In both preclinical models and two phase I clinical trials of non–small cell lung cancer with EGFR exon 20 mutations, the selective EGFR inhibitor sunvozertinib demonstrated manageable safety and antitumor activity.
Abstract
Epidermal growth factor receptor exon 20 insertion mutations (EGFRexon20ins) are detected in approximately 2% of patients with non–small cell lung cancer (NSCLC). Due to a lack of effective therapy, the prognosis of these patients is typically poor. Sunvozertinib (DZD9008) was designed as an oral, potent, irreversible, and selective EGFR tyrosine kinase inhibitor, showing activity against EGFRexon20ins and other mutations. In both cell lines and xenograft models, sunvozertinib shows potent antitumor activity. In the two ongoing phase I clinical studies, sunvozertinib was tolerated up to 400 mg once daily. The most common drug-related adverse events included diarrhea and skin rash. Antitumor efficacy was observed at the doses of 100 mg and above in patients with EGFRexon20ins NSCLC across different subtypes, with prior amivantamab treatment as well as with baseline brain metastasis. The median duration of response has not been reached.
Significance:
We report the discovery and early clinical development of sunvozertinib, a potential treatment option for the unmet medical need of EGFRexon20ins NSCLC.
This article is highlighted in the In This Issue feature, p. 1599
INTRODUCTION
Non–small cell lung cancer (NSCLC) remains one of the leading causes of cancer death worldwide. Genetic mutations, such as EGFR mutations, have been reported in NSCLC (1–4), and drugs have been approved for the treatment of NSCLC with EGFR sensitizing, T790M, and some types of uncommon mutations (5). For NSCLC with epidermal growth factor receptor exon 20 insertion mutations (EGFRexon20ins), which comprise about 2% of NSCLC (6), the development of targeted therapies has been more challenging. With available therapy, such as platinum-based chemotherapy, patients have a poor clinical prognosis with median progression-free survival (PFS) and overall survival rates of only around 6 months and 24 months, respectively (7–9). Recently, an EGFR and MET bispecific antibody, amivantamab, received accelerated approval from the FDA for treating this disease. However, the objective response rate (ORR) with amivantamab treatment was only around 40% (10). Mobocertinib, an EGFR tyrosine kinase inhibitor (TKI), has also recently received accelerated approval, and its ORR was only around 28% in a pivotal phase II study (11). Also, about 40% of patients experienced ≥grade 3 treatment-related adverse events (TEAE), and 21% of patients experienced ≥grade 3 diarrhea (11, 12), which required dose reduction or even discontinuation. Therefore, there is still room for improvement to treat EGFRexon20ins mutations.
Engineering one human EGFRexon20ins subtype 770_NPG into transgenic mice induced tumor formation in the lung (13), which suggests a potential oncogenic driver role for EGFRexon20ins. So far, more than 39 distinct subtypes of EGFRexon20ins have been reported in NSCLC (9). Each subtype might have different sensitivity to a particular EGFR TKI. Therefore, a compound with good tolerability that allows it to reach high enough doses to cover a wide spectrum of mutation subtypes is highly desirable.
Sunvozertinib (DZD9008) is an oral, potent, irreversible, and selective EGFR TKI targeting EGFRexon20ins as well as EGFR sensitizing, T790M, and uncommon mutations with weak activity against wild-type EGFR. In addition, sunvozertinib exhibits desirable drug metabolism and pharmacokinetic (DMPK) properties as an oral drug in both preclinical and clinical settings. Moreover, nonclinical toxicity studies demonstrated a favorable safety margin of sunvozertinib to support its clinical development. Currently, sunvozertinib is under phase I clinical development in the United States, Australia, South Korea, Taiwan (ClinicalTrials.gov: NCT03974022), and China (Chinadrugtrial: CTR20192097).
Here, we present the in vitro and in vivo activities of sunvozertinib on inhibiting EGFRexon20ins at the enzymatic and cellular levels and in xenograft and transgenic animal models. In addition, we also illustrate the mechanism of sunvozertinib on suppressing EGFRexon20ins through a structure–activity relationship. Finally, safety, pharmacokinetics (PK), and antitumor efficacy results from the two ongoing phase I clinical studies are also reported.
RESULTS
Design and Structure of Sunvozertinib that Can Selectively Bind to EGFRexon20ins Protein
We focused on designing inhibitors with an irreversible (covalent) binding mode with C797 similar to that of osimertinib. Without any clear structural rationale for attaining wild-type selectivity, we opted for using the osimertinib scaffold as the starting point for optimization of EGFRexon20ins potency and monitoring selectivity against wild-type EGFR using cellular assays. In addition, maintaining potent activity against EGFR sensitizing mutations and double mutations of T790M was also a key attribute in our inhibitor's design strategy. Based on the available structural information of EGFRexon20ins, we decided to replace the rotationally less flexible methylindole on osimertinib with a more flexible anilinophenyl moiety on C-4 of the pyrimidine hinge binding motif. Different substitution patterns on the phenyl ring were extensively investigated. Screening of the newly designed compounds began with the direct testing in EGFR cellular target engagement and antiproliferation assays, bypassing the unknown effect of ATP concentration on compound activity in biochemical assays with different EGFRexon20ins. A brief discussion of the structure–activity relationship culminating in the discovery of sunvozertinib (Fig. 1) follows. The initial screening campaign using cellular phosphorylated EGFR (pEGFR) assays (L858R/T790M, ASV, NPH, and wild-type) on our irreversible EGFR inhibitor library identified both triazine and pyrimidine hinge binding scaffolds as hits with EGFRexon20ins activities that were more potent than that of osimertinib (Supplementary Table S1). Initial lead optimization on the novel and more potent triazine series led to the identification of ortho-substituted anilinophenyl head groups as the preferred substructure for potent EGFRexon20ins potency. The enhanced potency of the inhibitors with EGFRexon20ins mutants can be envisioned as the ortho-substituent on the anilinophenyl group occupying part of the back pocket on the ATP-binding site and interacting favorably with the αC-helix. Among all the ortho-substituents investigated, we settled with the 2-hydroxypropan-2-yl moiety as in compound Cpd-5 for further optimization based on its potency and in vitro DMPK parameters. Incubation of compound Cpd-5 with human hepatocytes indicated oxidative liability that occurred predominantly on the head-group phenyl ring and the dimethylaminoethyl side chain. To this end, introducing both 4-fluoro and 5-chloro substitutions as in compound Cpd-6 successfully reduced human hepatocyte clearance from 34 to 7.1 (μL/min)/(106 cells). Additional modification of the flexible dimethylaminoethyl side chain in Cpd-6 into a conformational more rigid (dimethylamino) pyrrolidin-1-yl in compound Cpd-8 further decreased the human hepatocyte clearance to 2.8 (μL/min)/(106 cells) while maintaining the required EGFR mutant activity. Toward the end game, a match-pair direct comparison between compounds in the triazine and pyrimidine series was performed. Although the triazine series consistently showed better EGFRexon20ins potency, the pyrimidine core offered a more desirable selectivity against the wild-type EGFR. Finally, DZD9008 in the pyrimidine series was selected for further profiling in numerous in vivo efficacy and safety experiments and was eventually recommended as the development candidate of sunvozertinib. Nonclinical DMPK properties of sunvozertinib are summarized in Supplementary Table S2. Sunvozertinib is highly permeable with reasonable bioavailability in both rat and dog. At least 2-fold cellular selectivity between cells expressing major EGFRexon20ins subtypes and wild-type EGFR was observed. This is probably due to the different binding affinity of sunvozertinib against mutants and wild-type EGFR protein in a cytoplasmic environment with the presence of variable and high concentrations of ATP. The synthesis of sunvozertinib is described in Supplementary Methods, and the 1H and 13C nuclear magnetic resonance spectra of sunvozertinib are illustrated in Supplementary Fig. S1.
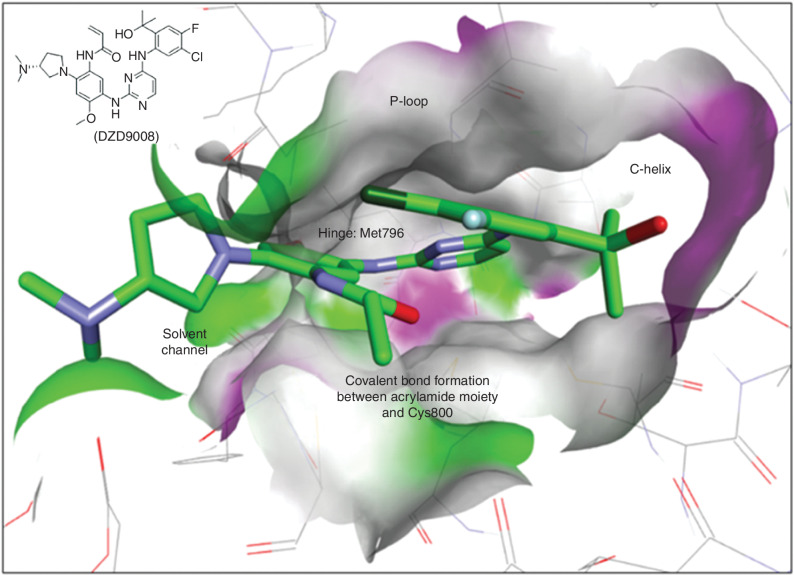
Figure 1.
Modeling of sunvozertinib with EGFRexon20ins 770_NPG (Protein Data Bank code: 4LRM). Key interactions of sunvozertinib with EGFR proteins include (i) bidentate interactions of aminopyrimidine with a hinge (Met796), (ii) acrylamide group forms an irreversible covalent bond with Cys800, (iii) 2-hydroxypropan-2-yl group occupies space next to the C-helix, and (iv) polar interaction of dimethylaminopyrrolidine with solvent channel residues. Green: carbon; purple: nitrogen; red: oxygen; dark green: chloride; light blue: fluorine. Colors on the protein surface represent the ATP-binding pocket and are for clarity only. DZD9008 = sunvozertinib.
Sunvozertinib Is an EGFRexon20ins Inhibitor Showing Good PK/Pharmacodynamic Correlation in Xenograft Models and Weak Activity against Wild-type EGFR
To test the enzymatic activity of sunvozertinib against EGFRexon20ins mutations, we utilized the commercially available recombinant kinase domain of EGFRexon20ins NPG enzyme to establish the enzymatic assay. Sunvozertnib showed potent inhibition with IC50 of 2.1 nmol/L against EGFRexon20ins NPG and slightly weaker inhibition against wild-type EGFR with IC50 of 2.4 nmol/L in an enzymatic assay performed with a compound in preincubation condition (Supplementary Fig. S2A).
To explore a broader kinome selectivity profile, sunvozertinib was evaluated by the in vitro kinase assay of 117 recombinant human kinases performed without compound preincubation condition (Supplementary Fig. S2B). At 1 μmol/L, sunvozertinib inhibited 15 of 117 kinases by >50%, including EGFR, HER2, HER4, and EGFR resistant mutation (Supplementary Table S3). Dose–response curves were established in these kinases, whose activity was inhibited by >50% in the single-point screening to determine IC50 (Supplementary Table S4). Of these 15 kinases, only one kinase (EGFR T790M mutation) was inhibited with IC50 <150 nmol/L, and two additional kinases (BTK, wild-type EGFR) were inhibited with IC50 <250 nmol/L. The IC50 of EGFR T790M is about 10-fold more potent than that of wild-type EGFR. Overall, sunvozertinib displayed potent kinase inhibitory activity against only the mutant EGFR, with limited off-target activity against the rest of the kinome.
To test the cellular activity of sunvozertinib, 14 different subtypes of EGFRexon20ins were engineered into Ba/F3 cell lines. In these cell lines, sunvozertinib showed potent activity in downregulating pEGFR with an IC50 of 6 to 40 nmol/L (Fig. 2A). In other cell lines expressing EGFR-sensitizing mutations, T790M resistant mutations, and uncommon mutations, sunvozertinib more potently downregulated pEGFR, with IC50 ranging from 1.1 to 12 nmol/L (Fig. 2A). The effect of sunvozertinib on wild-type EGFR was assessed by measuring pEGFR expression in the A431 cell line that overexpressed wild-type EGFR, which was used by other clinically approved EGFR TKIs (14, 15). In this cell line, sunvozertinib was less potent in downregulating pEGFR, with an IC50 of 58 nmol/L (Fig. 2A). Thus, sunvozertinib has 1.4- to 9.6-fold, 4.8-fold, and 52-fold selectivity on EGFRexon20ins, uncommon mutations, and sensitizing/resistant mutations, respectively, over wild-type EGFR. Notably, it has 2.5- to 3.1-fold selectivity on the most common subtypes of EGFRexon20ins (insASV, insSVD, and insNPH) versus wild-type EGFR. The antiproliferation potency GI50 (the concentration of the compound producing 50% proliferation inhibition) values in cells carrying different EGFR-mutant variants or wild-type EGFR are shown in Supplementary Fig. S2C. Sunvozertinib potently suppresses cell proliferation with a GI50 of 6 to 88 nmol/L in these cells. The wild-type pEGFR IC50 and antiproliferation GI50 were 52 nmol/L and 113 nmol/L in Ba/F3 cell clones, respectively, which were comparable with the data on the A431 cell line.
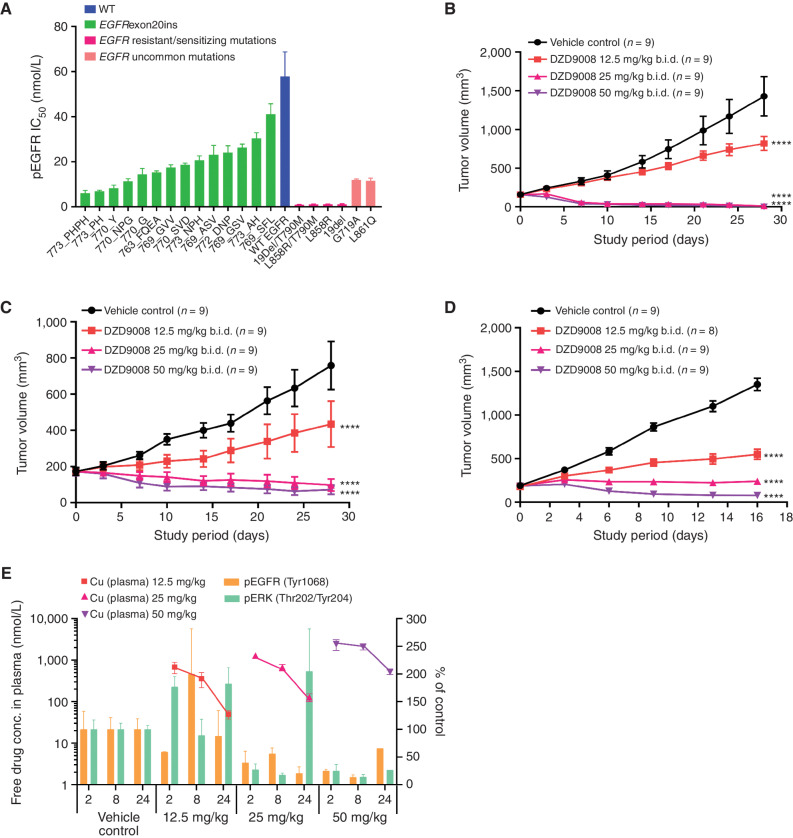
Figure 2.
In vitro and in vivo antitumor activity of sunvozertinib in EGFRexon20ins, sensitizing mutation, uncommon mutation, or resistant mutation cell lines and animal models. A, The cellular activity of sunvozertinib on EGFRexon20ins as well as sensitizing mutation, uncommon mutation, or resistant mutation versus wild-type (WT) EGFR, shown as pEGFR IC50. Cell lines carrying EGFRexon20ins were treated with sunvozertinib at a series of concentrations for 4 hours, and then pEGFR (Tyr1068) was measured with an MSD SECTOR Imager. In the A431 cell line carrying wild-type EGFR, after compound treatment for 4 hours, cells were stimulated with 100 ng/mL of recombinant human EGF for 10 minutes before lysis. The potency in each cell line was the average value from three biologically independent experiments. Data, mean ± SEM. One-way ANOVA test was used for comparison with wild-type EGFR. ****, P < 0.0001. B, Antitumor activity of sunvozertinib in the PDX model LU0387 carrying EGFRexon20ins insNPH. b.i.d., twice daily. C, Antitumor activity of sunvozertinib in the PDX model LU3075 carrying EGFRexon20ins 772_DNP. D, Antitumor activity of sunvozertinib in the A431 xenograft model expressing wild-type EGFR. Tumor volume in the different treatment groups at the endpoint was performed by two-way ANOVA. ****, P < 0.0001. E, PK/PD relationship of sunvozertinib in the PDX LU3075 model. The pEGFR (Tyr1068) and pERK (Thr202/Tyr204) expression in tumor tissues was detected by IHC and normalized to the vehicle control group. Each time point had tumor tissues from three mice to detect pEGFR or pERK signal except the time point of 24 hours in the 50 mg/kg group, which included only one mouse due to complete remission of tumor nodules in some mice. conc., concentration; pERK, phosphorylated ERK. DZD9008 = sunvozertinib.
In the EGFRexon20ins patient-derived xenograft (PDX) model LU0387 (EGFRexon20ins 773_NPH) or LU3075 (EGFRexon20ins 772_DNP), the oral administration of sunvozertinib demonstrated profound antitumor efficacy in a dose-dependent manner. At doses of ≥25 mg/kg, tumor regression was observed (Fig. 2B and C). In contrast, no tumor regression was observed at 25 mg/kg in the xenograft model carrying wild-type EGFR (Fig. 2D), suggesting its selectivity between mutant and wild-type EGFR. The waterfall plots of the antitumor activity of sunvozertinib in xenograft models are shown in Supplementary Fig. S3A and S3B to visually reflect such differences. In addition, the antitumor activity of sunvozertinib was also demonstrated in a transgenic mice model carrying one of the most prevalent EGFRexon20ins mutation subtypes, insASV (Supplementary Fig. S4A and S4B), which further confirmed its antitumor activity against EGFRexon20ins. Moreover, the antitumor activity of sunvozertinib was also assessed in a brain metastasis (BM) model carrying T790M mutation. As shown in Supplementary Fig. S5, sunvozertinib induced profound tumor regression in luci-H1975 BM models at the tested doses of 25 mg/kg twice daily (b.i.d.) or 50 mg/kg b.i.d. The antitumor effect of sunvozertinib was comparable with the effect of osimertinib at its clinically equivalent dose.
At the end of the efficacy study, blood samples and tumor tissues were collected to analyze the relationship between drug concentrations and pEGFR or phosphorylated ERK (pERK) inhibition in tumor tissues. As shown in Fig. 2E, an increased dose of sunvozertinib led to higher drug concentration in the plasma and more profound pEGFR inhibition. Its plasma concentrations at the dose of ≥25 mg/kg covered in vitro pEGFR IC50 of EGFRexon20ins 772_DNP-engineered Ba/F3 cells for more than 16 hours. At ≥25 mg/kg, sunvozertinib led to more than 50% pEGFR inhibition 2 hours after dosing, and the effect was maintained for about 24 hours. In addition, the dose-dependent modulation of pERK (Thr202/Tyr204) was detected in this model. Sunvozertinib at ≥25 mg/kg led to more than 70% pERK inhibition, and the effect lasted for ≥8 hours. These data suggest a good in vivo PK and pharmacodynamic (PD) relationship by sunvozertinib.
Phase I Clinical Studies of Sunvozertinib in NSCLC Patients with EGFR or HER2 Mutations
There are two ongoing phase I clinical studies for sunvozertinib (WU-KONG1, ClinicalTrials.gov identifier: NCT03974022, and WU-KONG2, Chinadrugtrial identifier: CTR20192097). WU-KONG1 is being conducted in the United States, Australia, South Korea, and Taiwan, and WU-KONG2 is being conducted in China. Patients were eligible for the studies if they had locally advanced or metastatic NSCLC harboring EGFR or HER2 mutations (including EGFR exon20 insertion mutation) and relapsed from a prior standard of treatment, with radiologically measurable disease. Patients with BM could be enrolled under stable and asymptomatic conditions. As the key study design of these two studies is similar, a pooled analysis was performed to assess safety, PK, and antitumor efficacy.
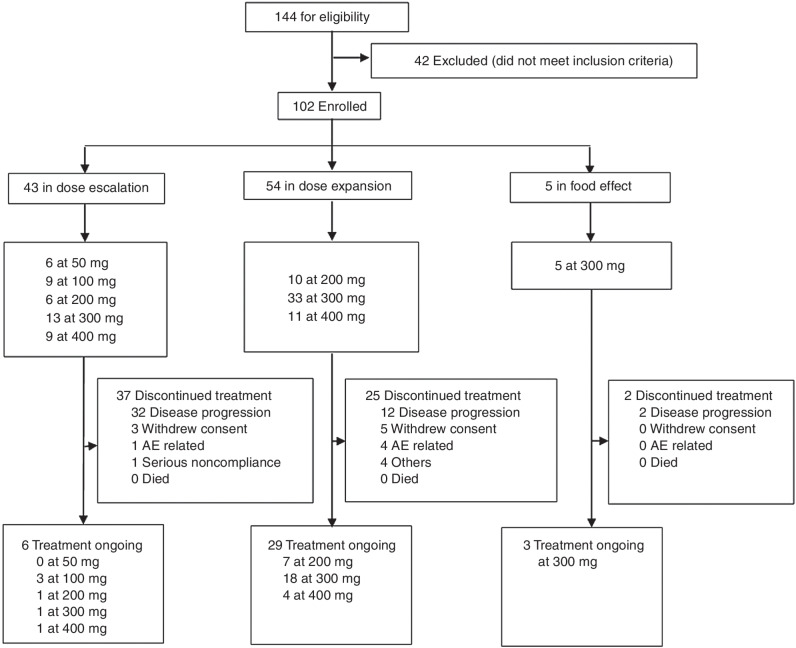
Between July 9, 2019, and April 3, 2021, sunvozertinib was administered to 102 patients with EGFR or HER2 mutant NSCLC, among whom 54 and 48 patients were from WU-KONG1 and WU-KONG2 studies, respectively (Fig. 3). Forty-three, 54, and 5 patients were enrolled into dose-escalation, dose-expansion, or food effect cohorts, respectively. Sixty-two of 102 patients (60.8%) carried EGFRexon20ins. As of April 3, 2021, the enrollment of the dose-escalation cohorts was completed, and the food effect and dose-expansion cohorts were still open. In the dose-escalation cohorts, five dose levels of sunvozertinib were explored: 50, 100, 200, 300, and 400 mg once daily. Based on safety, tolerability, PK, and efficacy signals in the dose-escalation cohorts, three dose levels (200, 300, and 400 mg) were selected for expansion. At the data cutoff date, 6, 28, and 3 patients in the dose escalation, dose-expansion, or food effect cohorts, respectively, are still receiving sunvozertinib treatment.
Figure 3.
Trial profiles. Pooled summary of WU-KONG1 and WU-KONG2 studies. Data cutoff date: April 3, 2021. Sunvozertinib (DZD9008) was dosed once daily. AE, adverse event.
Patient demographics are shown in Table 1. The median age was 59 years, 57 of 102 patients (55.9%) were women, and 101 patients (99%) had Eastern Cooperative Oncology Group (ECOG) score ≤1. The median lines of prior therapy were 3 (1–10), and 93 patients (91.2%) had received at least one line of prior chemotherapy. Mutation types of the 102 patients include EGFRexon20ins (61%, 62/102), EGFR sensitizing mutation (4%, 4/102), EGFR T790M (1%, 1/102), EGFR sensitizing/T790M double mutation (6%, 6/102), EGFR uncommon point mutation (1%, 1/102), and HER2exon20ins (28%, 28/102).
Table 1.
Patient demographics and baseline characteristics
| 50 mg | 100 mg | 200 mg | 300 mg | 400 mg | Total | |
|---|---|---|---|---|---|---|
| Characteristics | (n = 6) | (n = 9) | (n = 16) | (n = 51) | (n = 20) | (n = 102) |
| Age | ||||||
| Median | 48.5 | 60.0 | 60.5 | 62.0 | 53.5 | 59.0 |
| Min, max | 36, 72 | 48, 83 | 36, 83 | 32, 82 | 47, 85 | 32, 85 |
| Gender, n (%) | ||||||
| Female | 5 (83.3) | 5 (55.6) | 11 (68.8) | 26 (51.0) | 10 (50.0) | 57 (55.9) |
| Male | 1 (16.7) | 4 (44.4) | 5 (31.3) | 25 (49.0) | 10 (50.0) | 45 (44.1) |
| Race, n (%) | ||||||
| Asian | 5 (83.3) | 8 (88.9) | 15 (93.8) | 40 (78.4) | 17 (85.0) | 85 (83.3) |
| White | 1 (16.7) | 1 (11.1) | 1 (6.3) | 11 (21.6) | 3 (15.0) | 17 (16.7) |
| Baseline ECOG, n (%) | ||||||
| 0 | 5 (83.3) | 2 (22.2) | 11 (68.8) | 15 (29.4) | 5 (25.0) | 38 (37.3) |
| 1 | 1 (16.7) | 6 (66.7) | 5 (31.3) | 36 (70.6) | 15 (75.0) | 63 (61.8) |
| 2 | 0 (0.0) | 1 (11.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (1.0) |
| Baseline BM, n (%) | ||||||
| Yes | 2 (33.3) | 4 (44.4) | 4 (25.0) | 25 (49.0) | 9 (45.0) | 44 (43.1) |
| No | 4 (66.7) | 5 (55.6) | 12 (75.0) | 26 (51.0) | 11 (55.0) | 58 (56.9) |
| Number of prior cancer therapies | ||||||
| Median | 3.0 | 3.0 | 1.5 | 2.0 | 2.5 | 3.0 |
| Min, max | 1, 5 | 2, 6 | 1, 6 | 1, 10 | 1, 8 | 1, 10 |
| Mutation types, n (%) | ||||||
| EGFRexon20ins | 3 (50.0) | 2 (22.2) | 11 (68.8) | 36 (70.6) | 10 (50.0) | 62 (60.8) |
| EGFR sensitizing mutation | 0 (0.0) | 1 (11.1) | 1 (6.3) | 0 (0.0) | 2 (10.0) | 4 (3.9) |
| EGFR T790M | 0 (0.0) | 0 (0.0) | 1 (6.3) | 0 (0.0) | 0 (0.0) | 1 (1.0) |
| EGFR double mutation | 2 (33.3) | 2 (22.2) | 0 (0.0) | 2 (3.9) | 0 (0.0) | 6 (5.9) |
| HER2exon20ins | 1 (16.7) | 4 (44.4) | 3 (18.8) | 12 (23.5) | 8 (40.0) | 28 (27.5) |
| EGFR uncommon mutation | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (2.0) | 0 (0.0) | 1 (1.0) |
NOTE: Pooled analysis from WU-KONG1 and WU-KONG2 studies. Data cutoff date: April 3, 2021.
Abbreviation: ECOG, Eastern Cooperative Oncology Group.
In the dose-escalation cohorts, sunvozertinib was tolerated up to 400 mg. Two patients experienced dose-limiting toxicities (DLT), including one from the 300-mg cohort who experienced grade 3 diarrhea and later respiratory distress syndrome, and another one from the 400-mg cohort who experienced grade 3 cardiac arrythmia. Across all dose levels, 34 patients (33.3%) experienced ≥grade 3 drug-related adverse events (AE) assessed by investigators. Twenty-four (23.5%), 16 (15.7%), and 6 (5.9%) had dose interruption, reduction, and discontinuation due to drug-related TEAEs (Table 2). The most common TEAEs (>10%) are listed in Supplementary Table S5. All grade diarrhea and rash occurred in 53.9% (55/102) and 40.2% (41/102) of patients, respectively, whereas the incidence of ≥grade 3 diarrhea was only 4.9% (5/102), and no patients experienced ≥grade 3 rash. Based on safety and tolerability data in the dose-escalation cohorts, 400 mg was defined as the maximum tolerated dose (MTD), and 200 mg to 400 mg were selected for dose expansion.
Table 2.
Summary of safety profiles of sunvozertinib
| 50 mg | 100 mg | 200 mg | 300 mg | 400 mg | All | |
|---|---|---|---|---|---|---|
| (n = 6) | (n = 9) | (n = 16) | (n = 51) | (n = 20) | (n = 102) | |
| AE category | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) |
| Patients with any TEAE | 6 (100.0) | 9 (100.0) | 16 (100.0) | 51 (100.0) | 20 (100.0) | 102 (100.0) |
| Patients with any TEAE with CTCAE grade ≥3 | 2 (33.3) | 2 (22.2) | 2 (12.5) | 20 (39.2) | 14 (70.0) | 40 (39.2) |
| Patients with any drug-related AE | 6 (100.0) | 8 (88.9) | 16 (100.0) | 49 (96.1) | 20 (100.0) | 99 (97.1) |
| Patients with any drug-related AE with CTCAE grade ≥3 | 1 (16.7) | 1 (11.1) | 1 (6.3) | 17 (33.3) | 14 (70.0) | 34 (33.3) |
| Patients with any treatment-emergent SAE | 0 (0.0) | 2 (22.2) | 3 (18.8) | 14 (27.5) | 7 (35.0) | 26 (25.5) |
| Patients with any drug-related SAE | 0 (0.0) | 0 (0.0) | 1 (6.3) | 9 (17.6) | 6 (30.0) | 16 (15.7) |
| Patients with any TEAE leading to death | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (3.9) | 1 (5.0) | 3 (2.9) |
| Patients with any drug-related AE leading to death | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (2.0) | 0 (0.0) | 1 (1.0) |
| Patients with any TEAE leading to treatment discontinuation | 0 (0.0) | 1 (11.1) | 0 (0.0) | 4 (7.8) | 2 (10.0) | 7 (6.9) |
| Patients with any drug-related AE leading to treatment discontinuation | 0 (0.0) | 0 (0.0) | 0 (0.0) | 4 (7.8) | 2 (10.0) | 6 (5.9) |
| Patients with any TEAE leading to dose reduction | 0 (0.0) | 0 (0.0) | 0 (0.0) | 6 (11.8) | 10 (50.0) | 16 (15.7) |
| Patients with any drug-related AE leading to dose reduction | 0 (0.0) | 0 (0.0) | 0 (0.0) | 6 (11.8) | 10 (50.0) | 16 (15.7) |
| Patients with any TEAE leading to drug interruption | 0 (0.0) | 2 (22.2) | 2 (12.5) | 16 (31.4) | 8 (40.0) | 28 (27.5) |
| Patients with any drug-related AE leading to drug interruption | 0 (0.0) | 1 (11.1) | 1 (6.3) | 14 (27.5) | 8 (40.0) | 24 (23.5) |
NOTE: Pooled analysis of the WU-KONG1 and WU-KONG2 studies. Data cutoff date: April 3, 2021. Causality assessed by investigators.
Abbreviations: CTCAE, Common Terminology Criteria for Adverse Events version 5.0; SAE, serious adverse event.
Plasma samples were obtained from patients in the dose- escalation (n = 40) and dose-expansion (n = 24) cohorts for PK analysis. Across all dose levels (50 mg–400 mg), sunvozertinib was absorbed with a median tmax of 4 to 7 hours after single oral dosing and a tss,max of 5 to 6 hours after repeat dosing. With once-daily dosing, plasma concentrations of sunvozertinib appear to approach a steady state within 15 days. At the steady state, sunvozertinib exhibited approximately dose-proportional increases in exposures (Css,max and AUCss) across the dose range investigated. Comparing repeat dosing with single dosing, there was around 3-fold accumulation of drug exposure, probably due to its half-life of around 50 hours. At the doses of ≥200 mg, the geometric mean of Css,max and Css,min (Supplementary Fig. S6; Supplementary Table S6) was above pEGFR IC50 (Fig. 2A) of the majority of EGFRexon20ins subtypes.
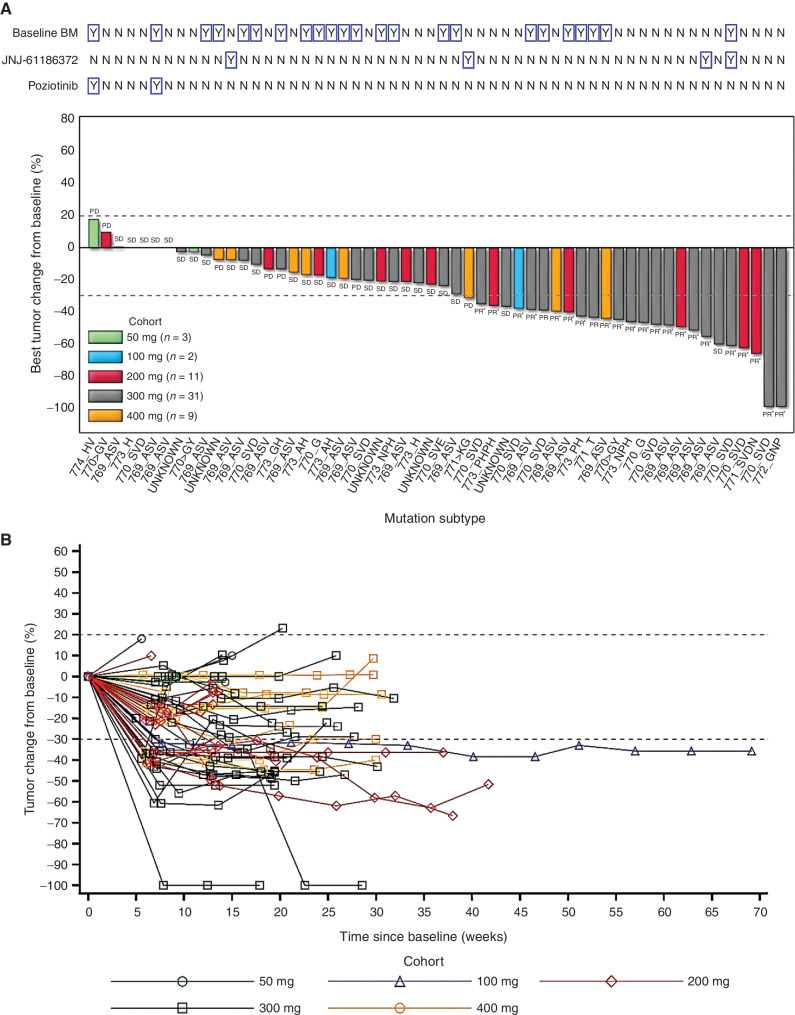
In 56 patients with EGFRexon20ins who were evaluable for efficacy assessment, partial response (PR) was observed at the doses of ≥100 mg. Across all dose levels, the best ORR was 41.1%, and the confirmed ORR was 37.5%. At the recommended phase II doses (PR2D), 200 and 300 mg, the ORRs were 45.5% (confirmed, 45.5%) and 48.4% (confirmed, 41.9%), respectively (Table 3). In dose-expansion cohorts, the best and confirmed ORRs were 47.4% and 44.7%, respectively, across 200 to 400 mg. Antitumor efficacy was observed in different EGFRexon20ins subtypes (Supplementary Table S7). PR was observed in patients with baseline BM or those with prior amivantamab treatment (Fig. 4A). By the data cutoff, with a median follow-up duration of 4.2 months, the median DoR was >3.5 months and had not been reached, the longest DoR was >8 months, and 15 of 23 patients (65.2%) were still receiving ongoing treatment and responding (Fig. 4B). With a median follow-up duration of 5.6 months, the median PFS was >4 months and had not been reached.
Table 3.
Antitumor activity of sunvozertinib in patients with previously treated EGFRexon20ins NSCLC
| 50 mg | 100 mg | 200 mg | 300 mg | 400 mg | Total | |
|---|---|---|---|---|---|---|
| (n = 3) | (n = 2) | (n = 11) | (n = 31) | (n = 9) | (n = 56) | |
| Best response, n (%) | ||||||
| PR (unconfirmed) | 0 (0.0) | 1 (50.0) | 5 (45.5) | 15 (48.4) | 2 (22.2) | 23 (41.1) |
| PR (confirmed) | 0 (0.0) | 1 (50.0) | 5 (45.5) | 13 (41.9) | 2 (22.2) | 21 (37.5) |
| Stable disease | 2 (66.7) | 1 (50.0) | 4 (36.4) | 15 (48.4) | 5 (55.5) | 27 (48.2) |
| Progressive disease | 1 (33.3) | 0 (0.0) | 2 (18.2) | 3 (9.7) | 2 (22.2) | 8 (14.3) |
| Confirmed ORR, n (%) | 0 (0.0) | 1 (50.0) | 5 (45.5) | 13 (41.9) | 2 (22.2) | 21 (37.5) |
| Disease control rate, n (%) | 2 (66.7) | 2 (100.0) | 9 (81.8) | 28 (90.3) | 7 (77.7) | 48 (85.7) |
NOTE: Pooled data from WU-KONG1 and WU-KONG2 studies. Data cutoff date: April 3, 2021. Tumor response was assessed by investigators.
Figure 4.
Clinical activity of sunvozertinib in EGFRexon20ins NSCLC patients with postbaseline target lesion assessments. A, Best percentage change from baseline in target lesions by dose level, molecular subtype, prior amivantamab or poziotinib treatment status, and baseline BM. B, Plot showing percentage change from baseline in target lesion by time on treatment and dose level. Pooled analysis of WU-KONG1 and WU-KONG2 studies was performed. Data cutoff date: April 3, 2021. Tumor response was assessed by investigators according to RECIST 1.1. PD, progressive disease; SD, stable disease. *, confirmed response. EGFRexon20ins subtypes were confirmed by next-generation sequencing using tumor tissue or/and plasma circulating tumor DNA. JNJ-61186372 = amivantamab. Note: Among the 56 patients, a total of 45 subjects had tumor tissue and/or plasma samples tested by a central laboratory using next-generation of sequencing; 41 of 45 were confirmed as EGFRexon20ins-positive, and the overall concordance rate was 91%. However, 31 subjects had only tumor tissue tested by the central laboratory, and the concordance rate between local and central laboratory testing was 97% (30/31).
In addition, the preliminary antitumor activity of sunvozertinib was also observed in patients with EGFR sensitizing mutation, EGFR sensitizing/T790M double mutation, and HER2exon20ins (Supplementary Table S8).
DISCUSSION
The discovery and development of EGFR TKIs, such as gefitinib and osimertinib, have substantially improved the clinical outcome of EGFR-mutated NSCLC. However, due to the unique structural features of EGFRexon20ins proteins, the current approved EGFR TKIs for sensitizing and T790M mutations were not active against them, and thus a potent and selective inhibitor is urgently needed. Exon20ins mutations occur mostly along a loop between αC and β4 on the kinase domain, with a noticeable exception occurring within the αC-helix (763_764insFQEA; ref. 16). Currently, only one exon20ins mutant (770_771insNPG) crystal protein structure is available in the public domain (Protein Data Bank codes: 4LRM, 7LGS; refs. 15, 17). Due to the scarcity of crystallographic data on different exon20ins mutants, computational modeling was used to study the conformation of various exon20ins mutants and to identify their differences from the wild-type protein (18–20). Most studies concluded that exon20ins mutations preferentially stabilize the αC-helix in the catalytic active conformation. Apart from the αC-helix orientation, it is also speculated that different exon20ins mutants may have an indirect effect on the conformation of the P-loop, hinge, or other β-sheet structures around the ATP-binding pocket, making the ATP-binding pocket smaller for the mutants. This rationale was used to explain the potency of poziotinib against EGFRexon20ins by Robichaux and colleagues (13). However, the modeling studies could not reveal any structural differentiation within the ATP-binding pockets between the EGFRexon20ins mutants and the wild-type protein, partly because all amino acid sequences within the ATP-binding pockets are the same, and the majority of the exon20ins mutations occur outside the ATP-binding pocket. Nevertheless, the differences between the mutants and wild-type ATP-binding pockets can have a subtle effect on ATP and inhibitor binding. Similar to the advantage previously being taken for the development of first-generation EGFR TKIs for exon 19 deletion and L858R substitution mutants, the decrease in ATP-binding Km with the exon 19 deletion and L858R mutants compared with that of the wild-type led to the successful identification of potent and selective EGFR inhibitors, such as gefitinib. Unfortunately, this ATP Km difference was decreased when T790M resistant mutation occurred after gefitinib treatment. The development of third-generation irreversible inhibitors, such as osimertinib (21), alleviated this detrimental effect of high ATP concentration in cells. Yasuda and colleagues have shown that exon20ins-mutant D770_N771insNPG having ATP Km = 36.8 μmol/L that is in between L858R (ATP Km = 68.5 μmol/L) and wild-type (ATP Km = 4.98 μmol/L; ref. 17). The lack of clear structural and biophysical data for most of the exon20ins mutants makes designing pan-exon20ins inhibitors with wild-type protein selectivity challenging. Recently, Gonzalvez and colleagues have reported the design of mobocertinib (TAK-788) targeting EGFRexon20ins (15). The research group focused on exploiting the unoccupied space around the gatekeeper Thr790 and osimertinib. The isopropyl ester substitution on C-5 of the pyrimidine core of osimertinib was found to be optimal. However, it is unclear how a larger molecule of mobocertinib interacts with predicted smaller ATP-binding pockets for EGFRexon20ins mutants. It is also interesting that mobocertinib can maintain its potency against EGFR T790M mutation. The added isopropyl ester on C-5 on the pyrimidine hinge moiety may interact unfavorably with the larger methionine. Furthermore, with the aforementioned unoccupied pocket lined in the proximity with the amino acid sequence on the far loop between αC and β4, we envisioned insertion mutations on exon 20, especially on the far loop, may have a profound effect on that region. Nevertheless, the data from mobocertinib collectively suggested that inhibitors with modified substitutions on the pyrimidine hinge binding core can gain potency against EGFRexon20ins with a certain level of wild-type selectivity. For a more thorough understanding of how EGFRexon20ins and wild-type proteins interact with inhibitors with different scaffolds, the availability of more crystallography data is essential.
In our EGFRexon20ins program, in contrast to mobocertinib, we decided to keep the C-5 position (which is near Thr790) on the pyrimidine open and directly replace the rotationally less flexible methylindole on osimertinib with a more flexible anilinophenyl moiety on C-4 of pyrimidine, hoping for flexible accommodation of the inhibitor among different mutants with slightly variable sizes of the ATP-binding pocket. Together with the suggestion from modeling studies that the C-helix in EGFRexon20ins mutants is more stabilized, we focused on designing inhibitors with substituents on the anilinophenyl moiety that may provide specific interactions with the adjacent C-helix and under the P-loop. After several design–make–test–analyze cycles, our discovery effort culminated in the identification of the clinical candidate sunvozertinib by optimizing the head group on C-4 of the pyrimidine hinge binder and the solvent-exposed amino side chain for both on-target activities against EGFR mutations with desirable drug-like properties (Supplementary Table S2).
The two phase I studies aimed to address the safety, tolerability, PK, and antitumor efficacy of sunvozertinib. The starting dose of 50 mg and the dose-escalation scheme were defined based on nonclinical toxicity studies and PK/PD modeling. Sunvozertinib was generally tolerated up to 400 mg, with 1 of 13 subjects in the 300-mg group and one out of nine subjects in the 400-mg group experienced dose-limiting toxicity. There was a trend showing increased incidence and severity of AEs with the dose increase. Given that 50% of subjects in the 400-mg group need a dose reduction, this dose was not selected for future development. The most common drug-related AEs were diarrhea and skin rash, which were consistent with the mechanism of sunvozertinib through inhibiting the EGFR pathway. Compared with other approved EGFR TKIs, such as afatinib (3) and dacomitinib (22), and EGFRexon20ins inhibitors, such as mobocertinib (11, 12), sunvozertinib showed a more favorable safety profile, with a few patients taking 200 mg and 300 mg experiencing ≥grade 3 drug-related TEAEs that required dose reduction or discontinuation. These data aligned with the original design on the selectivity of this compound.
From 50 mg to 400 mg, sunvozertinib showed a dose-proportional PK profile in humans, suggesting its good oral absorption. Due to its long half-life, sunvozertinib exhibited around 3-fold accumulation of drug exposure after repeat dosing versus single dosing. Similar to the PK profile of osimertinib (23), sunvozertinib exhibited a flat PK curve with small fluctuations between Cmax and Cmin at a steady state, which provides a stable and continuous coverage of the target. This PK pattern could potentially minimize drug-related AEs. These data suggest that sunvozertinib has a favorable PK profile in humans as an oral agent, and at ≥100-mg dose levels, free plasma concentrations of sunvozertinib were able to cover pEGFR IC50 of some subtypes of EGFRexon20ins, and the doses of ≥200 mg could cover the majority of subtypes (Supplementary Fig. S6). Indeed, at doses of ≥100 mg, antitumor efficacy was observed in phase I studies.
Noteworthily, the antitumor activity of sunvozertinib was observed in heavily pretreated patients, with a median three lines of prior systemic therapy. In addition, in four prior amivantamab-treated patients, two showed PR. With these data, sunvozertinib was granted Breakthrough Therapy designation by the FDA for the treatment of patients with locally advanced or metastatic NSCLC with EGFRexon20ins whose disease has progressed on or after platinum-based chemotherapy. A future study is warranted to confirm the antitumor activity of sunvozertinib in a larger cohort of patients with prior amivantamab treatment or prior EGFRexon20ins TKI treatment. Moreover, it is encouraging to see the antitumor activity of sunvozertininb in patients with baseline BM; however, as the intracranial lesions were not assessed as target lesions in this study, future studies are warranted to confirm this finding.
In conclusion, both preclinical and phase I clinical data suggest that sunvozertinib has a good safety/tolerability, PK, and efficacy profile, which warrants its further clinical development for treating NSCLC with EGFRexon20ins.
METHODS
Cell Lines
The A431 human epidermoid carcinoma cell line and the Ba/F3 murine pro-B cell line were obtained from ATCC. Human cell identity was confirmed by short-tandem repeat analysis (GenePrint 10 System, Promega). A431 cells were cultured in DMEM (Gibco) with 10% FBS. Ba/F3 cells were cultured in RPMI 1640 with 10% FBS and 10% conditioned medium of WEHI3B (ATCC) as a source of IL3. Ba/F3 cells carrying EGFRexon20ins 773_NPH were purchased from Crown Bioscience, Inc. All cells were maintained and propagated as monolayer cultures in a humidified incubator with 5% CO2 at 37°C.
Expression Vectors and Transfection
The cDNA sequence of the wild-type EGFR gene was obtained from GenBank (accession number NM005228.3). Full-length cDNAs of different types of human EGFRexon20ins were generated at Shanghai Sunbio Biotechnology Co., Ltd. and confirmed by Sanger sequencing. The cDNAs were then subcloned into the pMT143 lentiviral vector (Shanghai Sunbio Biotechnology Co., Ltd.). The lentivirus was packaged in 293T/17 cells (ATCC; cat. #CRL-11268, RRID: CVCL_1926) by transfection of lentiviral constructs and packaging mix (Shanghai Sunbio Biotechnology Co., Ltd.). Ba/F3 cells were infected by lentivirus with 5 μg/mL polybrene (Sigma-Aldrich), selected in 2 μg/mL puromycin (Invitrogen) as single-cell clones, and maintained in 1 μg/mL puromycin with IL3 depletion. Expression of exogenous EGFR in Ba/F3 cells was confirmed by Sanger sequencing at the mRNA level and Western blot at the protein level.
pEGFR MSD Assay
All cells were seeded into a 96-well plate in their corresponding culture medium containing 1% FBS. The seeding density for Ba/F3 cells and A431 was 50,000 cells/well and 20,000 cells/well, respectively. After overnight incubation, all cells were treated with compounds at a series of concentrations for 4 hours and then were lysed directly except A431 or the wild-type EGFR engineered Ba/F3 cell clone was stimulated with 100 ng/mL of recombinant human EGF for 10 minutes before lysis. pEGFR (Tyr1068) and total EGFR levels in the cell lysates were measured by an electrochemiluminescent method [MULTI-SPOT96 4-Spot HB Prototype EGFR Triplex ANALYTES: pEGFR (Tyr1068), pEGFR (Tyr1173), total EGFR; MESO SCALE DISCOVERY, cat. #N45ZB-1] with an MSD SECTOR Imager, which outputs the ratio of pEGFR/total EGFR for each well. The percentage of inhibition was calculated as follows: % Inhibition = 100 × [1 − (ratio of sample well − ratio of min. control well)/(ratio of max. control well − ratio of min. control well)]. The concentration of the compound producing 50% inhibition of EGFR phosphorylation was calculated in best-fit curves with GraphPad Prism (RRID:SCR_002798, GraphPad Software Inc.).
Cell Proliferation Assay
Ba/F3 cells expressing EGFR mutations were seeded in 384-well plates at 1,250 cells/well in RPMI 1640 medium containing 10% FBS. A431 cells were seeded in 384-well plates at 1,000 cells/well in DMEM containing 10% FBS. At the same time, a day 0 plate was prepared with duplicate rows of each cell line. After overnight incubation, the assay plates were dosed with compounds at a series of concentrations. Along with dosing the assay plates, the day 0 plate was processed using an MTS or a CellTiter-Glo assay to measure the number of viable cells (G0). The MTS assay is a colorimetric method for determining the number of viable cells using a Spark plate reader (Tecan). The assay plates were further incubated for 72 hours, and the number of viable cells (G3) was measured by the MTS or CellTiter-Glo assay. The percentage of proliferation was calculated as follows: % proliferation = 100 × (G3 value of sample well − G0 value)/(G3 value of DMSO control − G0 value). The concentration of the compound producing 50% proliferation inhibition (GI50) was further calculated in best-fit curves using XLFit software.
Generation of PDX Models in Mice and Compound Treatment
All studies involving animals were conducted according to the guidelines approved by Institutional Animal Care and Use Committees and the standard and local regulatory requirements of Dizal Pharmaceuticals or Crown Bioscience Inc. Six- to eight-week-old, specific pathogen–free immunocompromised female nude mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. or Nanjing Biomedical Research Institute of Nanjing University.
PDX models (LU0387 and LU3075) were established subcutaneously in immunocompromised female nude mice by implanting patients’ surgical tumor tissues and serially re-engrafting in vivo. When tumor nodules reached 500 to 600 mm3, tumor tissues were sliced into 3 mm × 3 mm × 3 mm fragments and implanted subcutaneously in female immune-compromised nude mice for compound testing.
Tumor nodules were measured in two dimensions with a caliper, and the tumor volume was calculated using the following formula: tumor volume = (length × width2) × 0.52. When the mean tumor volume reached 150 to 250 mm3, tumor-bearing mice were randomized into different treatment groups. Mice were then treated orally with either vehicle or drug twice daily from the day after randomization. The tumor volume and body weight of the mice were measured twice weekly, and the raw data were recorded according to their study number and measurement date in the assigned lab notebook. Tumor growth inhibition from the start of treatment was assessed by comparison of the mean change in tumor volume between the control and treatment groups and presented as tumor growth inhibition. The arithmetic mean tumor volume was used for efficacy calculation. The calculation was based on the arithmetic mean of relative tumor volume (RTV) in each group. RTV was calculated by dividing the tumor volume on the treatment day by the initial tumor volume. The efficacy of tumor growth inhibition on a specific day, for each treated group, was calculated by the following formula: inhibition % = (CG − TG) × 100/(CG-1), among which “CG” means the arithmetic mean of RTV of the control group, and “TG” means the arithmetic mean of RTV of the treated group.
Determination of Sunvozertinib Plasma Concentrations for PK/PD Study in the Animal Model
Blood samples from the LU3075 PDX mice model were collected at 0.5, 1, 2, 4, 8, and 24 hours on day 28 after the last dosing of sunvozertinib at 12.5, 25, and 50 mg/kg twice daily and put into different K2-EDTA–coagulated tubes, and then plasma was harvested after centrifugation. All plasma samples were stored at around −80°C prior to LC/MS-MS analysis.
Standards were prepared by spiking blank plasma with sunvozertinib covering 1.2 to 1200 ng/mL. Plasma samples were precipitated by adding 4-fold volume of acetonitrile containing internal standard (tolbutamide 30 ng/mL). After 2 minutes of vortex and 10 minutes of centrifugation at 4000 rpm, the supernatant was analyzed by LC/MS-MS (QTRAP5500, Applied Biosystems). Two sets of standard curves were run at the beginning and end of each batch from plasma sample analysis along with two sets of quality controls at different concentrations.
IHC Staining for pEGFR Expression in Tumor Tissue
Tumor tissues from the LU3075 PDX mice model were collected at 2, 8, and 24 hours on day 28 after the last dosing of sunvozertinib. Samples were harvested following formalin fixation and paraffin embedding (FFPE) for further study. IHC was performed on 3-μm FFPE sections using a Lab Vision autostainer (Thermo) for pERK (Thr202/Tyr204) staining and Ventana discovery XT automation (Roche) for pEGFR (Tyr1068) staining. Monoclonal rabbit anti-pEGFR (Tyr1068) antibody (cat. #2234) and pERK (Thr202/Tyr204) antibody (cat. #4376) were purchased from Cell Signaling Technology. The stained IHC slides were first reviewed and interpreted by a qualified pathologist and then quantified by the HALO system. H-score was also performed when the cases were illegible on the HALO platform. Statistical analysis was performed by a t test.
Phase I Clinical Studies
Study objectives
The primary objectives were to assess the safety and tolerability of sunvozertinib and define its MTD. Secondary objectives included the PK of sunvozertinib, and its antitumor efficacy was assessed by the investigator according to RECIST 1.1.
Study design and patients
WU-KONG1 (ClinicalTrials.gov: NCT03974022) is a phase I/II, open-label, multinational study being conducted at 17 centers in the United States, Australia, Taiwan, and South Korea. This study included two parts: part A (dose-escalation, food effect, and dose-expansion cohorts) and part B (dose-extension cohorts). This article reports the data from part A dose-escalation, food effect, and dose-expansion cohorts. Dose-escalation and food effect cohorts enrolled pretreated NSCLC patients with an EGFR or HER2 mutation, and dose-expansion cohorts enrolled only pretreated NSCLC patients with an EGFR or HER2 exon20ins. Key inclusion criteria included age 18 years or above; histologically or cytologically confirmed NSCLC with EGFR or HER2 mutation assessed in local laboratories; life expectancy of at least 3 months; an Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1; adequate organ system functions; and patients with BM can be enrolled only under the condition that BM is stable. For dose-expansion cohorts, patients should not have been previously treated with an EGFRexon20ins TKI. Key exclusion criteria included unsolved ≥grade 2 AEs, according to Common Terminology Criteria for Adverse Events (CTCAE) version 5.0, from previous treatment; a medical history of interstitial lung disease; spinal cord compression or leptomeningeal metastasis; and QTc >470 ms.
WU-KONG2 (Chinadrugtrial: CTR20192097) is a phase I, open-label, multicenter study being conducted at eight centers in China. This study included two parts: part A (dose escalation) and part B (dose expansion). A study design similar to that of the WU-KONG1 dose-escalation and dose-expansion cohorts is applied to this study.
All patients provided written informed consent before participating in this study. Before site activation, the study protocol was approved by the institutional review board or ethics committee at every center participating in the study. The study was undertaken in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines, as defined by the International Conference on Harmonisation. A Safety Review Committee, composed of investigators and core study team members, was formed for study monitoring, safety management, and decision-making.
Procedures
In dose-escalation cohorts, sunvozertinib was administered at 50, 100, 200, 300, and 400 mg once daily, respectively. In the food effect cohort, sunvozertinib was given at a 300-mg single dose, with crossover of fed and high-fat conditions. After PK sampling, patients continued with the repeat dosing of sunvozertinib. In dose-expansion cohorts, sunvozertinib was administered at 200 or 300 mg once daily. Patients received sunvozertinib treatment until disease progression, intolerable AEs, or withdrawal of consent. The investigators assessed safety at every scheduled visit. Categories of AEs were based on terms from the Medical Dictionary for Regulatory Activities (MedDRA), version 24.0. AEs were graded using CTCAE version 5.0, and investigators judged whether AEs were related to sunvozertinib. Tumor assessment was performed by investigators according to RECIST 1.1.
Plasma samples were collected at the scheduled visit and time points, after single or repeat dosing, to assess the sunvozertinib PK profile. Plasma concentrations of sunvozertinib were determined by validated the LC/MS-MS method following protein precipitation using acetonitrile with internal standard. The PK parameters were determined by the noncompartmental analysis method.
Archived tumor tissues or/and plasma samples at baseline were collected for retrospective confirmation of EGFRexon20ins mutation status.
Statistical Analysis
A Bayesian adaptive design was used to inform the dose-escalation and MTD estimation (24). The MTD was defined as the highest dose at which the predicted probability of dose-limiting toxicity was less than 30%. All patients who received at least one dose of sunvozertinib were included in the safety analysis, and patients with EGFRexon20ins NSCLC who had at least one posttreatment tumor assessment were included in the efficacy analysis.
Data Availability
The data generated in this study are available within the article and its supplementary data files.
Supplementary Material
Acknowledgments
We thank the patients, their families, and their caregivers; the investigators and their team members at each study site; and colleagues from Dizal Pharmaceuticals.This study was sponsored by Dizal Pharmaceuticals. Some of this work was supported in part by grants from the Wuxi Municipal Bureau on Science and Technology (WX03-02B0105-062000-01; to L. Zheng) on the WU-KONG2 study.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
M. Wang reports a consultant role with Dizal Pharmaceuticals. J.C.-H. Yang reports personal fees and other support from Daiichi Sankyo, Merck KGaA, Merck Sharp & Dohme, Novartis, Pfizer, Roche/Genentech, Takeda Oncology, Yuhan Pharmaceuticals, Amgen, Bayer, Boehringer Ingelheim, and Bristol Myers Squibb, grants, personal fees, and other support from AstraZeneca, other support from Puma Technology and Eli Lilly, and personal fees from Ono Pharmaceutical, Gilead, and GSK outside the submitted work. P.L. Mitchell reports personal fees from Roche, MSD, Amgen, AstraZeneca, Bristol Myers Squibb, Lilly, Novartis, Pfizer, and Puma Biotechnology outside the submitted work. D.R. Camidge reports institutional support for the Dizal Pharmaceuticals trial. C.-H. Chiu reports personal fees from AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Chugai Pharmaceutical, Eli Lilly, Janssen, Merck KGaA, Merck Sharp & Dohme, Novartis, Ono Pharmaceutical, Pfizer, Roche, Takeda, and Amgen outside the submitted work. V.W. Zhu reports other support from AstraZeneca, Blueprint, Nuvalent, Roche Foundation Medicine, Roche/Genentech, Takeda, TP Therapeutics, and Xcovery outside the submitted work. M. Millward reports other support from Dizal Pharmaceuticals during the conduct of the study, as well as personal fees from Merck Sharp & Dohme, Takeda Pharmaceuticals, Bristol Myers Squibb, Roche Products, Pfizer Australia, Novartis Pharma AG, Amgen Australia, Limbic, Merck Pte. Ltd., Beigene Australia, and Guardant Health and nonfinancial support from AstraZeneca outside the submitted work. L. Bazhenova reports AstraZeneca, Genentech, Beyondspring, Takeda, Bluprint Pharma, Novartis, Boehringer Ingelheim, Regeneron, Merck, Bristol Myers Squibb, Janssen, Daichi Sankyo, Neuvogen, Turning Point Therapeutics, Sanofi, Bayer, Mirati, Novovure, and ORIC outside the submitted work. C.K. Lee reports other support from Dizal Pharmaceuticals during the conduct of the study, as well as grants and personal fees from AstraZeneca, Roche, Amgen, and Merck KGaA, and personal fees from Takeda, Yuhan, Pfizer, MSD Oncology, GSK, Novartis, Janssen, and Boehringer Ingelheim outside the submitted work. Y. Xu reports grants from AstraZeneca outside the submitted work. W.-H. Hsu reports personal fees from AstraZeneca, Roche, Pfizer, Eli Lilly, Chugai, Takeda, Ono, Amgen, Jassen, ACT Genomics, MSD, and Guardant Health outside the submitted work. L. Zheng is an employee of and a shareholder in Dizal Pharmaceuticals. P.A. Jänne reports grants and personal fees from AstraZeneca, Boehringer Ingelheim, Pfizer, Roche/Genentech, Chugai, SFJ Pharmaceuticals, Voronoi, Biocartis, Novartis, Sanofi Oncology, Mirati Therapeutics, Transcenta, Silicon Therapeutics, Syndax, Nuvalent, Bayer, Esai, Allorion Therapeutics, Accutar Biotech, AbbVie, Eli Lilly, Daiichi Sankyo, and Takeda Oncology, and grants from Revolution Medicines and Puma outside the submitted work, as well as a patent for EGFR mutations issued, licensed, and with royalties paid from LabCorp. No disclosures were reported by the other authors.
Authors’ Contributions
M. Wang: Conceptualization, data curation, formal analysis, investigation, methodology, writing–original draft, project administration, writing–review and editing. J.C-H. Yang: Conceptualization, data curation, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. P.L. Mitchell: Data curation, investigation, writing–original draft, writing–review and editing. J. Fang: Conceptualization, data curation, investigation, writing–original draft, writing–review and editing. D.R. Camidge: Conceptualization, data curation, investigation, writing–original draft, writing–review and editing. W. Nian: Data curation, investigation, writing–original draft, writing–review and editing. C.-H. Chiu: Data curation, investigation, writing–original draft, writing–review and editing. J. Zhou: Data curation, investigation, writing–original draft, writing–review and editing. Y. Zhao: Data curation, investigation, writing–original draft, writing–review and editing. W.-C. Su: Data curation, investigation, writing–original draft, writing–review and editing. T.-Y. Yang: Data curation, investigation, writing–original draft, writing–review and editing. V.W. Zhu: Data curation, investigation, writing–original draft, writing–review and editing. M. Millward: Data curation, investigation, writing–original draft, writing–review and editing. Y. Fan: Data curation, investigation, writing–original draft, writing–review and editing. W.-T. Huang: Data curation, investigation, writing–original draft, writing–review and editing. Y. Cheng: Data curation, investigation, writing–original draft, writing–review and editing. L. Jiang: Data curation, investigation, writing–original draft, writing–review and editing. D. Brungs: Data curation, investigation, writing–original draft, writing–review and editing. L. Bazhenova: Data curation, investigation, writing–original draft, writing–review and editing. C.K. Lee: Data curation, investigation, writing–original draft, writing–review and editing. B. Gao: Data curation, investigation, writing–original draft, writing–review and editing. Y. Xu: Data curation, investigation, methodology, writing–original draft, writing–review and editing. W.-H. Hsu: Data curation, investigation, writing–original draft, writing–review and editing. L. Zheng: Conceptualization, data curation, formal analysis, methodology, writing–original draft, project administration, writing–review and editing. P.A. Jänne: Conceptualization, data curation, formal analysis, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non–small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380–8. [DOI] [PubMed] [Google Scholar]
- 2. Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239–46. [DOI] [PubMed] [Google Scholar]
- 3. Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang Y, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213–22. [DOI] [PubMed] [Google Scholar]
- 4. Moyer JD, Barbacci EG, Iwata KK, Arnold L, Boman B, Cunningham A, et al. Induction of apoptosis and cell cycle arrest by CP-358,774, an inhibitor of epidermal growth factor receptor tyrosine kinase. Cancer Res 1997;57:4838–48. [PubMed] [Google Scholar]
- 5. Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 2018;378:113–25. [DOI] [PubMed] [Google Scholar]
- 6. Arcila ME, Nafa K, Chaft JE, Rekhtman N, Lau C, Reva BA, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther 2013;12:220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oxnard GR, Lo PC, Nishino M, Dahlberg SE, Lindeman NI, Butaney M, et al. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol 2013;8:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Naidoo J, Sima CS, Rodriguez K, Busby N, Nafa K, Ladanyi M, et al. Epidermal growth factor receptor exon 20 insertions in advanced lung adenocarcinomas: clinical outcomes and response to erlotinib. Cancer 2015;121:3212–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang G, Li J, Xu H, Yang Y, Yang L, Xu F, et al. EGFR exon 20 insertion mutations in Chinese advanced non-small cell lung cancer patients: molecular heterogeneity and treatment outcome from nationwide real-world study. Lung Cancer 2020;145:186–94. [DOI] [PubMed] [Google Scholar]
- 10. Sabari JK, Shu CA, Park K, Leighl N, Mitchell P, Kim S, et al. Amivantamab in post-platinum EGFR Exon 20 insertion mutant non–small cell lung cancer. J Thorac Oncol 2021;16(3 Suppl S57–S698):S108–9. Abstract nr OA04.04. [Google Scholar]
- 11. Zhou C, Ramalingam S, Li B, Fang J, Kim TM, Kim S, et al. Mobocertinib in NSCLC with EGFR exon 20 insertions: results from EXCLAIM and pooled platinum-pretreated patient populations. J Thorac Oncol 2021;16(3 Suppl S57–S698):S108–9. Abstract nr OA04.03. [Google Scholar]
- 12. Riely GJ, Neal JW, Camidge DR, Spira AI, Piotrowska Z, Costa DB, et al. Activity and safety of mobocertinib (TAK-788) in previously treated non–small cell lung cancer with EGFR exon 20 insertion mutations from a phase I/II trial. Cancer Discov 2021;11:1688–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robichaux JP, Elamin YY, Tan Z, Carter BW, Zhang S, Liu S, et al. Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat Med 2018;24:638–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 2014;4:1046–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gonzalvez F, Vincent S, Baker TE, Gould AE, Li S, Wardwell SD, et al. Mobocertinib (TAK-788): a targeted inhibitor of EGFR exon 20 insertion mutants in non-small cell lung cancer. Cancer Discov 2021;11:1672–87. [DOI] [PubMed] [Google Scholar]
- 16. Friedlaender A, Subbiah V, Russo A, Banna GL, Malapelle U, Rolfo C, et al. EGFR and HER2 exon 20 insertions in solid tumours: from biology to treatment. Nat Rev Clin Oncol 2022;19:51–69. [DOI] [PubMed] [Google Scholar]
- 17. Yasuda H, Park E, Yun CH, Sng NJ, Lucena-Araujo AR, Yeo WL, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med 2013;5:216ra177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ikemura S, Yasuda H, Matsumoto S, Kamada M, Hamamoto J, Masuzawa K, et al. Molecular dynamics simulation-guided drug sensitivity prediction for lung cancer with rare EGFR mutations. Proc Natl Acad Sci U S A 2019;116:10025–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koivu MKA, Chakroborty D, Tamirat MZ, Johnson MS, Kurppa KJ, Elenius K. Structural basis for the functional changes by EGFR exon 20 insertion mutations. Cancers 2021;13:1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Di Gaetano S, Pirone L, Galdadas I, Traboni S, Iadonisi A, Pedone E, et al. Structural basis of the effect of activating mutations on the EGF receptor. eLife 2021;10:e65824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Finlay MR, Anderton M, Ashton S, Ballard P, Bethel PA, Box MR, et al. Discovery of a potent and selective EGFR inhibitor (AZD9291) of both sensitizing and T790M resistance mutations that spares the wild type form of the receptor. J Med Chem 2014;57:8249–67. [DOI] [PubMed] [Google Scholar]
- 22. Wu YL, Cheng Y, Zhou X, Lee KH, Nakagawa K, Niho S, et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol 2017;18:1454–66. [DOI] [PubMed] [Google Scholar]
- 23. Jänne PA, Yang JC, Kim DW, Planchard D, Ohe Y, Ramalingam SS, et al. AZD9291 in EGFR inhibitor–resistant non–small-cell lung cancer. N Engl J Med 2015;372:1689–99. [DOI] [PubMed] [Google Scholar]
- 24. Babb J, Rogatko A, Zacks S. Cancer phase I clinical trials: efficient dose escalation with overdose control. Stat Med 1998;17:1103–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files.