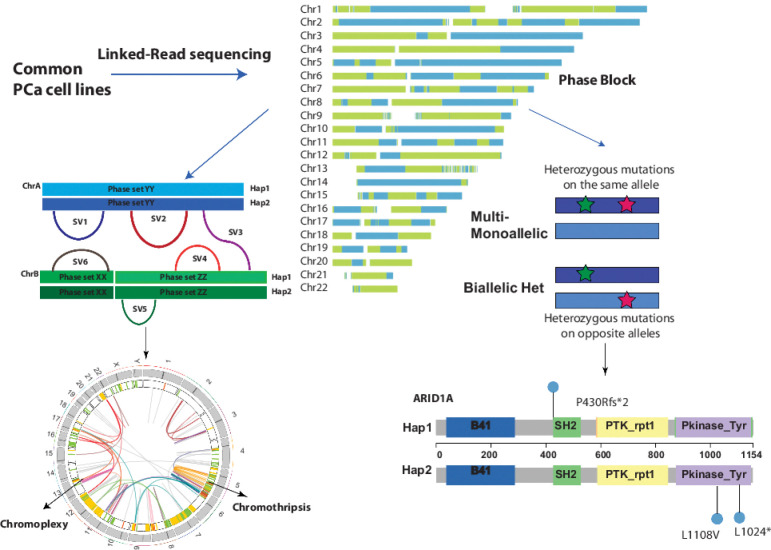
Abstract
A limited number of cell lines have fueled the majority of preclinical prostate cancer research, but their genomes remain incompletely characterized. Here, we utilized whole-genome linked-read sequencing for comprehensive characterization of phased mutations and rearrangements in the most commonly used cell lines in prostate cancer research including PC3, LNCaP, DU145, CWR22Rv1, VCaP, LAPC4, MDA-PCa-2b, RWPE-1, and four derivative castrate-resistant (CR) cell lines LNCaP_Abl, LNCaP_C42b, VCaP-CR, and LAPC4-CR. Phasing of mutations allowed determination of “gene-level haplotype” to assess whether genes harbored heterozygous mutations in one or both alleles. Phased structural variant analysis allowed identification of complex rearrangement chains consistent with chromothripsis and chromoplexy. In addition, comparison of parental and derivative CR lines revealed previously known and novel genomic alterations associated with the CR phenotype.
Implications:
This study therefore comprehensively characterized phased genomic alterations in the commonly used prostate cancer cell lines, providing a useful resource for future prostate cancer research.
Visual Overview
Introduction
Cancer cell line models are vital resources in cancer research. For the prostate cancer field, due to difficulties in establishing cell lines from cancer tissues, there are only about 20 unique parental cell lines established in the past half-century of research (1, 2). A query of PubMed revealed >23,000 publications with these cell lines, with seven of them (PC3, LNCaP, DU145, CWR22Rv1, VCaP, LAPC4, MDA-PCa-2b) accounting for >99% of the citations (Supplementary Materials and Methods). Early landmark efforts for molecular characterization of the prostate cancer cell lines revealed both chromosomal instability and gene mutations as key oncogenic drivers (2, 3).
Advances in next-generation sequencing (NGS) have fueled large-scale genomic studies of >1,000 prostate cancer genomes (4, 5). These studies identified recurrently mutated prostate cancer driver genes even when they were rare across individuals (4). Whole-genome sequencing studies also identified key patterns of structural variants (SV), including complex chained and clustered rearrangement breakpoints termed chromoplexy and chromothripsis, as drivers of prostate cancer (6–9). NGS has also allowed ever more comprehensive and deep characterizations of the ways in which the prostate cancer cell lines resemble or differ from primary human cancer tissues in their genetic alterations (10). However, it is still unclear to what extent the common prostate cancer cell lines recapitulate the genomic mutational and structural alterations found in patient samples.
Long-read sequencing and linked-read sequencing have allowed comprehensive assessment of haplotypes and even end-to-end assembly of human genomes (8, 11, 12). Combining the power of sequencing fidelity and throughput of conventional Illumina sequencing with emulsion-based barcoding of smaller fragments derived from very high molecular weight (HMW) DNA, linked-read sequencing allows phasing of variants, and greater power for detecting structural alterations.
Here we report a comprehensive analysis of phased mutations and SVs determined through linked-read sequencing of the commonly used prostate cancer cell lines, including castrate-resistant (CR) subclones of three of those cell lines. These analyses revealed that this collection of cell lines harbored a significant fraction of recurrent putative driver mutations and complex SVs observed in human prostate cancer.
Materials and Methods
Cell lines and culturing methods
Cell lines were obtained from ATCC (CWR22Rv1, DU145, LNCaP, MDA-PCa-2b, RWPE-1, VCaP), Johns Hopkins Biorepository (LAPC4-CR, VCaP-CR), NCI (PC3), UroCor, OK (LNCaP_C42b), Charles Sawyer's lab, UCLA (LAPC4), and Zoran Culig's lab, Innsbruck Medical University (LNCaP_Abl). All were cultured in media as instructed by providers and used at passages less than 10 (Supplementary Table S1). Frozen pellets were used for Mycoplasma testing (MycoDtect kit, Greiner Bio-One) and short tandem repeat (STR) genotyping (GenePrint 10 System, Promega) performed by the Johns Hopkins Genetic Resource Core Facility in July 2019 right before sequencing was performed. All cell lines were Mycoplasma free and passed the a priori defined STR genotype threshold of 70%. Sources, RRIDs, culture conditions, passage number, and STR genotype results are reported in Supplementary Table S1.
HMW DNA extraction
HMW DNA was extracted following the “salting out” protocol described by 10× Genomics (Pleasanton, CA). Briefly, 2 million cells were lysed overnight at 37°C in Lysis buffer containing 2 mmol/L EDTA, 300 mmol/L NaCl, 8 mmol/L Tris-HCl, 125 μg/mL proteinase K, and 0.5% SDS. HMW DNA was precipitated using 1 mol/L NaCl, washed in absolute ethanol, and resuspended overnight at 4°C in TE buffer. The TapeStation 4200 system (Agilent) was used to assess DNA quality. All samples contained more than 80% of DNA having greater than 60 kb in length (Supplementary Table S2).
Barcoding, library preparation, and sequencing
A total of 1 (±0.2) ng of DNA was quantified by Qubit Broad Range (Thermo Fisher Scientific, Q32850) and used for gel beads-in-emulsion (GEM) creation and barcoded library generation using the 10× Chromium Genome protocol. Resulting library fragment sizes were determined using the DNA 1000 and 2100 Kit BioAnalyzer (Agilent Technologies), and indexed with Chromium i7 Sample Index Plate (catalog no. 220103, 10× Genomics). The indexed libraries were sequenced to 30×–50× average coverage on an Illumina HiSeqX platform using paired-end 150 bp × 150 bp reads. The resulting sequencing BCL files were processed by the Long Ranger Pipeline (2.2; ref. 13; 10× Genomics) for alignment, variant discovery, and phasing.
Analysis for phased mutations and SVs
Long Ranger pipeline was used for phasing of small variants (SNP and micro-indels), phased SVs, and unphased copy-number variants (CNV). Passed variants from the Long Ranger phased_variants.vcf output files were annotated using the vcf2maf (14) pipeline and the Uniprot canonical isoform reference. Gene-level haplotype was inferred through a custom R (version 4.0.0) script that used the haplotype (column GT) and phase set (column PS) information provided by Long Ranger in phase_variants.vcf files. Any gene with at least one mutation with GT = “1|1” was categorized as “Hemi/homozygous,” and any gene with only one heterozygous (GT = “0|1” or “1|0”) mutation was categorized as “monoallelic.” For any gene with multiple heterozygous mutations, if all mutations were on the same phase set and belonged to the same haplotype, it was categorized as “multi-monoallelic.” If all mutations were on the same phase set and there was at least one mutation belonging to the opposite allele, it was categorized as “biallelic heterozygous.” In cases where multiple heterozygous mutations did not belong to the same phase set or were unphased (GT = “0/1”), the genes were categorized as “no info.”
Phased SVs were defined as large SV calls in large_sv_calls.bedpe file that had phased set information (PS1/PS2). A custom R script, termed ChainLink, allowed inference of phased SV breakpoints as follows: SVs were chained together based on both shared phased set and shared haplotype for each breakpoint. Each SV chain contained at least two SVs.
ChainFinder (9) was used in parallel using phased SVs from all 12 cell lines to determine background breakpoint frequency. “copy number type” was set as “Seq” and Deletion_threshold was set at 2. Other parameters were left as default. SnpEff 4.5.1 (RRID:SCR_005191) was used to annotate the closest gene to each breakpoint coordinate.
Graphs were prepared using 10× Genomics Loupe Browser 4.1.0, the R packages Rcircos (1.2.1; RRID:SCR_003310), and ggplot2 (3.3.5; RRID:SCR_014601), and assembled on Adobe Illustrator (24.1.1; RRID:SCR_010279).
Data availability statement
Raw data were made available at SRA with the following object ID: PRJNA751700. SVs and CNVs data were made available at dbVar under the accession number nstd213. Supplementary Data are included.
Results
Linked-read sequencing of human prostate cancer cell lines
We performed linked-read sequencing of HMW DNA from 11 prostate cancer cell lines and one immortalized nonmalignant prostatic epithelial cell line RWPE-1 (Supplementary Tables S1 and S2; Workflow: Supplementary Fig. S1A). The majority of cell lines were of low passage from the original source, and shared a majority of mutations found in driver genes from the Cancer Cell Line Encyclopedia (CCLE) project (Supplementary Tables S3 and S4). Whole-genome copy number and gene-level copy-number analysis also showed a high degree of correlation between related cell lines when comparing the current data to that from cells of later passages or with CCLE data, while unrelated cell lines showed lower correlation (Supplementary Fig. S1B; Supplementary Tables S3 and S4). As expected, we confirmed that cell lines with known microsatellite instability (MSI) and mismatch repair deficiency exhibited high mutation rates, and microsatellite stable (MSS) VCaP and PC3 cells exhibited lower mutational burdens (Supplementary Fig. S1C; Supplementary Table S5; ref. 15). In addition to MMR gene defects in some cell lines, many cell lines showed mutations in HDR genes, as indicated in Supplementary Table S6.
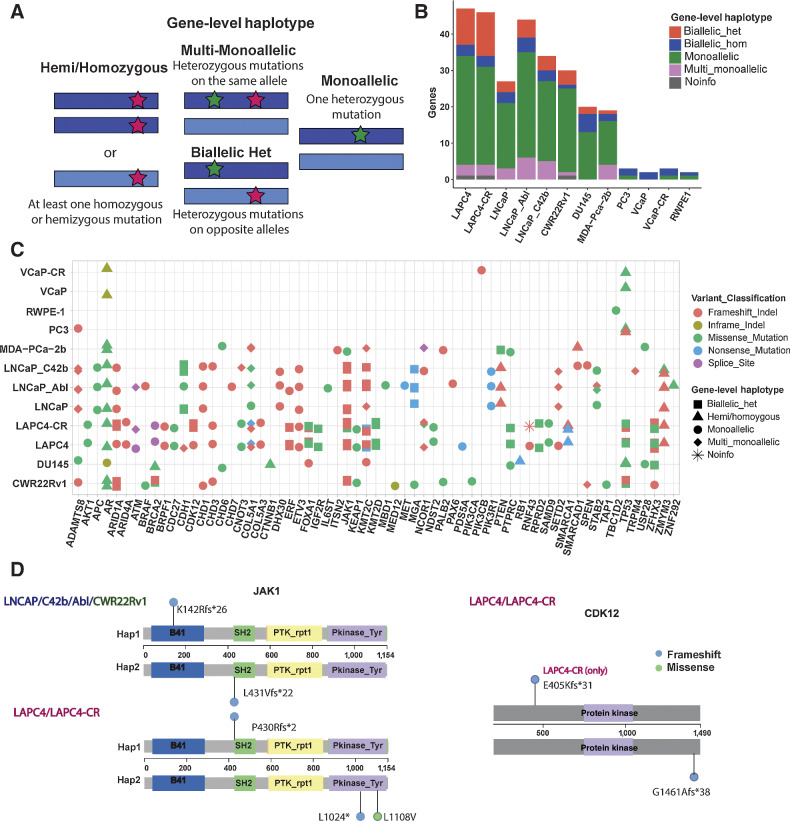
Gene-level phasing of somatic mutations to distinguish between monoallelic and biallelic gene mutations
Linked-read sequencing allowed phasing of heterozygous mutations, thus revealing allelic mutation status at the gene level. Using this phasing information, we developed the concept “gene-level haplotype” for each gene to determine whether a given gene is altered at a single or both alleles (Fig. 1A). We focused our analysis on 97 putative driver genes recurrently mutated in prostate cancer cataloged by Armenia and colleagues (4) that we called “Longtail” genes. Among these, we found 87 genes to be mutated in our panel of 12 cell lines (Fig. 1B; Supplementary Fig. S1D). MSI cell lines showed higher percentage of genes inactivated by biallelic heterozygous mutations, whereas MSS cell lines showed mostly monoallelic or hemi/homozygous mutations (Fig. 1B). To enrich missense mutations among potential cancer drivers, we filtered to those specific missense mutations documented in the Cancer Mutation Census (CMC) project under COSMIC (16), yielding 58 Longtail genes with putative driver mutations found in our panel of cell lines (Fig. 1C).
Figure 1.
Phasing of somatic mutations to determine gene-level haplotype. A, Schematic of gene-level haplotype, derived from phased linked read sequencing data, distinguishes between genes with multiple heterozygous mutations on the same or different alleles. Dark blue or light blue bars represent each allele for a given gene. Red and green stars indicate examples for the positions of mutations. B, Somatic mutations on “Longtail” genes previously found to be recurrently mutated in prostate cancer (4) stratified by gene-level haplotype. C, Cell line mutations in the Longtail panel of cancer driver genes that are recurrently mutated in human prostate cancer, with variant classification for each mutation, and gene-level haplotype for each gene annotated for each cell line. D, Lollipop plots showing locations of JAK1 and CDK12 biallelic heterozygous mutations relative to positions of Pfam domains. Colors of the lollipop heads indicate the variant classification for each mutation (blue = frameshift; green = missense).
Gene-level haplotypes allowed identification of several genes inactivated in a monoallelic versus biallelic manner (Supplementary Table S7). Illustrative examples of genes with biallelic alterations are shown for CDK12 and JAK1 (Fig. 1D). CDK12 mutation denotes a distinct subclass of prostate cancer characterized by tandem duplication and high neoantigen burden (6). In our cell lines, LAPC4 and its CR clone both had CDK12 G1461Afs*38 (COSM2837928) mutation. This somatic mutation is the most common frameshift CDK12 mutation documented in the CMC, recurrently found in human cancers (16). It is interesting to note that this frameshift mutation led to alteration of the last 30 amino acids of the protein, along with lengthening the protein by 9 amino acids. It is thus intriguing to speculate that it may have led to dominant negative function rather than just loss of function. LAPC4-CR lost the other copy of CDK12 from an E405Kfs*31 mutation, which truncated the remaining two thirds of the protein, and therefore had mutations in both alleles (Fig. 1D; Supplementary Table S7).
Another example to note was JAK1, a tyrosine kinase implicated in immune and apoptosis evasion (17). LNCaP, its CR clones, and CWR22Rv1 all had p.L431Vfs*22 (COSM41842) and p.K142Rfs*26 (COSM1639943), both of which are recurrent JAK1 mutations in human cancer (Fig. 1D). LAPC4 and LAPC4-CR shared p.P430Rfs*2 (COSM1560531) and LAPC4-CR had additional mutations at p.L1108V and p.L1024* on the kinase domain. Loss of function mutations in JAK1 is common among MSI cell lines and associated with reduced interferon response (17). Among the cell lines examined here, biallelic heterozygous mutations on JAK1 were observed in three MSI cell lines. We can speculate that these mutations may have been involved in evasion of apoptosis and immune surveillance in the original cancers from which these cell lines were derived (17).
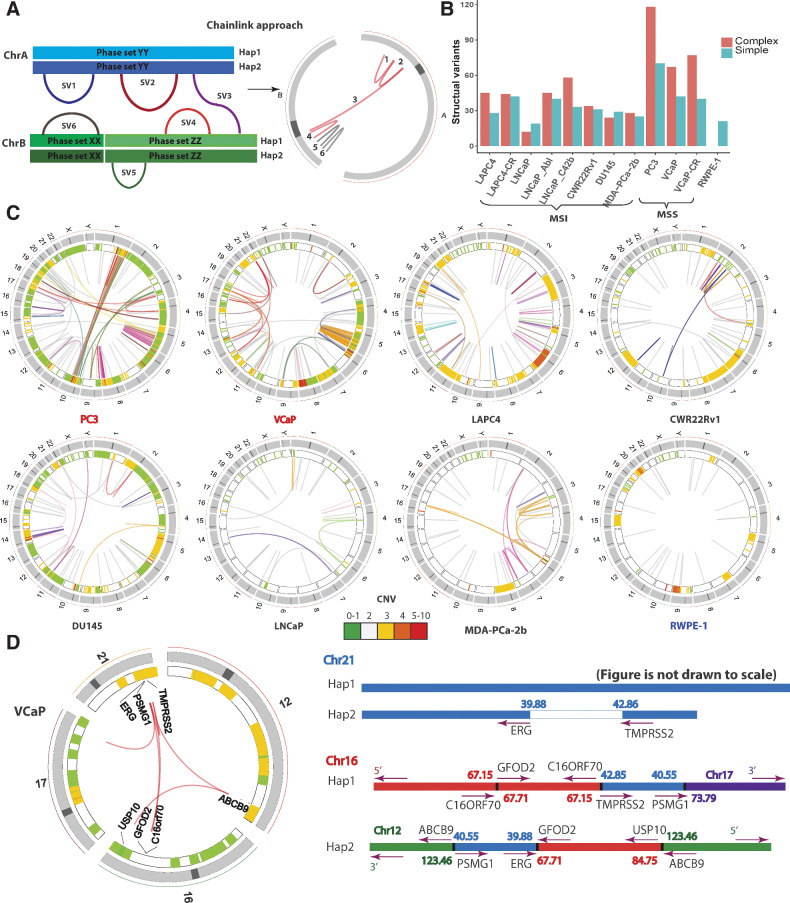
Phasing of structural alterations in prostate cancer cell lines
We next examined SVs in the prostate cancer cell lines. At a glance, in contrast with the high number of mutations (Supplementary Fig. S1C), MSI cell lines exhibit less SVs compared with MS stable cell lines (Supplementary Fig. S2A). By combining rearrangement breakpoint information with the associated haplotype information of each segment of the breakpoint, we phased multiple breakpoints to the same allele to identify complex rearrangements consistent with chromoplexy or chromothripsis. We developed a pipeline called ChainLink to infer clustered SVs by combining phase information and SV identification (Fig. 2A). We identified complex rearrangements in all prostate cancer cell lines, ranging from three chains in LNCaP to 11 chains in VCaP-CR (Fig. 2B and C; Supplementary Table S8). The modal number of SVs in one chain was 3. The non-cancer line RWPE-1 notably did not have any complex SVs (Fig. 2B and C; Supplementary Table S8), but did have evidence of having subclonal population(s) with potential aneuploidy (Supplementary Fig. S3A and S3B). Many of these complex rearrangements found in prostate cancer were consistent with chromoplexy and chromothripsis (Fig. 2C and D; Supplementary Fig. S2D and S2E).
Figure 2.
Identification and phasing of structural alterations. A, The ChainLink approach uses phase information for each breakpoint to chain together complex SVs. In this schematic, SV 1, 2, 3, 4 on two different chromosomes are chained together into a complex SV cluster based on phase information on each breakpoint; SV 5 and 6 are considered simple SVs. B, Number of SVs classified as simple or complex in each cell line. C, Circos plots representing CNV and SVs on the eight parental cell lines in this study: MSI stable (red), MSI high (black), and nonmalignant immortalized prostatic epithelium (navy). Heatmap track beneath chromosome ideograms represents CNV with colors representing copy number. Innermost link track represents large SVs: complex SVs are colored by chain; simple SVs are gray. D, Left: Circos plot showing the chromoplexy event associated with the TMPRSS2-ERG fusion gene formation in VCaP. Genes interrupted by these SVs are labeled on the innermost track. Right: genomic anatomy of the breakpoints involved in the chromoplexy event leading to the TMPRSS2-ERG fusion gene in VCaP and detailed rearrangement configuration of the 3 Mb sequence between the TMPRSS2-ERG genomic breakpoint. Genomic coordinates are in Mb per reference genome hg19. Arrows represent gene direction.
We compared the performance of ChainLink with a previous method called ChainFinder, which uses rearrangement breakpoints identified by conventional paired-end sequencing data to infer chained rearrangements based on comparing breakpoint locations with an expected distribution of breakpoints as if they had arisen independently (9). ChainLink identified all chains called by ChainFinder (Supplementary Fig. S2B and S2C). Moreover, ChainFinder identified fewer chains, and fewer SVs per chain, relative to ChainLink (Supplementary Fig. S2B and S2C). These analyses suggested that the use of phasing information from linked-read sequencing can increase the sensitivity of identifying rearrangements within complex chained rearrangements compared with more indirect statistical methods.
Using ChainLink, we were able to decipher the complete genomic anatomy of the complex SV underlying the TMPRSS2-ERG fusion gene in VCaP cells (Fig. 2D; Supplementary Table S9). It was previously known that the 3 Mbp sequence between the TMPRSS2 and ERG genes was rearranged to other genomic segments in this cell line, rather than being deleted (18); however, the precise genomic anatomy of rearrangements involving this 3 Mbp stretch was not well understood. Here we show that this 3 Mb piece was broken up into two parts, with one rearranged with sequences on chromosomes 16 and 17, and the other rearranged with sequences on chromosomes 16 and 12. Altogether, this rearrangement consisted of seven SV breakpoints in a highly complex genomic anatomy (Fig. 2D). Parallel ChainFinder analysis only called three of these SVs and could not fully decipher the complex anatomy of the rearrangements associated with this fusion gene (Supplementary Table S9).
In addition to the aforementioned chromoplexy event, VCaP also harbored a highly complex chromothripsis event on chromosome 5 characterized by clustered SVs with random orientation and alternating copy-number changes (Supplementary Fig. S2E). PC3 also showed evidence of chromothripsis, occurring on multiple chromosomes. In addition to two striking chromothripsis rearrangement clusters on chromosome 5q and 8q, we noted an unusual interchromosomal chromothripsis event bridging chromosomes 1 and 10 (Supplementary Fig. S2D; Supplementary Tables S9 and S10). This was suggestive of an interchromosomal rearrangement between chromosomes 1 and 10 in the PC3 cells, with subsequent catastrophic chromothripsis of the newly formed fused chromosome.
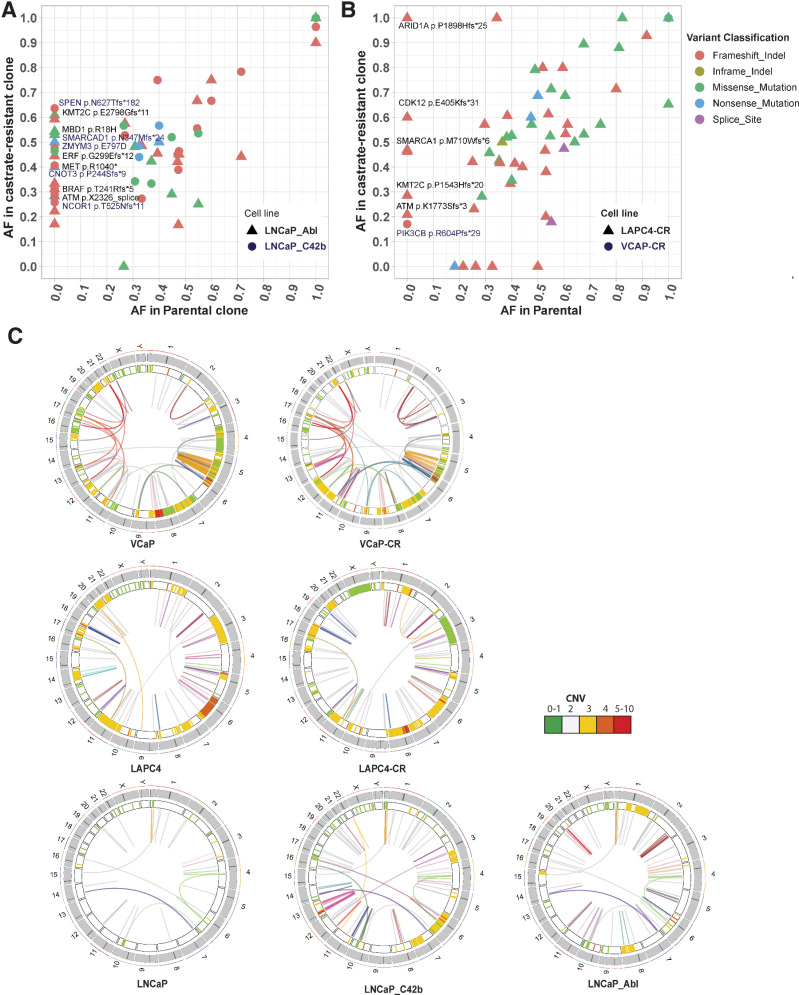
Somatic variants associated with resistance to androgen deprivation in prostate cancer cell lines
Understanding mechanisms of resistance to androgen receptor targeted therapy remains an important area of prostate cancer research. Among the cell lines with AR expression, it was interesting to note that there were several alterations in the AR gene itself, many of which have been previously documented (1, 2). CWR22Rv1, MDA-PCa-2b, VCaP, LAPC4, and LNCaP all have mutations in AR; CWR22Rv1 has duplication around the third exon of AR, and VCaP showed amplification of the entire AR locus (ref. 19; Supplementary Fig. S3C and S3D; Supplementary Table S11). While some of these alterations, such as the AR T878A mutation in LNCaP, have well-documented functional implications, the functional significance of other mutations are not well understood (20). To understand whether there are acquired mutations associated with progression to resistance to androgen deprivation, we next compared the somatic mutational and SV landscape in the parental LNCaP, LAPC4, and VCaP cell lines versus their previously established androgen deprivation–resistant subclones (refs. 2, 20, 21; Fig. 3). While the CR cell lines largely retained the mutations present in their parental cell line, confirming that they are truly subclones of those parental lines, they gained many additional mutations not present in the parental cell lines (Fig. 3A and B). LNCaP_Abl, LNCaP_C42b, and LAPC4-CR all gained mutations in genes encoding epigenetic machinery, including members of the SWI/SNF chromatin remodeling complex (ARID1A, ARID1B, SMARCA4, and SS18; Fig. 3A and B). Interrogating the cBioPortal database for mutations in 28 SWI/SNF subunits across five prostate cancer genomic studies (13, 22–25) found that 11% of prostate cancer samples (170/1,578 total patients) showed mutations in at least one SWI/SNF subunit (Supplementary Fig. S4A and S4B), with more mutations found in CR prostate cancer than castrate-sensitive prostate cancer (Supplementary Fig. S4B and S4C). These preliminary observations suggest SWI/SNF mutations potentially play a role in driving CR prostate cancer. Other epigenetic machinery proteins that gained mutations in the CR cell lines include BRD3, SMARCAD1, MBD1, KMT2C, and NCOR2 (Fig. 3A and B).
Figure 3.
Comparison of LNCaP, LAPC4 parental cell lines with their respective CR derivatives. A and B, Allele frequencies for each mutation in CR clones LNCaP_Abl, LNCaP_C42b, LAPC4-CR and their associated parental lines (LNCaP and LAPC4). Mutations seen along the y-axis represent new mutations arising in the CR lines not found in the parental cells. C, Juxtaposition of SVs in parental and CR clones. Heatmap track beneath chromosome band represents CNV with colors representing copy number. Innermost link track represents large SVs as defined by barcode overlap: complex SVs are colored by chain; simple SVs are gray.
CR clones also accumulated more SVs and CNVs, many of which may have contributed to their CR progression (Supplementary Fig. S5). Most notably, 8q24 amplification was found in both LAPC4-CR and LNCaP_Abl (Supplementary Fig. S5A and S5B). In LNCaP_Abl, there were three copy-number gains at 8q24 with one spanning the MYC enhancer (Supplementary Fig. S5A). The LAPC4-CR line, which harbored biallelic mutation of CDK12, showed multiple tandem duplications around both the MYC gene and its enhancer (Supplementary Fig. S5B and S5E); such an association between CDK12 mutation and tandem duplications around the MYC gene and its enhancer was recently reported in human prostate cancer (7). VCaP-CR has an AR enhancer amplification, a well-studied recently discovered SV driving castrate resistant phenotype (Supplementary Fig. S5C; ref. 8). The AR enhancer duplication in VCaP-CR occurred in addition to the AR amplification already present in VCaP. Finally, C42b gained a series of complex SVs on chromosome 10, 11, 12, 13, and 14 (Fig. 3C; Supplementary Fig. S5D; Supplementary Table S10). Most notable was a chromothripsis-like complex rearrangement on chromosome 13 (Supplementary Fig. S5D; Supplementary Table S10). These complex rearrangements involved multiple amplifications and translocations within chromosome 13, dysregulating genes within 13q12.11 to 13q13.2 and 13q31.3. Among these, LATS2 encodes a tumor suppressor protein kinase that functions as a positive regulator of TP53 and as a corepressor of androgen-responsive gene expression. Future mechanistic studies can examine the role of these mutations in driving resistance to androgen deprivation or prostate cancer progression.
Discussion
Linked-read sequencing allowed allelic phasing of both mutations and SVs in the commonly used prostate cancer cell lines. The phasing information of somatic mutations was used to derive “gene-level haplotypes,” which distinguished between multiple heterozygous mutations on the same or different copies of each gene. This allowed for the first time a genome-wide analysis of monoallelic versus biallelic driver gene alterations in the prostate cancer cell lines, providing a valuable resource for future studies (Supplementary Tables S6–S13). While it is certainly possible and likely that the cell lines can be prone to cell line drift, the high degree of similarity, with respect to mutations and copy-number alterations, between the cell lines used in this study and those reported in the CCLE project and in batches of cell lines from independent lab and across passage numbers, suggests that many of the alterations and findings reported here should be generalizable (Supplementary Fig. S1; Supplementary Tables S3 and S4).
The linked read sequencing data are well suited to identifying rearrangement breakpoints as overlap of barcodes from noncontiguous genomic regions. Furthermore, by developing a simple approach termed ChainLink, we leveraged the phasing/haplotype information from the linked read data to phase SV breakpoints and identify chained complex rearrangements, including those consistent with chromoplexy and chromothripsis (Supplementary Table S10). First, we were able to decipher for the first time the precise genomic anatomy of several complex rearrangement events, including the highly recurrent TMPRSS2-ERG rearrangement present in VCaP cells (Fig. 2D; Supplementary Table S9), as well as several chromoplexy and chromothripsis events in numerous cell lines that were previously unrecognized (Fig. 2C; Supplementary Fig. S2D and S2E). Second, by phasing all of the individual breakpoints that constituted these complex rearrangements, we showed that the breakpoints all occurred across a single phased allele, and did not represent rearrangements that were staggered independently in different alleles. This supports the model that chromoplexy and chromothripsis occur in a single concerted event, rather than through accumulation of multiple independent rearrangements.
These analyses also established the relevance of this set of prostate cancer cell lines to human prostate cancer. First, the cell lines captured a large fraction of recurrently mutated driver genes in human prostate cancer as have been reported in large-scale prostate cancer genome sequencing studies (4). In addition, integrating the phased mutation and SV information, we saw many associations between mutations and SV patterns that have been described in human cancers including: (i) the association of CDK12 mutations with tandem duplications as seen in the LAPC4/LAPC4-CR cell lines; (ii) the association of TP53 mutations with presence of chromothripsis (PC3 and VCaP cells); and (iii) correlation between pathogenic BRCA2 mutations and genome wide deletion in DU145 (Figs. 1C and 2C; ref. 7). Finally, comparing the CR cell lines to the castration sensitive parental cell lines, we found that several mutations in epigenetic pathways occurred in the CR lines.
One limitation of this study is that the approaches used for phasing variants and rearrangement breakpoints would not have accounted sufficiently for aneuploidy. As a result of this, we would be limited in determining the phasing of variants that arose after a specific allele gained a copy. Nonetheless, because we had such a high rate of phasing of the identified somatic mutations, it is likely that most of these mutations arose prior to any copy-number gains of those segments.
Taken together, this study serves as a comprehensive compilation of genomic features for the most commonly studied prostate cancer cell lines, provides important insights into the pathogenesis of chromoplexy, represents a valuable resource for the field, and presents genomic analysis frameworks for future long-read sequencing studies.
Supplementary Material
Acknowledgments
We would like to thank Jennifer Meyers from the SKCCC Experimental and Computational Genomics Core and Lisa Haley from the Pathology Molecular Diagnostics Lab for their help with sequencing runs and quality controls of DNA samples.
This work was supported by NIH/NCI grants P50CA058236, U01CA196390, R01CA183965, DOD CDMRP grant W81XWH-21-1-0295, and by the Prostate Cancer Foundation, Commonwealth Foundation, Maryland Cigarette Restitution Fund, and the Irving Hansen Foundation. The Experimental and Computational Genomics Core at the SKCCC was supported by the NIH/NCI Cancer Center Support Grant P30CA006973, and M.A. Garrison was supported by the National Science Foundation Graduate Research Fellowship under grant number DGE-1746891.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Disclosures
W.B. Isaacs reports being a coinventor on patent no. 9593380, Inst. related to the discovery of HOXB13 as a prostate cancer susceptibility gene. W.G. Nelson reports grants from NIH, Prostate Cancer Foundation, Department of Defense CDMRP, Commonwealth Foundation, Maryland Cigarette Restitution Fund, and Irving Hansen Foundation during the conduct of the study; grants and personal fees from Cepheid; personal fees from Digital Harmonics, Armis Biosciences, and Brahm Astra Therapeutics outside the submitted work; in addition, W.G. Nelson has a patent for LAPC-CR (castration-resistant) pending, a patent for VCaP-CR (castration-resistant) pending, a patent for Agents for reversing epigenetic silencing of genes issued to Option to Brahm-Astra Therapeutics, and a patent for Induction of synthetic lethality by epigenetic drugs pending. S. Yegnasubramanian reports grants from NIH, Prostate Cancer Foundation, Commonwealth Foundation, Maryland Cigarette Restitution Fund, Department of Defense CDMRP, and Irving Hansen Foundation during the conduct of the study; grants and personal fees from Cepheid; other support from Digital Harmonic, Brahm Astra Therapeutics; grants from Bristol Meyers Squibb and Janssen outside the submitted work; in addition, S. Yegnasubramanian has a patent for LAPC4-CR pending, a patent for VCaP-CR pending, a patent for Agents for reversing epigenetic silencing of genes issued, and a patent for Induction of synthetic lethality with epigenetic therapy pending. No disclosures were reported by the other authors.
Authors' Contributions
M.-T. Pham: Conceptualization, resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. A. Gupta: Resources, data curation, software, formal analysis, validation, methodology. H. Gupta: Formal analysis, validation, investigation, visualization, writing–original draft. A. Vaghasia: Data curation, funding acquisition, investigation, writing–original draft. A. Skaist: Software, formal analysis. M.A. Garrison: Formal analysis, writing–review and editing. J.B. Coulter: Conceptualization, writing–original draft. M.C. Haffner: Conceptualization, resources, funding acquisition, writing–original draft. S.L. Zheng: Data curation, software, formal analysis. J. Xu: Resources, data curation, formal analysis. C. DeStefano Shields: Writing–review and editing. W.B. Isaacs: Conceptualization, data curation, supervision, funding acquisition, validation, methodology, writing–original draft, writing–review and editing. S.J. Wheelan: Conceptualization, resources. W.G. Nelson: Conceptualization, resources, supervision, funding acquisition, writing–original draft. S. Yegnasubramanian: Conceptualization, resources, data curation, supervision, funding acquisition, investigation, methodology, writing–original draft, writing–review and editing.
References
- 1. Sobel RE, Sadar MD. Cell lines used in prostate cancer research: a compendium of old and new lines—part 1. J Urol 2005;173:342–59. [DOI] [PubMed] [Google Scholar]
- 2. van Bokhoven A, Varella-Garcia M, Korch C, Johannes WU, Smith EE, Miller HL, et al. Molecular characterization of human prostate carcinoma cell lines. Prostate 2003;57:205–25. [DOI] [PubMed] [Google Scholar]
- 3. van Bokhoven A, Caires A, Maria MD, Schulte AP, Lucia MS, Nordeen SK, et al. Spectral karyotype (SKY) analysis of human prostate carcinoma cell lines. Prostate 2003;57:226–44. [DOI] [PubMed] [Google Scholar]
- 4. Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018;50:645–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 2015;163:1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu Y, Cieślik M, Lonigro RJ, Vats P, Reimers MA, Cao X, et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell 2018;173:1770–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 2018;174:758–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Viswanathan SR, Ha G, Hoff AM, Wala JA, Carrot-Zhang J, Whelan CW, et al. Structural alterations driving castration-resistant prostate cancer revealed by linked-read genome sequencing. Cell 2018;174:433–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baca S, Prandi D, Lawrence M, Mosquera J, Romanel A, Drier Y, et al. Punctuated evolution of prostate cancer genomes. Cell 2013;153:666–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sienkiewicz K, Ratan A. Protocol for integrative subtyping of lower-grade gliomas using the SUMO pipeline. STAR Protoc 2022;3:101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marks P, Garcia S, Barrio AM, Belhocine K, Bernate J, Bharadwaj R, et al. Resolving the full spectrum of human genome variation using linked-reads. Genome Res 2019;29:635–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zheng GXY, Lau BT, Schnall-Levin M, Jarosz M, Bell JM, Hindson CM, et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol 2016;34:303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet 2013;45:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kandoth C. Vcf2maf. 1.6.19; 2020.
- 15. Sun X, Chen C, Vessella RL, Dong J. Microsatellite instability and mismatch repair target gene mutations in cell lines and xenografts of prostate cancer. Prostate 2006;66:660–6. [DOI] [PubMed] [Google Scholar]
- 16. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 2018;47:D941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Albacker LA, Wu J, Smith P, Warmuth M, Stephens PJ, Zhu P, et al. Loss of function JAK1 mutations occur at high frequency in cancers with microsatellite instability and are suggestive of immune evasion. PLoS One 2017;12:e0176181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun X, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005;310:644–8. [DOI] [PubMed] [Google Scholar]
- 19. Li Y, Hwang TH, Oseth LA, Hauge A, Vessella RL, Schmechel SC, et al. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene 2012;31:4759–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Culig Z, Hoffmann J, Erdel M, Eder IE, Hobisch A, Hittmair A, et al. Switch from antagonist to agonist of the androgen receptor bicalutamide is associated with prostate tumour progression in a new model system. Br J Cancer 1999;81:242–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haffner MC, Bhamidipati A, Tsai HK, Esopi DM, Vaghasia AM, Low JY, et al. Phenotypic characterization of two novel cell line models of castration-resistant prostate cancer. Prostate 2021;81:1159–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019;116:11428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Robinson D, Van Allen EM, Wu Y, Schultz N, Lonigro RJ, Mosquera J, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stopsack KH, Nandakumar S, Wibmer AG, Haywood S, Weg ES, Barnett ES, et al. Oncogenic genomic alterations, clinical phenotypes, and outcomes in metastatic castration-sensitive prostate cancer. Clin Cancer Res 2020;26:3230–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data were made available at SRA with the following object ID: PRJNA751700. SVs and CNVs data were made available at dbVar under the accession number nstd213. Supplementary Data are included.