription of antisense mRNA.
FIG. 3.
Northern blot analysis showing antisense rep2009 mRNA transcribed from pNZ44-rep2009rev. Lane 1, total RNA isolated from SMQ86 harboring pNZRep-3; lane 2, total RNA isolated from SMQ86 harboring pNZ44-rep2009rev. RNA samples were prepared and blotted as described in Materials and Methods. An 800-bp PCR DNA fragment representing rep2009 was used as a hybridization probe. Transcript size was estimated by using the 0.24- to 9.5-kb RNA ladder from Gibco BRL (Life Technologies Ltd., Paisley, United Kingdom). Sizes (in kilobases) are indicated on the left. The position of the main transcript representing antisense rep2009 mRNA is indicated on the right.
Antisense constructs targeted against replication module genes interfere with intracellular phage DNA replication.
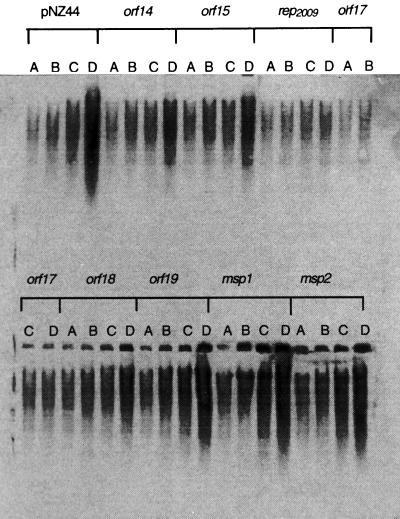
We have previously demonstrated that intracellular Tuc2009 DNA replication is severely inhibited in UC509.9 harboring a Per2009-conferring plasmid (31). In order to determine if the resistance phenotype conferred by antisense constructs was due to a similar mechanism, intracellular Tuc2009 DNA replication in strains harboring antisense constructs was monitored. As Fig. 4 shows, there was a direct correlation between the level of phage resistance conferred and the reduction in intracellular phage DNA replication for each of the antisense constructs targeted against replication module genes. The two constructs which conferred the most pronounced resistance phenotypes, pNZ44-rep2009rev and pNZ44-orf17rev, caused the most dramatic inhibition of DNA replication, whereas constructs targeting other genes of the replication module conferred much lower levels of resistance and had little or no effect on DNA replication. Constructs targeting the major structural proteins had no effect on DNA replication. These results indicate that transcription of antisense mRNA disrupts efficient translation of proteins essential for DNA replication, thus inhibiting phage proliferation. Antisense constructs that targeted orf14, orf15, rep2009, orf17, and orf18 of Tuc2009 caused some inhibition of phage DNA replication, resulting in the conferred Per phenotype, but only pNZ44-rep2009rev and pNZ44-orf17rev conferred resistance against phages Q30, Q33, and ul36. The latter result indicated that the rep2009 and orf17 homologues are also important for DNA replication in these phages.
FIG. 4.
Southern blot analysis showing the effects of antisense constructs on intracellular Tuc2009 DNA replication. Total cellular DNA was isolated at 20-min intervals from Tuc2009-infected UC509.9 harboring antisense constructs. Lanes A, zero time; lanes B, 20 min; lanes C, 40 min; lanes D, 60 min. The gene targeted by each antisense construct is indicated.
Isolation of phages that are not sensitive to engineered resistance mechanisms.
The ability of phages of the P335 species to overcome abortive infection or Per mechanisms has been reported previously, and it has been shown that Per- and Abi-insensitive phages can be isolated from large plaques in these instances (6, 15, 35). Large plaques, which were indicative of Per2009-insensitive (Per2009r) phages, were observed at a frequency of approximately 10−6 when phages Q30, Q33, and ul36 were titrated on L. lactis SMQ86 harboring Per2009-conferring plasmids (i.e., pNZRep-Cii, pSMG-1, and pSMG-2).
Plaque assays were performed with the three phages by using SMQ86 harboring pNZRep-Cii, and a single large plaque was picked in each case and propagated on SMQ86(pNZRep-Cii). The resulting three “new” phages, designated Q30r, Q33r, and ul36r, exhibited an EOP of 1 on strains harboring Per2009-conferring plasmids. It was notable that these Per2009r phages also exhibited an EOP of 1 when they were titrated on SMQ86 harboring antisense plasmid pNZ44-rep2009rev or pNZ44-orf17rev. Comparison of the genomes of the Per2009-sensitive and Per2009r phages by restriction analysis revealed significant differences in the genomic organizations of Q30r and ul36r and their parent phages (Fig. 5). Bouchard and Moineau (6) have recently described genomic reorganization in AbiK-insensitive ul36 mutant phages and demonstrated that these phages had exchanged DNA with phage sequences in the host chromosome via homologous recombination. No such genomic reorganization was observed for Q33r (Fig. 4B), and PCR and sequence analysis revealed that this phage still contained a rep2009 homologue identical to that of the Per2009-sensitive parent strain. Large plaques were also observed at a frequency of 10−6 when the parent phages were titrated on SMQ86 harboring antisense construct pNZ44-rep2009rev or pNZ44-orf17rev, which indicated that the large plaques also represented antisense-insensitive phages. Interestingly, the plaque morphology of Tuc2009 on UC509.9 harboring Per2009-conferring or antisense-producing plasmids was uniformly pinpoint, and Per2009r phage were never observed, indicating that evolution of engineered resistance-insensitive phages may depend on the lactococcal host.
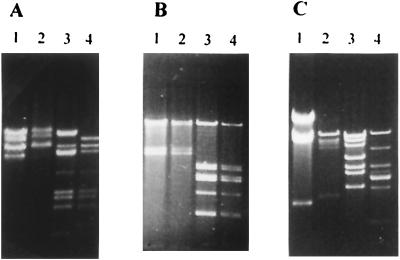
FIG. 5.
Restriction profiles of phage genomes comparing Per2009-sensitive and Per2009r phages. (A) Lanes 1 and 3, Per2009-sensitive phage Q30; lanes 2 and 4, Per2009-insensitive phage Q30r. (B) Lanes 1 and 3, Per2009-sensitive phage Q33; lanes 2 and 4, Per2009-insensitive phage Q33r. (C) Lanes 1 and 3, Per2009-sensitive phage ul36; lanes 2 and 4, Per2009-insensitive phage ul36r. In all panels lanes 1 and 2 contained preparations restricted with PstI and lanes 3 and 4 contained preparations restricted with EcoRV.
DISCUSSION
In this study we demonstrated that the Per2009 system may be used to inhibit proliferation of three other phages, Q30, Q33, and ul36. Furthermore, cloning of multiple copies of ori2009 on high-copy-number plasmid pNZ8048 was shown to significantly increase the efficiency of the Per2009 phenotype conferred against two of the four phages. This demonstrates that increasing the number of false phage origins of replication supplied in trans has the effect of titrating larger amounts of an essential replication factor. A dose-response relationship between the number of cloned ori2009 DNA fragments and the Per2009 phenotype conferred was observed for Tuc2009 (Table 3). A different relationship between the number of ori2009 DNA fragments cloned and the Per2009 phenotype was observed for the three other phages, Q30, Q33, and ul36 (Table 3). These results led us to conclude that these four phages employ similar means of DNA replication. However, it does appear that the overall replicative process is slightly different for Q33 and ul36 and significantly different for Q30. It may be that intracellular expression of phage replication factors is different for the four phages and that one copy of the ori2009 DNA fragment, supplied on plasmid pNZ8048, is sufficient to titrate limiting amounts of a replication factor for Q33 and ul36, whereas more copies are needed to achieve a similar effect for Tuc2009 and Q30. These results are in good agreement with those recently reported by Bouchard and Moineau (6). These authors described a Per system based on shuttle vector pMIG3. This Per system was significantly less effective than Per2009 at inhibiting Q33 and ul36 proliferation and actually conferred no resistance against Q30. pMIG3 has a low copy number in L. lactis (47), and therefore titration of essential phage DNA replication factors by ori sequences on this plasmid would be expected to be considerably less than titration of the same sequence supplied on the high-copy-number vector pNZ8048.
Five of the six antisense constructs targeted at the replication module open reading frames of Tuc2009 had an inhibitory effect on Tuc2009 proliferation and were shown to interfere with DNA replication, whereas no significant protection was provided by major structural protein-targeted constructs. In particular, pNZ44-rep2009rev and pNZ44-orf17rev were shown to be highly effective at inhibiting Tuc2009 DNA replication and provided significant protection against all four phages. To our knowledge, this represents the most efficient antisense strategy acting against bacteriophages infecting Lactococcus described to date. We have previously suggested that in addition to the Rep2009 protein, at least one other phage-encoded replication factor is essential for Tuc2009 DNA replication (31), and the results obtained in this study indicate that the protein product of orf17 is a very likely candidate. One of the possible functions of the orf17-encoded protein is as a helicase loader which facilitates delivery of the helicase to the origin of replication during DNA replication initiation (30). Genes coding for such helicase loader proteins have been identified immediately downstream of replisome organizer protein-encoding genes in lactococcal phage rlt (45) and Bacillus subtilis phage SPP1 (5).
Interestingly, it has been reported that ul36 is also sensitive to the φ50-derived Per50 system, and Moineau et al. (35) proposed that ul36 contains an origin of replication similar to that of φ50. A comparison of the sequences of the ul36 and Tuc2009 replication modules revealed extensive homology between the two sequences. However, no significant similarity was observed between the putative origins of replication for φ50 (ori50) and the ul36-Tuc2009 origin of replication sequence (ori2009). These observations led us to conclude that ul36 may in fact contain at least two functional origins of replication, a situation similar to that found in B. subtilis phage SPP1 (39) and S. thermophilus phage 7201 (42). In the latter case both origins of replication for 7201, ori7201A and ori7201B, were found to confer a Per phenotype when they were cloned independently.
It has been reported previously that when P335 phages are titrated on strains harboring Per or abortive infection plasmids, Perr and Abir mutant phages may be isolated (6, 15, 35, 38). Recently, Durmaz and Klaenhammer sequenced a 7.8-kb region of φ31.1, a recombinant Abir phage isolated after infection of NCK203 (Abi+) (15). This newly acquired region contained numerous regions of homology with temperate lactococcal bacteriophages, as well as homologues of lambda recombination protein BET and E. coli Holliday junction resolvase RUS, factors which may contribute to efficient recombination processes, as well as a new origin of replication.
Per2009r and antisense-resistant phages could be isolated when Q30, Q33, and ul36 were titrated on SMQ86 harboring Per2009 plasmids, and restriction analysis revealed that genomic reorganizations had taken place in Q30r and ul36r. The occurrence of these new recombinant phages was most likely due to acquisition of DNA from the host chromosome via homologous recombination, as recently described for ul36 when it was titrated on the AbiK+ Lactococcus strain SMQ88 (6). No obvious genomic reorganization appeared to have taken place in Q33r, indicating that the occurrence of this Per2009-insensitive phage was somewhat different than the occurrence of the other two phages. It may be that Q33r acquired a mutation which resulted in higher levels of expression of replication factors, thereby negating the effect of Per2009 and antisense plasmids. Interestingly, recombinant Per2009r phages were never isolated on UC509.9, indicating that prophages capable of exchanging DNA with Tuc2009 are not present in this strain. These observations add to the evidence that the presence of prophages on the chromosomes of Lactococcus strains may act as reservoirs for the evolution of new lytic phages (6, 15, 35).
In conclusion, by cloning multiple copies of ori2009, we improved the efficiency of the Per2009 system for preventing Tuc2009 proliferation, and we also identified three other phages against which the Per2009 system acts. We developed an optimized antisense strategy which is very effective at preventing proliferation of four P335 type phages. Data from experiments performed with antisense constructs further indicate that the Rep2009 protein and its homologues encoded by other P335 phages play an important role in DNA replication in these phages. Furthermore, a novel important DNA replication protein, Orf17, was identified for Tuc2009, and very similar proteins were present in other phages of the P335 species. Work is currently under way in our laboratory to elucidate the role that this protein plays in phage DNA replication.
ACKNOWLEDGMENTS
We thank Sylvain Moineau for supplying L. lactis SMQ86 and phages Q30, Q33, and ul36. We thank Michiel Kleerebezem for supplying plasmid pNZ8048. We also thank Jos Seegers for helpful discussions.
Stephen McGrath is the recipient of Forbairt research scholarship BR/96/196. This work was supported by a European Community Biotechnology grant (contract BIO4-CT96-0402).
REFERENCES
- 1.Alatossava T, Klaenhammer T R. Molecular characterization of three small isometric-headed bacteriophages which vary in their sensitivity to the lactococcal phage resistance plasmid pTR2030. Appl Environ Microbiol. 1991;57:1346–1353. doi: 10.1128/aem.57.5.1346-1353.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison G E, Klaenhammer T R. Phage resistance mechanisms in lactic acid bacteria. Int Dairy J. 1998;8:207–226. [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arendt E K, Daly C, Fitzgerald G F, van de Guchte M. Molecular characterization of lactococcal bacteriophage Tuc2009 and identification and analysis of genes encoding lysin, a putative holin, and two structural proteins. Appl Environ Microbiol. 1994;60:1875–1883. doi: 10.1128/aem.60.6.1875-1883.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayora S, Stasiak A, Alonso J C. The Bacillus subtilis bacteriophage SPP1 G39P delivers and activates the G40P DNA helicase upon interacting with the G38P-bound replication origin. J Mol Biol. 1999;288:71–85. doi: 10.1006/jmbi.1999.2662. [DOI] [PubMed] [Google Scholar]
- 6.Bouchard J D, Moineau S. Homologous recombination between a lactococcal bacteriophage and the chromosome of its host strain. Virology. 2000;270:65–75. doi: 10.1006/viro.2000.0226. [DOI] [PubMed] [Google Scholar]
- 7.Chung D K, Chung S K, Batt C A. Antisense RNA directed against the major capsid protein of Lactococcus lactis subsp. cremoris bacteriophage 4-1 confers partial resistance to the host. Appl Microbiol Biotechnol. 1992;37:79–83. doi: 10.1007/BF00174207. [DOI] [PubMed] [Google Scholar]
- 8.Chung D K, Kim J H, Batt C A. Cloning and nucleotide sequence of the major capsid protein from Lactococcus lactis ssp. cremoris bacteriophage F4–1. Gene. 1991;101:121–125. doi: 10.1016/0378-1119(91)90233-2. [DOI] [PubMed] [Google Scholar]
- 9.Coakley M, Fitzgerald G, Ros R P. Application and evaluation of the phage resistance- and bacteriocin-encoding plasmid pMRC01 for the improvement of dairy starter cultures. Appl Environ Microbiol. 1997;63:1434–1440. doi: 10.1128/aem.63.4.1434-1440.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffey A, Fitzgerald G F, Daly C. Identification and characterization of a plasmid encoding abortive infection from Lactococcus lactis subsp. lactis UC811. Neth Milk Dairy J. 1989;43:229–244. [Google Scholar]
- 11.Costello V A. Ph.D thesis. National University of Ireland. Cork, Ireland: University College Cork; 1988. [Google Scholar]
- 12.Daly C, Fitzgerald G F, Davis R. Biotechnology of lactic acid bacteria with special reference to bacteripophage resistance. Antoine Leeuwenhoek. 1996;70:99–110. doi: 10.1007/BF00395928. [DOI] [PubMed] [Google Scholar]
- 13.de Ruyter P G, Kuipers O P, de Vos W M. Controlled gene expression systems for Lactococcus lactis with the food- grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Djordjevic G M, O'Sullivan D J, Walker S A, Conkling M A, Klaenhammer T R. A triggered-suicide system designed as a defense against bacteriophages. J Bacteriol. 1997;179:6741–6748. doi: 10.1128/jb.179.21.6741-6748.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durmaz E, Klaenhammer T R. Genetic analysis of chromosomal regions of Lactococcus lactis acquired by recombinant lytic phages. Appl Environ Microbiol. 2000;66:895–903. doi: 10.1128/aem.66.3.895-903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foley S, Lucchini S, Zwahlen M C, Brussow H. A short noncoding viral DNA element showing characteristics of a replication origin confers bacteriophage resistance to Streptococcus thermophilus. Virology. 1998;250:377–387. doi: 10.1006/viro.1998.9387. [DOI] [PubMed] [Google Scholar]
- 17.Forde A, Daly C, Fitzgerald G F. Identification of four phage resistance plasmids from Lactococcus lactis subsp. cremoris HO2. Appl Environ Microbiol. 1999;65:1540–1547. doi: 10.1128/aem.65.4.1540-1547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garvey P, van Sinderen D, Twomey D P, Hill C, Fitzgerald G F. Molecular genetics of bacteriophage and natural phage defence systems in the genus Lactococcus. Int Dairy J. 1995;5:905–947. [Google Scholar]
- 19.Harrington A, Hill C. Construction of a bacteriophage-resistant derivative of Lactococcus lactis subsp. lactis 425A by using the conjugal plasmid pNP40. Appl Environ Microbiol. 1991;57:3405–3409. doi: 10.1128/aem.57.12.3405-3409.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill C. Bacteriophage and bacteriophage resistance in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:87–108. [Google Scholar]
- 21.Hill C, Miller L A, Klaenhammer T R. Cloning, expression, and sequence determination of a bacteriophage fragment encoding bacteriophage resistance in Lactococcus lactis. J Bacteriol. 1990;172:6419–6426. doi: 10.1128/jb.172.11.6419-6426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inouye M. Antisense RNA:its functions and applications in gene regulation—a review. Gene. 1988;72:25–34. doi: 10.1016/0378-1119(88)90124-2. [DOI] [PubMed] [Google Scholar]
- 23.Jarvis A W. Bacteriophages of lactic acid bacteria. J Dairy Sci. 1989;72:3406–3428. [Google Scholar]
- 24.Jarvis A W. Sources of lactic streptococcal phages in cheese plants. N Z J Dairy Sci. 1987;22:93–103. [Google Scholar]
- 25.Kim J H, Kim S G, Chung D K, Bor Y C, Batt C A. Use of antisense RNA to confer bacteriophage resistance in dairy starter cultures. J Ind Microbiol. 1992;10:71–78. doi: 10.1007/BF01583838. [DOI] [PubMed] [Google Scholar]
- 26.Kim S G, Batt C A. Antisense mRNA-mediated bacteriophage resistance in Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1991;57:1109–1113. doi: 10.1128/aem.57.4.1109-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S G, Bor Y C, Batt C A. Bacteriophage resistance in Lactococcus lactis ssp. lactis using antisense ribonucleic acid. J Dairy Sci. 1992;75:1761–1767. doi: 10.3168/jds.S0022-0302(92)77935-1. [DOI] [PubMed] [Google Scholar]
- 28.Klaenhammer T R, Fitzgerald G F. Bacteriophage and bacteriophage resistance. In: Gasson M J, de Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. Glasgow, United Kingdom: Blackie Academic and Professional; 1994. pp. 106–168. [Google Scholar]
- 29.Lillehaug D. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J Appl Microbiol. 1997;83:85–90. doi: 10.1046/j.1365-2672.1997.00193.x. [DOI] [PubMed] [Google Scholar]
- 30.Marians K J. Prokaryotic DNA replication. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 31.McGrath S, Seegers J F, Fitzgerald G F, van Sinderen D. Molecular characterization of a phage-encoded resistance system in Lactococcus lactis. Appl Environ Microbiol. 1999;65:1891–1899. doi: 10.1128/aem.65.5.1891-1899.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McIntyre K, Heap H A, Davey G P, Limsowtin G K Y. The distribution of lactococcal bacteriophage in the environment of a cheese manufacturing plant. Int Dairy J. 1991;1:183–197. [Google Scholar]
- 33.Moineau S, Borkaev M, Holler B J, Walker S J, Kondo J K, Vedamuthu E R, Vandenbergh P A. Isolation and characterization of lactococcal bacteriophages from cultured buttermilk plants in the United States. J Dairy Sci. 1996;79:2104–2111. [Google Scholar]
- 34.Moineau S, Fortier J, Ackermann H W, Pandian S. Characterization of lactococcal bacteriophages from Quebec cheese plants. Can J Microbiol. 1992;38:857–882. [Google Scholar]
- 35.Moineau S, Pandian S, Klaenhammer T R. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl Environ Microbiol. 1993;60:1832–1841. doi: 10.1128/aem.60.6.1832-1841.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moineau S, Pandian S, Klaenhammer T R. Restriction/modification systems and restriction endonucleases are more effective on lactococcal bacteriophages that have emerged recently in the dairy industry. Appl Environ Microbiol. 1992;59:197–202. doi: 10.1128/aem.59.1.197-202.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Sullivan D, Coffey A, Fitzgerald G F, Hill C, Ross R P. Design of a phage-insensitive lactococcal dairy starter via sequential transfer of naturally occurring conjugative plasmids. Appl Environ Microbiol. 1998;64:4618–4622. doi: 10.1128/aem.64.11.4618-4622.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Sullivan D J, Hill C, Klaenhammer T R. Effect of increasing the copy number of bacteriophage origins of replication in trans on incoming phage proliferation. Appl Environ Microbiol. 1993;59:2449–2456. doi: 10.1128/aem.59.8.2449-2456.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedre X, Weise F, Chai S, Luder G, Alonso J C. Analysis of cis and trans acting elements required for the initiation of DNA replication in the Bacillus subtilis bacteriophage SPP1. J Mol Biol. 1994;236:1324–1340. doi: 10.1016/0022-2836(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. .. [Google Scholar]
- 41.Sing W D, Klaenhammer T R. Conjugal transfer of bacteriophage resistance determinants on pTR2030 into Streptococcus cremoris strains. Appl Environ Microbiol. 1986;51:1264–1271. doi: 10.1128/aem.51.6.1264-1271.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stanley E, Walsh L, van der Zwet A, Fitzgerald G F, van Sinderen D. Identification of four loci isolated from two Streptococcus thermophilus phage genomes responsible for mediating bacteriophage resistance. FEMS Microbiol Lett. 2000;182:271–277. doi: 10.1111/j.1574-6968.2000.tb08907.x. [DOI] [PubMed] [Google Scholar]
- 43.van der Vossen J M B M, van der Leile D, Venema G. Isolation and characterization of Lactococcus lactis subsp. cremoris Wg2-specific promoters. Appl Environ Microbiol. 1987;53:2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Sinderen D, Karsens H, Kok J, Terpstra P, Ruiters M H, Venema G, Nauta A. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage rlt. Mol Microbiol. 1996;19:1343–1355. doi: 10.1111/j.1365-2958.1996.tb02478.x. [DOI] [PubMed] [Google Scholar]
- 45.Walker S A, Klaenhammer T R. An explosive antisense RNA strategy for inhibition of a lactococcal bacteriophage. Appl Environ Microbiol. 2000;66:310–319. doi: 10.1128/aem.66.1.310-319.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker S A, Klaenhammer T R. Molecular characterization of a phage-inducible middle promoter and its transcriptional activator from the lactococcal bacteriophage phi31. J Bacteriol. 1998;180:921–931. doi: 10.1128/jb.180.4.921-931.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wells J M, Wilson P W, Le Page R W. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol. 1993;74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]