Abstract
Background
Recent research has demonstrated that Type 2 Diabetes (T2D) risk is influenced by a number of common polymorphisms, including MC4R rs17782313, PPARG rs1801282, and TCF7L2 rs7903146. Knowledge of the association between these single nucleotide polymorphisms (SNPs) and body weight changes in different forms of prediabetes treatment is still limited. The aim of this study was to investigate the association of polymorphisms within the MC4R, PPARG, and TCF7L2 genes on the risk of carbohydrate metabolism disorders and body composition changes in overweight or obese patients with early carbohydrate metabolism disorders.
Methods and results
From 327 patients, a subgroup of 81 prediabetic female patients (48.7 ± 14.8 years) of Eastern European descent participated in a 3-month study comprised of diet therapy or diet therapy accompanied with metformin treatment. Bioelectrical impedance analysis and genotyping of MC4R rs17782313, PPARG rs1801282, and TCF7L2 rs7903146 polymorphisms were performed. The MC4R CC and TCF7L2 TT genotypes were associated with increased risk of T2D (OR = 1.46, p = 0.05 and OR = 2.47, p = 0.006, respectively). PPARG CC homozygotes experienced increased weight loss; however, no additional improvements were experienced with the addition of metformin. MC4R TT homozygotes who took metformin alongside dietary intervention experienced increased weight loss and reductions in fat mass (p < 0.05).
Conclusions
We have shown that the obesity-protective alleles (MC4R T and PPARG C) were positively associated with weight loss efficiency. Furthermore, we confirmed the previous association of the MC4R C and TCF7L2 T alleles with T2D risk.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11033-022-07254-y.
Keywords: Type 2 diabetes, Obesity, Diet, Single nucleotide polymorphism, Risk allele
Introduction
Type 2 Diabetes (T2D) develops as a result of a complex interaction between adverse environmental and certain genetic factors [1]. At present, over 700 DNA polymorphisms have been identified that are associated with altered risk of T2D [2–4]; as such, T2D disease development is polygenic in nature [5].
It is well established that T2D may develop through various different pathways, including insulin resistance (IR) and beta-cell function deficiency, suggesting that different gene polymorphisms may be involved in T2D pathogenesis. These genes include CDKAL1, CDKN2A, CDKN2B, TCF7L2, KCNJ11, UCP2, WFS1, and ABCC8, amongst many others [6]. Conversely, an increased T2D predisposition may be driven by severe insulin resistance, which itself can be modified through polymorphisms of the FTO, IRS1, PPARG, and PPARGC1A genes [6]. Most of the identified genes associated with T2D affect the insulin secretion [7]. In the Russian population, genes that influence insulin synthesis and secretion in β-cells of the pancreas also appear to be the main driver in the development of T2D [8].
Effective, timely treatment of early carbohydrate metabolism disorders in order to prevent the future development of T2D is one of the most significant practical problems facing modern diabetology. The excess of abdominal body fat is a relevant risk factor for the development of over-inflammation/oxidative stress, which worsens the prognosis for further development of carbohydrate metabolism disorders [9, 10]. Thus, alongside advocating glycemic control, recommendations from various endocrinological associations highlight the importance of a reduction in patient body weight by 5–10% from the initial presentation [11, 12]. Previously, patients were exclusively educated around the principles of a balanced diet, without prescribing concomitant drug therapy. Recent research suggests that drugs from the biguanide group, and, in particular, metformin, may be effective in the early treatment of T2D. Metformin has a hypoglycemic effect, can help reduce body weight, and also serve to normalize lipid profiles [13, 14]. However, not all patients effectively respond to biguanide drug therapy [15–17], with one of the key drivers of this inter-individual variation in treatment response being identified as polymorphisms in the genes regulating metabolism [18, 19].
The aim of the present study was to explore the association of polymorphisms within MC4R, PPARG, and TCF7L2 with the risk of different carbohydrate metabolism disorders and changes in the body composition in overweight or obese patients with early carbohydrate metabolism disorders.
Ethics statements
The study was approved by the Local Ethics Committee of Kazan State Medical University (No 10 of 18.12.2018) and was carried out in accordance with the Declaration of Helsinki as revised in 2000. All the subjects provided informed consent before participating in the study.
Participants
The study consisted of two parts: a case–control study and an intervention study. The case–control study involved 327 overweight or obese adults with T2D development risks (having first-degree relative with diabetes, history of cardiovascular diseases, hypertension ≥ 140/90 mmHg or undertaking therapy for hypertension, HDL cholesterol level < 35 mg/dL (0.90 mmol/L) and/or a triglyceride level > 250 mg/dL (2.82 mmol/L) in anamnesis, women with polycystic ovary syndrome, physical inactivity [11] and those who did not take medications that influence carbohydrate and fat metabolism. All of the patients underwent an oral glucose tolerance test and, as a result, 95 newly diagnosed prediabetic patients, 134 with the verified diagnosis of T2D, and 98 obese with normal glucose metabolism were identified. The average age at the time of the survey was 55.8 ± 12.7 years. Anthropometric parameters of all subgroups are shown in Supplementary Table 1. The control group included non-obese and non-diabetic healthy controls (all Caucasians of Eastern European descent and citizens of Russia).
Oral glucose tolerant test
At the beginning of the study all participants underwent an oral glucose tolerance test (OGTT) with a standard carbohydrate load of 75 g dry glucose diluted in 300–400 mL, as recommended by the American Diabetes Association (ADA) for diagnostics of T2D and prediabetes. The criteria for carbohydrate metabolism disorders were due to the Standards of Medical Care in Diabetes of ADA [11].
Intervention
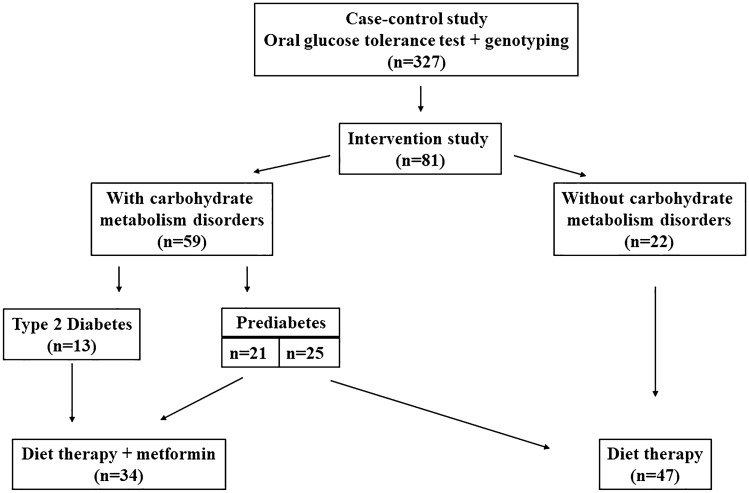
Out of the 327 patients, a subgroup of 81 female patients (48.7 ± 14.8 years) agreed to participate in a 3-month study comprised of further intervention (Fig. 1). Then the patients were divided into two groups utilising the simple randomization method. The first group comprised 47 patients (45.8 ± 14.8 years) undertaking diet therapy, which meant a balanced diet, with the exclusion of simple carbohydrate, and limiting complex carbohydrate and fat intake (the caloric intake was limited by 20 percent, and the macronutrient composition was comprised of 55% carbohydrate, 30% fat, and 15% protein). The second group comprised 34 patients (52.6 ± 14.2 years) who underwent the same diet therapy and 1500 mg/day metformin intake. The diet therapy guidelines and the dose of metformin were in accordance with Standards of Medical Care in Diabetes of ADA [11]. For the 3-month study period, once per week participant visits were performed, which involved food diaries checking.
Fig. 1.
Study design
Bioimpedancemetry
At the beginning of the study and at 3-month follow up, all participants underwent bioelectrical impedance analysis with “DIAMANT-AIST” body composition analyzer (Saint-Petersburg, Russia). Changes in body mass, BMI, waist and hip circumferences, fat mass, total water and body cell mass were characterized.
Genotyping
DNA was extracted from the blood samples of 327 participants using “AmpliPrime DNA-sorb-B” (NextBio, Moscow Russia), following the manufacturer’s recommended protocols. Nanodrop (ThermoFisher, USA) measurements were taken to measure the quality and quantity of the DNA. All samples were genotyped using allelic discrimination assays with TaqMan probes (Sintol, Moscow, Russia) on CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, California, USA). Assays were used for MC4R rs17782313 T/C, PPARG rs1801282 C/G, TCF7L2 rs7903146 C/T, including appropriate primers and fluorescently labeled probes to detect the alleles.
An initial genotyping study of MC4R rs17782313 in 304 DNA samples, of PPARG rs1801282 in 324 DNA samples, of TCF7L2 rs7903146 in 327 DNA samples was conducted. The distribution of genotypes and alleles of participants in the studied groups was compared with non-obese controls from the general (Russian) population: n = 172 for the MC4R rs17782313, n = 257 for the PPARG rs1801282, n = 404 for the TCF7L2 rs7903146.
Statistical analysis
Hardy–Weinberg equilibrium was tested by comparing the observed genotype frequencies with the expected frequencies using the Chi-square test with one degree of freedom in Microsoft Excel. Statistical analysis was conducted using GraphPad Instat. One-way analysis of variance (ANOVA) was applied to determine statistical significance among different groups. Paired t-tests were used to detect the significance of dynamic changes. Differences in phenotypes between groups were analyzed using unpaired t-tests. Body composition dynamics were calculated by percentage change of body composition parameters (% change from baseline) (Table 1). p < 0.05 was considered as statistically significant. Odds ratio with 95% confidence interval (CI) was used to assess the strength of the association of the investigated single nucleotide polymorphisms (SNPs). Association analysis was also performed assuming co-dominant, dominant, and recessive models.
Table 1.
Genotype and allelic frequencies of selected SNPs in patients and controls
| Gene, rs | Group | n | Genotypes | P1 | Risk allele, % | P2 | ||
|---|---|---|---|---|---|---|---|---|
| MC4R rs17782313 | TT | CT | CC | C | ||||
| T2D | 121 | 78 (64.5%) | 33 (27.3%) | 10 (8.3%) | 0.05* | 21.9 | 0.24 | |
| Prediabetes | 88 | 54 (61.4%) | 29 (33.0%) | 5 (5.7%) | 0.46 | 22.2 | 0.32 | |
| Overweight | 95 | 54 (56.8%) | 39 (41.1%) | 2 (2.1%) | 0.33 | 22.6 | 0.37 | |
| Controls | 172 | 92 (53.5%) | 70 (40.7%) | 10 (5.8%) | 1.00 | 26.2 | 1.00 | |
| PPARG rs1801282 | CC | CG | GG | C | ||||
| T2D | 131 | 93 (71.0%) | 29 (22.1%) | 9 (6.9%) | 0.003* | 82.1 | 0.07 | |
| Prediabetes | 94 | 69 (73.4%) | 23 (24.5%) | 2 (2.1%) | 0.39 | 85.6 | 0.65 | |
| Overweight | 99 | 69 (69.7%) | 25 (25.3%) | 5 (5.1%) | 0.03* | 82.3 | 0.11 | |
| Controls | 257 | 192 (74.7%) | 63 (24.5%) | 2 (0.8%) | 1.00 | 87.0 | 1.00 | |
| TCF7L2 rs7903146 | CC | CT | TT | T | ||||
| T2D | 131 | 73 (55.7%) | 41 (31.3%) | 17 (13.0%) | 0.02* | 28.6 | 0.04 | |
| Prediabetes | 91 | 44 (48.4%) | 32 (35.2%) | 15 (16.5%) | 0.001* | 34.1 | 0.001* | |
| Overweight | 105 | 58 (55.2%) | 32 (30.5%) | 15 (14.3%) | 0.01* | 29.5 | 0.03* | |
| Controls | 404 | 246 (60.9%) | 135 (33.4%) | 23 (5.7%) | 1.00 | 22.4 | 1.00 | |
*p < 0.05, statistically significant differences between participants with metabolic disorders and controls
Results
Case–control study
Genotype and allelic frequencies of three SNPs in patients and controls are shown in Table 1. The genotype distribution for each SNP was in agreement with the predicted Hardy–Weinberg equilibrium values (p > 0.05 in the T2D and control groups).
Genotype and allele frequencies of the TCF7L2 rs7903146 polymorphism differed significantly between type 2 diabetic patients and non-diabetic subjects (p = 0.02 and p = 0.04, respectively). The frequency of the risk (T) allele was 28.6% in T2D group and 22.4% in non-diabetic subjects, and this allele was significantly associated with T2D risk (OR = 1.39, 95% CI 1.01–1.90, p = 0.02). Moreover, the TT genotype was associated with a higher risk for T2D (OR = 2.47, 95% CI 1.28–4.78, p = 0.006). The T allele (OR = 1.66, 95% CI 1.05–2.63, p = 0.03) and TT genotype of the TCF7L2 rs7903146 (OR = 3.27, 95% CI 1.63–6.55, P = 0.0005) were associated with prediabetes. The TT genotype of TCF7L2 rs7903146 showed a significantly increased risk for obesity (OR = 2.76, 95% CI 1.38–5.50, p = 0.003). The risk genotype (CC) of MC4R rs17782313 SNP showed a significantly increased risk for T2D (OR = 1.46, 95% CI 0.59–3.62, p = 0.05). The C allele of PPARG rs1801282 SNP showed a significantly reduced risk for T2D (OR = 0.11, 95% CI 0.02–0.05, p = 0.0006) (Table 1).
Associations between MC4R, PPARG and TCF7L2 SNPs and weight loss efficiency
Participants in the diet therapy with metformin group showed a significant decrease in body weight (− 4.21 ± 0.67% vs. − 2.15 ± 0.48%; p = 0.01), BMI (− 1.77 ± 0.27% vs. − 0.87 ± 0.17%; p = 0.005), total water (− 0.38 ± 0.13% vs. + 0.02 ± 0.08%; p = 0.01) and body cell mass (− 0.44 ± 0.11% vs. − 0.15 ± 0.06%; p = 0.02) compared those participants in the dietary intervention only group (Table 2). There were no statistically significant differences in body composition changes between women with different menopausal status (Supplementary Table 2).
Table 2.
Changes in body composition in 81 women (% change from baseline)
| Parameter, % | Diet therapy | Diet therapy with metformin | p |
|---|---|---|---|
| Body weight | − 2.15 ± 0.48 | − 4.21 ± 0.67 | 0.01* |
| BMI | − 0.87 ± 0.17 | − 1.77 ± 0.27 | 0.005* |
| Waist circumference | − 4.25 ± 0.79 | − 4.36 ± 0.86 | 0.90 |
| Hip circumference | − 3.20 ± 0.87 | − 4.10 ± 0.88 | 0.34 |
| Waist/hip ratio | 1.49 ± 0.72 | − 0.51 ± 0.62 | 0.33 |
| Fat mass | − 0.90 ± 0.20 | − 1.20 ± 0.21 | 0.30 |
| Total water | + 0.02 ± 0.08 | − 0.38 ± 0.13 | 0.01* |
| Body cell mass | − 0.15 ± 0.06 | − 0.44 ± 0.11 | 0.02* |
Data are Mean ± SEM
*p < 0.05, statistically significant changes after interventions
Genotype and allele frequencies of three SNPs in the intervention groups are shown in Supplementary Table 3.
Carriers of the PPARG rs1801282 CC genotype experienced a more substantial decrease in body weight (− 2.92 ± 0.57% vs. − 0.33 ± 0.70%; p = 0.013), waist/hip ratio (− 2.78 ± 0.97% vs. 0.70 ± 1.52%; p = 0.05) and BMI (− 3.51 ± 0.61% vs. − 0.22 ± 0.87%; p = 0.01) compared with G allele carriers in the diet therapy group. No differences in the diet therapy with metformin group were found (p > 0.05) (Table 3).
Table 3.
The PPARG rs1801282 genotypes and body composition changes
| Parameter, % | Diet therapy | P | Diet therapy with metformin | P | ||
|---|---|---|---|---|---|---|
| CC (n = 33) | CG + GG (n = 14) | CC (n = 22) | CG + GG (n = 12) | |||
| Body weight | − 2.92 ± 0.57 | − 0.33 ± 0.70 | 0.013* | − 4.61 ± 0.83 | − 3.53 ± 1.15 | 0.45 |
| BMI | − 3.51 ± 0.61 | − 0.22 ± 0.87 | 0.01* | − 1.92 ± 0.94 | − 1.52 ± 1.39 | 0.49 |
| Waist circumference | − 4.82 ± 1.02 | − 2.49 ± 0.98 | 0.15 | − 3.83 ± 1.14 | − 5.25 ± 1.30 | 0.43 |
| Hip circumference | − 3.10 ± 0.99 | − 3.40 ± 1.8 | 0.86 | − 3.79 ± 1.13 | − 5.47 ± 1.41 | 0.37 |
| Waist/hip ratio | − 2.78 ± 0.97 | 0.70 ± 1.52 | 0.05* | − 0.51 ± 0.82 | − 0.53 ± 1.00 | 0.99 |
| Fat mass | − 1.10 ± 0.25 | − 0.36 ± 0.27 | 0.10 | − 1.25 ± 0.25 | − 1.15 ± 0.39 | 0.82 |
| Total water | − 0.05 ± 0.10 | + 0.25 ± 0.16 | 0.09 | − 0.48 ± 0.15 | − 0.21 ± 0.23 | 0.34 |
| Body cell mass | − 0.21 ± 0.07 | + 0.01 ± 0.08 | 0.10 | − 0.54 ± 0.14 | − 0.27 ± 0.20 | 0.29 |
Data are Mean ± SEM
*p < 0.05, statistically significant differences
Carriers of the MC4R rs17782313 TT genotype demonstrated a significantly greater reduction in body weight (− 5.35 ± 0.89% vs. − 2.5 ± 0.86%; p = 0.037), BMI (− 5.91 ± 0.95% vs. − 3.1 ± 1.22%; p = 0.044), hip circumference (− 5.98 ± 1.03% vs. − 2.07 ± 1.39%; p = 0.028) and fat mass (− 1.6 ± 0.28% vs. − 0.65 ± 0.26%; p = 0.027) compared with C allele carriers in the diet therapy with metformin group. No differences in diet therapy group were found (p > 0.05) (Table 4).
Table 4.
The MC4R rs17782313 genotypes and dynamics of body composition changes
| Parameter, % | Diet therapy | P | Diet therapy + metformin | P | ||
|---|---|---|---|---|---|---|
| TT (n = 30) | TC + CC (n = 14) | TT (n = 21) | TC + CC (n = 14) | |||
| Body weight | − 2.67 ± 0.59 | − 1.03 ± 0.76 | 0.11 | − 5.35 ± 0.89 | − 2.5 ± 0.86 | 0.037* |
| BMI | − 3.19 ± 0.65 | − 1.59 ± 0.87 | 0.16 | − 5.91 ± 0.95 | − 3.1 ± 1.22 | 0.04* |
| Waist circumference | − 4.56 ± 1.03 | − 3.59 ± 1.16 | 0.57 | − 5.48 ± 0.86 | − 2.66 ± 1.66 | 0.11 |
| Hip circumference | − 3.57 ± 1.09 | − 2.41 ± 1.45 | 0.54 | − 5.98 ± 1.03 | − 2.07 ± 1.39 | 0.028* |
| Waist/hip ratio | − 1.28 ± 0.84 | − 1.94 ± 1.40 | 0.67 | − 0.20 ± 0.74 | − 0.99 ± 1.11 | 0.54 |
| Fat mass | − 1.09 ± 0.26 | − 0.42 ± 0.26 | 0.12 | − 1.6 ± 0.28 | − 0.65 ± 0.26 | 0.027* |
| Total water | + 0.006 ± 0.11 | + 0.06 ± 0.12 | 0.78 | − 0.43 ± 0.19 | − 0.3 ± 0.15 | 0.61 |
| Body cell mass | − 0.17 ± 0.08 | − 0.10 ± 0.09 | 0.62 | − 1.03 ± 0.17 | − 1.33 ± 0.15 | 0.60 |
Data are mean ± SEM
*p < 0.05, statistically significant differences
No association between the TCF7L2 rs7903146 genotypes and body composition changes was found (Supplementary Table 4).
Discussion
In this study we explored the effect of a dietary intervention comprised of the exclusion of simple carbohydrates and limiting of complex carbohydrates and fats on body composition in overweight or obese patients with early carbohydrate metabolism disorders, and the influence of polymorphisms in genes related with the risk of T2D on these outcomes. We confirmed the association of MC4R rs17782313 C and TCF7L2 rs7903146 T alleles with the risk of T2D. Furthermore, we found that the protective alleles against obesity (MC4R rs17782313 T and PPARG rs1801282 C) were associated with weight loss effectiveness.
TCF7L2 encodes a transcription factor in the Wnt signaling pathway. Nucleotide substitution (C to T) in this gene increases the risk of T2D [20]. The association of TCF7L2 rs7903146 with the development of T2D is reproducible in many populations. In particular, in the present study of the Russian population, we revealed that the T allele of this polymorphism is more common in the participants with prediabetes, T2D and obesity. The same results were shown in a number of other studies, which demonstrate the association of TCF7L2 rs7903146 with T2D in across many ethnic populations [21–23]. However, there appears to be no significant association between the T allele of TCF7L2 rs7903146 and T2D risk in Chinese population [24].
The molecular mechanism of variation in TCF7L2 and risk of T2D and prediabetes is currently unknown. However, some studies demonstrate involvement of TCF7L2 in body weight regulation and the development of obesity and T2D due to impairment of β-cell function and then insulin secretion [25, 26]. The association of TCF7L2 rs7903146 with the outcome of metformin treatment also remains poorly studied, and represents a subject for active discussion by researchers in the future. A systematic review of 34 studies of the Cochrane Library and EMBASE demonstrated that this polymorphism is not associated with the effectiveness of metformin treatment [27]. We also didn’t find any significant differences in body composition changes in studied groups due to the variants of TCF7L2 rs7903146.
To date, most studies exploring the association of MC4R rs17782313 with risk of obesity and insulin resistance, high BMI and large waist circumference demonstrate an influence of the C allele [28–30]. It should be noted that melanocortin 4 receptor, which is encoded by MC4R, takes part in the regulation of insulin secretion [31], accordingly, nucleotide substitution (T to C) in the MC4R gene can lead to pathologies of lipid and carbohydrate metabolism.
This study demonstrates that the CC genotype of MC4R rs17782313 is more common in patients with T2D, but not prediabetes or obesity. Moreover, a number of studies have shown the association of polymorphisms within MC4R and the development of T2D [32]. Particularly, it was reported, that the MC4R rs17782313 C allele is most often associated with an increased T2D risk [33].
In our study, with adherence to diet therapy, C allele carriers and TT homozygotes of MC4R rs17782313 didn’t differ in body compositions changes, which is in agreement with the results of Diabetes Prevention Program (DPP) [34]. However, participants in our study who underwent metformin and diet therapy experienced a more substantial decrease in body weight and fat mass if there wereTT homozygotes. Although the effect of this polymorphism on the results of metformin therapy has not been previously studied, there is data demonstrating the association of MC4R rs9966412 and rs17066859 SNPs with loss of weight in patients while adding metformin to the treatment plan [35]. We suggest that it may be related with the action of melanocortin 4 on the production of insulin by the β-cells of the pancreas, as in case of monogenic form of obesity.
Among the known polymorphic markers of PPARG, rs1805192 is the most studied. The main role of the nuclear receptor superfamily PPARγ is the control of genetic expression, which influences the regulation of carbohydrate and lipid metabolism, adipogenesis, and synthesis of TNF-α, resistin and adiponectin.
At present, there is contradictory data on the association of PPARG rs1801282 with the development of metabolic syndrome (MS) and T2D. Accordingly, a number of studies demonstrated a decreased risk of MS and T2D in G allele carriers [36, 37]. In studies of other ethnic groups, this protective effect has not been found; indeed, the rs1801282 G allele was associated with increased risk of obesity in the populations of Spain, India and Mexico [38–41].
Cole SA et al. showed that the carriers of CG genotype are more predisposed to the increasing of BMI and development of obesity [40]. In contrast, Swarbrick et al., reported no association of CG genotype with obesity, arterial hypertension or T2D. Nevertheless, they made an interesting conclusion that lipid metabolism disorders are much more common among obese G allele carriers [42].
In our study, performed on a Russian population, we report results: the GG genotype was more common in patients with T2D, which is consistent with a number of studies on populations of Spain and India [38, 39]. However, the C allele was associated with a reduced risk of T2D. One potential explanation of this finding may be the relatively small number of GG genotype carriers.
We found that the C allele (protective against obesity) is associated with a more pronounced weight loss and decrease of waist/hip ratio (an indicator of abdominal fat tissue) among patients undertaking diet therapy, which is aligned with the results of earlier studies from Adamo et al. and Matsuo et al.; here, the G allele was more common among patients resistant to diet therapy [43, 44]. Furthermore, in DPP and the study of Frank et al., the PPARG rs1801282 G allele was associated with the short-term (up to 6 months) and long-term (up to 2 years) maintenance of achieved weight loss [34, 45]. We suggest that the resistance of G allele carriers to changes in body weight may be explained by the fact that the presence of this allele is associated with a decrease in the transcriptional activity of PPARγ [46, 47]. In this case, the PPARG rs1801282 C allele is associated with an increase in the transcriptional activity of PPARγ, which in turn leads to an increase in the sensitivity of cells to the action of insulin [48], promoting weight loss among patients. The absence of differences in weight loss between carriers of different alleles in the metformin plus diet therapy group may be explained by the suggestion that metformin may decrease the expression of PPARG [49].
The small number of subjects and the community setting, along with the exclusive use of female participants in the intervention study, all act as limitations to the present study. Also, it should be noted that other DNA polymorphisms and intervention methods such as exercise may potentially affect treatment outcomes [50]. In future, these limitations may be overcome by increasing of the number of participants, genetic markers, and including participants from both sexes.
Conclusions
We confirmed the association of MC4R rs17782313 and TCF7L2 rs7903146 SNPs with risk of T2D development. We also detected that MC4R rs17782313 and PPARG rs1801282 genotypes play an important role in body composition changes. CC homozygotes for PPARG rs1801282 appear to experience more pronounced weight loss; however, the addition of metformin to the treatment plan leveled any further changes in body weight. TT homozygotes for MC4R rs17782313 experienced increased weight loss and reductions in fat mass following the addition of metformin to diet therapy. According to our results, we suggest that these SNPs may be helpful in predicting the risk of T2D development and developing an optimal treatment plan in the early stages of the disorder. Further studies will show if the studied polymorphisms have a long-term effect on body composition changes.
Supplementary Information
Below is the link to the electronic supplementary material.
Funding
The authors have not disclosed any funding.
Declarations
Conflict of interest
The authors have no conflict of interest to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Elena V. Valeeva, Email: vevaleeva@ya.ru
Ildus I. Ahmetov, Email: i.akhmetov@ljmu.ac.uk
References
- 1.Park KS. The search for genetic risk factors of type 2 diabetes mellitus. Diabetes Metab J. 2011;35:12. doi: 10.4093/DMJ.2011.35.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krentz NAG, Gloyn AL. Insights into pancreatic islet cell dysfunction from type 2 diabetes mellitus genetics. Nat Rev Endocrinol. 2020;16:202–212. doi: 10.1038/S41574-020-0325-0. [DOI] [PubMed] [Google Scholar]
- 3.Hu M, Cherkaoui I, Misra S, Rutter GA. Functional genomics in pancreatic β cells: recent advances in gene deletion and genome editing technologies for diabetes research. Front Endocrinol. 2020;11:576632. doi: 10.3389/FENDO.2020.576632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vujcovic M, Keaton JM, Lynch JA, Miller DR. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat Genet. 2020;52:680–691. doi: 10.1038/S41588-020-0637-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingelsson E, McCarthy MI. Human genetics of obesity and Type 2 diabetes mellitus: past, present, and future. Circ Genom Precis Med. 2018;11:e002090. doi: 10.1161/CIRCGEN.118.002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruchat SM, Vohl MC, Weisnagel SJ, Rankinen T. Combining genetic markers and clinical risk factors improves the risk assessment of impaired glucose metabolism. Ann Med. 2010;42:196–206. doi: 10.3109/07853890903559716. [DOI] [PubMed] [Google Scholar]
- 7.Voight BF, Scott LJ, Steinthorsdottir V, Morris AP. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/NG.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikitin AG, Potapov VY, Brovkina OI, Koksharova EO. Association of polymorphic markers of genes FTO, KCNJ11, CDKAL1, SLC30A8, and CDKN2B with type 2 diabetes mellitus in the Russian population. PeerJ. 2017;5:e3414. doi: 10.7717/PEERJ.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Onofrio N, Pieretti G, Ciccarelli F, Gambardella A. Abdominal fat SIRT6 expression and its relationship with inflammatory and metabolic pathways in pre-diabetic overweight patients. Int J Mol Sci. 2019;20:1153. doi: 10.3390/ijms20051153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sardu C, Trotta MC, Pieretti G, Gatta G (2021) MicroRNAs modulation and clinical outcomes at 1 year of follow-up in obese patients with pre-diabetes treated with metformin vs. placebo. Acta Diabetol. 10.1007/s00592-021-01743-5 [DOI] [PubMed]
- 11.American Diabetes Association. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2021; Diabetes Care. 2021;44(Supplement 1):S15 LP-S33. 10.2337/dc21-S002 [DOI] [PubMed]
- 12.Garber AJ, Handelsman Y, Grunberger G, Einhorn D. Consensus statement by the american association of clinical endocrinologists and american college of endocrinology on the comprehensive type 2 diabetes management algorithm – 2020 executive summary. Endocr Pract. 2020;26:107–139. doi: 10.4158/CS-2019-0472. [DOI] [PubMed] [Google Scholar]
- 13.Donelly LA, Doney AS, Hattersley AT, Morris AD. The effect of obesity on glycaemic response to metformin or sulphonylureas in Type 2 diabetes. Diabet Med. 2006;23:128–133. doi: 10.1111/J.1464-5491.2005.01755.X. [DOI] [PubMed] [Google Scholar]
- 14.Jermendy G, Erdesz D, Nagy L, Yin D. Outcomes of adding second hypoglycemic drug after metformin monotherapy failure among type 2 diabetes in Hungary. Health Qual Life Outcomes. 2008;6:88. doi: 10.1186/1477-7525-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bankura B, Das M, Kumar Pattanayak A, Adhikary B. Inter-patient variability in clinical efficacy of metformin in type 2 diabetes mellitus patients in West Bengal. India J Metabolic Synd. 2016;5:2. doi: 10.4172/2167-0943.1000198. [DOI] [Google Scholar]
- 16.Cook MN, Girman CJ, Stein PP, Alexander CM. Glycemic control continues to deteriorate after sulfonylureas are added to metformin among patients with type 2 diabetes. Diabetes Care. 2005;28:995–1000. doi: 10.2337/DIACARE.28.5.995. [DOI] [PubMed] [Google Scholar]
- 17.Turner RC, Cull CA, Frighi V, Holman RR. Glycemic control with diet, sulfonylurea, metformin, or insulin in patients with type 2 diabetes mellitus: progressive requirement for multiple therapies (UKPDS 49). UK Prospective Diabetes Study (UKPDS) Group. JAMA. 1999;281:2005–2012. doi: 10.1001/JAMA.281.21.2005. [DOI] [PubMed] [Google Scholar]
- 18.Pawlyk AC, Giacomini KM, McKeon C, Shuldiner AR. Metformin pharmacogenomics: current status and future directions. Diabetes. 2014;63:2590–2599. doi: 10.2337/DB13-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang DD, Hu FB. Precision nutrition for prevention and management of type 2 diabetes. Lancet Diabetes Endocrinol. 2018;6:416–426. doi: 10.1016/S2213-8587(18)30037-8. [DOI] [PubMed] [Google Scholar]
- 20.Ding W, Xu L, Zhang L, Han Z. Meta-analysis of association between TCF7L2 polymorphism rs7903146 and type 2 diabetes mellitus. BMC Med Genet. 2018 doi: 10.1186/S12881-018-0553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsiao TJ, Lin E. A common rs7903146 variant of the transcription factor 7-like 2 gene is associated with type 2 diabetes mellitus and fasting glucose in a Taiwanese population. Diabetes Metab. 2017;43:83–85. doi: 10.1016/J.DIABET.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Zhou KC, Liu HW, Wang C, Fu YJ. Association of transcription factor 7-like 2 (TCF7L2) gene polymorphism with type 2 diabetes mellitus in Chinese Korean ethnicity population. Medicine. 2019;98:e14288. doi: 10.1097/MD.0000000000014288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mel’nikova ES, Rymar OD, Ivanova AA, Mustafina SV (2020) Association of polymorphisms of genes TCF7L2, FABP2, KCNQ1, ADIPOQ with the prognosisof the development of type 2 diabetes mellitus. Ter Arkh. 92:40–47. 10.26442/00403660.2020.10.000393 [DOI] [PubMed]
- 24.Zheng X, Ren W, Zhang S, Liu J. Association of type 2 diabetes susceptibility genes (TCF7L2, SLC30A8, PCSK1 and PCSK2) and proinsulin conversion in a Chinese population. Mol Biol Rep. 2012;39:17–23. doi: 10.1007/S11033-011-0705-6. [DOI] [PubMed] [Google Scholar]
- 25.Lyssenko V, Lupi R, Marchetti P, Del Guerra S. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007;117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loos RJ, Franks PW, Francis RW, Barosso I. TCF7L2 polymorphisms modulate proinsulin levels and beta-cell function in a British Europid population. Diabetes. 2007;56:1943–1947. doi: 10.2337/DB07-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maruthur NM, Gribble MO, Bennett WL, Bolen S. The pharmacogenetics of type 2 diabetes: a systematic review. Diabetes Care. 2014;37:876–886. doi: 10.2337/DC13-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyre D, Delplanque J, Chèvre JC, Lecoeur C. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/NG.301. [DOI] [PubMed] [Google Scholar]
- 29.Loos RJ, Lindgren CM, Li S, Wheeler E. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/NG.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sull JW, Lee M, Jee SH. Replication of genetic effects of MC4R polymorphisms on body mass index in a Korean population. Endocrine. 2013;44:675–679. doi: 10.1007/S12020-013-9909-Y. [DOI] [PubMed] [Google Scholar]
- 31.Yu K, Li L, Zhang L, Guo L. Association between MC4R rs17782313 genotype and obesity: a meta-analysis. Gene. 2020 doi: 10.1016/J.GENE.2020.144372. [DOI] [PubMed] [Google Scholar]
- 32.Xi B, Takeuchi F, Chandak GR, Kato N. Common polymorphism near the MC4R gene is associated with type 2 diabetes: data from a meta-analysis of 123,373 individuals. Diabetologia. 2012;55:2660–2666. doi: 10.1007/S00125-012-2655-5. [DOI] [PubMed] [Google Scholar]
- 33.Sull JW, Kim G, Jee SH (2020) Association of MC4R (rs17782313) with diabetes and cardiovascular disease in Korean men and women. BMC Med Genet. 21:1 21:1–6. 10.1186/S12881-020-01100-3 [DOI] [PMC free article] [PubMed]
- 34.Delahanty LM, Pan Q, Jablonski KA, Watson KE. Genetic predictors of weight loss and weight regain after intensive lifestyle modification, metformin treatment, or standard care in the diabetes prevention program. Diabetes Care. 2012;35:363–366. doi: 10.2337/DC11-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan Q, Delahanty LM, Jablonski KA, Knowler WC. Variation at the Melanocortin 4 Receptor gene and response to weight-loss interventions in the Diabetes Prevention Program. Obesity. 2013;21:E520. doi: 10.1002/OBY.20459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarhangi N, Sharifi F, Hashemian L, Doabsari MH. PPARG (Pro12Ala) genetic variant and risk of T2D: a systematic review and meta-analysis. Sci Rep. 2020;10(1):12764. doi: 10.1038/S41598-020-69363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Liu J, Ouyang Y, Fang M. The association between the Pro12Ala variant in the PPARγ2 gene and type 2 diabetes mellitus and obesity in a Chinese population. PLoS ONE. 2013;8(8):e71985. doi: 10.1371/JOURNAL.PONE.0071985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.González Sánchez JL, Serrano Ríos M, Fernández Perez C, Laakso M. Effect of the Pro12Ala polymorphism of the peroxisome proliferator-activated receptor gamma-2 gene on adiposity, insulin sensitivity and lipid profile in the Spanish population. Eur J Endocrinol. 2002;147:495–501. doi: 10.1530/EJE.0.1470495. [DOI] [PubMed] [Google Scholar]
- 39.Bhatt SP, Misra A, Sharma M, Luthra K. Ala/Ala genotype of Pro12Ala polymorphism in the peroxisome proliferator-activated receptor-γ2 gene is associated with obesity and insulin resistance in Asian Indians. Diabetes Technol Ther. 2012;14:828–834. doi: 10.1089/DIA.2011.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cole SA, Mitchell BD, Hsueh WC, Pineda P. The Pro12Ala variant of peroxisome proliferator-activated receptor-gamma2 (PPAR-gamma2) is associated with measures of obesity in Mexican Americans. Int J Obes Relat Metab Disord. 2000;24:522–524. doi: 10.1038/SJ.IJO.0801210. [DOI] [PubMed] [Google Scholar]
- 41.Mansoori A, Amini M, Kolahdooz F, Seyedrezazadeh E. Obesity and Pro12Ala polymorphism of peroxisome proliferator-activated receptor-gamma gene in healthy adults: a systematic review and meta-analysis. Ann Nutr Metab. 2015;67:104–118. doi: 10.1159/000439285. [DOI] [PubMed] [Google Scholar]
- 42.Swarbrick MM, Chapman CM, McQuillan BM, Hung J. A Pro12Ala polymorphism in the human peroxisome proliferator-activated receptor-gamma 2 is associated with combined hyperlipidaemia in obesity. Eur J Endocrinol. 2001;144:277–282. doi: 10.1530/EJE.0.1440277. [DOI] [PubMed] [Google Scholar]
- 43.Adamo KB, Dent R, Langfeld CD, Cox M. Peroxisome proliferator-activated receptor gamma 2 and acyl-CoA synthetase 5 polymorphisms influence diet response. Obesity. 2007;15:1068–1075. doi: 10.1038/OBY.2007.630. [DOI] [PubMed] [Google Scholar]
- 44.Matsuo T, Nakata Y, Katayama Y, Iemitsu M. PPARG genotype accounts for part of individual variation in body weight reduction in response to calorie restriction. Obesity. 2009;17:1924–1931. doi: 10.1038/OBY.2009.199. [DOI] [PubMed] [Google Scholar]
- 45.Franks PW, Jablonski KA, Delahanty L, Hanson RL. The Pro12Ala variant at the peroxisome proliferator-activated receptor gamma gene and change in obesity-related traits in the Diabetes Prevention Program. Diabetologia. 2007;50:2451–2460. doi: 10.1007/S00125-007-0826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meirhaeghe A, Amouyel P. Impact of genetic variation of PPARgamma in humans. Mol Genet Metab. 2004;83:93–102. doi: 10.1016/J.YMGME.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 47.Mazur II, Drozdovska S, Andrieieva O, Vinnichuk Y. PPARGC1A gene polymorphism is associated with exercise-induced fat loss. Mol Biol Rep. 2020;47:7451–7457. doi: 10.1007/S11033-020-05801-Z. [DOI] [PubMed] [Google Scholar]
- 48.Groop L, Pociot F. Genetics of diabetes–are we missing the genes or the disease? Mol Cell Endocrinol. 2014;382:726–739. doi: 10.1016/J.MCE.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 49.Choi RY, Ham JR, Lee HI, Cho HW. Scopoletin supplementation ameliorates steatosis and inflammation in diabetic mice. Phytother Res. 2017;31:1795–1804. doi: 10.1002/PTR.5925. [DOI] [PubMed] [Google Scholar]
- 50.Leońska-Duniec A, Ahmetov II, Zmijewski P. Genetic variants influencing effectiveness of exercise training programmes in obesity - an overview of human studies. Biol Sport. 2016;33(3):207–214. doi: 10.5604/20831862.1201052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.