Abstract
Tight junctions are essential for barrier integrity, inflammation, and cancer. Vitamin D and the vitamin D receptor (VDR) play important roles in colorectal cancer (CRC). Using the human CRC database, we found colonic VDR expression was low and significantly correlated with a reduction of Claudin-5 mRNA and protein. In the colon of VDRΔIEC mice, deletion of intestinal VDR led to lower protein and mRNA levels of Claudin-5. Intestinal permeability was increased in the VDR−/− colon cancer model. Lacking VDR and a reduction of Claudin-5 are associated with an increased number of tumors in the VDR−/− and VDRΔIEC mice. Furthermore, gain and loss functional studies have identified CLDN-5 as a downstream target of VDR. We identified the Vitamin D response element (VDRE) binding sites in a reporter system showed that VDRE in the Claudin-5 promoter is required for vitamin D3-induced Claudin-5 expression. Conditional epithelial VDR overexpression protected against the loss of Claudin-5 in response to inflammation and tumorigenesis in vivo. We also reported fecal VDR reduction in a colon cancer model. This study advances the understanding of how VDR regulates intestinal barrier functions in tumorigenesis and the possibility for identifying new biomarker and therapeutic targets to restore VDR-dependent functions in CRC.
Keywords: Claudin, barrier function, inflammation, colon cancer, colitis, tight junction, Vitamin D, vitamin D receptor
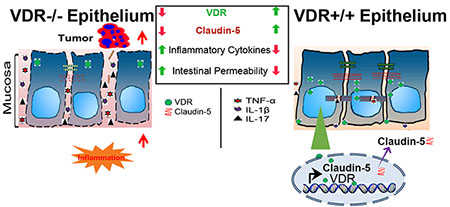
Graphical Abstract
Intestinal epithelial VDR is important for maintaining cellular and physiological levels of TJ protein Claudin-5 to prevent inflammation and tumorigenesis in the colon. At the molecular level, the CLDN-5 gene is a newly discovered downstream target of the transcriptional factor VDR. Lack of VDR led to a reduction of Claudin-5 and more tumors, whereas enhancing VDR increased Claudin-5 to protect the intestinal epithelial cells from tumorigenesis. Overall, we noted a link between VDR signaling and barrier functions in CRC.
Introduction
Tight junction structures are essential in intestinal innate immunity and barrier functions. The disruption of TJs is a common manifestation of various diseases, including chronic inflammation and cancers. Changes in expression and distribution of TJ proteins such as Claudin-2, −5, and −8 lead to discontinuous TJs and barrier dysfunction in active Crohn’s disease (CD), a type of inflammatory bowel disease 1. Claudin-5 is expressed in the epithelia and endothelia and form paracellular barriers and pores that determine permeability. This protein is downregulated in colon cancer 2, 3.
VDR is a nuclear receptor that mediates most known functions of the biologically active form of vitamin D 4–6. VDR possesses multiple critical roles in regulating innate and adaptive immunity, intestinal homeostasis, host response to microbiota, and tight junction structure 7–14. Vitamin D/VDR deficiency has been implicated in patients with inflammatory bowel disease and colon cancer 15–21. Our study demonstrated that VDR is essential for maintaining intestinal and microbial homeostasis 22, and protecting against intestinal tumorigenesis 23 24. Although vitamin D has been extensively studied, many critical questions about the biological functions of intestinal VDR in CRC remain unanswered. Although VDR and TJ proteins (e.g., Claudins) are involved in colon cancer, it remains unclear if they are closely related or function independently. Considering the multiple functional roles of VDR in the development of colon cancer 20, 24, it is important to dissect the cellular and molecular mechanisms by which VDR contributes to barrier function in protecting the host from tumorigenesis.
Here, we revisited the human CRC database and determined that colonic VDR expression is low and positively correlated with the reduction of the TJ protein Claudin-5 in CRC, including colitis-associated colon cancer. We investigate the novel role of VDR in regulating Claudin-5 expression using VDR−/− and intestinal epithelial VDR knockout mice (VDRΔIEC) in a colitis-associated colon cancer model. Human organoids, human colon cancer samples, VDR−/− mouse embryonic fibroblasts (MEF) cells, and cultured intestinal epithelial cells were used to determine the molecular mechanism. We determined that VDR is an important transcriptional regulator for maintaining physiological levels of the target gene Claudin-5 in the intestine. Furthermore, we generated a conditional intestinal epithelial VDR- overexpressed mouse model to study the protective role of VDR in the maintenance of TJs in the context of inflammation and tumorigenesis. Our goal is to provide a detailed understanding of how VDR status contributes to intestinal inflammation and cancer. Our finding may offer an additional avenue to treat colon cancer by restoring the barrier functions and developing a new protocol for risk assessment and prevention of cancer.
Results
Reduced VDR was positively correlated with low Claudin-5 expression in CRC patients
We first examined the gene expression levels of VDR and Claudin-5 in normal and human CRC samples by reviewing the GEO database GSE 4183 and GSE 8671 from Affymetrix data (human genome U133 Plus 2.0 arrays). Reduced VDR and Claudin-5 expression was observed in patients with CRC (Fig. 1A). To quantify and visualize the correlations between intestinal Claudin-5 and the VDR protein, we performed a correlation analysis between VDR and Claudin-5 and conducted a scatter plot with a regression line (Fig. 1B). We found significantly coordinated expression of VDR and Claudin-5 in biopsy samples collected from patients with CRC. We further analyzed data obtained from human colitis-associated colon cancer (Fig. 1C). VDR and Claudin-5 expression was significantly reduced in patients with colitis-associated CRC (GEO database GSE8671, GSE10714, and GSE37283) (Fig. 1C). We identified a positive correlation between VDR and Claudin-5 in biopsy samples collected from colitis-associated CRC patients and healthy controls (Fig. 1D). We then examined the protein levels of intestinal VDR in normal and CRC human colon samples using IHC. Compared to normal intestines, CRC patients with CRC possessed significantly lower VDR expression (Fig. 1E). Immunofluorescence (IF) staining of Claudin-5 revealed significantly lower Claudin-5 expression in CRC human colon samples (Fig. 1F). We performed correlation analysis and scatter plot of the staining intensity changes between the VDR protein and Claudin-5 in the colon. The results revealed that the staining intensity of Claudin-5 and intestinal VDR was positively associated with the Pearson correlation analysis (Fig. 1G). Thus, we revealed that colonic VDR expression is low and is correlated with the reduction of Claudin-5 at both mRNA and protein levels in human CRC, including colitis-associated colon cancer.
Fig. 1. Reduced VDR was correlated with low Claudin-5 expression in human colorectal cancer (CRC) patients.
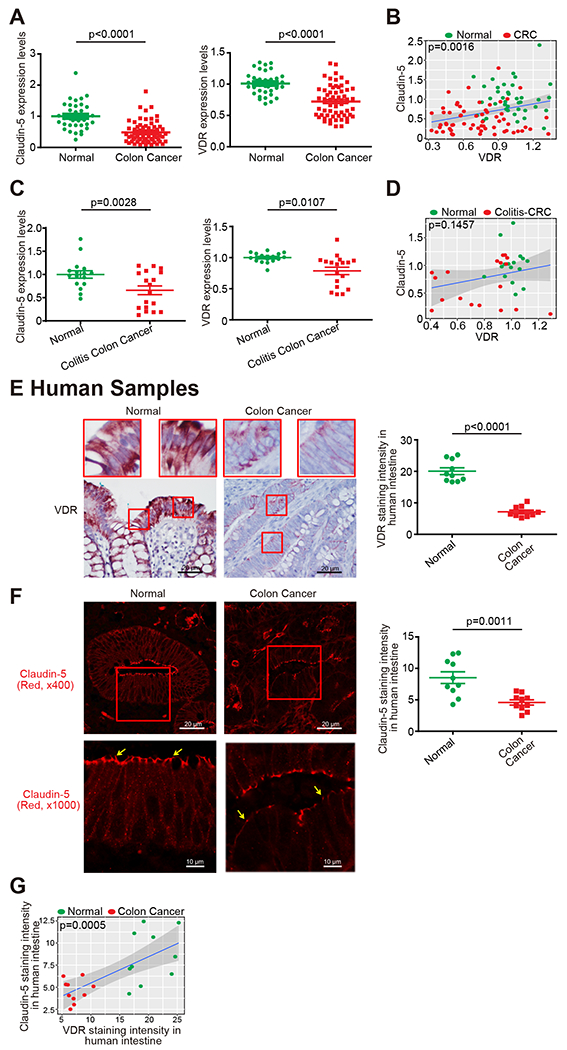
(A) Reduced VDR and Claudin-5 expression in patients with CRC (GEO database GSE4183 and GSE8671, (data were expressed as mean ± SD; Normal, n=40; CRC, n=62; Student’s t-test). All p values are shown in the figure. (B) Significantly coordinated (the Pearson correlation coefficient is 0.3083 with p-value = 0.001621) expression of VDR and Claudin-5 in biopsy samples collected from CRC patients. We performed a regression of VDR against Claudin-5 and conducted a scatter plot analysis with a regression line (GEO database GSE4183 and GSE8671, Normal, n=40; CRC, n=62; Intercept = 0.244; Slope = 0.5297). Values for healthy controls are presented in blue, and values for CRC patients are presented in red. (C) Reduced VDR and Claudin-5 expression in patients with Colitis-associated CRC (GEO database GSE8671, GSE10714 and GSE37283, data were expressed as mean ± SD; Normal, n=16; Colitis-associated CRC, n=18; Student’s t-test). (D) Coordinated expression of VDR and Claudin-5 in biopsy samples collected from Colitis-associated CRC patients. We performed a regression of VDR against Claudin-5 and conducted a scatter plot analysis with a regression line (GEO database GSE8671, GSE10714, and GSE37283 Normal, n=16; Colitis-associated CRC, n=18; the Pearson correlation coefficient is 0.2549 with p-value = 0.1457). (E) Intestinal VDR staining in normal and CRC human colon samples. Compared with normal intestines, the intestine from CRC patients possessed significantly lower VDR expression. (Images are representative of experiments performed in triplicate; Normal, n=10; Colorectal cancer, n=10; Student’s t-test). (F) IF staining of Claudin-5 in normal and CRC human colon samples. Compared to normal intestines, the intestines of CRC patients exhibited significantly lower Claudin-5 expression. (Images are representative of experiments in triplicate; Normal, n=10; Colon cancer, n=10; Student’s t-test). (G) The Pearson correlation analysis of staining intensity between intestinal Claudin-5 and VDR in human colon samples (the Pearson correlation coefficient is 0.7033 with p-value = 0.0005417. n = 10 for Normal and Colon cancer, respectively). All p values are shown in this figure.
Larger and more tumors developed in VDR deficient mice
Animal models have been developed to reflect the initiation and progression of human colon cancer 25. Azoxymethane (AOM) 26 mice develop hyperproliferative colonic mucosa, aberrant crypt foci (ACF), and eventually carcinomas 27. An AOM-dextran sulfate sodium 28 model is widely used to study colitis-associated colon cancer 29. We next investigated the role of VDR in regulating Claudin-5 expression in the development of cancer using an AOM/DSS treated mouse models (Fig. 2A). For wild-type VDR+/+ and whole body VDR knockout (VDR−/−) mice, representative colons with tumors are shown (Fig. 2B.) We observed that AOM/DSS-treated VDR−/− mice developed more tumors in the colon (Fig. 2C). The maximum tumor size was significantly larger in VDR−/− mice than in VDR+/+ mice (Fig. 2D). Furthermore, pathological analysis of colon samples indicated differences in tumor stage (carcinoma versus adenoma) between VDR−/− mice and the VDR+/+ AOM/DSS experimental groups (Fig. 2E). Epithelial hyperproliferation plays a critical role in the development of cancer. The IF data of the proliferative marker PCNA revealed that PCNA in the colon was significantly increased in the VDR−/− mice compared to that in the VDR+/+ mice (Fig. 2F). Chronic inflammation is one of the factors that contribute to CRC. We determined that serum cytokines TNF-α, IL-1β, and IL17 were significantly higher in the VDR−/− mice, compared to the VDR+/+ mice (Fig. 2G).
Fig. 2. VDR−/− mice developed a greater number of tumors compared to tumors in VDR+/+ mice.
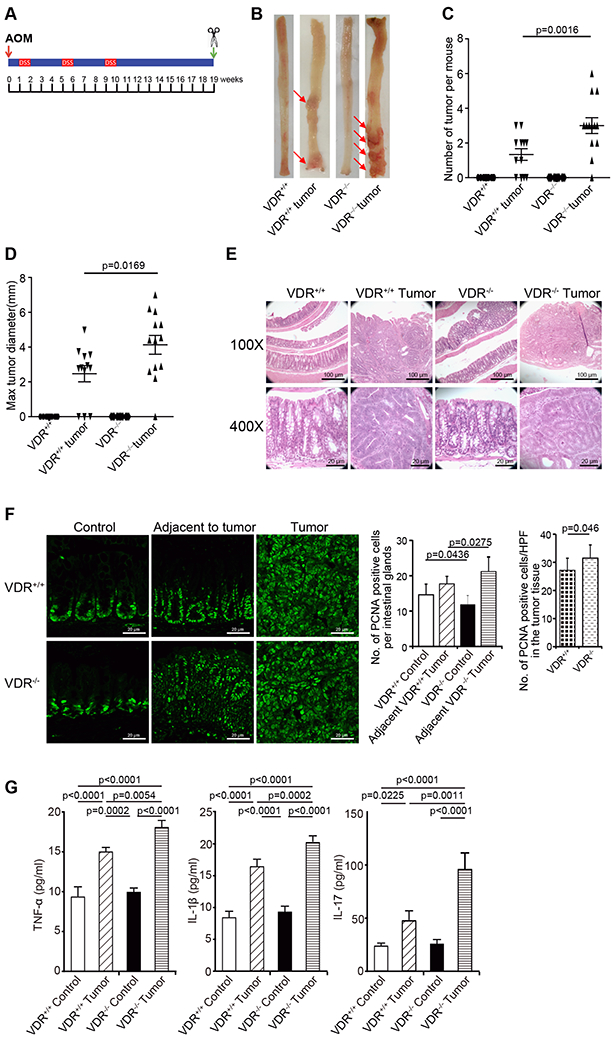
(A) Schematic overview of the AOM/DSS-induced colon cancer model. AOM (10 mg/kg) was injected on day 0. On Day 7, 2% DSS solution was administered to mice in drinking water. Seven days of DSS was followed by three weeks of drinking water free of DSS. An additional two cycles of DSS were administered prior to sacrifice at Week 19. (B) Colonic tumors in situ. Representative colons from different groups. Tumors were indicated by red arrows. (C) Tumor numbers in AOM-DSS induced colon cancer model: VDR+/+ and VDR−/− mice (data are expressed as mean ± SD. n = 10-13, one-way ANOVA test). (D) Max tumor size in AOM-DSS induced colon cancer model: VDR+/+ and VDR−/− mice (data are expressed as mean ± SD. n = 10-13, one-way ANOVA test). (E) Representative H&E staining of “Swiss rolls” of representative colons from the indicated groups. Images are from a single experiment and represent 10 mice per group. (F) Quantitation of PCNA-positive cells in control mucosa per intestinal glands or in the tumors tissue per high-power field. PCNA expression in the tumor tissue of VDR−/− mice was significantly higher than that in the VDR+/+ mice (data are expressed as mean ± SD. n = 5, Student’s t-test). (G) Serum cytokines such as TNF-α, IL-1β, and IL-17 were significantly increased, particularly in the AOM-DSS-induced VDR−/− mice colon cancer model. Each single experiment was assayed in triplicate. Data are expressed as mean ± SD. n = 6, one-way ANOVA test. All p values are shown in the figure.
VDR deletion leads to decreased Claudin-5 expression in tumor tissues
We examined changes in barrier function by testing intestinal permeability in mice with or without tumors. Mice were gavaged with fluorescein dextran (Molecular weight 3 kDa). After 4-h, blood samples were collected for fluorescence intensity measurement. Higher fluorescence intensity is indicative of higher intestinal permeability. As shown in Fig. 3A, AOM/DSS treatment increased intestinal permeability in both VDR+/+ and VDR−/−mice, while the VDR−/− mice exhibited significantly higher permeability post-treatment. Based on the in vivo intestinal permeability data, we hypothesized that the TJ proteins would be altered in the AOM/DSS mice. In the VDR−/− mice, we observed significant downregulation of Claudin-5 at the mRNA and protein levels in the colon (Fig. 3B & 3C). In the basal condition, Claudin-2 expression was decreased in VDR−/− mice, compared to the VDR+/+ mice. However, there was no difference in the tumor tissues between VDR+/+ and VDR−/− mice. The tight junction protein Claudin-7 did not show any changes either (Fig. 3C). Claudin-5 staining was observed at the crypt surface and at the lower portion of the intestine. Reduced Claudin-5 expression was confirmed through the immunostaining of colon tissues in the AOM/DSS mice (Fig. 3D & 3G). However, VDR deletion did not alter the expression of the TJ protein Claudin-7 in the colon of VDR−/− mice, compared to that of VDR+/+ mice (Fig. 3E). VDR expression was also decreased in mice with AOM/DSS-induced colon cancer (Fig. 3F & 3H). Moreover, we used our recently established method to measure VDR levels according to qPCR in fecal samples 30. We detected a significant reduction in VDR in fecal samples from mice with tumors (Fig. 3I). These data also suggest a decreased VDR in epithelial cells that are shed from mice with tumors.
Fig. 3. VDR deletion led to decreased Claudin-5 expression in tumor tissues.
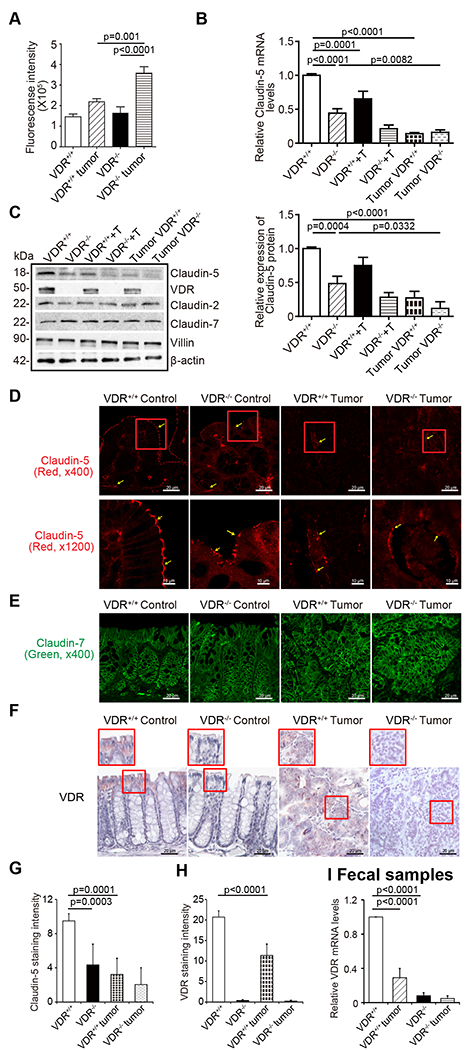
(A) Intestinal permeability increased in the AOM-DSS-induced VDR−/− mice colon cancer model. Fluorescein Dextran (Molecular weight 3 kDa, diluted in HBSS) was gavaged (50 mg/g mouse). Four hours later, mouse blood samples were collected for fluorescence intensity measurement (data are expressed as mean ± SD; n = 5 mice/group, 1-way ANOVA test). (B) VDR deletion decreased Claudin-5 at the mRNA level in the colon (data are expressed as mean ± SD. n = 5, one-way ANOVA test). (C) VDR deletion decreased Claudin-5 at the protein levels in the colon (data are expressed as mean ± SD. n = 5, one-way ANOVA test). Claudin-2 was decreased in the VDR−/− mice, compared to VDR+/+ mice in the basal condition, but there was no difference between the tumor tissue of VDR+/+ and VDR−/− mice. (D) and (G) Claudin-5 was decreased in the tumor tissue of VDR−/− mice, compared to levels in the tumor tissue of VDR+/+ mice according to immunofluorescence staining. Images are from a single experiment and represent 6 mice per group. (Data are expressed as mean ± SD. n = 6, one-way ANOVA test). (E) Claudin-7 was unchanged in the tumor tissue of VDR−/− mice, compared to levels in the tumor tissue of VDR+/+ mice according to immunofluorescence staining. Images are from a single experiment and represent 6 mice per group. (F) and (H) Intestinal VDR expression was decreased in the AOM-DSS-induced colon cancer model. Images are from a single experiment and represent 6 mice per group. (Data are expressed as mean ± SD. n = 6, one-way ANOVA test). (I) VDR levels in fecal samples were detected using RT-PCR. VDR expression was downregulated in the AOM-DSS-treated VDR+/+ mice (data are expressed as mean ± SD. n = 5, one-way ANOVA test). All p values are shown in this figure.
Conditional deletion of intestinal epithelial VDR led to increased permeability and reduced Clauidn-5 in the AOM/DSS cancer model.
To understand the tissue-specific role of Claudin-5 in colon cancer, we further studied intestinal permeability and Claudin-5 using a conditional intestinal epithelial VDR deletion VDRΔIEC model by fluorescein dextran measurement. We found that intestinal permeability indicated by the serum fluorescence intensity was significantly increased in VDRΔIEC mice (Fig. 4A). Intestinal epithelial VDR specific deletion led to significantly decreased Claudin-5 at the mRNA level in the colon (Fig. 4B) and further decreased in the mice with colon cancer. However, Claudin-2 was decreased in the VDR−/− mice, compared to the VDR+/+ mice at the basal condition, but not decreased in the tumor tissues of VDR+/+ and VDR−/− mice. Claudin-7 was not altered in the absence of VDR. At the protein level, we found reduced Claudin-5 in the VDRΔIEC mice (Fig. 4C). In tumor tissues of VDRΔIEC mice, epithelial Claudin-5 was disorganized and significantly decreased (Fig. 4D), compared to that in tumors of VDRloxp mice (Fig. 4F). In contrast, Claudin-7 was not altered in tumors from VDRΔIEC mice compared to the tumor tissue of VDRloxp mice (Fig. 4E). The VDR expression in fecal samples was downregulated in the AOM-DSS VDRloxp mice (Fig. 4G).
Fig. 4. VDR-specific deletion in mouse intestines lead to decreased Claudin-5 expression in tumor tissues.
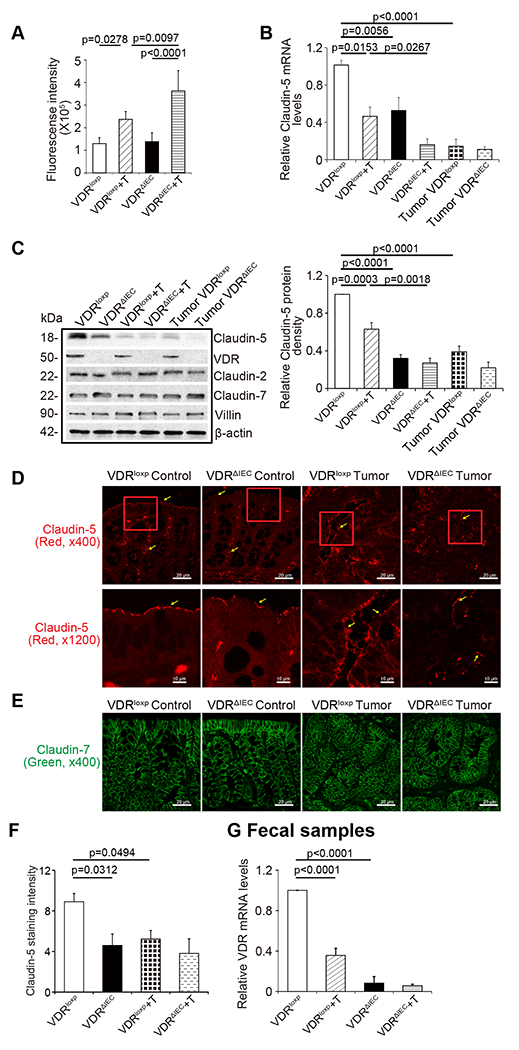
(A) Intestinal permeability was increased in the AOM-DSS-induce VDRΔIEC mice colon cancer model (data are expressed as mean ± SD; n = 5 mice/group, One-way ANOVA test). (B) VDR-specific deletion in mouse intestines decreased Claudin-5 at the mRNA level in the colon (data are expressed as mean ± SD. n = 5, one-way ANOVA test) (C) VDR-specific deletion in mouse intestines decreased Claudin-5 protein in the colon (data are expressed as mean ± SD. n = 5, one-way ANOVA test.) Claudin-2 was decreased in the VDRΔIEC mice, compared to the VDRloxp mice in the basal level, but not decreased in the tumor tissue of VDRΔIEC and VDRloxp mice. (D) Claudin-5 was decreased in the tumor tissue of VDRΔIEC mice compared to levels in the tumor tissue of VDRloxp mice according to immunofluorescence staining. Images are from a single experiment and represent 6 mice per group. (E) Claudin-7 expression was not changed in the AOM-DSS-induced VDRloxp mice colon cancer model. Images are from a single experiment and represent of 6 mice per group. (F) Intensity of the staining of Claudin-5. (Data are expressed as mean ± SD. n = 6, one-way ANOVA test). (G) VDR level in fecal samples was detected by RT-PCR. VDR expression was downregulated in the AOM-DSS-treated VDRloxp mice (data are expressed as mean ± SD. n = 3, one-way ANOVA test). All p values are shown in the figure.
Identification of the Vitamin D-response element (VDRE) in the Claudin-5 promoter
To confirm the direct regulation of VDR on Claudin-5, we examined various models at the basal level without any treatment in vivo and in vitro. In the VDR−/− mice, we observed that these mice possessed lower Claudin-5 protein levels in the colon than did VDR+/+ mice, and TJ Claudin-7 was not altered in the absence of VDR (Fig. S1A). We further detected significantly decreased mRNA levels of Claudin-5 in the intestine of VDR−/− mice (Fig. S1B). The density of Claudin-5 fluorescence staining was weaker in the VDR−/− mouse intestines (Fig. S1C). We also checked the specificity of intestinal VDR in regulating Claudin-5 expression in VDRΔIEC mice (Fig. S1D). Claudin-5 mRNA levels were significantly reduced in VDRΔIEC mice compared to those in VDR-lox mice (Fig. S1E). As expected, Claudin-7 expression remained unchanged. These data indicated that intestinal VDR specifically regulates the expression level of Claudin-5 in the colon. To confirm our finding in vitro, we used MEFs with VDR deletion. Lack of VDR led to a robust decrease of Claudin-5 at protein and mRNA levels in VDR−/− MEFs at the basal level (Fig. S1F & S1G). The density of Claudin-5 fluorescence staining was also weaker in the VDR−/− MEFs (Fig. S1H).
VDR acts as a transcription factor to regulate the expression of its target genes 31, 32. Activated VDR binds to the VDRE in the target gene promoter to regulate gene transcription 33. We reasoned that VDR might bind to the Claudin-5 promoter,thus altering the mRNA expression of the Claudin-5 gene. Further, we performed ChIP assay using the colon mucosal extract from VDR−/− mice and nonspecific IgG as a negative control to assess the binding of VDR to the Claudin-5 promoter. The samples were amplified by conventional PCR with Ikβα as positive control and Claudin-1 as a negative control as indicated in previous publications 34. CHIP-PCR demonstrated that VDR binds to the Claudin-5 promoter in the VDR+/+ mouse colon (Fig. 5A). The VDRE sequence (AGTTTGGTTGTGGGCT) within the Claudin-5 promoter region is shown in Figure 5B. We next assessed whether vitamin D3 could enhance Claudin-5 promoter activity through the VDRE binding sites. DNA sequences in the promoter region of vitamin D regulated genes were used in an in vitro reporter luciferase assay. A schematic drawing of transcriptional binding sites in the WT Claudin-5 promoter and its mutants are shown in Fig. 5C & Fig. S2. Plasmids with WT or deletions of VDRE, D2, or D3 in the Claudin-5 promoter binding were transfected into human SKCO15 cells, respectively followed by vitamin D3 treatment. We found Vitamin D3 enhanced WT-Claudin-5 promoter activity in cells. Deletions of D2 and D3 binding sites did not affect the Claudin-5 promoter activity. In contrast, deletions of VDRE binding sites clearly suppressed the promoter activity (Fig. 5D). These results demonstrate that the binding of VDRE binding sites in the Claudin-5 promoter is required for vitamin D3-induced Claudin-5 expression. However, siRNA-based Claudin-5 knock-down did not reduce VDR expression at the mRNA level (Fig. 5E). At the protein level, reduced Claudin-5 did not change the status of VDR protein or Claudin-7 at the protein level (Fig. 5F). Together, these results suggest that VDR transcriptionally regulates Claudin-5 at the mRNA level and that VDR is the upstream regulator of Claudin-5.
Fig 5. VDR binds to the Claudin-5 promoter in vivo and in vitro.
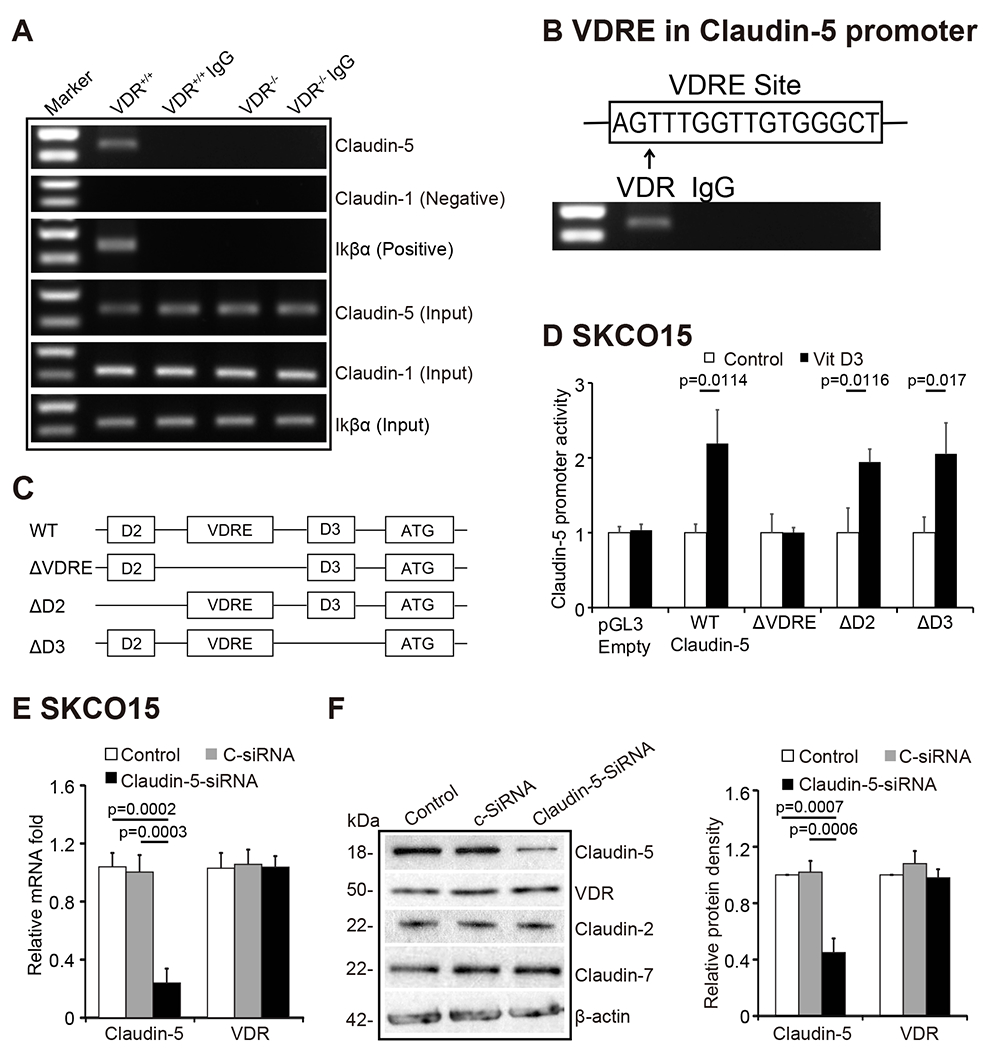
(A) CHIP-PCR amplification demonstrated that VDR binds to the promoter regions of Claudin-5 in mouse colons. PCR assays were performed and included input-positive controls and IgG/villin-negative controls. n = 3 separate experiments. (B) Claudin-5 promoter regions with VDRE sequence. (C) A schematic representation of transcriptional binding sites in the WT Claudin-5 promoter and deletion mutants. Plasmids include WT, binding site deletions of ΔVDRE, ΔD2, or ΔD3 in the Claudin-5 promoter. D2 and D3 domain were randomly selected upstream and downstream of the VDRE domain. (D) WT Claudin-5 reporter gene plasmids and the deletion mutant plasmids were transfected to SKCO15 cells. Luciferase activity was measured in the cell monolayers incubated in the absence or presence of vitamin D3 (20 nM) for 24 hours (data are expressed as mean ± SD. N =3, Student’s t-test). (E) Claudin-5 knockdown using siRNA (40 nM for 72 hours) did not reduce VDR expression at the mRNA level (data are expressed as mean ± SD. n = 3, one-way ANOVA test). (F) The protein expression in SKCO15 cells using siRNA (40 nM for 72 hours) (data are expressed as mean ± SD. n = 3, one-way ANOVA test). All p values are shown in the figure.
High VDR levels led to increased Claudin-5 protein and mRNA levels in vitro.
We then explored the possibility of enhancing VDR to maintain the physiological level of Claudin-5. Vitamin D3 is known to increase VDR expression and activate VDR signaling. We used the human colonic epithelial SKCO15 cell line that is widely used in studying TJs 35, 36. Claudin-5 mRNA level was significantly elevated in SKCO15 cells treated with Vitamin D3, whereas Claudin-7 mRNA was not altered by vitamin D3 treatment (Fig. 6A). The protein level of Claudin-5 and Claudin-2 were induced by Vitamin D3 (Fig. 6B). In vivo, Claudin-5 mRNA and protein levels were also increased in vitamin D3-treated mice (Fig. 6C & 6D). The protein level of Claudin-2 was also increased in vitamin D3-treated mice. (Fig. 6D). Colonoids are three-dimensional (3D) cell cultures that incorporate some of the key features of the represented organ 37. In this study, we developed human colonoids (Fig. 6E), and we observed vitamin D3 treatment significantly increased Claudin-5 mRNA level in these colonoids (Fig. 6F). Furthermore, vitamin D3 treatment significantly increased Claudin-5 and Claudin-2 protein levels in human colonoids, whereas there was no change of Claudin-7 after vitamin D3 treatment (Fig. 6G).
Fig 6. High VDR levels increased Claudin-5 at the protein and mRNA level in vitro.
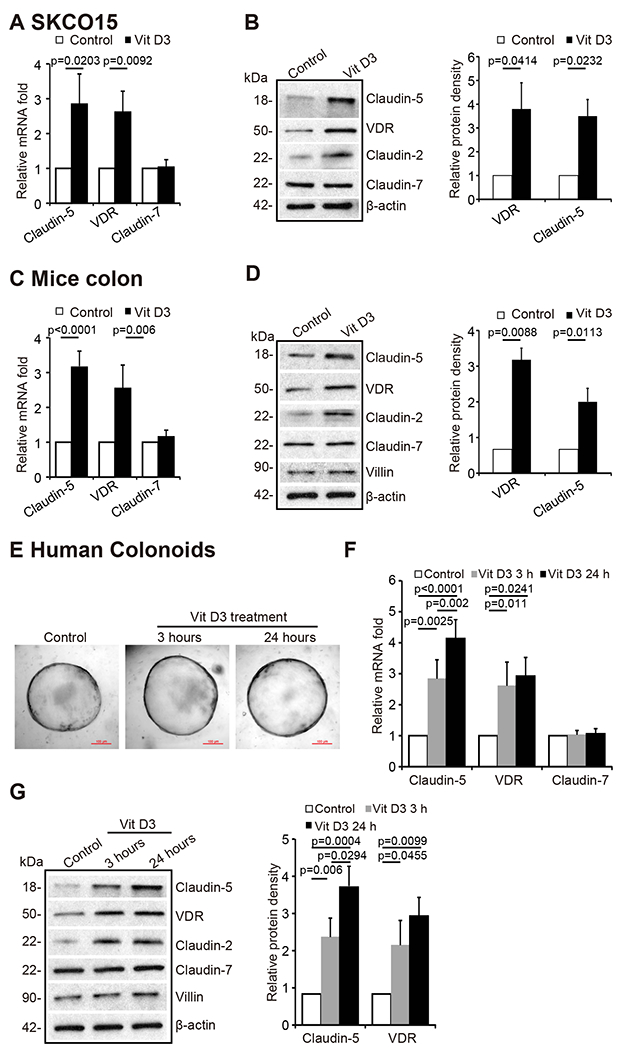
(A) Claudin-5 mRNA and (B) protein levels were increased after 24-hour vitamin D3 treatment at 20 nM in SKCO15 cells (data are expressed as mean ± SD. Student’s t-test, n = 3). (C) Claudin-5 mRNA and (D) protein levels were higher in vitamin D3- treated VDR+/+ mice. VDR+/+ mice (6-8 weeks) were gavaged by 0.2 μg vitamin D3 in 0.1 ml corn oil 3 times per week for 4 weeks (data are expressed as mean ± SD. Student’s t-test, n= 5 mice/group). (E) The micrographs showed representative human colonoids treated with Vit D3 (20 nM) for the indicated time points. (F) Claudin-5 mRNA and (G) protein levels were increased after vitamin D3 treatment in human colonoids (data are expressed as mean ± SD, n= 5, one-way ANOVA test). All p values are shown in the figure.
Intestinal epithelial VDR overexpression protected against the loss of Claudin-5 in respond to intestinal inflammation.
To further investigate the protective role of VDR in maintaining TJs in inflammation, we used a conditional intestinal epithelial VDR specific-overexpressed (O-VDR) mouse model generated in our previous study 38. Epithelial VDR overexpression in mouse intestines significantly increased Claudin-5 expression at both the mRNA and protein levels (Fig. 7A & 7B). Claudin-5 exhibited a minor decrease at the mRNA and protein levels in the colon of O-VDR mice treated with DSS, compared to that in the O-VDRloxp mice (Fig. 7C & 7D). Using IF staining, we determined that Claudin-5 was better preserved in the colon of O-VDR mice treated with DSS compared to that in the O-VDRloxp mice (Fig. 7E & 7G). As anticipated, Claudin-7 expression was unchanged in the intestinal tissue of O-VDR mice treated with DSS compared to that in the O-VDRloxp mice (Fig. 7F). VDR levels in fecal samples were detected using RT-PCR. VDR levels had a minor decrease in DSS-treated O-VDR mice, compared to that in O-VDRloxp mice treated with DSS (Fig. 7H). Moreover, there were fewer inflammatory cytokines such as IL-1β and IL-17 in the colons of DSS-induced O-VDR mice compared to O-VDRloxp mice (Fig. 7I).
Fig 7. Overexpressed intestinal epithelial VDR led to increased Claudin-5 and reduced inflammation in vivo.
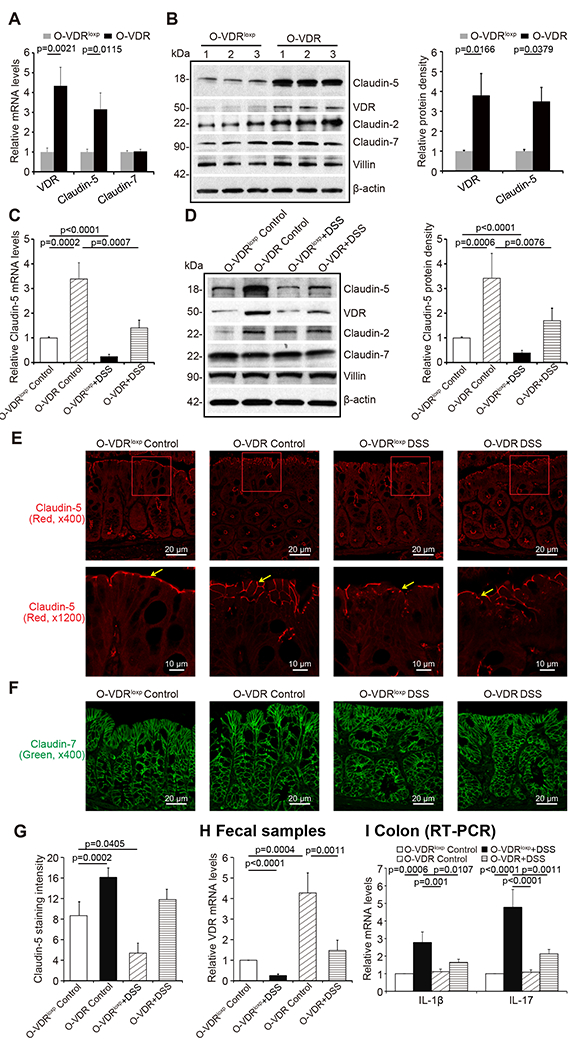
(A) VDR overexpression in mice intestines increased Claudin-5 expression in the colon at mRNA and (B) protein levels (data are expressed as mean ± SD. n = 3, one-way ANOVA test). (C) Claudin-5 was minorly decreased at the mRNA and (D) protein levels in the intestinal tissue of O-VDR mice treatment with DSS compared to levels in the O-VDRloxp mice (data are expressed as mean ± SD. n = 3, one-way ANOVA test). (E) Claudin-5 was minorly decreased in the intestinal tissue of O-VDR mice treated with DSS compared to levels in the O-VDRloxp mice, according to immunofluorescence staining. Images are from a single experiment and represent 5 mice per group. (F) Claudin-7 was unchanged in the intestinal tissue of O-VDR mice treated with DSS compared to levels in the O-VDRloxp mice according to immunofluorescence staining. Images are from a single experiment and represent 5 mice per group. (G) Intensity of the staining of Claudin-5. Images are from a single experiment and represent 5 mice per group. (Data are expressed as mean ± SD. n = 5, one-way ANOVA test). (H) VDR level in fecal samples was detected by RT-PCR. VDR expression was slightly decreased in O-VDR mice treated with DSS compared to O-VDRloxp mice treated with DSS (data are expressed as mean ± SD. n = 3, one-way ANOVA test). (I) The inflammatory cytokines IL-1β and IL-17 were less increased in the DSS-induced O-VDR mice colitis model compared to the levels in O-VDRloxp mice (data are expressed as mean ± SD. n = 3, one-way ANOVA test). All p values are shown in the figure.
Overexpressed intestinal epithelial VDR mice had less and smaller tumors in the AOM/DSS colon cancer model
The role of overexpression intestinal epithelial VDR was further examined in the development of colon cancer using an AOM/DSS model. Fig. 8A showed the tumors in the colon of representative O-VDRloxp and O-VDR mice. We observed that AOM/DSS-treated O-VDR mice developed fewer tumors than O-VDRloxp mice (Fig. 8B). The tumor size was significantly smaller in O-VDR mice than in O-VDRloxp mice (Fig. 8C). Claudin-5 exhibited a minor decrease at the mRNA and protein levels in the tumor tissue of O-VDR mice, compared to that in the tumors from O-VDRloxp mice (Fig. 8D & 8E). Moreover, there were fewer inflammatory cytokines, such as IL-1β and IL-17, in the tumor tissue of AOM/DSS-induced O-VDR mice, than those in O-VDRloxp mice (Fig. 8F).
Fig 8. Intestinal epithelial VDR overexpression mice have fewer and smaller tumors and show protection from decreased Claudin-5 and increased inflammation.
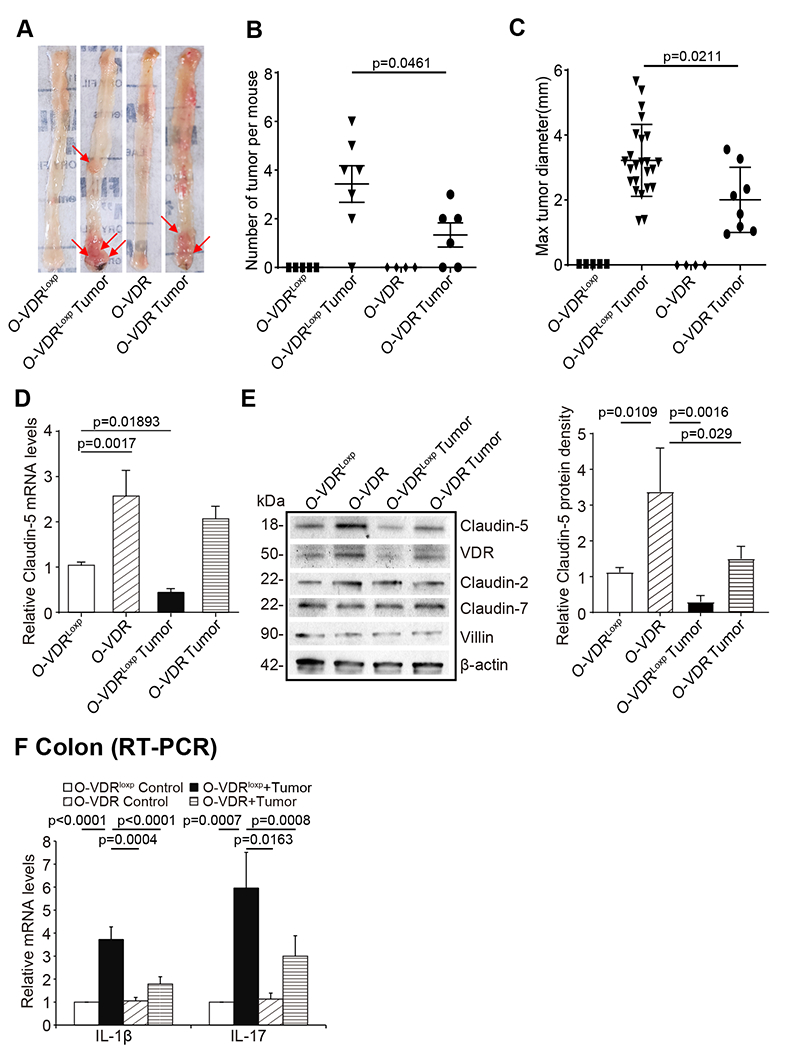
(A) Colonic tumors in situ. Representative colons from different groups. Tumors were indicated by red arrows. (B) Tumor numbers in AOM-DSS induced colon cancer model: O-VDRloxp and O-VDR mice (data are expressed as mean ± SD. n = 4-7, one-way ANOVA test,). (C) Max tumor size in AOM-DSS induced colon cancer model: O-VDRloxp and O-VDR mice (data are expressed as mean ± SD. n = 4-7, one-way ANOVA test). (D) Claudin-5 at the mRNA and (E) protein levels were decreased in the tumor tissue than in the control mice. O-VDRloxp mice tumor tissue had much more decrease than the O-VDR mice (data are expressed as mean ± SD. n = 3, one-way ANOVA test). (F) The inflammatory cytokines IL-1β and IL-17 were less increased in the AOM/DSS-induced O-VDR mice colon cancer model, than the levels in O-VDRloxp mice (data are expressed as mean ± SD. n = 3, one-way ANOVA test). All p values are shown in the figure.
Discussion
In the current study, we determined that low colonic VDR expression was significantly correlated with the reduction of Claudin-5 in human CRC. We demonstrated that VDR is important for maintaining cellular and physiological levels of TJ protein Claudin-5 in the colon to prevent inflammation and tumorigenesis. Our study further revealed a complex role for vitamin D/VDR regulation of CLDN-5 in the development of colon cancer. Lacking VDR reduced Claudin-5 in tumors, and enhanced VDR increased Claudin-5 to protect the intestinal epithelial cells from tumorigenesis. At the molecular level, our data have demonstrated that the CLDN-5 gene is a newly discovered downstream target of the transcriptional factor VDR. Overall, we noted a link between VDR signaling and barrier functions in CRC, thus suggesting a potential biomarker and target for a novel therapeutic strategy. Our study provides insight into how VDR signaling is involved in the tissue barrier related to tumorigenesis.
The intestinal barrier includes several elements that aid in its function as a physical and immunological barrier. These elements include the intestinal microbiota, secretory immunoglobulin A, antimicrobial peptides, the inner lamina propria, and epithelial cells. Epithelial cells play physical and physiological roles in health and disease at the cellular level. VDR signaling is involved in the epithelial barrier function related to various human diseases and remains largely unexplored 39. As a nuclear receptor, VDR mediates most known functions of 1,25-dihydroxyvitamin D (1,25(OH)2D3), the active form of vitamin D 4. However, the role of VDR has rarely been evaluated in studies examining human colon cancer. A recent study among patients with digestive tract cancer and vitamin D supplementation determined that when compared to placebo, this treatment did not result in significant improvement in relapse-free survival at 5 years 40. The dosage of vitamin D3 was insufficient among participants who possessed more severe deficiency at baseline. Therefore, the status of the VDR level must be considered over the course of many trials or as a biological measurement to clarify the underlying mechanisms. The traditional model of treatment using vitamin D that guided early vitamin studies should give way to a model incorporating more complex mechanisms of action of the Vitamin D/VDR system. Various methods have investigated the intestinal barrier, but the correlation of results across studies is difficult, representing a major shortcoming in the field 41.
The current study provides important insights into how VDR regulates Claudin-5 expression under normal physiological conditions and during tumor growth in the colon. We revealed a positive correlation between VDR and Claudin-5 at the mRNA and protein levels in healthy and tumor colons, thus suggesting the unique role of Claudin-5 in the intestine. There are 27 claudin family members that contribute to tight junctions 42, and not all Claudins are the same. Claudin-2 and Claudin-12 form paracellular Ca2+ channels in intestinal epithelia and are important for vitamin D-dependent calcium homeostasis 43. Our previous studies have shown that Claudin-2 is hyper regulated in colitis with VDR reduction 44, 45. Our current study has demonstrated the mechanism of the VDR-dependent function of Claudin-5 in the intestine. Interestingly, we found that the tight junction Claudin-7 was not altered in response to VDR-deficient status in the colon. In the lungs, VDR may play an important role in maintaining the pulmonary barrier integrity. We have reported that VDR deletion could increase lung permeability by altering the expression of TJ molecules, particularly Claudin-2, −4, −10, −12, and −18 46. Abnormal gut barrier function may serve as a biomarker for the risk of IBD onset 47. Our findings also suggest that the positively correlated status of VDR and Claudin-5 could be potentially applied to risk assessment, early detection, and prevention of CRC, including colitis-associated colon cancer.
Colorectal cancer is the second-leading cause of cancer-related death and is most curable in its early stages. Targeting barrier functions and microbiome 48 has been made regarding colon cancer therapy. Our study has provided a detailed understanding of how VDR status contributes to changes in TJs in the context of intestinal inflammation and colon cancer. Currently, there are no guidelines for monitoring vitamin D status, treating hypovitaminosis D, and maintaining optimal vitamin D stores in patients with IBD 49 or CRC. These tasks may prove particularly difficult due to malabsorption, gastrointestinal losses, and increased permeability associated with intestinal dysfunction. Based on the research progress regarding the novel roles of VDR in intestinal immunity and barrier functions, we expect that studies on VDR in intestinal barriers of colitis and colon cancer will have a marked impact on the prevention, diagnosis, and therapy of colitis and colon cancer patients.
Barrier function and VDR status are not only essential for the maintenance of intestinal homeostasis, but they are also critical for the development of chronic mucosal inflammation and cancer. Gut microbiome regulate both innate and adaptive immunity of the host. A conditional deletion of the VDR in intestinal epithelial cells or immune cells led to different changes of the intestinal virome and altered viral-bacterial interactions 50. CRC is a disease determimed by multiple factors, including host genetic backgound, immunity, environment, and microbiome 51–53. Malfunction of the innate immune system and barrier function may promote the development of CRC. Moreover, VDR is known to protects against dysbiosis and tumorigenesi 24. This knowledge can be used to develop intestinal VDR-associated mirobiome and Claudin-5 as clinical biomarkers for identifying patients who may benefit from currently available interventions and could also be used for the eventual development of novel strategies for the prevention and treatment of human CRC.
Materials and Methods
Human tissue samples
This study was performed in accordance with approval from the University of Rochester Ethics Committee (RSRB00037178) and UIC Ethics Committee (Institutional Review Board: 2017-0384). Colorectal tissue samples were obtained from 10 CRC patients with neoplasia and 10 patients without neoplasia (49-74years old). Human tissues for organoids are from healthy volunteers.
Gene expression datasets
For expression analyses, we used microarray data reported in the NCBI Gene Expression Omnibus database (GEO). To find the correlation between VDR and Claudin-5 at the gene expression level, we gathered data by searching the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) for expression profiling studies using colonic samples from colon cancer subjects. We randomly identified the GEO database reference series: GSE4183 54, GSE8671 55, GSE10714 56, and GSE37283 57. In these studies, the authors performed microarray analysis using colonic biopsy samples from healthy controls as well as from the inflamed and non-inflamed colonic mucosa from CRC subjects. From the databases, 40 healthy controls and 62 CRC patients were randomly selected for the CRC group, while 16 healthy controls and 18 colitis-associated CRC were randomly selected for the colitis-associated group. Both were subjected to further analyses.
Animals
VDR−/− mice on a C57BL/6 background were obtained by breeding heterozygous VDR+/− mice 45. VDRΔIEC mice were obtained by crossing the VDRLoxP-B mice, originally provided by Dr. Geert Carmeliet, with villin-cre mice (Jackson Laboratory, 004586), as we previously reported 23, 31, 58. We further backcrossed this strain with C57BL/6 mice for more than 10 generations after arriving our animal facility.
Intestinal-specific VDR-overexpressing (O-VDR) mice were generated in C57BL/6 mice strain background, as reported in our recent study 38. The mouse VDR (mVDR) sequence was cloned into the Stbl3 vector (size 6631 bp). The mVDR was cloned in (from ~2210 bp to ~3316bp) under EF1A promoter (1bp to 1105bp). A LoxP site was integrated after EF1A promoter (from 1105 bp to 2210 bp). VDR expression in O-VDR mice is Cre driven 38. This O-VDRloxp mouse line is labeled as O-VDRloxp in our gain of function study, distinct from the VDRloxP/loxP mouse made for VDRΔIEC mice.
Experiments were performed on 2-3 months old mice, including male and female. Mice were provided with water ad libitum and maintained in a 12 h dark/light cycle. The animal work was approved by the Rush University Animal Resources committee and UIC Office of Animal Care. The animal work was approved by the UIC Office of Animal Care (ACC 15-231,17-218, and 18-216).
Induction of colon cancer by AOM-DSS in mice
Mice were treated with 10mg/kg of AOM (Sigma-Aldrich, Milwaukee, WI, USA) by intraperitoneal injection as previously described 24. After a 7-day recovery period, mice received three cycles of 2% DSS in the drinking water. Tumor counts and measurements were performed in a blinded fashion under a stereo-dissecting microscope (Nikon SMZ1000, Melville, NY, USA). Microscopic analysis was performed for the severity of inflammation and dysplasia on hematoxylin and eosin-stained ‘Swiss rolled’ colons by a gastrointestinal pathologist blinded to treatment conditions. Mice were scarified under anesthesia.
Induction of colitis and experimental design
Eight to ten-week-old mice of a specific genetic background were grouped randomly into control and DSS treatment groups. Colitis was induced by adding 5% (weight/volume) dextran sodium sulfate 28 (mol. wt 36-50 kD; USB Corporation, Cleveland, OH, USA) to the drinking water for 7 days. Mice were monitored regularly, and their body weights were noted every day. All mice were provided a regular chow diet ad libitum. We checked the effect of DSS on both OVDR mice and compared them with their respective control group. On day 7, mice were sacrificed, and intestinal tissue and blood samples were harvested for RNA, protein, immunofluorescence, and cytokine analysis, as described in the results section. The samples were immediately frozen and kept at −80°C until use.
Vitamin D3 treatment in vivo
C57/BL/6 wild-type mice (6-8-week-old males and females) were gavaged with 1,25 D3 (0.2 μg/day in 100 μl of corn oil) 3 times a week for 4 weeks, as described in our previous study 59. Intestinal tissue was collected following euthanasia.
Cell culture
Mouse embryonic fibroblasts (MEF) were isolated from embryonic day 13.5 embryos generated from VDR+/− x VDR+/− mouse breeding as previously described 34. VDR+/+ and VDR−/− MEFs were used in experiments after more than 15 passages when they had been immortalized. MEFs and SKCO15 cells were grown in high glucose Dulbecco’s Modified Eagle Medium (DMEM) (Hyclone, SH30243.01) containing 10% (v/v) fetal bovine serum (GEMINI, 900-108), 50 μg/ml streptomycin, and 50 U/ml penicillin (Mediatech, Inc., 30-002CI), as previously described 60.
Colonoids cultures and treatment with Vit D3
Human colonoids were prepared and maintained as previously described 61. Mini gut medium (advanced DMEM/F12 supplemented with HEPES, L-glutamine, N2, and B27) was added to the culture, along with R-Spondin, Noggin, EGF, and Wnt-3a. On day 7 after passage, colonoids were treated by Vit D3 (20 nM) for indicated times.
Western blot analysis and antibodies
Mouse colonic epithelial cells were collected by scraping the tissue from the colon of the mouse, including the proximal and distal regions. The cells were sonicated in lysis buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, pH 8.0, 1% Triton X-100) with 0.2 mM sodium ortho-vanadate, and protease inhibitor cocktail. The protein concentration was measured using the BioRad Reagent (BioRad, Hercules, CA, USA). Cultured cells were rinsed twice with ice-cold HBSS, lysed in protein loading buffer (50 mM Tris, pH 6.8, 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol), and then sonicated. Equal amounts of protein were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and immunoblotted with primary antibodies. The following antibodies were used: anti-Claudin-5 (Invitrogen, 35-2500, Carlsbad, CA, USA), anti-Claudin-7 (Invitrogen, 34-9100, Carlsbad, CA, USA), anti-VDR (Santa Cruz Biotechnology, SC-13133, Dallas, TX, USA), anti-Villin (Santa Cruz Biotechnology, SC-7672 Dallas, TX, USA), or anti-β-actin (Sigma-Aldrich, A5316, St. Louis, MO, USA) antibodies and were visualized by ECL (Thermo Fisher Scientific, 32106, Waltham, MA, USA). Membranes that were probed with more than one antibody were stripped before re-probing.
Intestinal permeability
Fluorescein Dextran (Molecular weight 3 kDa, diluted in HBSS) was gavaged (50 mg/g mouse). Four hours later, mouse blood samples were collected for fluorescence intensity measurement, as previously reported 44.
Immunofluorescence
Colonic tissues were freshly isolated and embedded in paraffin wax after fixation with 10% neutral buffered formalin. Immunofluorescence was performed on paraffin-embedded sections (4 μm), after preparation of the slides as described previously 60 followed by incubation for 1 hour in blocking solution (2% bovine serum albumin, 1% goat serum in HBSS) to reduce nonspecific background. The tissue samples were incubated overnight with primary antibodies at 4°C. The following antibodies were used: anti-Claudin-5 and anti-Claudin-7. Slides were washed 3 times for 5 minutes each at room temperature in wash buffer. Samples were then incubated with secondary antibodies (goat anti-rabbit Alexa Fluor 488, Molecular Probes, CA; 1:200) for 1 hour at room temperature. Tissues were mounted with SlowFade Antifade Kit (Life technologies, s2828, Grand Island, NY, USA), followed by a coverslip, and the edges were sealed to prevent drying. Specimens were examined with a Zeiss laser scanning microscope LSM 710 (Carl Zeiss Inc., Oberkochen, Germany).
Immunohistochemistry (IHC)
After preparation of the slides, antigen retrieval was achieved by incubating the slides for 15 min in hot preheated sodium citrate (pH 6.0) buffer and 30 min of cooling at room temperature. Endogenous peroxidases were quenched by incubating the slides in 3% hydrogen peroxide for 10 min, followed by three rinses with HBSS, and incubation for 1 hour in 3% BSA + 1% goat serum in HBSS to reduce nonspecific background. Primary antibodies VDR (1:300) were applied for overnight in a cold room. After three rinses in HBSS, the slides were incubated in secondary antibody (1:100, Jackson ImmunoResearch Laboratories, Cat.No.115-065-174, West Grove, PA, USA) for 1 hour at room temperature. After washing with HBSS for 10 minutes, the slides were incubated with vectastain ABC reagent (Vector Laboratories, Cat.No. PK-6100, Burlingame, CA 94010, USA) for 1 hour. After washing with HBSS for five minutes, color development was achieved by applying a peroxidase substrate kit (Vector Laboratories, Cat.No. SK-4800, Burlingame, CA 94010) for 2 to 5 minutes, depending on the primary antibody. The duration of peroxidase substrate incubation was determined through pilot experiments and was then held constant for all of the slides. After washing in distilled water, the sections were counterstained with haematoxylin (Leica, Cat.No.3801570, Wetzlar, Germany), dehydrated through ethanol and xylene, and cover‐slipped using a permount (Fisher Scientific, Cat.No.SP15-100, Waltham, MA, USA ).
Real-Time quantitative PCR
Total RNA was extracted from epithelial cell monolayers or mouse colonic epithelial cells using TRIzol reagent (Fisher Scientific, 15596026, Waltham, MA, USA). RNA reverse transcription was done using the iScript cDNA synthesis kit (Bio-Rad Laboratories, 1708891) according to the manufacturer’s directions. The RT-cDNA reaction products were subjected to quantitative real-time PCR using the CFX96 Real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA) and iTaq™ Universal SYBR green supermix (Bio-Rad Laboratories, 1725121, Hercules, CA, USA) according to the manufacturer’s directions. All expression levels were normalized to β-actin levels of the same sample. Percent expression was calculated as the ratio of the normalized value of each sample to that of the corresponding untreated control cells. All real-time PCR reactions were performed in triplicate. Primer sequences were designed using Primer-BLAST or were obtained from Primer Bank primer pairs listed in Table 1.
Table 1:
Real-time PCR Primers
| Primers name | Sequence |
|---|---|
| mβ-actinF | 5′-TGTTACCAACTGGGACGACA-3′ |
| mβ-actinR | 5′-CTGGGTCATCTTTTCACGGT-3′ |
| mVDRF | 5′-GAATGTGCCTCGGATCTGTGG-3′ |
| mVDRR | 5′-ATGCGGCAATCTCCATTGAAG-3′ |
| mClaudin-5F | 5′-AGGCACGGGTAGCACTCACG-3′ |
| mClaudin-5R | 5′-CATAGTTCTTCTTGTCGTAATC-3′ |
| mClaudin-7F | 5′-GCGACAACATCATCACAGCC-3′ |
| mClaudin-7R | 5′-CCTTGGAGGAATTGGACTTGG-3′ |
| mTNF-α F | 5′-CCCTCACACTCAGATCATCTTCT-3′ |
| mTNF-α R | 5′-GCTACGACGTGGGCTACAG-3′ |
| mIL-1βF | 5′-GCAACTGTTCCTGAACTCAACT-3′ |
| mIL-1βR | 5′-ATCTTTTGGGGTCCGTCAACT-3′ |
| mIL-17F | 5′-TTTAACTCCCTTGGCGCAAAA-3′ |
| mIL-17R | 5′-CTTTCCCTCCGCATTGACAC-3′ |
| hβ-actinF | 5′-AGAGCAAGAGAGGCATCCTC-3′ |
| hβ-actinR | 5′-CTCAAACATGATCTGGGTCA-3′ |
| hVDRF | 5′-GGACTGCCGCATCACCAA-3′ |
| hVDRR | 5′-TCATCTCCCGCTTCCTCT-3′ |
| hClaudin-5F | 5′-TTCGCCAACATTGTCGTCC-3′ |
| hClaudin-5R | 5′-TCTTCTTGTCGTAGTCGCCG-3′ |
| hClaudin-7F | 5′-CATCGTGGCAGGTCTTGCC-3′ |
| hClaudin-7R | 5′-GATGGCAGGGCCAAACTCATAC-3′ |
Chromatin immunoprecipitation (CHIP) assay
Binding of VDR to the Claudin-5 promoter was investigated using the ChIP assay as described previously 62. Briefly, mouse colonic epithelial cells were collected by scraping the tissue from the colon of the mouse and the cells were treated with 1% formaldehyde for 10 min at 37°C. Cells were washed twice in ice-cold phosphate-buffered saline containing protease inhibitor cocktail tablets. Cells were scraped into conical tubes, pelleted, and lysed in SDS Lysis Buffer. The lysate was sonicated to shear DNA into fragments of 200-1000 bp (4 cycles of 10 s sonication, 10 s pausing, Branson Sonifier 250, USA). The chromatin samples were pre-cleared with salmon sperm DNA–bovine serum albumin-sepharose beads then incubated overnight at 4 °C with VDR antibody. Immune complexes were precipitated with salmon sperm DNA-bovine serum albumin-sepharose beads. DNA was prepared by treatment with proteinase K, extraction with phenol and chloroform, and ethanol precipitation.
Identification of functional VDRE
PCR was used to construct the deletion of entire VDRE binding sites (ΔVDRE) and negative control (ΔD2/ΔD3) These fragments were separately subcloned into the firefly luciferase reporter plasmid pGL3-basic (Sequence see supplement Figure S2). Deletions of different domains of the Claudin-5 promoter cloned into the in pGL3 vector, driving luciferase expression, were transfected into SKCO15 cells. Luciferase activity in cell lysates was assayed by the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Firefly luciferase activity was normalized to Renilla luminescence activity, and the activity was expressed as relative units.
Multiplex ELISA assay
A mouse-specific ProcartalPlex™ Multiplex Immunoassay plate from Invitrogen Thermo Fisher Scientific was used to detect serum cytokine levels. The assay was performed using the manufacturer’s instruction manual using proper standards. Eventually, the plate was read using a Megpix Luminex machine.
Test fecal VDR by PCR
Total RNA was extracted from mouse fecal samples, as previously described 63. Briefly, about 100 mg of frozen fecal pellet was used for RNA extraction using Trizol Reagent (Thermo Fisher Scientific, Cat.No.15596018, Waltham, MA, USA). To increase RNA yield in high quality, RNeasy minispin column (Qiagen, Cat No.217004, Hilden, Germany) was used by following the manufacturer’s instructions.
Statistical Analysis
All data were expressed as the mean ± SD. All statistical tests were 2-sided. All p-values < 0.05 were considered statistically significant. After the Shapiro-Wilk test confirmed that the data are normally distributed, the differences between samples were analyzed using unpaired Student’s t-test for two groups and one-way ANOVA for more than two groups as appropriate, respectively. The p-values in ANOVA analysis were adjusted using the Tukey method to ensure accurate results. Pearson correlation analyses and scatter plots were conducted for staining intensity changes between VDR protein and Claudin-5, using SAS version 9.4 (SAS Institute, Inc., Cary, NC, USA) and R software (R Core Team (2021), R Foundation for Statistical Computing, Vienna, Austria). Other statistical analyses were performed using GraphPad Prism 6 (GraphPad, Inc., San Diego, CA., USA).
Supplementary Material
A short summary.
1. What is already known about this subject?
Tight junction structures are essential in intestinal barrier integrity, inflammation, and cancer.
Vitamin D deficiency and the vitamin D receptor (VDR) play important roles in the development of colon cancer.
2. What are the new findings?
Our study is the first to link barrier function, a specific tight junction protein, and genetic susceptibility through intestinal epithelial VDR in human colorectal cancer.
Our study fills an existing gap by characterizing the mechanism of intestinal epithelial VDR in regulating barrier functions through alterations in TJs in tumorigenesis. VDR is important for maintaining the of physiological level of the TJ protein Claudin-5 in the colon. The CLDN-5 gene is a downstream target of the VDR signaling pathway. Lack of VDR led to a reduction of Claudin-5 in tumors, whereas enhancing VDR increased Claudin-5 to protect the intestinal epithelial cells from tumorigenesis.
We reported fecal VDR reduction in a colon cancer model. This introduces the possibility of identifying new biomarkers and therapeutic targets to restore VDR-dependent functions in CRC.
3. How might it impact clinical practice in the foreseeable future
Diagnosis of CRC considering VDR status
Barrier functions and regulator as direct or indirect biomarkers
Intestinal barriers in cancer prevention and treatment
Acknowledgments:
We would like to thank Dr. David Zhou for assisting with the CRC human samples, Drs. Shaoping Wu and Rong Lu for assisting with the AOM/DSS model, and Jason S. Xia for proofreading.
Funding:
This research was funded by the UIC Cancer Center, the NIDDK/National Institutes of Health grant R01 DK105118 and R01DK114126, VA Merit Award VA 1 I01 BX004824-01, and DOD BC160450P1 to Jun Sun. The study sponsors played no role in the study design, data collection, analysis, and interpretation of data. The contents do not represent the views of the United States Department of Veterans Affairs or the United States Government.
Footnotes
Conflicts of Interest: The authors declare no conflict of interest. The funders played no role in the study design, the collection, analyses, or interpretation of data, the writing of the manuscript, or the decision to publish the results.
Preprint: 10.1101/2021.04.29.441977 in bioRxiv: https://biorxiv.org/cgi/content/short/2021.04.29.441977v1
References
- 1.Zeissig S, Burgel N, Gunzel D, Richter J, Mankertz J, Wahnschaffe U et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn’s disease. Gut 2007; 56(1): 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu L, Han J, Li L, Wang Y, Li Y, Zhang S. Claudin Family Participates in the Pathogenesis of Inflammatory Bowel Diseases and Colitis-Associated Colorectal Cancer. Frontiers in immunology 2019; 10: 1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cherradi S, Martineau P, Gongora C, Del Rio M. Claudin gene expression profiles and clinical value in colorectal tumors classified according to their molecular subtype. Cancer management and research 2019; 11: 1337–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haussler MR, Whitfield GK, Haussler CA, Hsieh J-C, Thompson PD, Selznick SH et al. The nuclear vitamin D receptor: biological and molecular regulatory properties revealed. J Bone and Mineral Research 1998; 13: 325–349. [DOI] [PubMed] [Google Scholar]
- 5.Xue Y, Fleet JC. Intestinal vitamin D receptor is required for normal calcium and bone metabolism in mice. Gastroenterology 2009; 136(4): 1317–1327, e1311–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouillon R, Carmeliet G, Verlinden L, van Etten E, Verstuyf A, Luderer HF et al. Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev 2008; 29(6): 726–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogura M, Nishida S, Ishizawa M, Sakurai K, Shimizu M, Matsuo S et al. Vitamin D3 modulates the expression of bile acid regulatory genes and represses inflammation in bile duct-ligated mice. J Pharmacol Exp Ther 2009; 328(2): 564–570. [DOI] [PubMed] [Google Scholar]
- 8.Kamen DL, Tangpricha V. Vitamin D and molecular actions on the immune system: modulation of innate and autoimmunity. J Mol Med; 88(5): 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waterhouse JC, Perez TH, Albert PJ. Reversing bacteria-induced vitamin D receptor dysfunction is key to autoimmune disease. Ann N Y Acad Sci 2009; 1173: 757–765. [DOI] [PubMed] [Google Scholar]
- 10.Liu PT, Krutzik SR, Modlin RL. Therapeutic implications of the TLR and VDR partnership. Trends Mol Med 2007; 13(3): 117–124. [DOI] [PubMed] [Google Scholar]
- 11.Gombart AF, Borregaard N, Koeffler HP. Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up-regulated in myeloid cells by 1,25-dihydroxyvitamin D3. FASEB J 2005; 19(9): 1067–1077. [DOI] [PubMed] [Google Scholar]
- 12.Kong J, Zhang Z, Musch MW, Ning G, Sun J, Hart J et al. Novel role of the vitamin D receptor in maintaining the integrity of the intestinal mucosal barrier. Am J Physiol Gastrointest Liver Physiol 2008; 294(1): G208–216. [DOI] [PubMed] [Google Scholar]
- 13.Adams JS, Hewison M. Unexpected actions of vitamin D: new perspectives on the regulation of innate and adaptive immunity. Nat Clin Pract Endocrinol Metab 2008; 4(2): 80–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu S, Bruce D, Froicu M, Weaver V, Cantorna MT. Failure of T cell homing, reduced CD4/CD8alphaalpha intraepithelial lymphocytes, and inflammation in the gut of vitamin D receptor KO mice. Proceedings of the National Academy of Sciences of the United States of America 2008; 105(52): 20834–20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019; 569(7758): 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abreu MT, Kantorovich V, Vasiliauskas EA, Gruntmanis U, Matuk R, Daigle K et al. Measurement of vitamin D levels in inflammatory bowel disease patients reveals a subset of Crohn’s disease patients with elevated 1,25-dihydroxyvitamin D and low bone mineral density. Gut 2004; 53(8): 1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim WC, Hanauer SB, Li YC. Mechanisms of disease: vitamin D and inflammatory bowel disease. Nat Clin Pract Gastroenterol Hepatol 2005; 2(7): 308–315. [DOI] [PubMed] [Google Scholar]
- 18.Wang TT, Dabbas B, Laperriere D, Bitton AJ, Soualhine H, Tavera-Mendoza LE et al. Direct and indirect induction by 1,25-dihydroxyvitamin D3 of the NOD2/CARD15-defensin beta2 innate immune pathway defective in Crohn disease. The Journal of biological chemistry; 285(4): 2227–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rufo PA, Bousvaros A. Current therapy of inflammatory bowel disease in children. Paediatr Drugs 2006; 8(5): 279–302. [DOI] [PubMed] [Google Scholar]
- 20.Sun J Vitamin D and mucosal immune function. Curr Opin Gastroenterol 2010; 26(6): 591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song M, Chan AT, Jun J. Features of the Gut Microbiome, Diet, and Environment That Influence Risk of Colorectal Cancer. Gastroenterology 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu R, Zhang Y, Xia Y, Zhang J, Kaser A, Blumberg R et al. Paneth cell alertness to pathogens maintained by vitamin D receptors. Gastroenterology 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y-G, Lu R, Wu S, Chatterjee I, Zhou D, Xia Y et al. Vitamin D receptor protects against dysbiosis and tumorigenesis via the JAK/STAT pathway in intestine. Biorxiv 2020; 02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang YG, Lu R, Wu S, Chatterjee I, Zhou D, Xia Y et al. Vitamin D Receptor Protects Against Dysbiosis and Tumorigenesis via the JAK/STAT Pathway in Intestine. Cell Mol Gastroenterol Hepatol 2020; 10(4): 729–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology 2017; 153(6):1621–1633 e1626. [DOI] [PubMed] [Google Scholar]
- 26.Andoh A, Imaeda H, Aomatsu T, Inatomi O, Bamba S, Sasaki M et al. Comparison of the fecal microbiota profiles between ulcerative colitis and Crohn’s disease using terminal restriction fragment length polymorphism analysis. J Gastroenterol 2011; 46(4): 479–486. [DOI] [PubMed] [Google Scholar]
- 27.Bird RP, Good CK. The significance of aberrant crypt foci in understanding the pathogenesis of colon cancer. Toxicol Lett 2000; 112-113(8): 395–402. [DOI] [PubMed] [Google Scholar]
- 28.Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A et al. Large-scale whole-genome sequencing of the Icelandic population. Nature Genetics 2015; 47(5): 435–444. [DOI] [PubMed] [Google Scholar]
- 29.Kang X, Zhang R, Kwong TN, Lui RN, Wu WK, Sung JJ et al. Serrated neoplasia in the colorectum: gut microbiota and molecular pathways. Gut Microbes 2021; 13(1): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yongguo Zhang YX, Jun Sun, . . A simple and sensitive method to detect vitamin D receptor expression in various disease models using stool samples. Genes and Diseases 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu S, Zhang YG, Lu R, Xia Y, Zhou D, Petrof EO et al. Intestinal epithelial vitamin D receptor deletion leads to defective autophagy in colitis. Gut 2015; 64(7): 1082–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun J, Kong J, Duan Y, Szeto FL, Liao A, Madara JL et al. Increased NF-kappaB activity in fibroblasts lacking the vitamin D receptor. Am J Physiol Endocrinol Metab 2006; 291(2): E315–322. [DOI] [PubMed] [Google Scholar]
- 33.Kato S The function of vitamin D receptor in vitamin D action. J Biochem 2000; 127(5): 717–722. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y-g, Wu S, Lu R, Zhou D, Zhou J, Carmeliet G et al. Tight junction CLDN2 gene is a direct target of the vitamin D receptor. Scientific Reports 2015; 5: 10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capaldo CT, Koch S, Kwon M, Laur O, Parkos CA, Nusrat A. Tight function zonula occludens-3 regulates cyclin D1-dependent cell proliferation. Molecular biology of the cell 2011; 22(10): 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov AI, McCall IC, Babbin B, Samarin SN, Nusrat A, Parkos CA. Microtubules regulate disassembly of epithelial apical junctions. BMC cell biology 2006; 7: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang YG, Zhu X, Lu R, Messer JS, Xia Y, Chang EB et al. Intestinal epithelial HMGB1 inhibits bacterial infection via STAT3 regulation of autophagy. Autophagy 2019; 15(11): 1935–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chatterjee I, Zhang Y, Zhang J, Lu R, Xia Y, Sun J. Overexpression of Vitamin D Receptor in Intestinal Epithelia Protects Against Colitis via Upregulating Tight Junction Protein Claudin 15. Journal of Crohn’s and Colitis 05 March, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang YG, Wu S, Sun J. Vitamin D, Vitamin D Receptor, and Tissue Barriers. Tissue barriers 2013; 1(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urashima M, Ohdaira H, Akutsu T, Okada S, Yoshida M, Kitajima M et al. Effect of Vitamin D Supplementation on Relapse-Free Survival Among Patients With Digestive Tract Cancers: The AMATERASU Randomized Clinical Trial. JAMA 2019; 321(14): 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vancamelbeke M, Vermeire S. The intestinal barrier: a fundamental role in health and disease. Expert Rev Gastroenterol Hepatol 2017; 11(9): 821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mineta K, Yamamoto Y, Yamazaki Y, Tanaka H, Tada Y, Saito K et al. Predicted expansion of the claudin multigene family. FEBS Letters 2011; 585(4): 606–612. [DOI] [PubMed] [Google Scholar]
- 43.Fujita H, Sugimoto K, Inatomi S, Maeda T, Osanai M, Uchiyama Y et al. Tight junction proteins claudin-2 and −12 are critical for vitamin D-dependent Ca2+ absorption between enterocytes. Molecular biology of the cell 2008; 19(5): 1912–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang YG, Lu R, Xia Y, Zhou D, Petrof E, Claud EC et al. Lack of Vitamin D Receptor Leads to Hyperfunction of Claudin-2 in Intestinal Inflammatory Responses. Inflamm Bowel Dis 2019; 25(1): 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang YG, Wu S, Lu R, Zhou D, Zhou J, Carmeliet G et al. Tight junction CLDN2 gene is a direct target of the vitamin D receptor. Sci Rep 2015; 5: 10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, Lu R, Zhang YG, Sun J. Vitamin D Receptor Deletion Leads to the Destruction of Tight and Adherens Junctions in Lungs. Tissue barriers 2018; 6(4): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turpin W, Lee SH, Raygoza Garay JA, Madsen KL, Meddings JB, Bedrani L et al. Increased Intestinal Permeability is Associated with Later Development of Crohn’s Disease. Gastroenterology 2020. [DOI] [PubMed] [Google Scholar]
- 48.Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol 2019; 16(11): 690–704. [DOI] [PubMed] [Google Scholar]
- 49.Pappa HM, Grand RJ, Gordon CM. Report on the vitamin D status of adult and pediatric patients with inflammatory bowel disease and its significance for bone health and disease. Inflamm Bowel Dis 2006; 12(12): 1162–1174. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, Zhang Y, Xia Y, Sun J. Imbalance of the intestinal virome and altered viral-bacterial interactions caused by a conditional deletion of the vitamin D receptor. Gut Microbes 2021; 13(1): 1957408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song M, Chan AT, Sun J. Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology 2020; 158(2): 322–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun J, Kato I. Gut microbiota, inflammation and colorectal cancer. Genes Dis 2016; 3(2): 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun J Impact of bacterial infection and intestinal microbiome on colorectal cancer development. Chinese Medical Journal 2021; In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galamb O, Gyorffy B, Sipos F, Spisak S, Nemeth AM, Miheller P et al. Inflammation, adenoma and cancer: objective classification of colon biopsy specimens with gene expression signature. Disease markers 2008; 25(1): 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabates-Bellver J, Van der Flier LG, de Palo M, Cattaneo E, Maake C, Rehrauer H et al. Transcriptome profile of human colorectal adenomas. Molecular cancer research : MCR 2007; 5(12): 1263–1275. [DOI] [PubMed] [Google Scholar]
- 56.Galamb O, Sipos F, Solymosi N, Spisak S, Krenacs T, Toth K et al. Diagnostic mRNA expression patterns of inflamed, benign, and malignant colorectal biopsy specimen and their correlation with peripheral blood results. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2008; 17(10): 2835–2845. [DOI] [PubMed] [Google Scholar]
- 57.Pekow J, Dougherty U, Huang Y, Gometz E, Nathanson J, Cohen G et al. Gene signature distinguishes patients with chronic ulcerative colitis harboring remote neoplastic lesions. Inflamm Bowel Dis 2013; 19(3): 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y-g, Lu R, Xia Y, Zhou D, Petrof E, Claud EC et al. Lack of Vitamin D Receptor Leads to Hyperfunction of Claudin-2 in Intestinal Inflammatory Responses. Inflammatory Bowel Diseases 2018; 25(1): 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu R, Zhang YG, Xia Y, Sun J. Imbalance of autophagy and apoptosis in intestinal epithelium lacking the vitamin D receptor. FASEB J 2019: fj201900727R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu R, Wu S, Liu X, Xia Y, Zhang YG, Sun J. Chronic effects of a Salmonella type III secretion effector protein AvrA in vivo. PLoS One 2010; 5(5): e10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu R, Voigt RM, Zhang Y, Kato I, Xia Y, Forsyth CB et al. Alcohol Injury Damages Intestinal Stem Cells. Alcoholism, clinical and experimental research 2017; 41(4): 727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu S, Xia Y, Liu X, Sun J. Vitamin D receptor deletion leads to reduced level of IkappaBalpha protein through protein translation, protein-protein interaction, and post-translational modification. Int J Biochem Cell Biol 2010; 42(2): 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y-g, Xia Y, Sun J. A simple and sensitive method to detect vitamin D receptor expression in various disease models using stool samples. Genes & Diseases 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


