Abstract
Gene regulation in the human genome is controlled by distal enhancers that activate specific nearby promoters1. One model for this specificity is that promoters might have sequence-encoded preferences for certain enhancers, for example mediated by interacting sets of transcription factors or cofactors2. This “biochemical compatibility” model has been supported by observations at individual human promoters and by genome-wide measurements in Drosophila3-9. However, the degree to which human enhancers and promoters are intrinsically compatible has not been systematically measured, and how their activities combine to control RNA expression remains unclear. Here we designed a high-throughput reporter assay called ExP STARR-seq (enhancer x promoter self-transcribing active regulatory region sequencing) and applied it to examine the combinatorial compatibilities of 1,000 enhancer and 1,000 promoter sequences in human K562 cells. We identify simple rules for enhancer-promoter compatibility: most enhancers activated all promoters by similar amounts, and intrinsic enhancer and promoter activities combine multiplicatively to determine RNA output (R2=0.82). In addition, two classes of enhancers and promoters showed subtle preferential effects. Promoters of housekeeping genes contained built-in activating motifs for factors such as GABPA and YY1, which decreased the responsiveness of promoters to distal enhancers. Promoters of variably expressed genes lacked these motifs and showed stronger responsiveness to enhancers. Together, this systematic assessment of enhancer-promoter compatibility suggests a multiplicative model tuned by enhancer and promoter class to control gene transcription in the human genome.
Introduction
The extent to which distal enhancers might activate specific types of promoters has been an outstanding question in human gene regulation. Since their initial discovery, enhancers have been defined in part based on their ability to activate multiple non-cognate promoter sequences10,11. High-throughput reporter assays have now confirmed that many enhancer sequences derived from the human genome have the capability to activate various human, viral, and synthetic promoters12-18.
Yet, other observations have suggested that enhancers and promoters have some degree of intrinsic specificity. Early studies identified individual examples where particular enhancers or cofactors showed stronger activation with certain core promoters3-8. More recently, in Drosophila, studies using high-throughput reporter assays revealed that developmental and housekeeping gene promoters show >10-fold preferences for different classes of genomic enhancers9, have differing levels of sequence-encoded responsiveness to enhancer activation19, and respond differently to recruitment of various transcriptional cofactors20. Together, these studies have suggested a ‘biochemical compatibility’ model where different enhancers might have an intrinsic preference for activating different promoter sequences based on the transcription factors (TFs) and cofactors they can recruit2,21.
Despite these advances, the biochemical compatibility model has not been systematically tested for human enhancers and promoters. As such, it remains unclear whether compatibility classes of enhancers and promoters exist in the human genome, and, if so, how their enhancer and promoter activity combine and how such specificity is encoded.
Measuring enhancer-promoter compatibility
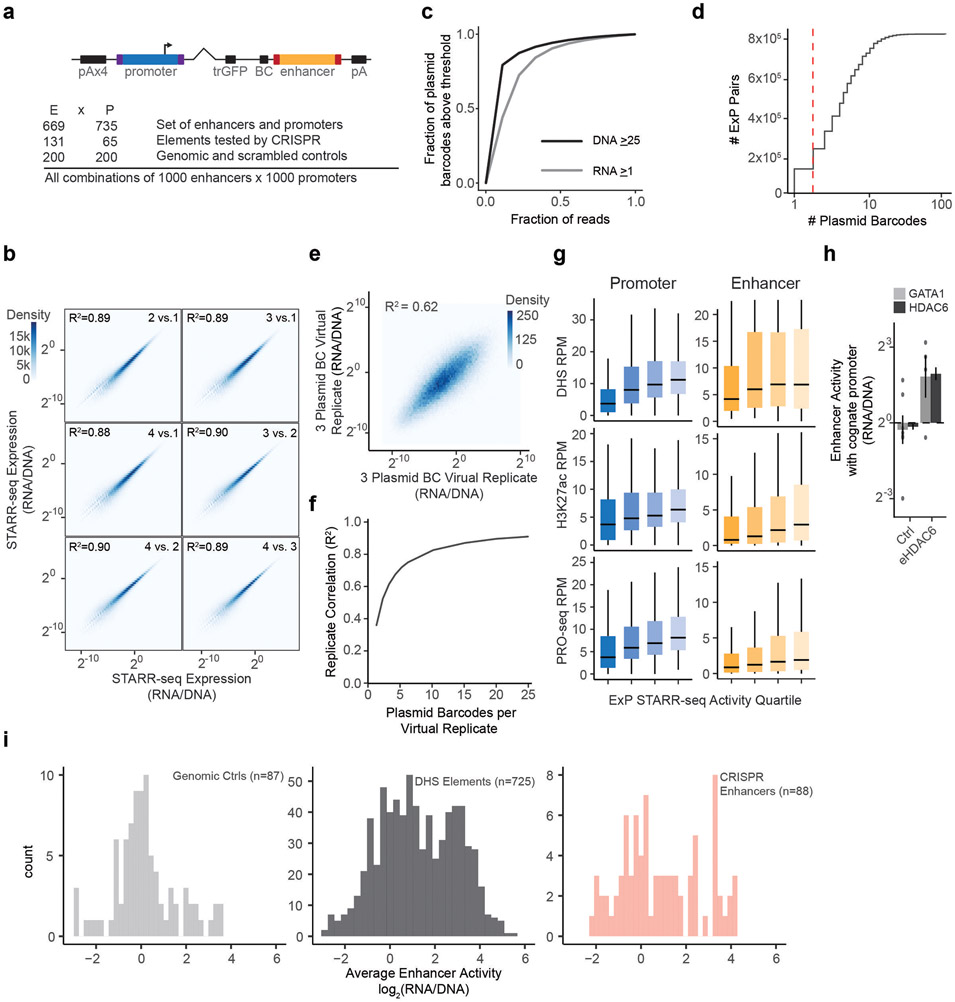
To investigate these questions, we developed an assay called enhancer x promoter (ExP) STARR-seq to test the ability of ~1,000 candidate enhancers to activate ~1,000 promoters (Fig. 1, Extended Data Fig. 1). In this assay, we synthesize pools of enhancer and promoter sequences (here, 264-bp) and clone them in all pairwise combinations located ~340-bp apart in the revised human STARR-seq plasmid-based reporter vector (Fig. 1a, Extended Data Fig. 1a)17. In STARR-seq assays, the enhancer sequence is transcribed and quantified using targeted RNA-seq to determine the level of expression of each plasmid13. For ExP STARR-seq, we introduce a unique 16-bp “plasmid barcode” adjacent to the enhancer sequence that allows us to determine which reporter transcripts are produced from which enhancer-promoter pairs. We transiently transfect this pool of plasmids into cells, measure the level of reporter transcripts produced, and calculate “STARR-seq expression” as the amount of RNA normalized to DNA input for each plasmid. This approach allows us to quantitatively measure the expression of hundreds of thousands of combinations of enhancer and promoter sequences, estimate the activities of individual enhancers and promoters, and test their compatibilities (see Methods).
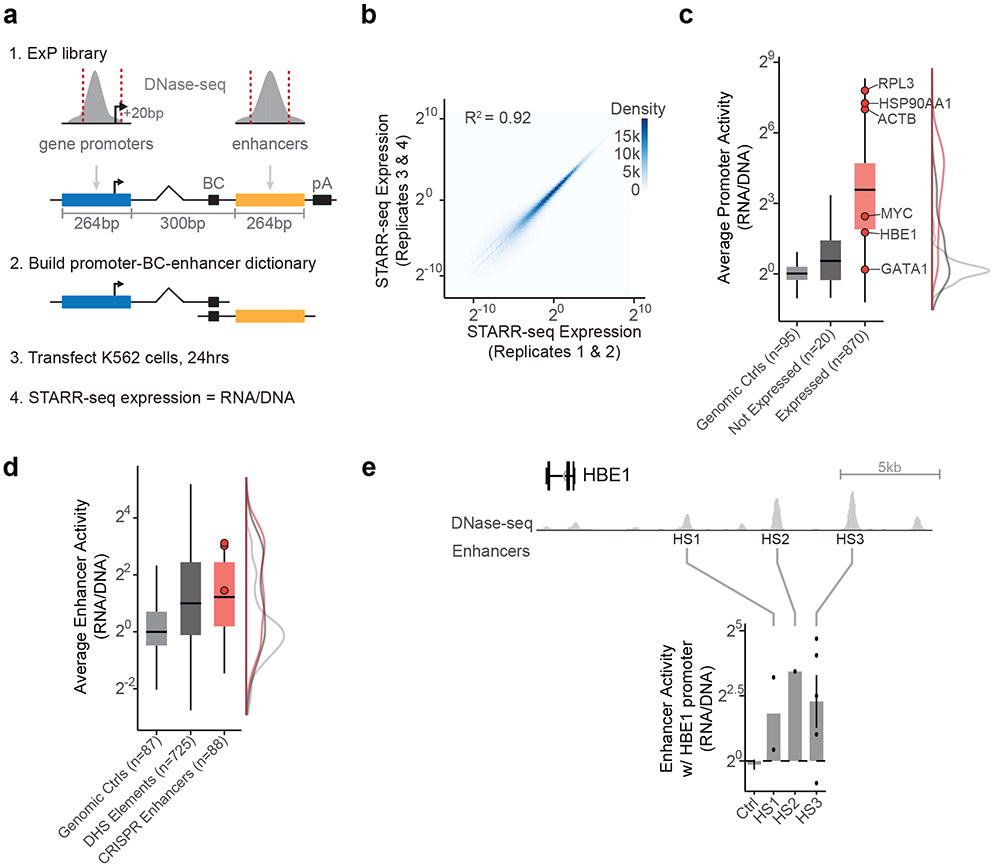
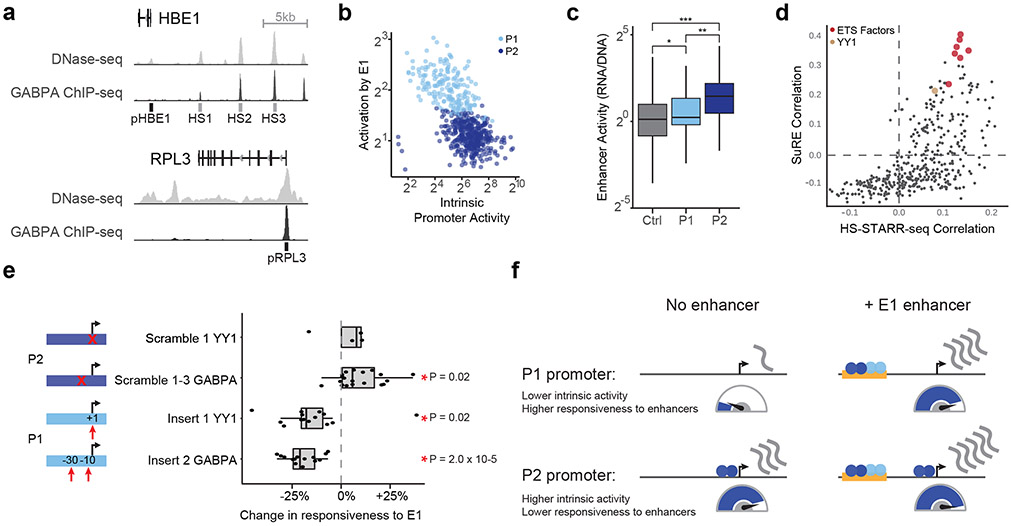
Fig. 1. Enhancer x Promoter STARR-seq.
a. ExP STARR-seq method for measuring the activities of enhancer and promoter sequences and testing their compatibilities. 264-bp sequences are selected and cloned in all pairwise combinations into the promoter and enhancer positions of a plasmid vector, together with a plasmid barcode (BC). We build a dictionary linking promoter-BC-enhancer triplets via sequencing (see Extended Data Fig. 1a). We then transfect the ExP STARR-seq plasmid pool into cells, where the promoter sequence on a given plasmid initiates transcription of a polyadenylated RNA containing the plasmid barcode and enhancer. We sequence these RNAs and calculate STARR-seq expression as the frequency of RNAs observed for each plasmid normalized by the frequency of that plasmid in the input DNA plasmid pool.
b. Correlation of ExP STARR-seq expression between biological replicate experiments, calculated for individual enhancer-promoter pairs with unique plasmid barcodes. Axes represent the average STARR-seq expression (RNA/DNA) of two biological replicates. Density: number of enhancer-promoter plasmids.
c. Average promoter activity (STARR-seq expression when paired with random genomic controls in the enhancer position) of promoter sequences derived from random genomic controls (set at 0), genes not expressed in K562s, and all other gene promoters. Box is median and interquartile range, whiskers are +/− 1.5 x IQR.
d. Average enhancer activity (STARR-seq expression of plasmids containing a given enhancer averaged across all promoters) of enhancer sequences derived from random genomic controls, accessible elements, and genomic enhancers validated in CRISPR experiments. Box and whiskers as in (c). Red dots represent three enhancers near HBE1 (see panel e).
e. Sequences derived from three genomic enhancers that regulate HBE1 in the genome (HS1-HS3) activate the HBE1 promoter in ExP STARR-seq. Ctrl: Average of 44 random genomic control sequences in the enhancer position that passed thresholds (see Methods). Error bars: 95% CI across plasmid barcodes, n=110 (ctrl), 2 (HS1), 1 (HS2), 5 (HS3).
Hereafter, for clarity, we use the terms “enhancer sequences” and “promoter sequences” to refer to sequences cloned into the enhancer and promoter positions in the ExP STARR-seq assay, and “genomic enhancers” and “genomic promoters” to refer to the corresponding elements in the genome.
We applied ExP STARR-seq to examine the combinatorial activities of 1,000 enhancer and 1,000 promoter sequences (Supplementary Table 1, Supplementary Table 2) in K562 erythroleukemia cells, which have been deeply profiled by the ENCODE Project1 and where we have previously collected data about which genomic enhancers regulate which genomic promoters using CRISPR interference (CRISPRi) screens22. Here, we selected promoter sequences to include (i) 65 genes studied in prior CRISPR screens; (ii) 735 additional genes sampled from across the genome to span a range of transcriptional activity (based on precision run-on sequencing (PRO-seq) data in K562 cells); and (iii) 200 control sequences including random genomic control sequences that are not accessible by ATAC-seq, and dinucleotide shuffled sequences (Extended Data Fig. 1a, see Methods). The promoter sequences were chosen to include approximately 20-bp downstream of the genomic transcription start site (as observed in capped analysis of gene expression (CAGE) data), and ~242-bp upstream (264 bp total, see Methods). In the enhancer position of ExP STARR-seq, we included (i) 131 accessible genomic elements we previously tested by CRISPRi; (ii) 669 other accessible genomic elements selected to span a range of quantitative histone 3 lysine 27 acetylation (H3K27ac) and DNase-seq signals (centered on the summit of the DNase-seq peak); and (iii) 200 controls including random genomic control sequences and dinucleotide shuffled sequences (Extended Data Fig. 1a, See Methods).
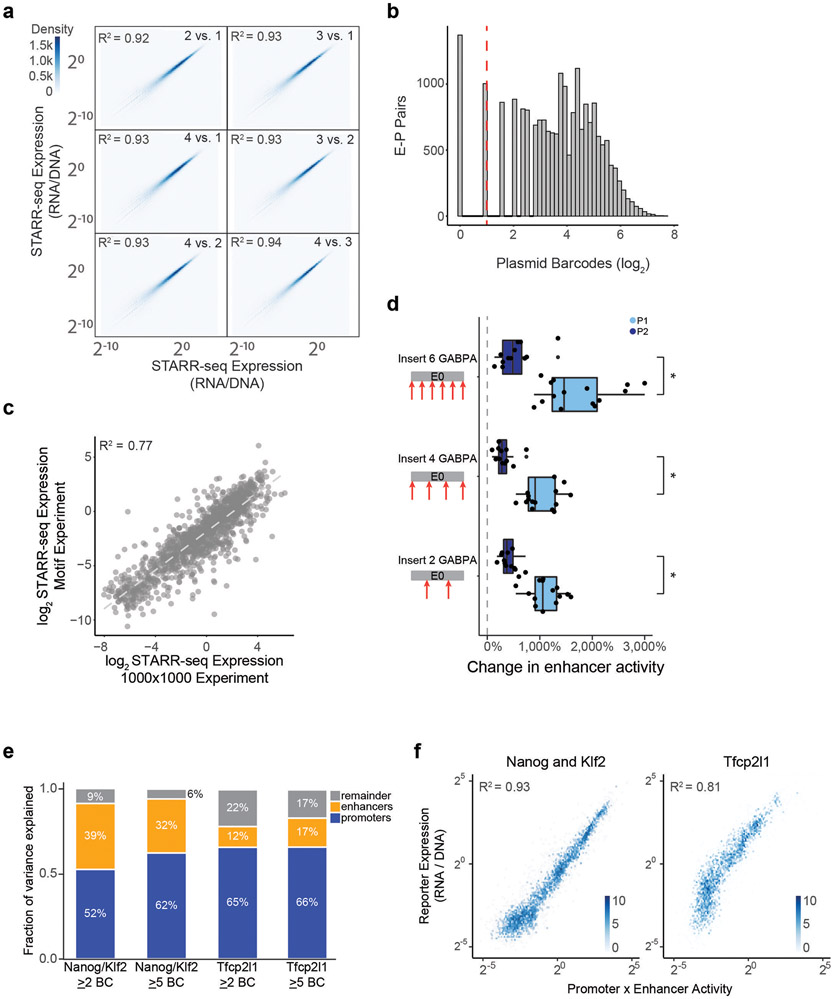
We cloned these 1,000 enhancer and 1,000 promoter sequences in all pairwise combinations, transfected the plasmid pool into K562 cells in 4 biological replicates of 50 million cells each, and sequenced each STARR-seq RNA and input DNA library to a depth of at least 2.6 billion and 470 million reads, respectively (Extended Data Fig. 1c). We focused our analysis on the 604,268 enhancer-promoter pairs where we obtained good coverage (see Methods). STARR-seq expression (RNA/DNA) varied over six orders of magnitude, and was highly reproducible, when comparing expression for individual plasmid barcodes between biological replicates (R2 = 0.92, Fig. 1b, Extended Data Fig. 1b), when comparing expression for an enhancer-promoter pair averaged across plasmid barcodes between biological replicates (R2 = 0.92), and when comparing expression for different plasmid barcodes for a given enhancer-promoter pair (R2 = 0.62, Extended Data Fig. 1d-f, see Methods).
Promoter sequences showed a very large (>1,500-fold) dynamic range of expression levels, similar to previous studies23 (“average promoter activity” = STARR-seq expression averaged across pairings with the 200 random genomic control sequences in the enhancer position). The strongest promoters in the dataset corresponded to housekeeping genes such as RPL3, HSP90AA1, and ACTB, and the weakest promoters included shuffled control sequences and non-expressed genes in K562 cells (Fig. 1c). Enhancer sequences also showed a wide (682-fold) range of STARR-seq expression in the dataset when averaged across promoters (“average enhancer activity”), and were on average 2-fold more active than random genomic control sequences (Fig. 1d, Extended Data Fig. 1i). Enhancer and promoter activity from ExP STARR-seq were correlated with biochemical features of activity at the corresponding genomic elements, including with levels of chromatin accessibility, H3K27ac, and nascent gene and eRNA transcription (Extended Data Fig. 1g).
We also found that sequences derived from known genomic enhancers activated their cognate promoters in the ExP STARR-seq assay. For example, we included 3 enhancers in the beta-like globin locus control region (HS1-HS3) that are known to coordinate expression of hemoglobin subunits during erythrocyte development24,25 and where CRISPRi perturbations in K562 cells reduce the expression of hemoglobin subunit epsilon 1 (HBE1) by 10-86%26,27. In ExP STARR-seq, each of these enhancers activated the HBE1 promoter (by 5.21-15.9-fold versus random genomic controls, Fig. 1e). Similarly, an enhancer that we previously showed to regulate GATA1 and HDAC6 in the genome28 led to 6.76 and 6.87-fold activation of the GATA1 and HDAC6 promoters in ExP STARR-seq, respectively (Extended Data Fig. 1h).
Taken together, these results show that ExP-STARR-seq produces quantitative and reproducible measurements of enhancer and promoter sequence activity over a large dynamic range.
Broad enhancer-promoter compatibility
We used this ExP STARR-seq dataset to test whether specific enhancers activate specific promoters. Surprisingly, virtually all active enhancer sequences activated all promoter sequences by similar amounts. For example, a small subset of 5 enhancers activated 5 promoters by a similar fold-change, even though the promoters spanned a 5.62-fold range of basal activities (Extended Data Fig. 2a-b; each enhancer-promoter pair had good coverage in the assay, median = 27 plasmid barcodes per pair). More generally, enhancers activated most promoters by similar fold-changes, with an average Spearman correlation across all pairs of promoters = 0.81 (Fig. 2a,c, Extended Data Fig. 2c), and pairs of enhancers showed similar proportional activation of promoters, with an average Spearman = 0.72 (Fig. 2b,d, Extended Data Fig. 2d-e). These observations indicate that, in this STARR-seq assay, there is broad compatibility between individual enhancer and promoter sequences — a striking difference from previous observations in Drosophila9,19.
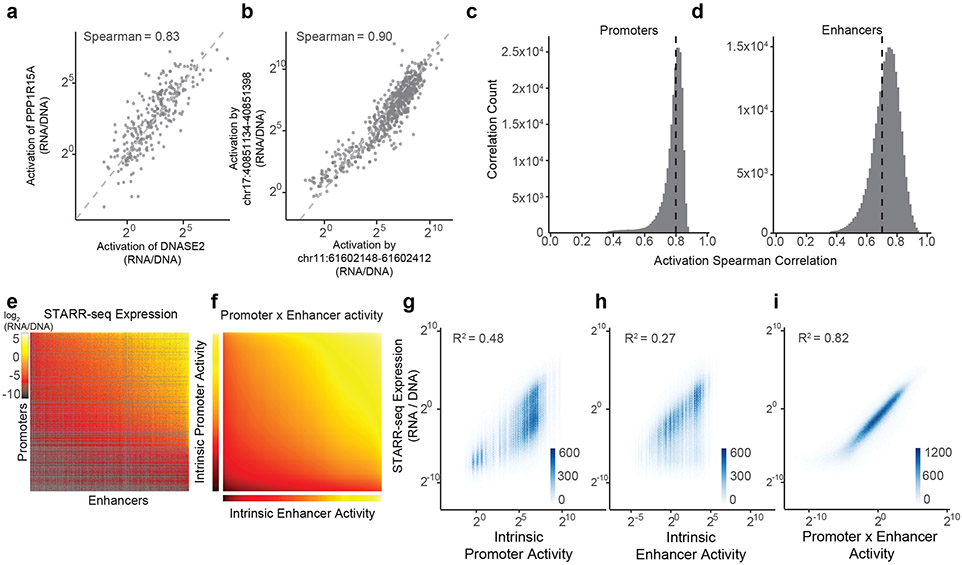
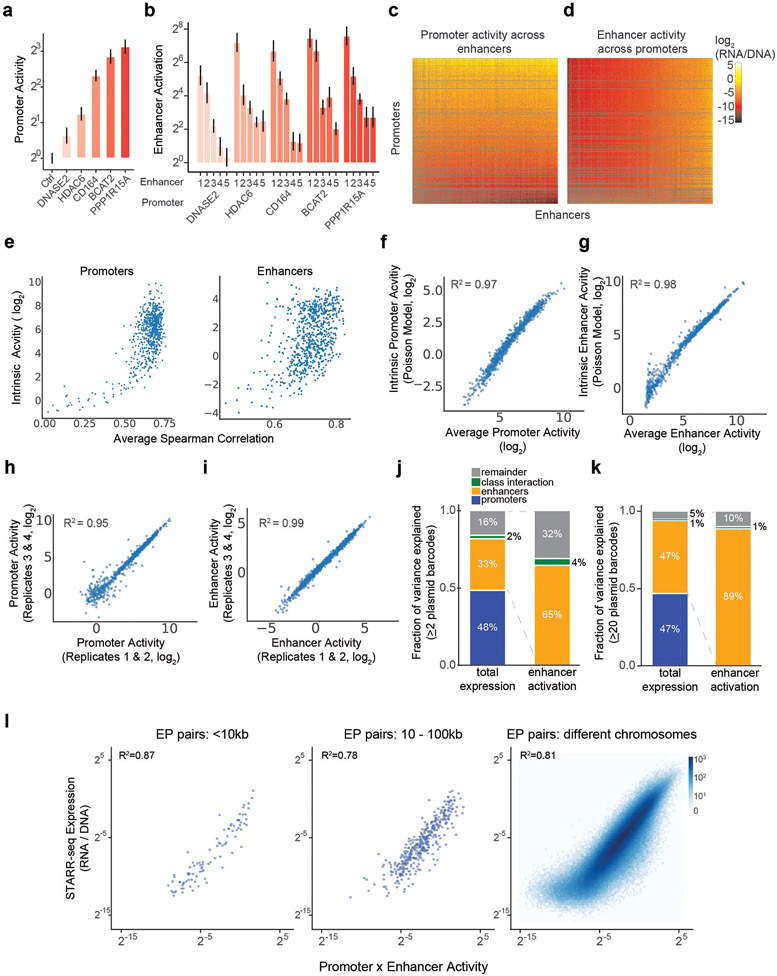
Fig. 2. Enhancer and promoter activities combine multiplicatively.
a. Correlation of enhancer activation for PPP1R15A and DNASE2 promoters. Each point is a shared enhancer sequence.
b. Correlation of enhancer activation by chr17:40851134-40851398 and chr11:61602148-61602412 enhancers. Each point is a shared promoter sequence.
c. Distribution of pairwise correlations of enhancer activation between promoter sequences, as in (a). Black dotted line = mean Spearman correlation.
d. Distribution of pairwise correlations of promoter activation between enhancer sequences, as in (b). Black dotted line = mean Spearman correlation.
e. Heatmap of ExP STARR-seq expression across all pairs of promoter (vertical) and enhancer sequences (horizontal). Axes are sorted by intrinsic promoter and enhancer activities. Grey: missing data.
f. Heatmap representing the multiplication of intrinsic promoter activity (vertical) with intrinsic enhancer activity (horizontal) from the Poisson model.
g-i. Correlation of ExP STARR-seq expression with intrinsic promoter activity (g), intrinsic enhancer activity (h), and the product of intrinsic promoter and enhancer activities (i). Density color scale: number enhancer-promoter pairs.
Activities combine multiplicatively
This pattern of effects — where enhancers showed similar fold-activation across many promoters, and promoters showed similar levels of activation by many enhancers — suggested that intrinsic enhancer and promoter activities combine multiplicatively to produce the RNA output in STARR-seq. To quantify this, we correlated expression in the STARR-seq assay with intrinsic enhancer activity, intrinsic promoter activity, and the multiplicative product of intrinsic enhancer and promoter activities.
To do so, we fit the following Poisson count model:
where RNA is RNA reads counts per plasmid, DNA is DNA read counts per plasmid, P is the intrinsic promoter activity, E is intrinsic enhancer activity, and k is a free intercept term used to scale the activities of promoters, enhancers, and their pairings relative to the average of random genomic control sequences (see Methods). This multiplicative model assumes that there is no sequence or biochemical specificity between individual pairs of enhancers and promoters, and that differences in expression are solely due to differences in intrinsic enhancer and promoter activities. Hereafter, we define “intrinsic enhancer activity” and “intrinsic promoter activity” as the fits from this model, which are similar to the “average activities” calculated above (Extended Data Fig. 2f-g) but better account for missing data and counting noise (see Methods). These estimates of activity were reproducible across replicate experiments and when comparing nonoverlapping plasmid barcodes (Extended Data Fig. 2h-i).
The multiplicative combination of intrinsic promoter and enhancer activities explained 82% of the total variance in STARR-seq expression, while intrinsic promoter and enhancer activities alone explained 48% and 27%, respectively (correlation with log2 STARR-seq expression across all enhancer-promoter pairs with at least 2 plasmid barcodes, Fig. 2e-i). The multiplicative model fit similarly well between enhancer-promoter pairs located nearby to one another in the genome (<10kb and <100kb), as it did for enhancer-promoter pairs located on different chromosomes (Extended Data Fig. 2j). From the point of view of ‘enhancer activation’ (fold-activation of an enhancer on a promoter, normalizing out promoter strength), intrinsic enhancer activity explained 65% of the variance, with 35% unexplained (Extended Data Fig. 2k). At least part of the remaining variance is likely due to experimental noise, because the proportion of variance explained by the multiplicative model increased when we examined E-P pairs with ≥20 barcodes (increasing from 82% to 94% variance explained for STARR-seq expression, and 65% to 89% explained by intrinsic enhancer activity for enhancer activation) (Extended Data Fig. 2l).
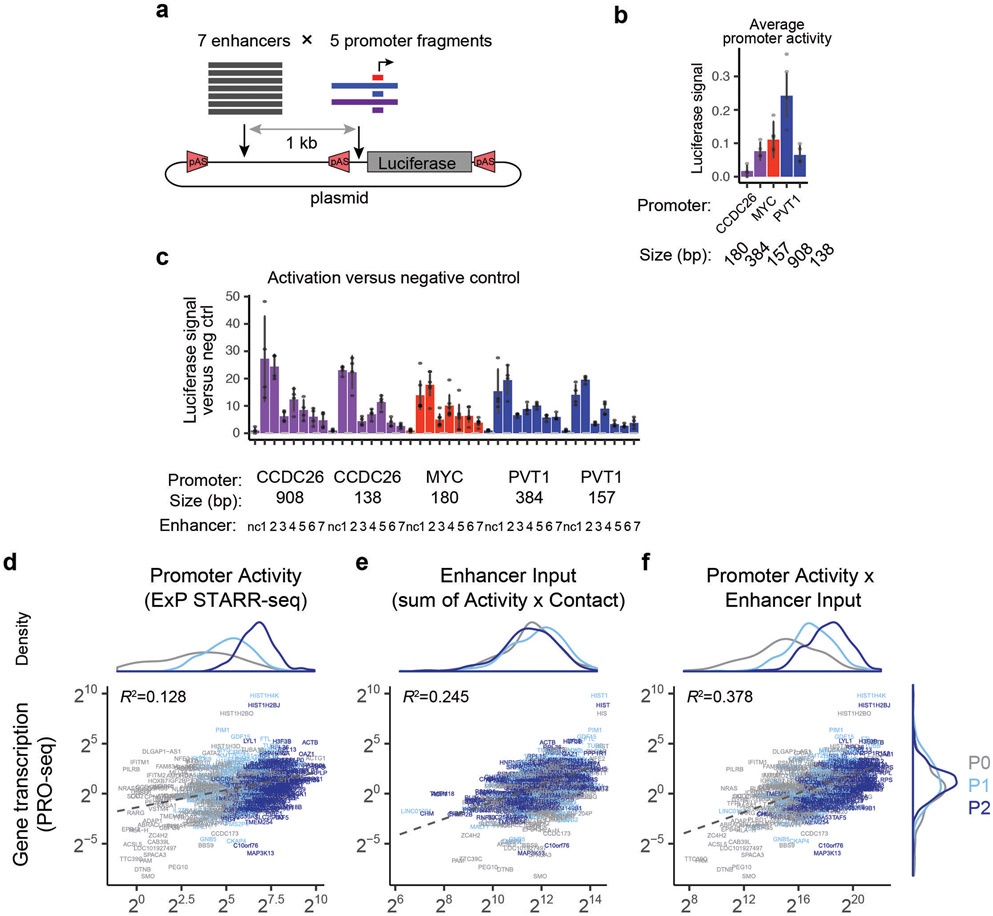
To confirm that this multiplicative relationship was not due to the specific design of our ExP STARR-seq assay, we cloned 7 enhancers from the MYC locus (1.0-2.2 kb) and 5 promoter sequences (138-908 bp, including the promoters of MYC and other nearby genes) in all combinations into a different reporter plasmid in which the enhancer is located 1 kb upstream of the promoter, and measured the expression of these constructs using a luciferase reporter assay (Extended Data Fig. 3a, Supplementary Table 3). Again, despite a range of intrinsic promoter activities (Extended Data Fig. 3b), all enhancer sequences activated all promoter sequences by a similar fold-change, and a multiplicative function of enhancer and promoter activities explained 84% of the total variance in the measurements (Extended Data Fig. 3c). We further tested whether gene transcription in the genome (as measured by PRO-seq) could be modeled as a multiplicative function of promoter activity (measured by STARR-seq) and enhancer inputs (here, calculated as the sum of Activity x Contact (ABC) scores22 for all nearby enhancers, which allowed us to include all enhancers in each locus including those not tested in ExP STARR-seq). We found indeed that gene transcription correlated with this promoter activity x enhancer input model (R2 = 0.378) much better than with either promoter activity or enhancer inputs alone (R2 = 0.128 and 0.245, respectively) (Extended Data Fig. 3d-f).
Thus, RNA expression in these reporter assays represents, to a first approximation, the multiplicative product of intrinsic enhancer activity and intrinsic promoter activity.
Classes of enhancers and promoters
Although we did not observe a strong degree of specificity among enhancer and promoter sequences, we asked whether there might exist classes with more subtle, quantitative preferences. To do so, we calculated, for each enhancer-promoter pair, its deviation from the multiplicative enhancer x promoter model (observed STARR-seq expression versus the product of intrinsic enhancer activity and intrinsic promoter activity, see Methods).
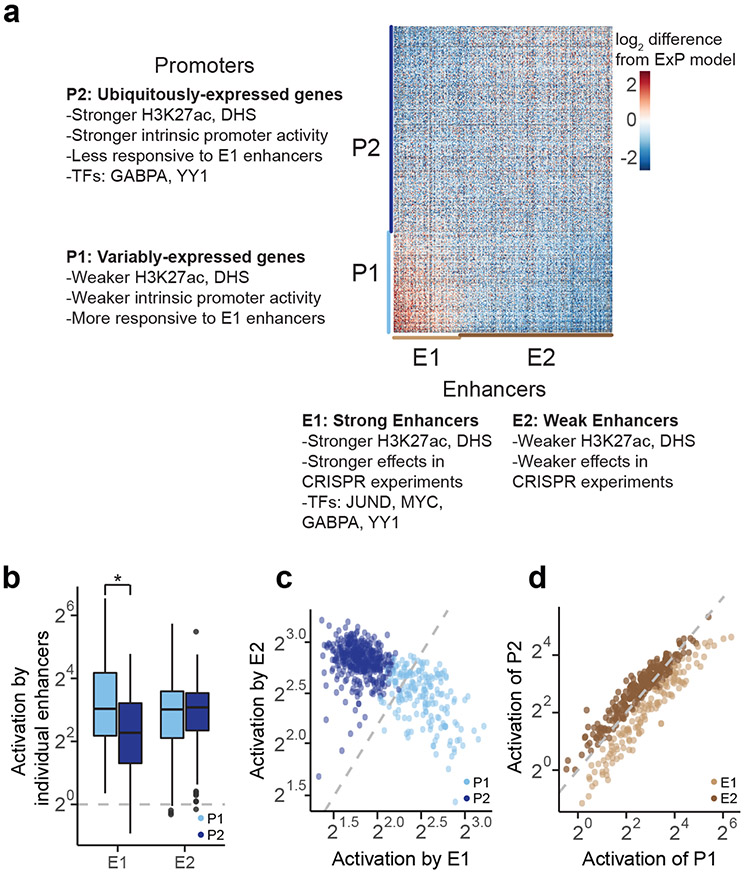
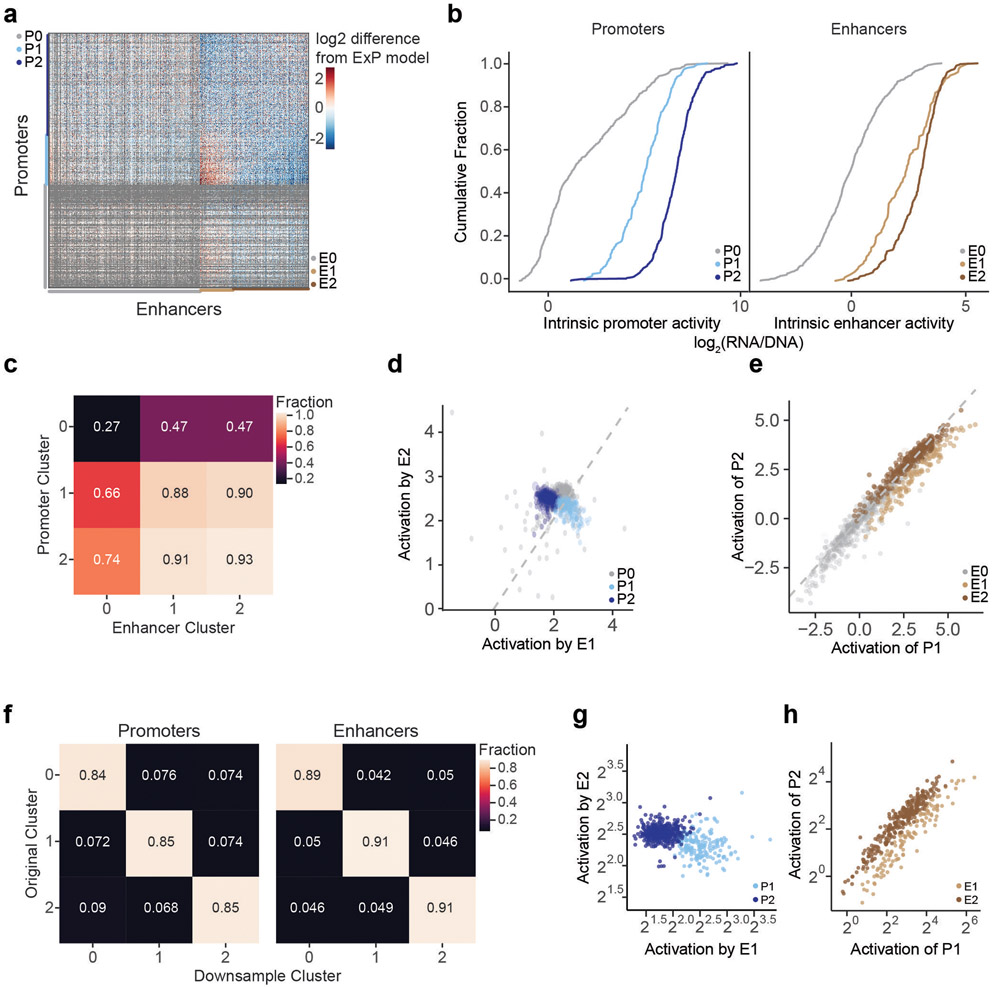
We identified two clusters of enhancer sequences (E1 and E2, n=126 and 290 respectively) that showed differential effects with respect to two sets of promoter sequences (P1 and P2, n=192 and 391 respectively) (Fig. 3a, Extended Data Fig. 4). In particular, E1 enhancer sequences activated P1 promoters more strongly than P2 promoters (by 1.93-fold, P = 4.19e-08, t-test), whereas E2 enhancer sequences activated promoters in both clusters approximately equally (1.05-fold stronger for P2 versus P1, P = 0.424, t-test; Fig. 3b). These sets of enhancers and promoters appeared to represent extremes of a graded scale: promoter responsiveness to E1 vs E2 enhancer sequences varied over a ~3-fold range (Fig. 3c, Extended Data Fig. 4d,g Extended Data Fig. 5a), and enhancer activation of P1 vs P2 promoters varied over a ~2-fold range (Fig. 3d, Extended Data Fig. 4e,h, Extended Data Fig. 5b). Cluster assignments were stable to down-sampling of promoter and enhancer sequences (Extended Data Fig. 4f, see Methods). Two additional clusters, P0 and E0, contained the remaining sequences, which had very weak activity and/or missing data and were excluded from analysis in subsequent sections (Extended Data Fig. 4a-c).
Fig. 3. Compatibility classes of enhancers and promoters.
a. Heatmap of deviations in enhancer-promoter STARR-seq expression from a multiplicative enhancer-promoter model (color scale: fold-difference between observed expression versus expression predicted by multiplicative model; gray: missing data). Vertical axis: promoter sequences grouped by class and sorted by responsiveness to E1 vs. E2 (see b); horizontal axis: enhancer sequences grouped by class and sorted by activation of P1 vs. P2 (see c).
b. Activation of P1 vs P2 promoters by E1 and E2 enhancer sequences (equivalently: Responsiveness to E1 vs E2 enhancer sequences). n=126 (E1) and 290 (E2). Boxes are median and interquartile range, whiskers are +/− 1.5*IQR. *P-value = 4.2 x 10−8, two-sample t-test.
c. For each promoter, the average activation by (responsiveness to) E1 enhancer sequences (x-axis) versus the average activation by E2 enhancer sequences (y-axis). P1 promoters (light blue) are activated more strongly by E1 versus E2 enhancers.
d. For each enhancer, the average fold-activation when paired with P1 promoters (x-axis) versus P2 promoters (y-axis). E1 enhancers (light brown) more strongly activate P1 promoters.
We quantified the additional variance explained by promoter and enhancer class by extending the multiplicative ExP model:
where PEClassInteraction is a weighted indicator variable for each of the 9 possible E-P class combinations. Promoter and enhancer class specificity explained an additional 2% of the total variation in STARR-seq expression, or, after normalizing for promoter activity, an additional 4% of the variance in enhancer activation (Extended Data Fig. 2k).
Together, these observations identify classes of enhancer sequences and classes of promoter sequences with subtle quantitative differences in compatibility. We next sought to characterize these classes of enhancer and promoter sequences and understand how such preferential effects might be encoded.
Properties of enhancer classes
To characterize the two classes of ExP STARR-seq enhancer sequences, we compared the classes with respect to biochemical features of their corresponding elements in the genome, sequence motifs, effects in CRISPR experiments, and other features.
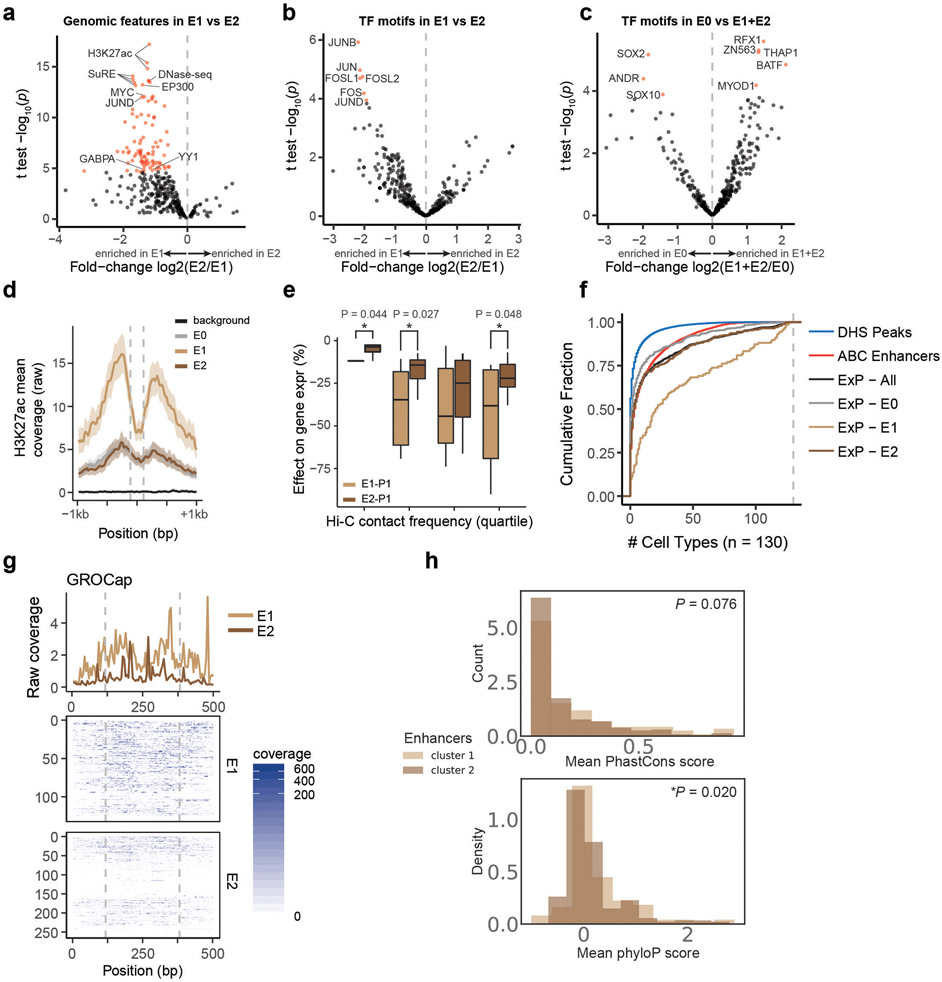
E1 and E2 classes showed biochemical features of strong and weak genomic enhancers, respectively. The features most strongly associated with E1 versus E2 sequences in the genome included H3K27ac, DNase I hypersensitivity, AP-1 factor binding (JUN, ATF3), and other known activating TFs (Fig. 3a, Extended Data Fig. 6a-d, Supplementary Table 4). E2 sequences in the genome were also DNase accessible and sometimes bound these factors, but to a significantly lesser degree. Consistent with these observations, E1 sequences had stronger effects on gene expression in CRISPR perturbation experiments, even when controlling for 3D contact with the target gene (Extended Data Fig. 6e). E1 sequences were more likely to be predicted to be enhancers in K562 cells (94% of E1 predicted to regulate a gene by the Activity-by-Contact (ABC) model, versus 49% of E2), and more likely to be broadly active in many cell types (32% of E1 predicted to be ABC enhancers in >50 of 130 other biosamples, versus 13% of E2, Extended Data Fig. 6f). Both classes contained a large fraction of sequences predicted to be an enhancer in at least one other related or unrelated cell type (90% of E1 and 70% of E2), suggesting that some E2 genomic elements may act as strong enhancers in other cell types. With regards to sequence features, E1 enhancer sequences were significantly enriched for FOS and JUN motifs, while E2 enhancer sequences were not significantly enriched for any particular motif (Benjamini-Hochberg corrected P < 0.05, Extended Data Fig. 6b-c, Supplementary Table 5). Both E1 and E2 genomic enhancers appeared to produce enhancer RNAs, as measured by GRO-cap (Extended Data Fig. 6g), and showed similar levels of sequence conservation (Extended Data Fig. 6h).
These observations suggest that the differences in how these classes of enhancer sequences activate different promoters in ExP-STARR-seq could be related to their ability to recruit activating TFs (see below). We note that, despite these clear differences in genomic activity, the two classes of enhancer sequences showed, on average, similar levels of activity in the ExP-STARR-seq assay (Extended Data Fig. 4b). This may reflect previous observations that sequences in STARR-seq might affect reporter expression by acting on steps other than transcriptional activation16, or that the episomal STARR-seq assay often detects activity for sequences that do not appear to be active in their endogenous chromosomal context17,29.
Properties of promoter classes
The two classes of promoter sequences also showed striking differences in their functional annotations, intrinsic promoter activity, and responsiveness to enhancers in the genome.
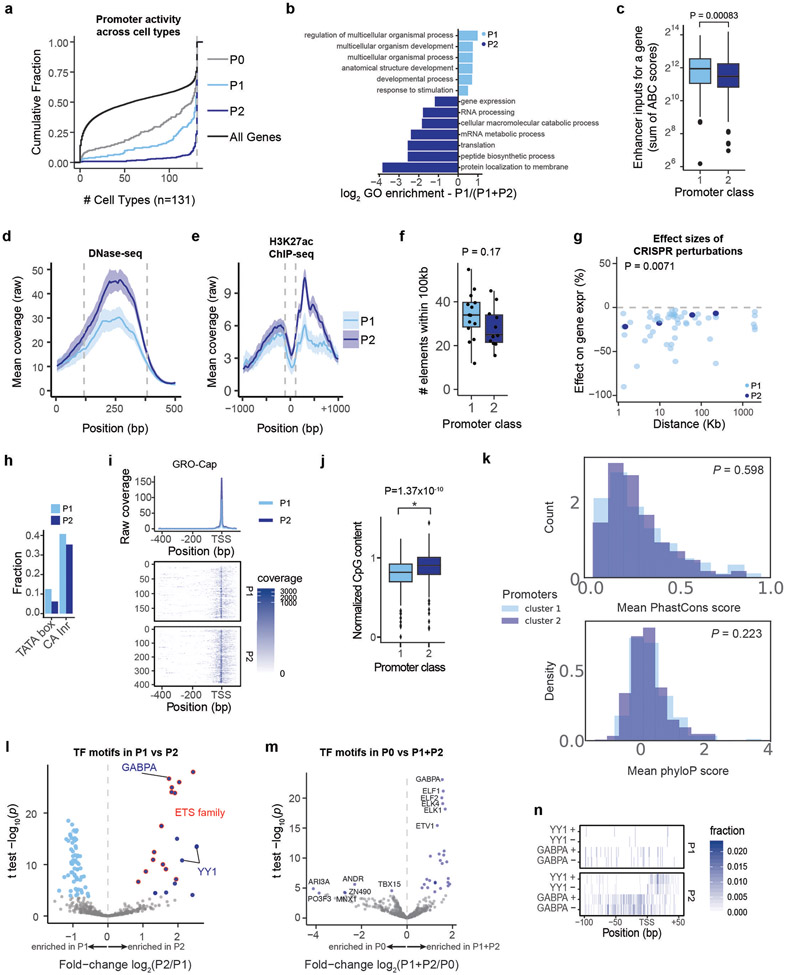
We found that many P2 promoter sequences corresponded to ubiquitously and uniformly expressed genes (often referred to as “housekeeping” genes), whereas P1 promoters largely corresponded to genes that were more variably expressed across cell types (Fig. 4a). For example, P2 promoters included beta actin (ACTB), all 37 tested ribosomal subunits (e.g., RPL13, RPS11), components of the electron transport chain (e.g., NDUFA2, ATP5B), and others (Supplementary Table 1). In contrast, P1 promoters included erythroid-specific genes (e.g., 3 hemoglobin genes) and variably expressed TFs (e.g., KLF1, JUNB, REL, MYC). Across a panel of 131 cell types and tissues (“biosamples”) most P2 promoters (76%) were active in all 131 biosamples, compared to only 45% of P1 promoters (Extended Data Fig. 7a), and P1 and P2 promoters were associated with developmental and housekeeping gene ontology terms, respectively (Extended Data Fig. 7b).
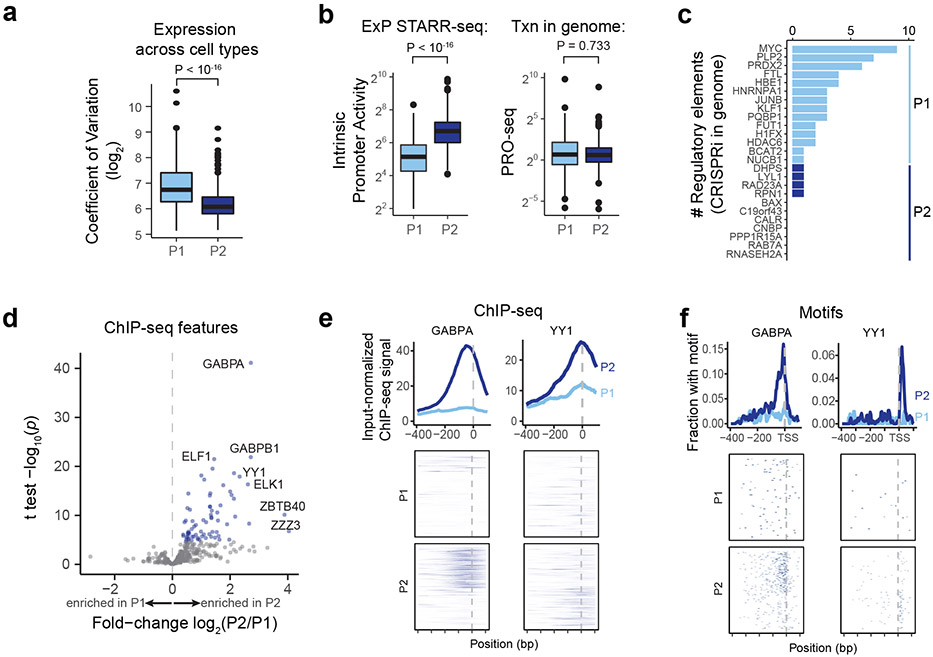
Fig. 4. Promoter classes correspond to enhancer-responsive versus ubiquitously expressed genes.
a. Variability of expression of genes corresponding to P1 and P2 promoters. Coefficient of variation is calculated across 1829 CAGE experiments from the FANTOM5 Consortium48. n=192 (P1) and 391 (P2). Boxes are median and interquartile range, whiskers are +/− 1.5*IQR. P-value is from two-sample t-test.
b. Intrinsic promoter activity for P1 vs P2 promoters (ExP STARR-seq) and genomic transcription level of genes corresponding to P1 vs P2 promoters (PRO-seq reads per kilobase per million in gene bodies). n=192 (P1) and 391 (P2). Boxes are median and interquartile range, whiskers are +/− 1.5*IQR.
c. Number of activating genomic regulatory elements identified in comprehensive CRISPRi screens for genes corresponding to P1 promoters (n=14) and P2 promoters (n=11)22.
d. Volcano plot comparing ChIP-seq and other biochemical features for P2 versus P1 promoters (see Supplementary Table 6). X-axis: ratio of average signal at P2 versus P1 promoters. Blue points: features with significantly higher signal at P2 promoters; no features have significantly higher signal at P1 promoters.
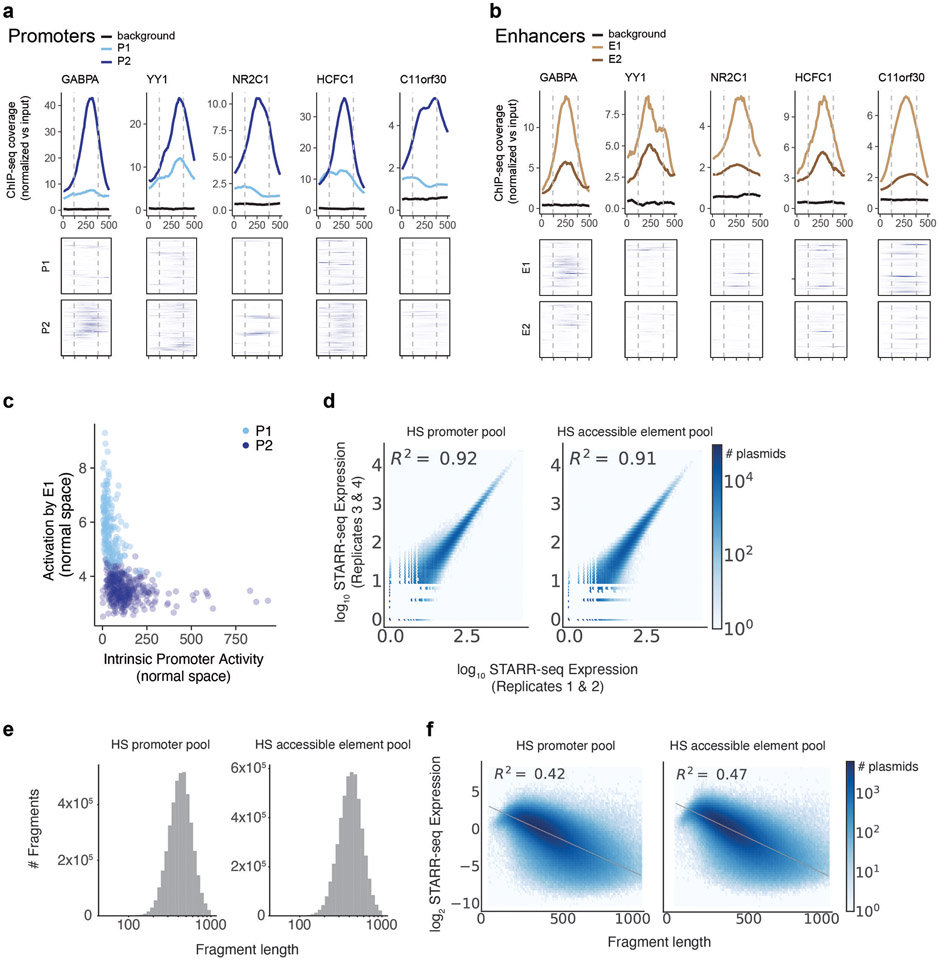
e. ChIP-seq signal for GABPA and YY1 in K562 cells at P1 and P2 promoters in the genome, aligned by TSS (see Methods). Top: average ChIP signal (normalized to input) +/− 95% c.i. Bottom: signal at individual genomic promoters.
f. Motif occurrences for GABPA and YY1 in P1 and P2 promoters, aligned by TSS.
P1 promoters had on average 3.2-fold weaker intrinsic promoter activity than P2 promoters, as measured by ExP-STARR-seq (P < 10−16, Mann-Whitney U-test; Fig. 4b, Extended Data Fig. 4b), but showed similar levels of transcription in their native genomic locations (as measured by PRO-seq in the gene body; P = 0.733, Mann-Whitney U-test; Fig. 4b), and had more activating chromatin environments based on predictions of enhancer input from the ABC model (P = 0.00083, Mann-Whitney U-test; Extended Data Fig. 7c-e). This suggests that P1 promoters may be more dependent on genomic context for their level of transcription in the genome.
Indeed, genes corresponding to P1 promoters had more genomic regulatory elements in CRISPR experiments. In data from previous studies in which CRISPRi was used to perturb every DNase-accessible element near selected promoters, the 14 genes corresponding to P1 promoters had an average of 3.6 (median: 3) distal enhancers in CRISPR experiments, whereas the 11 genes corresponding to P2 promoters had only 0.36 (median: 0, Fig. 4c, Extended Data Fig. 7f). Distal enhancers for P1 genes in the genome also had stronger effect sizes (P = 0.0071, t-test, Extended Data Fig. 7g).
Together, these observations suggest that P1 promoter sequences correspond to variably expressed genes and depend more on distal enhancers for their transcriptional activation both in ExP STARR-seq and in the genome, whereas P2 promoter sequences correspond to ubiquitously expressed genes that are relatively less sensitive to distal enhancers in both contexts.
TFs distinguish promoter classes
We next sought to identify sequence and chromatin features that distinguish P1 (more responsive) from P2 (less responsive) promoters.
We considered canonical core promoter motifs, which have been observed to differ between various subsets of promoters30-34, but did not find strong relationships. P1 and P2 promoter sequences had similar frequencies of the canonical ‘CA’ Initiator dinucleotide at the transcription start site (TSS) (40.1% vs 35.3%, Extended Data Fig. 7h), and corresponded to genes with similar patterns of dispersed versus focused TSSs in the genome (Extended Data Fig. 7i). Consistent with previous studies comparing features of housekeeping versus other gene promoters30-34, P2 promoters had a slightly higher frequency of CpG dinucleotides (median 0.90 vs 0.81 normalized CpG content for P2 and P1 promoters, Extended Data Fig. 7j), and P1 promoters had a 2-fold higher frequency of TATA box sequences upstream of the TSS (12.5% vs 6.1%), although only a small proportion of promoters contained this motif (Extended Data Fig. 7h). Both groups of promoters showed similar levels of sequence conservation (Extended Data Fig. 7k).
Accordingly, we explored which other sequence features or TF binding measurements distinguished P2 from P1 promoters. We examined 3,206 other features (including ChIP-seq measurements, TF motif predictions, and other features), and identified striking differences in the frequencies of certain transcription factor binding sites and motifs (Fig. 4d, Extended Data Fig. 7l-m, Supplementary Tables 6, 7), which in combination could classify the two promoter classes with 94% accuracy in 6-fold cross-validation (Supplementary Table 8, see Methods). The most significantly enriched features included ChIP-seq signal for ETS family factors (GABPA, ELK1, ELF1), YY1, HCFC1, NR2C1, and C11orf30 / EMSY (Fig. 4d, Extended Data Fig. 8a). For example, two of the top factors (GABPA and YY1) together showed strong binding to a total of 64% of P2 promoters in the genome: 50% of P2 promoters showed strong GABPA binding (vs 8% of P1 promoters; P = 9.9 x 10−22, BH-corrected Fisher’s exact test), and 29% of P2 promoters showed strong YY1 binding (vs 5% of P1 promoters, P = 9.4 x 10−9, BH-corrected Fisher’s exact test) (Fig. 4e). Notably, the sequence motifs for these factors showed positional preferences consistent with a function in regulating transcription initiation: the motif for GABPA was typically located 0-20 nucleotides upstream of the TSS (mode: −10), and for YY1 was often positioned at either +18 bp (both strands) or +2 bp (negative strand) from the TSS (Fig. 4f, Extended Data Fig. 7n). Consistent with these factors playing a functional role, previous studies have found that adding GABPA or YY1 motifs to promoters increases gene expression in various reporter assays and cell types35-38.
Together, these analyses suggest that P2 promoters can best be distinguished from P1 promoters by the presence of certain transcription factors including GABPA and YY1, rather than canonical core promoter motifs.
P2 promoters contain built-in enhancers
We considered how transcription factors such as GABPA and YY1 might contribute to the reduced enhancer responsiveness of P2 versus P1 promoters. Interestingly, we noticed that these same factors showed strong binding in the genome not only at P2 promoters (Fig. 4e,f), but also at some E1 enhancers (Extended Data Fig. 6a, Extended Data Fig. 8b). For example, 3 of the genomic enhancers for HBE1 (all classified as E1 in ExP STARR-seq) contained GABPA sequence motifs and showed strong GABPA binding by ChIP-seq, whereas the genomic promoter of HBE1 (classified as P1) lacked these features (Fig. 5a).
Fig. 5. P2 promoters contain built-in enhancer sequences.
a. DNase-seq and GABPA ChIP-seq binding at the HBE1 promoter (pHBE1, P1), HS1-HS3 enhancers (E1), and RPL3 promoter (pRPL3, P2).
b. Correlation between intrinsic promoter activity and responsiveness of promoters to E1 enhancers (average activation by E1 sequences, expressions vs. random genomic controls). Each point is one promoter.
c. Average enhancer activity in HS-STARR-seq (RNA/DNA) of random genomic background fragments (Ctrl, N = 3.9 million) and P1 (N = 192) and P2 (N = 391) promoters. *P =5.2*10−4, **P = 1.1*10−15, ***P = 1.4*10−66, two-sided t-test. Boxes are median and interquartile range, whiskers are +/− 1.5*IQR.
d. For each of 400 sequence motifs that appeared in at least 5% of HS-STARR-seq fragments, correlation (Pearson R) of motif occurrence with intrinsic promoter activity (SuRE signal, y-axis) and with intrinsic enhancer activity (HS-STARR-seq signal among fragments not overlapping TSS, x-axis).
e. Change in promoter responsiveness to E1 enhancers (average fold-activation by E1 enhancers) after scrambling YY1 or GABPA motifs in P2 promoters or inserting YY1 or GABPA motifs into P1 promoters. Each point is a promoter, *P < 0.05, two-sided t-test. Boxes are median and interquartile range, whiskers are +/− 1.5*IQR.
f. A model for enhancer-promoter compatibility. Enhancers multiplicatively scale the RNA output of promoters. P2 promoters contain built-in activating sequence motifs that both increase intrinsic promoter activity and reduce responsiveness to distal enhancers.
These observations suggested that P2 promoters may have reduced responsiveness to E1 enhancers because they contain some of the same motifs, potentially saturating some step in transcription. Accordingly, we explored the hypothesis that promoters contain ‘built-in’ E1 enhancer sequences that would increase promoter activity and decrease responsiveness to distal E1 enhancers.
Consistent with this hypothesis, we found that (i) across all promoters, responsiveness to E1 enhancers was inversely correlated with intrinsic promoter activity, in a way that appeared to saturate; (ii) P2 promoters had stronger enhancer activity than P1 promoters; (iii) nearly all of the TF motifs enriched in P2 promoters were predictive of both promoter activity and enhancer activity; and (iv) scrambling or inserting GABPA or YY1 motifs affected the responsiveness of promoters to E1 enhancers:
We first compared intrinsic promoter activity with responsiveness to E1 enhancers, and found that they were correlated both when considering all promoters in ExP STARR-seq (Pearson R = −0.62, log2 space; Fig. 5b) and when considering only P1 promoters (R = −0.51). As promoter activity increased, responsiveness to E1 enhancers decreased rapidly (for example, from ~9-fold average activation by E1 enhancers for the SNAI3 P1 promoter) and appeared to saturate at ~3-fold for most P2 promoter sequences (Extended Data Fig. 8c).
We next tested whether P2 promoters had stronger intrinsic enhancer activity. To do so, we generated a second STARR-seq dataset in which we measured the enhancer activity of >8.9 million sequences derived from DNase-accessible elements and promoters (by hybrid selection (HS)-STARR-seq, see Methods, Extended Data Fig. 8d-f). In this dataset, many promoter elements tested in ExP STARR-seq (along with thousands of other accessible elements) were densely tiled (an average of ~11 fragments each covering at least 90% of the promoters tested in the ExP assay), allowing us to test the enhancer activity of entire P1 and P2 promoter sequences. P2 promoters indeed showed ~2-fold higher intrinsic enhancer activity than P1 promoters in HS-STARR-seq (P = 1.14 x 10−16, t-test, Fig. 5c), supporting a model where these promoters contain built-in enhancers.
We examined whether the sequence motifs enriched in P2 promoters contribute to both enhancer activity and promoter activity. To do so, we examined data on enhancer activity from HS-STARR-seq along with another previous experiment that measured promoter activity for millions of random genomic fragments in K562 cells (SuRE23). 16 of the 17 motifs enriched in P2 promoters, including motifs for GABPA and YY1, were positively correlated with both enhancer activity and promoter activity (Fig. 5d, Supplementary Table 9, see Methods).
Finally, we conducted an ExP STARR-seq experiment in which we scrambled or inserted transcription factor motifs into promoter or enhancer sequences (Fig. 5e, Extended Data Fig. 9a-d). As predicted, inserting GABPA or YY1 motif instances into P1 promoter sequences significantly decreased responsiveness to E1 enhancers (GAPBA: average −19.8%, P = 2.0 x 10−5; YY1: average −14.8%%, P = 0.02; n = 14 insertions each). Conversely, mutating one or more motif instances in P2 promoter sequences usually increased responsiveness to E1 enhancers (GABPA: average +8.9%, P = 0.02, n = 20; YY1: average +2.6% P = 0.7, n = 4). We also tested inserting GABPA motifs into E0 (very weak) enhancer sequences, and found that they increased enhancer activity, and more so for P1 vs P2 promoters (average with P1: +1289%, average with P2: +417%, P = 7.9 x 10−14 (Extended Data Fig. 9d).
Together, these observations suggest a model for promoter sequence organization (Fig. 5f). Promoters encode binding motifs for activating factors, including GABPA and YY1, that act as ‘built-in’ enhancers for the promoter. This not only increases the autonomous activity of the promoter, but also reduces its responsiveness to distal enhancers. While P2 promoters have strong built-in enhancers, P1 promoters appear to have weaker or fewer built-in activating motifs, rendering them more sensitive to distal enhancers.
Compatibility rules in a second dataset
In a parallel study, Martinez-Ara et al. conducted a similar experiment to examine the compatibilities among hundreds of enhancer and promoter sequences in mouse embryonic stem cells (22,406 total enhancer-promoter pairs, measured in 2 separate experiments)39. This dataset provided an opportunity to assess the extent to which the compatibility rules we identified generalize to a second cell type, organism, and assay format. With regards to assay format, this study used a different plasmid design (MPRA format with the enhancer located just upstream of the promoter), method for element selection (densely sampled from 3 genomic loci), and enhancer and promoter sequence lengths (~400 bp).
The patterns of enhancer-promoter compatibility in this second dataset were highly similar to ExP STARR-seq. The multiplicative enhancer x promoter model explained 91% and 78% of the variance in RNA expression in the two experiments and 81% and 34% of the variance in enhancer activation (Extended Data Fig. 9e-f), with a fraction of variance unexplained that could result in part from additional specificity factors (see also Martinez-Ara et al. 202139). Promoters with stronger intrinsic activity were less sensitive to activation by enhancer sequences, and ETS-family motifs including GABPA were the strongest motifs positively correlated with enhancer activity and negatively correlated with enhancer responsiveness (see 39), consistent with features of P1 and P2 promoters identified in ExP STARR-seq. We note that analysis by Martinez-Ara et al. shows that most enhancers in both their MPRA and our ExP STARR-seq experiments show statistically significant deviations from the multiplicative model39, but the magnitude of such deviations is small and explain only a small fraction of the variance in reporter expression (Extended Data Fig. 9e-f). Together, both datasets indicate that, across multiple cell types and mammalian genomes, (i) enhancers and promoters are broadly compatible and (ii) there is an additional layer of selectivity in which specific motifs such as for GABPA and YY1 can tune enhancer-promoter activation.”
Discussion
Since the discovery of the first enhancers forty years ago10,11, many enhancer and promoter sequences have been combined and found to be compatible12-18. At the same time, studies of individual natural or synthetic core promoters have been found to have some degree of specificity when combined with various transcriptional cofactors or enhancer sequences3-8.
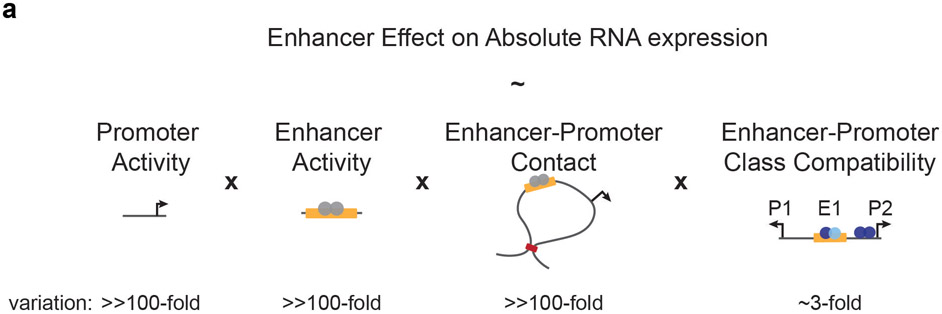
Here we develop and apply ExP STARR-seq to systematically quantify enhancer-promoter compatibility, and identify a simple rule for combining human enhancer and promoter activities. Enhancers are intrinsically compatible with many Pol II promoter sequences, and act multiplicatively to scale the RNA output of a promoter. As a result, independent control of intrinsic enhancer activity and intrinsic promoter activity can create significant variation in RNA expression: in our data, promoter activity and enhancer activity each vary over >3 orders of magnitude, with their multiplicative combination explaining much of the observed >250K-fold variation in STARR-seq expression (Fig. 2i, Extended Data Fig. 2k-l). This broad compatibility appears to be consistent with recent studies using other reporter approaches, which found that human core promoters or enhancers are similarly scaled when they are inserted into different genomic loci40,41 and that randomly generated enhancer and promoter sequences combine multiplicatively in STARR-seq experiments in three other cell lines36. This is also consistent with our previous finding that the effects of enhancers on nearby genes in the genome can be predicted with good accuracy using the Activity-by-Contact model, which assumes that all enhancers and promoters are equally compatible and that enhancer activity and 3D enhancer-promoter contact frequencies tune the relative effect of an enhancer on gene expression22.
We also identify two classes of enhancers and promoters that show subtle preferences in activation. One class of promoters, corresponding largely to ubiquitously expressed (housekeeping) genes, is less responsive to distal enhancers both in ExP STARR-seq and in the genome, while the second class of promoters, corresponding to variably expressed genes, is more responsive. Previous studies have identified numerous differences in sequence content and motifs between the promoters of housekeeping and context-specific genes30-34. We find that these promoters indeed show intrinsic differences in their levels of activity and responsiveness to enhancers. In particular, P2 promoters contain built-in activating sequences that increase both enhancer and promoter activity, which appears to reduce their responsiveness to distal enhancers. This model for human promoters appears to differ qualitatively from previous studies in Drosophila, which found that the promoters of housekeeping and developmentally regulated genes can both be highly responsive, but to distinct sets of enhancer sequences and cofactors9,20.
Together, these observations suggest a model where the effects of enhancers on nearby genes in the human genome is controlled by quantitative tuning of intrinsic promoter activity, intrinsic enhancer activity, and 3D enhancer-promoter contacts, with enhancer-promoter class compatibilities playing an additional but smaller role (Extended Data Fig. 10). Beyond these factors, further work will be required to identify and predict cases where promoters are responsive only to certain chromatin environments, cofactors, or enhancer sequences3-8. Regarding the latter possibility, other parallel studies have examined effects not explained by a multiplicative ExP model and found that combinations of transcription factors in enhancer and promoter sequences may mediate additional specificity36,39.
A remaining challenge will be to link the sequences that control enhancer and promoter activities with effects on particular biochemical steps in transcription. In this regard, we find that GABPA and YY1 bind both to P2 promoters and to distal enhancers, and are associated with increased enhancer activity, increased promoter activity, and reduced promoter responsiveness to distal enhancers. This suggests that distal enhancers may act, in part, on a particular rate-limiting step in transcription that can be saturated by inclusion of built-in activating sequences in a gene promoter. Indeed, a previous study found that adding GABPA and YY1 motifs to several promoters led to an increase in RNA expression that saturates at 2 or 5 copies of the motif, respectively.35 Given the preferred positions of these motifs within 20 bp of the TSS — as well as previous findings that these proteins physically interact with general transcription factors42,43 and/or influence transcriptional initiation and TSS selection37,44-46 — such a rate-limiting step might involve assembly of the preinitiation complex. In addition to this step, our data are consistent with a model in which enhancers and promoters control additional steps in transcription that combine multiplicatively and do not saturate in the dynamic range of our assay. Examples of such processes that could combine multiplicatively include control of burst frequency and burst size47. Further work will be required to investigate these possibilities.
Our study has several limitations that highlight areas for future work. First, the episomal STARR-seq assay does not capture all mechanisms that might influence transcriptional activation in the genome, and may capture effects of sequences on other mechanisms such as RNA stability16,17,29. Second, our experiments were not well powered to quantify possible compatibility among the weakest enhancers and promoters. Third, the exact proportions of variance explained by factors in the multiplicative model are influenced by the method of selecting enhancers and promoters for the experiment. Fourth, the extent to which promoter and enhancer classes might change across cell types is unclear. Further investigation with genome editing, orthogonal assays, and additional cell types will be required to resolve these outstanding questions.
Together, our findings identify simple rules for human enhancer-promoter compatibility, which will propel efforts to model gene expression, map the effects of human genetic variation, and design regulatory sequences for gene therapies.
Methods
Genome build
All analyses and coordinates are reported using human genome reference hg19.
Design of ExP STARR-seq
We designed ExP STARR-seq to systematically measure the intrinsic, sequence-encoded compatibility or specificity of many pairs of human enhancer and promoter sequences. The key design features we considered when developing this assay were the ability to measure the activity of individual enhancer-promoter sequence combinations, to precisely quantify the expression of each enhancer-promoter pair, and to test hundreds of thousands of combinations in order to identify patterns of compatibility or specificity across a large number of human sequences.
Accordingly, we designed a new variant of the STARR-seq high-throughput plasmid reporter assay called enhancer x promoter (ExP) STARR-seq. In both STARR-seq and ExP STARR-seq, enhancer sequences are cloned downstream of a promoter, transfected into cells, and transcribed to produce a reporter mRNA transcript, which is then sequenced to quantify the relative expression levels of plasmids containing different enhancer sequences13. In ExP STARR-seq, we modified the cloning and RNA sequencing strategy to enable testing different enhancer sequences in combination with different promoter sequences.
To clone combinations of enhancer and promoter sequences into a reporter plasmid, we synthesized 264-bp enhancer and promoter sequences in an oligo pool format, PCR amplified enhancer and promoter sequences separately, and inserted them into the hSTARR-seq_SCP1 vector_blocking 4 vector17 in the promoter position (replacing the original SCP1 promoter) or enhancer position in a single pooled cloning step using Gibson assembly to generate all pairwise combinations of chosen enhancer and promoter sequences (Fig. 1a, Extended Data Fig. 1a). We chose this specific STARR-seq vector with 4 polyA sequences upstream of the promoter position because it was specifically designed in order to avoid spurious transcription initiation from the origin of replication17, which would interfere with the STARR-seq signal from the cloned enhancer-promoter pairs. This STARR-seq vector also includes 5’ and 3’ splice sites upstream of the enhancer that allows using a PCR primer targeting the splice junction to specifically amplify cDNA derived from the reporter mRNA while avoiding amplifying the plasmid DNA sequence.
To quantify the reporter mRNA transcripts and determine which enhancer-promoter pair they correspond to, we further adapted the cloning and RNA sequencing design. In the standard STARR-seq assay, the reporter mRNA contains the enhancer sequence but not the full promoter sequence, and therefore cannot determine from which promoter a given reporter mRNA is derived. Accordingly, in ExP STARR-seq we introduced a random 16-mer sequence located just upstream of the enhancer sequence that we use as a “plasmid barcode” to identify which enhancer reporter mRNAs are derived from which enhancer-promoter pairs (Fig. 1a). After cloning the plasmid pool, we map which plasmid barcodes correspond to which promoters by applying Illumina high-throughput sequencing to a PCR amplicon containing the promoter sequence and plasmid barcode. From this, we build a dictionary to look up, for a given reporter mRNA containing a plasmid barcode and enhancer sequence, which enhancer-promoter-plasmid barcode construct that mRNA is derived from.
Finally, we selected the number of constructed tested (~1 million pairs of enhancer and promoter sequences cloned, with an average of 6.3 plasmid barcodes per pair) and sequencing depth (>1 billion reads per replicate) to enable highly precise measurements of expression for each enhancer-promoter pair. We obtained high reproducibility of enhancer-promoter expression levels between biological replicates (R2 = 0.92), allowing us to develop quantitative models of how enhancer and promoter activities combine.
Altogether, this approach enables precisely quantifying the expression levels of thousands of combinations of enhancer and promoter sequences.
Selection of enhancer and promoter sequences for ExP STARR-seq
To explore the compatibility of human enhancers and promoters, we selected 1000 promoter and 1000 enhancer sequences, including sequences from the human genome spanning a range of expression or activity levels, and dinucleotide shuffled controls. Based on available lengths of oligonucleotide pool synthesis, each sequence was 264bp.
Promoters: We selected the 1000 promoter sequences to include:
65 genes whose enhancers have previously been studied in CRISPR experiments in K562 cells22
715 genes sampled to span a range of potential promoter activities, including the 200 most highly expressed genes in K562 cells, based on CAGE signal at their TSS1 and a random sample of 515 other expressed genes (>1 TPM in RNA-seq data28).
20 genes that are not expressed or lowly expressed in K562 cells (<1 TPM), and that are expressed in both GM12878 and HCT-116 cells (in the top 70% of genes by TPM based on RNA-seq1.
100 random genomic sequences as negative controls (+ strand)
100 dinucleotide shuffles of these random genomic sequences
For the selected genes, we synthesized a 264-bp sequence including approximately 244 bp upstream and 20 bp downstream of the TSS. Here, we defined the TSS as the center of the 10-bp window with the most CAGE 5’ read counts within 1 Kbp of a RefSeq TSS. For lowly expressed genes (which lack clear CAGE signal), we used the hg19 RefSeq-annotated TSS. For genes studied in Fulco et al. 2019, we adjusted the assigned 10bp TSS window by manual examination of the CAGE if necessary.
Enhancers: We selected the 1000 enhancer sequences to include:
131 elements previously studied with CRISPR22, including (i) all distal elements (i.e., >1 Kb from an annotated TSS) with significant effects in previous CRISPRi tiling screens (activating or repressive), (ii) all distal elements predicted by the Activity-by-Contact model to regulate one of the tested genes in K562 cells22, and (iii) two promoter elements for PVT1 that also act as enhancers for MYC22. We selected 264-bp regions centered on the overlapping DHS narrow peak. For the small number of CRISPR elements that did not overlap a narrow peak, we tiled the corresponding element with 264-bp windows overlapping by 50 bp.
200 DNase peaks with the strongest predicted enhancer activity, and 351 other DNase peaks sampled evenly across the range of predicted enhancer activity. Here, we considered all distal DHS peaks in K562 cells (DHS narrow peaks22) and calculated predicted enhancer activity as the geometric mean of DNase I hypersensitivity and H3K27ac ChIP-seq read counts in K562 cells in the ~500-bp candidate enhancer regions used by the ABC model in Fulco et al. 201922. Some candidate ABC elements in this set span more than one DHS peak, in which case we divided the predicted enhancer activity equally among each overlapping peak. We downloaded introns from the UCSC Genome Browser ‘refGene’ track (version 2017-06-24), and removed any peaks overlapping an annotated splice donor or acceptor site. We then selected 264-bp regions centered on the remaining DHS narrow peaks.
100 random genomic sequences as negative controls
100 dinucleotide shuffles of these random genomic sequences
All enhancer sequences were taken from the hg19 reference in the + strand direction.
Library Cloning
We ordered 264bp sequences in an oligo array format from Twist Bioscience with separate pairs of 18bp adaptors (total length = 300bp) for enhancers (5’ = GCTAACTTCTACCCATGC, 3’ = GCAAGTTAAGTAGGTCGT) and promoters (5’ = TCATGTGGGACATCAAGC, 3’ = GCATAGTGAGTCCACCTT). We then PCR amplified enhancers and promoters separately from the same array using Q5 high-fidelity DNA polymerase (NEB M0492) (see Supplementary Table 10 for primer sequences). We amplified enhancers in four 50uL PCR reactions (98°C for 30 seconds; 15 cycles of 98°C for 15 seconds, 61°C for 15 seconds, and 72°C for 20 seconds) using primers (forward: TAGATTGATCTAGAGCATGCANNNNNNNNNNNNNNNNGAGTACTGGTATGTTCAGCTAACTTCTACCCATGC, reverse: TCGAAGCGGCCGGCCGAATTCGTCATTCCATGGCATCTCACGACCTACTTAACTTGC) which add an additional 17bp on either side, a 16bp N-mer plasmid barcode upstream, and homology arms for Gibson assembly on either side of the enhancer sequence. (For the motif ExP STARR-seq experiment, we used 18 cycles of PCR for amplifying the enhancers.) We amplified promoters in four 50uL PCR reactions (98°C for 30 seconds; 4 cycles of 98°C for 15 seconds, 61°C for 15 seconds, 72°C for 20 seconds; 11 cycles of 98°C for 15 seconds and 72°C for 20 seconds) using primers (forward: CTCTGGCCTAACTGGCCGGTACGAGTGAGCTCTCGTTCA TCATGTGGGACATCAAGC, reverse: CCCAGTGCCTCACGACCGGGCCTGGTAGCAAGCTTAGATAAGGTGGACTCACTATGC) which add an additional 17bp and homology arms for Gibson assembly on either side of the promoter sequence. (For the motif ExP STARR-seq experiment, we used 4 and 14 cycles of PCR for amplifying the promoters.) We purified the PCR products using 0.8X volume of AMPure XP beads (Beckman Coulter, A63881) and pooled the reactions together while keeping enhancers and promoters separate.
We digested the human STARR-seq screening vector (hSTARR-seq_SCP1 vector_blocking 4, Addgene #99319) with both Thermo SgrDI and BshTI (AgeI) (replaced with enhancer sequence), then NEB KpnI and ApaI (replaced with promoter sequence), with purification using 0.8X volume AMPure XP after each digestion. We then recombined 500ng of this digestion (including ~4.4kb of backbone vector and 250bp of filler sequence including a spliced region and truncated GFP ORF) with 150ng of both the purified enhancer and promoter products using Gibson assembly (NEB, E2611) for 1 hour at 50°C in a 40uL reaction and purified the reaction using 1X volume AMPure XP with 3 total ethanol washes.
We electroporated the assembled libraries into Lucigen Endura Electrocompetent cells (60242) using 0.1cm cuvettes (BioRad) using the Gene Pulser Xcell Microbial System (BioRad) (10 uF, 600 Ohms, 1800 Volts) following the manufacturer’s recommendations. We expanded the transformations for 12 hours in LB with carbenicillin while also estimating the number of transformed colonies by plating a serial dilution of transformation mixture as previously described49. We midiprepped the expanded transformations with ZymoPURE II Plasmid Midiprep (D4200), or with the Nucleobond Xtra Midiprep kit from Macherey-Nagel (for the motif ExP STARR-seq experiment).
Building the Barcode-Promoter Dictionary
We introduced a unique 16-bp “plasmid barcode” adjacent to the enhancer sequence to allow us to determine from which promoter each transcript originated, which, together with the self-transcribed enhancer, allow us to map each transcript to a promoter-enhancer pair.
To build the map from 16-bp plasmid barcodes to promoters we PCR-amplified a fragment containing both the promoter and plasmid barcode from the plasmid library (98°C for 1 minute and 16 cycles of 98°C for 10 seconds, 66°C for 15 seconds, and 72°C for 25 seconds, ExP_P1_fwd_I2: AATGATACGGCGACCACCGAGATCTACAC[index-2]GGGAGGTATTGGACAGGC, ExP_P3_rev: CAAGCAGAAGACGGCATACGAGATGCATGGGTAGAAGTTAGCTGAAC) and sequenced the promoter position with paired-end reads (using custom sequencing primers ExP_P1_fwd_seq_R1: GAGTGAGCTCTCGTTCATCATGTGGGACATCAAGC, ExP_P2_rev_seq_R2: TGGTAGCAAGCTTAGATAAGGTGGACTCACTATGC) and the plasmid barcode with an index read (using custom sequencing primer ExP_fwd_BC_seq: GTCCCAATTCTTGTTGAATTAGATTGATCTAGAGCATGCA). We mapped these sequences to a specially constructed index of the promoter sequences using bowtie2 (X: -q --met-stderr --maxins 2000 -p 4 --no-mixed --dovetail --fast). We dropped any BC-promoter pairs with singleton reads, then removed ambiguous pairings (more than one promoter for the same BC), and finally thresholded pairs with at least 5 reads to build the Barcode-Promoter dictionary.
Cell Culture
We maintained K562 erythroleukemia cells (ATCC) at a density between 100,000 and 1,000,000 cells per ml in RPMI-1640 (Thermo Fisher Scientific) with 10% heat-inactivated FBS, 2 mM L-glutamine and 100 units per ml streptomycin and 100 mg ml−1 penicillin by diluting cells 1:8 in fresh medium every 3 days. Cell lines were regularly tested for mycoplasma and authenticated through analysis of RNA-seq and ATAC-seq data.
Library Transfection
We nucleofected 10 million K562 cells with 15μg of the ExP plasmid library in 100μL cuvettes with the Lonza 4D-Nucleofector using settings and protocols specified by the manufacturer for K562 cells (T-016). We pooled 5 nucleofections together during recovery to form 50 million cell biological transfection replicates and generated 4 replicates for a total of 200 million total cells. After 24 hours, we harvested the cells in Qiagen buffer RLT (79216) and proceeded with STARR-seq library preparation. For the motif ExP STARR-seq experiment, we harvested 6.5-10 million nucleofected cells per replicate.
STARR-seq Library Preparation
We proceeded with STARR-seq library preparation using an adapted protocol from Arnold 201313. We split the 50 million-cell transfection replicates in half and extracted total RNA using 3 Qiagen RNeasy mini columns (74134), performing the on-column DNase step. We isolated polyA+ mRNA using the Qiagen Oligotex mRNA kit for the 1000 x 1000 ExP dataset (note this kit has been discontinued, we now use the Poly(A)Purist MAG kit from Thermo Fisher Scientific, AM1922). Following mRNA elution, we treated with TURBO DNase (Thermo Fisher Scientific, AM2238) in 100uL reactions at 37°C for 30 minutes, then added an additional 2uL of TURBO DNase and incubated at 37°C for 15 minutes. We purified the RNA following DNA digestion with Zymo RNA Clean & Concentrator 5 (R1013) (R1018 for the motif ExP STARR-seq experiment). We reverse transcribed the polyA+ mRNA using Thermo SuperScriptIV using the STARR_RT primer (CAAACTCATCAATGTATCTTATCATG) in 20uL reactions according to manufacturer’s recommendations. We included 1uL of ribonuclease inhibitor RNaseOUT (Invitrogen, 10777019). Following reverse transcription, we added 1uL of RNaseH (Thermo Fisher Scientific, EN0201) and incubated at 37°C for 20 minutes. We purified the cDNA with 1.8X volume of AMPure XP beads. We next selectively amplified the reporter transcript using intron-spanning junction primers with Q5 polymerase in 50uL reactions (98°C for 45 seconds and 15 cycles of 98°C for 15 seconds, 65°C for 30 seconds, and 72°C for 70 seconds, jPCR_fwd: TCGTGAGGCACTGGGCAG*G*T*G*T*C, jPCR_rev: CTTATCATGTCTGCTCGA*A*G*C, * = phosphorothioate bonds). Following purifications with 0.8X volume of AMPure XP beads, we performed a test final sequencing-ready PCR with a dilution of the junction PCR product to determine the optimal cycle number, then proceeded with the final PCR using Q5 polymerase in 50uL reactions (98°C for 45 seconds and ~9 cycles of 98°C for 10 seconds, 65°C for 30 seconds, and 72°C for 30 seconds, ExP_GFP_fwd_I2: AATGATACGGCGACCACCGAGATCTACAC[index-2]GGCTTAAGCATGGCTAGCAAAG, ExP_P4_rev: CAAGCAGAAGACGGCATACGAGATTCATTCCATGGCATCTCACG. For the 1000x1000 ExP STARR-seq experiment, we purified the final libraries with 2 rounds of 0.8X volume of SPRISelect (Beckman Coulter, B23318) (1 round of 0.7X SPRISelect for the motif ExP STARR-seq experiment).
Alignment and counting of STARR-seq data
To characterize activity in the STARR-seq assay, we define “STARR-seq expression” for a given plasmid (corresponding to a particular promoter, enhancer, and plasmid barcode) as the expression of the reporter RNA transcript normalized to the abundance of that plasmid in the input DNA pool.
To quantify STARR-seq expression, we sequenced the library of RNA transcripts produced from replicate transfections (described above) along with the DNA input with paired-end reads (using custom sequencing primers ExP_P3_fwd_seq_R1: GAGTACTGGTATGTTCAGCTAACTTCTACCCATGC, ExP_P4_rev_seq_R2: TCATTCCATGGCATCTCACGACCTACTTAACTTGC) and the plasmid barcode with an index read (using custom sequencing primer ExP_fwd_BC_seq: GTCCCAATTCTTGTTGAATTAGATTGATCTAGAGCATGCA). We aligned reads for both the RNA and DNA libraries to the designed enhancer sequences using bowtie2 (bowtie2 options: -q --met 30 --met-stderr --maxins 2000 -p 16 --no-discordant --no-mixed --fast).
We counted reads separately from PCR replicates derived from each biological replicate of 50M transfected cells, and scaled each of the PCR replicates within a biological replicate such that they had the same total normalized counts, equal to the maximum across all PCR replicates. We combined counts into per-biological replicate counts for further processing. We used the BC-promoter dictionary to identify the promoter associated with each transcript. We used the same mapping and BC-promoter assignment process for DNA.
For subsequent analysis, we discarded plasmids that had fewer than 25 DNA reads or fewer than 1 RNA transcript reads from further processing.
Computing technical reproducibility and influence of plasmid barcode sequences
To assess the technical reproducibility of ExP-STARR-seq, we first compared STARR-seq expression between biological replicate experiments. Specifically, we first combined data from biological replicates 1 & 2 and 3 & 4. Next, we correlated log2(RNA/DNA) for these groups before (Fig. 1b, Extended Data Fig. 1b) and after (Extended Data Fig. 1e,f) averaging across plasmid barcodes corresponding to the same enhancer-promoter pair.
We next assessed the variation between plasmids with the same enhancer and promoter sequences but different random 16-bp plasmid barcodes, because these 16 nucleotides of random sequence might contain transcription factor motifs or other sequences that affect STARR-seq expression. To do so, we combined data from all biological replicate experiments and created two “virtual replicates” for each enhancer-promoter pair by splitting the corresponding plasmid barcodes into two groups. For example, an enhancer-promoter pair with 6 plasmid barcodes was split into 2 virtual replicates each with 3 barcodes). We averaged log2 STARR-seq expression within enhancer-promoter pairs (across different barcodes) and correlated these virtual replicates. We compared versions of this analysis for increasing thresholds on the minimum number of barcodes in each virtual replicate (Extended Data Fig. 1e,f).
Estimating enhancer and promoter activities — naïve averaging approach
We sought to compare the intrinsic activities of different enhancer and promoter sequences in ExP STARR-seq — that is, the contribution of a given enhancer or promoter sequence to STARR-seq expression, relative to other sequences. We estimated enhancer activity and promoter activity in two ways: by a simple averaging method, and by fitting a multiplicative Poisson count model (see next section).
As a first approach to estimate promoter activity, we calculated, for each promoter sequence, the average log2 STARR-seq expression when that promoter is paired with random genomic sequences in the enhancer position (Fig. 1c). This quantity represents the “basal” or “autonomous” expression level of the promoter, in the absence of a strong activating sequence in the enhancer position.
As a first approach to estimate enhancer activity, we calculated, for each enhancer sequence, the average log2 STARR-seq expression of all pairs including that enhancer sequence (Fig. 1d).
As noted above, we fit this model on the set of plasmids with at least 25 DNA reads, and at least 1 RNA read. In addition, to reduce noise in our promoter and enhancer activity estimates, we required at least two separate plasmid barcodes per promoter-enhancer pair. These filters resulted in 604,268 promoter-enhancer pairs across 4,512,907 total unique plasmids (~ 7.5 plasmids per pair) that were used to estimate promoter and enhancer activity.
In practice, this averaging method of calculating enhancer and promoter activity was inaccurate and biased, for several reasons. First, the averaging method does not consider the variance introduced by sampling & counting noise in sequencing, which is significant because many promoter-enhancer pairs have low RNA read counts. Second, the averaging method does not account for differences introduced due to missing data. In the 1000 enhancer x 1000 promoter data matrix, many entries are missing either due to low RNA counts (resulting from counting and sampling noise, or low expression) or due to low DNA counts (resulting from variation introduced in cloning the plasmid library). As a result of these factors, the averaging method produces biased (inflated) estimates of activity for weaker enhancer and promoter sequences because the expression of plasmids containing these sequences is more likely to drop below the threshold of detection given our sequencing depth (Extended Data Fig. 2c-d).
Because this model explained the data well, we used this same model to estimate intrinsic enhancer and promoter activity.
Estimating intrinsic enhancer and promoter activities — multiplicative model
We fit a count-based Poisson model to address the limitations of using a simple averaging approach to estimate intrinsic enhancer and promoter activities (see previous section), and to quantify the extent to which the ExP STARR-seq data can be explained by a simple multiplicative function of intrinsic enhancer and promoter activities. In this multiplicative model, all enhancers are assumed to activate all promoters by the same fold-change, without enhancer-promoter interaction terms.
Specifically, we estimate enhancer and promoter activities from ExP STARR-seq data by fitting the observed RNA read counts to a multiplicative function of observed DNA input read counts, intrinsic enhancer activity, and intrinsic promoter activity:
In this formula, P is the intrinsic promoter activity of promoter sequence p, E is intrinsic enhancer activity of enhancer sequence e, and k is a global scaling/intercept term that accounts for factors that control the relative counts of DNA and RNA such as sequencing depth.
We fit these parameters using block coordinate descent on the negative log-likelihood of the distribution above, initially fixing k=0, then alternatively optimizing (i) promoter activities while holding enhancer activities constant, and (ii) enhancer activities while holding promoter activities constant.
We then re-normalized enhancer activities and promoter activities by the mean activity of random genomic sequences, and adjusted the scaling factor k accordingly.
In practice, this model produces similar estimates to simply taking the mean value of an enhancer sequence across all promoters, and vice versa, but accounts for missing data points in the 1000x1000 matrix, and provides a more robust estimate for very weak enhancers or promoters, which produce relatively little RNA and are therefore difficult to measure in this STARR-seq experiment except when paired with a strong element in the other group (Extended Data Fig. 2c-d).
Assessing a multiplicative model for gene expression in the genome
We tested whether gene transcription in the genome could be modeled as a multiplicative function of promoter activity and enhancer inputs. To measure gene transcription, we applied precision run-on sequencing (PRO-seq) as previously described56 to K562 cells to map transcriptionally engaged RNA polymerase, and assigned gene transcription as reads per kilobase per million in the gene body, excluding the region within 1 Kb of the annotated transcription start site. To estimate promoter activity, we used the intrinsic promoter activity estimate from ExP STARR-seq. To estimate enhancer input, we summed the Activity x Contact (ABC) scores for all nearby enhancers (within 5 Mb of the transcription start site). The ABC scores in turn are estimated based on multiplying enhancer activity (geometric mean of DHS and H3K27ac ChIP-seq read counts at an enhancer) by enhancer-promoter contact frequency (estimated from Hi-C data)50. We considered all genes with “active” promoters, defined as those with DHS and H3K27ac signals above the 40th percentile of all genes in the genome as used in the ABC Model50.
Computing and clustering residuals from the multiplicative model:
We explored whether enhancer-promoter compatibility could explain variation in STARR-seq expression beyond that explained by the multiplicative model. To do so, we looked for shared behaviors between groups of promoters and enhancers by clustering them according to their residual error from the Poisson model described above.
For each enhancer-promoter pair, we used the Poisson model above to compute predicted RNA given the input DNA counts and estimates of intrinsic enhancer and promoter activities. We then compute a transformed residual as
where pseudocount = 10 to stabilize variance of the estimates across the range of values for RNA51. We filtered to all enhancer-promoter pairs with at least two barcodes, and calculated the mean of the residuals across barcodes to form a (sparse) 1000x1000 matrix of residuals indexed by promoter and enhancers.
We clustered this matrix independently along rows and columns (treating missing pairs as having a residual of 0) using K-means with 3 clusters, labeling the clusters as 0,1, and 2 such that they had increasing mean activity estimates in the Poisson model. One cluster each of enhancers and promoters (E0 and P0) contained sequences that were missing many data points due to their weaker activity leading to dropout due to low RNA expression. The sparsity of data for the E0 and P0 clusters prevented accurate characterization of compatibility, and so we excluded these clusters from subsequent analysis.
Assessing reproducibility of the clusters:
We evaluated whether the clustering we observed in the residuals was a general trend of the data, or an artifact of a few promoters or enhancers. To test this possibility, we randomly downsampled the residual matrix to 25% of promoters and 25% of enhancers (6.25% of the total data) 100 times, and clustered the subsets. We found that the original (full-data) cluster identity of a promoter or enhancer predicted the downsampled cluster with greater than 80% accuracy (Extended Data Fig. 4f).
Estimating enhancer activity with specific promoter classes, and promoter responsiveness to specific enhancer classes:
We evaluated whether certain promoters were more responsive when paired with different enhancer classes, and whether certain enhancers had more activity when paired with promoters from different classes (Fig. 3c,d).
To explore differences in enhancer activity when paired with different promoter classes, we fit the Poisson model (described above) separately to two different subsets of the data: (i) all enhancer sequences paired with P1 or genomic background promoter sequences (yielding an estimate of the activity of an enhancer sequence on a P1 promoter), and (ii) all enhancer sequences paired with P2 or genomic background promoter sequences (yielding an estimate of the activity of an enhancer sequence on a P2 promoter).
Similarly, to estimate promoter responsiveness to either E1 or E2 enhancers, we fit the Poisson model to the subsets: (iii) all promoters paired with E1 or genomic background enhancer sequences (yielding an estimate of the responsiveness of a promoter sequence to E1 enhancers), and (iv) all promoters paired with E2 or genomic background enhancer sequences (yielding an estimate of the responsiveness of a promoter sequence to E2 enhancers).
We used the genomic background promoter sequences to set a common baseline.
Annotating enhancer and promoter sequences with genomic features and sequence motifs
To annotate enhancer and promoter sequences with features of transcription factor (TF) binding of the corresponding genomic elements, we downloaded list of Human TF ChIP-seq narrowpeak files from the ENCODE Project1, and annotated each enhancer or promoter sequence with the maximum signalValue column for any overlapping peak (or 0 signal, for no overlap). We then compared the fold-change in signal between classes of sequences (Fig. 4d, Extended Data Fig. 6a, Supplementary Table 11).
To annotate enhancer and promoter sequences with transcription factor motifs, we used FIMO52 (default parameters, including p-value threshold of 10−4) to identify matches for HOCOMOCO v11 CORE motifs53. We then compared the fold-change in motif counts between classes of sequences (Extended Data Fig. 6b,c, Extended Data Fig. 7l, Supplementary Table 5, Supplementary Table 7).
For comparing features between E1 and E2 enhancers, we compared motif, ChIP-seq, and other features between the E1 and E2 enhancer sequences that overlapped the summit of a DNase peak.
For analyzing the proportion of P2 promoters bound by various factors, we defined “strongly bound” as having ChIP-seq signal greater than 20% of maximum ChIP-seq signal among P1 and P2 promoters.
Logistic regression to classify P1 and P2 promoters
We trained a logistic regression classifier to distinguish P1 versus P2 promoters, and classify all promoters genome-wide as P1 or P2 (see Supplementary Table 8). We used as input sequence and genomic features as described above. To standardize features with varying distributions, we removed features with non-zero values in less than five promoters, normalized continuous TF binding and histone mark features with respect to DNase activity, and performed hyperbolic arcsine transformation on all continuous features.
Due to redundancy of ENCODE TF ChIP-seq data and highly similar motifs between certain TFs, many features are highly correlated. To address this, we constructed an undirected weighted graph where vertices are features and edges are defined by the Pearson correlation coefficient between two features. After removing edges with weight less than 0.8, we treated each connected component in the graph as a distinct feature. Most resulting connected components contained only one feature, while highly correlated redundant features were grouped into one connected component. For connected components with more than one feature, we collapsed the features into one by taking the average. In total, we used 2,535 features for model training.
We then trained an elastic net logistic regression model to classify P1 and P2 promoters using 80% of the data. We ranked features with non-zero model coefficients and selected top features based on elbow-point cutoff. We then retrained a model using this smaller set of features to mitigate overfitting, resulting in 145 final features. The model achieved 94% mean accuracy across 6-fold cross validation.
We applied this model to all gene promoters in the genome (see Supplementary Table 8). As expected, we observed striking functional difference between genes with predicted P1 and P2 promoters: nearly all ribosomal subunits (82 of 84) are predicted to be P2 promoters, and, more broadly, 78% of previously annotated housekeeping genes were predicted to have P2 promoters.
Assessing cell-type specificity of gene expression across cell types and tissues
We analyzed capped analysis of gene expression (CAGE) data from the FANTOM5 consortium across 1829 experiments (“biosamples”)30. We downloaded expression data (in transcripts per million) from https://fantom.gsc.riken.jp/5/datafiles/latest/extra/gene_level_expression/hg19.gene_phase1and2combined_tpm.osc.txt.gz on February 19, 2022. We defined ubiquitously expressed genes as genes that are expressed at >= 1 TPM in >=95% of biosamples. We defined uniformly versus variably expressed genes as genes whose maximum expression is less than (or greater than) 10 times the median expression across biosamples, respectively.
Comparison of CRISPR-derived regulatory elements for P1 vs P2 promoters
To compare the number and effect sizes of genomic regulatory elements for P1 and P2 promoters, we analyzed CRISPRi tiling screens from previous studies that perturbed all DNase accessible sites around selected genes22,27,28. We counted the number of activating distal regulatory elements — i.e., distal, non-promoter DNase accessible sites whose perturbation led to a significant reduction in gene expression (Fig. 4c). We also compared the effect sizes on gene expression for these same activating distal regulatory elements (Extended Data Fig. 6e, Extended Data Fig. 7e).
Luciferase assays
We tested the ability of each of 7 large MYC enhancer fragments to activate the promoters of 3 genes in the MYC locus — MYC, PVT1, and CCDC26 — using a classic plasmid luciferase-based enhancer assay. The 7 MYC enhancers were defined as the 1.0-2.2 kb sequences identified in our previous MYC proliferation-based CRISPRi screen28, and a 1 kb bacterial plasmid sequence was used as a negative control sequence. We cloned promoter fragments into plasmids in combination with each of these sequences (see Supplementary Table 3). The promoter fragments corresponded to the dominant transcription start site of each gene in K562 cells (as determined by CAGE). For each of PVT1 and CCDC26 — which do not appear to be regulated by most of the 7 MYC enhancers in the genome — we cloned two promoter fragments of different lengths to determine if nearby sequences might encode biochemical specificity. We designed an insertion site ~1 kb upstream of the promoter in the plasmid for inserting each enhancer sequence (Extended Data Fig. 3a), and we flanked this region with polyadenylation signals in either direction to avoid measuring luciferase activity driven from transcripts initiating from the enhancer elements themselves. Luciferase assays using the Dual-Luciferase Reporter Assay (Promega) were performed as previously described28 in 4-6 biological replicates. For each experiment, we calculated the fold-change in luciferase signal (Firefly / Renilla) for enhancer versus negative control (Extended Data Fig. 3c).
Assessing the cell-type specificity of E1 and E2 enhancers
We tested whether E1 and E2 enhancer sequences from ExP STARR-seq overlapped elements predicted to act as enhancers by the ABC model in K562 cells or in 128 other cell types and tissues. To do so, we intersected the E1 and E2 enhancer sequences with the ~200-bp regions predicted by the ABC model to act as enhancers for at least 1 nearby expressed gene, as previously defined50. The ABC enhancer-gene predictions from this previous study50 are available at https://www.engreitzlab.org/resources/.
Aligning promoters by transcription start site
For each 264-bp promoter sequence, we defined the primary transcription start site (TSS) as the nucleotide with the highest stranded 5’ signal in GRO-Cap data in K562 cells (GSM1480321)54. This primary TSS position was used for plotting genomic signals relative to TSS and in analyses of motif positioning (e.g., for GABPA and YY1).
Analysis of motif position relative to TSS
We used FIMO52 to scan for HOCOMOCO motifs in promoters including for GABPA (GABPA_HUMAN.H11MO.0.A), YY1 (TYY1_HUMAN.H11MO.0.A), and the TATA box (TBP_HUMAN.H11MO.0.A). We reported positional preferences as the distance between the primary transcription start site from GRO-cap (see above) and the center of the motif. For example, GABPA, the most common position was −10 relative to the TSS (i.e. with the second ‘G’ in the core ‘GGAA’ motif located at position −10).
Hybrid selection STARR-seq (HS-STARR-seq) to measure enhancer activity for millions of genomic sequences
We conducted two STARR-seq experiments to measure the enhancer activity of millions of long genomic sequences tiling across human enhancer and promoter sequences. To generate these tiling sequences, we used a hybrid selection strategy, similar to previous approaches55. Specifically, we purified genomic DNA from K562 cells, tagmented DNA using Tn5 and gel size selection to a size range of approximately 300-700 bp (Extended Data Fig. 8e), and conducted hybrid selection using RNA probes as previously described56 targeting either (i) all gene promoters (“HS promoter pool”) or (ii) all accessible elements (“HS accessible element pool”) in K562 cells (see Supplementary Table 12 and Supplementary Table 13 for probe sequences). We amplified these sequences using primers including a UMI (CapStarrFa_N10 primer: tagatTGAtCTAGAGCATGCACCGGCAAGCAGAAGACGGCATACGAGATNNNNNNNNNNATGTCTCGTGGGCTCGGAGATGT and CapStarrR primer: CGAAGCGGCCGGCCGAATTCGTCGATCGTCGGCAGCGTCAGATGTG) and cloned these selected sequences into the hSTARR-seq_ori vector17, which uses the bacterial origin of replication (ORI) sequence as the promoter for the reporter transcript, by Gibson assembly. In the final HS promoter and accessible element Pools, 9% and 12% of fragments mapped to their intended targets, respectively, and each element was tiled by a median of 20 and 55 sequences. We conducted the rest of the STARR-seq experiment as described above, transfecting 50 million cells per replicate for each of 4 replicates.
We sequenced the input DNA libraries to a depth of 880 million and 810 million reads (promoter and accessible element pools, respectively), and the RNA libraries to a depth of 1.1 billion reads (both pools). We aligned reads to the hg19 genome using bowtie2 (options: -q --met-stderr --maxins 1000 -p 4 --no-discordant --no-mixed). We discarded fragments with fewer than 25 aligned DNA reads. Biological replicates were highly correlated (R2 = 0.92 and 0.91 for promoter and accessible element pools) (Extended Data Fig. 8d).
We analyzed this data by computing a log2 activity per fragment equal to the log2(RNA / DNA). and correcting for a fragment-length bias. We noted that STARR-seq expression was highly inversely correlated with the length of the enhancer sequence, even among random genomic fragments that did not overlap putative regulatory elements, which could result from biases in library preparation and sequencing. To adjust for this, we fit a linear regression (separately for the two pools) and subtracted this regression from the log2(RNA / DNA) activity to give a bias-corrected activity. We then correlated motifs with bias-corrected activity. To estimate enhancer activity of promoters from the ExP, we found HS-STARR-seq fragments that overlapped at least 90% of an ExP promoter and averaged their activity scores.
Analysis of variance (ANOVA)
ANOVA results were computed based on intrinsic activities, and represent the sequential sum of squares (Type I ANOVA) analyses. For example, in Extended Data Fig. 2j-k, we calculated the proportion of variance explained by promoters, the proportion of variance explained by enhancers after controlling for promoter activity, the proportion of variance explained by class interactions after controlling for promoter and enhancer activity, and, finally, any remaining variance.
Motif ExP STARR-seq library design
For the motif-insertion experiment, we selected 15 promoters from the P1 and P2 promoter clusters that had also been studied in Fulco et al. 201922, and also with intrinsic activities between −2 and +2 (evenly-spaced sampling from promoters ordered by intrinsic activity). We included a weak promoter in each cluster (intrinsic activity < −3), and also included the promoters for GATA1 and RPL37A (two other weak promoters).
We selected 15 enhancers from each of the E1 and E2 enhancer clusters, from elements detected as endogenous enhancers in K562 cells in Fulco et al. 2019, with intrinsic activity between −2 and 2 (evenly-spaced sampling from enhancers ordered by intrinsic activity), and avoiding potential polyA signals in the enhancer sequence. We added one weak enhancer from each class (intrinsic activity < −3), and one weak scramble enhancer sequence from the original experiment.
To explore the effect of adding GABPA, YY1, or other motifs to promoters, in every promoter sequence selected above, we inserted 2 copies of the GABPA consensus motif, centered at −10 and −31 relative to the TSS (identified by CAGE) to generate a new promoter sequence. For each original sequence, we did the same with YY1 at +1 relative to the TSS. Consensus motifs were taken from the HOCOMOCO v11 CORE motifs53.
To explore the effect of breaking motifs in promoters, for each promoter sequence selected above, we identified any GABPA or YY1 motifs (FIMO52, default parameters, including p-value threshold of 10−4, motifs within 30 bases of the TSS), and swapped 2 bases in the core of the motifs (e.g., GGAAG to GAGAG in GABPA motifs, and ATGGC to AGTGC in YY1 motifs). For each original promoter sequence with more than one motif, we included a new promoter sequence with each individual motif modified, as well as a one new promoter sequence combining each of these modifications.
To explore the effect of adding GABPA motifs to enhancers, we inserted 2, 4, or 6 copies of the GAPBA consensus motif (GAACCGGAAGTGG) spaced by 21bp to a single random background enhancer.
In this experiment, each originally selected promoter was also included in the enhancer sequence pool, and all enhancer sequences were included in both orientations in the plasmid. Promoter and enhancer sequences used in this experiment are listed in Supplementary Table 14 and Supplementary Table 15.
Motif ExP STARR-seq analysis
For the Motif ExP STARR-seq experiment, we followed the methods detailed for the original STARR-seq experiment except where noted here. We sequenced the promoter DNA pool to a depth of 214.8M reads. For the construction of the Barcode-Promoter dictionary, we initially aligned the promoter sequence reads to a specially constructed index of the unedited promoter sequences, using bowtie2 as described for the original STARR-seq experiment but with a slightly relaxed alignment threshold (X:--score-min L,−0.7,−0.7). We then used a custom python script to check aligned reads at the sites of potential edits. The script reassigned a read to an edited promoter sequence if it perfectly matched the edited sequence at all edit positions and the immediate up and downstream base pair. If the read did not perfectly match an edited sequence but also did not match the wild type sequence at the edit positions, the read was discarded. Reads with <90% perfect match with the entire sequence to which they were assigned were discarded. From this point, we returned to the original STARR-seq methods, namely dropping BC-promoter pairs with singleton reads, removing ambiguous pairings, and thresholding pairs with at least 5 reads to build the Barcode-Promoter dictionary.
We sequenced the pool consisting of two replicates of the input DNA library as well as four replicates of the RNA library to a total of 345.5M reads. When quantifying STARR-seq expression, unlike with the original STARR-seq experiment, we sequenced two replicates of the input DNA library. Therefore, to quantify the DNA read count for the log2(RNA/DNA) calculation, we used the average count of the two replicates, weighted to the same total read count. STARR-seq expression was highly correlated across replicates, (min R2 = 0.92, Extended Data Fig. 9a). As with the original STARR-seq experiment, we only included in the final analysis plasmids that had greater than 25 DNA reads and greater than 1 RNA transcript, for a total of 362,905 plasmids across 19,019 Promoter-Enhancer pairs with an average of 19 barcodes per pair (Extended Data Fig. 9b). For unedited promoter and enhancer pairs, the STARR-seq expression was highly correlated with the expression in the original ExP STARR-Seq experiment (Extended Data Fig. 9c).
For edited promoters, to determine the change in responsiveness to E1 enhancers (Fig. 5e), we estimated unedited and edited responsiveness to E1 enhancers as described for the original experiment. We transformed the values out of log2 space and computed the percent change in the responsiveness as the percent difference between the responsiveness of the edited sequence and the responsiveness of the unedited sequence.
For edited enhancers, to determine change in enhancer activity with P1 and P2 promoters (Extended Data Fig. 9d), we calculated an inferred enhancer strength for each enhancer within a given enhancer-promoter pair as the ExP score for the pair minus the intrinsic promoter strength as calculated with the multiplicative model described above. We transformed these strength estimates out of log2 space and then determined the percent change in enhancer strength for each pair as the inferred strength of the edited enhancer with a given promoter minus the inferred strength of the unedited enhancer with the same promoter divided by the strength of the unedited enhancer. We then compared the change in enhancer strength for each edit type with all P1 promoters compared with P2 promoters using a two-tailed t-test.
Analysis of mouse embryonic stem cell MPRA experiments
Data from Martinez-Ara et al. 202139 was filtered to plasmids with at least 5 total DNA reads, and at least 1 RNA read in each of three replicates. Promoter-enhancer pairs were filtered to those with at least 2 barcodes. We estimated intrinsic promoter and enhancer activities using the same method as described above for the ExP-STARR multiplicative model, but without the final steps centering the activities of promoter and enhancer fragments from the genomic background to zero-strength.
Extended Data
Extended Data Fig. 1. Design and reproducibility of ExP STARR-seq.
a. ExP STARR-seq reporter construct (pA = polyadenylation signal; purple = promoter sequencing adaptors; angled = spliced sequence; trGFP = truncated GFP open reading frame with start and stop codon; BC = 16bp N-mer plasmid barcode; red = enhancer sequencing adaptors) and 1000x1000 K562 library contents.
b. Correlation of ExP STARR-seq expression between biological replicate experiments, calculated for individual enhancer-promoter pairs with unique plasmid barcodes. Axes represent the average STARR-seq expression (RNA/DNA) of individual biological replicates. Density: number of enhancer-promoter plasmids.
c. Fraction of remaining enhancer-promoter plasmids passing DNA (≥25) and RNA (≥1) threshold (y-axis) with downsampling of sequencing reads (x-axis).
d. Distribution of plasmid barcodes per enhancer-promoter pair, red dotted-line is threshold of two plasmid barcodes.
e. Correlation between virtual replicates, formed by sampling two nonoverlapping groups of three plasmid barcodes from pairs with at least 6 barcodes, and averaging log2(RNA/DNA) within groups.
f. Correlation between virtual replicates as in (c) for increasing numbers of plasmid barcodes per pair in virtual replicates.
g. DNase-seq, H3K27ac ChIP-seq, and PRO-seq (RPM) by increasing quartile of autonomous promoter activity and average enhancer activity in ExP STARR-seq (n = 800). Box: median and interquartile range (IQR). Whiskers: +/− 1.5 x IQR.
h. Activation in ExP STARR-seq (expression versus genomic controls in distal position) of GATA1 and HDAC6 promoters by eHDAC6 (chrX:48641342-48641606). Ctrl = activity of promoters with random genomic controls in enhancer position. Error bars: 95% CI across plasmid barcodes. n = 7 (GATA1-ctrl), 381 (HDAC6-ctrl), 4 (eHDAC6-GATA1), 37 (eHDAC6-HDAC6).
i. Average enhancer activity (STARR-seq expression of plasmids containing a given enhancer averaged across all promoters) of enhancer sequences derived from random genomic controls (n=87), accessible elements (n=725), and genomic enhancers validated in CRISPR experiments (n=89).
Extended Data Fig. 2. Comparison of methods of estimating enhancer and promoter activities and the multiplicative model.
a. Intrinsic promoter activity (expression versus random genomic controls in enhancer position) of five selected promoters. Error bars: 95% CI across plasmid barcodes (n=54-79). Promoter classes (see Methods): DNASE2 (P1), HDAC6 (P1), CD164 (P1), BCAT2 (P1), PPP1R15A (P2).
b. Activation (expression versus random genomic controls in enhancer position) of 5 selected promoters by 5 selected enhancers: 1 = chr11:61602148-61602412 (E1), 2 = chr19:49467061-49467325 (E1), 3 = chrX:48641342-48641606 (E1), 4 = chr19:12893216-12893480 (E2), 5 = chr17:40851134-40851398 (E1). Error bars: 95% CI across plasmid barcodes (n=12-56).
c-d. Heatmap of promoter activity (a, expression divided by intrinsic enhancer activity) or enhancer activity (b, expression divided by intrinsic promoter activity) across all pairs of promoter (vertical) and enhancer sequences (horizontal). Axes are sorted by intrinsic promoter and enhancer activities, as in Fig. 2j. Grey: missing data.
e. Intrinsic promoter and enhancer activity (y-axis, estimated by a Poisson count model) versus average pairwise Spearman correlation (as in Fig. 2c-d).
f-g. Correlation between two estimates of promoter (c) and enhancer (d) activities. One method (“average activity”, x-axis) estimates activity calculated by averaging across elements, and the other method (“intrinsic activity”, y-axis) estimates activity by using coefficients estimated by a Poisson count model (see Methods).
h-i. Correlation of intrinsic promoter (e) and enhancer (f) activity estimates from Poisson model using data from separate replicate experiments.
j-k. Fraction of variance explained by promoter activity, enhancer activity, class interaction from the perspective of expression (STARR-seq score) and enhancer activation (fold-activation of an enhancer on a promoter, normalizing out promoter strength) limited to pairs with 2 or more (c) or 20 or more (d) plasmid barcodes. Plot includes pairs with P0 promoters and E0 enhancers. Bar plots show sequential sum of squares (Type-I ANOVA).
l. Correlation of the multiplicative enhancer x promoter model with STARR-seq expression comparing enhancer-promoter pairs located within 10kb, 100kb, and pairs located on different chromosomes.
Extended Data Fig. 3. Validation of enhancer-promoter multiplication via luciferase assays and modeling gene transcription as a function of intrinsic promoter activity and enhancer inputs.
a. ExP luciferase reporter construct. Seven enhancer fragments, with flanking polyadenylation signals, were cloned upstream of five promoter fragments and measured via the dual luciferase assay.
b. Autonomous promoter activity of ExP luciferase (average luciferase signal of promoter with negative control) for 5 promoter sequences derived from 3 genes (MYC, PVT1, CCDC26). Error bars are 95% CI from 6 (MYC) or 4 (all other promoters) biological replicates.
c. Enhancer activation (luciferase signal versus negative control sequence in the enhancer position) of seven enhancers across five promoter fragments. Error bars are 95% CI from 6 (MYC) or 4 (all other promoters) biological replicates.
d-f. Gene transcription (y-axis): PRO-seq read counts in the gene body. a. Promoter Activity (x-axis, left): Intrinsic promoter activity, as measured by ExP STARR-seq. b. Enhancer Input (x-axis, center): enhancer activity (based on measurements of H3K27ac and DHS in the genome) multiplied by enhancer-promoter contact (based on Hi-C measurements), summed across all putative enhancers (DHS peaks) within 5 Mb of the gene promoter (excluding the promoter’s own peak), weighted by HiC contact as in the ABC Model22. c. Promoter Activity x Enhancer Input (x-axis, right). Labels: gene symbols for 741 promoters with sequence activity estimates from ExP STARR-seq and enhancer input estimates from ABC. Dotted lines: Line of best fit from linear regression in log2 space.
Extended Data Fig. 4. Enhancer and promoter cluster identification and reproducibility.
a. Heatmap of deviations in enhancer-promoter STARR-seq expression from a multiplicative enhancer-promoter model (color scale: fold-difference between observed expression versus expression predicted by multiplicative model; gray: missing data). Same as Fig 3a, except including clusters with weak sequences and missing data (E0 and P0). Vertical axis: promoter sequences grouped by class and sorted by responsiveness to E1 vs. E2; horizontal axis: enhancer sequences grouped by class and sorted by activation of P1 vs. P2.
b. Distribution of intrinsic enhancer and promoter activity (expression versus genomic controls) by cluster.
c. Fraction of enhancer-promoter pairs observed in ExP STARR-seq dataset (>= 2 plasmid barcodes) by cluster.
d. Correlation of average promoter activation (expression versus genomic controls in enhancer position) by E2 versus E1 enhancer sequences. Each point is one promoter sequence. Same as Fig. 3c, except including P0 promoter sequences.
e. Correlation of average activation of P2 versus P1 promoters. Each point is one enhancer sequence. Same as Fig. 3d, except including E0 enhancer sequences.
f. Robustness of enhancer and promoter cluster assignments to downsampling of enhancer and promoter sequences. Clustering was repeated in 100 random downsamplings to 25% of promoter sequences and 25% of enhancer sequences (6.25% of original matrix). Heatmap: Average fraction overlap between cluster assignments from the full and downsampled matrices.
g. Correlation of average promoter activation (expression versus genomic controls in enhancer position) by E2 versus E1 enhancer sequences using ‘average activity’ instead of model estimates. Each point is one promoter sequence.
h. Correlation of average activation of P2 versus P1 promoters using ‘average activity’ instead of model estimates. Each point is one enhancer sequence.
Extended Data Fig. 5. Classes of enhancer and promoter sequences show distinct patterns of activation and responsiveness.
a. For 6 representative enhancer sequences (3 E1 and 3 E2 sequences), the pairwise correlation of promoter activation (expression versus genomic controls in promoter position, averaged across plasmid barcodes). Each point is one promoter sequence.
b. For 6 representative promoter sequences (3 P2 and 3 P1 sequences), the pairwise correlation of activation by enhancers (expression versus genomic controls in enhancer position, averaged across plasmid barcodes). Each point is one enhancer sequence.
Extended Data Fig. 6. Classes of enhancer sequences correspond to strong and weak genomic enhancers.
a. Volcano plot comparing ChIP-seq and other genomic features for E2 versus E1 enhancer sequences (see Supplementary Table 4). X-axis: ratio of average signal at P2 versus P1 promoters. Red dots: features with significantly higher signal at E1; no features have significantly higher signal at E2 enhancer sequences.
b. Volcano plot comparing transcription factor motifs for E1 versus E2 enhancer sequences (see Supplementary Table 5). X-axis: ratio of average motif counts in E1 and E2 enhancer sequences. Red dots: Motifs significantly more frequent in E1 vs. E2 sequences.
c. Volcano plot comparing transcription factor motifs for E1 and E2 versus E0 enhancer sequences (see Supplementary Table 5). X-axis: ratio of average motif counts in E1 and E2 versus E0 sequences. Red dots: Motifs significantly more frequent in E1 and E2 versus E0 sequences (>0) or more frequent in E0 versus E1 and E2 (<0).
d. Mean H3K27ac ChIP-seq coverage of genomic elements corresponding to E0, E1, E2, or genomic control enhancer sequences (+/− 95% CI), aligned by DHS peak summit. Dotted lines mark bounds of the enhancer sequences used in ExP STARR-seq. E0 and E2 distributions are overlapping.
e. % effect of genomic elements corresponding to E1 vs. E2 enhancer sequences on expression of genes corresponding to P1 promoters in CRISPRi screens, separated by quartiles of 3D contact frequency measured by Hi-C (0.39-11.9 (n=9), 11.9-23.9 (n=31), 23.9-58.3 (36), 58.3-100(n=34)). *P < 0.05, two-sample, two-sided t-test. Boxes are median and interquartile range, whiskers are +/− 1.5*IQR.
f. Cumulative density plot showing the cell-type specificity of enhancer sequences selected for ExP STARR-seq, and DNase peaks or ABC enhancers in K562 cells. X-axis: # of cell types other than K562 in which the element is predicted to be an ABC enhancer.
g. GRO-Cap coverage of genomic enhancers used in ExP STARR-seq. Top: Mean coverage of enhancers corresponding to E1 vs. E2 classes. Bottom: Coverage across all individual enhancers.
h. Evolutionary conservation of enhancers separated by enhancer class, as measured by mean phastcon score (probability of each nucleotide belonging to a conserved element) and mean phyloP score (-log(p-value) under a null hypothesis of neutral evolution) across each element. P-value from KS test.
Extended Data Fig. 7. Properties of promoter classes.
a. Cumulative density plot showing the cell-type specificity of promoter chromatin activity (of promoters selected for ExP STARR-seq). X-axis: # of biosamples (cell types or tissues) other than K562 in which the promoter is active. Active = Top 50% of promoters by activity (geometric mean of H3K27ac and DHS signals, as used in the ABC model). All genes = all genes in the genome.
b. Gene ontology log2-enrichment for P1 promoters using P1 and P2 promoters as a background set.
c. Predicted enhancer inputs for each gene (sum of ABC scores for all candidate enhancers within 5 Mb of the TSS, excluding the promoter of the gene itself) for genes in the genome corresponding to P1 versus P2 promoters. P = 0.00083, Mann-Whitney U test. Boxes are median and interquartile range, whiskers are +/− 1.5*IQR.
d. DNase-seq signal in K562 cells at P1 and P2 promoters in the genome, aligned by boundaries of the 264-bp ExP STARR-seq promoter sequence (dotted gray lines, see Methods).
e. H3K27ac ChIP-seq signal in K562 cells at P1 and P2 promoters in the genome, aligned by boundaries of the 264-bp ExP STARR-seq promoter sequence (dotted grey lines, see Methods).
f. Number of nearby accessible elements (within 100 Kb of the gene promoter, considering top 150,000 DNase peaks in K562 cells as used in the ABC model22) for the 14 genes corresponding to P1 promoters and 11 genes corresponding to P2 promoters with comprehensive CRISPR tiling data. P = 0.17, Mann-Whitney U test. Boxes are median and interquartile range, whiskers are +/− 1.5*IQR.
g. % Effect of CRISPRi perturbations to genomic regulatory elements on genes corresponding to P1 vs. P2 promoters. P = 0.0071, t-test.
h. Fraction of promoter sequences containing TATA or CA initiator core promoter motifs.
i. GRO-Cap coverage of genomic promoters aligned by TSS. Top: Mean coverage of genomic promoters corresponding to P1 vs. P2 classes. Bottom: Coverage across all individual promoters.
j. Normalized CpG-content of P1 and P2 promoter sequences (n = 800), calculated as the ratio of observed to expected CpG = (CpG fraction) / ((GC content)2 / 2). Boxes are median and interquartile range, whiskers are +/− 1.5*IQR, P = 1.37*10−10, t-test.
k. Evolutionary conservation of promoters separated by promoter class, as measured by mean phastcon score (probability of each nucleotide belonging to a conserved element) and mean phyloP score (−log(p-value) under a null hypothesis of neutral evolution) across each element. P-value from KS test.
l. Volcano plot comparing frequency of transcription factor motifs in P2 versus P1 promoter sequences (see Supplementary Table 7). X-axis: ratio of average motif counts in P2 versus P1 promoter sequences. Light blue and dark blue dots: Motifs significantly more frequent in P1 or P2 promoter sequences, respectively. Red outline: significant motifs for ETS family TFs.
m. Volcano plot comparing frequency of transcription factor motifs in P2 and P1 versus P0 promoter sequences (see Supplementary Table 7). X-axis: ratio of average motif counts in P2 and P1 versus P0 promoter sequences. Dark blue dots: Motifs significantly more frequent in P2 and P1 vs. P0 promoter sequences.
n. Fraction of P2 promoter sequences with YY1 and GABPA binding motifs by nucleotide position, aligned by TSS and separated by strand (see Methods).
Extended Data Fig. 8. Transcription factors enriched at promoters and enhancers and hybrid-selection STARR-seq in K562 cells.
a. ChIP-seq signal for 5 transcription factors in K562 cells at P1 and P2 promoters in the genome, aligned by boundaries of the 264-bp ExP STARR-seq promoter sequence (see Methods). Top: average ChIP-seq signal normalized to input. Bottom: signal at individual genomic promoters. Black line: average for random genomic control sequences.
b. ChIP-seq signal at E1 and E2 enhancers in the genome. Black line: average for random genomic control sequences.
c. Correlation between intrinsic promoter activity and responsiveness of promoters to E1 enhancers (average activation by E1 sequences, expressions vs. random genomic controls). Each point is one promoter. Same as Fig. 5b, but in normal scale instead of log2 scale.
d. Correlation of HS-STARR-seq expression between biological replicate experiments for promoter and accessible element pools, calculated for individual elements with unique plasmid barcodes. Axes represent the average STARR-seq expression (RNA/DNA, log10 scale) of two biological replicates. Density: number of plasmids.
e. Fragment length distribution in HS-STARR-seq in promoter and accessible element pools, of fragments with at least 25 DNA counts.
f. STARR-seq expression (y-axis) and fragment length (x-axis) relationship in HS-STARR-seq. Density: number of plasmids.
Extended Data Fig. 9. Motif insertion and scramble ExP STARR-seq in K562 cells and generalizability of compatibility rules.
a. Correlation of ExP STARR-seq expression between biological replicate experiments, calculated for individual enhancer-promoter pairs with unique plasmid barcodes. Axes represent the average STARR-seq expression (RNA/DNA) of individual biological replicates. Density: number of enhancer-promoter plasmids.
b. Distribution of plasmid barcodes per enhancer-promoter pair. Red dotted-line: threshold of two plasmid barcodes.
c. STARR-seq expression in smaller-scale validation experiment (y-axis) vs. expression in the original ExP STARR-seq dataset (x-axis) for each enhancer-promoter pair included in both experiments. Dotted gray line: line of best fit from linear regression in log2 space.
d. Change in enhancer activity with P1 or P2 promoters (edited enhancer activity compared with unedited enhancer activity with a promoter) after inserting 2, 4, or 6 GABPA motifs into 1 E0 enhancer sequence. Each point represents one enhancer-promoter pair measured over 4 biological replicates. *P < 0.0001, two-tailed t-test. Boxes are median and interquartile range, whiskers are +/− 1.5*IQR.
e. Fraction of variance explained by intrinsic promoter activity and enhancer activity with respect to log2 reporter expression (reporter assay score) from Martinez-Ara et al. 202139. Left bars: experiment including promoters and enhancers from the Nanog and Klf2 loci. Right bars: experiment including promoters and enhancers from the Tfcp2l1 locus. For each experiment, values are shown for pairs with 2 or more, or 5 or more plasmid barcodes. Enhancer and promoter activities explain more of the variance when considering enhancer-promoter pairs with at least 5 vs. at least 2 barcodes. Bar plots show sequential sum of squares (Type-I ANOVA) for promoters, then enhancers.
f. Correlation of reporter assay expression with the product of intrinsic promoter and enhancer activities from two experiments from Martinez-Ara et al., 202139. Density color scale: number enhancer-promoter pairs.
Extended Data Fig. 10. Model of the effect of an enhancer on RNA expression.
a. Simple rules of enhancer and promoter compatibility. The effects of enhancers on nearby genes in the human genome are controlled by the quantitative tuning of intrinsic promoter activity, intrinsic enhancer activity, enhancer-promoter 3D contact, and enhancer-promoter class compatibility.
Supplementary Material
Acknowledgements
This work was supported by an NHGRI Genomic Innovator Award (R35HG011324 to J.M.E.); Gordon and Betty Moore and the BASE Research Initiative at the Lucile Packard Children’s Hospital at Stanford University (J.M.E.); an NIH Pathway to Independence Award (K99HG009917 and R00HG009917 to J.M.E.); the Harvard Society of Fellows (J.M.E.); the Novo Nordisk Foundation Center for Genomic Mechanisms of Disease (J.M.E.); the Broad Institute (E.S.L.); an AΩA Carolyn L. Kuckein Student Research Fellowship (D.T.B.); the NHGRI Ruth L. Kirschstein NRSA Predoctoral Institutional Research Training Grants (T32HG000044, V.L.); and by the National Institute of General Medical Sciences (T32GM007753, L.S.). We thank Bas van Steensel and Miguel Martinez-Ara for sharing data and discussing analysis. We thank C. Vockley, V. Subramanian, and members of the Engreitz and Lander labs for discussions and technical assistance.
Footnotes
Competing Interests
C.P.F. is now an employee and shareholder of Bristol Myers Squibb. J.M.E. is a shareholder of Illumina, Inc and other biotechnology companies. All other authors declare no competing interests.
Analysis and plotting tools
Data was processed and plotted using bowtie257 2.3.4.1, R58 4.0.2, python59 3.6.3, numpy60 1.17.4, scipy61 1.5.4, pandas62 1.1.5, matplotlib63 3.3.4, seaborn64 0.11.1, scikit-learn65 0.21.3, ncls66 0.0.51, statsmodels67 0.12.2.
Code Availability
Code for fitting the multiplicative ExP model is available at https://doi.org/10.5281/zenodo.6514733 or https://github.com/broadinstitute/ExP-model-fit.
Data Availability
Raw and processed data for ExP STARR-seq, motif ExP STARR-seq, HS STARR-seq, and K562 PRO-seq can be found in NCBI GEO under accession number GSE184426. Luciferase data can be found in Supplementary Table 3. Datasets used from the ENCODE Project are listed in Supplementary Table 10 and are available at https://www.encodeproject.org. Additional resources and protocols related to this study may be available at https://www.engreitzlab.org/resources/.
Main Text References
- 1.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Arensbergen J, van Steensel B & Bussemaker HJ In search of the determinants of enhancer-promoter interaction specificity. Trends Cell Biol. 24, 695–702 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emami KH, Navarre WW & Smale ST Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol. Cell. Biol 15, 5906–5916 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohtsuki S, Levine M & Cai HN Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 12, 547–556 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emami KH, Jain A & Smale ST Mechanism of synergy between TATA and initiator: synergistic binding of TFIID following a putative TFIIA-induced isomerization. Genes Dev. 11, 3007–3019 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler JEF Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes & Development vol. 15 2515–2519 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yean D & Gralla J Transcription reinitiation rate: a special role for the TATA box. Molecular and Cellular Biology vol. 17 3809–3816 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wefald FC, Devlin BH & Williams RS Functional heterogeneity of mammalian TATA-box sequences revealed by interaction with a cell-specific enhancer. Nature 344, 260–262 (1990). [DOI] [PubMed] [Google Scholar]
- 9.Zabidi MA, Arnold CD, Schernhuber K & Pagani M Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation. Nature (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerji J, Rusconi S & Schaffner W Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299–308 (1981). [DOI] [PubMed] [Google Scholar]
- 11.Banerji J, Olson L & Schaffner W A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33, 729–740 (1983). [DOI] [PubMed] [Google Scholar]
- 12.Melnikov A et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol 30, 271–277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnold CD et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–1077 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Kermekchiev M, Pettersson M, Matthias P & Schaffner W Every enhancer works with every promoter for all the combinations tested: could new regulatory pathways evolve by enhancer shuffling? Gene Expr. 1, 71–81 (1991). [PMC free article] [PubMed] [Google Scholar]
- 15.Tewhey R et al. Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay. Cell 172, 1132–1134 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Klein JC et al. A systematic evaluation of the design and context dependencies of massively parallel reporter assays. Nat. Methods 17, 1083–1091 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muerdter F et al. Resolving systematic errors in widely used enhancer activity assays in human cells. Nat. Methods 15, 141–149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen TA et al. High-throughput functional comparison of promoter and enhancer activities. Genome Res. 26, 1023–1033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnold CD et al. Genome-wide assessment of sequence-intrinsic enhancer responsiveness at single-base-pair resolution. Nat. Biotechnol 35, 136–144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haberle V et al. Transcriptional cofactors display specificity for distinct types of core promoters. Nature 570, 122–126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X & Noll M Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. EMBO J. 13, 400–406 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fulco CP et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Arensbergen J et al. Genome-wide mapping of autonomous promoter activity in human cells. Nat. Biotechnol 35, 145–153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wall L, deBoer E & Grosveld F The human beta-globin gene 3’ enhancer contains multiple binding sites for an erythroid-specific protein. Genes Dev. 2, 1089–1100 (1988). [DOI] [PubMed] [Google Scholar]
- 25.Tuan DY, Solomon WB, London IM & Lee DP An erythroid-specific, developmental-stage-independent enhancer far upstream of the human ‘beta-like globin’ genes. Proc. Natl. Acad. Sci. U. S. A 86, 2554–2558 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thakore PI et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klann TS et al. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol 35, 561–568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fulco CP et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y et al. Functional assessment of human enhancer activities using whole-genome STARR-sequencing. Genome Biol. 18, 219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haberle V & Stark A Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol 19, 621–637 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenhard B, Sandelin A & Carninci P Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet 13, 233–245 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Fan K, Moore JE, Zhang X-O & Weng Z Genetic and epigenetic features of promoters with ubiquitous chromatin accessibility support ubiquitous transcription of cell-essential genes. Nucleic Acids Res. 49, 5705–5725 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xi H et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 3, e136 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landolin JM et al. Sequence features that drive human promoter function and tissue specificity. Genome Res. 20, 890–898 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weingarten-Gabbay S et al. Systematic interrogation of human promoters. Genome Res. 29, 171–183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahu B, Hartonen T, Pihlajamaa P et al. Sequence determinants of human gene regulatory elements. Nat Genet 54, 283–294 (2022). doi: 10.1038/s41588-021-01009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu M et al. GA-binding protein-dependent transcription initiator elements. Effect of helical spacing between polyomavirus enhancer a factor 3(PEA3)/Ets-binding sites on initiator activity. J. Biol. Chem 272, 29060–29067 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Curina A et al. High constitutive activity of a broad panel of housekeeping and tissue-specific cis-regulatory elements depends on a subset of ETS proteins. Genes Dev. 31, 399–412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Ara M, Comoglio F, van Arensbergen J & van Steensel B (2022). Systematic analysis of intrinsic enhancer-promoter compatibility in the mouse genome. Mol Cell. 82, doi: 10.1101/2021.10.21.465269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maricque BB, Chaudhari HG & Cohen BA A massively parallel reporter assay dissects the influence of chromatin structure on cis-regulatory activity. Nat. Biotechnol (2018) doi: 10.1038/nbt.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong CKY & Cohen BA Genomic environments scale the activities of diverse core promoters. doi: 10.1101/2021.03.08.434469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiang CM & Roeder RG Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 267, 531–536 (1995). [DOI] [PubMed] [Google Scholar]
- 43.Austen M, Lüscher B & Lüscher-Firzlaff JM Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem 272, 1709–1717 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Sucharov C, Basu A, Carter RS & Avadhani NG A novel transcriptional initiator activity of the GABP factor binding ets sequence repeat from the murine cytochrome c oxidase Vb gene. Gene Expr. 5, 93–111 (1995). [PMC free article] [PubMed] [Google Scholar]
- 45.Carter RS & Avadhani NG Cooperative binding of GA-binding protein transcription factors to duplicated transcription initiation region repeats of the cytochrome c oxidase subunit IV gene. J. Biol. Chem 269, 4381–4387 (1994). [PubMed] [Google Scholar]
- 46.Usheva A & Shenk T YY1 transcriptional initiator: protein interactions and association with a DNA site containing unpaired strands. Proc. Natl. Acad. Sci. U. S. A 93, 13571–13576 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsson AJM et al. Genomic encoding of transcriptional burst kinetics. Nature 565, 251–254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.FANTOM Consortium and the RIKEN PMI and CLST (DGT) et al. A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang T, Lander ES & Sabatini DM Large-Scale Single Guide RNA Library Construction and Use for CRISPR-Cas9-Based Genetic Screens. Cold Spring Harb. Protoc 2016, db.top086892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nasser J et al. Genome-wide enhancer maps link risk variants to disease genes. Nature 593, 238–243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional References
- 51.Anscombe FJ THE TRANSFORMATION OF POISSON, BINOMIAL AND NEGATIVE-BINOMIAL DATA. Biometrika vol. 35 246–254 (1948). [Google Scholar]
- 52.Grant CE, Bailey TL & Noble WS FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kulakovskiy IV et al. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 46, D252–D259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Core LJ et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet 46, 1311–1320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vanhille L et al. High-throughput and quantitative assessment of enhancer activity in mammals by CapStarr-seq. Nat. Commun 6, 6905 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Engreitz JM et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R Core Team. R: A Language and Environment for Statistical Computing. (2014).
- 59.Van Rossum G & Drake FL Python 3 Reference Manual. (CreateSpace, 2009). [Google Scholar]
- 60.Harris CR et al. Array programming with NumPy. Nature 585, 357–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Virtanen P et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McKinney W & Others. Data structures for statistical computing in python. in Proceedings of the 9th Python in Science Conference vol. 445 51–56 (Austin, TX, 2010). [Google Scholar]
- 63.Hunter JD Matplotlib: A 2D Graphics Environment. Computing in Science Engineering 9, 90–95 (2007). [Google Scholar]
- 64.Waskom M seaborn: statistical data visualization. J. Open Source Softw 6, 3021 (2021). [Google Scholar]
- 65.Pedregosa F et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res 12, 2825–2830 (2011). [Google Scholar]
- 66.Stovner EB & Sætrom P PyRanges: efficient comparison of genomic intervals in Python. Bioinformatics 36, 918–919 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Seabold S & Perktold J Statsmodels: Econometric and statistical modeling with python. in Proceedings of the 9th Python in Science Conference vol. 57 61 (Austin, TX, 2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data for ExP STARR-seq, motif ExP STARR-seq, HS STARR-seq, and K562 PRO-seq can be found in NCBI GEO under accession number GSE184426. Luciferase data can be found in Supplementary Table 3. Datasets used from the ENCODE Project are listed in Supplementary Table 10 and are available at https://www.encodeproject.org. Additional resources and protocols related to this study may be available at https://www.engreitzlab.org/resources/.