Abstract
Diallyl trisulfide (DATS) is a promising small molecule phytochemical that exhibits in vitro and in vivo activity in multiple preclinical solid tumor models including breast cancer, but the underlying mechanism is not fully understood. We have shown previously that forkhead boxQ1 (FoxQ1) transcription factor is a novel target for breast cancer stem-like cells (bCSC) inhibition by DATS. Analysis of the breast TCGA (The Cancer Genome Atlas) data revealed that FoxQ1 expression was positively associated with that of SLC16A1/monocarboxylate transporter 1 (MCT1). Western blotting confirmed increased expression of MCT1 protein in SUM159 (basal-like) and MCF-7 cells (luminal-type) stably transfected to overexpress FoxQ1. Furthermore, FoxQ1 was recruited to the promoter of SLC16A1/MCT1. Treatment of SUM159 and MCF-7 cell lines with DATS resulted in suppression of MCT1 protein level that was accompanied by a decrease in intracellular and secreted levels of lactate. Overexpression or knockdown of MCT1 protein failed to alter DATS-mediated inhibition of colony formation or cell migration when compared to corresponding control cells. On the other hand, overexpression of MCT1 protein conferred partial but statistically significant protection against DATS-mediated inhibition of bCSC fraction (CD49fhigh/CD44high and aldehyde dehydrogenase 1 activity). The size of the mammospheres was relatively smaller in the DATS-treated group compared to control group. Inhibition of bCSC upon DATS treatment was augmented by knockdown of the MCT1 protein. In conclusion, the present study reveals that MCT1 is a novel target for bCSC inhibition by DATS treatment.
Keywords: MCT1, Diallyl Trisulfide, Breast Cancer, Chemoprevention
1. INTRODUCTION
Breast cancer is a heterogeneous disease that is a leading cause of cancer-related deaths in American women.1,2 More than 41,000 deaths are expected from breast cancer in 2021 in the United States according to the American Cancer Society. Edible plants like garlic and onions and their phytochemicals appear promising for prevention and treatment of cancer.3,4 Epidemiological studies have suggested that the risk of breast cancer is lowered by increased dietary intake of garlic.5,6 For example, a population-based case-control study in Puerto Rico (n=314 for primary breast cancer cases and n=346 for controls) noted an inverse association between breast cancer risk and moderate consumption of garlic/onion (odds ratio = 0.59; 95% confidence interval 0.35–1.01) or high garlic/onion consumption (odds ratio = 0.51; 95% confidence interval 0.30–0.87) when compared to low consumption cohort (Ptrend = 0.02).6 Similar conclusion was reached in a French case-control study.5 A double-blind and placebo-controlled interventional trial in patients with gastric cancer showed that administration of 200 mg of diallyl trisulfide (DATS) in combination with 100 μg of selenium for one month of each year during 1989–1991 was well tolerated.7 In the first five-year follow-up (1992–1996) after DATS administration was stopped, about 50% reduction in mortality rate and relative risk of cancer was also reported.8
The epidemiological observations were the foundation for multiple cellular mechanistic and in vivo efficacy studies of DATS in preclinical models of breast cancer. Various organosulfur compounds including fat-soluble diallyl disulfide and DATS as well as some water-soluble compounds are believed to be responsible for the anticancer benefit of garlic.9,10 The in vivo anticancer efficacy of DATS in a treatment setting has been investigated using xenograft models of human breast cancer cells.11–13 The MCF-7 xenograft growth was inhibited dramatically by administration of 5 μmol DATS/kg body twice/week for 1 month.11 After 12 weeks, none of vehicle-treated control mice were alive, but 50% of the DATS-treated mice were still alive.11 The in vivo growth of SUM159 human breast cancer xenograft was also inhibited by oral administration of 2 mg DATS/mouse three times/week.12 Daily oral administration of 25 and 50 mg DATS/kg body weight to MDA-MB-231 xenograft bearing nude mice significantly inhibited liver and lung metastasis that was associated with inhibition of thioredoxin system.13 However, oral DATS administration failed to prevent N-nitroso-N-methylurea-induced mammary cancer development in rats.14
Human breast cancer cell lines have been utilized to elucidate the mechanisms underlying anticancer effects of DATS.11–17 Inhibition of breast cancer cell proliferation, cell migration, self-renewal of breast cancer stem like cells (bCSC) are well-established anticancer effects of DATS.15–17 The DATS-treated breast cancer cells also exhibit G2/M phase cell cycle arrest and apoptosis induction.15,16,18 The DATS-induced apoptosis is triggered by generation of reactive oxygen species.15,16. A normal mammary epithelial cell line MCF-10A is resistant to growth inhibition and apoptosis induction by DATS treatment when compared to breast cancer cell lines MCF-7, MDA-MB-231, and BRI-JM04.16 At the molecular level, multiple targets of DATS have been identified, including c-Jun N-terminal kinase, forkhead box Q1 (FoxQ1), thiredoxin-1, signal transducer and activator of transcription 3, Wnt/β-catenin, α-secretases, lactate dehydrogenase, estrogen receptor-α, nuclear factor-κB, hypoxia inducible factor-1α, and mitogen-activated protein kinases, etc.11–24
We have shown previously that DATS decreases self-renewal of bCSC by inhibiting FoxQ1 transcription factor.12 Analysis of the breast TCGA (The Cancer Genome Atlas) revealed a positive association between expression of FoxQ1 and SLC16A1/monocarboxylate transporter 1 (MCT1) (data shown later). These observations prompted us to determine whether MCT1 is a novel target of regulation by FoxQ1 and if DATS inhibits MCT1.
2. MATHERIALS AND METHODS
2.1. Reagents and cell lines
DATS (99.2%) was purchased from LKT Laboratories (St. Paul, MN), diluted with dimethyl sulfoxide (DMSO; 28 mM stock), and stored at −80°C. Regents for cell culture including medium, fetal bovine serum, and antibiotics were from Life Technologies-Thermo Fisher Scientific (Waltham, MA). An antibody against MCT1 (for western blotting and confocal microscopy) and FoxQ1 (for western blotting) were from Proteintech Group (Rosemont, IL). Anti-MCT1 antibody for immunohistochemistry was from Invitrogen-Thermo Fisher Scientific and FoxQ1 antibody for Chromatin Immunoprecipitation (ChIP) assay was from Santa Cruz Biotechnology (Dallas, TX) An antibody against β-Actin was from Sigma (St. Louis, MO).A kit for measurement of lactate was purchased from Biovision (Milpitas, CA). The MCF-7 cell line was purchased from the American Association Type Culture Collection (Manassas, VA) and authenticated by us in 2017. The SUM159 cell line was purchased from Asterand and authenticated by us in 2017. Each cell line was maintained according to the supplier’s instructions. SUM159 cells were transfected with empty pCMV6 (Myc-FLAG-tagged) or the same vector encoding for MCT1 (OriGene, Rockville, MD). Stably transfected cells were generated by a 4-week culture in medium supplemented with 1.5 mg/mL of G418. The SUM159 cells were stably transfected with 4 μg of control small hairpin RNA (shRNA) or MCT1-targeted shRNA using transfection medium and reagents from Santa Cruz Biotechnology. The SUM159 clone with stable knockdown of MCT1 was selected with 2 μg/mL of puromycin for 5 weeks. Details of FoxQ1 overexpressing SUM159 and MCF-7 cells have been descried by us previously.12
2.2. TCGA data analysis
The RNA-seq data from breast cancer TCGA dataset (n=1097) was analyzed to determine the correlation between FoxQ1 and SLC16A1/MCT1 expression using the University of California, Santa Cruz Xena Browser (https://xena.ucsc.edu/public/). The correlation coefficient and statistical significance were determined by Pearson’s test.
2.3. Western blotting
Cell lysates were prepared as described by us previously.25 Western blotting was performed essentially as described by us previously.25 Blots were stripped and re-probed with anti-β-Actin for protein normalization. Protein level quantitation was done by densitometric scanning using UN-SCAN-IT gel analysis and graph digitizing software (Version 7.1; Silk Scientific Corporation, Orem, UT).
2.4. Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed following the manufacturers’ protocol (Magnetic Chip Kit; Pierce, Rockford, IL) using normal IgG (as a negative control) and FoxQ1 antibody. Other details of ChIP assay have been described by us previously.12 The FoxQ1 binding sites at the SLC16A1/MCT1 promoter were amplified (60°C, 1 minute, 40 cycles) with the following region-specific primers: site #1, 5′-TAAGTCCTAGCTGCCCCAAT3′ (forward) and 5′-CTGAGAGGTCCCATGTGCTT-3′ (reverse); site #2, ′TGGAAACACCAAAACTGTAACC-3′ (forward) and 5′-TTTCAGTTACTTCACCATTTCTGTG-3′ (reverse); site #3, 5′-TGTATGATCTGTGGAATCTAATTTTT -3′ (forward) and 5′-CCAATCTAATTGTCACTTTGTACCTG -3′ (reverse). Fold enrichment was normalized to the input.
2.5. Immunohistochemistry for MCT1 expression in triple negative breast cancer (TNBC)
Immunohistochemistry was performed to compare expression of MCT1 protein in tissue microarrays of TNBC (BR487a; US Biomax, Rockville, MD) and normal human mammary tissues (BRN801a; US Biomax). Immunohistochemistry for MCT1 was performed as described by us previously.12 At least 9 randomly selected non-overlapping and non-necrotic images were captured from each core of the tissue array and analyzed using plasma membrane algorithm of the Aperio Image Scope software (Leica Biosystems). Data are expressed as H-score that is based on intensity (0, 1+, 2+, and 3+) and % positivity (0%-100%) and calculated using the following formula: (% of negative cells × 0) + (% of 1+ cells × 1) + (% of 2+ cells × 2) + (% of 3+ cells × 3).
2.6. Confocal microscopy
Other details of confocal microscopy were described by us previously.26 Briefly, Cells were plated on coverslips in 12-well plates, allowed to attach by overnight incubation and then treated with DMSO or DATS for 24 h. The primary antibody (MCT1–1:300) was incubated overnight and then nuclei were stained with DAPI. Cells were observed under a Olympus FluoView FV1000 confocal microscope at 60x objective magnification.
2.7. Lactate measurement
Secreted level of lactate in the conditioned media of cells and intracellular level of lactate in cell lysates were determined using a kit from Biovison according to the manufacturer’s instructions. In brief, cells (SUM159– 3.5 × 105 cells/6-cm dish and MCF-7– 5 × 105 cells/6-cm dish) were plated in triplicate, incubated overnight, and then treated with DMSO or DATS (40 and 80 μM). After 24 hours, deproteinized medium and cell lysates were used for measurement of lactate level.
2.8. Colony forming assay
Cells (200 cells) were plated in six-well plates in triplicate. After overnight incubation, cells were treated with DMSO or the indicated concentrations of DATS. The medium containing DMSO or DATS was changed every third day. After 12 days, cells were fixed with methanol and stained with crystal violet for 30 min for colony visualization and quantitation.
2.9. Cell migration assay
Cell migration was determined as described by us previously using Transwell Boyden chambers from Corning (New York, NY) containing 8 μm pore size polycarbonate filter.27
2.10. Flow cytometry for determination of CD49fhigh/CD44high population and aldehyde dehydrogenase 1 (ALDH1) activity
Effect of DATS treatment on ALDH1 activity was determined using ALDEFLUOR™ kit from STEMCELL Technologies (Vancouver, BC) as recommended by the supplier. Diethylaminobenzaldehyde (DEAB), a specific ALDH1 inhibitor was used as a control. For analysis of CD49fhigh/CD44high population, cells were incubated with anti-CD49f (PE-conjugated) and anti-CD44 (FITC-conjugated) antibodies in the dark for 30 minutes at room temperature. The cells were washed with phosphate-buffered saline and analyzed using a Accuri™C6 flow cytometer.
2.11. Mammosphere formation assay
The mammosphere formation assay was performed as described by us previously.12 Briefly, one thousand cells were seeded in ultralow attachment plates in triplicate in medium containing penicillin/streptomycin, B27, insulin, hydrocortisone, epidermal growth factor, basic fibroblast growth factor, 2-mercaptoethanol and methylcellulose. The cells were treated with DMSO or DATS. After 7 days of incubation, the mammospheres were scored under an inverted microscope.
2.12. Statistical analysis
GraphPad Prism (version 8.0.0) was used to perform statistical analysis. Statistical analyses were done by one-way analysis of variance (ANOVA) followed by Dunnett’s adjustment (for dose-response comparison) or Bonferroni’s multiple comparisons test (for multiple group comparisons). Student’s t-test was used for binary comparisons.
3. RESULTS
3.1. FoxQ1 regulated MCT1 expression in breast cancer cells
Analysis of the breast cancer TCGA dataset revealed a statistically significant and positive association between expression of FoxQ1 and SLC16A1/MCT1 (Figure 1A). Overexpression of FoxQ1 in SUM159 and MCF-7 cell lines resulted in increased expression of MCT1 protein (Figure 1B). Three putative FoxQ1 occupancy sites were found at the promoter of SLC16A1/MCT1 (Figure 1C). The FoxQ1 was recruited at all 3 sites at the SLC16A1/MCT1 promoter in both SUM159 and MCF-7 cell lines (Figure 1D). These results indicate that MCT1 is a novel target of FoxQ1 in breast cancer cells.
Figure 1.
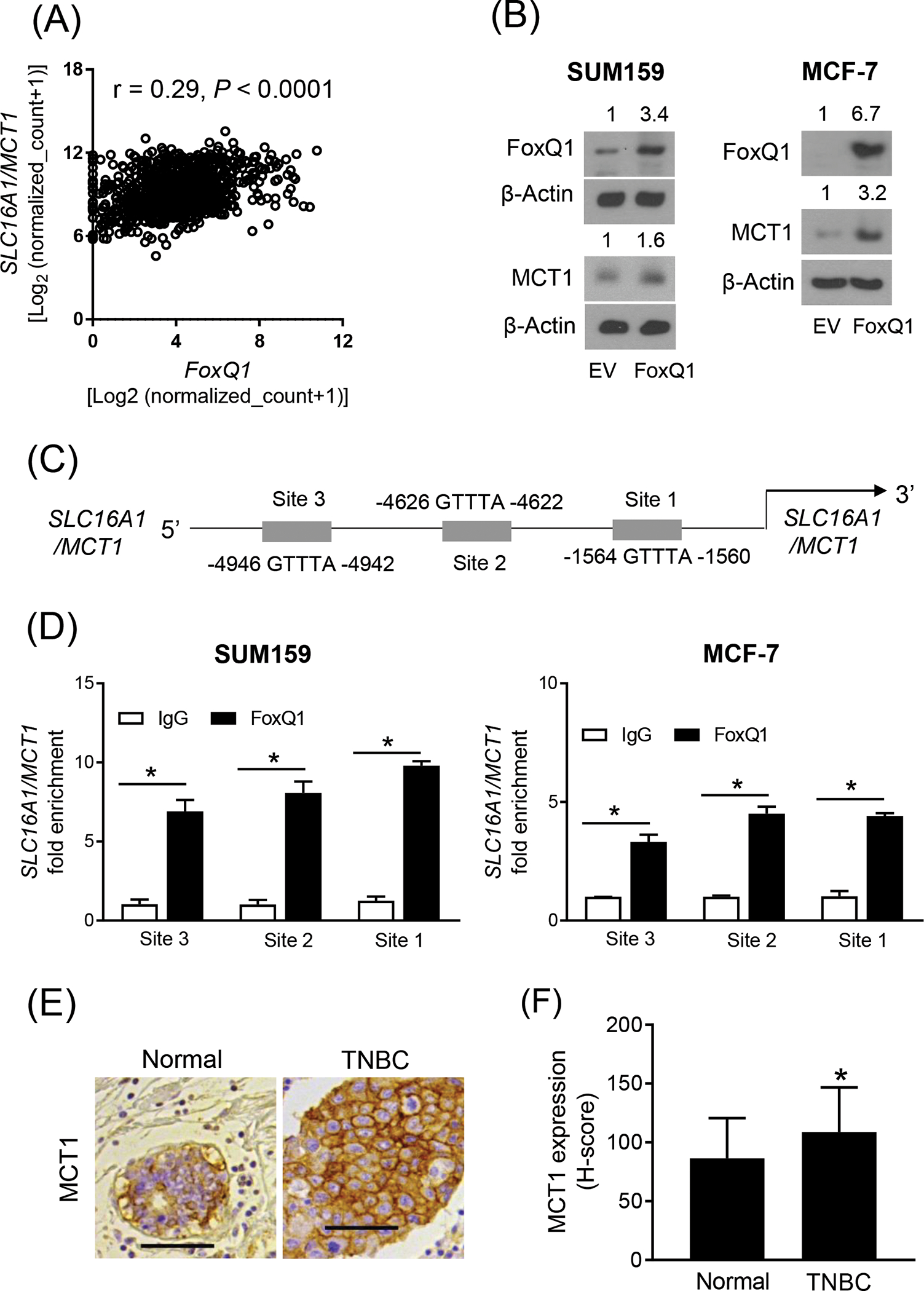
FoxQ1 is a novel regulator of MCT1. (A) Correlation between FoxQ1 and SLC16A1/MCT1 expression in breast tumors in TCGA (n=1097). Pearson’s test was used to determine the correlation coefficient and statistical significance. (B) Western blot analysis for FoxQ1 and MCT1 proteins using lysates from EV or FoxQ1 overexpressing SUM159 and MCF-7 cells. The numbers on top of the bands represent changes in protein level compared with corresponding EV cells. (C) Schematic diagram of putative FoxQ1 binding sites at the SCL16A1/MCT1 promoter. (D) Bar graphs show recruitment of FoxQ1 at the SLC16A1/MCT1 promoter in SUM159 and MCF-7 cells. Fold enrichment was calculated after normalization to input (mean ± SD; n = 3). *Statistical significance (P < 0.05) was determined by two-sided Student’s t test. (E) representative immunohistochemical image for MCT1 expression in a normal breast section and a TNBC section (20x objective, scale bar = 100 μm). (F) Quantitation of MCT1 protein expression (H-score) in normal breast (n= 50) and TNBC sections (n = 37). Results shown are mean ± SD. *Statistical significance was determined by two-sided Student’s t test. EV, empty vector
3.2. MCT1 expression was higher in TNBC when compared to normal mammary tissues
We have shown previously that expression of FoxQ1 protein is significantly higher in human TNBC in comparison with normal mammary tissue.12 Figure 1E shows immunohistochemistry for MCT1 protein in a representative section of normal human mammary tissue and a TNBC specimen. The expression of MCT1 was significantly higher in TNBC when compared to normal mammary tissues (Figure 1F). These results provided further evidence for regulation of MCT1 expression by FoxQ1 transcription factor.
3.3. DATS treatment downregulated MCT1 protein expression in breast cancer cells
We have shown previously that DATS treatment inhibits expression of FoxQ1 in SUM159 and MCF-7 cells.12 We raised the question of whether expression of MCT1 was also inhibited by DATS treatment. We explored this possibility by western blotting and immunocytochemistry. As can be seen in Figure 2A, the protein level of MCT1 was decreased by DATS treatment (Figure 2A,B). The DATS-mediated downregulation of MCT1 protein was confirmed by immunocytochemistry (Figure 2C). The intracellular level of lactate as well as its secretion into the conditioned media was also decreased by DATS treatment in both SUM159 and MCF-7 cells (Figure 2D). These results indicate that DATS downregulates MCT1 protein expression in breast cancer cells.
Figure 2.
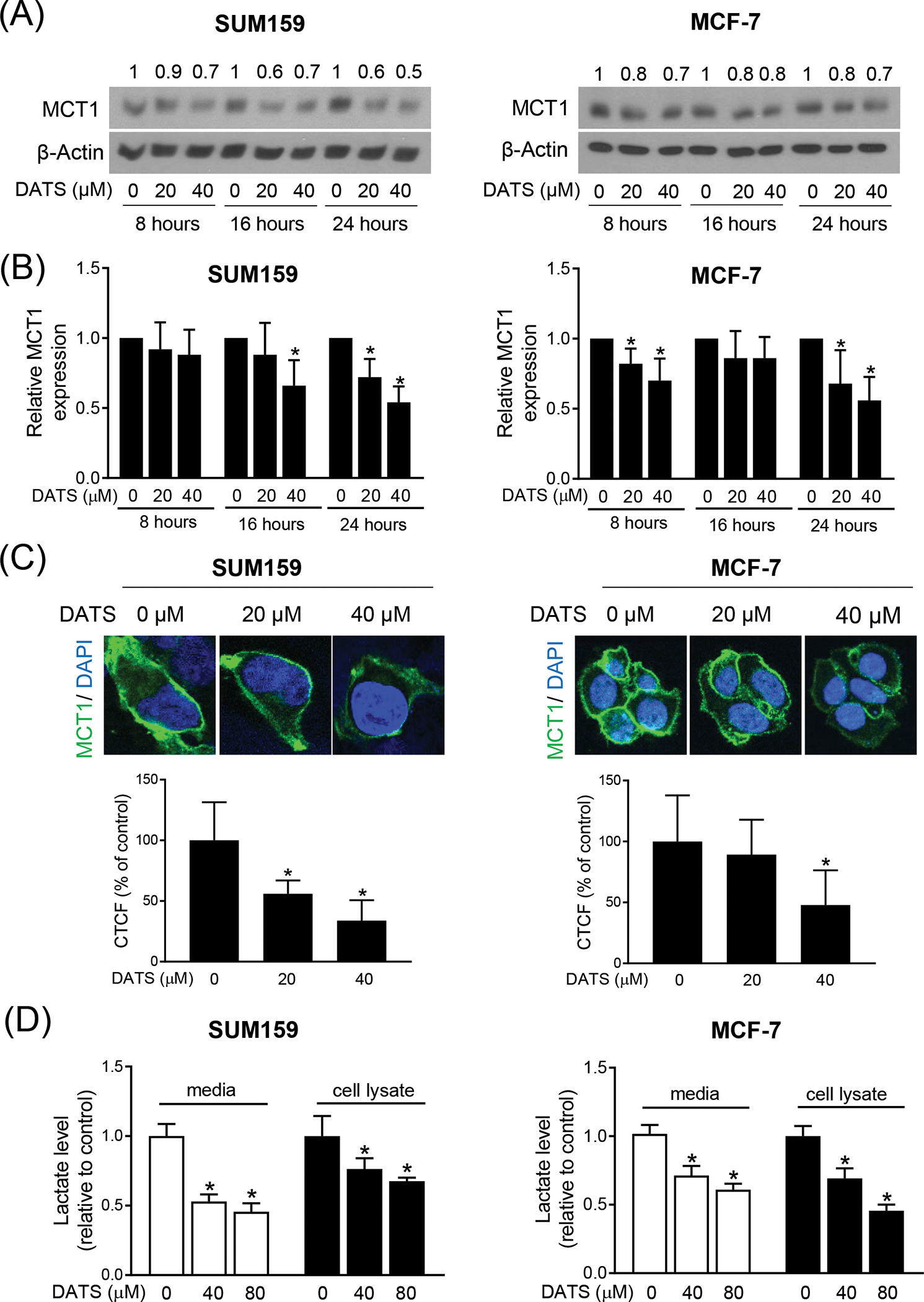
DATS treatment inhibited expression of MCT1 in Breast Cancer cells. (A) Immunoblotting for MCT1 using cell lysates from SUM159 and MCF-7 cells after treatment with DMSO or desired doses of DATS for the indicated time points. The numbers above the bands represent fold changes in expression relative to the corresponding DMSO-treated control. (B) Quantitation of the MCT1 protein expression from the western blot experiments. The results shown are mean ± SD from five independent experiments. *Significantly different (P < 0.05) compared with DMSO-treated control by one-way ANOVA followed by Dunnett’s test. (C) representative confocal microscopic images (60× oil objective) for MCT1 protein expression (green fluorescence) in SUM159 and MCF-7 cells after 24 h of treatment with DMSO or the indicated dose of DATS. DAPI (blue fluorescence) was used to stain nuclei. Bar graphs show the quantitation of corrected total cell fluorescence (CTCF) for MCT1 protein expression in SUM159 and MCF-7 cells treated with DMSO or the indicated dose of DATS. Results shown are mean ± SD (n = 10–16). *Significantly different (P < 0.05) compared with DMSO-treated control by one-way ANOVA followed by Dunnett’s adjustment. (D) Intracellular or Extracellular lactate levels in SUM159 and MCF-7 cells after treatment with DMSO or the indicated concentrations of DATS for 24 hours. Results were expressed as relative to DMSO-treated control. Results shown are mean ± SD (n = 3). *Significantly different (P < 0.05) compared with DMSO-treated control by one-way ANOVA followed by Dunnett’s adjustment. The results were consistent in two independent experiments.
3.4. MCT1 overexpression or knockdown failed to alter DATS-mediated inhibition of colony formation or cell migration in SUM159 cell line
We overexpressed MCT1 to determine its possible role in anticancer effects of DATS. The overexpression of MCT1 in transfected cells was confirmed by western blotting (Figure 3A). The colony formation by SUM159 cells was inhibited in both empty vector (EV) transfected control cells and MCT1 overexpressing cells (Figure 3B). However, MCT1 overexpression did not have any effect of DATS-mediated inhibition of colony formation (Figure 3B). Similar to colony formation assay, MCT1 overexpression did not affect inhibition of cell migration caused by DATS treatment (Figure 3C).
Figure 3.
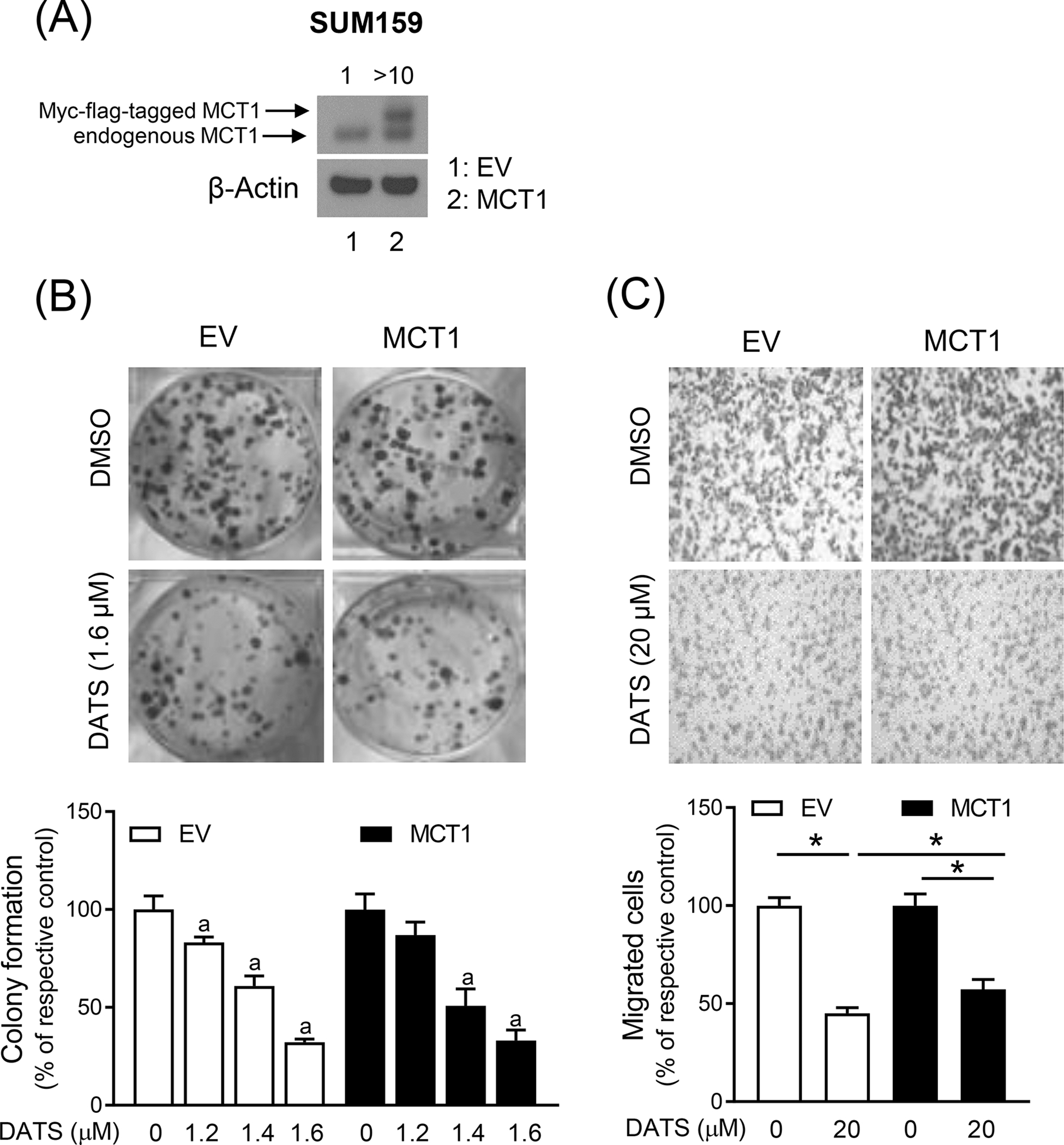
DATS-mediated inhibition of colony formation and migration is not affected by MCT1 overexpression (A) Immunoblotting for MCT1 protein using lysates from SUM159 cells stably transfected with the EV or the same vector encoding for Myc-flag-tagged MCT1 plasmid (MCT1). (B) Representative images of colony formation in EV and MCT1 overexpressing SUM159 cells after treatment with DMSO or DATS for 12 days. The graph shows the percentage of colony formation when compared to respective control. Results shown are mean ± SD (n = 3). Statistically significant (P < 0.05) compared with corresponding DMSO-treated control (a) by one-way ANOVA followed by Bonferroni’s multiple comparisons test. (C) Representative microscopic migration images (Boyden chamber assay) in EV and MCT1 overexpressing SUM159 cells after 24 h treatment with DMSO and DATS (100x magnification). Bar graph shows the percentage of cell migration when compared to respective control. Results shown are mean ± SD (n = 3). *Significantly different (P < 0.05) between the indicated groups by one-way ANOVA followed by Bonferroni’s multiple comparison test. The results were consistent in replicate experiments. EV, empty vector
We confirmed these observations using SUM159 cells stably transfected with a control shRNA or a MCT1-targeted shRNA. The level of MCT1 protein was decreased by about 90% in SUM159 cells transfected with MCT1-targeted shRNA when compared to control shRNA transfected cells (Figure 4A). The DATS treatment exhibited comparable effects on colony formation in cells transfected with a control shRNA and a MCT1-targeted shRNA (Figure 4B). The cell migration inhibition by DATS treatment was not affected by knockdown of MCT1 protein. Together, these results indicated that MCT1 downregulation did not contribute to DATS-mediated inhibition of colony formation or cell migration.
Figure 4.
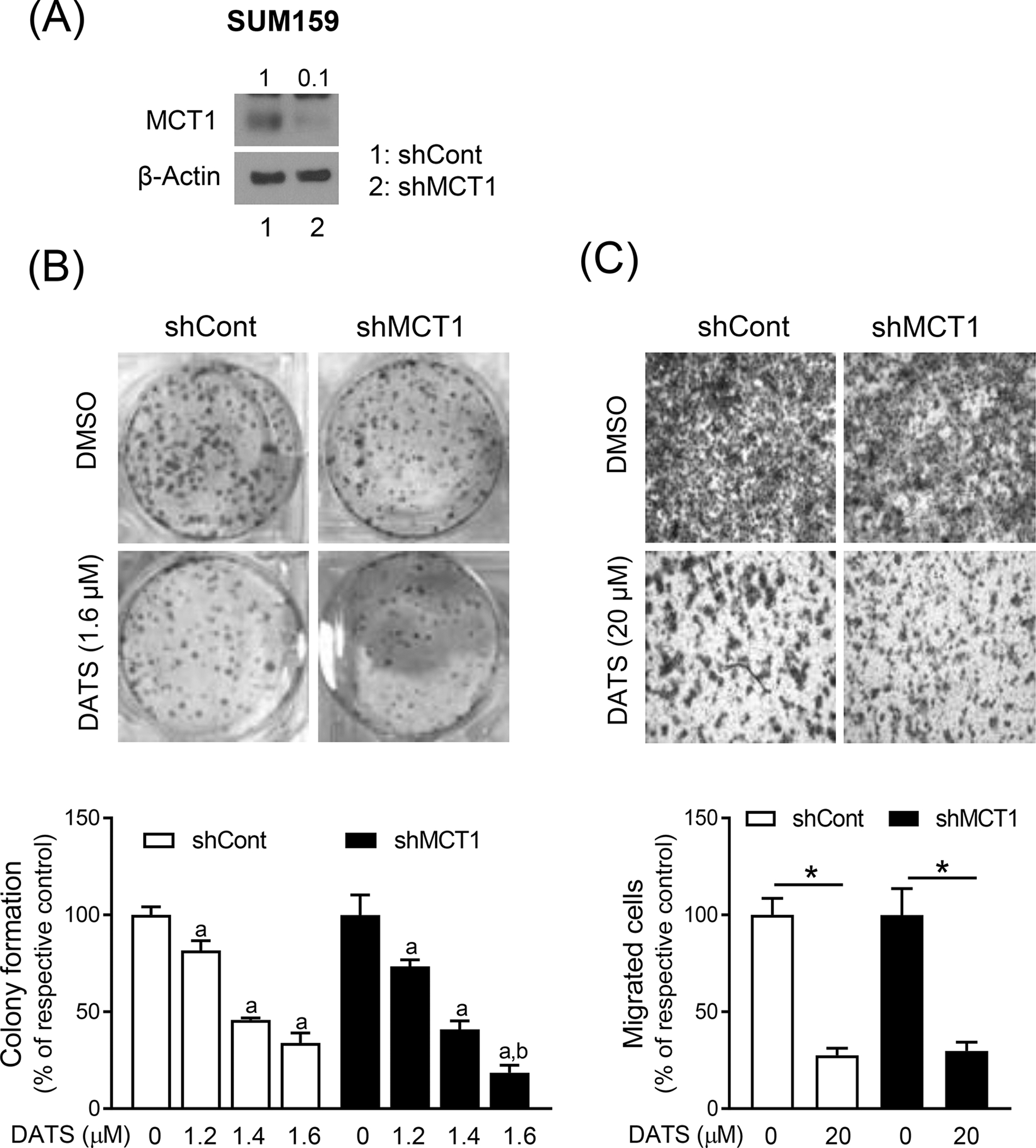
The effect of stable knockdown of MCT1 protein on DATS-mediated inhibition of colony formation and cell migration. (A) Immunoblotting for MCT1 protein using lysates from SUM159 cells stably transfected with a control shRNA (shCont) or a MCT1-targeted shRNA (shMCT1). (B) Representative images of colony formation in SUM159 cells stably transfected with a shCont or a shMCT1 after treatment with DMSO or DATS for 12 days. The graph shows the percentage of colony formation when compared to respective control. Results shown are mean ± SD (n = 3). Statistically significant (P < 0.05) compared with corresponding DMSO-treated control (a) or between shCont and shMCT1 transfected cells at the same dose of DATS (b) by one-way ANOVA followed by Bonferroni’s multiple comparisons test. (C) Representative microscopic migration images (Boyden chamber assay) in SUM159 cells stably transfected with a shCont and a shMCT1 after 24 h treatment with DMSO or DATS (100x magnification). Bar graph shows the percentage of cell migration when compared to respective control. Results shown are mean ± SD (n = 3). *Significantly different (P < 0.05) between the indicated groups by one-way ANOVA followed by Bonferroni’s multiple comparison test. Comparable results were observed in two independent experiments.
3.5. MCT1 overexpression attenuated DATS-mediated inhibition of bCSC
Next, we examined the effect of MCT1 overexpression on DATS-mediated inhibition of bCSC population. Figure 5A shows flow histograms for CD49fhigh/CD44high bCSC population in control and DATS-treated cells transfected with EV or MCT1 plasmid. The DATS treatment decreased CD49fhigh/CD44high population by 42% in EV cells (Figure 5B). Overexpression of MCT1 conferred partial but significant protection against DATS-mediated inhibition of CD49fhigh/CD44high population (Figure 5B). Flows histograms for ALDH1, which is another marker of bCSC, are shown in Figure 5C. Similar to analysis of CD49fhigh/CD44high population, the DATS-mediated inhibition of ALDH1 activity was partly but significantly attenuated by MCT1 overexpression (Figure 5D). The size and the number of mammospheres was decreased significantly by DATS treatment in both EV and MCT1 overexpressing SUM159 cells (Figure 5E). However, MCT1 overexpression did not affect DATS-mediated decrease in mammosphere number (Figure 5F).
Figure 5.
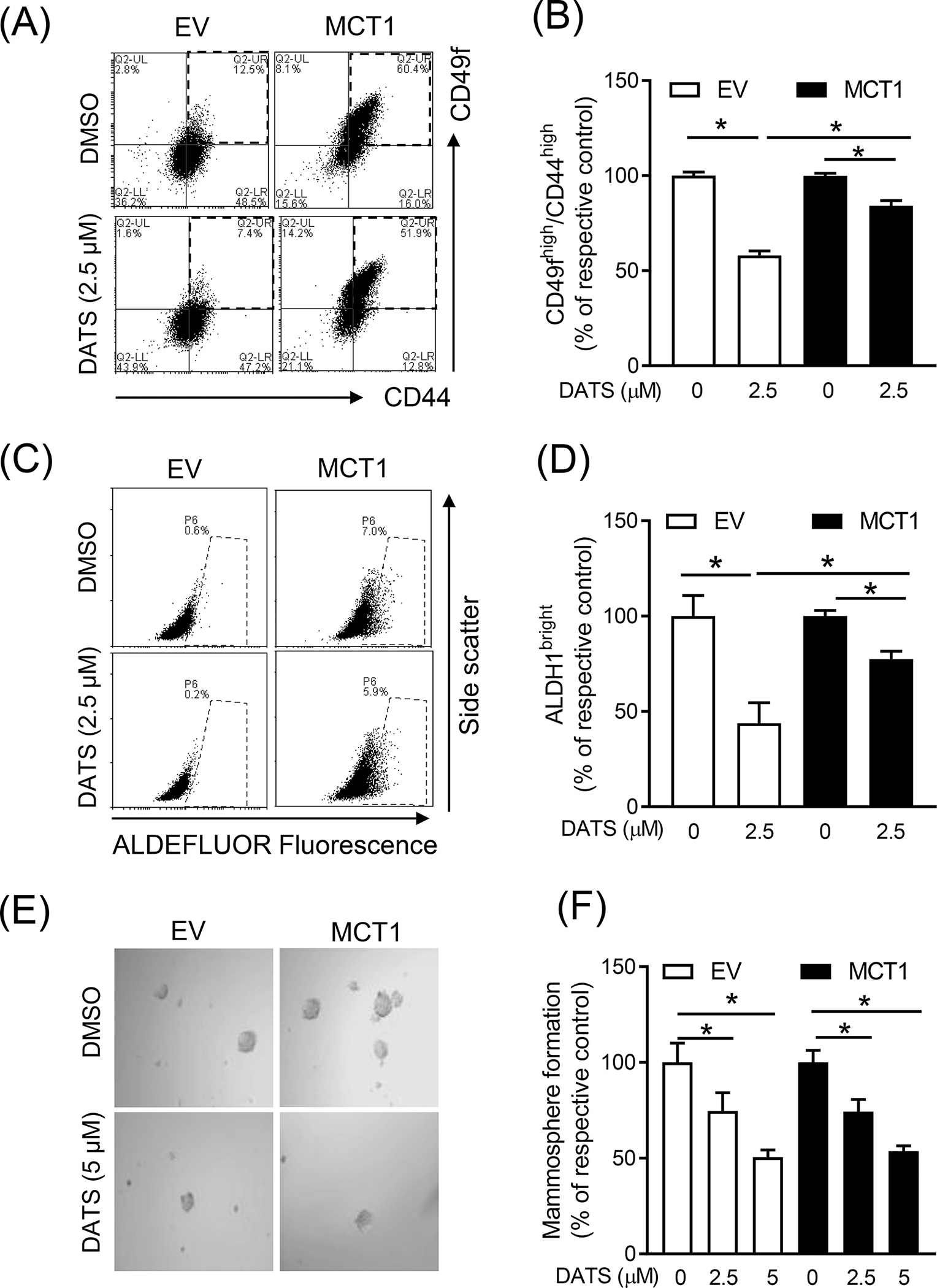
The role of MCT1 overexpression in DATS-mediated inhibition of bCSC (A) Representative flow histograms for the CD49fhigh/CD44high population in EV and MCT1 overexpressing SUM159 cells after treatment with DMSO or DATS (2.5 μM) for 72 hours. (B) Bar graph shows the percentage of the CD49fhigh/CD44high population when compared to respective control. (mean ± S.D., n = 3) (C) Representative flow histograms for ALDH1 activity in EV and MCT1 overexpressing SUM159 cells after 72-h treatment with DMSO or DATS (2.5 μM). (D) The graph shows the quantitation of ALDH1 activity when compared to respective control (mean ± S.D., n = 3). (E) Representative mammosphere images after 7 days of treatment with DMSO or DATS (5 μM) in EV and MCT1 overexpressing SUM159 (100x magnification). (F) Quantitation of mammosphere formation when compared to respective to control (mean ± S.D., n = 3). * P < 0.05 between the indicated groups by one-way ANOVA followed by Bonferroni’s multiple comparisons test. Each experiment was repeated at least twice with comparable results. EV, empty vector
Similar experiments were performed using cells transfected with a control shRNA and a MCT1-targeted shRNA. The DATS-mediated inhibition of CD49fhigh/CD44high population was significantly augmented by knockdown of MCT1 (Figure 6A, B). The DATS-mediated suppression of ALDH1 activity was also augmented by MCT1 knockdown but the difference was not significant (Figure 6C, D). However, the mammosphere frequency inhibition by DATS treatment was not affected by MCT1 knockdown (Figure 6E, F). Collectively, these results indicate that DATS-mediated inhibition of bCSC faction is partly regulated by FoxQ1-MCT1 axis.
Figure 6.
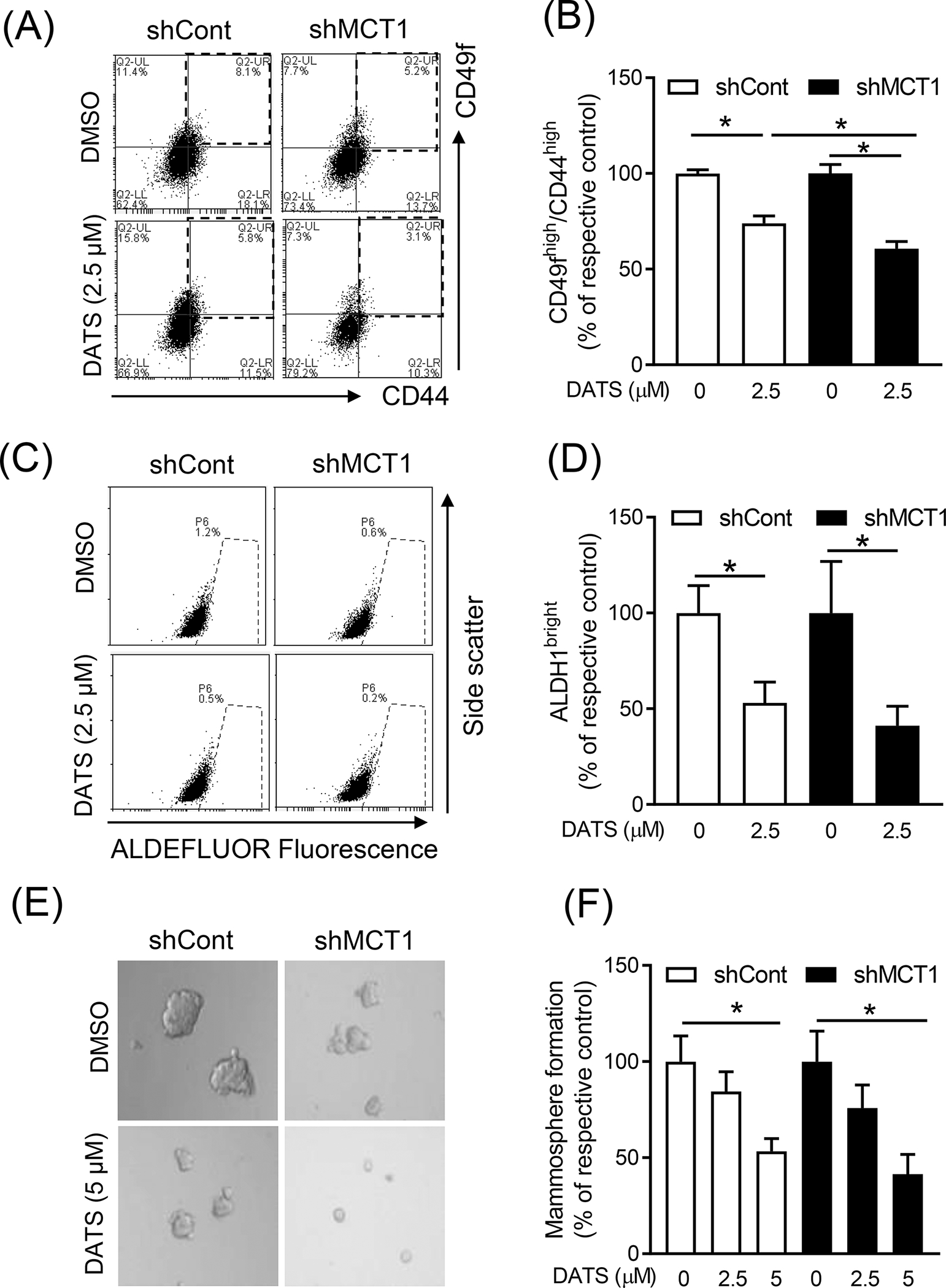
The effect of stable knockdown of MCT1 protein on DATS-mediated inhibition of bCSC. (A) Representative flow histograms for the CD49fhigh/CD44high population in SUM159 cells stably transfected with a shCont or a shMCT1 after treatment with DMSO or DATS (2.5 μM) for 72 hours. (B) Bar graph shows the percentage of the CD49fhigh/CD44high population when compared to respective control. (mean ± S.D., n = 3) (C) Representative flow histograms for ALDH1 activity in SUM159 cells stably transfected with a shCont or a shMCT1 after 72-h treatment with DMSO or DATS (2.5 μM). (D) The graph shows the quantitation of ALDH1 activity when compared to respective control (mean ± S.D., n = 3). (E) Representative mammosphere images after 7 days of treatment with DMSO or DATS (5 μΜ) in SUM159 cells stably transfected with a shCont or a shMCT1 (100x magnification). (F) Quantitation of mammosphere formation when compared to respective to control (mean ± S.D., n = 3). * P < 0.05 between the indicated groups by one-way ANOVA followed by Bonferroni’s multiple comparisons test. Similar results were observed in independent experiments.
4. DISCUSSION
The FoxQ1 transcription factor is best known for its role in regulation of epithelial to mesenchymal transition (EMT), bCSC maintenance, and metastasis.12,28,29 Overexpression of FoxQ1 in breast cancer cells increased migration and invasion and triggered EMT by repressing expression of epithelial marker E-cadherin.28 These investigators also showed that FoxQ1 expression was regulated by TGF-β1, and TGF-β1-induced EMT at both morphological and molecular level was blocked by knockdown of FoxQ1.28 Another study showed that platelet-derived growth factor receptor α/β were directly regulated by FoxQ1 and contribute to stemness and chemoresistance in breast cancer.29 Our own work revealed that tumor suppressor Dachshund homolog 1, whose expression is lost in invasive breast cancer, is negatively regulated by FoxQ1.12 The present study reveals that MCT1 is yet another novel target of regulation by FoxQ1 as evidenced by TCGA analysis and ChIP assay. MCT1 was shown to be overexpressed in hormone receptor-negative and high-proliferation subtypes, which was confirmed in the present study using TNBC specimens.30 High expression of MCT1 and MCT4 in breast cancer was associated with poor patient outcome.30
We have shown previously that FoxQ1 protein expression is significantly higher in TNBC and luminal type when compared to normal mammary tissues.12,31 Analysis of the breast TCGA dataset revealed significantly higher expression of FoxQ1 in black breast cancer patients compared with white women.31 Through RNA-seq comparison of FoxQ1 overexpressing SUM159 cells and corresponding EV transfected control cells, many previously reported transcriptional targets of FoxQ1 (e.g., E-cadherin, N-cadherin, fibronectin 1, etc.) were verified from the RNA-seq analysis.31 While majority of studies in breast cancer and other solid tumors support an oncogenic function of FoxQ1, one study reported significantly decreased expression of FoxQ1 mRNA in luminal type breast cancer and HER2 patients when compared to normal breast tissue samples.32,33 Low expression of FoxQ1 was associated with poor prognosis in breast cancer patients.33 The reasons for the discrepancy in oncogenic versus tumor suppressive functions of FoxQ1 are unclear and require further investigation.
We have shown previously that DATS treatment decreases bCSC faction in vitro and in vivo.12 Because DATS-mediated inhibition of bCSC is attenuated by FoxQ1 overexpression, we raised the question if MCT1 is also inhibited by DATS. MCT1 was shown to promote stemness in TNBC.34 The present study reveals that similar to FoxQ1, DATS treatment decreases protein level of MCT1 in both SUM159 and MCF-7 cells. The DATS-mediated inhibition of bCSC is only partially attenuated by ectopic expression of MCT1. These results suggest that inhibition of bCSC fraction by DATS treatment is likely mediated by MCT1 independent mechanisms as well. As an example, DATS was shown to inhibit bCSC by suppressing Wnt/β-catenin pathway.17 Interestingly, FoxQ1 was shown to be a target of Wnt in colorectal cancer.35
In conclusion, we demonstrate for the first time that MCT1 expression is directly regulated by FoxQ1 transcription factor and DATS-mediated inhibition of bCSC is partially caused by suppression of MCT1.
Abbreviations:
- ANOVA
analysis of variance
- bCSC
breast cancer stem-like cells
- DATS
diallyl trisulfide
- DMSO
dimethyl sulfoxide
- FoxQ1
forkhead box Q1
- MCT1
monocarboxylate transporter 1
- EV
empty vector
- shRNA
small hairpin RNA
- TCGA
The Cancer Genome Atlas
- TNBC
triple-negative breast cancer
REFERENCES
- 1.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 3.Tsubura A, Lai YC, Kuwata M, Uehara N, Yoshizawa K. Anticancer effects of garlic and garlic-derived compounds for breast cancer control. Anticancer Agents Med Chem. 2011;11:249–253. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Li S, Meng X, Gan RY, Zhang JJ, Li HB. Dietary natural products for prevention and treatment of breast cancer. Nutrients. 2017;9:728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Challier B, Perarnau JM, Viel JF. Garlic, onion and cereal fibre as protective factors for breast cancer: a French case-control study. Eur J Epidemiol. 1998;14:737–747. [DOI] [PubMed] [Google Scholar]
- 6.Desai G, Schelske-Santos M, Nazario CM, et al. Onion and garlic intake and breast cancer, a case-control study in Puerto Rico. Nutr Cancer. 2020;72:791–800. [DOI] [PubMed] [Google Scholar]
- 7.Li H, Li HQ, Wang Y, et al. An intervention study to prevent gastric cancer by micro-selenium and large dose of allitridum. Chin Med J (Engl). 2004;117:1155–1160. [PubMed] [Google Scholar]
- 8.Zheng GH, Li H, Fan WT, Li HQ. Study on the long-time effect on allitridum and selenium in prevention of digestive system cancers. Zhonghua Liu Xing Bing Xue Za Zhi (Chinese). 2005;26:110–112. [PubMed] [Google Scholar]
- 9.Antony ML, Singh SV. Molecular mechanisms and targets of cancer chemoprevention by garlic-derived bioactive compound diallyl trisulfide. Indian J Exp Biol. 2011;49:805–816. [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Liu X, Ruan J, Zhuang X, Zhang X, Li Z. Phytochemicals of garlic: Promising candidates for cancer therapy. Biomed Pharmacother. 2020;123:109730. [DOI] [PubMed] [Google Scholar]
- 11.Na HK, Kim EH, Choi MA, Park JM, Kim DH, Surh YJ. Diallyl trisulfide induces apoptosis in human breast cancer cells through ROS-mediated activation of JNK and AP-1. Biochem Pharmacol. 2012;84:1241–1250. [DOI] [PubMed] [Google Scholar]
- 12.Kim SH, Kaschula CH, Priedigkeit N, Lee AV, Singh SV. Forkhead box Q1 is a novel target of breast cancer stem cell inhibition by diallyl trisulfide. J Biol Chem. 2016;291:13495–134508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Zhao Y, Wei Z, et al. Targeting thioredoxin system with an organosulfur compound, diallyl trisulfide (DATS), attenuates progression and metastasis of triple-negative breast cancer (TNBC). Cell Physiol Biochem. 2018;50:1945–1963. [DOI] [PubMed] [Google Scholar]
- 14.Kim SH, Hahm ER, Singh KB, Singh SV. Diallyl trisulfide inhibits leptin-induced oncogenic signaling in human breast cancer cells but fails to prevent chemically-induced luminal-type cancer in rats. J Cancer Prev. 2020;25:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee BC, Park BH, Kim SY, Lee YJ. Role of Bim in diallyl trisulfide-induced cytotoxicity in human cancer cells. J Cell Biochem. 2011;112:118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandra-Kuntal K, Lee J, Singh SV. Critical role for reactive oxygen species in apoptosis induction and cell migration inhibition by diallyl trisulfide, a cancer chemopreventive component of garlic. Breast Cancer Res Treat. 2013;138:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Meng Y, Xie C, et al. Diallyl trisulfide inhibits breast cancer stem cells via suppression of Wnt/β-catenin pathway. J Cell Biochem. 2018;119:4134–4141. [DOI] [PubMed] [Google Scholar]
- 18.Malki A, El-Saadani M, Sultan AS. Garlic constituent diallyl trisulfide induced apoptosis in MCF7 human breast cancer cells. Cancer Biol Ther. 2009;8:2175–2185. [DOI] [PubMed] [Google Scholar]
- 19.Kiesel VA, Stan SD. Diallyl trisulfide, a chemopreventive agent from Allium vegetables, inhibits α-secretases in breast cancer cells. Biochem Biophys Res Commun. 2017;484:833–838. [DOI] [PubMed] [Google Scholar]
- 20.Cheng SY, Yang YC, Ting KL, et al. Lactate dehydrogenase downregulation mediates the inhibitory effect of diallyl trisulfide on proliferation, metastasis, and invasion in triple-negative breast cancer. Environ Toxicol. 2017;32:1390–1398. [DOI] [PubMed] [Google Scholar]
- 21.Hahm ER, Singh SV. Diallyl trisulfide inhibits estrogen receptor-α activity in human breast cancer cells. Breast Cancer Res Treat. 2014;144:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Zhu P, Wang Y, et al. Antimetastatic therapies of the polysulfide diallyl trisulfide against triple-negative breast cancer (TNBC) via suppressing MMP2/9 by blocking NF-κB and ERK/MAPK signaling pathways. PLoS One. 2015;10:e0123781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei Z, Shan Y, Tao L, et al. Diallyl trisulfides, a natural histone deacetylase inhibitor, attenuate HIF-1α synthesis, and decreases breast cancer metastasis. Mol Carcinog. 2017;56:2317–2331. [DOI] [PubMed] [Google Scholar]
- 24.Hahm ER, Kim SH, Mathan SV, Singh RP, Singh SV. Mechanistic targets of diallyl trisulfide in human breast cancer cells identified by RNA-seq analysis. J Cancer Prev. 2021;26:128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao D, Srivastava SK, Lew KL, et al. Allyl isothiocyanate, a constituent of cruciferous vegetables, inhibits proliferation of human prostate cancer cells by causing G2/M arrest and inducing apoptosis. Carcinogenesis. 2003;24:891–897. [DOI] [PubMed] [Google Scholar]
- 26.Singh KB, Hahm ER, Alumkal JJ, et al. Reversal of the Warburg phenomenon in chemoprevention of prostate cancer by sulforaphane. Carcinogenesis. 2019;40:1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Hahm ER, Singh SV. Withaferin A inhibits activation of signal transducer and activator of transcription 3 in human breast cancer cells. Carcinogenesis. 2010;31:1991–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Meng F, Liu G, et al. Forkhead transcription factor foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Cancer Res. 2011;71:1292–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng F, Speyer CL, Zhang B, et al. PDGFRα and β play critical roles in mediating Foxq1-driven breast cancer stemness and chemoresistance. Cancer Res. 2015;75:584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Wu Q, Sun S, et al. Monocarboxylate transporters in breast cancer and adipose tissue are novel biomarkers and potential therapeutic targets. Biochem Biophys Res Commun. 2018;501:962–967. [DOI] [PubMed] [Google Scholar]
- 31.Kim SH, Hahm ER, Singh KB, Singh SV. Novel mechanistic targets of forkhead box Q1 transcription factor in human breast cancer cells. Mol Carcinog. 2020;59:1116–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Zhang Y, Yao Z, Li S, Yin Z, Xu M. Forkhead box Q1: A key player in the pathogenesis of tumors (Review). Int J Oncol. 2016;49:51–58. [DOI] [PubMed] [Google Scholar]
- 33.Elian FA, Are U, Ghosh S, et al. FOXQ1 is differentially expressed across breast cancer subtypes with low expression associated with poor overall survival. Breast Cancer (Dove Med Press). 2021;13:171–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weng YS, Tseng HY, Chen YA, et al. MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol Cancer. 2019;18:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christensen J, Bentz S, Sengstag T, Shastri VP, Anderle P. FOXQ1, a novel target of the Wnt pathway and a new marker for activation of Wnt signaling in solid tumors. PLoS One. 2013;8:e60051. [DOI] [PMC free article] [PubMed] [Google Scholar]


